Abstract
Previous studies have shown that Rho guanine nucleotide exchange factor 7 (ARHGEF7) is implicated in cytoskeleton remodelling, which is important for cell motility and invasiveness, and exhibits frequent high-level genetic amplification in metastatic lesions of colorectal adenocarcinoma. Therefore, it was hypothesized that ARHGEF7 may be involved in the metastasis of colorectal adenocarcinoma. In the present study, it was demonstrated that the expression level of ARHGEF7 was significantly upregulated in colorectal adenocarcinoma tumor tissues compared with matched nontumorous tissues, and its expression level correlated with colorectal adenocarcinoma metastasis. In vitro assays showed that the overexpression of ARHGEF7 in CRC cells significantly enhanced cell migration and invasion, whereas the knockdown of ARHGEF7 in colorectal adenocarcinoma cells significantly decreased cell migration and invasion. In vivo assays showed that the overexpression of ARHGEF7 in CRC cells facilitated tumor metastasis, whereas the knockdown of ARHGEF7 in CRC cells significantly inhibited tumor metastasis. Furthermore, it was demonstrated that ARHGEF7 promoted cell motility by regulating the actin cytoskeleton. Finally, according to ReMARK guidelines for reporting prognostic biomarkers in cancer, it was found that a high expression of ARHGEF7 was significantly correlated with lymph node, mesenteric and distant metastasis. Patients with colorectal adenocarcinoma with a high expression of ARHGEF7 had shorter disease-free survival (DFS) and shorter overall survival (OS) rates, compared with those with a low expression of ARHGEF7, as determined by the Kaplan-Meier method with a log-rank test. Cox regression analysis showed that a high expression of ARHGEF7 was an independent risk factor for DFS and OS rates in colorectal adenocarcinoma.
Keywords: ARHGEF7, metastasis, coloretal cancer, cell motility
Introduction
Colorectal cancer (CRC) is the third most common type of cancer in men and women and is the fourth leading cause of cancer-associated mortality (1). The incidence of CRC is rapidly increasing in China, which is a serious threat to the health of the population (2). Colorectal adenocarcinoma accounts for 98% of cancer types among newly diagnosed cases. Advances in surgical technology and conception of treatment have been made, and optimization of treatment for patients with colorectal adenocarcinoma has led to an increase of survival rate at 5 and 10 years (3). However, metastasis remains the main cause of mortality and poor prognosis (4-6). The metastases produced by carcinomas are a result of a complex succession of the invasion-metastasis cascade (7). The fact that cancer cells gain cell motility and invasiveness is considered a critical step contributing to the metastasis of cancer (8). However, the mechanisms by which tumor cells become metastatic remain to be fully elucidated.
Rho guanine nucleotide exchange factor 7 (ARHGEF7), a guanine nucleotide exchange factor for Rho GTPases, functions in cell migration, attachment and cell spread (9-11). ARHGEF7 is implicated in cytoskeleton remodelling, which is important in cell migration (12,13). In other types of tumor, studies have shown that ARHGEF7 affects the motility of cells in vitro and invasion in vivo (14,15). Specially, it has been reported that the ARHGEF7 gene frequently exhibits high- level genetic amplification in metastatic lesions compared with primary sites in colorectal adenocarcinoma by SNP array (16). These data indicate that ARHGEF7 may be involved in colorectal adenocarcinoma metastasis. However, few investigations have focused on the role of ARHGEF7 in the metastasis of colorectal adenocarcinoma. Metastasis is the main contributor to the poor prognosis of patients (17). ARHGEF7 may be a prognostic biomarker and associated with colorectal adenocarcinoma metastasis.
The present study aimed to examine the expression of ARHGEF7 in colorectal adenocarcinoma. The role of ARHGEF7 in colorectal adenocarcinoma metastasis and its underlying molecular mechanism were examined using a series of in vitro and in vivo assays. Finally, whether the expression of ARHGEF7 is clinically relevant in patients with colorectal adenocarcinoma was determined according to ReMARK guidelines for the reporting of prognostic biomarkers in cancer (18).
Materials and methods
Colorectal adenocarcinoma samples
The present study was approved by the Ethics Committee of the Institutional Review Boards of the First Affiliated Hospital of Nanchang University (Nanchang, China) and Jiangxi Pingxiang People’s Hospital (Pingxiang, China). Prior informed consent was obtained from all participants, and the study was performed in accordance with the Declaration of Helsinki and current ethical guidelines. Firstly, 30 pairs of frozen fresh colorectal adenocarcinoma tumor tissues and corresponding nontumorous colorectal tissues (NCTs) from 30 patients were collected, and another five matched liver metastatic nodules (LMNs), tumor tissues and NCTs from five patients were collected following surgical resection at the Department of General Surgery, the First Affiliated Hospital of Nanchang University between July 2016 and January 2017. These tissues were used to screen the mRNA and protein expression of ARHGEF7. Secondly, another two sets of samples were used for prognostic analysis according to ReMARK guidelines for reporting prognostic biomarkers in cancer (18). Formalin-fixed, paraffin-embedded paired colorectal adenocarcinoma samples (including tumors and NCTs) obtained from 180 patients undergoing radical surgical resection at the Department of General Surgery, the First Affiliated Hospital of Nanchang University between January 2007 and December 2009 were designated as the training set. Another sample cohort containing 150 samples, including tumors and NCTs, from patients who underwent resection between July 2007 and July 2010 at the Department of General Surgery, Jiangxi Pingxiang People’s Hospital was designated as the validation set. The inclusion criteria for the samples enrolled in the study were as follows: Collection from patients with sporadic CRC, histopathologically diagnosed as adenocarcinoma by hematoxylin and eosin (H&E) staining; had completed clinicopathologic and follow-up data; were without neoadjuvant chemotherapies or distant metastasis prior to surgery. Patients with hereditary CRC were excluded from the study.
Prognostic evaluation
All patients were regularly followed-up by trained and experienced researchers. The follow-up period was defined as the interval between the date of surgery and that of the patient’s mortality or distant metastasis or the last follow-up. The median follow-up was 62.3 months (range 6.0-100.0 months) for the training set and 60.0 months (range 6.4-100.0 months) for the validation cohort. Patient mortality from other causes were treated as censored cases. Following surgery, all patients had regular clinical examination, serial monitoring of carcinoembryonic antigen (CEA) levels at 1-month interval, had computed tomography (CT) or magnetic resonance imaging (MRI) scan at a 3-month interval, and had investigation by colonoscopy at the 1-year interval. Recurrence or metastasis was diagnosed by clinical examination, serial CEA level, and CT or MRI or positron emission tomography-CT. Disease-free survival (DFS) was defined as the length of time following resection during which a patient survived without signs of recurrence or metastasis. Overall survival (OS) was defined as the interval between surgery and mortality, or between surgery and the last observation for surviving patients. Data for conventional clinical and pathological variables were also collected for analysis, including sex, age, serum CEA level, tumor differentiation, tumor site, tumor size, tumor grade, lymphatic vessel/vessel/neuron infiltration, mesenteric tumor deposit formation, and tumor-node-metastatsis (TNM) stage. The follow-up data were regularly updated in the database for each patient. Patients alive at the end of follow up or those who succumbed to mortality from causes without sign of recurrence or metastasis were censored.
Cell lines
The FHC normal colorectal mucosal cell line, and the HCT116, HT-29, SW480, SW620, LoVo cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). Short tandem repeat (STR) DNA fingerprinting was used to authenticate all cell lines prior to commencement of the study. All cell lines were routinely cultured with RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences, Logan, UT, USA), and maintained in a 5% CO2 humidified incubator at 37°C.
Vector construction and transfection
The lentiviral vector (LV) encoding short hairpin RNAs (shRNAs) for ARHGEF7 knockdown, and the LV encoding the ARHGEF7 gene were purchased from OriGene Technologies, Inc. (Rockville, MD, USA). The sequences of four shRNAs for ARHGEF7 knockdown were as follows: ARHGEF7-shRNA-Seq1, sense, 5′-ACAAGGTCCTCAGTTCCTTAGTGACTCTA-3′; ARHGEF7-shRNA-Seq2, sense, 5′-CCACCATAAAGCCTCATTCAGTGCCATCT-3′; ARHGEF7-shRNA-Seq3, sense, 5′-CCTGAACGGAAGCCTTCAGATGAGGAGTT-3′; and ARHGEF7-shRNA-Seq4, sense, 5′-TACGGCCATTGCAGACCAGTGAGAAGTTA-3′. The LoVo cells were transfected with LVs encoding the shRNAs, and the HCT116 cells were transfected with LVs encoding the human ARHGEF7 gene. An empty vector was used as the negative control and was designated as LV-control. The LV vectors were transfected into the CRC cells with an appropriate multiplicity of infection of 50. At 48 h post-transfection, 3.0 µg/ml puromycin (OriGene Technologies, Inc.) was added, and the cells were incubated for 2 weeks to select the stably transfected cells. The overexpression or downregulated expression of ARHGEF7 was confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analyses. The inhibitory efficiencies of the four shRNAs were validated and ARHGEF7-shRNA-Seq2 was used for subsequent experiments due to highly effective inhibition of the expression of ARHGEF7 in LoVo cells.
