Abstract
Bacillus circulans Jordan 32352 was isolated from decaying organic matter in the New Jersey soil in the early 1930s. This soil-dwelling bacterium produced an enzyme capable of degrading the type 3 capsular polysaccharide (Pn3P) of Streptococcus pneumoniae (Spn). Early reports of this enzyme, Pn3Pase, demonstrated its inducibility by, and specificity for Pn3P. We set out to identify and clone this enzyme for its recombinant expression and characterization. We first sequenced the genome of this bacterial species, and reclassified the Pn3Pase producing bacterium as Paenibacillus species 32352. We identified the putative protein of Pn3Pase through mass spectrometry-based proteomics and cloned the gene for recombinant expression. We then characterized the oligosaccharide products generated upon the enzymatic depolymerization of Pn3P. Sequence analysis suggests that this glycoside hydrolase belongs to a new carbohydrate-active enzyme GH family. To our knowledge, this is the only enzyme to demonstrate Pn3P depolymerization activity.
Keywords: capsular polysaccharide, enzyme processing, glycoside hydrolase, Paenibacillus
Introduction
In 1930, Avery and Dubos isolated an organism from soil taken from cranberry bogs, which was capable of depolymerizing type 3 capsular polysaccharide (CPS) of Streptococcus pneumoniae (Pn3P), a linear polymer of −3)βGlcA(1–4)βGlc(1– (Avery and Dubos 1930; Dubos and Avery 1931). The expression of this enzyme (Pn3Pase) was inducible in the presence of Pn3P, and the bacterium was able to grow with Pn3P as the sole carbon source. They described this bacterium as a sporulating, Gram-negative, aerobic bacillus with peritrichous flagella. A few years later, Sickles and Shaw isolated two additional similar strains demonstrating the same enzymatic activity targeting Pn3P (Sickles and Shaw 1934; Uetanabaro et al. 2003). These strains were designated as Bacillus palustris, before being accepted as synonymous to Bacillus circulans (Nakamura and Swezey 1983; Ash et al. 1993; Uetanabaro et al. 2003). Besides Pn3Pase, these strains demonstrated the ability to produce enzymes capable of depolymerizing S. pneumoniae capsular serotypes 2 and 8, although soluble protein in cell free extracts were inactive against these polysaccharides (Sickles and Shaw 1934; Torriani and Pappenheimer 1962). There are numerous possible practical applications for these strains and enzymes. For example, investigators have applied the Sickles and Shaw Pn3Pase enzyme while examining Pn3P biosynthesis as well as its immunological properties (Rice and Sickles 1946; Shaw and Sickles 1950; Forsee et al. 2006, 2009).
We obtained the “Bacillus circulans Jordan strain 32352” (i.e. the Sickles and Shaw strain) from American Type Culture Collection, and sequenced its genome (Middleton et al. 2017). 16 S rRNA analysis revealed that this bacterium belonged in the Paenibacillae genus, and it is now identified as Paenibacillus sp. 32352 (Pbac). Paenibacillus species are of growing interest since the genus was established in 1991 (Agard et al. 2006). These microbes are a rich source of extracellular enzymes that catalyze a variety of reactions, which have demonstrated utility in numerous agricultural and medical applications (Konishi and Maruhashi 2003; Guo et al. 2012; Chung et al. 2015; Knolhoff et al. 2015; Oliveira et al. 2015; Pasari et al. 2017). Database for carbohydrate-active enzyme (CAZy) annotation (dbCAN) of the Paenibacillus sp. 32352 genome indicates 665 carbohydrate-active entries out of 7200 predicted genes, 252 of those exhibiting glycoside hydrolase (GH)- or polysaccharide lyase-like architecture (Yin et al. 2012).
We set out to determine the identity of the gene producing Pn3Pase in order to express, utilize and study the enzyme’s unique Pn3P-specific activity. Here, we have identified the Paenibacillus Pn3Pase through proteomics of culture supernatant preparations with Pn3P supplemented minimal media growth. We have cloned the Pn3Pase gene and expressed the active enzyme in Escherichia coli. We have also identified kinetic parameters, characterized oligosaccharide products and determined optimal conditions for Pn3P depolymerization by Pn3Pase.
Results
Pn3P utilization induces expression of genes organized into locus
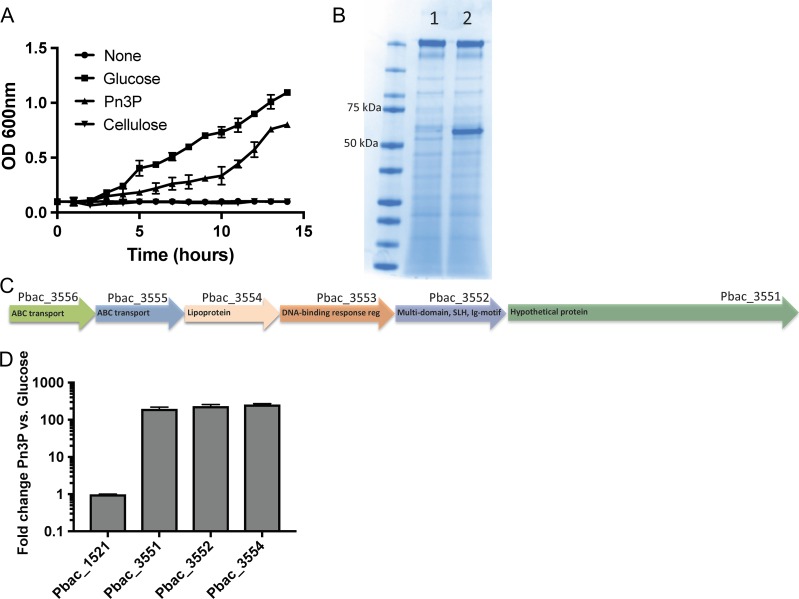
We began by culturing Pbac in minimal M9 media with either 2% glucose or Pn3P as the sole carbon source. Pbac growth was monitored by OD 600 nm for 14 h to achieve maximum Pn3Pase production (Figure 1A). Pbac was able to grow and utilize Pn3P and glucose, but was unable to utilize cellulose. Supernatants from these cultures were concentrated 20×, and proteins were visualized by coomassie staining. In the Pn3P growth conditions, a prominent band ~55 kDa was observed (Figure 1B). Proteomic analysis of these samples by LC–MS-MS identified numerous proteins that were present in both samples. Two proteins, however, were significantly enriched in the Pn3P growth condition as highlighted in Table I.
