Abstract
Objectives
To apply advanced diffusion MRI methods to the study of normal-appearing brain tissue in MS and examine their correlation with measures of clinical disability.
Methods
A multi-compartment model of diffusion MRI called neurite orientation dispersion and density imaging (NODDI) was used to study 20 patients with relapsing-remitting MS (RRMS), 15 with secondary progressive MS (SPMS), and 20 healthy controls. Maps of NODDI were analyzed voxel-wise to assess the presence of abnormalities within the normal-appearing brain tissue and the association with disease severity. Standard diffusion tensor imaging (DTI) parameters were also computed for comparing the 2 techniques.
Results
Patients with MS showed reduced neurite density index (NDI) and increased orientation dispersion index (ODI) compared with controls in several brain areas (p < 0.05), with patients with SPMS having more widespread abnormalities. DTI indices were also sensitive to some changes. In addition, patients with SPMS showed reduced ODI in the thalamus and caudate nucleus. These abnormalities were associated with scores of disease severity (p < 0.05). The association with the MS functional composite score was higher in patients with SPMS compared with patients with RRMS.
Conclusions
NODDI and DTI findings are largely overlapping. Nevertheless, NODDI helps interpret previous findings of increased anisotropy in the thalamus of patients with MS and are consistent with the degeneration of selective axon populations.
The factors triggering the transition from relapsing-remitting MS (RRMS) to secondary progressive MS (SPMS) are still largely unknown. The time to transition and the degree of long-term disability show only limited correlation with white matter (WM) lesion number or volume,1,2 suggesting that the accumulation of tissue damage outside of macroscopic lesions contributes to the determination of clinical disability, particularly in SPMS.1,3–5 Damage to the normal-appearing WM (NAWM) has been extensively studied using quantitative MRI techniques,3,6–8 and particularly diffusion MRI (dMRI).1,8 dMRI indirectly measures WM orientation and integrity by probing the random motion of water molecules within tissue.8 The most popular model of diffusion, diffusion tensor imaging (DTI),9 assumes a single water compartment and a dominant tissue orientation per voxel. The indices derived from this model, and in particular mean diffusivity (MD) and fractional anisotropy (FA), have been used as surrogate measures of microstructural tissue change, although their specificity is limited.8 Gray matter (GM) damage including lesions in the cortex and subcortical structures is a prominent aspect of MS pathology,2,10–13 which might be associated with disease progression. Because of the structure of GM, DTI is less sensitive to changes occurring within this tissue, where partial volume effects with CSF (in the cortex) can also act as a confound. Neurite orientation dispersion and density imaging (NODDI)14 was recently proposed to overcome the limitations of DTI and to capture the morphology of dendrites and of axons. The NODDI model is a simplified version of a diffusion model of neurite morphology,15 shown to correlate well with histologic analyses of neuropil orientation in vitro. NODDI was designed to work with data acquired within a clinically feasible scan time, thus enabling clinical applications. NODDI assumes that each MRI voxel may contain CSF and tissue and that the tissue itself comprises intra- and extracellular water. The main parameters derived from NODDI are neurite density index (NDI), quantifying neurite concentration, and neurite orientation dispersion index (ODI).14 The intracellular component of NODDI is designed to be representative of both axons and dendrites, thus providing an improved description of GM microstructure. In addition, NODDI enables changes to tissue density and tissue ODI (both expressed by FA changes when using DTI) to be disentangled, ultimately offering superior sensitivity to tissue changes. This ability to distinguish between neurite loss and changes to fiber arrangement can shed light on the mechanism underpinning the pathology observed in MS brains. A recent postmortem study16 highlighted significant changes in ODI within spinal cord lesions, supported by histopathology. Therefore, we hypothesize that reduced NDI should reflect decreased axonal and myelin density, whereas changes to ODI could reflect secondary fiber degeneration, but also morphological changes to single axons in the WM, and altered dendritic arborization in the GM. Initial observations in relatively small MS cohorts suggest that NDI is reduced in both lesions and NAWM, whereas changes in ODI can occur in both directions.17–19 This study aimed to identify MRI-derived hallmarks to distinguish between patients with RRMS and SPMS, exploiting the close relationship between NODDI metrics and brain microstructure. The main hypothesis was that SPMS would present more widespread changes in both NDI and ODI and that such changes would involve both the white and the GM. We first compared the ability of NODDI indices to characterize tissue abnormalities in MS brains with those of standard DTI indices, and we assessed the relationship between NODDI metrics in the GM and clinical scales. Ultimately, we tested NODDI for its potential ability in clarifying the mechanisms beyond relapsing-remitting and secondary progressive phenotypes and in providing clinical utility for biomarking MS progression. Our expectation was that specific patterns of ODI and NDI changes in the GM might help characterizing the pathophysiologic substrate for the transition from RR to SPMS.
Methods
Participants
Twenty patients with RRMS (mean ± SD, age 42.9 ± 6.1, male/female 8/12) and 15 patients with SPMS (mean ± SD, age 50.7 ± 7.2, male/female 8/7)20,21 were included. No patient experienced any clinical relapse or underwent any steroid treatment over the 3 months preceding recruitment. Twenty age- and sex-matched healthy participants (mean ± SD age 44.5 ± 11.7, male/female 8/12) were also recruited and served as healthy controls (HCs). All patients were rated with the Expanded Disability Status Scale (EDSS) score22 and the MS functional composite (MSFC) score.23 For each patient, the z-scores of the MSFC subtests were created with reference to the control participants. Patients were defined as impaired when their score was at least 2 SD below that of HC. Patients underwent MR scanning within 1 week after enrolment.
