Abstract
In a hepatitis B virus (HBV)-infected population, persistently low expression levels of serum HBV serum antigen (HBsAg) are present, particularly in chronic asymptomatic HBV carriers (ASCs). The present study sequenced the HBV Pre-S gene, and aimed to elucidate its features in ASCs with low HBsAg expression compared with in the established HBV Pre-S reference gene sequences from ASCs with high HBsAg expression. A total of 1,308 ASCs were grouped according to HBsAg serum levels (cut-off value, 10 IU/ml), and clinical characteristics were analyzed in detail. The HBV Pre-S gene was sequenced in 276 ASCs with low-level HBsAg; in addition, 100 of the remaining 1,032 ASCs with high-level HBsAg were randomly selected for HBV Pre-S gene sequencing on the basis of age matching with the low-level HBsAg group. Comparative analysis of the gene sequences from these groups was subsequently conducted. The major clinical features of the population with low-level HBsAg were as follows: Most were ASCs with chronic HBV infection; 97.1% were HBsAg/anti-HBe/anti-HBc-positive; 82.54% carried the B genotype; and 84.13% displayed the adw serotype. The results indicated that there were novel and meaningful mutations, including co-mutations, at numerous loci and sites in the Pre-S gene, as well as deletion mutations in the Pre-S2 gene. These mutations in the Pre-S1 and Pre-S2 gene frag ments accounted for 65.38% (68/104) of the 104 B genotype cases in the low-level HBsAg group and 90.91% (20/22) of the 22 C genotype cases in the low-level HBsAg group, respectively. In conclusion, Pre-S gene mutations may be associated with HBV replication defects, which may be the cause of the observed low expression levels of HBsAg.
Keywords: HBV, asymptomatic HBV carriers, low-level HBsAg, Pre-S gene, reference sequences, co-mutations
Introduction
Hepatitis B virus (HBV) infection is a global health issue, and China has a high incidence of HBV infection (1). HBV belongs to the Hepadnaviridae virus family, and its genome consists of partially double-stranded circular DNA, which is ~3.2 kb in length. HBV surface proteins include three components: Pre-S1, Pre-S2 and S, which are encoded by the Pre-S1, Pre-S2 and S open reading frames of the genome, respectively. The Pre-S1 and Pre-S2 proteins serve important roles in the viral invasion of hepatocytes, and in viral infection, viral assembly, viral replication and stimulation of immune responses in the body. The Pre-S1 protein fragment containing amino acid residues 21–47 is the most important region of the protein with regards to mediating attachment of HBV to hepatocytes. Pre-S2 contains the polymerized human serum albumin (PHSA) receptor; HBV can enter hepatocytes by pinocytosis mediated by PHSA (2,3).
It has previously been reported that Pre-S/S gene variations result in immune evasion and occult HBV infection (4,5), and serological detection of patients with Pre-S/S gene variations indicate the presence of both HBV surface antigen (HBsAg) and HBV surface antibody, or HBsAg negativity. Previous reports have also revealed that HBsAg expression is affected by high spontaneous error rates of HBV reverse transcriptase, with viral variants continuously arising during the infection course that are subject to endogenous and exogenous selection pressure (6,7). Pre-S (Pre-S1 and Pre-S2) gene mutations can affect HBsAg expression (3,8). However, with regards to the natural course of HBV infection in patients, few studies have analyzed the association between the HBV-infected population characterized by persistently low expression of serum HBsAg and low HBV DNA replication [the majority of these patients are chronic asymptomatic HBV carriers (ASCs)] and Pre-S gene mutations. Therefore, the present study sequenced the Pre-S (Pre-S1 and Pre-S2) genes in ASCs with persistently low HBsAg expression, and compared the results with the established Pre-S gene reference sequences from ASCs with persistently high HBsAg expression, in order to elucidate the features of the Pre-S gene and to investigate the mechanisms underlying the formation of persistently low HBsAg expression levels in ASCs. The present study has important clinical significance for the accumulation of clinical data, virology data and molecular epidemiological data. The HBV-infected population with low-level HBsAg exhibits low nucleic acid replication, which may be missed in HBV screening conducted after blood donation, or as a preoperative examination or traumatic examination; therefore, it has the potential to infect others. It is therefore important for research to focus on the prevention of HBV transmission in the HBV-infected population with low-level HBsAg.
Materials and methods
Specimen collection
The present study was approved by the Ethics Committee of the 117th Hospital of PLA (Hangzhou, China). A total of 1,308 serum specimens were obtained from ASCs from the specimen banks of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University (Hangzhou, China), the Clinical Laboratory Center of Hangzhou Infectious Disease Hospital (Hangzhou, China), and the Clinical Laboratory Center of the 117th Hospital of PLA. Serum samples were obtained from patients who were predominantly located in eastern China. All participants were recruited between February 2014 and December 2015, and provided written informed consent. Serum samples were collected from the patients between February 2014 and December 2015, and were stored at −70°C until future use. According to the laboratory results, 1-year follow-up information from patients, clinical information, and infection history or the stage of natural history (9–11), the following exclusion criteria were developed: i) Patients with combined infection with human immunodeficiency virus, hepatitis C virus and hepatitis D virus; ii) patients with acute HBV infection (acute hepatitis) or low HBsAg levels present in the early stage of infection; iii) patients with low HBsAg levels in the HBsAg/anti-HBs conversion stage during the recovery period; iv) patients with long-term coexistence of HBsAg/anti-HBs positivity; and v) patients whose medical histories indicated the use of liver protection agents, enzyme-lowering agents, immunomodulators or antiviral drugs during the 6-month period prior to specimen collection.
Sample grouping
ASCs were characterized by HBsAg positivity for >6 months, low or undetectable serum HBV DNA levels and normal serum alanine aminotransferase (ALT) levels (11,12).
HBV-infected patients with low-level HBsAg expression were characterized by more than three instances of serum HBsAg detection results with <10 IU/ml after >1 year of follow-up (the interval between specimen collections was >3 months) (13,14).
The 1,308 ASCs in the sample bank were grouped according to HBsAg level (13,14). There were 276 cases in the low-level HBsAg (≤10 IU/ml) group and 1,032 cases in the high-level HBsAg (>10 IU/ml) group. The mean age of the patients in the low-level HBsAg group was higher than that of the patients in the high-level HBsAg group (P<0.05), and the mean HBV DNA value of the patients in the low-level HBsAg group was lower than that of the patients in the high-level HBsAg group (P<0.05). The two groups did not exhibit statistically significant differences with regards to sex or ALT levels (P>0.05). The clinical information for these two groups is presented in Table I.
Table I.
