Abstract
Rationale
Biological significance of c-Kit as a cardiac stem cell marker and role(s) of c-Kit+ cells in myocardial development or response to pathologic injury remain unresolved due to varied and discrepant findings. Alternative experimental models are required to contextualize and reconcile discordant published observations of cardiac c-Kit myocardial biology and provide meaningful insights regarding clinical relevance of c-Kit signaling for translational cell therapy.
Objective
Demonstration of c-Kit myocardial biology through combined studies of both human and murine cardiac cells. Advancing understanding of c-Kit myocardial biology through creation and characterization of a novel, inducible transgenic c-Kit reporter mouse model that overcomes limitations inherent to knock-in reporter models, providing perspective to reconcile disparate viewpoints on c-Kit biology in the myocardium.
Methods and Results
In vitro studies confirm a critical role for c-Kit signaling in both cardiomyocytes and cardiac stem cells. Activation of c-Kit receptor promotes cell survival and proliferation in stem cells and cardiomyocytes of either human or murine origin. For creation of the mouse model, the cloned mouse c-Kit promoter drives Histone2B-EGFP (H2BEGFP) expression in a doxycycline inducible transgenic reporter line. The combination of c-Kit transgenesis coupled to H2BEGFP readout provides sensitive, specific, inducible, and persistent tracking of c-Kit promoter activation. Tagging efficiency for EGFP+/c-Kit+ cells is similar between our transgenic versus a c-Kit knock-in mouse line, but frequency of c-Kit+ cells in cardiac tissue from the knock-in model is 55% lower than our transgenic line. The c-Kit transgenic reporter model reveals intimate association of c-Kit expression with adult myocardial biology. Both cardiac stem cells and a subpopulation of cardiomyocytes express c-Kit in uninjured adult heart, upregulating c-Kit expression in response to pathologic stress.
Conclusions
c-Kit myocardial biology is more complex and varied than previously appreciated or documented, demonstrating validity in multiple points of coexisting yet heretofore seemingly irreconcilable published findings.
Subject Terms: Basic Science Research, Cell Signaling/Signal Transduction, Myocardial Biology, Stem Cells, Genetically Altered and Transgenic Models
Keywords: Cardiac, stem cell, c-Kit, cardiomyocyte, myocardium, signal transduction, molecular
INTRODUCTION
Cardiovascular research has struggled to advance the field of regenerative medicine, due in part to the lack of a unique and unequivocal marker for resident adult cardiac stem cells capable of mediating repair. In fact, the concept of myocardial regeneration that was initially advanced over a decade ago employed the stem cell marker c-Kit to identify presumptive cardiac stem cells 1, 2. Over the ensuing years the c-Kit+ cardiac stem cell population has become the focus of intense and diverse investigation by hundreds of laboratories worldwide at both the basic and clinical levels 3–10. At present, the presence and participation of c-Kit+ cells in myocardial development as well as response to pathological stress or injury is indisputable 2, 4, 8, 10–12. However, controversy persists regarding the roles(s) that cells expressing c-Kit play in cardiac biology. Such divergent perspectives suggest that fundamental understanding has not been achieved and that additional study is warranted to provide sorely needed novel insight(s). The present study was conceived and initiated to examine myocardial c-Kit biology with fresh perspective using a unique approach yielding novel findings.
Phenotypically, c-Kit serves as a marker of multiple cell types including stem cells, most often associated with hematopoietic progenitor cells, but in fact is present from early embryogenesis and persists in wide distribution throughout various adult tissues 13, 14. Functionally, c-Kit protein is a kinase that, upon activation by its cognate ligand stem cell factor (SCF), serves to induce cell proliferation, survival, and transcriptional activation 14. Variation in temporal appearance and expression level of c-Kit in multiple cell types and in response to environmental stimuli is indicative of a dynamically regulated protein that does not necessarily respect traditional nomenclature of a “stem cell marker”. Despite years of study, details of c-Kit signaling cascades and downstream targets remain obscure 15 and the respective activities for surface-expressed versus internal c-Kit remain poorly characterized 16. In the cardiac context, c-Kit expression has been documented in multiple cell types including (but not necessarily limited to) myocytes, endothelial cells, cardiac progenitor, and mesenchymal stem cells 17, 18. Indeed, observations of c-Kit protein induction suggest that stress responses and myocyte remodeling serve as inductive stimuli for expression of this “stem cell marker” in cardiomyocytes 4, 19.
Given the heterogeneous expression pattern for c-Kit that is not restricted solely to a canonical cardiac stem cell, the complex and varied findings derived from study of various genetically engineered c-Kit lineage-tracing models might indeed even be an expected outcome 18. Furthermore, every experimental approach to define c-Kit+ cell fate possesses inherent strengths and weaknesses to be considered when deriving conclusions. While the advantages of a genetically engineered knock-in model are well known 20, the inevitable silencing of one c-Kit allele perturbs endogenous c-Kit biology with potentially significant consequences for stem cell function. Furthermore, imperfect tagging efficiency in hemizygous knock-in mice allows for covert existence of an “invisible” c-Kit+ subpopulation in the myocardium comprising 25–30% of all c-Kit+ expressing cells 9 that cannot be followed. Consequently, c-Kit knock-in mice may harbor c-Kit dependent biological defects, impaired reporter sensitivity, as well as attendant uncertainty of the functional impact resulting from c-Kit allele hemizygosity that could affect proliferation and/or survival in culture.
An alternative approach to c-Kit+ tracking by “knock-in” techniques and associated allele inactivation is the use of transgenesis to introduce a cloned construct carrying a c-Kit promoter fragment driving expression of a long-lived reporter protein. The cloned c-Kit promoter employed as a driver for reporter gene expression has been extensively characterized both in vitro and in vivo 3, 21, 22, but has never been adopted to serve for inducible tracing of cell fate in the myocardial context. Transgenesis minimizes allele inactivation and potential deleterious consequences of impaired c-Kit activity that could influence interpretation of c-Kit expression and relevance in myocardial biology. For the current study, we employ a doxycycline-inducible transgenic mouse model to tag c-Kit+ cells with a long-lived, tetracycline responsive H2BEGFP (CKH2B) reporter 23. Endogenous c-Kit remains unperturbed and approximately 80% of nonmyocyte cardiac c-Kit+ cells express H2BEGFP reporter, similar to c-Kit coincidence with EGFP in a c-Kit/mER-Cre-mEr knock-in reporter model (CKmCm). A heterogeneous CKH2B cell population is revealed following induction with doxycycline including tagging of select adult cardiomyocytes (ACM) in vivo, a finding consistent with expression of c-Kit mRNA and protein in isolated ACMs. Increased expression of CKH2B in cultured ACM in response to stress is consistent with a role for c-Kit expression in cardiomyocyte survival, a finding recapitulated by acute pathologic injury in vivo. Similarly, c-Kit activation in cultured mouse and human cardiac progenitor cells (CPCs) enhances proliferation and survival, in line with with previous reports establishing the role of c-Kit signaling in human cardiac stem cells 19. Collectively, data presented here support validity of the c-Kit reporter model, pointing to heterogeneous and dynamic c-Kit expression in the postnatal myocardium that facilitates reconciliation of seemingly disparate observations from published literature and resolves multiple aspects of the controversial role for c-Kit in cardiac biology.
METHODS
The authors declare that all supporting data are available within the article [and its online supplementary files].
Mice
c-Kit/rtTA transgenic mice expressing the rtTA transactivator under control of the c-Kit promoter clone 2 21 were generated using standard genomics techniques and stabilized by breeding several generations into the FVB/N background. The CKH2B line was created by crossing c-Kit/rtTA mice with TRE-H2BEGFP mice purchased from Jaxmice (stock# 005104/pTRE-H2BEGFP). Copy number of the c-KitrtTA transgene per cell was determined by a qPCR method as described 24 with the following modifications. c-KitrtTA plasmid injected for transgenesis was used to establish the standard curve ranging in concentration from 5 fg to 5 ng of plasmid template per reaction. Genomic DNA (gDNA) was extracted from livers of mice hemizygous for the c-KitrtTA transgene or from non-rtTA controls. The qPCR reaction mix consisted of iQSYBR Green master mix, 50 ng gDNA, primers specific for rtTA and 18S, and was performed using a CFX Connect Bio-Rad cycler for 40 two-step cycles at 95C and 60C. A logarithmic standard curve was generated and copy number calculated per cell based on the assumption that each cell contains 6 pg genomic DNA, or that 1 ng DNA is the equivalent of 167 diploid cells. Genotyping of mice to detect rtTA and EGFP transgenic sequences in genomic tail DNA was performed by standard PCR using target specific primers. All primers for measuring copy number and genotyping are listed in Online Table I. c-Kit-MerCreMer/R26-NG (CKmCm) mice were created by crossing the Kit-MerCreMer driver line 9 to the FVB.Cg-Gt(ROSA)26Sortm1(CAG-lacZ,EGFP)Glh/J from Jaxmice (stock #012429, 25).
Animal Procedures
All procedures and experiments involving mice were approved by the SDSU Institutional Animal Care and Use Committee (IACUC). Acute diffuse injury was achieved by administering a single subcutaneous injection of 150 mg/kg isoproterenol. H2BEGFP reporter expression was induced in vivo with 0.2 mg/ml doxycycline in the drinking water for up to four days. CKmCm, three-month old male mice were fed normal chow or chow containing tamoxifen citrate for four weeks to induce EGFP reporter expression, after which hearts were retroperfused and fixed in formalin for paraffin processing. For flow cytometry analysis of heart and bone marrow, CKmCm mice were fed tamoxifen chow for four weeks followed by four weeks chow with no tamoxifen.
Cell isolation and expansion
Murine CPCs and ACMs were simultaneously isolated from transgenic hearts using a modified isolation protocol 26. Briefly, hearts were cannulated via the aortic arch and perfused for 15 minutes at 37C in digestion buffer containing Liberase, then physically dissociated in digestion buffer at 37C. Myocytes were filtered through a 100 micron nylon mesh and separated from nonmyocytes by gravity sedimentation. Fractions of myocytes and nonmyocytes were collected from three subsequent steps of digestion, sedimentation and filtration. Myocytes were plated on laminin coated slides or dishes. The nonmyocyte fraction was either analyzed for EGFP and c-Kit expression by flow cytometry, or filtered through a 40 micron nylon mesh and subjected to magnetic cell sorting to deplete the CD45+ population and enrich for c-Kit expressing cells using the Miltenyi magnetic sorting system (catalog# 130-090-312, mouse CD45 microbeads catalog# 130-052-301, mouse CD117 microbeads catalog# 130-091-224). CPCs were expanded in a humidified incubator at 37C, 5% CO2 in growth media consisting of 1:1 Dulbecco’s MEM/Ham’s F12 (DMEM/F12, LifeTechnologies) and Neurobasal Media (LifeTechnologies) containing 10% ES-FBS (LifeTechnologies), LIF (10ng/ml, Millipore), bFGF (10ng/ml, Peprotech), EGF (20ng/ml Peprotech), 1X Insulin-transferrin-selenium (ITS 500X, Lonza) 1% penicillin/streptomycin/2mM Glutamine (100X PSG, LifeTechnologies), supplemented with B27 and N2 (LifeTechnologies). Human CPCs were isolated from human LVAD discard tissue and expanded as previously described 27. Briefly, tissue was digested with collagenase, nonmyocyte cells were separated by low speed centrifugation (350g for 5 minutes) and filtered through sequential 100μcells were separated by lwere cultured for one day at 37°C, 5% CO2 in hCPC growth media (HAM’s F12 supplemented with 10% ES-FBS, 5mU/mL human EPO, 10ng/mL human bFGF, 0.2 mM Glutathione, 1X Penicillin/Streptomyocin/Glutamine), after which the c-Kit+ population was selected for by incubating with magnetic bead conjugated c-Kit antibodies (Miltenyi Biotec #130-091-332) and MAC sorting. Magnetically separated cells were expanded for at least 5 passages in hCPC growth media prior to use in experiments. Adult hCPCs were used at passage 6–10 in all experiments.
