Abstract
In the digital age of cardiovascular medicine, the rate of biomedical discovery can be greatly accelerated by the guidance and resources required to unearth potential collections of knowledge. A unified computational platform leverages metadata to not only provide direction but also empower researchers to mine a wealth of biomedical information and forge novel mechanistic insights. This review takes the opportunity to present an overview of the cloud-based computational environment, including the functional roles of metadata, the architecture schema of indexing and search, and the practical scenarios of machine learning-supported molecular signature extraction. By introducing several established resources and state-of-the-art workflows, we share with our readers a broadly-defined informatics framework to phenotype cardiovascular health and disease.
Keywords: Metadata, Indexing, Search, Molecular Signature, Machine Learning, Cardiovascular Medicine, cardiovascular disease, database, bioinformatics
Subject Terms: Genetics
Introduction
For an explorer on the high seas, discovery would not be remotely possible without a guiding compass and the resources required to unearth potential collections of wealth and knowledge. The hunt for biomedical data treasure troves is already well underway; many investigators have pioneered this exploratory adventure, and many more are already in pursuit of hidden hoards of knowledge. Defining an efficient, informatics-based path toward discovery is contingent upon providing aid and guidance to investigators sailing the sea of biomedical datasets, enabling them to succeed. Metadata transforms the landscape from a vast formidable ocean into more navigable waters. Indexing and search tools give data consumers multidimensional coordinates to access the resources they seek. Once investigators better understand their course and are equipped with the proper tools, they will be able to adequately extract information buried within the big data. In the digital age of cardiovascular medicine, a unified computational platform leverages metadata to not only provide direction but also empower data consumers to mine a wealth of biomedical information and unveil gems of knowledge.
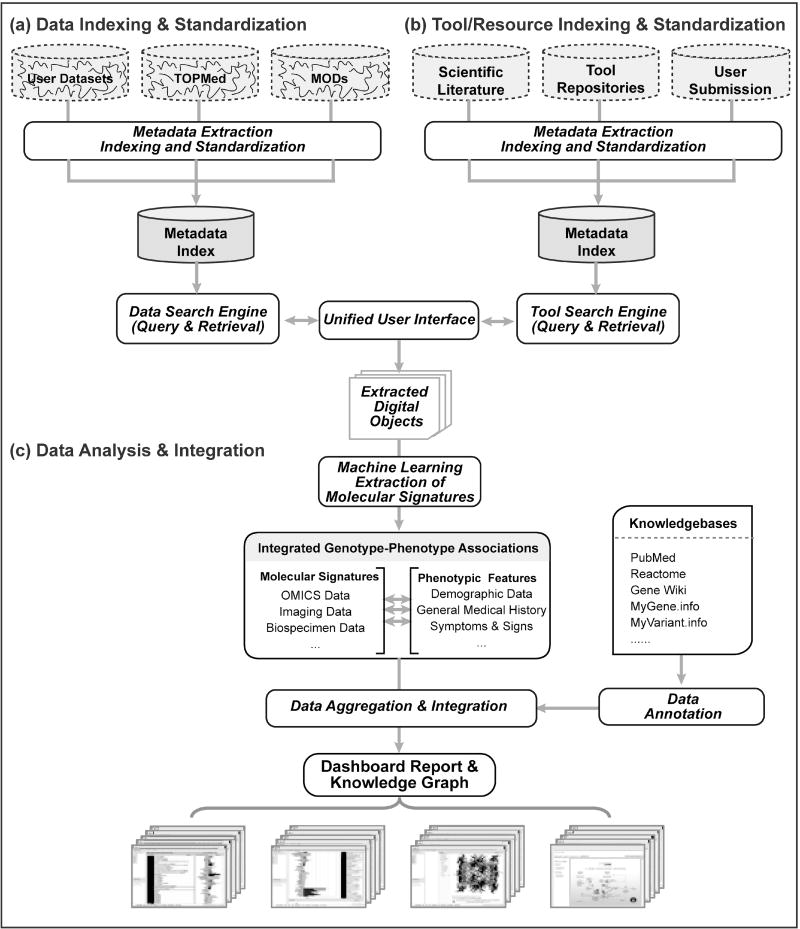
Unwilling to be constrained to the land and sea, modern scientists have opted to embark on a new frontier by launching their efforts into cloud computing, which has been envisioned as the next generation paradigm in biomedical informatics. As an internet-based computing solution, cloud computing provides shared resources on demand for data storage, processing, and dissemination that is reliable, cost effective, and customizable to suit individual user’s needs. These services provide users with a simple way to access databases, servers, storage, and a wide spectrum of software and applications through a web interface accessible from anywhere in the world. The maintenance of the hardware and infrastructure is managed by the cloud platform providers (e.g., Amazon Web Services, AWS1), giving users rapid access to flexible and low-cost computing resources. A user is able to instantly provision the type and size of the computing resources required, scaling up or down as desired, and is only responsible for the actual resource costs incurred. These features make cloud computing platforms a superior alternative to the previously dominant approach of maintaining a local high-performance computing infrastructure. This review takes the opportunity to present an overview of the cloud-based computational environment, including the functional roles of metadata, data indexing and search tools, and the entire platform in supporting computational efforts such as molecular phenotyping of cardiovascular disease. A conceptual overview of this framework is detailed in Figure 1.
Figure 1. A Cloud-based Computing Platform.
A unified computational platform leverages digital biomedical resources of many types. (a) To index and standardize datasets, metadata is extracted to create a metadata index. This information is utilized by the data search engine, which has both query and retrieval capabilities. (b) Similarly, indexing and standardization of tool and resource metadata employs the same framework to create a tool metadata index and corresponding resource search engine. The resulting metadata indices are then aggregated to empower a unified search interface composed of search processing engines optimized for each digital object type. The successful integration of disparate resources requires employing a unique persistent identifier such as digital object identifier (DOI) and standardized metadata representation. The entry point of the platform is a graphical user interface where users can query for digital objects. Through this interface, users can find relevant datasets and software tools with a single query. (c) This will facilitate data analysis, molecular signature extraction and building of new computational models using machine learning algorithms. To further the ingenuity from the extracted information and support collaborative creativity on the cloud, data harmonization and annotation are performed via existing biomedical computational resources. The amalgamation of multi-omics, imaging, and text data on the same unified platform allows users to efficiently establish genotype-phenotype associations, construct multidimensional knowledge graphs, and perform data enrichment through existing knowledgebases.
Section I Metadata
In 2016, a consortium of researchers, publishers, and research funders published the FAIR guiding principles to make data Findable, Accessible, Interoperable, and Re-usable2. The FAIR principles are about making data FAIR, but a key to achieving this is the metadata. Webster’s concisely defines metadata as “data that provides information about other data”3. Metadata serves as vital points of reference, adding a longitudinal and latitudinal perspective, laying out critical components to map and guide biomedical explorers with meaningful coordinates for features of interest in their daily dealing of data.
A classic example of metadata is bibliographic data. The large register card-based catalogues maintained by libraries in the 1970s illustrate the distinction between data and metadata at the physical level. The catalogues provided the metadata (author, title, keywords, etc.) as a searchable index, with each card pointing through a systematic identifier (e.g., “Journal Article”) to the data (e.g., the actual journal article) physically located in another part of the library. Significant resources went into the setup and maintenance of these catalogues; the quality of their indexing system, the frequency of updates, and their comprehensiveness were factors that could cost or save students and faculty many hours of work.
These roots are still clearly visible in current everyday research tools like PubMed. PubMed is a metadata catalogue; at the core, each record holds bibliographic information very similar to that typewritten on the library register cards of old. The data, the actual journal articles, are located elsewhere, such as at the sites of dozens of large publishers, or in PubMed Central. Of course, the identifiers pointing to the data today have to be unique not only in the context of the local library but also globally on the internet, creating the need for Globally Unique Identifiers (GUIDs), a system first utilized by Microsoft in 1999, and currently provided by systems like DOI.org4, 5or identifiers.org6.
