Summary
FBXW7 is well characterized as a tumor suppressor in many human cancers including melanoma; however, the mechanisms of tumor suppressive function have not been fully elucidated. We leveraged two distinct RNA sequencing datasets: human melanoma cell lines (n=10) with control versus silenced FBXW7 and a cohort of human melanoma tumor samples (n=51) in order to define the transcriptomic fingerprint regulated by FBXW7. Here, we report that loss of FBXW7 enhances a mitochondrial gene transcriptional program that is dependent on MITF in human melanoma and confers poor patient outcomes. MITF is a lineage-specific master regulator of melanocytes, and together with PGC-1alpha is a marker for melanoma subtypes with dependence for mitochondrial oxidative metabolism. We found that inactivation of FBXW7 elevates MITF protein levels in melanoma cells. In vitro studies examining loss of FBXW7 and MITF alone or in combination showed that FBXW7 is an upstream regulator for the MITF/PGC-1 signaling.
Keywords: FBXW7, MITF, PGC-1alpha, mitochondria, metabolism, melanoma
Melanoma is an aggressive cancer with high mutational load due to ultraviolet irradiation affecting several ‘driver’ oncogenes and tumor suppressors along with ‘non-driver’ or ‘passenger’ mutations (Reddy, Miller, & Tsao, 2017). While frequently altered driver genetic events are well characterized, their functional repertoire is not fully explored. BRAF and NRAS are highly mutated oncogenes in melanoma that cooperate with tumor suppressors such as CDKN2A, TP53, PTEN, NF1, and FBXW7 for tumor initiation and progression (Aydin et al., 2017; Dankort et al., 2009; Maertens et al., 2013). FBXW7 encodes an E3 ubiquitin ligase that is mutated or deleted in a variety of cancers including melanoma (Aydin et al., 2014; Davis, Welcker, & Clurman, 2014; Kourtis, Strikoudis, & Aifantis, 2015). Several oncoproteins are substrates of FBXW7 including MTOR, CYCLIN E1, NOTCH1, MYC, JUN, and PCG-1alpha rendering FBXW7 protein as an efficient tumor suppressor (Davis et al., 2014; Mao et al., 2008). Albeit as a proven tumor suppressor in cancer and its ability to regulate key signaling pathways during tumorigenesis, its functional roles in melanoma is not fully explored (Aydin et al., 2017; Aydin et al., 2014).
MITF is a lineage-specific master regulator of melanocyte development, survival, differentiation, and function (Hsiao & Fisher, 2014; King et al., 1999). It stimulates the melanin biosynthetic pathway by activating transcription of pigmentation genes (TYR, TYRP1, DCT, PMEL, and MLANA), and regulates melanocyte survival (BCL2, BCL2A1) and proliferation (CDK2) (Hsiao & Fisher, 2014; King et al., 1999). In melanoma, its role is complex as MITF acts as an oncogene in a subset of melanomas (MITF-high) whereas its down-regulation is well characterized in others (MITF-low). Recent studies broaden the functional roles of MITF by identifying it as a key regulator for tumor metabolism in BRAFV600E–driven melanomas (Haq et al., 2013; Vazquez et al., 2013). Shifts within the tumor’s energy dynamics from oxidative phosphorylation to aerobic glycolysis or vice versa are well recognized, a metabolic reprogramming that occurs to meet the tumor’s energy requirements (Vander Heiden, Cantley, & Thompson, 2009). Oxidative phosphorylation depends on the ability of functionally intact mitochondria to metabolize oxygen, whereas glycolysis can occur independent of mitochondria. PGC-1alpha (encoded by the PPARGC1A gene) controls mitochondrial biogenesis and oxidative phosphorylation (Vazquez et al., 2013). MITF directly regulates PGC-1alpha transcription. MITF-expressing melanomas have higher levels of oxidative genes (Haq et al., 2013). BRAFV600E suppresses MITF/PGC-1alpha axis, and enhances aerobic glycolysis, and its inhibition with the BRAF inhibitor vemurafenib re-routes away from glycolysis towards oxidative phosphorylation (McQuade & Vashisht Gopal, 2015). In this study, we describe that the FBXW7 tumor suppressor pathway is a bona fide regulator of MITF and the MITF/PGC-1alpha axis that associates with poor patient outcomes.