Clinical data of the low-level HBsAg and high-level HBsAg groups of chronic asymptomatic HBV carriers.
| Parameter | Low-level HBsAg group (n=276) | High-level HBsAg group (n=1,032) | P-value |
|---|---|---|---|
| Sex | |||
| Male | 169 | 671 | >0.05 |
| Female | 107 | 361 | |
| Age (years, means ± SD) | 55.09±16.45 | 43.63±10.95 | <0.05 |
| Laboratory results | |||
| ALT (U/l, means ± SD) | 23.13±9.60 | 25.32±10.54 | >0.05 |
| HBsAg (IU/ml, means ± SD) | 2.95±3.19 | 5,162.32±12,592.25 | <0.05 |
| HBV DNA (log10 IU/ml, means ± SD) | 1.32±1.60 | 2.93±2.83 | <0.05 |
| HBV DNA positive rate (%) | 45.65 (126/276) | 94.00 (94/100) | <0.05 |
ALT, alanine aminotransferase; HbsAg, HBV surface antigen; HBV, hepatitis B virus; SD, standard deviation.
Determination of biochemical and serological markers in chronic ASCs
ALT and HBV markers [HBsAg, anti-HBs, HBV e antigen (HBeAg), anti-HBe and anti-HBc] were measured using a C1600 biochemical analyzer (Abbott Laboratories, Lake Bluff, IL, USA) and an i2000 immunoassay instrument (Abbott Laboratories), respectively. ALT (cat. no. GP6040) and HBV marker [HBsAg (cat. no. 6C36-32), anti-HBs (cat. no. 7C18-30), HBeAg (cat. no. 6C32-20), anti-HBe (cat. no. 6C34-20) and anti-HBc (cat. no. 8L44-30)] kits were purchased from Beijing Leadman Biochemistry Co., Ltd. (Beijing, China) and Abbott Laboratories, respectively. The linear range of HBsAg was between 0.05 and 250 IU/ml. If the HBsAg level was >250.0 IU/ml, the samples were serially diluted 1:100 to obtain a value within the linear range of the assay, according to the manufacturer's protocol.
HBV DNA extraction and amplification
Viral DNA was extracted from 500 μl serum and eluted into 100 μl DNAse- and RNAse-free H2O using the nucleic acid extraction system NP968 (Tianlong Biotechnology Co., Ltd., Suzhou, China), according to the manufacturer's protocol. The level of HBV DNA in all samples was determined using a fluorescence quantitative polymerase chain reaction (PCR) detection kit [ACON Biotech (Hangzhou) Co., Ltd., Hangzhou, China] and an ABI StepOnePlus real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. The primer sequences were as follows: Forward, 5′-TTCCTCTTCATCCTGCTGCT-3′; reverse, 5′-AGTTTCCGTCCGAAGGTTTT-3′; fluorescence probe, 5′-FAM-ATCAACTACCAGCACGGGAC-BHQ1-3′. The amplification program used was as follows: Preheating at 95°C for 3 min; followed by 45 cycles of amplification, which consisted of heating at 94°C for 15 sec and 58°C for 30 sec; the fluorescence values of each tube were measured at the end of the 58°C step.
Sequencing of HBV Pre-S/S genomes
The target fragments of the HBV Pre-S/S genome in 276 serum samples from the low-level HBsAg group and 100 serum samples from the high-level HBsAg group, which were randomly selected on the basis of age matching with the low-level HBsAg group, were amplified using nested PCR (15). The patients from which the serum samples used for amplification were collected exhibited no statistically significant differences with regards to geographic region of origin, age or sex (P>0.05). The amplification and sequencing primer sequences are shown in Table II. The parameters and reaction systems used for amplification of the HBV Pre-S/S genes in nested PCR are presented in Table III. PCR amplification was performed using a Bio-Rad S1000 thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The positive products were purified, recovered and sent to Sangon Biotech Co., Ltd. (Shanghai, China) for sequencing using the ABI PRISM BigDye reagent kit and an ABI 3730 Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Table II.
Primers used for the amplification of the Pre-S/S genes in hepatitis B virus.
| Primer set and namea | Sequence (5′-3′) | Position (nt) | Fragment size (bp)b | Sequencing primer |
|---|---|---|---|---|
| IF | ACCWTATWCYTGGGAACAA | 2,819-2,837 | 1,554 | No |
| IR | TCAGCAAAYACTYGGCA | 1,190-1,174 | No | |
| IaF | ACCWTATWCYTGGGAACAA | 2,819-2,837 | 981 | Yes |
| IaR | GAYGAYGGGATGGGAATACA | 617-598 | Yes | |
| IbF | GACTYGTGGTGGACTTCTC | 251-269 | 939 | Yes |
| IbR | TCAGCAAAYACTYGGCA | 1,190-1,174 | Yes |
IF and IR refer to a pair of primers used in first-round amplification; Ia (IaF, IaR) and Ib (IbF, IbR) were used to amplify two fragments in the second-round amplification.
With reference to NC_003977 from National Center for Biotechnology Information. F, forward; R, reverse.
Table III.
Reaction parameters and systems used for the amplification of HBV Pre-S/S genes by nested PCR.
| Reaction step | Amplification parameters | Reaction system |
|---|---|---|
| First step | 95°C denaturation for 3 min followed by 5 cycles at 95°C for 30 sec. The Ta was then reduced from 57°C to 53°C over 4 cycles with a reduction of 1°C per cycle. The annealing time was 30 sec and extension was conducted at 72°C for 30 sec. Subsequently, 30 cycles were conducted at 95°C for 30 sec, Ta 53°C for 30 sec, 72°C for 30 sec, and a final extension step was performed at 72°C for 2 min. | Each 25-μl reaction tube contained 5 μl 5X KAPA2G buffer A, 5 μl 5X KAPA enhancer, 0.1 μl KAPA2G Robust HotStart DNA polymerase (Kapa Biosystems, Inc., Wilmington, MA, USA), 0.5 μl 10 μM dNTP Mix (Takara Bio, Inc., Otsu, Japan), l μl each 10 M primer, and 3 μl DNA template. The total volume was brought to 25 μl with PCR-specific water. |
| Second step | The same as the first step. | The same as the first step. The amplified DNA product obtained from the first step was used as the DNA template for the second step. |
PCR, polymerase chain reaction; Ta, annealing temperature.
The sequencing results were assembled using the SeqMan program in Lasergene software v7.1 (DNAstar, Inc., Madison, WI, USA). The comparison was performed using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Genotyping and serotyping
Pre-S/S genes assembled from multiple target fragments were compared and spliced using MEGA v6.0 software (16). The sequences obtained from sequencing and the National Center for Biotechnology Information (NCBI)-recommended representative genotypes B (AB033554, AF100309 and D00329) and C (AB014381, AY123041 and X04615) were used for construction of the phylogenetic tree and genotyping using the neighbor joining method (17) [≥96% homology of the S gene was considered the same genotype (18)] at a bootstrap setting of 1,000 times. The serotype was confirmed based on the amino acid expression at specific sites in the S gene sequence (19).
Establishment and validation of Pre-S gene reference sequences
Based on the successful sequencing results of the HBV Pre-S gene in 100 cases in the high-level HBsAg group, the base with the highest frequency at each position in the sequences of all major genotypes (B and C) was used as the reference base to establish a Pre-S reference sequence of genotype B in ASCs (ASC-R-Pre-S-B) and a Pre-S reference sequence of genotype C in ASCs (ASC-R-Pre-S-C) in eastern China.
