Abstract
Endoplasmic Reticulum aminopeptidase 1 (ERAP1) is an intracellular enzyme that can generate or destroy potential peptide ligands for MHC class I molecules. ERAP1 activity influences the cell-surface immunopeptidome and epitope immunodominance patterns but in complex and poorly understood manners. Two main distinct pathways have been proposed to account for ERAP1’s effects on the nature and quantity of MHCI-bound peptides: i) ERAP1 trims peptides in solution, generating the correct length for binding to MHCI or overtrimming peptides so that they are too short to bind, and ii) ERAP1 trims peptides while they are partially bound onto MHCI in manner that leaves the peptide amino terminus accessible. For both pathways, once an appropriate length peptide is generated it could bind conventionally to MHCI, competing with further trimming by ERAP1. The two pathways, although not necessarily mutually exclusive, provide distinct vantage points for understanding of the rules behind the generation of the immunopeptidome. Resolution of the mechanistic details of ERAP1-mediated antigenic peptide generation can have important consequences for pharmacological efforts to regulate the immunopeptidome for therapeutic applications, and for understanding association of ERAP1 alleles with susceptibility to autoimmune disease and cancer. We review current evidence in support of these two pathways and discuss their relative importance and potential complementarity.
Keywords: antigen processing, MHC class I, aminopeptidase, ERAP1, peptide, structure
MHCI peptide binding and presentation
The MHCI antigen processing and presentation pathway samples cellular proteins and presents peptide fragments bound to MHCI molecules for scrutiny by CD8+ T cells [1]. Many of these peptides are generated after protein degradation by the proteasome, a multi-subunit enzyme that degrades proteins to peptides 4–20 amino acids in length [2]. The majority of these peptides are rapidly destroyed by cytosolic aminopeptidases, but a small fraction escapes to the endoplasmic reticulum (ER) through the action of the transporter associated with antigen processing, TAP [3]. Most peptides translocated into the ER contain the same C-terminus as the antigenic epitopes finally presented by MHCI, due to similarly of proteasome cleavage sites and MHC peptide binding preferences, but many contain N-terminal extensions that have to be trimmed by ER resident aminopeptidases (ERAPs), before loading onto MHCI molecules [4]. MHCI molecules assemble in the ER with a polymorphic type I integral membrane glycoprotein heavy chain and an invariant soluble light chain (β2 microglobulin, β2m). The heavy chain forms the peptide binding site, a groove on the upper surface of the MHCI molecule [5]. MHCI residues within the binding groove interact via hydrogen bonds and salt bridges with the free N- and C-termini of peptides, thus limiting binding to (mostly) peptides of 8–9 residues. Side-chain binding pockets within the peptide binding site accommodate two or more residues (anchor residues) of the bound peptide [6, 7]. MHCI molecules are highly polymorphic with variation focused in side-chain binding pockets, so that a large variety of different peptides can be bound by various MHCI allelic variants. The loading of the peptide results in stabilization of MHCI-peptide complex, dissociation from the ER peptide loading machinery, and transit through Golgi to the cell surface to become available for immunosurveillance by receptors on CD8+T cells [8, 9].
Biological Function of ERAP1
N-terminally extended antigenic peptide precursors that survive cytosolic degradation and enter the ER, need to be further processed by ERAP1 and/or ERAP2, in order to acquire the right length required for MHCI binding. ERAP1 (ERAAP in mouse) is an IFN-γ-inducible, metalloaminopeptidase that trims N-terminal residues of extended peptide precursors down to 8-9 amino acids [10–12]. ERAP1 can also limit the production of antigenic peptides by overtrimming them [12, 13]. Several independent studies using RNA interference or knock-out animals confirmed this key involvement of ERAP1 in antigen presentation [10–13]. In addition these studies demonstrated that the absence of ERAP1 activity can lead to reduced number of peptide-MHCI complexes on the cell surface that also have a shorter half-life [14, 15]. In ERAP1-deficient mice the reduction in MHCI expression was proven to be due to faster dissociation of peptide-MHCI from the cell surface rather than to a slower rate of complex assembly in the ER [16]. Furthermore, ERAP1 plays roles in vivo in immune responses to viruses, either enhancing or reducing CTL responses to particular viral epitopes and thereby helping establish immunodominance hierarchy [17]. ERAP1 has also been shown to regulate tumor immunogenicity and cytotoxic response to cancer cells [18–20] and to play a role in regulating innate and inflammatory immune responses [21–23].
Effects of ERAP1 on the immunopeptidome
The collection of peptides that are presented by MHCI molecules on the cell surface define the immunopeptidome [24]. Although MHCI allelic variation is the most important determinant of the composition of the immunopeptidome, ERAP1 is an additional key editor that affects immunopeptidome quality and constitution [25, 26]. As noted above, reduction or loss of ERAP1 expression in cell lines or mouse models, led to presentation of unstable and structurally unique peptide-MHCI complexes to the cell surface. Many conventional complexes were lost, while concomitantly, new, highly immunogenic complexes emerged [14]. In other words, in the absence of ERAP1 trimming activity, some endogenous antigens were poorly presented, while presentation of other antigens was enhanced or remained unaffected, suggesting a selective effect on the repertoire of peptide-MHCI complexes [16, 17, 27, 28]. Additionally, a large fraction of the novel epitopes that arose, were longer in length and carried extended N-termini [4]. Taken together, it appears that ER proteolysis defines the composition and the structure of peptides presented to CD8+ cells.
Dimerization and synergism with ERAP2
Although ERAP1 is considered to be the dominant aminopeptidase in the ER for trimming peptide precursors, recent work has highlighted a second trimming enzyme, ERAP2, to be important for correct antigen processing. ERAP2 is highly homologous to ERAP1 (50% sequence identity) and can efficiently generate antigenic peptides in vitro [29, 30]. ERAP2 however, shows distinct trimming patterns compared to ERAP1, has a different specificity for N-terminal residues and appears to select peptides of different lengths [30, 31]. It has been proposed that ERAP2 activity is complementary to ERAP1 and several antigenic epitopes with complex or longer extensions seem to require both ERAP1 and ERAP2 for correct processing [29, 32]. ERAP1 and ERAP2 co-localize inside the cell, co-elute in chromatographic microsome fractionation and co-immunoprecipitate [29]. This has led to the suggestion that the human ER has a non-redundant system of physically associated ERAP1 and ERAP2 heterodimer that ensures highly efficient trimming of diverse precursor epitopes with complex N terminal extensions. A direct association of ERAP1 and ERAP2 has not however been demonstrated yet by biophysical methods. Recently, researchers used a leucine-zipper hetero-dimerization domain to construct an ERAP1/ERAP2 heterodimer and demonstrated that dimerization affected their enzymatic properties, suggesting that an ERAP1/ERAP2 dimer may be the biologically relevant antigen trimming entity in humans [33]. Structural analysis of ERAP2 by x-ray crystallography indicated a putative homo-dimerization domain in ERAP2 that utilizes residues that are largely conserved in ERAP1 [34]. Furthermore, both ERAP1 and ERAP2 are retained in the ER although they do not possess any ER retention signals, suggesting that they are retained there though protein-protein interactions that exist only inside the ER [35]. Indeed, disulfide bond formation between exon 10 of ERAP1 and ERp44, a factor involved in disulfide bond formation in the ER, has been suggested to be the main mechanism for ERAP1 retention in the ER and to regulate its release and thus the extracellular degradation of angiotensin II, although other protein-protein interactions with ER chaperones or the Peptide Loading Complex may also play roles [36] [37, 38]. ERAP2 contains an exon of similar length, which has little homology with the ERAP1 exon 10 but still retains the ERp44 interacting cysteine and has been found to fold in an appendix-like structure containing an internal disulfide bond and a helical segment [30]. It is currently not known if this exon is responsible for ERAP2 retention in the ER, although in an ERAP2 crystal structure it was found to make interactions with an adjacent ERAP2 molecule in the crystal, suggesting that it may constitute a protein-protein interaction structural motif [30]. IRAP, on the contrary, does not contain a similar exon, but has a transmembrane and a cytosolic domain that target it to endosomal vesicles or the cell membrane and have been suggested to be important for interactions and signaling [39–41].
