Abstract
Early-onset cardiovascular and renal diseases have a strong genetic component. In the present study, exome-wide association studies (EWASs) were performed to identify genetic variants that confer susceptibility to early-onset myocardial infarction (MI), hypertension, or chronic kidney disease (CKD) in Japanese individuals. A total of 8,093 individuals aged ≤65 years was enrolled in the study. The EWASs for MI, hypertension, and CKD were performed in 6,926 subjects (1,152 cases, 5,774 controls), 8,080 subjects (3,444 cases, 4,636 controls), and 2,556 subjects (1,051 cases, 1,505 controls), respectively. Genotyping of single nucleotide polymorphisms (SNPs) was performed with Illumina Human Exome-12 DNA Analysis BeadChip or Infinium Exome-24 BeadChip arrays. The associations of allele frequencies for 31,245, 31,276, or 31,514 SNPs that passed quality control to MI, hypertension, and CKD, respectively, was examined with Fisher's exact test. Bonferroni's correction for statistical significance of association was applied to compensate for multiple comparisons of genotypes with MI, hypertension, or CKD. The EWASs of allele frequencies revealed that 25, 11, and 11 SNPs were significantly associated with MI (P<1.60×10−6), hypertension (P<1.60×10−6), or CKD (P<1.59×10−6), respectively. Multivariable logistic regression analysis with adjustment for covariates showed that all 25, 11, and 11 SNPs were significantly associated with MI (P<0.0005), hypertension (P<0.0011), or CKD (P<0.0011), respectively. On examination of the results from previous genome-wide association studies and linkage disequilibrium of the identified SNPs, 11 loci (TMOD4, COL6A3, ADGRL3-CXCL8-MARCH1, OR52E4, TCHP-GIT2, CCDC63, 12q24.1, OAS3, PLCB2-VPS33B, GOSR2, ZNF77), six loci (MOB3C-TMOD4, COL6A3, COL6A5, CXCL8-MARCH1, NFKBIL1-6p21.3-NCR3, PLCB2-VPS33B), and seven loci (MOB3C-TMOD4, COL6A3, COL6A5, ADGRL3-CXCL8-MARCH1, MUC17, PLCB2-VPS33B, ZNF77) were identified as novel loci significantly associated with MI, hypertension, and CKD, respectively. Furthermore, six genes (TMOD4, COL6A3, CXCL8, MARCH1, PLCB2, VPS33B) were significantly associated with MI, hypertension and CKD; two genes (ADGRL3, ZNF77) with MI and CKD; and two genes (COL6A5, MOB3C) with hypertension and CKD. Therefore, 13 novel loci (MOB3C-TMOD4, COL6A3, ADGRL3-CXCL8-MARCH1, OR52E4, TCHP- GIT2, CCDC63, 12q24.1, OAS3, PLCB2-VPS33B, ZNF77, COL6A5, NFKBIL1-NCR3, MUC17) were identified that confer susceptibility to early-onset MI, hypertension, or CKD. The determination of genotypes for the SNPs at these loci may provide informative for assessment of the genetic risk for MI, hypertension, or CKD.
Keywords: myocardial infarction, hypertension, chronic kidney disease, genetics, exome-wide association study
Introduction
Coronary artery disease (CAD) and myocardial infarction (MI) are serious clinical conditions that remain the leading causes of mortality in the United States (1). Disease prevention is an important strategy for reducing the overall burden of these conditions, with the identification of biomarkers for disease risk being key for risk prediction and for potential intervention to reduce the chance of future coronary events.
In addition to conventional risk factors for CAD and MI, including hypertension, diabetes mellitus, and dyslipidemia, the importance of genetic factors has been highlighted (2). The heritability of CAD has been estimated to be 40-60% on the basis of family and twin studies (2,3). Genome-wide association studies (GWASs) in European-ancestry populations (4-10), African Americans (11), or Han Chinese (12,13) have identified various genes and loci that confer susceptibility to CAD or MI. A meta-analysis of GWASs for CAD among European-ancestry populations, which included low-frequency variants, identified 202 independent genetic variants at 129 loci with a false discovery rate of <5% (14). These genetic variants together account for ~28% of the heritability of CAD, showing that genetic susceptibility to this condition is largely determined by common variants with small effect sizes (2,14). A more recent meta-analysis for CAD in European-ancestry populations identified 304 independent genetic variants with a false discovery rate of <5%, with these variants explaining 21.2% of CAD heritability (15). Although several single nucleotide polymorphisms (SNPs) have been found to be significantly associated with MI in Japanese individuals (16,17), genetic variants that contribute to susceptibility to MI in the Japanese population remain to be fully elucidated.
Hypertension is a complex multifactorial disorder that results from an interaction between genetic background and both lifestyle and environmental factors (18). The heritability of hypertension has been estimated to be 84% and that of systolic blood pressure (BP) and diastolic BP has been estimated to be 53 and 49%, respectively (19). As hypertension is a major risk factor for CAD and stroke (20), personalized prevention of hypertension is an important public health goal.
GWASs have implicated various loci and genes as influencing BP or conferring susceptibility to hypertension in populations in Europe (6,21-24) and African (25,26) and in individuals from East Asia (27). A trans-ancestry GWAS in individuals of European and East or South Asian ancestry identified 12 loci associated with BP (28). A previous trans-ancestry meta-analysis in individuals of European and South Asian ancestry identified 30 susceptibility loci for BP or hypertension (29). Although an SNP in ADD2 was shown to be a susceptibility locus for hypertension in Japanese individuals (30), genetic variants that confer susceptibility to this condition in Japanese individuals await definitive identification.
Chronic kidney disease (CKD) is also a global public health problem, with affected individuals being at increased risk for end-stage renal disease (ESRD), cardiovascular disease and premature death (31,32). The identification of biomarkers for CKD risk is important to prevent progression to ESRD and to reduce the chance of future cardiovascular adverse events (33). In addition to conventional risk factors for CKD, including diabetes mellitus and hypertension (34), studies have shown the importance of genetic factors in renal function and in the development of CKD (35). The heritability of glomerular filtration rate for creatinine clearance has been estimated to be 52% (36).
GWASs have implicated several genes and loci in renal function or predisposition to CKD or ESRD in populations of European (37-42) or African (43) ancestry, and in renal function-related traits in East Asian populations (44). A meta-analysis of GWASs in European populations identified 53 loci that were significantly associated with estimated glomerular filtration rate (eGFR) (45). Although several SNPs have been shown to be associated with renal function in East Asian populations (44), genetic variants that contribute to predisposition to CKD in the Japanese population remain to be fully elucidated.
A previous study of monozygotic and dizygotic twins found that CAD-associated mortality rate at younger ages was significantly influenced by genetic factors in men and women, whereas the genetic effect was less pronounced at older ages (46). A family history of MI is also more apparent in individuals with early-onset MI compared with those with late-onset MI, suggestive of increased heritability in the former individuals (47,48). Similar to early-onset MI, early-onset forms of hypertension (49) and CKD (35) have been found to have a strong genetic component. As the genetic contribution may be higher in early-onset than in late-onset MI, hypertension and CKD, the statistical power of genetic association studies may be increased by focusing on subjects with early-onset disease (2,35,49).
In the present study, exome-wide association studies (EWASs) for MI, hypertension, and CKD were performed with the use of human exome array-based genotyping methods in order to identify genetic variants that confer susceptibility to these conditions in Japanese individuals. To increase the statistical power of the EWASs, subjects with early-onset disease were examined.
Materials and methods
Study subjects
In our previous EWASs, the median (mean) ages of subjects with MI, hypertension, and CKD were 67 (67.0), 68 (67.2), and 70 (69.9) years, respectively (50-52). Therefore patients with an age of ≤65 years were defined as individuals with early-onset forms of these conditions in the present study. A total of 8,093 Japanese individuals aged ≤65 years (subjects with MI, hypertension, or CKD and corresponding controls) was enrolled in the present study. In the EWASs for MI, hypertension, and CKD, 6,926 individuals (1,152 subjects with MI, 5,774 controls), 8,080 individuals (3,444 subjects with hypertension, 4,636 controls), and 2,556 individuals (1,051 subjects with CKD, 1,505 controls) were examined, respectively. The individuals were recruited as those who visited outpatient clinics or were admitted to participating hospitals in Japan (Gifu Prefectural Tajimi Hospital, Tajimi; Gifu Prefectural General Medical Center, Gifu; Japanese Red Cross Nagoya First Hospital, Nagoya, Japan; Northern Mie Medical Center Inabe General Hospital, Inabe, Japan; and Hirosaki University Hospital and Hirosaki Stroke and Rehabilitation Center, Hirosaki, Japan) due to the presence of various symptoms or for an annual health checkup between 2002 and 2014, or who were community-dwelling individuals recruited to a population-based cohort study in Inabe between 2010 and 2014 (53).
For the MI study, the diagnosis of MI was based on typical electrocardiographic changes and on increases in the serum activity of creatine kinase (MB isozyme) and in the serum concentration of troponin T. The diagnosis was confirmed by identification of the responsible stenosis in any of the major coronary arteries or in the left main trunk by coronary angiography. The control individuals had no history of MI, CAD, aortic aneurysm, or peripheral artery disease; of ischemic or hemorrhagic stroke; or of other atherosclerotic, thrombotic, embolic, or hemorrhagic disorders. Although certain control individuals had conventional risk factors for MI, including hypertension, diabetes mellitus, dyslipidemia, and CKD, they did not have cardiovascular complications.
In the hypertension study, the subjects with hypertension either had a systolic BP of ≥140 mmHg or diastolic BP of ≥90 mmHg (or both), or had taken antihypertensive medication. Individuals with severe valvular heart disease, congenital malformations of the heart or vessels, renal or endocrinologic diseases that cause secondary hypertension, or drug-induced hypertension were excluded from the study. The control individuals had a systolic BP of <140 mmHg and diastolic BP of <90 mmHg and no history of hypertension or of taking antihypertensive medication. BP was measured at least twice by a skilled physician or nurse, with subjects having first rested in the sitting position for >5 min.
For the CKD study, glomerular filtration rate was estimated with the use of the simplified prediction equation derived from a modified version of that described in the Modification of Diet in Renal Disease Study, as suggested by the Japanese Society of Nephrology (54): eGFR (ml min−1 1.73 m−2) = 194 × [age (years)] −0.287 × [serum creatinine (mg/dl)] −1.094 × [0.739 if female]. The National Kidney Foundation-Kidney Disease Outcomes Quality Initiative guidelines recommend a diagnosis of CKD if eGFR is <60 ml min−1 1.73 m−2 (55). Nonlinear relations between eGFR and the risk of adverse events, including mortality, cardiovascular episodes, and hospitalization, have been demonstrated, with an increased risk being associated with an eGFR of <60 ml min−1 1.73 m−2 and the risk markedly increasing further when values fall <45 ml min−1 1.73 m−2 (56). Therefore, the criterion of an eGFR of <60 ml min−1 1.73 m−2 was used for the diagnosis of CKD. The control individuals for the CKD study had an eGFR of ≥90 ml min−1 1.73 m−2 and had no functional or structural abnormalities of the kidneys or a history of renal disease. Although certain control individuals had hypertension, diabetes mellitus, dyslipidemia, or hyperuricemia, they had no renal complications.
A total of 785 subjects with both MI and hypertension and 3,946 controls overlapped between the corresponding studies, as did 324 subjects with both MI and CKD and 1,127 controls, and 750 subjects with both hypertension and CKD and 955 controls.
EWASs
Venous blood (5 or 7 ml) was collected into tubes containing 50 mmol/l ethylenediaminetetraacetic acid (disodium salt), peripheral blood leukocytes were isolated, and genomic DNA was extracted from these cells with the use of a Genomix DNA extraction kit (Talent Srl, Trieste, Italy) or SMITEST EX-R&D kit (Medical & Biological Laboratories, Co., Ltd., Nagoya, Japan). The EWASs for MI (1,152 cases, 5,774 controls), hypertension (3,444 cases, 4,636 controls), and CKD (1,051 cases, 1,505 controls) were performed with the use of a Human Exome-12 v1.2 DNA Analysis BeadChip or Infinium Exome-24 v1.0 BeadChip (Illumina, San Diego, CA, USA). These exome arrays include putative functional exonic variants selected from ~12,000 individual exome and whole-genome sequences. The exonic content consists of ~244,000 SNPs from European, African, Chinese, and Hispanic individuals (57). SNPs contained in only one of the exome arrays (~2.6% of all SNPs) were excluded from analysis. Quality control was performed (58) as follows: i) Genotyping data with a call rate of <97% were discarded, with the mean call rate for the remaining data being 99.9%; ii) sex specification was checked for each sample, and those for which sex phenotype in the clinical records was inconsistent with genetic sex chromosomes were discarded; iii) duplicate samples and cryptic relatedness were assessed by calculation of identity by descent; all pairs of DNA samples showing an identity by descent of >0.1875 were inspected and one sample from each pair was excluded; iv) the frequency of heterozygosity for SNPs was calculated for all samples, and those with low or high heterozygosity (>3 standard deviations from the mean) were discarded; v) SNPs in sex chromosomes or mitochondrial DNA were excluded from the analysis, as were nonpolymorphic SNPs or SNPs with a minor allele frequency of <1.0%; vi) SNPs with genotype distributions deviating significantly (P<0.01) from Hardy-Weinberg equilibrium in control individuals were discarded; vii) genotype data were examined for population stratification by principal components analysis (59), and population outliers were excluded from the analysis. A total of 31,245, 31,276, or 31,514 SNPs passed quality control for the EWASs of MI, hypertension, and CKD, respectively, and these SNPs were subjected to analyses.