RT-qPCR analysis
Total RNA was extracted from the cell lines (~1×107 cells) or fresh frozen tumor specimens (50-100 mg) using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. The RNA was then reverse transcribed to obtain cDNA using the universal cDNA synthesis kit (Toyobo Co., Ltd., Osaka, Japan) according to the manufacturer’s protocol. The aliquots of double-stranded cDNA were subjected to RT-qPCR. RT-qPCR analysis was performed using the SYBR®-Green Realtime PCR Master Mix assay kit (Toyobo Co., Ltd.) according to the manufacturer’s protocol; the assay was performed on a PRISM 7300 Sequence Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The cycling parameters were as follows: Initial denaturation at 95°C for 2 min, followed by 95°C for 15 sec, 55-60°C for 15 sec and 72°C for 15 sec for 50 cycles, and a final extension step at 72°C for 5 min. The primers for ARHGEF7 were as follows: Forward, 5′-CGCAAACCTGAACGGAAGC CTT-3′ and reverse, 5′-GTTTTGGCGCTGGTGCAGTAAG-3′. GAPDH was used as a control using the following primers: Forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′. The results were analyzed using the 2-ΔΔCq method with the following formula: ΔΔCq = ΔCqTumor - ΔCqNCT; ΔCq = CqARHGEF7-CqGAPDH (19).
Western blot analysis
Total proteins were extracted using RIPA lysis buffer. Protein concentration was then determined using the bicinchoninic acid method, and equal quantities of protein (20-60 µg) from each sample were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto PVDF membranes (EMD Millipore, Bedford, MA, USA). The blotted membranes were blocked with 5% skimmed milk for 30 min at 25°C and incubated with primary antibodies (mouse anti-ARHGEF7; 1:400; cat. no. sc-136035; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and then an appropriate HRP-conjugated goat anti-mouse secondary antibody (1;1,000; cat. no. 04-18-06; KPL, Inc., Gaithersburg, MD, USA) in order. Bands were detected with enhanced chemiluminescence regents (Thermo Fisher Scientific, Inc.). β-actin protein was also determined using the specific mouse anti-β-actin antibody (1:1,000; cat. no. A1978, Sigma; EMD Millipore) as a loading control. Protein expression levels were quantified using BandScan software 4.0 (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and defined as the ratio of target protein relative to β-actin.
Immunohistochemistry (IHC)
The detailed IHC procedures were performed as described previously (20). The paraffin- embedded tissues were sectioned into 4-µm slides. These slides were dewaxed, tissues were rehydrated, and antigen retrieval was performed using microwave-pretreated EDTA buffer (1 mM; pH 8.0) for 10 min. Following blocking, the slides were incubated for ARHGEF7 antibody (mouse anti-ARHGEF7 1:200; cat. no. sc-136035; Santa Cruz Biotechnology, Inc.) at 4°C overnight. The slides were incubated with biotin-labeled secondary and streptavidin-peroxidase (cat. no. SP-9002; Zhongshan Goldenbridge Biotechnology Co., Ltd., Beijing, China) for 30 min at 37°C. The samples were developed using 3,3′-diaminobenzidine substrate (cat. no. ZLI-9018) and counterstained with hematoxylin (cat. no. ZLI-9609) (both from Zhongshan Goldenbridge Biotechnology Co., Ltd.). Negative
controls were without primary antibody incubation during the procedure. The staining was determined in a blinded manner by two independent pathologists. The final immunostaining score (IS) was defined by the consistency of the grading by two pathologists. The expression levels of ARHGEF7 were scored based on staining intensity (SI) and percentage of positive cells (PP) using the IS as described previously (21). The SI was classified into four grades: 0, negative; 1, weak; 2, moderate; 3, strong. The PP was defined into five categories: 0, 0% positive cells; 1, 0-25% positive cells; 2, 25-50% positive cells; 3, 50-75% positive cells, and 4, 75-100% positive cells. IS = SI x PP.
Cell proliferation and colony formation assays
Methyl thiazolyl tetrazolium (MTT) assays were used to determine the level of cell proliferation. For the MTT assays, 5×103 cells were seeded into each well of 96-well plates. Three repeated wells for each group were detected every time. Fresh medium (100 µl; 0.5 mg/ml) containing MTT (Sigma; EMD Millipore) was added into each well and incubated at 37°C for 4 h. The medium was then replaced with 100 µl DMSO and shaken at room temperature for 10 min. The absorbance was measured at 570 nm. For the colony formation assays, 500 cells were seeded into 35-mm dishes (Corning Incorporated, Corning, NY, USA) and cultured in 5% CO2 for 2 weeks at 37°C. The number of colonies per dish was counted following staining with crystal violet (Beyotime Institute of Biotechnology, Jiangsu, China). Only positive colonies (diameter >40 µm) in the dishes were counted by inverted microscope (TE-2000S; Nikon Corporation, Tokyo, Japan) and compared (22). These experiments were performed in triplicate.
Transwell assays
Transwell migration and invasion assays were used to determine cell motility and invasion ability separately. For the Transwell invasion assays, the upper chamber of the insert was plated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). For the Transwell migration assays, the upper chamber of the insert was without Matrigel. Briefly, following preincubation with Mitomycin-C (10 µg/ml) for 1 h at 37°C to suppress cell proliferation, ~1×105 cells in serum- free medium were placed into the upper chamber of the insert. Following incubation in 5% CO2 at 37°C for 24 h, the cells in the upper chamber were removed with cotton swabs, fixed in 20% methanol, and then stained with a solution containing
0.1% crystal violet (Beyotime Institute of Biotechnology). The number of cells that adhered to the lower membrane of the inserts was counted. For each experimental group, the assays were performed in triplicate, and five random fields of view were selected for analysis.
Adhesion assay
Cell-extracellular matrix (ECM) adhesion and cell-cell adhesion assays were used to analyze the adhesive ability of the cells. For the cell-ECM adhesion assay, a 96-well plate was coated with fibronectin at 37°C for 1 h and washed twice with washing buffer (0.1% BSA in DMEM; HyClone; GE Healthcare Life Sciences). The plates were blocked in blocking buffer (0.5% BSA in DMEM) at 37°C in a CO2 incubator for 60 min. The cells (100 µl) at a density of ~1×105/ml were added into each well of a 96-well plate and cultured at 37°C. Five wells for each group were detected at 60, 90 or 120 min. The medium was entirely removed, and unbound cells were washed away with PBS. Fresh medium (100 µl; 0.5 mg/ml) containing MTT (Sigma; EMD Millipore) was added into each well and incubated at 37°C for 4 h. The medium was then replaced with 100 µl of DMSO and shaken at room temperature for 10 min. The absorbance was measured at 570 nm. For the cell-cell adhesion assay, subconfluent cell layers (~70-80%) were rinsed twice with Ca2- and Mg2-free PBS and detached by incubation in HBSS containing 1 mmol/l EDTA at 37°C for 20 min. Subsequently, single cells (100 µl) at density of ~1×105/ml were added into a 96-well plate (Costar; Corning Incorporated) with a fully confluent single cell layer, and cultured at 37°C for 0-120 min. The unbound cells were washed from the wells, and were collected and quantified using an inverted microscope (TE-2000S; Nikon Corporation). The adhesion rate was determined by counting representative aliquots from each sample on a hematocytometer. The percentage of adhesion was quantified at 60, 90 or 120 min as: N0-Nt/N0 × 100, where Nt is the total number of unbound cells at the incubation time t, and N0 is the total number of cells.
Cellular cytoskeleton analysis
Rhodamine-conjugated phalloidin was used to analyze cell cytoskeleton. The cells grown on cover slides were fixed, and then incubated with rhodamine-conjugated phalloidin (1:200; cat. no. CA1610; Solarbio Science and Technology Co., Ltd., Beijing, China). Following staining with DAPI (1:200; cat. no. C1002; Beyotime Institute of Biotechnology), images of the slides were captured using an inverted fluorescence microscope (TE-2000S; Nikon Corporation).
GTPase activity assays
In brief, the cells were grown to ~80% confluence in regular culture medium, were serum-starved for 24 h and were stimulated with 10% FBS for 5 min. GTP-bound Ras-related C3 botulinum toxin substrate 1 (Rac1), cell division cycle 42 (Cdc42) and total protein were detected using G-LISA Rac1 and G-LISA Cdc42 Activation Assay Biochem kits (Cytoskeleton, Inc., Denver, CO, USA) according to the manufacturer’s protocol.
Metastatic assays in an in vivo orthotopic model
The mice were provided by the Animal Institute of Nanchang University and were housed under specific pathogen-free conditions: Temperature, 25°C; relative humidity, ~40%; lighting, 10 h/day with fluorescent lights. The mice received ad libitum access to sterilized food and water. The experiments were performed according to the protocols approved by the Medical Experimental Animal Care Commission. For the in vivo metastatic assays, the orthotopic model in mice was constructed (23). Briefly, 1×106 cells were injected into the subserosal layer of the sigmoid colon of male BALB/c nude mice weighing ~16-20 g (5 weeks old). The mice were sacrificed following 7 weeks of cell inoculation. Following necropsy, tumors growing in the colon and peritoneum were excised. All livers and macroscopically enlarged mesenteric lymph nodes were harvested, fixed with 10% phosphate- buffered neutral formalin, sectioned serially, and stained with H&E for defining the presence of metastatic disease.
