Abstract
Specific response to the concurrent presence of two different inputs is one of the hallmarks of incorporating specificities in Nature. Artificial nanoassemblies that similarly respond to two very different inputs are of great interest in a variety of applications, especially in biomedicine. Here, we present a design strategy for amphiphilic nanoassemblies with such capabilities, enabled by photocaging a ligand moiety that is capable of binding to a specific protein. New molecular designs that offer nanoassemblies that respond to either of two inputs or only to the concurrent presence of two inputs are outlined. Such biomimetic nanoassemblies could find use in many applications, including drug delivery and diagnostics.
Keywords: stimulus responsive nanoassemblies, dual inputs, photoactivation of ligands, host-guest transformation, supramolecular disassembly
Graphical Abstract
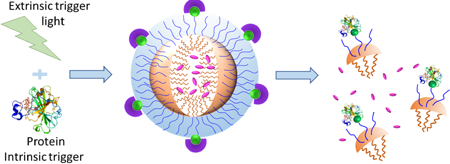
Nanoassemblies were engineered to respond to a combination of intrinsic and extrinsic inputs in AND/OR logic.
Supramolecular nanoassemblies that predictably respond to an environmental change have been of interest due to their implications in areas that range from material science to biomedicine.[1] When designing molecular assemblies that have the potential to impact biomedicine, the input triggers can be classified into two main categories: extrinsic and intrinsic inputs.[2] Extrinsic triggers have the advantage of offering external spatiotemporal control over the change in the properties of a molecular assembly, e.g. shining light at a specific location and time to disrupt a supramolecular assembly.[3] On the other hand, intrinsic triggers are directly correlated with an aberrant biological condition and therefore have the opportunity to be selective, e.g. lower pH at the extracellular space of disease tissues.[4] Although both these systems present complementary advantages, the specificity offered by either of these systems by itself is insufficient. Therefore, a viable strategy would involve systems that would respond to a specific combination of extrinsic and intrinsic stimuli. We present a simple, new supramolecular approach that responds to a specific combination of extrinsic and intrinsic stimuli.
We use proteins as the intrinsic trigger in our studies here, although the oft-targeted intrinsic triggers are pH, reducing conditions, and reactive oxygen species.[5] Proteins are challenging and interesting as inputs, because of their structural and functional fragility and because they are considered to be the primary cause of pathological imbalances in biology.[6] We use light as the extrinsic trigger in these studies. In engineering the combinations of these two triggers, we were inspired by the molecular logic gates proposed and studied over past couple of decades.[7] While there have been many reports on molecular logic gates involving small molecules,[8] such gated strategies in nanoscale assemblies are relatively limited.[9] We are particularly interested in developing systems that predictably respond to dual inputs, based on protein and light.
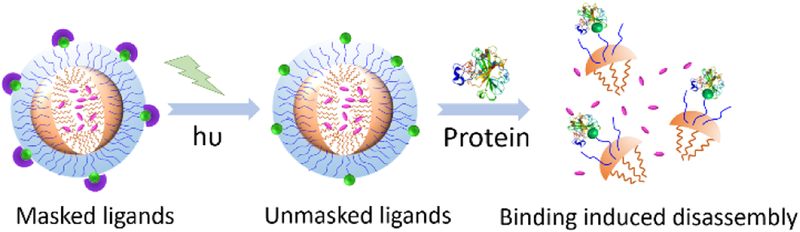
First, we targeted the design of a molecular assembly that would respond only in the presence of a specific protein and light, but not in the presence of either of these inputs by themselves or in their absence. Such a system is interesting, as they offer the best opportunity to be specific, because it requires the concurrent presence of two different stimuli. For the protein, we used bovine carbonic anhydrase (bCA). Primary aryl sulfonamides are well established ligands for this protein, where the active site zinc is known to be engaged with the sulfornamide moiety.[10] Examination of the structure of this binding interaction suggests that derivatizing the amino moiety of the sulfonamide group with an alkyl unit would cause this molecule to be not a good ligand for bCA. If such a substituent were to be removed in the presence of light, then the ligand is rendered activatable by light. Our design hypothesis is then that if such a functional group were to be then incorporated onto a protein-responsive assembly, then the assembly would respond only if there is both light and protein present, as shown in Figure 1.
Figure 1.
