Abstract
Background:
Hemophilia B is caused by genetic aberrations in the F9 gene. The majority of these are non-synonymous mutations that alter the primary structure of blood coagulation Factor IX (FIX). However, a synonymous mutation c.459G>A (Val107Val) was clinically reported to result in mild hemophilia B (FIX coagulant activity 15–20% of normal). The F9 mRNA of these patients showed no skipping or retention of introns and/or change in mRNA levels, suggesting that mRNA integrity does not contribute to the origin of the disease in affected individuals. The aim of this study is to elucidate the molecular mechanisms that can explain disease manifestations in patients with this synonymous mutation.
Methods:
We analyze the molecular mechanisms underlying the FIX deficiency through in silico analysis and reproducing the c.459G>A (Val107Val) mutation in stable cell lines. Conformation and non-conformation sensitive antibodies, limited trypsin digestion, activity assays for FIX, interaction with other proteins, and post-translation modifications were used to evaluate the biophysical and biochemical consequences of the synonymous mutation.
Results:
The Val107Val synonymous mutation in F9 was found to significantly diminish FIX expression. Our results suggest that this mutation slows FIX translation and affects its conformation resulting in decreased extracellular protein level. The altered conformation did not change the specific activity of the mutated protein.
Conclusions:
The pathogenic basis for one synonymous mutation (Val107Val) in the F9 gene associated with hemophilia B was determined. A mechanistic understanding of these synonymous variants yields potential for guiding and developing future therapeutic treatments.
Keywords: hemophilia, synonymous mutation, factor IX, protein folding, disease
INTRODUCTION
We and others have previously shown that synonymous mutations can affect protein folding, structure and function.[1] Synonymous mutations can affect protein biogenesis via multiple mechanisms, including affecting the efficiency of protein expression and/or protein folding.[2–7] Although no change in mRNA integrity was reported for these patients, a change in mRNA structure may have led to altered protein translation dynamics and reduced protein levels. Codon usage can affect translational speed; frequently used codons are translated more rapidly than infrequently used ones.[8–10] Modulation of local elongation rates through synonymous mutations has been shown to affect protein secretion,[11] thus it is possible that improper processing contributes to differences in protein expression, conformation and stability. It is likely that the multitude of mechanisms by which synonymous mutations alter protein structure and function have clinical consequences because a critical mass of literature shows that synonymous mutations are associated with numerous human diseases.[12–14]
Hemophilia B is a blood clotting disorder caused by mutations in the F9 gene, which encodes a serine protease in the blood coagulation system known as factor IX (FIX).[15] Mutations in F9 may lead to severe (FIX coagulant activity (CA) <1% of normal), moderate (CA 1–5%) or mild (CA 5–30%) hemophilia B.[16] A synonymous mutation, c.459G>A (GTG>GTA, p.Val153Val or Val107Val (amino acid number after the cleavage upon secretion)) recently identified in 5 non-related mild hemophilia B patients, resulted in a ~80% reduction in FIX coagulant activity.[15] Studies of lymphocyte F9 mRNA in these 5 individuals showed no skipping or retention of introns and/or change in mRNA levels, suggesting that mRNA integrity does not contribute to the origin of the disease.
FIX is a vitamin K-dependent blood coagulation factor that converts factor X to its active form. FIX is synthesized as a 461 amino acid precursor (primarily in the liver) and then secreted into plasma. FIX zymogen undergoes extensive co- and post-translational modifications, including but not limited to glycosylation (~17% carbohydrate by weight) and γ-carboxylation.[17] It has a 46 amino acid N-terminal pro-peptide (which includes a 28 amino acid signal sequence) that is cleaved. FIX circulates in the plasma as a single chain inactive zymogen of 415 amino acids. During blood clotting it is activated by two distinct mechanisms including either factor XIa (the intrinsic pathway) or factor VIIa/tissue factor (the extrinsic pathway). The activation of FIX results in the excision of the so-called activation peptide (aa 145–180) that converts FIX into its active form (factor FIXαβ) where the chains are linked together by a disulfide bond.
In the current study we have used a combination of in silico, in vitro and ex vivo (cellular) systems to understand the potential molecular mechanism(s) by which FIX c.459G>A (Val107Val) mutation causes reduction in FIX protein activity in the patient’s plasma.[15] The valine at position 107 is located at the beginning of the second β-sheet in the antiparallel among five tandem β- sheet structure of the second EFG-like domain with average conservation score [18] (http://consurftest.tau.ac.il/). This Val107Val mutation is one of ten identified synonymous variants that are clinically associated with hemophilia B. Our studies revealed that FIX c.459G>A (Val107Val) mutation affects FIX synthesis, maturation and conformation, therefore leading to FIX deficiency in the patient’s plasma.
MATERIALS AND METHODS
Computational analyses
The F9 mRNA sequence (RefSeq NM_000133.3) and 151 nucleotide fragment centered on the c.459G>A were analyzed with mfold (http://unafold.rna.albany.edu/?q=mfold),[19] Kinefold (http://kinefold.curie.fr/)[20] and NUPACK (http://www.nupack.org/)[21] software programs. Relative synonymous codon usage (RSCU) and codon adaptation index (CAI) were calculated as previously described.[22]
In vitro translation
In vitro translation of the in vitro transcribed F9 mRNAs was performed in the presence of [35S]Met following standard procedures with Rabbit Reticulocyte Lysate (RRL) system (Promega) supplemented with calf liver tRNAs.