Statistical analysis
For analysis of the characteristics of the study subjects, quantitative data were compared between subjects with MI, hypertension, or CKD and corresponding controls with an unpaired t-test. Categorical data were compared between the two groups with Pearson's χ2 test. Allele frequencies were estimated by the gene counting method, and Fisher's exact test was applied to identify departure from Hardy-Weinberg equilibrium. In the EWASs, the relation of allele frequencies of each SNP to MI, hypertension, or CKD was examined with Fisher's exact test. To compensate for multiple comparisons of genotypes with MI, hypertension, or CKD, Bonferroni's correction was applied for statistical significance of association. As 31,245, 31,276, and 31,514 SNPs that passed quality control were examined in the EWASs for MI, hypertension, and CKD, respectively, P<1.60×10−6 (0.05/31,245 and 31,276) for MI and hypertension or P<1.59×10−6 (0.05/31,514) for CKD was considered statistically significant. The inflation factor (λ) was 0.95 for MI, 1.03 for hypertension, and 1.07 for CKD. Multivariable logistic regression analysis was performed with MI as a dependent variable and independent variables included age, sex (0, woman; 1, man), the prevalence of hypertension, diabetes mellitus, and dyslipidemia (0, no history of these conditions; 1, positive history), and the genotype of each SNP. Similar analysis was performed with hypertension as a dependent variable and independent variables of age, sex, and the genotype of each SNP; and with CKD as a dependent variable and independent variables of age, sex, the prevalence of hypertension and diabetes mellitus, and the genotype of each SNP. The genotypes of the SNPs were assessed according to dominant [0, AA; 1, AB + BB (A, major allele; B, minor allele)], recessive (0, AA + AB; 1, BB), and additive genetic models, and the P-value, odds ratio, and 95% confidence interval were calculated. Additive models comprised additive 1 (0, AA; 1, AB; 0, BB) and additive 2 (0, AA; 0, AB; 1, BB) scenarios, which were analyzed simultaneously with a single statistical model. The association between the genotypes of SNPs and intermediate phenotypes of MI was examined with Pearson's χ2 test. The association between genotypes of SNPs and systolic or diastolic BP, the serum concentration of creatinine, or eGFR was examined by one-way analysis of variance (ANOVA). Bonferroni's correction was also applied to other statistical analysis as indicated. Statistical tests were performed with JMP Genomics version 9.0 software (SAS Institute, Cary, NC, USA).
Results
Characteristics of subjects
The characteristics of the 6,926 subjects enrolled in the MI study are shown in Table I. The age, the frequency of men, and the prevalence of obesity, hypertension, diabetes mellitus, dyslipidemia, CKD, and hyperuricemia, in addition to body mass index (BMI), systolic and diastolic BP, fasting plasma glucose (FPG) level, blood glycosylated hemoglobin (hemoglobin A1c) content, and the serum concentrations of triglycerides, creatinine, and uric acid were higher, whereas the serum concentration of high density lipoprotein (HDL)-cholesterol and eGFR were lower, in subjects with MI than in controls.
Table I.
Characteristics of subjects with MI and control individuals.
| Characteristic | Control | Myocardial infarction | P-value |
|---|---|---|---|
| Subjects (n) | 5,774 | 1,152 | |
| Age (years) | 50.6±10.2 | 55.6±7.4 | <0.0001 |
| Sex (men/women, %) | 52.1/47.9 | 85.4/14.6 | <0.0001 |
| Smoking (%) | 42.5 | 46.7 | 0.0105 |
| Obesity (%) | 31.0 | 42.7 | <0.0001 |
| Body mass index (kg/m2) | 23.2±3.5 | 24.6±3.5 | <0.0001 |
| Hypertension (%) | 31.7 | 68.9 | <0.0001 |
| Systolic BP (mmHg) | 121±18 | 139±26 | <0.0001 |
| Diastolic BP (mmHg) | 75±13 | 78±16 | <0.0001 |
| Diabetes mellitus (%) | 12.7 | 61.3 | <0.0001 |
| Fasting plasma glucose (mmol/l) | 5.66±1.78 | 7.66±3.39 | <0.0001 |
| Blood hemoglobin A1c (%) | 5.72±0.96 | 6.88±1.70 | <0.0001 |
| Dyslipidemia (%) | 56.9 | 84.9 | <0.0001 |
| Serum triglycerides (mmol/l) | 1.32±0.98 | 1.77±1.25 | <0.0001 |
| Serum HDL-cholesterol (mmol/l) | 1.65±0.45 | 1.17±0.34 | <0.0001 |
| Serum LDL-cholesterol (mmol/l) | 3.18±0.83 | 3.15±0.98 | 0.8593 |
| Chronic kidney disease (%) | 10.3 | 29.1 | <0.0001 |
| Serum creatinine (µmol/l) | 69.8±61.0 | 91.9±98.1 | <0.0001 |
| eGFR (ml min−1 1.73 m−2) | 78.7±17.1 | 71.2±27.8 | <0.0001 |
| Hyperuricemia (%) | 15.2 | 25.6 | <0.0001 |
| Serum uric acid (µmol/l) | 321±89 | 356±105 | <0.0001 |
Quantitative data are presented as the mean ± standard deviation and were compared between subjects with MI and controls using the unpaired t-test. Categorical data were compared between the two groups with Pearson's χ2 test. Based on Bonferroni's correction, P<0.0025 (0.05/20) was considered statistically significant. Obesity was defined as a body mass index of ≥25 kg/m2; hypertension as a systolic BP of ≥140 mmHg, diastolic BP of ≥90 mmHg, or the taking of anti-hypertensive medication; diabetes mellitus as a fasting plasma glucose level of ≥6.93 mmol/l, blood hemoglobin A1c content of ≥6.5%, or the taking of antidiabetic medication; dyslipidemia as a serum triglyceride concentration of ≥1.65 mmol/l, serum HDL-cholesterol <1.04 mmol/l, serum LDL-cholesterol ≥3.64 mmol/l, or the taking of anti-dyslipidemic medication; chronic kidney disease as an eGFR of <60 ml min−1 1.73 m−2; and hyperuricemia as a serum uric acid concentration of >416 µmol/l or the taking of uric acid-lowering medication. MI, myocardial infarction; BP, blood pressure; HDL, high density lipoprotein; LDL, low density lipoprotein; eGFR, estimated glomerular filtration rate.
The characteristics of the 8,080 subjects enrolled in the hypertension study are shown in Table II. The age, the frequency of men, and the prevalence of obesity, diabetes mellitus, dyslipidemia, CKD, and hyperuricemia, in addition to the BMI, FPG level, blood hemoglobin A1c content, and the serum concentrations of triglycerides, creatinine, and uric acid were higher, whereas the serum concentration of HDL-cholesterol and eGFR were lower, in subjects with hypertension than in controls.
Table II.
Characteristics of subjects with hypertension and control individuals.
| Characteristic | Control | Hypertension | P-value |
|---|---|---|---|
| Subjects (n) | 4636 | 3444 | |
| Age (years) | 49.2±10.3 | 55.8±7.6 | <0.0001 |
| Sex (men/women, %) | 52.0/48.0 | 67.8/32.2 | <0.0001 |
| Smoking (%) | 41.7 | 42.9 | 0.2829 |
| Obesity (%) | 24.3 | 44.4 | <0.0001 |
| Body mass index (kg/m2) | 22.6±3.3 | 24.5±3.7 | <0.0001 |
| Systolic BP (mmHg) | 114±12 | 144±24 | <0.0001 |
| Diastolic BP (mmHg) | 70±10 | 85±14 | <0.0001 |
| Diabetes mellitus (%) | 11.1 | 39.5 | <0.0001 |
| Fasting plasma glucose (mmol/l) | 5.61±1.78 | 6.72±2.83 | <0.0001 |
| Blood hemoglobin A1c (%) | 5.71±0.96 | 6.30±1.48 | <0.0001 |
| Dyslipidemia (%) | 51.3 | 75.5 | <0.0001 |
| Serum triglycerides (mmol/l) | 1.25±0.98 | 1.69±1.19 | <0.0001 |
| Serum HDL-cholesterol (mmol/l) | 1.63±0.46 | 1.41±0.45 | <0.0001 |
| Serum LDL-cholesterol (mmol/l) | 3.15±0.85 | 3.15±0.88 | 0.9206 |
| Chronic kidney disease (%) | 8.1 | 24.0 | <0.0001 |
| Serum creatinine (µmol/l) | 65.4±23.9 | 89.3±117.6 | <0.0001 |
| eGFR (ml min−1 1.73 m−2) | 80.4±18.8 | 72.5±21.3 | <0.0001 |
| Hyperuricemia (%) | 11.3 | 25.2 | <0.0001 |
| Serum uric acid (µmol/l) | 311±86 | 349±99 | <0.0001 |
Quantitative data are presented as the mean ± standard deviation and were compared between subjects with hypertension and controls with the unpaired t-test. Categorical data were compared between the two groups with Pearson's χ2 test. Based on Bonferroni's correction, P<0.0026 (0.05/19) was considered statistically significant. Obesity was defined as a body mass index of ≥25 kg/m2; hypertension as a systolic BP of ≥140 mmHg, diastolic BP of ≥90 mmHg, or the taking of antihypertensive medication; diabetes mellitus as a fasting plasma glucose level of ≥6.93 mmol/l, blood hemoglobin A1c content of ≥6.5%, or the taking of antidiabetic medication; dyslipidemia as a serum triglyceride concentration of ≥1.65 mmol/l, serum HDL-cholesterol <1.04 mmol/l, serum LDL-cholesterol ≥3.64 mmol/l, or the taking of anti-dyslipidemic medication; chronic kidney disease as an eGFR of <60 ml min−1 1.73 m−2; and hyperuricemia as a serum uric acid concentration of >416 µmol/l or the taking of uric acid-lowering medication. BP, blood pressure; HDL, high density lipoprotein; LDL, low density lipoprotein; eGFR, estimated glomerular filtration rate.
The characteristics of the 2,556 subjects enrolled in the CKD study are shown in Table III. The age, the frequency of men, and the prevalence of obesity, hypertension, diabetes mellitus, dyslipidemia, and hyperuricemia, in addition to the BMI, systolic and diastolic BP, FPG level, blood hemoglobin A1c content, and the serum concentrations of triglycerides and uric acid were higher, whereas the serum concentration of HDL-cholesterol was lower, in subjects with CKD than in controls.
Table III.
Characteristics of subjects with CKD and control individuals.
| Characteristic | Control | CKD | P-value |
|---|---|---|---|
| Subjects (n) | 1,505 | 1,051 | |
| Age (years) | 48.5±10.3 | 57.7±6.5 | <0.0001 |
| Sex (men/women, %) | 53.8/46.2 | 68.9/31.1 | <0.0001 |
| Smoking (%) | 43.2 | 37.3 | 0.0028 |
| Obesity (%) | 29.1 | 38.9 | <0.0001 |
| Body mass index (kg/m2) | 23.1±3.8 | 24.1±3.5 | <0.0001 |
| Hypertension (%) | 36.5 | 71.4 | <0.0001 |
| Systolic BP (mmHg) | 126±23 | 141±29 | <0.0001 |
| Diastolic BP (mmHg) | 76±14 | 80±16 | <0.0001 |
| Diabetes mellitus (%) | 24.8 | 45.2 | <0.0001 |
| Fasting plasma glucose (mmol/l) | 6.27±2.72 | 6.88±3.16 | <0.0001 |
| Blood hemoglobin A1c (%) | 6.08±1.51 | 6.42±1.51 | <0.0001 |
| Dyslipidemia (%) | 56.4 | 75.3 | <0.0001 |
| Serum triglycerides (mmol/l) | 1.35±1.21 | 1.69±1.07 | <0.0001 |
| Serum HDL-cholesterol (mmol/l) | 1.56±0.47 | 1.38±0.48 | <0.0001 |
| Serum LDL-cholesterol (mmol/l) | 3.08±0.85 | 3.13±0.93 | 0.0738 |
| Serum creatinine (µmol/l) | 51.7±9.3 | 150.3±198.0 | <0.0001 |
| eGFR (ml min−1 1.73 m−2) | 103.0±19.1 | 46.9±14.6 | <0.0001 |
| Hyperuricemia (%) | 8.5 | 40.6 | <0.0001 |
| Serum uric acid (µmol/l) | 297±80 | 381±106 | <0.0001 |
Quantitative data are presented as the mean ± standard deviation and were compared between subjects with CKD and controls with the unpaired t-test. Categorical data were compared between the two groups with Pearson's χ2 test. Based on Bonferroni's correction, P<0.0026 (0.05/19) was considered statistically significant. Obesity was defined as a body mass index of ≥25 kg/m2; hypertension as a systolic BP of ≥140 mmHg, diastolic BP of ≥90 mmHg, or the taking of antihypertensive medication; diabetes mellitus as a fasting plasma glucose level of ≥6.93 mmol/l, blood hemoglobin A1c content of ≥6.5%, or the taking of antidiabetic medication; dyslipidemia as a serum triglyceride concentration of ≥1.65 mmol/l, serum HDL-cholesterol <1.04 mmol/l, serum LDL-cholesterol ≥3.64 mmol/l, or the taking of anti-dyslipidemic medication; chronic kidney disease as an eGFR of <60 ml min−1 1.73 m−2; and hyperuricemia as a serum uric acid concentration of >416 µmol/l or the taking of uric acid-lowering medication. CKD, chronic kidney disease; BP, blood pressure; HDL, high density lipoprotein; LDL, low density lipoprotein; eGFR, estimated glomerular filtration rate.
