Abstract
Curcumin is an orange-yellow colored, lipophilic polyphenol substance derived from the rhizome of Curcuma longa that is widely used in many countries. Curcumin has many reported functions, including antioxidant and anti-inflammatory effects. Autophagy removes damaged organelles and protein aggregates in the cell. However, whether curcumin mediates its effects on neural stem cell (NSC) differentiation, cell cycle and apoptosis through autophagy is unknown. In the present study, the effects of curcumin and 3-methyladenine (3MA; an autophagy inhibitor, as a positive control) on the autophagy, differentiation, cell cycle progression and apoptosis of NSCs in different culture states were examined. In order to confirm the role of autophagy in these processes of NSC behavioral change, the protein expression level changes of markers of autophagy, such as autophagy-related protein 7 (Atg7), light chain (LC)3 and p62, were assessed. When NSCs were in an adherent state, 10 µM curcumin inhibited their differentiation into GFAP+ astrocytes or DCX+ immature neurons, while Atg7 and p62 protein expression were also reduced compared with the untreated control group. When NSCs were in a suspended state, 10 µM curcumin inhibited the cell cycle progression and apoptosis of NSCs as determined by western blotting, which was associated with a decreased autophagic flux and Atg7 expression. In addition, the curcumin-treated group trended in a similar direction to the 3MA-treated group. Thus, the data suggest that curcumin can inhibit differentiation, promote cell survival and inhibit cell cycle progression from G1 to S in NSCs, and that these effects are mediated through the regulation of Atg7 and p62.
Keywords: curcumin, cell cycle, neural stem cell, autophagy
Introduction
Curcumin (diferuloylmethane) is a natural product derived from the rhizome of Curcuma longa. Curcumin is one of the most common ingredients in Asian cuisine. Curcumin also has been found to have many potent effects, including anti-oxidative stress, anti-inflammatory, anti-proliferative, anticancer and neuroprotective properties (1–3). It was previously observed that curcumin enhanced the neurogenesis of neural progenitor cells by decreasing histone H3 and H4 acetylation (4) as well as stimulating developmental and adult hippocampal neurogenesis through the activation of ERK and p38 kinase (4,5). However, whether the effect of curcumin on NSCs is associated with autophagy has yet to be determined.
Autophagy is a highly regulated sequential process that delivers cytoplasmic macromolecules and damaged organelles to lysosomes for degradation, including mitochondria, endoplasmic reticulum, peroxisomes and misfolded proteins (6,7). Autophagy contributes to cell growth, cell development and cell homeostasis in normal conditions (7). Autophagic flux may be stimulated by multiple forms of cellular stress, including nutrient deprivation, energy limitations, hypoxia, toxins, radiation, DNA damage and intracellular pathogens (8,9).
In recent years, increasing evidence has supported that autophagy has a beneficial role in neurodegenerative disorders, including Huntington's, Alzheimer's, Parkinson's and amyotrophic lateral sclerosis (7,10,11). However, other evidence has indicated that excessive autophagy could contribute to neuronal death in cerebral ischemia (7,12–14). Furthermore, autophagy is also associated with cell aging, survival and proliferation (15–17).
In the present study, it was identified that curcumin actively suppressed the differentiation of NSCs into astrocytes and immature neurons while they were in adherent culture, and suppressed cell cycle progression and apoptosis in NSCs in suspension. Furthermore, transmission electron microscopy (TEM) revealed that the cytoplasm of the NSCs displayed autophagosomes following treatment with curcumin. Finally, it was identified that curcumin affected autophagy by inducing a decrease in autophagy-related gene (Atg)7 and p62 protein expression in NSCs in different culture states.
Materials and methods
Preparation of NSCs
NSCs were prepared from pregnant female Sprague-Dawley rats supplied by the Animal Breeding Center of Chinese Academy of Sciences (Shanghai, China) according to the method previously established by our group (18,19). All animal-related procedures were approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University (Wenzhou, China), and were conducted in accordance with the university's guidelines.
Briefly, embryonic cerebral cortices were collected and dissected from embryonic day 14.5 (n=8; E14.5) rats. The cells were isolated by mechanical pipetting with a fire-polished Pasteur pipette. The suspension was filtered through a 70-µm nylon mesh, seeded into a T25 Corning tissue culture flask (Corning Incorporated, Corning, NY, USA) at a density of 105 cells/ml and incubated at 37°C in a humidified, 5% CO2 atmosphere. The culture medium was composed of DMEM/F12, B27, N2 (all Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100× Penicillin-Streptomycin Solution (Beyotime Institute of Biotechnology, Haimen China), heparin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), HEPES and glutamine, with 20 ng/ml epidermal growth factor (EGF) and 20 ng/ml basic fibroblast growth factor (bFGF; Gibco; Thermo Fisher Scientific, Inc.) added, which is henceforth described as proliferation medium. At day 5, neurospheres were collected and passaged. Passage 2 NSCs were used for the subsequent assays.
To induce the differentiation of NSCs, dissociated cells in a single-cell suspension were seeded onto poly-L-lysine (cat. no. P-2636; Sigma-Aldrich; Merck KGaA) coated coverslips in a 24-well plate at a density of 5×104/coverslip. Differentiation medium contained 1% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) without EGF or bFGF. The cultures were allowed to differentiate for 6 days prior to being fixed for immunostaining.
The adherent and suspension cultures of NSCs were incubated in differentiation medium or proliferation medium, respectively, for 72 h prior to the preparation of protein extracts for western blotting. The medium was changed every 48 h.
Antibodies and reagents
Glial fibrillary acidic protein (GFAP) was detected by chicken polyclonal anti-GFAP (cat. no. AB5541; Merck KGaA). Other antibodies were as follows: Rabbit polyclonal anti-light chain (LC)3B (cat. no. 2775), rabbit monoclonal anti-Atg7 (D12B11; cat. no. 8558), anti-cyclin dependent kinase (CDK)2 (78B2; cat. no. 2546; Cell Signaling Technology, Inc.) and anti-p62 (cat. no. P0067), mouse monoclonal anti-nestin (cat. no. N5413) (both Sigma-Aldrich; Merck KGaA), anti-βIII-tubulin (cat. no. AB15708; Merck KGaA), anti-O4 (cat.no. O7139; Sigma-Aldrich; Merck KGaA), anti-β-actin (cat. no. sc-47778; Santa Cruz Biotechnology, Inc.), anti-tubulin βIII (cat. no. MAB1637; Merck KGaA), anti-doublecortin (DCX; cat. no. ab2253) and anti-caspase-3 antibody (cat. no. 9662; Cell Signaling Technology, Inc.). Curcumin (cat. no. C1386-5G) and monodansylcadaverine (MDC) were purchased from Sigma-Aldrich; Merck KGaA (cat. no. D4008-100MG). 3-Methyladenine (3MA, an inhibitor of phosphatidylinositol 3-kinases and autophagosome formation; cat. no. s2767) was purchased from Selleck Chemicals (Shanghai, China).
