Abstract
Background
Recent global progress in scaling up malaria control interventions has revived the goal of complete elimination in many countries. Decreasing transmission intensity generally leads to increasingly patchy spatial patterns of malaria transmission in elimination settings, with control programs having to accurately identify remaining foci in order to efficiently target interventions.
Findings
The role of connectivity between different pockets of local transmission is of increasing importance as programs near elimination since humans are able to transfer parasites beyond the limits of mosquito dispersal, thus re-introducing parasites to previously malaria-free regions. Here, we discuss recent advances in the quantification of spatial epidemiology of malaria, particularly Plasmodium falciparum, in the context of transmission reduction interventions. Further, we highlight the challenges and promising directions for the development of integrated mapping, modeling, and genomic approaches that leverage disparate datasets to measure both connectivity and transmission.
Conclusion
A more comprehensive understanding of the spatial transmission of malaria can be gained using a combination of parasite genetics and epidemiological modeling and mapping. However, additional molecular and quantitative methods are necessary to answer these public health-related questions.
Keywords: Malaria, Parasite genomics, Spatial modeling, Plasmodium falciparum
Background
The spatial dimensions of malaria control and elimination strategies
Assessing variation in spatial and temporal patterns of infection or in the distribution of a particular pathogen phenotype, such as drug resistance, is an important prerequisite for any infectious disease control effort. For malaria, these considerations are critical across the range of transmission settings (Fig. 1). In pre-elimination settings (e.g., E-2020 countries, including Swaziland, Costa Rica, China, and South Africa [1]), surveillance programs must locate and track imported infections, conduct contact tracing, and ensure that onward transmission resulting from importation events are rapidly extinguished. For countries with intermediate transmission (e.g., Bangladesh, Namibia, and Thailand), control programs must identify the transmission foci contributing to infections in the rest of the country and locate importation hotspots since these will require approaches focused on transmission reduction like vector control. Even in high transmission settings (e.g., Uganda, Nigeria, Democratic Republic of Congo, and Myanmar), which have traditionally focused on monitoring clinical cases and scaling up control and treatment strategies across the country, the renewed interest in measuring transmission has also raised the possibility of more effective program evaluation to assess the impact of interventions on transmission in different regions. Of particular importance in moderate to high transmission settings is the coordination between different regions when human mobility between them is frequent.
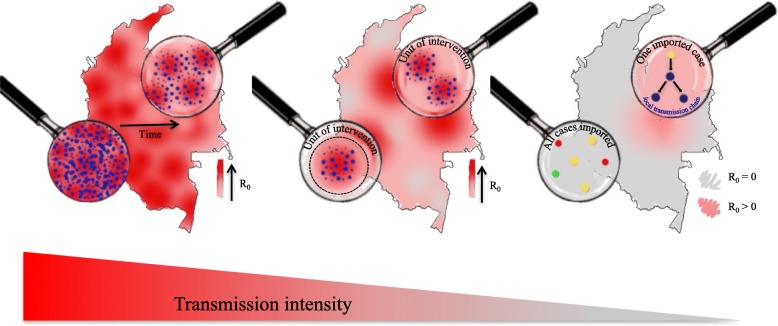
Fig. 1.