EWASs for MI, hypertension, and CKD
The association between allele frequencies for the 31,245, 31,276, and 31,514 SNPs that passed quality control to MI, hypertension, and CKD, respectively, were examined using Fisher's exact test. Following Bonferroni's correction, 25 and 11 SNPs were significantly (P<1.60×10−6) associated with MI (Table IV) or hypertension (Table V), respectively, and 11 SNPs were significantly (P<1.59×10−6) associated with CKD (Table VI).
Table IV.
25 SNPs significantly (P<1.60×10−6) associated with myocardial infarction in the exome-wide association study.
| Gene | SNP | Nucleotide substitutiona | Amino acid substitution | Chromosome | Position | MAF (%) | Allele OR | P-value (allele frequency) |
|---|---|---|---|---|---|---|---|---|
| PLCB2 | rs200787930 | C/T | E1106K | 15 | 40289298 | 1.2 | 0.01 | 8.81×10−26 |
| CXCL8 | rs188378669 | G/T | E31* | 4 | 73741568 | 1.2 | 0.01 | 1.33×10−25 |
| MARCH1 | rs61734696 | G/T | Q137K | 4 | 164197303 | 1.2 | 0.01 | 1.50×10−25 |
| VPS33B | rs199921354 | C/T | R80Q | 15 | 91013841 | 1.2 | 0.01 | 2.07×10−25 |
| TMOD4 | rs115287176 | G/A | R277W | 1 | 151170961 | 1.2 | 0.01 | 9.03×10−25 |
| COL6A3 | rs146092501 | C/T | E1386K | 2 | 237371861 | 1.2 | 0.02 | 1.27×10−24 |
| ZNF77 | rs146879198 | G/A | R340* | 19 | 2934109 | 1.2 | 0.02 | 1.27×10−24 |
| ADGRL3 | rs192210727 | G/T | R580I | 4 | 61909615 | 1.3 | 0.07 | 1.12×10−20 |
| ALDH2 | rs671 | G/A | E504K | 12 | 111803962 | 27.6 | 1.54 | 2.67×10−19 |
| ACAD10 | rs11066015 | G/A | 12 | 111730205 | 27.5 | 0.65 | 3.35×10−19 | |
| BRAP | rs3782886 | A/G | 12 | 111672685 | 29.3 | 1.49 | 5.52×10−17 | |
| HECTD4 | rs11066280 | T/A | 12 | 112379979 | 29.0 | 1.49 | 6.48×10−17 | |
| HECTD4 | rs2074356 | C/T | 12 | 112207597 | 25.4 | 1.48 | 4.24×10−15 | |
| OR52E4 | rs11823828 | T/G | F227L | 11 | 5884973 | 36.6 | 1.43 | 1.86×10−12 |
| rs12229654 | T/G | 12 | 110976657 | 22.5 | 1.39 | 1.45×10−10 | ||
| NAA25 | rs12231744 | C/T | R876K | 12 | 112039251 | 35.1 | 0.74 | 1.07×10−9 |
| ATXN2 | rs7969300 | T/C | N248S | 12 | 111555908 | 38.8 | 0.76 | 8.46×10−9 |
| GOSR2 | rs1052586 | T/C | 17 | 46941097 | 48.7 | 0.78 | 1.80×10−7 | |
| TCHP | rs74416240 | G/A | 12 | 109904793 | 13.3 | 1.37 | 2.47×10−7 | |
| GIT2 | rs925368 | A/G | N389S | 12 | 109953174 | 12.5 | 1.37 | 4.41×10−7 |
| rs2523644 | A/G | 6 | 31374707 | 8.1 | 1.49 | 4.75×10−7 | ||
| OAS3 | rs2072134 | C/T | 12 | 112971371 | 17.6 | 1.33 | 5.28×10−7 | |
| CCDC63 | rs10774610 | T/C | 12 | 110902439 | 23.7 | 1.30 | 5.42×10−7 | |
| rs2596548 | G/T | 6 | 31362769 | 5.4 | 1.60 | 6.01×10−7 | ||
| CCDC63 | rs10849915 | T/C | 12 | 110895818 | 23.6 | 1.28 | 7.71×10−7 |
Allele frequencies were analyzed using Fisher's exact test.
Major allele/minor allele. SNP, single nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio.
Table V.
11 SNPs significantly (P<1.60×10−6) associated with hypertension in the exome-wide association study.
| Gene | SNP | Nucleotide substitutiona | Amino acid substitution | Chromosome | Position | MAF (%) | Allele OR (allele frequency) | P-value |
|---|---|---|---|---|---|---|---|---|
| MARCH1 | rs61734696 | G/T | Q137K | 4 | 164197303 | 1.2 | 0.54 | 1.30×10−7 |
| PLCB2 | rs200787930 | C/T | E1106K | 15 | 40289298 | 1.2 | 0.55 | 3.33×10−7 |
| COL6A3 | rs146092501 | C/T | E1386K | 2 | 237371861 | 1.2 | 0.55 | 3.50×10−7 |
| COL6A5 | rs200982668 | G/A | E2501K | 3 | 130470894 | 1.3 | 0.56 | 3.52×10−7 |
| MOB3C | rs139537100 | C/T | R24Q | 1 | 46615006 | 1.2 | 0.56 | 4.76×10−7 |
| VPS33B | rs199921354 | C/T | R80Q | 15 | 91013841 | 1.2 | 0.55 | 4.84×10−7 |
| NCR3 | rs2515920 | T/A | 6 | 31594838 | 17.3 | 0.81 | 5.02×10−7 | |
| TMOD4 | rs115287176 | G/A | R277W | 1 | 151170961 | 1.2 | 0.55 | 5.49×10−7 |
| CXCL8 | rs188378669 | G/T | E31* | 4 | 73741568 | 1.2 | 0.56 | 6.93×10−7 |
| rs769177 | G/A | 6 | 31579834 | 17.2 | 0.81 | 1.35×10−6 | ||
| NFKBIL1 | rs2071593 | C/T | 6 | 31545022 | 18.8 | 0.82 | 1.59×10−6 |
Allele frequencies were analyzed using Fisher's exact test.
Major allele/minor allele. SNP, single nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio.
Table VI.
11 SNPs significantly (P<1.59×10−6) associated with chronic kidney disease in the exome-wide association study.
| Gene | SNP | Nucleotide substitutiona | Amino acid substitution | Chromosome | Position | MAF (%) | Allele OR | P-value (allele frequency) |
|---|---|---|---|---|---|---|---|---|
| COL6A5 | rs200982668 | G/A | E2501K | 3 | 130470894 | 1.3 | 0.21 | 5.08×10−9 |
| MARCH1 | rs61734696 | G/T | Q137K | 4 | 164197303 | 1.2 | 0.22 | 1.97×10−8 |
| MUC17 | rs78010183 | A/T | T1305S | 7 | 101035329 | 1.8 | 0.29 | 2.04×10−8 |
| MOB3C | rs139537100 | C/T | R24Q | 1 | 46615006 | 1.2 | 0.23 | 4.91×10−8 |
| PLCB2 | rs200787930 | C/T | E1106K | 15 | 40289298 | 1.2 | 0.23 | 4.94×10−8 |
| CXCL8 | rs188378669 | G/T | E31* | 4 | 73741568 | 1.2 | 0.23 | 7.83×10−8 |
| VPS33B | rs199921354 | C/T | R80Q | 15 | 91013841 | 1.2 | 0.23 | 7.86×10−8 |
| TMOD4 | rs115287176 | G/A | R277W | 1 | 151170961 | 1.2 | 0.23 | 1.25×10−7 |
| ADGRL3 | rs192210727 | G/T | R580I | 4 | 61909615 | 1.3 | 0.25 | 1.88×10−7 |
| ZNF77 | rs146879198 | G/A | R340* | 19 | 2934109 | 1.2 | 0.24 | 2.95×10−7 |
| COL6A3 | rs146092501 | C/T | E1386K | 2 | 237371861 | 1.2 | 0.24 | 2.99×10−7 |
Allele frequencies were analyzed using Fisher's exact test.
Major allele/minor allele. SNP, single nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio.
Multivariable logistic regression analysis of the association between SNPs and MI, hypertension, or CKD
The association of the 25 SNPs identified in the EWAS for MI with this condition was further examined by multivariable logistic regression analysis with adjustment for age, sex, and the prevalence of hypertension, diabetes mellitus, and dyslipidemia (Table VII). All 25 SNPs were significantly [P<0.0005 (0.05/100) in at least one genetic model] associated with MI. The association between the 11 SNPs identified in the EWAS for hypertension and this condition was examined by multivariable logistic regression analysis with adjustment for age and sex (Table VIII). All 11 SNPs were significantly [P<0.0011 (0.05/44)] associated with hypertension. The association between the 11 SNPs identified by the EWAS for CKD and this condition was also examined by multivariable logistic regression analysis with adjustment for age, sex, and the prevalence of hypertension and diabetes mellitus (Table IX). All 11 SNPs were significantly [P<0.0011 (0.05/44)] associated with CKD.
Table VII.
Association between SNPs and myocardial infarction as determined by multivariable logistic regression analysis.
| Gene | SNP | Dominant
|
Recessive
|
Additive 1
|
Additive 2
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | |||
| PLCB2 | rs200787930 | C/T | <0.0001 | 0.01 | 0.01-0.10 | <0.0001 | 0.01 | 0.01-0.10 | ||||||
| CXCL8 | rs188378669 | G/T | <0.0001 | 0.01 | 0.01-0.10 | <0.0001 | 0.01 | 0.01-0.10 | ||||||
| MARCH1 | rs61734696 | G/T | <0.0001 | 0.01 | 0.01-0.10 | <0.0001 | 0.01 | 0.01-0.10 | ||||||
| VPS33B | rs199921354 | C/T | <0.0001 | 0.01 | 0.01-0.10 | <0.0001 | 0.01 | 0.01-0.10 | ||||||
| TMOD4 | rs115287176 | G/A | <0.0001 | 0.01 | 0.01-0.11 | <0.0001 | 0.01 | 0.01-0.11 | ||||||
| COL6A3 | rs146092501 | C/T | <0.0001 | 0.01 | 0.01-0.10 | <0.0001 | 0.01 | 0.01-0.10 | ||||||
| ZNF77 | rs146879198 | G/A | <0.0001 | 0.01 | 0.01-0.11 | <0.0001 | 0.01 | 0.01-0.11 | ||||||
| ADGRL3 | rs192210727 | G/T | <0.0001 | 0.06 | 0.02-0.16 | 0.1736 | <0.0001 | 0.06 | 0.02-0.16 | 0.9960 | ||||
| ALDH2 | rs671 | G/A | <0.0001 | 2.00 | 1.56-2.55 | <0.0001 | 2.00 | 1.70-2.36 | 0.0040 | 1.46 | 1.13-1.90 | <0.0001 | 2.71 | 2.08-3.53 |
| ACAD10 | rs11066015 | G/A | <0.0001 | 0.51 | 0.39-0.65 | <0.0001 | 0.50 | 0.43-0.59 | 0.0049 | 0.69 | 0.53-0.89 | <0.0001 | 0.37 | 0.29-0.49 |
| BRAP | rs3782886 | A/G | <0.0001 | 1.98 | 1.67-2.34 | <0.0001 | 1.88 | 1.48-2.39 | <0.0001 | 1.84 | 1.55-2.20 | <0.0001 | 2.58 | 1.99-3.35 |
| HECTD4 | rs11066280 | T/A | <0.0001 | 2.02 | 1.71-2.38 | <0.0001 | 1.92 | 1.51-2.45 | <0.0001 | 1.88 | 1.58-2.24 | <0.0001 | 2.66 | 2.05-3.46 |
| HECTD4 | rs2074356 | C/T | <0.0001 | 1.85 | 1.57-2.17 | <0.0001 | 1.92 | 1.47-2.51 | <0.0001 | 1.73 | 1.46-2.06 | <0.0001 | 2.48 | 1.87-3.28 |
| OR52E4 | rs11823828 | T/G | <0.0001 | 1.56 | 1.30-1.87 | <0.0001 | 2.16 | 1.74-2.68 | 0.0152 | 1.28 | 1.05-1.56 | <0.0001 | 2.46 | 1.93-3.13 |
| rs12229654 | T/G | <0.0001 | 1.64 | 1.39-1.92 | <0.0001 | 1.89 | 1.41-2.53 | <0.0001 | 1.54 | 1.30-1.82 | <0.0001 | 2.27 | 1.67-3.07 | |
| NAA25 | rs12231744 | C/T | <0.0001 | 0.60 | 0.51-0.71 | <0.0001 | 0.49 | 0.37-0.64 | <0.0001 | 0.67 | 0.56-0.79 | <0.0001 | 0.40 | 0.30-0.53 |
| ATXN2 | rs7969300 | T/C | <0.0001 | 0.59 | 0.50-0.70 | <0.0001 | 0.55 | 0.43-0.71 | <0.0001 | 0.65 | 0.55-0.77 | <0.0001 | 0.44 | 0.34-0.57 |
| GOSR2 | rs1052586 | T/C | 0.0002 | 0.71 | 0.59-0.85 | <0.0001 | 0.61 | 0.50-0.75 | 0.0236 | 0.80 | 0.66-0.97 | <0.0001 | 0.53 | 0.42-0.67 |
| TCHP | rs74416240 | G/A | <0.0001 | 1.46 | 1.23-1.73 | 0.2849 | <0.0001 | 1.46 | 1.22-1.74 | 0.1399 | ||||
| GIT2 | rs925368 | A/G | <0.0001 | 1.48 | 1.24-1.76 | 0.5500 | <0.0001 | 1.49 | 1.25-1.79 | 0.3309 | ||||
| rs2523644 | A/G | <0.0001 | 1.74 | 1.41-2.14 | 0.5858 | <0.0001 | 1.76 | 1.42-2.17 | 0.4407 | |||||
| OAS3 | rs2072134 | C/T | <0.0001 | 1.45 | 1.23-1.71 | 0.0408 | 1.51 | 1.02-2.25 | <0.0001 | 1.42 | 1.20-1.69 | 0.0093 | 1.71 | 1.14-2.55 |
| CCDC63 | rs10774610 | T/C | <0.0001 | 1.38 | 1.18-1.62 | 0.0085 | 1.49 | 1.11-2.01 | 0.0009 | 1.33 | 1.12-1.58 | 0.0009 | 1.68 | 1.24-2.29 |
| rs2596548 | G/T | <0.0001 | 1.95 | 1.53-2.48 | 0.3820 | <0.0001 | 2.04 | 1.60-2.61 | 0.4739 | |||||
| CCDC63 | rs10849915 | T/C | <0.0001 | 1.38 | 1.18-1.63 | 0.0184 | 1.44 | 1.06-1.94 | 0.0006 | 1.34 | 1.14-1.59 | 0.0021 | 1.63 | 1.19-2.22 |
Multivariable logistic regression analysis was performed with adjustment for age, sex, and the prevalence of hypertension, diabetes mellitus, and dyslipidemia. Based on Bonferroni's correction, P<0.0005 (0.05/100) was considered statistically significant. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Table VIII.