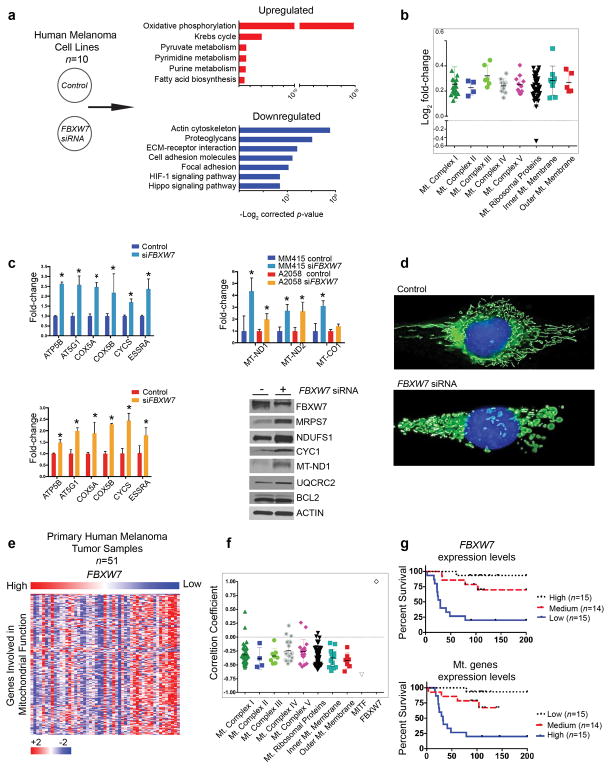
To gain insight into molecular mechanisms regulated by FBXW7 in melanoma, we examined transcriptional programs altered upon silencing of FBXW7 in ten human melanoma cell lines using RNA sequencing analysis. Functional gene groups involved in oxidative phosphorylation were found consistently elevated across the cell lines (Figure 1a). Specifically, a cohort of nuclear and mitochondrial-encoded genes encoding for components of the respiratory chain, inner and outer mitochondrial membrane factors, as well as those encoding for mitochondrial ribosomal proteins were found as significantly increased (Figure 1b). These results were validated using selected nuclear genes involved in regulating mitochondrial function and those encoded by mitochondria using qRT-PCR and western blotting (Figure 1c). We next examined morphological changes of mitochondria upon loss of FBXW7 in human melanoma cells. We noted that in control lines the mitochondria network demonstrates a normal distribution of fused and fragmented mitochondria, whereas in FBXW7 silenced cells an enhanced fragmented phenotype suggesting increased activity was noted (Figure 1d). To further validate these findings on human tumor samples, we interrogated our previously published RNA sequencing dataset (Badal et al., 2017) consisting human primary melanomas (n=51) as well as the TCGA melanomas. Our primary melanoma cohort consisted of treatment naïve melanomas representing early- (T1, ≤1.0 mm, n = 10), intermediate- (T2 and T3, 1.01–4.0 mm, n = 27), and late-category or advanced (T4, >4.0 mm, n = 14) tumors, with an overall survival of 90 months (mean) and 93.5 months (median) (Badal et al., 2017). We found that FBXW7 transcript levels anti-correlated with genes regulating mitochondrial function (Figure 1e, 1f, and Figure S1a). Down-regulation of FBXW7 transcripts and elevated levels of genes involved in mitochondrial function correlated with poor patient survival (Figure 1g). These results suggest that transcriptional silencing of FBXW7 associates with an active mitochondrial gene expression program and underlies an aggressive phenotype in melanoma. Intriguingly, we noted that FBXW7 was one of the genes with the highest inverse correlation with MITF (Figure 1f).
Figure 1. FBXW7 silencing enhances a mitochondrial gene transcription program.
(a) Human melanoma cell lines were transfected with either scrambled or FBXW7-specific pooled four distinct siRNAs (Dharmacon, Lafayette, CO), mRNA isolated, and subjected to RNA sequencing. Ten cell lines were examined: 501Mel, A2058, HT144, MeWo, MM415, MM485, SH4, SkMel3, WC119, and WM35. Differentially expressed genes between scrambled versus FBXW7 siRNA conditions were determined by using the limma software package (Law, Chen, Shi, & Smyth, 2014; Smyth, 2004). To identify functional pathways deregulated between the groups, the top 1000 up- and down-regulated genes were analyzed using DAVID (the Database for Annotation, Visualization, and Integrated Discovery). The x-axis represents the –log2 of the p-value and the y-axis the functional processes.