Actionable insight from genetic epidemiological studies of malaria across a range of transmission settings. This schematic depicts actionable insight that can be obtained from genetic epidemiological studies of malaria across a range of transmission settings, from high transmission (red) on the left to low transmission (gray) on the right. Here, both imported (stars) and local (points) infections, which may originate from different parasite lineages (various colors), are shown. In high transmission settings, parasites mix panmictically, polyclonal infections are common, and the goal is to evaluate the effectiveness of ongoing interventions. Genetic correlates of declining transmission (e.g., diversity) can provide sensitive indicators of the impact of an intervention. At intermediate transmission, parasites may cluster into interconnected populations. The goal is to delineate regions into units for targeted intervention and to identify the sources that seed transmission for maximally efficient resource allocation. In this setting, models incorporating human mobility and genetic measures of parasite relatedness can provide directional estimates of connectivity between parasite populations. At very low transmission, most infections are imported. The goal is to identify origins of imported parasites, quantify any onward transmission and, if onward transmission exists, the average length of local transmission chains. Models incorporating detailed case data, including genetic data and travel history, can reconstruct transmission chains to infer who acquires infection from who and how
Model of malaria spatial epidemiology
A variety of modeling approaches has been used to describe the spatial dynamics of malaria [2] and to effectively allocate resources. Geostatistical modeling approaches have been used to generate maps of epidemiological variables such as parasite prevalence [3] and intervention impact [4]. These maps derive from methods that interpolate across spatially idiosyncratic data sources, providing a spatially smoothed estimate of epidemiological metrics relevant for targeting of interventions. Nevertheless, certain important aspects of malaria epidemiology cannot be captured by interpolation methods. First, statistical methods may fail to distinguish between areas where cases reflect local transmission intensity versus regions with frequently imported infections; therefore, different assumptions about connectivity can lead to varying conclusions with regard to the capacity for local transmission and need for vector control [5]. Second, thinking beyond all but the most local scales, there is a myriad of ways to coordinate control efforts across different areas, for example, by grouping locations that naturally cluster together as larger units of transmission [6, 7]. Combined with transmission models that consider numerous non-linear feedbacks between control and transmission [8, 9] and are capable of accounting for location-specific intervention packages and their impacts [10, 11], these approaches could, theoretically, suggest an optimal elimination strategy. In practice, there are shortcomings in both the currently available data and models.
Quantifying connectivity is one of the most important aspects of characterizing the spatial dynamics of malaria, yet it can be one of the most vexing. Call data records routinely collected by mobile phone operators, as well as other novel data sources on human travel, have offered hope in recent years [5, 7, 12]. These data are not without their challenges, however, including variable cell tower densities, mobile phone market fragmentation, and possible disconnects between who is making calls and who is transmitting parasites [13]. Traditional travel survey data may be more directly related to known symptomatic individuals; however, these data are often limited in scope and accuracy [14]. Understanding which travel patterns are epidemiologically relevant further requires an understanding of vector distribution, identity, and abundance. The complex relationship between these ecological parameters of transmission and the epidemiology of disease, along with the lack of robust parasite strain markers, make it difficult to accurately identify the geographical source of particular infections, in turn hindering efforts to map the routes of parasite importation at the population level. Ultimately, models are necessary to appropriately combine information about human mobility with a variety of epidemiological data to arrive at an estimate of how parasite movement arises on different spatial scales. Indeed, recent work using mathematical models based on epidemiological data in Senegal showed that genetic data collected in parallel can provide consistent and confirmatory signals of significant transmission reductions followed by signatures of a rebound [15]; similar approaches in a spatial context may well be useful in other settings.
Parasite genetic signals may offer some of the richest information about these otherwise elusive patterns of parasite movement and, although this approach is still in its early stages, researchers have begun to assess the utility of molecular surveillance as a routine tool for the optimization of control and elimination strategies. We propose that the marriage of parasite genetic data and models in a spatial context may offer unique insights into the epidemiology of malaria. Below, we discuss the techniques, challenges, and promising applications of molecular surveillance.
Discussion
Applications of parasite genetics to spatial epidemiology of malaria
Molecular tools may be most valuable when epidemiological information is scarce and/or mobility data is unavailable. Genomic surveillance and phylogenetic analyses that relate the geographic distribution of genetic signals within and between populations have enabled near real-time estimation of transmission chains for non-sexually recombining, rapidly evolving pathogens (e.g., Ebola, influenza) [16, 17]. This nascent field of pathogen phylogeography has provided key insights into the routes of pathogen introductions and spread, particularly for viral diseases. However, directly extending these methods to a pathogen such as Plasmodium falciparum—a sexually recombining eukaryotic parasite with a complex lifecycle—requires both molecular and analytic advancements that are still at the early stages of development. In particular, the malaria parasite P. falciparum undergoes obligate sexual recombination and is often characterized by multi-genotype infections and low-density chronic blood-stage infections that can last for months in asymptomatic individuals. More complex still are the many challenges associated with the second most abundant cause of malaria, Plasmodium vivax [18]. Unlike P. falciparum parasites, P. vivax parasites can survive for months or years as dormant hypnozoites in the liver, where they are undetectable, and can relapse and cause blood-stage infection at any time. Since genetically diverse hypnozoites can build up in the liver, relapses lead to an even greater abundance of multi-genotype blood-stage infections and thus more frequent recombination between genetically diverse parasites. Moreover, in regions of ongoing transmission, relapses cannot be definitely distinguished from reinfections due to new mosquito bites, further complicating efforts to spatially track P. vivax infection. These complexities mean that standard population genetic or phylogenetic approaches do not effectively resolve relationships between malaria parasite lineages [19]. Therefore, new tools are needed for the effective molecular surveillance of both parasite species.
