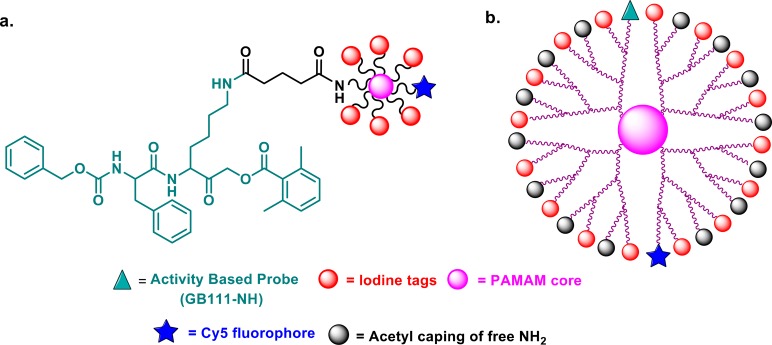
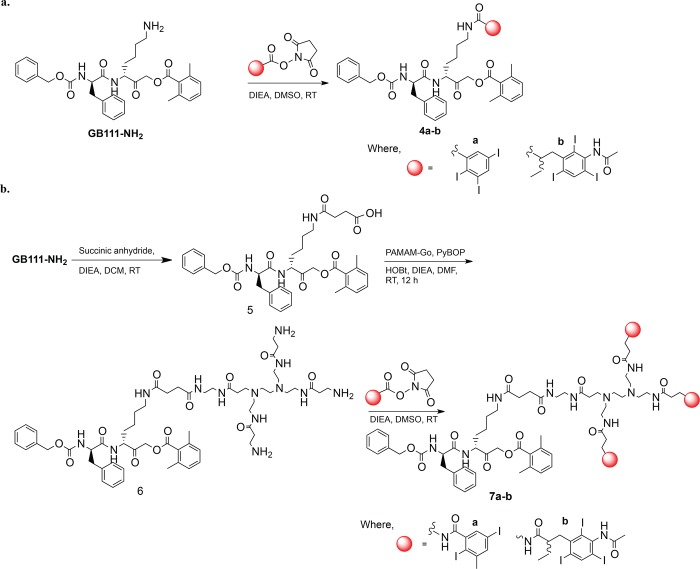
Abstract
X-ray computed tomography (CT) is a robust, precise, fast, and reliable imaging method that enables excellent spatial resolution and quantification of contrast agents throughout the body. However, CT is largely inadequate for molecular imaging applications due mainly to its low contrast sensitivity that forces the use of large concentrations of contrast agents for detection. To overcome this limitation, we generated a new class of iodinated nanoscale activity-based probes (IN-ABPs) that sufficiently accumulates at the target site by covalently binding cysteine cathepsins that are exceptionally highly expressed in cancer. The IN-ABPs are comprised of a short targeting peptide selective to specific cathepsins, an electrophilic moiety that allows activity-dependent covalent binding, and tags containing dendrimers with up to 48 iodine atoms. IN-ABPs selectively bind and inhibit activity of recombinant and intracellular cathepsin B, L, and S. We compared the in vivo kinetics, biodistribution, and tumor accumulation of IN-ABPs bearing 18 and 48 iodine atoms each, and their control counterparts lacking the targeting moiety. Here we show that although both IN-ABPs bind specifically to cathepsins within the tumor and produce detectable CT contrast, the 48-iodine bearing IN-ABP was found to be optimal with signals over 2.1-fold higher than its nontargeted counterpart. In conclusion, this study shows the synthetic feasibility and potential utility of IN-ABPs as potent contrast agents that enable molecular imaging of tumors using CT.
Keywords: Cathepsins, computed tomography, nanoscale activity-based probes, iodine contrast agent, PAMAM, molecular imaging
Computed tomography (CT) is an imaging technique widely used in a variety of research and clinical studies utilizing X-rays to create cross-section images of the body.1−5 CT’s high resolution, high efficiency, wide availability, and cost effectiveness makes it one of the most frequently used noninvasive clinical imaging modalities. CT imaging enables detailed three-dimensional (3D) visualization with excellent spatial resolution of internal body structures and is routinely used for diagnosis of diseases, treatment assessment, and prediction of therapy.6,7 It requires the presence of elements with high atomic weight and radiographic density higher than those of the surrounding biological tissue, thus iodine is often used to increase contrast.
Contrast agents are an essential part of radiology and are routinely applied in modern medicine, reported to have been used in 53% of all CT scans performed in the U.S. in 2011.8 Conventional, clinically used iodinated CT contrast agents, both ionic or nonionic, predominantly have a low molecular weight and accumulate nonspecifically in organs or tissues.9,10 Nevertheless, CT contrast requires high concentrations of contrast agents and the chemical properties of iodinated CT agents lead to a very rapid clearance from the body, limiting imaging to a time scale of minutes or even seconds for the majority of diagnostic applications.11
Nanoparticles have many advantages over conventional contrast agents, such as specific molecular targeting capability, prolonged blood circulation time, and controlled clearance pathways, thus allowing for molecular imaging and medical diagnostics.12,13 Nanoparticles incorporated with iodine, gold, and bismuth hold high electron density and therefore have been proposed as CT contrast agents, showing low rates of renal clearance and increased vascular residence time.14−16 In addition, nanoparticles and other macromoleculars, within the nanoscale range, accumulate preferentially in solid tumors due to the enhanced permeability and retention (EPR) effect.17,18 To enrich the number of contrasting atoms, we focused on polymeric dendrimers, planar hyperbranched nanostructures that can form spherical nanostructures; they possess chemical handles on which to graft therapeutics and contrast agents with great loading efficiency.19 As such, dendrimers have been reported as carriers of contrast agents for magnetic resonance imaging (MRI),20 electron paramagnetic resonance,21 and iodinated dendritic nanoparticles of CT.22 Indeed, nanoparticles and macromolecular agents are suitable for vascular imaging by CT because they exhibit longer half-lives as blood pool reagents.17,18 The majority of research for enhancing CT contrast on iodinated compounds such as nanosupentions,23 nanocapsules,24 nanoemulsions,25 and liposomes26 report macrophage targeting. Furthermore, covalent incorporation of iodine containing moieties to a macromolecular structure leads to high water solubility and good biocompatibility as potential blood pool contrast agents.15,22,27,28
The development of iodine-based CT probes for early disease detection is a highly attractive goal, however, the low sensitivity of CT scanners and minimum detectable signals make this a challenging task.10 Nevertheless, numerous groups have relied on highly expressed biological targets to direct nanoparticles, densely loaded with contrast molecules, to enable CT molecular imaging.29 For example, targeted contrast agents based on bismuth sulfide and gold nanoparticles targeted to high density lipoproteins that are specific for macrophages have been reported.30,31 Winter et al. reported on a nanoemulsion of iodinated oils modified with an antifibrin antibody attached to its surface,29,32 Hyafil et al. reported on iodinated polymeric nanoparticles targeted to macrophages in atherosclerotic plaque,15 and Montet’s group reported iodine-contrast media-loaded liposomes targeted to the E-selectin specific peptides.33 Additionally, Hill et al. developed a low density lipoprotein-like nanoparticle loaded with iodinated triglycerides targeted to HepG2 cancer cells.34 While diverse approaches leading to iodinated nanoparticles CT contrast media have been optimized, so far none of the compounds have been approved for clinical application.15,32−36 The several iodine blood-pool agents clinically used for enhancing CT contrast suggest that other highly iodinated compounds could be well tolerated.
