Abstract
Subcellular membrane-less assemblies are a reinvigorated area study in biology with spirited scientific discussions on the forces between the low-complexity protein domains within these assemblies. To illuminate these forces we determined atomic structures of five segments of protein low-complexity domains associated with membrane-less assemblies. Their common structural feature is the stacking of segments into kinked β-sheets which pair into protofilaments. Unlike steric zippers of amyloid fibrils, the kinked sheets interact weakly through polar atoms and aromatic sidechains. By computationally threading the human proteome on our kinked structures, we identified hundreds of low-complexity segments potentially capable of forming such interactions. These segments are found in proteins as diverse as RNA binders, nuclear pore proteins, and keratins, known to form networks and localize to membrane-less assemblies.
One Sentence Summary:
Threading reveals hundreds of low-complexity segments in the human proteome capable of forming kinked multivalent protofilaments.
Membrane-less organelles, such as P-bodies, nuclear paraspeckles and stress granules (SGs), form and redissolve in mammalian cells in response to stimuli (1, 2). Such phase separation is a property of macromolecules that are capable of multivalent interactions with each other, yielding a liquid phase having ~100 times the concentration of the macromolecule compared to the bulk liquid (3, 4). This type of phase separation is often seen with proteins that bind nucleic acids and contain low-complexity domains (LCDs) (1, 2, 5–8). For example, the SG associated proteins hnRNPA1, hnRNPA2, and FUS undergo liquid-liquid phase separation (9–12) and they contain LCDs which transition into reversible semi-solid phase hydrogels over time or at higher protein concentration (1, 5, 9). LCDs are common in the human proteome; they are largely intrinsically disordered (13), and dramatically underrepresented in the Protein Data Base (PDB) of known 3D structures (14).
Electron microscopy reveals that such hydrogels contain protein fibrils, and X-ray diffraction of the hydrogel yields a cross-β pattern (fig. S1C-E) (5, 15) reminiscent of amyloid. However, whereas the fibrils found in FUS hydrogels are heat and SDS-sensitive (5), amyloid fibrils resist denaturation by SDS and boiling. The spines of amyloid fibrils contain pairs of closely mating β-sheets along the fibril axis. Residue side-chains tightly interdigitate with sidechains of the opposing β-sheet to form a dry interface called a steric zipper, as seen in the structure of NKGAII from amyloid-beta (Aβ) (Fig. 1A) (16, 17). The steric zipper explains the extraordinary stability of some pathogenic amyloid. Apparently the relatively labile multivalent interactions of the hydrogel-forming proteins are different; their atomic-level details are largely unknown, although importantly ssNMR has shown that 57 of the 214 residue LCD of FUS form an ordered protofilament core with the remaining residues dynamically disordered (18).
Fig. 1. Structures of LARKS (B-F) compared to a steric zipper (A).
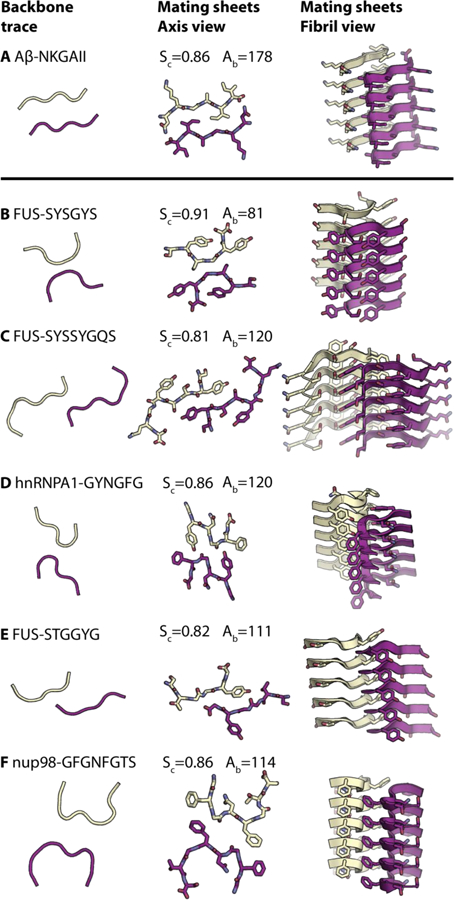
All structures composed of two mating β-sheets, one purple and the other yellow. The left-hand column shows the trace of the backbones of mating sheets to highlight kinks in the backbones of LARKS and the pleating of the classical β-sheets in steric zippers. The second column shows the atomic structures of mating sheets viewed down the fibril axes. The third column shows cartoons of the mating β-sheets viewed nearly perpendicular to the fibril axes. Each interface is characterized by the shape complementarity score (Sc=1.0 for perfect complementarity) and buried solvent-accessible surface area (Ab) in Ų between the mated sheets. Carbon atoms are colored purple or yellow, Nitrogen is blue, and Oxygen is red. Five layers of β-sheets are shown of the hundreds of thousands in the crystals. The kinked structures of LARKS are rare among mating β-sheets; dozens of other paired β-sheets form steric zippers (35).