Most national control programs are interested in spatial scales that are operationally relevant, namely within a given country or between countries if they are connected by migration. Population differentiation on international and continental geographic scales can be identified using principal component analysis, phylogenetic analysis, and the fixation index (FST) [20–24], yet these methods are not powered to detect finer-scale differentiation. This is because (1) recombination violates the assumptions underpinning classic phylogenetic analyses [25], and (2) principal component analysis based on a pairwise distance matrix and FST is influenced by drivers of genetic variation that act on a long time scale (i.e., the coalescent time of parasites) such that if migration occurs multiple times during this time frame, there will be little or no signal of differentiation among populations [26, 27]. In contrast, methods that exploit the signal left by recombination (rather than treating it as a nuisance factor) may have the power to detect geographic differentiation on spatial scales relevant for malaria control programs.
Recombination occurs in the mosquito midgut when gametes (derived from gametocytes) come together to form a zygote. If the gametes are genetically distinct, recombination will lead to the production of different, but highly related, sporozoites (and thus onward infections). These highly related parasites would tend to have genomes with a high degree of identity. Perhaps the simplest measure of this genetic similarity is “identity by state” (IBS), which is defined as the proportion of identical sites between two genomes and is a simple correlate of genetic relatedness between parasites. However, IBS makes no distinction between sites that are identical by chance and those that are identical due to recent shared ancestry, making it sensitive to the allele frequency spectrum of the particular population under study. Analyses that are probabilistic (e.g., STRUCTURE [28]) provide better resolution, but ultimately linkage disequilibrium-based methods, such as identity by decent (IBD) inferred under a hidden Markov model [29, 30] and chromosome painting [31], provide greater power. These IBD methods harness the patterns of genetic linkage disequilibrium that are broken down by recombination and are therefore sensitive to recent migration events and useful at smaller geographic scales. Additionally, they take advantage of the signals present in long contiguous blocks of genomic identity, which can be detected given a sufficient density of informative markers. The exact density required is a topic of current research and depends on the level of relatedness, required precision, and the nature of the genetic markers in question (e.g., the number and frequency of possible alleles for each marker).
In low transmission settings, such as Senegal and Panama, STRUCTURE as well as IBS (which approximates IBD, albeit with bias and more noise), can often be used to cluster cases and infer transmission patterns within countries [32–34]. In intermediate transmission settings, such as coastal regions of Kenya and border regions of Thailand, where genetic diversity is higher, IBS, IBD, and relatedness based on chromosome painting have been shown to recover genetic structure over populations of parasites on local spatial scales [27, 35]. However, due to dependence on allele frequency spectra, IBS is not as easily comparable across datasets and, as mentioned above, can be overwhelmed by noise due to identity by chance. Moreover, all of these methods currently have limited support for polyclonal samples. In high transmission settings, the complexity of infection is very high, making it difficult to calculate genetic relatedness between parasites within polyclonal infections or to estimate allele frequencies across polyclonal infections since the complexity entangles the signal from the genetic markers belonging to the individual clones, the number of which is unknown. Methods to disentangle (i.e., phase) parasite genetic data within polyclonal infections are being developed [36], while THE REAL McCOIL [37] has been developed to simultaneously infer allele frequencies and complexity of infection, allowing downstream calculation of FST. However, to fully characterize genetic structure at fine scales in high transmission settings, new methods that estimate IBD and other relatedness measures are needed to infer ancestry between polyclonal infections. Indeed, across all spatiotemporal scales and transmission intensities, we propose that rather than being defined by the transmission of discrete (clonal) parasite lineages, malaria epidemiology may be best characterized as the transmission of infection states, often comprised of an ensemble of parasites. Subsets of these ensembles are often transmitted together by a mosquito to another person, and therefore, the combination of alleles/parasites present in an infection state provides rich information about its origin(s) beyond the composition of individual parasites.