We took a unique approach of directing iodine CT contrast agents to cancerous tissue by targeting specific enzymatic activity rather than protein abundance using the activity-based-probes (ABP) methodology. A typical ABP includes a recognition element that drives selectivity to its protease, a contrast agent, and a “warhead”, usually an electrophile that enables an activity driven covalent linkage between the target and the contrast moiety.37−39 We targeted our probes to a subset of cysteine cathepsin proteases that are highly overexpressed and active in several pathologies characterized with high macrophage content such as cancer and atherosclerosis. With the extremely high abundance of our target cysteine cathepsins and the covalent nature of the probes we were able to overcome the challenges associated with the low sensitivity of CT imaging and generate iodine-based molecular imaging probes that enable cancer detection.
In an attempt to generate CT contrast agents for molecular imaging of cancer, we generated libraries of iodinated nanoscale activity-based probes (IN-ABPs) targeted to cathepsin proteases highly elevated in cancer.40−43 In general, the targeting moiety, based on the published ABP GB111-NH2,37 is highly selective to cathepsin B, L, and S. The short peptide portion of the probe (carbobenzoxy-phenylalanine-lysine) served as the recognition element to the cathepsins and the acyloxymethyl ketone as a warhead, enabling covalent linkage of the probe to its target. GB111-NH2 was tagged with various iodine-containing aromatic rings termed iodine tags (Scheme 1). To generate the iodine tags and enable attachment to the ABP core (GB111-NH2) we modified two commercial compounds; 2,3,5-triiodobenzoic acid (TBA) and iopanoic acid (IPA). The carboxylic acid of TBA was directly converted to succinimidyl ester (SE) while the IPA’s free amine was first acetylated and only then its carboxylic acid was converted to SE. The activated iodine tags were then simply attached to the free amine of GB111-NH2Scheme S1.
Scheme 1. Basic Library Design of Iodinated Nanoscale Activity Based Probes (IN-ABPs).
(a) Structure of G1 IN-ABP: PAMAM core labelled with six iodine tags, a Cy5 moiety, and a targeting moiety GB111-NH. (b) The general structure of PAMAM G3 IN-ABPs include 16 iodine tags, a targeting moiety (GB111-NH), 14 capping groups, and a Cy5 moiety.
To determine whether the attachment of the large iodine tags interferes with the probe binding to its target, we first generated IN-ABPs with only one or three tags using both TBA (1) and IPA (3) based tags. GB111-NH2 was either attached directly to a single tag resulting in 4a and 4b, (Scheme 2a) or reacted with succinic anhydride to enable PAMAM generation 0 (G0) binding through a succinic acid linker. The tags were then coupled via amide bond to form the three tagged compounds (7a, 7b) (Scheme 2b). These probes were then evaluated biochemically for their ability to bind and inhibit recombinant human cathepsins by a competitive inhibition assay.
Scheme 2. Synthesis of Iodine-Tagged ABPs (4a,b, 7a,b).
(a) Synthesis of single iodine tagged activity based probe. (b) Three iodide-tagged ABP (7a,b) were synthesized starting from GB111-NH2, which was reacted with succinic anhydride followed by coupling with ethylenediamine core PAMAM-Go and then reacted with SE of either 2,3,5-triiodobenzoic acid or iopanoic acid described in Scheme S1.
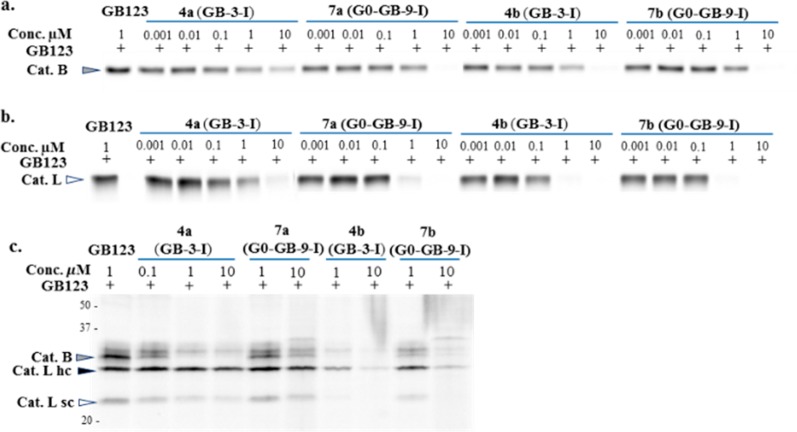
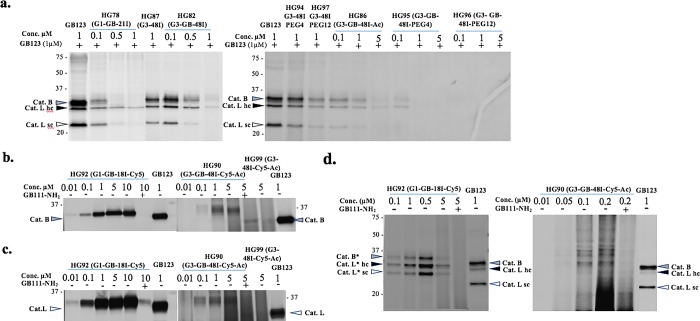
Recombinant human cathepsin B or L were incubated with an increasing concentration of the IN-ABPs, after which the residual cathepsin activity was detected by a Cy5 fluorescently labeled cathepsin ABP GB123.44 The free IN-ABPs were separated from the enzyme-probe complexes by SDS PAGE then detection of the probe-enzyme complex was carried out using a fluorescent scan of the gel. The single and three iodine tagged probes (4a, 4b, 7a, 7b) all showed cathepsin B and L inhibition with minor differences between the compounds, Figure 1a,b, indicated that the iodine tags do not interfere with binding of the probes to the protease.