Schematic representation of protein and light responsive nanoassembly.
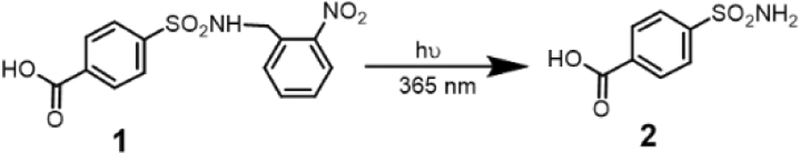
To test this hypothesis, we first tested whether small molecule sulfonamide ligand can be protected by an o-nitrobenzyl moiety, which can then be released in the presence of light. Accordingly, we synthesized the molecule 1 and evaluated the possibility of deprotection of the nitrobenzyl moiety due to light irradiation at 365 nm (Figure 1). Indeed, 1H NMR and LC-MS studies showed that the sulfonamide ligand was fully liberated to afford the sulfonamide ligand 2, in response to UV irradiation (Figure S1, S2). We also tested molecules 1 and 2 as the ligands for bCA using a 5-(Dimethylamino)-1-naphthalenesulfonamide (DNSA) in a competitive displacement assay, the fluorescence emission at 460 nm formed by DNSA-bCA complex indicates whether DNSA is replaced[11]. Our studies showed that when the ligand was masked in 1, it did not competitively remove DNSA, while the photo-cleaved product 2 was able to displace DNSA at a molar ratio of 1:1 for bCA and DNSA (Figure S3).
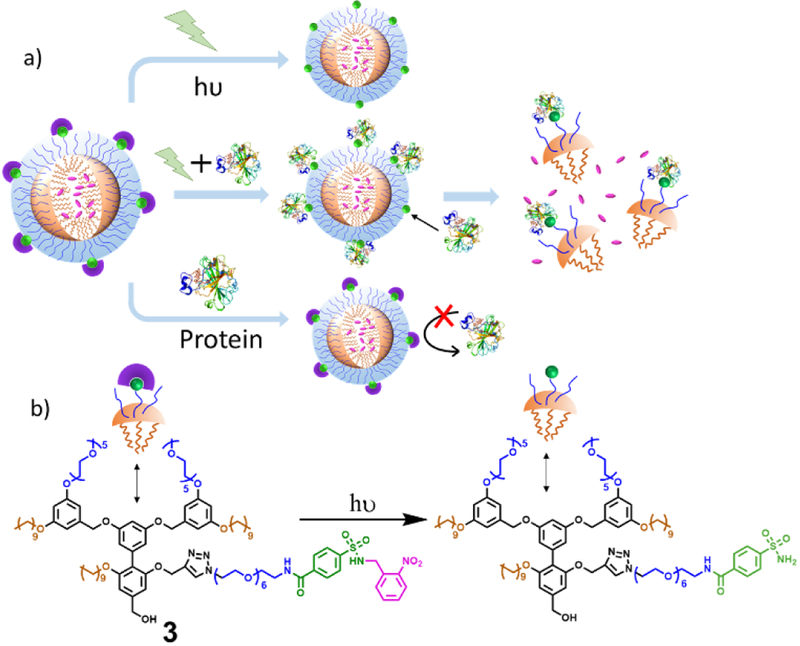
Next, we were interested in utilizing this to generate a nanoassembly that would predictably respond only to the concurrent presence of light irradiation and the protein. The molecular structure that potentially serves this purpose is shown in Figure 2b as 3. The facially amphiphilic trimer contains an alkyl chain as the hydrophobic moiety and an oligoethylene glycol (OEG) chain as the hydrophilic moiety in each of the repeat units. The key functional group, N-(o-nitrobenzyl) benzene sulfonamide, is clicked on to the central unit on the hydrophilic face of the amphiphile using the Huisgen cycloaddition reaction (see SI for synthetic details). This amphiphile is known to aggregate to form nanoassemblies, which could then disassemble in response to a ligand-protein binding because of the change in the hydrophilic-lipophilic balance (HLB) upon protein binding. We also hypothesized that this nanoassembly would disassemble only in response to both light and protein, but not to just one of these two inputs. When the assembly is irradiated with light, the sulfonamide moiety would be liberated; this change however would not be sufficient to change the HLB of the assembly. Similarly, since the ligand moiety is masked, it would be unavailable for the binding-induced disassembly in response to the protein. However, in the presence of both light and the protein, the nanoassembly should disassemble as the light would unmask the ligand, binding of which to the protein would cause a significant change in the HLB of the amphiphile.
