Abstract
The increased expression of sialyl-Lewisx (SLex) epitope on the surface of tumor cells has been known for decades. However, genetic manipulation of the expression of SLex and the role of SLex in cancer cell proliferation remains to be fully elucidated. The present study suggested that the monoclonal antibody of SLex (KM93) significantly inhibited the proliferation of human hepatocarcinoma (HCC) cells. The expression levels of three sialyl-Lewis oligosaccharide antigens, SLex, SLea and dimeric SLex (SDLex), were determined on the cell surface of the MHCC97 human HCC cell line. The expression of SLex was markedly higher in MHCC97 cells than in normal liver cells. The expression of SDLex was also relatively high, however, no significant difference was observed between normal liver cells and HCC cells. The expression of SLea was only detected in trace quantities. Fucosyltransferase (FUT) is the key enzyme of the fucosylation step in the biosynthesis of sialyl-Lewis oligosaccharide antigens. Therefore, the present study investigated the expression of FUTs. It was found that the mRNA and protein expression levels of FUT7 were high in the MHCC97 HCC cell line compared with levels in normal liver cells. FUT6 was also expressed at a high level, although the difference was not statistically significant between MHCC97 cells and normal liver cells. No expression of FUT3 was detected. The results were consistent with the change insialyl-Lewis antigens. The effects of FUT7 small interfering (si)RNA transfection on the expression of FUT7, expression of SLex and MHCC97 cell proliferation were also examined. Following FUT7 siRNA transfection, the expression of FUT7 was markedly downregulated, as determined by western blot and reverse transcription-quantitative polymerase chain reaction methods. The results from flow cytometry showed that the synthesis of SLex was also inhibited, which was consistent with the downregulated expression of FUT7. MHCC97 cell proliferation was also significantly inhibited following FUT7 siRNA transfection, which was correlated with suppression of the S-phase in cell cycle progression. By using inhibitors of various signaling pathways, it was found that the knockdown of FUT7 inhibited the activation of phospholipase Cγ (PLCγ) by inhibiting the translocation and phosphorylation of PLCγ. In conclusion, the results suggested that FUT7 has animportant functional role in human HCC cell proliferation by controlling cell cycle progression via the PLCγ/extracellular signal-regulated kinase signaling pathway. The inhibition of SLex and FUT7 siRNA transfection may provide a novel therapeutic methodology to treat tumors that express SLex glycoconjugates.
Keywords: human hepatocarcinoma cells, sialyl-Lewisx, fucosyltransferase 7, cell proliferation, signaling pathway
Introduction
The sialyl-Lewis oligosaccharide antigens (SLea and SLex) are blood group related antigens which are important in inflammation by initiating the leukocyte-endothelium interaction (1,2). To date, it has been found that sialyl-Lewis antigens are involvedin severalmolecular interactions, including differentiation, development and malignancy (3-5), and their expression is markedly enhanced in several types of carcinoma cells (6,7). It has also been reported that the expression of sialyl-Lewis oligosaccharide antigens on tumor cells is closely associated with tumor progression, and statistical relevance has been confirmed between the postoperative prognosis of tumor patients and the degree of sialyl-Lewis oligosaccharide antigen expression on tumor tissues (8,9). Therefore, sialyl-Lewis oligosaccharide antigens are widely considered as cancer markers. The present study determined the expression of three sialyl-Lewis oligosaccharide antigens, SLea, SLex, and dimeric SLex (SDLex) on the cell surface of the MHCC97 human hepatocarcinoma (HCC) cell line. The expression of SLex was higher in MHCC97 cells compared with its expression in normal liver cells. The expression of SDLex was also high, although did not differ significantly between the normal liver cells and MHCC97 cells. The expression of SLea was only detected in trace quantities. On using a monoclonal antibody of sialyl-Lewis antigens, the results indicated that the anti-SLex antibody KM93 significantly inhibited MHCC97 cell proliferation.
The synthesis of sialyl-Lewis antigens is regulated by a set of glycosyltransferases. Fucosylation is the final step in the process of sialyl-Lewis antigen synthesis and it is catalyzed by α1,3/1,4 fucosyltransferases (α1,3/1,4 FUTs). The α1,3/1,4FUTs catalyze the connection of fucose in a (1,3) and a (1,4) linkage to the sialylated precursors (10-15). Currently, several human α(1,3) FUT genes (αFUT-3, 4, 5, 6, 7 and 9) have been cloned, identified and characterized (15-24). Each enzyme has a unique substrate and tissue specificity (25). FUT4, FUT9, FUT7 and FUT3 efficiently fucosylate sialylated acceptors, whereas FUT4 and FUT9 preferentially fucosylate non-sialylated neutral acceptors (20,24). α1,3/1,4FUT-3 is the only αFUT with both α1,3 and α1,4 fucosylation activity, leading to the generation of α1,3-fucosyl-containing Lex and SLex, and its isomer α1,4-fucosyl-containing SLea, respectively (7). By contrast, α1,3FUT7 only catalyzes the synthesis of SLex (NeuAca2/3Galb1/4 (Fuca1-3) GlcNAcb1/3Galb1/R) by adding the distal fucose residue to α2,3-sialylated lactosamine (9). FUT7 is mainly expressed in tumor cells, endothelial cells and leukocytes. There is a rigid acceptor specificity for FUT7, which can only accept the distal N-acetylglucosamine on α2,3 sialylated lactosamines to form SLex antigen. Therefore, FUT7 is the key enzyme of SLex synthesis (9,22,23). It has been widely reported that the expression levels of α1,3/1,4FUTs are positively correlated with tumor progression and negatively correlated with patient prognosis (8,9). For example, tumor growth was significantly increased following transfection of the FUT3 gene into PC-3 prostate cancer cells (26). Additionally, cell proliferation was significantly inhibited following gtransfection of FUT3/6 antisense RNA into colon cancer cells (27). Until now, the role of FUT7 in cancer cell proliferation and growth has not been fully elucidated.
In the present study, the effects of the knockdown of FUT7 on the expression of SLex and human HCC cell proliferation were examined. The effects of FUT7 small interfering (si)RNAs on the expression and activation of signaling molecules were also investigated. It was found that the knockdown of FUT7 decreased the expression of SLex, inhibited the trans-location and phosphorylation of phospholipase Cγ (PLCγ), and arrested cell cycle progression, which subsequently led to the suppression of MHCC97 HCC cell proliferation. These findings indicated that SLex and FUT7 may be important in human HCC cell proliferation. Inhibition of the SLex oligosaccharide antigen may serve as a novel therapeutic method to treat tumors that express SLex glycoconjugates. In addition, FUT7 maybe a potential target and FUT7 RNA interference (RNAi) may be developed as a novel therapeutic approach for SLex-positive cancer therapy.
