Abstract
The altered expression of homeobox (HOX)A11 has been observed in various malignant tumor types, but it has remained to be determined in human lung adenocarcinoma (LUAD). In the present study, the expression of HOXA11 in LUAD and the potential associated mechanisms were assessed. Data from The Cancer Genome Atlas and Oncomine microarrays were gathered and in-house polymerase chain reaction data were produced to investigate the altered expression of HOXA11 in LUAD and its association with various clinicopathological characteristics. Genes co-expressed with HOXA11 were also identified by searching the cBioPortal and Multi Experiment Matrix databases, and performing a bioinformatics analysis, through which the potential molecular mechanisms of HOXA11 in LUAD were explored. The data analyses indicated that HOXA11 was overexpressed in the LUAD samples, and together with its co-expressed genes, it was indicated to participate in various key signaling pathways, including the focal adhesion, extracellular matrix-receptor interaction, axon guidance and small cell lung cancer signaling pathways. Furthermore, collagen type III α 1 chain (COL3A1), ephrin B2 (EFNB2), integrin subunit α 8 (ITGA8) and syndecan 2 (SDC2) were confirmed to be differentially expressed in LUAD vs. normal controls at the mRNA and protein level. Of note, LUAD patients with low expression of HOXA11 and ITGB1 had better overall survival rates. The present study indicated that HOXA11 may function as an oncogene in LUAD, and HOXA11 protein probably combines with ITGB1, COL3A1, EFNB2, ITGA8 and SDC2 to have a role in the focal adhesion pathway.
Keywords: homeobox A11, lung adenocarcinoma, co-expressed gene, bioinformatics
Introduction
Lung cancer is the second most commonly diagnosed malignant tumor type worldwide, and it causes a higher number of mortalities than any other cancer type, despite considerable progress in its early diagnosis and treatment over the past decades. The overall 5-year survival rate for lung cancer has not significantly changed over the past decade (1). In China, the morbidity and mortality associated with lung cancer have increased rapidly in recent years. Among the different lung cancer types, lung adenocarcinoma (LUAD) is the most common one; it is the most extensively elucidated subtype within the family of lung carcinomas and is particularly prevalent among Asian women (>70% in Japanese females) (2). Most LUADs are associated with a mutation in an oncogenic driver (3). The mortality rate of LUAD is high, as its early diagnosis is difficult and the disease progresses rapidly. Even in patients diagnosed at an early stage of the disease, when an initially curative surgery is performed, the 5-year survival rate remains <60% (4,5). Tobacco smoking is the major causative factor for lung cancer; however, although the etiological role of tobacco is crucial, up to 25% of lung cancers occur in non-smokers (6). In these cases, other risk factors associated to carcinogens appear to have an important role. Therefore, identification of novel targets in LUAD may offer novel viewpoints or ideas for a comprehensive management strategy for LUAD patients.
Homeobox (HOX)A11, also known as HOX1, HOX1I or radioulnar synostosis with amegakaryocytic thrombocytopenia 1, has been proven to have an essential role in numerous diseases, including renal cell carcinoma (RCC), T-cell acute lymphoblastic leukemia, glioblastoma and female reproductive system diseases (7-10). Growing evidence suggests that HOXA11 acts as a tumor suppressor in a wide range of tumor types. For instance, Wang et al (7) demonstrated that HOXA11 suppresses the proliferation, migration and invasion ability of RCC cells, while inducing cell apoptosis. Xia et al (11) reported that HOXA11 hypermethylation is relevant to the unfavorable prognosis of breast cancer patients, and the overexpression of HOXA11 suppresses cell growth. Se et al (8) indicated that a reduction of HOXA11 expression induces treatment resistance and leads to a poor prognosis in glioblastoma. Furthermore, Hwang et al (1) revealed that HOXA11 hypermethylation promotes non-small cell lung cancer development by enhancing cell proliferation or migration. However, the current knowledge on the association between HOXA11 and LUAD remains insufficient. To date, only one study has reported that HOXA11 may be an early diagnostic and independent prognostic marker for LUAD (12). The molecular mechanisms of HOXA11 in LUAD remain largely elusive and require further investigation.
With the rapid development of biological and computer technology, mass data have been emerging. More and more public databases have been established and applied for studying the genomic alterations in malignancies. At present, The Cancer Genome Atlas (TCGA) database (https://cancerge-nome.nih.gov/) is the biggest database of cancer-associated genomic alterations, and the analysis of its data may improve the current understanding of the genetic basis of various cancer types to then make it possible to diagnose and treat cancer at an early stage or even prevent it. Oncomine (https://www.oncomine.org) is the biggest database of information from oncogene chips in the world, representing an integrated data mining platform. These two databases collect information on genomic alterations via different methodologies. To gain in-depth knowledge regarding the role of HOXA11 in LUAD, the present study was performed to gather data from TCGA and Oncomine microarray chips, and produce in-house reverse transcription-quantitative polymerase chain reaction (RT-qPCR) data, with the aim of comprehensively investigating the clinical value of HOXA11 in LUAD in a large sample size and data produced with different research methods to evaluate whether HOXA11 has any diagnostic or prognostic value in LUAD. Genes co-expressed with HOXA11 were also identified by searching the cBioPortal (http://www.cbio-portal.org/) and Multi Experiment Matrix (MEM; http://biit.cs.ut.ee/mem/index.cgi) databases to determine the potential molecular mechanisms of HOXA11 in LUAD on a preliminary basis. The results revealed that HOXA11 may exert its function through the focal adhesion pathway.
Materials and methods
HOXA11 expression profile mining in TCGA
Relevant data from TCGA database (http://cancergenome.nih.gov) were downloaded using the UCSC Cancer Genomics Browser (https://genome-cancer.soe.ucsc.edu/). These data included the HOXA11 expression levels from 302 LUAD tissues and 22 tumor-adjacent normal control tissues, which were analysed to determine the diagnostic value of HOXA11 and its capacity to predict overall survival in LUAD. The HOXA11 values were carefully checked for each sample and values <1 counts were treated as missing values. Subsequently, the data were normalized via logarithmic transformation (log2) for further analysis. The corresponding clinical parameters of the LUAD patients were also extracted to investigate their association with HOXA11.
HOXA11 expression profile mining in the Oncomine database
To increase the reliability of the results, the online microarray database Oncomine was also used to analyze the transcript levels of HOXA11 in LUAD (13). The search terms used were as follows: Analysis type (cancer vs. normal), cancer types (non-small cell lung carcinoma; lung adenocarcinoma), sample type (clinical specimen) and data type (HOXA11); the other terms were the system defaults.
In-house HOXA11 RT-qPCR expression profiles
A total of 32 pairs of formalin-fixed paraffin-embedded tissues (tumor and adjacent non-cancerous) from LUAD patients who underwent curative surgical resection between January 2012 and February 2014 were collected at the Department of Pathology of the First Affiliated Hospital of Guangxi Medical University (Nanning, China). Among these patients, 23 were male and 9 were female, the median age was 56 years (range 33-90 years). TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to isolate total RNA following the manufacturer's protocol; next, the concentration and purity of the RNA was tested by measuring the absorbance at 260 and 280 nm, and the qualified RNA was reverse-transcribed in a large volume (20 µl) with the Prime Script RT Reagent Kit (Takara Bio Inc., Dalian, China) by using random primers under standard conditions. HOXA11 levels were detected using the SYBR Premix Ex Taq (Takara Bio Inc.) according to the manufacturer's protocol. GAPDH expression was used as the internal reference to normalize the results. Real-time RT-qPCR was performed using the 7900HT PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and the expression of HOXA11 and GAPDH were calculated using the 2−ΔΔCq method (14,15). The temperature protocol of the RT step was as follows: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. qPCR was performed using the following thermocycling conditions: Initial pre-denaturation for 5 min at 95°C, followed by 40 cycles of 95°C for 10 sec, 60°C for 10 sec and 72°C for 10 sec. All experiments were repeated in triplicate. The HOXA11 forward primer was 5′-CGC TTC AGA ACT CGT TGC TTT GC-3′ and the reverse primer was 5′-CGG AAG AAC TGG CAG TCT TTA CCT-3′. The GAPDH forward primer was 5′-TGA ACG GGA AGC TCA CTG G-3′, and the reverse primer was 5′-TCC ACC ACC CTG TTG CTG TA-3′.