To investigate relatively weak adhesion between LCDs of proteins recruited to SGs, we sought relevant atomic structures. Guided by studies of the LCDs of FUS and RBM14, which show that successive replacement of tyrosine residues by serine lowers their capacity to form hydrogels (1, 5), we scanned the LCD of FUS for tandem sequence motifs of the form [G/S]Y[G/S], finding two such segments: FUS-37SYSGYS42 and FUS-54SYSSYGQS61 (fig. S1A). Both segments crystallized as micron-sized needles and both atomic structures were determined, in addition to structures of three other segments identified by 3D profiling (see below): 243GYNGFG248 from protein hnRNPA1, 77STGGYG82 from FUS, and 116GFGNFGTS123 from nup98 (Fig. 1). Confirming the relevance of these structures to adhesion and multivalency of LCDs, a hydrogel is formed by a 26 residue synthetic peptide construct linking the three above segments of FUS (Fig. 2). Powder diffraction patterns of all 5 crystalline segments, this hydrogel, and the FUS-LCD hydrogel suggest they all share cross-β architecture (figs S2–3).
Fig. 2. Synthetic LARKS construct forms a labile hydrogel.
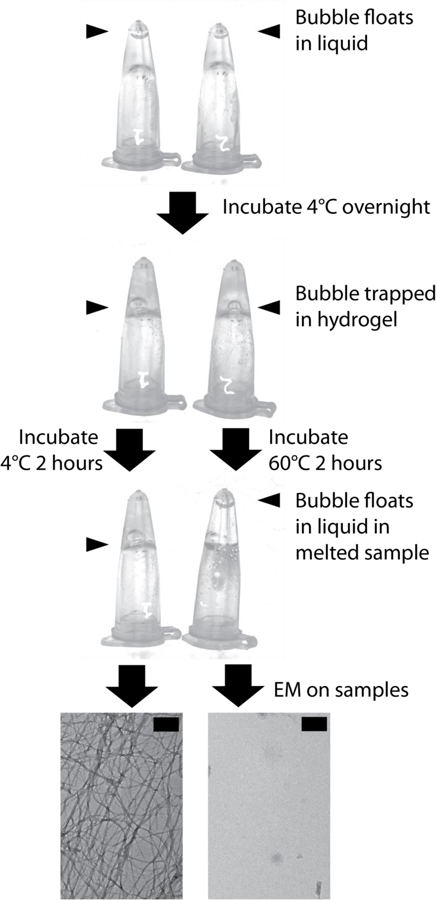
A synthetic LARKS construct with the sequence SYSGYSGDTSYSSYGQSNGPSTGGYG forms a labile hydrogel when dissolved in water at 50mg/ml and left overnight at 4°C. The hydrogel melts upon heating the sample to 60°C for two hours. A bubble (blue arrow) was introduced to the sample to show the difference between the liquid state (bubble rises) and hydrogel state (bubble does not rise). Electron microscopy confirms that fibrils were indeed melted. The hydrogel-forming property of this triple-LARKS sequence suggests that it is the multiple LARKS found in many LCDs that endow their unusual property of forming hydrogels. Scale bars are equal to 200nm.
All five segments crystalized as pairs of kinked β-sheets (Fig. 1). Each β-sheet runs the length of the crystal, formed from the stacking of about 300,000 segments and all structures show kinks at either glycines or aromatic residues instead of being extended (fig. S4). The structures share common adhesive features, including hydrogen bonds in-register to an identical segment below it (Fig. 1B-F, fig. S5). Aromatic residues predominate, both for inter-sheet stabilization and intra-sheet stabilization. Within sheets, the aromatic side-chains stack in an energetically favorable conformation, with the planes of the rings stacked parallel at a separation of 3.4 Å(19–21) (fig. S5). These aromatic “ladders” enhance the stability of each β-sheet. The kinks allow close approach of the backbones, providing favorable van der Waals or hydrogen-bond interactions between the sheets (fig. S5). These close interactions are quantified by the Structural Complementarity, Sc (Fig. 1), reflecting adhesion between the sheets. However the kinks prevent sidechains from interdigitating across the β-sheet interface so that the kinked interfaces bury smaller surface areas than found in pathogenic amyloid fibrils, and presumably have lower binding energies. Because of the distinction of the kinked structures from pathogenic steric zippers, we term them Low-complexity Aromatic-Rich Kinked Segments, or LARKS.