Figure 1.
Biochemical evaluations of Iodine-tagged ABPs (4a,b, 7a,b) by a competition assay. (a,b) Inhibition of recombinant cathepsin B and L, as described in Supporting Information. The reduction in intensity of the fluorescent band indicates inhibition of cathepsin activity. (c) Inhibition of endogenous cathepsin activity within intact NIH-3T3 cells, as described in Experimental Section. The iodine-tagged ABPs were found to be cell permeable and able to inhibit cathepsin activity.
Next, in order to evaluate the probe’s cell permeability and capability of labeling cellular cathepsins, we performed a competitive inhibition assay in intact NIH-3T3 cells. Probes were incubated with intact cells for 24 h then residual cathepsin activity was labeled by GB123. Cell lysates were separated on SDS PAGE, followed by scanning of the gel for fluorescence to detect residual cathepsin activity, Figure 1c. The inhibition of cellular cathepsins was clear and consistent with IPA tagged probes (4b and 7b). Interestingly, these probes exhibited better inhibition than the TBA probes (4a and 7a) most likely due to better cell permeability. We therefore generated our next probes with the IPA iodine tags only.
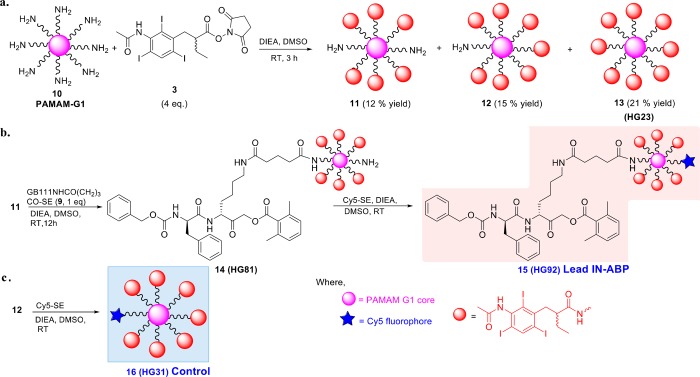
We moved to generating multiple tagged IN-ABPs with up to 48 iodine atoms (16 tags) based on PAMAM G1 and G3 cores. For the PAMAM G1 compounds, the PAMAM- core was reacted with a SE of IPA tag (3) in basic conditions, the mixture was then purified yielding six, seven, and eight iodine-tagged dendrimers (11, 12, 13, respectively), Scheme 3a. In parallel, GB111-NH2 was extended with glutaric acid and converted into a SE with NHS (9), Scheme S2. The six iodine-tagged ABP was generated by reacting 11 with GB111NHCO(CH2)3CO-SE (9) to give compound 14 (HG81), which was then reacted with Cy5-SE in basic conditions to give a Cy5-labeled six-iodine-tagged ABP 15 (HG92), Scheme 3b. A negative control compound containing 7 iodine tags without GB111-NH2, 16 (HG31) was synthesized by Cy5 attachment to compound 12, Scheme 3c. Compounds lacking Cy5 were also generated similarly, for example, the seven iodine-tagged ABP, 17 (HG78), by reacting compound 12 with GB111NHCO(CH2)3CO-SE (9), Scheme S3a. An ABP with two targeting peptides (18) was also generated by reacting 11 with two GB111NHCO(CH2)3CO-SE (9), Scheme S3b.
Scheme 3. Synthesis of IN-ABPs.
(a) Synthesis of six, seven and eight iodine-tagged contrast agents. The iodine tag IPA-SE (3) was attached to PAMAM-G1 generating 11, 12, 13 holding 6, 7, and 8 iodine tags respectively, each complex was purified with indicated yields. (b) Synthesis of Cy5 labelled IN-ABP (six tagged, 15) was achieved by coupling the six iodine-tagged PAMAM 11 with GB111NHCO(CH2)3CO-SE (9), then reacting with Cy5-SE. (c) Synthesis of seven tagged control 16 was accomplished by reacting the seven iodine tagged PAMAM 12 with Cy5-SE. A detailed procedure is described in Supporting Information.
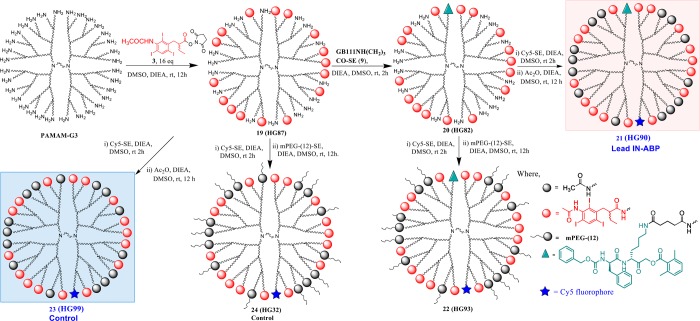
The design of targeted contrast reagent based on the dendrimer PAMAM G3 ((NH2)32) included a maximal usage of only 16 iodine tags due to solubility limitations. Since reports have found that highly cationic PAMAM dendrimers distort cellular membranes,45,46 the remaining amines were either capped by an acetyl group or with a polyethylene glycol (PEG) group, or left free. The synthetic route of the PAMAM G3 ABPs was similar to PAMAM G1 ABPs; the free amines of PAMAM G3 were first reacted with IPA tag 3 forming 19 (HG87), then with GB111NHCO(CH2)3CO-SE (9), resulting in 20 (HG82). HG82 was labeled with Cy5 which was then either acetylated or PEGylated to yield 27 (HG90) and 28 (HG93), respectively, Scheme 4. We aimed to control the stoichiometric ratio of the groups by adding precise molar ratios of GB111-NH2 and Cy5 onto each ABP, leading to an average of a single group on each probe. As negative controls, conjugates of PAMAM-G3 dendrimers with Cy5, acetylated or PEGylated but without GB111-NH, were synthesized resulting in 29, (HG99), and 30 (HG32), Scheme 4. Similar compounds lacking Cy5 were generated as described in Scheme S4. The synthesis, purification, and characterization of all IN-ABPs were performed using standard methods. 1H NMR was used to determine the average number of conjugates. For example, 16 IPA tags were found relying on the characteristic peaks at δ (ppm) 8.35 which belong to the phenyl proton (16 H’s) of the IPA aromatic ring. Similarly, GB111-NH, acetyl and PEG conjugates were determined as described in the experimental section. The number of conjugated Cy5 dyes per dendrimer was found to be one on average by spectrophotometry. A list of cathepsin IN-ABPs and controls synthesized with the number of iodine tags, atoms, and capping moieties is shown in Table 1.
