Abstract
Inhibition of proteasome activity blocks the degradation of dysfunctional proteins and induces cancer cell death due to cellular stress. Thus, proteasome inhibitors represent an attractive class of anticancer agents, and bortezomib, carfilzomib and ixazomib have been FDA-approved to treat multiple myeloma. However, cancer cells acquire resistance to these inhibitors through point mutations in the proteasome catalytic subunit or induction of alternative compensatory mechanisms. In this study, we identified a quinolin-chlorobenzothioate, QCBT7, as a new proteasome inhibitor showing cytotoxicity in a panel of cancer cell lines. QCBT7 is a more stable derivative of quinoline-8-thiol that targets the regulatory subunit instead of the catalytic subunit of the proteasome. QCBT7 caused the accumulation of ubiquitylated proteins in the cancer cells, indicating its proteasome inhibitory activity. Additionally, QCBT7 increased the expression of a set of genes (PFKFB4, CHOP, HMOX1 and SLC7A11) at both nascent RNA and protein levels, similarly to the known proteasome inhibitors MG132 and ixazomib. Together, QCBT7 induces proteasome inhibition, hypoxic response, endoplasmic reticulum stress and glycolysis, finally leading to cell death. Importantly, we have identified PFKFB4 as a potential biomarker of proteasome inhibitors that can be used to monitor treatment response.
Keywords: Quinolin-chlorobenzothioate, Proteasome inhibitor, Hypoxia, Endoplasmic reticulum stress, Glycolysis, Pancreatic cancer
1. Introduction
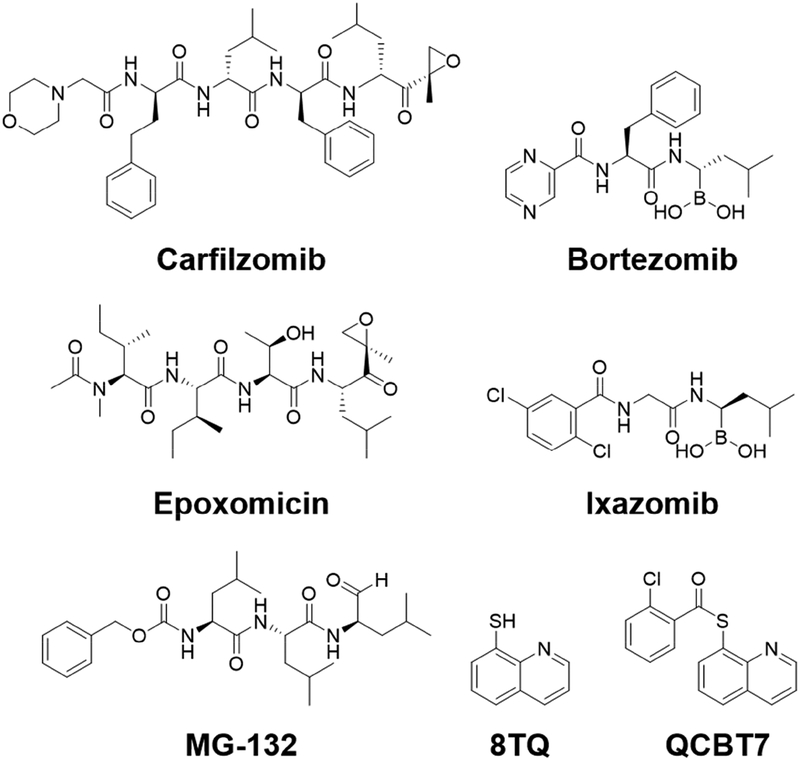
The human 26S proteasome is an essential protein degradation machinery designated to maintain cellular homeostasis via removal of unfolded and misfolded proteins, and it is involved in balancing cell survival and apoptotic cell death. Importantly, small molecule proteasome inhibitors activate unfolded protein response leading to cancer cell death and are attractive for cancer therapy [1, 2]. Bortezomib, ixazomib and carfilzomib were approved for treating multiple myeloma or mantle-cell lymphoma as proteasome inhibitors. They have boronic acid or epoxyketone scaffold, known to interact with the N-terminal threonine residues in the catalytic subunits of the proteasome thus blocking its activity (Figure 1) [1]. Derivatives of the approved proteasome inhibitors have shown efficacy in multiple cancers and are currently undergoing clinical trials in solid tumors, including pancreatic cancer [1–4]. Unfortunately, patients frequently develop resistance to these therapies due to either point mutations on the drug binding site or induction of alternative compensatory mechanisms such as the aggresome pathway [5]. Therefore, development of proteasome inhibitors with different scaffolds and mechanisms of action would be critical to overcome tumor cell resistance to first-line proteasome inhibitors.
Figure 1.
Structures of select proteasome inhibitors and the lead compound QCBT7. Carfilzomib and epoxomicin are epoxyketone-based compounds, and bortezomib and ixazomib are boronate-containing compounds. MG132 is a peptide aldehyde. 8TQ is 8-quinolinethiol hydrochloride. QCBT7 has a similar scaffold to 8TQ.
Pancreatic cancer is a difficult cancer to treat due to its rapid metastatic spread and late-stage diagnosis [6]. Pancreatic ductal adenocarcinoma (PDAC) is the major type of pancreatic cancer and is predicted to become the second most common cause of cancer-related death within the next decade in the United States [7, 8]. Pancreatic tumor cells can rapidly develop resistance to current chemotherapies, such as gemcitabine, nab-paclitaxel and FOLFIRINOX [9, 10]. Thus, it is essential to develop more effective therapeutics to better treat this dreadful disease. Inhibition of the ubiquitin-proteasome pathway contributes to apoptotic cell death in pancreatic cancer [11, 12]. Bortezomib sensitizes pancreatic cancer cells to endoplasmic reticulum (ER) stress and apoptosis [13]. Additionally, proteasome activator subunit 3 (PSME3) promotes the pancreatic cancer growth by activating the proteasome activity [14]. Therefore, the proteasome is an attractive target for therapeutics in pancreatic cancer.
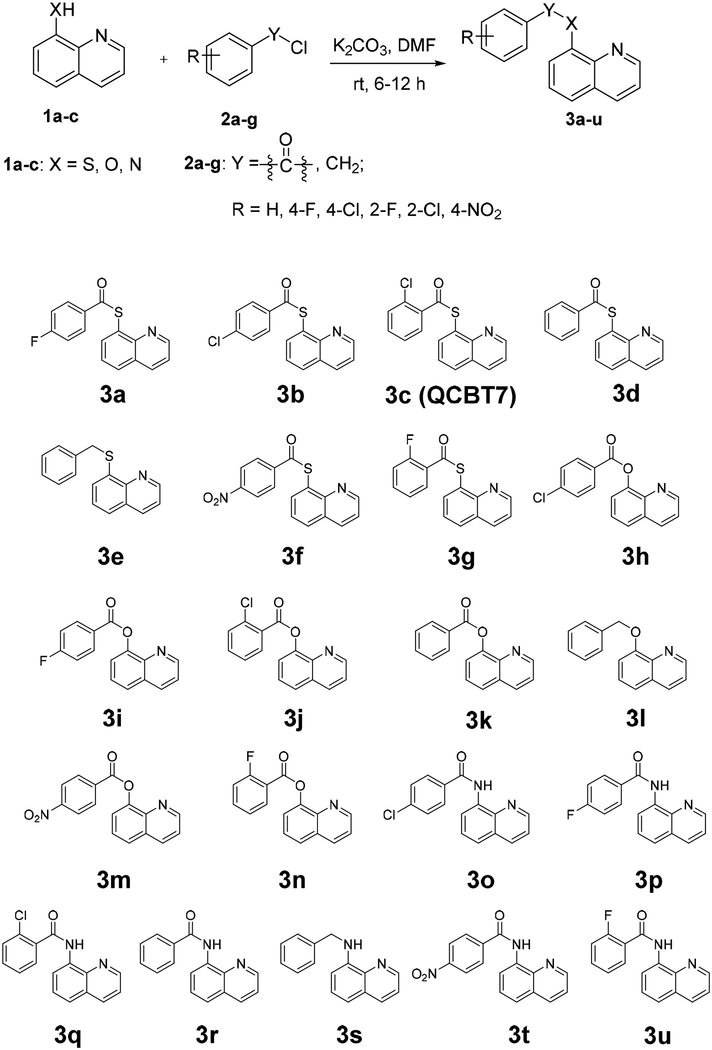
In a medium-throughput phenotypic screen of 20,000 diverse drug-like compounds, we identified a quinolin-chlorobenzothioate, QCBT7, with submicromolar cytotoxicity in the colon carcinoma cell line HCT 116. It was previously shown that the structurally similar compound 8-quinolinethiol hydrochloride (8TQ) inhibits the essential proteasome deubiquitinase Rpn11, instead of the catalytic subunit of the proteasome, suggesting that QCBT7 may also affect proteasome function (Figure 1) [15]. Quinoline derivatives possess antibacterial, antimalarial and anticancer activities. Thioester, amide and ester derivatives of quinoline have been used in antimicrobial and anticancer research [16–19]. To further identify new quinoline derivatives with potential proteasome inhibition and anticancer activities, we synthesized 21 analogs of QCBT7 to establish structural requirement for potency. We then profiled nascent RNA and protein expression in QCBT7-treated pancreatic cancer cells to understand how these cells respond to the treatment. Our results demonstrate that the induced transcription and protein patterns following QCBT7 treatment resemble those obtained for MG132 and ixazomib, indicating that QCBT7 blocks proteasome activity and induces hypoxic response, ER stress and glycolysis.