Fig. 1.
Culture of Paenibacillus sp. 32352 in minimal medium with Pn3P as carbon source. (A) Growth of Paenibacillus sp. 32352 (Pbac) on minimal medium plus 2% (w/v) glucose, Pn3P, cellulose or nothing as carbon source. (B) SDS-PAGE coomassie blue stained gel of concentrated culture supernatant of Pbac grown on glucose (1) or Pn3P (2). (C) Proposed locus of Pn3P utilization organization based on the Rapid Annotation Server (Aziz et al. 2008) (RAST) annotation. (D) Real-time PCR of select genes within putative locus in Pbac grown on 2% glucose or Pn3P, shown as fold change in expression in Pn3P culture versus glucose culture. SLH (surface layer homology) Ig-motif (immunoglobulin-like motif).
Table I.
Proteomic identification of culture supernatant proteins of Paenibacillus sp. 32352 grown in Pn3P. Highlighted are proteins unique to Pn3P grown culture.
| Gene | Description | Score | Coverage | #Unique peptides | # PSMsa | #AAs | MW (kDa) |
|---|---|---|---|---|---|---|---|
| Pbac_3554 | *Lipoprotein | 1301.24 | 82.02 | 49 | 597 | 506 | 55.9 |
| Pbac_1521 | Ig-like, group 2, surface layer protein | 450.46 | 50.17 | 44 | 218 | 1190 | 128.7 |
| Pbac_6539 | Hypothetical protein | 237.19 | 31.20 | 17 | 82 | 577 | 64.1 |
| Pbac_5871 | NLP/P60 family protein | 103.53 | 61.54 | 6 | 35 | 156 | 16.9 |
| Pbac_1659 | Alpha/beta hydrolase fold (EC 3.8.1.5) | 94.98 | 66.07 | 13 | 72 | 280 | 30.1 |
| Pbac_3551 | Hypothetical protein | 39.14 | 16.70 | 6 | 53 | 1545 | 168.0 |
| NCBI ref seq: WP_079,915,027 |
aPSM, peptide spectral matches, filtered at a 1% false discovery rate. Proteins considered with >30 PSM.
These two Pn3P induced Pbac proteins appear to be organized into a locus of Pn3P utilization (Figure 1C) consisting of ABC-type polysaccharide transport system, permease component (Pbac_3556), probable ABC transporter permease protein ytcP (Pbac_3555), lipoprotein (Pbac_3554), DNA-binding response regulator, AraC family (Pbac_3553), multidomain protein with a surface-layer homology region and immunoglobulin-like motif (Pbac_3552) and a hypothetical protein (Pbac_3551). Since two of these proteins are more abundant in the Pn3P growth conditions, we compared the transcription of three genes of this locus with the commonly expressed surface layer protein (Pbac_1521) in the glucose and Pn3P samples by RT-PCR. The transcription of genes 3551, 3552 and 3554 increase ~130-fold with Pn3P utilization, while mRNA expression of Pbac_1521 is unchanged between the two growth conditions (Figure 1D). The only major difference observed at the protein level, based on coomassie stain (Figure 1B) of the culture supernatant of Pn3P induced culture, is the dominant Pbac_3554 product. The sensitivity provided by mass spectrometry allowed us to identify Pbac_3551 in the supernatant as well, although at low abundance. While transcript levels are increased dramatically for all genes that were tested in this locus, some of these proteins may be closely associated with the cell surface and not efficiently shed similarly to Pbac_3554.
Pn3Pase identification and domain analysis
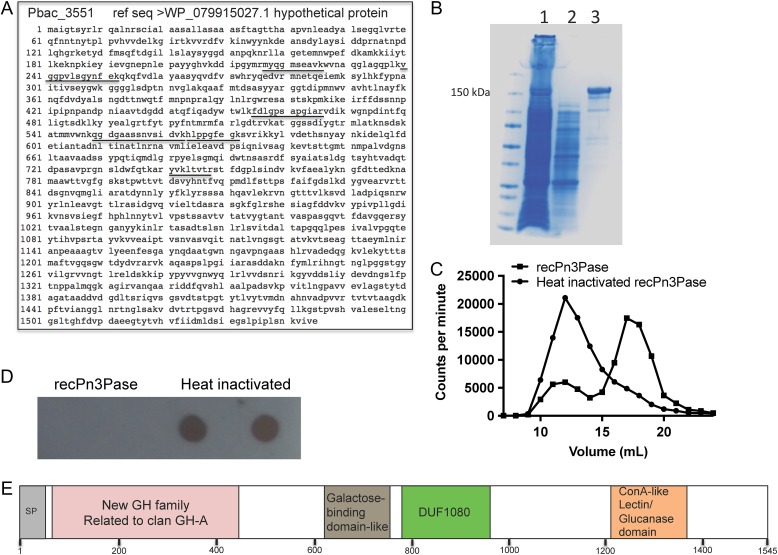
Pbac_3554, Pbac_3552 and Pbac_3551 were cloned and expressed in E. coli BL21 (DE3) cells to determine which upregulated gene product was responsible for Pn3P depolymerase activity. The hypothetical protein Pbac_3551 (Accession WP_079,915,027), with primary sequence shown in Figure 2A was expressed and purified by immobilized metal affinity chromatography (Figure 2B). Initial preparations have yielded ~2.5 mg/L of BL21 culture. Pbac_3551 (hypothetical protein) demonstrated rapid and efficient hydrolysis of tritium radioisotope labeled Pn3P, as shown by counts per min shift to lower molecular weight oligosaccharides when reaction products were separated by size exclusion chromatography (Figure 2C). Reactions of recombinant Pn3Pase with unlabeled Pn3P were performed, spotted on a PVDF membrane, and probed with a Pn3P monoclonal antibody. Reactivity to the monoclonal antibody was completely abolished after 4 h Pn3Pase treatment of Pn3P (Figure 2D).
Fig. 2.
