Abstract
Anorectal malformations (ARMs) are one of the most common congenital malformations of the digestive tract; however, the pathogenesis of this disease remains to be fully elucidated. MicroRNAs (miRNAs) are important in gastrointestinal development and may be involved in the pathogenesis of ARMs. The present study aimed to profile miRNAs and examine their potential functions in rats with ethylenethiourea (ETU)-induced ARMs. Pregnant Wistar rats (n=36) were divided randomly into ETU-treated and control groups. The rats in the ETU-treated group were gavage-fed 1% ETU (125 mg/kg) on gestational day 10 (GD10), whereas the control group rats received a corresponding dose of saline. Embryos were harvested by cesarean section on GD14, GD15 and GD16. Hindgut tissue was isolated from the fetuses for RNA extraction and microarray analysis, followed by bioinformatics analysis and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) validation. Overall, 38 miRNAs were differentially expressed (all upregulated) on GD14, 49 (32 upregulated and 17 downregulated) on GD15, and 42 (all upregulated) on GD16 in the ARM group compared with the normal group. The top 18 miRNAs with |log2(fold change)| >4.25 were selected for further bioinformatics analysis. Among these miRNAs, five were differentially expressed at two time-points and were involved in ARM-associated signaling pathways. The RT-qPCR analysis revealed that three miRNA (miR), miR-125b-2-3p, miR-92a-2-5p and miR-99a-5p, were significantly differentially expressed in rats with ARMs compared with the normal group. In conclusion, the results suggested that the differential expression of miR-125b-2-3p, miR-92a-2-5p and miR-99a-5p during key time-points of anorectal formation in rats may have functions in the pathogenesis of ARM.
Keywords: microRNA, anorectal malformation, gene expression, bioinformatics
Introduction
Anorectal malformations (ARMs) are common congenital malformations of the digestive tract manifesting as ectopic anus, anal stenosis and/or an abnormal fistula between the rectum and urinary tract, with an incidence of 2-6/10,000 (1-4). ARMs are usually combined with malformations of other systems, and although the majority of patients with ARMs require surgery, their quality of life is likely to be impaired postoperatively (5). Although earlier studies have revealed that multiple genes and signaling pathways are important in the pathophysiology of ARMs (3,6), the specific mechanisms remain to be fully elucidated. Therefore, it is necessary to investigate the pathogenesis of ARMs.
MicroRNAs (miRNAs) regulate gene expression at the post-transcriptional level by binding to the 3′-untranslated regions (3′-UTRs) of target mRNAs, and have been reported to be important in various congenital diseases and in the embryonic development of various systems (7-10). Previous studies have investigated the functions of miRNAs in gastrointestinal development in various species. For example, miRNA (miR)-143 was reported to regulate genes involved in the differentiation of connective tissue cells and to induce the proliferation and differentiation of smooth muscle cells, thus influencing bovine gut development (8). As the main effector of miR-17-92 cluster components, a high expression of miR-17-5p in the crypt progenitor compartment is potentially involved in controlling cell differentiation and proliferation in human colon development (11). However, current understanding of the functions of miRNAs in the development of ARMs remains limited. An increased understanding of miRNA expression profiles may reveal their functional roles in ARM development, thus providing a valuable basis for investigating the mechanisms responsible for this condition.
Ethylenethiourea (ETU)-induced ARM rat fetuses are the most commonly used animal model for investigating this disease, and the key abnormalities in the experimental embryos include the following: i) no fusion between the urorectal septum and cloacal membrane; ii) delay of tailgut regression; iii) abnormal apoptosis in the cloacal wall; iv) underdevelopment of the dorsal cloaca and its membrane (12,13). In the present study, miRNA expression patterns in the hindgut of rat fetuses with ETU-induced ARMs were profiled between gestational day (GD)14 and GD16, which are critical time-points during anorectal development (4,12-14). Additionally, advanced bioinformatics analysis was performed, including target gene prediction, construction of regulatory networks and functional enrichment analyses to clarify the molecular mechanisms involved in the development of ARMs.
Materials and methods
Ethics statement
The present study was approved by the China Medical University Ethics Committee (Shenyang, China; no. 2015 PS213K) and all procedures involving animals were performed in accordance with the guidelines for the care and use of laboratory animals (15).
Animal models and tissue preparation
Mature female Wistar rats (n=36; age, 7-9 weeks-old; body weight, 250-300 g) were provided by the Experimental Animal Center of Shengjing Hospital of China Medical University (Shenyang, China) and housed in a specific-pathogen-free animal laboratory (room temperature, 22±2°C; humidity, 55±5%; 12 h light/dark cycle and ad libitum access to water and food) at The Key Laboratory of Health Ministry for Congenital Malformations (Shenyang, China). The surgical procedures were performed following sacrifice by intraperitoneal injection of sodium pentobarbital, and all efforts were made to minimize animal suffering.
ARM was induced in fetal rats as described in earlier reports (4). Briefly, 18 pregnant rats were gavage-fed a single dose of 125 mg/kg of 1% ETU (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) on GD10, whereas the remaining 18 pregnant rats received corresponding doses of ETU-free saline as a control. The rat fetuses were obtained by cesarean delivery on GD14-16, and the presence of ARMs was determined by light microscopy.
No malformations were observed in the 258 embryos of the normal rats. Among the ETU-treated embryos, all 236 embryos had a short or absent tail, and 14 died in utero. The incidence of ARMs in the ETU-treated embryos was 85.6% (202/236).
miRNA profiling and microarray analysis
Assessment of RNA quality
Total RNA was isolated using an miRNeasy Mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. RNA quality was assessed using a K5500 micro-spectrophotometer (Shanghai Drawell Scientific Instrument Co., Ltd., Shanghai, China) and formaldehyde agarose gel electrophoresis. Values of absorbance (A)260/A280 ≥1.5 and A260/A230 ≥1 indicated acceptable RNA purity, and an RNA integrity number ≥7 from the Agilent 2200 RNA assay (Agilent Technologies, Inc., Santa Clara, CA, USA) indicated acceptable RNA integrity.
miRNA microarray assay
miRNA profiling was performed using the RiboArray platform (Ribobio Co., Ltd., Guangzhou, China). In brief, l μg of total RNA was labeled with Cy3 using the ULS™ microRNA labeling kit (Kreatech; Leica Microsystems, Inc., Buffalo Grove, IL, USA) and hybridized on the microarray. The microarray contained 761 specific oligos for rat miRNAs, based on the Sanger miRBase 21.0 database (www.mirbase.org/).