Scheme 4. Synthesis of IN-ABPs.
Synthesis of multiple tagged IN-ABPs labelled with Cy5 based on PAMAM G3 dendrimer core capped with acetyl or PEG groups. PAMAM G3 was first linked to 16 iodine tags by reacting with the IPA-SE (3) then a single GB111NHCO(CH2)3CO-SE (9) moiety was attached followed by reacting with Cy5-SE. The remaining free amines were then capped with acetyl, PEG, or left free, generating 21, 22, and 20, respectively. Control compounds lacking the GB111NHCO(CH2)3CO-SE (9) moiety were also generated with acetyl, PEG or with free amines, compound 23, 24, and 19 respectively.
Table 1. List of the PAMAM Dendrimer Core Multiple Tagged IN-ABPs.
| IN-ABP | PAMAM gen. | no. of iodine tagsa | no. of iodine atomsa | no. of GB111NH-a | no. of free NH2a | no. of acetyla | no. of PEG-4 (333)a | no. of PEG-12 (750)a | no. of Cy5b | molecular weightc |
|---|---|---|---|---|---|---|---|---|---|---|
| 4a | 1 | 3 | 1 | 1055.00 | ||||||
| 4b | 1 | 3 | 1 | 1168.08 | ||||||
| 7a | G0 | 3 | 9 | 1 | 2616.82 | |||||
| 7b | G0 | 3 | 9 | 1 | 2956.08 | |||||
| HG23 (13) | G1 | 8 | 24 | 6187.42 | ||||||
| HG81 (14) | G1 | 6 | 18 | 1 | 1 | 5653.11 | ||||
| HG92 (15) | G1 | 6 | 18 | 1 | 1 | 6305.34 | ||||
| HG31 (16) | G1 | 7 | 21 | 1 | 6230.83 | |||||
| HG78 (17) | G1 | 7 | 21 | 1 | 6247.91 | |||||
| HG81a (18) | G1 | 6 | 18 | 2 | 6308.4 | |||||
| HG87 (19) | G3 | 16 | 48 | 16 | ∼16426 | |||||
| HG82 (20) | G3 | 16.2 | 48 | 1.3 | 15 | ∼17415 | ||||
| HG90 (21) | G3 | 16.31 | 48 | 1.43 | 14 | 1.05 | ∼18826 | |||
| HG93 (22) | G3 | 16.23 | 48 | 1.78 | 14 | 0.94 | ∼27243 | |||
| HG99 (23) | G3 | 16.19 | 48 | 15 | 0.89 | ∼17737 | ||||
| HG32 (24) | G3 | 16.09 | 48 | 15 | 1.04 | ∼26668 | ||||
| HG86 (25) | G3 | 16.19 | 48 | 1.46 | 14 | ∼18104 | ||||
| HG95 (26) | G3 | 16.32 | 48 | 1.76 | 14 | ∼20846 | ||||
| HG96 (27) | G3 | 16.33 | 48 | 1.84 | 14 | ∼26743 | ||||
| HG33 (28) | G3 | 16 | 48 | 16 | ∼17098 | |||||
| HG94 (29) | G3 | 16.17 | 48 | 16 | ∼20878 | |||||
| HG97 (30) | G3 | 16 | 48 | 16 | ∼26585 |
Determined by 1H NMR spectrometry.
Number of Cy5 tags determined by using UV spectrophotometry.
Average molecular weight calculated.
We further analyzed all the iodinated dendrimers by zeta potential and a correlation was found between the changes in zeta potential of the dendrimers with a number of surface groups attached. For example, zeta potential of IPA and GB111-NH conjugated PAMAM HG82 was 34 ± 3 because of the partially positively charged NH2 surface groups, then a zeta potential decreased was detected after acetylation or PEGylation (HG86 and HG95, respectively). Table S1 shows the measured particle sizes and zeta potentials for all compounds generated. Most size distribution values were found to be higher than expected most likely due to water coating or aggregation of particles in the solution. TEM images of the selected G3 compounds are presented in Figure S1.
We turned to evaluate the inhibitory potency of the G1 IN-ABPs by competition assays of recombinant cathepsin B and L as well as of endogenous cathepsins within intact NIH-3T3 cells. Cathepsin inhibition was performed as described in Figure 1 by a competition assay. The G0 and G1 IN-ABPs, HG78, HG81, and HG81a, all inhibited cathepsin activity both in vitro and in intact cells with adequate potency in a dose response manner, Figure S2. Importantly, the inhibition potency of the G1 IN-ABPs was comparable to the single tag and tritag compounds shown in Figure S2. Furthermore, having two targeting moieties, GB111 attached to the same probe did not increase the potency (HG81a).
Finally, we evaluated potency toward cathepsins of the G3 probes, HG82, HG86, HG95, and HG96. All G3 compounds showed potent inhibition in a dose response manner of both recombinant cathepsin B and L as well as cellular cathepsins. These surprising results indicated that despite the large size of the compounds, there was no impairment of cell permeability or binding interaction with the cathepsin targets. Furthermore, HG82 with 15 free amines was slightly less potent than the acetylated and PEGylated G3 analogs in the cellular assay. To ensure that the binding was dependent on protease activity, control nanoscale compounds lacking the reactive moiety (HG87, HG94, and HG97) were tested with no detectable inhibition (Figure 2a and Figure S2d,e).
Figure 2.