Calculations and experiments support our structural inference that LARKS have smaller binding energies than steric zippers. We estimated energies of separation of the pairs of β-sheets in LARKS and steric zippers by applying atomic solvation parameters (22, 23) to our structures: the mean atomic solvation energy for separation of our LARKS interfaces is 567 ± 556 cal/mol/β-strand, whereas it is 1431 ± 685 cal/mol/β-strand for 75 steric zipper structures (fig. S6). These crude estimates suggest that the adhesive energy of one pair of β-strands in a LARKS is of the order of thermal energy, so that pairs of β-sheets adhere only through multivalent interactions of strands. In contrast the adhesive energy of one pair of strands in a steric zipper is several times that of thermal energy. Consistent with calculations, the synthetic muli-LARKS construct of Figure 2 dissolves when gently heated. Thus paired kinked β-sheets of LARKS are less strongly bound than the paired β-sheets in amyloid fibrils, yet still produce fibrils with the cross β-diffraction pattern of pathogenic amyloid.
To identify potential LARKS in the human proteome, we used computational 3D profiling, a method which tests the compatibility of query sequences with a template structure(24, 25). Here, we threaded human sequences onto the backbones of SYSGYS, GYNGFG, and STGGYG, placed and optimally re-packed side-chains, and then evaluated the Rosetta energy (Fig. 3A)(26). We advanced the threading by one residue and repeated the procedure until the end of the query sequence was reached. This 3D profiling predicted that nucleoporin proteins are enriched in LARKS (Fig. 3C). Our confidence in this prediction was bolstered by the success of earlier predictions that GYNGFG and STGGYG could form LARKS, based on threading with only the SYSGYS template. Here, again, we were able to validate our profiling algorithm by determining the structure of GFGNFGTS from the porin nup98, confirming LARKS architecture (Fig. 1F), and providing evidence that LARKS are present in a different type of membraneless organelle (27).
Fig. 3. 3D profiling to identify LARKS in LC domains of human proteins.
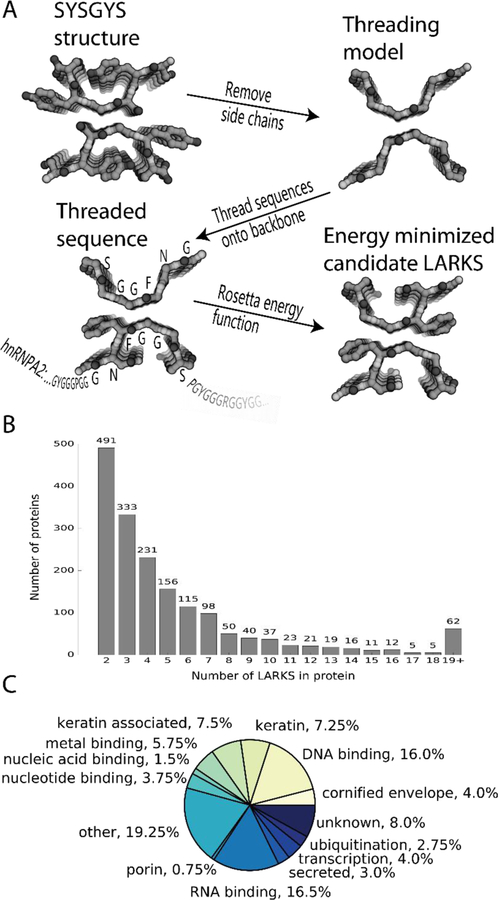
(A) Method: Sidechains are removed from the backbones of one of our atomic structures of a LARKS. Then the sequence of interest (hnRNPA1 shown) is threaded through the six residue template by placing the query sidechains on the template backbone. Sidechains are repacked and a Rosetta energy function is used to estimate if the structure is favorable for the threaded sequence. The sequence then advances through the template by one residue increments, producing successive models. (B) The frequency of the number of LARKS in 1725 human proteins predicted to house at least two LARKS. Proteins having two or more LARKS are predicted to have the capacity to form networks and possibly gels. (C) The annotated functions of the 400 proteins with the most predicted LARKS.
Analyzing the non-redundant human proteome of 20120 sequences from UniProt, we found 5867 proteins with LCDs. Of these, 2500 proteins contain at least one LARKS and 1725 proteins contain two or more LARKS, thus able to form multivalent interactions, and hence protein networks and gels. Hundreds of proteins house three or more LARKS (Fig. 3B). The 400 human LCDs most enriched in LARKS average 14 LARKS.