Drug treatment
Curcumin, MDC and 3MA stocks of 1 mg/ml were prepared in dimethylsulfoxide (DMSO) and stored at −20°C, in the dark. Subsequent to passaging, NSCs were treated with 10 µM curcumin or 10 µM 3MA for 72 h by adding the stock solutions to the medium. Then, they were fixed in 4% paraformaldehyde for 15 min, and processed for immunostaining as subsequently described. The 3MA group was used as a positive control.
Determination of cell viability
Cell viability was determined by a water-soluble tetrazolium salts (WST-1 Cell Proliferation and Cytotoxicity Assay kit; Beyotime Institute of Biotechnology) assay (20). Neural stem cells in suspension were seeded at a density of 2×104 cells/well into a 96-well plate. Following treatment with curcumin for 72 h, WST-1 was added to each well and incubated for 2 h at 37°C. The optical density was measured at 450 nm using a microplate reader. The data were presented as the mean from four independent experiments in quadruplicate.
Immunocytochemistry assay
Immunofluorescence was used to characterize the differentiation of NSCs in vitro, as described previously (18,19). Briefly, cells on poly-L-lysine coated cover-slips were fixed with 4% paraformaldehyde for 10 min at room temperature (RT), washed and stored in 0.01 mol/l PBS (pH 7.4). The sections of neurospheres or cell culture were blocked in 10% goat serum (cat. no. S26-100 ML; Merck KGaA) in PBS (for O4) or 0.3% Triton X-100-containing 10% goat serum in PBS (for GFAP, βIII-tubulin and nestin) for 1 h at RT, and incubated with the following primary antibodies overnight at 4°C: Monoclonal mouse antibodies against nestin (dilution, 1:800) for NSCs, βIII-tubulin (dilution, 1:400) for neurons, GFAP (dilution, 1:500) for astrocytes and O4 (dilution, 1:100) for oligodendrocytes. After washing three times with PBS, the cultures were incubated with rhodamine-conjugated goat anti-mouse or DyLight 488 (cat. no. 611-545-215) or 594-conjugated goat anti-rabbit antibodies (cat. no. 111-005-047) (dilution, 1:150; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 1 h at 37°C and washed three times with PBS. Subsequently, the cells were incubated with Hoechst 33258 (Beyotime Institute of Biotechnology) for 3 min at RT to stain the nuclei. Finally, the coverslips were mounted onto slides in 70% glycerol. Stained sections were observed and scanned under a fluorescence microscope (Olympus BX53; Olympus Corporation, Tokyo, Japan).
MDC staining
Neural stem cells were seeded on slides. Cells were incubated and treated with curcumin or 3MA for 72 h in proliferation medium, finally incubated with 0.05 mM MDC for 1 h at 37°C, and then washed four times with PBS (pH 7.4) (21). Cells were immediately visualized with a fluorescence microscope (Olympus BX53; Olympus Corporation). The fluorescence intensity values of all groups were digitally quantified using ImageJ image analyzer software (version 1.45; National Institutes of Health, Bethesda, MD, USA).
Transmission electron microscopy (TEM)
NSCs were seeded in 100-mm dishes. Cells were incubated and treated with curcumin or 3MA for 72 h in the proliferation medium. At the end of incubation, cell monolayers were washed with PBS and scraped gently with a plastic cell scraper. The harvested cells were pelleted by centrifugation at 13,000 × g for 10 min, and fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in cacodylate buffer. After rinsing with cacodylate buffer, the samples were post-fixed in 2% osmium tetroxide for 1 h. The samples were rinsed with water and dehydrated in a graded alcohol series (50, 75, 80 and 100%). The samples were then embedded in epoxy resin. Representative areas were chosen for ultra-thin sectioning, and viewed with a Hitachi 7000 STEM transmission electron microscope (Hitachi, Ltd., Tokyo, Japan) (6).
Western blot analysis
Cells were plated at a density of 1×106 cells per 100-mm culture dish and treated with curcumin or 3MA as previously described. Cells were washed twice with PBS and lysed in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1.0 mM Na3VO4, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 100 µg/ml phenylmethylsulfonyl fluoride, 30 µl/ml aprotinin and 4 µg/ml leupeptin, pH 7.5). Lysates were centrifuged and the supernatants diluted in sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 50 mM DTT and 0.1% bromophenol blue), then boiled for 5 min. Equal amounts of protein were resolved on 12% SDS-PAGE and transferred to polyvinylidene fluoride membranes. The membranes were blocked at RT for 1 h in 5% (w/v) dry skim milk in TBS plus 0.1% Tween-20 (TBST), rinsed in TBST and incubated with primary antibodies at 4°C overnight. The primary antibodies used were mouse antibodies specific for β-actin, βIII-tubulin, GFAP, caspase-3 DCX, p62, Atg7, LC3B and CDK2 (all dilution, 1:1,000). After rinsing, blots were incubated in TBST with peroxidase-conjugated secondary antibodies at RT for 1 h. The secondary antibodies used included goat anti-mouse IgG and goat anti-rabbit IgG (both dilution, 1:1,000). The peroxidase reaction was visual-ized with an enhanced chemiluminescence reagent. Films were digitized and densitometry was performed using ImageJ software.
Statistical analysis
Data were analyzed by one-way analysis of variance followed by Dunnett's post hoc test to determine whether there were significant differences between individual groups. All test assumptions regarding distribution and variance were met for each data set. All analyses were based on biological replicates (n=3) from the same independent experiment, not technical replicates or combined experiments. P<0.05 was considered to represent a statistically significant difference.
