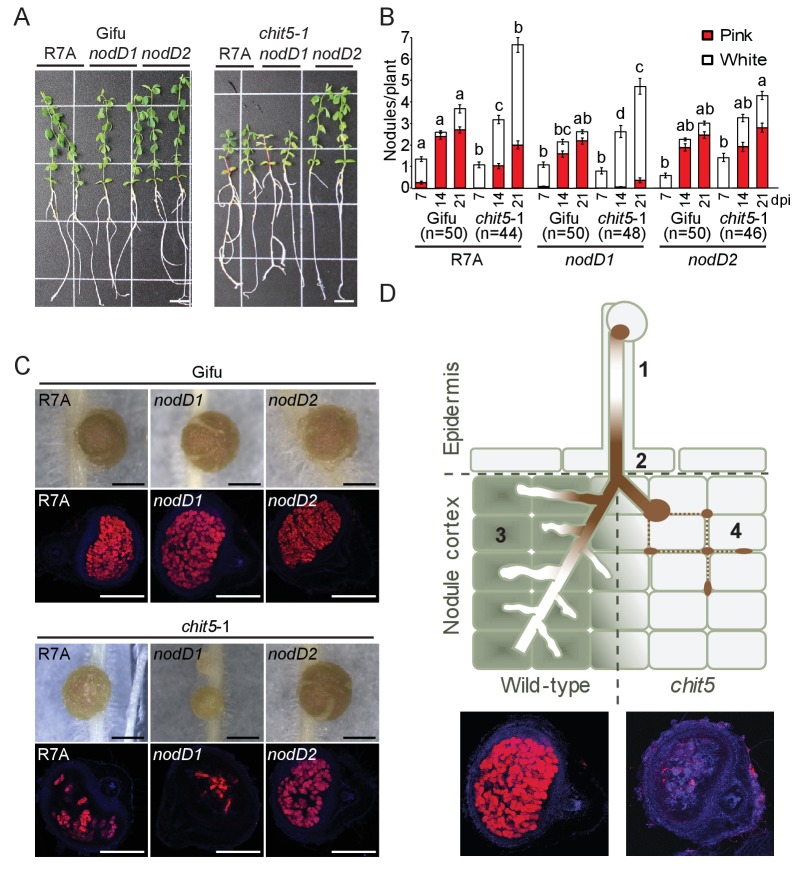
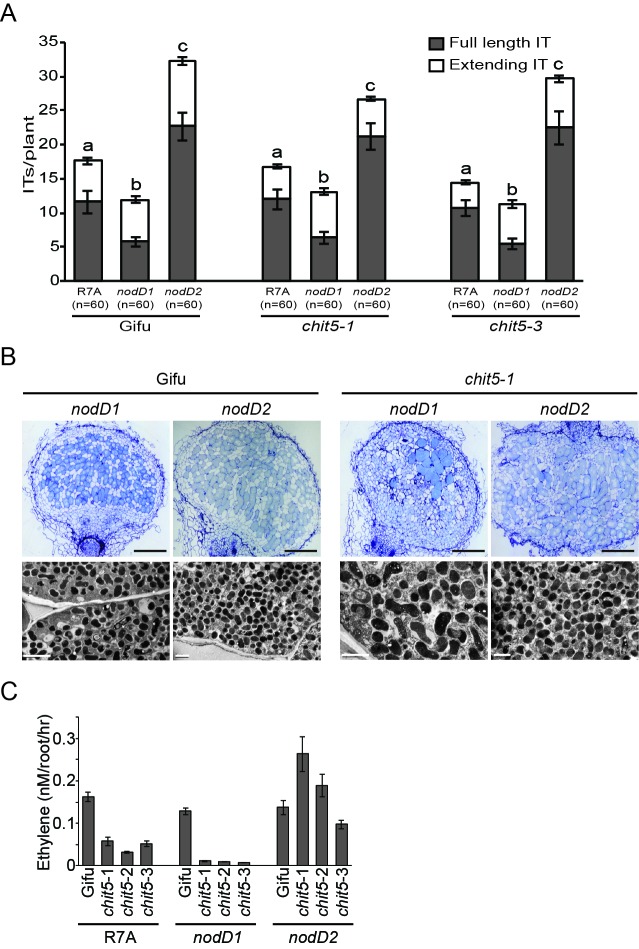
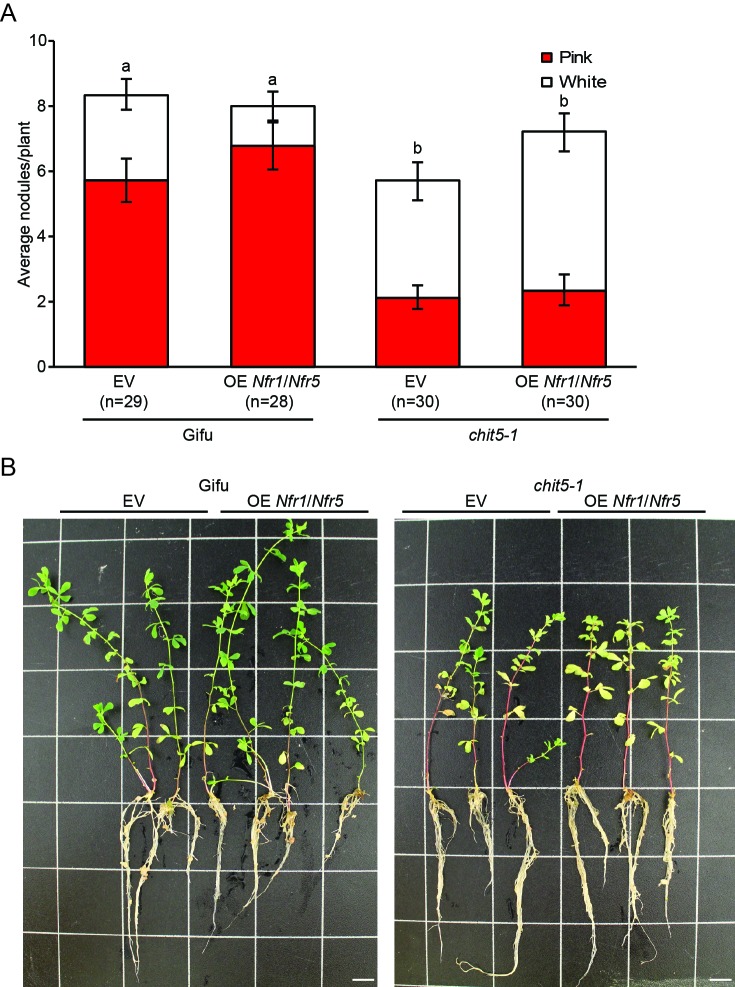
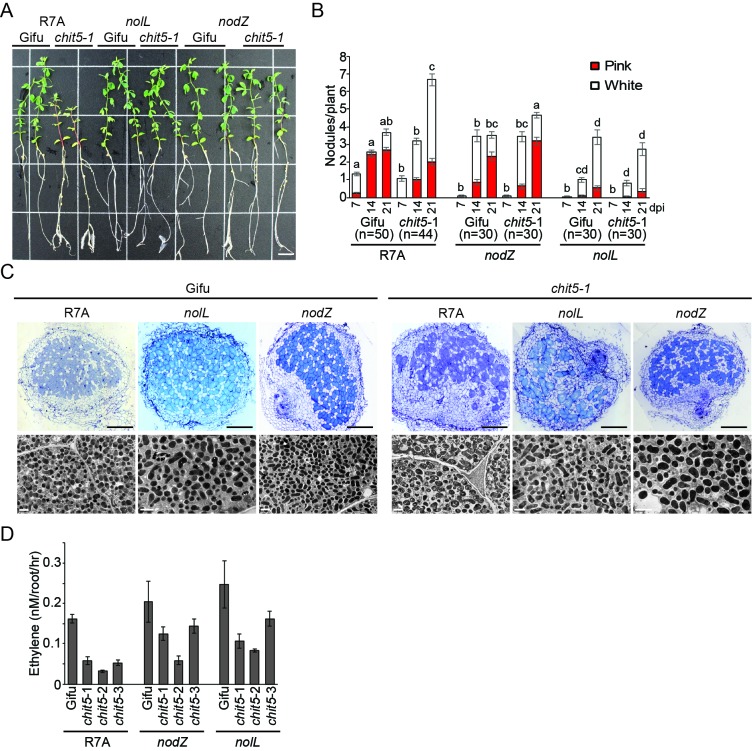
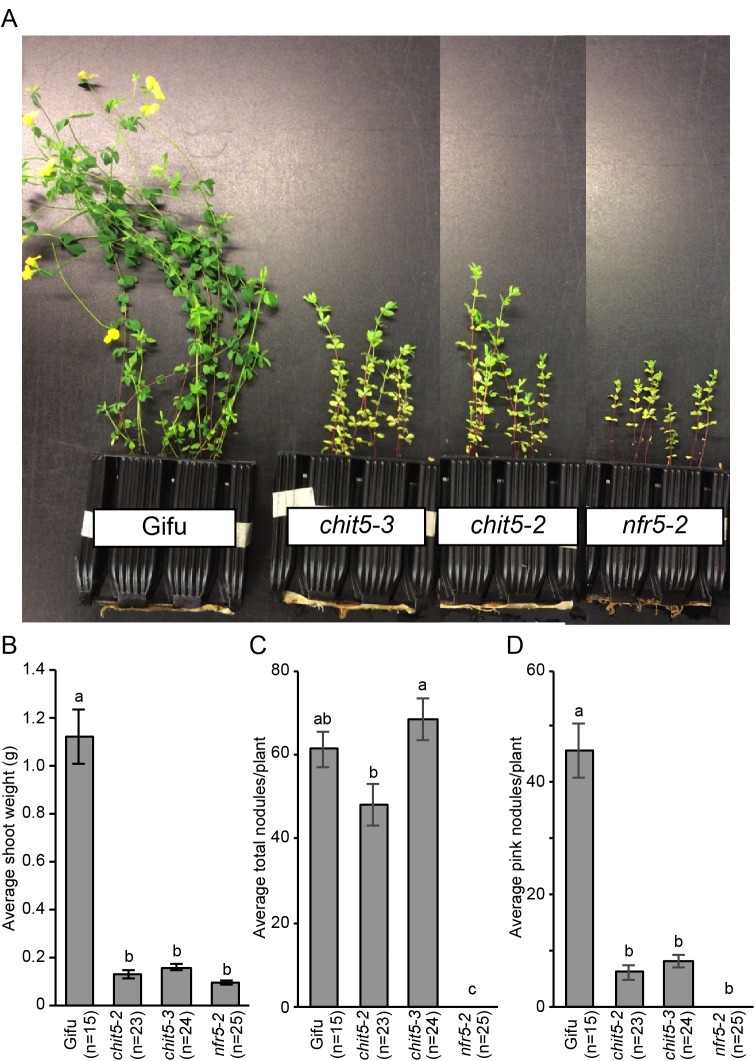
Figure 4. The symbiotic impairment of chit5 mutants can be overcome by an M.loti R7A mutant affected in the regulation of Nod factor biosynthesis.
(A) Representative images of Gifu and chit5-1 plants inoculated with the indicated M. loti R7A strains 7 wpi. Scale bars are 1 cm. (B) Average number of pink and white nodules formed on Gifu and chit5-1 by the indicated M. loti R7A strains over 3 weeks. Error bars represent SEM and statistical comparisons of the number of pink nodules formed between strains on each genotype at each time point are shown using ANOVA and Tukey post hoc testing with p values < 0.05 as indicated by different letters. (C) Light microscopy of whole nodules 3 wpi and confocal images of nodule sections from Gifu and chit5-1 plants inoculated with the indicated strains tagged with DsRed. Scale bars are 0.5 cm. (D) Proposed model of CHIT5 activity and representative nodule images of wild-type and chit5 mutant illustrating the observed phenotype. (1) Bacteria maintain a low level of Nod factors within root hair-traversing ITs by preferential activity of NodD1. (2) and (3) Bacterial amplification inside ITs, coupled with the switch for preferential activity of NodD2 leads to higher levels of fully decorated pentameric Nod factors (LCO-V). Chit5 expression (green filled cells in wild-type) is crucial for maintaining a balanced Nod factor level enabling IT extension inside primordia, efficient bacterial endocytosis and, ultimately, development of the nitrogen-fixing organ (3). (4) In the chit5 mutants (light grey filled cells) higher levels of Nod factors (LCO-V) impede IT elongation and branching inside primordia leading to bacteria accumulation in between the cells (dotted brown line) and scattered infection.