Standard protocol approvals, registrations, and patient consents
This study was approved by the local Ethics Committee of the Santa Lucia Foundation. All participants provided written informed consent before study initiation.
MRI acquisition
MRI data were acquired at 3T (Philips Achieva, Philips, Best, The Netherlands), including (1) dual-echo turbo spin echo (TSE) (TR = 2,000 ms, TE1 = 10 ms, TE2 = 80 ms; TSE factor = 10; matrix = 400 × 250 × 28; slice gap = 1 mm; field of view [FOV] = 230 × 184 × 139 mm3; slice thickness = 4 mm; total number of slices = 28); (2) fast fluid-attenuated inversion recovery (FLAIR) (repetition time [TR] = 11,000 ms, echo time [TE] = 125 ms, inversion time [TI] = 2,800 ms; TSE factor = 31; matrix = 288 × 211 × 28; slice gap = 1 mm; FOV = 230 × 192 × 167 mm3; slice thickness = 5 mm; total number of slices = 28); (3) T1-weighted inversion-recovery fast field echo (TR = 11 ms; TE = 5.3 ms; Flip angle = 8°; matrix = 256 × 228 × 190; slab thickness = 0.9 mm; FOV = 230 × 192 × 167 mm3); and (4) 2-shell high angular resolution diffusion imaging (HARDI) scheme, optimized for the NODDI protocol.14 The optimized protocol consists of 1 shell with 30 gradient directions at b = 711 s/mm2 and the other with 60 directions at b = 2,855 s/mm2. The HARDI shells were acquired using diffusion-weighted spin-echo echo-planar imaging (TR = 12.5 s, TE = 91 ms, isotropic resolution = 2.3 mm3, SENSE reduction factor = 2). Nine b = 0 images were also acquired, with the same imaging parameters as the dMRI. The scan time for the NODDI component of this protocol was approximately 20 minutes.
Lesion volume assessment
T2-hyperintense lesions were identified by the consensus of 2 expert observers (B.S. and M.B.) on FLAIR and T2-weighted images, for every patient, and outlined on FLAIR scans using semi-automated local thresholding contouring software (Jim 5.0; Xinapse System, West Bergholt, Essex, UK, xinapse.com/), yielding the total lesion volume. A lesion mask was then created for each patient by assigning a value of 1 to every voxel corresponding to a lesion and a value of 0 elsewhere. A diffeomorphic transformation was computed using the advanced normalization tools (ANTs)24 to warp FLAIR images into standard space, and the same transformation was then applied to the corresponding lesion mask. A probabilistic lesion map, indicating the percentage of patients with a lesion in a given area, was obtained by combining every patient's lesion mask in standard space (figure 1, A). Then, a population lesion mask was created by thresholding the probabilistic lesion mask at 10% and binarizing it (i.e., setting the value equal to 1 for voxels where at least 10% of the patients had a lesion and 0 elsewhere). This threshold was chosen to match with previous studies.25 The population lesion mask was subtracted from the statistical parametric mapping (fil.ion.ucl.ac.uk/spm/) brain mask, and the resulting image was used to confine the statistical analysis of quantitative data (i.e., NODDI) to the normal-appearing brain tissue, thus excluding the areas where at least 10% of patients had lesions. Figure 1, B shows the binary mask used for the analysis (thresholded at 10%) compared with the lesion distribution across all patients (i.e., thresholded at 1%).
Figure 1. Lesion distribution in patients.
(A) Probabilistic lesion map showing the absolute number and percentage (in brackets) of patients with MS with a lesion in a given area. The map is overlaid onto a T1-weighted image in Montreal Neurological Institute space. (B) Binarized lesion mask after thresholding at 10% (purple) and 1% (blue). This comparison shows the extent of tissue included in the analysis, which was partially affected by macroscopic lesions.
Computation of the NODDI maps
For every participant, dMRI data were corrected for eddy current effects and involuntary movement by affine coregistration using the FLIRT tool (part of the FMRIB Software Library; FSL21), as described elsewhere.26 NODDI fitting was then performed using the accelerated microstructure imaging via the convex optimization (github.com/daducci/AMICO/)27 toolbox, which is a linear implementation of the NODDI model. To obtain DTI maps comparable with the existing literature, we used the inner shell (b = 711) and the b0 images to estimate the tensor by multivariate linear regression in CAMINO (camino.cs.ucl.ac.uk). MD and FA maps were derived for every participant. The resulting NODDI (NDI and ODI) and DTI (FA and MD) maps were normalized into standard space, warping the skull-stripped mean b0 images to the skull-stripped T2 MNI atlas available in FSL (JHU-ICBM-T2-2 mm) and then applying the same nonlinear transformation to the NODDI and DTI maps. To compute this space transformation, we used the diffeomorphic transformations algorithm as implemented by ANTs,21 which is able to compensate for the spatial distortions shown by the diffusion-weighted images. Finally, the normalized images were smoothed using an 8 mm3 Gaussian kernel before statistical analysis.