To validate the accuracy and usability of ASC-R-Pre-S-B and ASC-R-Pre-S-C established in the present study, homology analyses were performed using the sequences of genotypes B and C of ASCs in NCBI (defined as the NCBI-ASC-Pre-S-B group and the NCBI-ASC-Pre-S-C group), and the reference sequences of genotypes B and C recommended by the NCBI (NCBI-R-Pre-S-B: AB033554, AF100309 and D00329; NCBI-R-Pre-S-C: AB014381, AY123041 and X04615) (17).
Comparative analyses of HBV Pre-S gene sequences
The HBV Pre-S gene sequences of ASCs in the high- and low-level HBsAg groups were compared with the established ASC-R-Pre-S-B and ASC-R-Pre-S-C reference sequences. Each case in which a base at a given site was replaced by another base and led to a change in the amino acid of the expression protein was defined as a mutation (nonsynonymous mutation) (20). In addition, cases in which mutations were present in more than two codons in the same Pre-S gene sequence were defined as multi-site co-mutations (21). The BLAST tool was used to search and compare the sequences in the NCBI database to determine whether the amino acid mutation sites, including single-site mutations, multiple-site co-mutations, deletion mutations and insertion mutations, in the Pre-S protein in the low- and high-level HBsAg groups were newly discovered or unreported mutation sites. The mutation sites and mutation frequencies reported in the NCBI database, including single-site mutations, multiple-site co-mutations, deletion mutations and insertion mutations, which displayed statistically significant differences in the low- and high-level HBsAg groups were defined as the meaningful mutations; mutations with a frequency of ≥10% were defined as hotspot mutations, and mutations with a frequency of <10% were defined as non-hotspot mutations (22).
Statistical analysis
SPSS version 12.01 (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. The scatter plots were produced using Graphpad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA), and the Venn diagrams were produced with Canvas 11 software (ACD Systems International, Inc., Victoria, BC, Canada). The results are presented as the means ± standard error of the mean, and HBV DNA levels (IU/ml) were logarithmically transformed for analysis. Continuous and categorical variables were compared between groups using the independent t-test, and the χ2 test or Fisher's exact test. The frequency of amino acid mutation sites was analyzed using the Cochran-Mantel-Haenszel test method. All P-values were two-tailed, and P<0.05 was considered to indicate a statistically significant difference.
Results
Basic serological characteristics of chronic ASCs
The HBV serum markers of 1,308 ASCs were examined. The HBV Pre-S/S genes of 276 cases in the low-level HBsAg group and those of 100 randomly selected cases in the high-level HBsAg group were sequenced, and genotyping and serotyping were performed. The results demonstrated that the distribution of HBV serological marker patterns, genotypes and serotypes between the low- and high-level HBsAg groups all exhibited statistically significant differences (P<0.05; Fig. 1).
Figure 1.
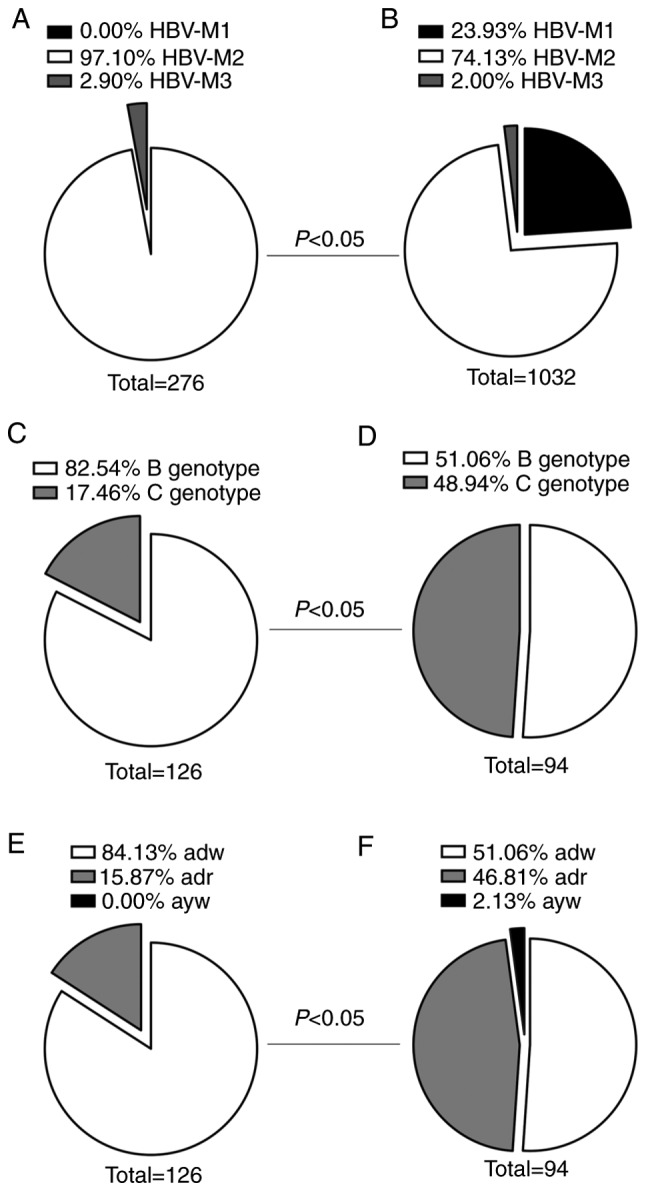
Distribution of HBV serological marker patterns, genotypes and serotypes in the low-level HBsAg and high-level HBsAg groups. Among the 276 cases in the low-level HBsAg group, Pre-S/S sequencing was successful in 126 cases. Among the 100 randomly selected cases in the high-level HBsAg group, Pre-S/S sequencing was successful in 94 cases. Distribution of serological patterns of HBV in the (A) low-level HBsAg group and (B) high-level HBsAg group (P<0.05). HBV-M1, HBsAg/HBV e antigen/anti-HBc-positive; HBV-M2, HBsAg/anti-HBe/Anti-HBc-positive; HBV-M3, HBsAg/anti-HBc-positive. Distribution of genotypes in the (C) low-level HBsAg group and (D) high-level HBsAg group (P<0.05). Distribution of serotypes in the (E) low-level HBsAg group and (F) high-level HBsAg group (P<0.05). HBsAg, HBV surface antigen; HBV, hepatitis B virus.