As an example, in the PubMed rendering of the publication “Percent emphysema, airflow obstruction, and impaired left ventricular filling” by Barr et al.7, the journal, publication date, pages, DOI, title, author list, and affiliations are all classical metadata attributes. Medical Subject Headings8, 9 (MeSH terms, see below) and the links to the full text are more recent additions to the list of typically captured metadata. The abstract is, on the one hand, metadata describing the main publication and, on the other hand, a substantial part of the publication itself, and thus illustrates the difficulties of clearly distinguishing between data and metadata. Previously, a card index could only be built and maintained for one or very few metadata attributes, usually author surname and perhaps keywords. Modern approaches have blurred the distinction between data and metadata and now allow us to create searchable indices over many attributes, even large chunks of text like abstracts and full text.
However, different metadata attributes need to be treated differently, as at the most basic level a keyword search is different from a numeric range like publication year. This can require a metadata schema, which is a logical outline of the metadata attributes and their relationships and is achieved by defining rules such as syntax and requirements for each individual attribute. For large systems like PubMed, which receives metadata from many different publishers, it is essential to clearly define which metadata attributes are required and in which precise form. For PubMed, this definition is applied through the Journal Article Tags Suite (JATS)10, first created in 2003 and iteratively improved ever since. In the past year, the Data Tag Suite (DATS)11 has been developed as a similar system to JATS to support the DataMed data discovery index. While both of these domain-specific metadata definition systems provide a rich, detailed description of metadata attributes in their domains, schema.org12, founded by Google, Microsoft, Yahoo, and Yandex, aims to develop an internet-wide metadata definition across many domains. Schema.org markup is embedded directly in HTML pages to support major search engines and to automatically generate short page “abstracts” to be displayed in “boxes” on search result pages. While already widely used on commercial sites, schema.org adoption is still low in the biomedical sciences, but it is expected to rapidly increase due to initiatives like bioschemas.org13 that develop specific extensions for the life sciences.
Computational power can easily handle some previously difficult problems, like whether to index John le Carré under ‘l’ or ‘c’, but fundamental challenges still remain. The diversity of our language allows us to use the keywords “adverse effect” or “side effect” almost synonymously, but for a search algorithm, they look quite different without further effort. The differences are exacerbated in large indices that include a broad range of data sources. Different journals, book editors, publishers, all have different ways of expressing similar meaning. This is why metadata harmonization is an essential activity that can be decisive for the utility of any metadata resource. As an example, the National Library of Medicine has used MeSH terms since 19548, 9 as a reference system for indexing articles for PubMed. In MeSH, more than 22,000 terms are arranged in a hierarchical controlled vocabulary, which provides reference terms to reduce the ambiguity of synonyms. Perhaps even more importantly, the hierarchical structure of MeSH and many other controlled vocabularies (or ontologies) makes searches aware of general-specific relationships between terms. Through the use of MeSH, PubMed can, for example, match a manuscript annotated with “Ectopia Cordis” when searching for “Cardiovascular Abnormalities”.
Zhou et al. recently analyzed the available PubMed metadata in the particularly challenging domain of clinical case reports14. By their nature, case reports describe unique, very rare, or novel patient phenotypes and treatment strategies; therefore the discovery of multiple, independently published case reports can be critical for the advancement of patient care in the domain. In a test sample of 700 reports, the study demonstrated an average improvement of 45% metadata coverage over current annotation in PubMed through careful manual annotation, but it also clearly acknowledges that this level of manual annotation is not feasible for the vast number of clinical case studies in PubMed and that advanced text mining methods will be required to improve metadata annotation in this and related document types.
In a different but similarly structured domain, the new AzTec catalogue of software tools and resources in the biomedical domain15, 16 uses the “Bioinformatics operations, data types, formats, identifiers and topics” (EDAM) ontology17 to specify the input and output formats for software tools. In addition to specifying for a human reader if a tool under consideration may be suitable for her/his needs, such well-structured metadata annotation may even support automated reasoning and the suggestions for complex data processing pipelines, chaining software tools to achieve desired workflows.
In supporting the FAIR principles, it is essential that the metadata is represented and harmonized using reference systems, which themselves are supporting the FAIR principles. While MeSH terms are free to use by all interested parties, some ontologies, such as the widely used SNOMED CT clinical healthcare terminology, require a license for use18.
To support efficient search engines, metadata from many different resources is often centrally aggregated while the actual data is widely distributed, as shown in our PubMed example. However, the separation of metadata and data supports another series of use cases, namely the distinction between Findable and Accessible. While most publishers have an interest in making their publications findable through open provision of metadata to public indices like PubMed, many publications are only accessible through license/subscription arrangements. In biomedical science, the open metadata/restricted data model is widely implemented to control access to studies involving human subjects. dbGaP19 and the European Genome-phenome Archive (EGA)20 are both providing open metadata catalogues of their studies to maximize their impact and reuse, but actual data access is only granted after validation of the scientific use case. This is an example of the critical importance of rich and precise metadata; based on the metadata, a researcher has to decide whether the data is likely to be worth his/her time in going through a potentially complex access authorization procedure. The Beacon project of the Global Alliance for Genomics and Health21 aims to enrich support for such decisions by providing minimal, privacy-preserving extracts of actual data, thus blurring the lines between data and metadata, in the genomic equivalent of indexing PubMed abstracts in addition to classic bibliographic data.
A combination of well structured, well indexed metadata and modern approaches like these advanced privacy-preserving query interfaces will be required for efficient discovery and reuse of data from a multitude of large scale studies like the NHLBI Program on Trans-Omics for Precision Medicine (TOPMed)22, which now envisions to sequence more than 100,000 individuals and provide a rich source of additional ‘omics data. For example, the capacity to filter, select, and construct on-demand synthetic cohorts for data (re)use in targeted research depends critically upon our ability to expose, structure, and exploit metadata optimally.
Section II Indexing and Search
Introduction
Indexing and search are long standing topics in data science, whose significant impact and wide utility are exemplified by search engines like Google. Without them, it would be impossible to find the information one desires when browsing the Web. The Google search engine uses web crawlers to automatically collect web pages on the Internet. It uses this aggregated information to create and maintain a large index of keywords, where these keywords may present in both web pages and the metadata of the web pages. Google’s search results are then ranked by the PageRank algorithm23, which uses the metadata and hyperlinks in web pages to evaluate the relevance and importance of web pages to the search keyword(s).
The same challenges are facing biomedical fields, in which data of all types is produced at an unprecedented rate. There are many types of digital objects that include but are not limited to datasets, data repositories, knowledgebases, reports, standalone software and tools, analytic pipelines, online services, and application programmable interfaces (APIs). Various efforts have been undertaken to make these digital objects FAIR, of which many focus on specific types of data. For example, PubMed contains more than 27 million citations and abstracts of biomedical publications from MEDLINE and life science journals, as well as books available on the NCBI Bookshelf. It stores and indexes metadata from these publications in a database and provides a Google-like search engine. A keyword-based query will be expanded to add appropriate metadata field names, e.g., Boolean operators, and relevant MeSH terms8, 9 to be posted against the metadata database. The results will include a list of publications ranked by their relevance to the queried keywords.
Computational Infrastructure Supporting Index and Search
An index structure is an auxiliary data structure built to organize the storage of data and to facilitate the search, enhancing the findability of the data. With the help of a well-designed indexing structure, the search algorithm aims to efficiently and accurately locate relevant information or data for the users. Cloud-based infrastructures provide the flexibility to scale on demand for support of fluctuating web traffic and workloads. The available cloud services support public, private, or hybrid storage depending on the security requirements and privacy concerns. Several of the household names are Amazon Web Services (AWS)1, Google Cloud Platform24, and Microsoft Azure25.