Results
NSCs can be successfully isolated while retaining their differentiation ability
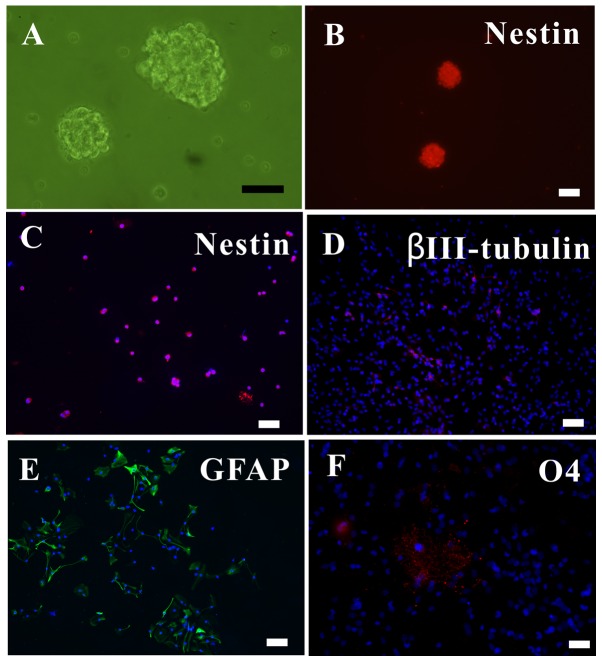
The in vitro cells proliferated to form neurospheres, which were observed under an inverted microscope (Fig. 1A). As nestin is a marker of NSCs (19), the nestin+ neurospheres and dissociated single cells were observed with immunofluorescence (Fig. 1B and C).
Figure 1.
Growth and fate of newly differentiated NSCs in vitro. (A) NSC neurospheres. (B) Neurospheres or (C) single NSCs in growth medium supplemented with 20 ng/ml EGF and bFGF, expressing Nestin. With 1% fetal bovine serum instead of EGF and bFGF, NSCs differentiated into (D) βIII-tubulin+ neurons, (E) GFAP+ astrocytes and (F) O4+ oligodendrocytes. Scale bar, 50 µm. NSC, neural stem cells; EGF, epidermal growth factor; bFGF, basic fibroblast growth factor.
To ascertain the differentiation ability of the NSCs, the NSCS were subjected to differentiating media for 5 days, after which they were immunolabeled for neurons (βIII-tubulin+; Fig. 1D), astrocytes (GFAP+; Fig. 1E) and oligodendrocytes (O4+; Fig. 1F). Thus, it was verified that the extracted NSCs could proliferate, self-renew and differentiate into the three major neural lineages.
Curcumin (10 µM) enhances NSC viability
In order to investigate the effect of different curcumin concentrations on NSCs in vitro, cell viability was determined by a WST-1 assay. NSCs were treated with 0, 5, 10, 20 and 50 µM curcumin. At 72 h, the viability of NSCs treated with 10 µM curcumin was significantly different compared with NSCs treated with 0 µM (P<0.05; Fig. 2), while differences among the 0, 5, 20 and 50 µM groups were all non-significant (P>0.05; Fig. 2). These results demonstrate that a 10-µM dose of curcumin can promote the proliferation of NSCs in vitro. Thus, a dose of 10 µM curcumin was selected for the following studies.
Figure 2.
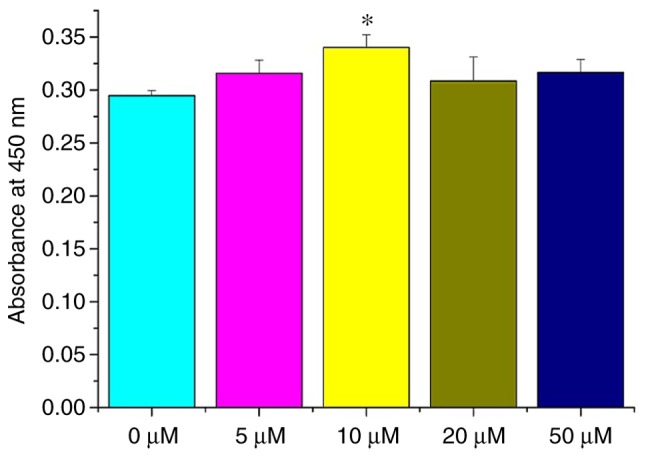
Effect of curcumin on NSC viability and cytotoxicity. At 72 h, 10 µM curcumin increased the viability of NSCs in vitro. *P<0.05 vs. 0 µM. NSC, neural stem cells.
Curcumin inhibits the differentiation of adherent NSCs by decreasing Atg7 and p62 expression
It was investigated whether treating with curcumin would result in changes in cell differentiation using immunocytochemistry and western blot analyses. After 3 days culture in differentiation medium, treatment with curcumin evidently decreased the number of NSCs differentiating into GFAP+ astrocytes (Fig. 3A–D) and DCX+ immature neurons compared with in the control group. However, βIII-tubulin+ expression could not be detected in all groups (Fig. 3D). The result of curcumin treatment was similar to that of 3MA treatment.
Figure 3.
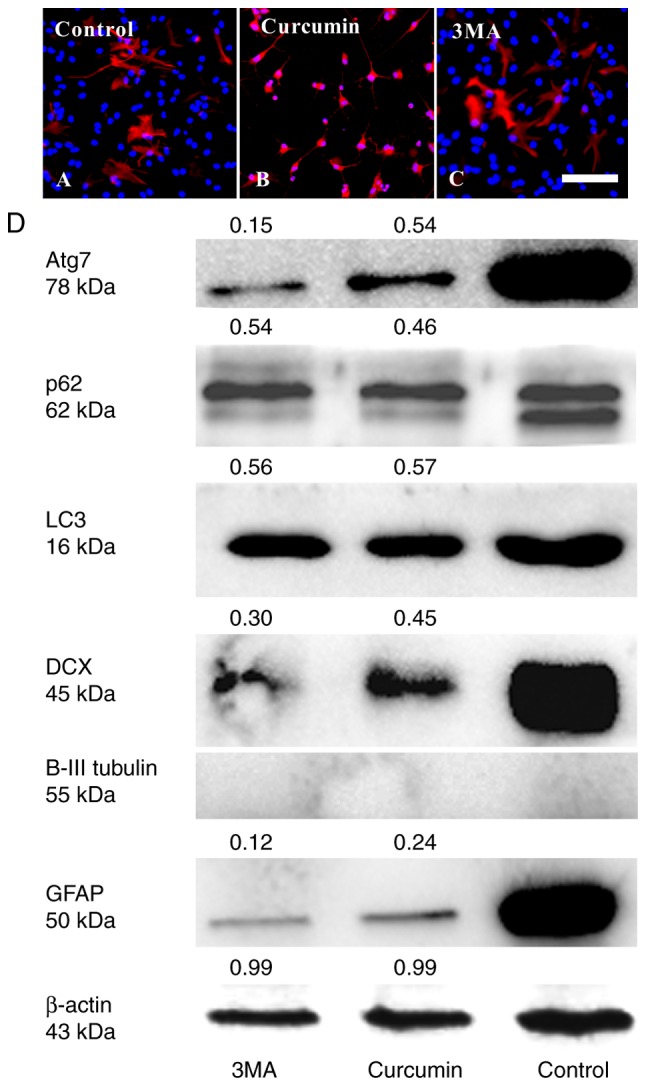
Effect of curcumin treatment on the differentiation of NSCs in adherent state. NSCs were treated as indicated for 72 h. Cells were stained with immunofluorescence and visualized by fluorescence microscopy, including (A) control, (B) curcumin and (C) 3MA-treated cells. (D) Cells were lysed and used for western blot analysis. β-actin was used as the loading control. The relative level of protein was quantified by ImageJ. The experiments were repeated three times. NSC, neural stem cells.
