Abstract
Atherosclerotic cardiovascular disease and acute myocardial infarction are the leading causes of mortality worldwide, and apoptosis is the major pathway of cardiomyocyte death under hypoxic conditions. Although studies have reported changes in the expression of certain pro-apoptotic and anti-apoptotic genes in hypoxic cardiomyocytes, genetic regulations are complex in human cardiomyocytes and there is much that remains to be fully elucidated. The present study aimed to identify differentially expressed genes in hypoxic human AC16 cardiomyocytes using next-generation sequencing and bioinformatics. A total of 24 genes (15 upregulated and 9 downregulated) with potential micro (mi)RNA-mRNA interactions were identified in the miRmap database. Utilising the Gene Expression Omnibus database of cardiac microvascular endothelial cells, tensin 1, B-cell lymphoma 2-interacting protein 3 like, and stanniocalcin 1 were found to be upregulated, and transferrin receptor and methyltransferase like 7A were found to be downregulated in response to hypoxia. Considering the results from miRmap, TargetScan and miRDB together, two potential miRNA-mRNA interactions were identified: hsa-miRNA (miR)-129-5p/CDC42EP3 and hsa-miR-330-3p/HELZ. These findings contribute important insights into possible novel diagnostic or therapeutic strategies for targeting cardiomyocytes under acute hypoxic stress in conditions, including acute myocardial infarction. The results of the present study also introduce an important novel approach in investigating acute hypoxic pathophysiology.
Keywords: hypoxia, AC16, cardiomyocyte, next-generation sequencing, bioinformatics
Introduction
Atherosclerotic cardiovascular disease is the leading cause of mortality worldwide (1,2). The cardiomyocytes (CMs) in patients with acute myocardial infarction (AMI) undergo cell death, alteration of cell cycle, and abrupt mitochondrial oxidative metabolism in either ischemic or hypoxic conditions (3-5). Therefore, antiplatelet agents, anticoagulation agents, and emergent coronary revascularization are often used to treat patients with acute coronary syndrome (6).
Apoptotic cell death is important in myocardial damage following AMI, which may lead to malignant arrhythmia, heart failure and cardiac death (7,8). During hypoxia and reoxygenation injury, CMs exhibit typical morphological characteristics of apoptotic nuclei, including membrane blebs, myofibillary disarrangement, chromatin condensation, and peripheral margination of mitochondria (3). Increased myocardial apoptosis has also been found in autopsies of patients who have succumbed to AMI and multi-vessel coronary artery diseases (9,10). Furthermore, changes in several pro-apoptotic and anti-apoptotic mediators have been noted in ischemic or hypoxic CMs in vivo, in isolated animal hearts, and in humans (5,7,8,11,12).
Unlike traditional Sanger sequencing technology, next-generation sequencing (NGS) provides rapid analyses of large quantities of genomic information, including DNAs, mRNAs, microRNAs (miRNAs), and non-coding RNAs (13,14). Using NGS in combination with bioinformatics analyses, whole exon sequencing has been performed in previous studies to identify a wide spectrum of genetic mutations in several scientific fields (13,14). NGS has also been used to identify novel genetic regulations in basic and clinical research (15,16), and it has been used in the genetic diagnosis of congenital heart diseases and inherited cardiac dysrhythmia (16,17).
Although several studies have investigated the signaling pathways and cellular responses of CMs in ischemic or hypoxic conditions, the regulation of genetic expression with regard to hypoxia-related apoptosis is complex and remains to be fully elucidated (18-20). Therefore, the present study was designed to perform a comprehensive investigation of various genetic expression changes in hypoxic human cardiomyocytes using NGS and bioinformatic analyses.
Materials and methods
Cell culture
The AC16 human cardiomyocyte cell line, purchased from EMD Millipore (Billerica, MA, USA), was cultured following standard manufacturer protocol. The cells (1×106 cells/well) were seeded in 10% FBS (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and DMEM/F-12 medium (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and allowed to grow overnight. The AC16 CMs were then incubated in either a normoxic condition (37°C, 20% O2 and 5% CO2) or hypoxic condition (37°C, 1% O2 and 5% CO2) for 24 h. The hypoxic condition was maintained in a physiological oxygen workstation (InvivO2 400; Baker Ruskinn, Sanford, ME, USA).
Flow cytometry and detection of apoptosis
The cells were analyzed using an Annexin V-FITC Early Apoptosis Detection kit (Cell Signaling Technology, Inc., Danvers, MA, USA) and a BD Accuri C6 Plus flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) as previously reported (21).
RNA sequencing
Total RNAs from the normoxic and hypoxic AC16 CMs were extracted with TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc.) following the manufacturer's standard protocol. The purified RNA was measured at OD260nm with an ND-1000 spectrophotometer (Nanodrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and qualitatively analyzed with an RNA 6000 LabChip kit (Agilent Technologies, Inc., Santa Clara, CA, USA) and the Bioanalyzer 2100 (Agilent Technologies, Inc.).
Library preparation and deep sequencing were performed according to the manufacturer's protocol (Illumina, Welgene Biotechnology Company, Taipei, Taiwan), as described in our previous reports (15,22). For small RNA sequencing, following reverse transcription of total RNA, cDNA with sizes indicating 18-40-nucleotide RNA fragments (140-155 nucleotides in length with both adapters) was extracted and sequenced on the Illumina instrument (75 single-end cycles). Following trimming or removing low-quality data using Trimmomatic (version 0.36) (23), the qualified reads were analyzed using miRDeep2 software (version 2.0.0.8) (24) and the human genome from the University of California Santa Cruz (UCSC) database (https://genome.ucsc.edu/). The miRNAs with low levels [<1 normalized read per million (rpm)] in both hypoxic and normoxic cells were excluded. For transcriptome sequencing, the library constructed with the SureSelect Strand Specific RNA Library Preparation kit (Agilent Technologies, Inc.) was sequenced with the TruSeq SBS kit on the Solexa platform (Illumina NextSeq 500; 75 cycles, single-end). Following trimming or removing low-quality data using Trimmomatics (23), the qualified reads were analyzed using Cufflinks (25) and the Ensembl database (https://asia.ensembl.org/index.html). The genes with low expression levels (<0.3 fragment per kilobase of transcript per million mapped reads) in both hypoxic and normoxic cells were excluded.
miRNA database analyses
The miRmap is a web database used to predict putative genes targeted by candidate miRNAs and is available from http://cegg.unige.ch/mirmap (26). In the present study, a search was performed for putative targeted miRNAs from human species and miRmap scores >99.0. A search was also performed for potential miRNA interactions in the miRmap, TargetScan (http://www.targetscan.org/vert_71/) and miRDB (http://www.mirdb.org/) databases.
Ingenuity® Pathway Analysis (IPA)
IPA software (Ingenuity Systems, Redwood City, CA, USA) integrates numerous results and performs multiple analyses providing a comprehensive interpretation of a large quantity of experimental data. In the present study, IPA analysis was performed for the network analyses of candidate genes.
Gene expression omnibus (GEO) database analysis
The GEO database (https://www.ncbi.nlm.nih.gov/geo/) is a useful web database containing raw gene expression data from microarray studies and NGS. The present study used a GEO array (GEO accession: GDS3483) investigating the gene responses to hypoxia in primary human pulmonary microvascular endothelial cells and cardiac microvascular endothelial cells (27). Data were collected from the cardiac microvascular endothelial cells under normoxia or hypoxia (1% O2, 5% CO2, and 94% N2) over various durations (3, 24 and 48 h). The raw data extracted from the GEO database were re-plotted and statistically analyzed using analysis of variance (ANOVA) followed by Dunnett's test using GraphPad Prism® 7 software (GraphPad Software, Inc., La Jolla, CA, USA).
Database for Annotation Visualization and Integrated Discovery (DAVID) analysis
The DAVID (https://david.ncifcrf.gov/) database (28) is a useful tool for gene functional classification. It integrates data from multiple functional annotation databases, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. A list of genes of interest is classified by clustering of related biological functions, signaling pathways, or diseases by calculating the similarity of global annotation profiles using an agglomeration algorithm method. The functions of differentially expressed genes were analyzed following methods used in our previous studies (15,22).
Statistical analysis
The expression levels of the genes were compared between cells treated with hypoxia for different lengths of time (3, 24 and 48 h) and normoxic cells using ANOVA, followed by Dunnett's test. P<0.05 (two-tailed) was considered to indicate a statistically significant difference.
Results
Gene expression profiling and miRNA changes in hypoxic AC16 CMs
As shown in Fig. 1A, RNAs were extracted from the cells and sent for NGS followed by bioinformatics analyses. The AC16 CMs incubated under hypoxic conditions exhibited increased apoptosis compared with the cells incubated under normoxic conditions (Fig. 1B).
Figure 1.
