Abstract
Cysteine sulfinic acid or S-sulfinylation is an oxidative post-translational modification (OxiPTM) that is known to be involved in redox-dependent regulation of protein function, but has been historically difficult to analyze biochemically. To facilitate the detection of S-sulfinylated proteins, we demonstrate that a clickable, electrophilic diazene probe (DiaAlk) enables capture and site-centric proteomic analysis of this OxiPTM. Using this workflow, we revealed a striking difference between sulfenic acid (S-sulfenylation) and S-sulfinylation dynamic response to oxidative stress, indicative of different roles for these OxiPTMs in redox regulation. We also identified >55 heretofore unknown protein substrates of the cysteine sulfinic acid reductase, sulfiredoxin (SRX), extending its function well beyond 2-Cys peroxiredoxins (2-Cys PRDX1–4), and offering new insight into the role of this unique oxidoreductase as a central mediator of ROS-associated diseases, particularly cancer. DiaAlk therefore provides a novel tool to profile S-sulfinylated proteins and study their regulatory mechanisms in cells.
The sulfur atom of cysteine has the ability to assume many different oxidation states, such as sulfhydryl (–SH, –2), sulfenic acid (–SOH, 0), sulfinic acid (–SO2H, +2) and sulfonic acid (–SO3H, +4), which exhibit distinct chemical properties and biological reactivity1 (Fig. 1a). Oxidative post-translational modifications (OxiPTMs), like sulfenic acid2, were long overshadowed by other canonical representatives, like disulfides (–S–S–, –1), but recent advances in chemoproteomics3,4 have enabled identification of this cysteine oxoform in a substantial number of proteins5,6, fundamentally changing our perspective on its role in physiology7–9 and disease10. The biological function of sulfinic acid has also been hotly debated, largely owing to a general consensus that this modification could not be reduced under physiological conditions11. This observation led many to conclude that sulfinic acids in proteins were irreversible or caused by random cysteine oxidation during isolation. However, this idea was challenged by the discovery that peroxiredoxin enzymes were reversibly inactivated by oxidation of the catalytic cysteine to sulfinic acid in cells12,13. Soon after, an ATP-dependent enzyme with sulfinic acid reductase activity, sulfiredoxin (SRX), was isolated from yeast14 and a human homolog was subsequently identified15. To date, peroxiredoxin isoforms 1–4 (PRDX1–4) are the only confirmed SRX substrates16, posing interesting questions concerning the possibility of other targets and broader role of this unique oxidoreductase in redox regulation. Individual studies have identified a functional role for sulfinic acid modification or S-sulfinylation in a growing list of proteins17, including the Parkinson’s disease protein, DJ-1 (refs. 18,19). Despite its regulatory potential20 and association with diseases21 linked to reactive oxygen species (ROS)22 generation, including cancer23–25, the scope, dynamics, and function of sulfinic acid is essentially unknown, owing to major challenges of detection.
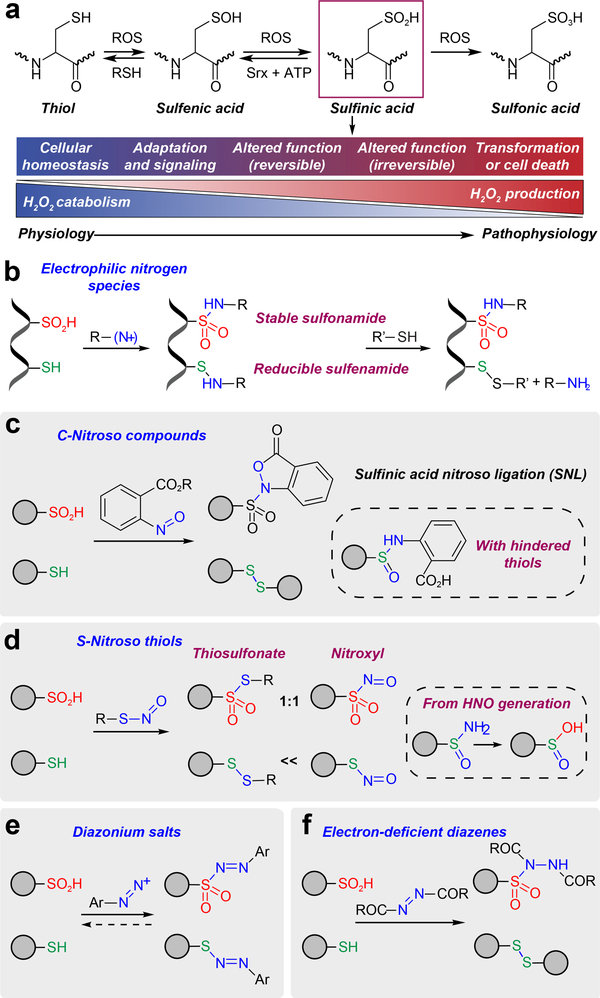
Figure 1 |. Development of electrophilic nitrogen species (ENS) for labeling sulfinic acids.
(a) The sulfur atom of cysteine has the ability to assume many different oxidation states. As the balance between H2O2 production and catabolism changes, adaption occurs with subsequent activation of signaling pathways via protein cysteine OxiPTM, which may be reversible or irreversible. An excessive imbalance can lead to cellular transformation or cell death. (b) General ENS approach for labeling of sulfinic acid in the presence of thiols. (c) Sulfinic acid nitroso ligation (SNL). (d) Reactivity of sulfinic acids and thiols with S-nitrosothiols. (e) Diazonium salts react with both sulfinic acids and thiols, generating a reversible adduct. (f) Electron-deficient diazenes promote covalent modification of sulfinic acids to give a stable sulfonamide adduct.
The weak nucleophilicity of the sulfur in sulfinic acid, compared to the more abundant and reactive thiol(ate), has limited the development of general methods for biochemical analysis. Antibodies have been generated for monitoring sulfinic acid, but have limited affinity and specificity26. Alternatively, a difference of +32 Da has been used to detect sulfinic acid by mass spectrometry, however this approach is plagued by oxidation during lengthy sample preparation27 and the existence of other modifications with the same nominal mass shift, like S-sulfhydration (–SSH). Acidic conditions can be used to favor the reaction of sulfinic acid with N-maleimides, but thiols and other bio-functional groups remain vulnerable to irreversible labeling28. To exploit the unique properties of S-sulfinylated proteins, chemical methods based on electrophilic nitrogen species (ENS) have also been developed29 (Fig. 1b). ENSs are soft electrophiles in which the nitrogen carries a partial positive charge. A soft nitrogen electrophile may react with both thiol and sulfinic acid, but the stability of the resulting adduct depends upon the nature of the sulfur nucleophile. Attack of a thiol on the electron deficient nitrogen center gives a labile sulfenyl amide, which is readily converted back to the thiol by common reducing agents, such as dithiothreitol (DTT). By contrast, the addition of sulfinic acid to the same species yields a stable sulfonamide. Our initial effort, termed sulfinic acid nitroso ligation or SNL (Fig. 1c) supported the validity of the ENS approach by demonstrating efficient addition of electron-deficient C-nitroso compounds to sulfinic acids29,30 (Supplementary Fig. 1a). SNL exhibits high selectivity toward sulfinic acid, but in cases where the thiol is sterically hindered, the labile sulfenamine intermediate can rearrange to give a nonreducible sulfoxide adduct (Supplementary Fig. 1b). This side-reaction can be ameliorated by pre-blocking thiols, and a biotinylated derivative, termed NO-Bio (Supplementary Fig. 1c) was successfully elaborated for several applications30, however complex mass spectrometric fragmentation patterns rendered the probe incompatible with site-centric proteomics. A subsequent study31 used S-nitrosothiols to profile sulfinic acid (Fig. 1d), however this reaction produces the aggressive electrophilic oxidant, nitroxyl (HNO) as a byproduct. HNO generation precludes quantitative conversion of sulfinic acid to the thiosulfonate and converts thiols to sulfinamides and sulfinic acid32 (Fig. 1d and Supplementary Fig. 2). Despite their limitations, these early approaches have revealed noteworthy candidate S-sulfinylated proteins associated with diverse cellular pathways, pointing toward a more complex role for this OxiPTM in biology, while also highlighting the need for more advanced methods to detect and quantify S-sulfinylated proteins from cells.
RESULTS
Electrophilic diazenes form stable sulfonamide adducts
Sulfinic acid has a pKa of ~2 and is deprotonated at physiological pH. Accordingly, the precise chemical target is the sulfinate anion (–SO2–), an ambident nucleophile. The key challenge in sulfinate detection is the stronger nucleophilicity of abundant, competing thiolates. ENS offer a novel solution to this problem because attack of a sulfinate on the electron-deficient nitrogen center yields a stable sulfonamide, whereas a thiolate gives a labile sulfenamide (Fig. 1b). However, as outlined above, C- and S-nitroso ENS suffer issues that limit their utility in chemoproteomics. We therefore examined two new classes of ENS, diazonium salts (Fig. 1e) and diazenes (Fig. 1f), as potential solutions for protein sulfinic acid detection.
We first selected 4-nitrobenzenediazonium tetrafluoroborate (NDF) as a prototype for an electron-poor diazonium salt. When mixed in equal amounts at pH 5, N-Fmoc cysteine sulfinic acid reacted rapidly to yield an orange diazosulfone product (Supplementary Fig. 3a). Under identical conditions, N-Fmoc cysteine also underwent facile reaction with NDF, forming a diazosulfenyl derivative (Supplementary Fig. 3b). At basic pH, the diazonium cation can also be conjugated to tyrosine residues33. Indeed, NDF formed adducts with a single-cysteine variant of thiol peroxidase, C64,82S Gpx334, even when the remaining thiol (C36) was pre-blocked by methyl methanethiosulfonate (MMTS; Supplementary Fig. 4). Known reversibility of sulfur adducts and cross-reactivity with tyrosines thus excluded application of diazonium salts to analysis of sulfinic acid.