Cathepsin binding of IN-ABPs assessed by competition assay and direct labeling. (a) Inhibition of endogenous cathepsin activity within intact NIH-3T3 cells, as described in Experimental Section. Decrease in band intensity indicates efficient cathepsin binding. (b,c) Direct labeling of recombinant cathepsin B and L by G1 and G3 Cy5 labeled IN-ABPs (HG92 and HG90). Indicated concentration of probes were incubated with enzymes with or without cathepsin inhibitor (GB111-NH2)36 pretreatment. Samples were run on gel that was scanned for fluorescence. Increase in band intensity indicates efficient cathepsin binding. (d) Intact NIH-3T3 cells with or without inhibitor pretreatment were treated with HG92, HG90 for 24 h in growth media, cells were lysed and separated by gel that was scanned for fluorescence. Clear selective binding of endogenous cathepsins are seen. Molecular weight shift of cathepsin-probe complex is marked with *.
Encouraged by the potency and cell permeability of the G1 and G3 IN-ABPs, we moved to compare the G1 and G3 specificity to cathepsin proteases in intact cells. We generated two Cy5 labeled IN-ABPs, a G1 probe 15 (HG92) and an acetylated G3 probe 21 (HG90) together with two corresponding controls, 16 (HG31) and 23 (HG99), respectively. First, we examined direct labeling of the Cy5 labeled IN-ABP to recombinant human cathepsins B and L by incubation with increasing concentrations of probes, and then by visualizing the IN-ABP-enzyme complex using a fluorescent scan of the SDS PAGE gel. Both HG92 and HG90 were found to bind cathepsin B and L in an activity-dependent manner as this binding was inhibited by pretreatment with the cathepsin inhibitor GB111-NH2. The smaller size of HG92 was more potent than HG90, and resulted in cleaner detectable bands probably due to the smearing of the large HG90 on the gel (Figure 2b,c). We evaluated cell permeability and selectivity toward cellular cathepsins by a direct labeling assay in which the Cy5-labeled IN-ABPs were added to the growth media of intact NIH-3T3 cells for 24 h, then cell lysates were analyzed by in-gel fluorescence. Encouragingly, both Cy5-labeled IN-ABPs tested showed high cell permeability and selective cathepsin binding with a detectable molecular weight shift of the probe-cathepsin complex as expected from the large weight of the probes (marked with asterisk, Figure 2d). We further generated a PEGylated Cy5 G3 IN-ABP 22 (HG93) that bound recombinant cathepsins very weakly making it difficult to detect the cathepsin binding in intact cells because of the probes’ resolution on gel, thus HG93 was neglected (Figure S3). All together our in vitro testing revealed suitable selectivity and potency of both HG92 and HG90 IN-ABPs to progress to in vivo imaging studies.
Prior to in vivo testing we assessed cellular cytotoxicity of the PAMAM dendrimeric compounds by a methylene blue cell viability assay. We found that the IN-ABPs, HG90 and HG92 were not toxic, thus, these compounds were selected for in vivo studies. Nevertheless, two compounds with multiple free amines, HG87 and HG82, and two of the acetylated compounds, HG99 and HG86, caused 15–35% reduced viability in the highest concentration of 10 μM after 48 h. Altogether, most of the compounds had negligible toxicity to the cells in the concentration range tested (Figure S4).
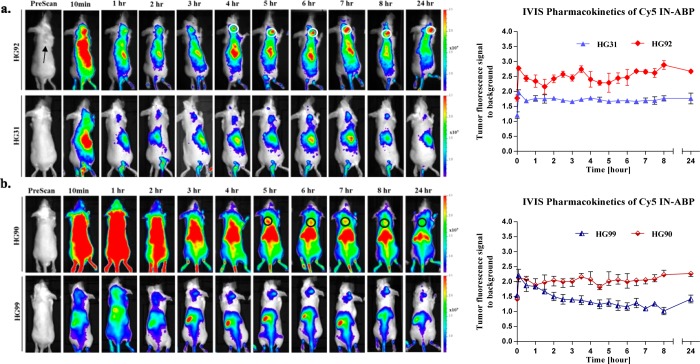
We then set out to investigate the CT imaging capabilities of the probes in tumor-bearing mice. We selected the two Cy5 IN-ABPs, G1 and G3, HG92 and HG90, together with their respective controls HG31 and HG99 lacking the targeting moiety. Prior to the CT analysis we explored the pharmacokinetics of the IN-ABP exploiting their fluorescent signal by noninvasive optical imaging to better time the CT scans. Following intravenous IN-ABP injections to tumor bearing mice, the fluorescent compounds rapidly circulated throughout the animal and high fluorescent signals could be seen in virtually all tissues, including the tumors (Figure 3a,b). The G1 IN-ABP, HG92, produced a clear tumor-specific signal that could be detected 4 h post injection, on average, as a result of sufficient tumor accumulation and partial clearance of the nonbound probe. This specific tumor signal increased over time and reached a maximum at 6–8 h post injection. Because of the covalent nature of the targeted IN-ABP a significant amount was retained in the tumor even 24 h post injection. As expected, the nontargeted contrast agent analog, HG31, only slightly accumulated in the tumor, most likely due to the EPR effect, as could be seen by the tumor fluorescence quantification (Figure 3a). Additionally, the G3 IN-ABP, HG90, showed similar kinetics to HG92 with a specific signal detected at 5 h post injection. Moreover, the nontargeted G3 contrast agent, HG99 showed monoexponential decay (Figure 3b). On the basis of these results, the optimal time for the CT scans was determined to be 5 and 24 h following administration of the probes.
Figure 3.
Noninvasive optical imaging of mice tumors. (a) Noninvasive fluorescent images of mice injected with Cy5-labeled G1 IN-ABP HG92 (top) and control HG31 (bottom). Left panel, tumor bearing mice were prescanned for fluorescence, and at indicated times post iv injection of compounds. Tumor fluorescence marked with white circles. Right panel, average signal to background fluorescence within tumors plotted over time. (b) Noninvasive fluorescent images and quantification from HG90, Cy5-labeled G3 IN-ABP, and control HG99 in tumors was acquired similarly. Tumor fluorescence is seen within the black circles. Fluorescent images were acquired with an in vivo imaging system (IVIS) equipped with a 630/690 nm excitation/emission filters, fluorescent scale bar is depicted on right of images in units of (p/sec/cm2/sr)/(μW/cm2).