Ex vivo expression vectors
Wild-type F9 ORF with a truncated intron 1, originating from the pCI-neo-hFIX1b vector bearing human F9 cDNA, was sub-cloned into pcDNA3.1/V5-His-TOPO (Invitrogen/Life Technologies). The c.459G>A (Val107Val) variant was constructed by site-directed mutagenesis using QuikChange II XL kit (Stratagene/Agilent Technologies).
Cell culture and transfection
Human HEK293 cells and Huh-7 cells were transformed using FuGENE 6 Transfection Reagent (Roche Diagnostics). Stable cell lines were created under selection pressure using Geneticin. The transfection conditions were tuned to present relatively low expression so slight differences between the wild-type and mutant will be detected.
RNA isolation and quantitative RT-PCR
Total RNA was isolated by Qiagen RNeasy Kit following manufacturer’s protocol with an additional 15 minutes on-column DNase incubation step. F9 mRNA expression levels were measured against the GAPDH reference gene using real-time quantitative RT-PCR. F9 cDNA was obtained by reverse transcription (Roche Diagnostics) followed by PCR-amplification.
PAGE, immunoblotting analysis, ELISA and activity measurements
Media concentration and cellular extracts were prepared as described earlier.[23] Extra- and intracellular protein expression were quantified via Western blotting under denaturing and native conditions[24] and/or FIX specific ELISA (Invitrogen/Thermo Fisher Scientific). Western blotting was performed using anti-V5 antibody (Catalog # R960-CUS, Invitrogen/Thermo Fisher Scientific), anti-human FIX antibodies (ESN1, American Diagnostica; AHIX-5041, Haematologic Technologies; 9D (Catalog #sc-59503), Santa Cruz Biotechnology, Inc.; and GMA-138 (Catalog #sc-65952), Santa Cruz Biotechnology, Inc.), anti-FIX pro-peptide antibody (a gift from Dr. Peters, Biogen) or anti-human GAPDH antibody (Catalog #sc-47742, Santa Cruz Biotechnology), horseradish peroxidase conjugated anti-mouse secondary antibody (Catalog #31340, Thermo Fisher Scientific) and horseradish peroxidase conjugated anti-rabbit secondary antibody (Catalog #611–1302, Rockland, Inc.). All the primary antibodies are derived from mouse monoclonal except for the pro-peptide antibody which is rabbit polyclonal.
Media was resolved by size-exclusion chromatography using a Superdex 200 column. aPTT assay was performed with TriniCLOT Automated aPTT (Trinity Biotech), FIX deficient plasma (HRF) and international standard FIX (#07/182, NIBSC) by an ACL Elite Pro system (Instrumentation Laboratory Company). Chromogenic assay was performed using a commercial kit (Biophen) and measured by Victor X3 plate reader (PerkinElmer) at 405 nm.
Protein purification, limited proteolysis and mass spectrometry analysis
Media from FIX expressing stable cell lines was collected for 3–4 days, concentrated 10-fold and subjected to His-tag FIX affinity purification using cobalt resin (Talon, Clontech). The eluted protein was dialyzed against 150 mM NaCl, 20 mM HEPES buffer. Supernatant was filtered through Pall Acropak filters (0.2 μm, Pall, Port). Cell culture harvest was directly loaded onto an IMAC HiTrap column. Proteins were eluted with 20 CV linear gradients (0% to 100%) of a 25 mM HEPES, 500 mM NaCl, and 250 mM Imidazole buffer. The eluate was then dialyzed using a 10 kDa membrane against the 10 mM HEPES, 150 mM NaCl buffer. Protein was separately purified for CD analysis with a V5-tagged purification kit (MBL International) per manufacturer’s protocol.
The integrity of purified protein was verified by SDS-PAGE followed by silver staining (Invitrogen/ Thermo Fisher Scientific) (figure 1a). Purified proteins were subjected to limited proteolysis with increasing trypsin concentrations (Sigma Aldrich) (0, 0.002, 0.005 and 0.009 mg/mL) for 5 minutes at 37 °C. Digestion was terminated by adding SDS-sample buffer and boiling the samples for 20 minutes at 100 °C. Samples were analyzed by western blotting with anti-human FIX antibody ESN1.
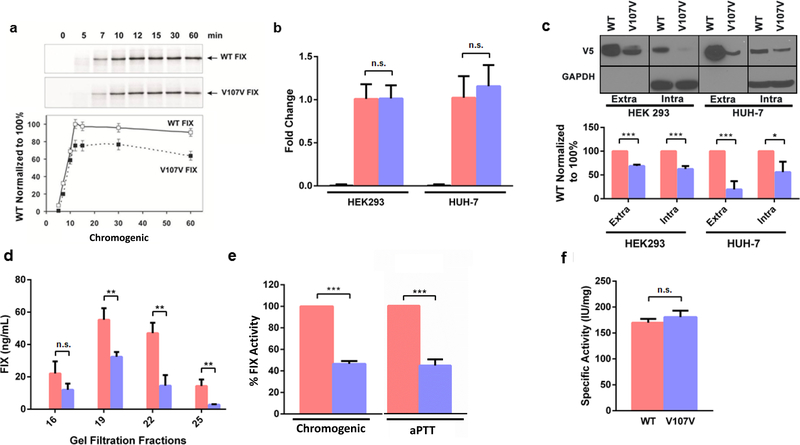
Figure 1. GTG>GTA (Val107Val) mutation reduces FIX protein expression levels and activity.