Statistical analysis
The statistical analyses on demographic, clinical, and conventional MRI data were performed using SPSS 21.0 for Windows (SPSS Inc). Chi-square tests and one-way analyses of variance with post hoc Tukey comparisons (to assess specific group differences) were used to compare all groups for sex and age, respectively. Disease duration, MSFC score, and T2-L volume were compared between patient groups using 2-sample t tests (2-tailed). Between-group EDSS score differences were assessed using a Kruskal-Wallis test. Voxel-wise statistical analyses of MRI data were performed using SPM8. Between-group comparisons of NODDI (NDI and ODI) and DTI (MD and FA) metrics were performed using 4 separate full factorial designs, in which 3 groups were modeled (HC, RRMS, and SPMS). Age and sex were always added as covariates of no interest. T-contrasts were used for testing between-group differences. Linear correlation was used to assess the potential relationship between each clinical score (EDSS and MSFC) and NODDI (NDI and ODI) and DTI (FA and MD) metrics in the brain. First, we tested for the presence of any group-by-score interaction, by modeling 2 groups (RRMS and SPMS). If no significant interaction was present, the analysis was repeated by grouping all patients together. If a significant result was found, scatterplots of the diffusion index averaged across all significant voxels vs the clinical scores were examined to support the interpretation. All voxel-wise analyses were adjusted for age and sex, explicitly masking for normal-appearing brain tissue (i.e., using the 10% thresholded mask). For detection of results on whole-brain level, we always used a cluster-forming threshold of p = 0.001, with cluster-level whole brain corrected (family wise error) p < 0.05 as the statistical significance threshold. These thresholds are based on current recommendations.28
Results
Demographic, clinical, and conventional MRI data
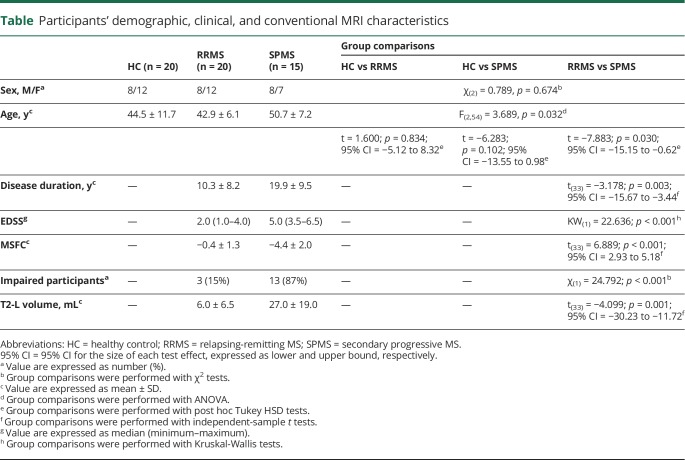
Participants' demographic, clinical, and conventional MRI (lesion volume) data are reported in table. There were no significant differences between HC and either patient group with respect to age and sex distribution. As expected, patients with SPMS were older and more disabled than those with RRMS and had higher T2-L volumes.
Table.
Participants' demographic, clinical, and conventional MRI characteristics
Voxel-wise NDI comparisons
Patients with RRMS compared with HC showed bilateral reductions of NDI in the tail of the hippocampus, fornix and fimbria, cingulate and lingual gyri, corpus callosum, thalamus, cerebellum, and brainstem (figure 2, A, upper panel, in red). More extensive NDI reductions were observed in patients with SPMS compared with HC involving the pre- and postcentral gyri, inferior and orbitofrontal gyri, operculum, superior, middle and inferior temporal gyri, supramarginal and angular gyri, cuneus and precuneus, insula, lingual gyrus, parahippocampal and fusiform gyri, hippocampus, fornix and fimbria, corpus callosum, paracingulate and cingulate gyri, thalamus, internal/external capsule, caudate, putamen, cerebellum, and brainstem (figure 2, B, upper panel, in red). Despite at a lesser extent, a similar pattern of NDI reductions was found when comparing patients with SPMS with patients with RRMS (figure 2, C, upper panel, in red). No areas of increased NDI were found in either patient group compared with HC and in patients with SPMS compared with patients with RRMS.
Figure 2. Voxel-wise differences across groups.
NODDI indices are shown in red, DTI indices in green, and the overlap in yellow. NDI/MD and ODI/FA are shown on the same overlay, respectively. (A) Differences between HC and RRMS. (B) Differences between HC and SPMS. (C) Differences between RRMS and SPMS. Significant clusters are shown at p value < 0.05 (FWE corrected), overlaid onto a template in Montreal Neurological Institute space. DTI = diffusion tensor imaging; FA = fractional anisotropy; FWE = family wise error; HC = healthy control; MD = mean diffusivity; NDI = neurite density index; NODDI = neurite orientation dispersion and density imaging; ODI = orientation dispersion index; RRMS = relapsing-remitting MS; SPMS = secondary progressive MS.