Statistical analysis
All data were analyzed using the statistical software SPSS 18.0 for Windows (SPSS, Inc., Chicago, IL, USA). The differences between groups were analyzed using Student’s t-test between two groups or by one-way analysis of variance in more than two groups when the variance was homogeneous. If the variance was not homogeneous, the differences between groups were analyzed using the Mann-Whitney U test between two groups or Kruskal-Wallis H test in more than two groups. χ2 analysis was used to analyze the correlation between the expression of ARHGEF7 and clinicopathologic features, and the presence of metastasis between two groups. Survival curves were constructed with the Kaplan-Meier method and compared using the log-rank test. The Cox proportional hazards regression model was established to identify independent factors for OS and DFS rates of patients. Receiver operating characteristics (ROC) curves were constructed to assess sensitivity, specificity, and respective areas under the curves with 95% CI. The cut-off value was assessed by the highest Youden index. All tests were two-tailed and P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of ARHGEF7 is significantly increased and associated with metastasis in colorectal adenocarcinoma
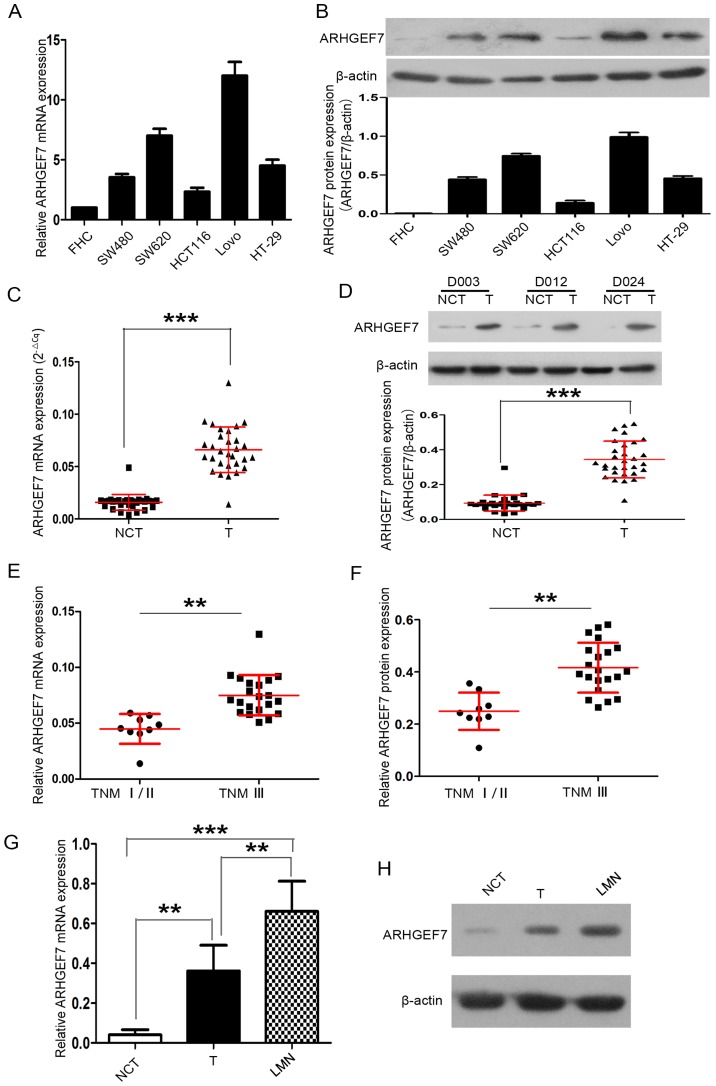
Firstly, the expression of ARHGEF7 in colorectal adenocarcinoma cell lines was detected. Compared with the FHC normal colorectal mucosal cell line, ARHGEF7 mRNA and protein were expressed at high levels in the colorectal adenocarcinoma cell lines, including the HCT116, HT-29, SW480, SW620 and LoVo cell lines (Fig. 1A and B). The expression of ARHGEF7 was also determined in 30 paired colorectal adenocarcinoma samples. RT-qPCR analysis revealed that the mRNA expression of ARHGEF7 was markedly higher in tumor tissues than in corresponding NCTs (P<0.001; Fig. 1C). Western blot analysis also showed that the protein expression of ARHGEF7 was markedly higher in tumor tissues than in NCTs (P<0.001; Fig. 1D).
Figure 1.
ARHGEF7 is overexpressed in colorectal adenocarcinoma and associated with metastasis. ARHGEF7 was significantly upregulated in colorectal adenocarcinoma cell lines. (A) RT-qPCR analysis of ARHGEF7 mRNA showed that, compared with the FHC normal colorectal mucosal cell line, mRNA expression of ARHGEF7 was elevated in HCT116, HT-29, SW480, SW620, LoVo colorectal adenocarcinoma cell lines. (B) Western blot results showed ARHGEF7 protein was overexpressed in HCT116, HT-29, SW480, SW620 and LoVo cells relative to FHC cells. Expression of ARHGEF7 was significantly upregulated in colorectal adenocarcinoma tissues. (C) RT-qPCR was used to analyze mRNA expression of ARHGEF7 in colorectal adenocarcinoma T tissues (n=30) and corresponding NCTs (n=30). RT-qPCR results showed that the mRNA expression level of ARHGEF7 was significantly higher in T tissues than in NCTs. (D) Western blot results showed that the expression of ARHGEF7 was higher in T tissues than in NCTs. Upper panel: Representative western blot of randomly selected samples (D003, D012 and D024)l; lower panel: Scatter plot for ARHGEF7 expression in T tissues and NCTs. (E) mRNA and (F) protein expression of ARHGEF7 in colorectal adenocarcinoma tissues from advanced stage (TNM III) patients were significantly higher than that in those from early stage patients (TNM I/II). Expression of ARHGEF7 progressively increased from NCTs, to T, to LMNs from the same patient (n=5). (G) RT-qPCR analysis of ARHGEF7 mRNA in NCTs, T tissues and LMNs. P=0.003 NCT vs. T; P<0.001 NCT vs. LMN; P=0.005 T vs. LMN (P-values determined by analysis of variance and further SNK test). (H) Representative images of western blot results showed that expression of ARHGEF7 progressively increased from NCTs, to T, to LMNs from the same patient. **P<0.01; ***P<0.001. ARHGEF7, Rho guanine nucleotide exchange factor 7; T, tumor; NCT nontumorous tissues; TNM, tumor-node-metastasis; LMN, liver metastatic nodule; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Subsequently, 30 primary tumors of different TNM stages were analyzed. The expression of ARHGEF7 in colorectal adenocarcinoma tissues was significantly higher in advanced stage patients (TNM stage III) than in early stage patients (TNM stage I/II) (P<0.01; Fig. 1E and F). The expression of ARHGEF7 was further analyzed in LMNs, primary tumors and corresponding NCTs from the same patient (n=5), and the results showed that the expression of ARHGEF7 was progressively increased from NCT to tumor to LMN at the mRNA (Fig. 1G) and protein (Fig. 1H) levels. The LMNs had the highest expression of ARHGEF7, whereas NCTs had the lowest expression (Fig. 1G and H). Taken together, these data confirmed that the expression of ARHGEF7 was significantly elevated in colorectal adenocarcinoma tissues and may be associated with metastasis in colorectal adenocarcinoma.
ARHGEF7 promotes colorectal adenocarcinoma cell motility and invasion in vitro
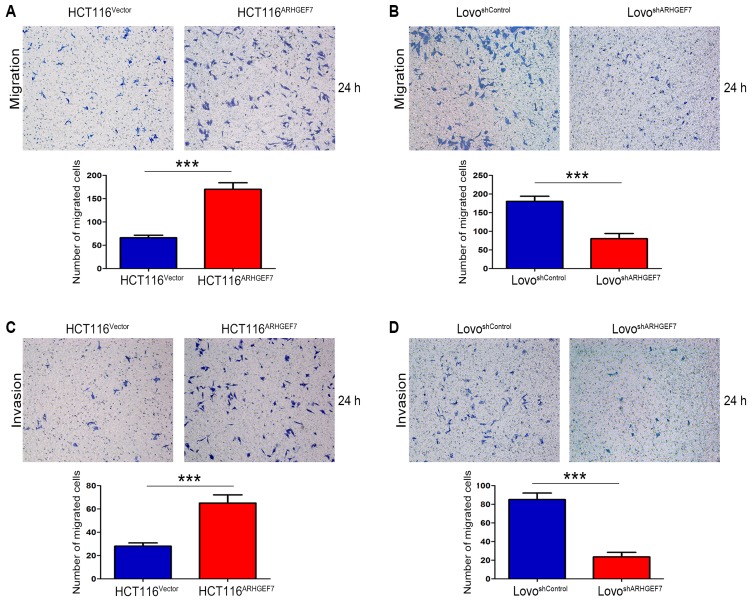
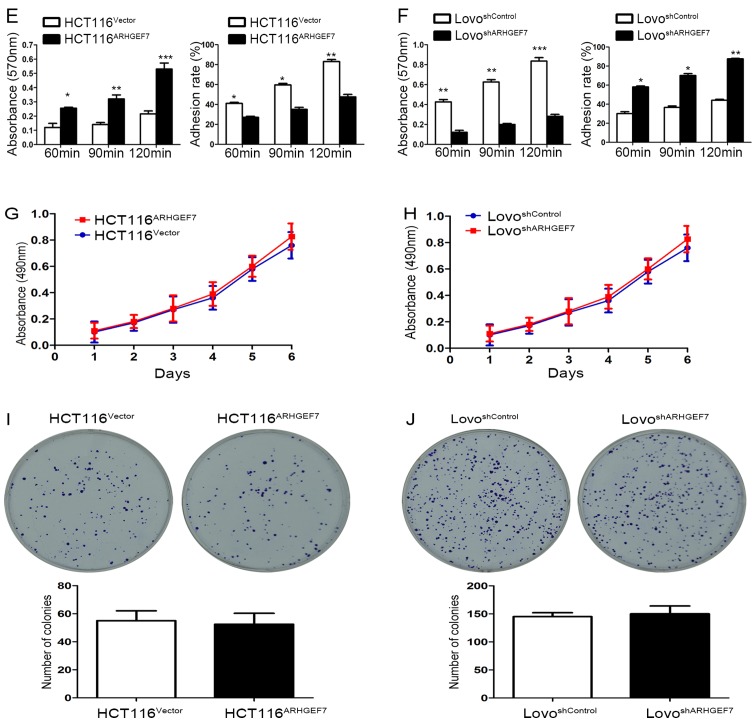
To determine the role of ARHGEF7 in the metastasis of colorectal adenocarcinoma, ARHGEF7 was overexpressed in HCT116 cells (Fig. 2A) and was stably knocked down in LoVo cells (Fig. 2B) according to the expression level of ARHGEF7 and biological characteristics of CRC cell lines. The Transwell migration assays showed that the overexpression of ARHGEF7 significantly increased motility of HCT116 cells (Fig. 3A), whereas the downregulation of ARHGEF7 decreased the motility of LoVo cells (Fig. 3B). Similar results were observed in the Transwell invasion assays (Fig. 3C and D). Adhesion assays showed that ARHGEF7 knockdown suppressed cell-ECM adhesion and enhanced cell-cell adhesion (Fig. 3E and F), whereas the overexpression of ARHGEF7 increased cell-ECM adhesion and inhibited cell-cell adhesion (Fig. 3E and F). Subsequently, the effect of ARHGEF7 on the proliferation of CRC cells was examined. The MTT assays showed that the overexpression of ARHGEF7 had minimal effect on HCT116 cell proliferation (Fig. 3G). Similarly, the downregulation of ARHGEF7 did not affect LoVo cell proliferation (Fig. 3H). The colony formation assays also showed that the capacity of colony formation of HCT116ARHGEF7 cells was almost equal to that of HCT116Vector cells (Fig. 3I). The colony formation capacity of the LoVoshSocrin cells was similar to that of the LoVoshControl cells (Fig. 3J). These data indicated that ARHGEF7 was involved in promoting colorectal adenocarcinoma cell motility and invasion in vitro.