Association between SNPs and hypertension as determined by multivariable logistic regression analysis.
| Gene | SNP | Dominant
|
Recessive
|
Additive 1
|
Additive 2
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | |||
| MARCH1 | rs61734696 | G/T | 0.0002 | 0.62 | 0.48-0.79 | 0.0002 | 0.62 | 0.48-0.79 | ||||||
| PLCB2 | rs200787930 | C/T | 0.0003 | 0.63 | 0.49-0.81 | 0.0003 | 0.63 | 0.49-0.81 | ||||||
| COL6A3 | rs146092501 | C/T | 0.0002 | 0.61 | 0.47-0.79 | 0.0002 | 0.61 | 0.47-0.79 | ||||||
| COL6A5 | rs200982668 | G/A | 0.0005 | 0.65 | 0.50-0.83 | 0.0005 | 0.65 | 0.50-0.83 | ||||||
| MOB3C | rs139537100 | C/T | 0.0006 | 0.64 | 0.50-0.83 | 0.0006 | 0.64 | 0.50-0.83 | ||||||
| VPS33B | rs199921354 | C/T | 0.0003 | 0.63 | 0.49-0.81 | 0.0003 | 0.63 | 0.49-0.81 | ||||||
| NCR3 | rs2515920 | T/A | <0.0001 | 0.74 | 0.67-0.82 | 0.0930 | <0.0001 | 0.74 | 0.66-0.82 | 0.0226 | 0.72 | 0.54-0.95 | ||
| TMOD4 | rs115287176 | G/A | 0.0004 | 0.63 | 0.49-0.81 | 0.0004 | 0.63 | 0.49-0.81 | ||||||
| CXCL8 | rs188378669 | G/T | 0.0004 | 0.63 | 0.49-0.81 | 0.0004 | 0.63 | 0.49-0.81 | ||||||
| rs769177 | G/A | <0.0001 | 0.74 | 0.67-0.82 | 0.0946 | <0.0001 | 0.74 | 0.67-0.83 | 0.0238 | 0.72 | 0.54-0.96 | |||
| NFKBIL1 | rs2071593 | C/T | <0.0001 | 0.74 | 0.67-0.82 | 0.2491 | <0.0001 | 0.74 | 0.67-0.82 | 0.0628 | ||||
Multivariable logistic regression analysis was performed with adjustment for age and sex. Based on Bonferroni's correction, P<0.0011 (0.05/44) was considered statistically significant. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Table IX.
Association between SNPs and chronic kidney disease as determined by multivariable logistic regression analysis.
| Gene | SNP | Dominant
|
Recessive
|
Additive 1
|
Additive 2
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | |||
| COL6A5 | rs200982668 | G/A | 0.0002 | 0.25 | 0.12-0.52 | 0.0002 | 0.25 | 0.12-0.52 | ||||||
| MARCH1 | rs61734696 | G/T | 0.0004 | 0.26 | 0.12-0.55 | 0.0004 | 0.26 | 0.12-0.55 | ||||||
| MUC17 | rs78010183 | A/T | 0.0008 | 0.35 | 0.19-0.65 | 0.0008 | 0.35 | 0.19-0.65 | ||||||
| MOB3C | rs139537100 | C/T | 0.0006 | 0.27 | 0.13-0.57 | 0.0006 | 0.27 | 0.13-0.57 | ||||||
| PLCB2 | rs200787930 | C/T | 0.0007 | 0.27 | 0.13-0.57 | 0.0007 | 0.27 | 0.13-0.57 | ||||||
| CXCL8 | rs188378669 | G/T | 0.0004 | 0.26 | 0.12-0.55 | 0.0004 | 0.26 | 0.12-0.55 | ||||||
| VPS33B | rs199921354 | C/T | 0.0007 | 0.27 | 0.13-0.58 | 0.0007 | 0.27 | 0.13-0.58 | ||||||
| TMOD4 | rs115287176 | G/A | 0.0007 | 0.27 | 0.13-0.58 | 0.0007 | 0.27 | 0.13-0.58 | ||||||
| ADGRL3 | rs192210727 | G/T | 0.0006 | 0.27 | 0.12-0.57 | 0.4876 | 0.0007 | 0.27 | 0.13-0.57 | 0.9964 | ||||
| ZNF77 | rs146879198 | G/A | 0.0009 | 0.28 | 0.13-0.59 | 0.0009 | 0.28 | 0.13-0.59 | ||||||
| COL6A3 | rs146092501 | C/T | 0.0010 | 0.28 | 0.13-0.60 | 0.0010 | 0.28 | 0.13-0.60 | ||||||
Multivariable logistic regression analysis was performed with adjustment for age, sex, and the prevalence of hypertension and diabetes mellitus. Based on Bonferroni's correction, P<0.0011 (0.05/44) was considered statistically significant. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Association between SNPs associated with MI and intermediate phenotypes
The present study examined the associations between the 25 SNPs associated with MI and intermediate phenotypes of this condition, including hypertension, diabetes mellitus, hypertriglyceridemia, hypo-HDL-cholesterolemia, hyper-low density lipoprotein (LDL)-cholesterolemia, CKD, obesity, and hyperuricemia, using Pearson's χ2 test. The SNPs rs200787930 of PLCB2, rs188378669 of CXCL8, rs61734696 of MARCH1, rs199921354 of VPS33B, rs115287176 of TMOD4, rs146092501 of COL6A3, rs146879198 of ZNF77, and rs192210727 of ADGRL3 were significantly [P<0.0003 (0.05/200)] associated with hypo-HDL-cholesterolemia, hyper-LDL-cholesterolemia, and CKD, with rs61734696 also being associated with hypertension. The SNPs rs671 of ALDH2, rs11066015 of ACAD10, and rs11066280 and rs2074356 of HECTD4 were significantly associated with hypo-HDL-cholesterolemia, hyper-LDL-cholesterolemia, and hyperuricemia. The SNPs rs3782886 of BRAP and rs12229654 at 12q24.1 were significantly associated with hypo-HDL-cholesterolemia and hyperuricemia. The SNP rs11823828 of OR52E4 was significantly associated with diabetes mellitus, rs12231744 of NAA25 with hypo-HDL-cholesterolemia, rs7969300 of ATXN2 with hyper-triglyceridemia, and rs10774610 and rs10849915 of CCDC63 with hyperuricemia (Table X).
Table X.
Association between SNPs associated with myocardial infarction and intermediate phenotypes.
| Gene | SNP | Hypertension | DM | Hyper-TG | Hypo-HDL | Hyper-LDL | CKD | Obesity | Hyperuricemia | |
|---|---|---|---|---|---|---|---|---|---|---|
| PLCB2 | rs200787930 | C/T | 0.0004 | 0.0026 | 0.5013 | <0.0001 | <0.0001 | <0.0001 | 0.0482 | 0.7764 |
| CXCL8 | rs188378669 | G/T | 0.0007 | 0.0039 | 0.8922 | <0.0001 | <0.0001 | <0.0001 | 0.0259 | 0.9412 |
| MARCH1 | rs61734696 | G/T | 0.0002 | 0.0013 | 0.5876 | <0.0001 | <0.0001 | <0.0001 | 0.0381 | 0.7835 |
| VPS33B | rs199921354 | C/T | 0.0006 | 0.0026 | 0.5939 | <0.0001 | <0.0001 | <0.0001 | 0.0398 | 0.9038 |
| TMOD4 | rs115287176 | G/A | 0.0006 | 0.0073 | 0.9087 | <0.0001 | <0.0001 | <0.0001 | 0.0578 | 0.8457 |
| COL6A3 | rs146092501 C | /T | 0.0004 | 0.0058 | 0.8385 | <0.0001 | <0.0001 | <0.0001 | 0.0459 | 0.8305 |
| ZNF77 | rs146879198 | G/A | 0.0014 | 0.0053 | 0.8625 | <0.0001 | <0.0001 | <0.0001 | 0.0245 | 0.9643 |
| ADGRL3 | rs192210727 | G/T | 0.0106 | 0.0262 | 0.6918 | <0.0001 | <0.0001 | <0.0001 | 0.0501 | 0.5669 |
| ALDH2 | rs671 | G/A | 0.0017 | 0.0231 | 0.0253 | <0.0001 | 0.0002 | 0.0297 | 0.0479 | <0.0001 |
| ACAD10 | rs11066015 | G/A | 0.0014 | 0.0300 | 0.0261 | <0.0001 | 0.0002 | 0.0376 | 0.0578 | <0.0001 |
| BRAP | rs3782886 | A/G | 0.0012 | 0.0675 | 0.0233 | <0.0001 | 0.0008 | 0.0329 | 0.0548 | <0.0001 |
| HECTD4 | rs11066280 | T/A | 0.0052 | 0.2161 | 0.0093 | <0.0001 | 0.0002 | 0.0172 | 0.1293 | <0.0001 |
| HECTD4 | rs2074356 | C/T | 0.0245 | 0.2148 | 0.0042 | <0.0001 | 0.0003 | 0.0218 | 0.2238 | <0.0001 |
| OR52E4 | rs11823828 | T/G | 0.0429 | <0.0001 | 0.0917 | 0.2341 | 0.8312 | 0.0161 | 0.0768 | 0.1053 |
| rs12229654 | T/G | 0.0200 | 0.4309 | 0.0996 | <0.0001 | 0.0290 | 0.0633 | 0.2134 | <0.0001 | |
| NAA25 | rs12231744 | C/T | 0.0913 | 0.5214 | 0.0151 | 0.0002 | 0.2175 | 0.8586 | 0.3543 | 0.0055 |
| ATXN2 | rs7969300 | T/C | 0.0368 | 0.6341 | 0.0001 | 0.0005 | 0.7189 | 0.3631 | 0.3155 | 0.0047 |
| GOSR2 | rs1052586 | T/C | 0.5175 | 0.0843 | 0.3162 | 0.0718 | 0.1779 | 0.6525 | 0.4307 | 0.5766 |
| TCHP | rs74416240 | G/A | 0.4244 | 0.0279 | 0.0630 | 0.0594 | 0.0142 | 0.0037 | 0.3409 | 0.1148 |
| GIT2 | rs925368 | A/G | 0.3492 | 0.0515 | 0.0337 | 0.0873 | 0.0105 | 0.0020 | 0.4399 | 0.0998 |
| rs2523644 | A/G | 0.4464 | 0.2581 | 0.5651 | 0.0797 | 0.0674 | 0.0397 | 0.4203 | 0.3215 | |
| OAS3 | rs2072134 | C/T | 0.6477 | 0.0936 | 0.1902 | 0.0004 | 0.0030 | 0.1862 | 0.2580 | 0.0185 |
| CCDC63 | rs10774610 | T/C | 0.3573 | 0.2133 | 0.2623 | 0.0030 | 0.0102 | 0.0055 | 0.5909 | <0.0001 |
| rs2596548 | G/T | 0.2749 | 0.0268 | 0.2615 | 0.2138 | 0.3223 | 0.0751 | 0.9065 | 0.2475 | |
| CCDC63 | rs10849915 | T/C | 0.3386 | 0.2256 | 0.2891 | 0.0025 | 0.0135 | 0.0060 | 0.5869 | <0.0001 |
Data are P-values. The association between genotypes of each SNP and intermediate phenotypes was examined with Pearson's χ2 test. Based on Bonferroni's correction, P<0.0003 (0.05/200) were considered statistically significant and are shown in bold. SNP, single nucleotide polymorphism; DM, diabetes mellitus; hyper-TG, hypertriglyceridemia; hypo-HDL, hypo-high density lipoprotein-cholesterolemia; hyper-LDL, hyper-low density lipoprotein-cholesterolemia; CKD, chronic kidney disease.