(a) FIX in vitro translation. Representative [35S]-autoradiogram of the wild-type FIX and Val107Val (GTG>GTA) mutant in vitro translation products in RRL system (top panel) and quantitation analysis of the band intensities of three independent experiments (bottom panel). The intensity of the FIX translation products of wild-type at 10 min was set to 100% in each case. (b) mRNA levels of FIX wild-type (pink) and FIX Val107Val (purple) mutant determined by qPCR in HEK293 and/or Huh-7 cell lines, respectively. Fold changes were normalized to GAPDH mRNA levels (left bar); cells transfected with empty vector were used as control. (c) Intra- and extracellular FIX protein levels measured by Western blotting with anti-V5 antibody (top panel) and quantitation analysis of the band intensities of three independent experiments (bottom panel). (d) FIX levels of FIX wild-type and FIX Val107Val determined by ELISA (pooled fractions of the HEK293 extracellular protein after size-exclusion chromatography of equal total amount of protein separated on Superdex-200). (e) FIX activity in media of Huh-7 cell line, stably expressing wild-type or Val107Val (GTG>GTA) mutant per total protein. FIX activity was measured using chromogenic method and one-stage clotting method (aPTT). Similar results were obtained using HEK293 cell line (data not shown). (f) Specific activity (IU/mg) of purified wild-type and Val107Val (GTG>GTA) protein was measured by chromogenic method (*** = p<0.005 by Student’s t-test (p = 0.00075–0.005436)). In all graphs, data represent the means ± SD of at least three independent experiments.
Activation by factor XIa
Purified FIX (1 μM) was incubated with factor XIa (2 nM) in 20 mM HEPES (pH 7.4), 150 mM NaCl, 0.2% polyethylene glycol (PEG) and 5 mM calcium. At timed intervals samples were removed and added to EDTA and lima bean trypsin inhibitor (LBTI, 250 μg/mL) to inhibit factor XIa. LBTI does not inhibit FIXa. The amount of FIXa was assessed as the ability to cleave Pefachrome FIXa. The relative rate of substrate cleavage is plotted against the time.
Factor IXa active site titration
Purified FIXa was incubated with the indicated concentration of antithrombin with 5 U/mL heparin in 20 mM HEPES, 150 mM NaCl and 0.2% PEG for 2 hours. The amount of FIXa was assessed as the ability to cleave Pefachrome FIXa. The rate of substrate cleavage is plotted against the concentration of antithrombin. Since FIXa forms a one to one complex with anithrombin, the amount of antithrombin required to completely inhibit FIXa activity is equal to the amount of FIXa.
Factor X activation by the factor IXa/VIIIa complex
Buffer for these studies was 20 mM HEPES (pH 7.4), 150 mM NaCl, 0.2% PEG, 5 mM calcium chloride. The indicated concentration of FIXa was incubated with 0.4 nM factor VIIIa, 40 μM phospholipid vesicles, and 170 nM factor X. Factor VIII was activated with thrombin; subsequently the thrombin was inhibited with hirudin. Phospholipid vesicles were phosphatidylcholine: phosphatidylethanolamine: phosphatidylserine (PC:PE:PS) 41:44:15, a composition designed to mimic the lipid composition of highly activated platelets. At timed intervals samples were removed and the amount of factor Xa was assessed as the ability to cleave Pefachrome FXa. The rates of substrate cleavage were plotted against time and the rate of factor X activation was determined as the slope of the data fit. The rate of factor X activation was plotted against the FIXa concentration.
Lys-C peptide mapping
Based on Peters et al., 2013,[25] 50 μg FIX samples were vacuum dried and denatured with 50 μL with 6 M guanidine-HCl-150 mM Tris-HCl-2.5 mM EDTA (pH 8.0). 1 μL of DTT (1 M) was added and incubated at 37 °C for 30 minutes to reduce the sample. 6 μL of iodoacetamide (0.5 M) was added and incubated in the darkness at room temperature for 30 minutes. FIX samples were then buffer exchanged into 50 mM Tris-HCl (pH 7.2) using micro bio-spin column (Bio-Rad). The sample was digested with 5 μL of 0.5 mg/mL Lys-C (Wako Chemicals USA) at 25 °C for 3 hours and then quenched by adding 1 μL of 10% trifluoroacetic acid. The 15 μg of digested sample was analyzed by LC-UV-MS/MS (Agilent 1290 UPLC coupled with Thermo LTQ linear ion trap mass spectrometer). The peptides were identified and confirmed by MS and MS/MS, for both sequence and posttranslational modifications.
Circular dichroism (CD)
CD was measured by JASCO J-810 CD spectropolarimeter, equipped with a Peltier temperaturecontrolled cell holder. Each spectrum (representing the sum of five separate accumulations) was measured with a 0.1-cm path length cell at 5 °C with a 10 nm/minute scanning speed and wavelength range (190–260 nm). CD spectrums were evaluated at a concentration range of 0.40.7 mg/mL. Thermal unfolding and refolding patterns of the FIX protein were conducted from 5 to 55 °C (measurements at 10 °C intervals; temperature shift speed of 1 °C/minute) and cooled to 5 °C. FIX secondary structure content was estimated by Dichroweb software (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml).