2. Results and discussion
2.1. Identification of QCBT7
In a medium-throughput screen of 20,000 drug-like compounds derived from a diverse library, QCBT7 was one of the most cytotoxic hits and had an IC50 value of 0.6 μM in HCT 116 cells [20]. QCBT7 was further tested in the pancreatic cancer cell lines, showing IC50 values of 2.6 μM and 1.1 μM in MIA PaCa-2 and PANC-1 cells, respectively (Table 1). Considering the stability of the thioesters in the cellular reducing environment, we synthesized ester and amide derivatives of QCBT7 to potentially increase the stability and maintain cytotoxicity.
Table 1.
Cytotoxicity of 8-thioester/ester/amide-quinoline derivatives of QCBT7 in a panel of cancer cell lines.
| Compound | IC50(μM) | |||
|---|---|---|---|---|
| MIA PaCa-2 | PANC-1 | HCT 116 | KYSE-70 | |
| 3a | 3.3 ± 0.2 | 1.5 ± 0.4 | 1.1 ± 0.4 | 1.1 ± 0.3 |
| 3b | 3.4 ± 0.3 | 1.5 ± 0.6 | 1.1 ± 0.6 | 1.3 ± 0.1 |
| 3c (QCBT7) | 2.6 ± 0.6 | 1.1 ± 0.1 | 0.6 ± 0.3 | 0.8 ± 0.1 |
| 3d | 2.4 ± 0.8 | 0.9 ± 0.3 | 0.5 ± 0.3 | 0.7 ± 0.1 |
| 3e | >20 | >20 | >20 | >20 |
| 3f | 4.9 ± 3.3 | 1.7 ± 0.1 | 1.5 ± 0.6 | 0.9 ± 0.2 |
| 3g | 3.6 ± 0.1 | 0.9 ± 0.4 | 0.7 ± 0.1 | 0.6 ± 0.1 |
| 3h | 10.3 ± 5.6 | 9.4 ± 4.4 | 4.7 ± 1.5 | 11.8 ± 0.5 |
| 3i | 6.5 ± 0.7 | 5.1 ± 2.1 | 4.0 ± 0.7 | 6.9 ± 0.1 |
| 3j | 6.5 ± 0.1 | 7.1 ± 1.1 | 3.0 ± 0.8 | 15.2 ± 5.7 |
| 3k | 5.4 ± 0.4 | 4.9 ± 1.4 | 3.7 ± 1.1 | 6.6 ± 0.2 |
| 3l | >20 | >20 | >20 | >20 |
| 3m | 7.0 ± 0.4 | 7.6 ± 1.2 | 4.2 ± 2.0 | 4.1 ± 0.3 |
| 3n | 5.8 ± 0.8 | 5.5 ± 0.2 | 2.9 ± 0.1 | 6.5 ± 0.3 |
| 3o | >20 | >20 | >20 | >20 |
| 3p | >20 | >20 | >20 | >20 |
| 3q | >20 | >20 | >20 | >20 |
| 3r | >20 | >20 | >20 | >20 |
| 3s | >20 | >20 | >20 | >20 |
| 3t | >20 | >20 | >20 | >20 |
| 3u | >20 | >20 | >20 | >20 |
Data are shown as mean ± standard deviation from at least three independent experiments.
2.2. Synthesis of QCBT7 and its derivatives
Derivatives of QCBT7 were prepared according to the general procedure shown in Scheme 1. Benzoyl chloride or benzyl chloride was reacted with 8-quinolinethiol hydrochloride (8TQ), 8-hydroxylquinoline or 8-aminoquinoline, respectively, in the presence of potassium carbonate in dimethylformamide (DMF) to afford final products 3a-3u in 72–95% yields (v).
2.3. 8-thioester and 8-ester quinoline derivatives are more cytotoxic than 8-amide quinoline derivatives
Cytotoxicity of QCBT7 and its derivatives was assessed using the colorimetric MTT assay in a panel of cancer cell lines (Table 1). The 8-thioester derivatives (3a-d, 3f, 3g) had similar IC50 values around 1 – 4 μM, while the 8-ester derivatives (3h-k, 3m, 3n) were less potent (IC50: 5–10 μM). Different substituents on the benzene ring (R group in Scheme 1) had little effect on the cytotoxicity of the compounds. The 8-amide derivatives (3o-r, 3t, 3u) were much less cytotoxic (IC50 > 20 μM) although the amide-quinoline was more stable than the ester-quinoline and thioester-quinoline in the cellular reducing environment. The ether analogs 3e and 3l were inactive. These results suggest that the thioester-quinoline and ester-quinoline scaffolds are responsible for the cytotoxicity of this series of compounds. Due to the substantial reduction in cytotoxicity of the amide-quinoline derivatives and similar high potency among 8-thioeter-quinoline analogs, we selected QCBT7 for further stability and mechanistic studies.
2.4. QCBT7 is more stable than 8TQ in solution
To assess the chemical stability of QCBT7, we performed liquid chromatography mass spectrometry (LCMS) of QCBT7 at 3 different time points after it was added to DMSO or to a 1:1 DMSO/water solution. N-acetylcysteine (NAC) was added in the solution to simulate a reducing environment and react with free thiols in the solution. 8TQ is the parent compound from which QCBT7 was synthesized, and is likely to be the functional scaffold for this series of compounds. QCBT7 was more stable than 8TQ under a range of conditions (Table 2, Supplementary Figure 2S). In 100% DMSO, QCBT7 was still intact after 7 days, while 8TQ formed dimers once it was dissolved in DMSO. Addition of NAC did not affect the results, and 8TQ also reacted with NAC forming a conjugate at 2 and 5 hours. In a 1:1 solution of DMSO and water, 8TQ formed dimers within one minute, while QCBT7 was stable and present in a monomeric form after 48 hours. Additionally, NAC reacted with 8TQ as early as one minute, but did not react with QCBT7 in the 1:1 solution. In conclusion, QCBT7 is more stable than 8TQ in DMSO and 1:1 solution of DMSO and water, and NAC has little effect on QCBT7 even after 48-hour incubation. Therefore, we selected QCBT7, the more stable compound, for mechanistic studies.
Table 2.
Stability of (a) QCBT7 and (b) 8TQ in DMSO and 1:1 DMSO/water solution with or without NAC.
| (a) Percent of QCBT7 remaining | ||||
| Time / h | DMSO | DMSO+NAC | 1:1 DMSO/water | 1:1 DMSO/water+NAC |
| 2 | N/A | 100 | 100 | 100 |
| 24 | N/A | 100 | 100 | 100 |
| 48 | 100 (> 7 days) | 97.8 | 97.6 | 100 |
| (b) Percent of 8TQ remaining | ||||
| Time / h | DMSO | DMSO+NAC | 1:1 DMSO/water | 1:1 DMSO/water+NAC |
| 0.017 | 74.9 | 72.8 | 78.7 | 72.2 |
| 0.5 | 34.6 | N/A | N/A | N/A |
| 2 | 0 | 13.1 | 13.3 | 7.6 |
| 5 | N/A | 0 | 0 | 0 |
N/A: not measured at the time point. LCMS spectrums are shown in the Supplementary Figure 2S.
2.5. QCBT7 upregulates gene sets related to proteasome inhibition, UPR, glycolysis and hypoxia
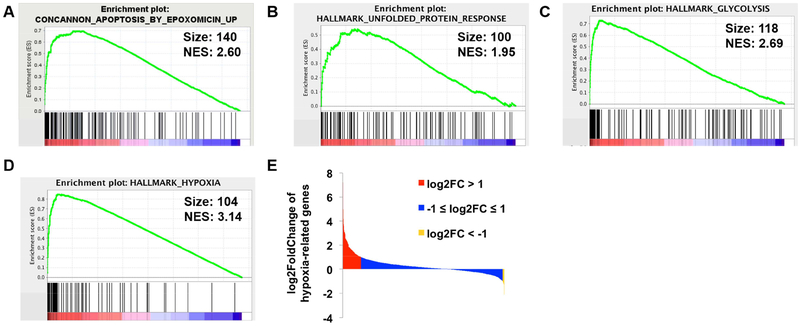
To better elucidate the potential mechanisms of action of QCBT7 in pancreatic cancer, we utilized Bru-seq technique to assess its effects on nascent transcription signatures in MIA PaCa-2 cells. After cells treated with 3.3 μM QCBT7 for 4 h, we observed 326 upregulated genes with fold change (FC) over 2 and 127 downregulated genes with FC below 0.5 (Table 3, Supplementary Figure 3S). In the set of upregulated genes, STRING analysis revealed three major biological processes: carbon metabolism, HIF-1 signaling pathway and glycolysis [21]. We performed gene set enrichment analysis (GSEA) using all the genes ranked by the log2-fold change from the Bru-seq results [22, 23]. The CONCANNON_APOPTOSIS_BY_EPOXOMICIN_UP gene set was ranked as the 13th enriched gene set of the C2 curated gene sets from the Molecular Signatures Database (MSigDB), suggesting that QCBT7 causes a similar transcriptional response to proteasome inhibitor epoxomicin (Figure 2A, Supplementary Table 1S). Epoxomicin is a potent, selective, irreversible and cell-permeable 20S proteasome inhibitor (Figure 1) [24]. Additionally, we observed the enrichment of genes involved in the unfolded protein response (UPR), glycolysis and hypoxia in the Hallmark gene sets (Figure 2B–D). Notably, a much higher number of hypoxia-related genes were upregulated (158) than downregulated (13) by QCBT7 (Figure 2E). Other gene sets indicated that QCBT7 also stimulated the transcription of genes involved in apoptotic cell death. Taken together, the results from STRING and GSEA are consistent, and we hypothesize that QCBT7 causes proteasome inhibition, hypoxic response, glycolysis and unfolded protein response.