Materials and methods
Cell culture and inhibitors treatment
The THLE-2 normal human liver cell line and HT29 human colorectal adenocarcinoma cell line were purchased from the American Type Culture Collection (Manassas, VA, USA). The MHCC97 human HCC cell line was purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). The THLE-2 cell line was grown in LONZA BEGM medium (Lonza/Clonetics Corporation, Walkersville, MD, USA). The MHCC97 cell line was cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FCS (Invitrogen, Thermo Fisher Scientific, Inc.). The cells were grown at 37°C in a humidified incubator under 5% CO2 and passaged when 70-80% confluent using 0.25% (w/v) trypsin solution in 0.04% (w/v) EDTA. At 1 h following passage of MHCC97 cells, PLCγ inhibitor (U73122; 10.5 mM; Merck KGaA, Darmstadt, Germany), protein kinase A (PKA) inhibitor (KT5720; 280 nM; Sigma-Aldrich; Merck KGaA), phosphoinositide 3-kinase (PI3K) inhibitor (LY294002; 16.5 mM; Sigma-Aldrich; Merck KGaA), extracellular signal-regulated kinase (Erk) inhibitor (ErkI; 50 mM; Merck KGaA) and CDC25 inhibitor (CDC25 inhibitor II; 1.05 mM; Merck KGaA) were added into the culture medium and the cells were cultured for a further 24 h. Cell survival was detected through MTT assay.
Detection of Lewis antigen with flow cytometric analysis
Cell surface Lewis antigen expression was detected by measuring the indirect immunofluorescence intensity using the FACS Calibur system (BD Biosciences, San Jose, CA, USA). Cell Quest Pro software (version 5.1; BD Biosciences) was used for cell acquisition and analysis. The cells were detached with 2 mM EDTA, washed twice with cold PBS and then prepared as a single cell suspension. A suspension of 106 cells was then incubated with mouse anti-SLex antibody (KM93, cat. no. 11-006, 1:100, GlycoTech, Gaithersburg, MD, USA), anti-SDLex antibody (FH6, cat. no. 11-007, 1:100, GlycoTech) and anti-SLea antibody (CA19-9, cat. no. 11-009, 1:100, GlycoTech) at 4°C for 1 h. Chilled PBS was used to wash the cells three times to remove unbound antibodies. The cells were then incubated with R-phycoerythin or peridinin chlorophyll protein (R-PE or Per-CP)-conjugated rabbit anti-mouse IgM (cat. no. 562033 and 553409, 1:200, BD Biosciences) at 4°C for 30 min in the dark. Chilled PBS was used to wash and remove unbound secondary antibodies. The cells were then fixed using 1% formaldehyde (Sigma; EMD Millipore, Billerica, MA, USA) for 30 min in the dark. A total of 1×104 cells from each sample tube were acquired for analysis using FACS Calibur. Cells incubated with secondary antibody only and unstained cells were used as controls.
Reverse-transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA from the cells were obtained using ab RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) following the manufacturer's protocol. Subsequently, 1 µg of total RNA was mixed with SYBR-Green PCR Master mix reagents of the PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) to synthesize the complementary DNA (cDNA) in the 7900HT real time PCR system (Applied Bio systems, Thermo Fisher Scientific, Inc.) according to the standard quantitative PCR protocol (25). Briefly, the RT-qPCR procedure was performed in a 20-µl reaction volume, including primers (0.25 µl, 10 pmol/l), 2X Mix SYBR-Green I (10 µl; Promega, Madison, WI, USA), template DNA (1 µl) and sterile water. The PCR steps constituted an initial denaturation step (95°C for 2 min), 50 cycles of 95°C for 15 sec and then 60°C for 45 sec. The fold change of target gene expression was calculated according to the following formula: Fold change = 22[[Cq (control) gene X-Cq (control) actin] [Cq (activated) gene X-Cq (activated) actin]] (28). The following primer sequences were used: Human FUT7, forward 5′-CCA CGA TCA CCA TCC TTG-3′ and reverse 5′-AGG CTT CGG TTG GCA CTC-3′; human FUT6, forward 5′-CAT TTC TGC TGC CTC AGG-3′ and reverse 5′-GGG CAA GTC AGG CAA CTC-3′; human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward 5′-GAA GGT GAA GGT CGG AGT C-3′ and reverse 5′-GAA GAT GGT GAT GGG ATT TC-3′. The relative mRNA expression levels of FUT7 and FUT6 were normalized to the endogenous mRNA expression of GAPDH.
Western blot analysis
RIPA buffer (Sigma; EMD Millipore) with protease inhibitor (Sigma; EMD Millipore) was used to lyse cells (26). The protein was collected, and a BCA Pierce Assay (Thermo Fisher Scientific, Inc.) was used for the quantification of protein concentration. Subsequently, 50 µg of protein from each sample was denatured and resolved on a 10% SDS-PAGE gradient gel (EMD Millipore), and then electro-blotted onto a PVDF nitrocellulose membrane (EMD Millipore). The PVDF membranes were then incubated with 5% non-fat milk for 1 h at room temperature, and then incubated with anti-FUT7 (1:1,000, cat. no. MAB64091), anti-PLCγ1 (1:500, cat. no. MAB8137) and anti-phosphorylated PLCγ1 (1:500, cat. no. MAB74541) which were purchased from Bio-Techne China (Shanghai, China), anti-FUT6 (1:1,000, cat. no. NBP1-57936; Novus Biologicals, LLC, Littleton, CO, USA), or anti-β-actin antibody (1:1,000, cat. no. MAB8929; Bio-Techne China) at 4°C overnight. Following washing with TBST, the membrane was incubated with HRP-conjugated secondary antibodies (1:3,000, cat. no. HAF008; Bio-Techne China) for 1.5 h at room temperature. Finally, the signals were developed by enhanced chemiluminescence (Pierce, Thermo Fisher Scientific, Inc.). Images of the results were captured and the images were scanned. The optical density of each protein band was quantified by a scanning densitometer and Quantity One software, version 4.4.1 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each lane of protein band density was normalized with corresponding β-actin density.
The cytosolic protein was isolated from particulate- conjugated protein using a digitonin separation method (29). The cells were collected and resuspended in 1 ml saline solution (1 mM EDTA, 150 mMNaCl, 1 mM PMSF, 2 mM EGTA, 1 µg/ml aprotinin, 10 µg/ml leupeptin, and 100 µg/ml digitonin) with occasional agitation for 10 min. The cells were then centrifuged at 13,000 × g for 5 min at 4°C and the resulting supernatant contained the cytosolic proteins. The cell pellet was dissolved in 1 ml lysis buffer (pH 7.4, 1 mM EDTA, 10 mM PBS, 1% Triton X-100, 2 mM EGTA, 1 mM PMSF, 0.1% SDS, 1 µg/ml aprotinin, and 10 µg/ml leupeptin) and contained the membrane protein (particulate-conjugated proteins). Subsequently, 80 µg of protein was separated by SDS-PAGE and transferred onto a PVDF membrane. The expression levels of PLCγ1 and phosphorylated PLCγ1 were detected by western blot analysis, as described above.