Pn3Pase identification and domain schematic of amino acid sequence. (A) Amino acid sequence and accession of Pn3Pase. Underlined are tryptic peptides identified with high confidence by proteome discoverer software. (B) Purification is His-tagged recombinant Pn3Pase by Nickel-NTA column. Lane (1) uncaptured flow-through, (2) 5× concentrated wash, (3) elution. (C) Separation of recombinant Pn3Pase depolymerized tritium radioisotope labeled Pn3P by size exclusion chromatography, measured by counts per minute in each 1 ml fraction. (D) Dot blot of recombinant Pn3Pase depolymerized cold Pn3P probed with Pn3P monoclonal antibody. (E) Schematic of predicted domains of Pn3Pase by InterPro.
The translated protein sequence of Pbac_3551 consists of 1545 amino acids. Predicted cleavage of the signal peptide by SignalP 4.1 server (Petersen et al. 2011) from residues 1 to 40 would yield a mature protein of 164.1 kDa. Protein sequence analysis and classification by InterPro online software (Finn et al. 2017) recognized homology to GH, family 39 (GH39) at the N-terminus, most similar to the β-xylosidase from Themoanaerobacterium saccharolyticum (beta/alpha)8 barrel region (Yang et al. 2004). Other regions related to galactose-binding domain-like related to carbohydrate-binding module, family 6 (CBM6) from 621 to 765, a domain of unknown function DUF1080 from 781 to 950 with structural similarity to an endo-1,3-1,4-beta glucanase belonging to GH, family 16 (GH16), and a concanavalin A-like lectin/glucanase domain from 1209 to 1348 (Figure 2E) were found. Based on CAZy database, although segments of this enzyme display some homology to numerous carbohydrate-active proteins, no overall homology to known GH families across the length of the enzyme exist (data not shown). This unique enzyme will potentially be sorted into a new dedicated GH family related to clan GH-A. Clan GH-A are retaining enzymes with a catalytic domain displaying a (beta/alpha)8 barrel fold (Henrissat and Bairoch 1996).
Characterization of oligosaccharide products
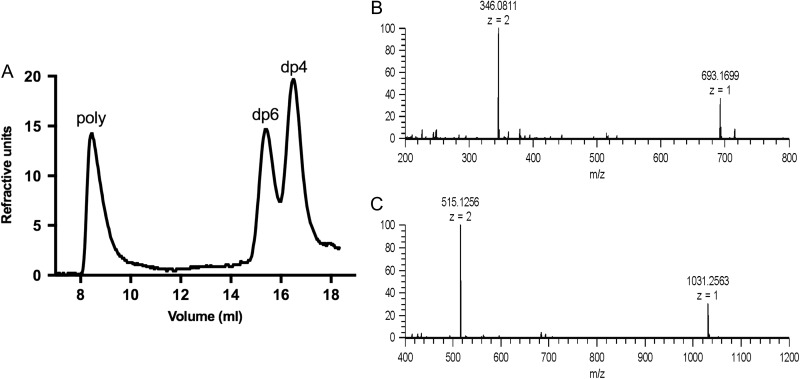
The oligosaccharide products of Pn3Pase hydrolysis were separated by size exclusion chromatography. This hydrolysis yielded two major product peaks eluting late in the Superdex peptide column corresponding to a tetrasaccharide and hexasaccharide (Figures 3A–C). The identities of these peaks were confirmed as tetrasaccharide (Figure 3B) and hexasaccharide (Figure 3C) by electrospray ionization mass spectrometry. Mass spectrometry data are summarized in Table II.
Fig. 3.
Identification of oligosaccharide products by electrospray ionization mass spectrometry. Separation of oligosaccharide products (A) and mass spectra of the tetrasaccharide (B) and the hexasaccharide (C). Experimental molecular weights and the accuracies are shown in Table II.
Table II.
Different charge states of the tetrasaccharide and hexasaccharide detected by mass spectrometry.
| m/z Observed | Charge state | Experimental M | Theoretical M | Error (ppm) | |
|---|---|---|---|---|---|
| Tetrasaccharide | 693.1699 | 1 | 694.1777 | 694.1804 | −3.85 |
| 346.0811 | 2 | 694.1779 | 694.1804 | −3.66 | |
| Hexasaccharide | 1031.2563 | 1 | 1032.2641 | 1032.2653 | −1.13 |
| 515.1256 | 2 | 1032.2669 | 1032.2653 | 1.51 |
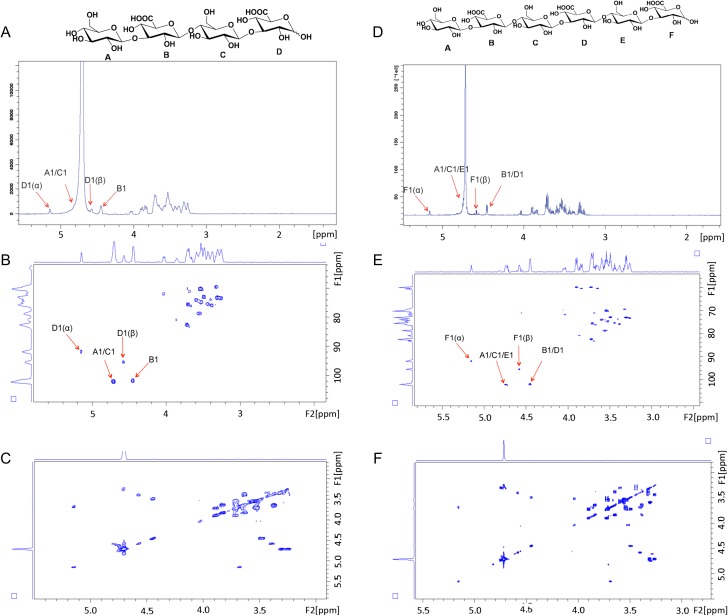
The characterization of oligosaccharide products by NMR spectroscopy is presented in Figure 4. All anomeric proton signals are assigned in representative 1H NMR spectra of tetra- and hexasaccharides (Figure 4). Anomeric proton signals of residue A, C and E at ~4.70 ppm were overlapped with HOD peak (Figure 4A and D) but showed up in 2D heteronuclear single quantum coherence (HSQC) spectrum (Figure 4B and E). The 3JHH coupling constants of B-1 and D-1 were 8.22 Hz, demonstrating β-linkages. The signals at 5.15 (3JHH = 3.69) and 4.58 (3JHH = 8.05) ppm correspond to α- and β-configuration of GlcA residue F, respectively. By combining HSQC and correlated spectroscopy (COSY) experiments (Figure 4C and F), we were able to identify that proton signal of F-5 possessed a chemical shift at a low field (~4.05 ppm) suggesting that GlcA residue (F) was at the reducing end of the carbohydrate chain. These data demonstrate that Pn3Pase cleaves the β(1–4) linkage between glucuronic acid and glucose in the polysaccharide chain.