Data normalization and analysis
Firstly, chip images were examined to exclude impurity, scratch marks and high background intensity. Following background adjustment by subtraction of background from foreground, the microarray data were normalized using a quantile (median) method and box-plots were generated based on the relative logarithm expression using the limma package in R software (version 3.3.0; www.r-project.org). Three-dimensional principal component analysis was performed using the scatter-plot3d package in R. Analysis of the differentially expressed miRNAs was performed using the limma package in R. Standard selection criteria for identifying the differentially expressed miRNAs were established as a fold change (FC) >2 or <−2, and P<0.05. Hierarchical clustering analysis of the miRNAs was performed and graphs were generated using the gplots package in R software.
Target gene prediction, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses
miRNAs with |log2(FC)| >4.25 were selected for further analysis. Target genes of these miRNAs were predicted using the target gene-prediction platforms miRWalk 3.0 (mirwalk.umm.uni-heidelberg.de/), TargetScan 7.1 (www.targetscan.org/vert_71/), miRBase 22 (www.mirbase.org/), miRDB (mirdb.org/miRDB/), and DIANA v5.0 (diana.imis.athena-innovation. gr/DianaTools/index.php?r=site/page&view=software). Those miRNAs predicted by at least three databases were considered as predicted target genes. Experimentally validated target genes were identified using miRWalk 3.0.
The biological functions of the key miRNAs were determined by GO clustering, including the three domains 'biological process' (BP), 'cellular component' (CC) and 'molecular function' (MF), and KEGG pathway analysis of the target genes using the Database for Annotation, Visualization and Integrated Discovery Bioinformatics Resources 6.7 (david-d.ncifcrf.gov/); corresponding results were displayed using the ggplot2 and gcookbook packages in R software.
Regulatory networks of the miRNA-target genes and miRNA-target gene signaling pathways were constructed and visualized using Cytoscape 3.4.0 (www.cytoscape.org/) in the Java Environment (16).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT was performed using a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). Stem-loop RT primers and PCR primers for each miRNA were purchased from Ribobio Co., Ltd. (cat. nos. U6 MQP-0202, rno-mir-125b-2-3p miRQ0004731-1-1, rno-mir-187-5p miRQ0017144-1-1, rno-mir-3542 miRQ0017796-1-1, rno-mir-92a-2-5p miRQ0017108-1-1 and rno-mir-99a-5p miRQ0000820-1-1). Fluorescence qPCR was performed on an ABI 7500 detection system (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Each 20 μl reaction mixture contained 2 μl template cDNA, 0.4 μM of each gene-specific primer, 10 μl SYBR Premix Ex Taq II (Tli RNaseH Plus, 2X), 0.4 μl ROX Reference Dye II (50X), and 6 μl sterilized Rnase-free water. The reactions were incubated at 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec, and 60°C for 60 sec. The reactions were incubated at 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec, and 60°C for 60 sec. With stable expression in the normal and ARM groups, U6 was selected as an endogenous reference gene for qPCR normalization purposes and the relative expression level of each miRNA was calculated using the 2−ΔΔCq method (17). RT-qPCR analysis was performed with three replicates.
Statistical analysis
Statistical analysis was performed using SPSS 21.0 software (IBM SPSS, Armonk, NY, USA). Statistically significant differences were identified using Student's t-test (two-tailed). All values are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Screening of differentially expressed miRNAs and clustering analysis
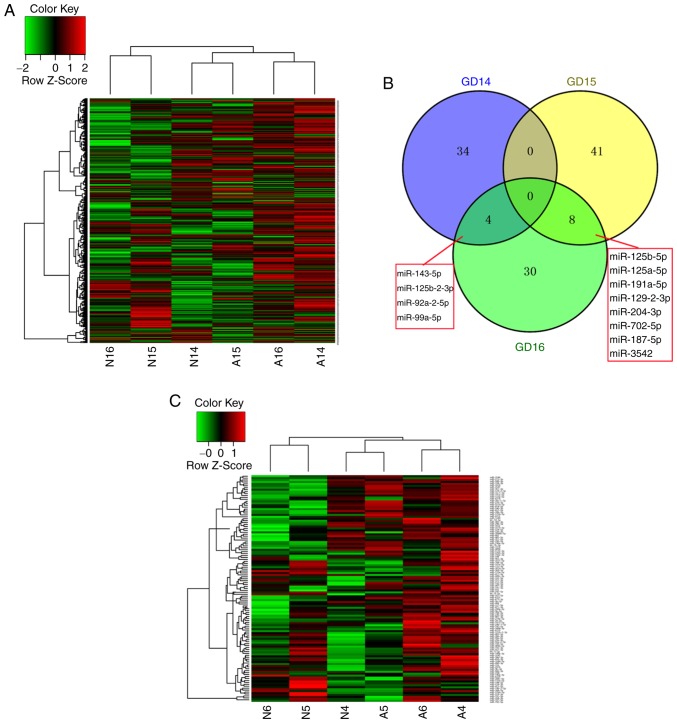
The miRNAs potentially involved in ARM development were investigated using microarray screening of differentially expressed miRNAs at key time-points during anus formation (GD14, GD15 and GD16). A heatmap of the 761 miRNAs at the three time-points is shown in Fig. 1A. The results of the microarray screening revealed different expression patterns between the normal and ARM groups, and the majority of miRNAs exhibited increased expression in the ARM group. In the normal group, a higher expression level of miRNAs was detected at GD15 compared with GD14 and GD16, and the expression trend was opposite to that in the ARM group.
Figure 1.
miRNA expression and miRNA-target gene regulatory network. In the heatmap, green and red indicate a decrease and increase of miRNA expression, respectively. The intensity of the color corresponds to the relative signal level on a logarithmic scale. (A) Hierarchical heatmap of the overall microarray screening. (B) Venn diagram of differentially expressed miRNAs at three time-points. (C) Hierarchical heatmap of differentially expressed miRNAs.