These platforms currently employ key technologies to render digital objects FAIR. Specifically, a digital object needs to have a unique persistent identifier (e.g., DOI4, 5), metadata that adequately describes the intrinsic properties of the object (e.g., data type), and the provenance of the object (e.g., source of the data, the time it was generated). Different repositories may use different metadata schema. How the metadata is physically represented and stored may vary as well: it may be in an extensible markup language (XML) file26, e.g., the mass spectral files for peptides in a proteomics study; a JavaScript object notation (JSON) file27, e.g., variation data representation and exchange28; a text document, e.g., a PDF file or an Word file; or held within the data structures of a relational graph or a document database. It is often made available via a representational state transfer API or as SPARQL Protocol29 and Resource Description Framework (RDF) Query Language endpoints30. The FAIRness (in particular, interoperability) of the data objects across different repositories therefore largely depends on how well these metadata could be accessed and understood by human and computer programs. An efficient strategy by a search engine is to transform these metadata into a harmonized metadata schema so that digital objects can be efficiently indexed and examined on a unified platform.
In the era of data science in cardiovascular medicine, the two most relevant questions to an investigator are (i) what are the datasets that harbor the necessary information to answer the biomedical questions of his/her interest, and (ii) what types of tools and resources are best suited to extract information from these datasets? With metadata acting as points of reference throughout the sea of biomedical digital objects, comprehensive indexing comprises the multidimensional coordinates of our explorers’ map, while search tools act as a navigation system to guide researchers to potential areas of novel biomedical discovery. Employing effective metadata templates and structures facilitates indexing of both datasets and tools. For example, one effective tool to locate datasets is OmicsDI31, an open-source cloud-based platform to discover, access, and disseminate omics datasets. Powered by an institutionally supported search engine (hosted on the EMBL-EBI private cloud), it currently harmonizes and indexes over 90,000 digital objects from 15 repositories in four continents. Metadata from these repositories are enriched and converted to a common XML format. The EBI search engine (running Apache Lucene32) creates indices on the XML documents stored in MongoDB, which in turn enables robust and scalable search capability. In parallel, AzTec (aztec.bio) is an open-source cloud-based platform for discovering and accessing biomedical tools and resources. Running on Amazon EC2, AzTec currently hosts over 10,000 resources spanning 17 domains, including imaging, gene ontology, text mining, data visualization, and various omics analyses. The metadata are converted into JSON format and indexed by Apache Lucene32, which are then enhanced by the semantic information retrieval engine called Aztec-IR.
Cardiovascular Use Case
In view of the rapid development of cloud-based technologies and the revolutionary advancements of data science in the biomedical field, we envision the wide utility of an indexing and search platform for cardiovascular researchers. Let us examine a use case scenario where application of indexing and search engine platform might be applied to support advancement in cardiovascular medicine. Several cardiovascular investigators are interested in the Multi-Ethnic Study of Atherosclerosis (MESA) dataset hosted by TOPMed22, which represents patient datasets, including various study goals, high-dimensional variables, and rich molecular information on phenotype and genotype. The study of any TOPMed dataset is contingent upon carefully guarded privacy and restricted access. The investigators wish to identify a dataset validating a molecular signature of their own patient cohorts of atherosclerosis (see Section III below for identification of molecular signatures). Furthermore, they are interested in learning about model systems that could enable further understanding of underlying molecular mechanisms, for which they turn to Model Organism Databases (MODs)33. To accomplish their tasks, it would require an effective search of properly indexed TOPMed datasets as well MODs, which are a cluster of organized molecular datasets with highly curated information and complete open data. These search tasks can be accomplished by creating a central entry point for targeted search activities in a data commons – from simple keyword searches to more advanced searches through privacy-preserving interfaces – facilitating the identification of a spectrum of relevant biomedical digital objects.
The central entry point will be a unified interface that integrates multiple current search engines (for example, AzTec and OmicsDI) designed for different types of digital objects. To harmonize the integration, one could employ a unique persistent identifier (such as DOI, which is already widely used and is adopted by both AzTec and OmicsDI) and construct a comprehensive and standardized metadata representation for all types of digital objects. Metadata harmonization requires employing controlled vocabularies and ontologies; normalizing the entities of different data objects using the existing metadata schemas; and standardizing the representations of clinical, technical, and analytical protocols. In particular, with respect to standardization of the metadata for clinical datasets, the integration of MeSH terms8, 9 and International Statistical Classification of Diseases (ICD-10)34 would be beneficial.
Once the datasets most useful to a particular study have been discovered and located, there remain challenges in transferring this data from its host source to the computational workspace, be that an individual computer or a cloud-based server. Many requirements must be met in order to carry out point-to-point transmission of large datasets in a secure fashion; security and encryption are especially important with regards to clinical health data. Currently, there exist software and infrastructure (see changes below) that researchers can utilize for these purposes with confidence that transmission will be successful and secure. Globus is designed as an infrastructure solution for all data management and transfer concerns for researchers, enabling them to securely store and share data without having significant back-end knowledge of data management35. Globus highlights their “fire and forget” approach, in which users may run a command, and Globus will handle any concerns that come up (e.g. connection interruptions) with minimal user interaction. More information on Globus and its options can be accessed at https://www.globus.org/. Similarly, bbcp allows for file copying from one point to another at higher speeds, requiring only that both the source and target have installed the software. This software and its documentation may be accessed at https://www.slac.stanford.edu/~abh/bbcp/. Researchers using AWS can use Snowball, which is capable of transferring data at the petabyte scale into and out of the AWS cloud. Snowball can be accessed at https://aws.amazon.com/snowball/. All of these options provide biomedical researchers, regardless of their knowledge of sophisticated data management architectures, with avenues to effectively gather and acquire data for their own investigations.
Section III Molecular Phenotyping of Cardiovascular Diseases
Introduction
Many modern cardiovascular research studies have focused on elucidating underlying molecular mechanisms of a particular biological process and/or clinically relevant question. To this end, data science offers great opportunities to support such investigative endeavors. Where traditional biological research may be akin to panning for gold, with access to only a small stream of data, data science provides access to the main vein of gold contained deep within the ever growing mountain of data. Recently, data science tools have been developed to surpass what human power may reach. For biomedical data with high complexity and heterogeneity, machine learning, as a burgeoning discipline in data science, offers powerful capacity to tackle such computational challenges. Below, we have selected a generalized cardiovascular case scenario to introduce computational resources available for their dataset interrogation and extraction, making new discoveries and advancing cardiovascular medicine.
Machine Learning-empowered Molecular Signature Extraction
Let us consider given molecular datasets based on two or more longitudinal cohorts in cardiovascular diseases, Control and Treatment groups. Machine learning-based analytical methods can be employed to (i) extract molecular signatures that differentiate the two cohorts, (ii) draw multilevel causal inference between clinically relevant variables and phenotypes, and (iii) build predictive models for clinical outcomes. The following three types of machine learning approaches can be used to build models that are suitable for such analyses: deep learning36, class imbalance learning37, and probabilistic graphical models38, 39; their key capabilities are summarized in Table 1.
Table 1.
Overview of Capabilities of Machine Learning Types.
| Computational Capabilities | Data Capabilities | |||||||
|---|---|---|---|---|---|---|---|---|
| Methods | Computation Efficiency |
Predictive Modeling |
Multilevel Causal Inference |
Temporal Analysis |
Rare Disease Diagnosis |
Imaging Data |
Text Data | Molecular/ Phenotype Data |
| CIL | * * * | * * | * | * * | * * * | * | * | * * * |
| DL | * * | * * * | * | * * | * | * * * | * * * | * * * |
| PGM | * * | * * | * * * | * * | * | * * | * * | * * * |
Class Imbalance Learning (CIL), Deep Learning (DL), and Probabilistic Graphical Modeling (PGM) methods are characterized according to computational capabilities (i.e., computation efficiency or cost-sensitive, predictive modeling, multilevel causal inference, temporal analysis, and rare disease diagnosis) and data capabilities, broken down by data type (i.e., imaging, text, and molecular/phenotypic data).