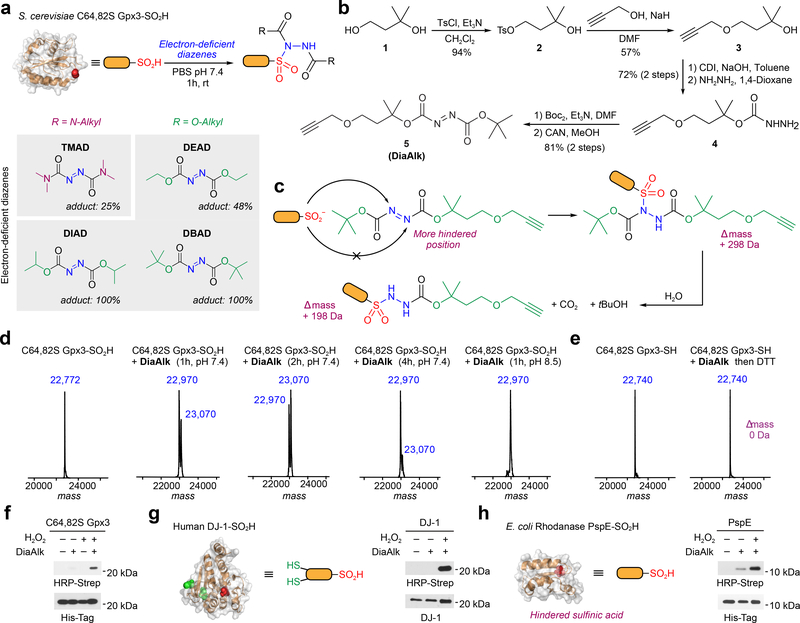
Next, we tested organic diazene compounds for their utility in sulfinic acid detection (Fig. 2). The first diazene examined was tetramethylazodicarboxamide (TMAD; Fig. 2a). To evaluate small-molecule or protein cysteine sulfinic acid labeling, reaction mixtures were analyzed by liquid chromatography tandem mass spectrometry (LC-MS, or intact MS for proteins). TMAD showed promising reactivity with N-Fmoc cysteine sulfinic acid (Supplementary Fig. 5) and a protein model (C64,82S Gpx3-SO2H, ref. 30; Supplementary Fig. 6), forming the expected sulfonamide adduct. However, the reaction of TMAD with Gpx3-SO2H was not quantitative (~25%; Fig. 2a and Supplementary Fig. 6a). Hypothesizing that this observation was due to the low electrophilicity of the nitrogen center, we tested the effect of electrophilic substitution on TMAD reactivity (Fig. 2a and Supplementary Fig. 6a). Diisopropyl azodicarboxylate (DIAD) and di-tert-butyl azodicarboxylate (DBAD) quantitatively converted Gpx3-SO2H to the sulfonamide adduct, whereas the yield of diethyl azodicarboxylate (DEAD) was only 48% (Fig. 2a and Supplementary Fig. 6a). The reaction of DBAD with N-Fmoc cysteine sulfinic acid gave full conversation to sulfonamide product in minutes (Supplementary Fig. 7a). Interestingly, we noted that a single tert-butyloxy carbonyl (BOC) group was lost from the sulfonamide adduct in a time-dependent manner, affording a mono-substituted product that was stable (Supplementary Fig. 7b); similar observations were made with Gpx3-SO2H (23,003 versus 22,903 m/z; Supplementary Fig. 6b,c). Due to its superior reactivity, DBAD was selected for further development.
Figure 2 |. Reactivity of electron-deficient diazenes toward recombinant protein models.
(a) Labeling efficiency of electron-deficient diazenes with recombinant C64,82S Gpx3-SO2H as measured by intact protein MS. (b) Synthesis of DiaAlk (5). (c) Sulfinic acid attacks the less hindered nitrogen of the diazene to generate the initial sulfonamide adduct (+ 298 Da). The BOC group undergoes hydrolysis to yield the final product (+ 198 Da). (d) Deconvoluted MS spectra of C64,82S Gpx3-SO2H before and after treatment with Dia-Alk at various times and pH. (e) ESI-MS spectra of C64,82S Gpx3-SH before and after Dia-Alk treatment, followed by DTT. (f-h) Immunoblot showing DiaAlk detection of sulfinic acid in C64,82S Gpx3 (f), DJ-1 (g) and PspE (h) by streptavidin–HRP. Equal protein loading was confirmed by re-probing immunoblots with the indicated antibody. Representative data from three independent experiments are shown. Uncropped scans of immunoblots are provided in Supplementary Fig. 22.
Validating DiaAlk and DiaFluo as sulfinic acid probes
To facilitate detection and enrichment of labeled targets we then synthesized a “clickable” alkyne derivative of DBAD, termed DiaAlk (5; Fig. 2b and Supplementary Note). With addition of the single alkyne tag, DBAD is converted into an asymmetrical molecule and, in principle, the sulfinic acid can attack at both positions of the N=N. Critically, analysis of the reaction between DiaAlk and sulfinic acid indicated that attack occurs at the less hindered nitrogen atom of the diazene to generate a sulfonamide adduct, leading to a mass increase of 298 Da (Fig. 2c and Supplementary Fig. 8a). The second-order rate constant for the reaction of DiaAlk and N-Fmoc cysteine sulfinic acid in neutral buffer was measured as 2.5×103 M−1min−1 (Supplementary Fig. 8b). Analogous to DBAD, hydrolysis of the initial DiaAlk sulfonamide adduct (“double BOC”) gave a mono-substituted product (“single BOC”) with retention of the alkyne tag (Supplementary Fig. 8a). Likewise, intact MS demonstrated quantitative labeling of Gpx3-SO2H by DiaAlk, followed by time- and pH-dependent cleavage of the BOC group (Fig. 2d). Analysis of C64,82S Gpx3-SH after treatment with DiaAlk and then DTT confirmed that the diazene-based probe does not form a stable adduct with Gpx3 C36 in its thiol form (Fig. 2e). The stability of the N-Fmoc cysteine- and Gpx3-DiaAlk adducts were also verified by exposure to conditions used during proteomic sample preparation (Supplementary Figs. 8c and 9). Gpx3 C36 was further confirmed as the site of adduct formation by tryptic digest and LC-MS/MS analysis, representing >95% of spectral counts from DiaAlk labeling (Supplementary Fig. 10).
The specificity of DiaAlk was also substantiated by MS and immunoblot analysis of DJ-1 (harboring a H2O2-sensitive C106 and two other redox-active cysteines, C46 and C53; Supplementary Fig. 11) and phage shock protein E (PspE; harboring a single hindered cysteine, C67; Supplementary Fig. 12). To visualize S-sulfinylation, proteins were exposed to H2O2 (or untreated), incubated with DiaAlk, subjected to Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) with azide-biotin and analyzed by streptavidin-horseradish peroxidase (Strep-HRP) immunoblot blot (Fig. 2f-h). When benchmarked against other methods, including commercially available antibodies26 that recognize sulfinic and sulfonic acid in select peptide sequences, and our earlier NO-Bio probe30, DiaAlk exhibited superior sensitivity (Supplementary Fig. 13). DiaAlk labeling of proteomes from HeLa lysates was redox-, dose- and time-dependent, as expected (Supplementary Fig. 14).
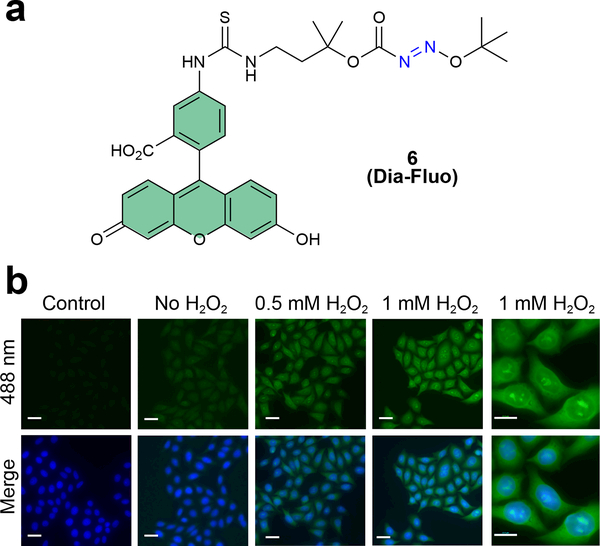
Finally, we also synthesized a fluorescent derivative of DiaAlk, DiaFluo (6, Fig. 3a and Supplementary Note), for visualization of protein S-sulfinylation in mammalian cells. HeLa cells were exposed to H2O2 (or untreated), fixed, blocked with N-ethylmaleimide (NEM), probed with DiaFluo, and imaged by fluorescence microscopy. A clear peroxide-dependent increase in fluorescence intensity was observed, which was localized throughout the cell, whereas controls minus DiaFluo exhibited only background signal (Fig. 3b). These results establish DiaAlk as a robust probe for labeling sulfinic acid in a variety of protein microenvironments (e.g., surface exposed, charged residues, and buried) and its complementary analog, DiaFluo, for its utility in fluorescence imaging.
Figure 3 |. Detection of protein S-sulfinylation from cells using DiaFluo.
(a) Structure of fluorescein-tagged DiaAlk analog, DiaFluo (6). (b) Fluorescence microscopy images of S-sulfinylation in HeLa cells before and after stimulation with H2O2. Cells were fixed, permeabilized, and blocked with NEM. Protein S-sulfinylation was visualized at 488 nm (green) after treatment with or without (control) DiaFluo. Nuclei were counterstained with DAPI (blue). Scale bars, 20 μm. Representative data from three independent experiments and more than 40 images are shown.
General strategy to discover new S-sulfinylated proteins
Having validated the selectivity of DiaAlk in small-molecule and protein sulfinic acids, we next set out to identify novel S-sulfinylation targets, using two approaches. First, we investigated S-sulfinylation of proteomes resulting from addition of exogenous H2O2 to A549 and HeLa cells using our established chemoproteomic workflow3,4. These analyses are designed to explicitly delineate proteomic cysteines capable of reacting with H2O2 to undergo sulfinic acid modification, an event known as “hyperoxidation”. Second, we used chemoproteomics to map differential S-sulfinylation in SRX deficient (Srx−/−) and SRX replete (Srx+/+) mouse embryonic fibroblast (MEF) cells35 which we classify as “SRX-regulated” cysteines. The latter comparison identifies SRX-dependent changes in S-sulfinylation, with the possibility to reveal new biology specific to this reductase.