Fig. 4.
Nuclear magnetic resonance characterization of oligosaccharide products. 1H NMR spectrum (A), 2D HSQC NMR spectrum (B), and 2D COSY NMR spectrum (C) of the Pn3 tetrasaccharide. 1H NMR spectrum (D), 2D HSQC NMR spectrum (E) and 2D COSY NMR spectrum (F) of the Pn3 hexasaccharide.
Pn3Pase activity analysis
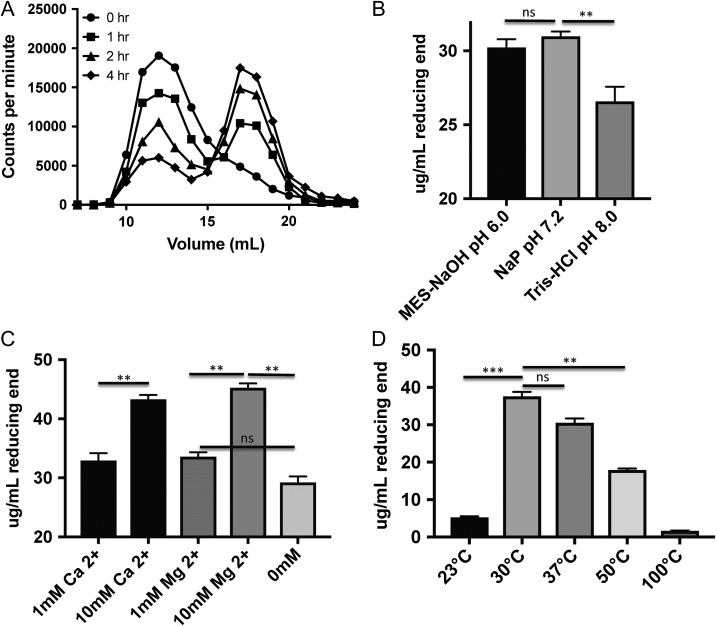
A time course experiment was performed using tritiated Pn3P and the recombinant Pn3Pase to understand whether this enzymatic degradation proceeds through endolytic or exolytic cleavage. Reaction products were separated by size exclusion chromatography after proceeding for the given time. Low molecular weight oligosaccharides are generated early on in the reaction (Figure 5A). Both an increase in CPM for the oligosaccharide elution volume, and corresponding decrease in CPM for the higher molecular weight polymer over time suggest an exolytic type cleavage (Figure 5A) that preferentially generates tetrasaccharides and hexasaccharides (Figure 3A). A gradual shift of the peak from left to right over time would be indicative of true random endolytic cleavage.
Fig. 5.
Activity assessment of Pn3Pase. (A) Time course assay of recombinant Pn3Pase depolymerized tritium radioisotope labeled Pn3P separated by size exclusion chromatography, measured by counts per min in each 1 ml fraction. (B) Optimization of recombinant Pn3Pase in three different buffer conditions, measured by concentration of glucuronic acid reducing end generated in μg/ml. (C) Metal dependence determination with Mg2+ and Ca2+ reaction supplements, measured by concentration of glucuronic acid reducing end generated in ug/ml. (D) Temperature dependence of recombinant Pn3Pase, measured by concentration of glucuronic acid reducing end generated in μg/ml. Statistical significance was determined with the two-tailed Student t-test, ***P < 0.001, **P < 0.01; ns, not significant.
Two-hour reactions were performed in three different buffers at pH 6.0, 7.2 and 8.0, detecting the concentration of reducing end glucuronic acid by the p-hydroxybenzoic acid hydrazide (PAHBAH) method (Blakeney and Mutton 1980) to determine the optimum reaction conditions for Pn3Pase. Pn3Pase displays slightly better activity in sodium phosphate buffer at pH 7.2 than in MES buffer pH 6.0, but performs significantly worse in Tris buffer at pH 8.0 (Figure 5B). Further optimization focusing on metal-ion dependence was performed with Mg2+ and Ca2+ in Tris–HCl pH 7.4. Pn3Pase displays a concentration-dependent increase in activity with addition of Ca2+ or Mg2+, as it produces a higher concentration of reducing end GlcA in presence of 10 mM of these divalent cations (Figure 5C). Surprisingly, Pn3Pase activity is minimal at 23°C, optimal at 30°C, and slightly less than optimal at 37°C, the temperature at which all other experiments are carried out (Figure 5D). Our initial kinetic analyses determined the Km and Vmax to be 108.4 ug/mL and 24.82 umole/min/mg, respectively.
Discussion
The Paenibacillus genus, literal Latin translation, “almost Bacillus,” was ruled distinct from true Bacillus species when phylogenetic analysis on 16 S rRNA gene sequences was performed for a number of strains previously defined as Bacillus (Ash et al. 1993). Sequence analysis showed that several bacterial strains sorted into this genus required reassignment. Species belonging to this genus have been obtained from diverse ecological niches (Grady 2016) from aquatic (Baik et al. 2011) to desert environments (Lim et al. 2006), and from hot springs (Bouraoui et al. 2013) to extreme cold regions (Kishore et al. 2010). Many Paenibacillus species are found in soil (Sickles and Shaw 1934; Shida et al. 1997) and plant root environments (Li et al. 2014); however, a number are isolated from human samples as well (Leão et al. 2010). Paenibacillus species are a rich source for a variety of agricultural, biomedical and industrial products (Grady et al. 2016). Extracellular enzymes demonstrating numerous activities (Hong and Meng 2003) have applications in production of a variety of industrially significant materials (Grady et al. 2016). A number of these species are efficient nitrogen fixers that have been applied agriculturally to promote crop growth (Xie et al. 2014). In addition, protective action of novel antimicrobial peptides and compounds obtained from Paenibacilli has been demonstrated (Allard et al. 2014).