Voxel-wise ODI comparisons
Across the brain, we observed areas of both increased and reduced ODI in patients with MS compared with HC. Patients with RRMS showed significant ODI increase in the left supramarginal and angular gyri, in the left cingulate and paracingulate gyri, in the left corpus callosum anteriorly, in the right cingulate and precuneus gyri, and in the fornix, thalamus, and cerebellum bilaterally (figure 2, A, bottom row, in red). Patients with SPMS showed a more widespread and bilateral pattern of ODI increase including all these anatomic regions plus the orbitofrontal gyrus, operculum, hippocampal, parahippocampal and fusiform gyri, insula, caudate, and brainstem (figure 2, B, middle row, in red). Patients with SPMS compared with HC showed also significant ODI reductions in the caudate nucleus, in the anterior and posterior limb of the internal capsule, and in the thalamus bilaterally (figure 2, B, bottom row). When looking at the differences between patient groups, increased ODI was observed in patients with SPMS in the hippocampus and parahippocampus, in the superior and middle temporal gyri, cingulate and paracingulate gyri, and in the anterior corpus callosum bilaterally (figure 2, C, middle row, in red). In addition, patients with SPMS compared with patients with RRMS showed significant ODI reductions in the right thalamus (figure 2, C, bottom row).
Voxel-wise DTI differences across groups
Between-group differences in FA and MD that are shown in figure 2 are overlaid onto the same sections as the NODDI parameters, to ease the comparison. For all the categorical comparisons, MD increases were broadly overlapping with NDI reductions, whereas reductions in FA-mapped areas were ODI was found to be increased. No areas of increased FA in either patients with RRMS or patients with SPMS compared with HC and in patients with SPMS compared with patients with RRMS were found.
NDI correlations
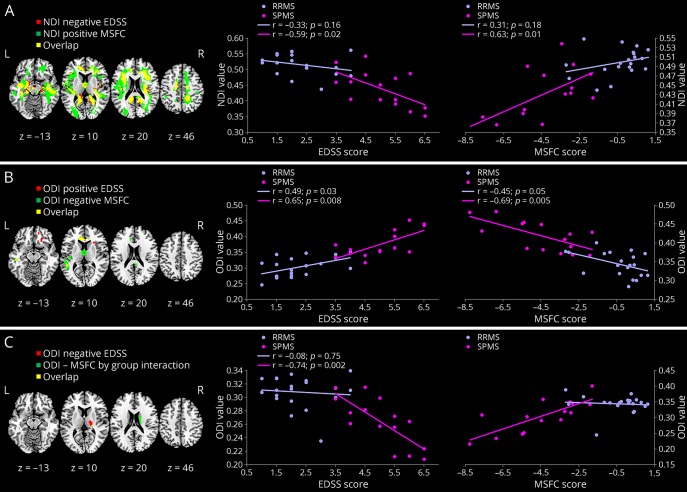
When looking at the relationship between EDSS or MSFC scores and NDI, no significant interaction for both EDSS and MSFC score by group (RRMS vs SPMS) was observed. By contrast, significant correlations were found between NDI and EDSS, as well as MSFC scores across all patients with MS. The areas of significant correlation are shown in figure 3, A (in red for EDSS and green for MSFC). NDI was found to correlate negatively with EDSS and positively with MSFC scores in several areas of the brain. These clusters, which were largely overlapping (yellow areas in the figure), but were larger for the MSFC scores, are localized in the pre- and postcentral gyri, inferior and orbitofrontal frontal gyri, operculum, superior, middle and inferior temporal gyri, supramarginal and angular gyri, cuneus and precuneus, insula, lingual gyrus, parahippocampal and fusiform gyri, hippocampus, fornix and fimbra, corpus callosum, paracingulate and cingulate gyri, thalamus, internal/external capsule, caudate, putamen, cerebellum, and brainstem. The scatterplots suggest that such correlations are driven by the SPMS group.
Figure 3. Areas of significant association.
Areas of significant association between NODDI metrics and EDSS (in red) and MSFC (in green). The overlap is shown in yellow. (A) Areas of significant association between NDI and EDSS/MSFC. (B) Areas of significant association between ODI and EDSS (positive) and MSFC (negative). (C) Areas of significant correlation between ODI and EDSS (negative) and MSFC (positive). For each panel, the scatterplot shows the trends for RRMS and SPMS separately. In all scatterplots NDI, ODI, and FA are averaged across all significant voxels, ranging from 0 to 1, and are dimensionless. Significant clusters are shown at p value < 0.05 (FWE corrected) overlaid onto a template in Montreal Neurological Institute space. EDSS = Expanded Disability Status Scale; FA = fractional anisotropy; FWE = family wise error; MSFC = MS functional composite; NDI = neurite density index; NODDI = neurite orientation dispersion and density imaging; ODI = orientation dispersion index; RRMS = relapsing-remitting MS; SPMS = secondary progressive MS.
ODI correlations
No areas of significant score by group interaction were found for EDSS and ODI. Increased EDSS scores were associated with increased ODI, bilaterally, in the cingulate gyrus, precuneus, insula, corpus callosum, cerebellum, and brainstem, other than the left superior and middle temporal gyri and the right orbito-frontal gyrus (figure 3, B). In addition, reduced ODI changes in the right caudate nucleus and thalamus were associated with increased EDSS scores (figure 3, C). To support the interpretation of these results, scatterplots of the average ODI from the areas of significant association vs the clinical scores are presented alongside. When looking at the relationship between MSFC scores and ODI, a significant interaction for the MSFC score by group (RRMS vs SPMS) was observed in the right thalamus, indicating an association between increased MSFC scores and increased ODI values in SPMS, which is not present in patients with RRMS (figure 3, C). By contrast, reduced ODI values were associated with increased MSFC across all patients in the anterior portion of caudate and cingulate gyri, as well as in the left insula, orbito-frontal, superior, and medial temporal gyri (figure 3, B). Again, the scatterplots indicate that these correlations are mainly driven by SPMS (figure 3, B).