Chemoproteomic analysis of H2O2-sensitive cysteines
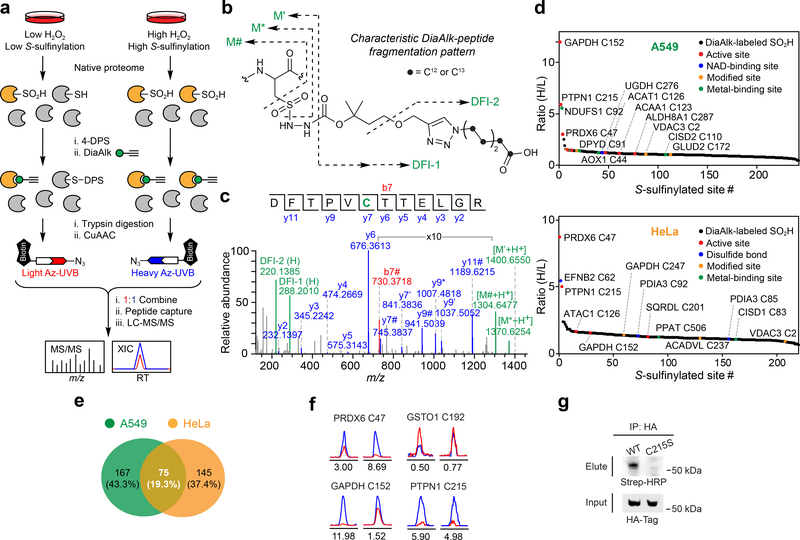
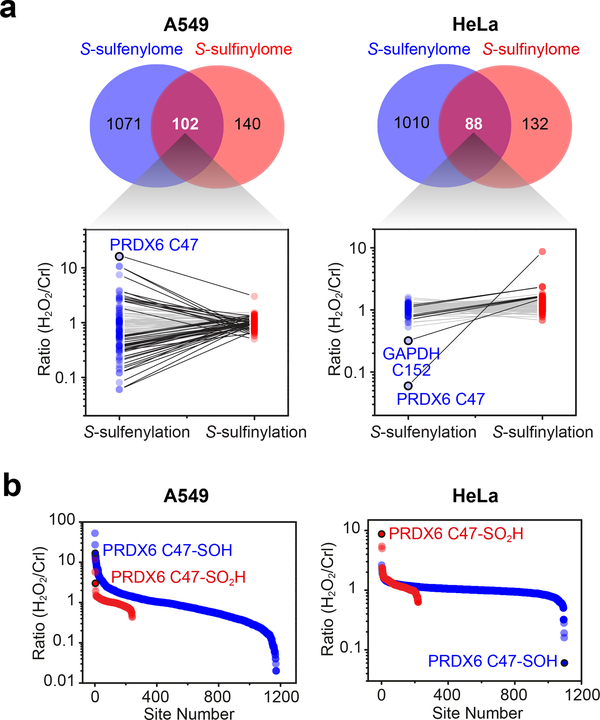
Focusing initially on cysteines that undergo “hyperoxidation”, we applied DiaAlk to profile S-sulfinylation caused by treatment of cells with H2O2 (1 mM, 15 min; Fig. 4a). Native proteomes obtained from A549 or HeLa cells were blocked with 4,4’dipyridyldisulfide (4-DPS) to prevent thiol-mediated probe consumption, labeled with DiaAlk (5 mM, 2 h), and then digested with trypsin. Tagged peptides from control and H2O2-treated samples were conjugated to light and heavy azido biotin functionalized with a photocleavable linker (Az-UV-biotin) via CuAAC, combined, purified by streptavidin, and released by photocleavage of the biotin linker. Liberated DiaAlk-triazohexanoic acid-modified peptides were analyzed by LC-MS/MS (Fig. 4b,c). In aggregate, we identified and quantified 387 probe-tagged S-sulfinylated sites on 296 proteins from A549 and HeLa cells, with 19.3% (75 sites) common to both cell lines (Fig. 4d,e and Supplementary Dataset 1). Identified among these hits were known sites of S-sulfinylation including PRDX636 (Cys47), GAPDH16 (Cys152), and PTPN137 (also known as PTP1B, Cys215), as were a cohort of novel S-sulfinylation events at functionally important sites, such as active sites (e.g., ALDH1L1 Cys707, SQRDL Cys201, and ACAT1 Cys126), metal-binding sites (e.g., NDUFS1 Cys92, AOX1 Cys44, and DPYD Cys82), and disulfide bonds (e.g., EFNB2 Cys62 and PDIA3 Cys85-Cys92). Bioinformatic analysis indicated their distribution across all major subcellular compartments (Supplementary Fig. 15a). Of note, we found that 47.3% of S-sulfinylated proteins were highly enriched in exosomes, which protect cells against stress by removal and/or transport of oxidized proteins38,39. S-sulfinylated proteins were also substantially enriched in diverse biological processes and pathways, including oxidation-reduction, cell-cell adhesion, RNA processing, glycolysis, protein import into the nucleus, and fatty acid beta-oxidation (Supplementary Fig. 15b,c). To examine the chemoselectivity of DiaAlk at the broadest possible level, we also searched our proteomic data for potential cross-reactivity with other nucleophilic residues (Supplementary Fig. 16a). Importantly, cysteine sulfinic acid accounted for >99.9% of all spectral counts (Supplementary Fig. 16b). A blind search for post-translational modifications (PTMs) also failed to identify unwanted DiaAlk-adducts. These data demonstrate that DiaAlk selectively maps known and novel S-sulfinylation sites of functional importance in human proteomes.
Figure 4 |. Site-centric and quantitative chemoproteomic profiling of protein S-sulfinylation.
(a) Schematic representation of site-centric quantitative chemoproteomic workflow for global profiling of S-sulfinylation in native proteomes. (b) Characteristic fragmentation of DiaAlk-triazohexanoic acid modified peptides. DFI: diagnostic fragment ions. (c) MS/MS spectrum of a DiaAlk-tagged peptide, which unequivocally identifies Cys47 of PRDX6 as a S-sulfinylated site. (d) Site-specific changes in S-sulfinylation in A549 (top) and HeLa (bottom) cells in response to H2O2 stress. Heavy (H2O2) to light (control) ratios are correlated with functional annotations from the UniProt database. (e) Venn diagram of A549- and HeLa-sulfinylomes labeled with DiaAlk. (f) Extracted ion chromatograms showing changes in DiaAlk-tagged peptides from PRDX6 (Cys47), GAPDH (Cys152), GSTO1 (Cys192), and PTPN1 (Cys215) from H2O2 stimulation of A549 (left) and HeLa cells (right). The profiles for light- and heavy-labeled peptides are shown in red and blue, respectively. Heavy (H2O2) to light (control) ratios were calculated from three independent experiments and are displayed below the individual chromatograms. (g) DiaAlk labels HA-tagged wild-type PTPN1, but not C215S, in HeLa cells. Representative data from three independent experiments are shown. Uncropped scans of immunoblots are provided in Supplementary Fig. 22.
As dynamic sites of S-sulfinylation may function as “sensors” of stress, we also quantified the H2O2-dependent fold-change in oxidation (RH2O2/Crl or RH/L; Fig. 4d, Supplementary Fig. 17 and Supplementary Dataset 1). Identified sites often exhibited differential responses to H2O2 depending on cell type (Fig. 4f). For example, S-sulfinylation of PRDX6 Cys47 dramatically increased in HeLa cells in response to H2O2 treatment (RH/L=8.69), while this change was relatively small in A549 cells (RH/L=3.00). By contrast, S-sulfinylation of GAPDH Cys152 exhibited higher sensitivity toward H2O2 in A549 cells (RH/L=11.98) compared to HeLa cells (RH/L=1.52). Distinct from these oxidation events, a small fraction of cysteine sulfinic acids (5.0% and 0.45% of the A549 and HeLa S-sulfinylomes, respectively), such as Cys192 of glutathione S-transferase omega 1 (GSTO1), exhibited a substantial decrease in S-sulfinylation after H2O2 treatment. These data suggest susceptibility to further oxidation to sulfonic acid, which would appear as a loss of signal. Though it often suggested that sulfenyl amide formation with the adjacent backbone nitrogen of Ser216 protects active site Cys215 against “hyperoxidation”, we observed PTPN1 as highly S-sulfinylated in both cell types (RH/L=5.90 and =4.98 in A549 and HeLa, respectively). To independently validate site-specific, H2O2-induced S-sulfinylation of PTPN1, epitope-tagged constructs (wild-type and C215S) were expressed in HeLa cells, labeled in situ with DiaAlk (5 mM, 2 h), immunoprecipitated, reacted with biotin-azide and visualized by streptavidin immunoblot. This analysis confirmed DiaAlk labeling of wild-type PTPN1, while mutation of Cys215 abrogated labeling, as expected (Fig. 4g).
H2O2 stress induces distinct OxiPTM fold-change dynamics
Considering the intrinsic relationship of protein S-sulfinylation and S-sulfenylation (–SOH)17, we next asked whether these oxiPTMs exhibit similar target profiles. Here, we conducted a quantitative analysis of the S-sulfenylome from both A549 and HeLa cells under identical H2O2 stress using our recently described chemoproteomic method3,4. In total, we identified and quantified 1,173 and 1,098 S-sulfenylation sites in A549 and HeLa cells, respectively (Fig. 5 and Supplementary Dataset 2). Approximately 40% of S-sulfinylated sites were identified in the corresponding S-sulfenylome dataset (Fig. 5a) which, considering the lineage of these OxiPTMs and comparatively transitory nature of sulfenic acids, represents excellent overlap. Of particular interest, we noted that oxidative stress induced distinct fold-change dynamics in both OxiPTMs. For example, most S-sulfinylation sites in A549 cells (91.7%) were unchanged after H2O2 stimulation (0.67≤RH/L≤1.5), whereas 60.3% of the corresponding S-sulfenylome underwent substantial fold-changes (>1.5; Fig. 5b). By contrast, the majority of S-sulfinylation (84.1%) and S-sulfenylation (95.1%) sites in HeLa were unchanged (0.67≤RH/L≤1.5; Fig. 5b), which is consistent with elevated basal levels of H2O2 production in this particular cancer cell line40. In HeLa cells, the most highly down-regulated S-sulfenylation event was the peroxidatic cysteine of PRDX6 (Cys47), consistent with “hyperoxidation” to sulfinic acid (Fig. 5a). Similar reciprocity was observed at the redox-sensitive cysteine of GAPDH Cys152 (Fig. 5a). The finding that S-sulfenylation is considerably more dynamic is supported by mechanistic studies41 which indicate that oxidation of a thiolate to sulfenic acid is more facile than oxidation of sulfenic to sulfinic acid (reflecting the intrinsic reduction in sulfur nucleophilicity) and cell-based studies that highlight the ability of antioxidant systems to protect cysteines from hyperoxidation7,13,15,35.