One star (*) denotes that the type has not yet demonstrated capability in that category;
two stars (* *) indicate the type has demonstrated capability under certain conditions;
three stars (* * *) mean that the type is preferred for the capability listed. CIL is the preferred method for rare disease diagnosis, and it is suitable for predictive modeling as well with molecular and phenotypic data. Moreover, in combination with incremental learning methods, CIL can built adaptive models on temporal data. Deep learning has uniquely high accuracy in predictive modeling and is preferable for all types of data, so long as they have a high number of features or dimensions. PGM is most suitable for performing multilevel causal inferences, such as determining meaningful relationships between factors and outcomes, and is best done on molecular and phenotypic data.
Deep learning36 is a branch of machine learning algorithms that revolutionized many fields including image recognition, speech recognition, natural language processing, and machine translation by delivering comparable or better performance than human experts. Deep learning uses multilayer artificial neural networks to learn feature representations of the data based on the assumption that the observed data were generated by the interactions of layered factors corresponding to levels of abstractions or compositions of features at varying resolutions. The specific network architecture and composition of artificial neurons at each layer may vary depending on the tasks to be accomplished. For supervised learning tasks, deep learning models obviate feature engineering by transforming observed data into intermediate features, as well as derive layered architectures that disentangle interactions between these features and discover the ones that are useful in improving outcome. Deep learning models have recently demonstrated superior performance in medical image segmentation and classification40, clinical decision support (using electronic health records)41, drug discovery42, and understanding gene regulation43; specific to cardiovascular research, deep learning has been employed to detect multiple types of cardiac arrhythmias from wearable heart rate monitor data44and build models to predict heart failure onset45, among other studies. Deep learning has recently been applied to rare disease datasets46; this currently is a burgeoning area of research in cardiovascular medicine47.
Rare disease cohorts pose a challenge for traditional machine learning methods in that the number of individuals available to be studied who bear the disease is far less than the number of individuals to be studied without the disease, thereby causing an imbalance. Class Imbalance Learning (CIL)37 has proven to be effective in building predictive models for biomedical applications, particularly when subjects with positive clinical outcomes greatly outnumber the subjects with negative clinical outcomes. The CIL algorithms include data sampling and cost-sensitive learning methods. Data sampling methods are model agnostic, allowing investigators to either use linear models for better interpretability or non-linear kernel/deep networks for high predictive accuracy. Cost-sensitive methods are computationally more efficient than sampling methods for large datasets. CIL has been effectively applied in the prediction of acute cardiac complications48 as well as for cardiovascular risk stratification49 using both data sampling and cost-sensitive-based approaches. In addition, CIL methods have recently been combined with incremental learning algorithms50 to address the challenges associated with learning from data streams and dynamic time series data51. Although CIL is very effective in predicting clinical outcomes and identifying the molecular signatures in rare diseases, it may not be able to infer multilevel causal relationships between clinically relevant factors and phenotypes.
To identify the factors (e.g., genes, proteins, medical history, environmental factors) most relevant to the target variable (e.g., phenotype) in a multilevel manner, probabilistic graphical models (PGM) can be used, as they can represent multilevel causal relationships between measured variables and disease phenotypes. Graphical models can reveal how a combination of factors impact the disease, partition cohorts in subpopulations of specific characteristics and associations to phenotypes, and lead to more personalized disease management protocols, as has been demonstrated on large scale gene expression datasets for Chronic Obstructive Pulmonary Disease (COPD) and Interstitial Lung Disease (ILD)52. Dynamic Bayesian networks, a type of probabilistic graphical model, are used in the integration of multiple high-dimensional datasets to create gene regulatory networks in cardiac differentiation53 and in the temporal abstraction of coronary disease outcomes using longitudinal clinical data54. Graphical models have the tendency to become very computationally expensive, particularly with high-dimensional datasets. They also impose some unrealistic assumptions like no presence of cycles in the underlying graph or the unimodal nature of variable types (i.e., all variables to be continuous or discrete only). More recently, new algorithms that learn graph structures over mixed data types have been proposed and used in chronic lung diseases52 and sickle cell55. Graphical model-based deep learning architectures also exist, but their predictive performance is generally inferior to those commonly used neural networks based deep learning methods. Moreover, drawing multilevel causal inference on temporal data using graphical models presents unique challenges and is an open avenue of research.
The advance of data acquisition technologies has exponentially expanded the scope and dimension of modern biomedical datasets, necessitating scalable, collaborative, community-based computational endeavors. With the molecular signatures acquired at hand, investigators could further pursue data harmonization, data annotation, and data mining via a number of existing computational resources; most of them are accessible on the cloud. Distributed cloud resources facilitate collaborative analysis by giving a better vantage point and better access to powerful tools. Below, we highlight several data science resources that have gained considerable user appreciation in the broad scientific community; we use them as examples to support the research efforts in understanding data harmonization, data annotation, and data mining.
Data Harmonization, Annotation, and Mining
The effective analysis of multi-omics datasets is contingent on the integration of these disparate data types; there are specific aspects of each data type that present unique challenges, making this task arduous and computationally complex. To address this, Harmonizome was created as an amalgamation of information from 114 datasets. Stored in a relational database, the web interface allows users to submit queries and download relevant data; a combination of statistical and machine learning methods have created over 71 million associations between close to 300,000 attributes and 55,000 genes, illustrating the clear utility of such a unified resource56. The creators of Harmonziome have provided a REST API for remote computation or integration into other programs. The platform can be accessed at: http://amp.pharm.mssm.edu/Harmonizome/.
Once specific datasets have been identified and accessed, the next goal is to acquire in-depth understanding of any information out there relevant to the datasets of interest. This type of data annotation effort can be supported by tools like Enrichr, which is an open-source, freely available tool for analysis and visualization of gene enrichment using unbiased lists of genes or proteins generated from genome wide, ChIP-seq, RNA-seq, microarray, and MS studies. These unbiased lists are used to compute gene enrichment against existing knowledge contained in ontologies and other annotated lists of gene set libraries organized by the functional groups of genes57. Legacy datasets are maintained to preserve past analyses and to address the issue of provenance, a key component of the FAIR doctrine. Enrichr utilizes the Data Driven Documents (D3) JavaScript library to present enrichment results. A REST API has also been provided for Enrichr, intended for users aiming to further enrich their own data programmatically. The analysis platform utilizes 114 libraries with over 210,000 annotated gene sets; over 7.2 million gene lists have been analyzed using Enrichr as of July, 2017. Accessible at http://amp.pharm.mssm.edu/Enrichr/.
The resources available to constantly aggregate relevant information for one specific molecule and to manually distill it into an accessible format is currently limited. The Gene Wiki portal, as part of the Molecular and Cellular Biology Wikiproject on Wikipedia, aims to leverage crowdsourcing efforts to annotate encyclopedic knowledge of individual genes and proteins58–60. Gene Wiki exists within Wikipedia as human gene and protein pages, which serve as living documents with an ever-increasing annotation network that both researchers and citizen scientists from a broad variety of backgrounds can contribute to by providing structured information. For example, embedded semantic linking within the Gene Wiki pages enables increased access to primary scientific knowledge by the general public. Over 10,000 individual gene pages are encompassed in Gene Wiki, among which there exist well annotated pages for the top 50 popularly studied cardiovascular proteins61, 62, including natriuretic peptide B, angiotensin-converting enzyme, sodium channel protein type 5 subunit alpha, potassium voltage-gated channel H2, and C-reactive protein. The Gene Wiki pages are also nicely clustered around organelle subproteomes; in the context of the mitochondrial subproteome, it covers all major metabolic pathways and over 550 mitochondrial proteins. The total pages are edited over 15,000 times and visited more than 50 million times in a single year. More information can be found at: https://en.wikipedia.org/wiki/Portal:Gene_Wiki.