Association between SNPs associated with hypertension and systolic or diastolic BP
The present study examined the associations between genotypes for the 11 SNPs associated with hypertension and systolic or diastolic BP by one-way ANOVA (Table XI). All 11 SNPs were significantly [P<0.0023 (0.05/22)] associated with systolic BP, whereas only rs2071593 of NFKBIL1 was associated with diastolic BP.
Table XI.
Association between SNPs associated with hypertension and systolic or diastolic BP.
| Gene | SNP | Systolic BP (mmHg) | P-value | Diastolic BP (mmHg) | P-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MARCH1 | rs61734696 | G/T | GG | GT | <0.0001 | GG | GT | 0.0239 | ||
| 128±24 | 119±16 | 77±14 | 75±12 | |||||||
| PLCB2 | rs200787930 C | /T | CC | CT | <0.0001 | CC | CT | 0.0482 | ||
| 128±24 | 119±16 | 77±14 | 75±12 | |||||||
| COL6A3 | rs146092501 C | /T | CC | CT | <0.0001 | CC | CT | 0.0584 | ||
| 128±24 | 119±15 | 77±14 | 75±12 | |||||||
| COL6A5 | rs200982668 | G/A | GG | GA | <0.0001 | GG | GA | 0.0341 | ||
| 128±24 | 119±16 | 77±14 | 75±12 | |||||||
| MOB3C | rs139537100 C | /T | CC | CT | <0.0001 | CC | CT | 0.0359 | ||
| 128±24 | 119±16 | 77±14 | 75±12 | |||||||
| VPS33B | rs199921354 C | /T | CC | CT | <0.0001 | CC | CT | 0.0348 | ||
| 128±24 | 119±16 | 77±14 | 75±12 | |||||||
| NCR3 | rs2515920 | T/A | TT | TA | AA | <0.0001 | TT | TA | AA | 0.0062 |
| 128±24 | 125±22 | 126±23 | 77±14 | 76±13 | 76±13 | |||||
| TMOD4 | rs115287176 | G/A | GG | GA | <0.0001 | GG | GA | 0.0793 | ||
| 128±24 | 119±16 | 77±14 | 75±12 | |||||||
| CXCL8 | rs188378669 | G/T | GG | GT | <0.0001 | GG | GT | 0.0703 | ||
| 128±24 | 119±16 | 77±14 | 75±12 | |||||||
| rs769177 | G/A | GG | GA | AA | 0.0001 | GG | GA | AA | 0.0094 | |
| 128±24 | 125±22 | 126±23 | 77±14 | 76±13 | 76±13 | |||||
| NFKBIL1 | rs2071593 | C/T | CC | CT | TT | <0.0001 | CC | CT | TT | 0.0012 |
| 128±24 | 125±22 | 127±23 | 77±14 | 76±13 | 76±13 | |||||
Data are presented as the mean ± standard deviation and were compared among genotypes by one-way analysis of variance. Based on Bonferroni's correction, P<0.0023 (0.05/22) was considered statistically significant and are shown in bold. SNP, single nucleotide polymorphism; BP, blood pressure.
Association between SNPs associated with CKD and the serum concentration of creatinine or eGFR
The present study examined the associations between the 11 SNPs associated with CKD and the serum concentration of creatinine or eGFR by one-way ANOVA (Table XII). All 11 SNPs were significantly [P<0.0023 (0.05/22)] associated with eGFR, however, none were associated with the serum concentration of creatinine.
Table XII.
Association between SNPs associated with chronic kidney disease and the serum concentration of creatinine and eGFR.
| Gene | SNP | Serum creatinine (µmol/l) | P-value | eGFR (ml min−1 1.73 m−2) | P-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| COL6A5 | rs200982668 | G/A | GG | GA | 0.0281 | GG | GA | <0.0001 | ||
| 93.6±138.5 | 61.8±34.4 | 79.3±32.9 | 95.2±20.2 | |||||||
| MARCH1 | rs61734696 | G/T | GG | GT | 0.0352 | GG | GT | <0.0001 | ||
| 93.5±138.3 | 62.4±35.0 | 79.3±32.9 | 94.7±20.3 | |||||||
| MUC17 | rs78010183 | A/T | AA | AT | 0.0068 | AA | AT | <0.0001 | ||
| 93.8±139.0 | 59.2±18.8 | 79.4±33.0 | 93.0±19.9 | |||||||
| MOB3C | rs139537100 C | /T | CC | CT | 0.0374 | CC | CT | <0.0001 | ||
| 93.5±138.4 | 62.4±35.5 | 79.3±32.9 | 94.9±20.8 | |||||||
| PLCB2 | rs200787930 C | /T | CC | CT | 0.0380 | CC | CT | <0.0001 | ||
| 93.5±138.3 | 62.5±35.4 | 79.3±32.9 | 94.8±20.8 | |||||||
| CXCL8 | rs188378669 | G/T | GG | GT | 0.0397 | GG | GT | <0.0001 | ||
| 93.5±138.3 | 62.6±35.6 | 79.3±32.9 | 95.0±20.9 | |||||||
| VPS33B | rs199921354 C | /T | CC | CT | 0.0406 | CC | CT | <0.0001 | ||
| 93.5±138.4 | 62.6±35.8 | 79.3±32.9 | 94.8±21.1 | |||||||
| TMOD4 | rs115287176 | G/A | GG | GA | 0.0426 | GG | GA | <0.0001 | ||
| 93.4±138.2 | 62.8±35.8 | 79.4±32.9 | 94.6±20.9 | |||||||
| ADGRL3 | rs192210727 | G/T | GG | GT | TT | 0.1294 | GG | GT | TT | 0.0002 |
| 93.2±137.5 | 63.0±35.8 | 64.5 | 79.3±32.9 | 94.4±21.4 | 94.3 | |||||
| ZNF77 | rs146879198 | G/A | GG | GA | 0.0491 | GG | GA | <0.0001 | ||
| 93.4±138.1 | 63.1±36.4 | 79.4±32.9 | 94.3±21.1 | |||||||
| COL6A3 | rs146092501 C | /T | CC | CT | 0.0466 | CC | CT | <0.0001 | ||
| 93.4±138.2 | 62.8±36.4 | 79.4±32.9 | 94.6±21.2 | |||||||
Data are presented as the mean ± standard deviation and were compared among genotypes by one-way analysis of variance. Based on Bonferroni's correction, P<0.0023 (0.05/22) was considered statistically significant and are shown in bold. SNP, single nucleotide polymorphism; eGFR, estimated glomerular filtration rate.
Linkage disequilibrium analyses
The linkage disequilibrium (LD) among SNPs associated with MI, hypertension, or CKD was examined. For the MI study, rs192210727 of ADGRL3, rs188378669 of CXCL8, and rs61734696 of MARCH1 were in LD [square of the correlation coefficient (r2), 0.907-0.972]. Two SNPs (rs2596548, rs2523644) at chromosome 6p21.3 were also in LD (r2=0.624). LD plots of the 13 SNPs located at chromosome 12q24.11 to 12q24.13 are shown in Fig. 1. There was significant LD between rs74416240 of TCHP and rs925368 of GIT2 (r2=0.937); between rs10849915 and rs10774610 of CCDC63 (r2=0.992); and among rs3782886 of BRAP, rs11066015 of ACAD10, rs671 of ALDH2, and rs2074356 and rs11066280 of HECTD4 (r2=0.814-0.994). A significant LD (r2=0.994) was also apparent between rs200787930 of PLCB2 and rs199921354 of VPS33B.
Figure 1.
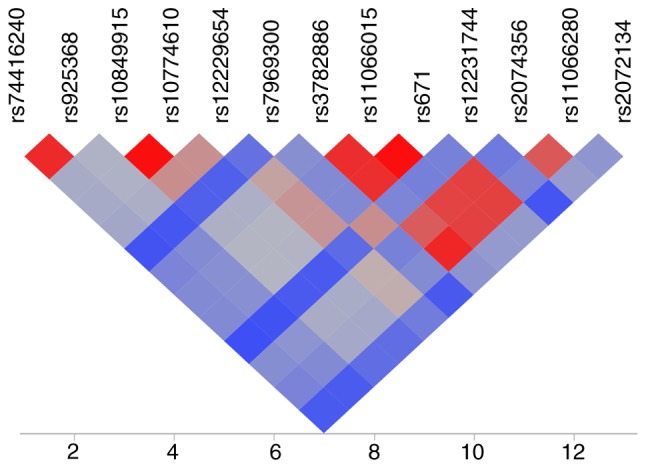
LD map of 13 SNPs at 12q24.11-12q24.13 associated with myocardial infarction. LD was calculated as the square of the correlation coefficient (r2), with the extent of LD increasing according to the color order of blue < gray < red. LD, linkage disequilibrium.
For the hypertension study, there was significant LD between rs139537100 of MOB3C and rs115287176 of TMOD4 (r2=0.984); between rs188378669 of CXCL8 and rs61734696 of MARCH1 (r2=0.972); among rs2071593 of NFKBIL1, rs769177 at 6p21.3, and rs2515920 of NCR3 (r2=0.883-0.980); and between rs200787930 of PLCB2 and rs199921354 of VPS33B (r2=0.994).
For the CKD study, complete LD (r2=1.000) was apparent between rs139537100 of MOB3C and rs115287176 of TMOD4, and between rs200787930 of PLCB2 and rs199921354 of VPS33B. Significant LD (r2=0.905-0.952) was also observed among rs192210727 of ADGRL3, rs188378669 of CXCL8, and rs61734696 of MARCH1.
Associations between the genes, chromosomal loci and SNPs identified in the present study and the phenotypes examined in previous GWASs
The present study examined the association of the genes, chromosomal loci and SNPs identified in the present study with the phenotypes previously investigated by GWASs available in the Genome-Wide Repository of Associations Between SNPs and Phenotypes Search database (https://grasp.nhlbi.nih.gov/Search.aspx) developed by the Information Technology and Applications Center at the National Center for Biotechnology Information (National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, USA) (60,61).
For the MI study, chromosomal region 6p21.3, ATXN2, BRAP, ACAD10, ALDH2, NAA25, and HECTD4 were previously shown to be associated with MI or CAD (Table XIII). The remaining 15 genes or loci identified in the present study have not been previously found to be associated with MI or CAD, although GOSR2 was previously found to be associated with systolic BP. For the hypertension study, none of the genes, chromosomal loci, or SNPs identified were found to be associated with systolic or diastolic BP or with hypertension in previous GWASs, although 6p21.3 was previously shown to be associated with CAD (Table XIV). For the CKD study, none of the genes or SNPs identified were shown to be associated with renal function-related traits or CKD in previous GWASs (Table XV).
Table XIII.
Association between genes, chromosomal loci, and SNPs associated with MI in the present study and previously examined cardiovascular disease-related phenotypes.
| Gene/chr. locus | SNP | Chr. | Position | Previously examined phenotypes |
|---|---|---|---|---|
| TMOD4 | rs115287176 | 1 | 151170961 | None |
| COL6A3 | rs146092501 | 2 | 237371861 | None |
| ADGRL3 | rs192210727 | 4 | 61909615 | None |
| CXCL8 | rs188378669 | 4 | 73741568 | Total cholesterol (23063622) |
| MARCH1 | rs61734696 | 4 | 164197303 | Adiponectin concentrations (20887962) |
| 6p21.3 | rs2596548 | 6 | 31362769 | MI (21971053) |
| rs2523644 | 31374707 | |||
| OR52E4 | rs11823828 | 11 | 5884973 | None |
| TCHP | rs74416240 | 12 | 109904793 | None |
| GIT2 | rs925368 | 12 | 109953174 | None |
| CCDC63 | rs10849915 | 12 | 110895818 | None |
| rs10774610 | 110902439 | |||
| 12q24.1 | rs12229654 | 12 | 110976657 | HDL-cholesterol (21909109) |
| ATXN2 | rs7969300 | 12 | 111555908 | MI (19820697, 19198610), CAD (19820697, 23202125, 21060863) |
| BRAP | rs3782886 | 12 | 111672685 | MI (21971053, 19820697, 21107343), CAD (21971053, 21572416, 23202125, 19820697, 23364394, 21060863) |
| ACAD10 | rs11066015 | 12 | 111730205 | CAD (23364394, 23202125) |
| ALDH2 | rs671 | 12 | 111803962 | MI (21971053), CAD (21971053, 21572416, 23202125) |
| NAA25 | rs12231744 | 12 | 112039251 | MI (19820697, 23202125), CAD (21971053, 19820697) |
| HECTD4 | rs2074356 | 12 | 112207597 | MI (19820697), CAD (21971053, 21572416, 22751097, 19820697, 23364394, 23202125) |
| rs11066280 | 112379979 | |||
| OAS3 | rs2072134 | 12 | 112971371 | HDL-cholesterol (21909109) |
| PLCB2 | rs200787930 | 15 | 40289298 | Triglycerides (23063622) |
| VPS33B | rs199921354 | 15 | 91013841 | Type 2 diabetes (22885922) |
| GOSR2 | rs1052586 | 17 | 46941097 | Systolic BP (21909110, 21909115) |
| ZNF77 | rs146879198 | 19 | 2934109 | None |
Data were obtained from the Genome-Wide Repository of Associations Between SNPs and Phenotypes Search database (https://grasp.nhlbi.nih.gov/Search.aspx) with P<1.0×10−6. Numbers in parentheses are PubMed IDs. SNP, single nucleotide polymorphism; MI, myocardial infarction; Chr., chromosome; HDL, high density lipoprotein; CAD, coronary artery disease; BP, blood pressure.