Polymorphic variation
ERAP1 is polymorphic and several single nucleotide polymorphisms (SNPs) in its gene that encode amino acid changes have been associated with predisposition to a variety of diseases, ranging from viral infections to cancer and autoimmunity [26, 42]. Functional studies on ERAP1 alleles have suggested that disease-associated SNPs affect enzymatic function and specificity [43–45]. Indeed, experiments comparing cell lines harboring different ERAP1 allelic forms have discovered distinct effects on the cellular immunopeptidome and provided hints on the pathogenesis of HLA-associated autoimmunity [46–48]. ERAP2 also harbors common coding SNPs although fewer than for ERAP1. The ERAP2 SNP rs2248374 has been found to regulate the expression levels of the enzyme via nonsense RNA mediated decay [49] and SNP rs2549782 codes for a substitution near the catalytic site of the enzyme, affecting both activity and specificity [50]. ERAP2 SNPs have been associated with predisposition to autoimmunity, preeclampsia and resistance to HIV infection, possibly due to changes in antigen processing and presentation [51–54].
Basic and unique enzymatic properties
ERAP1 is a member of the M1 family of aminopeptidases, which utilize a bound zinc ion to hydrolyze the amino terminal peptide bond of a substrate [55, 56]. This class of enzyme is found in bacteria and eukaryotes, with myriad substrate specificities and physiological roles, ranging from metabolism to neurotransmitter and hormone degradation, to immune function and antigen processing [57–60]. This versatility is possible due to the large size of the substrate binding pocket, which extends away from the active site and can accommodate substrates up to 20 amino acids long in the case of ERAP1 [10]. Different M1 family members have different preferences for substrate N-terminal residues, with ERAP1 exhibiting broad specificity for aliphatic residues [31]. Along the length of the substrate, ERAP1 exhibits preferences for particular side chains at particular positions, but in a more degenerate pattern than other more specific proteases. Two notable preferences are for a bulky aliphatic or aromatic group at S1’ (amino-terminal to the cleavage site), and for either a large hydrophobic group or a basic group at Somega, (carboxy-terminal side chain) [31, 61].
Unlike other M1 aminopeptidases, ERAP1 has a strong preference for trimming substrates longer than eight or nine amino acids [10, 12]. This activity is well suited for efficient generation of peptides of length suitable for loading onto MHC-I proteins. A mechanism for peptide-length dependent regulation of ERAP1 aminopeptidase activity has been proposed, based on cooperativity and allosteric activation observed for short but not full-length peptide substrates [62]. The model postulates a regulatory site distinct from the active site that can be accessed by long substrates, with binding to this site favoring adoption of a closed conformation with increased catalytic activity [62]. Crystal structures of a complex between the C-terminal domain of ERAP1 and a C-terminus of peptides presented in fusion with the same domain suggested a possible location of this regulatory site [63].
Known crystal structures
Crystal structures have been reported for ERAP1, ERAP2, and IRAP, the members of the oxytocinase subfamily of M1 aminopeptidases, all of which have roles in antigen presentation. The structures confirmed that these enzymes closely resemble the other M1 aminopeptidases structurally. They consist of four domains. Domain I is a mixed all-beta sandwich present in other aminopeptidases that caps off the amino-terminal side of active site. Domain II contains the active-site zinc in a thermolysin-like α/β fold. Domain III is an Ig-like domain unique to the M1 family. Domain IV contains 8 Armadillo-like helix-turn-helix repeats forming a bowl-like structure [64].
ERAP1 has been crystallized in two different conformations, wherein the C-terminal domain IV is either closely interacting with domains I/II and enclosing the active site (termed ‘closed’), or is rotated away in a hinge motion which exposes the active site (‘open’). This large motion correlates with a reorganization of the active site, with Tyr438 oriented in a catalytically active orientation only in the closed state. In the open structure, Tyr438 has rotated such that the hydroxyl group is displaced 5.5 angstroms relative to the closed structure, and is no longer oriented to interact with a putative bound substrate [65] [62]. This correlation between ERAP1 conformation and active site organization suggests that the enzyme might cycle between these states in order to bind substrate, catalyze hydrolysis, and release product. This is notable within the entire M1 aminopeptidase family, of which several members have been crystallized in the closed conformation, including the homologous ERAP2 and IRAP [30, 34, 66].
Conformational change and conformational dynamics
Conversion between structural conformations is a widely studied feature of many enzymes. Such structural alterations can range from small loop reorganizations to large domain motions [67]. One longstanding model of enzyme function assumes a structural change upon substrate binding referred to as a ‘induced fit’ [68]. If these conversions are required for catalysis and occur slowly relative to the enzyme catalytic rate, small perturbations in conformational dynamics can have notable effects on enzyme’s function [69]. The identification of two ERAP1 conformations by crystallography raises the possibility that conformational interconversion may occur for ERAP1 and possibly for other members of the M1 aminopeptidase family. Using molecular dynamics simulations, an energy landscape connecting the open and closed structures has been identified with several local minima. Simulations of polymorphic variants of ERAP1 indicate that Q730E and K528R can each alter the population of conformations [45]. The relative stability of the closed conformation requires electrostatic interactions between domains I/II and IV [70]. The molecular dynamics model was supported by concordance with small angle X-ray scattering data of mutants designed to disrupt these interdomain interactions [70].