In the context of cellular processes, most molecules do not function in isolation; rather, a cascade of molecules work together to achieve a biological outcome. Through its intuitive interface and computational analysis tools, Reactome enables users to extract information from multidimensional datasets with high complexity63, 64. Users can supply their own datasets and employ highly optimized, in-memory pathway analysis tools to visualize carefully crafted, enriched pathways65. Moreover, this analysis tool provides the capability to explore the visualization of pathways in the Pathway Browser, which supports such features as zooming and event highlighting, as demonstrated in Figure 266–75. Reactome presents curated information on proteins, complexes, reactions, and pathways from 19 species, including 10,684 human proteins and isoforms, and 66 cardiovascular pathways. Reactome’s data as well as software tools are freely available for download at: http://reactome.org/.
Figure 2. Overview of Pathways Involved in Protein Metabolism as Annotated in the Reactome Pathways Database66.
Terms in italics reference the labels in the figure. Metabolism of proteins, as annotated here, covers the full life cycle of a protein from its synthesis to its posttranslational modification and degradation. Protein synthesis is accomplished through Translation of an mRNA sequence into a polypeptide chain. Protein folding is achieved through the function of molecular chaperones that recognize and associate with proteins in their non-native state; as well as facilitate their folding by stabilizing the conformation of productive folding intermediates67. Following translation, many newly formed proteins undergo Post-translational modification, essentially irreversible covalent modifications critical for their mature locations and functions68, including Ubiquitination, Methionine oxidation, Carboxyterminal post-translational modifications, Neddylation, and Phosphorylation. Most mitochondrial proteins are encoded in the nucleus, synthesized in the cytosol and then by the process of Mitochondrial protein import sorted and targeted to four locations, outer membrane, intermembrane space, inner membrane, and matrix69–72. Peptide hormones are synthesized as parts of larger precursor proteins whose cleavage in the secretory system (endoplasmic reticulum, Golgi apparatus, secretory granules) is annotated in Peptide hormone metabolism. After secretion, peptide hormones are modified and degraded by extracellular proteases73. Two responses to protein damage are annotated in Reactome. The Unfolded Protein Response (UPR) is a regulatory system that protects the Endoplasmic Reticulum (ER) from overload. First, the UPR is provoked by the accumulation of improperly folded protein in the ER during times of unusually high secretory activity74. Second, Protein repair enables the reversal of damage to some amino acid side chains caused by reactive oxygen species. Pulmonary surfactants are lipids and proteins that are secreted by the alveolar cells of the lung that decrease surface tension at the air/liquid interface within the alveoli to maintain the stability of pulmonary tissue75. Nuclear regulation, transport, metabolism, reutilization, and degradation of surfactant are described in the Surfactant metabolism pathway. Amyloid fiber formation, the accumulation of mostly extracellular deposits of fibrillar proteins, is associated with tissue damage observed in numerous diseases including late phase heart failure (cardiomyopathy) and neurodegenerative diseases such as Alzheimer's, Parkinson's, and Huntington's. The figure has been copied under CC-BY with permission from Reactome, an interactive version with links to more detailed sub-pathway representations is accessible at [http://reactome.ncpsb.org/PathwayBrowser/#/R-HSA-392499]
A cloud-based virtual space can provide an environment as a sandbox to accommodate datasets, computational tools, and analytic pipelines to work synergistically. Galaxy is an open-source platform with a wide range of analytical tools to perform biomedical research on open-source large datasets. By uploading their own data to the computational infrastructure of Galaxy, users can answer biomedical questions surrounding omics datasets by utilizing the countless tools on the site; informatics experience is not a requirement for performing these high throughput analyses, as the platform allows users to deploy preexisting computational workflows and create their own pipelines76. The project currently has over 4,500 publications that cite, mention, or discuss Galaxy, demonstrating its broad use in many fields of biological research. The platform can be accessed, along with in-depth tutorials, at https://usegalaxy.org/. Another popular open-source software platform is Cytoscape, used for analysis, visualization, integration, and annotation of complex networks such as molecular interaction networks and biological pathways77. The platform enables enhancement of the network data by integrating a wide variety of metadata formats using APIs from external sources and databases into the network structure. Two powerful aspects of Cytoscape are its extensibility and its active user community; using the Java-based open API, over 320 applications have been authored by third party developers to create added functionality and interoperability, with nearly 1 million App Store downloads as of July 2017. Cytoscape is accessible at http://cytoscape.org/. In a similar fashion, more recent project such as Apache Taverna and Synapse of Sage Bionetwork aim to empower users with limited programmatic experience to perform biomedical computation. Taverna, now part of the Apache incubator, is designed as a workflow management system where users have ultimate customization of analytical workflows (accessible at: https://taverna.incubator.apache.org/). Sage Bionetworks created Synapse with the intention of providing a collaborative platform for researchers to track their investigations; they have multiple API clients for a variety of programming frameworks and languages (available at: https://www.synapse.org/).
With the recent advancement of cloud-based computational technologies, many software applications have been engineered for portability in parallel to interoperability. Utilizing open-source data APIs MyGene.info and MyVariant.info serve to compile biomedical information regarding genes and variants into structured annotations; MyGene.info contains information for multiple species, whereas MyVariant.info contains information solely regarding human variations. The platforms benefit from an Elastic search-based indexing engine that clusters data objects across multiple repositories of data, which enables high-performance querying and scalability78, 79. Both tools, on average, receive more than 3 million requests per month and cover information regarding 19 billion genes and variants spanning. The APIs can be accessed at http://mygene.info/ and http://myvariant.info/.
In addition to these tools with widespread biomedical scope, there are tools and platforms that have been developed to specifically tackle cardiovascular research questions. The Cardiac Atlas Project was created as a database for the open sharing of cardiovascular imaging data. Including the MESA study among its epidemiological cohorts, the project houses this data for widespread access in order to catalyze computational modeling and high-througput statistical analyses80, 81. More information can be found at http://www.cardiacatlas.org/. Moreover, the American Heart Association recently began collaboration with Amazon Web Services (AWS) to launch the AHA Institute for Precision Cardiovascular Medicine. Leveraging the computational infrastructure in AWS, the AHA Precision Medicine Platform was created as a cloud-based source of openly accessible data and state-of-the-art tools to foster synergistic investigations among researchers. Details may be accessed at https://precision.heart.org/about.
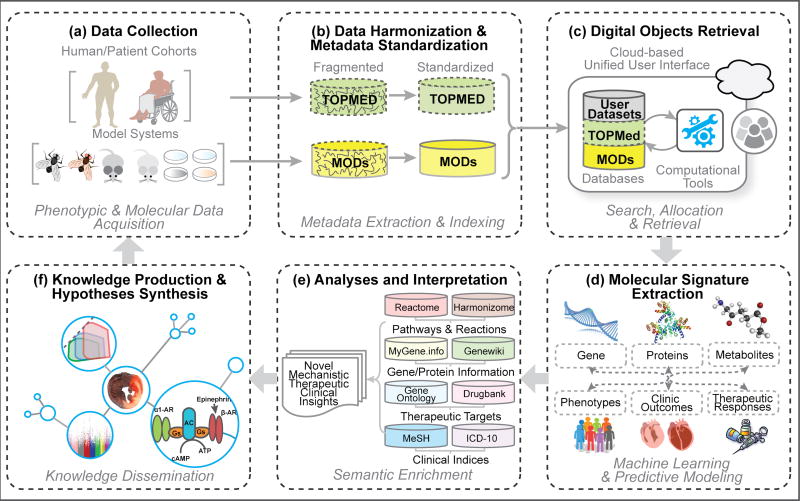
A typical biomedical research workflow inspired by modern informatics would involve well thought-out steps and the integration of many resources to draw together, cluster, and aggregate all necessary information, producing new knowledge. An example of such a workflow is detailed in Figure 3. Employing cutting-edge machine learning technologies in conjunction with aggregated knowledgebases, analytical workspaces, and other digital resources can be tailored to the needs of individual investigators and the specific question they have at hand. As cloud-based technologies render these resources more accessible than ever before, their utilities and benefits are increasingly appreciated by the scientific community. With the field of data science rapidly transforming, especially in the context of cardiovascular medicine, the sustainability of the domain and the community hinges on deploying FAIR principles wherever possible.