ALLN treatment
Stable FIX expressing clones were treated with cycloheximide (10 μg/mL) (Sigma Aldrich) and subsequently by N-acetyl-leucyl-leucyl-norleucinal (ALLN), an inhibitor of neutral Ca2+ dependent cysteine proteases, calpain and the proteasome [26] (Sigma Aldrich) at a 20 μg/mL concentration, 30 hours prior to harvesting (control wells were treated with DMSO). Protein expression was analyzed by Western blotting and ELISA.
Glycosylation assessment of purified FIX
The wild-type and Val107Val purified FIX proteins (1.5 μg/reaction) were treated with the commercial mix of PNGase F, Endo-O-Glycosidase, alfa-2-Neuraminidase, beta-1, 4-Galactosidase and beta-N-Acetylglucosaminidase at 37 °C for 15 minutes (EDEGLY Enzymatic Protein Deglycosylation Kit, Sigma). After termination of the reaction by adding loading dye solution, the reaction mixtures were run on 8% Bis-Tris SDS-PAGE Western blotting using anti- human FIX antibody (9D). The commercially available plasma derived FIX (Mononine®, CSL Behring, gift from Dr. Mikhail V. Ovanesov, FDA) and recombinant FIX (BeneFIX® Wyeth Pharmaceuticals Inc.) were used as positive controls.
RESULTS
The ~80% reduction of FIX coagulant activity observed in plasma of hemophilia B patients with the c.459G>A (Val107Val) mutation can be explained by a number of plausible mechanisms, which could include effects on both mRNA and protein levels. Analysis of F9 mRNA in affected individuals showed that changes in mRNA expression levels and/or its integrity do not contribute to FIX deficiency in affected individuals[15]. These data suggest that FIX deficiency in patients likely arise due to i) reduced translation of mRNA, caused by a c.459G>A (Val107Val) mutation, leading to reduced protein expression levels ii) reduced protein stability, secretion efficiency and/or activity due to changes in FIX conformation and/or altered post translational modifications and/or all of the above. We addressed these possibilities by employing a combination of in silico, in vitro and ex vivo approaches.
Effects of synonymous FIX Val107Val mutation on F9 mRNA structure, codon usage and thermodynamic stability
Three computational algorithms (mfold, Kinefold and NUPACK) were used to assess the effect of the c.459G>A (Val107Val) mutation on mRNA structure, showed a moderate reduction in mRNA thermodynamic stability (supplementary figure S1a-c). Although decreased mRNA thermodynamic stability generally leads to increased protein expression levels, we found that in vitro translation involving the c.459G>A mutant yields 25–30% less protein (figure 2a). This suggests that the mutation moderately reduces global translation rates; however, this result does not clarify the significance of its decreased mRNA thermodynamic stability. Measures of codon usage, the Relative Synonymous Codon Usage (RSCU) and Codon Adaptation Index (CAI),[10] show that the GTA codon is substantially less frequently used in humans than GTG (table 1).
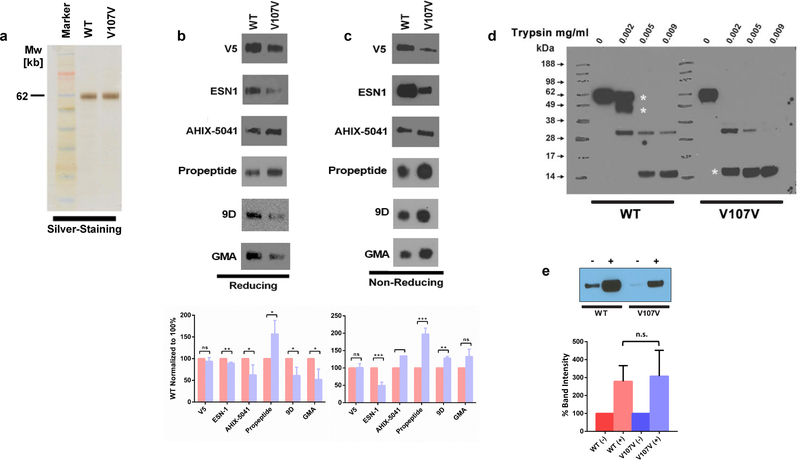
Figure 2. FIX wild-type and GTG>GTA (Val107Val) variants reveal differential structural properties.
(a) Equal amounts of purified FIX protein quantified based on PAGE silver staining were loaded on PAGE and detected under (b) Denaturing and (c) native conditions by Western blot analysis using various antibodies. (d) Limited proteolysis of FIX with trypsin. Samples were analyzed by Western blotting under PAGE denaturing conditions. Asterisks point to the main differences in the pattern. The data are representative of three independent experiments. (e) The effect of ALLN on FIX expression. Huh-7 cells stably expressing FIX wild-type and the FIX Val107Val variants were treated with either DMSO or with 20 μg/mL ALLN for 36 hours. Intracellular protein extracts from these cells were served for the Western blotting with anti-V5 antibody. Data are representative of the mean ± SD for three individual experiments with normalization to wild-type and Val107Val FIX without ALLN. In all graphs, data represent the means ± SD of at least three independent experiments.
Table 1.
RSCU and CAI values for wild-type (GTG) and mutant (GTA) valine codons based on the entire human genome or F9 codon usage.
| Codon for valine | RSCU (genome)a | ΔRSCU (genome)b | RSCU (F9)a | ΔRSCU (F9) | CAIa |
|---|---|---|---|---|---|
| GTG | 1.853 | -- | 0.757 | -- | 1 |
| GTA | 0.466 | −1.387 | 0.540 | −0.216 | 0.242 |
The RSCU values were calculated using both human genome codon usage and codon usage of human F9 wild-type open reading frame (RefSeq NM_000133.3); CAI was calculated using the human genome.