Meta-analysis
To strengthen the reliability of the results, all the datasets included were used together to perform a meta-analysis. The mean value, standard deviation and sample size of the tumor and normal control groups were calculated using SPSS Statistics software version 24.0 (IBM Corp., Armonk, NY, USA). The true positives (TPs), false positives (FPs), false negatives (FNs), and true negatives (TNs) were also calculated according to the following equation:
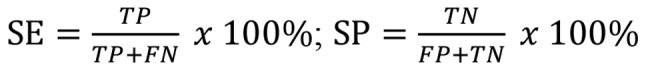 |
Where SE represents sensitivity; SP represents specificity; TP+FN is the sample size of the tumour; and FP+TN is the sample size of the normal control. The meta-analysis was performed using STATA 12.0 (StataCorp LP, College Station, TX, USA) for pooling the standardized mean difference (SMD), sensitivity, specificity and summary ROC of HOXA11 expression (16,17). An I2 test was used to evaluate heterogeneity, when I2 <50%, a fixed-effects model was used; while I2 ≥50% the random-effects model was selected (18).
Identification of co-expressed genes and pathways associated with HOXA11
The genes co-expressed with HOXA11 and their associated pathways and functions were then investigated. Using two online tools, the MEM database and cBioPortal, the genes co-expressed with HOXA11 were retrieved and extracted. The candidate co-expressed genes obtained with the two online tools were then overlapped and displayed in Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/). Bioinformatics analyses, including determination of gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and the generation of the Protein-Protein Interaction (PPI) network, were performed to investigate the underlying molecular mechanisms. The GO and KEGG analyses were performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics tool (version 6.8; https://david.ncifcrf.gov/), whereas PPI analysis was performed using the search tool for the retrieval of interacting genes/proteins (STRING) version 10.5 (http://www.string-db.org/). The GO analysis comprised the following three categories: Biological process (BP), cellular component (CC) and molecular function (MF). In addition, Cytoscape version 3.4.0 (http://cytoscape.org) provided a visualization of the functional network between HOXA11 and these co-expressed genes. Furthermore, mRNA and protein levels of the molecules in the top three KEGG pathways were confirmed with TCGA data (http://gepia.cancer-pku.cn/) and immunohistochemical staining results (https://www.protein-atlas.org/). In addition, an overall survival analysis of these genes in LUAD was performed using TCGA data from the GEPIA website (gepia.cancer-pku.cn/), which is a TCGA project based interactive web server used to analyse RNA sequencing expression and prognosis.
Statistical analysis
The expression levels of HOXA11 are presented as the mean ± standard deviation. The unpaired, two-tailed Student's t-test was used for comparison of data between two groups from TCGA and Oncomine. The paired t-test was applied for the in-house RT-qPCR data. Analysis of variance was utilized for identifying significant differences among three groups. The diagnostic value of HOXA11 in LUAD was identified by drawing the receiver operating characteristic (ROC) curve. The correlation between HOXA11 expression and co-expressed genes was analyzed by Pearson correlation analysis. To assess the ability of HOXA11 to predict patient survival, Kaplan-Meier survival curves were drawn and significant differences were assessed using the log-rank test. All of the abovementioned statistical analyses were performed with SPSS Statistics software version 24.0 (IBM Corp., Armonk, NY, USA). GraphPad Prism 7 software (GraphPad Software, Inc., La Jolla, CA, USA) was used for graphic presentation of data. In addition, meta-analysis was performed using STATA 12.0 for pooling the SMD, sensitivity, specificity and summary ROC of HOXA11 expression from the three data sources. P<0.05 was considered to indicate a statistically significant difference.
Results
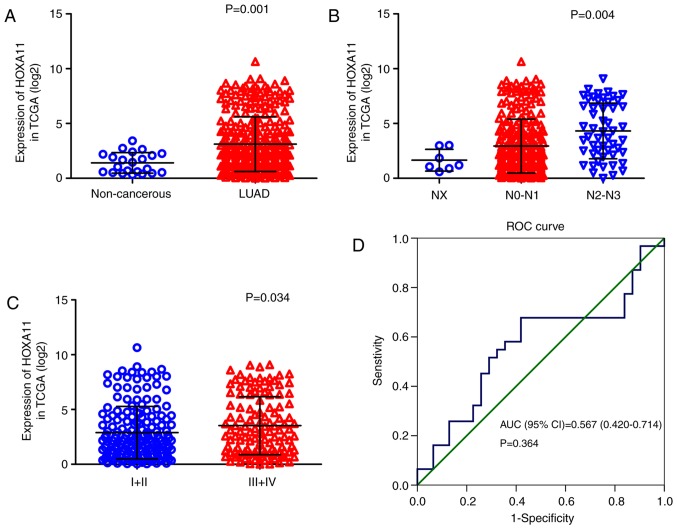
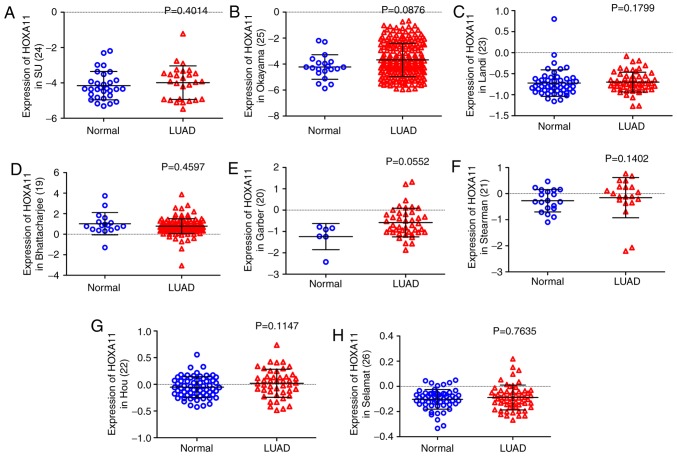
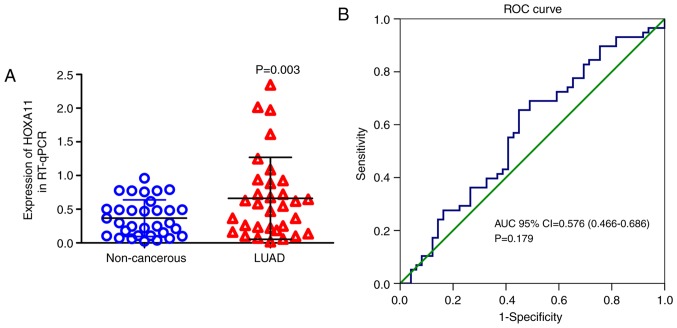
Overexpression of HOXA11 in LUAD
The clinical significance of HOXA11 in LUAD was validated using data from three different sources. First, the calculation of HOXA11 expression values with the data from TCGA (Table I) revealed that HOXA11 was significantly overexpressed in LUAD (3.15±2.506) compared with that in adjacent, non-tumorous tissue samples (1.41±0.937, P=0.001; Fig. 1A) and that the level of HOXA11 in stage III/IV LUAD tissues was significantly higher than that in stage I/II LUAD tissues (P=0.034; Fig. 1B). The number of lymph node metastases was positively correlated with the expression levels of HOXA11 (P<0.001; Fig. 1C). In addition, the area under the curve (AUC) for upregulated HOXA11 in LUAD diagnosis was 0.706 [95% confidence interval (CI): 0.620-0.793, P=0.0012; Fig. 1D], with a cutoff value of 2.747 (sensitivity 44.2% and specificity 95.5%). Furthermore, except from the data of Bhattacharjee et al (19), all of the eight datasets obtained from the Oncomine database indicated that HOXA11 expression in LUAD tissues was higher than that in the normal controls (20-26), although all the P-values were >0.05 (Fig. 2). Among the eight datasets, that by Garber et al (20) revealed that HOXA11 had a moderate diagnostic value for LUAD, while it was poor in the other studies. In the present study, HOXA11 expression was also detected in 32 pairs of formalin-fixed, paraffin-embedded tumors and adjacent non-cancerous tissues by RT-qPCR. These in-house data suggested that HOXA11 was significantly overexpressed in the LUAD vs. normal tissue samples (P=0.003) (Table II). Furthermore, HXOA11 was indicated to be of diagnostic value for LUAD, as the AUC was 0.632 (95% CI: 0.495-0.770, P=0.069; Fig. 3), with a cutoff value of 0.5236 (sensitivity 53.1% and specificity 78.1%).