Current sampling and sequencing strategies for genomic epidemiology of malaria
The use of genetic approaches described above will depend on the routine generation of parasite genetic data since any molecular surveillance system will improve with more data and must be tailored to the sampling framework and sequencing approach. To date, many studies attempting to obtain epidemiologic information from genomic data have taken advantage of existing samples rather than having sampling tailored to the questions and public health interventions of interest. This is understandable given that a number of these studies have been exploratory and that informed decisions regarding sampling require a priori empiric data on parasite population structure (unavailable in most places) and a predetermined analysis plan (difficult when analytical approaches are actively in development). A more direct/tailored study design should be possible as more parasite genomic data become available and analytical methods mature. However, in general, a greater sampling of infections will be required to answer fine-scale questions regarding transmission (e.g., whether infections are local versus imported, determining the length of transmission chains) than for larger-scale questions such as relative connectivity of parasite populations between distinct geographic regions. Now that sequencing can be performed from blood spots collected on filter papers or even rapid diagnostic tests, collecting samples from passively detected symptomatic cases at health facilities offers the most efficient means of collecting large numbers of infected cases, often with high parasite densities, thus making them easier to genotype. Nevertheless, while this may be sufficient to characterize the underlying parasite population in some settings and for some questions, in others, the capture of asymptomatic cases through active case detection may be essential to understand transmission epidemiology, e.g., to determine the contribution of the asymptomatic reservoir in sustaining local transmission.
The discriminatory power of the genotyping method will depend on the local epidemiology and transmission setting. The two most common genotyping approaches, namely relatively small SNP barcodes and panels of microsatellite markers [38], have been extensively used to monitor the changes in the diversity and structure of the parasite population. However, signals in these markers may not be sufficient to distinguish geographic origin and have limited resolution in certain transmission settings [37, 39, 40]. Increasing the number of loci and/or discrimination of each locus may be necessary to answer the questions relevant to elimination. Further, increasing discrimination by using multiallelic loci has particular advantages since these may provide more information content than biallelic loci [41]. This is particularly true in polyclonal infections, frequent even in areas close to elimination, because heterozygous genotypes of biallelic loci contain little information (all possible alleles are present), whereas detecting, for example, 3 out of 20 potential alleles in an infection, still allows informative comparisons between infecting strains. In addition, some genotypable multiallelic loci contain extremely high diversity, which can be combined in relatively small numbers to create high-resolution genotypes. Targeting specific regions of the genome for sequencing after amplification by PCR (amplicon sequencing) or other methods, such as molecular inversion probes [42], offers efficient approaches to genotyping multiallelic short-range haplotypes, SNPs, and/or microsatellites, providing a flexible platform for deeper and more consistent coverage of regions of interest at lower cost than whole genome sequencing. Amplicon sequencing may be of particular interest for genotyping minor strains in polyclonal infections and/or low-density samples, whereas molecular inversion probes may excel for more highly multiplexed marker assays where capturing low-density samples is not critical. Identifying a panel of optimally informative genetic markers to address a specific question remains a major challenge that must balance the cost, throughput, and discriminatory power. For example, at fine geographic scales, larger numbers of more closely spaced markers with representative coverage of the genome may be required in contrast to studies comparing distant parasite populations; the density at which infected individuals are sampled and the underlying diversity and genetic structure will also affect the number and type of loci required.
With proper consideration, a parsimonious set of genetic targets may be identified as useful to answer a number of general questions regarding malaria genomics. Nonetheless, the development of a marker toolbox and genotyping methods tailored to answering questions relevant for transmission at different spatial scales is an important goal. To this end, several ambitious sequencing studies have begun, and over 4000 P. falciparum genomes have been sequenced from different transmission settings around the globe (such as the Pf3K Project, https://www.malariagen.net/data/pf3k-pilot-data-release-3) [40, 43, 44]. These genetic data are all publicly available, providing a crucial framework to build upon when designing more local, sequence-based epidemiological studies that balance the trade-off between the number of genetic loci evaluated and the quality of the data (e.g., depth of sequence coverage) for each parasite sample. Genomic sequencing methods are evolving rapidly towards high-throughput and low-cost, deep sequencing approaches that can be performed on routinely collected patient samples, allowing for evaluation of even asymptomatic low-density infections, e.g., by selective enrichment of parasite DNA [45, 46]. These enrichment methods can exacerbate the non-uniformity of sequencing coverage variation across the parasite genome and can require specialized filters to remove erroneous heterozygous calls, yet they generally produce genotypes exhibiting very high concordance with those from samples sequenced via alternate means [46, 47]. Preferential amplification of dominant strains in a polyclonal infection (i.e., missing minority clones) and the inability to detect copy number variation have also been described as potential limitations of these selective enrichment methods [47]. Nevertheless, despite these limitations, these methods are enabling cost-effective whole genome sequences from routinely collected blood samples. Moving forward, we must ensure that rich metadata are made easily available in the context of genome sequences, so that links can be made to experimental, epidemiological, and ecological variables and models.