To further validate the role of autophagy in the effect of curcumin on NSC differentiation, changes in the expression of autophagy markers, including LC3, Atg7 and p62, were determined (Fig. 3D). LC3, is an essential component of autophagosomes widely used as autophagy marker. The degree of conversion of cytosolic LC3I to membrane bound LC3II indicates the level of autophagic activity (6). It was identified that the total LC3 (16 kDa) in the curcumin and 3MA groups was lower than in the control group. In addition, the LC3II isoform was not detected in any of the groups. As total LC3 is not a good marker for autophagy (22), it was not possible to ascertain whether NSC differentiation was associated with LC3. Another approach is to detect the Atg7 and p62 expression levels. Atg7 is essential for the early elongation and closure of the autophagosomal membrane (9,23). The level of p62 degradation is used to detect autophagic flux (24); p62 accumulates when autophagy is inhibited, and decreased levels are observed when autophagy is induced (25). Therefore, the reduction in the number of GFAP+ astrocytes and DCX+ immature neurons may be due to differences in the protein levels of Atg7 and p62.
Following exposure to 3MA or curcumin, Atg7 and p62 levels were much lower than in the untreated control group (Fig. 3D). 3MA is a known inhibitor of type III phosphatidylinositol 3-kinase (PI3K) and autophagy induction. Thus, the results preliminarily indicated that curcumin may have inhibited the differentiation of NSCs through PI3K inhibition (26,27) or decreasing the protein levels of Atg7 and p62.
Curcumin inhibits the cell cycle progression and apoptosis of suspended NSCs by decreasing Atg7 and increasing p62
The effect of treatment with curcumin on the cell cycle and apoptosis of NSCs was then considered. As NSCs also express GFAP (28), there was no change in the expression of GFAP protein between the three groups (Fig. 4).
Figure 4.
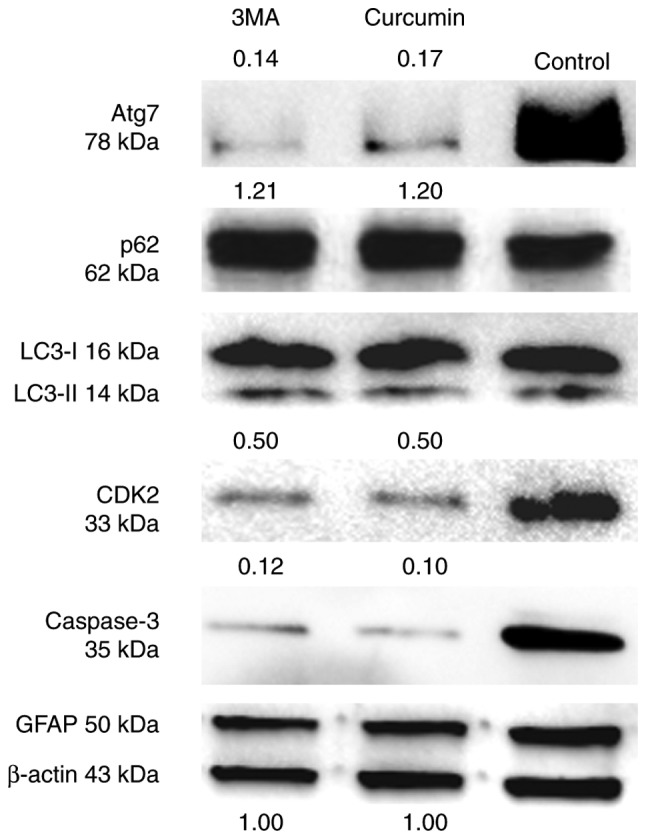
Effect of curcumin treatment on the cell cycle and apoptosis of NSCs in suspended state. NSCs were treated with control, curcumin and 3MA for 72 h. β-actin was used as the loading control. The relative level of protein was quantified by ImageJ. Experiments were repeated three times. NSC, neural stem cells; 3MA, 3-methyladenine.
CDK2 is a marker of the cell cycle required during S phase (29), and caspase-3 is a critical executioner of apoptosis. The results suggested that curcumin or 3MA could markedly decrease the protein level of these two markers compared with untreated cells (Fig. 4). Additionally, the changes in the expression of autophagy markers in suspended NSCs was assessed. As identified through western blot analysis, LC3I and II protein levels were unchanged between the three groups. However, Atg7 protein levels were much lower following treatment with curcumin or 3MA compared with the control group. The level of p62 protein was slightly increased in the curcumin and 3MA treatment groups compared with the control group (Fig. 4). Taken together, these findings indicate that curcumin can affect the progression of NSCs from G1 to S phase, and prevent their apoptosis. Furthermore, they suggest that increased autophagic flux and decreased Atg7 expression are involved in the process of NSC S-phase arrest and reduced apoptosis.