Establishment and validation of Pre-S reference sequences
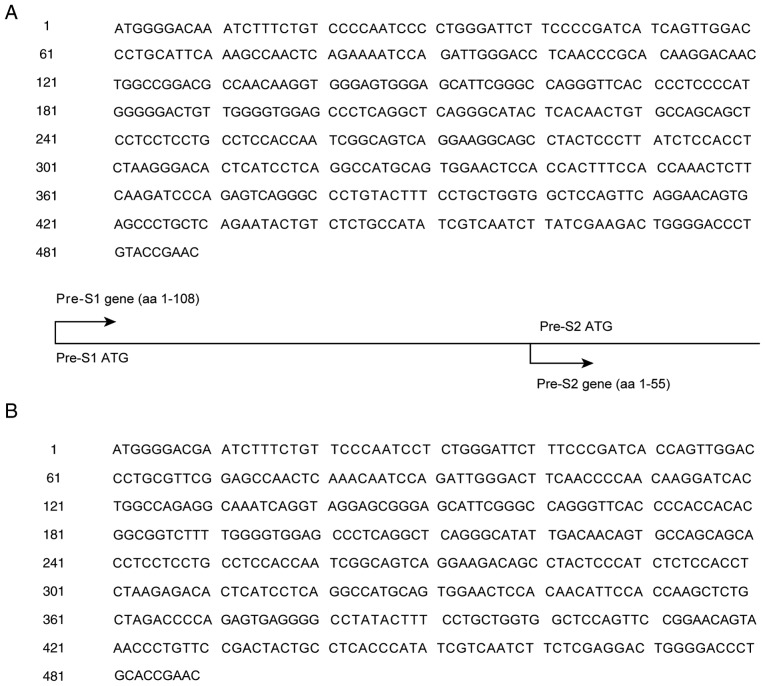
Genotyping was performed on the 94 cases in the high-level HBsAg group for which successful sequencing results were obtained. The major genotypes were the B and C genotypes. The base in the same position with the highest frequency in the B and C sequences was used as the reference to establish the Pre-S reference sequence of the B genotype in ASCs (ASC-R-Pre-S-B) and the Pre-S reference sequence of the C genotype in ASCs (ASC-R-Pre-S-C). The results are shown in Fig. 2.
Figure 2.
Pre-S reference sequences of genotype B and C in ASCs. (A) Pre-S reference sequences of genotype B in ASCs (ASC-R-Pre-S-B). (B) Pre-S reference sequences of genotype C in ASCs (ASC-R-Pre-S-C). ASC, asymptomatic hepatitis B virus carriers.
Homology analyses of the nucleotides and amino acid residues in the Pre-S sequences of the B and C genotypes in ASCs in NCBI (NCBI-ASC-Pre-S-B and NCBI-ASC-Pre-S-C) and the Pre-S reference sequences of genotypes B and C recommended by NCBI (NCBI-R-Pre-S-B and NCBI-R-Pre-S-C) were performed. The ASC-R-Pre-S-B and ASC-R-Pre-S-C sequences established in the present study displayed high homology (98.5–100%) with the Pre-S sequences of genotypes B and C in ASCs reported in most areas in China, high homology with the majority of the Pre-S reference sequences of genotypes B and C in Asia recommended by NCBI (98.2–99.5%), and lower homology (96.4–96.9%) with those in Canada and Indonesia (Table IV).
Table IV.
Analysis of the homology of Pre-S gene sequences between ASC-R-Pre-S-B and ASC-R-Pre-S-C, and NCBI-ASC-Pre-S-B, NCBI-ASC-Pre-S-C, NCBI-R-Pre-S-B and NCBI-R-Pre-S-C (%).
| Reference sequence | Comparison sequence | Accession number | Area | Homology (%) nucleotide | Amino acid |
|---|---|---|---|---|---|
| ASC-R-Pre-S-B | NCBI-ASC-Pre-S-B | JX661484 | Shanghai | 99.4 | 100 |
| JX661473 | Shanghai | 99.5 | 99.2 | ||
| JN406371 | Wuhan | 99.5 | 99.0 | ||
| KY470957 | Yunnan | 99.6 | 99.2 | ||
| KY470963 | Yunnan | 99.4 | 98.7 | ||
| DQ463800 C | anada | 96.5 | 96.9 | ||
| DQ463801 C | anada | 96.4 | 96.4 | ||
| NCBI-R-Pre-S-B | D00329 | Japan | 98.7 | 99.2 | |
| AF100309 | Shanghai | 99.4 | 98.7 | ||
| AB033554 | Indonesia | 96.5 | 96.4 | ||
| ASC-R-Pre-S-C | NCBI-ASC-Pre-S-C | EU306721 | Yunnan | 99.2 | 99.0 |
| EU439007 | Yunnan | 98.9 | 98.5 | ||
| JX661496 | Shanghai | 99.4 | 99.2 | ||
| JX661498 | Shanghai | 99.1 | 98.7 | ||
| AY641558 | South Korea | 99.2 | 99.5 | ||
| AY641560 | South Korea | 98.9 | 99.0 | ||
| AB033556 | Japan | 98.8 | 99.2 | ||
| AB222715 | Japan | 99.3 | 99.2 | ||
| NCBI-R-Pre-S-C | AB014381 | Japan | 99.1 | 98.7 | |
| AY123041 | Japan | 98.8 | 98.2 | ||
| X04615 | Japan | 98.9 | 98.7 |
ASC-R-Pre-S-B and ASC-R-Pre-S-C, Pre-S reference sequences of genotypes B and C in ASCs established in the present study; NCBI-ASC-Pre-S-B and NCBI-ASC-Pre-S-C, Pre-S sequences of genotypes B and C in ASCs uploaded to NCBI; NCBI-R-Pre-S-B and NCBI-R-Pre-S-C, Pre-S reference sequences of genotypes B and C recommended by NCBI. ASC, asymptomatic hepatitis B virus carriers; NCBI, National Center for Biotechnology Information.
Comparative analyses of HBV Pre-S gene sequences
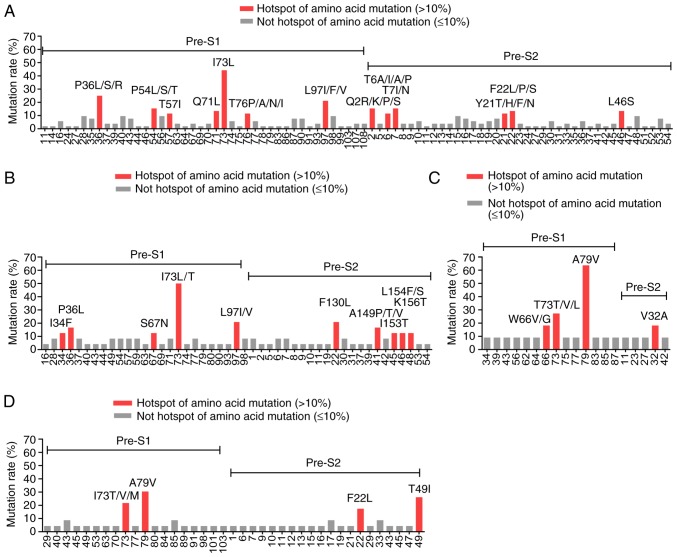
The ASC-R-Pre-S-B and ASC-R-Pre-S-C reference sequences established in the present study were compared with the Pre-S gene sequences of genotypes B and C in the low- and high-level HBsAg groups. For genotype B, the number of amino acid mutation sites in the Pre-S protein in the low-level HBsAg group was higher than that in the high-level HBsAg group (P<0.05), whereas the number of hotspot mutations was not significantly different in the two groups (P>0.05). The distributions and frequencies of mutation sites in the two groups are shown in Fig. 3A and B. In genotype C, the number of amino acid mutation sites and the number of hotspot mutation sites in the Pre-S protein was not significantly different in the two groups (P>0.05). The distributions and frequencies of mutation sites in these two groups are presented in Fig. 3C and D.