DTI correlations
Significant associations with clinical scores were found also for DTI indices. In particular, disability was mainly associated with FA of the corpus callosum, inferior longitudinal fasciculus, and orbito-frontal WM. MD showed broader areas of association, including medial-temporal structures, insular cortex, thalamus, and parietal lobe. These are shown in the figure e-1 (links.lww.com/NXI/A76).
Data availability
All relevant data are available on request addressed to the corresponding author.
Discussion
So far, NODDI has been applied to several neurologic and psychiatric conditions, including Alzheimer disease,29 focal cortical dysplasia,30 idiopathic Parkinson disease,31 and psychosis.32 One of the main advantages of NODDI over DTI is in its ability to disentangle the contributions of axonal/dendritic density and fiber orientation to microscopic changes, by contrast both represented by FA changes. As discussed below, NODDI thus allows a better definition of the neurobiological substrate underlying brain tissue damage in MS and, possibly, its relationship with clinical aspects of the disease. Our data indicate the presence of diffuse brain abnormalities in patients with MS, with RRMS showing only localized changes and patients with SPMS showing a significantly more widespread involvement, particularly in the GM. Both DTI and NODDI indices have proven to be sensitive to these changes; however, NODDI provides valuable insights into the likely nature of these abnormalities. GM pathology (including demyelinated cortical lesions and diffuse cortical and deep GM degeneration) has emerged as an important feature of long-term disability and disease progression in MS.33 Indeed, pathologic evidence suggests that the WM is predominantly involved in inflammatory demyelination of RRMS, whereas cortical and subcortical GM is predominantly involved in neurodegenerative processes of SPMS.33 We found that patients with SPMS exhibited a widespread loss of neurite integrity (i.e., decrease of NDI) along with a loss of fiber coherences (i.e., an increase of ODI) in the GM (cortical and subcortical). Conversely, neurite/fiber abnormalities in the GM of patients with RRMS were less extensive. This fits with the hypothesis that GM pathology is subclinical in the early RR phase of MS, but dominates the clinical picture of SPMS by causing irreversible disability.5,12,34 In concert with our results, a recent postmortem study35 evaluated axonal and dendritic alterations in different cortical brain regions (i.e., insular, fronto-temporal, and occipital lobe) of patients with MS and long disease duration. The authors found a widespread loss of dendritic spines across the whole GM and of cortical axons within the demyelinated GM.35 Changes to the status of local microglia have also been proposed as a possible contributor to the observed dMRI changes in the GM.12,36 This is still a matter of debate because a recent postmortem study has failed to demonstrate a significant presence of activated microglia in the cortex, thus suggesting that inflammation is not prominent in GM lesions.37 Consistent with that, a recent study combining 7T postmortem MRI data with histopathologic examination38 was designed specifically to test the hypothesis that microglial activation may explain the increased FA in cortical GM lesions and concluded that such an explanation is unlikely. Based on comparison with histology, it was recently shown that NDI reflects both axonal and myelin loss,16 whereas ODI reflects morphologic changes to axons and dendrites. Our results suggest that the changes to ODI are confined to specific regions of the brain and might have a relationship with disease progression. We found both increased and decreased ODI in different areas of the brain. This apparently counterintuitive result can be explained by the fact that ODI reflects a complex combination of structural changes, which range from axonal loss, dendritic pruning, and tissue reorganization. Some of these changes will result in an increase, whereas others in a decrease of this parameter. This will also be partially determined by the original tissue configuration (i.e., whether the area of the change is characterized by a high or low dispersion in the healthy brain). In other words, areas that in the normal brain exhibit a high ODI, such as the GM, are more likely to show a decrease in ODI, if, for example, they experience a loss of dendrites. By contrast, highly coherent WM fibers might show an increase in ODI after tissue disruption. A range of other scenarios can also occur. Indeed, we observed a significantly reduced ODI in the caudate nuclei and thalami in patients with SPMS when compared with both HC and patients with RRMS. More importantly, in the areas in which ODI was reduced, this same parameter was strictly associated with patients' EDSS and MSFC scores. Both these associations were significant in patients with SPMS but not in those with RRMS (in the case of MSFC, with a significant interaction). Although we can only speculate on the pathology underpinning these findings, we argue that the lower neurite ODI in the caudate nuclei and thalamus of patients with SPMS can be due to targeted degeneration of a single axon population, which would result in diminished dispersion. Although we did not find evidence of increased FA in the same area, this might be due to the fact that FA reflects both loss of NDI and changes in dispersion, which might cancel each other out.39 Other authors have previously reported increased FA in the thalamus and basal ganglia of patients with MS,12,40 as discussed below. The presence of profound neuronal loss in the thalamus and other deep GM nuclei has already been previously implicated in MS pathophysiology, often in association with clinical measures of disability.2 This neuronal loss in the deep GM could, in part, be explained by anterograde and/or retrograde neuronal degeneration due to lesions within connecting fiber tracts.34,41 With