Two pathways for peptide processing by ERAP1: solution versus on-MHC trimming
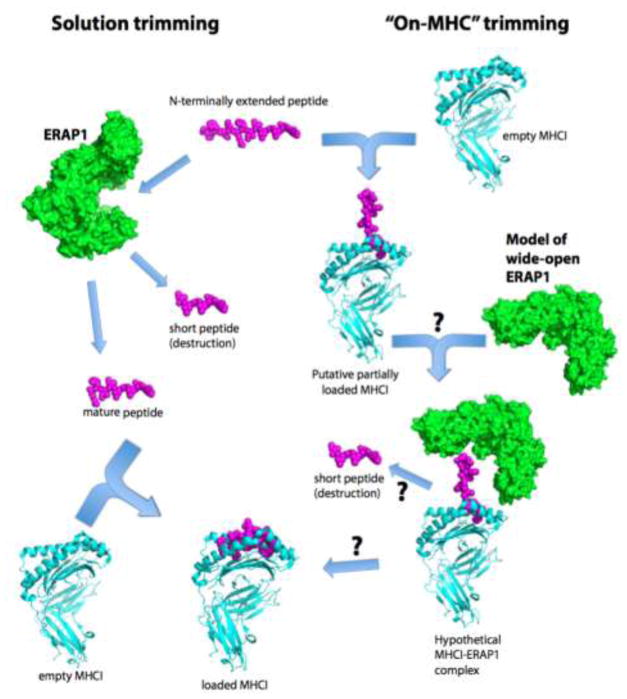
Although the importance for ERAP1 mediated peptide generation has been well established in the literature, the physiological substrate for ERAP1 trimming is controversial. As early as 1990, Falk et al. proposed several models to explain MHCI-restricted generation of antigenic peptides from extended precursors [71], generally encompassed by two main ideas: that the peptide is generated by proteolysis in solution before binding to MHCI, or it is generated while bound onto MHCI, using the latter as a template. Thus, in one possible pathway ERAP1 trims precursor peptides in solution and relies on its unique enzymatic properties to optimize them for MHCI; mature peptides then can bind onto MHCI at which point they are protected from any further ERAP1 trimming (Figure 1, left pathway). Alternatively, peptide precursors can bind onto the MHCI with N-terminal extensions such that ERAP1 can trim them while still bound onto the MHCI (Figure 1, right pathway). Competition between ERAP1 trimming and optimal binding of the peptide onto MHCI is the main driving force behind MHCI cargo optimization in either pathway. Below, we describe each pathway in more detail and outline the main supporting experimental evidence.
Figure 1.
Pathways for ERAP1-restricted peptide trimming. Left pathway, ERAP1 trims N-terminally extended peptides in solution down to mature, correct-length peptides that can then proceed to bind onto MHCI. Overtrimming to short peptides equates to epitope destruction. Right pathway, N-terminally extended peptides bind onto MHCI in a sub-optimal manner. Formation of a ternary complex with ERAP1 allows N-terminal trimming while the peptide is still bound on the MHCI. Optimal interactions between the generated mature epitope and the MHCI allow for ERAP1 dissociation.
Pathway 1: Peptide generation in solution – experimental evidence
Members of the M1 family of aminopeptidases, including ERAP1, trim peptidic substrates in solution [72]. Many studies have demonstrated that ERAP1 has this activity and can efficiently trim peptides in solution, generating appropriate epitopes for MHCI binding [10, 32, 43, 73–75]. Still, the catalytic efficiency of ERAP1 is not particularly high as suggested by a study comparing trimming of peptides by ERAP1 and Leucine aminopeptidase (LAP), which found that ERAP1 was about 50-fold slower [45]. The same study however, showed a lack of length preference by LAP, whereas a strong preference of ERAP1 for longer peptides, consistent with earlier reports [10, 45, 62]. The solution ability of ERAP1 to prefer to trim peptides longer than 9 residues and spare many shorter peptides has been proposed as a mechanism to specifically generate correct-length peptides for binding onto MHCI [10, 76].
Pathway 2: Peptide trimming while bound onto MHCI – experimental evidence
The unique biological function of ERAP1 and its indispensability for generating MHCI cargo suggests that this aminopeptidase may have specific properties suited for its biological purpose. Although currently there is no direct biophysical evidence that ERAP1 can interact with the MHCI, some studies have suggested that ERAP1 can trim peptides while they are bound onto MHCI. Specifically, Kanaseki et al. found that ERAAP (the mouse homologue of ERAP1) generated the correct H2-Ld ligand only when both ERAAP1 and H2-Ld were present in the ER of cells and otherwise overtrimmed the peptide, suggesting the MHC is indispensible for protecting the mature epitope from ERAAP [77]. In the same study, the authors detected ERAAP-generated, N-terminally extended peptides resistant to further trimming (due to a proline amino acid insertion), bound onto cell-surface MHC and theorized that ERAAP was trimming the initial precursor while bound onto the MHC. In a following study however, Infantes et al. found that H-2Ld actually protected N-terminally extended peptide precursors from ERAP1 and the 9mer epitope was only generated when the precursor was free in solution, not while bound to MHC [74]. In another study supportive of the on-MHC cleavage pathway, Reeves et al. found that ERAP1 was able to trim the N-terminal amino acid from a precursor inside the ER when the peptide was covalently tethered onto the H2-Kb MHC using a disulfide trap single-chain MHCI construct [44]. Finally, recently Chen et al. used a leucine zipper ERAP1-ERAP2 dimer construct to demonstrate in vitro trimming of N-terminally elongated epitope precursors bound by MHCI [78]. Curiously however, this ERAP1-ERAP2 construct appeared to trim through a Pro residue, in contrast to the inability of ERAAP to trim through Pro in the Kanaseki et al. study and others [11, 77, 79].
Interplay of trimming models with crystal structures and conformational dynamics
To date, the known experimental structures of ERAP1 and MHC-I are more consistent with pathway #1 than pathway #2. Crystal structures of ERAP1, even in the open conformation, cannot dock onto MHC-I such that a peptide N-terminus could sit/synonym? at the ERAP1 active site [62]. The same appears to apply for ERAP2 and IRAP [30, 80]. If ERAP1 has the ability to open further, however, this may allow docking onto MHC to form a ternary structure [81]. Indeed, molecular dynamics simulations have suggested a continuous but rugged energy terrain between different conformations of ERAP1, suggesting that other conformations are possible, albeit of high energy [70]. Even if such a “wide-open” structure could form however, then for efficient processing, Tyr438 would have to properly orient for catalysis when ERAP1 is in the open state. Interestingly, in a recent “semi-closed” crystal structure of the homologous IRAP, the catalytic Tyr was found to properly orient towards a transition-state analogue bound in the catalytic site [80, 82].
MHCI peptide editing, N-terminal extensions and the peptide loading complex
Conventionally, peptide-MHCI binding is thought to require both amino- and carboxyl-termini for stable interaction [83, 84]. Peptides with longer than optimal length (10–13 residues) have been observed to bind, but in a bulged conformation with buried termini and exposed central regions [85]. This might be thought to preclude on-MHC processing by ERAP, which requires a free amino terminus. Recently two groups have reported peptide-MHCI structures with protruding N-termini, for an HIV gag-derived epitope binding/bound to HLA-B57 [86] or HLA-B58 [87]. These add to an earlier report of a LCMV gp33-derived epitope binding to H2-Kb with an exposed N-terminal residue [88]. In each of these cases the peptide is bound throughout the full length of the peptide binding site, occupying with a small Ser or Ala side chain the “A pocket” that usually accommodates the amino terminus, making a sharp kink stabilized by several hydrogen bonds that orient one or two amino terminal residues outside the groove. Although these structures reveal an exposed amino-terminal residue, trimming by ERAP1 to generate a conventional 9mer-residue peptide would appear to require breaking many peptide-MHC interactions so that the active site could access the scissile bond. Even for the “wide-open” ERAP1 conformation more “wide-open” than currently observed by X-ray crystallography or SAXS, ERAP1 trimming has been estimated to require at least four residues protruding beyond the last MHC interaction sites [76, 89]. Thus these N-extended conformations are not likely to represent ERAP1 substrates without considerable additional structural rearrangement or stabilization of the MHC with partially-occupied peptide.