Statistics
Two-tailed t-tests were used to compare to two groups for all analyses other than the linear mixed model for Figure 5 as described below. p<0.05 is considered statistically significant.
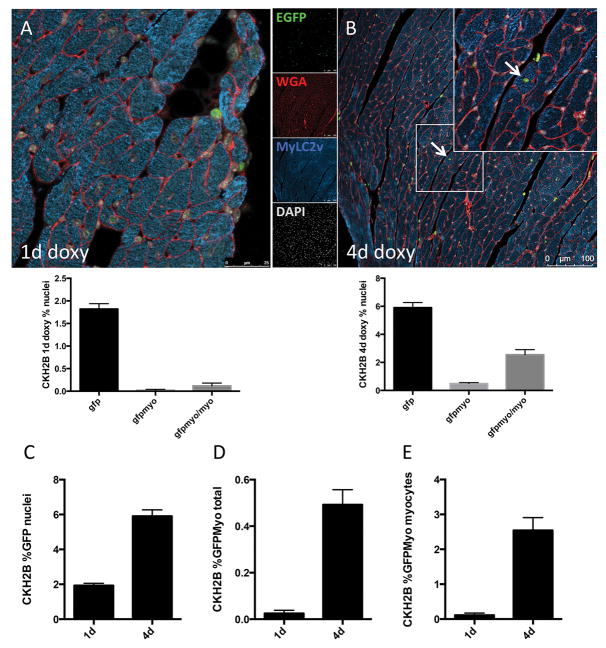
Figure 5. H2BEGFP reporter accumulates in cardiac myocytes in vivo.
EGFP is detected and quantitated in nuclei of immunolabeled CKH2B hearts treated for one day (A) or four days (B) with doxycycline (red=wheat germ agglutinin, green=H2BEGFP, blue=myosin light chain 2v (MYLC2V), gray=DAPI). Quantitation of the increase in EGFP+ cells per total nuclei (C), EGFP+ myocytes per total nuclei (D) and EGFP+ myocytes per total myocytes (E) represented in histograms. n=3 hearts, minimum 10 fields of view per heart.
To formally test the difference in the means of the knock-in versus transgenic, the repeated measures of c-Kit for each animal are modeled using a linear mixed model. A post-hoc Tukey two-sided test for comparison of means indicates some evidence to suggest a significant difference in means of CKH2B+Doxy vs CKmCm+Tx (P =0.0838). (See Supplement Material for description and details of the statistical analyses.)
To formally test the difference of c-Kit density per unit area, the repeated measures of c-Kit for each animal are modeled using a linear mixed model. Based on the significant results of the mice strain factor, a post-hoc Tukey two-sided test for multiple comparison of means indicates a significant difference in means of CKmCm-Tx vs CKH2B+Doxy (P=0.00769), CKH2B+Doxy vs CKmCm+Tx (P= 0.00542), and no significant difference in means of CKmCm-Tx vs CKmCm+Tx (P= 0.90577). (See Supplement Material for description and details of statistical analyses.) Online Table III provides values for efficiency of co-expression assessed with c-Kit+ / GFP+ coincidence in three experiments using CKH2B and CKmCm mouse heart samples.
RESULTS
c-Kit signaling promotes proliferation and survival in CPCs
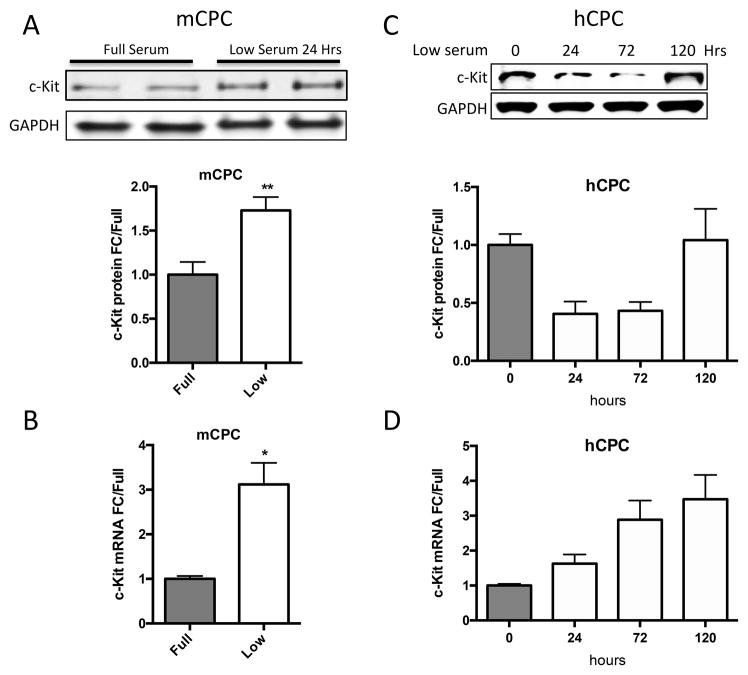
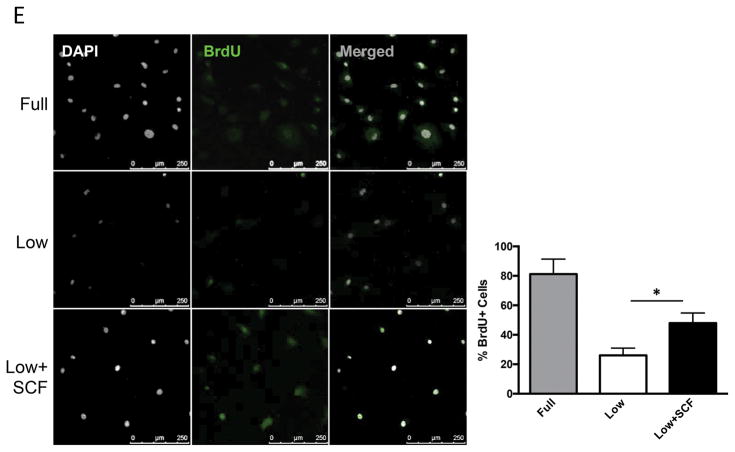
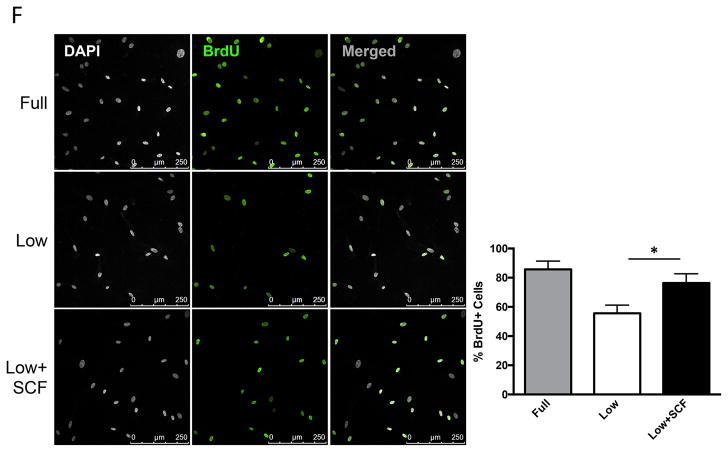
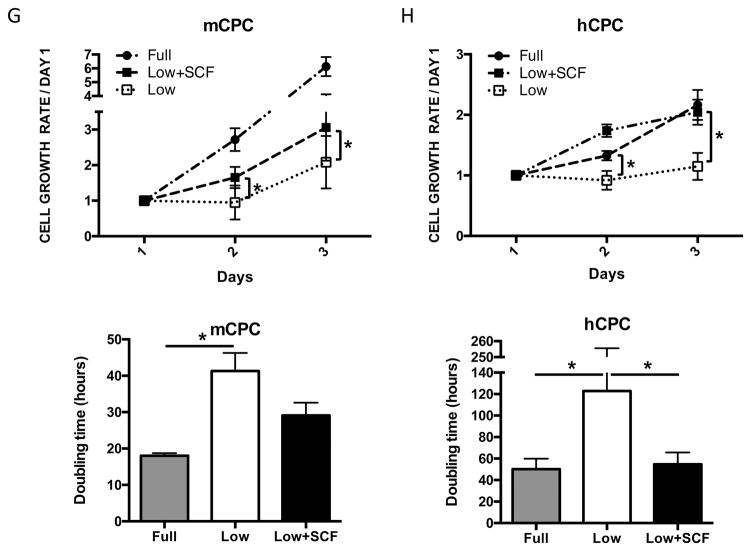
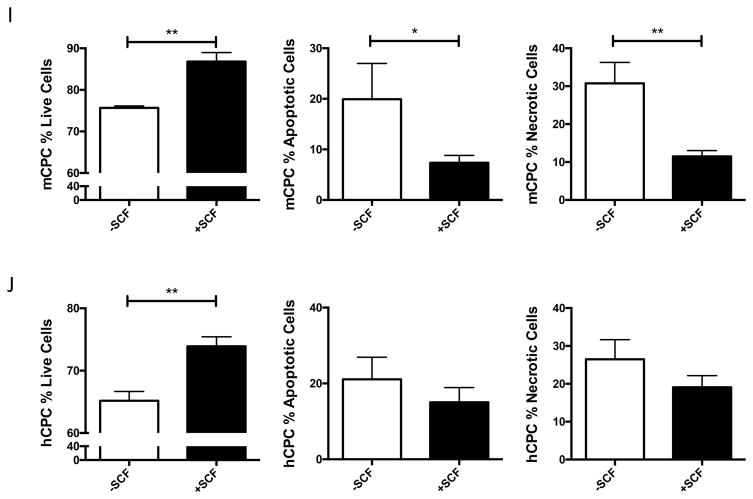
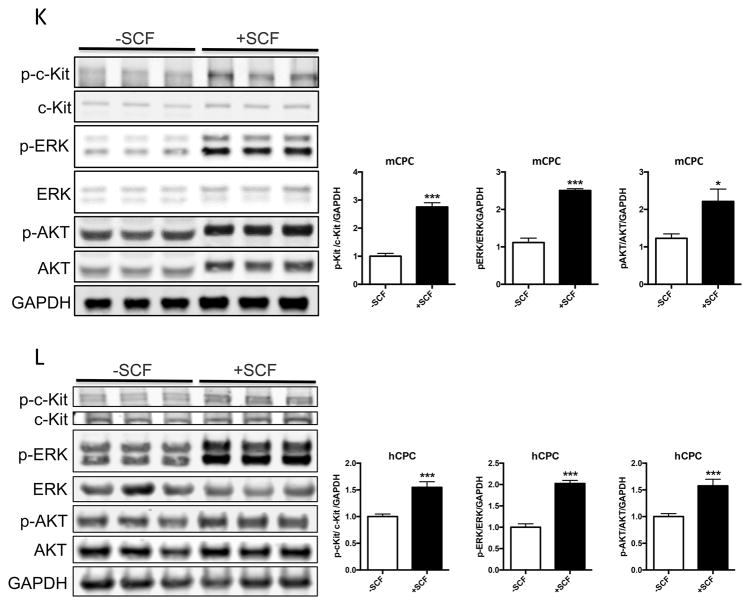
c-Kit signaling stimulates proliferation and enhances survival in multiple cell types 28,29,30, including human cardiac stem cells 19. Mouse and human CPCs were assayed for c-Kit expression, proliferation and cell death in response to low serum culture conditions. Serum starvation for 24 hours induces a significant 73% increase in c-Kit protein and threefold higher levels of c-Kit mRNA in mCPC (Figure 1A,B), while c-Kit protein levels in hCPC initially decrease by 60% at 24 hours and return to full serum levels after five days of starvation, as measured in three distinct lines (Figure 1C). Interestingly, c-Kit mRNA levels increase by 63% in hCPCs after 24 hours, and rise to more than threefold by five days in low serum (Figure 1D). Treatment of serum starved mCPC and hCPC with c-Kit ligand SCF significantly enhances BrdU incorporation by 84% and 37%, respectively (Figure 1E, F). Similarly, SCF treatment increases cell growth rate and shortens doubling time in serum starved cells (Figure 1G, H), confirming the pro-proliferative activity of c-Kit signaling in CPCs. Viability in serum starved mCPCs is significantly higher and cell death significantly blunted in cells treated with SCF, as measured by a 14% increase in cell survival and 63% decrease in both apoptosis and necrosis using flow cytometry, indicative of protective c-Kit signaling in these cells. Likewise, SCF treated hCPC exhibit a significant 13% increase in viability, and 29% and 28% downward trends in apoptosis and necrosis, respectively (Figure 1I,J). Activation of c-Kit by SCF is verified in mouse and human CPCs as evidenced by significant upregulation of phospho-c-Kit and of downstream targets phospho-ERK1/2 and phospho-AKT (Figure 1K,L). Inhibition of c-Kit with Imanitib or with c-Kit specific antibody blunts phosphorylation of c-Kit, ERK and AKT, demonstrating specificity of SCF mediated c-Kit pathway activation in mCPCs (Online Figure IA,B) and hCPCs (Online Figure IC). To determine if macrophages or mast cells contribute to c-Kit signaling in mCPCs, freshly isolated NMCs and cultured mCPCs were analyzed for macrophage (F4/80, CD206) mast cell (FceR1) and hematopoietic (CD45) markers by flow cytometry (Online Figure ID,E). These populations comprise between 7 and 12% of the whole NMC population (Online Figure ID), but are absent above baseline in mCPCs (Online Figure IE), indicating that cardiac progenitors, not cultured mast or macrophage cells, drive c-Kit signaling in these cells. Consistent with previous reports, live cultured mCPCs express stromal markers (CD105, CD90.1, PDGFRα). Surface c-Kit expression does decrease with culture as measured by flow cytometry in unfixed mCPCs (Online Figure IF), nevertheless, c-Kit is detected in the majority of fixed mCPCs assessed by flow cytometry (Online Figure IF) and immunofluorescence (Online Figure X). Collectively, these data demonstrate that the c-Kit pathway is active in mouse and human CPCs, and that c-Kit signaling promotes CPC proliferation and survival in low serum conditions.