Combining data layers to map malaria
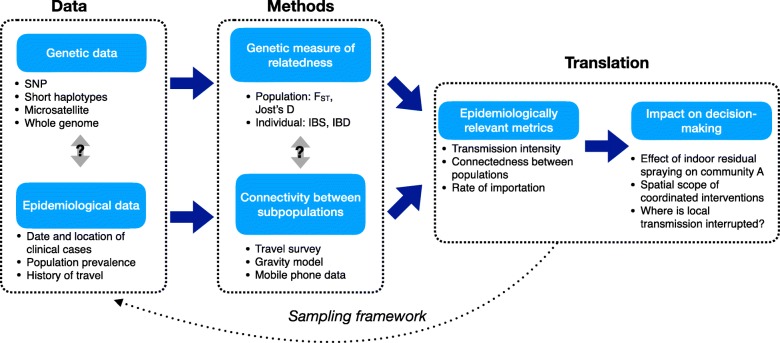
In concrete terms, we want to be able to clearly identify if two locations are epidemiologically linked. However, given the current methods available and in development, the complicated life cycle of the parasite, and the epidemiology of malaria, any single data source or method is unlikely to produce a complete picture of the spatial dynamics of malaria parasites. Figure 2 illustrates an analytical pipeline linking different spatially explicit datasets to methods and ultimately interventions, highlighting current uncertainties and the need to consider policy-relevant metrics when designing sampling frameworks. In particular, we believe that future development should focus on identifying how these different types of data can be combined and integrated to provide a more complete picture of connectivity and transmission dynamics. If we view this problem in terms of a simplified traditional medical statistic, malaria parasite data have a high false-negative rate (the analysis mostly underestimates relatedness between parasites), whereas connectivity data inferred from mobile phone data or other proxy measures of travel have a high false-positive rate (the analysis mostly overestimates the number of epidemiologically relevant connections). Ideally, joint inference methods that combine these data sources would help improve the type I (false-positivity rate) and type II (false-negativity rate) errors in each type of data.
Fig. 2.
The analysis pipeline. Both genetic and epidemiological data can be collected and analyzed in order to understand the parasite flow (with example datasets and methods listed above). To identify how these two methods can be combined, directly related to policy-relevant questions, and translated to control measures will require the development of novel inference frameworks and the design of studies across a range of transmission settings
Conclusions
These new data streams therefore offer great potential, but understanding how to effectively combine them in ways that consider the biases and strengths of each data type will require significant research investment. Furthermore, making these methods relevant for implementation is a consideration that must be at the forefront of research efforts. For example, the ongoing availability of each data stream, the feasibility of implementing these analytical approaches in the context of national control programs as well as the capacity-building required to do so, will ultimately determine their impact. This means that tools must provide clearly communicated estimates of uncertainty and will need to be straightforward for their use in different contexts, easy to communicate, and generalizable.
Acknowledgements
This work is supported by Maximizing Investigators’ Research Award for Early Stage Investigators, R35GM124715 (COB, AW, ART), a Wellcome Trust Sustaining Health Grant (106866/Z/15/Z to COB, AW, ART; https://wellcome.ac.uk/), the Models of Infectious Disease Agent Study program, cooperative agreement U54GM088558 (to COB; https://www.nigms.nih.gov/Research/specificareas/MIDAS/Pages/default.aspx), and the Bill and Melinda Gates Foundation OPP 1132226 (to TAP, BG, ST) and OPP 1110495 (to TAP). BG is a Chan Zuckerberg Biohub investigator. AW is supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. RV is funded by a Skills Development Fellowship, jointly funded by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement and is also part of the EDCTP2 programme supported by the European Union.