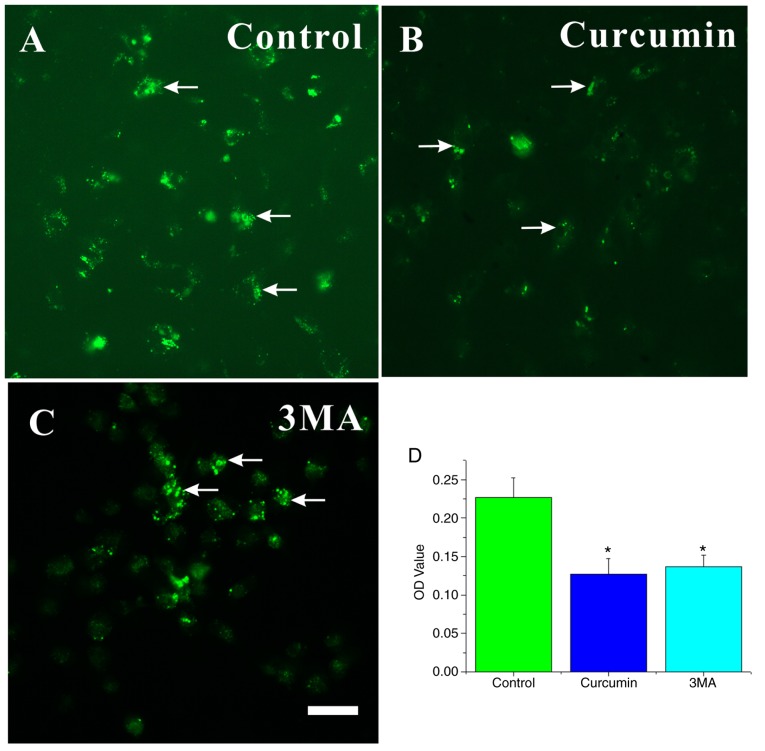
Autophagic vacuoles (AVOs) decreased in curcumin-treated NSCs
MDC is a specific marker for autolysosomes and AVOs, which appear as spherical structures in the cytoplasm and the perinuclear region (30). Thus, the MDC staining of AVOs was used to assess the extent of autophagy in the cells. Large dots indicative of AVOs appeared in the cytoplasm of NSCs in the control group (Fig. 5A). The fluorescence intensity of MDC in NSCs significantly decreased in the curcumin- or 3MA-treated groups compared with the control group (P<0.05; Fig. 5B–D).
Figure 5.
MDC-labeled AVOs occur in curcumin- and 3MA treated NSCs. Characterization of the MDC-labeled vacuoles. NSCs were seeded on slides. Cells were incubated in proliferation medium and (A) left untreated or treated with 10 µM (B) curcumin or (C) 3MA for 72 h, and then stained with 0.05 mM MDC. (D) Cells were mounted and analyzed by fluorescence microscopy. The fluorescence intensity values (OD value) of all groups were digitally quantified .using ImageJ. The results are presented as the mean ± standard deviation (n=3). Scale bar, 25 µm. *P<0.05 compared with control. MDC, monodansylcadaverine; AVO, autophagic vacuoles; 3MA, 3-methyladenine; NSC, neural stem cells.
TEM demonstrated autophagosome formation in curcumin-treated NSCs
In order to study the ultrastructural changes of NSCs treated with curcumin or 3MA, TEM was performed to identify AVOs, as previously described (31). AVOs containing extensively degraded organelles, including mitochondria and endoplasmic reticulum, were detected in the cytoplasm of NSCs (Fig. 6A). In contrast, NSCs treated with curcumin or 3MA had relatively few autophagosomes in the cytoplasm (Fig. 6B and C). These results suggest that curcumin treatment affected autophagy initiation in NSCs.
Figure 6.
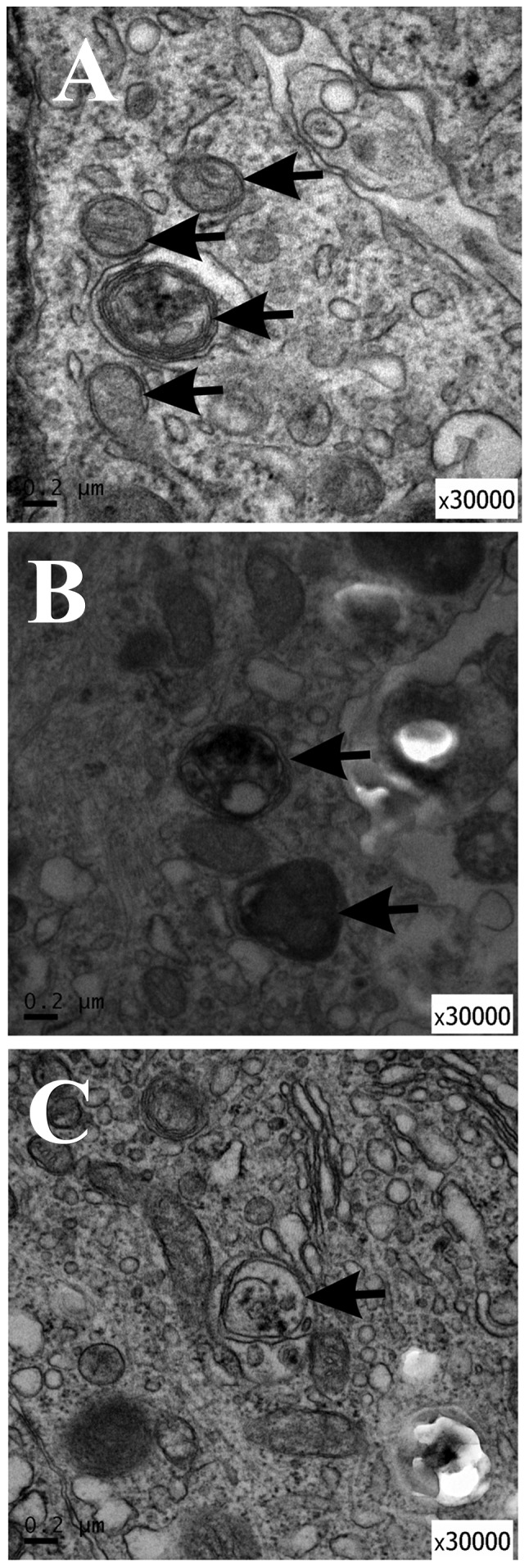
Ultrastructural features of autophagy in NSCs treated with curcumin or 3MA for 72 h. (A) Negative control (untreated) cells; (B) NSCs treated with 10 µM curcumin; (C) NSCs treated with 10 µM 3MA, as a positive control. The autophagic vacuoles are indicated by arrowheads. Scale bar, 0.2 µM; magnification, ×30,000. NSC, neural stem cells; 3MA, 3-methyladenine.