Knockdown of FUT7 in MHCC97 cells by RNAi
The expression of FUT7 in MHCC97 cells was silenced using specific siRNAs (Silencer siRNA transfection, Ambion, Thermo Fisher Scientific, Inc.). Scramble siRNA (siNC; Silencer siRNA trans-fection, Ambion, Thermo Fisher Scientific, Inc.) was used to confirm the specificity of FUT7 siRNAs. Untransfected cells were used as acontrol. The siRNA transfection into MHCC97 cells was performed following the manufacturer's protocol. A total of 1×105 cells/ml of MHCC97 cells were re-suspended in DMEM. The transfection complexes were prepared by mixing RNAi MAX Lipofectamine transfection agent (Ambion; Thermo Fisher Scientific, Inc.) and siRNA (20 nM) in DMEM. The MHCC97 cells and transfection complexes were mixed and then incubated for 24 h at 37°C in 6-well (2×105 cells/well) plates (Nalge Nunc International, Penfield, NY, USA). The cells were collected at 48 h for RNA analysis and 72 h for protein analysis. The knockdown of FUT7 was confirmed by RT-qPCR and western blot analyses.
Cell survival MTT assay
To investigate the effect of FUT7 silencing on cell proliferation and viability, MTT was added to the MHCC97 cells at 0, 24, 48, 72, 96 and 120 h following transfection with FUT7 siRNA. In detail, 5×104 cells were seeded into 96-well plates and, the following day, FUT7 was knocked down as described above. MTT (20 µl, 5 mg/ml) was added into each well at different time points. Following culture for 4 h at 37°C the DMEM was gently aspirated and 100 µl of dimethyl sulfoxide was added into each well. The absorbance of formazan at a 490 nm wavelength was measured using a Bio-Rad Microplate Reader 550 (Bio-Rad Laboratories, Inc.) and a cell growth curve was plotted.
Analysis of cell cycle
Ice-cold PBS was used to wash the collected control cells and transfected cells, and 1×106 cells were fixed in 70% ethanol for 24 h at -20°C. The cells were then washed twice with chilled PBS, and stained withpropidium iodide (PI; 20 µg/ml, Sigma; EMD Millipore) including RNase (100 µg/ml) for 30 min at room temperature. A BD FACScan flow cytometer was used to analyze the DNA content of cell cycle phases. A total of 2×104 cells per sample were collected and analyzed using BD FACSDiva™ analysis software (version 6.2; BD Biosciences).
Statistical analysis
Statistical analysis was performed using SPSS 20.0 software (IBM SPSS, Armonk, NY, USA). All values are expressed as the mean ± standard error of the mean and are representative of at least three independent experiments. The data were analyzed using one-way analysis of variance. When the results were significant, post hoc testing of differences between each group was performed using Bonferroni's correction. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of sialyl-Lewis oligosaccharide antigens and FUTs in the MHCC97 human HCC cell line
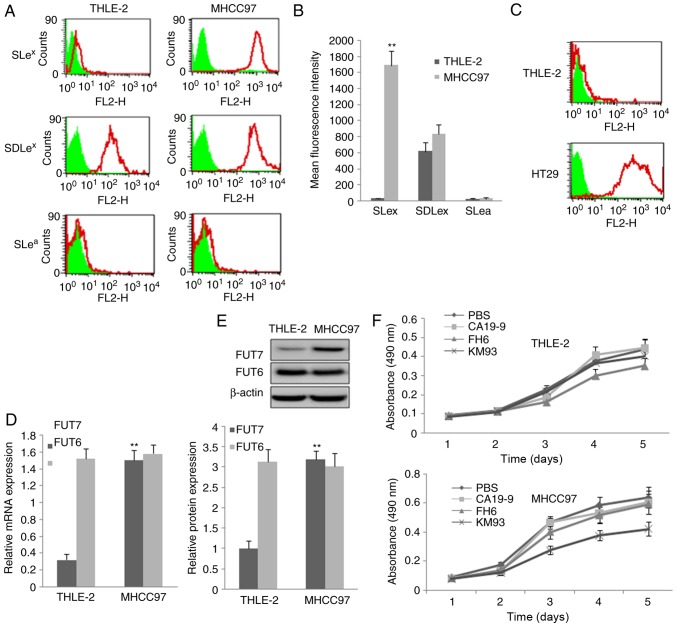
The expression of three sialyl-Lewis antigens, SLea, SDLex and SLex, were determined and the results are shown in Fig. 1A and B. The expression of SLex was significantly increased in the MHCC97 human HCC cells compared with that in THLE-2 normal human liver cells. The expression of SDLex was high, however, no significant difference was observed between the MHCC97 cells and THLE-2 cells. The expression of SLea was absent or negligible. The high expression of SLea in the HT29 cells confirmed the high affinity of the anti-Slea antibody (Fig. 1C). RT-qPCR and western blot methods were used to detect the mRNA and protein levels of FUT7 and FUT6, respectively. The mRNA and protein expression levels of FUT7 were high in the MHCC97 cells compared with expression levels in the THLE-2 normal liver cells. FUT6 was also expressed at high levels, however, no significant difference was found between the MHCC97 cells and THLE-2 cells (Fig. 1D and E). No FUT3 was detected.
Figure 1.
Expression of three sialyl-Lewis oligosaccharides and relative FUTs and the inhibition effect of monoclonal antibody on cell proliferation. (A) MHCC97 human hepatocellular carcinoma cells and THLE-2 normal liver cells were assayed by flow cytometry using three monoclonal antibodies targeting sialyl-Lewis antigens. PBS, cells treated with PBS and without antibodies as a control; KM93, cells treated with mAb against SLex; FH6, cells treated with mAb against SDLex; CA19-9, cells treated with mAb against SLea. In the flow cytometry histograms, the areas in green show the number of unstained cells and the areas outlined in red represent cells binding to mAb. (B) Quantitative analysis of the expression of sialyl-Lewis oligosaccharides by calculation of mean fluorescence intensity. THLE-2 cells were used as a control. (C) THLE-2 cells and HT29 cells were assayed by flow cytometry using monoclonal antibody against SLea antigen to confirm the high affinity of the SLea antibody. (D) mRNA expression of FUT7 and FUT6 were detected by reverse transcription-quantitative polymerase chain reaction analysis. The relative gene mRNA level was normalized to the respective glyceraldehyde-3-phosphate dehydrogenase level. THLE-2 cells were used asa control. (E) Protein expression levels of FUT7 and FUT6 protein were evaluated by western blot analysis (above) and quantitative analysis of relative protein levels was performed (below). THLE-2 cells were used as a control. **P<0.01 (n=3), vs. THLE-2 Control. (F) Effect of monoclonal antibodies on the proliferation of THLE-2 cells and MHCC97 cells. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H tetrazolium bromide assay showed that MHCC97 cell proliferation was significantly suppressed by KM93, a mAb against SLex. FUT, fucosyltransferase; SLex, sialyl-Lewis X, SLea, sialyl-Lewis a; SDLe X, dimeric SLe; mAb, monoclonal antibody.