Table I.
Expression of HOXA11 in LUAD based on the The Cancer Genome Atlas database.
| Clinicopathological feature | N (%) | HOXA11 expression (log2) | T- or F-value | P-value |
|---|---|---|---|---|
| Tissue type | ||||
| Normal | 22 (6.8) | 1.41±0.937 | T=3.239 | 0.001 |
| LUAD | 302 (93.2) | 3.15±2.506 | ||
| Age (years) | ||||
| <60 | 96 (31.8) | 3.12±2.644 | T=−0.140 | 0.888 |
| ≥60 | 206 (68.2) | 3.16±2.446 | ||
| Sex | ||||
| Male | 138 (45.7) | 3.20±2.558 | T=0.318 | 0.750 |
| Female | 164 (54.3) | 3.11±2.469 | ||
| Ethnicity | ||||
| Caucasian | 226 (87.9) | 3.06±2.526 | F=0.212 | 0.809 |
| Black | 26 (10.1) | 3.29±2.270 | ||
| Asian | 5 (1.9) | 2.55±1.461 | ||
| Tumor stage | ||||
| T1/T2 | 255 (85.0) | 3.13±2.507 | T=−0.422 | 0.673 |
| T3/T4 | 45 (15.0) | 3.30±2.561 | ||
| N stage | ||||
| NX | 7 (2.3) | 1.65±0.992 | F=8.111 | <0.001 |
| N0/N1 | 243 (80.5) | 2.94±2.456 | ||
| N2/N3 | 52 (17.2) | 4.32±2.537 | ||
| M stage | ||||
| MX | 81 (27.0) | 3.12±2.449 | F=0.142 | 0.867 |
| M0 | 202 (67.3) | 3.12±2.532 | ||
| M1 | 17 (5.7) | 3.45±2.655 | ||
| Clinical stage | ||||
| I/II | 177 (59.0) | 2.89±2.395 | T=−2.127 | 0.034 |
| III/IV | 123 (41.0) | 3.53±2.648 | ||
| Survival status | ||||
| Dead | 121 (40.1) | 3.48±2.683 | T=1.827 | 0.069 |
| Alive | 181 (59.9) | 2.93±2.363 | ||
| Recurrence | ||||
| Distant metastasis | 53 (58.2) | 3.15±2.559 | F=1.362 | 0.261 |
| Loco-regional recurrence | 35 (38.5) | 2.61±2.526 | ||
| New primary tumor | 3 (3.3) | 0.97±1.053 |
Values are expressed as the mean ± standard deviation. LUAD, lung adenocarcinoma; N, Number; SD, standard deviation; T, Student's t-test; F, one-way analysis of variance; TNM, tumor-nodes-metastasis; HOXA11, homeobox A11.
Figure 1.
HOXA11 expression in LUAD tissues and adjacent non-cancerous lung tissues from TCGA. (A) Relative levels of HOXA11 in LUAD tissues and adjacent non-cancerous lung tissues. (B) Nodal stage. (C) Tumor-nodes-metastasis stage. (D) ROC curve. TCGA, The Cancer Genome Atlas; HOX, homeobox; AUC, area under curve; CI, confidence interval; ROC, receiver operating characteristic; LUAD, lung adenocarcinoma.
Figure 2.
HOXA11 expression in Oncomine database microarrays. (A) Su et al (24), (B) Okayamal et al (25), (C) Landi et al (23), (D) Bhattacharjee et al (19), (E) Garber et al (20), (F) Stearman et al (21), (G) Hou et al (22) and (H) Selamat et al (26). HOX, homeobox.
Table II.
Expression of HOXA11 in LUAD tissues based on RT-qPCR in house.
| Clinicopathological features | N (%) | HOXA11 expression (2 −ΔΔCq)
|
P-value | |
|---|---|---|---|---|
| Mean ± SD | T- or F-value | |||
| Tissues | 0.003 | |||
| Non-tumor | 32 (50.0) | 0.3667±0.27144 | T=3.201 | |
| LUAD | 32 (50.0) | 0.6616±0.60821 | ||
| Size | 0.877 | |||
| ≤3 cm | 9 (28.1) | 0.6888±0.71826 | T=0.156 | |
| >3 cm | 23 (71.9) | 0.6509±0.57727 | ||
| TNM | 0.615 | |||
| I-II | 19 (59.4) | 0.7073±0.70770 | T=0.508 | |
| III-IV | 13 (40.6) | 0.5947±0.44298 | ||
| Sex | 0.786 | |||
| Male | 23 (71.9) | 0.6429±0.65476 | T=−0.274 | |
| Female | 9 (28.1) | 0.7093±0.50092 | ||
| Age | 0.659 | |||
| <60 years | 20 (62.5) | 0.6240±0.52890 | T=−0.446 | |
| ≥60 years | 12 (37.5) | 0.7243±0.74329 | ||
| Smoking | 0.793 | |||
| No | 18 (56.3) | 0.6361±0.43031 | T=−0.265 | |
| Yes | 14 (43.7) | 0.6943±0.79871 | ||
| Vascular invasion | 0.154 | |||
| No | 30 (93.8) | 0.6217±0.57800 | T=−1.463 | |
| Yes | 2 (6.2) | 1.2598±1.00785 | ||
| LNM | 0.920 | |||
| No | 18 (56.3) | 0.6518±0.73299 | T=−0.102 | |
| Yes | 14 (43.7) | 0.6742±0.42335 | ||
| Grade | 0.383 | |||
| I | 5 (15.6) | 0.4371±0.24929 | F=0.991 | |
| II | 24 (75.0) | 0.7480±0.67319 | ||
| III | 3 (9.4) | 0.3440±0.17608 | ||
LUAD, lung adenocarcinoma; N, number; T, Student's t-test; SD, standard deviation; F, one-way analysis of variance; LNM, lymph node metastasis.
Figure 3.
HOXA11 expression determined in LUAD tissues and adjacent non-cancerous tissues obtained at our hospital by using reverse transcription- quantitative polymerase chain reaction. (A) Relative levels of HOXA11 in LUAD tissues and adjacent non-cancerous lung tissues; (B) ROC curve. HOX, homeobox; AUC, area under curve; CI, confidence interval; ROC, receiver operating characteristic; LUAD, lung adenocarcinoma.