Discussion
Curcumin has been used for many centuries as a traditional agent in treating inflammatory diseases and other ailments. Curcumin is reported to contribute to the proliferation and neurogenesis of NSCs (5). Additionally, curcumin can promote the differentiation of glioma-initiating cells by inducing autophagy (32) and induce glioma stem-like cell formation (33). However, the role and mechanism of curcumin in NSCs remains to be characterized. In the present study, curcumin treatment prevented the differentiation of NSCs in the adherent state via a reduction in p62 expression. Curcumin treatment impeded cell cycle progression and reduced the rate of apoptosis by decreasing the Atg7 protein level and increasing the autophagic flux of p62. Taken together, these data strongly suggest that curcumin inhibits the differentiation and cell cycle of NSCs by adjusting Atg7 and p62 protein levels. It thus appears that the outcomes of curcumin treatment may differ depending on the conditions of culture.
In the present study, the effect of curcumin on the survival of NSCs in vitro was assessed using the WST method. The viability of NSCs was higher following treatment with 10 µM curcumin compared with the control group. No cytotoxicity was observed following treatment with 10 or 20 µM curcumin. Additionally, the caspase-3 protein level decreased in suspended NSCs treated with curcumin as detected by western blotting. Caspase-3 is activated in apoptotic cells. Therefore, the data showed that curcumin may prevent apoptosis and promote NSC survival.
However, CDK2, a regulator of cell cycle progression from G1 to S phase, was also reduced in suspended NSCs. This is not contrary to the WST results, as cellular proliferation may be not associated with CDK2 (34–36). Autophagy exerts a major influence on the G1 and S phases of the cell cycle (37). For example, a previous study has indicated that Atg7 is required for the p53-dependent expression of p21CDKN1A and the cell cycle arrest of mouse embryonic fibroblasts starved of serum and amino acids (38). However, another study reported that the knockdown of Atg7 specifically increased p27 protein abundance; p27 is a CDK inhibitor that prevents cell proliferation (39). These two studies suggest opposing roles for Atg7 in the cell cycle. Accordingly, we hypothesize that curcumin promotes NSC survival through reducing Atg7 to decrease the expression of CDK inhibitors.
Five CDKs active in the cell cycle have been identified so far, including during G1 (CDK4, 6 and 2), S (CDK2), G2 and M (CDK1). The present study has demonstrated that curcumin treatment may inhibit G1-to-S progression by downregulating CDK2. However, whether other factors affecting the cell cycle, including CDK1, 4 and 6, and CDK inhibitors, are also involved in the effect of curcumin on the NSC cell cycle is unclear. Further research in this area is required to fully understand the mechanisms of the effect.
Accumulating data have confirmed that the association between autophagy and apoptosis is complex. Caspases can cleave various autophagy-related proteins, and the cleaved fragments generated have different functional activities and cellular localization (40). Caspase-8 contributes to the cleavage of Atg3, preventing its pro-autophagic activity (41), whereas caspase-9 can interact with Atg7 to facilitate autophagy (42). A recent study has also indicated that caspase-3 has both anti- and pro-autophagic effects (43). The knockdown of Atg12 leads to a marked inhibition in caspase activity, including that of caspase-3 (44). The present study revealed that curcumin could decrease Atg7 protein level and downregulate caspase-3 expression, indicating that a decrease in Atg7 may have led to the inhibition of caspase-3, potentially impairing Atg7-mediated autophagosome formation. These data are in line with a previous study (42). It was hypothesized that an increase in p62 level may decrease the sensitivity of NSCs to caspase-3.
A previous study identified that the inhibition of autophagy through the deletion of Atg5, Atg16L1 or Atg7 did not impair the maintenance and differentiation of postnatal NSCs, whereas p62 accumulation promoted the apoptosis of autophagy-deficient NSCs by increasing the superoxide concentration (45). In the present study, it was shown that curcumin treatment prevented the differentiation of NSCs into astrocytes or immature neurons accompanied by a reduction in the Atg7 and p62 protein levels. Therefore, we hypothesize that Atg7 or p62 may be involved in the effect on NSC differentiation mediated by curcumin. p62 expression can prevent oxidative stress (46,47) and be used to detect the state of reactive oxygen species buffering systems (48). On the other hand, H2O2 exposure can increase the neurogenesis and oligodendrogenesis of NSCs (49), and curcumin has anti-oxidative effects (50); thus, it was hypothesized that the oxidative stress and p62 protein expression decrease induced by curcumin treatment may result in the dysfunction of NSC differentiation.
Consistent with previous research (9), for NSCs in a suspended culture state, the levels of p62 increased, whereas the Atg7 levels decreased, compared with untreated control cells. An increase in LC3-II is not a measure of autophagic flux per se, since it can also indicate the inhibition of autophagosome clearance (51). However, there were no differences in LC3-II expression among the control and the curcumin- and 3MA-treated groups. Thus, the results demonstrated that curcumin could enhance the autophagic flux of p62 and suppress the Atg7 protein level in suspended NSCs.
The MDC-labeled vesicles were also assessed in the groups. A previous study has indicated that MDC-labeled vesicles are not exclusively autolysosomes, and that MDC labels any acidic compartment (21). In addition, MDC dots can still be detected in Atg5−/− mouse embryonic stem cells (52). Therefore, TEM was also applied; the TEM results were consistent with the MDC-labeling.
It was previously identified that NSC dynamics can be modulated by different ion channels, such as K+, Na+, Cl+ and TRP channels (53). Moreover, curcumin also affects the functions of these channels (54,55). Thus, curcumin may also affect NSC differentiation, proliferation and apoptosis through an effect on these channels.
In summary, to the best of our knowledge, it was demonstrated for the first time that curcumin inhibited the differentiation, cell cycle progression and apoptosis of NSCs through modulating the expression of Atg7 and p62 in vitro. However, the results also suggest that effect of curcumin may be dependent on the cell culture state. p62, a marker of autophagic flux, was evidently decreased in adherent NSCs and increased in suspended NSCs. 3MA, used as a positive control as an inhibitor of autophagy, induced similar effects to curcumin. Thus, we hypothesize that curcumin may also affect PI3K in NSCs. However, the results of the present study were preliminary; it is not yet possible to be certain whether Atg7 modulation mediated the effects of curcumin intervention. Employing gene overexpression and knockdown would further demonstrate the roles of Atg7 and p62 in the effects of curcumin treatment. Furthermore, the connections between Atg7 and p53 in the effect of curcumin on NSCs will need to be considered in the future.