Figure 2.
(a) Schematic representation of protein AND light gated disassembly and guest release, (b) Molecular structure of 3.
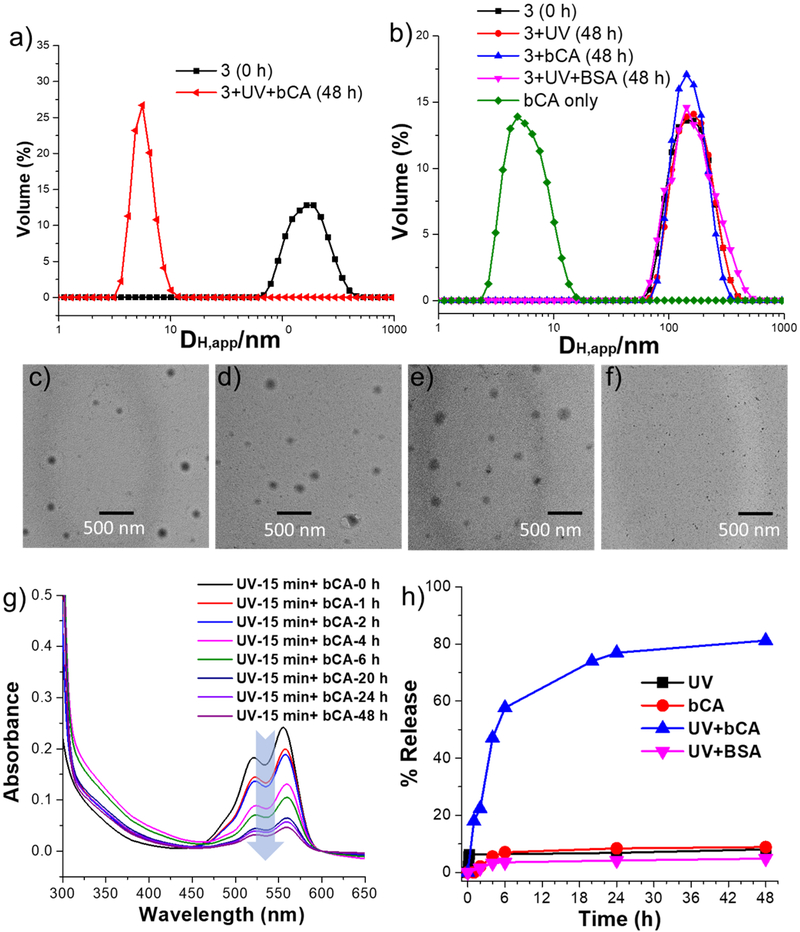
Prior to testing this hypothesis, we characterized the nanoassembly, formed from molecule 3. Synthetic details and the molecular characterization are shown in the SI. Since 3 contains, the nanoassembly formed would be an amphiphilic assembly, the critical aggregation concentration (CAC) can be estimated using the possibility of incorporating a hydrophobic molecule within the interiors of the assembly. The CAC for 3 was found to be ~36 μM (Figure S4). To assess the size of the nanoassembly formed, an aqueous solution of 3 was assessed using dynamic light scattering (DLS), at a concentration above its CAC (50 μM). The amphiphilic nanoassembly was found to have an apprarent hydrodynamic diameter of >120 nm (Figure 3a). The spherical morphology of the assembly was ascertained using transmission electron microscopy (TEM) (Figure 3c) and atomic force microscopy (AFM) (Figure S6). Size from TEM images showed that the observed aggregates are slightly lower than those from DLS, this difference is likely due to the shrinkage of the particles in the dry state or due to overestimation of the size of the particles in DLS as it also includes hydration shells around the particles.
Figure 3.