(b) Genes involved in mitochondrial function found as differentially expressed between scrambled versus FBXW7 siRNA (adjusted p-value ≤ 0.05) were grouped based on functional category. The x-axis represents mitochondrial functional groups and the y-axis the log2 fold-change.
(c) Two melanoma cell lines, A2058 and MM415, were transfected with scrambled or FBXW7-specific siRNA in triplicates, followed by total RNA isolation and qRT-PCR of genes for mitochondrial function, including those encoded by mitochondria (MT-ND1, MT-ND2, and MT-CO1). Selected proteins were validated by western blot using MM415 whole cell lysates. Densitometry analysis is indicated in Figure S1b.
(d) MM415 cells were transfected with scrambled or FBXW7-specific siRNA for 48 hours. Cells were seeded on rat-tail collagen I coated plates for 24 hours prior to the following indicated treatments. Mitochondria and nuclei were labeled with MitoTracker Green (100 nM) and Hoechst 33342 (20 mM) for 30 minutes at 37°C, respectively. Phenol red free media supplemented with 10% FBS, 2 mM L-glutamine, and antibiotics was used for all imaging performed on a Zeiss Imager.Z1 equipped with a N-Achroplan 40×/0.75 water immersion lens and an AxioCAM MRm digital camera. Images were captured using AxioVision 4.8 and Zeiss Zen software.
(e) Our previously published (Badal et al., 2017) RNA-Seq dataset of human primary melanomas (n=51) was queried for FBXW7 and mitochondrial genes. Heatmap represents supervised clustering based on FBXW7 mRNA expression levels (high to low, x-axis) and mitochondrial genes (y-axis).
(f) Pearson correlation coefficient values (+1 to −1, y-axis) for FBXW7 and genes regulating mitochondrial function (x-axis) were calculated from the RNA-Seq dataset, and are shown in a dot plot graph that also includes MITF.
(g) Kaplan-Meier survival curves of the patient cohort were examined for FBXW7 and genes regulating mitochondrial function. Survival curves showing clinical outcome of melanoma patients based on FBXW7 (top) and mitochondrial gene (bottom) expression levels are indicated. Samples are divided into three groups of similar size with high (n=15), medium (n=14), and low (n=15) expression levels.
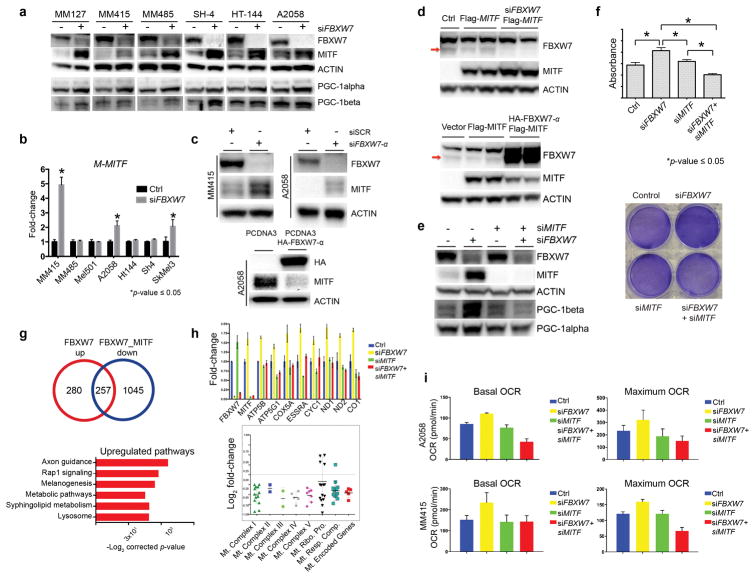
The MITF/PGC-1alpha axis regulates mitochondrial oxidative phosphorylation and detoxification of reactive oxygen species (Haq et al., 2013; Vazquez et al., 2013). While PGC-1alpha is a substrate of FBXW7 (Olson et al., 2008), to our knowledge, modulation of MITF by FBXW7 and FBXW7/MITF/PGC-1alpha in the context of tumor metabolism has not been previously described. Based on our findings in human melanoma samples, we next examined the relationship between FBXW7 and MITF by silencing FBXW7 using FBXW7-specific siRNA in six melanoma cell lines. We observed consistent and significant increase of MITF protein levels upon loss of FBXW7 in these cells that harbored either oncogenic BRAFV600E or NRASQ61 (Figure 2a). As expected, PGC-1alpha and PGC-1beta showed uniform increase. To determine whether this regulation is at the transcriptional level, we examined MITF mRNA levels in a panel of FBXW7-silenced melanoma cell lines by qRT-PCR. MITF mRNA was elevated in some, but not all cell lines (Figure 2b). Isoforms of FBXW7 are expressed in different cellular compartments; the alpha isoform is expressed in the nucleus (Davis et al., 2014). We next confirmed that FBXW7-α regulates MITF by loss-of-function or gain-of-function approaches using alpha isoform-specific-siRNA and -expression vector, respectively (Figure 2c). These results were validated in 293T cells where MITF is not constitutively expressed (Figure 2d). When FBXW7 and MITF were silenced together versus individually, we found that PGC-1alpha levels