Figure 1. c-Kit signaling promotes proliferation and survival in CPCs.
c-Kit protein and RNA were quantified in mouse and human CPCs cultured in full or low serum media. c-Kit protein levels increase twofold in mCPCs cultured for 24 hours in low serum versus full growth media as measured by immunoblotting (A). c-Kit RNA levels in mCPCs exposed to low serum increase threefold as measured by qPCR (B). hCPC culture for five days in low serum exhibit an initial decrease in c-Kit protein levels which return to full serum levels by five days (C), while c-Kit mRNA increases steadily over time (D). Decreased proliferation in mCPCs and hCPCs cultured in low serum is partially rescued by SCF treatment as indicated by BrdU incorporation (green=BrdU, E,F) and daily cell counts (G,H). Doubling time of CPCs calculated from growth rates increases following serum depletion but is restored with addition of SCF (G,H). Viability is higher, while apoptosis and necrosis are lower in SCF treated CPCs as measured by AnnexinV/PI flow cytometry analysis (I,J). Specific activation of c-Kit in mCPCs and hCPCs is confirmed by increased phosphorylation of c-Kit, ERK and AKT following SCF treatment (K,L). n=3 for all experiments. p<0.05 denotes statistical significance. Statistical significance between two groups was determined by unpaired t-test for all data except J, which was analyzed by paired t-test. *=p<0.05, **=p<0.01, ***=p<0.001.
Cardiac myocytes express active c-Kit and upregulate c-Kit in response to stress in vitro
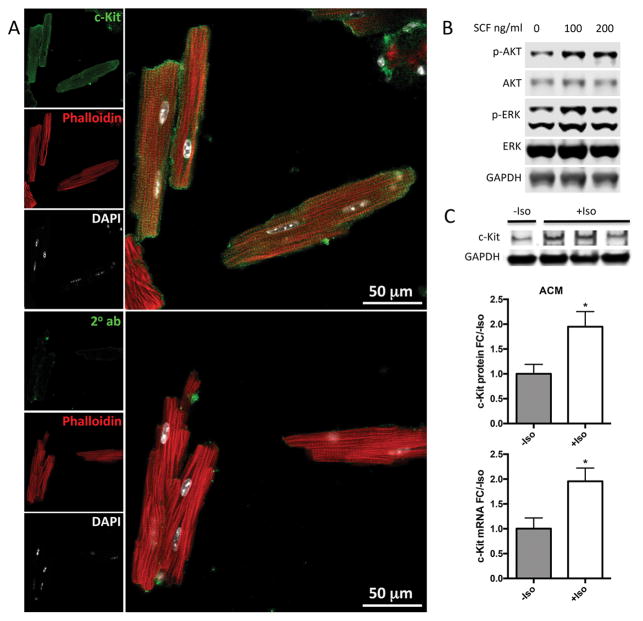
Prior studies have reported increased cardiac c-Kit expression in myocytes following myocardial injury 4. Isolated ACM immunolabeled for c-Kit confirm myocyte specific c-Kit protein expression (Figure 2A, Online Figure IIB, C). c-Kit was detected in most adult cardiac myocytes (ACM), whether fixed immediately after isolation or plated for two hours and then fixed. Using either method, c-Kit staining was found in the majority of ACM (79.25%, or 42 out of 53 immediately fixed ACM). Upregulation of phospho-ERK and phoshpo-AKT in ACM treated with SCF verifies activation of c-Kit activity in these cells (Figure 2B, Online Figure IIA). Finally, c-Kit protein and mRNA levels are elevated twofold in isoproterenol treated ACMs (Figure 2C). Taken together, these results indicate that mouse ACMs express c-Kit mRNA and protein, exhibit c-Kit pathway activation by SCF, and upregulate c-Kit expression in response to cellular stress in vitro.
Figure 2. Cardiac myocytes express active c-Kit and upregulate c-Kit expression in response to stress in vitro.
Adult mouse cardiac myocytes (ACM) were isolated, fixed and immunolabeled for c-Kit (green) colocalized with phalloidin (red) and DAPI (white) (A). Activation of c-Kit by SCF was confirmed by immunoblotting for phospho-ERK and phospho-AKT (B). ACM were treated with isoproterenol to induce cellular stress. c-Kit mRNA and protein levels were measured by qPCR and immunoblotting, respectively. c-Kit protein and mRNA levels increased significantly in isoproterenol treated ACMs as measured by immunoblotting and qPCR, respectively (C). n=3 experiments for A,B. *=p<0.05 as measured by t-test.
H2BEGFP reporter is expressed in transgenic cardiac and noncardiac c-Kit+ cells in vivo
An inducible transgenic c-Kit reporter mouse was created to further probe cardiac c-Kit biology in vivo. The rtTA transcription factor was cloned downstream of a previously characterized 14 kb fragment of the mouse c-Kit promoter 3, 21 to generate the c-KitrtTA mouse line (Online Figure III). c-KitrtTA mice were crossed with mice harboring the tetracycline responsive H2BEGFP reporter construct (TRE-H2BJ/eGFP, Online Figure IIIB)23 to create a double transgenic line in which doxycycline treatment induces H2BEGFP expression in cells harboring an active c-KitrtTA construct (CKH2B, Online Figure IIIC, D). c-KitrtTA transgene copy number was calculated to be approximately 4 copies per diploid genome in hemizygous animals, as measured by quantitative PCR of genomic DNA using a previously described method (Online Figure IV)24. Transgene expression was validated in CKH2B mice treated with doxycycline (Figure 3). Cardiac c-Kit+ cells from doxycycline treated CKH2B mice express the H2BEGFP reporter in the nucleus, as illustrated by immunolabeling of paraffin sections (Figure 3A) and freshly isolated, unfixed nonmyocytes (NMC) immunolabeled for c-Kit and analyzed by flow cytometry (Figure 3B,D). Likewise, noncardiac tissues exhibit H2BEGFP induction in c-Kit+ cells as measured by flow cytometry of bone marrow (Figure 3C,E) and immunostaining in c-Kit+ rich tissues such as intestinal crypts and cryptopatches, and pancreatic Islet of Langerhans (Online Figure VIIIA-C). Furthermore, H2BEGFP is expressed in vascular and stromal cardiac cells, as indicated by colocalization with CD31 and vimentin by IHC-P (Online Figure VA,B) and with CD105, CD90.1 and PDGFRα as measured by flow cytometry (Online Figure VE, Online Figure XI). Vascular smooth muscle does not appear to express H2BEGFP (Online Figure VD), and CD45+/EGFP+ cells were rarely observed in uninjured tissue sections, however flow cytometry reveals a higher percentage of CD45+/H2BEGFP NMC cells (Online Figure VE).
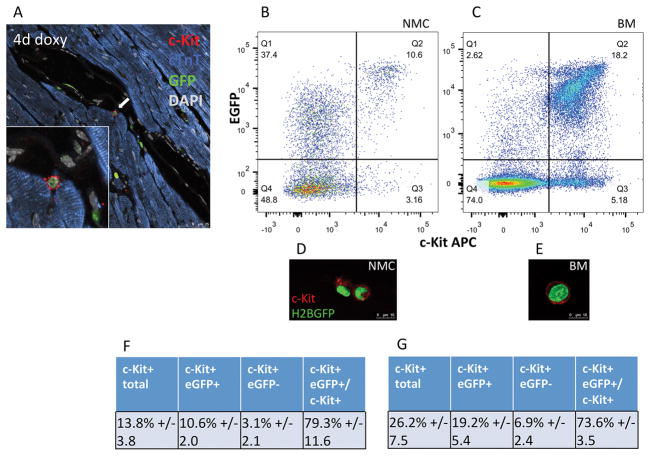
Figure 3. H2BEGFP reporter expression is induced in c-Kit+ cells in vivo.