Table 3.
Top 50 upregulated genes in QCBT7-treated MIA PaCa-2 cells.
| Gene | FC | Gene | FC | Gene | FC | Gene | FC | Gene | FC |
|---|---|---|---|---|---|---|---|---|---|
| HMOX1 | 149 | BNIP3L | 15 | BTG1 | 9 | ENO2 | 8 | ZNF395 | 6 |
| LOC344887 | 45 | ADM | 14 | PGK1 | 9 | GLA | 7 | GBE1 | 6 |
| OSGIN1 | 36 | GCLC | 13 | IL8 | 9 | TXNRD1 | 7 | PAM | 6 |
| PFKFB4 | 32 | PDK3 | 12 | ANGPTL4 | 8 | EID3 | 7 | HK2 | 6 |
| MIR210HG | 30 | TNFSF9 | 12 | NQO1 | 8 | SRXN1 | 7 | PGD | 6 |
| SLC7A11 | 27 | FAM162A | 12 | EGLN1 | 8 | P4HA1 | 6 | INSIG2 | 5 |
| DOK3 | 24 | PDK1 | 11 | DDIT4 | 8 | JUNB | 6 | DUSP5 | 5 |
| SCAND2 | 23 | ARRDC3 | 10 | DNAJB4 | 8 | PIR | 6 | CCNG2 | 5 |
| ANKRD37 | 21 | RNF122 | 9 | HILPDA | 8 | UPRT | 6 | RNF24 | 5 |
| P4HA2 | 19 | CHAC1 | 9 | C3orf58 | 8 | WSB1 | 6 | ERO1L | 5 |
FC is fold change, defined as treatment over control. HMOX1, PFKFB4, PDK1, PGK1, EGLN1, ENO2 and HK2 are involved in HIF-1 signaling pathway. PGK1, ENO2, HK2 and PGD are related to carbon metabolism. PGK1, ENO2 and HK2 affect glycolysis.
Figure 2.
QCBT7 upregulated the gene sets related to proteasome inhibition, unfolded protein response (UPR), glycolysis and hypoxic response. (A-D) GSEA enrichment plots for top enriched gene sets matched with upregulated genes from QCBT7 Bru-seq sample (FDR <= 0.001). (A) Epoxomicin, a proteasome inhibitor. (B) UPR. (C) Glycolysis. (D) Hypoxia. Size is the number of genes, and NES is the normalized enrichment score for each gene set. (E) Log2FoldChange of 1387 hypoxia-related genes from QCBT7 Bru-seq results. 1 and −1 are used as the cutoff values for upregulated and down-regulated genes.
2.6. QCBT7 has similar expression signatures to proteasome inhibitors and hypoxia inducers
To further test our hypothesis that QCBT7 inhibits proteasome activity and induces hypoxia, glycolysis and ER stress, we performed connectivity map (CMAP) analysis of QCBT7 Bru-seq results (Table 4) [25]. In the top 50 correlated perturbagens (small molecules and genetic reagents causing gene expression changes in cell lines), knockdown of PSMD1, PSMB5 and PSMA1 are listed, which encode different proteasome subunits. Additionally, proteasome inhibitors, such as MG132 and MLN-2238 (ixazomib), display similar expression signatures to QCBT7. Hypoxia-inducible factor (HIF) activators, such as VU-0418947–2 and VU-0418946–1, also show high similarity (high connectivity score), suggesting that QCBT7 induces hypoxic response. Taken together, CMAP analysis supports the hypothesis that QCBT7 causes proteasome inhibition and elicits a hypoxic response.
Table 4.
Connectivity map analysis of QCBT7. Top compound perturbagens (CP), gene over-expression perturbagens (OE), gene knock-down perturbagens (KO) and perturbational class member (PCL) are listed.
| Type | Name | Description | Pc_selection |
|---|---|---|---|
| CP | VU-0418947–2 | hypoxia inducible factor activator | 99.89 |
| CP | VU-0418946–1 | hypoxia inducible factor activator | 99.86 |
| CP | MG-132 | proteasome inhibitor | 99.58 |
| CP | MLN-2238 | proteasome inhibitor | 99.3 |
| OE | NFE2L2 | basic leucine zipper proteins, nuclear factor (erythroid-derived 2)-like 2 | 99.91 |
| KD | PSMD1 | Proteasome subunits, proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 99.82 |
| KD | PSMB5 | Proteasome subunits, proteasome (prosome, macropain) subunit, beta type, 5 | 99.61 |
| KD | PSMA1 | Proteasome subunits, proteasome (prosome, macropain) subunit, alpha type, 1 | 99.45 |
| PCL | HIF activator | N/A | 100 |
| PCL | Proteasome inhibitor | N/A | 99.72 |
Pc_selection is the connectivity score (from −100 to 100). A positive higher score means more positive connection between the perturbagen and QCBT7.
2.7. QCBT7 and MG132 have similar transcriptional profiles
MG132, a peptide-aldehyde proteasome inhibitor, blocks the proteolytic activity of the 26S proteasome by covalently binding to the 20S catalytic subunit [26]. Due to its lack of specificity, it has only been used as a research tool to study the ubiquitin-proteasome pathway. In a separate study, we performed Bru-seq of MG132-treated HeLa cancer cells. Although a different cell line, we still identified similarity in the CONCANNON_APOPTOSIS_BY_EPOXOMICIN_UP gene set and WINTER_HYPOXIA_METAGENE gene set between QCBT7 and MG132.
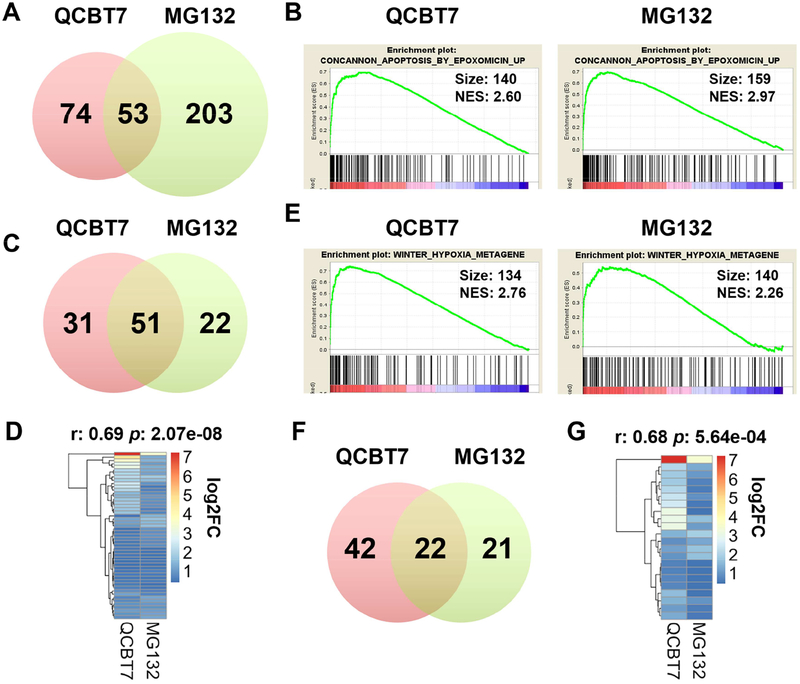
QCBT7 and MG132 share 53 common upregulated enriched gene sets (FDR < 0.001) from C2 curated gene sets (Figure 3A, Supplementary Table 2S). Treatments with QCBT7 and MG132 induced 51 common genes contributing to the enrichment of the CONCANNON_APOPTOSIS_BY_EPOXOMICIN_UP gene set (Figure 3B–C). Moreover, their transcript expressions were strongly correlated (r = 0.69, p = 2.07e-08) (Figure 3D). STRING analysis indicates that the 51 common upregulated genes are involved in apoptotic signaling pathway, unfolded protein response and ER stress. Furthermore, we observed 22 common genes in the WINTER_HYPOXIA_METAGENE gene set from both QCBT7- and MG132-treated samples, and their nascent transcript expressions also had a strong correlation (r = 0.68, p = 5.64e-04) (Figure 3E–G). These 22 genes are related to HIF-1 signaling pathway, glycolysis and cellular stress.
Figure 3.
QCBT7 and MG132 have common enriched gene sets in proteasome inhibition pathway and hypoxic response. (A) Venn diagram of all upregulated enriched gene sets between QCBT7 and MG132 (FDR < 0.001). (B) GSEA enrichment plots of CONCANNON_APOPTOSIS_BY_EPOXOMICIN_UP gene set in QCBT7 and MG132 Bru-seq samples. (C) 51 genes in common from the EPOXOMICIN gene set. (D) Heatmap of the 51 common genes using log2FC (r = 0.69, p = 2.07e-08). (E) GSEA enrichment plots of WINTER_HYPOXIA_METAGENE gene set in two samples. (F) 22 genes in common from HYPOXIA gene set. (G) Heatmap of the 22 common genes using log2FC (r = 0.68, p = 5.64e-04).
Together, these results demonstrate that QCBT7 is similar to the proteasome inhibitor MG132 at the transcription level and regulates redox signaling and ER stress. GCLM, HMOX1, NQO1 and SQSTM1, from the 51 common genes in the EPOXOMICIN gene set, are involved in redox signaling pathways. GCLM encodes a glutamate-cysteine ligase modifier subunit, which is a rate-limiting enzyme for glutathione synthesis. HMOX1 (heme oxygenase 1) responds to cellular oxidative stress and catalyzes cleavage of heme in order to maintain redox balance [27]. NQO1 (NAD(P)H quinone dehydrogenase 1) can also prevent oxidative stress via reducing quinones to hydroquinones. In addition, SQSTM1 (sequestosome 1) mediates the activation of NF-κB signaling together with TNF receptor-associated factor 6, and is a positive regulator of Nrf2 signaling [28]. CEBPB, DDIT3 and PPP1R15A are three ER stress-related genes induced by both QCBT7 and MG132. Proteasome inhibitors may stimulate CEBPB, a transcription factor that interacts with DDIT3/CHOP, an ER stress biomarker, to induce cellular stress and cause cancer cell death [27, 29]. PPP1R15A (GADD34) sensitizes cells to proteasome inhibitors by promoting ER stress, reactive oxygen species production and autophagy [30].
