Abstract
Dental follicle stem/progenitor cells have the potential to undergo osteogenesis. naked cuticle homolog 2 (Nkd2) is a signal-inducible feedback antagonist of the canonical Wnt signaling pathway. The purpose of the present study was to investigate the function of Nkd2 in the differentiation of dental follicle stem/progenitor cells (DFSCs) into osteoblasts. Immunohistochemistry, reverse transcription-quantitative polymerase chain reaction and western blotting were employed to detect Nkd2 expression in rat DFSCs. In addition, rat DFSCs (rDFSCs) were transfected with small interfering RNAs to examine the effect of Nkd2 on the differentiation of these cells into osteoblasts. Furthermore, the function of Nkd2 in the Wnt/β-catenin pathway in rDFSCs was investigated using β-catenin/T-cell factor luciferase activity assays and western blotting. It was revealed that the expression of Nkd2 was upregulated during the differentiation of rDFSCs into osteoblasts. Furthermore, osteoblast differentiation ability and Wnt/β-catenin pathway activity were significantly decreased in Nkd2-silenced rDFSCs compared with the si-NC group (P<0.05 and P<0.001, respectively). The results suggest that Nkd2 promotes the differentiation of rDFSCs into osteoblasts through Wnt/β-catenin signaling.
Keywords: naked cuticle homolog 2, dental follicle stem/progenitor cells, differentiation, osteoblasts, Wnt/β-catenin
Introduction
The dental follicle (DF) is a loose connective tissue sac surrounding the enamel organ and dental papilla of a developing tooth germ prior to eruption. This tissue is believed to contain DF stem/progenitor cells (DFSCs) that are able to give rise to cementoblasts, periodontal ligament cells and osteoblasts (1). In vitro, DFSCs have the potential to differentiate into osteoblasts (2). The Wnt/β-catenin signaling pathway is critical for normal bone and tooth formation and development (3). Multiple pathways, including bone morphogenetic protein (BMP), Notch, insulin-like growth factor and Wnt signaling, are involved in the differentiation of DFSCs into osteoblasts (3). Previous studies have revealed that the expression of Wnts has spatial and temporal specificity during tooth development (4-6) and that the Wnt/β-catenin pathway participates in the differentiation of DFSCs into osteoblasts (7-11). The enhanced and reduced activity of the Wnt/β-catenin pathway may result in abnormalities in tooth development (12-14).
Drosophila naked cuticle (Nkd), a Drosophila segment polarity gene, encodes an inducible antagonist for the Wnt signaling molecule Wingless (Wg) (15) and Nkd functions as a signal-inducible feedback antagonist of Wg in fly embryos (16). Nkd homolog 2 (Nkd2) is one of the mammalian homologs of Nkd, and has been demonstrated to negatively regulate the canonical Wnt signaling pathway by interacting with Dishevelled (Dvl) (17,18). In 293T cells, myristoylated Nkd2 interacts with Dvl-1 at the plasma membrane, which results in the ubiquitin-mediated proteasomal degradation of the two proteins, thus attenuating Wnt signaling (19). Nkd2 also suppresses canonical Wnt/β-catenin signaling during multiple stages of early development in zebrafish (20). An additional role for Nkd2 is to escort transforming growth factor-α (TGF-α)-containing vesicles to the basolateral surface of polarized epithelial cells (21). Nkd2 may regulate Wnt and epidermal growth factor receptor signaling via its function in TGF-α delivery and Dvl stabilization (22). Nevertheless, the functions and mechanisms of Nkd2 during the differentiation of rat DFSCs (rDFSCs) into osteoblasts remain largely unknown.
In the present study, it was hypothesized that Nkd2 regulates the Wnt/β-catenin pathway during the differentiation of rDFSCs into osteoblasts. Therefore, the function of Nkd2 during rDFSC differentiation into osteoblasts was examined, and its function in Wnt/β-catenin signaling was assessed. It was demonstrated that Nkd2 is able to promote the differentiation of rDFSCs into osteoblasts through the Wnt/β-catenin pathway, results that may provide novel insight into the molecular mechanism of regeneration of tooth root and alveolar bone.
Materials and methods
Immunohistochemistry
All animal procedures were ethically approved by the Ethics Committee of the Guanghua College of Stomatology, Sun Yat-sen University [Guangzhou, China; approval no. ERC-(2014)-21]. Sprague-Dawley rats (Laboratory Animal Center, Sun Yat-sen University) were housed in a specific pathogen-free laboratory animal center at a temperature of 20-26°C, 35 pa higher than the standard atmosphere, light-dark cycle 12/12 h, and fed using standard rodent chow (ad libitum access to food and water). A total of 70 rats (1, 3, 5, 7, 9, 11 and 13 days old; 10 rats for each group, nonsexual selection with 38 female mice and 32 male mice, weight 5-20 g) were obtained from the Laboratory Animal Center and euthanized by cervical dislocation immediately.
Mandibles containing the first molars were dissected, fixed in 4% paraformaldehyde at 4°C for 24 h, decalcified in methanoic acid for 3-7 days and embedded in paraffin. Furthermore, serial 5-mm sections were then cut, collected on 3-aminopropyltriethoxysilane-coated slices and air dried. Mesiodistal sections of mandibular tissues were deparaffinized with two washes in dimethyl benzene for 10 min succeeded by rehydration with serial dilutions (95, 90, 80 and 70%) of ethanol for 5 min respectively. Sections were heated in a water bath (10 min at 95°C) in citrate buffer for antigen retrieval. Following incubation with blocking buffer consisting of 1% bovine serum albumin (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at room temperature for 1 h, the slides were incubated overnight at 4°C with the primary antibody Nkd2 (1:100; cat. no. sc163145; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or with PBS as a negative control. Subsequent to washing using PBS, the slides were incubated with the horseradish peroxidase (HRP)-conjugated secondary antibody rabbit anti-goat IgG (1:200; cat. no. A21030; Abbkine Scientific Co., Ltd., Wuhan, China) at room temperature for 1 h and images were obtained using an electron optical microscope (magnification, x200; Zeiss AG, Oberkochen, Germany) as previously described (23). Subsequent to image acquisition, image analysis was performed using ImageJ 1.48 version (National Institutes of Health, Bethesda, MD, USA), as previously described (24). Quantification of the immunoscore of all images was blinded for histopathological diagnosis. A specific brown color was selected as the standard for all positive image scores as previously described (24). The mean gray value of the negative group was used to set the threshold adjustment, and the mean gray value of each image was obtained. The results were evaluated using SPSS software version 16.0 (SPSS, Inc., Chicago, IL, USA) with a paired Student's t-test (each group was compared with the negative group), and the data are expressed as the mean ± standard deviation, with P<0.05 considered to indicate a statistically significant difference.
Cell culture and induction of osteoblast differentiation
DFs were gently isolated from the first mandibular molars of Sprague-Dawley rats (5 to 7 days old) and digested with 0.1% collagenase type I (Collagen-I) and 10 U/ml dispase (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 30 min at 37°C to obtain rDFSCs, as previously described (7). To ensure a uniform cell population, the rDFSCs were cultured in α-modified Eagle's medium (α-MEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin for 3 to 4 passages in a 37°C incubator with 5% CO2. rDFSCs at passages 3-5 were used for all experiments.
rDFSCs were cultured until sub-confluent (>80%) in standard cell culture medium prior to the induction of cementoblast/osteoblast differentiation, and then stimulated with osteoblast induction medium containing 10 mmol β-glycerophosphate (Sigma-Aldrich; Merck KGaA), 0.2 mmol ascorbic acid (Sigma-Aldrich; Merck KGaA) and 100 nmol dexamethasone (Sigma-Aldrich; Merck KGaA). Differentiation of rDFSCs was determined by Alizarin Red staining and the expression of osteoblast markers.