respect to diffusion anisotropy, the caudate nucleus and the thalamus are characterized by differing structures: the cells in the caudate (cell bodies and dendrites) tend to show near-isotropy at the voxel scale, whereas the presence of myelinated fibers between thalamic nuclei tend to result in higher anisotropy of the latter structure. The caudate nucleus receives information from multiple cortical regions and transmits it via the globus pallidus and the thalamus back to the neocortex,40,42,43 thus exhibiting a large ODI. If the fibers that connect cortical areas with these subcortical structures are selectively affected by Wallerian degeneration or neuronal dysfunction (diaschisis), this could result in decreased ODI. Such an interpretation has been previously suggested by Ciccarelli et al.,40 who showed a paradoxical increase in FA of the normal-appearing basal ganglia (caudate and putamen) along with a MD reduction in patients with MS. These authors excluded a contribution from gliosis, which would have caused a more marked tissue disorganization (i.e., reduced anisotropy and increased T2) and attributed this apparently paradoxical finding to axonal degeneration due to fiber transection in remote focal MS lesions. Later on, other authors also reported paradoxical increases of tissue anisotropy in the basal ganglia and in the thalamus of patients with MS, together with correlations with scores of disease severity.2,10,12,36 A recent study12 speculated that an increase in FA of both caudate and thalamic nuclei might reflect progressive GM degeneration in patients with RRMS and SPMS and that this measure might constitute a sensitive biomarker of specific pathologic processes, such as the loss of dendrites. The increased specificity of NODDI confirms these earlier speculations and suggests that such pathologic processes might be closely associated with disability, specifically in SPMS. As a consequence, deep GM ODI might provide a valuable biomarker, and its ability to predict the risk of transition from RRMS to SPMS should be evaluated with longitudinal designs.
This study also suffers from a series of limitations. First, we chose to exclude from our analysis voxels where at least 10% of the patient cohort had a lesion. On the one hand, this led to the exclusion of a large proportion of the WM, which might explain the specific location of the changes we found. On the other hand, by not excluding all the lesions, we are unable to draw conclusions about the independent contributions of lesions and NAWM to the abnormalities we detected. One possible way to assess this would be to include the total lesion load as an additional covariate to our clinical correlation analysis. However, in this heterogeneous patient cohort, lesion load and disability are highly likely to be correlated (SPMS tends to have a higher lesion load), and therefore, adding T2L volume as a covariate is bound to account for a large part of the variance without necessarily informing the underlying processes. Another potential confound is atrophy. Partial volume with CSF can bias the voxel-wise comparison of dMRI parameters; however, as NODDI models separately the isotropic component of diffusion (free water), it should be less sensitive to atrophy than DTI. To ensure that atrophy was not the main driver for our findings, we repeated the analysis adjusting for GM volume. As expected, the results were virtually identical (figure e-2, links.lww.com/NXI/A76). Although NODDI arguably provides advantages compared with DTI, its limitations must also be considered. Similar to all models, it provides a simplified representation of tissue complexity, and to enable the use of a clinically friendly acquisition time, it makes a series of assumptions, which might not always hold true.44 Despite this, increasing evidence supports the concordance of NODDI with histology.16 Our data show widespread microscopic abnormalities in the GM of MS brains and support the notion that different substrates (i.e., neuropil, axons, dendrite proliferation, and connections) are likely to contribute to neurodegenerative progression of disease and global disability of the patients. GM abnormalities are more prominent in SPMS than RRMS and correlate strictly with patients' clinical disability. Although confirmation from larger studies is needed, this indicates that NODDI may provide reliable information of prognostic value in MS and might in future be used for clinical trial monitoring and, hopefully, for clinical routine. This will be facilitated by the advent of increasingly faster acquisition schemes, such as those based on simultaneous multislice.45 The changes we observed in the basal ganglia and in the thalamus might result from the selective degeneration of a single axonal population. Future work should focus on relating NODDI changes directly with histopathologic findings and comparing them with other, complementary, MRI contrasts.
Glossary
- ANT
advanced normalization tool
- DTI
diffusion tensor imaging
- EDSS
Expanded Disability Status Scale
- FA
fractional anisotropy
- FLAIR
fluid-attenuated inversion recovery
- FOV
field of view
- GM
gray matter
- HARDI
high angular resolution diffusion imaging
- HC
healthy control
- MD
mean diffusivity
- MSFC
MS functional composite
- NAWM
normal appearing WM
- NDI
neurite density index
- NODDI
neurite orientation dispersion and density imaging
- ODI
orientation dispersion index
- RRMS
relapsing-remitting MS
- SPMS
secondary progressive MS
- TE
echo time
- TI
inversion time
- TR
repetition time
- TSE
turbo spin echo
- WM
white matter
Authors contributions
B. Spanò, M. Bozzali, C. Caltagirone, and M. Cercignani were responsible for study design and conception and drafting of the manuscript. B. Spanò, G. Giulietti, V. Pisani, M. Morreale, E. Tuzzi, U. Nocentini, and A. Francia contributed to data collection and analysis.
Study funding
This work was funded by a grant from the Italian Ministry of Health (RF-2013-02358409) awarded to M. Cercignani.