Proteasome inhibitors can regulate redox homeostasis and induce ER stress [1, 27]. MG132 has been shown to activate the Nrf2-ARE signaling pathway that protects cells from oxidative stress [31]. The inhibition of proteasome activity caused by MG132 leads to the unfolded protein response and apoptosis [26, 32]. The findings in our study are consistent with previous reports on the mechanisms of action of MG132. Moreover, they are similar to those of QCBT7 implied from the STRING and GSEA results.
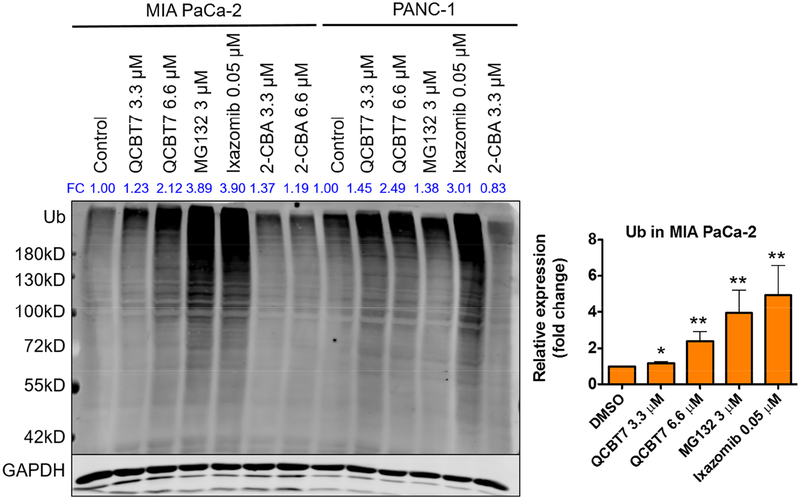
2.8. QCBT7 induces the accumulation of ubiquitylated proteins
To investigate whether QCBT7 inhibits proteasome activity as suggested from Bru-seq analysis, immunoblot was performed using the antibody against the ubiquityl group of ubiquitylated proteins in QCBT7-treated pancreatic cancer cells. Ubiquitylated proteins are normally subjected to proteasome-mediated degradation, but inhibition of the proteasome results in the accumulation of slower-migrating ubiquitylated proteins. QCBT7 treatment resulted in the dose-dependent accumulation of ubiquitylated proteins in both MIA PaCa-2 and PANC-1 cells (Figure 4). Similarly, both MG132 and ixazomib caused a significant accumulation of ubiquitylated proteins (p < 0.005). QCBT7 showed a stronger accumulating effect than MG132 in PANC-1, while ixazomib was the most potent inducer of ubiquitylated proteins in both cell lines. These results support the hypothesis that QCBT7 has proteasome inhibitory activity. Ixazomib, which reversibly and selectively bind to the proteasome beta 5 subunit (PSMB5) of the 20S catalytic subunit, has been approved as an orally bioavailable proteasome inhibitor for the treatment of multiple myeloma and is currently being tested in combination with other standard-of-care treatments in clinical trials for solid tumors [33, 34].
Figure 4.
QCBT7 causes the accumulation of ubiquitylated proteins in MIA PaCa-2 and PANC-1 cell lines, similar to MG132 and ixazomib. Cells were treated with QCBT7, MG132, ixazomib and 2-chlorobenzoic acid (2-CBA) for 24 h. MG132 and ixazomib were used as positive controls. FC: fold change, relative expression of the protein normalized to GAPDH expression. Data shown are representative of 3 independent experiments. Error bars represent standard deviation. * denotes p < 0.05, ** denotes p < 0.005 and *** denotes p < 0.0005 compared with DMSO control.
We also tested 2-chlorobenzoic acid (2-CBA), a potential by-product of QCBT7 in the cellular reducing environment (Supplementary Scheme 1S). 2-CBA did not cause accumulation of ubiquitylated proteins, suggesting that QCBT7 or 8TQ is the active proteasome inhibitory molecules.
2.9. QCBT7 increases the protein expression of genes related to glycolysis, ER stress and hypoxia in pancreatic cancer cells
To validate proteins and signaling pathways identified with the Bru-seq analysis, we performed immunoblot of select genes related to glycolysis, ER stress and hypoxia in MIA PaCa-2 and PANC-1 cells.
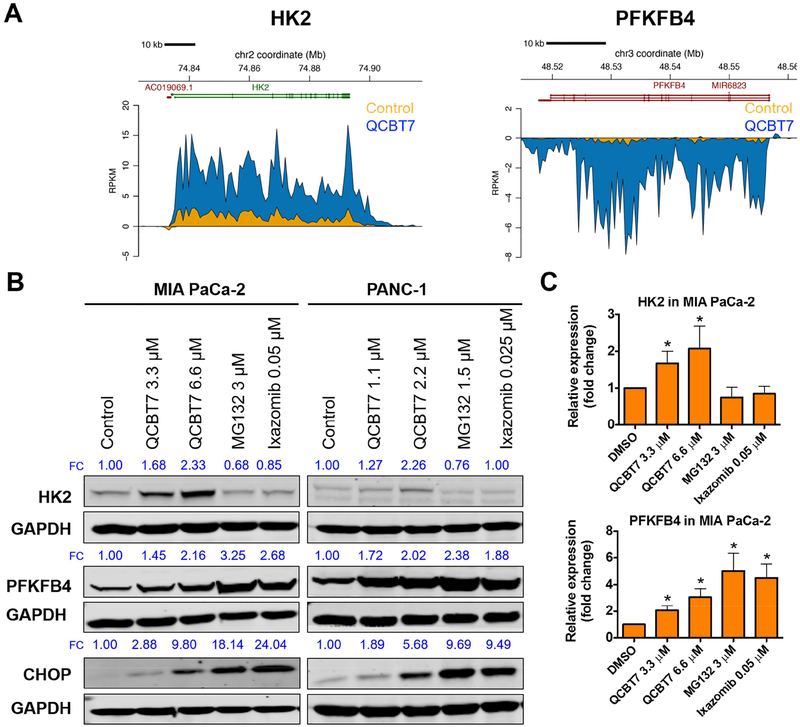
HK2 (hexokinase 2) and PFKFB4 (6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 4) are two glycolytic genes among the top 50 upregulated genes (Figure 5A, Table 3). QCBT7 elevated HK2 and PFKFB4 protein expression dose-dependently in both MIA PaCa-2 and PANC-1 cells (Figure 5B), and it significantly increased their protein expression at 3.3 μM (Figure 5C, p < 0.05). HK2 and PFKFB4 are related to hexose metabolic processes and their protein expression increases under hypoxic conditions [35].
Figure 5.
QCBT7 increases nascent RNA and protein expression of genes related to glycolysis and ER stress in pancreatic cancer cells. (A) Trace diagrams of nascent RNA expression of HK2 and PFKFB4. Blue represents QCBT7 treatment and yellow represents the DMSO control. (B) Immunoblot of HK2, PFKFB4 and CHOP in MIA PaCa-2 and PANC-1 cells after 24 h treatment. FC: fold change, relative expression of the protein normalized to GAPDH expression. Data shown are representative of 3 independent experiments. (C) Quantification of relative expression of each protein in 3 independent experiments. Data are reported as mean ± standard deviation. * denotes p < 0.05 compared with DMSO control.
QCBT7 also induced CHOP expression, which is a protein biomarker for ER stress (Figure 5B). Moreover, MG132 and ixazomib significantly increased PFKFB4 and CHOP expression, but not HK2, suggesting that they also have some differences in the downstream signaling due to potential distinct binding sites of the proteasome.