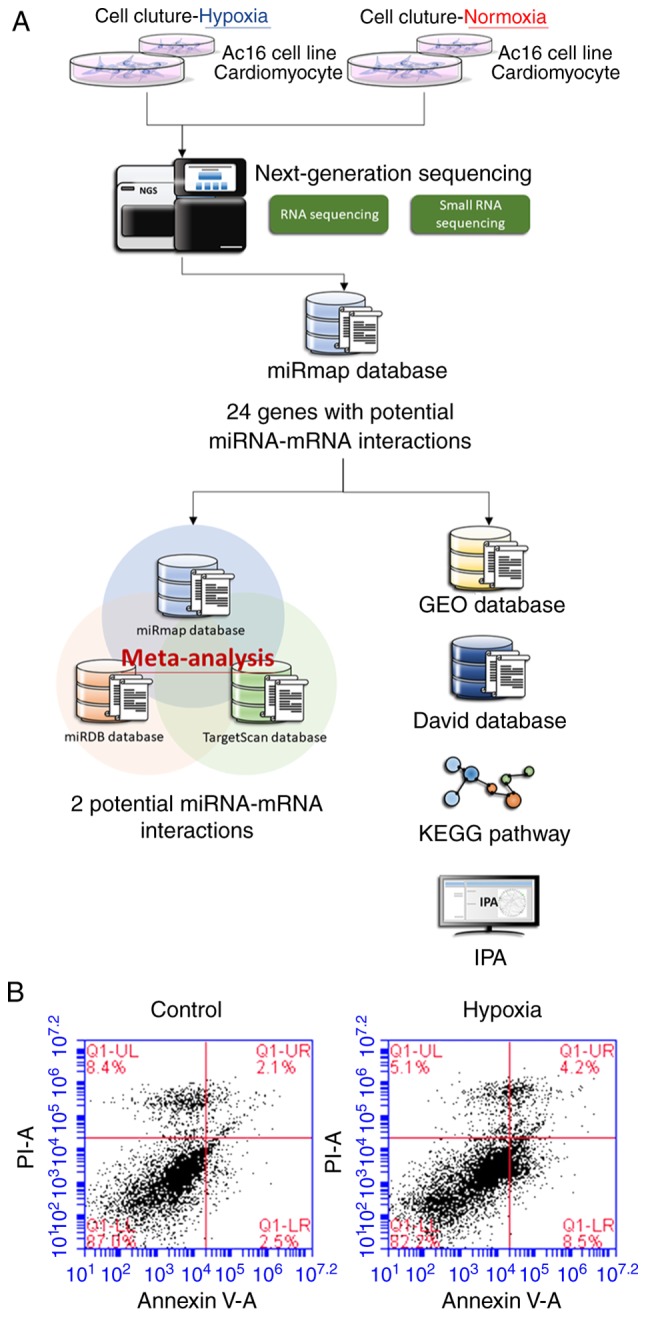
Investigating hypoxia-related changes in AC16 CMs. (A) Flowchart showing the methods for investigating various genetic expression changes in hypoxic AC16 CMs using next-generation sequencing and bioinformatics approaches. (B) AC16 CMs (5×105) were incubated in either a normoxic (control) or hypoxic condition for 24 h, and analyzed using a flow cytometer following staining with fluorescein isothiocyanate-conjugated Annexin V and propidium iodide. Increased apoptosis was noted in the hypoxic AC16 CMs. miRNA, microRNA; CMs, cardiomyocytes; DAVID, Database for Annotation, Visualization and Integrated Discovery; GEO, Gene expression omnibus; KEGG, Kyoto Encyclopedia of Genes and Genomes; IPA, Ingenuity® Pathway Analysis.
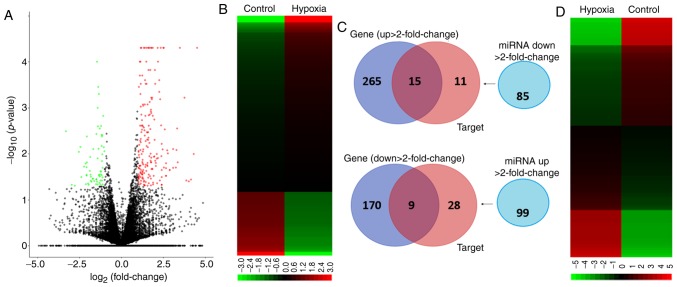
A volcano plot of differentially upregulated (right panel) and downregulated (left panel) gene expression in hypoxic, vs. normoxic AC16 CMs is shown in Fig. 2A. Genes with −log10 (P-value) >1.3 and >2-fold changes (Fig. 2B) were selected for further analyses. The small RNA-sequencing data generated with NGS were analyzed to identify potentially significant changes in miRNA profiles in the AC16 CMs under hypoxia, vs. normoxia. As shown in Fig. 2C and D, 184 miRNAs were identified with >2-fold changes (99 upregulated and 85 downregulated). Following selection of those with thresholds of >1 rpm in both hypoxic and normoxic cells, 62 miRNAs were identified, including 30 miRNAs with >2-fold upregulation and 32 miRNAs with >2-fold downregulation (Table I).
Figure 2.
Potential miRNA-mRNA interactions in hypoxic AC16 CMs. (A) Volcano plot of −log10 (P-value), vs. log2 (fold change) shows differentially upregu-lated and downregulated genes in hypoxic, vs. normoxic AC16 CMs; genes with −log10 (P-value) >1.3 and ≥2-fold changes are plotted in red (upregulated) and green (downregulated). (B) RNA-sequencing heatmap analysis showing differentially expressed genes (total 459 genes) with fold changes >2.0. (C) Genes Venn diagram shows that 280 genes were upregulated and 179 genes were downregulated in hypoxic AC16 CMs, vs. normoxic AC16 CMs; the miRNA Venn diagram showed that there were 85 downregulated miRNAs and 99 upregulated miRNAs in hypoxic AC16 CMs, vs. normoxic AC16 CMs; the Targets Venn diagram revealed the putative gene targets of the miRNAs with >2-fold changes predicted with miRmap database with threshold of miRmap scores >99.0. The intersection Venn diagram between Genes and Targets revealed 24 genes with potential miRNA-mRNA interactions. (D) Small RNA-sequencing heatmap analysis showing differentially expressed miRNAs (total 184 miRNAs) with fold changes >2. miRNA, microRNA; CMs, cardiomyocytes.
Table I.
miRNAs with significant change in hypoxic, vs. normoxic AC16 CMs.