Figure 3.
Distribution of Pre-S1 and Pre-S2 mutation sites in the amino acid sequence of the B and C genotypes in the low- and high-level HBsAg groups. (A) Mutation sites in the amino acid sequences of the Pre-S1 and Pre-S2 proteins in the B genotype in the low-level HBsAg group. (B) Mutation sites in the amino acid sequences of the Pre-S1 and Pre-S2 proteins in the B genotype in the high-level HBsAg group. (C) Mutation sites in the amino acid sequences of the Pre-S1 and Pre-S2 proteins in the C genotype in the low-level HBsAg group. (D) Mutation sites in the amino acid sequences of the Pre-S1 and Pre-S2 proteins in the C genotype in the high-level HBsAg group. HBsAg, hepatitis B virus surface antigen.
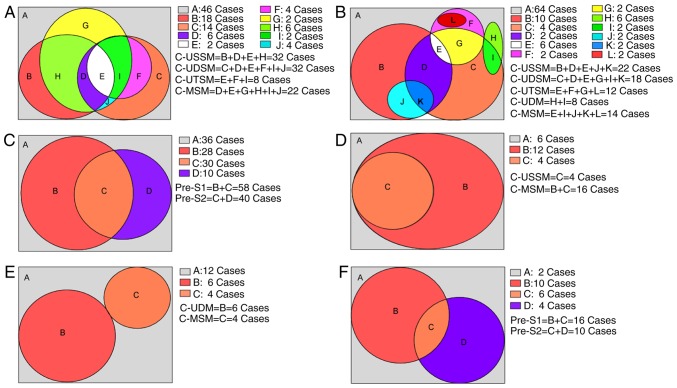
The distribution of the characteristic amino acid substitutions due to mutations found in the Pre-S1 and Pre-S2 proteins of different genotypes from individuals in different groups is presented in Table V. In genotype B, the Pre-S1 gene had 14 single-site mutations, eight double-site co-mutations and two triple-site co-mutations; these mutations were detected only in the low-level HBsAg group. One single-site mutation (L97V) was present in both groups. BLAST comparison of these mutation sites indicated that none of them had been reported in NCBI or in the literature (unreported mutation sites). In addition, BLAST comparison revealed that five single-site mutations were reported in NCBI or in the literature; the frequency of these mutations differed significantly in the two groups (meaningful site mutations, P<0.05). The characteristic single-site mutations and multiple-site co-mutations that resulted in amino acid substitutions accounted for 55.77% (58/104) of the 104 cases in the low-level HBsAg group (Fig. 4A). The Pre-S2 gene had 14 single-site mutations, nine double-site co-mutations, three triple-site mutations and three deletion mutations; these were only found in the low-level HBsAg group. Comparison of these mutation sites using BLAST indicated that none of them had been reported in NCBI or in the literature (unreported mutation sites). In addition, BLAST comparison revealed that two of the single-site mutations were reported in NCBI or in the literature; the frequencies of these mutations differed significantly between the two groups (meaningful site mutations, P<0.05). These characteristic amino acid single-site mutations and multiple-site co-mutations accounted for 38.46% (40/104) of the 104 cases in the low-level HBsAg group (Fig. 4B). In total, the characteristic amino acid single-site mutations and multiple-site co-mutations in the Pre-S1 and Pre-S2 gene fragments accounted for 65.38% (68/104) of the 104 cases in the low-level HBsAg group (Fig. 4C).
Table V.
Characteristic amino acid mutation sites in genotypes B and C in the low- and high-HBsAg groups.
| Genotype | Gene name | Mutation type | Low-level HBsAg group (B: n=104; C: n=22) | High-level HBsAg group (B: n=48; C: n=46) |
|---|---|---|---|---|
| B | Pre-S1 | Unreported mutation sitea Meaningful mutation siteb | 14 single-site mutations, 8 double-site mutations, 2 triple-site mutationsc F56I/V(10), T76A/N/P(12) | L97V(2), S67N+Q71L(4), S67N+L97I/V(4) L34F(6), V49A(4), P59S/L(4) |
| Pre-S2 | Unreported mutation sitea | 14 single-site mutations, 8 double-site combined mutations, 3 triple-site combined mutationsd Pre-S2Δ2-5(4), Pre-S2Δ12-14(2), Pre-S2Δ5-22(2) | F8Y(2), I42T(4), T6A/ S+F22L(4), F22L+A41P/S(2) | |
| Meaningful mutation siteb | P15L/S/T(10), Y21T/F/H/N(12) | Not detected | ||
| C | Pre-S1 | Unreported mutation sitea | D39A(2), W66V(2) | S98C(2) |
| Meaningful mutation siteb | W66V/G(4), A79V(14) | A79V(14) | ||
| Pre-S2 | Unreported mutation sitea | Pre-S2Δ9-22(4), Pre-S2Δ12-22(2) | V17A/G+F22L(4), Pre-S2Δ15-22(4), Pre-S2Δ8-15(2) | |
| Meaningful mutation siteb | V32A(4) | T49I(12) |
Searching and comparing using BLAST did not identify Pre-S gene sequences in the same genotype (B and C) containing this single-site mutation or did not identify Pre-S gene sequences in the same genotype (B and C) containing this multiple-site mutation (2 or 3 sites had mutations at the same time).
Searching and comparing using BLAST revealed that the mutation was already reported in NCBI or in the literature; however, the frequency of the mutation in the high- and low-level HBsAg groups was significantly different (P<0.05). The underlined and bold mutation sites had significantly greater mutation frequencies in the high-level HBsAg group than in the low-level HBsAg group.
Single-site mutations (14): S27A(2), N35S/D(4), P36R(2), D43H(2), K46T(2), L74K/S(4), T75P(2), T86H(2), N87I(2), Q93H/L(4), L97F/V(4), P99L(2), Q107H(4), and A108V(4); double-site combined mutations (8): E28K+D43N(4), P36L/S/R+F56V(6), N40I+F56V(2), P54L+V77L/M(4), T57I+S90T(4), A70T+L97I(4), Q71L+N87T(4), and T76P/I+S98T(4); and triple-site combined mutations (2): N35K+N40T+F56I(4) and Q71L+N87T+L97I(4).