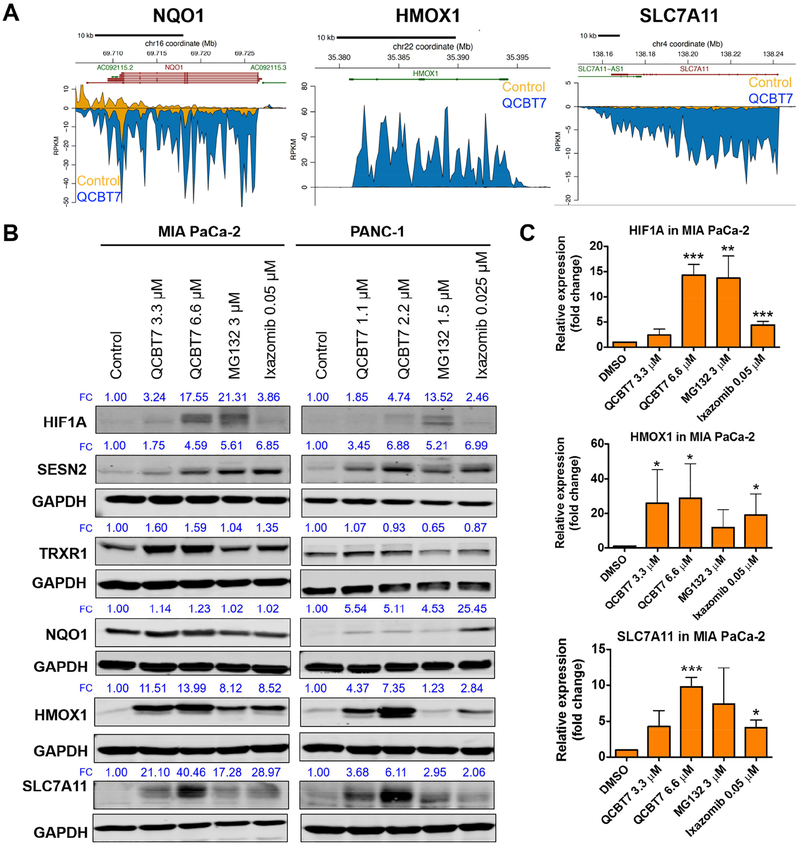
Furthermore, QCBT7 increased the expression of hypoxia-related proteins, including HIF1A, SESN2, TRXR1, NQO1, HMOX1 and SLC7A11 (Figure 6). At the nascent transcript level, NQO1, HMOX1 and SLC7A11 were dramatically upregulated by QCBT7 (Figure 6A). HMOX1 and SLC7A11 protein expression also increased significantly by 6.6 μM QCBT7, and NQO1 showed dramatic increase in protein expression in PANC-1 but not MIA PaCa-2 cells (Figure 6B–C). HIF1A, SESN2 and TRXR1 were upregulated by QCBT7 in both cell lines, and HIF1A and SESN2 showed a dose-dependent increase (Figure 6B). MG132 and ixazomib had similar effects on HIF1A, SESN2, HMOX1 and SLC7A11 protein levels (Figure 6B–C). SESN2 (sestrin 2) is inducible by hypoxia and oxidative stress conditions to balance metabolic homeostasis [36]. HMOX1 is a downstream target of HIF1A, and is sensitive to cellular redox signaling [37]. In addition, hypoxia affects glutathione metabolism [35]. TRXR1 (thioredoxin reductase 1) was elevated by QCBT7 to possibly balance the increase of HIF1A and maintain redox homeostasis. SLC7A11 (solute carrier family 7 member 11), which regulates glutamate transport, also showed an increase expression with the treatment [38].
Figure. 6.
QCBT7 increases nascent RNA and protein expression of genes related to hypoxia in pancreatic cancer cells. (A) Trace diagrams of nascent RNA expression of NQO1, HMOX1 and SLC7A11. Blue represents QCBT7 treatment and yellow represents the DMSO control. (B) Immunoblot of HIF1A, SESN2, TRXR1, NQO1, HMOX1 and SLC7A11 in MIA PaCa-2 and PANC-1 cells after 24 h treatment. FC: fold change, relative expression of the protein normalized to GAPDH expression. Data shown are representative of 3 independent experiments. (C) Quantification of relative expression of each protein in 3 independent experiments. Data are reported as mean ± standard deviation. * denotes p < 0.05, ** denotes p < 0.005 and *** denotes p < 0.0005 compared with DMSO control.
Our immunoblot results of MG132 and ixazomib corroborate the previously reported mechanisms of proteasome inhibitors in inducing hypoxia and ER stress. MG132 increases the protein level of SESN2 due to the accumulation of ubiquitylated SESN2, and it is dependent on Nrf-2 [36]. MG132, ixazomib and bortezomib increase HMOX1 and CHOP expression level due to Nrf2 signaling and ER stress [27, 31–34, 39]. Compared with MG132 and ixazomib, QCBT7 also induces the expression of most proteins, such as PFKFB4, CHOP, HIF1A and HMOX1, supporting their similar mechanisms (Figure 5–6). The signaling pathways affected by QCBT7 have a strong correlation with its proteasome inhibition activity.
Overall, HK2, PFKFB4, CHOP, HIF1A, SESN2, TRXR1, NQO1, HMOX1 and SLC7A11 proteins are robustly upregulated in pancreatic cancer cells by QCBT7. Most proteins show a dose-dependent increase in response to QCBT7, MG132 and ixazomib. These results indicate that QCBT7 induces hypoxia, ER stress and glycolysis, and finally leads to cancer cell death, similar to MG132 and ixazomib.
2.10. PFKFB4 is a potential biomarker to study proteasome inhibition
We identified that protein expression of PFKFB4 was significantly elevated by MG132, ixazomib and QCBT7. Additionally, QCBT7 increased PFKFB4 protein level back in the PFKFB4 knockdown MIA PaCa-2 cells (Supplementary Figure 6S). Our results show for the first time that PFKFB4 transcript and protein levels can be used to evaluate the response to proteasome inhibitors in pancreatic cancer. PFKFB4 is essential for cellular response to hypoxia, glucose metabolism and cell survival, and its depletion in p53-deficient prostate cancer cells inhibits tumor growth and cell survival [35, 40–42]. PFKFB4 is also recognized as an autophagy regulator, relating to cancer cell death [43]. Most importantly, it may be used as a new response regulatory protein to study the downstream signaling of MG132, ixazomib and other proteasome inhibitors.
3. Conclusions
In this study, the STRING, GSEA and CMAP analyses of the novel compound QCBT7 reveal that it induces transcriptional responses related to proteasome inhibition, hypoxia, ER stress and glycolysis. Moreover, QCBT7 blocked the degradation of ubiquitylated proteins, similar to MG132 and ixazomib. We demonstrated that PFKFB4, HK2, CHOP, HIF1A, SESN2, TRXR1, NQO1, HMOX1 and SLC7A11 protein levels increased upon QCBT7 treatment. MG132 and ixazomib also elevated PFKFB4, CHOP, HIF1A, SESN2, HMOX1 and SLC7A11 protein levels. Additionally, for the first time we identify PFKFB4 as a potential biomarker to monitor proteasome inhibition. In conclusion, QCBT7 shares similarity with proteasome inhibitors at both the transcript and protein levels, and blocks the protein degradation pathway. Importantly, our study reveals and confirms the association between hypoxia, glycolysis, ER stress and proteasome inhibition in pancreatic cancer cells.
4. Experimental section
4.1. Chemical synthesis
All commercial reagents and anhydrous solvents were purchased and used without purification, unless specified. Column chromatography was performed using a Biotage chromatography system on Biotage or Silicycle normal phase silica gel columns. NMR spectra were recorded on a Bruker instrument (300 or 500 MHz). Chemical shifts (δ) are reported in parts per million (ppm) units relative to residual undeuterated solvent. Mass spectra were obtained on a Shimadzu LCMS-2020 liquid chromatography mass spectrometer using the electron spray ionization (ESI) method. HPLC was used to determine purity of biologically tested compounds with a Shimadzu HPLC Test Kit C18 column (3 μm, 4.6 × 50 mm) under the following gradient elution condition: mobile phase A of acetonitrile/water (10–95%) or mobile phase B of methanol/water (10–95%). The purity was established by integration of the areas of major peaks detected at 254 nm and all final products were >95% pure as determined by HPLC/MS and 1H NMR.
8-Quinolinethiol hydrochloride (1 mmol), 8-hydroxylquinoline (1 mmol) or 8-aminoquinoline (1 mmol) was dissolved in 5 ml of DMF, then substituted benzoyl chloride (1 mmol) or substituted benzyl chloride (1 mmol), and potassium carbonate (2 mmol) were added sequentially. The mixed solution was stirred for 6–12 h at room temperature. Upon completion of the reaction, DMF was evaporated under reduced pressure. The residue was extracted with ethyl acetate (3 × 10 mL). The combined organic layers were washed with saturated aqueous NH4Cl solution and brine, dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure. The resulting mixture was purified by silica column chromatography to give products 3a-3u.
4.1.1. S-(quinolin-8-yl) 4-fluorobenzothioate (3a)
Light green solid; yield: 91%; 1H NMR (300 MHz, Chloroform-d) δ 9.12 – 8.98 (m, 1H), 8.31 – 8.22 (m, 1H), 8.17 (dd, J = 8.6, 5.4 Hz, 2H), 8.11 (d, J = 7.2 Hz, 1H), 7.65 (t, J = 7.8 Hz, 1H), 7.49 (dd, J = 8.2, 4.2 Hz, 1H), 7.16 (t, J = 8.5 Hz, 2H), 7.09 (d, J = 8.5 Hz, 1H); 13C NMR (126 MHz, Chloroform-d) δ 187.92, 167.08, 165.12, 150.95, 147.72, 137.66, 136.79, 133.23, 130.38, 130.29, 129.08, 127.68, 126.53, 121.75, 115.88, 115.70. HRMS (ESI): m/z calculated for C16H10FNOS [M+H]+, 284.0540; found, 284.0539. LCMS: m/z 284 [M+H]+, purity: 100%.
4.1.2. S-(quinolin-8-yl) 4-chlorobenzothioate (3b)
Light green solid; yield: 89%; 1H NMR (500 MHz, Chloroform-d) δ 9.00 (s, 1H), 8.07 (ddt, J = 75.2, 60.4, 13.9 Hz, 5H), 7.69 – 7.32 (m, 4H); 13C NMR (126 MHz, Chloroform-d) δ 188.26, 150.95, 147.66, 139.95, 137.62, 136.79, 135.24, 130.44, 129.09, 128.95, 127.52, 126.54, 121.77. HRMS (ESI): m/z calculated for C16H10ClNOS [M+H]+, 300.0244; found, 300.0244. LCMS: m/z 300 [M+H]+, purity: 100%.