and cell survival characteristics regulated by FBXW7 were dependent on MITF (Figure 2e and 2f). To further explore the MITF-dependency in the context of FBXW7 inactivation, we silenced FBXW7 and MITF alone or in combination together with scrambled siRNA, and performed RNA sequencing. These experiments confirmed that genes regulating mitochondrial function and energy homeostasis are mediated by inactivation of the FBXW7 tumor suppressor that is dependent on MITF, delineating a previously unknown signaling link between FBXW7/MITF/PGC-1alpha (Figure 2g and 2h). To determine the functional impact of our findings, specifically on the mitochondrial respiration, we next examined oxygen consumption rate (OCR) in control versus silenced FBXW7 and MITF alone or in combination in two melanoma cell lines using the Seahorse Mito Stress platform. We noted significant increase of mitochondrial respiration upon inactivation of FBXW7 that was dependent on MITF, providing further evidence on the regulatory role of FBXW7 on the function of mitochondria (Figure 2i).
Figure 2. FBXW7 tumor suppressor modulates MITF, and MITF-dependent mitochondrial gene transcriptional program and oxidative metabolism.
(a) MITF protein levels were assayed using Western blotting (Pierce, Waltham, MA, USA) following transient transfection of either scrambled or FBXW7-specific siRNA in a panel of human melanoma cell lines. MM127, MM415, and MM485 harbor an NRASQ61 mutation whereas SH4, HT144, and A2058 melanoma lines have the BRAFV600E mutation. PGC-1alpha (Santa Cruz Biotechnology, Inc. Dallas, TX, USA) and PGC-1beta (Bethyl Laboratories, Inc. Montgomery, TX, USA) levels are shown. β-actin (Cell Signaling Technology, Inc., Danvers, MA, USA) was used as loading control. Densitometry is depicted in Figure S2.
(b) A panel of human melanoma cell lines were transfected with scrambled or FBXW7-specific siRNA and at 72h was processed for total RNA isolation and qRT-PCR for M-MITF. Expression levels of M-MITF were normalized to β-actin. The data plotted represent means ± standard deviations of three independent experiments. Significance is indicated by p-value ≤ 0.05.
(c) MITF levels were examined by Western blotting after silencing FBXW7 with FBXW7-α specific siRNA in MM415 and A2058 lines (top panel) and overexpressing FBXW7-α using HA-tagged FBXW7-α expression plasmid (bottom panel) in the A2058 human melanoma cell line. Experiments were performed at 72h. Densitometry is depicted in Figure S2.
(d) MITF levels were examined upon silencing of FBXW7 using FBXW7-specific siRNA in the presence of flag-tagged MITF expression vector or expression vector alone (top panel). Empty vector and flag-tagged MITF expression plasmid alone or together with HA-tagged FBXW7-α were transiently transfected in 293T cells. MITF protein levels were examined using Western blotting (bottom panel). β-actin was used as loading control. Densitometry is depicted in Figure S2.
(e) FBXW7 and MITF were silenced alone or in combination using gene-specific siRNAs in the MM415 human melanoma cell line (siMITF from Dharmacon, Lafayette, CO) followed by Western blotting. MITF, PGC-1alpha, and PGC-1beta levels were analyzed. β-actin was used as loading control. Densitometry is depicted in Figure S2. Densitometry is depicted in Figure S2.
(f) FBXW7 and MITF were silenced alone or in combination using gene-specific siRNAs in MM415 melanoma cells followed by XTT assay (Cell Signaling Technology, Inc., Danvers, MA, USA) to analyze cell viability using manufacturer’s instructions (top panel). The absorbance was read at 450nm in a 96-well plate reader. Representative crystal violet stainings are depicted in the bottom panel showing cellular density in the different experimental conditions.