Figure 5 |. Comparison of S-sulfenylome and S-sulfinylome sites and dynamic fold-changes.
(a) Venn diagram showing overlap between the S-sulfenylome and S-sulfinylome. Ratios (H2O2 versus untreated control) obtained from common sites in the S-sulfenylome and S-sulfinylome dataset are plotted as line series and displayed on a log10 scale on the y-axis. Two-fold or smaller changes between –SOH and –SO2H are depicted with light grey lines; fold-changes greater than two are depicted in black. (b) Ratio (H2O2 versus untreated control) of S-sulfenylated and S-sulfinylated sites identified in the same cell line are plotted as a scatter graph and displayed on a log10 scale on the y-axis.
Chemoproteomics reveals new substrates of sulfiredoxin
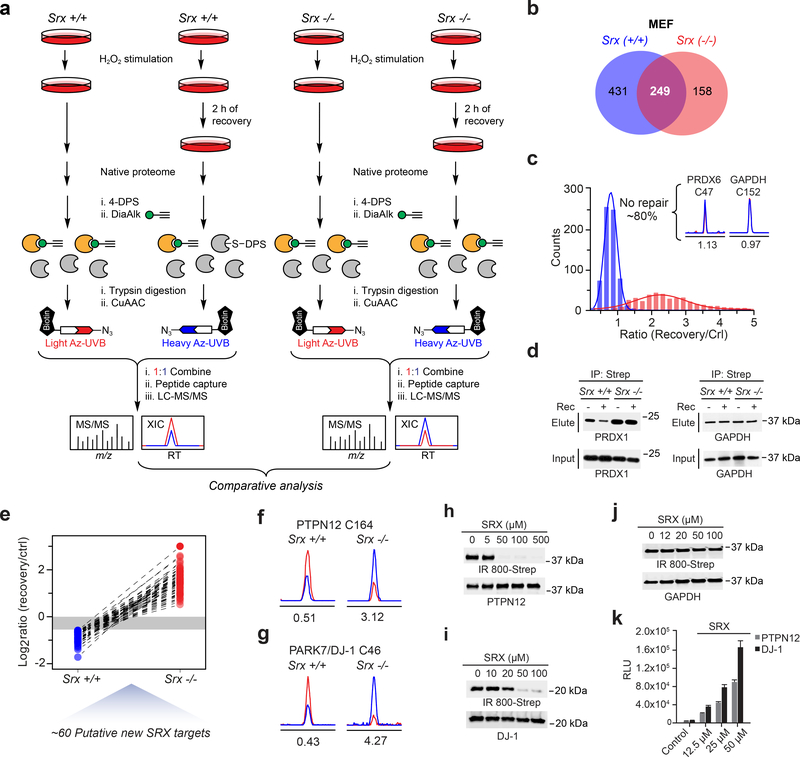
To complement our H2O2-sensitivity data, we next examined SRX-regulated changes in S-sulfinylation. For these studies, we evaluated turnover of the S-sulfinylome in Srx+/+ and Srx−/− MEFs35 using an established “recovery” method14,16. Cells were exposed to H2O2 stress in order to induce Srx expression in wild-type MEFs; Supplementary Fig. 18) and harvested for chemoproteomic analysis or placed in H2O2-free culture medium to recover for 2 h (Fig. 6a). In principle, SRX-targeted sulfinic acids should be turned over (i.e., reduced) in Srx+/+, but not Srx−/− cells. Native proteomes obtained from Srx+/+ and Srx−/− MEFs were DPS-blocked, labeled with DiaAlk (5 mM, 2 h), and digested into tryptic peptides. The resulting peptide mixtures, with and without recovery, were conjugated to heavy and light Az-UV-biotin reagents, respectively. DiaAlk-tagged peptides were then affinity purified on streptavidin, photo-released, and analyzed by LC-MS/MS. In total, we identified and quantified 680 S-sulfinylated sites (521 proteins) and 407 sites (339 proteins) in Srx+/+ and Srx−/− MEFs, respectively (Fig. 6b and Supplementary Dataset 3). As shown in Figure 6c, ~80% of protein sulfinic acids could not be turned over in Srx+/+ MEFs (0.67<RH/L<1.5). For example, sulfinic acid forms of PRDX6 (Cys47) and GAPDH (Cys152) remained unchanged in Srx+/+ MEFs during the recovery period (Fig. 6c, inset), indicating that these sites are not reduced by SRX. The observation that PRDX6 and GAPDH S-sulfinylation is not repaired by SRX is consistent with earlier biochemical findings16 and independently validate our method. The overall level of S-sulfinylation was also increased in Srx−/− MEFs during the recovery period (Supplementary Dataset 3), affirming the susceptibility of Srx-deficient cells to acute oxidative stress21,35. As tryptic peptides bearing the catalytic cysteines of established SRX substrates PRDX 1–4 were too long and hydrophobic for robust chemoproteomic analysis, we next examined whether DiaAlk could detect changes in S-sulfinylation in one of these sentinel peroxidases, PRDX1, by immunoblot. H2O2-stimulated Srx+/+ and Srx−/− MEFs were subject to the “recovery” workflow, labeled with DiaAlk (5 mM, 2 h), proteomes clicked with biotin-azide, captured by streptavidin, and visualized by immunoblot. This analysis verified the ability of DiaAlk to detect SRX-dependent changes in S-sulfinylation of PRDX1, whereas GAPDH, as a negative control, showed no change, as expected (Fig. 6d).
Figure 6 |. Proteome-wide analysis of SRX-regulated changes in S-sulfinylation.
(a) Chemoproteomic workflow to map differential S-sulfinylation in SRX deficient (Srx−/−) and SRX replete (Srx+/+) MEFs, classified as “SRX-regulated” cysteines. (b) Venn diagram showing overlap between the Srx+/+ and Srx−/− S-sulfinylomes in MEFs. (c) Distribution of dynamic (measured ratios) of protein S-sulfinylation in Srx+/+ (blue) and Srx−/− (red) MEFs. Ratios between heavy (recovery) and light (without recovery, or control) were measured as the turnover rates for each S-sulfinylation event. Inset, extracted ion chromatograms (XIC) of DiaAlk-modified peptides from PRDX6 (Cys47) and GAPDH (Cys152) from Srx +/+ MEFs. The profiles for light- and heavy-labeled peptides are shown in red and blue, respectively. Heavy (recovery) to light (control) ratios were calculated from two independent experiments and are displayed below the individual chromatograms. (d) S-sulfinylation of PRDX1, but not GAPDH, is reversible and Srx-dependent, as detected by DiaAlk. Representative data from three independent experiments are shown. Uncropped scans of immunoblots are provided in Supplementary Fig. 22. (e) Line series plot showing the turnover rates of the same S-sulfinylation sites identified in both Srx+/+ and Srx−/− MEFs. (f,g) XIC of DiaAlk-modified peptides from PTPN12 (Cys164) (f) and DJ-1 (Cys46) (g) from Srx+/+ and Srx−/− MEFs. Heavy (recovery) to light (control) ratios were calculated from two independent experiments and are displayed below the individual chromatograms. (h-j) Recombinant SRX1 reduces S-sulfinylation of PTPN12 (h) or DJ-1 (i), but not GAPDH (j), as detected by BioDiaAlk (7). Uncropped scans of immunoblots are provided in Supplementary Fig. 22. (k) Luminescence assay measures SRX1 ATPase activity in the presence of PTPN12 or DJ-1. Signal in the absence of SRX1 was subtracted as background; control reaction lacks ATP. Representative data from three independent experiments are shown, each performed in triplicate; error bars represent the standard deviation (s.d).
In total, our chemoproteomic analyses enabled site-specific and quantitative comparison of turnover at 249 S-sulfinylation sites among Srx+/+ and Srx−/− MEF cells (Fig. 6b). Of these, 59 sulfinic acids on 55 proteins exhibited substantial turnover in Srx+/+ MEFs (RH/L<0.67) while remaining unchanged or increased (RH/L≥1) in Srx−/− MEFs (Fig. 6e; Supplementary Dataset 3), suggesting that they might be substrates of SRX. For example, S-sulfinylation at Cys164 of the protein tyrosine phosphatase non-receptor type 12 (PTPN1242, also known as PTP-PEST) decreased by half in Srx+/+ MEF cells during the recovery period, while turnover was not observed in the Srx knockout (Fig. 6f); analogous observations were made for DJ-1 Cys46 (Fig. 6g). To substantiate our proteomic discovery of new SRX substrates, S-sulfinylated proteins were incubated with increasing amounts of recombinant SRX43, labeled with a biotinylated DiaAlk derivative, BioDiaAlk (7, Supplementary Note), and then visualized by streptavidin blot (Fig. 6h-j). PTPN12 and DJ-1 exhibited an SRX-dependent decrease in S-sulfinylation, whereas the GADPH control showed no change. To further characterize this biochemical reaction, we used a luminescent assay, ADP-Glo™, to monitor the ADP product formed by SRX reduction of sulfinic acid. ATP hydrolysis was observed by SRX in a dose-dependent manner in reactions containing S-sulfinylated PTPN12 and DJ-1 substrates (Fig. 6k), but not GAPDH (Supplementary Fig. 19a) further substantiating proteomic and immunoblot analyses. Control experiments verified PRDX2-SO2H as an SRX substrate in BioDiaAlk and ATP-hydrolysis assays (Supplementary Fig. 19a,b). Still further, among three S-sulfinylated cysteines mapped onto the actin-regulating redox sensor, cofilin 144,45 (CFL1), only Cys39-SO2H could be recovered in Srx+/+ MEFs (Supplementary Fig. 20a). To substantiate this finding, we expressed an epitope-tagged version of wild-type CFL1 or the C39A mutant in HEK293T cells and observed that this mutation partially reversed turnover of S-sulfinylation during recovery (Supplementary Fig. 20b). We also observed that H2O2 treatment increased the association of wild-type CFL1 and SRX (Supplementary Fig. 20c), further supporting S-sulfinylated CFL1 as a substrate of SRX. Collectively, these results establish SRX-mediated reduction of S-sulfinylation in new targets, indicating that substrate scope extends well beyond 2-Cys PRDX1–4. Finally, another interesting observation from our chemoproteomic analyses is that S-sulfinylation of evolutionarily conserved Cys92 in the mitochondrial NADH-ubiquinone oxidoreductase, NDUS1, was recovered in both Srx+/+ and Srx−/− MEFs, while no protein turnover was observed (Supplementary Fig. 21). These findings suggest the possibility that the sulfinic acid at this site might be reduced by a yet-to-be discovered reductase in cells, which merits exploration in future studies.