| miRNA | Precursor | AC16 CMs hypoxia
|
AC16 CMs normoxia
|
Fold change | Up/down | |
|---|---|---|---|---|---|---|
| seq (norm) | seq (norm) | |||||
| hsa-miR-10b-3p | hsa-mir-10b | 2.81 | 1.03 | 2.73 | Up | |
| hsa-miR-1276 | hsa-mir-1276 | 4.84 | 1.84 | 2.63 | Up | |
| hsa-miR-142-5p | hsa-mir-142 | 6.08 | 2.18 | 2.79 | Up | |
| hsa-miR-181b-2-3p | hsa-mir-181b-2 | 8.33 | 3.33 | 2.50 | Up | |
| hsa-miR-210-3p | hsa-mir-210 | 392.50 | 122.50 | 3.20 | Up | |
| hsa-miR-210-5p | hsa-mir-210 | 23.52 | 8.49 | 2.77 | Up | |
| hsa-miR-212-5p | hsa-mir-212 | 6.19 | 1.95 | 3.17 | Up | |
| hsa-miR-224-3p | hsa-mir-224 | 4.61 | 1.15 | 4.01 | Up | |
| hsa-miR-26a-2-3p | hsa-mir-26a-2 | 7.43 | 2.75 | 2.70 | Up | |
| hsa-miR-26b-3p | hsa-mir-26b | 10.80 | 4.47 | 2.42 | Up | |
| hsa-miR-299-5p | hsa-mir-299 | 27.57 | 10.09 | 2.73 | Up | |
| hsa-miR-3200-3p | hsa-mir-3200 | 10.01 | 3.67 | 2.73 | Up | |
| hsa-miR-330-3p | hsa-mir-330 | 9.79 | 4.70 | 2.08 | Up | |
| hsa-miR-33a-3p | hsa-mir-33a | 3.15 | 1.26 | 2.50 | Up | |
| hsa-miR-34b-3p | hsa-mir-34b | 13.39 | 5.28 | 2.54 | Up | |
| hsa-miR-365b-5p | hsa-mir-365b | 3.60 | 1.15 | 3.13 | Up | |
| hsa-miR-3944-5p | hsa-mir-3944 | 2.48 | 1.15 | 2.16 | Up | |
| hsa-miR-454-5p | hsa-mir-454 | 14.40 | 5.73 | 2.51 | Up | |
| hsa-miR-485-3p | hsa-mir-485 | 22.84 | 5.51 | 4.15 | Up | |
| hsa-miR-486-3p | hsa-mir-486-1 | 8.78 | 2.64 | 3.33 | Up | |
| hsa-miR-486-3p | hsa-mir-486-2 | 8.89 | 2.29 | 3.88 | Up | |
| hsa-miR-494-3p | hsa-mir-494 | 18.00 | 8.83 | 2.04 | Up | |
| hsa-miR-548e-3p | hsa-mir-548e | 6.86 | 3.21 | 2.14 | Up | |
| hsa-miR-550a-3p | hsa-mir-550a-1 | 2.48 | 1.03 | 2.41 | Up | |
| hsa-miR-550a-3p | hsa-mir-550a-2 | 2.48 | 1.03 | 2.41 | Up | |
| hsa-miR-550a-3p | hsa-mir-550a-3 | 2.48 | 1.03 | 2.41 | Up | |
| hsa-miR-5690 | hsa-mir-5690 | 5.51 | 2.41 | 2.29 | Up | |
| hsa-miR-582-3p | hsa-mir-582 | 9.45 | 3.67 | 2.57 | Up | |
| hsa-miR-590-3p | hsa-mir-590 | 15.42 | 7.34 | 2.10 | Up | |
| hsa-miR-641 | hsa-mir-641 | 8.21 | 3.90 | 2.11 | Up | |
| hsa-miR-766-3p | hsa-mir-766 | 10.69 | 5.05 | 2.12 | Up | |
| hsa-miR-92b-5p | hsa-mir-92b | 11.59 | 5.51 | 2.10 | Up | |
| hsa-miR-98-3p | hsa-mir-98 | 8.78 | 4.36 | 2.01 | Up | |
| hsa-miR-1249-3p | hsa-mir-1249 | 2.48 | 5.28 | −2.13 | Down | |
| hsa-miR-1262 | hsa-mir-1262 | 1.35 | 3.56 | −2.64 | Down | |
| hsa-miR-1292-5p | hsa-mir-1292 | 1.24 | 2.52 | −2.03 | Down | |
| hsa-miR-1303 | hsa-mir-1303 | 2.59 | 5.51 | −2.13 | Down | |
| hsa-miR-1306-5p | hsa-mir-1306 | 1.69 | 4.01 | −2.37 | Down | |
| hsa-miR-134-3p | hsa-mir-134 | 1.58 | 3.44 | −2.18 | Down | |
| hsa-miR-1908-3p | hsa-mir-1908 | 1.01 | 2.29 | −2.27 | Down | |
| hsa-miR-23a-5p | hsa-mir-23a | 5.63 | 14.68 | −2.61 | Down | |
| hsa-miR-296-3p | hsa-mir-296 | 1.35 | 3.67 | −2.72 | Down | |
| hsa-miR-29a-5p | hsa-mir-29a | 2.48 | 6.54 | −2.64 | Down | |
| hsa-miR-3130-3p | hsa-mir-3130-1 | 1.13 | 4.59 | −4.06 | Down | |
| hsa-miR-3130-3p | hsa-mir-3130-2 | 1.13 | 4.59 | −4.06 | Down | |
| hsa-miR-3167 | hsa-mir-3167 | 2.93 | 6.08 | −2.08 | Down | |
| hsa-miR-3176 | hsa-mir-3176 | 9.34 | 22.94 | −2.46 | Down | |
| hsa-miR-33b-3p | hsa-mir-33b | 2.14 | 4.82 | −2.25 | Down | |
| hsa-miR-3615 | hsa-mir-3615 | 4.95 | 15.71 | −3.17 | Down | |
| hsa-miR-3619-5p | hsa-mir-3619 | 1.13 | 4.01 | −3.55 | Down | |
| hsa-miR-377-5p | hsa-mir-377 | 6.53 | 13.30 | −2.04 | Down | |
| hsa-miR-4454 | hsa-mir-4454 | 3.04 | 6.31 | −2.08 | Down | |
| hsa-miR-483-5p | hsa-mir-483 | 1.69 | 5.05 | −2.99 | Down | |
| hsa-miR-487a-5p | hsa-mir-487a | 1.35 | 3.10 | −2.30 | Down | |
| hsa-miR-503-5p | hsa-mir-503 | 4.28 | 10.44 | −2.44 | Down | |
| hsa-miR-541-3p | hsa-mir-541 | 2.14 | 7.46 | −3.49 | Down | |
| hsa-miR-548d-5p | hsa-mir-548d-1 | 2.59 | 11.70 | −4.52 | Down | |
| hsa-miR-548d-5p | hsa-mir-548d-2 | 3.26 | 13.65 | −4.19 | Down | |
| hsa-miR-548n | hsa-mir-548n | 2.81 | 5.73 | −2.04 | Down | |
| hsa-miR-548o-3p | hsa-mir-548o | 13.73 | 27.87 | −2.03 | Down | |
| hsa-miR-548o-3p | hsa-mir-548o-2 | 13.73 | 27.87 | −2.03 | Down | |
| hsa-miR-551b-3p | hsa-mir-551b | 1.13 | 2.87 | −2.54 | Down | |
| hsa-miR-6840-5p | hsa-mir-6840 | 1.46 | 2.98 | −2.04 | Down | |
| hsa-miR-6852-5p | hsa-mir-6852 | 2.03 | 4.36 | −2.15 | Down | |
| hsa-miR-877-5p | hsa-mir-877 | 9.00 | 18.24 | −2.03 | Down | |
| hsa-miR-887-5p | hsa-mir-887 | 1.01 | 3.10 | −3.07 | Down | |
| hsa-miR-92a-1-5p | hsa-mir-92a-1 | 1.35 | 4.01 | −2.97 | Down | |
| hsa-miR-93-3p | hsa-mir-93 | 1.58 | 7.57 | −4.79 | Down | |
miRNA/miR, microRNA; Up, upregulated; Down, downregulated.
Identification of potential miRNA-mRNA interactions in hypoxic AC16 CMs
The present study aimed to determine the potential miRNA-mRNA interactions in hypoxic AC16 CMs. To achieve this, the putative targets of the miRNAs with >2-fold changes were searched (Fig. 2D) from the NGS results using a miRmap database search for those with threshold miRmap scores >99.0. The genes showing >2-fold changes (Fig. 2B) were matched with these putative targets, and, as shown in the intersection Venn diagram in Fig. 2C, 24 genes were potentially involved in miRNA-mRNA interactions (15 upregulated and nine downregulated) (Table II).
Table II.
Genes selected by intersection between RNA sequencing candidates and microRNA putative targets.
| Official gene symbol | Gene name | Log2 ratio (hypoxia/control) | Gene expression | miRNA with putative interaction |
|---|---|---|---|---|
| SLC6A13 | Solute carrier family 6 member 13 | 11.77 | Up | hsa-miR-3619-5p |
| HIF3A | Hypoxia inducible factor 3 α subunit | 3.29 | Up | hsa-miR-1254, |
| hsa-miR-377-5p, | ||||
| hsa-miR-615-5p | ||||
| EPB49 | Dematin actin binding protein | 2.60 | Up | hsa-miR-1254 |
| APLN | Apelin | 2.16 | Up | hsa-miR-3619-5p, |
| hsa-miR-503-5p | ||||
| ACVRL1 | Activin A receptor like type 1 | 2.11 | Up | hsa-miR-3619-5p |
| SLC2A5 | Solute carrier family 2 member 5 | 1.98 | Up | hsa-miR-3619-5p |
| STC1 | Stanniocalcin 1 | 1.77 | Up | hsa-miR-541-3p, |
| hsa-miR-615-5p | ||||
| SLC6A8 | Solute carrier family 6 member 8 | 1.64 | Up | hsa-miR-541-3p |
| TNS1 | Tensin 1 | 1.41 | Up | hsa-miR-3176 |
| CAMK1D | Calcium/calmodulin dependent protein kinase ID | 1.24 | Up | hsa-miR-1254 |
| C1QL1 | Complement C1q like 1 | 1.20 | Up | hsa-miR-541-3p |
| GPR68 | G protein-coupled receptor 68 | 1.14 | Up | hsa-miR-4741 |
| CD4 | CD4 molecule | 1.13 | Up | hsa-miR-4741 |
| MME | Membrane metalloendopeptidase | 1.09 | Up | hsa-miR-1303 |
| BNIP3L | BCL2 interacting protein 3 like | 1.08 | Up | hsa-miR-93-3p |
| APOL6 | Apolipoprotein L6 | −1.09 | Down | hsa-miR-4421 |
| METTL7A | Methyltransferase like 7A | −1.13 | Down | hsa-miR-4421 |
| TFRC | Transferrin receptor | −1.17 | Down | hsa-miR-296-5p |
| BCL2 | BCL2, apoptosis regulator | −1.26 | Down | hsa-miR-296-5p |
| CHAC1 | Chac glutathione specific | −1.44 | Down | hsa-miR-3918 |
| γ-glutamylcyclotransferase 1 | ||||
| DIO2 | Iodothyronine deiodinase 2 | −1.48 | Down | hsa-miR-4717-3p |
| SLC7A11 | Solute carrier family 7 member 11 | −1.50 | Down | hsa-miR-3129-3p, |
| hsa-miR-4768-5p, | ||||
| hsa-miR-3074-5p, | ||||
| hsa-miR-590-3p | ||||
| TMEM144 | Transmembrane protein 144 | −1.84 | Down | hsa-miR-582-3p |
| HDAC11 | Histone deacetylase 11 | −2.11 | Down | hsa-miR-3918, |
| hsa-miR-766-3p |
miR, microRNA.