Figure 3. Workflow Detailing Data Science Elements in Research Experimental Design or Clinical Study Design.
This regenerative workflow overviews the experimental design that drives biomedical research forward in the age of data science. (a) Data can be collected from either human cohorts or model systems. Through techniques and methods tailored to the individual dataset, phenotypic and/or molecular data are acquired that may comprise one or more variables. (b) As there are many types of data and many features that can describe the data, e.g., sequencing technology used for a transcriptomics dataset, we will begin by first conducting data harmonization; subsequently, we will extract the metadata pertaining to the dataset in order to enable indexing and standardization. (c) With this data transformed into an accessible format and integrated into a unified interface, the investigator can then search for and retrieve relevant digital objects, i.e., datasets or computational tools most appropriate for the proposed study. (d) These resources can be deployed to perform state-of-the-art analyses, such as machine learning and predictive modeling, to discover robust genotype-phenotype associations and establish molecular signatures for the cohort. (e) Once we have molecular signatures at hand, they will be processed and further analyzed to gain novel mechanistic, therapeutic, and clinical insights. (f) Armed with these new insights, investigators can contribute to the network of biomedical knowledge and inform new hypotheses to continue the upward spiral of cardiovascular research. Notably, this is not a closed cycle; this workflow demonstrates two potential scenarios. First, an investigator can charter through the course of experimental design and repeat, forging their continued path onto their next projects. Second, by fomenting and inspiring other investigations to be conducted in parallel, the data collected from one set of studies may foster and cultivate joint analyses and further propagate collaborative discovery among the biomedical community.
Concluding Remarks
In the immediate future, we foresee cloud-based solutions making substantial contributions to advancing precision cardiovascular health. Regardless of what computational solutions the long-term future holds, we believe a computational environment built on the guiding principles of collaborative teamwork as well as shared common resources will prevail. As summarized, biomedical researchers are benefiting from collaborative analytical environments, such as Sage Bionetwork’s infrastructure, where multiple teams in different geographical locations can work on model system or cohort study analyses. Fundamentally, these platforms facilitate innovation and discovery by bring people together across the world. From a patient-care perspective, one prospective use of cloud-based technologies is enabling real-time virtual patient and physician communications on secure, individual health commons, thereby empowering patients to approach their care in unique ways which best suit their needs. These advances can be furthered through the use of mobile health devices to monitor cardiovascular parameters (e.g., heart rate, blood pressure) and lifestyle (e.g., physical activity, social media), collecting and analyzing these types of data to gain personal insights into one’s health.
As the digital paradigm has shifted from individually produced, isolated digital objects to those available on the cloud, cardiovascular investigators potentially have myriad resources at their fingertips. Employing FAIR principles in an integrated, cloud computing interface can drive both biomedical research and clinical practice forward by encouraging and inspiring users to perform data-based research without needing in-depth information about the infrastructural capacity required to perform the computational analyses best suited to their study.
Having said that, there are some initial costs in deploying a cloud computing environment. First, data (and metadata) that are typically stored on local databases and public data repositories need to be migrated to cloud storage. Importing these legacy data into the cloud often requires data cleaning and harmonization, which may entail disk file transfers from one system to another, custom software, human annotation and curation, and other methods. The specific method depends entirely on the systems involved and the nature and state of the data being migrated. Deploying existing software and applications on the cloud may also be non-trivial, depending on their current implementation. In some rare cases where a software or application was not implemented in a programming language that is supported by the cloud infrastructure and/or requires system configurations that are not compatible with the cloud infrastructure, reimplementation of (part of) the software or application may be needed. Similar challenges present themselves if one wishes to migrate from one cloud to another or to build a cloud environment that bridges multiple cloud computing platforms. Serious considerations need to be given to ensure compatibility, interoperability, and portability. The success of deploying a cloud computing environment also depends on initial user acceptability and degree of adaptation. This requires a change of mindset for many users to embrace a new and more collaborative research environment and computing platform. Training activities and onsite technical supports may be needed to facilitate the transition.
In a cloud environment, the data physically reside in remote locations, aggravating concerns about data security and privacy protection. A number of techniques have been proposed by researchers for data protection and to attain the highest level of data security in the cloud. However, there are still many gaps to be filled by making these techniques more effective. These have been the focus of data governance that concerns the entire lifecycle of data management, including its organization, integrity, confidentiality, availability, privacy, and security. Better data governance policy and practice may not only support data privacy, security, and user trust but also play a key role in metadata generation and management; preserving data provenance and lineage; allowing simplified methods for tracing errors and correcting them; and enhancing reproducibility in data aggregation, analytics, and integration. This will also enable proper credit attribution and encourage good scientific practice and user engagement.
The sea of biomedical data is rising, and the scientific community demands tools to not only stay afloat, but also navigate the waters to their final destinations. The fundamental paths to biomedical discovery have been forged, thanks largely in part to the trailblazing investigators that have created analytical tools and repositories. Looking forward, there abounds opportunities for efficient extraction of the rich information hidden in untapped datasets; a community effort involving novel approaches to cloud-based computing will empower investigators to seize the gems of biomedical research and propel the transformation of cardiovascular medicine into a new era.
Acknowledgments
We thank Drs. Ding Wang and Bilal Mirza, as well as Anders Garlid, Chelsea Ju, Jessica Lee, Zeyu Li, Patrick Tan, and Justin Wood at the University of California, Los Angeles (UCLA) for their critical input on the content. We would also like to thank Dr. Lisa Matthews, at New York University Medical Center at Cornell University; and Cristoffer Sevilla, Steven Jupe, and Peter D’Eustachio at the European Bioinformatics Institute (EBI) in the United Kingdom, for their contributions regarding Reactome enlisted pathways and other related components.
Sources of Funding:
This work was supported in part by National Institutes of Health U54 GM114833 (to P. Ping, H. Hermjakob, and W. Wang), U41 HG003751 (to H. Hermjakob), U01 HL137159 (to P. Benos), R01 LM012087 (to P. Benos, R35 HL135772 (to P. Ping), and the T.C. Laubisch endowment at UCLA (to P. Ping).
Non-standard Abbreviations and Acronyms
- API
Application Programming Interface
- AWS
Amazon Web Services
- CC-BY
Creative Commons Attribution
- ChIP-seq
Chromatin Immunoprecipitation Sequencing
- CIL
Class Imbalance Learning
- DATS
Data Tag Suite
- dbGaP
database of Genotypes and Phenotypes
- DOI
Digital Object Identifier
- EBI
European Bioinformatics Institute
- EDAM
EMBRACE Data and Methods
- EGA
European Genome-phenome Archive
- ER
Endoplasmic Reticulum
- FAIR
Findable, Accessible, Interoperable, Resuable
- GUID
Globally Unique Identifier
- ILD
Interstitial Lung Disease
- JATS
Journal Article Tag Suite
- JSON
JavaScript Object Notation
- MESA
Multi-ethnic Study of Atherosclerosis
- MOD
Model Organism Database
- MS
Mass Spectrometry
- PGM
Probabilistic Graphical Models
- RDF
Resource Description Framework
- SPARQL
SPARQL Protocol and RDF Query Language
- TOPMed
Trans-Omics for Precision Medicine
- UPR
Unfolded Protein Response
- XML
Extended Markup Language
Footnotes
Disclosures:
None.