DISCUSSION
Protein S-sulfinylation is an emergent and complex OxiPTM in biology that still requires chemical biology methods for functional studies. Here we demonstrate that cysteine sulfinic acid labeling with the alkyne-functionalized electrophilic diazene reporter, DiaAlk, enables stable covalent modification of S-sulfinylated proteins from mammalian cells. Our chemoproteomic workflow permits site-specific quantification of dynamic fold-changes in sulfinylome modifications. The observation that MS/MS spectra of DiaAlk/click-modified peptides contain diagnostic fragment ions (DFIs) further increases the confidence of S-sulfinylated peptide identification. DiaAlk trapping coupled with our chemoproteomic workflow enabled us to robustly quantify sites of protein S-sulfinylation in normal and H2O2-stressed cells, confirming many known and revealing new S-sulfinylated proteins.
Increasing evidence indicates that “hyperoxidation” of a cysteine thiolate is not as rare or unregulated as previously believed. For example, analysis of soluble proteins from rat liver by gel focusing electrophoresis estimated that ≥5% of cysteine residues exist as –SO2H46. Our study reports hundreds of previously unknown sites of protein S-sulfinylation. Interestingly, the majority of sites identified in this study did not undergo substantial fold-changes in extent of modification during stress, suggesting that S-sulfinylation may exist at high occupancy under basal conditions, ostensibly accumulating over a protein’s lifetime, with reduction/degradation as the rate-limiting step in this cycle. Consistent with this hypothesis, we found that ~50% of S-sulfinylated proteins were enriched in exosomes, which sort and transport oxidized proteins. By contrast, our previous studies5 and present data indicate a dynamic S-sulfenylome, with >60% undergoing substantial fold-changes during H2O2 stress in A549 cells. Regarding the lineage between sulfenic and sulfinic acid states, our findings can be rationalized from several standpoints: From a chemical perspective, the reaction between a thiolate and H2O2 is more facile, compared to the less nucleophilic sulfur atom in sulfinic acid and this difference in rates constants has been well documented2,41. From a biological viewpoint, S-sulfinylation may accumulate due to the moderate intrinsic catalytic activity of SRX11,15 and the observation that not all protein sulfinic acids appear to be viable substrates for SRX-targeted reduction. With more available biological routes for reduction2,7, the S-sulfenylation cycle has the capacity to be more dynamic or adaptive, supporting different roles for these OxiPTMs in thiol-based redox-regulation and signaling. Of note, not all sites of S-sulfinylation may stem from non-enzymatic oxidation. For example, cysteine oxidase enzymes in plants can oxidize cysteines to sulfinic acid using oxygen as a cofactor47, although it is currently unknown whether a similar enzyme function in animals.
On the other hand, we also identified numerous S-sulfinylation sites with substantial fold-changes in modification. S-sulfinylation events undergoing the greatest alterations were found to be enriched among functional cysteines (e.g., metal-binding, active site, disulfide bonds) and were distributed across all major compartments. S-sulfinylated proteins were also substantially enriched in various biological processes and pathways. For example, we identified enzymes involved in cell adhesion, which has been reported to be regulated by H2O248, as highly enriched in the S-sulfinylome. In this regard, filamin B (FLNB), an actin-binding protein that connects cell membrane constituents to the actin cytoskeleton, was extensively S-sulfinylated at Cys2501 and Cys2115. Glycolysis represents another important cellular pathway targeted by S-sulfinylation. One particularly intriguing case was identified in phosphoglycerate kinase 1 (PGK1), which coordinates ATP production with one-carbon metabolism and cellular redox regulation. A recent study indicates that endogenous H2O2 production and oxidative stress leads to mitochondrial translocation of PGK1, where it phosphorylates alternative substrates49. Among the seven cysteines in PGK1, we found that Cys50 exclusively underwent S-sulfinylation during stress. Given this correlation, it is tempting to speculate that oxidation of Cys50 mediates PGK1 translocation and/or modulates kinase activity, however, the precise role of S-sulfinylation remains to be uncovered.
The initial discovery of SRX, an ATP-dependent oxidoreductase that reduces cysteine sulfinic acid14, ignited interest in S-sulfinylation as a “redox-switch” or regulatory modification11. Until the present work, 2-Cys PRDX1–4 have remained the only known substrates of SRX, which reduces sulfinic acid at their peroxidatic cysteine with concomitant restoration of catalytic activity. Measuring the site-specific turnover of the cellular S-sulfinylome, in the presence and absence of SRX, was made possible by the development of DiaAlk and application to our quantitative chemoproteomic workflow. In total, 59 sites exhibited substantial turnover in Srx+/+ MEFs (RH/L<0.67) while remaining unchanged or increased (RH/L≥1) in Srx−/−MEFs, providing a strong indication that they might be SRX substrates. Additional labeling and biochemical studies with recombinant proteins provided evidence for SRX-dependent of reduction of S-sulfinylation in both PTPN12 and DJ-1. Our proteomic data are further supported by cell-based studies, which pinpoint Cys39 as the site of S-sulfinylation in CFL1 and show H2O2-dependent association of wild-type CFL1 with SRX. This collective work, thus identifies new targets of SRX beyond 2-Cys PRDX1–4. This discovery should dramatically accelerate our understanding the role of this antioxidant enzyme in regulating redox balance, circadian rhythmn, protein stability, and how its overexpression facilitates survival of cancer cells. The enzymatic reversibility of S-sulfinylation among a cohort of proteins vis-à-vis SRX also reveals a deeper layer of thiol-based redox regulation, previously unknown. For example, PTPN12, a recently characterized tumor suppressor, is pivotal for regulation of EGFR signaling and required for embryonic development50. S-sulfinylated Cys164 is directly adjacent to the ß3-loop-ß4 of PTPN12, a structurally diverse region among PTP-PEST family members, which dictates substrate specificity42. Likewise, S-sulfinylation mediates a redox-switch that enables DJ-1 to participate in cytoprotective signaling under stress18,19. These, and numerous other instances in our datasets, indicate a broader role for individual S-sulfinylation sites in cells.
Finally, it is important to acknowledge some limitations of the current study. In particular, one drawback of our chemoproteomic method is that it is not yet optimized to identify S-sulfinylated cysteines at a global level directly in cells, but rather in native proteomes supplemented with antioxidants, including catalase. Therefore, an important future goal will be elaboration of chemoproteomic analysis of S-sulfinylated proteins in situ, including exploration of thiol-blocking tactics with reduced cytotoxicity and/or modulation of probe reactivity. Despite these “NextGen” challenges, the current study details a suite of highly enabling and novel chemical probes for unambiguous mapping of S-sulfinylated cysteines and discloses unprecedented biology through the identification and validation of new SRX substrates. As such, these studies broaden the scope for our understanding of cellular “redox circuitry” and pave the way for future directions of research that expand knowledge of cysteine “hyperoxidation” as a regulatory modification in signaling and as pathological hallmarks in human disease.
ONLINE METHODS
General materials and methods.
N-Fmoc-cysteine, 4-nitrobenzenediazonium tetrafluoroborate, tetramethylazodicarboxamide (TMAD), diethyl azodicarboxylate (DEAD), di-isopropyl azodicarboxylate (DIAD), di-tertbutyl azodicarboxylate (DBAD), 4,4’-dipyridyl disulfide (4-DPS), methyl methanethiosulfonate (MMTS) and iodoacetyl-LC-biotin (IAM-Bio) were purchased from Thermo Fisher. BTD, a clickable probe for detecting S-sulfenylation, and UV-cleavable azido-biotin reagents were synthesized, as previously described2. Recombinant PTPN12 was purchased from Sino Biological. GAPDH was purchased from Sigma. Reagents and solvents were purchased from Sigma and were used without further purification. For chemical synthesis, all reactions were performed under a nitrogen atmosphere in oven-dried glassware. Analytical thin layer chromatography (TLC) was carried out using Analtech Uniplate silica gel plates and visualized using a combination of UV and potassium permanganate staining. Flash chromatography was performed using silica gel (32–63 μM, 60 Å pore size) from Sorbent Technologies Incorporated. NMR spectra were obtained on a Bruker Avance 400 (400 MHz for 1H; 100 MHz for 13C). 1H and 13C NMR chemical shifts are reported in parts per million (ppm) referenced to the residual solvent peak. Low-resolution electrospray ionization (ESI) mass spectra were obtained with an Agilent 6120 Single Quadrupole LC-MS.
Antibodies.