Table XIV.
Association between genes, chromosomal loci, and SNPs associated with hypertension in the present study and previously examined cardiovascular disease-related phenotypes.
| Gene/chr. locus | SNP | Chr. | Position | Previously examined phenotypes |
|---|---|---|---|---|
| MOB3C | rs139537100 | 1 | 46615006 | None |
| TMOD4 | rs115287176 | 1 | 151170961 | None |
| COL6A3 | rs146092501 | 2 | 237371861 | None |
| COL6A5 | rs200982668 | 3 | 130470894 | None |
| CXCL8 | rs188378669 | 4 | 73741568 | Total cholesterol (23063622) |
| MARCH1 | rs61734696 | 4 | 164197303 | Adiponectin concentrations (20887962) |
| NFKBIL1 | rs2071593 | 6 | 31545022 | Total cholesterol (20686565) |
| 6p21.3 | rs769177 | 6 | 31579834 | Total cholesterol (20686565), triglycerides (20686565), CAD (21971053) |
| NCR3 | rs2515920 | 6 | 31594838 | Total cholesterol (20686565) |
| PLCB2 | rs200787930 | 15 | 40289298 | Triglycerides (23063622) |
| VPS33B | rs199921354 | 15 | 91013841 | Type 2 diabetes (22885922) |
Data were obtained from the Genome-Wide Repository of Associations Between SNPs and Phenotypes Search database (https://grasp.nhlbi.nih.gov/Search.aspx) with a P<1.0×10−6. Numbers in parentheses are PubMed IDs. SNP, single nucleotide polymorphism; Chr., chromosome; CAD, coronary artery disease.
Table XV.
Association between genes and SNPs associated with chronic kidney disease in the present study and previously examined cardiovascular and renal phenotypes.
| Gene | SNP | Chr. | Position | Previously examined phenotypes |
|---|---|---|---|---|
| MOB3C | rs139537100 | 1 | 46615006 | None |
| TMOD4 | rs115287176 | 1 | 151170961 | None |
| COL6A3 | rs146092501 | 2 | 237371861 | None |
| COL6A5 | rs200982668 | 3 | 130470894 | None |
| ADGRL3 | rs192210727 | 4 | 61909615 | None |
| CXCL8 | rs188378669 | 4 | 73741568 | Total cholesterol (23063622) |
| MARCH1 | rs61734696 | 4 | 164197303 | Adiponectin concentrations (20887962) |
| MUC17 | rs78010183 | 7 | 101035329 | None |
| PLCB2 | rs200787930 | 15 | 40289298 | Triglycerides (23063622) |
| VPS33B | rs199921354 | 15 | 91013841 | Type 2 diabetes (22885922) |
| ZNF77 | rs146879198 | 19 | 2934109 | None |
Data were obtained from the Genome-Wide Repository of Associations Between SNPs and Phenotypes Search database (https://grasp.nhlbi.nih.gov/Search.aspx) with a P<1.0×10−6. Numbers in parentheses are PubMed IDs. SNP, single nucleotide polymorphism; Chr., chromosome.
Gene Ontology (GO) analysis of genes identified in the present study
The potential biological functions of the 18 genes (MOB3C, TMOD4, COL6A3, ADGRL3, CXCL8, MARCH1, OR52E4, TCHP, GIT2, CCDC63, OAS3, PLCB2, VPS33B, ZNF77, COL6A5, NFKBIL1, NCR3, MUC17) identified in the present study were examined with the use of the QuickGO database of Gene Ontology and GO Annotations (https://www.ebi.ac.uk/QuickGO; European Bioinformatics Institute, European Molecular Biology Laboratory, Hinxton, UK) (62,63). The GO analysis predicted various biological functions for the 18 genes, including those associated with muscle contraction (TMOD4), cell adhesion (COL6A3, COL6A5), G protein-coupled receptor signaling (ADGRL3, OR52E4, PLCB2), the inflammatory response (CXCL8), the immune response (MARCH1, OAS3 NCR3), apoptosis (TCHP), GTPase activity (GIT2), protein transport (VPS33B), NF-κB signaling (NFKBIL1), and lectin receptor signaling (MUC17) (Table XVI).
Table XVI.
Gene Ontology analysis of the 18 genes identified in the present study.
| Gene | Function | Biological process |
|---|---|---|
| MOB3C | Protein binding, metal ion binding | Uncharacterized |
| TMOD4 | Actin binding, tropomyosin binding | Muscle contraction, actin filament organization, myofibril assembly, pointed-end actin filament capping |
| COL6A3 | Serine-type endopeptidase inhibitor activity, extracellular matrix structural constituent conferring tensile strength, collagen-containing extracellular matrix | Cell adhesion, muscle organ development, negative regulation of endopeptidase activity, extracellular matrix organization |
| ADGRL3 | G protein-coupled receptor activity, calcium ion binding, protein binding, carbohydrate binding | Neuron migration, signal transduction, G protein-coupled receptor signaling pathway, positive regulation of synapse assembly, cell-cell adhesion via plasma-membrane adhesion molecules |
| CXCL8 | Cytokine activity, interleukin-8 receptor binding, protein binding | Positive regulation of angiogenesis, induction of positive chemotaxis, defense response, inflammatory response, immune response, cell cycle arrest, negative regulation of G protein-coupled receptor protein signaling pathway, negative regulation of cell proliferation, cytokine-mediated signaling pathway, calcium-mediated signaling, regulation of cell adhesion, positive regulation of neutrophil chemotaxis, receptor internalization, response to endoplasmic reticulum stress, intracellular signal transduction, PKR-like endoplasmic reticulum kinase-mediated unfolded protein response, neutrophil activation, cellular responses to fibroblast growth factor stimulus, lipopolysaccharide, interleukin-1, and tumor necrosis factor |
| MARCH1 | Ubiquitin-protein transferase activity, zinc ion binding, MHC protein binding, ubiquitin protein ligase activity | Protein polyubiquitination, immune response, antigen processing and presentation of peptide antigen via MHC class II |
| OR52E4 | G protein-coupled receptor activity, olfactory receptor activity | Signal transduction, G protein-coupled receptor signaling pathway, sensory perception of smell, response to stimulus, detection of chemical stimulus involved in sensory perception of smell |
| TCHP | Protein binding | Apoptotic process, negative regulation of cell growth, negative regulation of cilium assembly |
| GIT2 | GTPase activator, kinase activity, metal ion binding | Phosphorylation, positive regulation of GTPase activity |
| CCDC63 | Cell differentiation | Spermatogenesis, spermatid development, cilium movement, outer dynein arm assembly |
| OAS3 | Nucleotide binding, 2′-5′-oligoadenylate synthetase activity, RNA binding, protein binding, ATP binding, nucleotidyltransferase activity, metal ion binding | Immune response, nucleobase-containing compound metabolic process, defense response to virus, negative regulation of viral genome replication, interferon-g-mediated signaling pathway, regulation of ribonuclease activity |
| PLCB2 | Phosphatidylinositol phospholipase C activity, calcium ion binding, phosphoric diester hydrolase activity | Phospholipid metabolic process, intracellular signal transduction, G protein-coupled receptor signaling pathway, activation of phospholipase C activity, Wnt signaling pathway, calcium-modulating pathway, inositol phosphate metabolic process |
| VPS33B | Protein binding, AP-3 adaptor complex, clathrin complex | Vesicle docking involved in exocytosis, endosome organization, protein transport, vesicle-mediated transport, peptidyllysine hydroxylation, melanosome localization, lysosome localization, collagen metabolic process, membrane fusion, platelet a granule organization |
| ZNF77 | RNA polymerase II transcription factor activity, sequence-specific DNA binding, metal ion binding | Regulation of DNA-templated transcription, regulation of transcription by RNA polymerase II |
| COL6A5 | Collagen-containing extracellular matrix, protein binding, extracellular matrix structural constituent conferring tensile strength | Cell adhesion |
| NFKBIL1 | Protein binding | Inhibitor of NF-κB kinase/NF-κB signaling, cytoplasmic sequestering of transcription factor, negative regulation of lipopolysaccharide-mediated signaling pathway, NF-κB transcription factor activity, tumor necrosis factor production, Toll-like receptor signaling pathway |
| NCR3 | Signaling receptor binding, protein binding, protein homodimerization activity, cell adhesion molecule binding | Regulation of immune response, homophilic and heterophilic cell-cell adhesion via plasma-membrane adhesion molecules, cell recognition, positive regulation of natural killer cell-mediated cytotoxicity, susceptibility to natural killer cell-mediated cytotoxicity and T cell-mediated cytotoxicity |
| MUC17 | Protein binding, PDZ domain binding, extracellular matrix constituent, lubricant activity | Stimulatory C-type lectin receptor signaling pathway, O-glycan processing, cellular homeostasis |
Data for predicted functions and biological processes for the genes were obtained from the database of Gene Ontology and GO Annotations (QuickGO; https://www.ebi.ac.uk/QuickGO). GO, Gene Ontology; MHC, major histocompatibility complex; NF-κB, nuclear factor-κB.
Network analysis of genes identified in the present study
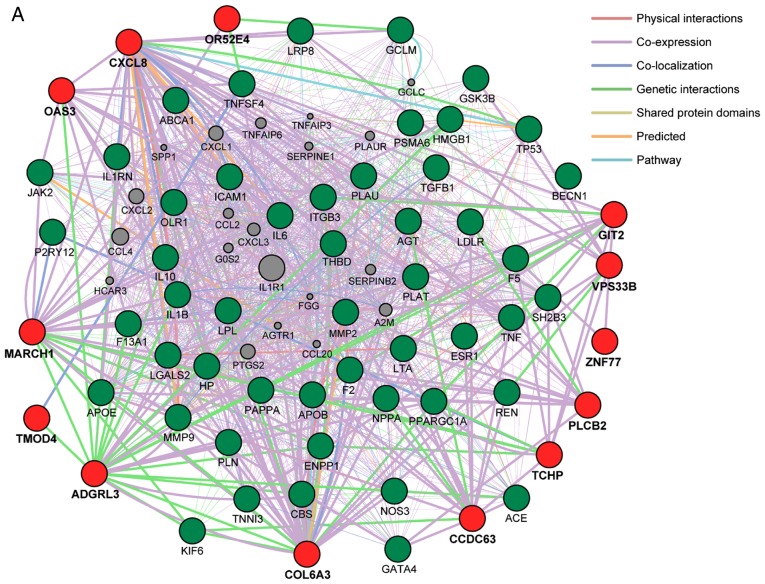
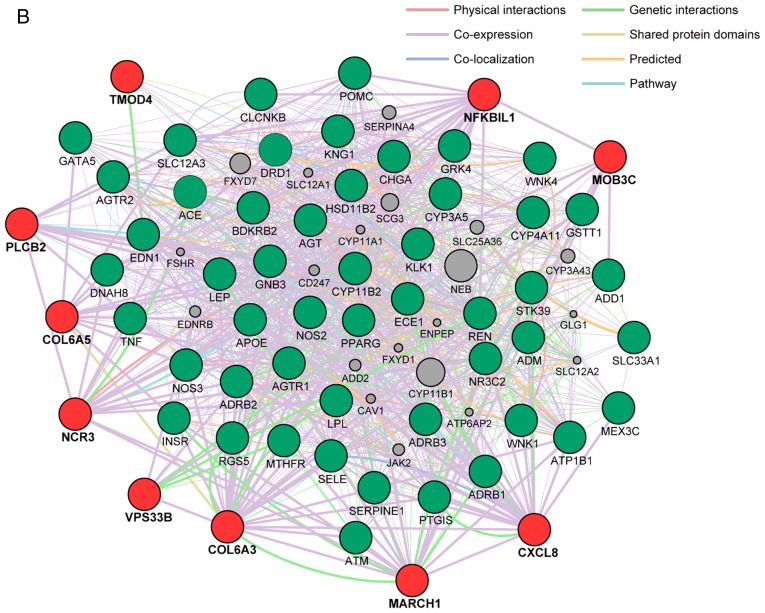
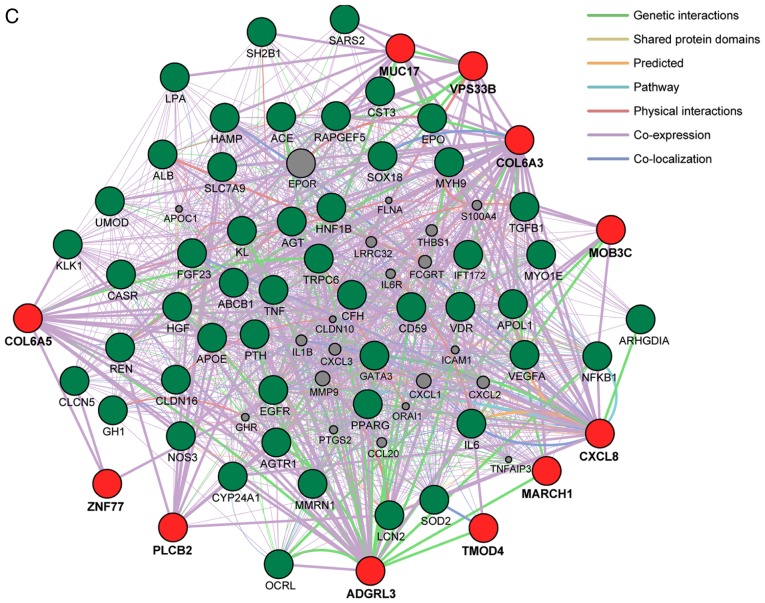
Network analysis of the 18 genes identified in the present study was performed to predict functional gene-gene interactions with the use of the GeneMANIA Cytoscape plugin (http://apps.cytoscape.org/apps/genemania; Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, ON, Canada) (64-66) and Cytoscape v3.4.0 software (http://www.cytoscape.org; The Cytoscape Consortium, San Diego, CA, USA) (67). Three sets of 50 genes were selected from the DisGeNET database (http://www.disgenet.org/web/DisGeNET; Integrative Biomedical Informatics Group, Research Programme on Biomedical Informatics, Barcelona Biomedical Research Park, Barcelona, Spain) (68,69) according to the rank order (high to low) of scores for association with MI, hypertension, or CKD (Fig. 2). The network analysis revealed that the 13 (Fig. 2A), 10 (Fig. 2B), and 11 (Fig. 2C) genes found to be associated with MI, hypertension, and CKD, respectively, in the present study have potential direct or indirect interactions with the corresponding sets of 50 genes previously shown to be associated with these conditions.