References
- 1.Amazon Web Services. [Accessed August 1, 2017];What is Cloud Computing? Available at: https://aws.amazon.com/what-is-cloud-computing/
- 2.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJ, Groth P, Goble C, Grethe JS, Heringa J, t Hoen PA, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merriam-Webster, Inc. [Accessed August 1, 2017];The definition of "metadata". Available at: https://www.merriam-webster.com/dictionary/metadata.
- 4. [Accessed August 1, 2017];The International DOI Foundation. Available at: https://www.doi.org.
- 5.Paskin N. Digital Object Identifier (DOI®) System. Encyclopedia of Library and Information Sciences, Third Edition. 2009:1586–1592. [Google Scholar]
- 6.Juty N, Le Novere N, Laibe C. Identifiers.org and MIRIAM Registry: community resources to provide persistent identification. Nucleic Acids Res. 2012;40:D580–586. doi: 10.1093/nar/gkr1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barr RG, Bluemke DA, Ahmed FS, Carr JJ, Enright PL, Hoffman EA, Jiang R, Kawut SM, Kronmal RA, Lima JA, Shahar E, Smith LJ, Watson KE. Percent emphysema, airflow obstruction, and impaired left ventricular filling. N Engl J Med. 2010;362:217–227. doi: 10.1056/NEJMoa0808836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. [Accessed August 1, 2017];U.S. National Library of Medicine. Available at: https://www.nlm.nih.gov/mesh/
- 9.Lipscomb CE. Medical Subject Headings (MeSH) Bull Med Libr Assoc. 2000;88:265–266. [PMC free article] [PubMed] [Google Scholar]
- 10. [Accessed August 1, 2017];National Center for Biotechnology Information, U.S. National Library of Medicine. Available at: https://jats.nlm.nih.gov.
- 11.Sansone SA, Gonzalez-Beltran A, Rocca-Serra P, Alter G, Grethe JS, Xu H, Fore IM, Lyle J, Gururaj AE, Chen X, Kim HE, Zong N, Li Y, Liu R, Ozyurt IB, Ohno-Machado L. DATS, the data tag suite to enable discoverability of datasets. Sci Data. 2017;4:170059. doi: 10.1038/sdata.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. [Accessed August 1, 2017];The Schema.org Community Group. Available at: https://schema.org.
- 13. [Accessed August 1, 2017];The Bioschemas Community Group. Available at: https://bioschemas.org.
- 14.Zhou Y, Liem DA, Lee JM, Bleakley B, Caufield JH, Murali S, Wang W, Zhang L, Bui AT, Sun Y, Watson KE, Han J, Ping P. Uncovering medical insights from vast Amounts of biomedical data in clinical case reports. bioRxiv. 2017:172460v1. [Google Scholar]
- 15. [Accessed August 1, 2017];Project Supported by NIH Big Data to Knowledge award U54 GM114833. Available at: https://aztec.bio.
- 16.Wang W, Bleakley B, Ju C, Kyi V, Tan P, Choi H, Huang X, Zhou Y, Wood J, Wang D, Bui A, Ping P. Aztec: A platform to Render Biomedical Software Findable, Accessible, Interoperable, and Reusable. arXiv. 2017:1706.06087v1. [Google Scholar]
- 17. [Accessed August 1, 2017];EDAM Community Project. Available at: http://edamontology.org/
- 18. [Accessed August 1, 2017];SNOMED International. Available at: http://www.snomed.org/snomed-ct/get-snomed-ct.
- 19. [Accessed August 1, 2017];National Center for Biotechnology Information, U.S. National Library of Medicine. Available at: https://www.ncbi.nlm.nih.gov/gap.
- 20. [Accessed August 1, 2017];European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) Available at: https://www.ebi.ac.uk/ega/
- 21. [Accessed August 1, 2017];Beacon Project. Available at: https://genomicsandhealth.org/work-products-demonstration-projects/beacon-project-0.
- 22. [Accessed August 1, 2017];The National Heart, Lung, and Blood Institute. Trans-Omics for Precision Medicine (TOPMed) Program. doi: 10.1093/aje/kwab115. Available at: https://www.nhlbi.nih.gov/research/resources/nhlbi-precision-medicine-initiative/topmed. [DOI] [PMC free article] [PubMed]
- 23.Page L, Brin S, Motwani R, Winograd T. The PageRank citation ranking: Bringing order to the web. Stanford InfoLab. 1999 [Google Scholar]
- 24.Google Inc. [Accessed August 1, 2017];Google Cloud Platform. Available at: https://cloud.google.com/
- 25.Microsoft Corporation. [Accessed August 1, 2017];Microsoft Azure. Available at: http://www.microsoft.com/azure/default.mspx.
- 26.Bray T, Paoli J, Sperberg-McQueen CM. Extensible markup language (XML) World Wide Web Journal. 1997;2:29–66. [Google Scholar]
- 27.Bray T. The javascript object notation (json) data interchange format. 2014 [Google Scholar]
- 28.Byrne M, Fokkema IF, Lancaster O, Adamusiak T, Ahonen-Bishopp A, Atlan D, Beroud C, Cornell M, Dalgleish R, Devereau A, Patrinos GP, Swertz MA, Taschner PE, Thorisson GA, Vihinen M, Brookes AJ, Muilu J. VarioML framework for comprehensive variation data representation and exchange. BMC Bioinformatics. 2012;13:254. doi: 10.1186/1471-2105-13-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris S, Seaborne A, Prud’hommeaux E. SPARQL 1.1 query language. W3C recommendation. 2013 [Google Scholar]
- 30.Pan JZ. Handbook on ontologies. Springer; Berlin Heidelberg: 2009. Resource description framework; pp. 71–90. [Google Scholar]
- 31.Perez-Riverol Y, Bai M, da Veiga Leprevost F, Squizzato S, Park YM, Haug K, Carroll AJ, Spalding D, Paschall J, Wang M, Del-Toro N, Ternent T, Zhang P, Buso N, Bandeira N, Deutsch EW, Campbell DS, Beavis RC, Salek RM, Sarkans U, Petryszak R, Keays M, Fahy E, Sud M, Subramaniam S, Barbera A, Jimenez RC, Nesvizhskii AI, Sansone SA, Steinbeck C, Lopez R, Vizcaino JA, Ping P, Hermjakob H. Discovering and linking public omics data sets using the Omics Discovery Index. Nat Biotechnol. 2017;35:406–409. doi: 10.1038/nbt.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Apache Jakarta Project. Apache Software Foundation. Apache Lucene; 2004. Apache Lucene-a high-performance, full-featured text search engine library. [Google Scholar]
- 33.National Human Genome Research Insititute. [Accessed August 1, 2017];Compuptaional Genomics and Data Science Program: Model Organism Databases (MODs) Available at: https://www.genome.gov/10001837/model-organism-databases/
- 34.World Health Organization. ICD-10: international statistical classification of diseases and related health problems: tenth revision. Geneva: 2004. [Google Scholar]
- 35.Ananthakrishnan R, Chard K, Foster I, Tuecke S. Globus Platform-as-a-Service for Collaborative Science Applications. Concurr Comput. 2015;27:290–305. doi: 10.1002/cpe.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 37.He H, Garcia EA. Learning from Imbalanced Data. IEEE Transactions on Knowledge and Data Engineering. 2009;21:1263–1284. [Google Scholar]
- 38.Pearl J. Causality: Models, Reasoning and Inference. Cambridge University Press; 2009. [Google Scholar]
- 39.Spirtes P, Glymour C, Scheines R. Causation, Prediction, and Search. 2. Cambridge, MA: The MIT Press; 2000. [Google Scholar]
- 40.Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miotto R, Li L, Kidd BA, Dudley JT. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Sci Rep. 2016;6:26094. doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma J, Sheridan RP, Liaw A, Dahl GE, Svetnik V. Deep neural nets as a method for quantitative structure-activity relationships. J Chem Inf Model. 2015;55:263–274. doi: 10.1021/ci500747n. [DOI] [PubMed] [Google Scholar]