ΔRSCU = RSCUmutant – RSCUWT
Owing to a decrease in codon usage frequency, a decrease in translation efficiency would be expected in the mutant, but a single codon substitution generally would not be expected to lead to a drastic change in translation and it cannot account for the ~80% lower FIX activity in patients. These data suggest that additional mechanism(s) are responsible for decreased FIX activity in patients.
Ex vivo analysis of FIX Val107Val mutation on FIX expression
To further investigate changes in protein structure/ function due to the c.459G>A mutation, we compared the levels and activity of wild-type and variant proteins expressed in HEK293 and Huh-7 stably transfected cell lines, which were constructed to have equivalent levels of mRNA (figure 1b). mRNA was examined for its integrity and sequencing reveals no unexpected splicing events in these systems (supplementary figure S2). Expression of FIX was monitored using a set of specific antibodies. Compared to wild-type, expression of mutant FIX was significantly lower in both cell lines regardless of whether FIX was identified in the medium (extracellular) or in the cell lysate (intracellular). The c.459G>A (Val107Val) mutation resulted in a 40 to 80% reduction in the amount of secreted protein (figure 1c). Moreover, equal amounts of the total secreted protein from media collected from an equal number of cells were subjected to size exclusion chromatography followed by ELISA. Lower levels of mutant FIX (in comparison with wildtype) were found in all fractions (figure 1d). The decreased level of Val107Val FIX protein in the media was reflected by activity measurements, showing diminished levels (~45% of wildtype FIX, figure 1e) of activity, resulting in comparable specific activity (figure 1f).
A “silently” mutated FIX Val107Val protein exhibits an altered conformation
We showed above that the c.459G>A (Val107Val) mutation results in lower expression of FIX protein. Non-synonymous mutations (altering protein structure) in the second EGF-like domain (where Val107Val mutation is located) were shown to cause impaired intracellular processing and secretion.[27] Current knowledge about the effects of synonymous mutations on protein function suggests that they can also alter protein structure. Therefore, we further decided to probe protein structure and investigate protein processing.
First we investigated the global secondary structure of the mutant protein by circular dichroism. Both wild-type and Val107Val were found to have similar secondary structure content, with minor variation seen among replicate measurements (supplementary figure S3, S4). Thermal unfolding and refolding experiments also indicated that both wild-type and mutant FIX share similar thermal stability (supplementary figure S3). These results are consistent with the prevailing consensus that synonymous mutations are usually not associated with a change in secondary protein structure of mRNA.
We next investigated binding of conformation sensitive antibodies to equal amounts of purified protein, because antibodies are often sensitive to proteins’ conformation changes. Immunoblots on equal amounts of purified FIX using six antibodies are depicted in figure 2. Under denaturing conditions, immunoblotting with monoclonal anti-FIX antibody ESN1 showed comparable FIX bands for both variants (figure 2b); however a stronger signal for the wild-type protein was observed under native conditions (figure 2c). The use of antibodies 9D and GMA-138 revealed weaker signal for Val107Val variant under denaturing conditions while weaker signal for the wild-type variant was observed under native conditions. A high molecular band was always observed in the native gel systems,[24] but the intensities of the high molecular weight complexes (upper band) constantly correlate with the intensities of the lower band regardless of ratio of the lower bands between wild-type and Val107Val FIX variants (data not shown). These results suggest that conformational changes in the variant may alter the accessibility of the antiFIX epitopes and are consistent with limited trypsin proteolysis which shows that the mutant has a greater sensitivity to trypsin (figure 2d).
Enhanced sensitivity to trypsin digestion also suggests that the Val107Val FIX protein may be prone to increased intracellular degradation. However, the use of a cellular proteasome inhibitor (ALLN) resulted in similar stabilization of both proteins (figure 2e).
Detailed functional characterization
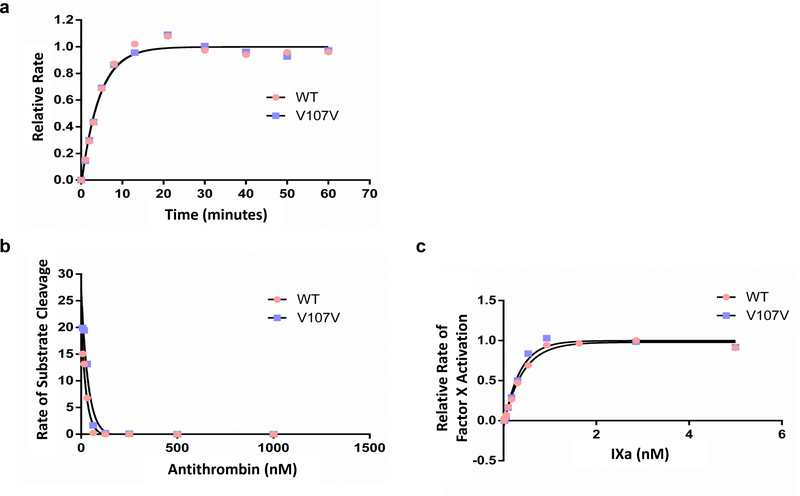
While we found the specific activity of purified wild-type and Val107Val FIX to be similar, the Val107Val mutant protein clearly exhibits altered conformation as shown through differential immunoreactivity. Interestingly the crystal structure of FIX (2WPJ.pdb) indicates that valine 107 is found in a region critical for an interaction with FVIII [28–30] and Val107Ala nonsynonymous mutation reduces FIX specific activity due to elevated Km to FVIII.[31] Moreover, five of ten clinically associated synonymous FIX mutations, in three amino acid codons, are in the second EGF domain (figure 3). Valine 107 may therefore be positioned in a region sensitive to changes in translation rate, and subsequently to a subtle change in the structure. Therefore, we tested whether interaction with other proteins in the coagulation cascade were affected. The various properties examined included (a) the rate of activation by factor XIa and (b) the relative effect of the cofactor FVIIIa on the FIXa activity. The results obtained from these assays are presented in figure 4, and in all cases, they yielded similar value for the wild-type and mutant FIX, suggesting that any difference in the conformation is subtle.