A total of 38 miRNAs were upregulated on GD14 in the ARM group compared with the normal group; 32 were upregulated and 17 were downregulated at GD15 in the ARM group compared with the normal group; and 42 were upregulated at GD16 in the ARM group compared with the normal group. The overlap of these miRNAs between the three time-points is shown in Fig. 1B. The detailed information and expression patterns of the differentially expressed miRNAs at the three time-points are shown in Table I and Fig. 1C.
Table I.
Differentially expressed miRNAs at GD14, GD15 and GD16.
| miRNA | lgFC | FDR |
|---|---|---|
| G14; N vs. A (38) | ||
| miR-125b-2-3p | 5.6523 | 0.0181 |
| miR-92a-2-5p | 4.9863 | 0.0033 |
| miR-99a-5p | 4.7777 | 0.0207 |
| miR-331-3p | 4.7257 | 0.0171 |
| miR-410-3p | 4.5907 | 0.0124 |
| miR-26b-3p | 4.2617 | 0.0215 |
| miR-190a-3p | 4.2564 | 0.0052 |
| miR-212-5p | 4.1900 | 0.0495 |
| miR-802-5p | 4.1618 | 0.0230 |
| miR-3586-3p | 4.1185 | 0.0394 |
| miR-143-5p | 4.1014 | 0.0133 |
| miR-132-3p | 4.0487 | 0.0068 |
| miR-200a-3p | 3.9410 | 0.0020 |
| miR-6319 | 3.8708 | 0.0209 |
| miR-497-3p | 3.8449 | 0.0105 |
| miR-29a-3p | 3.7200 | 0.0008 |
| miR-133a-5p | 3.6877 | 0.0338 |
| miR-501-5p | 3.4335 | 0.0379 |
| miR-369-5p | 3.4053 | 0.0480 |
| miR-322-5p | 3.3726 | 0.0006 |
| miR-146a-3p | 3.1359 | 0.0068 |
| miR-9b-3p | 3.0811 | 0.0077 |
| miR-31b | 3.0555 | 0.0216 |
| miR-301b-3p | 3.0376 | 0.0278 |
| let-7i-3p | 3.0366 | 0.0087 |
| miR-291b | 3.0339 | 0.0077 |
| miR-490-3p | 2.9198 | 0.0040 |
| miR-30e-3p | 2.8834 | 0.0112 |
| miR-421-3p | 2.8269 | 0.0162 |
| miR-6329 | 2.6684 | 0.0479 |
| miR-411-5p | 2.4638 | 0.0261 |
| miR-344i | 2.2869 | 0.0288 |
| miR-326-3p | 1.8954 | 0.0491 |
| miR-376b-5p | 1.8833 | 0.0128 |
| miR-666-3p | 1.8752 | 0.0217 |
| miR-192-3p | 1.7802 | 0.0307 |
| miR-3557-5p | 1.6589 | 0.0399 |
| miR-376b-3p | 1.0623 | 0.0007 |
| G15; N vs. A (49) | ||
| miR-741-3p | 4.9521 | 0.0013 |
| miR-935 | 4.9112 | 0.0077 |
| miR-542-5p | 3.3498 | 0.0025 |
| miR-1249 | 3.2639 | 0.0007 |
| miR-5132-3p | 2.1773 | 0.0133 |
| miR-129-2-3p | 2.0751 | 0.0011 |
| miR-409b | 1.9867 | 0.0165 |
| miR-7a-1-3p | 1.8004 | 0.0432 |
| miR-3550 | 1.7798 | 0.0096 |
| miR-349 | 1.6717 | 0.0172 |
| miR-874-5p | 1.6689 | 0.0232 |
| miR-3547 | 1.6623 | 0.0007 |
| miR-551b-5p | 1.6278 | 0.0027 |
| miR-361-3p | 1.5764 | 0.0205 |
| miR-3546 | 1.5650 | 0.0199 |
| miR-455-3p | 1.5239 | 0.0484 |
| miR-212-3p | 1.5230 | 0.0169 |
| miR-743a-3p | 1.4840 | 0.0116 |
| miR-196a-3p | 1.3656 | 0.0000 |
| miR-28-5p | 1.3441 | 0.0328 |
| miR-194-3p | 1.3365 | 0.0083 |
| miR-15b-5p | 1.3145 | 0.0170 |
| miR-191b | 1.2964 | 0.0156 |
| miR-540-3p | 1.2545 | 0.0328 |
| let-7b-3p | 1.2488 | 0.0363 |
| miR-191a-5p | 1.2430 | 0.0180 |
| miR-6315 | 1.1330 | 0.0307 |
| miR-125b-5p | 1.1254 | 0.0038 |
| miR-125a-5p | 1.1246 | 0.0073 |
| miR-210-3p | 1.1245 | 0.0308 |
| miR-652-5p | 1.0856 | 0.0302 |
| miR-20b-5p | 1.0384 | 0.0139 |
| miR-144-5p | −6.6610 | 0.0130 |
| miR-539-3p | −5.9386 | 0.0043 |
| miR-708-5p | −5.4602 | 0.0468 |
| miR-412-5p | −5.1433 | 0.0053 |
| miR-187-5p | −4.9449 | 0.0021 |
| miR-3542 | −4.7610 | 0.0261 |
| miR-224-3p | −4.2339 | 0.0112 |
| miR-101a-5p | −4.2198 | 0.0133 |
| miR-200c-5p | −4.0638 | 0.0208 |
| miR-19b-2-5p | −3.4133 | 0.0476 |
| miR-384-5p | −3.2683 | 0.0244 |
| miR-135a-5p | −2.8032 | 0.0037 |
| miR-204-3p | −2.7952 | 0.0009 |
| miR-741-5p | −2.7242 | 0.0060 |
| let-7f-5p | −2.2251 | 0.0124 |
| miR-702-5p | −1.4132 | 0.0039 |