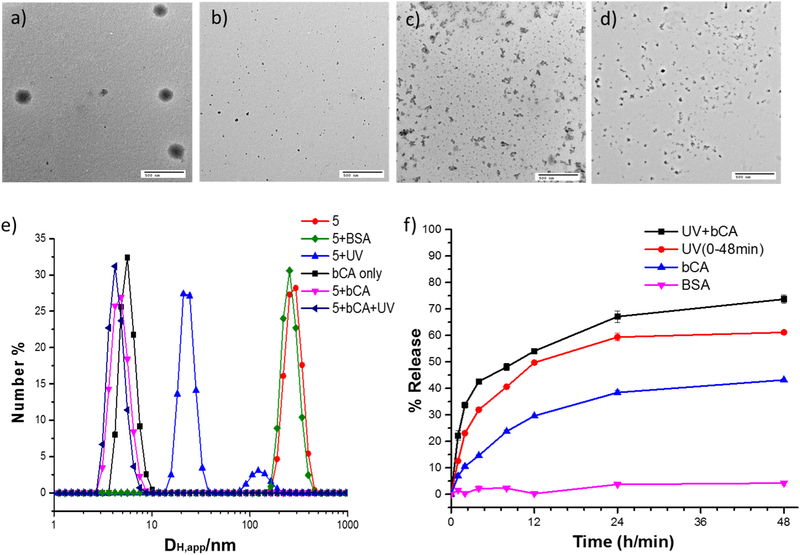
(a) Apparent hydrodynamic diameter (DH,app) of nanoassembly formed by 3 (50 μM) determined by one-angle dynamic light scattering, and 3 in presence of bCA and UV after 48h, (b) DH, app of nanoassembly 3 in presence of UV, bCA, UV and BSA, TEM images of 3 (50 μM) in presense of (c) no inputs, (d) UV light, (e) bCA, (f) bCA and UV light, (g) DiI release from 50 μM 3 solution in response to UV and bCA, (h) Plot of % release of DiI.
Next, to test our hypothesis that the nanoassembly from 3 would be sensitive to the concurrent presence of both light and proteins, we treated a 50 μM solution of 3 with 365 nm light irradiation for 15 minutes and 60 μM bCA. We were gratified to find that the size of the assembly reduced from >120 nm to <10 nm (Figure 3a). To fully test whether this is indeed a response to the combination of these two inputs, effects of the light irradiation and the presence of bCA were tested independently. In both these cases, there was no discernible change in the size of the assembly, compared to the assembly of 3 itself (Figure 3b). The size change in the presence of both stimuli, and lack thereof in the presence of either of these stimuli, were also confirmed by TEM (Figure 3c-3f) and AFM (Figure S6a-S6d). These results provided the first indicator that the system is only responsive to the presence of both stimuli.
To test these findings further, we utilized the host-guest properties of the nanoassembly. Since 3 forms amphiphilic aggregates with a hydrophobic interior in the aqueous phase, it can function as a nanocontainer to host water-insoluble guest molecules. We envisaged that by taking advantage of this container-like feature and employing AND logic inputs to the nanoassemblies, we will be able to regulate the guest release profile. Here, we use 1,1’-dioctadecyl-3,3,3’3’-tetramethyl-indocarbocyanine perchlorate (DiI), as the guest molecule to be entrapped inside the hydrophobic interior of 3. Encapsulation of DiI in this assembly was found to be quite stable with time, where there was a <10% change in the characteristic absorption of DiI over 48 hours (Figure S7). Similarly, when the 50 μM solution of 3 was irradiated with light at 365 nm or when it was treated with 60 μM concentration of bCA, the change in absorption peak was small and indistinguishable from the assembly in the absence of any stimulus (Figure 3h). Interestingly however, a rather dramatic decrease in DiI absorption was observed in the presence of both light and bCA, where ~60% of the guest molecules were released from the assembly in ~6 hours and >80% of the molecules were released in 48 hours (Figure 3g, h). These data are all consistent with our hypothesis that our nanoassembly is programmed to respond only in the presence of both stimuli. However, it is important to show that the presumed disassembly and guest release is indeed due to specific protein-ligand binding. To test the specificity of the protein-ligand binding, we applied UV irradiation and bovine serum albumin (BSA), a protein that has no specific interaction with sulfonamide, as the simultaneous inputs to investigate the size transformation and guest release. Indeed, there was neither any change in the size of the nanoassembly nor was there any discernible guest release over 48 h. These results further validate that the assembly is specific in response to bCA.