Meta-analysis
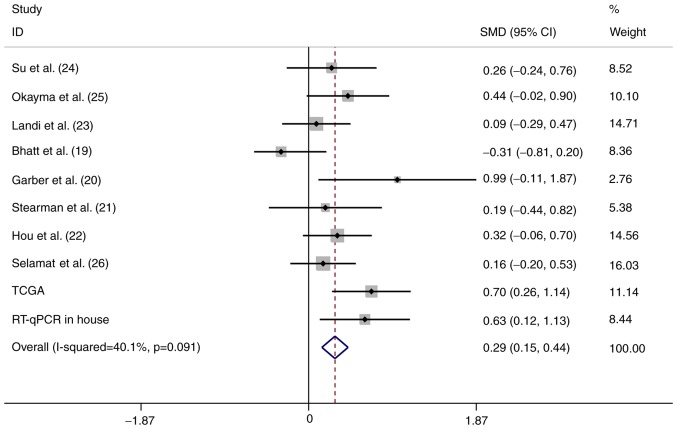
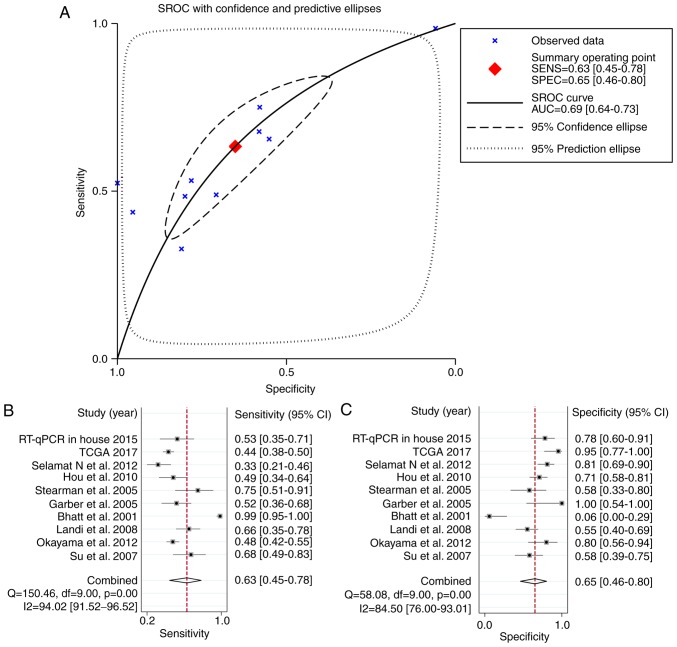
To strengthen the reliability of the results, a meta-analysis of the data from the three different sources was performed. In total, 934 LUAD and 319 normal control samples (159 were normal tissues from healthy subjects and 160 were tumor-adjacent tissues) were included. A fixed-effects model was selected and the pooled SMD of the 10 studies was 0.29 (95% CI: 0.15-0.44). A significant difference was identified in the expression of HOXA11 between LUAD and normal tissues, and the heterogeneity among the individual datasets was low (I2=40.1%, P=0.091; Fig. 4). This result further proved that HOXA11 was overexpressed in LUAD. In addition, the potential diagnostic value of HOXA11 in LUAD was analyzed using the merged data, and the meta-analysis revealed that the pooled AUC for the diagnostic value of HOXA11 for LUAD was 0.69 (95% CI: 0.64-0.73; Fig. 5A). The combined sensitivity and specificity were 0.63 (95% CI: 0.45-0.78) and 0.65 (95% CI: 0.46-0.80), respectively (Fig. 5A and B), indicating that HOXA11 probably has a role in the tumorigenesis of LUAD.
Figure 4.
Forest plot of the meta-analysis for HOXA11 expression in patients with lung adenocarcinoma within the 10 datasets included. The fixed-effects model was applied when combining the SMD. The grey squares represent the SMD value of each dataset; the different size of the squares reflect the weight of each study in the meta-analysis. The horizontal lines indicate the 95% confidence intervals of each study. The diamond represents the effect size (pooled SMD). SMD, standardized mean difference; CI, confidence interval; PCR, polymerase chain reaction; TCGA, The Cancer Genome Atlas; HOX, homeobox.
Figure 5.
Diagnostic meta-analysis. (A) SROC of HOXA11 on the early diagnosis of LUAD within the 10 datasets included. (B) Sensitivity and (C) specificity analysis of the early diagnostic value of HOXA11 for LUAD with 10 datasets included. The black squares represent the sensitivity/specificity value of each dataset. The horizontal lines indicate the 95% confidence intervals of each study. The diamonds represent the effect size (pooled sensitivity/specificity). HOX, homeobox; CI, confidence interval; PCR, polymerase chain reaction; TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma; df, degrees of freedom; SENS, sensitivity; SPEC, specificity; SROC, summary receiver operating characteristic.
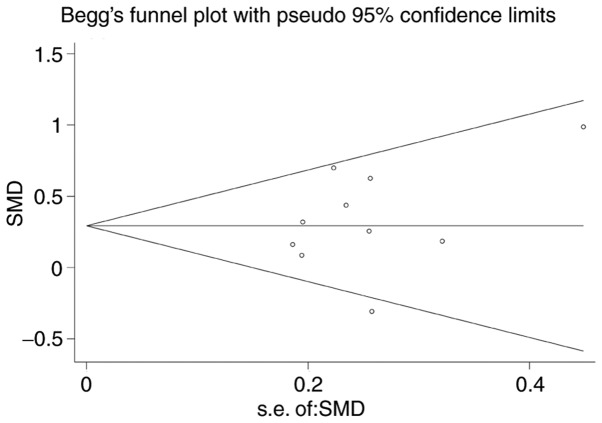
Begg's funnel plot was performed for all of the datasets (19-26) to assess publication bias, and the results yielded P=0.138. This result suggested the absence of publication bias in the present study (Fig. 6).
Figure 6.
Publication bias.
Genes co-expressed with HOXA11
A total of 3,972 genes co-expressed with HOXA11 were extracted from the MEM database with two independent gene probe tests. According to the co-expression analysis in the cBioPortal database, the mRNAs of 5,393 genes were identified as being co-expressed with HOXA11. Ultimately, to more accurately obtain genes co-expressed with HOXA11, the co-expressed genes extracted from the MEM and cBioPortal databases were overlapped and 911 were thereby identified for further analysis (Fig. 7).
Figure 7.
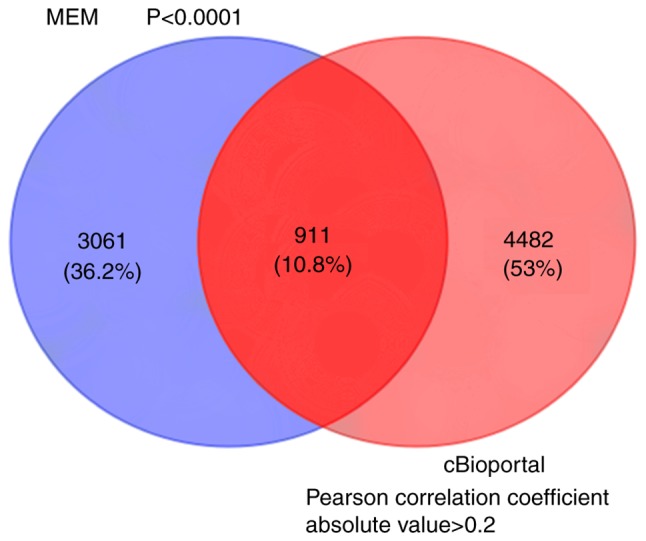
Venn diagram for the integration between MEM and cBioPortal co-expression genes. MEM, Multi Expression Matrix.