Figure 2.
Validation of the expression of ARHGEF7 in colorectal adenocarcinoma cell lines with ARHGEF7 overexpression or knockdown. (A) ARHGEF7 was upregulated by ARHGEF7-expression LV particles in HCT116 cells. RT-qPCR and western blot analyses were used to evaluate the upregulation of exogenous ARHGEF7 in HCT116 cells. (B) ARHGEF7 was downregulated by shRNA-ARHGEF7 LV particles in LoVo cells. RT-qPCR and western blot analyses were used to assess the inhibitory efficiency of four designed ARHGEF7 shRNA LV particles and control. Expression of ARHGEF7 in control cell was set to 1. Relative expression levels of ARHGEF7 in these cells are shown in the diagram. ARHGEF7-shRNA-Seq2 reduced ARHGEF7 expression by >90% and was used for subsequent experiments due to its highly effective inhibitory efficiency. ARHGEF7, Rho guanine nucleotide exchange factor 7; shRNA, short hairpin RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; LV, lentiviral.
Figure 3.
ARHGEF7 promotes colorectal adenocarcinoma cell migration and invasion in vitro. ARHGEF7 promoted colorectal adenocarcinoma cell migration in vitro. Transwell migration assays of (A) HCT116 cells with ARHGEF7 overexpression (HCT116ARHGEF7) or control (HCT116Vector), and (B) LoVo cells with ARHGEF7 knockdown (LoVoshARHGEF7) or control (LoVoshControl). ARHGEF7 promoted colorectal adenocarcinoma cell invasion in vitro. Original magnification, ×100. Transwell invasion assays of (C) HCT116ARHGEF7 and HCT116Vector, and (D) LoVoshARHGEF7 and LoVoshControl. Original magnification, ×100. Adhesion assays were used to determine the effect of ARHGEF7 on cell adhesion.
ARHGEF7 enhances the invasion and metastasis of colorectal adenocarcinoma cells in vivo
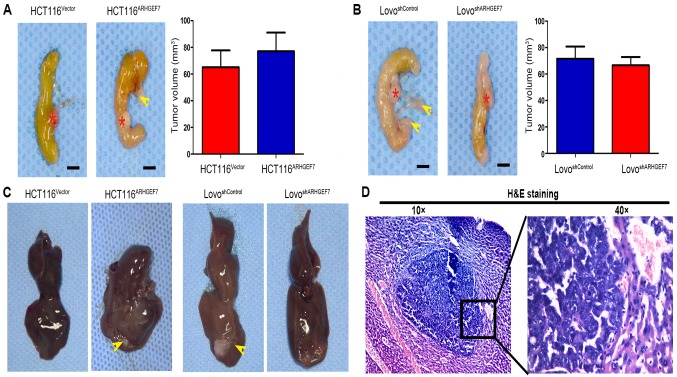
The role of ARHGEF7 in colorectal adenocarcinoma was further evaluated using in situ xenograft metastasis mice models. Following 7 weeks of cell inoculation, the tumor volume of orthotopic mice colon tumors was analyzed. Compared with orthotopic implantation of HCT116Vector cells, mice with implantation of HCT116ARHGEF7 cells had the similar tumor incidence (Table I) and tumor volume in the colon (P>0.05; Fig. 4A). However, mice with implantation of HCT116ARHGEF7 cells had increased peritoneum incidence of regional mesenteric lymph node metastasis (P=0.031; Fig. 4A; Table I) and liver metastasis (P=0.038; Fig. 4C and D; Table I) than mice with implantation of HCT116Vector cells. In the mice implanted with LoVo cells, the tumor incidence and volume of orthotopic colon tumors derived from the LoVoshSocrin cells was not significantly smaller than that of orthotopic colon tumors derived from LoVoshControl cells (P>0.05; Fig. 4B; Table I). However, mice receiving implantation of LoVoshSocrin cells showed lower incidence of regional mesenteric lymph node metastasis (P=0.010; Fig. 4B; Table I) and liver metastasis (P=0.031; Fig. 4C and D; Table I) than the control. Taken together, these results demonstrated that ARHGEF7 promoted colorectal adenocarcinoma metastasis in vivo.
Table I.
Human colon cancer cells implanted in the colon of nude mice.
| Group | Tumor incidence | P-value | Incidence of lymph node metastasis | P-value | Hepatic metastasis | P-value |
|---|---|---|---|---|---|---|
| HCT116Vector | 6/8 | 0.450 | 0/8 | 0.031 | 0/8 | 0.038 |
| HCT116ARHGEF7 | 8/8 | 5/8 | 4/8 | |||
| LoVoshControl | 8/8 | 0.500 | 8/8 | 0.010 | 8/8 | 0.031 |
| LoVoshARHGEF7 | 7/8 | 2/8 | 3/8 |
ARHGEF7, Rho guanine nucleotide exchange factor 7; sh, short hairpin RNA.
Figure 3.
Continued. (E) ARHGEF7 knockdown suppressed cell-ECM adhesion and enhanced cell-cell adhesion. (F) Overexpression of ARHGEF7 increased cell-ECM adhesion and inhibited cell-cell adhesion. Methyl thiazolyl tetrazolium assays were performed to assess the effect of ARHGEF7 on (G) HCT116 and (H) LoVo cell proliferation. Colony formation assays were used to determine the proliferation of (I) HCT116 and (J) LoVo cells. Neither overexpression of ARHGEF7 in HCT116 cells or ARHGEF7 knockdown in LoVo cells affected colony formation. *P<0.05; **P<0.01; ***P<0.001. Data are presented as the mean ± standard deviation. ARHGEF7, Rho guanine nucleotide exchange factor 7; shRNA, short hairpin RNA; ECM, extracellular matrix.
ARHGEF7 promotes colorectal adenocarcinoma metastasis by regulating actin cytoskeleton rearrangements
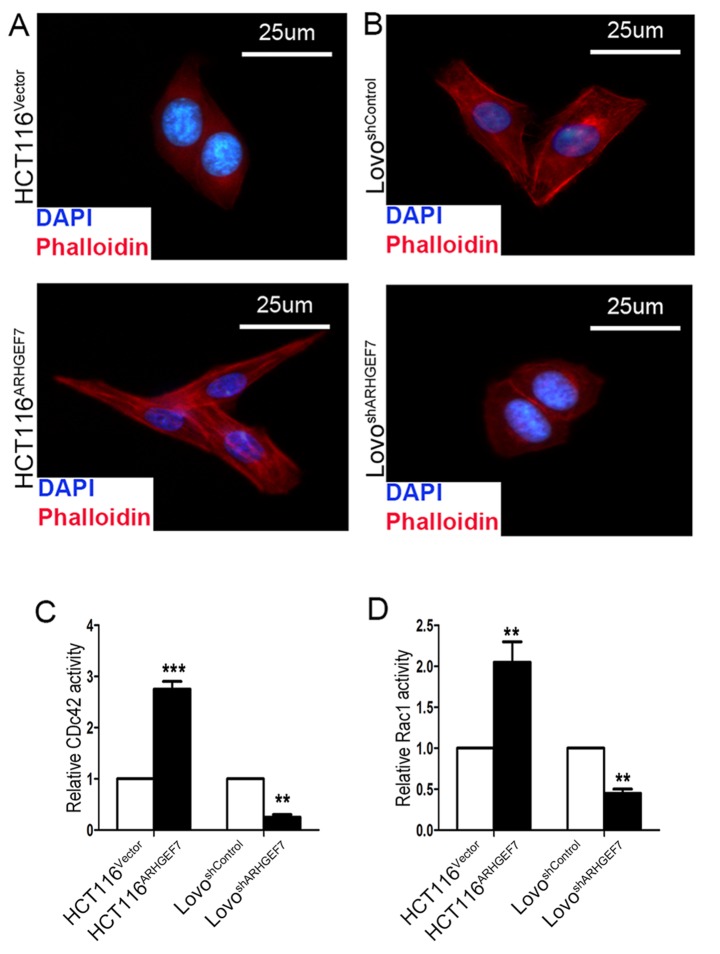
The above results showed that ARHGEF7 promoted colorectal adenocarcinoma metastasis by facilitating cell motility, which is associated with cytoskeletal reorganization by polymerization of filamentous actin (F-actin) (24). Therefore, the present study investigated the effect of ARHGEF7 on regulating the actin cytoskeleton in CRC cell motility by staining F-actin. F-actin immunofluorescence staining was used to analyze the cytoskeleton. The immunofluorescence analysis showed that, compared with the HCT116Vector cells, the appearance of F-actin fibers was present in the HCT116ARHGEF7 cells, which had an increase in filopodia and lamellipodia (Fig. 5A). By contrast, the LoVoshARHGEF7 cells exhibited loosely organized F-actin and shrinkable parallel bundles stress fibers relative to the LoVoshControl cells (Fig. 5B). ARHGEF7 is an identified guanine nucleotide exchange factor for Cdc42/Rac (10), which is involved in cell migration by the extension and maintenance of filopodia (25,26). The effect of ARHGEF7 on the GTP/GDP-binding status of Rac1 and Cdc42 was determined. The results showed that GTP-Cdc42 and GTP-Rac1 activation was significantly increased in the ARHGEF7-overexpressing HCT116 cells (Fig. 5C and D). By contrast, in the LoVo cells, basal Rac1 and Cdc42 activities were marked reduced by ARHGEF7 knockdown (Fig. 5C and D). Therefore, these data indicated that ARHGEF7 may be associated with colorectal adenocarcinoma metastasis by regulating actin cytoskeleton rearrangements.