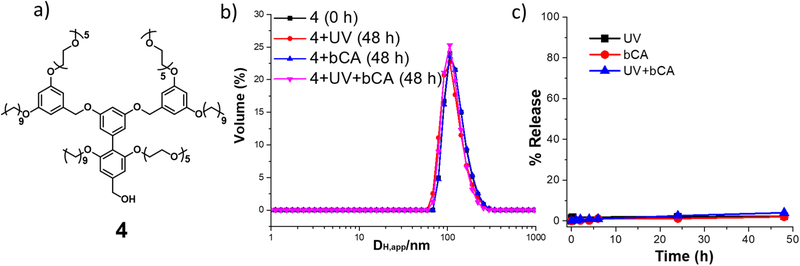
Also, we were interested in another control experiment, where we utilize a structurally related amphiphile forms a similar nanoassembly, but lacks the features that respond to light or to the specific protein. In this case, we prepared the trimeric amphiphile, 4, in which every unit contains both hydrophobic alkyl chains and hydrophilic PEG moieties without any light sensitive moieties or protein-binding ligand functionalities. This molecule, too, forms a similarly sized nanoassembly in aqueous phase. Similar to the methods above, we studied the effects of individual and concurrent orthogonal inputs of UV light and bCA protein. No size transition or discernible guest release were observed, independent of whether a single input, no input, or both inputs were applied (Figure 4b and 4c). These results validate that the introduction of N-(o-nitrobenzyl) benzene sulfonamide ligand is critical for realizing the observed AND-gated disassembly and guest release.
Figure 4.
(a) Molecular structure of 4, (b) DH,app of 4 nanoassembly (concentration of 4 = 50 μM); (c) Plot of % release of DiI from 50 μM 4 solution.
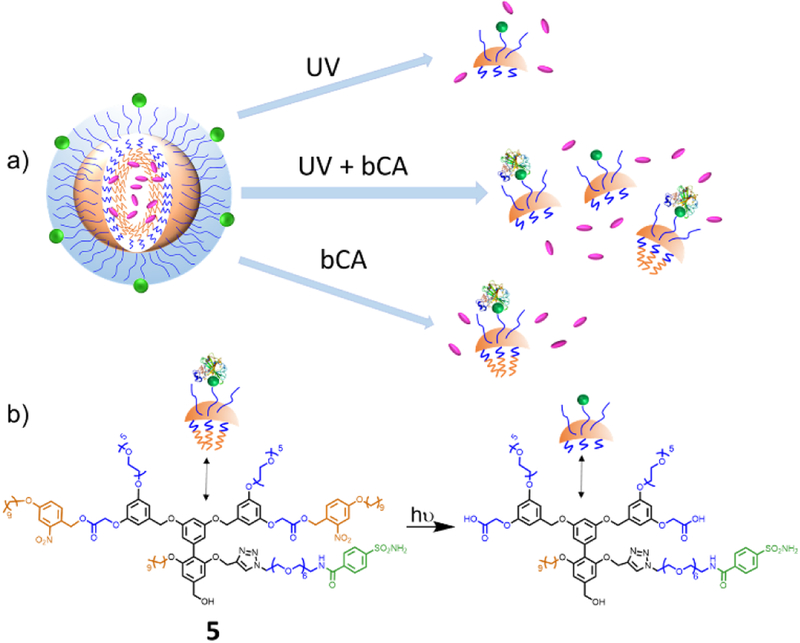
In dual responsive logic-gated systems, the next challenge in designing nanoscopic systems involves the OR gate, where a nanoassembly can respond to either of the inputs. To address this design challenge, we designed and synthesized the amphiphile 5, shown in Figure 5. This molecule contains a sulfonamide moiety in the middle repeat unit on the hydrophilic face of the amphiphile, similar to 3, but the bCA-ligand is present here in its unmasked form. At the two terminal units, the hydrophobic decyl chain is linked to the trimeric scaffold using a photo-responsive o-nitrobenyzl ester linker. Synthetic details and are shown in the SI. We envisage here that when 5 is exposed to UV light, photo-induced cleavage of the nitrobenzyl ester will disconnect the long hydrophobic chain from the amphiphilic oligomer, while concurrently generating a carboxylic acid moiety. This transformation should render the entire oligomer much more hydrophilic, thus triggering disassembly. On the other hand, when treated with bCA, the already unmasked and available sulfonamide ligand should bind to the protein efficiently, causing a change in the HLB of the amphiphile to result in disassembly. To test these design hypotheses, the size transformation of a solution of 5 was evaluated using DLS. As shown in Figure 6e, either UV light or the bCA protein inputs induce a size change in the nanoassembly from ~150 nm to ~10 nm. TEM and AFM images of D2 before and after applying one or both inputs further confirm the disassembly event (Figure 6a–d, Figure S14a–d). We also tested the host-guest properties of the assembly in the presence of these stimuli. Indeed, the DiI guest encapsulated in the D2 nanoassembly was released, when exposed to the bCA protein or the UV irradiation (Figure 6f). Note that the extent of molecular release with the protein binding is smaller than that of unmasked 3. This is expected, because the overall hydrophobicity of the interior of the assembly from 5 is significantly higher than that from 3, because of the introduction of additional aromatic units in the two of the three hydrophobic units. In fact, aromatic-aromatic interactions have been shown to have a substantial effect on the stability of encapsulation of molecules in these nanoassemblies.[12] Removal of these hydrophobic units, followed by treatment with the protein brings the guest release profile, comparable to that found with the unmasked 3.