Bioinformatics analysis
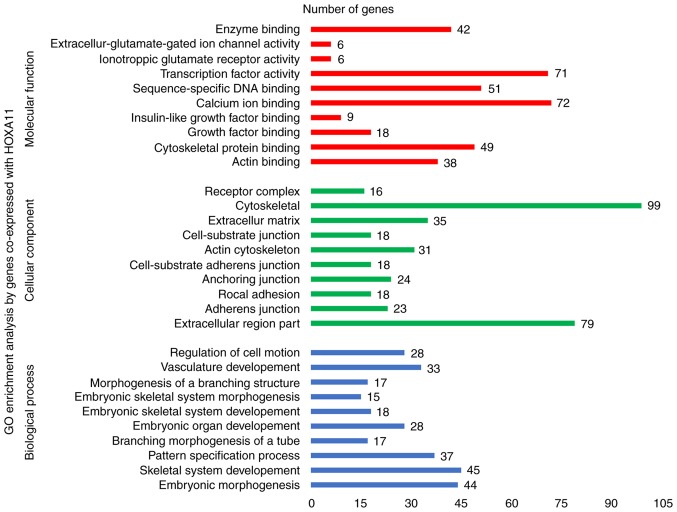
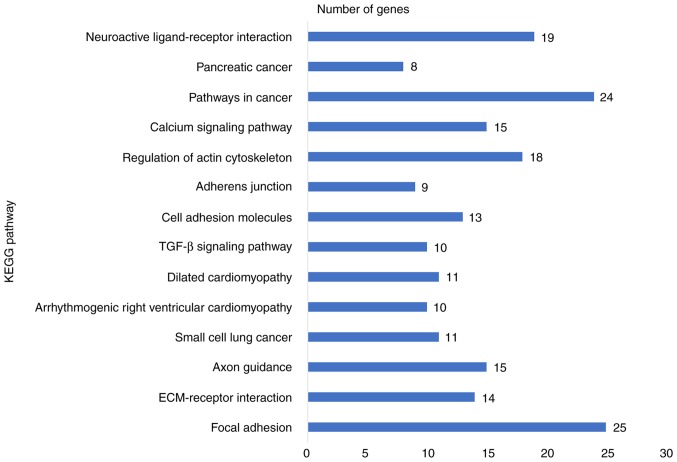
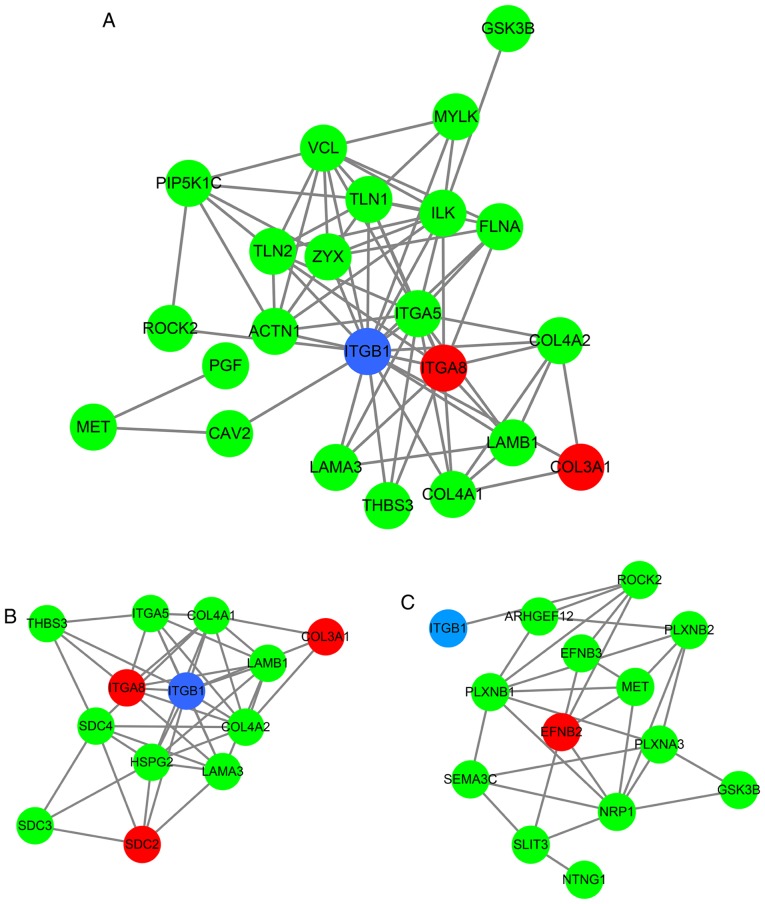
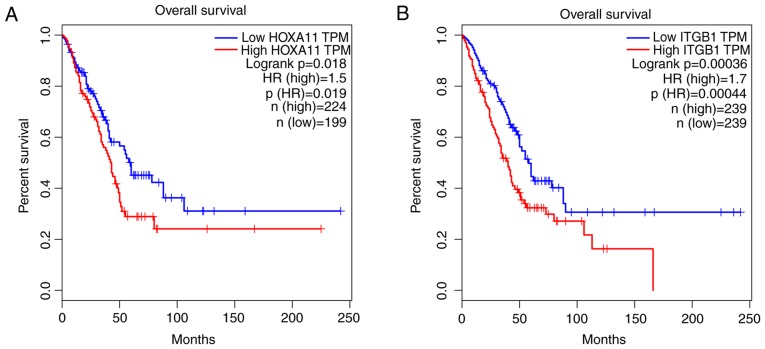
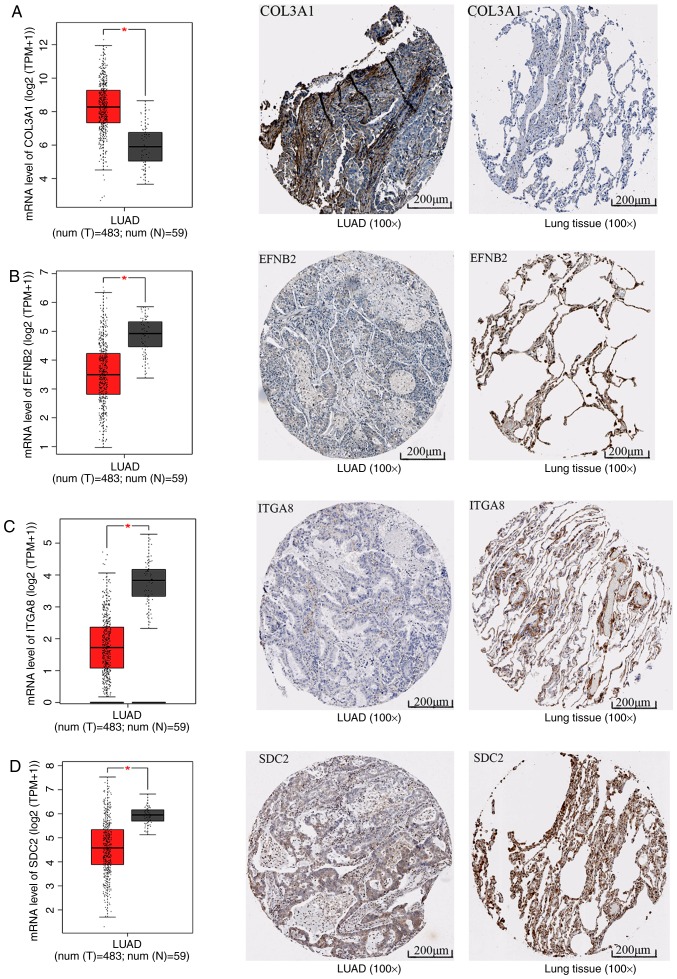
To explore the mechanisms and pathways of HOXA11 and its co-expressed genes, the 911 co-expressed genes selected were subjected to in silico analysis using the DAVID and STRING online tools. According to the GO enrichment analysis, these co-expressed genes were mainly enriched in the terms 'vasculature development' (GO:0001944, P=9.3×10−7), 'regulation of cell motion' (GO:0051270, P=1.08×10−6), 'cell adhesion' (GO:0007155, P=6.52×10−6) and 'biological adhesion' (GO:0022610, P=6.58×10−6) in the category BP; 'adherens junction' (GO:0005912, P=7×10−6), 'anchoring junction' (GO:0070161, P=1.2×10−5), 'receptor complex' (GO:0043235, P=5.62×10−4) and 'cell junction' (GO:0030054, P=1.86×10−3) in the category CC; and 'actin binding' (GO:0003779, P=2×10−6), 'growth factor binding' (GO:0019838, P=1.3×10−5), 'calcium ion binding' (GO:0005509, P=8.8×10−5) and 'transcription regulator activity' (GO:0030528, P=9.5×10−3) in the category MF. The GO functional annotation results are presented in Table III and Fig. 8 with the top 10 significantly enriched terms by the co-expressed genes provided for each category. KEGG pathway analysis revealed a significant enrichment of the co-expressed genes in the pathways 'focal adhesion' (hsa04510; P=6.5×10−5), 'extracellular matrix (ECM)-receptor interaction' (hsa04512; P=2.7×10−4), 'axon guidance' (hsa04360; P=5.4×10−3) and 'small-cell lung cancer' (hsa05222; P=9.5×10−3; Table IV and Fig. 9). To investigate the interaction of the genes enriched in the top three pivotal KEGG pathways (focal adhesion, ECM-receptor interaction and axon guidance), a PPI network was constructed (Fig. 10). The network analysis revealed that integrin β1 (ITGB1) and neuropilin 1 (NRP1) are the most important hub genes in the three pivotal pathways mentioned above. Pearson analysis of the correlation between HOXA11 and these two hub genes was performed, and ITCB1 was identified to be negatively correlated with HOXA11. Thus, an analysis of the influence of HOXA11 and ITGB1 on the overall survival of LUAD patients was then performed. Of note, LUAD patients with low expression levels of HOXA11 or ITGB1 in the tumour tissues had better overall survival rates than those with high expression (Fig. 11). Furthermore, high-throughput sequencing data and immunohistochemical staining confirmed that the mRNA and protein levels of several molecules in the top three KEGG pathways were dysregulated. Collagen type III α1 chain (COL3A1) was significantly upregulated in the 'ECM-receptor interaction' pathway, ephrin B2 (EFNB2) and syndecan 2 (SDC2) were separately downregulated in the 'axon guidance' and 'ECM-receptor interaction' pathways, and ITGA8 was significantly and concurrently downregulated in the 'focal adhesion' and 'ECM-receptor interaction' pathways (Fig. 12). The aberrant expression of these genes may cause abnormal changes in the above three signaling pathways to induce the tumorigenesis and progression of LUAD.