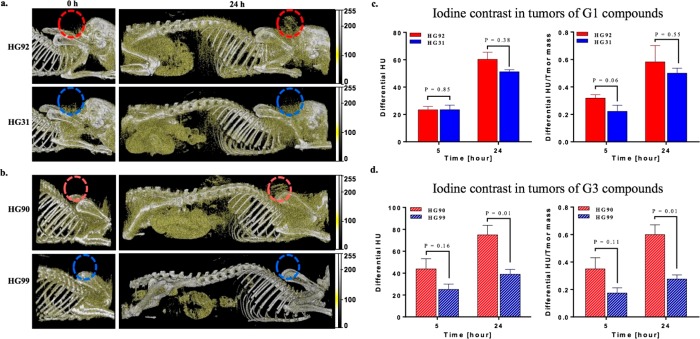
The use of iodine compounds as X-ray CT contrast agents is challenging since both tissue and iodine attenuates the CT beam similarly, thus clinically, extremely high amounts of iodine are administered to gain contrast.7 Here we set out to determine whether the CT scanner is capable of detecting tumor protease activity in vivo using relative low doses of our IN-ABPs. Additionally, we tested whether the covalent nature of the IN-ABPs was advantageous over the nontargeted controls. In our setting, tumor-bearing mice were scanned by CT prior to compound injection and again 5 and 24 h post administration as suggested by the optical imaging experiments. The CT iodine signal was optimized relative to soft tissue by adjusting the setup from a 0.2 mm aluminum filter, a tube voltage of 40 kVp and 500 mA to 60 kVp and 350 mAs with a 0.5 mm aluminum filter. This setting enables detection of small amounts of iodine in tissue since a higher fraction and total number of photons with energies between 33 keV and ∼45 keV, just above iodine’s K-edge (33.2 keV), reach the detector. The scanner equipped with 64 detectors was used for all scans to detect the iodine signal, enabling detection of a faint signal from the tumors even 5 h post injection of the IN-ABPs, using only 0.5 mg/mouse equivalent iodine (∼20 mg/kg). Nevertheless, at the 5 h scan the iodine was still distributed throughout the body making it difficult to distinguish the tumor from the body, Figure S5. Representative images of CT scans obtained 24h post IN-ABP injection (G3-HG90 and G1-HG92) revealed detectable accumulation in the tumor, liver, and stomach. While the tumor was clearly detected in the IN-ABP-injected mice it was barely detected in nontargeted G1-HG31 and G3-HG99 treated mice, Figure 4a,b. The signal in the tumor was due to retention of the probes because of both high cathepsin activity and the EPR effect. Liver signals most likely resulted from the physiological high cathepsin activity together with the compound’s retention due to the liver detoxification function. The stomach signal, however, resulted from the combined high cathepsin activity and signals from the chow that was also detected in the preinjection scans. While other organs were visible by CT, the IN-ABPs accumulation in the tumor enabled its clear detection.
Figure 4.
In vivo X-ray computed tomography. (a,b) Representative sagittal images of tumor-bearing mice that were scanned for CT contrast by a micro CT prior to and 24 h post iv injection of IN-ABP HG92, HG90 and respective controls HG31, HG99. Tumors placed on the back of the mice are marked by colored circles. Gold color marks contrast from iodine, gray color marks contrast from bone. (c,d) Differential CT tumor contrast in HU (above background at t = 0), left, and normalized to each tumor mass [80–160 mg], right, of IN-ABP HG92, HG90 and respective control HG31, and HG99. Red bars represent targeted IN-ABPs, blue bars represent control compounds.
Quantitative analysis of the tumor CT contrast was presented as the increase in Hounsfield Units (HU) above baseline (Differential HU), Figure 4c,d left, and relative to tumor mass, Differential HU/Tumor mass, Figure 4c,d on the right. These presentations indicate that tumor accumulation of the targeted IN-ABP, G3-HG90 and G1-HG92, are higher than their nontargeted analogs, G3-HG99 and G1-HG31. Nevertheless, the difference in HU of targeted versus control compounds became significant only in the 24 h G3-HG90 scans where sufficient accumulation of the heavily iodinated probe occurred. In the G1-HG92 scans, more probe accumulated than its control though the difference was not significant most likely because of the lower iodine content of the probe, Figure 4c,d. We assume that more G1-HG92 accumulates in the tumor than G3-HG90, however, the larger number of iodine atoms in G3-HG90 results in similar CT contrasts of these IN-ABPs.
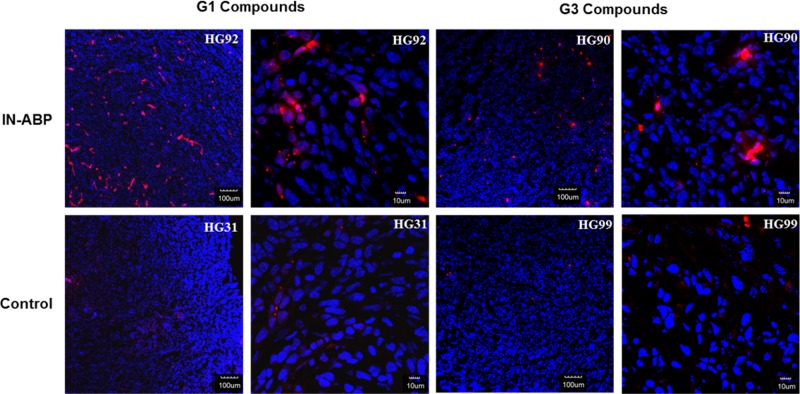
Tumors from injected animals were also analyzed by fluorescent microscopy. Significantly enhanced Cy5 fluorescence was detected in tumors from targeted IN-ABP injected mice, G3-HG90 and G1-HG92, versus their controls, G3-HG99 and G1-HG31. This increase in fluorescence is a reflection of the high levels of IN-ABP bound to the excessive amount of activity cysteine cathepsin in the tumors (Figure 5). These data demonstrate an excellent correlation with the CT in vivo data.
Figure 5.
Tumors from animals 24 h post injection with indicated compounds were frozen in OCT, sectioned, and stained with DAPI. Representative fluorescent scans acquired with a confocal microscope are presented, red- Cy5 from IN-ABPs, blue- DAPI. High Cy5 fluorescence was detected only in tumors from targeted IN-ABPs HG92 and HG90.