Figure 4.
ARHGEF7 promotes colorectal adenocarcinoma cell metastasis in vivo. Representative orthotopic tumors from (A) HCT116ARHGEF7 and (B) LoVoshARHGEF7 cells and their control cells are shown. Red stars indicated orthotopic tumor; yellow arrow indicated metastatic lymph node. Tumor volumes of each group are shown and compared in the bar charts; ARHGEF7 exhibited minimal effect on colorectal adenocarcinoma cell growth in vivo. Representative images of (C) macrographic and (D) micrographic metastasis in the liver. ARHGEF7, Rho guanine nucleotide exchange factor 7; shRNA, short hairpin RNA; H&E, hematoxylin and eosin.
High expression of ARHGEF7 correlates with aggressive clinicopathological characteristics and predicts poor prognosis in patients with colorectal adenocarcinoma
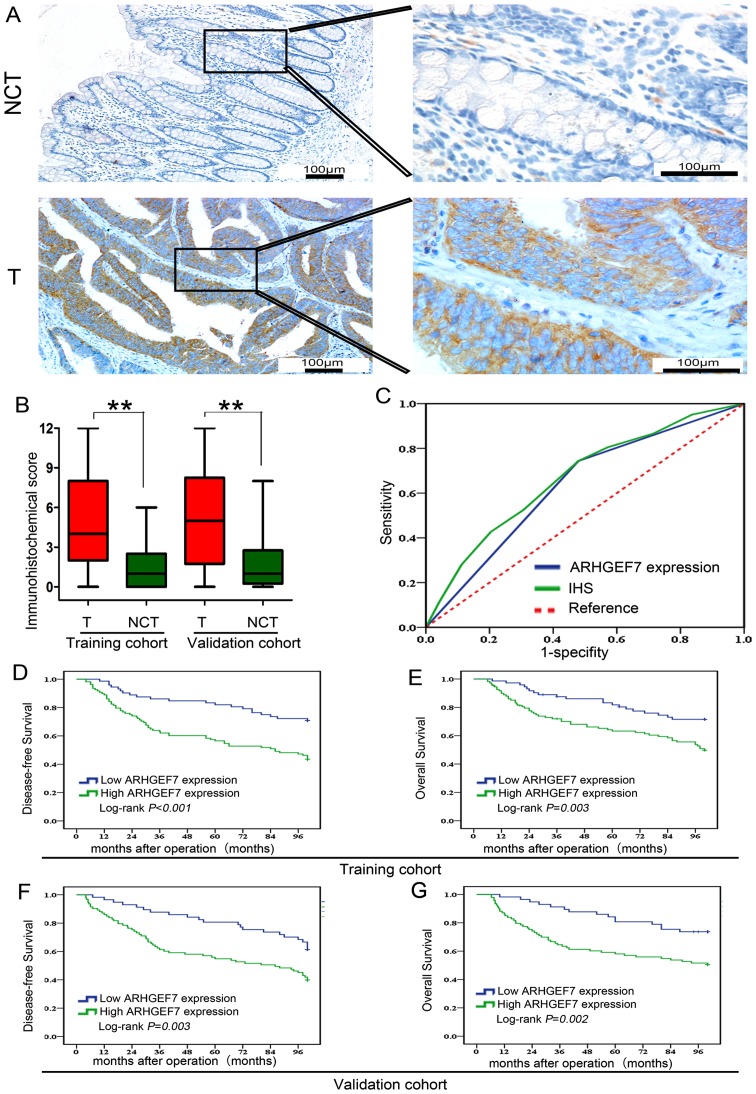
It has been demonstrated that ARHGEF7 is involved in colorectal adenocarcinoma metastasis, which is the main causative factor for poor prognosis in colorectal adenocarcinoma. Therefore, the present study further determined whether the expression of ARHGEF7 was clinically relevant to colorectal adenocarcinoma according to ReMARK guidelines for reporting prognostic biomarkers in cancer (18). An IHC method was used to assay two sets of colorectal adenocarcinoma samples from two independent cohorts, including training and validation cohorts (Fig. 6). The clinicopathologic characteristics of the patients in these two sets are shown in Table II. Based on the training cohort, the results showed that the expression of ARHGEF7 was mainly located in the cytoplasm (Fig. 7A), and the expression level of ARHGEF7 was significantly higher in tumor tissues than in adjacent NCTs (Fig. 7B). An ROC curve for metastasis was utilized to determine the cut-off value according to the results of the IHC staining analysis (Fig. 7C). The highest Youden index was the cut-off value, defined as 4, thus an IS of 4 was selected as a cut-off value for low expression of ARHGEF7. In the training cohort, a high expression of ARHGEF7 was found in 60.0% (108/180) of the tumor tissues, compared with only 20% (36/180) of the adjacent NCTs (P<0.001; Table III). A high expression of ARHGEF7 was significantly correlated with lymph node, mesenteric and distant metastasis (Table IV). Following analysis with the Kaplan-Meier method and log- rank test, patients with colorectal adenocarcinoma with a high expression of ARHGEF7 had either shorter DFS (P<0.001; Fig. 7D) or shorter OS (P=0.003; Fig. 7E), compared with patients with a low expression of ARHGEF7. Finally, to determine whether high expression of ARHGEF7 was an independent prognostic factor for colorectal adenocarcinoma, univariate analysis was first performed followed by multivariate Cox proportional hazards analysis. The data showed a high expression of ARHGEF7 as an independent risk factor for DFS (HR 3.541, 95% CI 1.959-6.400, P<0.001; Table V) and OS (HR 2.050, 95% CI 1.157-3.633, P=0.012; Table V) in colorectal adenocarcinoma.
Figure 5.
ARHGEF7 promotes colorectal adenocarcinoma metastasis by regulating actin cytoskeleton rearrangements Representative images of (A) HCT116 and (B) LoVo cell cytoskeletons show that ARHGEF7 affected the polymerization of filamentous actin in the colorectal adenocarcinoma cells. Original magnification, ×40. The effects of ARHGEF7 on the GTP/GDP-binding status of (C) Cdc42 and (D) Rac1 and were determined. GTP-Cdc42 and GTP-Rac1 activation was significantly increased in ARHGEF7-overexpressing HCT116 cells. In LoVo cells, basal Rac1 and Cdc42 activity was markedly reduced by ARHGEF7 knockdown. ARHGEF7, Rho guanine nucleotide exchange factor 7; shRNA, short hairpin RNA; Rac1, Ras-related C3 botulinum toxin substrate 1; Cdc42, cell division cycle 42.
Table II.
Clinicopathologic characteristics of patients with colorectal adenocarcinoma in the training cohort and validation cohort.
| Clinicopathologic variable | Training cohort (n) (n=180) | Validation cohort (n) (n=150) | P-value |
|---|---|---|---|
| Sex | |||
| Female | 75 | 65 | 0.760 |
| Male | 105 | 85 | |
| Age (years) | |||
| ≤60 | 89 | 70 | 0.615 |
| >60 | 91 | 80 | |
| CEA (ng/ml) | |||
| ≤5 | 110 | 98 | 0.429 |
| >5 | 70 | 52 | |
| Tumor differentiation | |||
| I/II | 83 | 67 | 0.454 |
| III/IV | 97 | 83 | |
| Tumor site | |||
| Colon | 78 | 64 | 0.903 |
| Rectum | 102 | 86 | |
| Tumor size | |||
| ≤5 cm | 94 | 88 | 0.241 |
| >5 cm | 86 | 62 | |
| pT stage | |||
| T1/T2 | 56 | 34 | 0.086 |
| T3/T4 | 124 | 116 | |
| pN stage | |||
| N0 | 85 | 63 | 0.342 |
| N+ | 95 | 87 | |
| Lymphatic/microvascular/ | |||
| nerve invasion | |||
| Negative | 81 | 61 | 0.429 |
| Positive | 99 | 89 | |
| Mesenteric tumor | |||
| deposit formation | |||
| Negative | 114 | 91 | 0.619 |
| Positive | 66 | 59 | |
| Distant metastasis | |||
| Negative | 98 | 83 | 0.872 |
| Positive | 82 | 67 |
Distant metastasis was identified during postoperative follow-up. CEA, carcinoembryonic antigen.