A search was also performed for the potential miRNA-mRNA interactions of the miRNAs with >2-fold changes in various miRNA target predicting databases. On combining the results from miRmap, TargetScan and miRDB, two miRNA-mRNA interactions were identified: hsa-miR-129-5p/CDC42EP3 and hsa-miR-330-3p/HELZ (Fig. 3). However, although hsa-miR-129-5p and hsa-miR-330-3p were significantly upregulated in hypoxic CMs, no significant differences in the expression levels of CDC42EP3 or HELZ were noted between the hypoxic and normoxic CMs.
Figure 3.

Potential miRNA-mRNA interactions from miRmap, TargetScan and miRDB databases. miRmap (selection criteria: miRmap score >99.0), TargetScan and miRDB (selection criteria: miRDB score >99.0) were used to identify genes with potential miRNA-mRNA interactions of the miRNAs with >2-fold changes. The intersection of three databases revealed two potential miRNA-mRNA interactions: hsa-miR-129-5p/CDC42EP3 and hsa-miR-330-3p/HELZ. miRNA, microRNA.
Validation of the differentially expressed genes in response to hypoxia
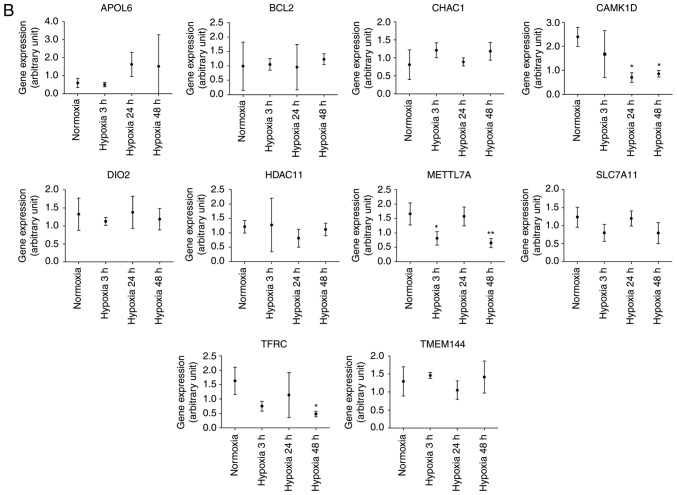
The 24 genes with potential miRNA-mRNA interactions (Table II) were further compared against associated array data obtained from the GEO database. Briefly, the gene expression levels in cardiac microvascular endothelial cells were extracted from the GEO database (GEO accession: GDS3483) (27). The expression levels of the genes were compared between cells treated with hypoxia over different lengths of time (3, 24 and 48 h) and those of normoxic cells. The findings were partially similar to those from the NGS CM data. In hypoxic cardiac microvascular endothelial cells, tensin 1 (TNS1), B-cell lymphoma 2 (BCL2)/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3L), and stanniocalcin 1 (STC1) were significantly upregulated (Fig. 4A) and transferrin receptor (TFRC) and methyltransferase like 7A (METTL7A) were significantly downregulated (Fig. 4B). However, unlike the findings based on NGS CM data, Calcium/calmodulin dependent protein kinase 1D (CAMK1D) was significantly downregulated.
Figure 4.
GEO database analysis of genes with potential miRNA-mRNA interactions. The 24 genes with potential miRNA-mRNA interactions, including (A) upregulated and (B) downregulated genes in hypoxic cardiomyocytes are listed in Table II. They were further validated using related array data from the GEO database. The gene expression levels in cardiac microvascular endothelial cells were extracted from the GEO database (GEO accession: GDS3483), and the expression levels of the genes were compared between cells treated with hypoxia for different lengths of time (3, 24 and 48 h) and normoxic cells. *P<0.05; **P<0.01; ***P<0.001. miRNA, microRNA; GEO, Gene Expression Omnibus.
GO annotations and KEGG pathway analyses of DEGs in hypoxic AC16 CMs
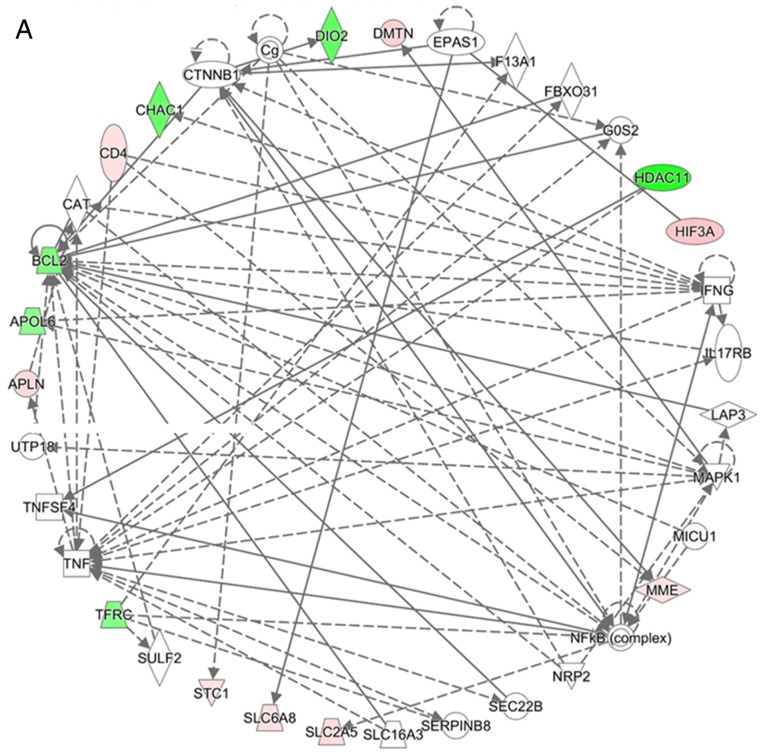
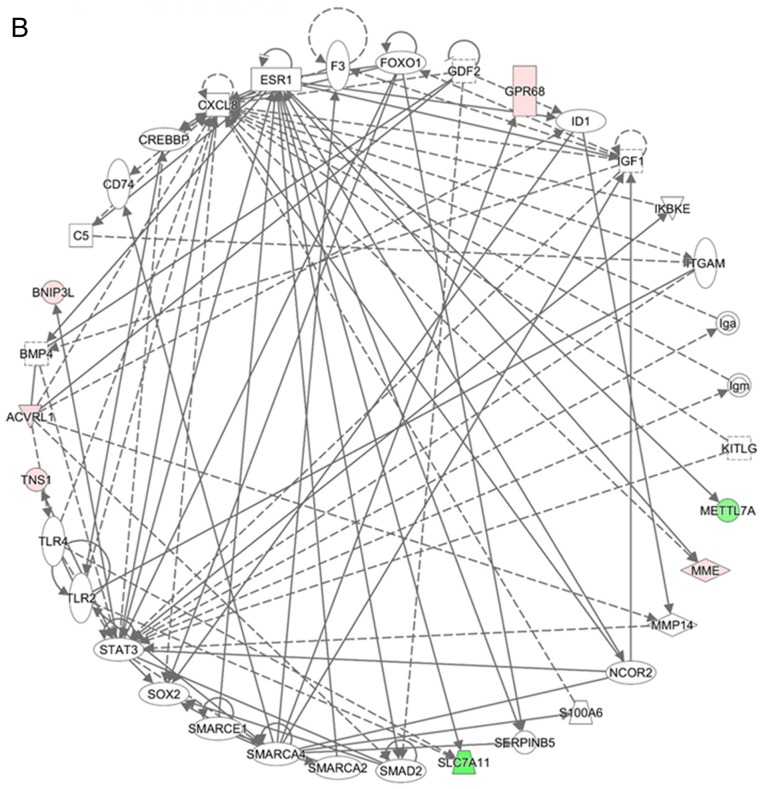
Network analyses of the 24 genes with potential miRNA-mRNA interactions were performed using IPA software. The top three networks associated with genes targeted by miRNAs differentially expressed in hypoxic AC16 CMs are listed in Table III. In network 1 (Fig. 5A), 14 targeted genes (iodothyronine deiodinase 2, solute carrier family 2 member 5, apolipoprotein L6, histone deacetylase 11, STC1, membrane metalloendopeptidase (MME), chac glutathione specific γ-glutamylcyclotransferase 1, apelin, TFRC, CD4 molecule, hypoxia inducible factor 3 α subunit, BCL2, dematin actin binding protein, and solute carrier family 6 member 13) were associated with free radical scavenging, small molecule biochemistry, and carbohydrate metabolism. In network 2 (Fig. 5B), seven targeted genes (solute carrier family 7 member 11, activin A receptor like type 1, BNIP3L, MME, METTL7A, TNS1, and G protein-coupled receptor 68) were associated with cancer, organismal injury and abnormalities, and cell death and survival. In network 3, only CAMK1D was associated with carbohydrate metabolism, small molecule biochemistry, and digestive system development and function. Two genes (complement C1q like 1 and transmembrane protein 144) were not involved in these networks.