- 43.Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 44.Rajpurkar P, Hannun AY, Haghpanahi M, Bourn C, Ng AY. Cardiologist-Level Arrhythmia Detection with Convolutional Neural Networks. arxiv. 2017:1707.01836. [Google Scholar]
- 45.Choi E, Schuetz A, Stewart WF, Sun J. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc. 2017;24:361–370. doi: 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Shen D. Computational medicine: A cybernetic eye for rare disease. Nat Biomed Eng. 2017;1:0032. doi: 10.1038/s41551-017-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial Intelligence in Precision Cardiovascular Medicine. J Am Coll Cardiol. 2017;69:2657–2664. doi: 10.1016/j.jacc.2017.03.571. [DOI] [PubMed] [Google Scholar]
- 48.Liu N, Koh ZX, Chua EC, Tan LM, Lin Z, Mirza B, Ong ME. Risk scoring for prediction of acute cardiac complications from imbalanced clinical data. IEEE J Biomed Health Inform. 2014;18:1894–1902. doi: 10.1109/JBHI.2014.2303481. [DOI] [PubMed] [Google Scholar]
- 49.Singh A, Guttag JV. A comparison of non-symmetric entropy-based classification trees and support vector machine for cardiovascular risk stratification; Conf Proc IEEE Eng Med Biol Soc; 2011. pp. 79–82. [DOI] [PubMed] [Google Scholar]
- 50.Gepperth A, Hammer B. Incremental learning algorithms and applications; Proceedings of the European Symposium on Artificial Neural Networks (ESANN 2016) on "Computational Intelligence and Machine Learning"; 2016. pp. 357–368. [Google Scholar]
- 51.Mirza B, Lin Z. Meta-cognitive online sequential extreme learning machine for imbalanced and concept-drifting data classification. Neural Netw. 2016;80:79–94. doi: 10.1016/j.neunet.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Sedgewick AJ, Shi I, Donovan RM, Benos PV. Learning mixed graphical models with separate sparsity parameters and stability-based model selection. BMC Bioinformatics. 2016;17(Suppl 5):175. doi: 10.1186/s12859-016-1039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong W, Koyano-Nakagawa N, Li T, Garry DJ. Inferring dynamic gene regulatory networks in cardiac differentiation through the integration of multi-dimensional data. BMC Bioinformatics. 2015;16:74. doi: 10.1186/s12859-015-0460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orphanou K, Stassopoulou A, Keravnou E. DBN-Extended: A Dynamic Bayesian Network Model Extended With Temporal Abstractions for Coronary Heart Disease Prognosis. IEEE J Biomed Health Inform. 2016;20:944–952. doi: 10.1109/JBHI.2015.2420534. [DOI] [PubMed] [Google Scholar]
- 55.Bae H, Monti S, Montano M, Steinberg MH, Perls TT, Sebastiani P. Learning Bayesian Networks from Correlated Data. Sci Rep. 2016;6:25156. doi: 10.1038/srep25156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, Ma'ayan A. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016;2016 doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huss JW, 3rd, Lindenbaum P, Martone M, Roberts D, Pizarro A, Valafar F, Hogenesch JB, Su AI. The Gene Wiki: community intelligence applied to human gene annotation. Nucleic Acids Res. 2010;38:D633–639. doi: 10.1093/nar/gkp760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su AI, Good BM, van Wijnen AJ. Gene Wiki Reviews: marrying crowdsourcing with traditional peer review. Gene. 2013;531:125. doi: 10.1016/j.gene.2013.08.093. [DOI] [PubMed] [Google Scholar]
- 60.Tsueng G, Good BM, Ping P, Golemis E, Hanukoglu I, van Wijnen AJ, Su AI. Gene Wiki Reviews: Raising the quality and accessibility of information about the human genome. Gene. 2016;592:235–238. doi: 10.1016/j.gene.2016.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lam MP, Venkatraman V, Cao Q, Wang D, Dincer TU, Lau E, Su AI, Xing Y, Ge J, Ping P, Van Eyk JE. Prioritizing Proteomics Assay Development for Clinical Translation. J Am Coll Cardiol. 2015;66:202–204. doi: 10.1016/j.jacc.2015.04.072. [DOI] [PubMed] [Google Scholar]
- 62.Lam MP, Venkatraman V, Xing Y, Lau E, Cao Q, Ng DC, Su AI, Ge J, Van Eyk JE, Ping P. Data-Driven Approach To Determine Popular Proteins for Targeted Proteomics Translation of Six Organ Systems. J Proteome Res. 2016;15:4126–4134. doi: 10.1021/acs.jproteome.6b00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D'Eustachio P. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D'Eustachio P, Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, D'Eustachio P, Stein L, Hermjakob H. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics. 2017;18:142. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, Matthews L, May B, Milacic M, Rothfels K, Shamovsky V, Webber M, Weiser J, Williams M, Wu G, Stein L, Hermjakob H, D'Eustachio P. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44:D481–487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 68.Knorre DG, Kudryashova NV, Godovikova TS. Chemical and functional aspects of posttranslational modification of proteins. Acta Naturae. 2009;1:29–51. [PMC free article] [PubMed] [Google Scholar]
- 69.Kutik S, Guiard B, Meyer HE, Wiedemann N, Pfanner N. Cooperation of translocase complexes in mitochondrial protein import. J Cell Biol. 2007;179:585–591. doi: 10.1083/jcb.200708199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milenkovic D, Muller J, Stojanovski D, Pfanner N, Chacinska A. Diverse mechanisms and machineries for import of mitochondrial proteins. Biol Chem. 2007;388:891–897. doi: 10.1515/BC.2007.097. [DOI] [PubMed] [Google Scholar]
- 71.Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Endo T, Yamano K. Multiple pathways for mitochondrial protein traffic. Biol Chem. 2009;390:723–730. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- 73.Chertow BS. The role of lysosomes and proteases in hormone secretion and degradation. Endocr Rev. 1981;2:137–173. doi: 10.1210/edrv-2-2-137. [DOI] [PubMed] [Google Scholar]
- 74.Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium. 2002;32:235–249. doi: 10.1016/s0143416002001823. [DOI] [PubMed] [Google Scholar]
- 75.Griese M. Pulmonary surfactant in health and human lung diseases: state of the art. Eur Respir J. 1999;13:1455–1476. doi: 10.1183/09031936.99.13614779. [DOI] [PubMed] [Google Scholar]
- 76.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, Gruning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu C, Macleod I, Su AI. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xin J, Mark A, Afrasiabi C, Tsueng G, Juchler M, Gopal N, Stupp GS, Putman TE, Ainscough BJ, Griffith OL, Torkamani A, Whetzel PL, Mungall CJ, Mooney SD, Su AI, Wu C. High-performance web services for querying gene and variant annotation. Genome Biol. 2016;17:91. doi: 10.1186/s13059-016-0953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fonseca CG, Backhaus M, Bluemke DA, Britten RD, Chung JD, Cowan BR, Dinov ID, Finn JP, Hunter PJ, Kadish AH, Lee DC, Lima JA, Medrano-Gracia P, Shivkumar K, Suinesiaputra A, Tao W, Young AA. The Cardiac Atlas Project--an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics. 2011;27:2288–2295. doi: 10.1093/bioinformatics/btr360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suinesiaputra A, Medrano-Gracia P, Cowan BR, Young AA. Big heart data: advancing health informatics through data sharing in cardiovascular imaging. IEEE J Biomed Health Inform. 2015;19:1283–1290. doi: 10.1109/JBHI.2014.2370952. [DOI] [PMC free article] [PubMed] [Google Scholar]