It is possible that cellular antigen presentation machinery can stabilize a complex between MHC and partially-bound peptide in a conformation that is suitable for on-MHC trimming. Nascent MHCI molecules without peptides are kept in a receptive state, through the formation of a complex (peptide loading complex, PLC) with TAP and other ER chaperone proteins, including tapasin, calnexin, calreticulin, ERp57 and protein disulfide isomerase, PDI [90, 91]. In this complex, tapasin interacts directly with MHCI molecules and is required for optimal peptide loading [92] via a peptide editing process [93], calnexin and calreticulin stabilize the heavy chain before β2-microglobulin association, and ERp57 is a thioreductase that supports the formation of a disulfide bridge that connects the walls of the MHCI peptide-binding groove with its base [91]. A recently determined structure of the PLC by cryo-EM has shed light on the structural composition of the complex but did not contain ERAP1, although some room for additional interactions is available [38]. The exact manner in which peptide enters the peptide loading complex is not clear, and it is possible that a complex of stabilized MHC with partially bound peptide is suitable for ERAP processing. ERAP1 has not been shown to directly interact with this complex, but thiol-dependent interactions similar to those observed for ERp44 could be envisioned that might promote processing of transient states.
Role of ERAP1 in antigenic peptide destruction
Whereas the two pathways described above present distinct strategies to generating MHCI peptide ligands, they also may affect the ability of ERAP1 to overtrim peptides to lengths too short for forming stable complexes with MHCI. Several studies have suggested an important role of ERAP1 in destroying peptides by overtrimming, effectively eradicating a pool of peptides from the antigenic peptide repertoire [12, 14, 27, 28, 94]. MHCI have also been demonstrated to be able to protect good peptide ligands from ERAP1 degradation [74, 77]. In Pathway #1, peptide destruction can occur in solution, determined by the affinity of the peptide for ERAP1. Conversely in Pathway #2, destruction can occur on-MHC, determined by a combination of peptide affinity for ERAP1 and MHCI. A combination of both processes however is also possible, with weakly-binding peptides dissociating off the MHCI to be trimmed by ERAP1 in solution. Currently however, the relative importance of destruction versus generation for shaping the immunopeptidome is not clear.
Functional repercussions of each trimming pathway
A main difference between the two pathways presented in Figure 1 lies in enzyme kinetics and selectivity. In pathway #1, the substrate for ERAP1 is free peptide, and so both kinetics and selectivity are determined by interactions between the peptide and ERAP1. In pathway #2 however, the ERAP1 substrate is the MHCI-peptide complex, and it is reasonable to argue that in this case both kinetics and selectivity are determined not only by interactions between the peptide and ERAP1, but also by as well as interactions between MHCI and ERAP1. Although a direct ERAP1/MHCI interaction has not been demonstrated up to date, parallels between the formation of a transient MHCI-ERAP1 complex and the formation of Peptide Loading Complex can be easily drawn [91]. Several studies have analyzed the kinetics and specificity of ERAP1 trimming peptides in solution and in general have found good, albeit not absolute, agreement between solution trimming and cell-based experiments [31, 44, 95]. On-MHCI ERAP1 trimming kinetic analysis has not been reported, although it may be a powerful tool to compare these two modes of action.
Differential effects on generation of immunopeptidome
A key distinction between the two pathways is the shift of the burden from determining selectivity of the antigen generation process from ERAP1 (pathway #1) to MHCI (pathway #2), a distinction that could strongly alter our understanding of how the immunopeptidome is generated. According to the on-MHC trimming pathway, the peptide is still partially bound onto MHC and as a result these peptide-MHCI interactions could guide peptide generation. Such a model reduces the importance of ERAP1 in shaping the immunopeptidome and highlights the importance of peptide-MHCI interactions, which are already well studied [96]. The effects of ERAP1 on the immunopeptidome are now well-documented by multiple studies, including specific effects on sequence [27, 28, 46, 97] making it difficult to imagine its role being limited to that of a general aminopeptidase activity. However, solution overtrimming could account for significant effects on the immunopeptidome, even if in the case of an on-MHC trimming pathway. More high-resolution proteomic studies may be necessary to discern the exact effect of ERAP1 selectivity in shaping the immunopeptidome.
Effects of trimming models on inhibitor design
Crystallographic analysis of ERAP1 has shown that the active site is reconfigured between the two known conformations and based on those reconfigurations it has been proposed that the closed conformation is the one that is enzymatically active [62, 65]. As noted above, the experimentally determined open conformation has a poorly structured S1 specificity pocket and a catalytic Tyr438 facing away from the Zn(II) atom, casting doubt on its catalytic efficiency. These considerations make structure-based or mechanism-based inhibitor design based on the open conformation difficult and as a result, the closed conformation of ERAP1 has been used for such approaches till now [98–100]. However, if the on-MHC trimming pathway with a “wide-open” conformation [89] is dominant in the cell, such design efforts may have to be redirected to using open conformations of ERAP1 for efficient targeting. Indeed, in a recent study, ERAP1 variants with weakened domain-closure interactions were found to be much more weakly inhibited by a mechanism-based transition-state analog inhibitor [70]. It should be noted however that the same compound as well as similar structure-inspired inhibitors have been found to be active in cells, indirectly suggesting that the closed ERAP1 conformation is indeed relevant in the cellular context [21, 31, 101] [99].
Possible complementarity of trimming pathways
Determining the exact pathway for the generation or destruction of MHCI peptide ligands by ERAP1 is important for our understanding of the shaping of the immunopeptidome and for designing inhibitors that can manipulate it. It is also possible however that both pathways operate in parallel depending on the peptide and its MHC-binding kinetics and affinity. Slow-binding or weaker binding peptides may spend more time in solution to be trimmed or destroyed by ERAP1, whereas some peptides may associate with MHCI immediately after they are translocated in the ER and can only be trimmed in that context. This notion adds an additional level of complexity in the generation of the immunopeptidome. The effects of ERAP1 deletion on the immunopeptidome are however complex [27, 28, 46] suggesting that our current understanding of the underlying mechanisms may not yet be sufficient to fully explain the physiological process.
On-MHC Trimming by ERAP2 and IRAP?