Inhibition of SLex by KM93 monoclonal antibody inhibits MHCC97 cell proliferation
Different monoclonal antibodies of sialyl-Lewis antigens were added to inhibit the sialyl-Lewis antigens on the cell surface of MHCC97 cells and normal liver cells, and cell growth curves were plotted using the data obtained from the MTT method. It was found that the anti-SLex monoclonal antibody KM93 significantly suppressed MHCC97 cell proliferation, but not that of normal liver cells. The anti-SDLex antibody FH6 had a minimalsuppression effect, which was not statistically significant. The anti-SLea antibody CA19-9 did not suppress MHCC97 cell proliferation (Fig. 1F).
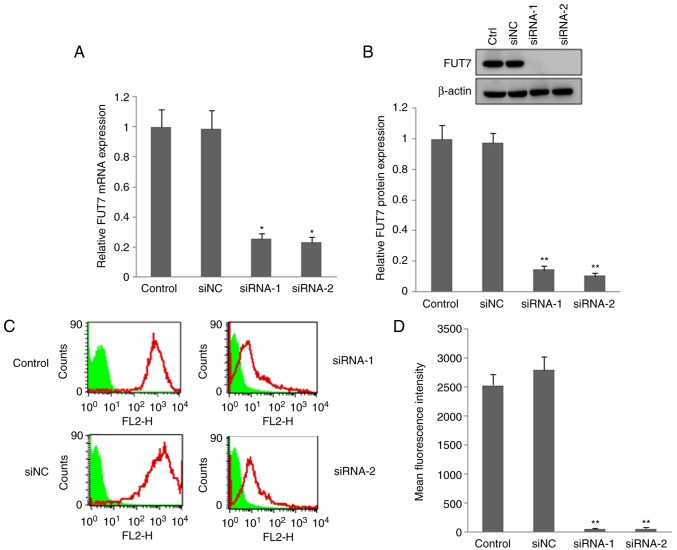
FUT7 siRNAs transfection reduced the expression of FUT7 and SLex
RT-qPCR, western blot and flow cytometry methods were used to investigate the effect of FUT7 RNAi on the expression of FUT7 and SLex. The RT-qPCR and western blot results showed that the mRNA and protein levels of FUT7 (Fig. 2A and B) were markedly lower in the FUT7 siRNA-transfected MHCC97 cells, compared with those in the untransfected wild-type cells (control). The FUT7 siRNAs effectively inhibited the expression of FUT7. The inhibitory effect of FUT7 siRNA on the expression of SLex was also investigated by flow cytometry. The fluorescence intensity of SLex in the utransfected control cells was more marked than that in the FUT7-knockdown cells (Fig. 2C and D). siNC-transfected cells were used to confirm the specificity of FUT7 siRNA. No significant difference in the expression of FUT7 or SLex was found between the siNC cells and untransfected control cells. The above results suggested that siRNAs of FUT7 suppressed the expression of FUT7 and markedly inhibited the synthesis of SLex oligosaccharide antigen.
Figure 2.
Changes in the expression of FUT7 and SLex of MHCC97 cells following transfection with FUT7 siRNAs. siRNA targeting FUT7 was transfected into MHCC97 cells and the interference rate was confirmed by RT-qPCR and western blot analyses. Flow-cytometric analysis was used to detect the expression of SLex. (A) mRNA level of FUT7 detected by RT-qPCR analysis following FUT7-siRNA transfection. (B) Protein level of FUT7 detected by western blot analysis following FUT7-siRNA transfection (above) and quantitative analysis of the protein level of FUT7 following FUT7-siRNA transfection below). (C) Flow cytometric analysis of cell surface expression of SLex following FUT7-siRNA transfection. (D) Quantitative analysis of the expression of SLex by calculating the mean fluorescence intensity. *P<0.05 (n=3), vs. untransfected control cells; **P<0.01 (n=3), vs. untransfected control cells. FUT, fucosyltransferase; SLex, sialyl-Lewis X; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA; siNC, scramble-siRNA transfected cells; Control/Ctrl, untransfected cells.
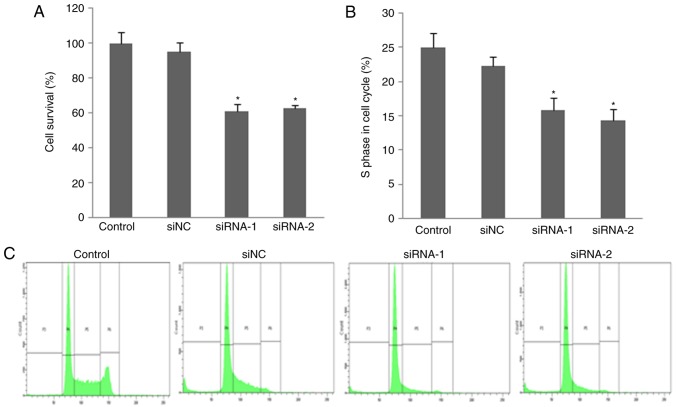
Knockdown of FUT7 inhibits human HCC cell proliferation
An MTT assay was used to investigate the impact of FUT7 knockdown in HCC cell proliferation. The number of proliferated cells was measured by detecting the absorbance at a wavelength of 490 nm. The MTT assay showed that cell proliferation of the FUT7-knockdown cells was suppressed compared with that of control cells. No significant difference was found between the cell proliferation rate of siNC cells and control cells (Fig. 3A). This indicated that the silencing of FUT7 may significantly suppress HCC cell proliferation.
Figure 3.
Cell proliferation and cell cycle assay after knockdown of FUT7. MHCC97 cells were transfected with FUT7 siRNAs and siNC. Cell proliferation was analyzed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H tetrazolium bromide assay and cell cycle was analyzed by flow cytometry. (A) Cell proliferation assay following knockdown of FUT7. (B) Cell cycle assay following knockdown of FUT7. (C) Flow cytometry profiles of MHCC97 cells following transfection. *P<0.05 (n=3), vs. untransfected control cells. FUT, fucosyltransferase; SLex, sialyl-Lewis X; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA; siNC, scramble-siRNA transfected cells; Control, untransfected cells.