Cardiac c-Kit+ cells from CKH2B mice express H2BEGFP reporter (white arrow) following administration of doxycycline, as visualized by immunolabeling (A, red=c-Kit, green=EGFP, blue=cardiac troponin T(cTnT), gray=DAPI). Coexpression of c-Kit and H2BEGFP is detected in fresh isolates of nonmyocyte cardiac cells (NMC, B) and whole bone marrow (BM, C) from doxycycline treated CKH2B mice as measured by flow cytometry, and illustrated by corresponding representative confocal images (red=c-Kit, green=H2BEGFP). Tabulation of flow cytometry analysis of c-Kit+ and H2BEGFP in CKH2B NMC (F) and BM (G). n=4 animals.
c-Kit+ cells comprise 13.8%+/−3.8 of all freshly isolated NMC, most of which (79.3%+/−11.6) co-express the H2BEGFP reporter in CKH2B doxycycline treated hearts (Figure 3F). Freshly isolated, unfixed whole bone marrow from the same animals exhibit a higher percentage of c-Kit cells in the overall population (26.2%+/−7.5), 82.4% of the H2BEGFP BM, more c-Kit+EGFP+ cells overall (19.2%+/−5.4 in total BM vs 10.6%+/−2.0 in total NMC, and 82.4% of H2BEGFP+ BM vs 29.5% of the H2BEGFP NMC population Online Figure VE), but a similar proportion of c-Kit+EGFP+ cells within the total c-Kit population (73.6%+/−3.5), reflective of higher overall endogenous c-Kit expression levels in bone marrow versus NMC, but similar tagging efficiency in the two c-Kit+ tissues (Figure 3G). To correlate EGFP+ signal with c-Kit mRNA expression in c-Kit− NMC, EGFP+ nonmyocytes were FACS sorted for positive or negative c-Kit protein expression and analyzed for c-Kit mRNA levels by qPCR (Online Figure VG). c-Kit mRNA was detected above negative control levels in the EGFP+/c-Kit− population, and at one third the level seen in EGFP+/c-Kit+ cells. These results serve as additional evidence that the H2BEGFP reporter reflects c-Kit promoter activity and further validates the CKH2B model.
Differences in coincidence of c-Kit / EGFP tagging as well as c-Kit+ cell frequency assessed by comparative analysis of transgenic versus knock-in reporter mouse models
Various tagging strategies have been developed to identify the c-Kit cell population in vivo using mouse models 4, 9, 10, 21, 31. An established c-Kit / EGFP knock-in reporter construct from a Cre-mediated inducible knock-in line (CKmCm) 9 was compared to the CKH2B model for both colocalization of c-Kit with EGFP, as well as frequency of c-Kit+ cells in myocardial tissue sections. Paraffin sections were immunolabeled for c-Kit, EGFP and DAPI, along with WGA to mark cell boundaries and cardiac troponin T (cTnT) to identify myocytes (representative images, Online Figure VIA,B). The total number of c-Kit+ cells was counted per area of tissue, and percentage of c-Kit cells expressing reporter determined by colocalization of c-Kit, EGFP and DAPI signals. Efficiency of coincident tagging between c-Kit and EGFP in cardiac c-Kit+/EGFP+ cells exhibited a trend but was not significantly higher in heart tissue sections from CKH2B+Doxy versus CKmCm+Tx mice (Figure 4A, p=0.08; 75.7%+/−6.9 SD vs 62%+/−4.5 SD, respectively, Online Table III), consistent with flow cytometry data for CKH2B+Doxy nonmyocyte (Figure 3F, G) or CKmCm+Tx bone marrow cells 9. Moreover, density of c-Kit+ cells per unit area is significantly higher in CKH2B versus CKmCm heart tissue sections (Figure 4B). This rigorous, blinded analysis reveals that the transgenic (CKH2B) reporter mice possess 2.2 fold higher cardiac c-Kit+ cell population density and a trend toward higher EGFP tagging of c-Kit+ cells relative to the knock-in (CKmCm) mice. However, neither model successfully tags all c-Kit+ cells with EGFP, demonstrating a similarity consistent with heterogeneity of the endogenous cardiac c-Kit cell population. Additionally, NMCs were isolated from tamoxifen treated CKmCm mice and analyzed for c-Kit and EGFP+ by flow cytometry. Consistent with findings in paraffin sections, the percentage of c-Kit+ NMCs is approximately half of the level observed in CKH2B NMCs (5.8%+/−1.1 SD versus 13.8%+/−3.8 SD, respectively, Figure 4C). Interestingly, the EGFP+ population was substantially lower in CKmCm BM and NMC than in CKH2B (0.04%+/−.02 SD versus 4.7%+/−2.7 SD NMC, 2.9% versus 20.4% BM in CKmCM and CKH2B respectively Online Figure VIC). Discrepanices in reporter expression may be due in part to differences in mouse strain and reporter induction protocol, however the lower proportion of EGFP+ cells in CKmCm is also consistent with the c-Kit haploinsufficiency.
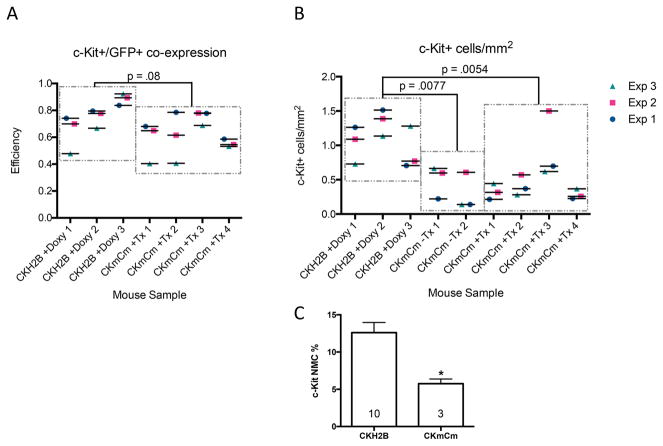
Figure 4. Similar c-Kit+/EGFP+ co-localization but decreased total cardiac c-Kit+ cells per area in CKmCm knock-in mice versus CKH2B transgenic mice.
Paraffin sections were immunolabeled for c-Kit and EGFP to quantitate reporter coexpression in cardiac c-Kit+ cells in situ. EGFP co-localization with c-Kit+ cells in hearts of CKmCm and CKH2B mice is comparable (p=0.0838)(A). Significantly fewer c-Kit+ cells per area were present in CKmCm+Tx and CKmCm-Tx mice compared to doxycycline treated CKH2B mice (p=0.0054 and p=0.0077, respectively). No statistical differences were found between CKmCm-Tx and CKmCm+Tx mice (0.9058)(B). CKH2B hearts n=3, CKmCm-Tx hearts n=2, CKmCm+Tx hearts n=4. Significance determined by mixed linear modeling as described in the Statistics section and the Supplemental Statistical Analysis. p<0.05 denotes significance. (C). Comparative flow cytometry analysis of nonmyocytes isolated from CKmCm+Tx versus CKH2B+doxycycline hearts reveals significantly lower percentage of c-Kit+ NMCs in CKmCm hearts. n=10 CKH2B, n=3 CKmCm, statistical significance determined by two-tailed t-test, *=p<0.05.
H2BEGFP reporter is expressed in CKH2B adult cardiac myocytes
c-Kit expression has been reported in neonatal mouse cardiomyocytes and genetically tagged mouse ACMs 4, 9, 31, 32, and measured in ACM (Figure 2). Therefore, expression of H2BEGFP reporter in myocytes was tested by immunolabeling for EGFP and cardiac myosin light chain in heart tissue from CKH2B mice treated with doxycycline for 1 or 4 days (Figure 5A, B). Quantitation of H2BEGFP signal in all nuclei and in myocyte nuclei reveals that approximately 1.8% of all cardiac nuclei express H2BEGFP after one day of induction, increasing to 6% by four days of treatment (Figure 5C). Cardiac myocyte nuclei positive for H2BEGFP comprise less that 0.1% of all nuclei at 1 day, and 0.5% of all nuclei by 4 days of induction (Figure 5D), while the proportion of cardiac myocytes expressing H2BEGFP reporter increases from 0.1% to 2.5% between 1 and 4 days, respectively (Figure 5E). ACMs isolated from doxycycline treated CKH2B hearts exhibit discreet nuclear H2BEGFP fluorescent signal in 9.4% of total myocytes (Online Figure VIIA, Figure 7F). Likewise, naïve CKH2B ACM treated with doxycycline post isolation express H2BEGFP (Online Figure VIIB). Taken together, these data confirm c-Kit expression in mouse ACM (Figure 2A) and demonstrate sensitive detection of c-Kit expression in the cardiomyocyte population as revealed by cumulative H2BEGFP expression over time with doxycycline induction. Additionally, paraffin sections of tamoxifen induced CKmCm were immunolabeled for EGFP and a cardiomyocyte marker to identify EGFP+ myocytes in the knock-in hearts. Between 1 and 5 EGFP+ myocytes were detected per heart section in three individual CKmCm hearts (Online Figure VID). Collectively, these data provide further evidence that c-Kit is expressed in adult cardiac myocytes.
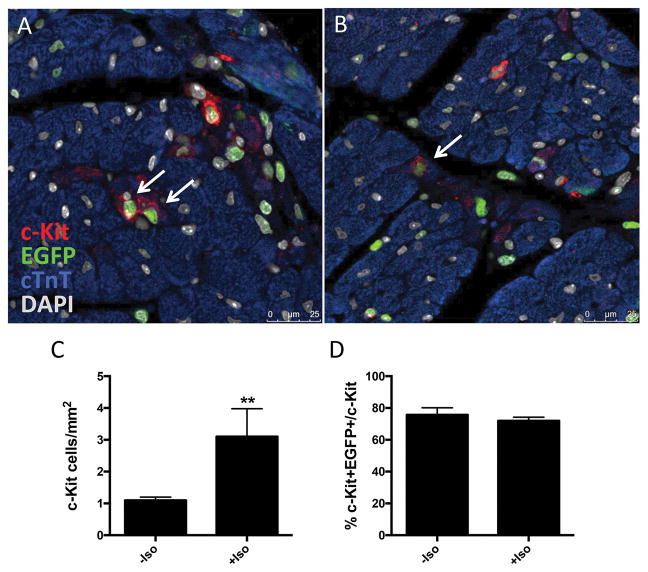
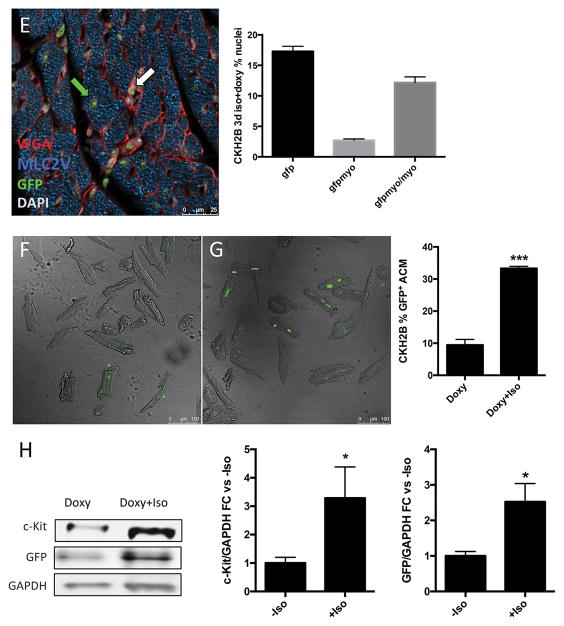
Figure 7. Cardiac c-Kit+ expression increases following diffuse injury in vivo.