While Paenibacillus sp. 32352 was isolated from a soil source (Sickles and Shaw 1934), it is appropriate to question the evolutionary pressure to obtain enzymes capable of acting on a human pathogen Spn CPS. Based on earlier studies, this particular strain demonstrates the ability to degrade three distinct pneumococcal CPSs (Sickles and Shaw 1934; Shaw and Sickles 1950). Whether these are the natural substrates for the enzymes, or whether other soil-dwelling microbes or plant matter possess similar glycan residues and linkages remains to be investigated.
However originally acquired, Paenibacillus sp. 32352 has adapted the Pn3Pase described here to play an important functional role in its metabolism. Early reports by Torriani and Pappenheimer on this species indicated the ability to induce Pn3Pase activity in culture supernatant by addition of Pn3P in the growth medium (Torriani and Pappenheimer 1962). The “inducibility” was in contrast to the Avery and Dubose findings that Pn3Pase formation only occurred in conditions where Pn3P is present as a sole carbon source (Dubos and Avery 1931). Here, we demonstrate that while Pn3P is not required for bacterial growth, it can serve as the sole nutrition source. Moreover, our data indicate that presence of Pn3P in the Pbac culture medium induces the expression of Pn3Pase.
Our attempts to purify multimilligram quantities of active, native Pn3Pase from an induced culture of this strain were unsuccessful, though Pn3P depolymerizing activity can be detected in these culture supernatant preparations. In this study, we have identified and cloned the Pn3Pase gene from Paenibacillus sp. 32352. We have fully characterized the oligosaccharide products. Kinetic values found in this manuscript will serve as initial parameters towards future structure–activity relationship studies of this enzyme. Future structural studies of Pn3Pase in complex with Pn3-oligosaccharides will determine factors of substrate specificity, active site residues, and catalytic mechanism of this unique GH. In addition, future experiments will assess this enzyme’s utility as a treatment for serotype 3 Spn infections by stripping the CPS from the bacterial surface. The capsular polysaccharide is a major virulence factor for type 3 strains, as non-encapsulated mutants fail to colonize (Magee and Yother 2001). The virulence mechanisms of CPS is to help Spn evade the immune system through resisting or inhibiting its phagocytosis by host macrophages while also limiting mucus-mediated clearance (Nelson et al. 2007). The Pn3P component of the current 13-valent vaccine (PCV13) induces variable immune responses to serotype 3 (Gruber et al. 2012; Dagan et al. 2013). Individuals vaccinated with PCV13 require higher opsonophagocytosis assay serum titers for serotype 3 in comparison with other serotypes (Kieninger et al. 2010). Despite current vaccination programs against Spn, it remains one of the world’s most lethal pathogens. Inefficiencies in current vaccination and antibiotic administration necessitate the discovery of alternative therapeutic approaches for controlling these and other encapsulated pathogens.
Materials and methods
Bacterial strains and growth conditions
Paenibacillus sp. 32352 (ATCC 14,175) was cultured aerobically with shaking at 37°C on Tryptic Soy Agar with 5% sheep blood (Hardy Diagnostics), or in minimal medium (M9 Teknoba) culture containing 1 mM MgSO4, 1 mM biotin, 1 mM thiamin and 2% glucose (Sigma Aldrich) or Pn3P powder (ATCC 172-X) as sole carbon source. Streptococcus pneumoniae type 3 (WU2 strain) and acapsular derivative (JD908), generous gifts from Moon Nahm (University of Alabama at Birmingham), were cultured aerobically without shaking at 37°C on Tryptic Soy Agar with 5% sheep blood (TSAB), or in Todd Hewitt Broth plus 0.5% yeast extract (THY) (BD Biosciences).
Proteomics of culture supernatants
Paenibacillus sp. 32352 was cultured in 5 ml of minimal medium M9 containing 2% (w/v) carbon source, as described above. Bacterial cultures were harvested at mid-log phase (OD 600 nm, 0.6). Culture supernatants were passed through a 0.45-μm syringe filter and concentrated to 1/20th culture volume using a microsep advance centrifugal device with 10 K molecular weight cutoff (Pall). Protein concentration was determined by bicinchoninic acid assay. An in-solution trypsin digestion was performed as described previously (Lim et al. 2008). Briefly, 20 μg protein from culture supernatant protein was reduced by incubation with 10 mM DTT for 1 h at 56°C, followed by carboxyamidomethylation with 55 mM iodoacetamide in the dark at room temperature for 45 min, and then digested with 1 μg of sequencing grade trypsin (Promega) in 40 mM ammonium bicarbonate overnight at 37°C. Trypsin digest was stopped with 1% trifluoroacetic acid and incubation on ice for 30 min. The resulting peptides were cleaned up using C18 spin columns (G Biosciences), dried down and reconstituted in 0.1% formic acid. The peptides were analyzed on a Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) interfaced with an UltiMate 3000 RSLCnano HPLC system (Thermo Fisher Scientific). Peptides were resolved on an Acclaim™ PepMap™ RSLC C18 column (75 μm ID × 15 cm; 2 μm particle size) at a flow rate of 200 nL/min using a linear gradient of 1–100% solvent B (0.1% formic acid in 80% acetonitrile) over 60 min and a total run time of 90 min. Data-dependent acquisition was carried out using the Orbitrap mass analyzer collecting full scans of 200–2000 m/z range at 60,000 mass resolution. Most intense precursor ions were selected using top speed mode with a maximum cycle time of 3 s. Precursor ions with charge state 2–5 were selected with dynamic exclusion set to exclude precursors for 20 s following a third selection within 10 s. Selected precursors were fragmented using collision-induced dissociation set to 38%, and resulting MS/MS ions were scanned out in the ion trap. The raw MS/MS spectra were searched against the Rapid Annotation Server (Aziz et al. 2008) annotated genome database for Paenibacillus sp. 32352 using SEQUEST HT in Proteome Discoverer 1.4 (Thermo Fisher Scientific) with precursor mass tolerance of 10 ppm and fragment tolerance of 0.3 Da. Static modification of +57.021 Da (carbamidomethylation of cysteine residues), and dynamic modification of +15.995 Da (oxidation of methionine residues) were allowed in the search parameters. Results were filtered at a 1% false discovery rate for peptide assignments.