While the covalent nature of the IN-ABP led to prolonged retention of the IN-ABPs within tissues that resulted in detected CT signals, it also enabled biochemical analysis of in vivo labeled tissue. We analyzed the tumor tissues as well as livers, kidneys, and spleens from IN-ABP (HG92) and nontargeted (HG31) treated mice. Tissue lysates were prepared and separated by SDS-PAGE that was scanned for Cy5 fluorescence. Pronounced labeling of tissue cathepsins was detected from the IN-ABP-cathepsin enzyme complex revealing differences in cathepsin B, L, and S activities within the different organs as seen by the indicative cathepsin bands between 22 and 35 kDa. As expected, no cathepsin labeling was detected in the nontargeted compound-treated mice, Figure S6.
The data shown here are consistent with the increasing ability of IN-ABP to label tumors that express elevated levels of cathepsin activity; this theme may be extended for new classes of targeted X-ray contrast agents targeted to different proteases. A disadvantage of the IN-ABP methodology is the lack of signal amplification, together with the low sensitivity of CT scanners, making signal detection challenging. Nevertheless, the very high levels of cathepsins within tumors enable sufficient accumulation of the targeted contrast agent, which is sufficient for tumor detection by CT.
In conclusion, we developed a new class of ABPs attached to iodinated polymeric dendrimers for detection of solid tumors using X-ray CT. Initially, we synthesized and characterized a library of IN-ABPs and tested them in vitro. We then studied the in vivo pharmacokinetics by optical imaging that set the basis for the in vivo kinetic CT imaging experiment. Specific accumulation of the IN-ABPs enabled molecular CT imaging due to their covalent nature, showing the increase in specific signals detecting cathepsin activity. Additionally, the signal detection was done using dramatically low iodine concentrations of approximately 20 mgI/kg, as compared to clinically used iodine containing reagents that are used at approximately 300 mgI/kg.47 The technique brought here describes a methodology that after further development may have extensive clinical diagnosis applications for detecting cancer and other pathologies with elevated cathepsin activity such as vulnerable atherosclerotic plaques with commonly used CT instruments.
Acknowledgments
This work was funded by the starting European Research Council (ERC) grant (Grant 337238) (GB), Grass Center for Drug Design and Synthesis of Novel Therapeutics (GB) Rothschild Caesarea Foundation (DT). We thank Eduard Berenshtein from the electron microscopy lab of the Hebrew University Core Research Facility for aiding with TEM analysis.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.7b03813.
Chemistry, experimental section, 1H NMR data of IN-ABP, biochemical evaluation, TEM images, and complementary characterizations (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Webb W. R.; Brant W.; Major N.. Fundamentals of body CT, 3rd ed.; Saunders Elsevier: Philadelphia, PA, 2005. [Google Scholar]
- Schenkman L. Science 2011, 331, 1002. 10.1126/science.331.6020.1002. [DOI] [PubMed] [Google Scholar]
- Schambach S. J.; Bag S.; Schilling L.; Groden C.; Brockmann M. A. Methods 2010, 50, 2. 10.1016/j.ymeth.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Badea C. T.; Drangova M.; Holdsworth D. W.; Johnson G. A. Phys. Med. Biol. 2008, 53, R319. 10.1088/0031-9155/53/19/R01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalender W. A. Phys. Med. Biol. 2006, 51, R29. 10.1088/0031-9155/51/13/R03. [DOI] [PubMed] [Google Scholar]
- Scally P.Medical Imaging; Oxford University Press: Oxford, England, 1999. [Google Scholar]
- Lusic H.; Grinstaff M. W. Chem. Rev. 2013, 113, 1641. 10.1021/cr200358s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMV Medical Information Division, Inc. IMV. 2012 CT Market outlook report; IMV Medical Information Division: Des Plaines, IL, 2012. [Google Scholar]
- Singh J.; Daftary A. J. J. Nucl. Med. Technol. 2008, 36, 69. 10.2967/jnmt.107.047621. [DOI] [PubMed] [Google Scholar]
- Krause W. Adv. Drug Delivery Rev. 1999, 37, 159–173. 10.1016/S0169-409X(98)00105-7. [DOI] [PubMed] [Google Scholar]
- Canty J. M. Jr.; Judd R. M.; Brody A. S.; Klocke F. J. Circulation 1991, 84, 2071. 10.1161/01.CIR.84.5.2071. [DOI] [PubMed] [Google Scholar]
- Popovtzer R.; Agrawal A.; Kotov N. A.; Popovtzer A.; Balter J.; Carey T. E.; Kopelman R. Nano Lett. 2008, 8, 4593. 10.1021/nl8029114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormode D. P.; Skajaa T.; Fayad Z. A.; Mulder W. J. M. Arterioscler., Thromb., Vasc. Biol. 2009, 29, 992. 10.1161/ATVBAHA.108.165506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormode D. P.; Naha P. C.; Fayad Z. A. Contrast Media Mol. Imaging 2014, 9, 37. 10.1002/cmmi.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyafil F.; Cornily J. C.; Feig J. E.; Gordon R.; Vucic E.; Amirbekian V.; Fisher E. A.; Feldman L.; Fayad Z. A.; et al. Nat. Med. 2007, 13, 636. 10.1038/nm1571. [DOI] [PubMed] [Google Scholar]
- Hallouard F.; Anton N.; Choquet P.; Constantinesco A.; Vandamme T. Biomaterials 2010, 31, 6249. 10.1016/j.biomaterials.2010.04.066. [DOI] [PubMed] [Google Scholar]