HRP-streptavidin (GE Healthcare, RPN1231, 1:80,000), His-tag (Cell Signaling Technologies, 2365, 1:1,000), anti-Actin (Santa Cruz Biotechnology, sc-1616 1:1,000), anti-PARK7/DJ-1 (Abcam, ab18257 1:1,000 or Proteintech, 11681–1-AP, 1:500), anti-oxDJ-1 (Millipore, MABN1773, 1:500), anti-PTPN12 (Invitrogen, MA1–12377, 1:2,000), anti-SRX (Santa Cruz Biotechnology, sc-166786, 1:100); anti-NDUFS1 (Abcam, ab169540, 1:5,000); anti-V5 (Invitrogen, R960–25, 1:1,000), anti-HA (Invitrogen, 71–5500, 1:1,000), anti-GAPDH (Abnova, H00002597-M01, 1:1000), anti-PRDX1 (Abnova, MAB7559, 1:1,000), anti-PRDX2 (Abnova, H00007001-M01, 1:200), rabbit anti-goat IgG-HRP (Invitrogen, 31402, 1:20,000 – 1:50,000), and rabbit anti-mouse IgG-HRP (Invitrogen, 61–6520, 1:20,000 – 1:50,000).
Cell lines and culture.
A549, HeLa, and HEK293T cells were sourced from the ATCC (Manassas, VA). Srx +/+ and Srx −/− MEFs were kindly provided by Dr. Sue Goo Rhee and Dr. Michael Toledano. A549, HeLa, HEK293T, and MEFs were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Corning) supplemented with 10% (v/v) fetal bovine serum (FBS, Invitrogen), 1% (v/v) penicillin-streptomycin (Invitrogen) and 1% of Glutagro® (Corning). Cells were maintained at 37 °C in a 5% CO2 humidified atmosphere.
Expression and purification of recombinant proteins.
C64,82S Gpx3 was expressed and purified, as described51. WT DJ-1, E18N DJ-1 and C106S DJ-1 were expressed and purified, as described19. PspE was expressed and purified, as described52. SRX and PRDX2 were expressed and purified, as described43.
Generation of protein sulfinic acids in vitro.
Recombinant proteins were reduced with dithiothreitol (DTT, 5 mM) for 30 min at 0 °C. Reactions were then passed through one Micro Bio-Spin P-30 column (BioRad) equilibrated with 50 mM HEPES or 100 mM PBS, pH 7.4. DTT-free proteins were then treated with hydrogen peroxide (H2O2, 2–5 eq.) at rt for 1 h. Reactions were quenched by passage through one Micro Bio-Spin P-6 column (BioRad). For C64,82S Gpx3, the reduction/oxidation cycle was repeated to ensure complete reduction of the homodimer. Clarified samples were supplemented with DTT (5 mM), snap-frozen in liquid N2, and stored at −80 °C. Formation of the sulfinic acid species was confirmed by intact LC-MS analysis, as described below.
Sample preparation for analysis of S-sulfinylation of recombinant proteins.
S-sulfinylated recombinant proteins incubated with DiaAlk (1 mM) in the dark at rt for 1 h. Excess DiaAlk was quenched by incubation with DTT (2 mM) for 30 min and then passed through one Micro Bio-Spin P-30 column pre-equilibrated with 100 mM PBS, pH 7.4. Click chemistry was performed on buffer exchanged samples by the addition of 0.1 mM biotin-PEG4-N3, 0.25 mM BTTP-CuSO4 (2:1) ratio, and 2.5 mM ascorbate in 50 mM PBS, pH 7.4 with 100 mM NaCl. After 1 h at rt, reactions were quenched by EDTA. Samples were analyzed by immunoblot, as described below. Equal loading was verified by antibodies against the indicated proteins.
Sample preparation for analyzing protein S-sulfinylation in cell lysates.
HeLa cells were treated with or without H2O2 (1 mM, 15 min), washed with PBS (3X) and lysed in modified RIPA buffer [50 mM triethanolamine, pH 7.4, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 200 U/mL catalase (C4963, Sigma), 1x EDTA-free complete mini protease inhibitors (Roche)]. After 20 min on ice (with frequent mixing), lysates were clarified by centrifugation at 14,000 − g at 4 °C for 20 min. Protein concentration was determined using a standard BCA assay (Pierce). Cell lysates were treated with 4,4′-dithiodipyridine (4-DPS, 200 mM) at rt for 1 h to block free thiols and subsequently buffer exchanged using one Micro Bio-Spin P-30 column pre-equilibrated with 100 mM HEPES, pH 8.5, 100 mM NaCl. DPS-free lysates were then reacted with DiaAlk (1 mM) or iodoacetyl-LC-biotin (250 μM in DMSO) in the presence of 0.5% of SDS in the dark at rt with rotation for 1 h. Reactions were quenched by incubation with DTT (1 mM) for 30 min and passed through one detergent removal spin column (87777, Pierce) pre-equilibrated with 100 mM PBS, pH 7.4. Click chemistry was performed on the buffer exchanged samples, as described above.
Immunoblotting.
Recombinant proteins or cell lysates were separated by SDS-PAGE followed by semi-dry transfer to polyvinylidene difluoride (PVDF, 1620177, BioRad). After transfer, the PVDF membrane was blocked with 5% BSA in TBST for 1 h at rt. The membrane was washed with TBST (3X) and incubated with indicated primary antibodies overnight at 4 °C. After incubation, the membrane was washed (3X) with TBST and incubated with the appropriate HRP-conjugated secondary antibody. The PVDF membrane was then washed with TBST (3X), visualized by chemiluminescence (ECL Plus, Pierce), exposed to film and analyzed by ImageJ (v1.52) software. SRX reduction of S-sulfinylated proteins was analyzed by transfer onto PVDF-FL (LI-COR). The membrane was then blocked with Odyssey Blocking Buffer (LI-COR) at rt for 1 h and probed with the indicated primary antibodies overnight at 4 °C. After incubation, membranes were washed with TBST (3X) and incubated with secondary antibodies: streptavidin 680LT (1:15,000, 925–32230, LI-COR) and goat-anti-mouse/rabbit 800CW (1:15,000, LI-COR, 925–32210/925–32211) in the dark at rt. The membrane then was then washed with TBST (3X) and analyzed using an Odyssey Imaging System (LI-COR). For analyzing cellular SRX and S-sulfinylation of DJ-1 and CFL1 from lysates and immunoprecipitants, samples were resolved by SDS-PAGE and transferred to a PVDF membrane (Merck Millipore, IPVH00010), blocked with 5% milk in TBST at rt for 1 h and probed with the indicated primary antibodies overnight at 4 °C. After incubation, the membrane was washed TBST (3X) and incubated with the appropriate HRP-conjugated secondary antibody. The PVDF membrane was then washed with TBST (3X), visualized on a Tanon 5200 scanner (Shanghai, China), and analyzed by GelCap software v 5.6 using ECL chemiluminescence (CWBIO, CW0049S).
Fluorescence imaging.
In a 4-well chamber slide (Thermo Scientific), 3 × 104 HeLa cells were seeded in each compartment. Cells were incubated at 37 °C in a 5% CO2 atmosphere for 24 h. Cell were exposed to H2O2 (0, 0.5 or 1 mM) for 15 min, and then washed with cold PBS (3X) containing 200 U/mL of catalase. Cells were then fixed with methanol at −30 °C for 15 min, permeabilized with 0.2% of Triton X-100 in PBS with iodoacetamide (IAM; 40 mM) in the dark for 1 h. The media was removed, and each compartment washed with 0.2% Triton X-100 in PBS (1 mL, 3X). Cells were incubated with DiaFluo (500 μL, 25 μM in DMSO) in PBS supplemented with 0.2% of Triton X-100 in the dark at rt for 30 min. Subsequently, the media was removed, and each compartment was washed with PBS containing 0.2% Triton X-100 (1 mL, 4X). After fixation and permeabilization, cells were incubated with a solution containing DAPI (1 mL, 5 μg/mL) in PBS in the dark at rt for 5 min. The media was removed, each compartment was washed with PBS (1 mL, 2X), mounted in Fluoromount-G (Thermo Fisher) and imaged by fluorescence microscopy (Olympus FV1000).
Sample preparation for chemoproteomic S-sulfinylome analysis.
Cell pellets from A549, HeLa or MEF cells treated with or without H2O2 (1 mM, 15 min), were washed with PBS (3X), and lysed on ice for 20 min in four volumes of NETN buffer (50 mM HEPES pH 7.5, 150 mM NaCl, and 1% Igepal) supplemented with 10 μL/mL of HALT protease and phosphatase inhibitors (Life Technologies, 78444) with 200 U/mL catalase. After 20 min incubation on ice with frequent mixing, the resulting native proteomes were incubated with 4-DPS (200 mM) at rt for 1 h. Excess small-molecules were removed by methanol/chloroform precipitation, as described previously4. The resulting protein pellets were resuspended in 0.5% SDS and incubated with DiaAlk (5 mM) in the dark at 37 °C for 2 h. The DiaAlk-labeled proteome was then treated with DTT (8 mM) at rt for 1 h. Reduced cysteines then were alkylated with IAM (32 mM) in the dark at rt for 30 min. Proteins were then precipitated with methanol-chloroform, as described4. Protein pellets were resuspended in 50 mM ammonium bicarbonate, analyzed by BCA assay and adjusted to a concentration of 2 mg/mL. Resuspended protein was digested with sequencing grade trypsin (Promega) at a 1:50 (enzyme/substrate) ratio for 8h at 37 °C. The tryptic digests were desalted with HLB extraction cartridges (Waters) and desalted samples were evaporated to dryness under vacuum. Desalted tryptic digests were reconstituted in a solution of 30% ACN at pH ~6. Click chemistry was performed by the addition of 1 mM light or heavy Azido-UV-biotin, 10 mM sodium ascorbate, 1 mM TBTA, and 10 mM CuSO4. Samples were allowed to react with rotation in the dark at rt for 2 h. Light and heavy isotopic tagged samples were mixed immediately following click chemistry. Samples were then purified by strong cation exchange (SCX) chromatography, as described4, and enriched with pre-washed streptavidin sepharose beads at rt for 2 h. After capture, the streptavidin sepharose beads were sequentially washed with 50 mM NaAc, pH 4.5, 50 mM NaAc, pH 4.5 containing 2 M NaCl, then ddH2O (2X each) with vortexing and/or rotation to remove non-specifically bound peptides and reconstituted in 25 mM ammonium bicarbonate. The streptavidin sepharose suspension was transferred to several glass tubes (VWR), irradiated with 365 nm UV light (Entela, Upland, CA) at rt with stirring for 2 h. The supernatant was collected, evaporated to dryness under vacuum, and stored at −20 °C until LC-MS analysis.