Table III.
Top 10 significantly enriched GO terms in three categories by genes co-expressed with homeobox A11.
| Category/term | Count | P-value | FDR |
|---|---|---|---|
| Biological process | |||
| GO:0048598-Embryonic morphogenesis | 44 | 6.79×10−10 | 0.000001 |
| GO:0001501-Skeletal system development | 45 | 7.03×10−10 | 0.000001 |
| GO:0007389-Pattern specification process | 37 | 4.66×10−8 | 0.000084 |
| GO:0048754-Branching morphogenesis of a tube | 17 | 7.79×10−8 | 0.000140 |
| GO:0048568-Embryonic organ development | 28 | 1.02×10−7 | 0.000183 |
| GO:0048706-Embryonic skeletal system development | 18 | 1.71×10−7 | 0.000306 |
| GO:0048704-Embryonic skeletal system morphogenesis | 15 | 5.22×10−7 | 0.000938 |
| GO:0001763-Morphogenesis of a branching structure | 17 | 5.32×10−7 | 0.000955 |
| GO:0001944-Vasculature development | 33 | 9.30×10−7 | 0.001669 |
| GO:0051270-Regulation of cell motion | 28 | 1.08×10−6 | 0.001945 |
| Cellular component | |||
| GO:0044421-Extracellular region part | 79 | 7.0×10−6 | 0.009522 |
| GO:0005912-Adherens junction | 23 | 7.0×10−6 | 0.010227 |
| GO:0005925-Rocal adhesion | 18 | 9.0×10−6 | 0.013065 |
| GO:0070161-Anchoring junction | 24 | 1.2×10−6 | 0.017438 |
| GO:0005924-Cell-substrate adherens junction | 18 | 1.6×10−6 | 0.022116 |
| GO:0015629-Actin cytoskeleton | 31 | 2.5×10−6 | 0.035229 |
| GO:0030055-Cell-substrate junction | 18 | 3.2×10−6 | 0.046211 |
| GO:0031012-Extracellular matrix | 35 | 9.3×10−6 | 0.133109 |
| GO:0005856-Cytoskeleton | 99 | 1.1×10−5 | 0.161576 |
| GO:0043235-Receptor complex | 16 | 5.6×10−5 | 0.798459 |
| Molecular function | |||
| GO:0003779-Actin binding | 38 | 2.0×10−6 | 0.002752 |
| GO:0008092-Cytoskeletal protein binding | 49 | 8.0×10−6 | 0.012857 |
| GO:0019838-Growth factor binding | 18 | 1.3×10−6 | 0.020705 |
| GO:0005520-Insulin-like growth factor binding | 9 | 1.7×10−6 | 0.026080 |
| GO:0005509-Calcium ion binding | 72 | 8.8×10−6 | 0.135240 |
| GO:0043565-Sequence-specific DNA binding | 51 | 2.3×10−5 | 0.357942 |
| GO:0003700-Transcription factor activity | 71 | 8.2×10−5 | 1.256619 |
| GO:0004970-Ionotropic glutamate receptor activity | 6 | 1.4×10−4 | 2.177531 |
| GO:0005234-Extracellular-glutamate-gated ion channel activity | 6 | 1.8×10−4 | 2.828013 |
| GO:0019899-Enzyme binding | 42 | 2.2×10−4 | 3.271727 |
GO, Gene Ontology; FDR, false discovery rate.
Figure 8.
GO enrichment analysis (top 10 entries) by genes co-expressed with HOXA11. GO, gene ontology; HOX, homeobox.
Table IV.
Kyoto Encyclopedia of Genes and Genomes pathways enriched by genes co-expressed with homeobox A11.
| Pathway | n | P-value | FDR |
|---|---|---|---|
| hsa04510: Focal adhesion | 25 | 6.50×10−6 | 0.077743 |
| hsa04512: ECM-receptor interaction | 14 | 2.68×10−5 | 0.318914 |
| hsa04360: Axon guidance | 15 | 5.41×10−4 | 6.249372 |
| hsa05222: Small cell lung cancer | 11 | 9.48×10−4 | 10.714508 |
| hsa05412: Arrhythmogenic right ventricular cardiomyopathy | 10 | 1.40×10−3 | 15.453584 |
| hsa05414: Dilated cardiomyopathy | 11 | 1.74×10−3 | 18.830973 |
| hsa04350: TGF-β signaling pathway | 10 | 3.12×10−3 | 31.416058 |
| hsa04514: Cell adhesion molecules | 13 | 3.47×10−3 | 34.317283 |
| hsa04520: Adherens junction | 9 | 4.01×10−3 | 38.569481 |
| hsa04810: Regulation of actin cytoskeleton | 18 | 4.43×10−3 | 41.646034 |
| hsa04020: Calcium signaling pathway | 15 | 6.18×10−3 | 53.213195 |
| hsa05200: Pathways in cancer | 24 | 6.82×10−3 | 56.834134 |
| hsa05212: Pancreatic cancer | 8 | 7.11×10−3 | 58.417821 |
| hsa04080: Neuroactive ligand-receptor interaction | 19 | 9.72×10−3 | 70.392280 |
FDR, false discovery rate; ECM, extracellular matrix; TGF, transforming growth factor; hsa, Homo sapiens.
Figure 9.
KEGG pathways enriched by genes co-expressed with HOXA11. KEGG, Kyoto Encyclopedia of Genes and Genomes; HOX, homeobox; TGF, transforming growth factor.
Figure 10.
Protein-protein interaction network of the genes co-expressed with HOXA11 enriched in the top three KEGG pathways. (A) Focal adhesion, (B) extracellular matrix-receptor interaction and (C) axon guidance. Blue circle, survival associated gene in lung adenocarcinoma; red circle, gene differentially expressed between lung adenocarcinoma and normal controls; green circle, gene clustered in top three KEGG pathways; HOX, homeobox.
Figure 11.