- Maeda H.; Nakamura H.; Fang J. Adv. Drug Delivery Rev. 2013, 65, 71. 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Maeda H.; Wu J.; Sawa T.; Matsumura Y.; Hori K. J. Controlled Release 2000, 65, 271. 10.1016/S0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- Lee C. C.; Mackay J. A.; Frechet J. M. J.; Szoka F. C. Nat. Biotechnol. 2005, 23, 1517. 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- Kobayashi H.; Sato N.; Hiraga A.; Saga T.; Nakamoto Y.; Ueda H.; Konishi J.; Togashi K.; Brechbiel M. W. Magn. Reson. Med. 2001, 45, 454.. [DOI] [PubMed] [Google Scholar]
- Yordanov A. T.; Yamada K.-I.; Krishna M. C.; Mitchell J. B.; Woller E.; Cloninger M.; Brechbiel M. W. Angew. Chem. 2001, 113, 2762.. [DOI] [PubMed] [Google Scholar]
- Yordanov A. T.; Lodder A. L.; Woller E.; Cloninger M.; Patronas N.; Milenic D.; Brechbiel M. W. Nano Lett. 2002, 2, 595. 10.1021/nl020246x. [DOI] [Google Scholar]
- Rabinow B. E. Nat. Rev. Drug Discovery 2004, 3, 785. 10.1038/nrd1494. [DOI] [PubMed] [Google Scholar]
- Kong W. H.; Lee W. J.; Cui Z. Y.; Bae K. H.; Park T. G.; Kim J. H.; Park K.; Seo S. W. Biomaterials 2007, 28, 5555. 10.1016/j.biomaterials.2007.08.044. [DOI] [PubMed] [Google Scholar]
- Solans C.; Izquierdo P.; Nolla J.; Azemar N.; Garcia-Celma M. J. Curr. Opin. Colloid Interface Sci. 2005, 10, 102. 10.1016/j.cocis.2005.06.004. [DOI] [Google Scholar]
- Mukundan S. Jr.; Ghaghada K. B.; Badea C. T.; Kao C. Y.; Hedlund L. W.; Provenzale J. M.; Johnson G. A.; Chen E.; Bellamkonda R. V.; Annapragada A. AJR, Am. J. Roentgenol. 2006, 186, 300. 10.2214/AJR.05.0523. [DOI] [PubMed] [Google Scholar]
- Fu Y.; Nitecki D. E.; Maltby D.; Simon G. H.; Berejnoi K.; Raatschen H. J.; Yeh B. M.; Shames D. M.; Brasch R. C. Bioconjugate Chem. 2006, 17, 1043. 10.1021/bc060019c. [DOI] [PubMed] [Google Scholar]
- Raatschen H. J.; Fu Y.; Brasch R. C.; Pietsch H.; Shames D. M.; Yeh B. M. Invest. Radiol. 2009, 44, 265. 10.1097/RLI.0b013e31819f1b60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter P.; Shukla H. P.; Caruthers S. D.; Scott M. J.; Fuhrhop R. W.; Robertson J. D.; Gaffney P. J.; Wickline S. A.; Lanza G. M. Acad. Radiol. 2005, 12, S9. 10.1016/j.acra.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Kinsella J. M.; Jimenez R. E.; Karmali P. P.; Rush A. M.; Kotamraju V. R.; Gianneschi N. C.; Ruoslahti E.; Stupack D.; Sailor M. J. Angew. Chem., Int. Ed. 2011, 50, 12308. 10.1002/anie.201104507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormode D. P.; Skajaa T.; van Schooneveld M. M.; Koole R.; Jarzyna P.; Lobatto M. E.; Calcagno C.; Barazza A.; Gordon R. E.; Zanzonico P.; Fisher E. A.; Fayad Z. A.; Mulder W. J. M. Nano Lett. 2008, 8, 3715. 10.1021/nl801958b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D.; Williams T. A.; Senpan A.; Allen J. S.; Scott M. J.; Gaffney P. J.; Wickline S. A.; Lanza G. M. J. Am. Chem. Soc. 2009, 131, 15522. 10.1021/ja906797z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C.; Schaefer S. C.; Juillerat-Jeanneret L.; Lagopoulos L.; Lehr H. A.; Becker C. D.; Montet X. Eur. Radiol 2009, 19, 2487. 10.1007/s00330-009-1434-2. [DOI] [PubMed] [Google Scholar]
- Hill M. L.; Corbin I. R.; Levitin R. B.; Cao W. G.; Mainprize J. G.; Yaffe M. J.; Zheng G. Acad. Radiol 2010, 17, 1359. 10.1016/j.acra.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Bogdanov A. A.; Weissleder R. Jr.; Brady T. J. Adv. Drug Delivery Rev. 1995, 16, 335. 10.1016/0169-409X(95)00034-5. [DOI] [Google Scholar]
- Krause W.; Hackmann-Schlichter N.; Maier F. K.; Muller R. Top. Curr. Chem. 2000, 210, 261. 10.1007/3-540-46577-4_6. [DOI] [Google Scholar]
- Blum G.; Mullins S. R.; Keren K.; Fonovic M.; Jedeszko C.; Rice M. J.; Sloane B. F.; Bogyo M. Nat. Chem. Biol. 2005, 1, 203. 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- Ren G.; Blum G.; Verdoes M.; Liu H.; Syed S.; Edgington L. E.; Gheysens O.; Miao Z.; jiang H.; Gambhir S. S.; Bogyo M.; Cheng Z. PLoS One 2011, 6, e28029. 10.1371/journal.pone.0028029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato D.; Boatright K. M.; Berger A. B.; Nazif T.; Blum G.; Ryan C.; Chehade K. A. H.; Salvesen G. S.; Bogyo M. Nat. Chem. Biol. 2005, 1, 33. 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- Jedeszko C.; Sloane B. F. Biol. Chem. 2004, 385, 1017. 10.1515/BC.2004.132. [DOI] [PubMed] [Google Scholar]
- Joyce J. A.; Baruch A.; Chehade K.; Meyer-Morse N.; Giraudo E.; Tsai F. Y.; Greenbaum D. C.; Hager J. H.; Bogyo M.; Hanahan D. Cancer Cell 2004, 5, 443. 10.1016/S1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Salpeter S. J.; Pozniak Y.; Merquiol E.; Ben-Nun Y.; Geiger T.; Blum G. Oncogene 2015, 34, 6066. 10.1038/onc.2015.51. [DOI] [PubMed] [Google Scholar]
- Ben-Nun Y.; Merquiol E.; Brandis A.; Turk B.; Scherz A.; Blum G. Theranostics 2015, 5, 847. 10.7150/thno.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G.; von Degenfeld G.; Merchant M. J.; Blau H. M.; Bogyo M. Nat. Chem. Biol. 2007, 3, 668. 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- Leroueil P. R.; Berry S. A.; Duthie K.; Han G.; Rotello V. M.; McNerny D. Q.; Baker J. R.; Orr B. G.; Banaszak Holl M. M. Nano Lett. 2008, 8, 420. 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]
- Jain K.; Kesharwani P.; Gupta U.; Jain N. K. Int. J. Pharm. 2010, 394, 122. 10.1016/j.ijpharm.2010.04.027. [DOI] [PubMed] [Google Scholar]
- DailyMed, NIH U.S. National Library of Medicine, https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=764f5085-7ee8-fdba-2c5b-ae6721014a94, accessed January 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.