Figure 5.
(a) Schematic representation of OR logic gated disassembly and guest release, (b) Molecular structure of 5.
Figure 6.
TEM images of 5 (50 μM) in presense of (a) no inputs, (b) UV light, (c) bCA, (d) bCA and UV light; (e) DH,app of 5 nanoassembly in response to UV and bCA, (f) Plot of % release of DiI.
In summary, we have demonstrated a set of amphiphilic supramolecular assemblies that can disassemble in the presence of an extrinsic physical stimulus (light) and an intrinsic biological stimulus (protein). Since these nanoassemblies are capable of sequestering hydrophobic guest molecules, the host-guest properties of the assemblies are also compromised in the presence of these inputs. We outline molecular designs that can respond to the presence of either one or both of these stimuli, as well as that would respond only to the concurrent presence of both stimuli. The latter system was developed by caging a protein-specific ligand with a photo-protecting group that masks the ligand from being available for protein binding and thus preventing binding-induced disassembly. Therefore, the nanoassembly requires the concurrent presence of both light and the specific protein for programmed disassembly. In the former scenario, where the nanoassembly responds to either of the inputs, the disassembly was achieved by strategically placing the light-responsive moieties and the protein-responsive moiety in two different parts of the amphiphilic building block. As controlled responses to the concurrent presence of two different stimuli present the possibility of substantially increasing specificity in responses, the design insights provided here will find use in the design of novel protein-responsive drug delivery and controlled-release systems.
Supplementary Material
Scheme 1.
Photo-induced cleavage of compound 1 to expose sulphonamide ligand 2.
Acknowledgements
We thank NIGMS of the NIH for support (GM-065255). X.L. gratefully acknowledges the support from the China Scholarship Council. H.S. thanks for the support from The Scientific and Technological Research Council of Turkey (TUBITAK).
References
- [1].Zhuang J, Gordon MR, Ventura J, Li L, Thayumanavan S, Chem. Soc. Rev 2013, 42, 7421–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee SM, Nguyen ST, Macromolecules 2013, 46, 9169–9180. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hu X, Zhang Y, Xie Z, Jing X, Bellotti A, Gu Z, Biomacromolecules 2017, 18, 649–673. [DOI] [PubMed] [Google Scholar]; d) Blum AP, Kammeyer JK, Rush AM, Callmann CE, Hahn ME, Gianneschi NC, J. Am. Chem. Soc 2015, 137, 2140–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Yu G, Jie K, Huang F, Chem. Rev 2015, 115, 7240–7303. [DOI] [PubMed] [Google Scholar]