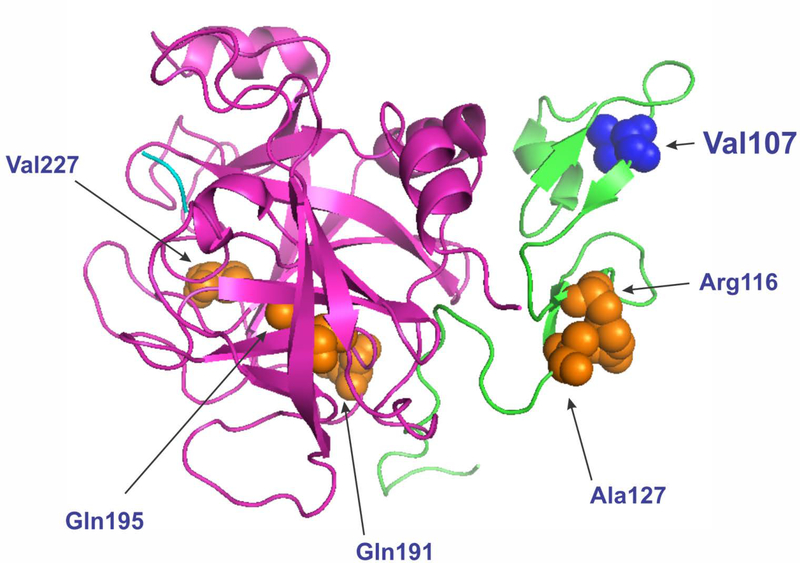
Figure 3. Structure of human FIXαβ: ribbon diagram (PDB ID 2WPJ).
Coagulation factor IXαβ: light chain fragment (residues 87–145; second EGF-like domain) is shown in green; heavy chain fragment (residues 181–415; catalytic domain) is in pink. Val107 under study is in blue (side chain is shown). Residues with clinically associated synonymous mutations are also shown (side chains are in orange). Residue numbers correspond to that of the mature/processed protein.
Figure 4. Effect of FVIIIa and FIXa.
FIXa functional studies. (a) FIX (1 μM) was incubated with 2 nM FXIa and FIX activation was monitored. FIXa was plotted against time. Data was fit to an exponential curve. Wild-type and Val107Val showed the same time course for activation. (b) The concentration of FIXa active sites was determined by incubating FIXa with a known concentration of antithrombin (AT) in the presence of heparin. FIXa activity was plotted against the AT concentration. Both wild-type and Val107Val variant were inhibited by AT/heparin. The concentration of AT required to inhibit both wild-type and Val107Val FIXa was the same indicating that both proteins have the same active concentration. (c) FIXa was incubated with FX in the presence of FVIIIa and lipid vesicles. The rate of factor X activation was monitored. The rate of activation was plotted against the concentration of FIXa. Data was fit to a quadratic equation. Both wild-type and Val107Val bound factor VIIIa equally well and activated factor X to the same extent.
Val107Val FIX mutant does not exhibit significant difference in its composition and the extent of post-translational modifications, specifically γ-carboxylation
Differences in conformation of the protein variants may arise as a result of the altered processing. It is also possible that critical FIX co- and post-translational modifications such as γ-carboxylation and glycosylation, could be altered in the mutant FIX protein, thus leading to differences in protein conformations. Mass spectrometric analysis of the purified FIX variants did not reveal any significant differences between wild-type and mutant proteins. The only notable difference was the relative proportion of pro-peptide (pro-FIX) in the wild-type and mutant proteins: 9.5% and 20.7% respectively (supplementary table S1a, supplementary figure S5). Supplementary table S2 lists the detected Gla peptides and the peptide containing Val107al (K9) in Lys-C peptide maps. The N- and O-glycosylation in the activation peptide of FIX were not characterized in detail due to its complexity. From these results, it is likely that moderate differences in translation rates between the two variants may lead to the moderately altered processing and subsequent changes in the conformation of FIX. Glycosylation status of intracellular wild-type and Val107Val FIX proteins was also examined (supplementary figure S6). No apparent difference was observed in deglycosylation pattern of these FIX proteins although this method detects overall molecular weight and is not sensitive to detect distinct glycosylation.