Table III.
Top three networks associated with genes targeted by microRNAs differentially expressed in hypoxic AC16 cardiomyocytes.
| Network | Top diseases and functions | Score | Focus molecules | Molecules in network |
|---|---|---|---|---|
| 1 | Free radical scavenging, small molecule biochemistry, carbohydrate metabolism | 32 | 14 | aAPLN, aAPOL6, aBCL2, CAT, aCD4, Cg, aCHAC1, CTNNB1, aDIO2, aDMTN, EPAS1, F13A1, FBXO31, G0S2, aHDAC11, aHIF3A, IFNG, IL17RB, LAP3, MAPK1, MICU1, aMME, NFkB(complex), NRP2, SEC22B, SERPINB8, SLC16A3, aSLC2A5, aSLC6A8, aSTC1, SULF2, aTFRC, TNF, TNFSF4, UTP18 |
| 2 | Cancer, organismal injury and abnormalities, cell death and survival | 13 | 7 | aACVRL1, BMP4, aBNIP3L, C5, CD74, CREBBP, CXCL8, ESR1, F3, FOXO1, GDF2, aGPR68, ID1, Iga, IGF1, Igm, IKBKE, ITGAM, KITLG, aMETTL7A, aMME, MMP14, NCOR2, S100A6, SERPINB5, aSLC7A11, SMAD2, SMARCA2, SMARCA4, SMARCE1, SOX2, STAT3, TLR2, TLR4, aTNS1 |
| 3 | Carbohydrate metabolism, small molecule biochemistry, digestive system development and function | 2 | 1 | aCAMK1D, ERG, GCG, INS |
Focus molecules.
Figure 5.
Networks analysis of the 24 differentially expressed genes with potential miRNA-mRNA interactions in hypoxic AC16 CMs. The network analyses of the 24 differentially expressed genes with potential miRNA-mRNA interactions in hypoxic AC16 CMs were investigated using Ingenuity® Pathway Analysis software. (A) In network 1, 14 targeted genes (DIO2, SLC2A5, APOL6, HDAC11, STC1, MME, CHAC1, APLN, TFRC, CD4, HIF3A, BCL2, DMTN, and SLC6A8) were associated with free radical scavenging, small molecule biochemistry, and carbohydrate metabolism. (B) In network 2, seven targeted genes (SLC7A11, ACVRL1, BNIP3L, MME, METTL7A, TNS1, and GPR68) were associated with cancer, organismal injury and abnormalities, and cell death and survival. In network 3, only CAMK1D was associated with carbohydrate metabolism, small molecule biochemistry, and digestive system development and function. CMs, cardiomyocytes; miRNA, microRNA. Diamonds, horizontal ovals, vertical ovals, squares, rectangles, trapezoids, and inverted triangles represent enzymes, transcription regulators, transmembrane receptors, cytokines, ligand-dependent nuclear receptors, transporters, and kinases, respectively. Solid and dashed lines represent direct and indirect action, respectively. Red and green symbols represent upregulated and downregulated molecules, respectively.
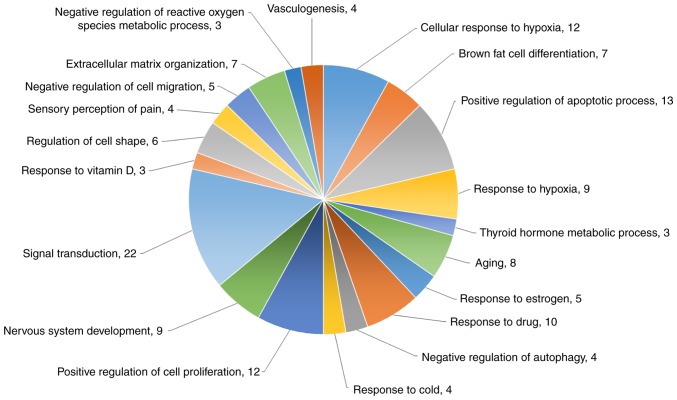
Functional analysis of the DEGs from mRNA sequencing was also performed by DAVID biological process analysis (Fig. 6). The top four biological processes of these genes were cellular response to hypoxia (12 genes), brown fat cell differentiation (seven genes), positive regulation of apoptotic process (13 genes), and response to hypoxia (nine genes). The protein-coding genes specifically associated with apoptosis, cell proliferation inhibition, and cell cycle arrest were also analyzed with a heatmap (Fig. 7).
Figure 6.
Top 20 biological processes of the DEGs in hypoxic AC16 CMs. The top 20 biological processes of DEGs in hypoxic AC16 CMs are presented in a pie chart. numbers represent numbers of genes in the biological process. The top four biological processes of these genes were cellular response to hypoxia (12 genes), brown fat cell differentiation (seven genes), positive regulation of apoptotic process (13 genes), and response to hypoxia (nine genes). CMs, cardio-myocytes; DEGs, differentially expressed genes.
Figure 7.

Heatmap of the differential expression of genes specifically associated with apoptosis, cell proliferation inhibition, and cell cycle arrest in hypoxic, vs. normoxic AC16 cardiomyocytes. The upregulated and down-regulated genes are shown in red and green, respectively.
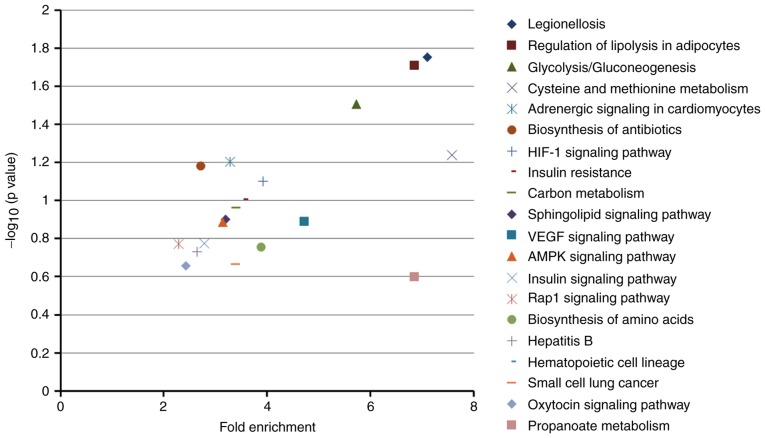
The top 20 KEGG pathways of dysregulated genes were identified using mRNA sequencing data (Table IV and Fig. 8). The legionellosis (fold enrichment=7.11), regulation of lipolysis in adipocytes (fold enrichment=6.86), and glycolysis/gluco-neogenesis (fold enrichment=5.73) pathways were significantly enriched in the hypoxic CMs.
Table IV.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of the dysregulated genes (top 20) identified from mRNA sequencing.
| Description | Count P | -value | Gene upregulated | Gene downregulated | Fold enrichment |
|---|---|---|---|---|---|
| Legionellosis | 4 | 0.02 | ITGB2, EEF1A2, BNIP3 | CXCL2 | 7.11 |
| Regulation of lipolysis in adipocytes | 4 | 0.02 | AQP7, PTGS2 | ADORA1, PIK3R3 | 6.86 |
| Glycolysis/gluconeogenesis | 4 | 0.03 | LDHA, TPI1, PGK1 | LDHC | 5.73 |
| Cysteine and methionine metabolism | 3 | 0.06 | LDHA | SDSL, LDHC | 7.58 |
| Adrenergic signaling in cardiomyocytes | 5 | 0.06 | CACNG6, RAPGEF4, | BCL2, PIK3R3 | 3.29 |
| PPP2R5B | |||||
| Biosynthesis of antibiotics | 6 | 0.07 | LDHA, TPI1, PGK1, RGN | SDSL, LDHC | 2.72 |
| HIF-1 signaling pathway | 4 | 0.08 | VEGFA | TFRC, BCL2, PIK3R3 | 3.92 |
| Insulin resistance | 4 | 0.10 | PPP1R3B, PPP1R3C | PPARGC1A, PIK3R3 | 3.55 |
| Carbon metabolism | 4 | 0.11 | TPI1, PGK1, RGN | SDSL | 3.40 |
| Sphingolipid signaling pathway | 4 | 0.13 | PPP2R5B | ADORA1, BCL2 PIK3R3 | 3.20 |
| VEGF signaling pathway | 3 | 0.13 | VEGFA, PTGS2 | PIK3R3 | 4.72 |
| AMPK signaling pathway | 4 | 0.13 | PFKFB4, PPP2R5B | PPARGC1A, PIK3R3 | 3.15 |
| Insulin signaling pathway | 4 | 0.17 | PPP1R3B, PPP1R3C | PPARGC1A, PIK3R3 | 2.78 |
| Rap1 signaling pathway | 5 | 0.17 | ITGB2, VEGFA, RAPGEF4, | PIK3R3 | 2.29 |
| Biosynthesis of amino acids | 3 | 0.18 | TPI1, PGK1 | SDSL | 3.89 |
| Hepatitis B | 4 | 0.19 | BCL2, TLR3, PIK3R3, IFIH1 | 2.65 | |
| Hematopoietic cell lineage | 3 | 0.22 | MME, CD4 | TFRC | 3.39 |
| Small cell lung cancer | 3 | 0.22 | PTGS2 | BCL2, PIK3R3 | 3.39 |
| Oxytocin signaling pathway | 4 | 0.22 | PTGS2, CACNG6, CAMK1D | PIK3R3 | 2.43 |
Significant values are shown in bold.