Figure 6.
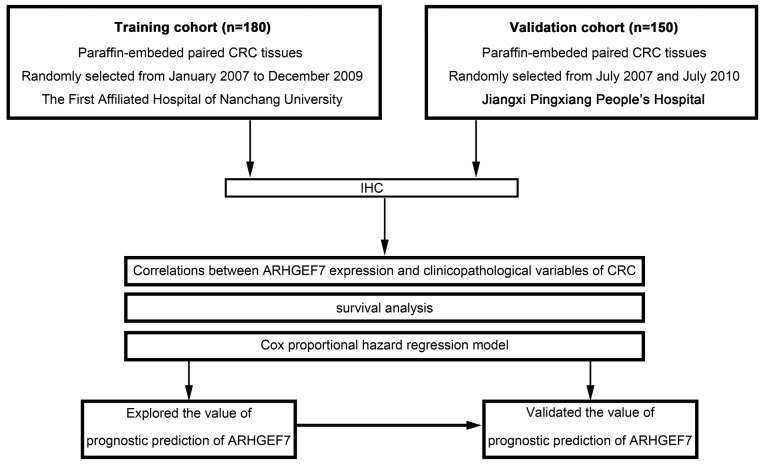
Flowchart of clinical experimental design in two independent cohorts of patients with CRC. ARHGEF7, Rho guanine nucleotide exchange factor 7; CRC, colorectal adenocarcinoma; IHC, immunohistochemistry.
Table III.
Immunohistochemical analysis of the protein expression of ARHGEF7 in tumor tissues and NCTs from the training and validation cohort.
| Training cohort
|
Validation cohort
|
|||||
|---|---|---|---|---|---|---|
| ARHGEF7 expression
|
P-valuea | ARHGEF7 expression
|
P-valuea | |||
| Type | High | Low | High | Low | ||
| Tumor | 108 | 72 | <0.001 | 93 | 57 | <0.001 |
| NCT | 36 | 144 | 45 | 105 | ||
Calculated by χ2 analysis. ARHGEF7, Rho guanine nucleotide exchange factor 7.
Table IV.
Correlations between expression of ARHGEF7 in CRC tissues and clinicopathologic variables of patients with CRC in the training cohort.
| Clinicopathologic variable | n | ARHGEF7 expression
|
P-value | |
|---|---|---|---|---|
| Low (n=72) | High (n=108) | |||
| Sex | ||||
| Female | 75 | 35 | 40 | 0.123 |
| Male | 105 | 37 | 68 | |
| Age (years) | ||||
| ≤60 | 89 | 38 | 51 | 0.465 |
| >60 | 91 | 34 | 57 | |
| CEA (ng/ml) | ||||
| ≤5 | 110 | 49 | 61 | 0.119 |
| >5 | 70 | 23 | 47 | |
| Tumor differentiation | ||||
| I/II | 83 | 43 | 40 | 0.003 |
| III/IV | 97 | 29 | 68 | |
| Tumor site | ||||
| Colon | 78 | 31 | 47 | 0.951 |
| Rectum | 102 | 41 | 61 | |
| Tumor size | ||||
| ≤5 cm | 94 | 40 | 54 | 0.465 |
| >5 cm | 86 | 32 | 54 | |
| pT stage | ||||
| T1/T2 | 56 | 33 | 23 | 0.001 |
| T3/T4 | 124 | 39 | 85 | |
| pN stage | ||||
| N0 | 85 | 47 | 38 | <0.001 |
| N+ | 95 | 25 | 70 | |
| Lymphatic/microvascular/nerve invasion | ||||
| Negative | 81 | 42 | 39 | 0.003 |
| Positive | 99 | 30 | 69 | |
| Mesenteric tumor deposit formation | ||||
| Negative | 114 | 54 | 60 | 0.008 |
| Positive | 66 | 18 | 48 | |
| Distant metastasis | ||||
| Negative | 98 | 51 | 47 | <0.001 |
| Positive | 82 | 21 | 61 | |
CRC, colorectal cancer; CEA, carcinoembryonic antigen; ARHGEF7, Rho guanine nucleotide exchange factor 7.
Table V.
Cox proportional hazard regression analyses for DFS and OS in the training cohort.
| Variable | n | DFS
|
OS
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis
|
Multivariate analysis
|
Univariate analysis
|
Multivariate analysis
|
||||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| Sex | |||||||||
| Female | 75 | Reference | Reference | ||||||
| Male | 105 | 1.051 (0.609-1.814) | 0.298 | NA | 1.176 (0.739-2.833) | 0.271 | NA | ||
| Age (years) | |||||||||
| ≤60 | 89 | Reference | Reference | NA | |||||
| >60 | 91 | 1.192 (0.715-1.987) | 0.180 | NA | 1.207 (0.572-2.137) | 0.170 | |||
| CEA (ng/ml) | |||||||||
| ≤5 | 110 | Reference | Reference | Reference | Reference | ||||
| >5 | 70 | 2.782 (1.948-4.083) | 0.003 | 1.615 (1.271-2.052) | 0.039 | 2.064 (1.356-3.142) | 0.008 | 1.432 (1.083-2.348) | 0.043 |
| Tumor differentiation | |||||||||
| I/II | 83 | Reference | Reference | Reference | Reference | ||||
| III/IV | 97 | 1.521 (1.034-2.416) | 0.045 | 1.007(0.634-1.629) | 0.102 | 1.773 (1.349-3.082) | 0.039 | 1.275 (0.802-2.027) | 0.218 |
| Tumor site | |||||||||
| Colon | 78 | Reference | Reference | ||||||
| Rectum | 102 | 1.206 (0.803-1.811) | 0.127 | NA | 1.216 (0.835-2.238) | 0.103 | NA | ||
| Tumor size | |||||||||
| ≤5 cm | 94 | Reference | Reference | Reference | Reference | ||||
| >5 cm | 86 | 2.033 (1.080-3.824) | 0.028 | 1.594 (0.914-2.780) | 0.073 | 2.078 (1.123-3.845) | 0.026 | 1.602 (0.923-2.781) | 0.057 |
| pT stage | |||||||||
| T1/T2 | 56 | Reference | Reference | Reference | Reference | ||||
| T3/T4 | 124 | 2.175 (1.532-4.782) | 0.012 | 1.876 (1.068-3.296) | 0.024 | 2.179 (1.723-3.835) | 0.010 | 1.832 (1.214-2.907) | 0.035 |
| pN stage | |||||||||
| N0 | 85 | Reference | Reference | Reference | Reference | ||||
| N+ | 95 | 4.309 (2.908-6.385) | <0.001 | 2.524 (1.715-3.715) | 0.001 | 4.204 (2.841-6.221) | <0.001 | 1.918 (1.534-3.235) | 0.022 |
| Lymphatic/microvascular/nerveinvasion | |||||||||
| Negative | 81 | Reference | Reference | Reference | Reference | ||||
| Positive | 99 | 3.034 (1.790-5.145) | 0.005 | 2.175 (1.532-4.782) | 0.010 | 4.914 (3.009-8.025) | <0.001 | 3.190 (1.209-3.967) | 0.001 |
| Mesenteric tumor deposit formation | |||||||||
| Negative | 114 | Reference | Reference | Reference | Reference | ||||
| Positive | 66 | 4.057 (2.485-6.623) | <0.001 | 2.323 (1.219-4.427) | 0.007 | 4.309 (2.908-6.385) | <0.001 | 2.875 (1.733-4.769) | 0.006 |
| ARHGEF7 expression | |||||||||
| Low | 72 | Reference | Reference | Reference | Reference | ||||
| High | 108 | 5.854 (3.193-10.733) | <0.001 | 3.541 (1.959-6.400) | <0.001 | 3.377 (2.135-5.342) | 0.003 | 2.050 (1.157-3.633) | 0.012 |
DFS, disease-free survival; OS, overall survival; CEA, carcinoembryonic antigen; ARHGEF7, Rho guanine nucleotide exchange factor 7.
To validate the clinical significance of ARHGEF7 in colorectal adenocarcinoma, IHC assays of another cohort of CRC samples from the validation cohort showed that the expression level of ARHGEF7 was significantly higher in the tumor tissues than in the adjacent NCTs (Fig. 7B). A high expression of ARHGEF7 was found in 62.0% (93/150) of the tumor tissues, compared with only 30% (45/150) of the NCTs (P<0.001; Table III) from the validation cohort. A high expression of ARHGEF7 was significantly correlated with lymph node, mesenteric and distant metastasis (Table VI). Analysis with the Kaplan-Meier method and log-rank test showed that patients with colorectal adenocarcinoma with a high expression of ARHGEF7 had either shorter DFS (P=0.003; Fig. 7F) or shorter OS (P=0.002; Fig. 7H), compared with those with a low expression of ARHGEF7. Finally, Cox proportional hazards analysis showed that a high expression of ARHGEF7 was an independent risk factor for DFS (HR 3.128, 95% CI 2.536-3.858, P=0.011; Table VII) and OS (HR 2.801, 95% CI 1.503-5.219, P=0.015; Table VII) in colorectal adenocarcinoma. Collectively, these data suggested that ARHGEF7 was a potent prognosticator in addition to promoting colorectal adenocarcinoma metastasis.
Table VI.