H2BEGFP expression in cardiac c-Kit+ cells visualized by immunostaining and confocal microscopy (A,B), red=c-Kit, green=EGFP, blue=cTnT, gray=DAPI, white arrows indicate c-Kit+/EGFP+ cells. Quantitation of c-Kit+ immunolabeled cells per area of tissue (C), and percentage of c-Kit+ cells expressing H2BEGFP reporter in uninjured versus injured CKH2B hearts (D). H2BEGFP expression in adult cardiac myocytes in hearts from doxycycline and isoproterenol treated CKH2B revealed and quantitated by immunostaining (E) (red=WGA, green=EGFP, blue=MyLC2V, gray=DAPI, white arrow indicates H2BEGFP in nonmyocyte, green arrow indicates H2BEGFP in cardiomyocyte). ACM isolated from CKH2B mice treated with doxycycline only (F) or doxycycline and isoproterenol (G) for three days as imaged and quantitated by confocal microscopy. Protein levels of c-Kit and EGFP quantitated in ACM isolated from Doxy or Doxy+Iso treated CKH2B hearts (H). Bright field=ACM, green=H2BEGFP fluorescence. n=3 mice (F–H), n=4 isolations (I).
H2BEGFP is expressed in CKH2B bone marrow stem cells in vitro
Bone marrow is an established source of c-Kit expressing stem cells. c-Kit+ bone marrow stem cells (BMSC) were isolated as previously described 33 from CKH2B mice, expanded and treated with doxycycline in vitro to assess transgene expression in a c-Kit-enriched stem cell system. As shown by live fluorescence imaging, untreated BMSC express no fluorescent signal (Online Figure IXA), while cells exposed to doxycycline exhibit induction of robust, chromatin-associated H2BEGFP fluorescence consistent with expected histone distribution (Online Figure IXB,C). Flow cytometry and confocal microscopy analyses of fixed, immunolabeled BMSCs further confirm low baseline reporter signal in untreated BMSCs (Online Figure IXD,H) and robust induction of H2BEGFP in doxycycline treated CKH2B (Online Figure IXE, F, I) but not in single transgenic c-KitrtTA BMSC (Online Figure IXG). Coexpression of c-Kit with the H2BEGFP signal is detected in approximately 60% of the total c-Kit+ BMSC population, as measured by flow cytometry (Online Figure IXF). BMSCs labeled for flow cytometry and imaged by confocal microscopy exhibit membrane c-Kit and nuclear H2BEGFP signals (Online Figure IXI). Cumulatively, these data demonstrate robust and appropriate induction of the H2BEGFP reporter in a cultured stem cell population enriched for c-Kit expression, consistent with previous publications characterizing the CKH2B promoter construct 21.
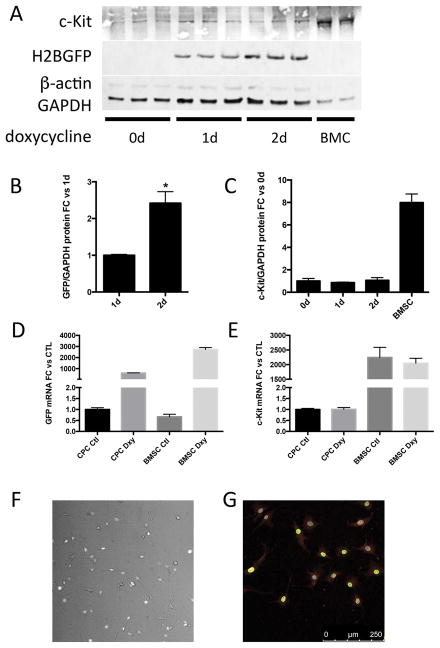
CKH2B CPCs express c-Kit and H2BEGFP reporter in vitro
Nonmyocyte c-Kit+ cardiac cells are an important source of stem and progenitor cells for cardiac regenerative therapy. c-Kit cardiac progenitor cells (CPCs) isolated from CKH2B hearts were induced with doxycycline and probed for c-Kit and H2BEGFP protein expression. H2BEGFP protein is undetectable in untreated CPCs and clearly induced by doxycycline treatment, as indicated by immunoblot (Figure 6A), fluorescence imaging of live and immunostained fixed cells (Figure 6F,G, Online Video I, Online Figure X). c-Kit protein and mRNA levels are substantially lower in CPCs than in BMSCs, and H2BEGFP levels increase twofold in CPCs between 1 and 2 days of exposure to doxycycline, whereas endogenous c-Kit levels remain unchanged (Figure 6C,E). Quantitative PCR confirms that mRNA levels of H2BEGFP but not endogenous c-Kit are augmented following doxycycline induction (Figure 6B,D), and live imaging verifies nuclear localization of the fluorescent H2BEGFP signal in CPCs (Figure 6F, Online Figure X). Collectively, these results demonstrate that the CKH2B reporter is induced in two cultured adult c-Kit stem cell populations and reflects the heterogeneity of c-Kit expression inherent in the parent tissue from which these cells are derived.
Figure 6. H2BEGFP reporter is induced in CKH2B c-Kit+ CPCs in vitro.
CKH2B CPCs express c-Kit and accumulate H2BEGFP after treatment with doxycycline (A). BMC=c-KitrtTA whole bone marrow. Quantitation of H2BEGFP protein levels in A relative to 1 day of doxycycline treatment (B) and of c-Kit relative to untreated CPCs (C). Quantitation of mRNA levels of EGFP (D) and c-Kit (E) relative to untreated CPCs. H2BEGFP fluorescence in c-Kit+ CKH2B CPCs following doxycycline induction in live cells (F) and fixed cells immunolabeled for c-Kit (G, red=c-Kit, green=EGFP, gray=DAPI).
Cardiac c-Kit expression increases following diffuse injury in vivo
Diffuse injury induces transient proliferation of CPCs in vivo 8. Mice received a single injection of 150mg/kg isoproterenol subcutaneously and hearts were analyzed by immunostaining or flow cytometry three days later. Consistent with previous reports, the c-Kit+ NMC population increased (12.6% to 20.1% in control versus injured hearts) as measured by flow cytometry (Online Figure XIC). CKH2B mice received a single injection of isoproterenol and treated with doxycycline for three days, and c-Kit colocalization with H2BEGFP was measured by immunolabeling in paraffin sections or flow cytometry on isolated NMC to assess the impact of diffuse injury on transgene expression. The H2BEGFP+ NMC population increased by 63% (4.9% to 8.0% of ungated NMC), while the proportion of c-Kit+ H2BEGFP NMCs declined from 29.5% to 20.7% in uninjured versus injured hearts, respectively (Online Figure XIC). The proportion of mesenchymal/stromal cells, as marked by CD105, CD90.1 and PDGFRα, all increased within the total NMC and EGFP+ NMC populations (Online Figure XID,E), supported by clear colocalization of vimentin with EGFP+ in damaged regions of cardiac CKH2B tissue sections (Online Figure XIA) The percentage of c-Kit+ cells labeled for H2BEGFP in injured heart was comparable to uninjured controls (75.7% versus 71.9%, respectively, Figure 7A,B,D). Cells coexpressing CD45 and EGFP were detected in injured areas of cardiac tissue sections (Online Figure XIB), while flow cytometry analysis indicates a decrease in the CD45+/H2BEGFP+ NMC population from 27% to 11% in uninjured versus injured hearts. The total number of c-Kit cells per area was 3.1 versus 1.1 per mm2 in injured versus uninjured heart sections (Figure 7C), consistent with flow cytometry data and previous reports of elevated c-Kit cell numbers in isoproterenol treated hearts 3 days after injury 8.
Prior studies have reported evidence for c-Kit expression in myocytes 4, 9, 10, 31, 32. Therefore, expression of H2BEGFP was assessed in cardiac myocytes isolated from hearts of CKH2B mice receiving a single injection of isoproterenol and treated with doxycycline for three days. H2BEGFP signal increased overall in isoproterenol treated hearts as measured by immunolabeling for EGFP and cardiac myosin light chain (Figure 7E). Total EGFP positive nuclei rose from 6% in unchallenged hearts to 17% following injury, the EGFP positive myocytes increased from 0.5% to 3% of total nuclei, and the proportion of EGFP labeled myocytes was 13% of the myocyte population versus 3% in control hearts (Figure 3E, Figure 7F), consistent with the upregulation of c-Kit in ACMs treated with isoproterenol in vitro (Figure 2C). Furthermore, the percentage of H2BEGFP+ ACM isolated from isoproterenol injured CKH2B hearts was more than threefold higher than those from uninjured controls (33.3% versus 9.4%, respectively, Figure 7F,G), and protein levels of c-Kit and H2BEGFP were also elevated in ACM from isoproterenol treated animals (Figure 7H). These data strongly support evidence for c-Kit expression in adult cardiac myocytes in vivo and in vitro, and emphasize a previously underappreciated and potentially important role for c-Kit signaling in the cardiomyocyte response to stress.
DISCUSSION
Over a decade of cumulative research supports the important biological role of c-Kit+ cells in cardiac development, homeostasis and repair 2, 8, 22, 34, 35 although recent studies using genetically modified knock-in mice have challenged certain aspects of c-Kit-mediated contribution to myocardial regeneration 9, 36. Knock-in mouse models thus far commonly focus upon the property of c-Kit as a canonical stem cell marker originally identified in the hematopoietic context, but devote relatively little attention to the critical biological role of c-Kit-dependent signal transduction. Regarding c-Kit as a perfunctory marker of stem cells overlooks substantial evidence that c-Kit signaling is an important determinant of myocardial biology and the molecular response to pathologic injury 19, 37. Findings presented here report biologically relevant c-Kit function in cardiac progenitor cells and myocytes, with important implications for the role of c-Kit expression and signaling in the cardiac injury response.
c-Kit is a type III receptor tyrosine kinase comprised of an extracellular ligand binding domain, transmembrane region and intracellular kinase signaling domain. Binding of c-Kit ligand Stem Cell Factor (SCF) initiates receptor dimerization, autophosphorylation, and activation of intracellular signaling cascades such as PI3K/AKT and Ras/MAPK/ERK. AKT and ERK signaling downstream of c-Kit confers powerful protective and proliferative effects in the heart as well as other tissues 14, 15, 37, 38. In the myocardial context, SCF-mediated activation of c-Kit or overexpression of constitutively active c-Kit protects against cardiomyopathic challenge 38, while defective c-Kit signaling leads to compromised cardiac function in response to age and stress 39–41. Consistent with published literature, findings obtained using isolated ACM demonstrate that c-Kit protein is functionally present and responsive to SCF exposure (Figure 2A–C, Online Figure II). Furthermore, c-Kit expression increases in ACMs following adrenergic stress (Figure 2C), potentially contributing to a protection of the myocardium from pathological injury.