Figure 8.
Enrichment analysis of KEGG pathways from differentially expressed genes in hypoxic AC16 CMs. In the graph, the top 20 KEGG pathways (details listed in Table IV) were plotted with −log10 (P-value), vs. fold enrichment. Higher fold enrichment values and −log10 (P-value) indicate that the pathway is expressed at a higher level and is more reliable. Legionellosis, regulation of lipolysis in adipocytes, glycolysis/gluconeogenesis pathways were significantly enriched in hypoxic AC16 CMs. KEGG, Kyoto Encyclopedia of Genes and Genomes; CMs, cardiomyocytes.
Discussion
Differential gene expression with altered signaling pathways has been reported in CMs responding to hypoxic/ischemic conditions (3,29,30). The reciprocal regulation of miRNAs and
mRNA in human CMs from patients with heart failure has also been investigated using microarray analysis (31). The present study comprehensively surveyed the differential expression of mRNAs and miRNAs and potential mRNA-miRNA interactions using NGS techniques and bioinformatics.
miRNAs are short, small non-coding RNAs, which regulate gene expression via several post-transcriptional processes (32). Hypoxia is a stress condition that may provoke multiple biological and molecular regulatory networks associated with apoptosis, cell proliferation, and alterations in metabolism (32). Previous studies have suggested that miRNAs may potentially be used as novel diagnostic makers and therapeutic targets for hypoxic CMs in clinical scenarios, including AMI (33). The present study found 62 miRNAs with >2-fold changes in hypoxic CMs. Based on the miRNA-mRNA interactions predicted using miRmap, TargetScan and miRDB databases, hsa-miR-129-5p and hsa-miR-330-3p were identified as having the highest potential of becoming such biomarkers. The overexpression of miR-129-5p inhibits cell proliferation in smooth muscle cells, glioblastoma multiforme cells, gastric cancer cells, and H9C2 CMs (34-37). Previous experimental studies have noted wingless-type MMTV integration site family member 5A, collagen, type I, a1, and cyclin-dependent kinase 6 to be the targets of miR-129-5p (34-37). The overexpression of miR-330 has been found to inhibit cell proliferation through B cell-specific Moloney murine leukemia virus integration site 1, E2F transcription factor 1 and musashi RNA binding protein 1 in osteosarcoma cells, gastric cells, and prostate cancer cells, respectively (38-40). However, miR-330 has also found to increase cell proliferation by regulating SH3-domain GRB2-like 2 and WNT signaling pathways in human glioblastoma cells and vascular endothelial cells, respectively (41,42). Although the expression levels of these genes exhibited no significant changes in AC16 CMs exposed to hypoxia, the downstream regulatory mechanisms of hsa-miR-129-5p and hsa-miR-330-3p in hypoxic CMs warrant further investigation.
The present study identified 24 genes with significant changes and potential miRNA-mRNA interactions. These genes were associated with important cellular functions, including cell proliferation, apoptosis, and carbohydrate metabolism. Using the gene expression profiles of cardiac microvascular endothelial cells obtained from the GEO database, it was possible to validate the significant upregulation of TNS1, BNIP3L and STC1, and the significant downregulation of TFRC and METTL7A in response to hypoxia. CAMK1D exhibited paradoxical changes in different cells.
TNS1 is a focal adhesion molecule which serves as a scaffold for cell mobility and adhesion through binding to fibronectin and β1-integrin (43,44). The overexpression of TNS1 has been found to contribute to the invasion and metastasis of breast cancer cells (45). TNS1 may also be involved in the micro-environmental changes that occur in the myocardium during hypoxia.
Programmed cell death may occur in CMs via the extrinsic cytokine death pathway or the intrinsic mitochondrial pruning pathway (46,47). The BNIP3 and BNIP3-like (BNIP3L, also known as NIP3-like protein X, NIP3L, or NIX), belong to the pro-apoptotic Bcl-2 protein family and are mitochondrial stress sensors (48). They can promote mitophagy and autophagy in response to hypoxia (49,50). Upregulated BNIP3 and BNIP3L trigger apoptosis in conditions which include myocardial hypertrophy and are important in cardiac remodelling (51,52). Therefore, these pathways have been investigated for their potential role in inhibiting ischemic CM apoptosis (53). Similarly, the present study found BNIP3L to be upregulated in hypoxic CMs and cardiac microvascular endothelial cells.
STC1, an endogenous glycoprotein, has anti-inflammatory and anti-oxidative effects through the uncoupling protein (UCP) family in mice kidney and lung injuries (54,55). However, its main role in CMs remains to be elucidated. STC1 has been found to be significantly increased in patients with dilated cardiomyopathy, but normalizes following treatment using a left ventricular assist device (56). STC1 inhibits super-oxide generation via the induction of UCP3 in CMs, which may ameliorate angiotensin II-mediated cardiac injury (57). However, the cardiotoxic effects of STC1 also appear to be mediated via mitochondrial injuries through loss of integrity of the mitochondrial membrane, increased mitochondrial calcium levels, and reactive oxygen species production (58). In the present study, STC1 was upregulated in hypoxic CMs and cardiac microvascular endothelial cells. Whether this up regulation of STC1 was associated with anti-oxidative effects or pro-oxidative effects warrants further investigation.
The present study revealed significant downregulation of TFRC and METTL7A in hypoxic CMs and cardiac microvascular endothelial cells. Previous studies have shown that either iron overloading or deficiency in CMs were associated with cardiotoxicity and heart failure. In mice, the lack of TFRC results in poor cardiac and mitochondrial function, and early death (59). Decreased expression of TFRC has been associated with reduced pulmonary smooth muscle cell proliferation in hypoxia (60). METTL7A has been recognized as a novel tumor suppressor gene (61). The actual role of the downregulation of METTL7A in CMs and cardiac microvascular endothelial cells in response to hypoxia remains uncertain and requires further investigation.
CAMK1D is a key regulator of granulocyte function and has been associated with newly-developed type 2 diabetes mellitus in Japan (62,63). The overexpression of CAMK1D results in increased cell proliferation and epithelial-mesenchymal transition activity in breast epithelial cells (64). In the present study, it was found that CAMK1D was significantly upregulated in hypoxic CMs but significantly downregulated in hypoxic cardiac microvascular endothelial cells. Although the reason for this paradoxical finding remains to be fully elucidated, it was hypothesized that the overexpression of CAMK1D in hypoxic CMs may be involved in alterations in carbohydrate metabolism and cell detachment.
In conclusion, the present study reports on the differentially upregulated/downregulated mRNAs and miRNAs in hypoxic human CMs (Fig. 9). An improved understanding of the mRNA and miRNA profiles, in addition to the potential miRNA-mRNA interactions, in hypoxic CMs, enables the development of novel diagnostic tools and therapeutic strategies for hypoxic human CMs as is found in AMI.
Figure 9.
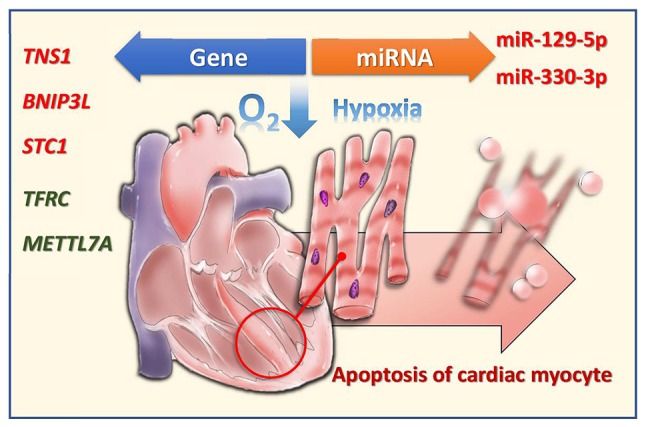
Changes in miRNAs and mRNAs in hypoxic AC16 cardiomyocytes. miRNA/miR, microRNA; TNS1, tensin 1; BNIP3L, B-cell lymphoma 2-interacting protein 3 like; STC1, stanniocalcin 1; TFRC, transferrin receptor; METTL7A, methyltransferase like 7A.