Immunofluorescence
rDFSCs were fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 20 min. The cells were blocked with 1% bovine serum albumin at 4°C for 1 h and then incubated overnight at 4°C with an anti-rat Nkd2 antibody (1:100; cat. no. sc163145; Santa Cruz Biotechnology, Inc.). Fluorescein isothiocyanate-conjugated immunoglobulin G secondary antibody (1:1,000; cat. no. E031230; EarthOx, LLC, Millbrae, CA, USA) was added to the cells at 37°C for 2 h as the secondary antibody prior to nuclear staining with 10 µg/ml DAPI at 37°C for 2 min. The cells were finally washed with PBS 3 times and examined under a confocal laser-scanning microscope (magnification, ×400; Zeiss AG).
Small interfering RNA (siRNA) transfection
rDFSCs (passages 3-5) were seeded in 6-well culture plates with a density of 105/ml and 200 ml cell suspension per well, and cultured until sub-confluent (>80%) prior to transfection with 50 nmol/l siRNA targeting the rat Nkd2 gene (si-Nkd2) using Lipofectamine RNAiMax (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 8 h according to the manufacturer's protocol. Cells transfected with non-targeting siRNA (si-NC) or cells incubated without siRNA (mock transfection) were used as negative controls. The oligonucleotides si-Nkd2 (forward, 5′ GCACUCCAGUGUGAUGUCUdTdT 3' and reverse, 3′ dTdTCGUGAGGUCACACUACAGA 5′) and si-NC were synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
Cell Counting Kit-8 (CCK-8) assay for proliferation
DFSCs were seeded in 96-well culture plates at a density of 4×103/ml, 200 µl cell suspension per well and transfected with si-Nkd2 or si-NC for 24, 48 or 72 h. Mock-transfected cells were used as negative controls. CCK-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) reagent was added to each well at 10% of the volume, and the samples were incubated at 37°C in the dark for 2 h. Optical density was measured at 450 nm using a microplate reader (Tecan Group, Ltd., Mannedorf, Switzerland).
Cell cycle distribution and apoptosis analyses
DFSCs transfected with si-Nkd2 or si-NC for 72 h were used for cell cycle distribution and apoptosis analyses. For the cell cycle distribution assay, 1×106 treated cells were fixed in 70% pre-chilled ethanol overnight at -20°C. Subsequently, the cells were labeled with Forevergen Cell Cycle kits (Forevergen; Guangzhou Yongnuo Biotechnology Co., Ltd., Guangzhou, China) according to the manufacturer's protocol. The Annexin V/propidium iodide apoptosis assay was performed using the PE Annexin V Apoptosis Detection kit I (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer's protocols. All samples were analyzed using an Accuri C6 Flow Cytometer and CFlow Plus 1.0.264.15 Software (BD Biosciences).
Cell migration assay
si-Nkd2- or si-NC-transfected rDFSCs were trypsinized with 0.25% Trypsin-EDTA solution (cat. no. 25200-056; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 1 min and resuspended in serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.) at a density of 3×105/ml, and 200 ml of the cell suspension was seeded in the upper chamber of a Costar Transwell System (Corning Incorporated, Corning, NY, USA). The lower chamber was filled with 600 ml α-MEM containing 10% FBS. The cells were allowed to migrate through 8-mm pores for 16 h at 37°C and the cells in the upper chamber were removed with a cotton swab. The cells present beneath the membrane were stained with 10 µg/ml DAPI at 37°C for 2 min. Fluorescence microscopy was performed with a electron fluorescence microscope (magnification, x100; Axiovert; Zeiss AG, Oberkochen, Germany). Ten fields per sample were randomly selected for the quantitative analysis of cell migration by counting cell nuclei.
Alkaline phosphatase activity (ALP) assay
si-Nkd2- or si-NC-transfected rDFSCs seeded at a density of 2×104/ml and 100 µl cell suspension per well in 96-well culture plates were grown in conditioned medium for 7 days. The plates were washed twice, and 55 µl 1% Triton X-100 was added to each well. Subsequent to incubation overnight at 4°C, 30 µl cell lysate per well was subjected to an ALP activity assay using an ALP kit (Nanjing Jiancheng Bioengingeering Institute, Nanjing, China). To normalize enzyme activity, the protein concentration was measured with a BCA protein assay kit (Boster Biological Technology, Pleasanton, CA, USA).
Alizarin Red staining
si-Nkd2- or si-NC-transfected rDFSCs were seeded at a density of 2×104/ml and 100 µl cell suspension per well in 96-well culture plates. Subsequent to osteogenic induction for 7 days, si-Nkd2- or si-NC-transfected rDFSCs were fixed with 4% paraformaldehyde at 4°C for 24 h and stained with 1% Alizarin Red S (Sigma-Aldrich; Merck KGaA) at 37°C for 15 min. Following the addition of 10% cetylpyri-dine, the cells were incubated at 37°C for 60 min. The extent of mineralization was detected at 562 nm using a microplate reader (Tecan Group, Ltd.).
Western blotting
Control and induced (0, 1, 2, 3 and 4 weeks), and si-Nkd2- or si-NC-transfected cells (≥106 cells per group) were harvested with radioimmunoprecipitation assay (RIPA) lysis buffer at 0°C for 30 min (10 mmol/l Tris-HCl, 1 mmol/l EDTA, 1% sodium dodecyl sulfate, 1% Nonidet P-40, 1:1,000 proteinase inhibitor cocktail, 50 mmol/l β-glycerophosphate and 50 mmol/l sodium fluoride) to obtain total proteins. Nuclear proteins were extracted using a CelLytic™ NuCLEAR™ Extraction kit (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocol. Proteins were quantified using a BCA Protein Assay kit (cat. no. P0012S; Beyotime Institute of Biotechnology, Shanghai, China), and 30 µg protein per lane was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto 0.22-mm nitrocellulose membranes (EMD Millipore, Billerica, MA, USA). The nitrocellulose membranes were blocked using 1% bovine serum albumin for 20 min at 37°C. Subsequent to washing with tris-buffered saline (TBS; 10 mM Tris, 154 mM NaCl, pH=7.5 with 0.1% Tween-20) for 5 min at room temperature 3 times, the membranes were incubated overnight at 4°C with goat anti-rat Nkd2 (1:100; cat. no. sc163145; Santa Cruz Biotechnology, Inc.), rabbit anti-rat β-catenin (1:1,000; cat. no. ab6302; Abcam, Cambridge, MA, USA), rabbit anti-rat runt-related transcription factor 2 (Runx2; 1:1,000; cat. no. ab23981; Abcam), rabbit anti-rat osteocalcin (OCN; 1:200; cat. no. AB10911; EMD Millipore), rabbit anti-rat β-actin (1:1,000; cat. no. G3202; Wuhan Saiwei Biotechnology Co., Ltd., Wuhan, China), rabbit anti-rat Cyclin D1 (1:1,000; cat. no. #2978; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit anti-rat c-Myc (1:1,000; cat. no. #5605; Cell Signaling Technology, Inc.), mouse anti-rat GAPDH (1:1,000; cat. no. #5174; Cell Signaling Technology, Inc.) and rabbit anti-rat histone H3 (1:500; cat. no. ab8580; Abcam) antibodies. The membranes were washed with TBS for 5 min at room temperature 3 times. Proteins were detected by incubated with HRP-conjugated secondary antibody rabbit anti-goat immunoglobulin G (IgG; 1:3,000; cat. no. A21030; Abbkine Scientific Co., Ltd.), goat anti-rabbit IgG (1:3,000; cat. no. A0208; Beyotime Institute of Biotechnology) and goat anti-mouse IgG (1:3,000; cat. no. A0216; Beyotime Institute of Biotechnology) at 37°C for 2 h and according to the enhanced chemiluminescence method (EMD Millipore). Densitometric analysis of the protein bands was performed using ImageJ software 1.48 version (National Institutes of Health, Bethesda, MD, USA).