Single-site mutations (14): S5A(2), Q10N(2), L12H/V(4), D14E(2), V17I(4), R18M(2), Y21F(2), P23H(2), T31K(2), A35/V(2), Q36R/P(6), N37H(2), I42T(2), and I45N(2); double-site combined mutations (8): Q2R/K+Y21T/D(4), Q2K+F22L(2), Q2R/K+T7I/N(8), T7I/N+F22L/P(6), F8L+D14E/N(4), Q13P/H+F22L(4), P15L+L20A/V/(2), and P15L+Y21H(2); and triple-site combined mutations (3): Q2R/K+T6A/S+F22L(6), P15L/S+R16G/K/S+A19T/P(6), and P15L/S/T+L20T/Q/R+Y21T/N(6).
Figure 4.
Distribution of cases according to mutation type in the Pre-S1 and Pre-S2 proteins of the B and C genotypes in the low-level hepatitis B virus surface antigen group. Within the Venn diagram, letters are used to represent region; capitalized A represents no CMs (unreported and meaningful mutations), the remaining capitals refer to CMs. (A) B genotype Pre-S1. (B) B genotype Pre-S2. (A and B) Of the total 104 cases, 58 and 40 cases were identified as CMs (all regions except the gray A region) in (A) and (B), respectively. (C) B genotype Pre-S1 and Pre-S2. Of the total 104 cases, 68 cases were identified as CMs (all regions in the rectangular except the gray A region) either in Pre-S1 or Pre-S2, with a total CM rate of 65.38% (68/104). (D) C genotype Pre-S1. (E) C genotype Pre-S2. (D and E) Of the total 22 cases, 16 and 10 cases were identified as CMs (all regions except the gray A region) in (D) and (E), respectively. (F) C genotype Pre-S1 and Pre-S2. Of the total 22 cases, 20 cases were identified as CMs (all regions except the gray A region) either in Pre-S1 or Pre-S2, with a total CM rate of 90.91% (20/22). CMs, characteristic mutations consisting of unreported and meaningful mutations; C-MSM, cases of meaningful site mutations; C-UDM, cases of unreported deletion mutations; C-UDSM, cases of unreported double-site co-mutations; C-USSM, cases of unreported single-site mutations; C-UTSM, cases of unreported triple-site co-mutations.
In genotype C, the Pre-S1 gene had two single-site mutations; these were only found in the low-level HBsAg group. Comparison of these mutations sites using BLAST indicated that they had not been reported in NCBI or in the literature (unreported mutation sites). In addition, comparison of the two single-site mutations in both groups using BLAST revealed that they had previously been reported in NCBI or in the literature; however, their mutation frequencies were significantly different in the two groups (meaningful site mutations, P<0.05). These characteristic amino acid single-site mutations and multiple-site co-mutations accounted for 72.73% (16/22) of the 22 cases in the low-level HBsAg group (Fig. 4D). The Pre-S2 gene had two locations at which deletion mutations occurred in the low-level HBsAg group, whereas the high-level HBsAg group had one double-site mutation and two locations at which deletion mutations occurred. Comparison of these mutation sites and deletion mutations using BLAST indicated that none of them had been previously reported in NCBI or in the literature. In addition, comparison of two single-site mutations in both groups using BLAST revealed that they had already been reported in NCBI or in the literature; however, the frequencies of these mutations were significantly different in the two groups (meaningful site mutations, P<0.05). These characteristic amino acid single-site mutations, multiple-site co-mutations and deletion mutations accounted for 45.45% (10/22) of the 22 cases in the low-level HBsAg group (Fig. 4E). In total, the characteristic amino acid single-site mutations and multiple-site co-mutations in the Pre-S1 and Pre-S2 gene fragments accounted for 90.91% (20/22) of the 22 cases in the low-level HBsAg group (Fig. 4F).
Discussion
Serum HBsAg is not only an important serum marker for the screening and diagnosis of HBV infection, but also an important reference indicator for the effects of HBV treatment (23). Jaroszewicz et al (24) and Nguyen et al (25) reported differences in serum HBsAg levels in HBV-infected patients in Asia and Europe at different stages during the natural course of infection. Serum HBsAg levels can also be used in auxiliary diagnosis to determine the stage of HBV infection with respect to the natural history of the virus (26). In our previous study, a survey was performed, and the distribution features, immune function and S gene sequences in an HBV-infected population with persistently low levels of serum HBsAg expression (<10 IU/ml) were analyzed (13,14). The present study investigated the clinical features of 276 HBV-infected patients with low HBsAg levels. The results demonstrated that the majority of individuals in the studied population with low HBsAg levels were chronic ASCs. The population with low HBsAg levels were not significantly different with regards to sex composition and ALT levels when compared with a population of 1,032 ASCs with high HBsAg levels; however, age, HBV serological marker pattern composition, HBV genotype composition and HBV serotype composition were significantly different in these two populations. The serological pattern HBsAg/anti-HBe/anti-HBc was present in 97.1% of the individuals in the low-level HBsAg group, and the group had low viral replication and low positive rates of HBV DNA that were similar to those reported in the literature (13,14). Therefore, chronic asymptomatic HBV infection, HBsAg/Anti-HBe/Anti-HBc positivity (97.1%), B genotype (82.54%) and adw serotype (84.13%) were considered major clinical features of the ASC population with low-level HBsAg.
Under normal circumstances, the large surface proteins encoded by the Pre-S1 gene are closely associated with the virus cycle, and with the interaction between viruses and hepatocyte receptors. The middle surface proteins encoded by the Pre-S2 gene are associated with the extracellular secretion of viruses. Mutations in the Pre-S region might alter the phenotype of HBsAg (2,3,8). Therefore, when base substitution, deletion or insertion mutations occur in the Pre-S gene, the infectivity of HBV, HBV nucleic acid replication, HBV serological markers, viral protein expression and secretion, immunogenicity and antigen-antibody binding capacity may be affected (3,8,27). The present study investigated Pre-S1 and Pre-S2 gene sequences in 276 HBV-infected patients with low HBsAg levels, compared with an established reference sequence. The results demonstrated that 28 unreported single-site mutations (14 in Pre-S1 and 14 in Pre-S2), 17 unreported double-site co-mutations (eight in Pre-S1 and nine in Pre-S2), five unreported triple-site co-mutations (two in Pre-S1 and three in Pre-S2), three unreported Pre-S2 deletion mutations, and seven meaningful single-site mutations (five in Pre-S1 and two in Pre-S2) were present in the Pre-S sequence of genotype B. The mutation frequencies of some unreported mutation sites and the frequencies of deletion mutations in the low- and high-level groups did not display statistically significant differences, which may be caused by natural substitution (to be further verified by protein expression); therefore, no statistical inferences were made regarding the observed unreported mutation sites and deletion mutations. In addition, the number of amino acid mutation sites in the Pre-S protein was higher in the low-level HBsAg group than in the high-level HBsAg group; however, the number of hotspot mutations in these two groups did not show a statistically significant difference.