The other oxytocinase subfamily members ERAP2 and IRAP also have been proposed to play important roles in antigen processing in specific different cellular contexts [102]. ERAP2 has been proposed to act as an accessory aminopeptidase and supplement ERAP1 specificity while IRAP to operate in a separate pathway of cross-presentation [29, 103]. Interestingly ERAP2 and IRAP do not share the same enzymatic properties with ERAP1 regarding length preference and rather prefer to trim peptides of intermediate length [30, 45, 80]. Although ERAP2 was proposed to trim on-MHC bound peptide precursors in the context of an ERAP1/ERAP2 dimer, neither ERAP2 nor IRAP alone is known if they can perform the same function. Still, multiple conformations have been suggested to be accessible for these two enzymes also, opening up the possibility that they also employ similar mechanisms to peptide trimming as ERAP1 [30, 34, 80, 81].
Future directions – insights
It has been 15 years since the first description of the role of ERAP1 in antigenic peptide generation [11–13] and although many studies have demonstrated a key role in immune homeostasis and in immune dysfunction, significant controversy remains regarding its molecular mechanism and physiological substrate. The two pathways described in this review can serve as a distillation of current literature and as a starting point to design experiments to discern the relative importance and contributions of each proposed pathway in vivo. Given the importance of ERAP1 in shaping immune responses, a deeper understanding of the relative importance of solution versus on-MHC processing is necessary to allow both prediction of the repercussions of ERAP1 activity modulation on immune function and also the optimization of small molecule inhibitors of ERAP1 for pharmacological applications.
HIGHLIGHTS.
ERAP1 is an intracellular enzyme that generates peptide ligands for MHC-I molecules
It’s molecular mechanism and physiological substrate are still controversial
We review current knowledge regarding ERAP1’s structure, function and mechanism
We discuss the two main proposed pathways for peptide generation by ERAP1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Lawrence J. Stern, Email: lawrence.stern@umassmed.edu.
Efstratios Stratikos, Email: stratos@rrp.demokritos.gr, stratikos@gmail.com.
References
- 1.Rock KL, Reits E, Neefjes J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol. 2016;37:724–737. doi: 10.1016/j.it.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saunders PM, van Endert P. Running the gauntlet: from peptide generation to antigen presentation by MHC class I. Tissue Antigens. 2011;78:161–170. doi: 10.1111/j.1399-0039.2011.01735.x. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard N, Kanaseki T, Escobar H, Delebecque F, Nagarajan NA, Reyes-Vargas E, Crockett DK, Raulet DH, Delgado JC, Shastri N. Endoplasmic reticulum aminopeptidase associated with antigen processing defines the composition and structure of MHC class I peptide repertoire in normal and virus-infected cells. J Immunol. 2010;184:3033–3042. doi: 10.4049/jimmunol.0903712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hewitt EW. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology. 2003;110:163–169. doi: 10.1046/j.1365-2567.2003.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neefjes J, Ovaa H. A peptide’s perspective on antigen presentation to the immune system. Nat Chem Biol. 2013;9:769–775. doi: 10.1038/nchembio.1391. [DOI] [PubMed] [Google Scholar]
- 7.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 9.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 10.Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc Natl Acad Sci U S A. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 12.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 13.Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL, Tsujimoto M, Goldberg AL. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 14.Hammer GE, Gonzalez F, James E, Nolla H, Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat Immunol. 2007;8:101–108. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- 15.Yan J, Parekh VV, Mendez-Fernandez Y, Olivares-Villagomez D, Dragovic S, Hill T, Roopenian DC, Joyce S, Van Kaer L. In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J Exp Med. 2006;203:647–659. doi: 10.1084/jem.20052271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammer GE, Gonzalez F, Champsaur M, Cado D, Shastri N. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat Immunol. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 17.York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci U S A. 2006;103:9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James E, Bailey I, Sugiyarto G, Elliott T. Induction of Protective Antitumor Immunity through Attenuation of ERAAP Function. J Immunol. 2013;190:5839–5846. doi: 10.4049/jimmunol.1300220. [DOI] [PubMed] [Google Scholar]
- 19.Keller M, Ebstein F, Burger E, Textoris-Taube K, Gorny X, Urban S, Zhao F, Dannenberg T, Sucker A, Keller C, Saveanu L, Kruger E, Rothkotter HJ, Dahlmann B, Henklein P, Voigt A, Kuckelkorn U, Paschen A, Kloetzel PM, Seifert U. The proteasome immunosubunits, PA28 and ER-aminopeptidase 1 protect melanoma cells from efficient MART-126-35 -specific T-cell recognition. Eur J Immunol. 2015;45:3257–3268. doi: 10.1002/eji.201445243. [DOI] [PubMed] [Google Scholar]
- 20.Cifaldi L, Lo Monaco E, Forloni M, Giorda E, Lorenzi S, Petrini S, Tremante E, Pende D, Locatelli F, Giacomini P, Fruci D. Natural killer cells efficiently reject lymphoma silenced for the endoplasmic reticulum aminopeptidase associated with antigen processing. Cancer Res. 2011;71:1597–1606. doi: 10.1158/0008-5472.CAN-10-3326. [DOI] [PubMed] [Google Scholar]
- 21.Aldhamen YA, Pepelyayeva Y, Rastall DP, Seregin SS, Zervoudi E, Koumantou D, Aylsworth CF, Quiroga D, Godbehere S, Georgiadis D, Stratikos E, Amalfitano A. Autoimmune disease-associated variants of extracellular endoplasmic reticulum aminopeptidase 1 induce altered innate immune responses by human immune cells. J Innate Immun. 2015;7:275–289. doi: 10.1159/000368899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goto Y, Ogawa K, Nakamura TJ, Hattori A, Tsujimoto M. TLR-mediated secretion of endoplasmic reticulum aminopeptidase 1 from macrophages. J Immunol. 2014;192:4443–4452. doi: 10.4049/jimmunol.1300935. [DOI] [PubMed] [Google Scholar]
- 23.Cui X, Rouhani FN, Hawari F, Levine SJ. Shedding of the type II IL-1 decoy receptor requires a multifunctional aminopeptidase, aminopeptidase regulator of TNF receptor type 1 shedding. J Immunol. 2003;171:6814–6819. doi: 10.4049/jimmunol.171.12.6814. [DOI] [PubMed] [Google Scholar]