Simple western size assay
Cells (si-Nkd2- or si-NC-transfected, and control or treated by Wnt3a) at a density of ≥106 per group were harvested with RIPA lysis buffer (10 mmol/l Tris-HCl, 1 mmol/l EDTA, 1% sodium dodecyl sulfate, 1% Nonidet P-40 and 1:1,000 proteinase inhibitor cocktail) for 30 min at 0°C for protein extraction and western blot analysis. Cellular extracts were prepared with Lysis kit-RIPA buffer (ProteinSimple, San Jose, CA, USA). Wes Antibody Diluent2 included in the respective Wes Master kit was used as the blocking buffer and used undiluted. Cellular extracts were blocked with the Wes Antibody Diluent2 for 30 min at room temperature. Immunoreactive proteins were detected using Wes-Rabbit (12-230 kDa) Master kit (cat. no. PS-MK01; ProteinSimple) and Wes-Mouse (12-230 kDa) Master kit (cat. no. PS-MK02; ProteinSimple) by the Wes™ Simple Western System (ProteinSimple) according to the manufacturer's protocol. Rabbit anti-rat Runx2 (1:20; cat. no. ab23981; Abcam), Collagen-I (1:10; cat. no. ab90395; Abcam), ALP (1:10; cat. no. ab83259; Abcam) and β-catenin (1:50; cat. no. ab23981; Abcam) antibodies were used as primary antibodies for detecting Runx2, Collagen-I and ALP, respectively. Cellular extracts were incubated with the primary antibodies for 30 min at room temperature. As the loading control, β-tubulin was detected using a mouse anti-rat β-tubulin monoclonal antibody (1:50; cat. no. AF7011; Affinity Biosciences, Cincinnati, OH, USA). The secondary antibodies were included in the respective Wes Master kit and used undiluted. Cellular extracts were incubated with the secondary antibodies for 30 min at room temperature. Quantitation of the detected proteins and image preparation were performed with Compass Software 2.5.8.0 (ProteinSimple). Parameters: Sample separation time, 25 min; separation voltage, 375 V.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The total RNA of control and induced (0, 1, 2, 3 and 4 weeks), and si-Nkd2- or si-NC-transfected cells was isolated with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. mRNA quantification was performed using an ABI 7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). Complementary DNA (cDNA) was synthesized with a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) in a total volume of 20 µl. RT was performed at 37°C for 1 h followed by incubation for 10 min at 70°C to inactivate the reverse transcriptase, following which 5 µl of the reaction mixture was incubated with FastStart Universal SYBR Green Master (Roche Diagnostics, Basel, Switzerland) in a total volume of 10 µl. The primers used for qPCR are listed in Table I. All reactions were performed with a hot-start preincubation step of 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C, 60 sec at 60°C and a final 5 min step at 60°C. The amount of template was quantified using the comparative cycle quantification cycle method according to the ABI 7300 Real-Time PCR System manufacturer's protocol. Relative gene expression was calculated with GAPDH as an internal control using the formula 2−ΔΔCq (25).
Table I.
Reverse transcription-quantitative polymerase chain reaction primer sequences.
| Gene | Primer sequences |
|---|---|
| Runt related transcription factor 2 | Forward: 5′-TAGAGGGGATGCCTTAGTG |
| Reverse: 5′-GAGGATGGAGGGAAACAA | |
| Osteocalcin | Forward: 5′-AGCAGGAGGGCAGTAAGG |
| Reverse: 5′-TCCAGGGGATCTGGGTAG | |
| β-catenin | Forward: 5′-TGGTGAAAATGCTTGGGTCG |
| Reverse: 5′-TCTGAAGGCAGTCTGTCGTAATAG | |
| Naked cuticle homolog 2 | Forward: 5′-ATCTGCCGCTGTGTGAGTGT |
| Reverse: 5′-CGTTTGTTTCCCATCTCTTTAGGT | |
| β-actin | Forward: 5′-TGCTATGTTGCCCTAGACTTCG |
| Reverse: 5′-GTTGGCATAGAGGTCTTTACGG |
Luciferase assay
Activation of the Wnt/β-catenin signaling pathway was measured using the activity of the transcription factor T-cell factor (TCF)/lymphoid enhancer factor (LEF), a direct downstream modulator of Wnt signaling, as previously described (26). The luciferase assay was performed following the RT protocol described by the manufacturer of TCF/LEF Reporter Assay kit (Qiagen GmbH, Hilden, Germany), which contains a TCF/LEF luciferase reporter construct in addition to a negative control (Cignal negative control; non-inducible firefly luciferase reporter construct) and a positive control (Cignal positive control; constitutively expressed firefly lucif-erase construct). The pGL4 Luciferase Reporter Vectors were used as the plasmid vectors as described by the manufacturer of TCF/LEF Reporter Assay kit. The reporter was a mixture of inducible transcription factor responsive construct and constitutively expressing Renilla luciferase construct (40:1). The inducible transcription factor-responsive construct encodes the firefly luciferase reporter gene under the control of a basal promoter element (TATA box) joined to tandem repeats of a specific Transcriptional Response Element. The negative control was a mixture of non-inducible reporter construct and constitutively expressing Renilla luciferase construct (40:1). The noninducible reporter construct encodes firefly luciferase under the control of a basal promoter element (TATA box), without any additional transcriptional response elements. The positive control was a constitutively expressing GFP construct, pre-mixed with a constitutively expressing firefly luciferase construct, and a constitutively expressing Renilla luciferase construct (40:1:1).
For co-transfection with si-Nkd2, rDFSCs were seeded at a density of 4×105 cells/ml and cultured in α-MEM containing 5% FBS without antibiotics. The constructs were diluted in 25 ml Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.), mixed with Lipofectamine RNAiMax reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and incubated at room temperature for 20 min to allow formation of the transfection complex. Subsequently, the mixture was added to cells in 96-well culture plates. After 24 h of transfection at RT, signals were assessed using Dual-Luciferase® Reporter Assay System (Promega Corporation, Madison, WI, USA). Luciferase activity in control (si-NC-transfected) cells was used for calibration.
Rescue experiment
si-Nkd2-transfected rDFSCs were seeded in 6-well culture plates at a density of 105/ml and 200 ml cell suspension per well. si-Nkd2-transfected rDFSCs were treated with 100 ng/ml Wnt3a or mock-treated and osteoblast induction medium at 37°C for 7 days. rDFSCs cultured in osteoblast induction medium at 37°C for 7 days served as the control. A simple western size assay was performed to assess the levels of ALP, Runx2 and β-catenin proteins.
Statistical analysis
All experiments were performed at least 3 times. Unpaired Student's t-test was used to compare the means of two groups. One-way analysis of variance was applied to compare more than two means and Bonferroni's test was performed to compare the means between the groups. The Kruskal-Wallis test was used to assess the variance of heterogeneity. All results were evaluated using SPSS software version 16.0 (SPSS, Inc., Chicago, IL, USA). The data were expressed as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significance difference.
Results
Expression of Nkd2 in rDFs
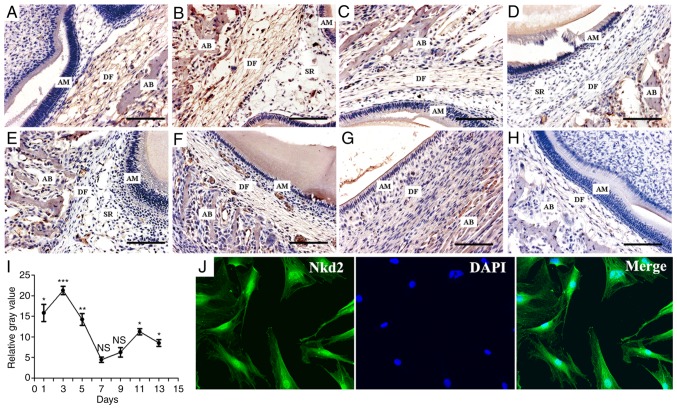
Nkd2 staining was positive on postpartum day 1 in the DF of newborn rats (Fig. 1A). Subsequently, Nkd2 expression peaked on day 3 (Fig. 1B) and became weaker on day 5 (Fig. 1C). Nkd2 staining was not detected on days 7 and 9 (Fig. 1D and E) but was detected again on days 11 (Fig. 1F) and 13 (Fig. 1G). The expression of Nkd2 in the DF at postpartum days 1, 3, 5, 11 and 13 were significantly higher compared with the respective days in the negative control group (Fig. 1H) (P<0.05, P<0.001, P<0.01, P<0.05 and P<0.05, respectively. Fig. 1I). However, at postpartum days 7 and 9, the difference was not significant (P>0.05). In addition, primary rDFSCs were isolated from the first mandibular molars of Sprague-Dawley rats, and the localization of Nkd2 in the cytomembrane, cytoplasm and nucleus of rDFSCs was observed by immunofluorescence and confocal laser-scanning microscopy (Fig. 1J).