Neither the B nor C genotype virus present in the two groups exhibited Pre-S1 deletion mutations or Pre-S insertion mutations. The Pre-S1 promoter deletion mutation (Pre-S1Δ1-8) that was previously reported to be associated with high HBsAg expression (8) was not observed in the present study. In addition, the Pre-S2 deletion mutation (Pre-S2Δ3-25) that was previously shown to be associated with defective viral replication (8) was not observed in this study. It has been reported (3) that Pre-S1 region 3′-end deletion mutations and Pre-S2 region 5′-end deletion mutations affect numerous functional sites in the S gene promoter and the CCAAT-binding factor (CBF) binding site required for S gene expression; such mutations usually result in abnormal HBV synthesis and secretion, and affect the binding of neutralizing antibodies. We aim to conduct further research on whether the Pre-S2 deletion mutations discovered in the present study are closely associated with persistently low HBsAg expression. In the low-level HBsAg group, the percentages of mutations (unreported mutations and meaningful mutations) in the Pre-S1 gene (B: 55.77% and C: 72.72%) were higher than in the Pre-S2 gene (B: 38.46% and C: 45.45%). However, no significant difference in the mutation rates of Pre-S1 and Pre-S2 between the B and C genotypes was detected. These findings suggested that Pre-S1 gene mutations may serve a leading role in the HBV-infected population with persistently low HBsAg expression.
To ensure the accuracy of analysis of the HBV Pre-S gene sequence in the low-level HBsAg population (28), Pre-S reference sequences of the B and C genotypes (ASC-R-Pre-S-B and ASC-R-Pre-S-C) suitable for research on eastern Chinese populations were established. The results indicated that ASC-R-Pre-S-B and ASC-R-Pre-S-C can be used as HBV S gene reference sequences for ASCs in China. These results also revealed that the establishment of reference sequences should consider both the nature of the study subjects (ASCs in the present study) and regional differences. In addition, the established reference sequences should be validated from different perspectives. Therefore, the established reference sequences will have representativeness, comparability and authority, and the obtained study results based on reference sequences will have creditability and scientific importance.
Previous research has addressed four possible mechanisms underlying the low expression levels of serum HBsAg in some individuals. i) S gene variations, particularly 'a' antigenic determinant mutations, which may cause alterations in the spatial conformation of the antigenic determinants of HBsAg, thereby affecting antigen detection and/or resulting in reduction of HBsAg secretion. In addition, the mutations rtA181T/sW172*, rtV191I/sW182* and rtM204I/sW196* in the S gene cause early termination of transcription; therefore, viruses cannot successfully synthesize and secrete HBsAg (2,7,29,30). ii) Pre-S/S gene variations result in differences between the HBV DNA level and the HBsAg concentration (3,8,27). iii) Either there is immune tolerance of unknown cause between HBV and hosts (13) or HBsAg mutations induced by the use of immu-nosuppressive agents enhance immune tolerance in the body; therefore, HBsAg cannot be completely cleared from the body, resulting in the presence of low HBsAg levels (31). iv) S gene methylation has a regulatory role in HBsAg secretion (32). The aforementioned findings are mainly associated with Pre-S/S gene variations, occult hepatitis, the use of immunosuppressive agents, and S gene methylation resulting from antiviral therapy. Very few studies have reported the features of HBV S gene sequences or the mechanisms by which ASC populations with low HBsAg levels are generated. Therefore, the present study used established genotype B and C reference sequences of HBV-infected ASCs with high HBsAg levels as a reference in comparative analyses of Pre-S gene sequences of HBV-infected ASCs with low HBsAg levels. The results indicated that the amino acid mutation rate and the number of mutation sites in the Pre-S protein in the low-level HBsAg group, which was mainly characterized by the B genotype, were higher than those in the high-level HBsAg group.
In conclusion, during immune clearance of HBV in the host, novel mutations at numerous sites (including co-mutations), meaningful mutations (including co-mutations) and deletion mutations in the HBV Pre-S gene may be induced; these might be associated with defects in HBV virus replication and might be one of the reasons underlying low-level HBsAg expression.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Medical Science Foundation of Nanjing Military Command (grant no. 12MA117) and the Natural Science Foundation of Zhejiang Province (grant no. Y15H200001).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JC participated in the project design and research, performed the statistical analysis, and was responsible for drafting and revising the manuscript. CS participated in the project design and coordination, assisted in writing the manuscript and helped with the statistical analysis. DC was responsible for sample collection. YD and MZ performed the virologic analysis and helped draft the manuscript. XX performed the molecular genetic analysis and sample collection. TW performed the virologic analysis and sample collection, and participated in the statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Prior to enrolment of the subjects, all subjects provided written informed consent to participate in the present study. The present study was approved by the Medical Ethics Committee of The 117th Hospital of the PLA (protocol no: PLA-117-20160309; Hangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ott JJ, Horn J, Krause G, Mikolajczyk RT. Time trends of chronic HBV infection over prior decades-A global analysis. J Hepatol. 2017;66:48–54. doi: 10.1016/j.jhep.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 2.Xiang KH, Michailidis E, Ding H, Peng YQ, Su MZ, Li Y, Liu XE, Dao Thi VL, Wu XF, Schneider WM, et al. Effects of amino acid substitutions in hepatitis B virus surface protein on virion secretion, antigenicity, HBsAg and viral DNA. J Hepatol. 2017;66:288–296. doi: 10.1016/j.jhep.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollicino T, Amaddeo G, Restuccia A, Raffa G, Alibrandi A, Cutroneo G, Favaloro A, Maimone S, Squadrito G, Raimondo G. Impact of hepatitis B virus (HBV) preS/S genomic variability on HBV surface antigen and HBV DNA serum levels. Hepatology. 2012;56:434–443. doi: 10.1002/hep.25592. [DOI] [PubMed] [Google Scholar]
- 4.Zhang ZH, Wu CC, Chen XW, Li X, Li J, Lu MJ. Genetic variation of hepatitis B virus and its significance for pathogenesis. World J Gastroenterol. 2016;22:126–144. doi: 10.3748/wjg.v22.i1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CH, Changchien CS, Lee CM, Tung WC, Hung CH, Hu TH, Wang JH, Wang JC, Lu SN. A study on sequence variations in pre-S/surface, X and enhancer II/core promoter/precore regions of occult hepatitis B virus in non-B, non-C hepatocellular carcinoma patients in Taiwan. Int J Cancer. 2009;125:621–629. doi: 10.1002/ijc.24416. [DOI] [PubMed] [Google Scholar]