4.1.3. S-(quinolin-8-yl) 2-chlorobenzothioate (3c)
Light yellow solid; yield: 76%; 1H NMR (400 MHz, Chloroform-d) δ 9.01 (dd, J = 4.3, 1.8 Hz, 1H), 8.27 – 8.14 (m, 2H), 8.00 (dd, J = 7.6, 1.7 Hz, 1H), 7.94 (dd, J = 8.2, 1.3 Hz, 1H), 7.69 – 7.61 (m, 1H), 7.52 – 7.39 (m, 3H), 7.36 (td, J = 7.2, 1.4 Hz, 1H); 13C NMR (101 MHz, Chloroform-d) ō 188.24, 149.79, 146.14, 136.05, 135.77, 131.38, 130.18, 129.84, 129.26, 128.65, 127.97, 126.94, 125.64, 125.56, 120.76. HRMS (ESI): m/z calculated for C16H10ClNOS [M+H]+, 300.0244; found, 300.0246. LCMS: m/z 300 [M+H]+, purity: 100%.
4.1.4. S-(quinolin-8-yl) benzothioate (3d)
Light yellow solid; yield: 83%; 1H NMR (300 MHz, Chloroform-d) δ 9.04 (d, J = 4.6 Hz, 1H), 8.30 – 8.20 (m, 1H), 8.15 (q, J = 6.9, 6.1 Hz, 2H), 7.95 (dd, J = 12.4, 7.8 Hz, 1H), 7.73 – 7.55 (m, 2H), 7.56 – 7.46 (m, 3H), 7.43 (d, J = 8.4 Hz, 1H); 13C NMR (126 MHz, Chloroform-d) ō 189.40, 150.91, 147.78, 137.62, 136.73, 133.53, 130.25, 129.05, 128.63, 127.76, 126.52, 121.70. HRMS (ESI): m/z calculated for C16H11NOS [M+H]+, 266.0634; found, 266.0634. LCMS: m/z 266 [M+H]+, purity: 95.0%.
4.1.5. 8-(Benzylthio)quinoline (3e)
Light yellow solid; yield: 94%; 1H NMR (300 MHz, Chloroform-d) ō 8.98 (d, J = 4.2 Hz, 1H), 8.24 – 8.11 (m, 1H), 7.60 (d, J = 8.0 Hz, 1H), 7.52 (d, J = 7.2 Hz, 1H), 7.49 – 7.40 (m, 4H), 7.31 (t, J = 6.9 Hz, 2H), 7.26 (d, J = 6.2 Hz, 1H), 4.33 (s, 2H). LCMS: m/z 252 [M+H]+, purity: 99.8%.
4.1.6. S-(quinolin-8-yl) 4-nitrobenzothioate (3f)
Yellow solid; yield: 96%; 1H NMR (300 MHz, Chloroform-d) ō 8.91 (dq, J = 4.2, 2.1 Hz, 1H), 8.36 – 8.18 (m, 4H), 8.15 (q, J = 3.2, 2.8 Hz, 1H), 8.09 – 7.93 (m, 1H), 7.68 – 7.56 (m, 1H), 7.47 (dd, J = 8.5, 4.5 Hz, 1H), 7.36 (d, J = 8.2 Hz, 1H); 13C NMR (126 MHz, Chloroform-d) ō 210.25, 161.22, 160.09, 151.07, 137.60, 136.99, 131.22, 130.86, 128.73, 126.64, 123.88, 123.60, 121.97. HRMS (ESI): m/z calculated for C16H10N2O3S [M+H]+, 311.0485; found, 311.0485. LCMS: m/z 311 [M+H]+, purity: 99.6%.
4.1.7. S-(quinolin-8-yl) 2-fluorobenzothioate (3g)
Light green solid; yield: 78%; 1H NMR (300 MHz, Chloroform-d) ō 9.04 (s, 1H), 8.22 (d, J = 8.3 Hz, 1H), 8.07 (s, 1H), 7.94 (d, J = 7.5 Hz, 1H), 7.66 (d, J = 8.0 Hz, 1H), 7.60 (s, 1H), 7.52 (dd, J = 8.4, 4.2 Hz, 1H), 7.44 (d, J = 7.7 Hz, 1H), 7.27 (s, 1H), 7.19 (s, 1H); 13C NMR (126 MHz, Chloroform-d) ō 186.30, 159.53, 150.94, 147.59, 137.59, 136.75, 134.51, 130.47, 130.15, 129.05, 126.52, 124.19, 121.74, 116.98, 116.80. HRMS (ESI): m/z calculated for C16H10FNOS [M+H]+, 284.0540; found, 284.0538. LCMS: m/z 285 [M+H]+, purity: 98.8%.
4.1.8. Quinolin-8-yl 4-chlorobenzoate (3h)
White solid; yield:83%; 1H NMR (300 MHz, Chloroform-d) ō 8.94 (s, 1H), 8.27 (t, J = 11.8 Hz, 3H), 7.80 (d, J = 7.3 Hz, 1H), 7.60 (d, J = 7.5 Hz, 2H), 7.53 (d, J = 8.7 Hz, 3H). LCMS: m/z 284 [M+H]+, purity: 99.6%.
4.1.9. Quinolin-8-yl 4-fluorobenzoate (3i)
White solid; yield: 80%; 1H NMR (300 MHz, Chloroform-d) ō 8.90 (d, J = 4.1 Hz, 1H), 8.49 – 8.33 (m, 2H), 8.22 (d, J = 8.3 Hz, 1H), 7.79 (d, J = 6.2 Hz, 1H), 7.68 – 7.53 (m, 2H), 7.45 (dd, J = 8.6, 4.3 Hz, 1H), 7.32 – 7.13 (m, 2H). LCMS: m/z 268 [M+H]+, purity: 100%.
4.1.10. Quinolin-8-yl 2-chlorobenzoate (3j)
White solid; yield: 71%; 1H NMR (300 MHz, Chloroform-d) ō 8.93 (d, J = 4.6 Hz, 1H), 8.40 (d, J = 7.7 Hz, 1H), 8.30 – 8.16 (m, 1H), 7.79 (d, J = 7.1 Hz, 1H), 7.69 – 7.52 (m, 4H), 7.51 – 7.40 (m, 2H). LCMS: m/z 284 [M+H]+, purity: 97.0%.
4.1.11. Quinolin-8-yl benzoate (3k)
White solid; yield: 84%; 1H NMR (300 MHz, Chloroform-d) ō 8.92 (s, 1H), 8.37 (d, J = 7.7 Hz, 2H), 8.23 (d, J = 8.5 Hz, 1H), 7.79 (d, J = 6.8 Hz, 1H), 7.66 (d, J = 7.4 Hz, 2H), 7.57 (q, J = 7.5, 6.8 Hz, 4H), 7.45 (dd, J = 8.5, 4.1 Hz, 1H). LCMS: m/z 250 [M+H]+, purity: 100%.
4.1.12. 8-(Benzyloxy)quinoline (3l)
White solid; yield: 86%; 1H NMR (300 MHz, Chloroform-d) ō 8.88 (s, 1H), 8.04 (s, 1H), 7.42 (m, 8H), 6.94 (s, 1H), 5.49 – 5.25 (m, 2H). LCMS: m/z 236 [M+H]+, purity: 100%.
4.1.13. Quinolin-8-yl 4-nitrobenzoate (3m)
Yellow solid; yield: 90%; 1H NMR (300 MHz, Chloroform-d) ō 8.89 (s, 1H), 8.54 (d, J = 8.3 Hz, 2H), 8.41 (d, J = 8.6 Hz, 2H), 8.25 (d, J = 8.3 Hz, 1H), 7.83 (s, 1H), 7.62 (s, 2H), 7.55 – 7.44 (m, 1H). LCMS: m/z 295 [M+H]+, purity: 98.4%.
4.1.14. Quinolin-8-yl 2-fluorobenzoate (3n)
White solid; yield:76%; 1H NMR (300 MHz, Chloroform-d) ō 9.01 (d, J = 4.1 Hz, 1H), 8.38 (t, J = 7.7 Hz, 1H), 8.29 (d, J = 8.3 Hz, 1H), 7.85 (d, J = 6.0 Hz, 1H), 7.66 (d, J = 4.9 Hz, 3H), 7.60 – 7.47 (m, 1H), 7.45 – 7.28 (m, 2H). LCMS: m/z 268 [M+H]+, purity: 97.6%.
4.1.15. 4-Chloro-N-(quinolin-8-yl)benzamide (3o)
White solid; yield: 80%; 1H NMR (300 MHz, Chloroform-d) ō 10.75 (s, 1H), 8.93 (dd, J = 7.1, 2.0 Hz, 1H), 8.88 (dd, J = 4.3, 1.7 Hz, 1H), 8.23 (dd, J = 8.2, 1.7 Hz, 1H), 8.10 – 8.00 (m, 2H), 7.63 (s, 1H), 7.61 – 7.58 (m, 1H), 7.58 – 7.53 (m, 2H), 7.53 – 7.48 (m, 1H). LCMS: m/z 283 [M+H]+, purity: 99.5%.
4.1.16. 4-Fluoro-N-(quinolin-8-yl)benzamide (3p)
White solid; yield: 83%; 1H NMR (300 MHz, Chloroform-d) ō 10.74 (s, 1H), 8.93 (d, J = 7.8 Hz, 1H), 8.88 (dd, J = 4.4, 1.6 Hz, 1H), 8.29 – 8.19 (m, 1H), 8.13 (dd, J = 8.6, 5.4 Hz, 2H), 7.72 – 7.56 (m, 2H), 7.52 (dd, J = 8.3, 4.2 Hz, 1H), 7.36 – 7.20 (m, 2H). LCMS: m/z 267 [M+H]+, purity: 99.4%.