Figure 1.
Immunohistochemistry results of Nkd2 expression in rDFs. (A) Nkd2 staining was positive in DFs on postpartum day 1. (B) Peak staining appeared on day 3. (C) Nkd2 staining was weaker on day 5. (D) Nkd2 staining was absent in DFs on day 7. (E) Nkd2 staining remained absent on day 9. (F) Positive staining was detected again on day 11. (G) Positive staining was detected on day 13. (H) Control sections revealed no specific immunoreaction. Scale bar, 100 µm. (I) Expression trend of Nkd2 in DFs. Statistical analysis was performed with a Student's t-test; the results are expressed as the mean ± standard deviation. *P<0.05, **P<0.01 and ***P<0.001 vs. the negative group (values at week 0). (J) Immunofluorescence using a confocal laser-scanning microscope revealed that rDFs at passage 3 were positive for Nkd2 (magnification, ×400). Nkd2, naked cuticle homolog 2; rDFs, rat dental follicles; AM, adamantoblast; AB, alveolar bone; SR, stellate reticulum; NS, not significant.
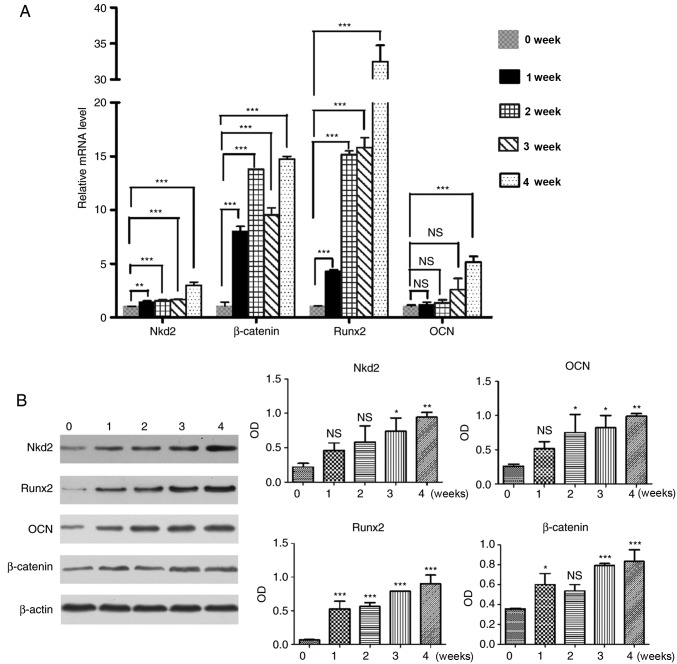
Nkd2 expression is upregulated in rDFSCs following osteogenic induction
Subsequent to culturing for 4 weeks in osteoblast induction medium, RT-qPCR (Fig. 2A) and western blotting (Fig. 2B) revealed that Nkd2 expression was upregulated in rDFSCs following osteogenic induction. This result was consistent with the observed expression of β-catenin, Runx2 and OCN. The expression of Nkd2 mRNA was significantly increased at week 1, 2, 3 and 4 compared with the control group (P<0.01, P<0.001, P<0.001 and P<0.001, respectively), which was the similar to the mRNA expression of β-catenin (P<0.001, P<0.001, P<0.001 and P<0.001, respectively), Runx2 (P<0.001, P<0.001, P<0.001 and P<0.001, respectively) and OCN (P>0.05, P>0.05, P>0.05 and P<0.001, respectively) compared with the control group. The expression of Nkd2 protein was also significantly upregulated on week 3 and 4 compared with the control group (P<0.05 and P<0.01) similar to the protein levels of β-catenin (P<0.001 and P<0.001), Runx2 (P<0.001 and P<0.001) and OCN (P<0.05 and P<0.01).
Figure 2.
Reverse transcription-quantitative polymerase chain reaction and western blotting were performed in rDFSCs following 1, 2, 3 and 4 weeks of culturing in osteogenic induction medium to detect the expression of Nkd2, β-catenin, Runx2 and OCN. rDFSCs harvested prior to the addition of osteogenic induction medium (at 80% confluence) served as controls. (A) Expression of Nkd2 mRNA was upregulated in rDFSCs following culturing in osteogenic induction medium, consistent with the expression of β-catenin, Runx2 and OCN. (B) Expression of Nkd2 protein was upregulated in rDFSCs following culturing in osteogenic induction medium, consistent with expression of β-catenin, Runx2 and OCN. *P<0.05, **P<0.01 and ***P<0.001 vs. values at week 0. rDFSC, rat dental follicle stem/progenitor cell; Nkd2, naked cuticle homolog 2; Runx2, runt-related transcription factor 2; OCN, osteocalcin; OD, optical density; NS, not significant.
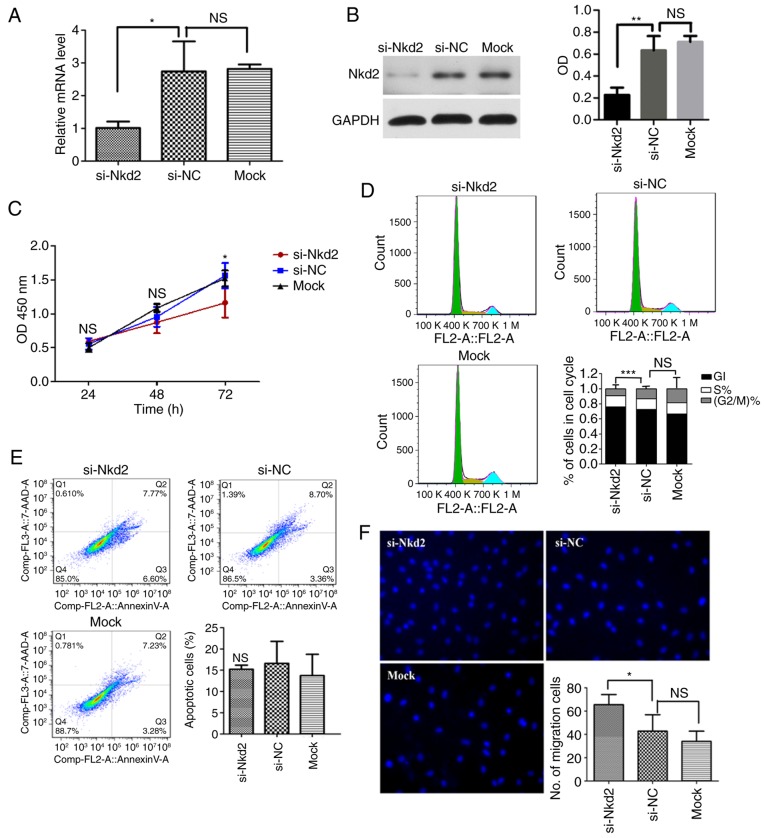
Involvement of Nkd2 in rDFSC proliferation and migration abilities
RT-qPCR and western blotting were employed to investigate the efficiency of si-Nkd2 transfection in rDFSCs. The mRNA and protein levels of Nkd2 were significantly reduced compared with the si-NC transfected group (P<0.05 and P<0.01, respectively; Fig. 3A and B).
Figure 3.