Abbreviations
- FST
Fixation index
- IBD
Identical by descent
- IBS
Identical by state
Authors’ contributions
AW and COB conceived the study and participated in its design and coorination. AW, ART, HHC, RV, ST, JAB, TAP, DEN, BG, and COB drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Amy Wesolowski, Email: awesolowski@jhu.edu.
Caroline O Buckee, Email: cbuckee@hsph.harvard.edu.
References
- 1.World Health Organization . World malaria report 2017. Geneva: WHO; 2017. [Google Scholar]
- 2.Reiner RC, Jr, Perkins TA, Barker CM, Niu T, Chaves LF, Ellis AM, George DB, Le Menach A, Pulliam JR, Bisanzio D, et al. A systematic review of mathematical models of mosquito-borne pathogen transmission: 1970–2010. J R Soc Interface. 2013;10(81):20120921. doi: 10.1098/rsif.2012.0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalrymple U, Mappin B, Gething PW. Malaria mapping: understanding the global endemicity of falciparum and vivax malaria. BMC Med. 2015;13:140. doi: 10.1186/s12916-015-0372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, Battle K, Moyes CL, Henry A, Eckhoff PA, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526(7572):207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruktanonchai NW, DeLeenheer P, Tatem AJ, Alegana VA, Caughlin TT, Zu Erbach-Schoenberg E, Lourenco C, Ruktanonchai CW, Smith DL. Identifying malaria transmission foci for elimination using human mobility data. PLoS Comput Biol. 2016;12(4):e1004846. doi: 10.1371/journal.pcbi.1004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatem AJ, Smith DL. International population movements and regional Plasmodium falciparum malaria elimination strategies. Proc Natl Acad Sci U S A. 2010;107(27):12222–12227. doi: 10.1073/pnas.1002971107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wesolowski A, Eagle N, Tatem AJ, Smith DL, Noor AM, Snow RW, Buckee CO. Quantifying the impact of human mobility on malaria. Science. 2012;338(6104):267–270. doi: 10.1126/science.1223467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerra CA, Reiner RC, Jr, Perkins TA, Lindsay SW, Midega JT, Brady OJ, Barker CM, Reisen WK, Harrington LC, Takken W, et al. A global assembly of adult female mosquito mark-release-recapture data to inform the control of mosquito-borne pathogens. Parasit Vectors. 2014;7:276. doi: 10.1186/1756-3305-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith DL, Dushoff J, Snow RW, Hay SI. The entomological inoculation rate and Plasmodium falciparum infection in African children. Nature. 2005;438(7067):492–495. doi: 10.1038/nature04024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker PG, Griffin JT, Ferguson NM, Ghani AC. Estimating the most efficient allocation of interventions to achieve reductions in Plasmodium falciparum malaria burden and transmission in Africa: a modelling study. Lancet Glob Health. 2016;4(7):e474–e484. doi: 10.1016/S2214-109X(16)30073-0. [DOI] [PubMed] [Google Scholar]
- 11.Nikolov M, Bever CA, Upfill-Brown A, Hamainza B, Miller JM, Eckhoff PA, Wenger EA, Gerardin J. Malaria elimination campaigns in the Lake Kariba region of Zambia: a spatial dynamical model. PLoS Comput Biol. 2016;12(11):e1005192. doi: 10.1371/journal.pcbi.1005192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wesolowski A, Buckee CO, Engo-Monsen K, Metcalf CJE. Connecting mobility to infectious diseases: the promise and limits of mobile phone data. J Infect Dis. 2016;214(suppl_4):S414–S420. doi: 10.1093/infdis/jiw273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall JM, Toure M, Ouedraogo AL, Ndhlovu M, Kiware SS, Rezai A, Nkhama E, Griffin JT, Hollingsworth TD, Doumbia S, et al. Key traveller groups of relevance to spatial malaria transmission: a survey of movement patterns in four sub-Saharan African countries. Malar J. 2016;15:200. doi: 10.1186/s12936-016-1252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wesolowski A, Stresman G, Eagle N, Stevenson J, Owaga C, Marube E, Bousema T, Drakeley C, Cox J, Buckee CO. Quantifying travel behavior for infectious disease research: a comparison of data from surveys and mobile phones. Sci Rep. 2014;4:5678. doi: 10.1038/srep05678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels RF, Schaffner SF, Wenger EA, Proctor JL, Chang HH, Wong W, Baro N, Ndiaye D, Fall FB, Ndiop M, et al. Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc Natl Acad Sci U S A. 2015;112(22):7067–7072. doi: 10.1073/pnas.1505691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudas G, Carvalho LM, Bedford T, Tatem AJ, Baele G, Faria NR, Park DJ, Ladner JT, Arias A, Asogun D, et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature. 2017;544(7650):309–315. doi: 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, Baele G, Russell CA, Smith DJ, Pybus OG, Brockmann D, et al. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 2014;10(2):e1003932. doi: 10.1371/journal.ppat.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira MU, de Oliveira TC. Challenges for Plasmodium vivax malaria elimination in the genomics era. Pathog Glob Health. 2015;109(3):89–90. doi: 10.1179/2047772415Z.000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang HH, Moss EL, Park DJ, Ndiaye D, Mboup S, Volkman SK, Sabeti PC, Wirth DF, Neafsey DE, Hartl DL. Malaria life cycle intensifies both natural selection and random genetic drift. Proc Natl Acad Sci U S A. 2013;110(50):20129–20134. doi: 10.1073/pnas.1319857110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 2013;45(6):648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright S. The genetical structure of populations. Annal Eugenics. 1951;15(4):323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 23.Mu J, Awadalla P, Duan J, McGee KM, Joy DA, McVean GA, Su XZ. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3(10):e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neafsey DE, Schaffner SF, Volkman SK, Park D, Montgomery P, Milner DA, Jr, Lukens A, Rosen D, Daniels R, Houde N, et al. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol. 2008;9(12):R171. doi: 10.1186/gb-2008-9-12-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frost SD, Pybus OG, Gog JR, Viboud C, Bonhoeffer S, Bedford T. Eight challenges in phylodynamic inference. Epidemics. 2015;10:88–92. doi: 10.1016/j.epidem.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang HH, Dordel J, Donker T, Worby CJ, Feil EJ, Hanage WP, Bentley SD, Huang SS, Lipsitch M. Identifying the effect of patient sharing on between-hospital genetic differentiation of methicillin-resistant Staphylococcus aureus. Genome Med. 2016;8(1):18. doi: 10.1186/s13073-016-0274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor AR, Schaffner SF, Cerqueira GC, Nkhoma SC, Anderson TJC, Sriprawat K, Pyae Phyo A, Nosten F, Neafsey DE, Buckee CO. Quantifying connectivity between local Plasmodium falciparum malaria parasite populations using identity by descent. PLoS Genet. 2017;13(10):e1007065. doi: 10.1371/journal.pgen.1007065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaffner SF, Taylor AR, Wong W, Wirth DF, Neafsey DE. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar J. 2018;17(1):196. doi: 10.1186/s12936-018-2349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henden L, Lee S, Mueller I, Barry A, Bahlo M. Detecting selection signals in Plasmodium falciparum using identity-by-descent analysis. bioRxiv. 2016; 10.1101/088039.
- 31.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8(1):e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang HH, Park DJ, Galinsky KJ, Schaffner SF, Ndiaye D, Ndir O, Mboup S, Wiegand RC, Volkman SK, Sabeti PC, et al. Genomic sequencing of Plasmodium falciparum malaria parasites from Senegal reveals the demographic history of the population. Mol Biol Evol. 2012;29(11):3427–3439. doi: 10.1093/molbev/mss161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniels R, Chang HH, Sene PD, Park DC, Neafsey DE, Schaffner SF, Hamilton EJ, Lukens AK, Van Tyne D, Mboup S, et al. Genetic surveillance detects both clonal and epidemic transmission of malaria following enhanced intervention in Senegal. PLoS One. 2013;8(4):e60780. doi: 10.1371/journal.pone.0060780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obaldia N, 3rd, Baro NK, Calzada JE, Santamaria AM, Daniels R, Wong W, Chang HH, Hamilton EJ, Arevalo-Herrera M, Herrera S, et al. Clonal outbreak of Plasmodium falciparum infection in eastern Panama. J Infect Dis. 2015;211(7):1087–1096. doi: 10.1093/infdis/jiu575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omedo I, Mogeni P, Bousema T, Rockett K, Amambua-Ngwa A, Oyier I, C Stevenson J, Y Baidjoe A, de Villiers EP, Fegan G, et al. Micro-epidemiological structuring of Plasmodium falciparum parasite populations in regions with varying transmission intensities in Africa Wellcome. Open Res. 2017;2:10. doi: 10.12688/wellcomeopenres.10784.