Sample preparation for chemoproteomic S-sulfenylome analysis.
Cell pellets from A549 or HeLa cells treated with or without H2O2 (1 mM, 15 min), were washed with PBS (3X), and lysed in the presence of BTD (5 mM) on ice for 20 min in NETN buffer with inhibitor cocktail with 200 U/mL catalase. After 20 min incubation on ice, native proteomes were clarified by centrifugation and further incubated at rt for 2 h. The BTD-labeled proteome was then processed for chemoproteomic analysis, as described above.
LC-MS.
BTD-based S-sulfenome and DiaAlk-based S-sulfinylome analyses were performed on a Q Exactive plus (Thermo Scientific) operated with an Easy-nLC1000 system (Thermo Scientific). Samples were reconstituted in 0.1% formic acid and pressure-loaded onto a 2 cm microcapillary precolumn packed with C18 (3 μm, 120 Å, SunChrom, USA). The pre-column was connected to a 12 cm 150-μm-inner diameter microcapillary analytical column packed with C18 (1.9 μm, 120 Å, Dr. Maisch GebH, Germany) and equipped with a homemade electrospray emitter tip. The spray voltage was set to 2.0 kV and the heated capillary temperature to 320 °C. LC gradient consisted of 0 min, 7% B; 14 min, 10% B; 51 min, 20% B; 68 min, 30% B; 69–75 min, 95% B (A = water, 0.1% formic acid; B = ACN, 0.1% formic acid) at a flow rate of 600 nL/min. HCD MS/MS spectra were recorded in the data-dependent mode using a Top-20 method. MS1 spectra were measured with a resolution of 70,000, an AGC target of 3e6, a max injection time of 20 ms, and a mass range from m/z 300 to 1400. HCD MS/MS spectra were acquired with a resolution of 17,500, an AGC target of 1e6, a max injection time of 60 ms, a 1.6 m/z isolation window and normalized collision energy of 30. Peptide m/z that triggered MS/MS scans were dynamically excluded from further MS/MS scans for 18 s. For intact protein analysis, the sample was analyzed on an electrospray linear ion trap mass spectrometer (LTQ-XL, Thermo Scientific) after separation on an Agilent Eclipse XDB-C8 2.1 mm x 15 mm trap with mobile phases A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile) which was used to trap, desalt, and elute proteins onto a Varian 2.1 mm x 50 mm 5 μm PLRP-S C18 column with a gradient of 5% to 100% B in 14 min at a flow rate of 200 μL/min.
Peptide identification and quantification.
Raw data files were searched against Homo sapiens Uniprot canonical database (Dec 2, 2016, 20,130 entries) using pFind 3.1.2 software (http://pfind.ict.ac.cn/software/pFind3/index.html)53–55. Precursor ion mass and fragmentation tolerance were 10 ppm and 20 ppm, respectively for the database search. A specific-tryptic search was employed with a maximum of three missed cleavages allowed. The maximum number of modifications allowed per peptide was three. For DiaAlk-based chemoproteomic S-sulfinylome analysis, modifications of +15.9949 Da (methionine oxidation, M), + 57.0214 Da (IAM alkylation, C), +387.1754 (C15H25N5O7, DiaAlk-triazohexanoic acid, C) were searched as dynamic modifications. For BTD-based chemoproteomic S-sulfenylome analysis, modifications of +15.9949 Da (methionine oxidation, M), + 57.0214 Da (IAM alkylation, C), +418.1312 (C19H22N4O5S, BTD-triazohexanoic acid, C). No fixed modifications were searched for both S-sulfenylome and S-sulfinylome analyses. A differential modification of 6.0201 Da on probe-derived modifications was used for all analyses. False discovery rates (FDRs) at spectrum, peptide, and protein level were < 1%. Quantification of heavy to light ratios (RH/L) was performed using pQuant as previously described56–58, which directly uses the RAW files as the input. pQuant calculates RH/L values based on each identified MS scan with a 15 ppm-level m/z tolerance window and assigns an interference score (Int. Score, also known as confidence score σ) to each value from zero to one. In principle, the lower the calculated Int. Score, the less co-elution interference signal was observed in the extracted ion chromatograms. In this regard, the median of peptide ratios with sigma less than or equal 0.5 were considered to calculate site-level ratios.
Srx-dependent reduction of sulfinic acid in recombinant proteins by BioDiaAlk.
S-sulfinylated proteins were incubated with recombinant SRX at the indicated concentrations in 100 mM HEPES containing 100 mM NaCl, pH 8. The reaction was performed in a 40 μL-reaction volume in the presence of 2 mM DTT, 1 mM ATP and 2 mM MgCl2 at 37 ºC. After 1 h, excess small molecules were removed by size-exclusion (Zeba spin column, 7K cutoff, 0.5 mL, Pierce), and labeled with BioDiaAlk (100 μM) in the dark at rt for 1 h.
Luminescence assay for Srx activity.
ATP-dependent SRX activity was measured by incubating recombinant S-sulfinylated protein with SRX at the indicated concentrations in an assay buffer containing 100 mM Hepes, pH 8, 100 mM NaCl, 2 mM DTT, 1 mM ATP and 2 mM MgCl2 at 37 º C for 2 h. The ADP by-product generated during Srx-mediated reduction of cysteine sulfinic acid was detected using the ADP-Glo™ assay (Promega) following the manufacturer’s protocol. Briefly, 5-μL of each reaction was incubated with 5 μL ADP-Glo™ reagent in a white 384-well plate at rt for 40 min, followed by dispensing of 10 μL of ADP-Glo detection reagent, and incubation at rt for 30 min. The plates were then read for luminescence signal using an Envision plate reader (PerkinElmer).
Expression of epitope-tagged proteins.
Full-length cDNA encoding human CFL1 (NM_005507) in pDONR223 was purchased from YouBao and subcloned into pLX304 (Addgene). The C39A mutant was generated by QuikChange mutagenesis using the primer CAAGAAGGCGGTGCTCTTCGCCCTGAGTGAGGACAAGAAG and cloned into the pLX304 vector containing the V5 epitope. HEK293T cells were transformed by incubating 30 μg of each of plasmid and 270 μL of polyethylenimine with 80%-confluent HEK293T cells on a 10-cm dish. After 6 h, cells were cultured in DMEM supplemented with 10% FBS for another 48 h. Full-length pJ3H-PTPN1 and -C215S PTPN1 mammalian expression vectors containing the HA-epitope have been described previously.59 HeLa cells were transformed at 85% confluency on a 10-cm dish and cultured, as described for HEK293T cells above.
Sample preparation for analyzing S-sulfinylation of epitope-tagged proteins.
HEK293T cells expressing V5-tagged wild-type and C39A CFL1 were washed twice with ice cold PBS and harvested. Cells then were lysed in NETN lysis buffer (50 mM HEPES, 150 mM NaCl, 1% Igepal, pH 7.5) containing inhibitor cocktail, 200 U/mL catalase, and 4-DPS (200 mM). Reagents were removed by precipitating with chloroform/methanol. The protein precipitate was resuspended in 500 μL 0.4% SDS and labeled with DiaAlk (5 mM) at 37 °C for 2 h. Proteins were precipitated a second time, resuspended in 500 μL 0.4% SDS and incubated with 0.2 mM azide-biotin, 1 mM sodium ascorbate, 0.1 mM TBTA and 1 mM CuSO4 at rt for 2 h. Excess reagents were removed by methanol/chloroform precipitation and the resulting protein pellets were dissolved in 0.4% SDS and enriched by streptavidin sepharose. After incubation at 4 °C for 16 h, the streptavidin resin was collected and washed with 1% SDS (2X), 4 M urea (2X), PBS (2X) and ddH2O (2X), resuspended in loading buffer, boiled to release the biotin-conjugated proteins, and analyzed by immunoblot. HeLa cells expressing HA-tagged wild-type and C215S PTPN1 were incubated with DiaAlk (5 mM) in serum-free media at 37 °C for 2 h. After labeling, cells were washed with PBS (3X) and lysed in NP-40 lysis buffer [50 mM Tris-HCl pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40, 50 mM NaF, 10 mM β-glycerolphosphate, 1 mM sodium vanadate, 1× EDTA-free protease cocktail inhibitors (Roche), and 200 U/mL catalase]. After 20 min on ice, cell debris was removed by centrifugation at 14,000 rpm at 4 °C for 15 min. After enrichment, HA-agarose resin was incubated with 100 μM azide-biotin, 500 μM BTTP, 250 μM CuSO4, and 2.5 mM sodium ascorbate in 50 mM HEPES, 100 mM NaCl, pH 7.4 in a final volume of 20 μL. After 1 h, the reaction was terminated with 10 μL Laemmli sample buffer, boiled to release biotin-conjugated proteins, and analyzed by immunoblot.
Immunoprecipitation.
HEK293T cells expressing V5-tagged WT and C39A CFL1 were washed with cold PBS (2X) and harvested. Cells were lysed in HEPES lysis buffer (50 mM HEPES, 150 mM NaCl, 1% Igepal, pH 7.5) containing inhibitor cocktail and 200 U/ml catalase. Lysates were incubated with V5 antibody (diluted at 1:50) at 4 °C for 6 h. Lysates were incubated with protein A/G magnetic beads (50 μL, Biotool, B23202) with rotation at rt for 1 h. Beads were washed with lysis buffer, eluted with 2X loading buffer and analyzed by immunoblot. HA-tagged wild-type and C215S PTPN1 were immunoprecipitated from 500 μg of lysate with 20 μL of anti-HA agarose (Pierce, 26181). After incubation at 4 °C for 16 h, the agarose resin was collected and washed with TBST (3X) and 50 mM HEPES, 100 mM NaCl, pH 7.4 (2X).
Statistics and reproducibility.