| miR-449c-3p | −1.2088 | 0.0303 |
| G16; N vs. A (42) | ||
| miR-409a-5p | 6.3869 | 0.0271 |
| miR-92a-2-5p | 4.8504 | 0.0128 |
| miR-484 | 4.7546 | 0.0084 |
| miR-673-3p | 4.5965 | 0.0110 |
| miR-125b-2-3p | 4.3396 | 0.0171 |
| miR-3542 | 4.2864 | 0.0007 |
| miR-127-5p | 4.2316 | 0.0246 |
| miR-6325 | 4.1901 | 0.0126 |
| miR-23b-5p | 4.1463 | 0.0001 |
| miR-3068-3p | 3.9677 | 0.0370 |
| miR-99a-5pa | 3.8542 | 0.0053 |
| miR-143-5p | 3.7453 | 0.0142 |
| miR-6318 | 3.7171 | 0.0420 |
| miR-9a-5p | 3.6453 | 0.0070 |
| miR-871-3p | 3.5678 | 0.0002 |
| miR-29b-1-5p | 3.5150 | 0.0137 |
| miR-219a-5p | 3.4518 | 0.0295 |
| miR-1912-5p | 3.2806 | 0.0171 |
| miR-187-5pa | 3.2137 | 0.0074 |
| miR-9a-3p | 3.1949 | 0.0292 |
| miR-296-5p | 2.8674 | 0.0025 |
| miR-483-3p | 2.6111 | 0.0454 |
| miR-500-3p | 2.5249 | 0.0157 |
| miR-881-3p | 2.4781 | 0.0438 |
| miR-665 | 2.4316 | 0.0108 |
| miR-3552 | 2.3654 | 0.0418 |
| miR-351-5p | 2.1768 | 0.0025 |
| miR-742-5p | 2.0011 | 0.0080 |
| miR-708-3p | 1.9967 | 0.0259 |
| miR-324-5p | 1.9943 | 0.0130 |
| miR-3573-3p | 1.9430 | 0.0298 |
| miR-29b-5p | 1.8790 | 0.0485 |
| miR-125a-5p | 1.8771 | 0.0059 |
| miR-485-5p | 1.7577 | 0.0154 |
| miR-129-2-3p | 1.6690 | 0.0001 |
| miR-191a-5p | 1.6207 | 0.0372 |
| miR-204-3p | 1.6186 | 0.0182 |
| let-7d-5p | 1.6180 | <0.0001 |
| miR-3084b-5p | 1.5984 | 0.0362 |
| miR-125b-5p | 1.2629 | 0.0247 |
| miR-702-5p | 1.2500 | 0.0419 |
| miR-30c-1-3p | 1.1605 | 0.0216 |
miRNAs marked with dashed lines are those with |log2(FC)| >4.25, and were selected for further analysis.
miR-99a-5p and miR-187-5p on GD16 had |log2(FC)| >4.25 on GD14 or GD15. miR/miRNA, microRNA; GD, gestational day; N, normal; A, anorectal malformation; lgFC, log2; FC, fold change.
Predicted and validated target genes of miRNAs
There were 18 differentially expressed miRNAs with |log2(FC)| >4.25 (miR-125b-2-3p, miR-144-5p, miR-187-5p, miR-190a-3p, miR-26b-3p, miR-331-3p, miR-3542, miR-409a-5p, miR-410-3p, miR-412-5p, miR-484, miR-539-3p, miR-673-3p, miR-708-5p, miR-741-3p, miR-92a-2-5p, miR-935 and miR-99a-5p), which were selected for further analysis. The numbers of predicted target genes for the above miRNAs were 229, 27, 104, 81, 177, 54, 219, 19, 57, 1, 112, 66, 17, 43, 143, 642, 148 and 5, respectively. Bone morphogenetic protein receptor type 2 (BMPR2) was the only experimentally validated target gene of miR-99a-5p in rats (18). The predicted and experimentally validated target genes of the 18 miRNAs were subjected to further analysis. The regulatory network between the miRNAs and their target genes is presented in Fig. 1D.
GO and KEGG enrichment analyses, and construction of the regulatory network
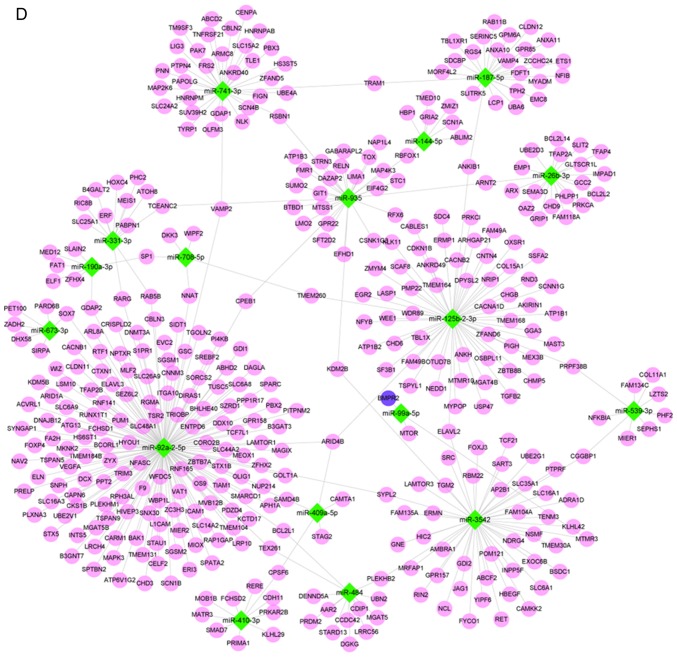
According to GO analysis of the target genes of the 18 miRNAs, the functions of DNA binding and gene regulation, metabolic and biosynthetic process of biomolecule, differentiation and projection of neuron were enriched. The top 15 enriched terms in the BP, CC and MF terms are presented in Fig. 2A.
Figure 2.
GO and KEGG enrichment analyses ranked by enriched factor, which was calculated by -log(q-value). The q-value is a modified Fisher exact P-value provided by Database for Annotation, Visualization and Integrated Discovery enrichment analysis. (A) GO enrichment results; the top 15 GO categories are listed in the diagram. (B) Top 30 enriched signaling pathways from KEGG analysis. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; CC, cellular component; MF, molecular function.