DISCUSSION
Here we have investigated a synonymous mutation in F9 that is clinically associated with hemophilia B phenotype.[15] Our studies suggest that the observed decrease in FIX activity in affected individuals can be explained by moderately decreased efficiency of protein translation, leading to altered pro-peptide processing, causing subsequent changes in protein integrity and conformation and thus decreasing overall protein expression. These changes are not caused by a change in mRNA integrity, but rather originate from a change in the efficiency of mutant mRNA translation. Whereas numerous non-synonymous mutations in the F9 gene have been linked to hemophilia B,[32] this report provides insight into the mechanism(s) by which a single synonymous mutation can sufficiently disrupt the protein properties to cause the clinical manifestation of a bleeding disorder (figure 5). Early studies that explored the underpinnings of synonymous mutation mediated effects on protein function [4] focused on single unique mechanisms. In this study we demonstrate that a synonymous mutation can have a multitude of effects. As the clinical effect of this GTG>GTA mutation can be quantified vis-à-vis decrease in FIX-activity, we show that the moderate decrease in protein translation alone is not sufficient to explain the substantial decrease in protein expression levels/activity and the clinical outcome.
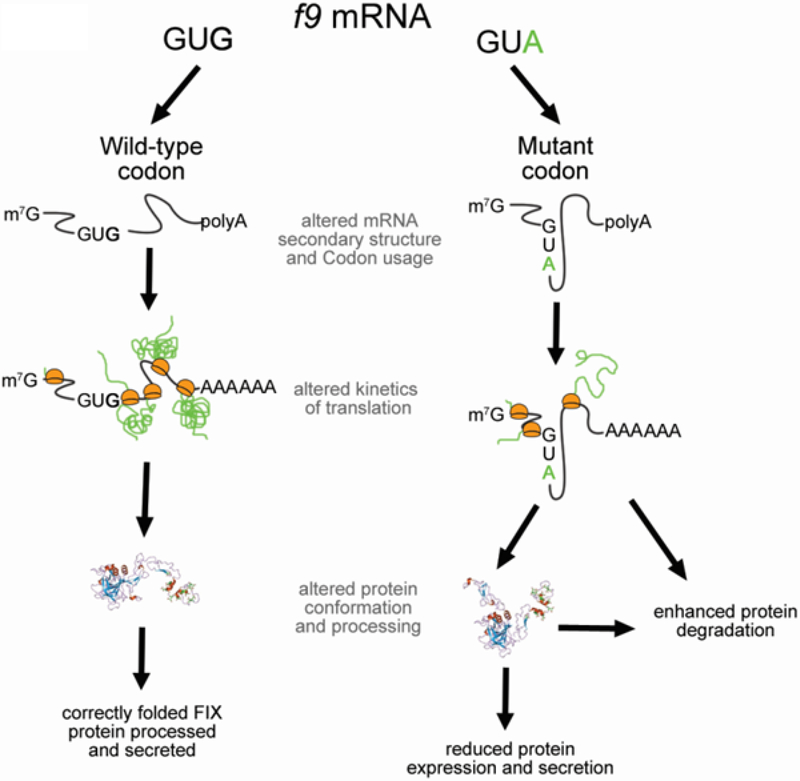
Figure 5.
Mechanism by which a single synonymous mutation can disrupt protein properties.
We also suggest that a specific location of the valine codon is one of the reasons, why the underlying Val107Val (GTG>GTA) mutation causes the disease. Effects of the synonymous codons are believed to be position dependent. In this particular case, the specific location of the mutation in the second EGF-like domain may be one of the major contributors to impaired post-translational modification(s) and overall extracellular protein levels. These are associated with subtle changes in its structure caused by altered translation dynamics (figure 3). Note that nonsynonymous mutations in the second EGF-like domain were previously shown to cause FIX impaired intracellular processing and secretion.[27] We have previously presented an extensive review of the multiple mechanisms by which synonymous mutations affect protein structure and function and the methods that may be used to study these.[4] This study shows that these mechanisms may act in concert and it is important to exhaustively (and where possible quantitatively) characterize the mechanistic basis of clinical outcomes driven by synonymous changes. As a final note it is important to point out that the synonymous change characterized in this study is not unique. At least nine additional synonymous mutations associated with hemophilia B are reported in the Centers for Disease Control and Prevention (CDC) F9 mutation database (http://www.cdc.gov/ncbddd/hemophilia/champs.html).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds from the laboratory of Hemostasis, Center for Biologics Evaluation and Research, FDA (CK-S), The American Heart Association [13GRNTI7070025 to AAK] and The National Institutes of Health [HL121779-01A1 to AAK]. We thank Puja Nanavaty, Cleveland State University, for technical assistance.