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu SJ, Almagro-Garcia J, McVean G. Deconvolution of multiple infections in Plasmodium falciparum from high throughput sequencing data. Bioinformatics. 2018;34(1):9–15. doi: 10.1093/bioinformatics/btx530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang HH, Worby CJ, Yeka A, Nankabirwa J, Kamya MR, Staedke SG, Dorsey G, Murphy M, Neafsey DE, Jeffreys AE, et al. THE REAL McCOIL: a method for the concurrent estimation of the complexity of infection and SNP allele frequency for malaria parasites. PLoS Comput Biol. 2017;13(1):e1005348. doi: 10.1371/journal.pcbi.1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Escalante AA, Ferreira MU, Vinetz JM, Volkman SK, Cui L, Gamboa D, Krogstad DJ, Barry AE, Carlton JM, van Eijk AM, et al. Malaria molecular epidemiology: lessons from the International Centers of Excellence for Malaria Research Network. Am J Trop Med Hyg. 2015;93(3 Suppl):79–86. doi: 10.4269/ajtmh.15-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sisya TJ, Kamn’gona RM, Vareta JA, Fulakeza JM, Mukaka MF, Seydel KB, Laufer MK, Taylor TE, Nkhoma SC. Subtle changes in Plasmodium falciparum infection complexity following enhanced intervention in Malawi. Acta Trop. 2015;142:108–114. doi: 10.1016/j.actatropica.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerqueira GC, Cheeseman IH, Schaffner SF, Nair S, McDew-White M, Phyo AP, Ashley EA, Melnikov A, Rogov P, Birren BW, et al. Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 2017;18(1):78. doi: 10.1186/s13059-017-1204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baetscher DS, Clemento AJ, Ng TC, Anderson EC, Garza JC. Microhaplotypes provide increased power from short-read DNA sequences for relationship inference. Mol Ecol Resour. 2018;18(2):296–305. doi: 10.1111/1755-0998.12737. [DOI] [PubMed] [Google Scholar]
- 42.Aydemir O, Janko M, Hathaway NJ, Verity R, Mwandagalirwa MK, Tshefu AK, Tessema SK, Marsh PW, Tran A, Reimonn T, et al. Drug resistance and population structure of Plasmodium falciparum across the Democratic Republic of Congo using high-throughput molecular inversion probes. J Infect Dis. 2018;218(6):946–955. doi: 10.1093/infdis/jiy223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Mudeppa DG, Sharma A, Mascarenhas A, Dash R, Pereira L, Shaik RB, Maki JN, White J, 3rd, Zuo W, et al. Distinct genomic architecture of Plasmodium falciparum populations from South Asia. Mol Biochem Parasitol. 2016;210(1–2):1–4. doi: 10.1016/j.molbiopara.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parobek CM, Parr JB, Brazeau NF, Lon C, Chaorattanakawee S, Gosi P, Barnett EJ, Norris LD, Meshnick SR, Spring MD, et al. Partner-drug resistance and population substructuring of artemisinin-resistant Plasmodium falciparum in Cambodia. Genome Biol Evol. 2017;9(6):1673–1686. doi: 10.1093/gbe/evx126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larremore DB, Sundararaman SA, Liu W, Proto WR, Clauset A, Loy DE, Speede S, Plenderleith LJ, Sharp PM, Hahn BH, et al. Ape parasite origins of human malaria virulence genes. Nat Commun. 2015;6:8368. doi: 10.1038/ncomms9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oyola SO, Ariani CV, Hamilton WL, Kekre M, Amenga-Etego LN, Ghansah A, Rutledge GG, Redmond S, Manske M, Jyothi D, et al. Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification. Malar J. 2016;15(1):597. doi: 10.1186/s12936-016-1641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowell AN, Loy DE, Sundararaman SA, Valdivia H, Fisch K, Lescano AG, Baldeviano GC, Durand S, Gerbasi V, Sutherland CJ, et al. Selective whole-genome amplification is a robust method that enables scalable whole-genome sequencing of Plasmodium vivax from unprocessed clinical samples. MBio. 2017;8(1). [DOI] [PMC free article] [PubMed]