- [2].a) Torchilin VP, J. Controlled Release 2001, 73, 137–172. [DOI] [PubMed] [Google Scholar]; b) Soussan E, Cassel S, Blanzat M, Rico‐Lattes I, Angew. Chem., Int. Ed 2009, 48, 274–288. [DOI] [PubMed] [Google Scholar]; c) Raghupathi KR, Guo J, Munkhbat O, Rangadurai P, Thayumanavan S, Acc. Chem. Res 2014, 47, 2200–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Hartgerink JD, Beniash E, Stupp SI, Science 2001, 294, 1684–1688. [DOI] [PubMed] [Google Scholar]; e) Cabral H, Nishiyama N, Kataoka K, Acc. Chem. Res 2011, 44, 999–1008. [DOI] [PubMed] [Google Scholar]
- [3].a) Mal NK, Fujiwara M, Tanaka Y, Nature 2003, 421, 350. [DOI] [PubMed] [Google Scholar]; b) Goodwin AP, Mynar JL, Ma Y, Fleming GR, Fréchet JM, J. Am. Chem. Soc 2005, 127, 9952–9953. [DOI] [PubMed] [Google Scholar]; c) Ferris DP, Zhao Y-L, Khashab NM, Khatib HA, Stoddart JF, Zink JI, J. Am. Chem. Soc 2009, 131, 1686–1688. [DOI] [PubMed] [Google Scholar]; d) Yesilyurt V, Ramireddy R, Thayumanavan S, Angew. Chem., Int. Ed 2011, 50, 3038–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Karimi M, Sahandi Zangabad P, Baghaee-Ravari S, Ghazadeh M, Mirshekari H, Hamblin MR, J. Am. Chem. Soc 2017, 139, 4584–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Yu G, Yu W, Mao Z, Gao C, Huang F, Small, 2015, 11, 919–925. [DOI] [PubMed] [Google Scholar]; g) Amir RJ, Pessah N, Shamis M, Shabat D, Angew. Chemie 2003, 115, 4632–4637. [DOI] [PubMed] [Google Scholar]; h) Amir RJ, Shabat D, Chem. Commun 2004, 1614–1615. [DOI] [PubMed] [Google Scholar]; i) Kostiainen MA, Smith DK, Ikkala O, Angew. Chemie Int. Ed 2007, 46, 7600–7604. [DOI] [PubMed] [Google Scholar]; j) Kostiainen MA, Kotimaa J, Laukkanen M-L, Pavan GM, Chem. - A Eur. J 2010, 16, 6912–6918. [DOI] [PubMed] [Google Scholar]; k) Nazemi A, Schon TB, Gillies ER, Org. Lett 2013, 15, 1830–1833. [DOI] [PubMed] [Google Scholar]; l) Nazemi A, Gillies ER, Chem. Commun 2014, 50, 11122–11125. [DOI] [PubMed] [Google Scholar]
- [4].a) Gillies ER, Jonsson TB, Fréchet JM, J. Am. Chem. Soc 2004, 126, 11936–11943. [DOI] [PubMed] [Google Scholar]; b) Du J, Tang Y, Lewis AL, Armes SP, J. Am. Chem. Soc 2005, 127, 17982–17983. [DOI] [PubMed] [Google Scholar]; c) Li Y, Du W, Sun G, Wooley KL, Macromolecules 2008, 41, 6605–6607. [Google Scholar]; d) Peng HS, Stolwijk JA, Sun LN, Wegener J, Wolfbeis OS, Angew. Chem., Int. Ed 2010, 122, 4342–4345. [DOI] [PubMed] [Google Scholar]; e) Li Y, Zhao T, Wang C, Lin Z, Huang G, Sumer BD, Gao J, Nat. Commun 2016, 7, 13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Chen T, Hu Y, Cen Y, Chu X, Lu Y, J. Am. Chem. Soc 2013, 135, 11595–11602. [DOI] [PubMed] [Google Scholar]; b) Yao Y, Wang Y, Huang F, Chem. Sci, 2014, 4312–4316. [Google Scholar]; c) Wang M, Sun S, Neufeld CI, Perez‐Ramirez B, Xu Q, Angew. Chem., Int. Ed 2014, 53, 13444–13448. [DOI] [PubMed] [Google Scholar]; d) Deng Z, Qian Y, Yu Y, Liu G, Hu J, Zhang G, Liu S, J. Am. Chem. Soc 2016, 138, 10452–10466. [DOI] [PubMed] [Google Scholar]; e) Napoli A, Valentini M, Tirelli N, Muller M, Hubbell JA, Nat. Mater 2004, 3, 183. [DOI] [PubMed] [Google Scholar]; f) Li C, Madsen J, Armes SP, Lewis AL, Angew. Chem., Int. Ed 2006, 45, 3510–3513. [DOI] [PubMed] [Google Scholar]; g) Ryu J, Chacko RT, Jiwpanich S, Bickerton S, Babu RP, Thayumanavan S, J. Am. Chem. Soc 2010, 132, 17227–17235. [DOI] [PubMed] [Google Scholar]; h) de Vries WC, Grill D, Tesch M, Ricker A, Nüsse H, Klingauf J, Studer A, Gerke V, Ravoo BJ, Angew. Chem., Int. Ed 2017, 56, 9603–9607. [DOI] [PubMed] [Google Scholar]