Concluding that c-Kit expression in cardiomyocytes is a normal biological process is reinforced by studying genetically engineered mouse models using three distinct c-Kit promoter systems including transgenic c-Kit-BAC reporter 4 and c-Kit locus knock-in systems 9, 18, 31, 42. “Instant on” c-Kit expression tagging in a c-Kit knock-in model demonstrated reporter expression in adult cardiac myocytes 31, consistent with H2BEGFP tagging of myocytes in CKH2B hearts after four days of doxycycline treatment (Figure 4). In the wake of pathologic damage to the myocardium, GFP signal in c-Kit-BAC border zone myocytes following cryoinjury indicates activation of c-Kit expression in stressed cardiomyocytes 4 is consistent with higher incidence of H2BEGFP in ACMs isolated from isoproterenol injured CKH2B hearts (Figure 7E–H) and elevated c-Kit protein levels in isoproterenol treated ACMs (Figure 2C). Sample preparation and detection method influence outcomes of quantitative assessments, as exemplified by comparing findings using immunolabeled paraffin sections versus freshly isolated ACM. The percentage of EGFP+ cardiomyocytes in paraffin sections (2.5%) represents an in situ view of the heart in which tissue content and context are preserved, yet epitope availability to antibodies may be diminished due to tissue processing. In comparison, the percentage of freshly isolated EGFP+ ACM from CKH2B hearts (9%) represents the end product of a multistep preparation that selects for cardiomyocytes that survive myocardial digestion and enrichment procedures. Furthermore, c-Kit protein is not detected in all EGFP+ cells in CKH2B cardiac cells because levels of c-Kit protein versus mRNA expression do not necessarily directly correlate (Online Figure VG). Another contributing factor in this observed discrepancy is sensitivity of c-Kit detection. For example, directly conjugated c-Kit antibody is used for detection of c-Kit in unfixed EGFP+ cells (Figures 3D,E). However, fixation and permeabilization of cells increases the proportion of c-Kit+ positive cells in the population since some c-Kit protein is intracellular16. Furthermore, applying unconjugated c-Kit primary antibody followed by a fluorescently tagged secondary yields higher sensitivity for detection of c-Kit protein because of increased fluorophore labeling. These methodological considerations can also contribute to variation in correlation between c-kit protein and EGFP reporter detection. Thus, derivation of quantitative findings using multiple approaches as performed in this study marginalizes bias in experimental design and helps to reconcile discrepant observations from prior publications. Consistent documentation of c-Kit in adult cardiomyocytes supports a regulatory role in biological processes and validates appearance of the CKH2B reporter in the present study as a reasonable and relevant indicator for c-Kit expression in ACMs. Differential induction of the two mouse models is an important discussion point. Although tagging efficiency of c-Kit+ cells is comparable in the two mouse models, the underlying genetics and induction protocols are dramatically different. To generate the EGFP+ tag in c-Kit+ cells in the knock-in model, CKmCm mice ingest tamoxifen in their feed for four weeks to effect permanent Cre-mediated recombination, whereas CKH2B animals imbibe doxycycline in their water for four days to drive expression of the H2BEGFP+ reporter. In the first system, EGFP+ cannot accumulate after recombination, unlike the tet-inducible system directly driving expression of H2BEGFP. These differences in reporter expression mechanisms influence reporter induction dynamics between the two models, in addition to the distinct c-Kit promoters used for tissue specific expression.
Induction and activation of c-Kit in CPCs echo findings in ACMs. c-Kit signaling confers protective, proliferative, and pro-migratory effects through induction of ERK and AKT in adult human and rat CPCs 19, 41. Likewise, the SCF/c-Kit pathway promotes proliferation in CPCs stimulated by co-culture with MSCs 43. Consistently, c-Kit levels increase in mouse CPCs following cellular stress (Figure 1A,B). Interestingly, in human CPCs, c-Kit protein levels decrease and recover after five days in low serum, whereas c-Kit mRNA increases steadily from 1 to 5 days in low serum (Figure 1C,D). Furthermore, SCF-mediated activation of c-Kit promotes CPC proliferation and blunts cell death in both mCPC and hCPC (Figure 1E–J), demonstrating the functional biological role of c-Kit in CPCs. Again, as is true for ACM, genetically engineered fluorescent tag reporter models contribute support for relevance of c-Kit cell biology through visualization and tracking of cardiac c-Kit+ cells in vivo and in vitro 3, 4, 9, 10, 31. The tetracycline-responsive H2BEGFP reporter was deliberately chosen for our study based upon literature precedents as a long-lived, easily identifiable chromatin-associated fluorescent tag originally designed for stem cell tracking studies 23, 44. Combined with the transgenic c-KitrtTA promoter, the resultant CKH2B reporter system presented in this report provides a sensitive, inducible readout for c-Kit expression without impacting endogenous c-Kit function consequential to targeted knock-in approaches that take advantage of the endogenous promoter but, by inherent necessity of experimental design, inactivate normal transcriptional activity of the c-Kit locus.
Another critical aspect of further validating the H2BEGFP model as a tool for investigating cardiac c-Kit cell biology is consistency with numerous reports detailing phenotypic and morphologic properties of CPCs 2, 3, 8, 22, 45, 46. H2BEGFP-tagged cells in transgenic hearts exhibit typical morphology of small, round c-Kit+ cells embedded in the myocardium (Figure 3A). Induction of H2BEGFP expression revealed by confocal analyses of paraffin sections and flow cytometry of freshly isolated nonmyocyte cardiac cells (Figure 3B–E) matches reports with other c-Kit reporter models 9. Likewise, CPCs isolated and expanded from CKH2B hearts exhibit classic CPC spindle morphology, express c-Kit, and upregulate H2BEGFP following doxycycline treatment (Figure 6, Online Video I, Online Figure X). These fundamental characterizations together with concurrence between our findings and prior publications 2, 46 reinforce the fidelity and authenticity of c-Kit properties as revealed in the H2BEGFP transgenic model.
Cardiac c-Kit cell expansion occurs in diffuse injury models in vivo 47, 48. Analysis of c-Kit+ cells in isoproterenol injured CKH2B hearts revealed sites of c-Kit+/H2BEGFP+ clusters and, interestingly, significantly more H2BEGFP+ cardiomyocytes in vivo and in vitro (Figure 7), consistent with elevated c-Kit expression in isoproterenol treated ACMs (Figure 2C) and reminiscent of EGFP positive myocytes in border zone myocytes of c-Kit-BAC-GFP hearts 4. Upregulation of c-Kit in CKH2B myocytes in response to stress further substantiates a previously unrecognized or underappreciated biologically relevant role for c-Kit signaling in cardiac myocytes.
c-Kit+ cells constitute a highly heterogeneous population with broad distribution and diverse function throughout the body postulated to possess a range of phenotypic and biological properties 35. The cardiac c-Kit+ cell subpopulation likewise comprises a diverse cell pool purported to participate in both direct contribution to tissue formation by lineage commitment as well as indirect paracrine-mediated signaling 35, 49–51. Tagging efficiency of such a heterogenous cell population will inevitably vary with promoter expression levels, whether endogenous or transgenic, and may be affected by the hemizygous state of current c-Kit knock-in reporter models. Interestingly, up to 35% of cardiac c-Kit+ cells do not express a genetic reporter tag in either system, as measured by flow cytometry and immunolabeing of paraffin sections (Figure 3, 4)9 further highlighting the complexity of c-Kit expression dynamics. Although approximately 70% of H2BEGFP+ nonmyocytes are negative for c-Kit protein by flow cytometry (Online Figure VE,F), these EGFP+/c-Kit− nonmyocytes possess c-Kit mRNA by RT-qPCR analysis (Online Figure VG) consistent with CKH2B transgenic promoter activity reflecting endogenous c-Kit mRNA expression. EGFP+ nonmyocytes diversity is readily apparent from immunoprofiling that shows heterogeneity of immune, interstitial cell and mesenchymal/stromal markers including FceR1, F4/80, CD206, vimentin, CD105, CD90.1 and PDGFRα (Online Figure ID and Online Figure VE). Such heterogeneity warrants cautious interpretation of c-Kit+ cell biology consistent with the existence of multiple distinct subpopulations possessing nuanced differential characteristics that are lost upon imposition of assessments focused upon average measurements.
Genetic knock-in models that disrupt the c-Kit locus by necessity confer a hemizygous genotype for c-Kit. Given the proliferative and protective roles of c-Kit signaling in CPCs, and evidence for survival signaling in ACMs, it is important to consider the potential impact of the hemizygous c-Kit phenotype on cardiac function and repair. Indeed, frequency of c-Kit cells decreased approximately 50% in myocardial tissue sections from knock-in versus transgenic samples (Figure 4B), perhaps a consequence of differences in c-Kit gene dosage (hemizygosity in the CKmCm knock-in mice), strain (FVB for CKH2B and C57Bl/6 for CKmCm), or reporter system (direct TRE-mediated induction in CKH2B versus Cre-mediated recombination in CKmCm). One explanation for the difference in cell labeling between the knock-in and the transgenic models could be a feed-forward mechanism whereby disruption of one copy of c-Kit expression in the knock-in model leads to reduced c-Kit levels in all cells that express c-Kit. Impairment of c-Kit function leads to unequivocal phenotypic consequences in Wv c-Kit mutant mice that exhibit impaired cardiac recovery after infarction, diminished cardiac function with advanced age, and compromised c-Kit cell terminal differentiation into cardiomyocytes 32, 39–41. Circumspection is reasonable when considering the myocardial response to injury in c-Kit hemizygous mice that could compromise reparative capacity of c-Kit+ cells in myocardial regeneration. Reporter models that do not disrupt the endogenous locus, such as the previously described c-Kit-BAC-GFP transgenic 4 or the inducible CKH2B system described here, bypass interference with endogenous c-Kit expression and associated changes in cellular function. Given the preponderance of evidence as cited in preceding paragraphs to substantiate the biological role for c-Kit in the normal and injured myocardium, inescapable c-Kit allele inactivation should not be readily dismissed or deemed irrelevant from interpretation of results using genetically engineered knock in models.
Together with the advantage of preserving normal endogenous c-Kit regulation, the CKH2B tagging system employs a transgenic c-Kit promoter to express rtTA transcription factor combined with a tet-responsive ubiquitous H2BEGFP reporter line. Doxycycline-mediated transgene induction can be performed at any stage of development without the potential adverse side effects of tamoxifen treatment in pregnant dams 52–57. Protein stability of H2BEGFP, with a reported half-life of several months in vivo 44, 58, provides long term tagging of c-Kit+ cells in CKH2B mice, while allowing for reversibility in mitotically active systems. Permanent tagging of c-Kit cells allowing for long-term lineage tracking of cells has been initiated through combining the c-KitrtTA transgenic construct with a tetracycline-responsive Cre line 59 and the ROSAmT/mG reporter mouse 60. Future studies using various c-KitrtTA transgenic models will establish distribution of c-Kit+ cells in the heart and clarify participation of c-Kit+ cells in response to cardiac injury.