4.1.17. 2-Chloro-N-(quinolin-8-yl)benzamide (3q)
White solid; yield: 72%; 1H NMR (300 MHz, Chloroform-d) ō 10.51 (s, 1H), 8.98 (dd, J = 6.9, 2.1 Hz, 1H), 8.82 (dd, J = 4.2, 1.7 Hz, 1H), 8.21 (dd, J = 8.2, 1.7 Hz, 1H), 7.89 – 7.80 (m, 1H), 7.62 (d, J = 7.1 Hz, 1H), 7.61 (s, 1H), 7.56 – 7.39 (m, 4H). LCMS: m/z 283 [M+H]+, purity: 100%.
4.1.18. N-(quinolin-8-yl)benzamide (3r)
White solid; yield: 83%; 1H NMR (300 MHz, Chloroform-d) ō 10.79 (s, 1H), 8.97 (dd, J = 7.4, 1.6 Hz, 1H), 8.88 (dd, J = 4.3, 1.7 Hz, 1H), 8.22 (dd, J = 8.3, 1.7 Hz, 1H), 8.19 – 8.05 (m, 2H), 7.69 – 7.55 (m, 5H), 7.51 (dd, J = 8.3, 4.2 Hz, 1H). LCMS: m/z 249 [M+H]+, purity: 100%.
4.1.19. N-benzylquinolin-8-amine (3s)
White solid; yield: 90%; 1H NMR (300 MHz, Chloroform-d) ō 8.66 (dd, J = 4.3, 1.8 Hz, 1H), 8.02 (dd, J = 8.4, 1.7 Hz, 1H), 7.45 (dd, J = 8.2, 1.8 Hz, 1H), 7.41 – 7.18 (m, 7H), 6.74 (dd, J = 7.5, 1.8 Hz, 1H), 4.42 (d, J = 7.4 Hz, 2H). LCMS: m/z 235 [M+H]+, purity: 100%.
4.1.20. 4-Nitro-N-(quinolin-8-yl)benzamide (3t)
Yellow solid; yield: 92%; 1H NMR (300 MHz, Chloroform-d) ō 10.85 (s, 1H), 8.97 – 8.91 (m, 1H), 8.91 – 8.85 (m, 1H), 8.42 (d, J = 8.3 Hz, 2H), 8.26 (d, J = 8.6 Hz, 3H), 7.64 (d, J = 4.9 Hz, 2H), 7.54 (dd, J = 8.3, 4.2 Hz, 1H). LCMS: m/z 294 [M+H]+, purity: 98.8%.
4.1.21. 2-Fluoro-N-(quinolin-8-yl)benzamide (3u)
White solid; yield: 74%; 1H NMR (300 MHz, Chloroform-d) ō 10.81 (d, J = 12.2 Hz, 1H), 8.63 (dd, J = 7.0, 2.1 Hz, 1H), 8.52 (dd, J = 4.2, 1.7 Hz, 1H), 7.84 (td, J = 8.3, 1.8 Hz, 2H), 7.26 – 7.21 (m, 2H), 7.19 (s, 1H), 7.12 (dd, J = 8.3, 4.2 Hz, 1H), 7.02 – 6.94 (m, 1H), 6.89 (d, J = 3.1 Hz, 1H). LCMS: m/z 267 [M+H]+, purity: 99.2%.
4.2. Biological evaluation
4.2.1. Cell Culture
MIA PaCa-2, PANC-1, HCT 116 and KYSE-70 cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% FBS (Atlanta Biologicals). Cells were grown at 37 °C in a humidified atmosphere of 5% CO2. All cell lines used were maintained in culture under 35 passages and tested regularly for mycoplasma contamination using PlasmoTest Kit (InvivoGen, San Diego, CA).
4.2.2. MTT assay
Cytotoxicity of compounds was determined by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as previously described [44]. Briefly, cells were seeded in 96-well tissue culture plates and treated with compounds or vehicle for 72 h after overnight attachment. 20 μL MTT (3 mg/mL) was added, and the cells were incubated with MTT for 3 h. DMSO was added after removing the media and absorbance was measured at 570 nm.
4.2.3. Immunoblot
After overnight attachment in 6-well tissue culture plates, cells were treated with QCBT7 (3.3 μM or 6.6 μM), MG132 (3 μM), ixazomib (0.05 μM), or 2-CBA (3.3 μM or 6.6 μM). After 24 h treatment, cells were washed with 1×DPBS and lysed using RIPA lysis buffer in the presence of 1×protease inhibitor cocktail (Sigma-Aldrich) and 1×phosphatase inhibitor cocktail (VWR International). Cell lysates were vortexed and centrifuged at 12,000×g for 15 min at 4 °C. Protein concentration of the samples was measured using a BCA protein assay (Thermo Fisher Scientific) and equal amounts of total proteins were resolved on 10% or 8% polyacrylamide via SDS-PAGE. The separated proteins were transferred onto PVDF membranes (Thermo Fisher Scientific, 0.45 μm) and blocked in 5% milk for 1 h at room temperature. The membranes were probed with primary antibodies in 5% milk or 5% BSA at 4 °C overnight with recommended dilution (HK2: Cell Signaling Technology (CST) #2106, PFKFB4: Thermo Fisher PA5–15475, CHOP: CST #2895, HIF1A: CST #3716, SESN2: CST #8487, TRXR1: CST #15140, NQO1: CST #3187, HMOX1: CST #5061, SLC7A11: CST #12691, Ub: CST #3936). Secondary antibodies were added at 1:6000 dilution (Thermo Fisher Scientific, DyLight 800, #SA5–35571 and # SA5–35521) and membranes were incubated for 1 h at room temperature. Finally, membranes were imaged using Odyssey Imaging Systems (LI-COR Biosciences).
4.2.4. PFKFB4 siRNA knock down study
siRNA targeting PFKFB4 was purchased from Thermo Fisher Scientific (#4427038). 8 million MIA PaCa-2 cells were seeded in 6-well tissue culture plates. After overnight attachment, 30 pmol and 60 pmol siPFKFB4 were added to MIA PaCa-2 cells following the manufacturer’s protocol. QCBT7 (3.3 μM) was added to the cells at the same time. Cells were collected after 24 h treatment for later immunoblot.
4.3. Bru-seq experiment and bioinformatics analysis
Bru-seq experiment for nascent RNA measurement was performed as previously reported [44]. Briefly, MIA PaCa-2 cells were treated with QCBT7 for 4 h. Bromouridine was added to the cells at a final concentration of 2 mM in the last 30 min of the treatment. Cells were then collected in Trizol and total RNA was isolated. The bromouridine-containing RNA population was further isolated and sequenced. Sequencing reads were mapped to the hg19 reference genome. Further analysis was conducted using DESeq, GSEA, STRING and CMAP. R (version 3.3.2) was used to calculate Pearson correlation and make related figures.
4.4. Statistics
Results were shown as mean ± standard deviation as stated in the figure or table legends. Unpaired t-test was performed for data analysis.
Supplementary Material
Scheme 1.
Preparation of QCBT7 derivatives.
Highlights.
QCBT7 was identified as a lead proteasome inhibitor among the 21 novel analogs.
QCBT7 shares a similar transcriptomic profile with MG132 and epoxomicin.
QCBT7 blocks the degradation of ubiquitylated proteins.
QCBT7 induces hypoxia, protein stress and glycolysis at both RNA and protein levels.
PFKFB4 is a potential new biomarker of response to proteasome inhibitors.
Acknowledgement
This work was supported by NIH grant R01 [CA188252] and a grant from the University of Michigan Forbes Institute for Cancer Discovery. We thank A. Shergalis for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interest.
Appendix A. Supplementary data
Supplementary information is in a separate file.