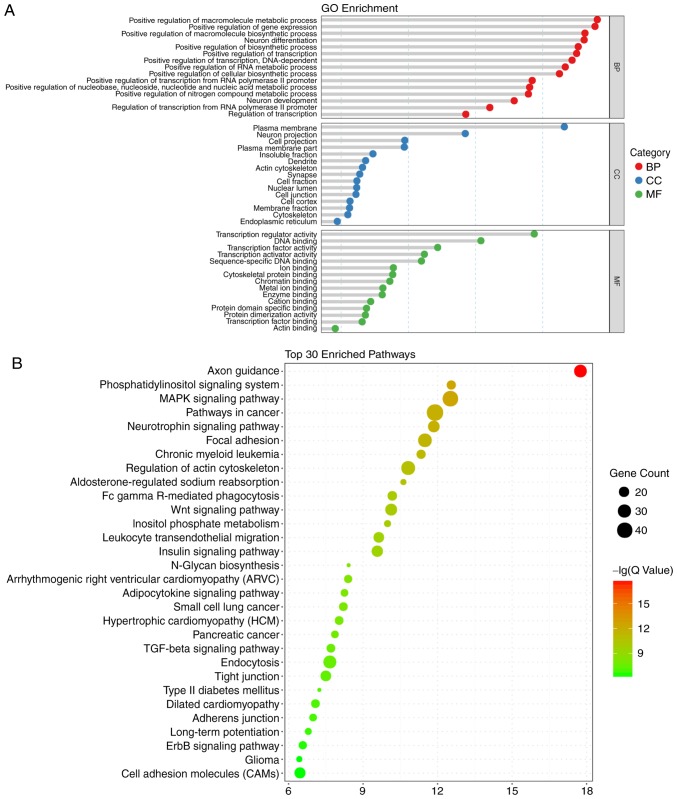
In terms of the KEGG analysis, a total of 49 signaling pathways were enriched, and the top 30 are listed in Fig. 2B. Among those pathways, 'axon guidance' was enriched the most, followed by 'phosphatidylinositol signaling system'. Notably, the enriched pathways included the Wingless Type MMTV integration site family (Wnt), mitogen-activated protein kinase (MAPK), and transforming growth factor-β (TGF-β) pathways, which are known to be closely associated with the process of embryonic development and ARM (6,19-21). Therefore, miRNA target gene-pathway networks were constructed for the miRNAs and target genes contributing to these three signaling pathways (Fig. 3A). A total of 15 core miRNAs associated with the signaling pathways of interest were screened out: miR-92a-2-5p, miR-125b-2-3p, miR-935, miR-190a-3p, miR-741-3p, miR-26b-3p and miR-539-3p were potentially associated with all three pathways; miR-484, miR-3542, miR-708-5p and miR-187-5p were potentially associated with two pathways; and miR-410-3p, miR-331-3p, miR-673-3p and miR-99a-5p were potentially associated with only one pathway. As demonstrated by the miRNA target gene-signaling pathway network, miR-92a-2-5p exhibited the most extensive connection with target genes in the relevant pathways.
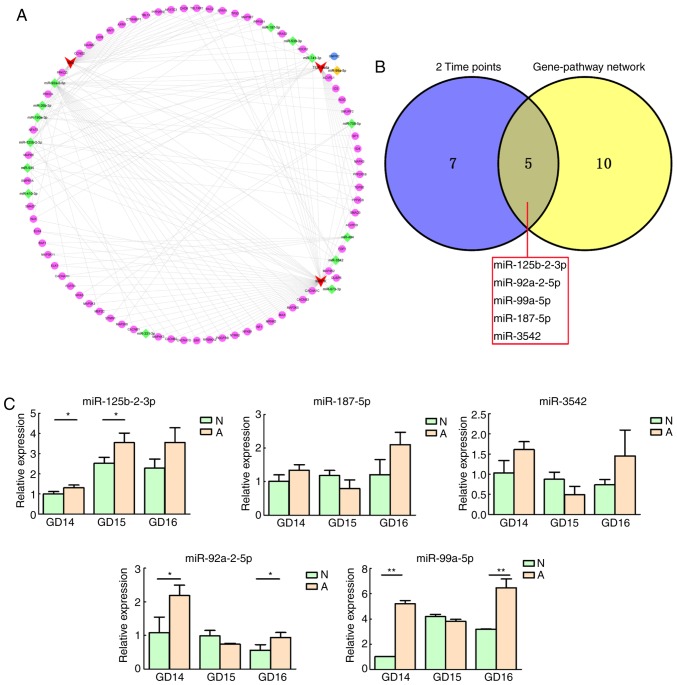
Figure 3.
miRNA-target gene-signaling pathway regulatory network and PCR validation. (A) miRNA-target gene-signaling pathway regulatory network. Diamonds and nodes represent miRNAs and target genes, among which the orange diamond and blue nodes represent an miRNA-mRNA pair validated by a dual luciferase experiment. Red polygons represent signaling pathways. Grey lines represent potential regulatory associations between miRNAs and their target genes. (B) Venn diagram of miRNAs differentially expressed at two time-points and included in the miRNA-target gene-signaling pathway regulatory network. Overlap between the two circles represent five miRNAs included in the network that are differentially expressed at two time-points and validated by PCR. (C) Relative expression levels of selected miRNAs in N and A groups. Relative miRNA expression is presented as the mean ± standard deviation. *P<0.05; **P<0.01. miR/miRNA, microRNA; PCR, polymerase chain reaction; N, normal; A, anorectal malformations; GD, gestational day.
Validation of important miRNAs
Among the 15 core differentially expressed miRNAs, five (miR-125b-2-3p, miR-92a-2-5p, miR-99a-5p, miR-187-5p and miR-3542) were differentially expressed at two time-points, and were selected for RT-qPCR validation (Fig. 3B). For all miRNAs, the relative expression levels were consistent with the microarray results (Fig. 3C). There were significant differences between the normal and ARM group for three miRNAs (miR-125-2-3p, miR-92a-2-5p and miR-99a-5p).
Discussion
The role of miRNAs in embryonic development and congenital deformities of gastrointestinal system have been described, however, the association between miRNAs and ARMs remains to be fully elucidated (8,10,11,22-24). Examining miRNA expression patterns may assist in identifying the pathogenesis of ARM. In the present study, miRNAs that may be involved in the course of ARM were profiled and bioinformatics analyses were performed to reveal the potential mechanism.
According to previous morphological investigations, rectourethal fistula and a common cloaca were present in all ARM rat fetuses, as a result of failed fusion of the urorectal septum and cloacal membrane at GD15 (4,13,25). Therefore, the present study detected aberrantly expressed miRNAs in ARM fetuses compared with normal rat fetuses at key time-points for anus formation (GD14, GD15 and GD16). The number of differentially expressed miRNAs at each time-point was 38, 49 and 42, respectively. Hierarchical analysis revealed that the increases of miRNAs in the ARM group were predominant, suggesting that increases in miRNAs may be one of the factors causing ARMs. In the normal group, there were differences in miRNA expression between the three time-points, which may be critical for embryonic development and organogenesis.