Effect of si-Nkd2 transfection on rDFSCs. (A) Efficiency of si-Nkd2 transfection in rDFSCs determined by reverse transcription-quantitative polymerase chain reaction. A total of 48 h after si-Nkd2 transfection, the levels of Nkd2 mRNA in the si-Nkd2 group were significantly lower compared with that in the si-NC group, but there was no difference between the si-NC and Mock groups. (B) Efficiency of si-Nkd2 transfection in rDFSCs determined by western blotting. A total of 72 h after si-Nkd2 transfection, the Nkd2 protein levels in the si-Nkd2 group were significantly lower compared with that in the si-NC group, but there was no difference between the si-NC and Mock groups. (C) Cell Counting Kit-8 assay results revealed that the proliferation of si-Nkd2-transfected rDFSCs was significantly lower compared with that of si-NC-transfected rDFSCs at 72 h after transient transfection. (D) A total of 72 h after transient transfection, cells were arrested in the G1 phase, and their rate of entry into G2 phase was diminished. (E) Silencing of Nkd2 had no significant effect on apoptosis rates in rDFSCs. (F) Transwell assay results demonstrated significantly increased migration in Nkd2-silenced rDFSCs. *P<0.05 and **P<0.01 with comparisons shown by lines. si-, small interfering RNA; Nkd2, naked cuticle homolog 2; NC, negative control; NS, not significant; OD, optical density; rDFSC, rat dental follicle stem/progenitor cell.
Silencing of Nkd2 expression significantly decreased the proliferation of rDFSCs after 72 h of transfection compared with the si-NC group (P<0.05; Fig. 3C). To assess whether si-Nkd2-transfected rDFSCs exhibited decreased proliferation or whether these cells underwent apoptosis, the cell cycle distribution and apoptosis rate were investigated. It was revealed that Nkd2 silencing arrested cells in the G1 phase and significantly reduced their rate of entry into the G2 phase compared with the si-NC group (P<0.001; Fig. 3D). Apoptosis analysis revealed that fewer cells underwent apoptosis due to si-Nkd2 transfection compared with the si-NC group, however this difference was not significant (P>0.05; Fig. 3E). Therefore, si-Nkd2 inhibited cell cycle progression instead of affecting survival. Furthermore, a Transwell assay revealed the significantly increased migration of rDFSCs silenced for Nkd2 compared with the si-NC group (P<0.05; Fig. 3F).
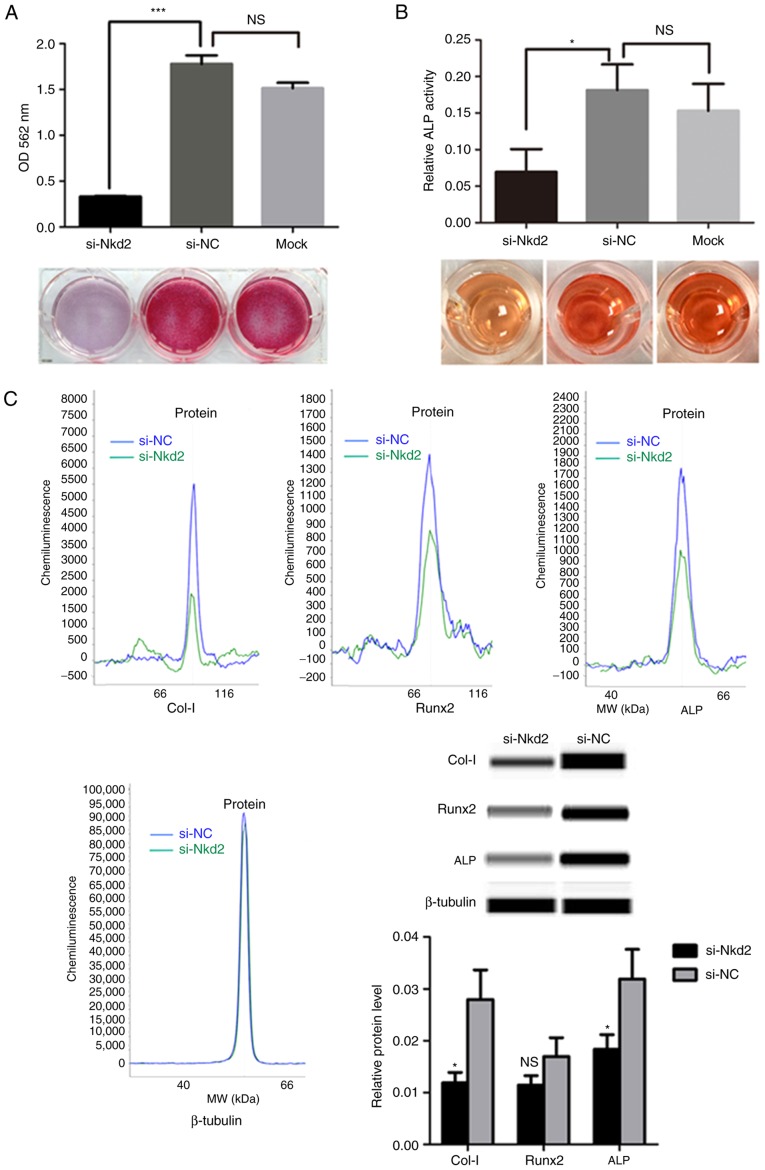
siRNA-mediated depletion of nkd2 reduces differentiation of rDFSCs to osteoblasts
Subsequent to culturing in osteoblast induction medium for 7 days, Alizarin Red staining demonstrated a significant reduction in the formation of mineralized nodules for Nkd2-silenced rDFSCs compared with the si-NC group (P<0.001; Fig. 4A). Furthermore, ALP activity was significantly lower in si-Nkd2-transfected cells compared with in si-NC- and Mock-transfected cells (P<0.05; Fig. 4B). As an indication of osteoblast differentiation, the expression of osteoblast-associated markers were examined using a simple western size assay. Collagen-I and ALP protein levels were significantly lower in si-Nkd2-transfected rDFSCs compared with in si-NC-transfected rDFSCs subsequent to culturing in osteoblast induction medium for 7 days (P<0.05). Runx2 expression was also reduced in Nkd2-silenced rDFSCs, albeit in a non-significant manner (P>0.05; Fig. 4C).
Figure 4.
Osteogenic differentiation ability of Nkd2-silenced rDFSCs. (A) Alizarin Red staining revealed the reduced formation of mineralized nodules by Nkd2-silenced rDFSCs following culturing in osteogenic induction medium for 7 days. (B) ALP activity was blocked in Nkd2-silenced rDFSCs. (C) A simple western size assay revealed that Col-I and ALP protein levels were significantly lower in si-Nkd2-transfected rDFSCs compared with in si-NC-transfected rDFSCs subsequent to culturing in osteogenic induction medium for 7 days. *P<0.05 and ***P<0.001 with comparisons shown by lines or vs. si-NC. OD, optical density; si-, small interfering RNA; Nkd2, naked cuticle homolog 2; NC, negative control; NS, not significant; ALP, alkaline phosphatase; Col-I, collagen type I; Runx2, runt-related transcription factor 2; rDFSC, rat dental follicle stem/progenitor cell.