Acknowledgments
The authors thank the Center for Research Resources and Development of Kaohsiung Medical University (Kaohsiung, Taiwan).
Funding
The present study was supported by research grants from the Ministry of Science and Technology (grant no. MOST 107-2320-B-037-011-MY3), Kaohsiung Medical University Hospital (grant nos. KMUHS10701 and KMUHS10712), and the Kaohsiung Medical University (grant no. KMU-DK108003).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HMS and PLK conceived the study. WHL, MJT, WAC, KFC, and PLK analyzed and interpreted the data. LYW and HYW performed the cell culture and flow cytometry. WHL, MJT and PLK were the major contributors in writing the manuscript. WAC produced the illustration. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors confirm that they have no competing interests.
References
- 1.Lee WH, Hsu PC, Chu CY, Su HM, Lee CS, Yen HW, Lin TH, Voon WC, Lai WT, Sheu SH. Cardiovascular events in patients with atherothrombotic disease: A population-based longitudinal study in taiwan. PLoS One. 2014;9:e92577. doi: 10.1371/journal.pone.0092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soler EP, Ruiz VC. Epidemiology and risk factors of cerebral ischemia and ischemic heart diseases: Similarities and differences. Curr Cardiol Rev. 2010;6:138–149. doi: 10.2174/157340310791658785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang PM, Haunstetter A, Aoki H, Usheva A, Izumo S. Morphological and molecular characterization of adult cardiomyocyte apoptosis during hypoxia and reoxygenation. Circ Res. 2000;87:118–125. doi: 10.1161/01.RES.87.2.118. [DOI] [PubMed] [Google Scholar]
- 4.Sun T, Dong YH, Du W, Shi CY, Wang K, Tariq MA, Wang JX, Li PF. The Role of MicroRNAs in myocardial infarction: From molecular mechanism to clinical application. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18040745. pii: E745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adachi S, Ito H, Tamamori-Adachi M, Ono Y, Nozato T, Abe S, Ikeda Ma, Marumo F, Hiroe M. Cyclin A/cdk2 activation is involved in hypoxia-induced apoptosis in cardiomyocytes. Circ Res. 2001;88:408–414. doi: 10.1161/01.RES.88.4.408. [DOI] [PubMed] [Google Scholar]
- 6.Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli-Ducci C, Bueno H, Caforio ALP, Crea F, Goudevenos JA, Halvorsen S, et al. 2017 ESC Guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation: The Task Force for the management of acute myocardial infarction in patients presenting with ST-segment elevation of the European Society of Cardiology (ESC) Eur Heart J. 2018;39:119–177. doi: 10.1093/eurheartj/ehx393. [DOI] [PubMed] [Google Scholar]
- 7.Krijnen PA, Nijmeijer R, Meijer CJ, Visser CA, Hack CE, Niessen HW. Apoptosis in myocardial ischaemia and infarction. J Clin Pathol. 2002;55:801–811. doi: 10.1136/jcp.55.11.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takemura G, Fujiwara H. Morphological aspects of apoptosis in heart diseases. J Cell Mol Med. 2006;10:56–75. doi: 10.1111/j.1582-4934.2006.tb00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biondi-Zoccai GG, Abbate A, Vasaturo F, Scarpa S, Santini D, Leone AM, Parisi Q, De Giorgio F, Bussani R, Silvestri F, et al. Increased apoptosis in remote non-infarcted myocardium in multivessel coronary disease. Int J Cardiol. 2004;94:105–110. doi: 10.1016/j.ijcard.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Abbate A, Melfi R, Patti G, Baldi F, D'Ambrosio A, Manzoli A, Baldi A, Di Sciascio G. Apoptosis in recent myocardial infarction. Clin Ter. 2000;151:247–251. [PubMed] [Google Scholar]
- 11.Saraste A, Pulkki K, Kallajoki M, Henriksen K, Parvinen M, Voipio-Pulkki LM. Apoptosis in human acute myocardial infarction. Circulation. 1997;95:320–323. doi: 10.1161/01.CIR.95.2.320. [DOI] [PubMed] [Google Scholar]
- 12.Abbate A, Bussani R, Biondi-Zoccai GG, Santini D, Petrolini A, De Giorgio F, Vasaturo F, Scarpa S, Severino A, Liuzzo G, et al. Infarct-related artery occlusion, tissue markers of ischaemia, and increased apoptosis in the peri-infarct viable myocardium. Eur Heart J. 2005;26:2039–2045. doi: 10.1093/eurheartj/ehi419. [DOI] [PubMed] [Google Scholar]
- 13.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Mardis ER. Next-generation sequencing platforms. Annu Rev Anal Chem (Palo Alto Calif) 2013;6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 15.Chen SC, Chen FW, Hsu YL, Kuo PL. Systematic analysis of transcriptomic profile of renal cell carcinoma under long-term hypoxia using next-generation sequencing and bioinformatics. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18122657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blue GM, Kirk EP, Giannoulatou E, Dunwoodie SL, Ho JW, Hilton DC, White SM, Sholler GF, Harvey RP, Winlaw DS. Targeted next-generation sequencing identifies pathogenic variants in familial congenital heart disease. J Am Coll Cardiol. 2014;64:2498–2506. doi: 10.1016/j.jacc.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 17.Lubitz SA, Ellinor PT. Next-generation sequencing for the diagnosis of cardiac arrhythmia syndromes. Heart Rhythm. 2015;12:1062–1070. doi: 10.1016/j.hrthm.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia P, Liu Y, Cheng Z. Signaling Pathways in Cardiac Myocyte Apoptosis. Biomed Res Int. 2016;2016:9583268. doi: 10.1155/2016/9583268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiong M, Wang ZV, Pedrozo Z, Cao DJ, Troncoso R, Ibacache M, Criollo A, Nemchenko A, Hill JA, Lavandero S. Cardiomyocyte death: Mechanisms and translational implications. Cell Death Dis. 2011;2:e244. doi: 10.1038/cddis.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regula KM, Ens K, Kirshenbaum LA. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res. 2002;91:226–231. doi: 10.1161/01.RES.0000029232.42227.16. [DOI] [PubMed] [Google Scholar]
- 21.Yen MC, Shih YC, Hsu YL, Lin ES, Lin YS, Tsai EM, Ho YW, Hou MF, Kuo PL. Isolinderalactone enhances the inhibition of SOCS3 on STAT3 activity by decreasing miR-30c in breast cancer. Oncol Rep. 2016;35:1356–1364. doi: 10.3892/or.2015.4503. [DOI] [PubMed] [Google Scholar]
- 22.Sheu CC, Tsai MJ, Chen FW, Chang KF, Chang WA, Chong IW, Kuo PL, Hsu YL. Identification of novel genetic regulations associated with airway epithelial homeostasis using next-generation sequencing data and bioinformatics approaches. Oncotarget. 2017;8:82674–82688. doi: 10.18632/oncotarget.19752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vejnar CE, Zdobnov EM. MiRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012;40:11673–11683. doi: 10.1093/nar/gks901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costello CM, Howell K, Cahill E, McBryan J, Konigshoff M, Eickelberg O, Gaine S, Martin F, McLoughlin P. Lung-selective gene responses to alveolar hypoxia: Potential role for the bone morphogenetic antagonist gremlin in pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2008;295:L272–L284. doi: 10.1152/ajplung.00358.2007. [DOI] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kacimi R, Chentoufi J, Honbo N, Long CS, Karliner JS. Hypoxia differentially regulates stress proteins in cultured cardiomyocytes: Role of the p38 stress-activated kinase signaling cascade, and relation to cytoprotection. Cardiovasc Res. 2000;46:139–150. doi: 10.1016/S0008-6363(00)00007-9. [DOI] [PubMed] [Google Scholar]