Figure 2.
(A) Network analysis of the 13 genes (TMOD4, COL6A3, ADGRL3, CXCL8, MARCH1, OR52E4, TCHP, GIT2, CCDC63, OAS3, PLCB2, VPS33B, ZNF77) associated with MI in the present study (red circles) was performed to predict functional gene-gene interactions with the use of the GeneMANIA Cytoscape plugin (http://apps.cytoscape.org/apps/genemania) and Cytoscape v3.4.0 software (http://www.cytoscape.org). A total of 50 genes (green circles) selected from the DisGeNET database (http://www.disgenet.org/web/DisGeNET) according to the rank order (high to low) of scores for association with MI were applied to the analysis. Interactions between red circles or between red and green circles are indicated by bold lines. Molecules represented by gray circles are putative mediators of interactions between genes. (B) Network analysis of the 10 genes (MOB3C, TMOD4, COL6A3, COL6A5, CXCL8, MARCH1, NFKBIL1, NCR3, PLCB2, VPS33B) associated with hypertension in the present study (red circles) was performed in the same manner; 50 genes (green circles) selected from the DisGeNET database according to the rank order of scores for association with hypertension were applied to the analysis. MI, myocardial infarction. (C) Network analysis of the 11 genes (MOB3C, TMOD4, COL6A3, COL6A5, ADGRL3, CXCL8, MARCH1, MUC17, PLCB2, VPS33B, ZNF77) associated with CKD in the present study (red circles) was performed in the same manner; 50 genes (green circles) selected from the DisGeNET database according to the rank order of scores for association with CKD were applied to the analysis. CKD, chronic kidney disease.
Discussion
As the prevalence of cardiovascular and renal diseases is increasing and is therefore a key public health concern (1,20,31,32), the identification of genetic variants that confer susceptibility to MI, hypertension, and CKD is important to prevent these conditions. In the present study, EWASs for MI, hypertension, and CKD were performed in subjects with early-onset forms of these conditions, with genetic factors likely to be more important in these individuals compared with those with late-onset forms.
In the MI study, it was found that 25 SNPs of 20 genes and two chromosomal loci were significantly associated with early-onset MI. Among these genes and loci, 6p21.3 (16), ATXN2 (9), BRAP (16), ACAD10 (9), ALDH2 (9), NAA25 (9), and HECTD4 (9) have previously been shown to be associated with MI or CAD. Therefore, 16 SNPs of 14 genes and one locus were newly identified. However, significant LD was detected among three SNPs in ADGRL3, CXCL8, and MARCH1; between two SNPs in TCHP and GIT2; between two SNPs in CCDC63; and between two SNPs in PLCB2 and VPS33B. Therefore, 11 novel loci (TMOD4, COL6A3, ADGRL3-CXCL8-MARCH1, OR52E4, TCHP-GIT2, CCDC63, 12q24.1, OAS3, PLCB2-VPS33B, GOSR2, ZNF77) were identified that confer susceptibility to MI.
In the hypertension study, 11 SNPs of 10 genes and one chromosomal locus were significantly associated with early-onset hypertension. None of these genes or loci have been found to be associated with systolic or diastolic BP or with hypertension in previous GWASs. However, there was significant LD between two SNPs in MOB3C and TMOD4; between two SNPs in CXCL8 and MARCH1; among three SNPs of NFKBIL1, 6p21.3, and NCR3; and between two SNPs in PLCB2 and VPS33B. Therefore six novel loci (MOB3C-TMOD4, COL6A3, COL6A5, CXCL8-MARCH1, NFKBIL1-6p21.3-NCR3, PLCB2-VPS33B) were identified that confer susceptibility to hypertension.
In the CKD study, 11 SNPs in 11 genes were significantly associated with early-onset CKD. None of these genes have been shown to be associated with renal function-related traits or CKD in previous GWASs. However, there was significant LD between two SNPs in MOB3C and TMOD4; between two SNPs in PLCB2 and VPS33B; and among three SNPs in ADGRL3, CXCL8, and MARCH1. Therefore, seven new loci (MOB3C-TMOD4, COL6A3, COL6A5, ADGRL3-CXCL8-MARCH1, MUC17, PLCB2-VPS33B, ZNF77) were identified that confer susceptibility to CKD.
Furthermore, rs115287176 of TMOD4, rs146092501 of COL6A3, rs188378669 of CXCL8, rs61734696 of MARCH1, rs200787930 of PLCB2, and rs199921354 of VSP33B were significantly associated with all three diseases (MI, hypertension, and CKD); rs192210727 of ADGRL3 and rs146879198 of ZNF77 were associated with both MI and CKD; and rs200982668 of COL6A5 and rs139537100 of MOB3C were associated with both hypertension and CKD. In addition, GOSR2, which was associated with MI, has previously been shown to be associated with systolic BP (23); and 6p21.3, which was associated with hypertension, has previously been shown to be associated with CAD (16). Therefore, 13 loci (MOB3C-TMOD4, COL6A3, ADGRL3-CXCL8-MARCH1, OR52E4, TCHP- GIT2, CCDC63, 12q24.1, OAS3, PLCB2-VPS33B, ZNF77, COL6A5, NFKBIL1-NCR3, MUC17) were newly identified that confer susceptibility to MI, hypertension, or CKD. Genes, chromosomal loci, and SNPs identified in the present study, in particular, those significantly associated with two or three diseases, may prove to be informative for clinical practice.
In the MI study, associations of CXCL8 to plasma total cholesterol (70), of MARCH1 to serum adiponectin concentration (71), of 12q24.1 and OAS3 to plasma HDL-cholesterol (72), of PLCB2 to plasma triglycerides (70), of VPS33B to type 2 diabetes mellitus (73), and of GOSR2 to systolic BP (23) have been shown in previous GWASs. In the present study, eight loci (TMOD4, COL6A3, ADGRL3-CXCL8-MARCH1, OR52E4, CCDC63, 12q24.1, PLCB2-VPS33B, ZNF77) newly associated with MI were significantly associated with one or more intermediate phenotypes; in particular, five loci (TMOD4, COL6A3, ADGRL3-CXCL8-MARCH1, PLCB2-VPS33B, ZNF77) associated with two or three diseases were associated with three or four intermediate phenotypes. As these intermediate phenotypes are important risk factors for MI, the association between these loci and MI may be attributable, at least in part, to their effects on intermediate phenotypes. By contrast, three loci (TCHP-GIT2, OAS3, and GOSR2) associated with MI were not associated with intermediate phenotypes. The molecular mechanisms underlying the association between these loci and MI remain to be elucidated.
In the hypertension study, although the six loci (MOB3C-TMOD4, COL6A3, COL6A5, CXCL8-MARCH1, NFKBIL1-6p21.3-NCR3, PLCB2-VPS33B) identified in the present study were all significantly associated with systolic BP, they have not been shown to be associated with BP or hypertension by previous GWASs. The associations of NFKBIL1 and NCR3 to plasma total cholesterol (74), of 6p21.3 to plasma total cholesterol and triglycerides (74), and of CXCL8 (70), MARCH1 (71), PLCB2 (70), and VPS33B (73) to lipid or glucose metabolism have been shown in previous GWASs. The effects of these loci on lipid or glucose metabolism may influence BP or hypertension, although the functional relevance of these loci to the pathogenesis of hypertension remains to be fully elucidated.
In the CKD study, although the seven loci (MOB3C-TMOD4, COL6A3, COL6A5, ADGRL3-CXCL8-MARCH1, MUC17, PLCB2-VPS33B, ZNF77) identified in the present study were all significantly associated with eGFR, they have not been shown to be associated with renal function-related traits or CKD in previous GWASs. The associations of CXCL8 (70), MARCH1 (71), PLCB2 (70), and VPS33B (73) to lipid or glucose metabolism have been demonstrated in previous GWASs. The effects of these loci on lipid or glucose metabolism may contribute, at least in part, to renal function or the development of CKD, although the molecular mechanisms underlying the association between these loci and CKD remain to be elucidated.
Previous GWASs have identified potential biological pathways underlying the associations between genetic loci and CAD (75,76). Network analysis of functional gene-gene interactions may be informative with regard to the clarification of biological processes underlying CAD and to the identification of therapeutic targets for this condition (77). Therefore the present study performed GO and network analyses to predict biological processes associated with the identified genes, and the interactions between these genes and those previously shown to be associated with MI, hypertension, or CKD.
GO analysis of the 13 genes associated with MI suggested that the functions of TMOD4 (actin filament organization); COL6A3 (cell adhesion and extracellular matrix organization); ADGRL3, OR52E4, and PLCB2 (G protein-coupled receptor signaling); CXCL8 (angiogenesis and inflammatory response); MARCH1 and OAS3 (immune response); TCHP (apoptotic process); GIT2 (regulation of GTPase activity); and VPS33B (vesicle-mediated transport and platelet α-granule organization) may be associated with the pathogenesis of MI. For the 10 genes associated with hypertension, the functions of TMOD4, COL6A3, COL6A5 (cell adhesion), CXCL8, MARCH1, NFKBIL1 (NF-κB signaling), NCR3 (immune response), PLCB2, and VPS33B may be associated with the pathogenesis of hypertension. Among the 11 genes associated with CKD, the functions of COL6A3, COL6A5, ADGRL3, CXCL8, MARCH1, MUC17 (cellular homeostasis), PLCB2, and VPS33B may be associated with the pathogenesis of CKD. Network analysis showed that the 13, 10, and 11 genes found to be associated with MI, hypertension, and CKD, respectively, in the present study had direct or indirect interactions with the corresponding sets of 50 genes selected from the DisGeNET database (68,69). However, the underlying molecular mechanisms of these interactions remain to be elucidated.
In our previous studies, it was shown that nine, nine, and four SNPs were associated with MI (P<0.01), hypertension (P<0.01), and CKD (P<0.05), respectively, as determined by multivariable logistic regression analysis with adjustment for covariates following an initial EWAS screening of allele frequencies among subjects with early-onset and late-onset forms of these conditions (50-52). The associations between five of the nine SNPs [rs202103723 (P=0.0107), rs188212047 (P=0.0039), rs1265110 (P=0.0004), rs9258102 (P=0.0373), rs439121 (P=0.0063)] and MI were replicated (P<0.05) in the present study. The associations between six of the nine SNPs [rs150854849 (P=0.0123), rs2867125 (P=0.0111), rs202069030 (P=6.40×10−10), rs139012426 (P=5.50×10−6), rs12229654 (P=0.0003), rs201633733 (P=0.0356)] and hypertension were replicated (P<0.05) in the present study. By contrast, none of the associations between the four SNPs and CKD were replicated in the present study, indicative of the importance of age in the development of CKD. These results suggested that genetic variants associated with MI, hypertension, and CKD differ, in part, between individuals with early-onset and late-onset disease.
There were several limitations to the present study: i) As that the results were not replicated, their validation is necessary in independent study populations or in other ethnicities; ii) it is possible that SNPs identified in the present study are in LD with other genetic variants in the same gene or in other nearby genes that are actually responsible for the development of MI, hypertension, or CKD; iii) the functional relevance of identified SNPs to the pathogenesis of MI, hypertension, and CKD remains to be elucidated. Due to the lack of experiments for functional analyses, the association of the SNPs identified in the present study with MI, hypertension, and CKD requires careful interpretation.
In conclusion, the present study identified 13 loci (MOB3C-TMOD4, COL6A3, ADGRL3-CXCL8-MARCH1, OR52E4, TCHP- GIT2, CCDC63, 12q24.1, OAS3, PLCB2-VPS33B, ZNF77, COL6A5, NFKBIL1-NCR3, MUC17) that confer susceptibility to MI, hypertension, or CKD. Determination of the genotypes for the SNPs at these loci may provide informative for assessment of the genetic risk for MI, hypertension, and CKD.
Acknowledgments
Not applicable.