- 6.Cento V, Van Hemert F, Neumann-Fraune M, Mirabelli C, Di Maio VC, Salpini R, Bertoli A, Micheli V, Gubertini G, Romano S, et al. Anti-HBV treatment induces novel reverse transcriptase mutations with reflective effect on HBV S antigen. J Infect. 2013;67:303–312. doi: 10.1016/j.jinf.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Kim H, Lee SA, Won YS, Lee H, Kim BJ. Occult infection related hepatitis B surface antigen variants showing lowered secretion capacity. World J Gastroenterol. 2015;21:1794–1803. doi: 10.3748/wjg.v21.i6.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim BK, Choi SH, Ahn SH, Chung AR, Park YK, Han KH, Kim S, Kim HS, Park JH, Kim KS, et al. Pre-S mutations of hepatitis B virus affect genome replication and expression of surface antigens. J Gastroenterol Hepatol. 2014;29:843–850. doi: 10.1111/jgh.12415. [DOI] [PubMed] [Google Scholar]
- 9.Chinese Society of Hepatology, Chinese Medical Association; Chinese Society of Infectious Diseases, Chinese Medical Association. Hou JL, lai W. The guideline of prevention and treatment for chronic hepatitis B: A 2015 update. Zhonghua Gan Zang Bing Za Zhi. 2015;23:888–905. doi: 10.3760/cma.j.issn.1007-3418.2015.12.002. In Chinese. [DOI] [PubMed] [Google Scholar]
- 10.European Association for the Study of the Liver. Electronic address easloffice@easloffice.eu; European Association for the Study of the Liver: EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 11.Guidelines for the Prevention . Care and treatment of persons with chronic hepatitis B infection WHO guidelines approved by the guidelines review committee. World Health Organization; Geneva: 2015. [PubMed] [Google Scholar]
- 12.Kumar M, Sarin SK, Hissar S, Pande C, Sakhuja P, Sharma BC, Chauhan R, Bose S. Virologic and histologic features of chronic hepatitis B virus-infected asymptomatic patients with persistently normal ALT. Gastroenterology. 2008;134:1376–1384. doi: 10.1053/j.gastro.2008.02.075. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Wu W. Determination of low level HBsAg in serum by microparticle enzyme immunoassay. Hepatobiliary Pancreat Dis Int. 2002;1:262–264. [PubMed] [Google Scholar]
- 14.Cheng J, Sun CG, Chen Y, Xu ZL, Dai YZ, Sun GZ, Li XJ. Relationship between the quantitative results of HBV DNA and HBV markers (HBV M) in population with low-level HBsAg. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2009;25:631–633. 636. In Chinese. [PubMed] [Google Scholar]
- 15.Chook JB, Teo WL, Ngeow YF, Tee KK, Ng KP, Mohamed R. Universal primers for detection and sequencing of hepatitis B virus genomes across genotypes A to G. J Clin Microbiol. 2015;53:1831–1835. doi: 10.1128/JCM.03449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao H, Liu Y, Chen J, Ding W, Li X, Xu Z, Yang Y, Chen R, Si L, Xu X, et al. Characterization of hepatitis B virus (HBV) preS/S gene mutations in blood donors with occult HBV infection in the Baoji area of North China. Transfusion. 2017;57:857–866. doi: 10.1111/trf.14046. [DOI] [PubMed] [Google Scholar]
- 18.Tong S, Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64(Suppl 1):S4–S16. doi: 10.1016/j.jhep.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: Genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 20.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: A molecular epidemiologic study. J Infect Dis. 2015;211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong YW, Di FL, Liu C, Zhang XC, Bi JF, Li YL, Wu SQ, Dong H, Liu LM, He J, et al. Hepatitis B virus basal core promoter/precore mutants and association with liver cirrhosis in children with chronic hepatitis B virus infection. Clin Microbiol Infect. 2016;22:379.e1–379.e8. doi: 10.1016/j.cmi.2015.10.033. [DOI] [PubMed] [Google Scholar]
- 22.Yin J, Xie J, Zhang H, Shen Q, Han L, Lu W, Han Y, Li C, Ni W, Wang H, Cao G. Significant association of different preS mutations with hepatitis B-related cirrhosis or hepatocellular carcinoma. J Gastroenterol. 2010;45:1063–1071. doi: 10.1007/s00535-010-0253-1. [DOI] [PubMed] [Google Scholar]
- 23.Martinot-Peignoux M, Marcellin P. Virological and sero-logical tools to optimize the management of patients with chronic hepatitis B. Liver Int. 2016;36(Suppl 1):S78–S84. doi: 10.1111/liv.12998. [DOI] [PubMed] [Google Scholar]
- 24.Jaroszewicz J, Calle Serrano B, Wursthorn K, Deterding K, Schlue J, Raupach R, Flisiak R, Bock CT, Manns MP, Wedemeyer H, Cornberg M. Hepatitis B surface antigen (HBsAg) levels in the natural history of hepatitis B virus (HBV)-infection: A European perspective. J Hepatol. 2010;52:514–522. doi: 10.1016/j.jhep.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen T, Thompson AJ, Bowden S, Croagh C, Bell S, Desmond PV, Levy M, Locarnini SA. Hepatitis B surface antigen levels during the natural history of chronic hepatitis B: A perspective on Asia. J Hepatol. 2010;52:508–513. doi: 10.1016/j.jhep.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Karra VK, Chowdhury SJ, Ruttala R, Polipalli SK, Kar P. Clinical significance of quantitative HBsAg titres and its correlation with HBV DNA levels in the natural history of hepatitis B virus infection. J Clin Exp Hepatol. 2016;6:209–215. doi: 10.1016/j.jceh.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Liu Y, Zhao J, Xu Z, Chen R, Si L, Lu S, Li X, Wang S, Zhang K, et al. Characterization of novel hepatitis B virus PreS/S-Gene mutations in a patient with occult hepatitis B virus infection. PLoS One. 2016;11:e0155654. doi: 10.1371/journal.pone.0155654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Q, Zhu H, Zhang Y, Li X, Zhang Z. Hepatitis B virus genotype A: Design of reference sequences for sub-genotypes. Virus Genes. 2016;52:325–333. doi: 10.1007/s11262-016-1307-0. [DOI] [PubMed] [Google Scholar]
- 29.Osiowy C. Detection of HBsAg mutants. J Med Virol. 2006;78(Suppl 1):S48–S51. doi: 10.1002/jmv.20607. [DOI] [PubMed] [Google Scholar]
- 30.Wang ML, Tang H. Nucleos(t)ide analogues causes HBV S gene mutations and carcinogenesis. Hepatobiliary Pancreat Dis Int. 2016;15:579–586. doi: 10.1016/S1499-3872(16)60064-4. [DOI] [PubMed] [Google Scholar]
- 31.Salpini R, Colagrossi L, Bellocchi MC, Surdo M, Becker C, Alteri C, Aragri M, Ricciardi A, Armenia D, Pollicita M, et al. Hepatitis B surface antigen genetic elements critical for immune escape correlate with hepatitis B virus reactivation upon immu-nosuppression. Hepatology. 2015;61:823–833. doi: 10.1002/hep.27604. [DOI] [PubMed] [Google Scholar]
- 32.Graumann F, Churin Y, Tschuschner A, Reifenberg K, Glebe D, Roderfeld M, Roeb E. Genomic methylation inhibits expression of hepatitis B virus envelope protein in transgenic mice: A non-infectious mouse model to study silencing of HBV surface antigen genes. PLoS One. 2015;10:e0146099. doi: 10.1371/journal.pone.0146099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.