FUT7 knockdown decreases HCC cell distribution into the S-phase
To further investigate the suppression of cell proliferation in FUT7-knockdown cells, cell cycle distribution was analyzed by flow cytometry. As shown in Fig. 3B and C, flow cytometry with PI-stained cells showed that untransfected control cells and siNC cells were presented in the G0/G1 (58.78±6.45 and 60.52±6.17%), S (25.20±2.17 and 22.18±2.87%) and G2/M (2.97±0.60 and 3.10±0.77%) phases. In the FUT7 siRNA-transfected MHCC97 cells, the S-phase fraction was lower (16.40±2.10% for FUT7 siRNA-1 and 13.27±2.75% for FUT7 siRNA-2 cells, P<0.05) and the G0/G1 fraction was higher (69.62±6.84% for FUT7 siRNA-1 and 67.72±6.53% for FUT7 siRNA-2 cells, respectively, P<0.05). This indicated that the fractions of cells in the S-phase of cell cycle, which represents a population of dividing cells, decreased following the knockdown of FUT7. A concomitant increase was observed in the number of cells at the G0/G1 phases. These results showed that FUT7 knockdown suppressed cell proliferation by inhibiting cell cycle progression into the S-phase. These results further emphasized the importance of the expression of FUT7 on cell proliferation.
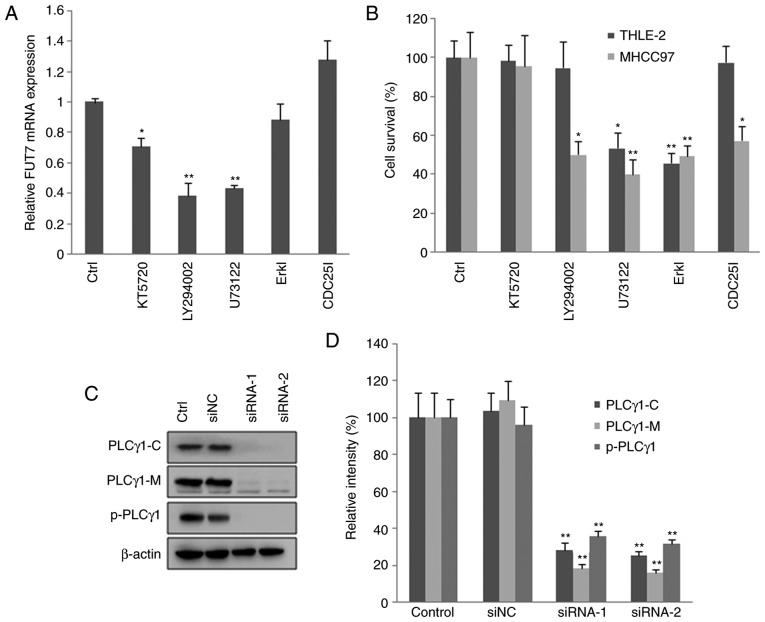
Effects of inhibiting PKA, PI3K/AKT and PLCγ/Erk signaling pathways on the expression of FUT and MHCC97 cell proliferation
To understand the mechanisms involved, the present study examined the effects of inhibitors of signaling pathways on the mRNA levels of FUT7. The RT-qPCR results showed that, in MHCC97 cells, PLCγ inhibitor, PKA inhibitor and PI3K inhibitor significantly reduced the mRNA levels of FUT7. By contrast, the Erk inhibitor and CDC25 inhibitor had no effect on the mRNA level of FUT7 (Fig. 4A). The MTT results showed that inhibitors of PLCγ and Erk reduced cell survival in normal liver cells. CDC25, PI3K, PLCγ and Erk inhibitors decreased the proliferation of MHCC97 cells, with the most marked effect observed with PLCγ inhibitor (Fig. 4B). These results indicated that the PLCγ inhibitor significantly reduced the mRNA level of FUT7, and inhibited MHCC97 HCC cell proliferation and survival.
Figure 4.
Knocking down FUT7 affects cell survival via various intracellular signaling mediators. MHCC97 cells were transfected with FUT7 siRNAs. After 1 h, inhibitors of signaling molecules were added to the culture medium, and the cells were cultured for a further 48 h for RNA analysis and 72 h for protein analysis. (A) mRNA levels of FUT7 were quantified by reverse transcription-quantitative polymerase chain reaction following treatment with inhibitors. (B) Following treatment with the inhibitors, cell survival was quantified using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H tetrazolium bromide assay in the THLE-2 normal liver cells and MHCC97 hepatocarcinoma cells. (C) Cells were permeabilized and centrifuged to isolate the cytosolic proteins and particulate-conjugated proteins. Results of western blot analysis of the target proteins are shown with profiles. (D) Quantitative analysis of relative protein levels. *P<0.05 (n=3), vs. Control; **P<0.01 (n=3), vs. Control. FUT, fucosyltransferase; Ctrl, control (no inhibitor); ErkI, extracellular signal-regulated kinase inhibitor; siRNA, small interfering RNA; siNC, scramble-siRNA transfected cells; PLCγ, phospholipase Cγ; PLCγ1-C, PLCγ1 in cytoplasm; PLCγ1-M, PLCγ1 conjugated with plasma membrane; p-PLCγ1, phosphorylated PLCγ1 conjugated with plasma membrane; Control, untransfected cells.
To observe the effect of FUT7 siRNA transfection on the activation of PLCγ, the proteins in the cytoplasm were isolated from particulate-conjugated proteins. The contents of PLCγ1 and phosphorylated PLCγ1 were detected by western blot analysis. The results (Fig. 4C and D) showed that FUT7 siRNA transfection decreased the content of PLCγ1 in the cytoplasm, and decreased the content of PLCγ1 and phosphorylated PLCγ1 conjugated on the plasma membrane. The results demonstrated that FUT7 siRNA transfection inhibited the translocation and phosphorylation of PLCγ1, and thus inhibited the activation of PLCγ1. Therefore, the aforementioned results indicated that FUT7 may affect cell proliferation through the activation of PLCγ. As Erk is the downstream signaling molecule of PLCγ (30), the results indicated that FUT7 modulated cell proliferation via the PLCγ/Erk signaling pathway in MHCC97 cells.
Discussion
Cell surface glycoconjugates have important functional roles in cancer cell proliferation, growth, metastasis and invasiveness. It has been reported that the changing expression and components of glycoconjugates are correlated with carcinogenesis in severaltyped of cancer (3-9). Abnormal elevation in the expression of sialyl-Lewis oligosaccharide antigens (SLea and SLex) has been correlated with the malignancy of a wide range of tumors (6-9). Several antibodies targeting the sialyl-Lewis oligosaccharide have been developed as potential therapeutic approaches for the treatment of sialyl-Lewis oligosaccharide-positive tumors (31-33). However, there has been limited investigation of methods to effectively inhibit the synthesis of sialyl-Lewis oligosaccharide and its effects on tumor cell proliferation and growth.