Disclosure
B. Spano, G. Giulietti, V. Pisani, M. Morreale, and E. Tuzzi report no disclosures. U. Nocentini served on the scientific advisory board of Biogen and received speaker honoraria from Merck Serono. A. Francia reports no disclosures; C. Caltagirone served on the editorial board of Neurological Sciences. M. Bozzali served as an associate editor for the Journal of Alzheimer's Disease and Frontiers Cellular Neuroscience and received research support from the Italian Ministry of Health. M. Cercignani served on the editorial board of Magnetic Resonance in Medicine and Frontiers in Cellular Neuroscience and received research support from the Italian Ministry of Health, the Wellcome Trust, Academy of Medical Science, Arthritis Research UK, and MND Association. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NN.
References
- 1.Bozzali M, Spanò B, Parker GJ, et al. . Anatomical brain connectivity can assess cognitive dysfunction in multiple sclerosis. Mult Scler 2013;19:1161–1168. [DOI] [PubMed] [Google Scholar]
- 2.Cavallari M, Ceccarelli A, Wang GY, et al. . Microstructural changes in the striatum and their impact on motor and neuropsychological performance in patients with multiple sclerosis. PLoS One 2014;9:e101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cercignani M, Bozzali M, Iannucci G, Comi G, Filippi M. Intra-voxel and inter-voxel coherence in patients with multiple sclerosis assessed using diffusion tensor MRI. J Neurol 2002;249:875–883. [DOI] [PubMed] [Google Scholar]
- 4.Spanò B, Cercignani M, Basile B, et al. . Multiparametric MR investigation of the motor pyramidal system in patients with “truly benign” multiple sclerosis. Mult Scler 2010;16:178–188. [DOI] [PubMed] [Google Scholar]
- 5.Calabrese M, Romualdi C, Poretto V, et al. . The changing clinical course of multiple sclerosis: a matter of gray matter. Ann Neurol 2013;74:76–83. [DOI] [PubMed] [Google Scholar]
- 6.Londoño AC, Mora CA. Nonconventional MRI biomarkers for in vivo monitoring of pathogenesis in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm 2014;1:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller TR, Mohan S, Choudhri AF, Gandhi D, Jindal G. Advances in multiple sclerosis and its variants: conventional and newer imaging techniques. Radiol Clin North Am 2014;52:321–336. [DOI] [PubMed] [Google Scholar]
- 8.Enzinger C, Barkhof F, Ciccarelli O, et al. . Nonconventional MRI and microstructural cerebral changes in multiple sclerosis. Nat Rev Neurol 2015;11:676–686. [DOI] [PubMed] [Google Scholar]
- 9.Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B 1994;103:247–254. [DOI] [PubMed] [Google Scholar]
- 10.Hasan KM, Halphen C, Kamali A, Nelson FM, Wolinsky JS, Narayana PA. Caudate nuclei volume, diffusion tensor metrics, and T(2) relaxation in healthy adults and relapsing-remitting multiple sclerosis patients: implications for understanding gray matter degeneration. J Magn Reson Imaging 2009;29:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutta R, Chang A, Doud MK, et al. . Demyelination causes synaptic alterations in hippocampi from multiple sclerosis patients. Ann Neurol 2011;69:445–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannoun S, Durand-Dubief F, Confavreux C, et al. . Diffusion tensor-MRI evidence for extra-axonal neuronal degeneration in caudate and thalamic nuclei of patients with multiple sclerosis. AJNR Am J Neuroradiol 2012;33:1363–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minagar A, Barnett MH, Benedict RH, et al. . The thalamus and multiple sclerosis: modern views on pathologic, imaging, and clinical aspects. Neurology 2013;80:210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 2012;61:1000–1016. [DOI] [PubMed] [Google Scholar]
- 15.Jespersen SN, Leigland LA, Cornea A, Kroenke CD. Determination of axonal and dendritic orientation distributions within the developing cerebral cortex by diffusion tensor imaging. IEEE Trans Med Imaging 2012;31:16–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grussu F, Schneider T, Tur C, et al. . Neurite dispersion: a new marker of multiple sclerosis spinal cord pathology? Ann Clin Transl Neurol 2017;4:663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider T, Brownlee W, Zhang H, Ciccarelli O, Miller DH, Wheeler-Kingshott CG. Sensitivity of multi-shell NODDI to multiple sclerosis white matter changes: a pilot study. Funct Neurol 2017;32:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Santis S, Bastiani M, Droby A, et al. . Characterizing microstructural tissue properties in multiple sclerosis with diffusion MRI at 7 T and 3 T: the impact of the experimental design. Neuroscience Epub 2018 Apr 7. [DOI] [PubMed]
- 19.Granberg T, Fan Q, Treaba CA, et al. . In vivo characterization of cortical and white matter neuroaxonal pathology in early multiple sclerosis. Brain 2017;140:2912–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lublin FD, Reingold SC. Defining the clinical course of multiple sclerosis: results of an international survey. National multiple sclerosis society (USA) advisory committee on clinical trials of new agents in multiple sclerosis. Neurology 1996;46:907–911. [DOI] [PubMed] [Google Scholar]