Overall survival analysis of lung adenocarcinoma patients stratified according to low or high HOXA11 or ITGB1 expression. (A) HOXA11 and (B) ITGB1. HOX, homeobox; HR, hazard ratio; ITGB1, integrin subunit β1; TPM, transcript per kilobase million. The median value was selected for splitting the high-expression and low-expression cohorts.
Figure 12.
mRNA and protein levels of molecules in the top three Kyoto Encyclopedia of Genes and Genomes pathways. The box plots represent the mRNA levels of the molecules; the red boxes represent the LUAD group and the gray boxes the normal controls. The five horizontal lines in the box plots from top to bottom represent the upper limit, upper quartile, median value, lower quartile and lower limit, respectively. Each black dot represents a tissue sample. In the immunohistochemical staining images, the left panel represents LUAD and the right panel represents the normal control. (A) COL3A1, (B) EFNB2, (C) ITGA8 and (D) SDC2. *P<0.05. LUAD, lung adenocarcinoma; COL3A1, collagen type III α1 chain; EFNB2, ephrin B2; ITGA8, integrin subunit α8; SDC, syndecan.
Discussion
HOX genes have been reported to encode for transcription factors that have essential roles in the embryonic development and differentiation of adult cells, and their altered expression has also been identified in cancer (27-29). Several studies also reported that HOXA11 was hypermethylated in lung cancer (1,30,31). However, the expression of HOXA11 in cancer and whether altered HOXA11 expression may contribute to the genesis of lung cancer remains to be fully elucidated. In the present study, the expression of HOXA11 in LUAD patients was determined using data obtained from TCGA database, revealing that HOXA11 was overexpressed in LUAD samples compared with its expression in adjacent, non-tumorous tissues. To gain further insight into the role of HOXA11 in LUAD, a total of 8 microarrays collected from the Oncomine database, as well as in-house RT-qRCR data, were used to validate the results from TCGA. Most of the microarray data indicated that HOXA11 expression in LUAD was higher than that in the normal samples, but only the results of two microarrays exhibited statistical significance. This may be for two possible reasons: The sample size of LUAD patients and tumor-adjacent tissues was small, while another likely cause is that, for certain microarrays, the sample size was unbalanced between the two compared groups (tumor and normal groups). However, the RT-qRCR data of the present study also suggested that HOXA11 was significantly upregulated in LUAD. Furthermore, to verify whether HOXA11 was differentially expressed between LUAD and normal lung tissue, comprehensive data produced by various research methods were used and a huge amount of data was analyzed to enhance the reliability of the results.
The meta-analysis of data gathered from three sources, with 934 LUAD and 319 normal controls, also revealed that HOXA11 was significantly overexpressed in LUAD [SMD=0.29 and 95%CI: 0.15-0.44], which indicated that HOXA11 has a role in the genesis of LUAD. In addition, analysis of the association between HOXA11 expression and clinical parameters indicated that the levels of HOXA11 were significantly higher in patients with lymphoid metastasis and advanced clinical stage, two important clinical parameters in cancer progression. For the other parameters, including tumor size, metastasis and survival status, no statistical significance was identified for their association with HOXA11 expression, while a higher level of HOXA11 was noted in risk groups (large tumor size, distant metastasis and deceased patients). The uneven sample size between the two compared groups may be the major cause for the insignificant difference (P>0.05). A further limitation of the present study is that the normal controls in included tissues from healthy individuals and tumor-adjacent samples. The tumor-adjacent samples of LUAD patients are located by inflammatory lesions, which may affect gene expression. Therefore, sample selection may be another reason for the insignificant differences observed. However, the results of the present study still suggested that HOXA11 may act as an oncogene in the initiation and progression of LUAD. HOXA11 may be used as a clinical predictor for the survival of LUAD patients. In future studies, it is required to further verify the role of HOXA11 expression in LUAD development/progression with a larger sample size and to identify the underlying regulatory mechanisms.
Due to the insufficient number of studies investigating the role of HOXA11 in cancer, the modes and molecular pathways via which upregulated HOXA11 exerts its function in LUAD have remained to be elucidated. Hence, the present study attempted to predict the potential mechanisms of the role of HOXA11 in LUAD in silico. With the development of high-throughput sequencing and novel computational approaches, accumulating evidence has proven that multiple genes always function together and then regulate tumor initiation and progression (32). To investigate the function of HOXA11, the cBioPortal and MEM databases were used in the present study to identify the genes co-expressed with HOXA11. The 911 co-expressed genes obtained were inputted into the DAVID online tool for predicting the pathways through which these genes are involved in LUAD progression. The KEGG pathway analysis results suggested that HOXA11 and its co-expressed genes were significantly enriched in the focal adhesion, ECM-receptor interaction and axon guidance pathways. The focal adhesion pathway has been reported to be correlated with cellular behavior, including migration and metastasis (33). From these results it may be deduced that HOXA11 promotes the development and progression of LUAD by affecting cancer cell adhesion and migration.
The genes enriched in the above pathways (focal adhesion, ECM-receptor interaction and axon guidance) were used to construct PPI networks, in which ITGB1 and NRP1 were identified as the hub genes. ITGB1 is a member of the integrin family and forms various heterodimeric receptors for cell adhesion to ECM proteins. Integrins are known to have important roles in regulating proliferation, cell migration, invasion and survival (34). Previous studies reported that the expression of ITGB1 is associated with the prognosis of patients with lung cancer (35), colorectal cancer (34) and breast cancer (36). However, its value as a prognostic marker, as well as its correlation with the expression of other genes in LUAD patients, has rarely been studied. In the present study, a Pearson correlation analysis was performed, which indicated that ITGB1 expression is positively correlated with HOXA11 in LUAD. Further investigation of the prognostic value with Kaplan-Meier analysis and a log-rank test also revealed that low expression levels of HOXA11 and ITGB1 were associated with a better overall survival rate of LUAD patients. However, logistic regression analysis has not been performed in the current study, it would be assessed in a future study. The mRNA (analysed using TCGA data) and protein levels (from the human protein atlas database) of COL3A1 were identified to be upregulated in LUAD tissues, while those of EFNB2, ITGA8 and SDC2 were significantly downregulated in LUAD. Therefore, it may be speculated that HOXA11 probably binds with ITGB1, COL3A1, EFNB2, ITGA8 and SDC2, which then get involved in the focal adhesion pathway to promote the initiation and progression of LUAD. Further experimental verification is required to investigate whether these potential targets were transcriptionally activated by HOXA11 protein binding with their activation sequence in the 3′UTR, which may promote the development and progression of LUAD.
In the present study the expression and potential clinical value of HOXA11 in LUAD patients was comprehensively analyzed using various data sources and methods. The results revealed that HOXA11 is overexpressed in LUAD. In addition, the bioinformatics analysis suggested that HOXA11 probably combines with ITGB1, COL3A1, EFNB2, ITGA8 and SDC2, which get involved in the focal adhesion pathway and then regulate the emergence and development of LUAD. However, the present study has certain limitations. First, the expression of HOXA11 was detected in only 32 pairs of LUAD and adjacent non-tumorous tissues. The normal controls in the current study included tissues from healthy individual combined with tumor-adjacent samples. Further investigations with an even sample size and even distribution of data are required to confirm the value of HOXA11 as a potential target for LUAD. In addition, the mechanism of HOXA11 in LUAD was predicted in silico. Further studies are required to experimentally investigate the molecular mechanisms of HOXA11 in LUAD.
Funding
The study was supported by funds from the National Natural Science Foundation of China (grant nos. NSFC81560469 and NSFC81360327), the Natural Science Foundation of Guangxi, China (grant nos. 2017GXNSFAA198016 and 2015GXNSFCA139009), the Guangxi Medical University Training Program for Distinguished Young Scholars (Gang Chen) and a Medical Excellence Award funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University (Gang Chen).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XY and YD designed the study, performed the experiments, analyzed and interpreted the data and wrote the manuscript. They contributed equally to the present study. XY, RQH and XJL performed the statistical analysis, and designed and completed the figures and tables. YD and XJL recruited the patients to obtain their specimens and performed the mRNA isolation and RT-qPCR. JM, GC and XHH participated in the design of the study, supervised the experiments and corrected the manuscript. All of the authors read and approved the final manuscript.