It has been reported that the SLea oligosaccharide is mainly expressed in tumor cells derived from digestive organs, including the pancreas, rectum, biliary tract and colon (34). In addition, it has been reported that the SLex oligosaccharide ismainly expressed in ovarian, breast and pulmonary tumor cells (35). It has also been reported that the level of SLex oligo-saccharide was significantly higher in serum of patients with HCC than in serum of normal controls and patients with liver cirrhosis (36). It has also been shown that the expression of SLex oligosaccharide is closely associated with the growth of HCC (37-39). The results of the present study also indicated that the expression of SLex in HCC tissues was higher than in adjacent normal liver tissues. In addition, the findings indicated that the expression of SLex was significantly increased in MHCC97 human HCC tumor cells compared with that in normal liver cells. The expression of SDLex was unchanged and the expression of SLea was only detected in trace quantities.
FUT7 is the key enzyme for the synthesis of the SLex oligosaccharide, therefore, the present study investigated the expression of FUT7 in HCC. The results indicated that the expression of FUT7 was markedly increased in MHCC97 cells compared with normal liver cells. Following transfection with FUT7 siRNAs, the expression of SLex was significantly decreased in the MHCC97 cells. It has been reported that FUT7 has high substrate specificity and that FUT7 is the only FUT responsible for the synthesis of SLex Lewis oligosaccharide antigen (22,23). FUT3/6 is responsible for the synthesis of SLex and SDLex, and FUT6 is the major enzyme which is responsible for the synthesis of SDLex from SLex (20). The results of the present study indicated that the expression of FUT6 was also high, however, no statistically significant difference was found between the MHCC97 cells and normal liver cells. The mRNA and protein levels of FUT7 and FUT6 were correlated with the expression of sialyl-Lewis antigens (SLex and SDLex) in the MHCC97 cells. The expression of FUT7 has been reported to be altered in several types of tumor (26,27). It has also been documented that the expression of glycan-related gene in human HCC cells, which is responsible for the synthesis of N-glycan and glycolipids, particularly the sialyl-Lewis antigen, was significantly increased (40). However, the mechanism of FUT7 in cancer cell growth has not been investigated fully in human HCC cells.
The results of the present study showed that FUT7 is not only responsible for SLex synthesis, but is also involved in cell proliferation. siRNAs of FUT7 suppressed the expression of FUT7 and markedly inhibited the synthesis of SLex oligosaccharide antigen and MHCC97 HCC cell proliferation. The results also indicated that anti-SLex monoclonal antibody KM93 significantly suppressed MHCC97 HCC cell proliferation. Therefore, the suppression of cell proliferation following FUT7 RNAi may be due to the decreased synthesis of SLex. The detailed functional role of SLex is being investigated in subsequent investigations.
Following the binding of hepatocyte growth factor (HGF) ligand to the HGF receptor (receptor tyrosine kinase (MET)) on the liver cell surface, the kinase catalytic activity of MET is activated, which triggers trans-phosphorylation of the tyrosines of MET, including Tyr 1234 and Tyr 1235, Tyr 1349 and Tyr 1356. These tyrosines recruit and engage a number of signaling transducers, including PI3K, Src homology 2-containing phosphotyrosine phosphatase2, and PLCγ, therefore initiating a whole spectrum of biological activities, including cell survival, proliferation, migration and invasion. The PLC/Erk signal transduction pathway is one of the most important signaling pathways in regulating cell proliferation, growth and differentiation (30). PLC can be divided into three subclasses: PLCβ, PLCγ and PLCδ, according to their different structures (41). PLCγ is one of the direct downstream signal transducers of growth factor receptors and has an important functional role in regulating cell growth and proliferation. PLCγ1 is widely distributed in various tissues and cells, whereas PLCγ2 is mainly distributed in the lungs, spleen and thymus. Therefore, PLCγ1 was selected as the main signaling target for the present study. The activation of PLCγ1 involves a series of molecular events (42). First, ligands conjugate with growth factor receptors that have receptor protein tyrosine kinase activity. The receptors are then dimerized and autophosphorylated. The intracellular domain of receptors is autophosphorylated to form the phosphorylated tyrosine residues, and these residues of the receptors form the binding site of the SH2 domain of PLCγ1. Subsequently, PLCγ1 is phosphorylated at certain tyrosine residues and is activated by receptor kinase. The activated PLCγ1 then separates from its receptors and trans-locates to the plasma membrane by binding phospholipid phosphatidylinositol 3,4,5 triphosphate (PIP3) through its PH structure. Subsequently, PLCγ1 can hydrolyze phospholipid phosphatidylinositol 3,4,5 biphosphate (PIP2) to produce diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3). The phosphorylation and translocation of PLCγ1 from the cytosol to the plasma membrane are necessary for its activation and function. Therefore, the activation of PLCγ1 can be detected by its phosphorylation and translocation. The experimental results showed that the proliferation of MHCC97 cells was suppressed following FUT7 siRNA transfection. The relative content of PLCγ1 conjugated with the plasma membrane of MHCC97 cells transfected with FUT7 siRNAs was also lower than that of the control cells, and the phosphorylated PLCγ1 conjugated with the plasma membrane was also decreased. This indicates that FUT7 knockdown inhibited the phosphorylation and translocation of PLCγ1 from the cytosol to the plasma membrane, and thus, inhibited the activation of PLCγ1. In addition, siRNA targeting FUT7 reduced cell proliferation and survival. The PLCγ inhibitor U73122 significantly reduced the mRNA levels of FUT7 and inhibited the proliferation and survival of the MHCC97 cells. The experiment using PLCγ inhibitor supported the hypo thesis that the expression level of FUT7 is important to sustain MHCC97 HCC cell proliferation and survival. The mechanistic mechanism of PLCγ/ERK pathway regulating the mRNA expression of FUT7 remains to be fully elucidated. It was hypothesized that activated PLCγ1 cleaves the PIP2 into DAG and IP3. DAG and calcium work together to activate protein kinase C. This leads to a series of phosphorylation events downstream in the mitogen-activated protein kinase (MAPK) cascade. First, the activated PKC activates RAF, and RAF kinase phosphorylates and activates MAPK kinase. This ultimately results in the phosphorylation and activation of ERKs. ERK regulates the activities of several transcription factors, including c-Jun, activating transcription factor 2, ELK1 and heat shock factor 1. By altering the levels and activities of transcription factors, ERK alters the transcription of genes that are involved in regulation of cell proliferation and cell cycle, including FUT7. The detailed signaling mechanism and the factors involved in PLCγ1 regulating the gene transcription of FUT7 remain fully elucidated.