We assigned cellular function to these 400 proteins based on their Uniprot annotations (Fig. 3C): 16% are DNA binding, 17% are RNA binding, and 4% are nucleotide binding, consistent with reports of nucleotide binding proteins in membraneless organelles (2, 8). Keratins (5%), keratin-associated (9%), and cornified envelope proteins (4%) are also enriched in LARKS. The finding of keratins is consistent with experiments (28) showing that keratin granules are trafficked to the cell cortex where they merge and eventually mature into filaments. Also rich in LARKS are proteins found in ribonucleoprotein particles such as the spliceosome or nucleolus (Fig. 4). Nucleoporins including nup54 and nup98 with FG repeats are enriched in predicted LARKS and purified FG repeats form a hydrogel (27, 29). The possibility that the FG repeats of nucleoporins may form LARKS in the diffusion barrier of the pore is supported by our structure of GFGNFGTS from nup98. We assigned additional cellular functions to these 400 proteins from their associated gene ontology (GO) terms. We found GO terms enriched in the human proteome for RNA transport, processing localization, SG assembly, and epithelial cell differentiation due to the numerous keratins enriched in LARKS. Therefore we propose 3D profiling for LARKS as a tool to identify proteins that may form networks and gels by multivalent interactions and may participate in membraneless organelles (fig. S10).
Fig. 4.
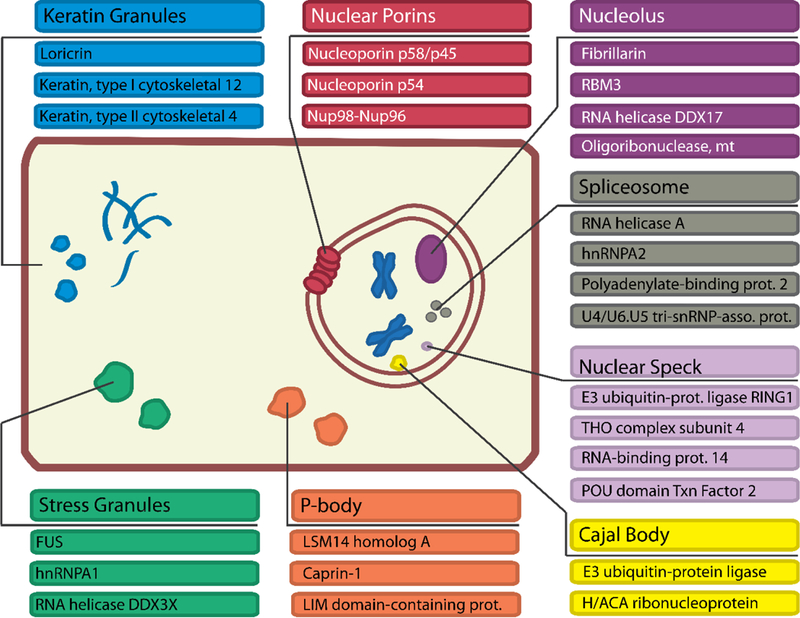
Functions of proteins among the 400 proteins most enriched in LARKS and dynamic intracellular bodies they are known to be a part of.
Conclusion:
The prevalence of LCDs within eukaryotic proteomes has long been recognized (30), but the role of these domains has not been fully defined. Previous discoveries include: LCDs can “functionally aggregate” (31); proteins with LCDs typically form more protein-protein interactions (32, 33) ; and proteins can interact homotypically and heterotypically through LC domains (1, 5, 34). Our atomic structures support the hypothesis that LC domains have the capacity to form gel-like networks. LARKS possess three properties that are consistent with their functioning as adhesive elements in protein gels formed from LC domains: i) High aqueous solubility contributed by their high proportion of hydrophilic residues: serine, glutamine, and asparagine; ii) Flexibility ensured by their high glycine content; iii) Multiple interaction motifs per chain (Fig. 3B), endowing them with multivalency, enabling them to entangle, forming networks as found in gels (Fig. 2). That each LARKS provides adhesion only comparable to thermal energy suggests that numerous LARKS must cooperate in gel formation, and that the interactions must be concentration dependent and may be transient. If steric zippers act as molecular glue, then LARKS in LCDs act as Velcro. These properties are compatible with the hypothesis that LARKS are a protein interaction motif that provides adhesion of LCDs in protein gels and perhaps in membrane-less assemblies (fig S10).