- [6].a) Chaudhuri TK, Paul S, FEBS J. 2006, 273, 1331–1349. [DOI] [PubMed] [Google Scholar]; b) Ross CA, Poirier MA, Nat. Med 2004, 10, S10–S17. [DOI] [PubMed] [Google Scholar]; c) Chiti F, Dobson CM, Annu. Rev. Biochem 2006, 75, 333–366. [DOI] [PubMed] [Google Scholar]; d) Dobson CM, Nature 2002, 418, 729–730. [DOI] [PubMed] [Google Scholar]; e) Takaoka Y, Sakamoto T, Tsukiji S, Narazaki M, Matsuda T, Tochio H, Shirakawa M, Hamachi I, Nat. Chem 2009, 1, 557–561. [DOI] [PubMed] [Google Scholar]
- [7].a) De Silva PA, Gunaratne NHQ, McCoy CP, Nature 1993, 364, 42–44. [Google Scholar]; b) Aviram A, J. Am. Chem. Soc 1988, 110, 5687–5692. [Google Scholar]; c) De Silva AP, Nat. Mater 2005, 4(1), 15. [Google Scholar]; d) De Silva AP, Uchiyama S Nat. Nanotech 2007, 2(7), 399–410. [DOI] [PubMed] [Google Scholar]
- [8].a) Collier CP, Wong EW, Belohradský M, Raymo FM, Stoddart JF, Kuekes PJ, Williams RS, Heath JR, Science, 1999, 285, 391–394. [DOI] [PubMed] [Google Scholar]; b) Kou S, Lee HN, van Noort D, Swamy KEE, Kim SH, Soh JH, Park S, Angew. Chem., Int. Ed 2008, 47, 872–876. [DOI] [PubMed] [Google Scholar]; c) Erbas‐Cakmak S, Akkaya EU, Angew. Chem., Int. Ed 2013, 52, 11364–11368. [DOI] [PubMed] [Google Scholar]; c) Margulies D, Melman G, Felder CE, Arad-Yellin R, Shanzer A, J. Am. Chem. Soc 2004, 126, 15400–15401. [DOI] [PubMed] [Google Scholar]
- [9].a) De Silva AP, James MR, McKinney BOF, Pears DA, Weir SM, Nat. Mater 2006, 5, 787. [DOI] [PubMed] [Google Scholar]; b) Margulies D, Hamilton AD, J. Am. Chem. Soc 2009, 131, 9142. [DOI] [PubMed] [Google Scholar]; c) Chen J, Fang Z, Lie P, Zeng L, Anal Chem. 2012, 84, 6321. [DOI] [PubMed] [Google Scholar]; d) Nikitin MP, Shipunova VO, Deyev SM, & Nikitin PI Nat. Nanotech, 2014, 9, 716–722. [DOI] [PubMed] [Google Scholar]; e) You M, Peng L, Shao N, Zhang L, Qiu L, Cui C, Tan W, J. Am. Chem. Soc 2014, 136, 1256–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL,Weibel DB, Whitesides GM, Chem. Rev 2008, 108, 946–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Alterio V, Di Fiore A, D’Ambrosio K, Supuran CT, De Simone G, Chem. Rev 2012, 112, 4421–4468. [DOI] [PubMed] [Google Scholar]
- [11].Li HN, Ci YX, Analytica chimica acta 1995, 317, 353–357. [Google Scholar]
- [12].a) Munkhbat O, Garzoni M, Raghupathi KR, Pavan GM, Thayumanavan S, Langmuir, 2016, 32, 2874–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tanford C, C. 2nd ed.; J. Wiley and Sons: New York, 1980. [Google Scholar]; c). Israelachvili JN, Mitchell DJ, Ninham BW, J. Chem. Soc., Faraday Trans. 1976, 72, 1525–1568. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.