In conclusion, the findings of the present study revealed that the suppression of PLCγ/Erk signaling pathway inhibited human HCC cell proliferation, which was, at least partially, mediated by the decreased expression of FUT7 and its product, SLex. The results demonstrated that FUT7 siRNAs downregulated SLex synthesis, inhibited PLCγ activation, arrested cell cycle progression and effectively reduced MHCC97 HCC cell proliferation. These results suggest that FUT7 siRNAs maybe a potential therapeutic agent to treat human SLex-positive cancer. Therefore, targeting FUT7 for inactivation with RNAi technology is an attractive approach for controlling HCC cell proliferation.
Funding
This study was supported by the National Natural Science Foundation (grant ns 81373173 and 30371359) and the Research Fund for the Doctoral Program of Higher Education (grant no. 20132104120002).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
DL and HL conceived and designed the experiments; DL, HS, ZB and GB Performed the experiments; WW, Ml and JL analyzed the data; DL and HL wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethical Committee of China Medical University (Shenyang, Liaoning, China). Written informed consent was obtained from each patient recruited.
Patient consent for publication
Written informed consent was obtained from each patient.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
References
- 1.Lowe JB. Glycan-dependent leukocyte adhesion and recruitment in inflammation. Curr Opin Cell Biol. 2003;15:531–538. doi: 10.1016/j.ceb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: The multistep paradigm. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 3.Kannagi R, editor. Molecular mechanism for cancer-associated induction of sialyl Lewis X and sialyl Lewis A expression-the Warburg effect revisited. Glycoconj J. 2004;20:353–364. doi: 10.1023/B:GLYC.0000033631.35357.41. [DOI] [PubMed] [Google Scholar]
- 4.Roseman S. Reflections on glycobiology. J Biol Chem. 2001;276:41527–41542. doi: 10.1074/jbc.R100053200. [DOI] [PubMed] [Google Scholar]
- 5.Liu YC, Yen HY, Chen CY, Chen CH, Cheng PF, Juan YH, Chen CH, Khoo KH, Yu CJ, Yang PC, et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc Natl Acad Sci USA. 2011;108:11332–11337. doi: 10.1073/pnas.1107385108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kannagi R. Carbohydrate-mediated cell adhesion involved in hematogenous metastasis of cancer. Glycoconj J. 1997;14:577–584. doi: 10.1023/A:1018532409041. [DOI] [PubMed] [Google Scholar]
- 7.Takada A, Ohmori K, Yoneda T, Tsuyuoka K, Hasegawa A, Kiso M, Kannagi R. Contribution of carbohydrate antigens sialyl Lewis A and sialyl Lewis X to adhesion of human cancer cells to vascular endothelium. Cancer Res. 1993;53:354–361. [PubMed] [Google Scholar]
- 8.Ito H, Hiraiwa N, Sawada-Kasugai M, Akamatsu S, Tachikawa T, Kasai Y, Akiyama S, Ito K, Takagi H, Kannagi R. Altered mRNA expression of specific molecular species of fucosyl- and sialyltransferases in human colorectal cancer tissues. Int J Cancer. 1997;71:5560–5564. doi: 10.1002/(SICI)1097-0215(19970516)71:4<556::AID-IJC9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 9.Ogawa JI, Inoue H, Koide S. α-2,3-Sialyltransferase type 3N and α-1,3-fucosyltransferase type VII are related to sialyl Lewis x synthesis and patient survival from lung carcinoma. Cancer. 1997;79:1678–1685. doi: 10.1002/(SICI)1097-0142(19970501)79:9<1678::AID-CNCR7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 10.Kuijipers TW. Terminal glycosyltransferase activity: A selective role in cell adhesion. Blood. 1993;81:873–882. [PubMed] [Google Scholar]
- 11.Candelier JJ, Mollicone R, Mennesson B, Bergemer AM, Henry S, Couillin P, Oriol R. Alpha-3-fucosyltransferases and their glycoconjugate antigen products in the developing human kidney. Lab Invest. 1993;69:449–459. [PubMed] [Google Scholar]
- 12.Stroup GB, Anumula KR, Kline TF, Caltabiano MM. Identification and characterization of two distinct a-1-3-L-fucosyltransferase activities in human colon carcinoma. Cancer Res. 1990;50:6787–6792. [PubMed] [Google Scholar]
- 13.Mollicone R, Candelier JJ, Mennesson B, Couillin P, Venot AP, Oriol R. Five specificity patterns of (1→3)-α-L-fucosyltransferase activity defined by use of synthetic oligosaccharide acceptors. Differential expression of the enzymes during human embryonic development and in adult tissues. Carbohydr Res. 1992;228:265–276. doi: 10.1016/S0008-6215(00)90564-0. [DOI] [PubMed] [Google Scholar]
- 14.Macher BA, Holmes EH, Swiedler SJ, Stults CM, Srnka CA. Human alpha 1-3 fucosyltransferases. Glycobiology. 1991;6:577–584. doi: 10.1093/glycob/1.6.577. [DOI] [PubMed] [Google Scholar]
- 15.Kukowska-Latallo JF, Larsen RD, Nair RP, Lowe JB. A cloned human cDNA determines expression of a mouse stage-specific embryonic antigen and the Lewis blood group alpha (1,3/1,4) fucosyltransferase. Genes Dev. 1990;4:1288–1303. doi: 10.1101/gad.4.8.1288. [DOI] [PubMed] [Google Scholar]
- 16.Goelz SE, Hession C, Goff D, Griffiths B, Tizaed R, Newman B, Chi-Rosso G, Lobb R. ELFT: A gene that directs the expression of an ELAM-1 ligand. Cell. 1990;63:1349–1356. doi: 10.1016/0092-8674(90)90430-M. [DOI] [PubMed] [Google Scholar]
- 17.Lowe JB, Kukowska-Latallo JF, Nair RP, Larsen RD, Marks RM, Macher BA, Kelly R, Ernst LK. Molecular cloning of a human fucosyltransferase gene that determines expression of the Lewis x and VIM-2 epitopes but not ELAM-1-dependent cell adhesion. J Biol Chem. 1991;266:17467–17477. [PubMed] [Google Scholar]
- 18.Kumar R, Potvin B, Muller WA, Stanley P. Cloning of a human alpha(1,3)-fucosyltransferase gene that encodes ELFT but does not confer ELAM-1 recognition on Chinese hamster ovary cell transfectants. J Biol Chem. 1991;266:21777–21783. [PubMed] [Google Scholar]
- 19.Weston BW, Nair RP, Larsen RD, Lowe JB. Isolation of a novel human alpha (1,3)fucosyltransferase gene and molecular comparison to the human Lewis blood group alpha (1,3/1,4) fucosyltransferase gene. Syntenic, homologous, nonallelic genes encoding enzymes with distinct acceptor substrate specificities. J Biol Chem. 1992;267:4152–4160. [PubMed] [Google Scholar]