Supplementary Material
Acknowledgments:
Our X-ray diffraction data were collected at the Northeastern Collaborative Access Team beamline 24-ID-E which is funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Atomic coordinates and structure factors have been deposited in the PDB with the following accession codes: SYSGYS (6BWZ), SYSSYGQS (6BXV), STGGYG (6BZP), GYNGFG (6BXX), and GFGNFGTS (6BZM). We thank NSF MCB-1616265, NIH AG-054022, DOE, and HHMI for support.
Abbreviations:
- LARKS
Low-complexity Aromatic-Rich Kinked Segments
- LCD
low-complexity domain
- Sc
Structural Complementarity
- SG
stress granule
- ssNMR
solid-state NMR
References and Notes:
- 1.Hennig S et al. , Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol 210, 529–539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguzzi A, Altmeyer M, Phase Separation: Linking Cellular Compartmentalization to Disease. Trends Cell Biol 26, 547–558 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Banani SF, Lee HO, Hyman AA, Rosen MK, Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P et al. , Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato M et al. , Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbaum-Garfinkle S, Brangwynne CP, Liquids, Fibers, and Gels: The Many Phases of Neurodegeneration. Dev. Cell 35, 531–532 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Lin Y, Protter DSW, Rosen MK, Parker R, Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison AF, Shorter J, RNA-binding proteins with prion-like domains in health and disease. Biochem. J 474, 1417–1438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami T et al. , ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 88, 678–690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel A et al. , A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Molliex A et al. , Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang S et al. , The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell 163, 829–839 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumari B, Kumar R, Kumar M, Low complexity and disordered regions of proteins have different structural and amino acid preferences. Mol. Biosyst 11, 585–594 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Wootton JC, Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput. Chem 18, 269–285 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Schwartz JC, Cech TR, Parker RR, Biochemical Properties and Biological Functions of FET Proteins. Annu. Rev. Biochem 84, 355–379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson R et al. , Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawaya MR et al. , Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Murray DT et al. , Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 171, 615–627.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinnokrot MO, Valeev EF, Sherrill CD, Estimates of the ab initio limit for pi-pi interactions: the benzene dimer. J. Am. Chem. Soc 124, 10887–10893 (2002). [DOI] [PubMed] [Google Scholar]
- 20.McGaughey GB, Gagné M, Rappé AK, pi-Stacking interactions. Alive and well in proteins. J. Biol. Chem 273, 15458–15463 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Arunan E, Gutowsky HS, The rotational spectrum, structure and dynamics of a benzene dimer. J. Chem. Phys 98 (1992). [Google Scholar]
- 22.Eisenberg D, McLachlan AD, Solvation energy in protein folding and binding. Nature 319, 199–203 (1986). [DOI] [PubMed] [Google Scholar]
- 23.Eisenberg DE, Wesson M, Yamashita M, Interpretation of Protein Folding and Binding with Atomic Solvation Parameters. Chem. Scr 29A, 217–221 (1989). [Google Scholar]
- 24.Bowie JU, Lüthy R, Eisenberg D, A method to identify protein sequences that fold into a known three-dimensional structure. Science 253, 164–170 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Goldschmidt L, Teng PK, Riek R, Eisenberg D, Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. U. S. A 107, 3487–3492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leaver-Fay A et al. , ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol 487, 545–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frey S, Richter RP, Görlich D, FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Windoffer R, Wöll S, Strnad P, Leube RE, Identification of novel principles of keratin filament network turnover in living cells. Mol. Biol. Cell 15, 2436–2448 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ader C et al. , Amyloid-like interactions within nucleoporin FG hydrogels. Proc. Natl. Acad. Sci. U. S. A 107, 6281–6285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim KL, Creamer TP, Abundance and distributions of eukaryote protein simple sequences. Mol. Cell. Proteomics MCP 1, 983–995 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Toretsky JA, Wright PE, Assemblages: functional units formed by cellular phase separation. J. Cell Biol 206, 579–588 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coletta A et al. , Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol 4, 43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uversky VN, Oldfield CJ, Dunker AK, Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu. Rev. Biophys 37, 215–246 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Kwon I et al. , Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenberg DS, Sawaya MR, Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem 86, 69–95 (2017). [DOI] [PubMed] [Google Scholar]
- 36.McCoy AJ et al. , Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ, Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Sheldrick GM, A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W, XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hattne J et al. , MicroED data collection and processing. Acta Crystallogr. Sect. Found. Adv 71, 353–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanc E et al. , Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr 60, 2210–2221 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winn MD et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The UniProt Consortium, UniProt: the universal protein knowledgebase. Nucleic Acids Res 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gene Ontology Consortium, Gene Ontology Consortium: going forward. Nucleic Acids Res 43, D1049–1056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halfmann R, A glass menagerie of low complexity sequences. Curr. Opin. Struct. Biol 38, 18–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