- 21.Polman CH, Reingold SC, Banwell B, et al. . Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 2011;69:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 1983;33:1444–1452. [DOI] [PubMed] [Google Scholar]
- 23.Cutter GR, Baier ML, Rudick RA, et al. . Development of a multiple sclerosis functional composite as a clinical trial outcome measure. Brain 1999;122:871–882. [DOI] [PubMed] [Google Scholar]
- 24.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011;54:2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodini B, Khaleeli Z, Cercignani M, Miller DH, Thompson AJ, Ciccarelli O. Exploring the relationship between white matter and gray matter damage in early primary progressive multiple sclerosis: an in vivo study with TBSS and VBM. Hum Brain Mapp 2009;30:2852–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cercignani M, Embleton K, Parker GJ, Bozzali M. Group-averaged anatomical connectivity mapping for improved human white matter pathway visualisation. NMR Biomed 2012;25:1224–1233. [DOI] [PubMed] [Google Scholar]
- 27.Daducci A, Canales-Rodríguez EJ, Zhang H, Dyrby TB, Alexander DC, Thiran JP. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage 2015;105:32–44. [DOI] [PubMed] [Google Scholar]
- 28.Woo CW, Krishnan A, Wager TD. Cluster-extent based thresholding in fMRI analyses: pitfalls and recommendations. Neuroimage 2014;91:412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colgan N, Siow B, O'Callaghan JM, et al. . Application of neurite orientation dispersion and density imaging (NODDI) to a tau pathology model of Alzheimer's disease. Neuroimage 2016;125:739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winston GP, Micallef C, Symms MR, Alexander DC, Duncan JS, Zhang H. Advanced diffusion imaging sequences could aid assessing patients with focal cortical dysplasia and epilepsy. Epilepsy Res 2014;108:336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamagata K, Hatano T, Okuzumi A, et al. . Neurite orientation dispersion and density imaging in the substantia nigra in idiopathic Parkinson disease. Eur Radiol 2016;26:2567–2577. [DOI] [PubMed] [Google Scholar]
- 32.Rae CL, Davies G, Garfinkel SN, et al. . Deficits in neurite density underlie white matter structure abnormalities in first-episode psychosis. Biol Psychiatry 2017;15:716–725. [DOI] [PubMed] [Google Scholar]
- 33.van Munster CE, Jonkman LE, Weinstein HC, Uitdehaag BM, Geurts JJ. Gray matter damage in multiple sclerosis: impact on clinical symptoms. Neuroscience 2015;303:446–461. [DOI] [PubMed] [Google Scholar]
- 34.Steenwijk MD, Daams M, Pouwels PJ, et al. . Unraveling the relationship between regional gray matter atrophy and pathology in connected white matter tracts in long-standing multiple sclerosis. Hum Brain Mapp 2015;36:1796–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jürgens T, Jafari M, Kreutzfeldt M, et al. . Reconstruction of single cortical projection neurons reveals primary spine loss in multiple sclerosis. Brain 2016;139:39–46. [DOI] [PubMed] [Google Scholar]
- 36.Calabrese M, Rinaldi F, Seppi D, et al. . Cortical diffusion-tensor imaging abnormalities in multiple sclerosis: a 3-year longitudinal study. Radiology 2011;261:891–898. [DOI] [PubMed] [Google Scholar]
- 37.Haider L, Zrzavy T, Hametner S, et al. . The topograpy of demyelination and neurodegeneration in the multiple sclerosis brain. Brain 2016;139:807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonkman LE, Klaver R, Fleysher L, Inglese M, Geurts JJ. The substrate of increased cortical FA in MS: a 7T post-mortem MRI and histopathology study. Mult Scler 2016;22:1804–1811. [DOI] [PubMed] [Google Scholar]
- 39.Filippi M, Bozzali M, Comi G. Magnetization transfer and diffusion tensor MR imaging of basal ganglia from patients with multiple sclerosis. J Neurol Sci 2001;183:69–72. [DOI] [PubMed] [Google Scholar]
- 40.Ciccarelli O, Werring DJ, Wheeler-Kingshott CA, et al. . Investigation of MS normal-appearing brain using diffusion tensor MRI with clinical correlations. Neurology 2001;56:926–933. [DOI] [PubMed] [Google Scholar]
- 41.Haider L, Simeonidou C, Steinberger G, et al. . Multiple sclerosis deep grey matter: the relation between demyelination, neurodegeneration, inflammation and iron. J Neurol Neurosurg Psychiatry 2014;85:1386–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Postuma RB, Dagher A. Basal ganglia functional connectivity based on a meta-analysis of 126 positron emission tomography and functional magnetic resonance imaging publications. Cereb Cortex 2006;16:1508–1521. [DOI] [PubMed] [Google Scholar]
- 43.Kotz SA, Anwander A, Axer H, Knösche TR. Beyond cytoarchitectonics: the internal and external connectivity structure of the caudate nucleus. PLoS One 2013;8:e70141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lampinen B, Szczepankiewicz F, Mårtensson J, van Westen D, Sundgren PC, Nilsson M. Neurite density imaging versus imaging of microscopic anisotropy in diffusion MRI: a model comparison using spherical tensor encoding. Neuroimage 2017;147:517–531. [DOI] [PubMed] [Google Scholar]
- 45.Moeller S, Yacoub E, Olman CA, et al. . Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med 2010;63:1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are available on request addressed to the corresponding author.