In summary, the biological role of cardiac c-Kit is an important one that may be underappreciated due to an overemphasis upon the use of c-Kit as a “marker” of stem cells. Pro-proliferative and protective effects of c-Kit signaling in cardiac cells are central to their regenerative potential and should be considered when designing experimental models to assess their role in cardiac formation and repair. All genetic lineage-tracing models have inherent limitations. Knock-ins frequently disrupt expression of the gene of interest and impair normal biological regulation, whereas transgenic promoter constructs may lack regulatory elements of the endogenous gene. Multi-component lineage tracing models designed to mark cells temporally and spatially harbor inducible or regulatable elements, or rely upon enzymatic recombination for tagging, introducing further layers of complexity and inescapable variables in efficiency, specificity, or even toxicity. In the final analysis all experimental models are tools, with even the most sophisticated promoter-driven reporter models unable to precisely capture post-transcriptional regulation of c-Kit expression. For example, recent studies reveal that c-Kit translation, but not transcription, is regulated by Pim1 kinase in the hematopoetic stem cell pool 61. Similarly, retinoic acid controls c-Kit translation through PI3K/Akt/mTOR during spermatogonial differentiation 62, 63. Additionally, microRNA 221 downregulates c-Kit protein expression in melanoma, hyperglycemic HUVECs, and PDGF-stimulated vascular smooth muscle cells 64–67. Multiple layers of c-Kit protein regulation dependent upon posttranslational modification, ubiquitination, internalization, and degradation rate all contribute to dynamic c-Kit expression beyond the reach of any one reporter system to reflect endogenous c-Kit biology. Acknowledging that there is much still to learn and embracing variations in perspectives rooted in divergent experimental models will resolve discrepancies, unify the cardiovascular research community, and provide new avenues for exploration moving forward with a renewed respect for the complexity of myocardial c-Kit biology.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
The mammalian heart has a limited capacity for self-renewal and repair. The identity of cells responsible for this regenerative activity remains an area of active investigation.
c-Kit is a receptor tyrosine kinase used to identify cardiac stem and progenitor cell populations. c-Kit signaling promotes cell proliferation and survival.
The cardiac c-Kit cell population is heterogeneous; it contributes to myocardial development, and participates in the myocardial response to injury.
Experimental animal models designed to identify and study the biological function of c-Kit expressing cells produce varying and discrepant results likely in part due to variations in promoter and reporter design.
What New Information Does This Article Contribute?
c-Kit expression increases in mouse and human cardiac progenitor cells in response to cellular stress.
c-Kit is expressed in adult cardiac myocytes and it abundance increases following cellular stress.
c-Kit signaling promotes proliferation and survival in mouse and human cardiac progenitor cells.
The novel, transgenic, inducible c-Kit reporter mouse model characterized in this study reveals previously unrecognized heterogeneous, dynamic c-Kit expression and biology in the postnatal myocardium
The biological role of cardiac c-Kit has been largely overlooked in the cardiovascular research community due to overemphasis on the use of c-Kit as a “marker” for stem cells. The protective and pro-proliferative effects of c-Kit signaling in cardiac cells are central to their regenerative potential and their role in cardiac formation and repair. Heterogeneity of the cardiac c-Kit+ cell population including c-Kit activity in cardiac myocytes, confirmed with our novel transgenic reporter mouse, reconcile and extend prior divergent reports as well as point toward new horizons for c-kit biology in the myocardium. The biological role(s) for cardiac progenitor cells, including those expressing c-kit, in maintaining cardiac homeostasis and promoting repair is clearly more nuanced and complex than previously represented in the evolving field of myocardial regeneration.
Acknowledgments
We gratefully acknowledge Dr. Roland Wolkowicz and Cameron Smurthwaite from the San Diego State University flow cytometry core facility, and Sharp Memorial Hospital staff for providing the Sussman lab with the human heart biopsy samples used for our cell isolations.
SOURCES OF FUNDING
M.A. Sussman is supported by NIH grants: R01HL067245, R37HL091102, R01HL105759, R01HL113647, R01HL117163, P01HL085577, and R01HL122525, as well as an award from the Fondation Leducq. N.A. Gude is supported by the National Institutes of Health (NIH) grants R37HL091102, R01HL117163, R01HL105759, U54CA132384. M.M. Monsanto is supported by NIH grant R01HL122525, Rees Stealy Research Foundation, Achievement Rewards for College Scientists (ARCS), and Elliott Family Fund Scholarship. K.M.Broughton is supported by NIH grant F32HL136196, F.G. Khalafallah is supported by NIH grant F32HL131299, J.van Berlo is supported by NIH grants R00HL112852 and R01HL130072.
Nonstandard Abbreviations and Acronyms
- ACM
Adult cardiac myocytes
- mCPC
Mouse cardiac progenitor cell
- hCPC
Human cardiac progenitor cell
- CKH2B
c-KitrtTA/H2BEGFP mouse line
- CKmCm
c-Kit-MerCreMer /R26-NG mouse line
- TRE
Tet-Responsive Element
Footnotes
DISCLOSURES
MA Sussman is a founding member of CardioCreate, Inc.
References
- 1.Orlic D, Kajstura J, Chimenti S, Limana F, Jakoniuk I, Quaini F, Nadal-Ginard B, Bodine DM, Leri A, Anversa P. Mobilized bone marrow cells repair the infarcted heart, improving function and survival. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10344–10349. doi: 10.1073/pnas.181177898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 3.Fransioli J, Bailey B, Gude NA, Cottage CT, Muraski JA, Emmanuel G, Wu W, Alvarez R, Rubio M, Ottolenghi S, Schaefer E, Sussman MA. Evolution of the c-kit-positive cell response to pathological challenge in the myocardium. Stem cells. 2008;26:1315–1324. doi: 10.1634/stemcells.2007-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tallini YN, Greene KS, Craven M, Spealman A, Breitbach M, Smith J, Fisher PJ, Steffey M, Hesse M, Doran RM, Woods A, Singh B, Yen A, Fleischmann BK, Kotlikoff MI. c-kit expression identifies cardiovascular precursors in the neonatal heart. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1808–1813. doi: 10.1073/pnas.0808920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyamoto S, Kawaguchi N, Ellison GM, Matsuoka R, Shin’oka T, Kurosawa H. Characterization of long-term cultured c-kit+ cardiac stem cells derived from adult rat hearts. Stem Cells and Development. 2010;19:105–116. doi: 10.1089/scd.2009.0041. [DOI] [PubMed] [Google Scholar]
- 6.Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Fischer KM, Cottage CT, Konstandin MH, Volkers M, Khan M, Sussman MA. Pim-1 kinase inhibits pathological injury by promoting cardioprotective signaling. Journal of Molecular and Cellular Cardiology. 2011;51:554–558. doi: 10.1016/j.yjmcc.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellison GM, Vicinanza C, Smith AJ, Aquila I, Leone A, Waring CD, Henning BJ, Stirparo GG, Papait R, Scarfo M, Agosti V, Viglietto G, Condorelli G, Indolfi C, Ottolenghi S, Torella D, Nadal-Ginard B. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 2013;154:827–842. doi: 10.1016/j.cell.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 9.van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin SC, Middleton RC, Marban E, Molkentin JD. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatzistergos KE, Takeuchi LM, Saur D, Seidler B, Dymecki SM, Mai JJ, White IA, Balkan W, Kanashiro-Takeuchi RM, Schally AV, Hare JM. cKit+ cardiac progenitors of neural crest origin. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:13051–13056. doi: 10.1073/pnas.1517201112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Zhu W, Bender I, Gong W, Kwak IY, Yellamilli A, Hodges TJ, Nemoto N, Zhang J, Garry DJ, van Berlo JH. Pathologic Stimulus Determines Lineage Commitment of Cardiac C-kit+ Cells. Circulation. 2017;136:2359–2372. doi: 10.1161/CIRCULATIONAHA.117.030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vicinanza C, Aquila I, Scalise M, Cristiano F, Marino F, Cianflone E, Mancuso T, Marotta P, Sacco W, Lewis FC, Couch L, Shone V, Gritti G, Torella A, Smith AJ, Terracciano CM, Britti D, Veltri P, Indolfi C, Nadal-Ginard B, Ellison-Hughes GM, Torella D. Adult cardiac stem cells are multipotent and robustly myogenic: c-kit expression is necessary but not sufficient for their identification. Cell Death and Differentiation. 2017;24:2101–2116. doi: 10.1038/cdd.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison-Graham K, Takahashi Y. Steel factor and c-kit receptor: from mutants to a growth factor system. Bioessays. 1993;15:77–83. doi: 10.1002/bies.950150202. [DOI] [PubMed] [Google Scholar]
- 14.Ronnstrand L. Signal transduction via the stem cell factor receptor/c-Kit. Cellular and Molecular Life Sciences. 2004;61:2535–2548. doi: 10.1007/s00018-004-4189-6. [DOI] [PubMed] [Google Scholar]
- 15.Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiological Review. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 16.Shi HL, Drummond CA, Fan XM, Haller ST, Liu J, Malhotra D, Tian J. Hiding inside? Intracellular expression of non-glycosylated c-kit protein in cardiac progenitor cells. Stem cell research. 2016;16:795–806. doi: 10.1016/j.scr.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hesse M, Fleischmann BK, Kotlikoff MI. Concise review: The role of C-kit expressing cells in heart repair at the neonatal and adult stage. Stem cells. 2014;32:1701–1712. doi: 10.1002/stem.1696. [DOI] [PubMed] [Google Scholar]
- 18.Hatzistergos KE, Hare JM. Murine Models Demonstrate Distinct Vasculogenic and Cardiomyogenic cKit(+) Lineages in the Heart. Circulation Research. 2016;118:382–387. doi: 10.1161/CIRCRESAHA.115.308061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vajravelu BN, Hong KU, Al-Maqtari T, Cao P, Keith MC, Wysoczynski M, Zhao J, Moore JBt, Bolli R. C-Kit Promotes Growth and Migration of Human Cardiac Progenitor Cells via the PI3K-AKT and MEK-ERK Pathways. PloS one. 2015 Oct 16;10:e0140798. doi: 10.1371/journal.pone.0140798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta SK, Shukla P. Gene editing for cell engineering: trends and applications. Critical Reviews in Biotechnology. 2016:1–13. doi: 10.1080/07388551.2016.1214557. [DOI] [PubMed] [Google Scholar]