Effect of Nkd2 on Wnt/β-catenin signaling in rDFSCs
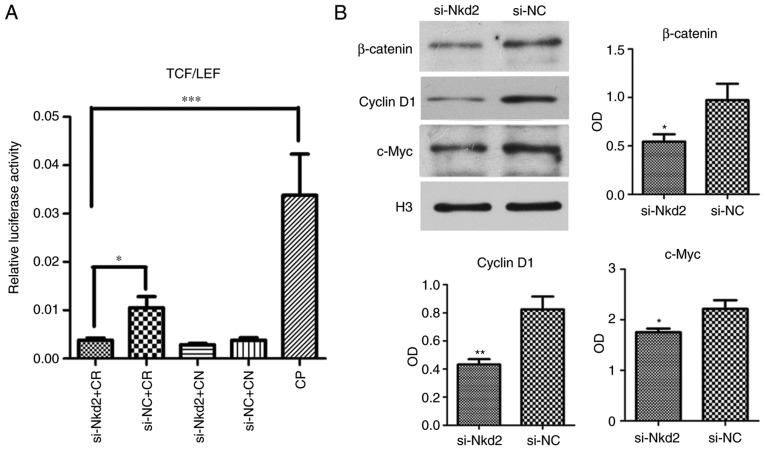
A luciferase activity assay at 72 h after si-Nkd2 transfection revealed that TCF/LEF activity was significantly reduced in Nkd2-silenced rDFSCs with Cignal reporter compared with the si-NC transfected group with the Cignal reporter (P<0.05) or compared with the Cignal positive control (P<0.001; Fig. 5A). Furthermore, western blotting of the nuclear proteins revealed that the expression of β-catenin, cyclin D1 and c-Myc was significantly lower in si-Nkd2-transfected rDFSCs compared with in si-NC-transfected rDFSCs (P<0.05, P<0.01 and P<0.05, respectively; Fig. 5B).
Figure 5.
si-Nkd2 transfection reduced activity of the Wnt/β-catenin pathway. (A) Luciferase activity at 72 h after transfection was significantly lower in si-Nkd2-transfected rDFSCs compared with in si-NC-transfected rDFSC. (B) Western blotting revealed that the expression of β-catenin, cyclinD1 and c-Myc was signficantly lower in si-Nkd2-transfected rDFSCs compared with in si-NC-transfected rDFCs. *P<0.05, **P<0.01 or ***P<0.001 with comparisons shown by lines or vs. the si-NC group. TCF, LEF, si-, Nkd2, CR, Cignal reporter; CN, Cignal negative control; NC, negative control; CP, Cignal positive control; rDFSC, rat dental follicle stem/progenitor cell.
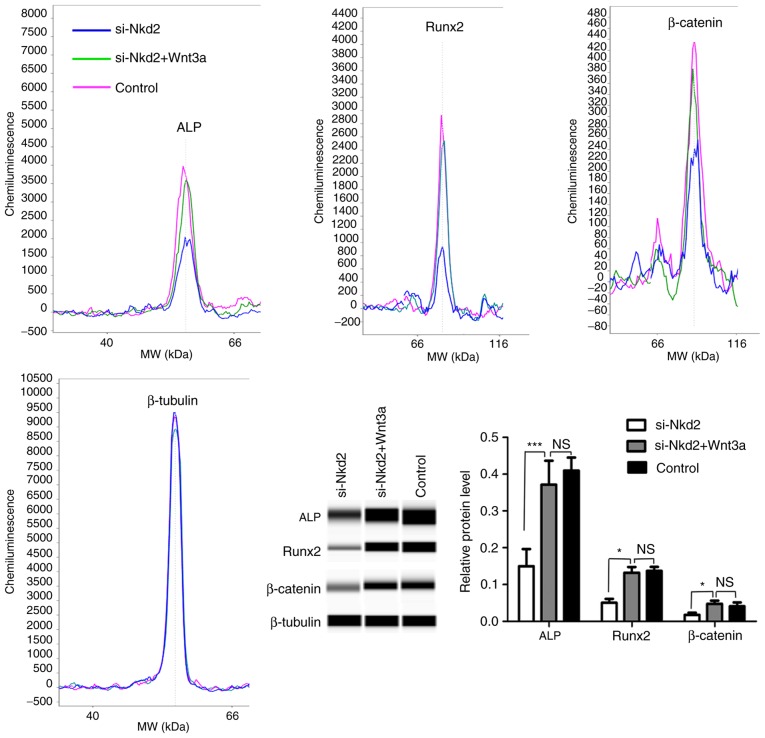
Next, si-Nkd2-transfected rDFSCs were treated with Wnt3a or mock-treated and cultured in osteoblast induction medium for 7 days. According to the results of the simple western size assay, the protein levels of ALP, Runx2 and β-catenin were significantly increased in Wnt3a-treated si-Nkd2-transfected rDFSCs compared with si-Nkd2-transfected rDFSCs (P<0.001, P<0.05 and P<0.05, respectively; Fig. 6).
Figure 6.
A simple western size assay demonstrating the levels of ALP, Runx2 and β-catenin proteins in Wnt3a- or mock-treated si-Nkd2-transfected rDFSCs. si-Nkd2-transfected rDFSCs were treated with Wnt3a or mock-treated and cultured in osteoblast induction medium for 7 days. Cells were harvested, and the protein levels of ALP, Runx2 and β-catenin proteins were analyzed by a simple western size assay. Significantly lower levels of ALP, Runx2 and β-catenin proteins were observed in si-Nkd2-transfected rDFSCs, and higher levels of proteins were observed in Wnt3a-treated cells compared with the control. rDFSCs cultured in osteoblast induction medium served as the control. *P<0.05 and ***P<0.001 with comparisons shown by lines. NS, not significant; si-, small interfering RNA; Nkd2, naked cuticle homolog 2; ALP, alkaline phosphatase; Runx2, runt-related transcription factor 2; rDFSC, rat dental follicle stem/progenitor cell.
Discussion
During tooth development, DFSCs give rise to the cementoblasts of the tooth and osteoblasts of the alveolar bone (27). Previously, rDFSCs with multipotential differentiation abilities were produced using tissue explants combined with enzymatic digestion (7). Nkd2 is an inhibitor of the Wnt/β-catenin pathway and is transcriptionally regulated by β-catenin. This signal-inducible feedback antagonist serves an important role during embryogenesis; in addition, Nkd2 is associated with the suppressive effects of Wnt proteins on development (16) and is differentially expressed in different tissues (28).
Using in situ hybridization, Wang et al (5) identified similar expression patterns for Wnt/β-catenin signaling components, including Wnt ligands (Wnt3 and Wnt5A), receptors [Frizzled class receptor (FZD)4, FZD6 and LDL receptor related protein (LRP)5], transducers (β-catenin), transcription factors (transcription factor 4 and lymphoid enhancer binding factor 1) and antagonists (Dickkopf WNT signaling pathway inhibitor 1 and sclerostin domain containing 1) in the development of human and mouse tooth germ. These results suggest an essential role of Wnt/β-catenin signaling in the regulation of mammal tooth development. β-catenin is a key component of the canonical Wnt pathway, and in a previous study, the expression of this protein was absent in dental follicles on days 1 and 3 in vivo, but progressively increased from days 5 to 13 (7). In the present study, the expression of Nkd2 was high at an early stage, unlike β-catenin, but was similar at later stages, suggesting its involvement in the development of teeth.
Studies have revealed that Wnt/β-catenin signaling may be involved in the osteogenic differentiation of DFSCs. In mice, the Wnt/β-catenin pathway regulates the bone morphogenetic protein 2-mediated differentiation of DFSCs (9), and the protein and mRNA levels of ALP, Runx2 and osterix are increased by Wnt3a treatment (11). Hertwig's epithelial root sheath cells regulate osteogenic differentiation of DFSCs through the Wnt pathway (10). Additionally, a previous study revealed that LiCl treatment upregulated the levels of total β-catenin, nuclear β-catenin, OCN, Runx2 and Collagen-I and suggested that Wnt/β-catenin signaling pathway positively regulates cement-oblast/osteoblast differentiation of DFCs (7). Runx2 and OCN are important markers of osteoblast differentiation (29-31). Runx2 is an important transcription factor that mediates osteogenesis. As the expression of Runx2 in the mesenchyme occurs prior to formation of bone, it is known as an early regulator of osteoblast differentiation. Gaur et al (32) revealed that the Wnt/β-catenin pathway promotes osteogenesis by directly stimulating Runx2 gene expression. In the present study, Nkd2 expression was upregulated in rDFSCs following osteogenic induction in vitro, which was consistent with the expression of β-catenin, Runx2 and OCN. Further analyses demonstrated that fewer mineralized nodules formed in Nkd2-silenced rDFSCs compared with si-NC transfected cells. Overall, the lower ALP activity (P<0.05) of si-NKd2-transfected cells combined with the significantly lower protein expression of osteoblast-associated markers Collagen-I and ALP (P<0.05 and P<0.05, respectively) in si-Nkd2-transfected rDFSCs indicated that the silencing of Nkd2 inhibited the differentiation of rDFSCs into osteoblasts. Runx2 expression was also reduced in Nkd2-silenced rDFSCs, albeit in a non-significant manner. Thus, it was hypothesised that Nkd2 is involved in the differentiation of rDFSCs into osteoblasts through Wnt/β-catenin signaling.