- 24.Admon A, Bassani-Sternberg M. The Human Immunopeptidome Project, a suggestion for yet another postgenome next big thing. Mol Cell Proteomics. 2011;10:O111 011833. doi: 10.1074/mcp.O111.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Georgiadou D, Stratikos E. Cellular mechanisms that edit the immunopeptidome. Curr Proteomics. 2009;6:13–24. [Google Scholar]
- 26.Alvarez-Navarro C, Lopez de Castro JA. ERAP1 structure, function and pathogenetic role in ankylosing spondylitis and other MHC-associated diseases. Mol Immunol. 2014;57:12–21. doi: 10.1016/j.molimm.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Nagarajan NA, de Verteuil DA, Sriranganadane D, Yahyaoui W, Thibault P, Perreault C, Shastri N. ERAAP Shapes the Peptidome Associated with Classical and Nonclassical MHC Class I Molecules. J Immunol. 2016;197:1035–1043. doi: 10.4049/jimmunol.1500654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnea E, Melamed Kadosh D, Haimovich Y, Satumtira N, Dorris ML, Nguyen MT, Hammer RE, Tran TM, Colbert RA, Taurog JD, Admon A. The Human Leukocyte Antigen (HLA)-B27 Peptidome in Vivo, in Spondyloarthritis-susceptible HLA-B27 Transgenic Rats and the Effect of Erap1 Deletion. Mol Cell Proteomics. 2017;16:642–662. doi: 10.1074/mcp.M116.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 30.Mpakali A, Giastas P, Mathioudakis N, Mavridis IM, Saridakis E, Stratikos E. Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (ER) Aminopeptidase 2. J Biol Chem. 2015;290:26021–26032. doi: 10.1074/jbc.M115.685909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zervoudi E, Papakyriakou A, Georgiadou D, Evnouchidou I, Gajda A, Poreba M, Salvesen GS, Drag M, Hattori A, Swevers L, Vourloumis D, Stratikos E. Probing the S1 specificity pocket of the aminopeptidases that generate antigenic peptides. Biochem J. 2011;435:411–420. doi: 10.1042/BJ20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorente E, Barriga A, Johnstone C, Mir C, Jimenez M, Lopez D. Concerted in vitro trimming of viral HLA-B27-restricted ligands by human ERAP1 and ERAP2 aminopeptidases. PLoS One. 2013;8:e79596. doi: 10.1371/journal.pone.0079596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evnouchidou I, Weimershaus M, Saveanu L, van Endert P. ERAP1-ERAP2 dimerization increases peptide-trimming efficiency. J Immunol. 2014;193:901–908. doi: 10.4049/jimmunol.1302855. [DOI] [PubMed] [Google Scholar]
- 34.Birtley JR, Saridakis E, Stratikos E, Mavridis IM. The crystal structure of human endoplasmic reticulum aminopeptidase 2 reveals the atomic basis for distinct roles in antigen processing. Biochemistry. 2012;51:286–295. doi: 10.1021/bi201230p. [DOI] [PubMed] [Google Scholar]
- 35.Hattori A, Tsujimoto M. Endoplasmic reticulum aminopeptidases: biochemistry, physiology and pathology. J Biochem. 2013;154:219–228. doi: 10.1093/jb/mvt066. [DOI] [PubMed] [Google Scholar]
- 36.Hattori A, Goto Y, Tsujimoto M. Exon 10 coding sequence is important for endoplasmic reticulum retention of endoplasmic reticulum aminopeptidase 1. Biol Pharm Bull. 2012;35:601–605. doi: 10.1248/bpb.35.601. [DOI] [PubMed] [Google Scholar]
- 37.Hisatsune C, Ebisui E, Usui M, Ogawa N, Suzuki A, Mataga N, Takahashi-Iwanaga H, Mikoshiba K. ERp44 Exerts Redox-Dependent Control of Blood Pressure at the ER. Mol Cell. 2015;58:1015–1027. doi: 10.1016/j.molcel.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, Moeller A, Tampe R. Structure of the human MHC-I peptide-loading complex. Nature. 2017;551:525–528. doi: 10.1038/nature24627. [DOI] [PubMed] [Google Scholar]
- 39.Saveanu L, van Endert P. The role of insulin-regulated aminopeptidase in MHC class I antigen presentation. Front Immunol. 2012;3:57. doi: 10.3389/fimmu.2012.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chai SY, Fernando R, Peck G, Ye SY, Mendelsohn FA, Jenkins TA, Albiston AL. The angiotensin IV/AT4 receptor. Cell Mol Life Sci. 2004;61:2728–2737. doi: 10.1007/s00018-004-4246-1. [DOI] [PubMed] [Google Scholar]
- 41.Keller SR. The insulin-regulated aminopeptidase: a companion and regulator of GLUT4. Front Biosci. 2003;8:s410–420. doi: 10.2741/1078. [DOI] [PubMed] [Google Scholar]
- 42.Stratikos E, Stamogiannos A, Zervoudi E, Fruci D. A role for naturally occurring alleles of endoplasmic reticulum aminopeptidases in tumor immunity and cancer pre-disposition. Front Oncol. 2014;4:363. doi: 10.3389/fonc.2014.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evnouchidou I, Kamal RP, Seregin SS, Goto Y, Tsujimoto M, Hattori A, Voulgari PV, Drosos AA, Amalfitano A, York IA, Stratikos E. Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J Immunol. 2011;186:1909–1913. doi: 10.4049/jimmunol.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeves E, Edwards CJ, Elliott T, James E. Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J Immunol. 2013;191:35–43. doi: 10.4049/jimmunol.1300598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamogiannos A, Koumantou D, Papakyriakou A, Stratikos E. Effects of polymorphic variation on the mechanism of Endoplasmic Reticulum Aminopeptidase 1. Mol Immunol. 2015;67:426–435. doi: 10.1016/j.molimm.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Martin-Esteban A, Sanz-Bravo A, Guasp P, Barnea E, Admon A, Lopez de Castro JA. Separate effects of the ankylosing spondylitis associated ERAP1 and ERAP2 aminopeptidases determine the influence of their combined phenotype on the HLA-B*27 peptidome. J Autoimmun. 2017 doi: 10.1016/j.jaut.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Guasp P, Barnea E, Gonzalez-Escribano MF, Jimenez-Reinoso A, Regueiro JR, Admon A, Lopez de Castro JA. The Behcet’s disease-associated variant of the aminopeptidase ERAP1 shapes a low-affinity HLA-B*51 peptidome by differential subpeptidome processing. J Biol Chem. 2017;292:9680–9689. doi: 10.1074/jbc.M117.789180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez de Castro JA, Alvarez-Navarro C, Brito A, Guasp P, Martin-Esteban A, Sanz-Bravo A. Molecular and pathogenic effects of endoplasmic reticulum aminopeptidases ERAP1 and ERAP2 in MHC-I-associated inflammatory disorders: Towards a unifying view. Mol Immunol. 2016;77:193–204. doi: 10.1016/j.molimm.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Andres AM, Dennis MY, Kretzschmar WW, Cannons JL, Lee-Lin SQ, Hurle B, Program NCS, Schwartzberg PL, Williamson SH, Bustamante CD, Nielsen R, Clark AG, Green ED. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evnouchidou I, Birtley J, Seregin S, Papakyriakou A, Zervoudi E, Samiotaki M, Panayotou G, Giastas P, Petrakis O, Georgiadis D, Amalfitano A, Saridakis E, Mavridis IM, Stratikos E. A common single nucleotide polymorphism in endoplasmic reticulum aminopeptidase 2 induces a specificity switch that leads to altered antigen processing. J Immunol. 2012;189:2383–2392. doi: 10.4049/jimmunol.1200918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson MP, Roten LT, Dyer TD, East CE, Forsmo S, Blangero J, Brennecke SP, Austgulen R, Moses EK. The ERAP2 gene is associated with preeclampsia in Australian and Norwegian populations. Hum Genet. 2009;126:655–666. doi: 10.1007/s00439-009-0714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanhille DL, Hill LD, Hilliard DD, Lee ED, Teves ME, Srinivas S, Kusanovic JP, Gomez R, Stratikos E, Elovitz MA, Romero R, Strauss JF., 3rd A Novel Haplotype Structure in a Chilean Population: Implications for ERAP2 Protein Expression and Preeclampsia Risk. Mol Genet Genomic Med. 2013;1:98–107. doi: 10.1002/mgg3.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cagliani R, Riva S, Biasin M, Fumagalli M, Pozzoli U, Lo Caputo S, Mazzotta F, Piacentini L, Bresolin N, Clerici M, Sironi M. Genetic diversity at endoplasmic reticulum aminopeptidases is maintained by balancing selection and is associated with natural resistance to HIV-1 infection. Hum Mol Genet. 2010;19:4705–4714. doi: 10.1093/hmg/ddq401. [DOI] [PubMed] [Google Scholar]
