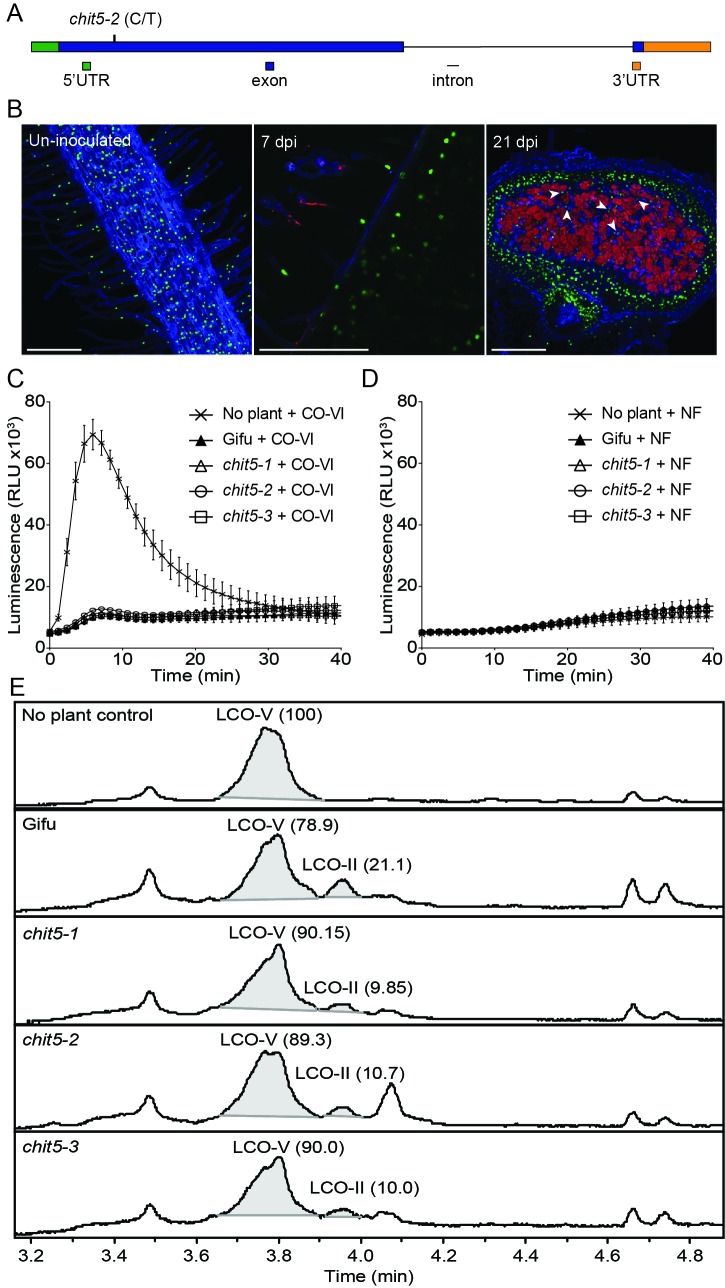
Figure 3. Chit5 encodes a root expressed class V chitinase with Nod factor hydrolase activity.
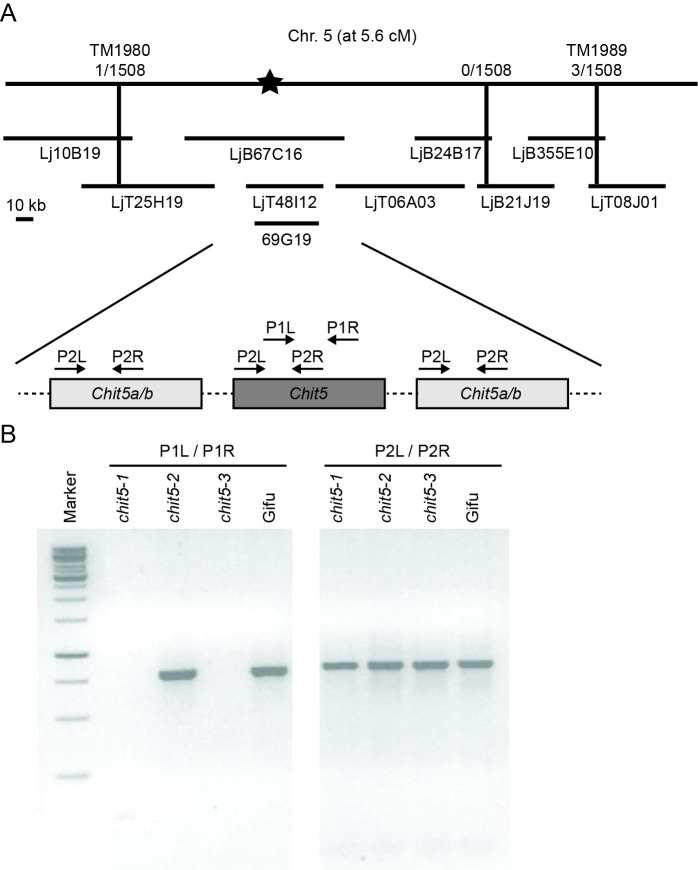
(A) Chit5 gene structure. Mutation in chit5-2 allele is shown. Chit5 gene is deleted in chit5-1 and chit5-3 alleles. (B) Chit5 promoter activity was monitored in Gifu roots transformed with a Chit5 promoter-tYFP-NLS (green nuclei) in uninoculated and M. loti R7A + DsRed inoculated roots. White arrows highlight examples of uninfected cells showing Chit5 promoter activity. Scale bars are 200 µm. (C) ROS induced by exudates of CO-VI-treated plants. (D) Absence of ROS induction by exudates of M. loti R7A Nod factor-treated plant exudates. (E) M. loti R7A Nod factor hydrolysis in the presence of the indicated plant genotypes measured by HPLC-MS. The average relative percentage of LCO-V and LCO-II fractions from two biological replicates (Figure 3—source data 1) determined from peak-peak integration is indicated in brackets. The peak eluting near 4.1 min is present in all plant treated samples but was not detected in the control sample. It lacks the characteristic fragmentation pattern of LCOs, and appears to be an aromatic, small molecule unrelated to chitinase activity.