Ethics approval and consent to participate
All experiments using patient tissues were authorized by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Guangxi, China).
Patient consent for publication
All subjects gave their informed consent prior to their inclusion in the study. Details that may disclose their identity were omitted.
Competing interests
The authors declare that they have no competing of interest.
Acknowledgments
Not applicable.
References
- 1.Hwang JA, Lee BB, Kim Y, Park SE, Heo K, Hong SH, Kim YH, Han J, Shim YM, Lee YS, Kim DH. HOXA11 hypermethylation is associated with progression of non-small cell lung cancer. Oncotarget. 2013;4:2317–2325. doi: 10.18632/oncotarget.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calvayrac O, Pradines A, Pons E, Mazières J, Guibert N. Molecular biomarkers for lung adenocarcinoma. Eur Respir J. 2017;49:1601734. doi: 10.1183/13993003.01734-2016. [DOI] [PubMed] [Google Scholar]
- 3.Sholl LM. Biomarkers in lung adenocarcinoma: A decade of progress. Arch Pathol Lab Med. 2015;139:469–480. doi: 10.5858/arpa.2014-0128-RA. [DOI] [PubMed] [Google Scholar]
- 4.Wallerek S, Sørensen JB. Biomarkers for efficacy of adjuvant chemotherapy following complete resection in NSCLC stages I-IIIA. Eur Respir Rev. 2015;24:340–355. doi: 10.1183/16000617.00005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi L, Li Y, Qin Y, Shi G, Li T, Wang J, Chen L, Gu Y, Zhao W, Guo Z. An individualised signature for predicting response with concordant survival benefit for lung adenocarcinoma patients receiving platinum-based chemotherapy. Br J Cancer. 2016;115:1513–1519. doi: 10.1038/bjc.2016.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalela R, Curull V, Enriquez C, Pijuan L, Bellosillo B, Gea J. Lung adenocarcinoma: From molecular basis to genome-guided therapy and immunotherapy. J Thorac Dis. 2017;9:2142–2158. doi: 10.21037/jtd.2017.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Cui Y, Sheng J, Yang Y, Kuang G, Fan Y, Jin J, Zhang Q. Epigenetic inactivation of HOXA11, a novel functional tumor suppressor for renal cell carcinoma, is associated with RCC TNM classification. Oncotarget. 2017;8:21861–21870. doi: 10.18632/oncotarget.15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Se YB, Kim SH, Kim JY, Kim JE, Dho YS, Kim JW, Kim YH, Woo HG, Kim SH, Kang SH, et al. Underexpression of HOXA11 is associated with treatment resistance and poor prognosis in glioblastoma. Cancer Res Treat. 2017;49:387–398. doi: 10.4143/crt.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitcomb BP, Mutch DG, Herzog TJ, Rader JS, Gibb RK, Goodfellow PJ. Frequent HOXA11 and THBS2 promoter methylation, and a methylator phenotype in endometrial adenocarcinoma. Clin Cancer Res. 2003;9:2277–2287. [PubMed] [Google Scholar]
- 10.Speleman F, Cauwelier B, Dastugue N, Cools J, Verhasselt B, Poppe B, Van Roy N, Vandesompele J, Graux C, Uyttebroeck A, et al. A new recurrent inversion, inv(7)(p15q34), leads to transcriptional activation of HOXA10 and HOXA11 in a subset of T-cell acute lymphoblastic leukemias. Leukemia. 2005;19:358–366. doi: 10.1038/sj.leu.2403657. [DOI] [PubMed] [Google Scholar]
- 11.Xia B, Shan M, Wang J, Zhong Z, Geng J, He X, Vu T, Zhang D, Pang D. Homeobox A11 hypermethylation indicates unfavorable prognosis in breast cancer. Oncotarget. 2017;8:9794–9805. doi: 10.18632/oncotarget.14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Chen C, Ren X, Sun W. DNA methylation profiling identifies the HOXA11 gene as an early diagnostic and prognostic molecular marker in human lung adenocarcinoma. Oncotarget. 2017;8:33100–33109. doi: 10.18632/oncotarget.16528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Chen G, Kronenberger P, Teugels E, Umelo IA, De Grève J. Targeting the epidermal growth factor receptor in non-small cell lung cancer cells: The effect of combining RNA interference with tyrosine kinase inhibitors or cetuximab. BMC Med. 2012;10:28. doi: 10.1186/1741-7015-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo YH, Tang W, Zhang X, Tan Z, Guo WL, Zhao N, Pang SM, Dang YW, Rong MH, Cao J. Promising significance of the association of miR-204 5p expression with clinicopathological features of hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e7545. doi: 10.1097/MD.0000000000007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Pang YY, He RQ, Lin P, Cen JM, Yang H, Ma J, Chen G. Diagnostic value of strand-specific miRNA-101 3p and miRNA-101 5p for hepatocellular carcinoma and a bioinformatic analysis of their possible mechanism of action. FEBS Open Bio. 2017;8:64–84. doi: 10.1002/2211-5463.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X, Zeng Z, Hou Y, Yuan T, Gao C, Jia W, Yi X, Liu M. MicroRNA-92a as a potential biomarker in diagnosis of colorectal cancer: A systematic review and meta-analysis. PLoS One. 2014;9:e88745. doi: 10.1371/journal.pone.0088745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Jr, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CY. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 26.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al. Genome-scale analysis of DNA methylation in lung adenocar-cinoma and integration with mRNA expression. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 28.Javed S, Langley SE. Importance of HOX genes in normal prostate gland formation, prostate cancer development and its early detection. BJU Int. 2014;113:535–540. doi: 10.1111/bju.12269. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Romero R, Marrero-Rodriguez D, Romero-Morelos P, Villegas V, Valdivia A, Arreola H, Huerta-Padilla V, Salcedo M. The role of developmental HOX genes in cervical cancer. Rev Med Inst Mex Seguro Soc. 2015;53(Suppl 2):S188–S193. In Spanish. [PubMed] [Google Scholar]
- 30.Nelson HH, Marsit CJ, Christensen BC, Houseman EA, Kontic M, Wiemels JL, Karagas MR, Wrensch MR, Zheng S, Wiencke JK, Kelsey KT. Key epigenetic changes associated with lung cancer development: Results from dense methylation array profiling. Epigenetics. 2012;7:559–566. doi: 10.4161/epi.20219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu W, Zhao S, Wang Y, Zhao BN, Zhao W, Zhou X. Identification of cancer prognosis-associated functional modules using differential co-expression networks. Oncotarget. 2017;8:112928–112941. doi: 10.18632/oncotarget.22878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Liu QZ, Gao XH, Chang WJ, Gong HF, Fu CG, Zhang W, Cao GW. Expression of ITGB1 predicts prognosis in colorectal cancer: A large prospective study based on tissue microarray. Int J Clin Exp Pathol. 2015;8:12802–12810. [PMC free article] [PubMed] [Google Scholar]
- 35.Oshita F, Kameda Y, Ikehara M, Tanaka G, Yamada K, Nomura I, Noda K, Shotsu A, Fujita A, Arai H, et al. Increased expression of integrin beta1 is a poor prognostic factor in small-cell lung cancer. Anticancer Res. 2002;22:1065–1070. [PubMed] [Google Scholar]
- 36.Yao ES, Zhang H, Chen YY, Lee B, Chew K, Moore D, Park C. Increased beta1 integrin is associated with decreased survival in invasive breast cancer. Cancer Res. 2007;67:659–664. doi: 10.1158/0008-5472.CAN-06-2768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.