- 20.Weston BW, Smith PL, Kelly RJ, Lowe JB. Molecular cloning of a fourth member of a human alpha (1,3) fucosyltransferase gene family Multiple homologous sequences that determine expression of the Lewis x, sialyl Lewis x, and difucosyl sialyl Lewis x epitopes. J Biol Chem. 1992;267:24575–24584. [PubMed] [Google Scholar]
- 21.Koszdin KL, Bowen BR. The cloning and expression of a human alpha-1,3 fucosyltransferase capable of forming the E-selectin ligand. Biochem Biophys Res Commun. 1992;187:152–157. doi: 10.1016/S0006-291X(05)81472-X. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki K, Kurata K, Funayama K, Nagata M, Watanabe E, Ohta S, Hanai N, Nishi T. Expression cloning of a novel alpha 1,3-fucosyltransferase that is involved in biosynthesis of the sialyl Lewis x carbohydrate determinants in leukocytes. J Biol Chem. 1994;269:14730–14737. [PubMed] [Google Scholar]
- 23.Natsuka S, Gersten KM, Zenita K, Kannagi R, Lowe JB. Molecular cloning of a cDNA encoding a novel human leukocyte alpha-1,3-fucosyltransferase capable of synthesizing the sialyl Lewis x determinant. J Biol Chem. 1994;269:16789–16794. [PubMed] [Google Scholar]
- 24.Kudo K, Ikehara Y, Togayachi A, Kaneko M, Hiraga T, Sasaki K, Narimatsu H. Expression, cloning and characterization of a novel murine alpha1,3-fucosyltransferase, mFuc-IX, that synthesis the Lewis X (CD15) epitope in brain and kidney. J Biol Chem. 1998;273:26729–26738. doi: 10.1074/jbc.273.41.26729. [DOI] [PubMed] [Google Scholar]
- 25.Narimatsu H. Human fucosyltransferases: Tissue distribution of blood group antigens, cancer associated antigens and fucosyltransferase. Tanpakushitsu Kakusan Koso. 1998;43(Suppl 16):S2394–S2403. In Japanese. [PubMed] [Google Scholar]
- 26.Albrethsen J, Bøgebo R, Gammeltoft S, Olsen J, Winther B, Raskov H. Upregulated expression of human neutrophil peptides 12 and 3 (HNP13) in colon cancer serum and tumours: A biomarker study. BMC Cancer. 2005;5:8. doi: 10.1186/1471-2407-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiller KM, Mayben JP, Bendt KM, Manousos GA, Senqer K, Cameron HS, Weston BW. Transfection of alpha (1,3) fucosyltransferase antisense sequences impairs the proliferative and tumorigenic ability of human colon carcinoma cells. Mol Carcinog. 2000;27:280–288. doi: 10.1002/(SICI)1098-2744(200004)27:4<280::AID-MC6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Cordeiro C, Freire AP. Digiton in permeabilization of Saccharomyces cerevisiae cells for in situ enzyme assay. Anal Biochem. 1995;229:145–148. doi: 10.1006/abio.1995.1394. [DOI] [PubMed] [Google Scholar]
- 30.Berridge MJ. Inositol triphosphate and diacylycerol as second messenger. Biochem J. 1984;220:345–360. doi: 10.1042/bj2200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klinger M, Farhan H, Just H, Drobny H, Himmler G, Loibner H, Mudde GC, Freissmuth M, Sexl V. Antibodies directed against Lewis -Y antigen inhibit signaling of Lewis-Y modified ErbB receptors. Cancer Res. 2004;64:1087–1093. doi: 10.1158/0008-5472.CAN-03-2435. [DOI] [PubMed] [Google Scholar]
- 32.Farhan H, Schuster C, Klinger M, Weisz E, Waxenecker G, Schuster M, Sexl V, Mudde GC, Freissmuth M, Kircheis R. Inhibition of xenograft tumor growth and down-regulation of ErbB receptors by an antibody directed against Lewis Y antigen. J Pharmacol Exp Ther. 2006;319:1459–1466. doi: 10.1124/jpet.106.107318. [DOI] [PubMed] [Google Scholar]
- 33.Scott AM, Geleick D, Rubira M, Clarke K, Nice EC, Smyth FE, Stockert E, Richards EC, Carr FJ, Harris WJ, et al. Construction, production, and characterization of humanized anti-Lewis Y monoclonal antibody 3S193 for targeted immunotherapy of solid tumors. Cancer Res. 2000;60:3254–3261. [PubMed] [Google Scholar]
- 34.Kannagi R. Carbohydrate antigen sialyl Lewis a-its pathophysiological significance and induction mechanism in cancer progression. Chang Gung Med J. 2007;30:189–209. [PubMed] [Google Scholar]
- 35.Kannagi R, Izawa M, Koike T, Miyazaki K, Kimura N. Carbohydrate-mediated cell adhesion in cancer metastasis and angiogenesis. Cancer Sci. 2004;95:377–384. doi: 10.1111/j.1349-7006.2004.tb03219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakai N, Okada Y, Tsuji T. Increased serum levels of the carrier molecules of the carbohydrate antigen sialyl Lewis X in liver diseases. Acta Med Okayama. 1997;51:79–85. doi: 10.18926/AMO/30781. [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara Y, Shimada M, Takenaka K, Kajiyama K, Shirabe K, Sugimachi K. The Sialyl Lewis X expression in hepatocarcinogenesis: Potential predictor for the emergence of hepatocellular carcinoma. Hepatogastroenterology. 2002;49:213–217. [PubMed] [Google Scholar]
- 38.Okada Y, Jin-no K, Ikeda H, Sakai N, Sotozono M, Yonei T, Nakanishi S, Moriwaki S, Tsuji T. Changes in the expression of sialyl-Lewisx, a hepatic necroinflammation-associated carbohydrate neoantigen, in human hepatocellular carcinomas. Cancer. 1994;73:1811–1816. doi: 10.1002/1097-0142(19940401)73:7<1811::AID-CNCR2820730707>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Torii A, Nakayama A, Harada A, Nakao A, Nonami T, Sakamoto J, Watanabe T, Ito M, Takagi H. Expression of the CD15 antigen in hepatocellular carcinoma. Cancer. 1993;71:3864–3867. doi: 10.1002/1097-0142(19930615)71:12<3864::AID-CNCR2820711212>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 40.Kang X, Wang N, Pei C, Sun L, Sun R, Chen J, Liu Y. Glycan-related gene expression signatures in human metastatic hepatocellular carcinoma cells. Exp Ther Med. 2012;3:415–422. doi: 10.3892/etm.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delmas P, Crest M, Brown DA. Functional organization of PLC signaling microdomains in neurons. Trends Neurosci. 2004;27:41–47. doi: 10.1016/j.tins.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 42.Liu B, Wu D. Analysis of G protein-mediated activation of phospholipase C in cultured cells. Methods Mol Biol. 2004;237:99–102. doi: 10.1385/1-59259-430-1:99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.