- 54.Kuiper JJ, Van Setten J, Ripke S, Van’t Slot R, Mulder F, Missotten T, Baarsma GS, Francioli LC, Pulit SL, De Kovel CG, Ten Dam-Van Loon N, Den Hollander AI, Veld PH, Hoyng CB, Cordero-Coma M, Martin J, Llorenc V, Arya B, Thomas D, Bakker SC, Ophoff RA, Rothova A, De Bakker PI, Mutis T, Koeleman BP. A genome-wide association study identifies a functional ERAP2 haplotype associated with birdshot chorioretinopathy. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hattori A, Matsumoto H, Mizutani S, Tsujimoto M. Molecular cloning of adipocyte-derived leucine aminopeptidase highly related to placental leucine aminopeptidase/oxytocinase. J Biochem (Tokyo) 1999;125:931–938. doi: 10.1093/oxfordjournals.jbchem.a022371. [DOI] [PubMed] [Google Scholar]
- 56.Schomburg L, Kollmus H, Friedrichsen S, Bauer K. Molecular characterization of a puromycin-insensitive leucyl-specific aminopeptidase, PILS-AP. Eur J Biochem. 2000;267:3198–3207. doi: 10.1046/j.1432-1327.2000.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danziger RS. Aminopeptidase N in arterial hypertension. Heart Fail Rev. 2008;13:293–298. doi: 10.1007/s10741-007-9061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peer WA. The role of multifunctional M1 metallopeptidases in cell cycle progression. Ann Bot. 2011;107:1171–1181. doi: 10.1093/aob/mcq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhosale M, Kumar A, Das M, Bhaskarla C, Agarwal V, Nandi D. Catalytic activity of Peptidase N is required for adaptation of Escherichia coli to nutritional downshift and high temperature stress. Microbiol Res. 2013;168:56–64. doi: 10.1016/j.micres.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem J. 1993;290(Pt 1):205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evnouchidou I, Momburg F, Papakyriakou A, Chroni A, Leondiadis L, Chang SC, Goldberg AL, Stratikos E. The internal sequence of the peptide-substrate determines its N-terminus trimming by ERAP1. PLoS ONE. 2008;3:e3658. doi: 10.1371/journal.pone.0003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen TT, Chang SC, Evnouchidou I, York IA, Zikos C, Rock KL, Goldberg AL, Stratikos E, Stern LJ. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol. 2011;18:604–613. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sui L, Gandhi A, Guo HC. Crystal structure of a polypeptide’s C-terminus in complex with the regulatory domain of ER aminopeptidase 1. Mol Immunol. 2016;80:41–49. doi: 10.1016/j.molimm.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gandhi A, Lakshminarasimhan D, Sun Y, Guo HC. Structural insights into the molecular ruler mechanism of the endoplasmic reticulum aminopeptidase ERAP1. Sci Rep. 2011;1:186. doi: 10.1038/srep00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kochan G, Krojer T, Harvey D, Fischer R, Chen L, Vollmar M, von Delft F, Kavanagh KL, Brown MA, Bowness P, Wordsworth P, Kessler BM, Oppermann U. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7745–7750. doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mpakali A, Saridakis E, Harlos K, Zhao Y, Kokkala P, Georgiadis D, Giastas P, Papakyriakou A, Stratikos E. Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. J Med Chem. 2017 doi: 10.1021/acs.jmedchem.6b01890. [DOI] [PubMed] [Google Scholar]
- 67.Gutteridge A, Thornton J. Conformational change in substrate binding, catalysis and product release: an open and shut case? FEBS Lett. 2004;567:67–73. doi: 10.1016/j.febslet.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 68.Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc Natl Acad Sci U S A. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Z, Xing J. Functional roles of slow enzyme conformational changes in network dynamics. Biophys J. 2012;103:1052–1059. doi: 10.1016/j.bpj.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stamogiannos A, Maben Z, Papakyriakou A, Mpakali A, Kokkala P, Georgiadis D, Stern LJ, Stratikos E. Critical Role of Interdomain Interactions in the Conformational Change and Catalytic Mechanism of Endoplasmic Reticulum Aminopeptidase 1. Biochemistry. 2017;56:1546–1558. doi: 10.1021/acs.biochem.6b01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Falk K, Rotzschke O, Rammensee HG. Cellular peptide composition governed by major histocompatibility complex class I molecules. Nature. 1990;348:248–251. doi: 10.1038/348248a0. [DOI] [PubMed] [Google Scholar]
- 72.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Georgiadou D, Hearn A, Evnouchidou I, Chroni A, Leondiadis L, York IA, Rock KL, Stratikos E. Placental leucine aminopeptidase efficiently generates mature antigenic peptides in vitro but in patterns distinct from endoplasmic reticulum aminopeptidase 1. J Immunol. 2010;185:1584–1592. doi: 10.4049/jimmunol.0902502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Infantes S, Samino Y, Lorente E, Jimenez M, Garcia R, Del Val M, Lopez D. Cutting Edge: H-2L(d) Class I Molecule Protects an HIV N-Extended Epitope from In Vitro Trimming by Endoplasmic Reticulum Aminopeptidase Associated with Antigen Processing. J Immunol. 2010;184:3351–3355. doi: 10.4049/jimmunol.0901560. [DOI] [PubMed] [Google Scholar]
- 75.Kim S, Lee S, Shin J, Kim Y, Evnouchidou I, Kim D, Kim YK, Kim YE, Ahn JH, Riddell SR, Stratikos E, Kim VN, Ahn K. Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat Immunol. 2011;12:984–991. doi: 10.1038/ni.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stratikos E, Stern LJ. Antigenic peptide trimming by ER aminopeptidases--insights from structural studies. Mol Immunol. 2013;55:212–219. doi: 10.1016/j.molimm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP Synergizes with MHC Class I Molecules to Make the Final Cut in the Antigenic Peptide Precursors in the Endoplasmic Reticulum. Immunity. 2006;25:795–806. doi: 10.1016/j.immuni.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen H, Li L, Weimershaus M, Evnouchidou I, van Endert P, Bouvier M. ERAP1-ERAP2 dimers trim MHC I-bound precursor peptides; implications for understanding peptide editing. Sci Rep. 2016;6:28902. doi: 10.1038/srep28902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Serwold T, Gaw S, Shastri N. ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat Immunol. 2001;2:644–651. doi: 10.1038/89800. [DOI] [PubMed] [Google Scholar]