Funding
The present study was supported by the CREST (grant no. JPMJCR1302), Japan Science and Technology Agency to YYamada, JS, and IT).
Availability of data and materials
All data underlying the findings described are available upon request from the corresponding author.
Authors' contributions
YYamada contributed to conception and design of the study; to acquisition, analysis, and interpretation of the data; and to drafting of the manuscript. KK, MO, HH, and TF each contributed to acquisition of the data and to revision of the manuscript. YYasukochi, IT, and JS contributed to analysis and interpretation of the data and to revision of the manuscript. All authors read and approved the final manuscript
Ethics approval and consent to participate
The study protocol complied with the Declaration of Helsinki and was approved by the Committees on the Ethics of Human Research of Mie University Graduate School of Medicine, Hirosaki University Graduate School of Medicine, and participating hospitals (Gifu Prefectural Tajimi Hospital, Gifu Prefectural General Medical Center, Japanese Red Cross Nagoya First Hospital, Northern Mie Medical Center Inabe General Hospital, and Hirosaki Stroke and Rehabilitation Center). Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, et al. Heart disease and stroke statistics-2018 update: A report from the american heart association. Circulation. 2018;137:e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 2.McPherson R, Tybjaerg-Hansen A. Genetics of coronary artery disease. Circ Res. 2016;118:564–578. doi: 10.1161/CIRCRESAHA.115.306566. [DOI] [PubMed] [Google Scholar]
- 3.Peden JF, Farrall M. Thirty-five common variants for coronary artery disease: The fruits of much collaborative labour. Hum Mol Genet. 2011;20:R198–R205. doi: 10.1093/hmg/ddr384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 6.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CARDIoGRAMplusC4D Consortium. Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, Bennett F, Bowden DW, Chakravarti A, Dreisbach A, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 african americans: The NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the chinese han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 13.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, Cheng J, Zhang L, Gu CC, Huang J, et al. Genome-wide association study in han chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi F, Yokota M, Yamamoto K, Nakashima E, Katsuya T, Asano H, Isono M, Nabika T, Sugiyama T, Fujioka A, et al. Genome-wide association study of coronary artery disease in the japanese. Eur J Hum Genet. 2012;20:333–340. doi: 10.1038/ejhg.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirokawa M, Morita H, Tajima T, Takahashi A, Ashikawa K, Miya F, Shigemizu D, Ozaki K, Sakata Y, Nakatani D, et al. A genome-wide association study identifies PLCL2 and AP3D1-DOT1L-SF3A2 as new susceptibility loci for myocardial infarction in japanese. Eur J Hum Genet. 2015;23:374–380. doi: 10.1038/ejhg.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padmanabhan S, Caulfield M, Dominiczak AF. Genetic and molecular aspects of hypertension. Circ Res. 2015;116:937–959. doi: 10.1161/CIRCRESAHA.116.303647. [DOI] [PubMed] [Google Scholar]
- 19.Byars SG, Ewbank D, Govindaraju DR, Stearns SC. Natural selection in a contemporary human population. Proc Natl Acad Sci USA. 2010;107(Suppl 1):S1787–S1792. doi: 10.1073/pnas.0906199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rapsomaniki E, Timmis A, George J, Pujades-Rodriguez M, Shah AD, Denaxas S, White IR, Caulfield MJ, Deanfield JE, Smeeth L, et al. Blood pressure and incidence of twelve cardiovascular diseases: Lifetime risks, healthy life-years lost, and age-specific associations in 1.25 million people. Lancet. 2014;383:1899–1911. doi: 10.1016/S0140-6736(14)60685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Consortium for Blood Pressure Genome-Wide Association Studies. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, Ntalla I, Surendran P, Liu C, Cook JP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, Rotimi C. A genome-wide association study of hypertension and blood pressure in african americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang J, Le TH Edwards, DRV Tayo, BO Gaulton, KJ Smith, JA Lu Y, Jensen RA, Chen G, Yanek LR, et al. Single-trait and multi-trait genome-wide association analyses identify novel loci for blood pressure in african-ancestry populations. PLoS Genet. 2017;13:e1006728. doi: 10.1371/journal.pgen.1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east asians. Nat Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, Kelly TN, Saleheen D, Lehne B, Mateo Leach I, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surendran P, Drenos F, Young R, Warren H, Cook JP, Manning AK, Grarup N, Sim X, Barnes DR, Witkowska K, et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato N, Miyata T, Tabara Y, Katsuya T, Yanai K, Hanada H, Kamide K, Nakura J, Kohara K, Takeuchi F, et al. High-density association study and nomination of susceptibility genes for hypertension in the japanese national project. Hum Mol Genet. 2008;17:617–627. doi: 10.1093/hmg/ddm335. [DOI] [PubMed] [Google Scholar]
- 31.Webster AC, Nagler EV, Morton RL, Masson P. Chronic kidney disease. Lancet. 2017;389:1238–1252. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 32.Weiner DE, Tighiouart H, Amin MG, Stark PC, MacLeod B, Griffith JL, Salem DN, Levey AS, Sarnak MJ. Chronic kidney disease as a risk factor for cardiovascular disease and all-cause mortality: A pooled analysis of community-based studies. J Am Soc Nephrol. 2004;15:1307–1315. doi: 10.1097/01.ASN.0000123691.46138.E2. [DOI] [PubMed] [Google Scholar]
- 33.James MT, Hemmelgarn BR, Tonelli M. Early recognition and prevention of chronic kidney disease. Lancet. 2010;375:1296–1309. doi: 10.1016/S0140-6736(09)62004-3. [DOI] [PubMed] [Google Scholar]
- 34.Yamagata K, Ishida K, Sairenchi T, Takahashi H, Ohba S, Shiigai T, Narita M, Koyama A. Risk factors for chronic kidney disease in a community-based population: A 10-year follow-up study. Kidney Int. 2007;71:159–166. doi: 10.1038/sj.ki.5002017. [DOI] [PubMed] [Google Scholar]
- 35.Vivante A, Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol. 2016;12:133–146. doi: 10.1038/nrneph.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bochud M, Elston RC, Maillard M, Bovet P, Schild L, Shamlaye C, Burnier M. Heritability of renal function in hypertensive families of African descent in the Seychelles (Indian Ocean) Kidney Int. 2005;67:61–69. doi: 10.1111/j.1523-1755.2005.00055.x. [DOI] [PubMed] [Google Scholar]
- 37.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chambers JC, Zhang W, Lord GM, van der Harst P, Lawlor DA, Sehmi JS, Gale DP, Wass MN, Ahmadi KR, Bakker SJ, et al. Genetic loci influencing kidney function and chronic kidney disease. Nat Genet. 2010;42:373–375. doi: 10.1038/ng.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Böger CA, Gorski M, Li M, Hoffmann MM, Huang C, Yang Q, Teumer A, Krane V, O'Seaghdha CM, Kutalik Z, et al. Association of eGFR-related loci identified by GWAS with incident CKD and ESRD. PLoS Genet. 2011;7:e1002292. doi: 10.1371/journal.pgen.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pattaro C, Köttgen A, Teumer A, Garnaas M, Böger CA, Fuchsberger C, Olden M, Chen MH, Tin A, Taliun D, et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet. 2012;8:e1002584. doi: 10.1371/journal.pgen.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorski M, van der Most PJ, Teumer A, Chu AY, Li M, Mijatovic V, Nolte IM, Cocca M, Taliun D, Gomez F, et al. 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep. 2017;7:46835. doi: 10.1038/srep46835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu CT, Garnaas MK, Tin A, Kottgen A, Franceschini N, Peralta CA, de Boer IH, Lu X, Atkinson E, Ding J, et al. Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLoS Genet. 2011;7:e1002264. doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada Y, Sim X, Go MJ, Wu JY, Gu D, Takeuchi F, Takahashi A, Maeda S, Tsunoda T, Chen P, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east asian populations. Nat Genet. 2012;44:904–909. doi: 10.1038/ng.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, Garnaas M, Tin A, Sorice R, Li Y, et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun. 2016;7:10023. doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 47.Nora JJ, Lortscher RH, Spangler RD, Nora AH, Kimberling WJ. Genetic-epidemiologic study of early-onset ischemic heart disease. Circulation. 1980;61:503–508. doi: 10.1161/01.CIR.61.3.503. [DOI] [PubMed] [Google Scholar]
- 48.Roncaglioni MC, Santoro L, D'Avanzo B, Negri E, Nobili A, Ledda A, Pietropaolo F, Franzosi MG, La Vecchia C, Feruglio GA, et al. Role of family history in patients with myocardial infarction. An italian case-control study. GISSI-EFRIM investigators. Circulation. 1992;85:2065–2072. doi: 10.1161/01.CIR.85.6.2065. [DOI] [PubMed] [Google Scholar]
- 49.Niiranen TJ, McCabe EL, Larson MG, Henglin M, Lakdawala NK, Vasan RS, Cheng S. Heritability and risks associated with early onset hypertension: Multigenerational, prospective analysis in the framingham heart study. BMJ. 2017;357:j1949. doi: 10.1136/bmj.j1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada Y, Sakuma J, Takeuchi I, Yasukochi Y, Kato K, Oguri M, Fujimaki T, Horibe H, Muramatsu M, Sawabe M, et al. Identification of STXBP2 as a novel susceptibility locus for myocardial infarction in Japanese individuals by an exome-wide association study. Oncotarget. 2017;8:33527–33535. doi: 10.18632/oncotarget.16536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada Y, Sakuma J, Takeuchi I, Yasukochi Y, Kato K, Oguri M, Fujimaki T, Horibe H, Muramatsu M, Sawabe M, et al. Identification of polymorphisms in 12q24.1, ACAD10, and BRAP as novel genetic determinants of blood pressure in Japanese by exome-wide association studies. Oncotarget. 2017;8:43068–43079. doi: 10.18632/oncotarget.17474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada Y, Sakuma J, Takeuchi I, Yasukochi Y, Kato K, Oguri M, Fujimaki T, Horibe H, Muramatsu M, Sawabe M, et al. Identification of C21orf59 and ATG2A as novel determinants of renal function-related traits in Japanese by exome-wide association studies. Oncotarget. 2017;8:45259–45273. doi: 10.18632/oncotarget.16696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamada Y, Matsui K, Takeuchi I, Oguri M, Fujimaki T. Association of genetic variants with hypertension in a longitudinal population-based genetic epidemiological study. Int J Mol Med. 2015;35:1189–1198. doi: 10.3892/ijmm.2015.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuo S, Imai E, Horio M, Yasuda Y, Tomita K, Nitta K, Yamagata K, Tomino Y, Yokoyama H, Hishida A, Collaborators developing the Japanese equation for estimated GFR Revised equations for estimated GFR from serum creatinine in japan. Am J Kidney Dis. 2009;53:982–992. doi: 10.1053/j.ajkd.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 55.National Kidney Foundation K/DOQI clinical practice guidelines for chronic kidney disease: Evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(Suppl 1):S1–S266. [PubMed] [Google Scholar]
- 56.Go AS, Chertow GM, Fan D, McCulloch CE, Hsu CY. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med. 2004;351:1296–1305. doi: 10.1056/NEJMoa041031. [DOI] [PubMed] [Google Scholar]
- 57.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, et al. Best practices and joint calling of the HumanExome BeadChip: The CHARGE Consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 60.Leslie R, O'Donnell CJ, Johnson AD. GRASP: Analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics. 2014;30:i185–i194. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eicher JD, Landowski C, Stackhouse B, Sloan A, Chen W, Jensen N, Lien JP, Leslie R, Johnson AD. GRASP v2.0: An update on the genome-wide repository of associations between SNPs and phenotypes. Nucleic Acids Res. 2015;43:D799–D804. doi: 10.1093/nar/gku1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R. QuickGO: A web-based tool for gene ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huntley RP, Binns D, Dimmer E, Barrell D, O'Donovan C, Apweiler R. QuickGO: A user tutorial for the web-based gene ontology browser. Database (Oxford) 2009;2009:bap010. doi: 10.1093/database/bap010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F, Furlong LI. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz F, Furlong LI. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Asselbergs FW, Guo Y, van Iperen EP, Sivapalaratnam S, Tragante V, Lanktree MB, Lange LA, Almoguera B, Appelman YE, Barnard J, et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am J Hum Genet. 2012;91:823–838. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jee SH, Sull JW, Lee JE, Shin C, Park J, Kimm H, Cho EY, Shin ES, Yun JE, Park JW, et al. Adiponectin concentrations: A genome-wide association study. Am J Hum Genet. 2010;87:545–552. doi: 10.1016/j.ajhg.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, Hwang JY, Oh JH, Kim DJ, Kim NH, et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet. 2011;43:990–995. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- 73.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, Leed A, Weale ME, Spencer CCA, Aguet F, et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49:1392–1397. doi: 10.1038/ng.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Howson JMM, Zhao W, Barnes DR, Ho WK, Young R, Paul DS, Waite LL, Freitag DF, Fauman EB, Salfati EL, et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet. 2017;49:1113–1119. doi: 10.1038/ng.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lempiäinen H, Brænne I, Michoel T, Tragante V, Vilne B, Webb TR, Kyriakou T, Eichner J, Zeng L, Willenborg C, et al. Network analysis of coronary artery disease risk genes elucidates disease mechanisms and druggable targets. Sci Rep. 2018;8:3434. doi: 10.1038/s41598-018-20721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data underlying the findings described are available upon request from the corresponding author.