- 21.Cairns LA, Moroni E, Levantini E, Giorgetti A, Klinger FG, Ronzoni S, Tatangelo L, Tiveron C, De Felici M, Dolci S, Magli MC, Giglioni B, Ottolenghi S. Kit regulatory elements required for expression in developing hematopoietic and germ cell lineages. Blood. 2003;102:3954–3962. doi: 10.1182/blood-2003-04-1296. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira-Martins J, Ogorek B, Cappetta D, Matsuda A, Signore S, D’Amario D, Kostyla J, Steadman E, Ide-Iwata N, Sanada F, Iaffaldano G, Ottolenghi S, Hosoda T, Leri A, Kajstura J, Anversa P, Rota M. Cardiomyogenesis in the developing heart is regulated by c-kit-positive cardiac stem cells. Circulation Research. 2012;110:701–715. doi: 10.1161/CIRCRESAHA.111.259507. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi MU, Pittman HK, Haisch CE, Verbanac KM. Real-time PCR to determine transgene copy number and to quantitate the biolocalization of adoptively transferred cells from EGFP-transgenic mice. Biotechniques. 2008;45(3):247–258. doi: 10.2144/000112913. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, Goldhamer DJ. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009;47:107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubli DA, Cortez MQ, Moyzis AG, Najor RH, Lee Y, Gustafsson AB. PINK1 Is Dispensable for Mitochondrial Recruitment of Parkin and Activation of Mitophagy in Cardiac Myocytes. PloS one. 2015;10:e0130707. doi: 10.1371/journal.pone.0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monsanto MM, White KS, Kim T, Wang BJ, Fisher K, Ilves K, Khalafalla FG, Casillas A, Broughton K, Mohsin S, Dembitsky WP, Sussman MA. Concurrent Isolation of Three Distinct Cardiac Stem Cell Populations from a Single Human Heart Biopsy. Circulation Research. 2017;121:113–124. doi: 10.1161/CIRCRESAHA.116.310494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young SM, Cambareri AC, Ashman LK. Role of c-KIT expression level and phosphatidylinositol 3-kinase activation in survival and proliferative responses of early myeloid cells. Cell Signal. 2006;18:608–620. doi: 10.1016/j.cellsig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal S, Kazi JU, Ronnstrand L. Phosphorylation of the activation loop tyrosine 823 in c-Kit is crucial for cell survival and proliferation. Journal of Biological Chemistry. 2013;288:22460–22468. doi: 10.1074/jbc.M113.474072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, Wu Z, Yin G, Liu H, Guan X, Zhao X, Wang J, Zhu J. Stem cell factor (SCF) protects osteoblasts from oxidative stress through activating c-Kit-Akt signaling. Biochemical and Biophysical Research Communications. 2014;455:256–261. doi: 10.1016/j.bbrc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Liu Q, Yang R, Huang X, Zhang H, He L, Zhang L, Tian X, Nie Y, Hu S, Yan Y, Zhang L, Qiao Z, Wang QD, Lui KO, Zhou B. Genetic lineage tracing identifies in situ Kit-expressing cardiomyocytes. Cell research. 2016;26:119–130. doi: 10.1038/cr.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Naqvi N, Yahiro E, Liu K, Powell PC, Bradley WE, Martin DI, Graham RM, Dell’Italia LJ, Husain A. c-kit is required for cardiomyocyte terminal differentiation. Circulation Research. 2008;102:677–685. doi: 10.1161/CIRCRESAHA.107.161737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quijada P, Toko H, Fischer KM, Bailey B, Reilly P, Hunt KD, Gude NA, Avitabile D, Sussman MA. Preservation of myocardial structure is enhanced by pim-1 engineering of bone marrow cells. Circulation Research. 2012;111:77–86. doi: 10.1161/CIRCRESAHA.112.265207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anversa P, Kajstura J, Rota M, Leri A. Regenerating new heart with stem cells. The Journal of clinical investigation. 2013;123:62–70. doi: 10.1172/JCI63068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Keith MC, Bolli R. “String theory” of c-kit(pos) cardiac cells: a new paradigm regarding the nature of these cells that may reconcile apparently discrepant results. Circulation Research. 2015;116:1216–30. doi: 10.1161/CIRCRESAHA.116.305557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sultana N, Zhang L, Yan JY, Chen JQ, Cai WB, Razzaque S, Jeong D, Sheng W, Bu L, Xu MJ, Huang GY, Hajjar RJ, Zhou B, Moon A, Cai CL. Resident c-kit(+) cells in the heart are not cardiac stem cells. Nature Communications. 2015;6:8701. doi: 10.1038/ncomms9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Siena S, Gimmelli R, Nori SL, Barbagallo F, Campolo F, Dolci S, Rossi P, Venneri MA, Giannetta E, Gianfrilli D, Feigenbaum L, Lenzi A, Naro F, Cianflone E, Mancuso T, Torella D, Isidori AM, Pellegrini M. Activated c-Kit receptor in the heart promotes cardiac repair and regeneration after injury. Cell Death & Disease. 2016;7:e2317. doi: 10.1038/cddis.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa K, Fish K, Aguero J, Yaniz-Galende E, Jeong D, Kho C, Tilemann L, Fish L, Liang L, Eltoukhy AA, Anderson DG, Zsebo K, Costa KD, Hajjar RJ. Stem cell factor gene transfer improves cardiac function after myocardial infarction in swine. Circulation:Heart Failure. 2015;8:167–174. doi: 10.1161/CIRCHEARTFAILURE.114.001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye L, Zhang EY, Xiong Q, Astle CM, Zhang P, Li Q, From AH, Harrison DE, Zhang JJ. Aging Kit mutant mice develop cardiomyopathy. PloS one. 2012;7:e33407. doi: 10.1371/journal.pone.0033407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fazel S, Cimini M, Chen L, Li S, Angoulvant D, Fedak P, Verma S, Weisel RD, Keating A, Li RK. Cardioprotective c-kit+ cells are from the bone marrow and regulate the myocardial balance of angiogenic cytokines. The Journal of clinical investigation. 2006;116:1865–1877. doi: 10.1172/JCI27019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cimini M, Fazel S, Zhuo S, Xaymardan M, Fujii H, Weisel RD, Li RK. c-kit dysfunction impairs myocardial healing after infarction. Circulation. 2007;116:I77–182. doi: 10.1161/CIRCULATIONAHA.107.708107. [DOI] [PubMed] [Google Scholar]
- 42.Wallner M, Duran JM, Mohsin S, Troupes CD, Vanhoutte D, Borghetti G, Vagnozzi RJ, Gross P, Yu DH, Trappanese DM, Kubo H, Toib A, Sharp TE, Harper SC, Volkert MA, Starosta T, Feldsott EA, Berretta RM, Wang T, Barbe MF, Molkentin JD, Houser SR. Acute Catecholamine Exposure Causes Reversible Myocyte Injury Without Cardiac Regeneration. Circulation Research. 2016;119:865–879. doi: 10.1161/CIRCRESAHA.116.308687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatzistergos KE, Saur D, Seidler B, Balkan W, Breton M, Valasaki K, Takeuchi LM, Landin AM, Khan A, Hare JM. Stimulatory Effects of Mesenchymal Stem Cells on cKit+ Cardiac Stem Cells Are Mediated by SDF1/CXCR4 and SCF/cKit Signaling Pathways. Circulation Research. 2016;119:921–930. doi: 10.1161/CIRCRESAHA.116.309281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, Hock H. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nature Biotechnology. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gude N, Muraski J, Rubio M, Kajstura J, Schaefer E, Anversa P, Sussman MA. Akt promotes increased cardiomyocyte cycling and expansion of the cardiac progenitor cell population. Circulation Research. 2006;99:381–388. doi: 10.1161/01.RES.0000236754.21499.1c. [DOI] [PubMed] [Google Scholar]
- 46.Smith AJ, Lewis FC, Aquila I, Waring CD, Nocera A, Agosti V, Nadal-Ginard B, Torella D, Ellison GM. Isolation and characterization of resident endogenous c-Kit+ cardiac stem cells from the adult mouse and rat heart. Nature Protocols. 2014;9:1662–1681. doi: 10.1038/nprot.2014.113. [DOI] [PubMed] [Google Scholar]
- 47.Ellison GM, Torella D, Karakikes I, Purushothaman S, Curcio A, Gasparri C, Indolfi C, Cable NT, Goldspink DF, Nadal-Ginard B. Acute beta-adrenergic overload produces myocyte damage through calcium leakage from the ryanodine receptor 2 but spares cardiac stem cells. The Journal of Biological Chemistry. 2007;282:11397–11409. doi: 10.1074/jbc.M607391200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nadal-Ginard B, Ellison GM, Torella D. The cardiac stem cell compartment is indispensable for myocardial cell homeostasis, repair and regeneration in the adult. Stem cell research. 2014;13:615–630. doi: 10.1016/j.scr.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 49.Chamuleau SA, Vrijsen KR, Rokosh DG, Tang XL, Piek JJ, Bolli R. Cell therapy for ischaemic heart disease: focus on the role of resident cardiac stem cells. Netherlands Heart Journal. 2009;17:199–207. doi: 10.1007/BF03086247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellison GM, Galuppo V, Vicinanza C, Aquila I, Waring CD, Leone A, Indolfi C, Torella D. Cardiac stem and progenitor cell identification: different markers for the same cell? Frontiers in Bioscience (Scholar Edition) 2010;2:641–652. doi: 10.2741/s91. [DOI] [PubMed] [Google Scholar]
- 51.Nigro P, Perrucci GL, Gowran A, Zanobini M, Capogrossi MC, Pompilio G. c-kit(+) cells: the tell-tale heart of cardiac regeneration? Cellular and Molecular Life Sciences. 2015;72:1725–1740. doi: 10.1007/s00018-014-1832-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Current Biology. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 53.Halakivi-Clarke L, Cho E, Onojafe I, Liao DJ, Clarke R. Maternal exposure to tamoxifen during pregnancy increases carcinogen-induced mammary tumorigenesis among female rat offspring. Clinical Cancer Research. 2000;6:305–308. [PubMed] [Google Scholar]
- 54.Park EJ, Sun X, Nichol P, Saijoh Y, Martin JF, Moon AM. System for tamoxifen-inducible expression of cre-recombinase from the Foxa2 locus in mice. Developmental Dynamics. 2008;237:447–453. doi: 10.1002/dvdy.21415. [DOI] [PubMed] [Google Scholar]
- 55.Huh WJ, Khurana SS, Geahlen JH, Kohli K, Waller RA, Mills JC. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology. 2012;142:21–24e7. doi: 10.1053/j.gastro.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S, Lee MS, Park J, Zhang JY, Jin DI. Oxidative stress in the testis induced by tamoxifen and its effects on early embryo development in isogenic mice. The Journal of Toxicological Sciences. 2012;37:675–679. doi: 10.2131/jts.37.675. [DOI] [PubMed] [Google Scholar]
- 57.Lizen B, Claus M, Jeannotte L, Rijli FM, Gofflot F. Perinatal induction of Cre recombination with tamoxifen. Transgenic Research. 2015;24:1065–1077. doi: 10.1007/s11248-015-9905-5. [DOI] [PubMed] [Google Scholar]
- 58.Busche A, Schmitz S, Fleige H, Robbins SH, Walzer T, Stewart CA, Forster R, Messerle M, Prinz I. Genetic labeling reveals altered turnover and stability of innate lymphocytes in latent mouse cytomegalovirus infection. The Journal of Immunology. 2011;186:2918–2925. doi: 10.4049/jimmunol.1003232. [DOI] [PubMed] [Google Scholar]
- 59.Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 61.An N, Cen B, Cai H, Song JH, Kraft A, Kang Y. Pim1 kinase regulates c-Kit gene translation. Experimental Hematology & Oncology. 2016;5:31. doi: 10.1186/s40164-016-0060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L, Tang J, Haines CJ, Feng H, Lai L, Teng X, Han Y. c-kit expression profile and regulatory factors during spermatogonial stem cell differentiation. BMC Developmental Biology. 2013;13:38. doi: 10.1186/1471-213X-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Busada JT, Chappell VA, Niedenberger BA, Kaye EP, Keiper BD, Hogarth CA, Geyer CB. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Developmental Biology. 2015;397:140–149. doi: 10.1016/j.ydbio.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Igoucheva O, Alexeev V. MicroRNA-dependent regulation of cKit in cutaneous melanoma. Biochemical and Biophysical Research Communications. 2009;379:790–794. doi: 10.1016/j.bbrc.2008.12.152. [DOI] [PubMed] [Google Scholar]
- 65.Godshalk SE, Paranjape T, Nallur S, Speed W, Chan E, Molinaro AM, Bacchiocchi A, Hoyt K, Tworkoski K, Stern DF, Sznol M, Ariyan S, Lazova R, Halaban R, Kidd KK, Weidhaas JB, Slack FJ. A Variant in a MicroRNA complementary site in the 3′ UTR of the KIT oncogene increases risk of acral melanoma. Oncogene. 2011;30:1542–1550. doi: 10.1038/onc.2010.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Song YH, Li F, Yang T, Lu YW, Geng YJ. MicroRNA-221 regulates high glucose-induced endothelial dysfunction. Biochemical and Biophysical Research Communications. 2009;381:81–83. doi: 10.1016/j.bbrc.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. The Journal of Biological Chemistry. 2009;284:3728–3738. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.