From the microarray results, 18 miRNAs with the highest significant FC values were selected for further bioinformatics analysis. By predicting target genes and searching for experimentally validated targets, an miRNA-target gene regulatory network was constructed. In a previous study by Jin et al (9), aberrantly expressed miRNAs in the terminal hindgut following the formation of ARM in Sprague-Dawley rat fetuses were profiled by the miRNA microarray, and miR-193 was screened out with |FC| >2. The present study focused on differentially expressed miRNAs prior to ARM having fully developed (GD14 and GD15), and the inclusion criteria of miRNAs selected for further bioinformatics analysis was |log2(FC)| >4.25; therefore, none of the miR-193 family members, including miR-193a-3p, miR-193a-5p, miR-193b-3p and miR-193b-5p, were included in the miRNAs selected for further analysis. Differences in the chip array and the samples used may also be factors in the discrepancy between these two studies.
Further functional analysis indicated that the target genes of the differentially expressed miRNAs were enriched in processes, including gene regulation and neuron development, and KEGG pathway analysis indicated that 49 significant signaling pathways were enriched, including the Wnt, MAPK and TGF-β signaling pathways, which have also previously been reported to be closely associated with ARM (6,19-21).
The neuron-associated terms that were enriched in the GO and KEGG functional analyses indicated that these miRNAs, were not only potentially associated with the development of ARMs, but may also be involved in neurode-velopmental abnormalities, which often appear concurrently with ARMs (26,27). In addition to the Wnt, MAPK and TGF-β pathways, the BMP signaling pathway is also important in ARMs (28-30). Wnt/β-catenin signal transduction dysregulation can lead to an ARMs phenotype (31). Studies have revealed that this may be caused by abnormal BMP signaling, as BMP4 and BMP7 were abnormally increased in the β-catenin-induced ARM phenotype, and furthermore, knockout of the BMP receptor, BMPR1A, partially rescued the malformation (32,33). However, in the previous study, only the BMRP1A receptor was knocked out, whereas the other BMP receptor, BMPR2, was not altered. Therefore, whether or not the knockout of BMPR2 and BMPR1A together is able to rescue the deformity to a greater degree, remains to be elucidated.
In the present study, the potential regulatory roles of the miRNAs and their corresponding target genes included in the enriched Wnt, MAPK and TGF-β signaling pathways were examined by constructing miRNA-target gene-pathway regulatory networks, which identified 15 core miRNAs indirectly associated with the these three signaling pathways. Among the 15 core miRNAs, five miRNAs (miR-125b-2-3p, miR-92a-2-5p, miR-99a-5p, miR-187-5p and miR-3542) were selected for RT-qPCR validation, as they were differentially expressed at two time-points in the microarray. The results revealed that the relative expression levels of miR-125b-2-3p, miR-92a-2-5p and miR-99a-5p were consistent with the microarray, with statistical significance.
miR-92 is a member of the miR-17-92 cluster, which can regulate fetal development at the early stages, and may promote proliferation and inhibit differentiation during embryogenesis (11,34). miR-125, which is a highly conserved miRNA family, can regulate early embryonic development by affecting cell fate and differentiation (35-37). Previously, GO analysis demonstrated that miR-125 controls axon guidance pathways, synaptic plasticity and catabolic processes (38). This GO and KEGG analysis was similar to the functional enrichment results in the present study, which suggests that miR-125 may be crucial in the pathogenesis of ARM. Regarding the miR-99a/100-125b cluster, different members can exert homogenous functions. RNAs of the miR-125 family can directly bind to the 3′-UTR of p53, B cell lymphoma-2 (Bcl-2), Bcl-2-like 12 and Mcl-1, and regulate cell proliferation and apoptosis (39,40). Similarly, miR-99 family members can reduce the expression of homeobox A1, which downregulates the expression of the downstream Bcl-2 gene, and leads to reduced cell survival and enhanced apoptosis (41). In hematopoietic stem and progenitor cells, miR-99a/100-125b can regulate homeostasis by shifting the balance between TGF-β and Wnt signaling (42); and in early chondrogenic differentiation in rats, miR-99a and miR-125b also have critical regulatory roles, with miR-99a being particularly important and negatively regulating chon-drogenic differentiation by directly targeting BMPR2 (18). These previous studies revealed that the dysregulation of apoptosis-associated genes, BMP family genes and homeobox genes are associated with ARM; however, the functions exerted by miRNAs in the disease remain to be fully elucidated, and further investigation is required.
In conclusion, the dynamic changes in differentially expressed miRNAs were investigated in normal and ARM rat fetuses at key time-points during anorectal development (GD14-16). The aberrant expression of miR-125b-2-3p, miR-92a-2-5p and miR-99a-5p during this period suggested that these miRNAs may be involved in ARM in rats. However, the result of this investigation into ARMs are preliminary, and further investigations are required to investigate the potential target genes which may contribute to ARM pathogenesis. In addition, normal and ARM fetuses were compared to screen miRNAs that may function in the course of this disease, however, the study did not evaluate miRNA expression in fetuses without the ARM phenotype following ETU treatment, which may assist in revealing the protective mechanisms that occur to avoid ARM following ETU stimulation. Further in vivo and in vitro investigations are also required to clarify the specific mechanisms responsible for the development of ARMs.
Acknowledgments
The authors would like to thank Professor Jie Liu (Ph.D., Science Experiment Center of China Medical University. Shenyang, China) and Mr. Liangcai Wu (M.M., Shanghai First Maternity and Infant Hospital, First Maternity and Infant Hospital Affiliated to Tongji University, Shanghai, China) for their assistance in discussions during the course of the investigations. Additionally, they are acknowledged for their critical and insightful comments on the manuscript received from reviewers.