Acknowledgments
Not applicable.
Funding
The study was supported by the Zhejiang Provincial Natural Science Foundation of China (grant nos. LY18H090013 and LY14F020035) and Wenzhou Municipal Science and Technology Bureau Fund (grant no. Y20170165).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JJW and CJX analyzed the data; JLW, ZNC, CJX performed the experiments and data analysis; CJX wrote the manuscript; JJW and CJX designed the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal-related procedures were approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University (Wenzhou, China), and were conducted in accordance with the university's guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: Role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72:29–39. doi: 10.1124/mol.106.033167. [DOI] [PubMed] [Google Scholar]
- 2.Wang R, Li Y, Li Y, Xu Y, Wu H, Li X. Curcumin protects against glutamate excitotoxicity in rat cerebral cortical neurons by increasing brain-derived neurotrophic factor level and activating. TrkB Brain Res. 2008;1210:84–91. doi: 10.1016/j.brainres.2008.01.104. [DOI] [PubMed] [Google Scholar]
- 3.Ramadan G, Al-Kahtani MA, El-Sayed WM. Anti-inflammatory and anti-oxidant properties of curcuma longa (turmeric) versus zingiber officinale (ginger) rhizomes in rat adjuvant-induced arthritis. Inflammation. 2011;34:291–301. doi: 10.1007/s10753-010-9278-0. [DOI] [PubMed] [Google Scholar]
- 4.Kang SK, Cha SH, Jeon HG. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. Stem Cells Dev. 2006;15:165–174. doi: 10.1089/scd.2006.15.165. [DOI] [PubMed] [Google Scholar]
- 5.Kim SJ, Son TG, Park HR, Park M, Kim MS, Kim HS, Chung HY, Mattson MP, Lee J. Curcumin stimulates proliferation of embryonic neural progenitor cells and neurogenesis in the adult hippocampus. J Biol Chem. 2008;283:14497–14505. doi: 10.1074/jbc.M708373200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long DX, Hu D, Wang P, Wu YJ. Induction of autophagy in human neuroblastoma SH-SY5Y cells by tri-ortho-cresyl phosphate. Mol Cell Biochem. 2014;396:33–40. doi: 10.1007/s11010-014-2139-7. [DOI] [PubMed] [Google Scholar]
- 7.Shi R, Weng J, Zhao L, Li XM, Gao TM, Kong J. Excessive autophagy contributes to neuron death in cerebral ischemia. CNS Neurosci Ther. 2012;18:250–260. doi: 10.1111/j.1755-5949.2012.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez-Puerto MC, Folkerts H, Wierenga ATJ, Schepers K, Schuringa JJ, Coffer PJ, Vellenga E. Autophagy proteins ATG5 and ATG7 are essential for the maintenance of human CD34+ hematopoietic stem-progenitor cells. Stem Cells. 2016;34:1651–1663. doi: 10.1002/stem.2347. [DOI] [PubMed] [Google Scholar]
- 10.Ouyang L, Zhang L, Zhang S, Yao D, Zhao Y, Wang G, Fu L, Lei P, Liu B. Small-molecule activator of UNC-51-like kinase 1 (ULK1) that induces cytoprotective autophagy for Parkinson's disease treatment. J Med Chem. 2018;61:2776–2792. doi: 10.1021/acs.jmedchem.7b01575. [DOI] [PubMed] [Google Scholar]
- 11.Karabiyik C, Lee MJ, Rubinsztein DC. Autophagy impairment in Parkinson's disease. Essays Biochem. 2017;61:711–720. doi: 10.1042/EBC20170023. [DOI] [PubMed] [Google Scholar]
- 12.Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu, Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281:30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 13.Fornai F, Longone P, Ferrucci M, Lenzi P, Isidoro C, Ruggieri S, Paparelli A. Autophagy and amyotrophic lateral sclerosis: The multiple roles of lithium. Autophagy. 2008;4:527–530. doi: 10.4161/auto.5923. [DOI] [PubMed] [Google Scholar]
- 14.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: An immunoelectron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 15.Cianfanelli V, Cecconi F. AMBRA1: When autophagy meets cell proliferation. Autophagy. 2015;11:1705–1707. doi: 10.1080/15548627.2015.1053681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Revuelta M, Matheu A. Autophagy in stem cell aging. Aging Cell. 2017;16:912–915. doi: 10.1111/acel.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Codogno P, Meijer AJ. Autophagy and signaling: Their role in cell survival and cell death. Cell Death Differ. 2005;2(Suppl 12):S1509–S1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C, Wu JM, Liao M, Wang JL, Xu CJ. The ROCK/GGtase pathway are essential to the proliferation and differentiation of neural stem cells mediated by simvastatin. J Mol Neurosci. 2016;60:474–485. doi: 10.1007/s12031-016-0811-y. [DOI] [PubMed] [Google Scholar]
- 19.Xu CJ, Xu L, Huang LD, Li Y, Yu PP, Hang Q, Xu XM, Lu PH. Combined NgR vaccination and neural stem cell transplantation promote functional recovery after spinal cord injury in adult rats. Neuropathol Appl Neurobiol. 2011;37:135–155. doi: 10.1111/j.1365-2990.2010.01117.x. [DOI] [PubMed] [Google Scholar]
- 20.Peng ZW, Xue F, Wang HN, Zhang RG, Chen YC, Wang Y, Zhang LY, Fan J, Tan QR. Paroxetine up-regulates neurogenesis in hippocampus-derived neural stem cell from fetal rats. Mol Cell Biochem. 2013;375:105–113. doi: 10.1007/s11010-012-1533-2. [DOI] [PubMed] [Google Scholar]
- 21.Vázquez CL, Colombo MI. Chapter 6 assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol. 2009;452:85–95. doi: 10.1016/S0076-6879(08)03606-9. [DOI] [PubMed] [Google Scholar]