- 30.Koeppen M, Lee JW, Seo SW, Brodsky KS, Kreth S, Yang IV, Buttrick PM, Eckle T, Eltzschig HK. Hypoxia-inducible factor 2-alpha-dependent induction of amphiregulin dampens myocardial ischemia-reperfusion injury. Nat Commun. 2018;9:816. doi: 10.1038/s41467-018-03105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., II Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nallamshetty S, Chan SY, Loscalzo J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radic Biol Med. 2013;64:20–30. doi: 10.1016/j.freeradbiomed.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ong SB, Katwadi K, Kwek XY, Ismail NI, Chinda K, Ong SG, Hausenloy DJ. Non-coding RNAs as therapeutic targets for preventing myocardial ischemia-reperfusion injury. Expert Opin Ther Targets. 2018;22:247–261. doi: 10.1080/14728222.2018.1439015. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu Z, Zhou M, Liu C. MicroRNA-129-5p inhibits vascular smooth muscle cell proliferation by targeting Wnt5a. Exp Ther Med. 2016;12:2651–2656. doi: 10.3892/etm.2016.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng A, Yin J, Li Y, Li R, Wang Z, Zhou X, Jin X, Shen F, Yan W, You Y. miR-129-5p targets Wnt5a to block PKC/ERK/NF-kappaB and JNK pathways in glioblastoma. Cell Death Dis. 2018;9:394. doi: 10.1038/s41419-018-0343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majumdar G, Raghow R. Trichostatin A induces a unique set of microRNAs including miR-129-5p that blocks cyclin-dependent kinase 6 expression and proliferation in H9c2 cardiac myocytes. Mol Cell Biochem. 2016;415:39–49. doi: 10.1007/s11010-016-2675-4. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol. 2018;96:19–25. doi: 10.1139/bcb-2016-0254. [DOI] [PubMed] [Google Scholar]
- 38.Zheng Z, Bao F, Chen X, Huang H, Zhang X. MicroRNA-330-3p Expression Indicates Good Prognosis and Suppresses Cell Proliferation by Targeting Bmi-1 in Osteosarcoma. Cell Physiol Biochem. 2018;46:442–450. doi: 10.1159/000488612. [DOI] [PubMed] [Google Scholar]
- 39.Guan A, Wang H, Li X, Xie H, Wang R, Zhu Y, Li R. MiR-330-3p inhibits gastric cancer progression through targeting MSI1. Am J Transl Res. 2016;8:4802–4811. [PMC free article] [PubMed] [Google Scholar]
- 40.Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT, Goan YG, Lu PJ. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene. 2009;28:3360–3370. doi: 10.1038/onc.2009.192. [DOI] [PubMed] [Google Scholar]
- 41.Qu S, Yao Y, Shang C, Xue Y, Ma J, Li Z, Liu Y. MicroRNA-330 is an oncogenic factor in glioblastoma cells by regulating SH3GL2 gene. PLoS One. 2012;7:e46010. doi: 10.1371/journal.pone.0046010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Ren J, Ma R, Zhang ZB, Li Y, Lei P, Men JL. Effects of microRNA-330 on vulnerable atherosclerotic plaques formation and vascular endothelial cell proliferation through the WNT signaling pathway in acute coronary syndrome. J Cell Biochem. 2018;119:4514–4527. doi: 10.1002/jcb.26584. [DOI] [PubMed] [Google Scholar]
- 43.Shih YP, Sun P, Wang A, Lo SH. Tensin1 positively regulates RhoA activity through its interaction with DLC1. Biochim Biophys Acta. 2015;1853:3258–3265. doi: 10.1016/j.bbamcr.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernau K, Torr EE, Evans MD, Aoki JK, Ngam CR, Sandbo N. Tensin 1 Is essential for myofibroblast differentiation and extracellular matrix formation. Am J Respir Cell Mol Biol. 2017;56:465–476. doi: 10.1165/rcmb.2016-0104OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhan Y, Liang X, Li L, Wang B, Ding F, Li Y, Wang X, Zhan Q, Liu Z. MicroRNA-548j functions as a metastasis promoter in human breast cancer by targeting Tensin1. Mol Oncol. 2016;10:838–849. doi: 10.1016/j.molonc.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorn GW., II Mitochondrial pruning by Nix and BNip3: An essential function for cardiac-expressed death factors. J Cardiovasc Transl Res. 2010;3:374–383. doi: 10.1007/s12265-010-9174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ney PA. Mitochondrial autophagy: Origins, significance, and role of BNIP3 and NIX. Biochim Biophys Acta. 2015;1853:2775–2783. doi: 10.1016/j.bbamcr.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 48.Chinnadurai G, Vijayalingam S, Gibson SB. BNIP3 subfamily BH3-only proteins: Mitochondrial stress sensors in normal and pathological functions. Oncogene. 2008;27(Suppl 1):S114–S127. doi: 10.1038/onc.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. 2015;43:331–342. doi: 10.1016/j.immuni.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazure NM, Pouyssegur J. Atypical BH3-domains of BNIP3 and BNIP3L lead to autophagy in hypoxia. Autophagy. 2009;5:868–869. doi: 10.4161/auto.9042. [DOI] [PubMed] [Google Scholar]
- 51.Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, Aronow BJ, Lorenz JN, Dorn GW., II Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 52.Dorn GW, II, Kirshenbaum LA. Cardiac reanimation: Targeting cardiomyocyte death by BNIP3 and NIX/BNIP3L. Oncogene. 2008;27(Suppl 1):S158–S167. doi: 10.1038/onc.2009.53. [DOI] [PubMed] [Google Scholar]
- 53.Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan JS, Huang L, Belousova T, Lu L, Yang Y, Reddel R, Chang A, Ju H, DiMattia G, Tong Q, Sheikh-Hamad D. Stanniocalcin-1 inhibits renal ischemia/reperfusion injury via an AMP-activated protein kinase-dependent pathway. J Am Soc Nephrol. 2015;26:364–378. doi: 10.1681/ASN.2013070703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang SE, Wu CP, Wu SY, Peng CK, Perng WC, Kang BH, Chu SJ, Huang KL. Stanniocalcin-1 ameliorates lipopolysac-charide-induced pulmonary oxidative stress, inflammation, and apoptosis in mice. Free Radic Biol Med. 2014;71:321–331. doi: 10.1016/j.freeradbiomed.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 56.Sheikh-Hamad D, Bick R, Wu GY, Christensen BM, Razeghi P, Poindexter B, Taegtmeyer H, Wamsley A, Padda R, Entman M, et al. Stanniocalcin-1 is a naturally occurring L-channel inhibitor in cardiomyocytes: Relevance to human heart failure. Am J Physiol Heart Circ Physiol. 2003;285:H442–H448. doi: 10.1152/ajpheart.01071.2002. [DOI] [PubMed] [Google Scholar]
- 57.Liu D, Huang L, Wang Y, Wang W, Wehrens XH, Belousova T, Abdelrahim M, DiMattia G, Sheikh-Hamad D. Human stanniocalcin-1 suppresses angiotensin II-induced superoxide generation in cardiomyocytes through UCP3-mediated anti-oxidant pathway. PLoS One. 2012;7:e36994. doi: 10.1371/journal.pone.0036994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan J, Mishra S, Shi J, Plovie E, Qiu Y, Cao X, Gianni D, Jiang B, Del Monte F, Connors LH, et al. Stanniocalcin1 is a key mediator of amyloidogenic light chain induced cardiotoxicity. Basic Res Cardiol. 2013;108:378. doi: 10.1007/s00395-013-0378-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu W, Barrientos T, Mao L, Rockman HA, Sauve AA, Andrews NC. Lethal cardiomyopathy in mice lacking transferrin receptor in the heart. Cell Rep. 2015;13:533–545. doi: 10.1016/j.celrep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naito Y, Hosokawa M, Sawada H, Oboshi M, Hirotani S, Iwasaku T, Okuhara Y, Morisawa D, Eguchi A, Nishimura K, et al. Transferrin receptor 1 in chronic hypoxia-induced pulmonary vascular remodeling. Am J Hypertens. 2016;29:713–718. doi: 10.1093/ajh/hpv163. [DOI] [PubMed] [Google Scholar]
- 61.Qi L, Song Y, Chan THM, Yang H, Lin CH, Tay DJT, Hong H, Tang SJ, Tan KT, Huang XX, et al. An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer. Nucleic Acids Res. 2017;45:10436–10451. doi: 10.1093/nar/gkx667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng J, Tang L, Hong Q, Ye H, Xu X, Xu L, Bu S, Wang Q, Dai D, Jiang D, Duan S. Investigation into the promoter DNA methylation of three genes (CAMK1D, CRY2 and CALM2) in the peripheral blood of patients with type 2 diabetes. Exp Ther Med. 2014;8:579–584. doi: 10.3892/etm.2014.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imamura M, Iwata M, Maegawa H, Watada H, Hirose H, Tanaka Y, Tobe K, Kaku K, Kashiwagi A, Kawamori R, et al. Genetic variants at CDC123/CAMK1D and SPRY2 are associated with susceptibility to type 2 diabetes in the Japanese population. Diabetologia. 2011;54:3071–3077. doi: 10.1007/s00125-011-2293-3. [DOI] [PubMed] [Google Scholar]
- 64.Bergamaschi A, Kim YH, Kwei KA, La Choi Y, Bocanegra M, Langerød A, Han W, Noh DY, Huntsman DG, Jeffrey SS, et al. CAMK1D amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol Oncol. 2008;2:327–339. doi: 10.1016/j.molonc.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.