Gene expression
Comparison of the levels of transcript expression from the Pn3P induced locus was performed by RT-PCR. Paenibacillus sp. 32352 was cultured in 5 ml of minimal medium containing 2% (w/v) carbon source, as described above. Triplicate bacterial cultures were harvested at mid-log phase (OD 600 nm, 0.6), RNA was purified using E.N.Z.A. Bacterial RNA Kit, followed by TRIzol (Thermo Fisher Scientific) extraction of RNA from contaminating genomic DNA as described previously (Rio et al. 2010). RNA purity was assessed with nanodrop, and 1 μg of RNA was used for reverse transcription reaction using iscript CDNA synthesis kit (BioRad). Quantitative real time-PCR was performed in a 96-well plate on a MyiQ system (BioRad) with iQ SYBR green mastermix. Primers used in RT-PCR are listed in Table III. The reactions were carried out in 20 μl, consisting of 10 μl of SYBR Green mix, 20 ng of cDNA and 1 μM primer mix. The reaction conditions were 95°C 180 s, followed by 45 cycles of 95°C for 10 s, 55°C for 20 s and 72°C for 30 s. The data were normalized to 16 S rRNA transcript levels, and changes in expression level were calculated as fold change compared with cultures of minimal medium with glucose supplement.
Table III.
Oligonucleotides used in this study.
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| Pbac_3551F-RT | gcacccgtgaatctggaagc |
| Pbac_3551R-RT | ctccataggaataagcgagagatagg |
| Pbac_3552F-RT | gagccgtcaggcaccatac |
| Pbac_3552R-RT | ctctaatgcaggctgcgg |
| Pbac_3554F-RT | ggctgcggcggcacta |
| Pbac_3554R-RT | tcccaaaacatcccggatcg |
| Pbac_1521F-RT | gatggcgtgaaagacgatac |
| Pbac_1521R-RT | acagttttgtcgttaacgcc |
| Pbac_16sF-RT | catgagggaatcatgaaacacg |
| Pbac_16sR-RT | gggctttcttctcaggtacc |
| Pbac_3551 F | ggggacaagtttgtacaaaaaagcaggcttcgaaggagatagaaccatggcacccgtgaatctggaagc |
| Pbac_3551 R | ggggaccactttgtacaagaaagctgggtgctccacgatcaccttattcgataacg |
| Pbac_3552 F | ggggacaagtttgtacaaaaaagcaggcttcgaaggagatagaaccatggagccgtcaggcaccatac |
| Pbac_3552 R | ggggaccactttgtacaagaaagctgggtgcatccctcctgcgatgtg |
| Pbac_3554 F | ggggacaagtttgtacaaaaaagcaggcttcgaaggagatagaaccatgggctgcggcggcacta |
| Pbac_3554 R | ggggaccactttgtacaagaaagctgggtgcttctttgcgctcttgttatactc |
Production of recombinant Pn3Pase
The coding region of Pbac_3551 (ref seq WP_079,915,027), Pbac_3552 (ref seq WP_079,915,028) and Pbac_3554 (ref seq WP_079,915,030.1) (minus predicted signal peptide and stop codon) was amplified from Paenibacillus sp. 32352 genomic DNA (DNeasy blood and tissue kit, Qiagen) using 2× platinum superfi mastermix (Thermo Fisher Scientific) with overhang containing B-sites to facilitate gateway cloning (Walhout et al. 2000) via BP reaction (Thermo Fisher Scientific) into pDONR221. Primers for cloning are listed in Table III. After DH5α transformation and DNA sequence confirmation, an LR-clonase reaction was performed to insert the gene into the pET-DEST42 (Thermo Fisher Scientific) destination vector for the expression of a carboxy-terminal His6-tagged fusion protein in E. coli BL21(DE3) cells. BL21(DE3) cells transformed with the pET-DEST42-“Pn3Pase” plasmid were grown in LB medium supplemented with 100 μg/ml ampicillin at 37°C, and cell density was monitored by absorbance at 600 nm. Once the OD 600 nm reached 0.6, the cells were transferred into 25°C, protein expression was induced by the addition of Isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mM and the cell culture was allowed to incubate for 8 h (until A600 reached ~1.1). Cells were harvested by centrifugation. Cells were then resuspended in phosphate-buffered saline (PBS, pH 7.2) with 1 mg/ml lysozyme for 20 min at 30°C, probe sonicated for 2 min (four cycles of 20 s on, 10 s off), clarified by centrifugation at 17,000 × g for 1 h at 4°C, and passed through a 0.45-μm syringe filter. Recombinant Pn3Pase was purified by Ni2+-NTA resin at 4°C, eluted with 300 mM imidazole and buffer exchanged into PBS pH 7.2. Protein concentration was determined by bicinchoninic acid assay. Purity was assessed by visualizing proteins on stain free tris-glycine gel (BioRad) using gel doc EZ imager (BioRad).
Enzyme assays
Tritiated Pn3P assays: Recombinant enzyme activity against type 3 capsular polysaccharide was assayed by incubation of 0.2 μg/ml recombinant protein, or heat killed control, with 10 μg/ml 3H-Pn3P in PBS. The reaction was stopped after 2 h by heating at 100°C for 5 min. Reaction mixture was separated on a superdex peptide 10/300 GL column (GE) on an NGC discoverer FPLC system (BioRad). Fractions of 1 ml were collected, and counts per min in each fraction were counted in a Tri-Carb 2910 TR liquid scintillation analyzer (Perkin Elmer). Time course experiments were analyzed by the same method.
Reducing end sugar assays: Recombinant Pn3Pase hydrolysis activity was determined by measuring the increase in reducing ends using the p-hydroxybenzoic acid hydrazide (PAHBAH) method (Blakeney and Mutton 1980). A reaction mixture (200 μl) containing 20 μg Pn3P, and 1 μg recombinant Pn3Pase in either 50 mM MOPS-NaOH (pH 6.0), 50 mM sodium phosphate buffer (pH 7.2) or 20 mM Tris–HCl (pH 8.0), was incubated at 37°C for 1 h and then heated at 100°C for 5 min to stop. Reaction mixture (40 μl) was mixed with 120 μl of 1% (w/v) PAHBAH–HCL solution, heated at 100°C for 5 min. Absorbance at 405 nm was measured on a Biotek synergy H1 microplate reader in a clear bottom 96-well microplate. Concentrations of reducing sugars were calculated based on GlcA standard curves generated by the same method in respective buffers. Metal dependence assays were performed similarly with the addition of MgCl2 or CaCl2 in Tris buffer (pH 7.4).