Footnotes
COMPETING INTERESTS
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 1999;462(3):387–91. [DOI] [PubMed] [Google Scholar]
- 2.Buhr F, Jha S, Thommen M, Mittelstaet J, Kutz F, Schwalbe H, Rodnina MV, Komar AA. Synonymous Codons Direct Cotranslational Folding toward Different Protein Conformations. Mol Cell 2016;61(3):341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SJ, Yoon JS, Shishido H, Yang Z, Rooney LA, Barral JM, Skach WR. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science 2015;348(6233):444–8. [DOI] [PubMed] [Google Scholar]
- 4.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007;315(5811):525–8. [DOI] [PubMed] [Google Scholar]
- 5.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci 2009;34(1):16–24. [DOI] [PubMed] [Google Scholar]
- 6.Yu CH, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, Liu Y. Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding. Mol Cell 2015;59(5):744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 2009;16(3):274–80. [DOI] [PubMed] [Google Scholar]
- 8.Chaney JL, Clark PL. Roles for Synonymous Codon Usage in Protein Biogenesis. Annu Rev Biophys 2015;44:143–66. [DOI] [PubMed] [Google Scholar]
- 9.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 2011;12(1):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 1987;15(3):1281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pechmann S, Chartron JW, Frydman J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol 2014;21(12):1100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, Harel-Bellan A, Mograbi B, Darfeuille-Michaud A, Hofman P. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet 2011;43(3):242–5. [DOI] [PubMed] [Google Scholar]
- 13.Gartner JJ, Parker SC, Prickett TD, Dutton-Regester K, Stitzel ML, Lin JC, Davis S, Simhadri VL, Jha S, Katagiri N, Gotea V, Teer JK, Wei X, Morken MA, Bhanot UK, Program NCS, Chen G, Elnitski LL, Davies MA, Gershenwald JE, Carter H, Karchin R, Robinson W, Robinson S, Rosenberg SA, Collins FS, Parmigiani G, Komar AA, KimchiSarfaty C, Hayward NK, Margulies EH, Samuels Y. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci U S A 2013;110(33):13481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt RC, Simhadri VL, Iandoli M, Sauna ZE, Kimchi-Sarfaty C. Exposing synonymous mutations. Trends Genet 2014;30(7):308–21. [DOI] [PubMed] [Google Scholar]
- 15.Knobe KE, Sjorin E, Ljung RC. Why does the mutation G17736A/Val107Val (silent) in the F9 gene cause mild haemophilia B in five Swedish families? Haemophilia 2008;14(4):723–8. [DOI] [PubMed] [Google Scholar]
- 16.Giangrande P Haemophilia B: Christmas disease. Expert Opin Pharmacother 2005;6(9):1517–24. [DOI] [PubMed] [Google Scholar]
- 17.Peters RT, Low SC, Kamphaus GD, Dumont JA, Amari JV, Lu Q, Zarbis-Papastoitsis G, Reidy TJ, Merricks EP, Nichols TC, Bitonti AJ. Prolonged activity of factor IX as a monomeric Fc fusion protein. Blood 2010;115(10):2057–64. [DOI] [PubMed] [Google Scholar]
- 18.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 2010;38(Web Server issue):W529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuker M Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003;31(13):3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xayaphoummine A, Bucher T, Isambert H. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res 2005;33(Web Server issue):W605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem 2011;32(1):170–3. [DOI] [PubMed] [Google Scholar]
- 22.Sharp PM, Li W-H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic acids research 1987;15(3):1281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards NC, Hing ZA, Perry A, Blaisdell A, Kopelman DB, Fathke R, Plum W, Newell J, Allen CE, S G, Shapiro A, Okunji C, Kosti I, Shomron N, Grigoryan V, Przytycka TM, Sauna ZE, Salari R, Mandel-Gutfreund Y, Komar AA, Kimchi-Sarfaty C. Characterization of coding synonymous and non-synonymous variants in ADAMTS13 using ex vivo and in silico approaches. PLoS ONE 2012;7(6):e38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simhadri VL, Hamasaki-Katagiri N, Tseng SC, Bentley AA, Zichel R, Hershko AY, Komar AA, Kimchi-Sarfaty C. Factor IX oligomerization underlies reduced activity upon disruption of physiological conditions. Haemophilia 2014;20(2):e157–63. [DOI] [PubMed] [Google Scholar]
- 25.Peters RT, Toby G, Lu Q, Liu T, Kulman JD, Low SC, Bitonti AJ, Pierce GF. Biochemical and functional characterization of a recombinant monomeric factor VIII-Fc fusion protein. J Thromb Haemost 2013;11(1):132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. Journal of Biological Chemistry 1997;272(14):9086–92. [DOI] [PubMed] [Google Scholar]
- 27.Enjolras N, Plantier JL, Rodriguez MH, Rea M, Attali O, Vinciguerra C, Negrier C. Two novel mutations in EGF-like domains of human factor IX dramatically impair intracellular processing and secretion. J Thromb Haemost 2004;2(7):1143–54. [DOI] [PubMed] [Google Scholar]
- 28.Brandstetter H, Bauer M, Huber R, Lollar P, Bode W. X-ray structure of clotting factor IXa: active site and module structure related to Xase activity and hemophilia B. Proc Natl Acad Sci U S A 1995;92(21):9796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngo JC, Huang M, Roth DA, Furie BC, Furie B. Crystal structure of human factor VIII: implications for the formation of the factor IXa-factor VIIIa complex. Structure 2008;16(4):597–606. [DOI] [PubMed] [Google Scholar]
- 30.Stoilova-McPhie S, Villoutreix BO, Mertens K, Kemball-Cook G, Holzenburg A. 3Dimensional structure of membrane-bound coagulation factor VIII: modeling of the factor VIII heterodimer within a 3-dimensional density map derived by electron crystallography. Blood 2002;99(4):1215–23. [DOI] [PubMed] [Google Scholar]
- 31.Chang YJ, Wu HL, Hamaguchi N, Hsu YC, Lin SW. Identification of functionally important residues of the epidermal growth factor-2 domain of factor IX by alanine-scanning mutagenesis. Residues Asn(89)-Gly(93) are critical for binding factor VIIIa. J Biol Chem 2002;277(28):25393–9. [DOI] [PubMed] [Google Scholar]
- 32.Hamasaki-Katagiri N, Salari R, Simhadri VL, Tseng SC, Needlman E, Edwards NC, Sauna ZE, Grigoryan V, Komar AA, Przytycka TM, Kimchi-Sarfaty C. Analysis of F9 point mutations and their correlation to severity of haemophilia B disease. Haemophilia 2012;18(6):933–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.