In the Wnt/β-catenin pathway, following the canonical Wnt ligand binding to the LRP5/LRP6 complex, the Dvl protein is recruited to the membrane, which results in the disassembly of the 'Wnt destruction complex', which includes Axin, APC, WNT signaling pathway regulator, casein kinase 1 α 1 and glycogen synthase kinase 3 β (GSK-3β). Inactivation of GSK-3β is the central step of canonical Wnt signaling, allowing β-catenin stabilization and translocation to the nucleus (33). Nkd2 has been reported to be expressed in the cytomembrane of epithelial cells; myristoylated Nkd2 was demonstrated to interact with Dvl-1, resulting in their mutual ubiquitin-mediated proteasomal degradation, which antagonizes Wnt signaling (17,19). However, in the present study, Nkd2 was observed in the cytomembrane, cytoplasm and nucleus of rDFSCs, suggesting that the function of Nkd2 in rDFSCs may be different from that in epithelial cells. Further investigation demonstrated that the activity of TCF/LEF and the expression of β-catenin, cyclin D1 and c-Myc were decreased in Nkd2-silenced rDFSCs, which validated the hypothesis that Nkd2 sustains the Wnt/β-catenin pathway in rDFSCs, despite being reported as a Wnt inhibitor (17,19). These characteristics of Nkd2 are similar to those of APC downregulated 1 (APCDD1), which is a membrane-bound glycoprotein. In hair follicle cells, APCDD1 inhibits the canonical Wnt/β-catenin pathway, and its inactivation is associated with an autosomal dominant form of hair loss (26). However, during the osteogenic differentiation of DFSCs, APCDD1 sustains the expression and activation of β-catenin (26).
The expression levels of Cyclin D1, a protein required for progression through the G1 phase of the cell cycle (34), were decreased in the nuclei of Nkd2-silenced rDFSCs in the present study. Cyclin D1 dimerizes with cyclin-dependent kinase 4/6 to regulate G1/S-phase transition and entry into S phase (35). In the present study, Nkd2-silenced rDFSCs were arrested in the G1 phase, and few cells underwent apoptosis. Therefore, Nkd2 silencing inhibited cell cycle progression as opposed to affecting cell survival. The effect of Nkd2 on cell migration remains controversial. Adamowicz et al (36) utilized a DNA microarray analysis to reveal the high expression of Nkd2 in subregions of malignant peripheral nerve sheath tumor types, suggesting that Nkd2 is able to reduce the adhesion of tumor cells, resulting in tumor metastasis. Other studies have reported that Nkd2 suppresses tumor growth and metastasis in gastric (37) and breast cancer (38), and osteosarcoma (39). In the present study, the increased migratory ability of si-Nkd2 rDFSCs was observed, which may have been caused by a reduction in β-catenin levels and cell adhesion.
The Wnt3a ligand has been recognized as a canonical Wnt activator. ALP is another marker of osteoblast differentiation (40). The present study detected the protein expression of ALP, Runx2 and β-catenin in Nkd2-silenced rDFSCs treated with Wnt3a or mock-treated and revealed the higher levels of these proteins in Nkd2-silenced rDFSCs treated with Wnt3a, which is able to activate the Wnt/β-catenin pathway. This result suggested that the repression of rDFSC differentiation into osteoblasts by si-Nkd2 transfection may be rescued by Wnt3a; thus, Nkd2 regulates the differentiation of DFSCs into osteoblasts, potentially via Wnt/β-catenin signaling.
It was concluded that Nkd2 serves a role in DFSCs during osteoblast differentiation; it promotes the differentiation of DFSCs into osteoblasts, potentially through the activation of Wnt/β-catenin signaling. However, further studies are required in order to reveal the mechanism of Wnt/β-catenin signaling activation by Nkd2 in DFSCs.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81170932 and 81700967), the Natural Science Foundation of Guangdong Province (grant nos. 2017A030310595 and 2015A030313048) and the Science and Technology Research Foundation of Shenzhen (grant no. JCYJ20170303155435885).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
CC and JL conceived and designed the study. CC, JZ, YD and YH performed the experiments. CC and JZ wrote the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
All animal procedures were approved by the Ethics Committee of the Guanghua College of Stomatology, Sun Yat-sen University [Guangzhou, China; approval no. ERC-(2014)-21].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Felthaus O, Gosau M, Ettl T, Prantl L, Morsczeck C. Migration of human dental follicle cells in vitro. J Periodontal Res. 2014;49:205–212. doi: 10.1111/jre.12096. [DOI] [PubMed] [Google Scholar]
- 2.Pan K, Sun Q, Zhang J, Ge S, Li S, Zhao Y, Yang P. Multilineage differentiation of dental follicle cells and the roles of Runx2 over-expression in enhancing osteoblast/cement-oblast-related gene expression in dental follicle cells. Cell Prolif. 2010;43:219–228. doi: 10.1111/j.1365-2184.2010.00670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morsczeck C, Schmalz G. Transcriptomes and proteomes of dental follicle cells. J Dent Res. 2010;89:445–456. doi: 10.1177/0022034510366899. [DOI] [PubMed] [Google Scholar]
- 4.Liu F, Millar SE. Wnt/β-catenin signaling in oral tissue development and disease. J Dent Res. 2010;89:318–330. doi: 10.1177/0022034510363373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang B, Li H, Liu Y, Lin X, Lin Y, Wang Y, Hu X, Zhang Y. Expression patterns of WNT/β-CATENIN signaling molecules during human tooth development. J Mol Histol. 2014;45:487–496. doi: 10.1007/s10735-014-9572-5. [DOI] [PubMed] [Google Scholar]
- 6.Xiang L, Chen M, He L, Cai B, Du Y, Zhang X, Zhou C, Wang C, Mao JJ, Ling J. Wnt5a regulates postnatal dental follicle stem/progenitor cells. Stem Cell Res Ther. 2014;5:135. doi: 10.1186/scrt525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y, Ling J, Wei X, Ning Y, Xie N, Gu H, Yang F. Wnt/β-catenin signaling participates in cementoblast/osteoblast differentiation of dental follicle cells. Connect Tissue Res. 2012;53:390–397. doi: 10.3109/03008207.2012.668980. [DOI] [PubMed] [Google Scholar]
- 8.Nemoto E, Koshikawa Y, Kanaya S, Tsuchiya M, Tamura M, Somerman MJ, Shimauchi H. Wnt signaling inhibits cement-oblast differentiation and promotes proliferation. Bone. 2009;44:805–812. doi: 10.1016/j.bone.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 9.Silvério KG, Davidson KC, James RG, Adams AM, Foster BL, Nociti FH, Jr, Somerman MJ, Moon RT. Wnt/β-catenin pathway regulates bone morphogenetic protein (BMP2)-mediated differentiation of dental follicle cells. J Periodontal Res. 2012;47:309–319. doi: 10.1111/j.1600-0765.2011.01433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Ge Y, Chen G, Yan Z, Yu M, Feng L, Jiang Z, Guo W, Tian W. Hertwig's epithelial root sheath cells regulate osteogenic differentiation of dental follicle cells through the Wnt pathway. Bone. 2014;63:158–165. doi: 10.1016/j.bone.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Nemoto E, Sakisaka Y, Tsuchiya M, Tamura M, Nakamura T, Kanaya S, Shimonishi M, Shimauchi H. Wnt3a signaling induces murine dental follicle cells to differentiate into cementoblastic/osteoblastic cells via an osterix-dependent pathway. J Periodontal Res. 2016;51:164–174. doi: 10.1111/jre.12294. [DOI] [PubMed] [Google Scholar]