- 80.Mpakali A, Saridakis E, Harlos K, Zhao Y, Papakyriakou A, Kokkala P, Georgiadis D, Stratikos E. Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing. J Immunol. 2015;195:2842–2851. doi: 10.4049/jimmunol.1501103. [DOI] [PubMed] [Google Scholar]
- 81.Papakyriakou A, Stratikos E. The Role of Conformational Dynamics in Antigen Trimming by Intracellular Aminopeptidases. Frontiers in Immunology. 2017;8 doi: 10.3389/fimmu.2017.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hermans SJ, Ascher DB, Hancock NC, Holien JK, Michell BJ, Chai SY, Morton CJ, Parker MW. Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides. Protein Sci. 2015;24:190–199. doi: 10.1002/pro.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 84.Bouvier M, Wiley DC. Importance of peptide amino and carboxyl termini to the stability of MHC class I molecules. Science. 1994;265:398–402. doi: 10.1126/science.8023162. [DOI] [PubMed] [Google Scholar]
- 85.Tynan FE, Burrows SR, Buckle AM, Clements CS, Borg NA, Miles JJ, Beddoe T, Whisstock JC, Wilce MC, Silins SL, Burrows JM, Kjer-Nielsen L, Kostenko L, Purcell AW, McCluskey J, Rossjohn J. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6:1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 86.Pymm P, Illing PT, Ramarathinam SH, O’Connor GM, Hughes VA, Hitchen C, Price DA, Ho BK, McVicar DW, Brooks AG, Purcell AW, Rossjohn J, Vivian JP. MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape. Nat Struct Mol Biol. 2017;24:387–394. doi: 10.1038/nsmb.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li X, Lamothe PA, Walker BD, Wang JH. Crystal structure of HLA-B*5801 with a TW10 HIV Gag epitope reveals a novel mode of peptide presentation. Cell Mol Immunol. 2017;14:631–634. doi: 10.1038/cmi.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Achour A, Michaelsson J, Harris RA, Odeberg J, Grufman P, Sandberg JK, Levitsky V, Karre K, Sandalova T, Schneider G. A structural basis for LCMV immune evasion: subversion of H-2D(b) and H-2K(b) presentation of gp33 revealed by comparative crystal structure.Analyses. Immunity. 2002;17:757–768. doi: 10.1016/s1074-7613(02)00478-8. [DOI] [PubMed] [Google Scholar]
- 89.Papakyriakou A, Stratikos E. The Role of Conformational Dynamics in Antigen Trimming by Intracellular Aminopeptidases. Front Immunol. 2017;8:946. doi: 10.3389/fimmu.2017.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hulpke S, Tampe R. The MHC I loading complex: a multitasking machinery in adaptive immunity. Trends Biochem Sci. 2013;38:412–420. doi: 10.1016/j.tibs.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 91.Cresswell P, Bangia N, Dick T, Diedrich G. The nature of the MHC class I peptide loading complex. Immunol Rev. 1999;172:21–28. doi: 10.1111/j.1600-065x.1999.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 92.Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–520. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 93.Thomas C, Tampe R. Proofreading of Peptide-MHC Complexes through Dynamic Multivalent Interactions. Front Immunol. 2017;8:65. doi: 10.3389/fimmu.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seregin SS, Rastall DP, Evnouchidou I, Aylsworth CF, Quiroga D, Kamal RP, Godbehere-Roosa S, Blum CF, York IA, Stratikos E, Amalfitano A. Endoplasmic reticulum aminopeptidase-1 alleles associated with increased risk of ankylosing spondylitis reduce HLA-B27 mediated presentation of multiple antigens. Autoimmunity. 2013 doi: 10.3109/08916934.2013.819855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hearn A, York IA, Rock KL. The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J Immunol. 2009;183:5526–5536. doi: 10.4049/jimmunol.0803663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ayres CM, Corcelli SA, Baker BM. Peptide and Peptide-Dependent Motions in MHC Proteins: Immunological Implications and Biophysical Underpinnings. Front Immunol. 2017;8:935. doi: 10.3389/fimmu.2017.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guasp P, Alvarez-Navarro C, Gomez-Molina P, Martin-Esteban A, Marcilla M, Barnea E, Admon A, Lopez de Castro JA. The Peptidome of Behcet’s Disease-Associated HLA-B*51:01 Includes Two Subpeptidomes Differentially Shaped by Endoplasmic Reticulum Aminopeptidase 1. Arthritis Rheumatol. 2016;68:505–515. doi: 10.1002/art.39430. [DOI] [PubMed] [Google Scholar]
- 98.Zervoudi E, Saridakis E, Birtley JR, Seregin SS, Reeves E, Kokkala P, Aldhamen YA, Amalfitano A, Mavridis IM, James E, Georgiadis D, Stratikos E. Rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses. Proc Natl Acad Sci U S A. 2013;110:19890–19895. doi: 10.1073/pnas.1309781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Papakyriakou A, Zervoudi E, Tsoukalidou S, Mauvais FX, Sfyroera G, Mastellos DC, van Endert P, Theodorakis EA, Vourloumis D, Stratikos E. 3,4-diaminobenzoic acid derivatives as inhibitors of the oxytocinase subfamily of M1 aminopeptidases with immune-regulating properties. J Med Chem. 2015;58:1524–1543. doi: 10.1021/jm501867s. [DOI] [PubMed] [Google Scholar]
- 100.Kokkala P, Mpakali A, Mauvais FX, Papakyriakou A, Daskalaki I, Petropoulou I, Kavvalou S, Papathanasopoulou M, Agrotis S, Fonsou TM, van Endert P, Stratikos E, Georgiadis D. Optimization and Structure-Activity Relationships of Phosphinic Pseudotripeptide Inhibitors of Aminopeptidases That Generate Antigenic Peptides. J Med Chem. 2016;59:9107–9123. doi: 10.1021/acs.jmedchem.6b01031. [DOI] [PubMed] [Google Scholar]
- 101.Chen L, Ridley A, Hammitzsch A, Al-Mossawi MH, Bunting H, Georgiadis D, Chan A, Kollnberger S, Bowness P. Silencing or inhibition of endoplasmic reticulum aminopeptidase 1 (ERAP1) suppresses free heavy chain expression and Th17 responses in ankylosing spondylitis. Ann Rheum Dis. 2016;75:916–923. doi: 10.1136/annrheumdis-2014-206996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tsujimoto M, Hattori A. The oxytocinase subfamily of M1 aminopeptidases. Biochim Biophys Acta. 2005;1751:9–18. doi: 10.1016/j.bbapap.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 103.Saveanu L, Carroll O, Weimershaus M, Guermonprez P, Firat E, Lindo V, Greer F, Davoust J, Kratzer R, Keller SR, Niedermann G, van Endert P. IRAP Identifies an Endosomal Compartment Required for MHC Class I Cross-Presentation. Science. 2009;325:213–217. doi: 10.1126/science.1172845. [DOI] [PubMed] [Google Scholar]