Correlations between the expression of ARHGEF7in CRC tissues and clinicopathologic variables of patients with CRC in the validation cohort.
| Clinicopathologic variable | n | ARHGEF7 expression
|
P-value | |
|---|---|---|---|---|
| Low (n=57) | High (n=93) | |||
| Sex | ||||
| Female | 65 | 29 | 36 | 0.144 |
| Male | 85 | 28 | 57 | |
| Age (years) | ||||
| ≤60 | 70 | 30 | 40 | 0.139 |
| >60 | 80 | 27 | 53 | |
| CEA (ng/ml) | ||||
| ≤5 | 98 | 38 | 60 | 0.788 |
| >5 | 52 | 19 | 33 | |
| Tumor differentiation | ||||
| I/II | 67 | 37 | 30 | <0.001 |
| III/IV | 83 | 20 | 63 | |
| Tumor site | ||||
| Colon | 64 | 23 | 41 | 0.653 |
| Rectum | 86 | 34 | 52 | |
| Tumor size | ||||
| ≤5 cm | 88 | 34 | 54 | 0.848 |
| >5 cm | 62 | 23 | 39 | |
| pT stage | ||||
| T1/T2 | 34 | 24 | 10 | <0.001 |
| T3/T4 | 116 | 33 | 83 | |
| pN stage | ||||
| N0 | 63 | 31 | 32 | 0.016 |
| N+ | 87 | 26 | 61 | |
| Lymphatic/microvascular/ | ||||
| nerve invasion | ||||
| Negative | 61 | 40 | 21 | <0.001 |
| Positive | 89 | 17 | 72 | |
| Mesenteric tumor deposit formation | ||||
| Negative | 91 | 35 | 56 | 0.885 |
| Positive | 59 | 22 | 37 | |
| Distant metastasis | ||||
| Negative | 83 | 39 | 44 | 0.012 |
| Positive | 67 | 18 | 49 | |
CRC, colorectal cancer; ARHGEF7, Rho guanine nucleotide exchange factor 7; CEA, carcinoembryonic antigen.
Table VII.
Cox proportional hazard regression analyses for DFS and OS in the validation cohort.
| Variable | n | DFS
|
OS
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis
|
Multivariate analysis
|
Univariate analysis
|
Multivariate analysis
|
||||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| Sex | |||||||||
| Female | 65 | Reference | Reference | ||||||
| Male | 85 | 1.316 (0.782-1.894) | 0.141 | NA | 1.617 (0.883-2.308) | 0.190 | NA | ||
| Age (years) | |||||||||
| ≤60 | 70 | Reference | Reference | ||||||
| >60 | 80 | 1.196 (0.616-2.322) | 0.342 | NA | 1.104 (0.804-1.623) | 0.239 | NA | ||
| CEA (ng/ml) | |||||||||
| ≤5 | 98 | Reference | Reference | Reference | Reference | ||||
| >5 | 52 | 1.702 (1.301-2.227) | 0.010 | 1.201 (1.009-1.430) | 0.047 | 1.875 (1.290-2.725) | 0.008 | 1.287 (1.100-1.505) | 0.039 |
| Tumor differentiation | |||||||||
| I/II | 67 | Reference | Reference | Reference | Reference | ||||
| III/IV | 83 | 1.349 (1.072-1.698) | 0.037 | 1.064 (0.804-1.801) | 0.094 | 1.381 (1.012-2.783) | 0.044 | 1.109 (0.803-1.532) | 0.141 |
| Tumor site | |||||||||
| Colon | 64 | Reference | Reference | ||||||
| Rectum | 86 | 1.332 (0.680-2.609) | 0.108 | NA | 1.206 (0.660-2.153) | 0.204 | NA | ||
| Tumor size | |||||||||
| ≤5 cm | 88 | Reference | Reference | Reference | Reference | ||||
| >5 cm | 62 | 1.603 (1.108-2.319) | 0.042 | 1.180 (0.904-1.540) | 0.089 | 1.506 (1.041-2.198) | 0.036 | 1.145 (0.937-1.399) | 0.109 |
| pT stage | |||||||||
| T1/T2 | 34 | Reference | Reference | Reference | Reference | ||||
| T3/ T4 | 116 | 3.054 (1.625-5.154) | 0.007 | 1.795 (1.187-2.753) | 0.031 | 3.634 (2.327-5.675) | 0.004 | 1.837 (1.351-2.985) | 0.027 |
| pN stage | |||||||||
| N0 | 63 | Reference | Reference | Reference | Reference | ||||
| N+ | 87 | 4.725 (3.051-7.317) | <0.001 | 3.314 (2.191-5.013) | 0.005 | 4.982 (3.087-8.040) | <0.001 | 3.526 (2.354-5.282) | 0.006 |
| Lymphatic/microvascular/nerve invasion | |||||||||
| Negative | 61 | Reference | Reference | Reference | Reference | ||||
| Positive | 89 | 3.004 (2.172-7.624) | 0.007 | 1.934 (1.072-3.489) | 0.044 | 3.582 (2.723-4.712) | 0.009 | 2.036 (1.288-4.732) | 0.20 |
| Mesenteric tumor deposit formation | |||||||||
| Negative | 91 | Reference | |||||||
| Positive | 59 | 3.904 (2.605-5.851) | <0.001 | Reference | 0.019 | Reference | <0.001 | Reference | 0.008 |
| ARHGEF7 | 2.524 (1.715-3.715) | 3.972 (1.433-6.587) | 2.832 (2.054-3.905) | ||||||
| Low | 57 | Reference | |||||||
| High | 93 | 3.834 (1.972-7.624) | 0.003 | Reference | 0.011 | Reference | 0.002 | Reference | 0.015 |
DFS, disease-free survival; OS, overall survival; CEA, carcinoembryonic antigen; ARHGEF7, Rho guanine nucleotide exchange factor 7; NA, not applicable.
Discussion
Colorectal adenocarcinoma is a frequently life-threatening disease with heterogeneous outcomes (27). Surgical resection remains the most important therapy used for patients with colorectal adenocarcinoma (3). However, cancer metastasis presents an obstacle in improving the clinical outcome, which affects up to 60% of patients with colorectal adenocarcinoma (28). The detailed molecular mechanism of metastasis remains to be fully elucidated. The present study revealed that the expression of ARHGEF7 was increased in colorectal adenocarcinoma tissues compared with that in NCTs. Increased expression of ARHGEF7 was found to be positively correlated with the metastatic potential of colorectal adenocarcinoma cells, suggesting that ARHGEF7 was involved in colorectal adenocarcinoma metastasis.
To define the role of ARHGEF7 in metastasis, a serial of in vitro assays showed that that overexpression of ARHGEF7 in colorectal adenocarcinoma cells significantly enhanced cell migration and invasion, whereas the knockdown of ARHGEF7 in colorectal adenocarcinoma cells significantly decreased cell migration and invasion. Furthermore, the in vivo assays showed that the overexpression of ARHGEF7 in colorectal adenocarcinoma cells facilitated tumor metastasis, whereas the knockdown of ARHGEF7 in colorectal adenocarcinoma cells significantly inhibited tumor metastasis. Although the effect of ARHGEF7 on the proliferation of colorectal adenocarcinoma cells was not observed, the results were consistent with the involvement of ARHGEF7 in cell migration and metastasis in other malignancies (14,29), suggesting that ARHGEF7 possesses oncogenic properties in promoting metastasis in colorectal adenocarcinoma.
Cancer metastasis occurs via a process involving abnormal cell migration (30). Cell migration, a dynamic physical process, is controlled by the cytoskeletal system, which includes the dynamics of actin organization (30). A previous study showed that ARHGEF7 had a direct regulatory role in promoting dense actin networks to direct cell migration (29). In the present study, the immunofluorescences analysis showed that ARHGEF7 promoted the polymerization of F-actin in colorectal adenocarcinoma cells and facilitated the formation of filopodia and lamellipodia. The results further showed that the activation of GTP-Cdc42 and GTP-Rac1 was significantly increased in the ARHGEF7-overexpressing colorectal adenocarcinoma cells. By contrast, basal Rac1 and Cdc42 activity was markedly reduced by ARHGEF7 knockdown in colorectal adenocarcinoma cells, supporting the hypothesis that ARHGEF7 serves as a GEF protein with activity towards Rac1 and Cdc42 in cell migration (26).
The prognosis for patients with metastatic disease remains poor (31), therefore, the identification of more potent molecular biomarkers for identifying patient subgroups with high-risk metastasis is urgently required. The present study also suggested that ARHGEF7 may be a promising prognostic biomarkers associated with survival rates. The prognostic significance of the expression of ARHGEF7 was validated in two independent cohorts according to ReMARK guidelines for reporting prognostic biomarkers in cancer (18). It was found that a high expression of ARHGEF7 was significantly correlated with metastasis, and shorter DFS or shorter OS. A high expression of ARHGEF7 was found to be an independent prognostic factor in colorectal adenocarcinoma. However, complex pathways contribute to colorectal adenocarcinoma progression, and the prediction of efficacy of ARHGEF7 and outcome in colorectal adenocarcinoma requires validation for clinical use.
In conclusion, the present study identified the frequently increased expression of ARHGEF7 in colorectal adenocarcinoma tissues and this expression pattern was associated with colorectal adenocarcinoma metastasis. Furthermore, it was demonstrated that ARHGEF7 promoted metastasis by regulating actin cytoskeleton rearrangements. Finally, it was shown that a high expression of ARHGEF7 was clinically relevant, with aggressive clinicopathological characteristics and poor prognosis in patients with colorectal adenocarcinoma, when consulting ReMARK guidelines for reporting prognostic biomarkers in cancer. Collectively, the data indicated that ARHGEF7 was important in colorectal adenocarcinoma metastasis and may be an independent potential prognostic marker for predicting clinical outcome in colorectal adenocarcinoma.