References
- [1].Manasanch EE, Orlowski RZ, Proteasome inhibitors in cancer therapy, Nat Rev Clin Oncol, 14 (2017) 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kaplan GS, Torcun CC, Grune T, Ozer NK, Karademir B, Proteasome inhibitors in cancer therapy: Treatment regimen and peripheral neuropathy as a side effect, Free Radic Biol Med, 103 (2017) 1–13. [DOI] [PubMed] [Google Scholar]
- [3].Goldberg AL, Development of proteasome inhibitors as research tools and cancer drugs, J Cell Biol, 199 (2012) 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Johnson DE, The ubiquitin-proteasome system: Opportunities for therapeutic intervention in solid tumors, Endocr Relat Cancer, 22 (2015) T1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cottini F, Guidetti A, Prada CP, Hideshima T, Maglio M, Varga C, Chauhan D, Laubach J, Anderson KC, Richardson PG, Resistance to proteasome inhibitors in multiple myeloma, in: Dou QP (Ed.) Resistance to proteasome inhibitors in cancer: Molecular mechanisms and strategies to overcome resistance, Springer International Publishing, Cham, 2014, pp. 47–80. [Google Scholar]
- [6].Li D, Xie K, Wolff R, Abbruzzese JL, Pancreatic cancer, Lancet, 363 (2004) 1049–1057. [DOI] [PubMed] [Google Scholar]
- [7].Semaan A, Maitra A, Pancreatic cancer in 2017: Rebooting pancreatic cancer knowledge and treatment options, Nat Rev Gastroenterol Hepatol, (2018). [DOI] [PubMed] [Google Scholar]
- [8].Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, Neale RE, Tempero M, Tuveson DA, Hruban RH, Neoptolemos JP, Pancreatic cancer, Nat Rev Dis Primers, 2 (2016) 16022. [DOI] [PubMed] [Google Scholar]
- [9].Aroldi F, Bertocchi P, Savelli G, Rosso E, Zaniboni A, Pancreatic cancer: New hopes after first line treatment, World J Gastrointest Oncol, 8 (2016) 682–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lambert A, Gavoille C, Conroy T, Current status on the place of folfirinox in metastatic pancreatic cancer and future directions, Therap Adv Gastroenterol, 10 (2017) 631–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Belalcazar A, Shaib WL, Farren MR, Zhang C, Chen Z, Yang L, Lesinski GB, El-Rayes BF, Nagaraju GP, Inhibiting heat shock protein 90 and the ubiquitin-proteasome pathway impairs metabolic homeostasis and leads to cell death in human pancreatic cancer cells, Cancer, 123 (2017) 4924–4933. [DOI] [PubMed] [Google Scholar]
- [12].Li X, Zhu F, Jiang J, Sun C, Zhong Q, Shen M, Wang X, Tian R, Shi C, Xu M, Peng F, Guo X, Hu J, Ye D, Wang M, Qin R, Simultaneous inhibition of the ubiquitin-proteasome system and autophagy enhances apoptosis induced by er stress aggravators in human pancreatic cancer cells, Autophagy, 12 (2016) 1521–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nawrocki ST, Carew JS, Pino MS, Highshaw RA, Dunner K Jr., Huang P, Abbruzzese JL, McConkey DJ, Bortezomib sensitizes pancreatic cancer cells to endoplasmic reticulum stress-mediated apoptosis, Cancer Res, 65 (2005) 11658–11666. [DOI] [PubMed] [Google Scholar]
- [14].Guo J, Hao J, Jiang H, Jin J, Wu H, Jin Z, Li Z, Proteasome activator subunit 3 promotes pancreatic cancer growth via c-myc-glycolysis signaling axis, Cancer Lett, 386 (2017) 161–167. [DOI] [PubMed] [Google Scholar]
- [15].Li J, Yakushi T, Parlati F, Mackinnon AL, Perez C, Ma Y, Carter KP, Colayco S, Magnuson G, Brown B, Nguyen K, Vasile S, Suyama E, Smith LH, Sergienko E, Pinkerton AB, Chung TDY, Palmer AE, Pass I, Hess S, Cohen SM, Deshaies RJ, Capzimin is a potent and specific inhibitor of proteasome isopeptidase rpn11, Nat Chem Biol, 13 (2017) 486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gorka AP, de Dios A, Roepe PD, Quinoline drug-heme interactions and implications for antimalarial cytostatic versus cytocidal activities, J Med Chem, 56 (2013) 5231–5246. [DOI] [PubMed] [Google Scholar]
- [17].Jain S, Chandra V, Kumar Jain P, Pathak K, Pathak D, Vaidya A, Comprehensive review on current developments of quinoline-based anticancer agents, Arab J Chem, (2016). Epub Oct 26, 2016 DOI: 10.1016/j.arabjc.2016.10.009. [DOI] [Google Scholar]
- [18].Liu XL, Shi Y, Kang JS, Oelschlaeger P, Yang KW, Amino acid thioester derivatives: A highly promising scaffold for the development of metallo-beta-lactamase l1 inhibitors, ACS Med Chem Lett, 6 (2015) 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou H-J, Wustrow D, Compositions and methods for JAMM protein inhibition, WO2014066506 (A2) (2014). [Google Scholar]
- [20].Kyani A, Tamura S, Yang S, Shergalis A, Samanta S, Kuang Y, Ljungman M, Neamati N, Discovery and mechanistic elucidation of a class of protein disulfide isomerase inhibitors for the treatment of glioblastoma, ChemMedChem, 13 (2018) 164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C, String 8--a global view on proteins and their functional interactions in 630 organisms, Nucleic Acids Res, 37 (2009) D412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles, Proc Natl Acad Sci USA, 102 (2005) 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC, Pgc-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes, Nat Genet, 34 (2003) 267–273. [DOI] [PubMed] [Google Scholar]
- [24].Meng LH, Mohan R, Kwok BHB, Elofsson M, Sin N, Crews CM, Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity, Proc Natl Acad Sci USA, 96 (1999) 10403–10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, Lahr DL, Hirschman JE, Liu Z, Donahue M, Julian B, Khan M, Wadden D, Smith IC, Lam D, Liberzon A, Toder C, Bagul M, Orzechowski M, Enache OM, Piccioni F, Johnson SA, Lyons NJ, Berger AH, Shamji AF, Brooks AN, Vrcic A, Flynn C, Rosains J, Takeda DY, Hu R, Davison D, Lamb J, Ardlie K, Hogstrom L, Greenside P, Gray NS, Clemons PA, Silver S, Wu X, Zhao WN, Read-Button W, Wu X, Haggarty SJ, Ronco LV, Boehm JS, Schreiber SL, Doench JG, Bittker JA, Root DE, Wong B, Golub TR, A next generation connectivity map: L1000 platform and the first 1,000,000 profiles, Cell, 171 (2017) 1437–1452 e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Guo N, Peng Z, Mg132, a proteasome inhibitor, induces apoptosis in tumor cells, Asia Pac J Clin Oncol, 9 (2013) 6–11. [DOI] [PubMed] [Google Scholar]
- [27].Nerini-Molteni S, Ferrarini M, Cozza S, Caligaris-Cappio F, Sitia R, Redox homeostasis modulates the sensitivity of myeloma cells to bortezomib, Br J Haematol, 141 (2008) 494–503. [DOI] [PubMed] [Google Scholar]
- [28].Riz I, Hawley TS, Marsal JW, Hawley RG, Noncanonical sqstm1/p62-nrf2 pathway activation mediates proteasome inhibitor resistance in multiple myeloma cells via redox, metabolic and translational reprogramming, Oncotarget, 7 (2016) 66360–66385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Barakat DJ, Mendonca J, Barberi T, Zhang J, Kachhap SK, Paz-Priel I, Friedman AD, C/ebpbeta regulates sensitivity to bortezomib in prostate cancer cells by inducing redd1 and autophagosome-lysosome fusion, Cancer Lett, 375 (2016) 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu L, Ito S, Nishio N, Sun Y, Chen N, Tanaka Y, Isobe K, Gadd34 facilitates cell death resulting from proteasome inhibition, Anticancer Res, 35 (2015) 5317–5324. [PubMed] [Google Scholar]
- [31].Cui W, Bai Y, Luo P, Miao L, Cai L, Preventive and therapeutic effects of mg132 by activating nrf2-are signaling pathway on oxidative stress-induced cardiovascular and renal injury, Oxid Med Cell Longev, 2013 (2013) 306073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Park HS, Jun do Y, Han CR, Woo HJ, Kim YH, Proteasome inhibitor mg132-induced apoptosis via er stress-mediated apoptotic pathway and its potentiation by protein tyrosine kinase p56lck in human jurkat t cells, Biochem Pharmacol, 82 (2011) 1110–1125. [DOI] [PubMed] [Google Scholar]
- [33].Fan Q, Liu B, Identification of the anticancer effects of a novel proteasome inhibitor, ixazomib, on colorectal cancer using a combined method of microarray and bioinformatics analysis, Onco Targets Ther, 10 (2017) 3591–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Augello G, Modica M, Azzolina A, Puleio R, Cassata G, Emma MR, Di Sano C, Cusimano A, Montalto G, Cervello M, Preclinical evaluation of antitumor activity of the proteasome inhibitor mln2238 (ixazomib) in hepatocellular carcinoma cells, Cell Death Dis, 9 (2018) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Leiherer A, Geiger K, Muendlein A, Drexel H, Hypoxia induces a hif-1 alpha dependent signaling cascade to make a complex metabolic switch in sgbs-adipocytes, Molecular and Cellular Endocrinology, 383 (2014) 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim MG, Yang JH, Kim KM, Jang CH, Jung JY, Cho IJ, Shin SM, Ki SH, Regulation of toll-like receptor-mediated sestrin2 induction by ap-1, nrf2, and the ubiquitin-proteasome system in macrophages, Toxicol Sci, 144 (2015) 425–435. [DOI] [PubMed] [Google Scholar]
- [37].Sheng XJ, Tu HJ, Chien WL, Kang KH, Lu DH, Liou HH, Lee MJ, Fu WM, Antagonism of proteasome inhibitor-induced heme oxygenase-1 expression by pink1 mutation, PLoS One, 12 (2017) e0183076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Martin L, Gardner LB, Stress-induced inhibition of nonsense-mediated rna decay regulates intracellular cystine transport and intracellular glutathione through regulation of the cystine/glutamate exchanger slc7a11, Oncogene, 34 (2015) 4211–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shin DH, Chun YS, Lee DS, Huang LE, Park JW, Bortezomib inhibits tumor adaptation to hypoxia by stimulating the fih-mediated repression of hypoxia-inducible factor-1, Blood, 111 (2008) 3131–3136. [DOI] [PubMed] [Google Scholar]
- [40].Kumar P, Gullberg U, Olsson I, Ajore R, Myeloid translocation gene-16 co-repressor promotes degradation of hypoxia-inducible factor 1, PLoS One, 10 (2015) e0123725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M, Zamboni N, Schulze A, Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival, Cancer Discov, 2 (2012) 328–343. [DOI] [PubMed] [Google Scholar]
- [42].Ros S, Floter J, Kaymak I, Da Costa C, Houddane A, Dubuis S, Griffiths B, Mitter R, Walz S, Blake S, Behrens A, Brindle KM, Zamboni N, Rider MH, Schulze A, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 is essential for p53-null cancer cells, Oncogene, 36 (2017) 3287–3299. [DOI] [PubMed] [Google Scholar]
- [43].Strohecker AM, Joshi S, Possemato R, Abraham RT, Sabatini DM, White E, Identification of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as a novel autophagy regulator by high content shrna screening, Oncogene, 34 (2015) 5662–5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kuang Y, Sechi M, Nurra S, Ljungman M, Neamati N, Design and synthesis of novel reactive oxygen species inducers for the treatment of pancreatic ductal adenocarcinoma, J Med Chem, 61 (2018) 1576–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.