- 12.Bae CH, Lee JY, Kim TH, Baek JA, Lee JC, Yang X, Taketo MM, Jiang R, Cho ES. Excessive Wnt/β-catenin signaling disturbs tooth-root formation. J Periodontal Res. 2013;48:405–410. doi: 10.1111/jre.12018. [DOI] [PubMed] [Google Scholar]
- 13.Kim TH, Lee JY, Baek JA, Lee JC, Yang X, Taketo MM, Jiang R, Cho ES. Constitutive stabilization of β-catenin in the dental mesenchyme leads to excessive dentin and cementum formation. Biochem Biophys Res Commun. 2011;412:549–555. doi: 10.1016/j.bbrc.2011.07.116. [DOI] [PubMed] [Google Scholar]
- 14.Zhang R, Yang G, Wu X, Xie J, Yang X, Li T. Disruption of Wnt/β-catenin signaling in odontoblasts and cementoblasts arrests tooth root development in postnatal mouse teeth. Int J Biol Sci. 2013;9:228–236. doi: 10.7150/ijbs.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez Arias A, Baker NE, Ingham PW. Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development. 1988;103:157–170. doi: 10.1242/dev.103.1.157. [DOI] [PubMed] [Google Scholar]
- 16.Zeng W, Wharton KA, Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. Naked cuticle encodes an inducible antagonist of Wnt signalling. Nature. 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- 17.Rousset R, Mack JA, Wharton KA, Jr, Axelrod JD, Cadigan KM, Fish MP, Nusse R, Scott MP. Naked cuticle targets dishevelled to antagonize Wnt signal. Genes Dev. 2001;15:658–671. doi: 10.1101/gad.869201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan D, Wallingford JB, Sun TQ, Nelson AM, Sakanaka C, Reinhard C, Harland RM, Fantl WJ, Williams LT. Cell autonomous regulation of multiple Dishevelled-dependent pathways by mammalian Nkd. Proc Natl Acad Sci USA. 2001;98:3802–3807. doi: 10.1073/pnas.071041898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu T, Li C, Cao Z, Van Raay TJ, Smith JG, Willert K, Solnica-Krezel L, Coffey RJ. Myristoylated Naked2 antago-nizes Wnt-beta-catenin activity by degrading Dishevelled-1 at the plasma membrane. J Biol Chem. 2010;285:13561–13568. doi: 10.1074/jbc.M109.075945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Raay TJ, Coffey RJ, Solnica-Krezel L. Zebrafish Naked1 and Naked2 antagonize both canonical and non-canonical Wnt signaling. Dev Biol. 2007;309:151–168. doi: 10.1016/j.ydbio.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Franklin JL, Graves-Deal R, Jerome WG, Cao Z, Coffey RJ. Myristoylated Naked2 escorts transforming growth factor alpha to the basolateral plasma membrane of polarized epithelial cells. Proc Natl Acad Sci USA. 2004;101:5571–5576. doi: 10.1073/pnas.0401294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu T, Li C. Convergence between Wnt-β-catenin and EGFR signaling in cancer. Mol Cancer. 2010;9:236. doi: 10.1186/1476-4598-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Wei X, Ling J, Xie N. Expression of matrilin-2 and -4 in human dental pulps during dentin-pulp complex wound healing. J Endod. 2011;37:642–649. doi: 10.1016/j.joen.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Mane DR, Kale AD, Belaldavar C. Validation of immunoexpression of tenascin-C in oral precancerous and cancerous tissues using ImageJ analysis with novel immunohistochemistry profiler plugin: An immunohistochemical quantitative analysis. J Oral Maxillofac Pathol. 2017;21:211–217. doi: 10.4103/jomfp.JOMFP_234_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC T method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Viale-Bouroncle S, Klingelhöffer C, Ettl T, Morsczeck C. The WNT inhibitor APCDD1 sustains the expression of β-Catenin during the osteogenic differentiation of human dental follicle cells. Biochem Biophys Res Commun. 2015;457:314–317. doi: 10.1016/j.bbrc.2014.12.107. [DOI] [PubMed] [Google Scholar]
- 27.Morsczeck C, Götz W, Schierholz J, Zeilhofer F, Kühn U, Möhl C, Sippel C, Hoffmann KH. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005;24:155–165. doi: 10.1016/j.matbio.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, Cagatay T, Amanai M, Zhang M, Kline J, Castrillon DH, Ashfaq R, Oz OK, Wharton KA., Jr Viable mice with compound mutations in the Wnt/Dvl pathway antagonists nkd1 and nkd2. Mol Cell Biol. 2007;27:4454–4464. doi: 10.1128/MCB.00133-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitagawa M, Tahara H, Kitagawa S, Oka H, Kudo Y, Sato S, Ogawa I, Miyaichi M, Takata T. Characterization of established cementoblast-like cell lines from human cementumlining cells in vitro and in vivo. Bone. 2006;39:1035–1042. doi: 10.1016/j.bone.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Lengner CJ, Drissi H, Choi JY, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Activation of the bone-related Runx2/Cbfa1 promoter in mesenchymal condensations and developing chondrocytes of the axial skeleton. Mech Dev. 2002;114:167–170. doi: 10.1016/S0925-4773(02)00050-3. [DOI] [PubMed] [Google Scholar]
- 31.An J, Yang H, Zhang Q, Liu C, Zhao J, Zhang L, Chen B. Natural products for treatment of osteoporosis: The effects and mechanisms on promoting osteoblast-mediated bone formation. Life Sci. 2016;147:46–58. doi: 10.1016/j.lfs.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 32.Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 33.Libro R, Bramanti P, Mazzon E. The role of the Wnt canonical signaling in neurodegenerative diseases. Life Sci. 2016;158:78–88. doi: 10.1016/j.lfs.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 34.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 35.Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299:35–55. doi: 10.1016/S0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 36.Adamowicz M, Radlwimmer B, Rieker RJ, Mertens D, Schwarzbach M, Schraml P, Benner A, Lichter P, Mechtersheimer G, Joos S. Frequent amplifications and abundant expression of TRIO, NKD2, and IRX2 in soft tissue sarcomas. Genes Chromosomes Cancer. 2006;45:829–838. doi: 10.1002/gcc.20343. [DOI] [PubMed] [Google Scholar]
- 37.Jia Y, Cao B, Yang Y, Linghu E, Zhan Q, Lu Y, Yu Y, Herman JG, Guo M. Silencing NKD2 by promoter region hypermethylation promotes gastric cancer invasion and metastasis by up-regulating SOX18 in human gastric cancer. Oncotarget. 2015;6:33470–33485. doi: 10.18632/oncotarget.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong Y, Cao B, Zhang M, Han W, Herman JG, Fuks F, Zhao Y, Guo M. Epigenetic silencing of NKD2, a major component of Wnt signaling, promotes breast cancer growth. Oncotarget. 2015;6:22126–22138. doi: 10.18632/oncotarget.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao S, Kurenbekova L, Gao Y, Roos A, Creighton CJ, Rao P, Hicks J, Man TK, Lau C, Brown AM, et al. NKD2, a negative regulator of Wnt signaling, suppresses tumor growth and metastasis in osteosarcoma. Oncogene. 2015;34:5069–5079. doi: 10.1038/onc.2014.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manzano-Moreno FJ, Ramos-Torrecillas J, Melguizo-Rodríguez L, Illescas-Montes R, Ruiz C, García-Martínez O. Bisphosphonate modulation of the gene expression of different markers involved in osteoblast physiology: Possible implications in bisphosphonate-related osteonecrosis of the Jaw. Int J Med Sci. 2018;15:359–367. doi: 10.7150/ijms.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.