(g) FBXW7 and MITF were silenced alone or in combination using gene-specific siRNAs in MM415 cells followed by total RNA isolation, library preparation (TruSeq RNA Library Prep Kit Illumina, San Diego, CA), and sequencing (Illumina NextSeq500, San Diego, CA), and analyzed as described previously (Badal et al., 2017). Differentially expressed genes for each condition were analyzed. Functional pathways and genes regulating mitochondrial function deregulated upon FBXW7 silencing that are dependent on MITF are selected and displayed.
(h) Validation experiment was performed using qRT-PCR on the same conditions using a selected number of genes involved in mitochondrial function.
(i) Oxygen consumption rate (OCR) was examined through the Seahorse technology using the manufacturer’s guidelines (Santa Clara, CA, USA). A2058 and MM415 melanoma cells were transfected with either scrambled, or gene-specific siRNAs for FBXW7 and MITF alone or in combination. At 72 hours the Mito Stress Test was performed and the data was normalized by a colorimetric assay.
In summary, we characterized the transcriptomic signature of human melanomas expressing low levels of FBXW7 that associated with poor patient outcomes. We identified FBXW7 tumor suppressor as a regulator of MITF. Specifically, a global up-regulation of a mitochondrial gene transcriptional program and mitochondrial function dependent on MITF/PGC-1alpha axis was defined. Our data suggested that FBXW7 regulates MITF via post-transcriptional mechanisms. Our studies expand the functional repertoire of FBXW7 tumor suppressor in melanoma.
Supplementary Material
Significance.
The functional roles of FBXW7 in melanoma are not fully elucidated. Our study provide evidence of a novel mechanism of FBXW7 by regulating MITF and the MITF/PGC-1alpha axis to support the metabolic requirements of the tumor, and underscore a subtype of melanomas with elevated oxidative phosphorylation and poor prognosis.
Acknowledgments
This work has been supported in part by the Icahn School of Medicine at Mount Sinai (J.T.C.), a pilot developmental research grant from the Tisch Cancer Institute (J.T.C. and J.E.C), a gift from the Dow Family Charitable Fund (J.T.C.), and research grants from the National Institutes of Health (CA158557 and CA177940 (J.T.C), CA206005 (J.E.C.), and the Tisch Cancer Institute Cancer Center Support Grant P30 CA196521).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- Aydin IT, Abbate F, Rajan GS, Badal B, Aifantis I, Desman G, Celebi JT. FBXW7 inactivation in a BrafV600E-driven mouse model leads to melanoma development. Pigment Cell Melanoma Res. 2017 doi: 10.1111/pcmr.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin IT, Melamed RD, Adams SJ, Castillo-Martin M, Demir A, Bryk D, … Celebi JT. FBXW7 mutations in melanoma and a new therapeutic paradigm. J Natl Cancer Inst. 2014;106(6):dju107. doi: 10.1093/jnci/dju107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badal B, Solovyov A, Di Cecilia S, Chan JM, Chang LW, Iqbal R, … Celebi JT. Transcriptional dissection of melanoma identifies a high-risk subtype underlying TP53 family genes and epigenome deregulation. JCI Insight. 2017;2(9) doi: 10.1172/jci.insight.92102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, … Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26(4):455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, … Widlund HR. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23(3):302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao JJ, Fisher DE. The roles of microphthalmia-associated transcription factor and pigmentation in melanoma. Arch Biochem Biophys. 2014;563:28–34. doi: 10.1016/j.abb.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R, Weilbaecher KN, McGill G, Cooley E, Mihm M, Fisher DE. Microphthalmia transcription factor. A sensitive and specific melanocyte marker for MelanomaDiagnosis. Am J Pathol. 1999;155(3):731–738. doi: 10.1016/S0002-9440(10)65172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, Strikoudis A, Aifantis I. Emerging roles for the FBXW7 ubiquitin ligase in leukemia and beyond. Curr Opin Cell Biol. 2015;37:28–34. doi: 10.1016/j.ceb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, … Cichowski K. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3(3):338–349. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, Balmain A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321(5895):1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade JL, Vashisht Gopal Y. Counteracting oxidative phosphorylation-mediated resistance of melanomas to MAPK pathway inhibition. Mol Cell Oncol. 2015;2(3):e991610. doi: 10.4161/23723556.2014.991610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22(2):252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BY, Miller DM, Tsao H. Somatic driver mutations in melanoma. Cancer. 2017;123(S11):2104–2117. doi: 10.1002/cncr.30593. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, … Puigserver P. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23(3):287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.