Enzyme kinetics
The Michaelis–Menten constant (Km) and the maximum velocity (Vmax) of Pn3Pase were measured using Pn3P as substrate (average molecular weight: 400,000 Da). The substrate was used at eight concentrations (3200, 1600, 800, 400, 200, 100, 50 and 25 nM) in phosphate-buffered saline at pH 7.4. Pn3Pase was added at 1ug/mL and the reactions were incubated at 37°C. Reactions were stopped at 0, 4, 8, 12, 16 and 20 min (corresponding to approximately 10% of total depolymerization yielding mostly tetrasaccharides) by boiling the reaction at 100°C for 5 min. The amount of product formed was measured using PAHBAH–HCl, as described above, with tetrasaccharides obtained from enzymatic depolymerization to generate a standard curve for data fitting. Initial velocity was calculated using the amount of product formed in the linear region of absorbance. Initial velocities of each substrate concentration were inserted into the Michaelis–Menten equation to determine Km and Vmax.
Oligosaccharide analysis
Pn3P powder (2 mg) was incubated with 100 μg of Pn3Pase at 37°C for 48 h. The reaction was stopped by heating at 100°C for 5 min, and loaded onto Superdex peptide 10/300 GL column (GE). Products were separated in phosphate-buffered saline at a flow rate of 1 ml/min and monitored by refractive index. Fractions (0.5 ml) were collected and oligosaccharide peaks were purified, desalted into water on a packed fine P2 column (Biorad). Desalted oligos were lyophilized and subject to ESI-MS for mass determination and NMR for structural and reducing end identification.
NMR: Oligosaccharides were dissolved in 400 μL 2H2O (99.9%, Sigma-Aldrich, St. Louis, MO) and lyophilized three times to remove the exchangeable protons. The samples were re-dissolved in 400 μl 99.96% 2H2O and transferred to NMR microtubes. 1H spectroscopy, 13C spectroscopy, 1H–1H COSY and 1H–13C HSQC spectroscopy experiments were all performed at 298 K on Bruker 600 or 800 MHz spectrometer with Topspin 2.1.6 software.
Acknowledgements
We thank Dr. Bernard Henrissat for providing important feedback on CAZY classification of Pn3Pase.
Funding
National Institutes of Health Grant (R01AI123383 to F.A.), (R01-HL125371 to R.J.L.) and National Center for Biomedical Glycomics (P41GM103490 to L.W.).
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Author contributions
D.R.M. and F.Y.A. conceptualized, designed and coordinated the study, and wrote the article. D.R.M., X.Z., P.L.W., A.O., X.L., R.L., N.G., R.B., L.W., R.J.L. and F.Y.A. performed, and/or analyzed the experiments presented. All authors reviewed and approved the final submitted version of the article.
References
- Agard N, Baskin J, Prescher J, Lo A, Bertozzi C. 2006. A comparative study of bioorthogonal reactions with azides. ACS Chem Biol. 1:644–648. [DOI] [PubMed] [Google Scholar]
- Allard S, Enurah A, Strain E, Millner P, Rideout SL, Brown EW, Zheng J. 2014. In situ evaluation of Paenibacillus alvei in reducing carriage of Salmonella enterica serovar Newport on whole tomato plants. Appl Environ Microbiol. 80:3842–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash C, Priest FG, Collins MD. 1993. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek. 64:253–260. [DOI] [PubMed] [Google Scholar]
- Avery OT, Dubos R. 1930. The specific action of a bacterial enzyme on pneumococci of type III. Science. 72:151–152. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M et al. . 2008. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik KS, Choe HN, Park SC, Kim EM, Seong CN. 2011. Paenibacillus wooponensis sp. nov., isolated from wetland freshwater. Int J Syst Evol Microbiol. 61:2763–2768. [DOI] [PubMed] [Google Scholar]
- Blakeney A, Mutton L. 1980. A simple colorimetric method for the determination of sugars in fruit and vegetables. J Sci Food Agric. 31:889–897. [Google Scholar]
- Bouraoui H, Rebib H, Ben Aissa M, Touzel JP, O’donohue M, Manai M. 2013. Paenibacillus marinum sp. nov., a thermophilic xylanolytic bacterium isolated from a marine hot spring in Tunisia. J Basic Microbiol. 53:877–883. [DOI] [PubMed] [Google Scholar]
- Rice CE, Sickles GR. 1946. Studies of anti-pneumococcal serum; complement-fixing activity of fractionated rabbit serum. J Immunol. 54:267–274. [PubMed] [Google Scholar]
- Chung J, Kim S, Choi K, Kim JO. 2015. Degradation of polyvinyl alcohol in textile waste water by Microbacterium barkeri KCCM 10507 and Paenibacillus amylolyticus KCCM 10508. Environ Technol. 37:4, 452–458. [DOI] [PubMed] [Google Scholar]
- Dagan R, Patterson S, Juergens C, Greenberg D, Givon-Lavi N, Porat N, Gurtman A, Gruber WC, Scott DA. 2013. Comparative immunogenicity and efficacy of 13-valent and 7-valent pneumococcal conjugate vaccines in reducing nasopharyngeal colonization: a randomized double-blind trial. Clin Infect Dis. 57:952–962. [DOI] [PubMed] [Google Scholar]
- Dubos R, Avery OT. 1931. Decompostion of the capsular polysaccharide of pneumococcus type III by a bacterial enzyme. J Exp Med. 54:51–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztányi Z, El-Gebali S, Fraser M et al. . 2017. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsee WT, Cartee RT, Yother J. 2006. Role of the carbohydrate binding site of the Streptococcus pneumoniae capsular polysaccharide type 3 synthase in the transition from oligosaccharide to polysaccharide synthesis. J Biol Chem. 281:6283–6289. [DOI] [PubMed] [Google Scholar]
- Forsee WT, Cartee RT, Yother J. 2009. Characterization of the lipid linkage region and chain length of the cellubiuronic acid capsule of Streptococcus pneumoniae. J Biol Chem. 284:11826–11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady E, MacDonald J, Liu L, Richman A, Yuan Z. 2016. Current knowledge and perspectives of Paenibacillus: A review. Microb Cell Fact. 15:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber WC, Scott DA, Emini EA. 2012. Development and clinical evaluation of Prevnar 13, a 13-valent pneumocococcal CRM197 conjugate vaccine. Ann N Y Acad Sci. 1263:15–26. [DOI] [PubMed] [Google Scholar]