- 22.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 23.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 26.Hamzehzadeh L, Atkin SL, Majeed M, Butler AE, Sahebkar A. The versatile role of curcumin in cancer prevention and treatment: A focus on PI3K/AKT pathway. J Cell Physiol. 2018;233:6530–6537. doi: 10.1002/jcp.26620. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Zhu R. Curcumin suppresses the progression of laryngeal squamous cell carcinoma through the upregulation of miR-145 and inhibition of the PI3K/Akt/mTOR pathway. Onco Targets Ther. 2018;11:3521–3531. doi: 10.2147/OTT.S159236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed AI, Shtaya AB, Zaben MJ, Owens EV, Kiecker C, Gray WP. Endogenous GFAP-positive neural stem/progenitor cells in the postnatal mouse cortex are activated following traumatic brain injury. J Neurotrauma. 2012;29:828–842. doi: 10.1089/neu.2011.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2010;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munafó DB, Colombo MI. A novel assay to study autophagy: Regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114:3619–3629. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- 31.Gukovskaya AS, Gukovsky I. Autophagy and pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2012;303:G993–G1003. doi: 10.1152/ajpgi.00122.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuang W, Long L, Zheng B, Ji W, Yang N, Zhang Q, Liang Z. Curcumin promotes differentiation of glioma-initiating cells by inducing autophagy. Cancer Sci. 2012;103:684–690. doi: 10.1111/j.1349-7006.2011.02198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi L, Wang Z, Sun G. Curcumin induces glioma stem-like cell formation. Neuroreport. 2015;26:167–172. doi: 10.1097/WNR.0000000000000320. [DOI] [PubMed] [Google Scholar]
- 34.Méndez J. Cell proliferation without cyclin E-CDK2. Cell. 2003;114:398–399. doi: 10.1016/S0092-8674(03)00649-4. [DOI] [PubMed] [Google Scholar]
- 35.Tetsu O, Mccormick F. Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell. 2003;3:233–245. doi: 10.1016/S1535-6108(03)00053-9. [DOI] [PubMed] [Google Scholar]
- 36.Kelly TJ, Brown GW. Regulation of chromosome replication. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 37.Tasdemir E, Maiuri MC, Tajeddine N, Vitale I, Criollo A, Vicencio JM, Hickman JA, Geneste O, Kroemer G. Cell cycle-dependent induction of autophagy, mitophagy and reticulophagy. Cell Cycle. 2007;6:2263–2267. doi: 10.4161/cc.6.18.4681. [DOI] [PubMed] [Google Scholar]
- 38.Lee IH, Kawai Y, Fergusson MM, Rovira II, Bishop AJ, Motoyama N, Cao L, Finkel T. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–228. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J, Yang L, Tian Z, Hua X, Gu J, Li J, Liu C, Jin H, Wang Y, Jiang G, et al. ATG7 overexpression is crucial for tumorigenic growth of bladder cancer in vitro and in vivo by targeting the ETS2/miRNA196b/FOXO1/p27 axis. Mol Ther Nucleic Acids. 2017;7:299–313. doi: 10.1016/j.omtn.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ojha R, Ishaq M, Singh SK. Caspase-mediated crosstalk between autophagy and apoptosis: Mutual adjustment or matter of dominance. J Cancer Res Ther. 2015;11:514–524. doi: 10.4103/0973-1482.163695. [DOI] [PubMed] [Google Scholar]
- 41.Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, Gozuacik D. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis. 2012;17:810–820. doi: 10.1007/s10495-012-0735-0. [DOI] [PubMed] [Google Scholar]
- 42.Han J, Hou W, Goldstein LA, Stolz DB, Watkins SC, Rabinowich H. A complex between atg7 and caspase-9: A novel mechanism of cross-regulation between autophagy and apoptosis. J Biol Chem. 2014;289:6485–6497. doi: 10.1074/jbc.M113.536854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang JL, Chow JM, Chang JH, Wen YC, Lin YW, Yang SF, Lee WJ, Chien MH. Quercetin simultaneously induces G0/G1-phase arrest and caspase-mediated crosstalk between apoptosis and autophagy in human leukemia HL-60 cells. Environ Toxicol. 2017;32:1857–1868. doi: 10.1002/tox.22408. [DOI] [PubMed] [Google Scholar]
- 44.Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The autophagy protein atg12 associates with antiapoptotic BCL-2 family members to promote mitochondrial apoptosis. Mol Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Wang C, Chen S, Yeo S, Karsli-Uzunbas G, White E, Mizushima N, Virgin HW, Guan JL. Elevated p62/SQSTM1 determines the fate of autophagy-deficient neural stem cells by increasing superoxide. J Cell Biol. 2016;212:545–560. doi: 10.1083/jcb.201507023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taniguchi K, Yamachika S, He F, Karin M. p62/SQSTM1-Dr. Jekyll and Mr. Hyde that prevents oxidative stress but promotes liver cancer. FEBS Lett. 2016;590:2375–2397. doi: 10.1002/1873-3468.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Cano M, Handa JT. p62 provides dual cytoprotection against oxidative stress in the retinal pigment epithelium. Biochim Biophys Acta. 2014;1843:1248–1258. doi: 10.1016/j.bbamcr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carroll B, Otten EG, Manni D, Stefanatos R, Menzies FM, Smith GR, Jurk D, Kenneth N, Wilkinson S, Passos JF, et al. Oxidation of SQSTM1/p62 mediates the link between redox state and protein homeostasis. Nat Commun. 2018;9:256. doi: 10.1038/s41467-017-02746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez Estrada C, Covacu R, Sankavaram SR, Svensson M, Brundin L. Oxidative stress increases neurogenesis and oligodendrogenesis in adult neural progenitor cells. Stem Cells Dev. 2014;23:2311–2327. doi: 10.1089/scd.2013.0452. [DOI] [PubMed] [Google Scholar]
- 50.Panchal HD, Vranizan K, Lee CY, Ho J, Ngai J, Timiras PS. Early anti-oxidative and anti-proliferative curcumin effects on neuroglioma cells suggest therapeutic targets. Neurochem Res. 2008;33:1701–1710. doi: 10.1007/s11064-008-9608-x. [DOI] [PubMed] [Google Scholar]
- 51.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 52.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Yasuda T, Adams DJ. Physiological roles of ion channels in adult neural stem cells and their progeny. J Neurochem. 2010;114:946–959. doi: 10.1111/j.1471-4159.2010.06822.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Chen Q, Wang Y, Peng W, Cai H. Effects of curcumin on ion channels and transporters. Front Physiol. 2014;5:94. doi: 10.3389/fphys.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nalli M, Ortar G, Schiano Moriello A, Di Marzo V, De Petrocellis L. Effects of curcumin and curcumin analogues on TRP channels. Fitoterapia. 2017;122:126–131. doi: 10.1016/j.fitote.2017.09.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.