Abbreviations
- miRNAs
microRNAs
- ARMs
anorectal malformations
- ETU
ethylenethiourea
- GD
gestational day
- FC
fold change
- 3′-UTRs
3′-untranslated regions
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genome
- Wnt
Wingless type MMTV integration site family
- MAPK
mitogen-activated protein kinase
- TGF-β
transforming growth factor-β
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- BMP
bone morphogenetic protein
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81470788 and 81770511), the Project of Key Laboratory of the Education Department of Liaoning Province (grant no. LS201601) and the Outstanding Scientific Fund of Shengjing Hospital (grant no. 201502).
Availability of data and materials
Data sharing is not applicable, as no datasets were generated or analyzed during the study.
Authors' contributions
YB and XT conceived and designed the experiments; CL performed the experiments; CL analyzed the data; ZY and WW contributed to data interpretation; CL wrote the manuscript.
Ethics approval and consent to participate
The present study was approved by the China Medical University Ethics Committee (no. 2015 PS213K).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cuschieri A, EUROCAT Working Group Descriptive epidemiology of isolated anal anomalies: A survey of 4.6 million births in Europe. Am J Med Genet. 2001;103:207–215. doi: 10.1002/ajmg.1532. [DOI] [PubMed] [Google Scholar]
- 2.de Blaauw I, Wijers CH, Schmiedeke E, Holland-Cunz S, Gamba P, Marcelis CL, Reutter H, Aminoff D, Schipper M, Schwarzer N, et al. First results of a European multi-center registry of patients with anorectal malformations. J Pediatr Surg. 2013;48:2530–2535. doi: 10.1016/j.jpedsurg.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 3.Wijers CH, van Rooij IA, Marcelis CL, Brunner HG, de Blaauw I, Roeleveld N. Genetic and nongenetic etiology of nonsyndromic anorectal malformations: A systematic review. Birth Defects Res C Embryo Today. 2014;102:382–400. doi: 10.1002/bdrc.21068. [DOI] [PubMed] [Google Scholar]
- 4.Bai Y, Chen H, Yuan ZW, Wang W. Normal and abnormal embryonic development of the anorectum in rats. J Pediatr Surg. 2004;39:587–590. doi: 10.1016/j.jpedsurg.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Grano C, Bucci S, Aminoff D, Lucidi F, Violani C. Quality of life in children and adolescents with anorectal malformation. Pediatr Surg Int. 2013;29:925–930. doi: 10.1007/s00383-013-3359-8. [DOI] [PubMed] [Google Scholar]
- 6.Draaken M, Prins W, Zeidler C, Hilger A, Mughal SS, Latus J, Boemers TM, Schmidt D, Schmiedeke E, Spychalski N, et al. Involvement of the WNT and FGF signaling pathways in non-isolated anorectal malformations: Sequencing analysis of WNT3A, WNT5A, WNT11, DACT1, FGF10, FGFR2 and the T gene. Int J Mol Med. 2012;30:1459–1464. doi: 10.3892/ijmm.2012.1124. [DOI] [PubMed] [Google Scholar]
- 7.Dong R, Shen Z, Zheng C, Chen G, Zheng S. Serum microRNA microarray analysis identifies miR-4429 and miR-4689 are potential diagnostic biomarkers for biliary atresia. Sci Rep. 2016;6:21084. doi: 10.1038/srep21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang G, Malmuthuge N, McFadden TB, Bao H, Griebel PJ, Stothard P, Guan le L. Potential regulatory role of microRNAs in the development of bovine gastrointestinal tract during early life. PLoS One. 2014;9:e92592. doi: 10.1371/journal.pone.0092592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin S, Wang J, Chen H, Xiang B. Differential miRNA expression analysis during late stage terminal hindgut development in fetal rats. J Pediatr Surg. 2017;52:1516–1519. doi: 10.1016/j.jpedsurg.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Wang S, Guo Z, Wu H, Jin X, Wang Y, Li X, Liang S. miRNA profiling reveals dysregulation of RET and RET-regulating pathways in Hirschsprung's disease. PLoS one. 2016;11:e0150222. doi: 10.1371/journal.pone.0150222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monzo M, Navarro A, Bandres E, Artells R, Moreno I, Gel B, Ibeas R, Moreno J, Martinez F, Diaz T, et al. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. 2008;18:823–833. doi: 10.1038/cr.2008.81. [DOI] [PubMed] [Google Scholar]
- 12.Macedo M, Martins JL, Meyer KF. Evaluation of an experimental model for anorectal anomalies induced by ethyl-enethiourea. Acta Cir Bras. 2007;22:130–136. doi: 10.1590/S0102-86502007000200010. [DOI] [PubMed] [Google Scholar]
- 13.Qi BQ, Beasley SW, Frizelle FA. Clarification of the processes that lead to anorectal malformations in the ETU-induced rat model of imperforate anus. J Pediatr Surg. 2002;37:1305–1312. doi: 10.1053/jpsu.2002.34996. [DOI] [PubMed] [Google Scholar]
- 14.Faria DJ, Simões Mde J, Martins JL. Is it possible folic acid reduce anorectal malformations ethylenethiourea induced in rats? Acta Cir Bras. 2015;30:517–522. doi: 10.1590/S0102-865020150080000001. [DOI] [PubMed] [Google Scholar]
- 15.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals . Guide for the Care and Use of Laboratory Animals. 8th edition. National Academies Press; Washington, DC: 2011. [Google Scholar]
- 16.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X, Wang J, Sun H, Qi Y, Xu W, Luo D, Jin X, Li C, Chen W, Lin Z, et al. MicroRNA-99a regulates early chondrogenic differentiation of rat mesenchymal stem cells by targeting the BMPR2 gene. Cell Tissue Res. 2016;366:143–153. doi: 10.1007/s00441-016-2416-8. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, Li L, Cheng W. Anorectal malformation: The etiological factors. Pediatr Surg Int. 2015;31:795–804. doi: 10.1007/s00383-015-3685-0. [DOI] [PubMed] [Google Scholar]
- 20.Wong EH, Ng CL, Lui VC, So MT, Cherny SS, Sham PC, Tam PK, Garcia-Barceló MM. Gene network analysis of candidate loci for human anorectal malformations. PLoS One. 2013;8:e69142. doi: 10.1371/journal.pone.0069142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura T, Tsuchiya K, Watanabe M. Crosstalk between Wnt and Notch signaling in intestinal epithelial cell fate decision. J Gastroenterol. 2007;42:705–710. doi: 10.1007/s00535-007-2087-z. [DOI] [PubMed] [Google Scholar]