- Guo Y, Huang E, Yuan C, Zhang L, Yousef AE. 2012. Isolation of a Paenibacillus sp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Appl Environ Microbiol. 78:3156–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 316(Pt 2):695–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong TY, Meng M. 2003. Biochemical characterization and antifungal activity of an endo-1,3-beta-glucanase of Paenibacillus sp. isolated from garden soil. Appl Microbiol Biotechnol. 61:472–478. [DOI] [PubMed] [Google Scholar]
- Kieninger DM, Kueper K, Steul K, Juergens C, Ahlers N, Baker S, Jansen KU, Devlin C, Gruber WC, Emini EA et al. . 2010. Safety, tolerability, and immunologic noninferiority of a 13-valent pneumococcal conjugate vaccine compared to a 7-valent pneumococcal conjugate vaccine given with routine pediatric vaccinations in Germany. Vaccine. 28:4192–4203. [DOI] [PubMed] [Google Scholar]
- Kishore KH, Begum Z, Pathan AA, Shivaji S. 2010. Paenibacillus glacialis sp. nov., isolated from the Kafni glacier of the Himalayas, India. Int J Syst Evol Microbiol. 60:1909–1913. [DOI] [PubMed] [Google Scholar]
- Knolhoff AM, Zheng J, McFarland MA, Luo Y, Callahan JH, Brown EW, Croley TR. 2015. Identification and structural characterization of naturally-occurring broad-spectrum cyclic antibiotics isolated from Paenibacillus. J Am Soc Mass Spectrom. 26:1768–1779. [DOI] [PubMed] [Google Scholar]
- Konishi J, Maruhashi K. 2003. 2-(2’-Hydroxyphenyl)benzene sulfinate desulfinase from the thermophilic desulfurizing bacterium Paenibacillus sp. strain A11-2: Purification and characterization. Appl Microbiol Biotechnol. 62:356–361. [DOI] [PubMed] [Google Scholar]
- Leão RS, Pereira RH, Ferreira AG, Lima AN, Albano RM, Marques EA. 2010. First report of Paenibacillus cineris from a patient with cystic fibrosis. Diagn Microbiol Infect Dis. 66:101–103. [DOI] [PubMed] [Google Scholar]
- Li QQ, Zhou XK, Dang LZ, Cheng J, Hozzein WN, Liu MJ, Hu Q, Li WJ, Duan YQ. 2014. Paenibacillus nicotianae sp. nov., isolated from a tobacco sample. Antonie Van Leeuwenhoek. 106:1199–1205. [DOI] [PubMed] [Google Scholar]
- Lim JM, Jeon CO, Lee JC, Xu LH, Jiang CL, Kim CJ. 2006. Paenibacillus gansuensis sp. nov., isolated from desert soil of Gansu Province in China. Int J Syst Evol Microbiol. 56:2131–2134. [DOI] [PubMed] [Google Scholar]
- Lim JM, Sherling D, Teo CF, Hausman DB, Lin D, Wells L. 2008. Defining the regulated secreted proteome of rodent adipocytes upon the induction of insulin resistance. J Proteome Res. 7:1251–1263. [DOI] [PubMed] [Google Scholar]
- Magee AD, Yother J. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect Immun. 69:3755–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton DR, Lorenz W, Avci FY. 2017. Complete genome sequence of the bacterium Bacillus circulans Jordan strain 32352. Genome Announc. 5:e00289–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura L, Swezey J. 1983. Taxonomy of Bacillus circulans Jordan 1890-base composition and reassociation of deoxyribonucleic acid. Int J Syst Bacteriol. 33:46–52. [Google Scholar]
- Nelson AL, Roche AM, Gould JM, Chim K, Ratner AJ, Weiser JN. 2007. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect Immun. 75:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A, Leite M, Kluskens LD, Santos SB, Melo LD, Azeredo J. 2015. The first Paenibacillus larvae bacteriophage endolysin (PlyPl23) with high potential to control american foulbrood. PLoS One. 10:e0132095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasari N, Adlakha N, Gupta M, Bashir Z, Rajacharya GH, Verma G, Munde M, Bhatnagar R, Yazdani SS. 2017. Impact of module-X2 and carbohydrate binding module-3 on the catalytic activity of associated glycoside hydrolases towards plant biomass. Sci Rep. 7:3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 8:785–786. [DOI] [PubMed] [Google Scholar]
- Rio DC, Ares M, Hannon GJ, Nilsen TW. 2010. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc. 2010(6), pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- Shaw M, Sickles GM. 1950. Production of specific pneumococcus carbohydrate-splitting enzymes in media to which the specific substrate was not added. J Immunol. 64:27–32. [PubMed] [Google Scholar]
- Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K. 1997. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol. 47:289–298. [DOI] [PubMed] [Google Scholar]
- Sickles GM, Shaw M. 1934. A systematic study of microörganisms which decompose the specific carbohydrates of the pneumococcus. J Bacteriol. 28:415–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani A, Pappenheimer AM. 1962. Inducible polysaccharide depolymerases of Bacillus palustris. J Biol Chem. 237:3–13. [PubMed] [Google Scholar]
- Uetanabaro AP, Wahrenburg C, Hunger W, Pukall R, Spröer C, Stackebrandt E, de Canhos VP, Claus D, Fritze D. 2003. Paenibacillus agarexedens sp. nov., nom. rev., and Paenibacillus agaridevorans sp. nov. Int J Syst Evol Microbiol. 53:1051–1057. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. 2000. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328:575–592. [DOI] [PubMed] [Google Scholar]
- Xie JB, Du Z, Bai L, Tian C, Zhang Y, Xie JY, Wang T, Liu X, Chen X, Cheng Q et al. . 2014. Comparative genomic analysis of N2-fixing and non-N2-fixing Paenibacillus spp.: Organization, evolution and expression of the nitrogen fixation genes. PLoS Genet. 10:e1004231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JK, Yoon HJ, Ahn HJ, Lee BI, Pedelacq JD, Liong EC, Berendzen J, Laivenieks M, Vieille C, Zeikus GJ et al. . 2004. Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase. J Mol Biol. 335:155–165. [DOI] [PubMed] [Google Scholar]
- Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40:W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]