Funding
This study was supported by the Natural Science Foundation of Jiangxi, China (grant no. 20142BAB205055) and also partly supported by the National Natural Science Foundation of China (grant no. 81702922).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
TL, XL and BX designed the experiments. XL, LD, JL, ZW, DL, SL, HD, GH, CT and CX performed experiments and analyzed data. TL, BX, XL and LD provided patient samples and collected data. TL, XL, JL, ZW wrote and revised the paper.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Institutional Review Boards of the First Affiliated Hospital of Nanchang University and Jiangxi Pingxiang People’s Hospital, and was performed in accordance with the Declaration of Helsinki and current ethical guidelines. Prior informed consent was obtained from all participants.
Patient consent for publication
Patients provided written informed consent for publication.
Competing interests
The authors confirm that they have no competing interests.
Figure 7.
ARHGEF7 is a potential prognosticator for colorectal adenocarcinoma clinical outcome following radical surgery. (A) Representative IHC staining of ARHGEF7 protein in tumor tissues and NCTs from the training and validation cohort. (B) Tumor tissues exhibited higher protein expression of ARHGEF7 than corresponding NCTs. A box-plot was used to show the differential expression and a Mann-Whitney U test was used to analyze the difference between tumor tissues and NCTs from the training and validation cohort. (C) ROC curve analysis. The sensitivity and 1-specificity of IS in colorectal adenocarcinoma tumor tissues was plotted (green line). The AUC and the P-value, as calculated by ROC curve analysis, were AUC=0.668 and P<0.001 respectively. The highest Youden index cut-off value was defined as 4; an IS of 4 was selected as a cut-off value for low expression of ARHGEF7. The sensitivity and 1-specificity of ARHGEF7 expression in colorectal adenocarcinoma tissues were plotted (blue line). The AUC and the P-value as calculated by ROC curve analysis were AUC=0.632 and P=0.002, respectively. Kaplan-Meier analysis of the correlation between the expression of ARHGEF7 and (D) DFS and (E) OS of patients with colorectal adenocarcinoma in the training cohort. Kaplan-Meier analysis of the correlation between the expression of ARHGEF7 and (F) DFS and (G) OS of patients with colorectal adenocarcinoma in the validation cohort. ARHGEF7, Rho guanine nucleotide exchange factor 7; T, tumor; NCTs, nontumorous tissues; ROC, receiver operating characteristic; AUC, area under the curve; DFS, disease-free survival; OS, overall survival; IHC, immunohistochemistry; IS, immunostaining score.
Acknowledgments
The authors would like to thank Dr Jian Lei (Department of Pathology, Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University) for providing technical support for histology.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi H, Ueno H, Hashiguchi Y, Mochizuki H. Distribution of lymph node metastasis is a prognostic index in patients with stage III colon cancer. Surgery. 2006;139:516–522. doi: 10.1016/j.surg.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Reddy GK. Prognostic significance of increasing lymph node count in stage III colorectal cancer. Clin Colorectal Cancer. 2006;5:403–404. doi: 10.1016/S1533-0028(11)70215-8. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg R, Engel J, Bruns C, Heitland W, Hermes N, Jauch KW, Kopp R, Pütterich E, Ruppert R, Schuster T, et al. The prognostic value of lymph node ratio in a population-based collective of colorectal cancer patients. Ann Surg. 2010;251:1070–1078. doi: 10.1097/SLA.0b013e3181d7789d. [DOI] [PubMed] [Google Scholar]
- 7.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Koh CG, Manser E, Zhao ZS, Ng CP Lim L. Beta1PIX, the PAK-interacting exchange factor, requires localization via a coiled-coil region to promote microvillus-like structures and membrane ruffles. J Cell Sci. 2001;114:4239–4251. doi: 10.1242/jcs.114.23.4239. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, Eom M, Lee SJ, Kim S, Park H, Park D. βPix-enhanced p38 Activation by Cdc42/Rac/PAK/MKK3/6-mediated Pathway. J Biol Chem. 2001;276:25066–25072. doi: 10.1074/jbc.M010892200. [DOI] [PubMed] [Google Scholar]
- 11.Kuo JC, Han X, Hsiao CT, Yates JR, III, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campa F, Machuy N, Klein A, Rudel T. A new interaction between Abi-1 and betaPIX involved in PDGF-activated actin cytoskeleton reorganisation. Cell Res. 2006;16:759–770. doi: 10.1038/sj.cr.7310091. [DOI] [PubMed] [Google Scholar]
- 13.Heidary Arash E, Song KM, Song S, Shiban A, Attisano L. Arhgef7 promotes activation of the Hippo pathway core kinase Lats. EMBO J. 2014;33:2997–3011. doi: 10.15252/embj.201490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Han M, Whetsell W, Jr, Wang J, Rich J, Hallahan D, Han Z. Tax-interacting protein 1 coordinates the spatiotemporal activation of Rho GTPases and regulates the infiltrative growth of human glioblastoma. Oncogene. 2014;33:1558–1569. doi: 10.1038/onc.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu YH, Lin WL, Hou YT, Pu YS, Shun CT, Chen CL, Wu YY, Chen JY, Chen TH, Jou TS. Podocalyxin EBP50 ezrin molecular complex enhances the metastatic potential of renal cell carcinoma through recruiting Rac1 guanine nucleotide exchange factor ARHGEF7. Am J Pathol. 2010;176:3050–3061. doi: 10.2353/ajpath.2010.090539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muñoz-Bellvis L, Fontanillo C, González-González M, Garcia E, Iglesias M, Esteban C, Gutierrez ML, Abad MM, Bengoechea O, De Las Rivas J, et al. Unique genetic profile of sporadic colorectal cancer liver metastasis versus primary tumors as defined by high- density single-nucleotide polymorphism arrays. Mod Pathol. 2012;25:590–601. doi: 10.1038/modpathol.2011.195. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Davis C, Shah S, Hughes D, Ryan JC, Altomare D, Peña MM. IL-33 promotes growth and liver metastasis of colorectal cancer in mice by remodeling the tumor microenvironment and inducing angiogenesis. Mol Carcinog. 2017;56:272–287. doi: 10.1002/mc.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Lei X, Liang Y, Chen J, Xiao S, Lei J, Li J, Duanmu J, Jiang Q, Liu D, Tang C, et al. Sorcin predicts poor prognosis and promotes metastasis by facilitating epithelial-mesenchymal transition in hepatocellular carcinoma. Sci Rep. 2017;7:10049. doi: 10.1038/s41598-017-10365-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Liu Su, Chen SQ, Chen J, Chen J, He F, Huang C, Wu D, Lin W, Huang LW, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25:605–620. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 22.García-Echeverría C, Pearson MA, Marti A, Meyer T, Mestan J, Zimmermann J, Gao J, Brueggen J, Capraro HG, Cozens R, et al. In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell. 2004;5:231–239. doi: 10.1016/S1535-6108(04)00051-0. [DOI] [PubMed] [Google Scholar]
- 23.Yokoi K, Thaker PH, Yazici S, Rebhun RR, Nam DH, He J, Kim SJ, Abbruzzese JL, Hamilton SR, Fidler IJ. Dual inhibition of epidermal growth factor receptor and vascular endothelial growth factor receptor phosphorylation by AEE788 reduces growth and metastasis of human colon carcinoma in an orthotopic nude mouse model. Cancer Res. 2005;65:3716–3725. doi: 10.1158/0008-5472.CAN-04-3700. [DOI] [PubMed] [Google Scholar]
- 24.Insall RH, Machesky LM. Actin dynamics at the leading edge: From simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Reddy PN, Radu M, Xu K, Wood J, Harris CE, Chernoff J, Williams DA. p21-activated kinase 2 regulates HSPC cytoskeleton, migration, and homing via CDC42 activation and interaction with β-Pix. Blood. 2016;127:1967–1975. doi: 10.1182/blood-2016-01-693572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ten Klooster JP, Jaffer ZM, Chernoff J, Hordijk PL. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J Cell Biol. 2006;172:759–769. doi: 10.1083/jcb.200509096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahani DV, Bajwa MA, Andrabi Y, Bajpai S, Cusack JC. Current status of imaging and emerging techniques to evaluate liver metastases from colorectal carcinoma. Ann Surg. 2014;259:861–872. doi: 10.1097/SLA.0000000000000525. [DOI] [PubMed] [Google Scholar]
- 29.Yu HW, Chen YQ, Huang CM, Liu CY, Chiou A, Wang YK, Tang MJ, Kuo JC. β-PIX controls intracellular viscoelasticity to regulate lung cancer cell migration. J Cell Mol Med. 2015;19:934–947. doi: 10.1111/jcmm.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang B, Wu N, Guan R, Pang L, Li X, Li S, Tang L, Guo Y, Chen J, Sun D, et al. Overexpression of RCC2 enhances cell motility and promotes tumor metastasis in lung adenocarcinoma by inducing epithelial-mesenchymal transition. Clin Cancer Res. 2017;23:5598–5610. doi: 10.1158/1078-0432.CCR-16-2909. [DOI] [PubMed] [Google Scholar]
- 31.Sanoff HK, Sargent DJ, Campbell ME, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC, Goldberg RM. Five-year data and prognostic factor analysis of oxaliplatin and irinotecan combinations for advanced colorectal cancer. J Clin Oncol. 2008;26:5721–5727. doi: 10.1200/JCO.2008.17.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.