- 22.Lei H, Tang J, Li H, Zhang H, Lu C, Chen H, Li W, Xia Y, Tang W. MiR-195 affects cell migration and cell proliferation by downregulating DIEXF in Hirschsprung's disease. BMC Gastroenterol. 2014;14:123. doi: 10.1186/1471-230X-14-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park C, Yan W, Ward SM, Hwang SJ, Wu Q, Hatton WJ, Park JK, Sanders KM, Ro S. MicroRNAs dynamically remodel gastrointestinal smooth muscle cells. PLoS One. 2011;6:e18628. doi: 10.1371/journal.pone.0018628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang W, Tang J, He J, Zhou Z, Qin Y, Qin J, Li B, Xu X, Geng Q, Jiang W, et al. SLIT2/ROBO1-miR-218-1-RET/PLAG1: A new disease pathway involved in Hirschsprung's disease. J Cell Mol Med. 2015;19:1197–1207. doi: 10.1111/jcmm.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumaru D, Murashima A, Fukushima J, Senda S, Matsushita S, Nakagata N, Miyajima M, Yamada G. Systematic stereoscopic analyses for cloacal development: The origin of anorectal malformations. Sci Rep. 2015;5:13943. doi: 10.1038/srep13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Jia H, Zhang H, Chen Q, Zhang T, Bai Y, Yuan Z. Abnormal innervation patterns in the anorectum of ETU-induced fetal rats with anorectal malformations. Neurosci Lett. 2011;495:88–92. doi: 10.1016/j.neulet.2011.02.057. [DOI] [PubMed] [Google Scholar]
- 27.Guan K, Li H, Fan Y, Wang Y, Yuan Z. Defective development of sensory neurons innervating the levator ani muscle in fetal rats with anorectal malformation. Birth Defects Res A Clin Mol Teratol. 2009;85:583–587. doi: 10.1002/bdra.20576. [DOI] [PubMed] [Google Scholar]
- 28.Mandhan P, Quan QB, Beasley S, Sullivan M. Sonic hedgehog, BMP4, and Hox genes in the development of anorectal malformations in Ethylenethiourea-exposed fetal rats. J Pediatr Surg. 2006;41:2041–2045. doi: 10.1016/j.jpedsurg.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Tang XB, Wang WL, Yuan ZW, Bai YZ. Spatiotemporal expression of BMP7 in the development of anorectal malformations in fetal rats. Int J Clin Exp Pathol. 2015;8:3727–3734. [PMC free article] [PubMed] [Google Scholar]
- 30.Pyati UJ, Cooper MS, Davidson AJ, Nechiporuk A, Kimelman D. Sustained Bmp signaling is essential for cloaca development in zebrafish. Development. 2006;133:2275–2284. doi: 10.1242/dev.02388. [DOI] [PubMed] [Google Scholar]
- 31.Ng RC, Matsumaru D, Ho AS, Garcia-Barceló MM, Yuan ZW, Smith D, Kodjabachian L, Tam PK, Yamada G, Lui VC. Dysregulation of Wnt inhibitory factor 1 (Wif1) expression resulted in aberrant Wnt-β-catenin signaling and cell death of the cloaca endoderm, and anorectal malformations. Cell Death Differ. 2014;21:978–989. doi: 10.1038/cdd.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyagawa S, Harada M, Matsumaru D, Tanaka K, Inoue C, Nakahara C, Haraguchi R, Matsushita S, Suzuki K, Nakagata N, et al. Disruption of the temporally regulated cloaca endodermal β-catenin signaling causes anorectal malformations. Cell Death Differ. 2014;21:990–997. doi: 10.1038/cdd.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens ML, Chaturvedi P, Rankin SA, Macdonald M, Jagannathan S, Yukawa M, Barski A, Zorn AM. Genomic integration of Wnt/β-catenin and BMP/Smad1 signaling coordinates foregut and hindgut transcriptional programs. Development. 2017;144:1283–1295. doi: 10.1242/dev.145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jevnaker AM, Khuu C, Kjøle E, Bryne M, Osmundsen H. Expression of members of the miRNA17-92 cluster during development and in carcinogenesis. J Cell Physiol. 2011;226:2257–2266. doi: 10.1002/jcp.22562. [DOI] [PubMed] [Google Scholar]
- 35.Ouchi Y, Yamamoto J, Iwamoto T. The heterochronic genes lin-28a and lin-28b play an essential and evolutionarily conserved role in early zebrafish development. PLoS One. 2014;9:e88086. doi: 10.1371/journal.pone.0088086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim KH, Seo YM, Kim EY, Lee SY, Kwon J, Ko JJ, Lee KA. The miR-125 family is an important regulator of the expression and maintenance of maternal effect genes during preimplantational embryo development. Open Biol. 2016;6:160181. doi: 10.1098/rsob.160181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ambros V. MicroRNAs and developmental timing. Cur Opin Genet Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malmevik J, Petri R, Klussendorf T, Knauff P, Åkerblom M, Johansson J, Soneji S, Jakobsson J. Identification of the miRNA targetome in hippocampal neurons using RIP-seq. Sci Rep. 2015;5:12609. doi: 10.1038/srep12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin H, Sun Y, Wang X, Park J, Zhang Y, Li M, Yin J, Liu Q, Wei M. Progress on the relationship between miR-125 family and tumorigenesis. Exp Cell Res. 2015;339:252–260. doi: 10.1016/j.yexcr.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 40.Tong Z, Liu N, Lin L, Guo X, Yang D, Zhang Q. miR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomed Pharmacother. 2015;75:129–136. doi: 10.1016/j.biopha.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 41.Chen D, Chen Z, Jin Y, Dragas D, Zhang L, Adjei BS, Wang A, Dai Y, Zhou X. MicroRNA-99 family members suppress Homeobox A1 expression in epithelial cells. PLoS One. 2013;8:e80625. doi: 10.1371/journal.pone.0080625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emmrich S, Rasche M, Schöning J, Reimer C, Keihani S, Maroz A, Xie Y, Li Z, Schambach A, Reinhardt D, Klusmann JH. miR-99a/100-125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFbeta and Wnt signaling. Genes Dev. 2014;28:858–874. doi: 10.1101/gad.233791.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable, as no datasets were generated or analyzed during the study.