For intact protein MS, representative data from three independent experiments with one LC-MS run for each are shown. For immunoblotting DiaAlk and BioDiaAlk-tagged proteins, representative data from three independent experiments are shown. Raw scans of immunoblots are provided in Supplementary Fig. 22. For luminescence assays, representative data from three independent experiments are shown, each performed in triplicate, and error bars represent the standard deviation (SD). For A549 and HeLa cells, each DiaAlk-based chemoproteomic analysis was performed in three independent experiments with one LC-MS/MS run for each, and each BTD-based chemoproteomic analysis was performed in two independent experiments with two LC-MS/MS runs for each. The profiles for light- and heavy-labeled peptides are shown in red and blue, respectively. Heavy (H2O2) to light (control) ratios were calculated from three independent experiments and are displayed below the individual chromatograms. For MEF Srx +/+ and MEF Srx −/− cells, each DiaAlk-based chemoproteomic analysis was performed in two independent experiments with two LC-MS/MS runs for each. The profiles for light- and heavy-labeled peptides are shown in red and blue, respectively. Heavy (Recovery) to light (control) ratios were calculated from two independent experiments and are displayed below the individual chromatograms. All independent quantitative chemoproteomic experiments were started from cell cultures. The quality of these quantitative datasets is assessed by standard deviation (SD) value of all the measured ratios (H2O2 versus control or recovery versus non-recovery) and are provided in each Supplementary Dataset. The medium coefficient of variation for each quantitative dataset is provided in each Supplementary Dataset legend.
DATA AVAILABILITY STATEMENT
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files.
Supplementary Material
Acknowledgements
This work was supported by the National Key R&D Program of China (2016YFA0501303), the National Natural Science Foundation of China (31770885), and Beijing Nova Program (Z171100001117014) to J. Y., and the US National Institutes of Health (R01 GM102187 and R01 CA174864 to K.S.C) and (R01 GM072866 to W.T.L.). This work was also supported by the Wake Forest Baptist Comprehensive Cancer Center (P30CA012197) to W.T.L. We thank Q. Zhou and W. Leng (National Center for Protein Sciences • Beijing, China) for expert technical assistance, C. Liu and H. Chi (Institute of Computing Technology, CAS, Beijing, China) for helpful discussions in proteomic informatics, K. Tallman and N. Porter (Vanderbilt University, Nashville, TN) for providing light and heavy azido-UV-biotin reagents, P. Wu (The Scripps Research Institute, La Jolla, CA) for providing the BTTP click ligand, M. Wilson (University of Nebraska, Lincoln) for providing recombinant DJ-1, S. G. Rhee (Yonsei University College of Medicine, Seoul, Korea) and M. Toledano (Institut des Science du Vivant Frédérique Joliot, Gif-sur-Yvette, France) for providing Srx +/+ and Srx −/− MEF cells.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Paulsen CE & Carroll KS Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev 113, 4633–4679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta V & Carroll KS Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta 1840, 847–875 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta V, Yang J, Liebler DC & Carroll KS Diverse redoxome reactivity profiles of carbon nucleophiles. J. Am. Chem. Soc 139, 5588–5595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J et al. Global, in situ, site-specific analysis of protein S-sulfenylation. Nat. Prot 10, 1022–1037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Gupta V, Carroll KS & Liebler DC Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat. Comm 5, 4776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould NS et al. Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Cell Chem. Biol 22, 965–975 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Depuydt M et al. A periplasmic reducing system protects single cysteine residues from oxidation. Science 326, 1109–1111 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Paulsen CE et al. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol 8, 57–64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulathu Y et al. Regulation of A20 and other OTU deubiquitinases by reversible oxidation. Nat. Comm 4, 1569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seo YH & Carroll KS Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc. Natl. Acad. Sci. USA 106, 16163–16168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacob C, Holme AL & Fry FH The sulfinic acid switch in proteins. Org. Biomol. Chem 2, 1953–1956 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Woo HA et al. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J. Biol. Chem 278, 47361–47364 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Wood ZA, Poole LB & Karplus PA Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300, 650–653 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Biteau B, Labarre J & Toledano MB ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425, 980–984 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Chang TS et al. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem 279, 50994–51001 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Woo HA et al. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J. Biol. Chem 280, 3125–3128 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Lo Conte M & Carroll KS The redox biochemistry of protein sulfenylation and sulfinylation. J. Biol. Chem 288, 26480–26488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canet-Aviles RM et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 101, 9103–9108 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackinton J et al. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J. Biol. Chem 284, 6476–6485 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kil, et al. Circadian oscillation of sulfiredoxin in the mitochondria. Mol. Cell 59, 651–663 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Ramesh A et al. Role of sulfiredoxin in systemic diseases influenced by oxidative stress. Redox Biol. 2, 1023–1028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickinson BC & Chang CJ Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol 7, 504–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei Q, Jiang H, Matthews CP & Colburn NH Sulfiredoxin is an AP-1 target gene that is required for transformation and shows elevated expression in human skin malignancies. Proc. Natl. Acad. Sci. USA 105, 19738–19743 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Q et al. Sulfiredoxin-Peroxiredoxin IV axis promotes human lung cancer progression through modulation of specific phosphokinase signaling. Proc. Natl. Acad. Sci. USA 108, 7004–7009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H et al. Sulfiredoxin inhibitor induces preferential death of cancer cells through reactive oxygen species-mediated mitochondrial damage. Free Rad. Biol. Med 91, 264–274 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Woo HA & Rhee SG in Methods in Redox Signaling (ed. Dipak Das) Ch. 4, 19 (Mary Ann Liebert, Inc, 2010). [Google Scholar]
- 27.Lee CF, Paull TT & Person MD Proteome-wide detection and quantitative analysis of irreversible cysteine oxidation using long column UPLC-pSRM. J. Prot. Res 12, 4302–4315 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo YH et al. Profiling protein S-sulfination with maleimide-linked probes. Chembiochem 18, 2028–2032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo Conte M & Carroll KS Chemoselective ligation of sulfinic acids with aryl-nitroso compounds. Angew. Chem. Int. Ed 51, 6502–6505 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo Conte M, Lin J, Wilson MA & Carroll KS A chemical approach for the detection of protein sulfinylation. ACS Chem. Biol 10, 1825–1830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majmudar JD et al. Harnessing redox cross-reactivity to profile distinct cysteine modifications. J. Am. Chem. Soc 138, 1852–1859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitroka S et al. Direct and nitroxyl (HNO)-mediated reactions of acyloxy nitroso compounds with the thiol-containing proteins glyceraldehyde 3-phosphate dehydrogenase and alkyl hydroperoxide reductase subunit C. J. Med. Chem 56, 6583–6592 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlick TL, Ding Z, Kovacs EW & Francis MB Dual-surface modification of the tobacco mosaic virus. J. Am. Chem. Soc 127, 3718–3723 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Delaunay A, Pflieger D, Barrault MB, Vinh J & Toledano MB A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471–481 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Baek JY et al. Sulfiredoxin protein is critical for redox balance and survival of cells exposed to low steady-state levels of H2O2. J. Biol. Chem 287, 81–89 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim KH, Lee W & Kim EE Crystal structures of human peroxiredoxin 6 in different oxidation states. Biochem. Biophys. Res. Comm 477, 717–722 (2016). [DOI] [PubMed] [Google Scholar]
- 37.van Montfort RL, Congreve M, Tisi D, Carr R & Jhoti H Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 423, 773–777 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Mullen L, Hanschmann EM, Lillig CH, Herzenberg LA & Ghezzi P Cysteine oxidation targets peroxiredoxins 1 and 2 for exosomal release through a novel mechanism of redox-dependent secretion. Mol. Med 21, 98–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szabo-Taylor K et al. Oxidative and other posttranslational modifications in extracellular vesicle biology. Semin. Cell Dev. Biol 40, 8–16 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Porta C, Moroni M, Guallini P, Torri C & Marzatico F Antioxidant enzymatic system and free radicals pathway in two different human cancer cell lines. Anticancer. Res 16, 2741–2747 (1996). [PubMed] [Google Scholar]
- 41.Chauvin JR & Pratt DA On the reactions of thiols, sulfenic acids, and sulfinic acids with hydrogen peroxide. Angew. Chem. Int. Ed 56, 6255–6259 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Li H et al. Crystal Structure and Substrate Specificity of PTPN12. Cell Rep 15, 1345–1358, doi: 10.1016/j.celrep.2016.04.016 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Jönsson TJ et al. Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin. Biochemistry 44, 8634–8642 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klamt F et al. Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat. Cell Biol 11, 1241–1246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cameron JM et al. Polarized cell motility induces hydrogen peroxide to inhibit cofilin via cysteine oxidation. Curr. Biol 25, 1520–1525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamann M, Zhang T, Hendrich S & Thomas JA Quantitation of protein sulfinic and sulfonic acid, irreversibly oxidized protein cysteine sites in cellular proteins. Meth. Enzymol 348, 146–156 (2002). [DOI] [PubMed] [Google Scholar]
- 47.White MD et al. Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat. Comm 8, 14690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroder K NADPH oxidases in redox regulation of cell adhesion and migration. Antioxid. Redox Signal 20, 2043–2058 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Li X et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol. Cell 61, 705–719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun T et al. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell 144, 703–718, doi: 10.1016/j.cell.2011.02.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paulsen CE & Carroll KS Chemical dissection of an essential redox switch in yeast. Chem. Biol 16, 217–225 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Cheng H, Donahue JL, Battle SE, Ray WK & Larson TJ Biochemical and genetic characterization of PspE and GlpE, two single-domain sulfurtransferases of Escherichia coli. Open Microbiol. J 2, 18–28 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chi H et al. pFind-Alioth: A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. J. Proteomics 125, 89–97 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Li D et al. pFind: a novel database-searching software system for automated peptide and protein identification via tandem mass spectrometry. Bioinformatics 21, 3049–3050 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Wang LH et al. pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrom 21, 2985–2991 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Tan D et al. Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu C et al. pQuant improves quantitation by keeping out interfering signals and evaluating the accuracy of calculated ratios. Anal. Chem 86, 5286–5294 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Ma Y, McClatchy DB, Barkallah S, Wood WW & Yates JR 3rd. HILAQ: A novel strategy for newly synthesized protein quantification. J. Proteome Res 16, 2213–2220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia FJ & Carroll KS Redox-based probes as tools to monitor oxidized protein tyrosine phosphatases in living cells. Eur. J. Med. Chem 17, 28–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files.