The human gut microbial community is associated strongly with host physiology and human diseases. This observation has prompted research on pre- and probiotics, two concepts enabling specific changes in the composition of the human gut microbiome that result in beneficial effects for the host. Here, we show how fructooligosaccharide-inulin prebiotics are fermented by commercial probiotic bacterial strains involving specific sets of enzymes and transporters. Cross-feeding strains such as Lactobacillus paracasei W20 may thus act as keystone strains in the degradation of prebiotic inulin in the human gut, and this strain–exo-inulinase combination may be used in commercial Lactobacillus-inulin synbiotics.
KEYWORDS: cross-feeding, Lactobacillus paracasei, exo-inulinase, inulin, probiotics
ABSTRACT
Probiotic gut bacteria employ specific metabolic pathways to degrade dietary carbohydrates beyond the capabilities of their human host. Here, we report how individual commercial probiotic strains degrade prebiotic (inulin type) fructans. First, a structural analysis of commercial fructose oligosaccharide-inulin samples was performed. These β-(2-1)-fructans differ in termination by either glucose (GF) or fructose (FF) residues, with a broad variation in the degrees of polymerization (DPs). The growth of individual probiotic bacteria on short-chain inulin (sc-inulin) (Frutafit CLR), a β-(2-1)-fructan (DP 2 to DP 40), was studied. Lactobacillus salivarius W57 and other bacteria grew relatively poorly on sc-inulin, with only fractions of DP 3 and DP 5 utilized, reflecting uptake via specific transport systems followed by intracellular metabolism. Lactobacillus paracasei subsp. paracasei W20 completely used all sc-inulin components, employing an extracellular exo-inulinase enzyme (glycoside hydrolase family GH32 [LpGH32], also found in other strains of this species); the purified enzyme converted high-DP compounds into fructose, sucrose, 1-kestose, and F2 (inulobiose). The cocultivation of L. salivarius W57 and L. paracasei W20 on sc-inulin resulted in cross-feeding of the former by the latter, supported by this extracellular exo-inulinase. The extent of cross-feeding depended on the type of fructan, i.e., the GF type (clearly stimulating) versus the FF type (relatively low stimulus), and on fructan chain length, since relatively low-DP β-(2-1)-fructans contain a relatively high content of GF-type molecules, thus resulting in higher concentrations of GF-type DP 2 to DP 3 degradation products. The results provide an example of how in vivo cross-feeding on prebiotic β-(2-1)-fructans may occur among probiotic lactobacilli.
IMPORTANCE The human gut microbial community is associated strongly with host physiology and human diseases. This observation has prompted research on pre- and probiotics, two concepts enabling specific changes in the composition of the human gut microbiome that result in beneficial effects for the host. Here, we show how fructooligosaccharide-inulin prebiotics are fermented by commercial probiotic bacterial strains involving specific sets of enzymes and transporters. Cross-feeding strains such as Lactobacillus paracasei W20 may thus act as keystone strains in the degradation of prebiotic inulin in the human gut, and this strain–exo-inulinase combination may be used in commercial Lactobacillus-inulin synbiotics.
INTRODUCTION
Probiotics are live microorganisms that, when administered in adequate amounts, confer a health benefit to the host (1). Probiotic bacteria are typically found in the genera Bifidobacterium and Lactobacillus; however, some strains of Enterococcus and Escherichia also exhibit interesting probiotic properties. Providing bacteria with prebiotic compounds enables the selective stimulation and enrichment of desirable bacterial strains important for sustaining human health. Prebiotics are defined as the substrates that are selectively utilized by host microorganisms conferring a health benefit (2). A prominent class of prebiotics is constituted by dietary nondigestible carbohydrates. These nondigestible carbohydrates escape metabolism by human digestive enzymes and are therefore available as carbon and energy sources to bacteria residing in the lower gastrointestinal tract (3).
There are several classes of prebiotic carbohydrates, including arabino xylanooligosaccharides (AXOS), maltooligosaccharides (MOS), galactooligosaccharides (GOS), and fructooligosaccharides (FOS, such as inulin and oligofructose [OF]) (4). Fructose-based prebiotics are the most-well-studied nondigestible carbohydrates considered for their prebiotic effects. These include the β-(2-1)-fructans derived by enzyme synthesis from sucrose and inulin or oligofructose obtained from plant sources. β-(2-1)-Fructans exhibit structural differences in both their degrees of polymerization (DPs), which may vary between 2 and 60, and the constituent terminal nonreducing end sugar, being glucose for GF-type fructans and fructose for FF-type fructans (5, 6).
Bacteria that metabolize prebiotics and other nondigestible carbohydrates produce short-chain fatty acids (SCFA), such as acetate, propionate, and butyrate. These biologically important compounds mediate crucial health effects in the human body at the molecular level (7). Prebiotics thus serve as selective nutritional sources for probiotic bacteria, and the two independently defined dietary concepts of pre- and probiotics may synergistically come to action in the human gut.
Probiotic bacteria may benefit selectively from the presence of prebiotic carbohydrates through their set of carbohydrate-active enzymes (CAZymes), which include glycoside hydrolases (GHs), enabling carbohydrate utilization as carbon and energy sources (8). GHs encoded in bacterial genomes often outnumber the number of human GH genes by several fold (9). Bacterial species strongly vary in the set of GH enzymes encoded. Different carbohydrates are thus fermented by different bacterial species, resulting in the selective growth stimulation of species that benefit from a certain carbohydrate structure. The modulation of the human gut microbiota by prebiotic carbohydrates thus may occur at the species level (10). This provides clear opportunities to supply combinations of probiotic strains and prebiotic compounds with beneficial effects. In practice, however, pre- and probiotic treatments are often still given independently from each other. It remains largely unknown how the increased beneficial effects of novel probiotic bacterial strains can be potentiated through the addition of selected prebiotics. Further studies into the physiology and growth of strain-specific probiotic bacteria on prebiotic oligosaccharides are therefore clearly needed.
The utilization of certain β-(2-1)-fructans has been investigated with various strains from the genera Lactobacillus and Bifidobacterium (11–14). These studies mostly focused on the selective utilization of fructans with a certain length per strain. For instance, Bifidobacterium longum and Bifidobacterium animalis grew better on short-chain fructooligosaccharide (scFOS) Actilight 950P (low DP) than on inulin from dahlia tubers (high DP) (11). A study of oligofructose and inulin utilization by 18 Bifidobacterium strains from 10 different species revealed 4 groups, reflecting their individual catabolic abilities, but none of the strains consumed inulin completely (12). The ability to utilize specific fructan components differed even among strains from the same Lactobacillus or Bifidobacterium species (13, 14). No single species used FOS/inulin completely. Besides being directly stimulated by prebiotic substrates, probiotic bacteria may indirectly benefit from prebiotic substrates via cross-feeding mediated by smaller carbohydrate degradation products or by SCFA as the end products of bacterial fermentation. While the structural identity of SCFA for cross-feeding in other bacteria often is clear (15–17), little is known about the structures of the extracellular degradation products of prebiotic fructans that may accumulate through bacterial GH enzyme activity. The elucidation of such structural features is essential for the further unraveling of the cross-feeding mechanisms that may occur among human gut microbiota and will enable the determination of the ecological roles that individual members play and show how members contribute to the gut microbiome as a whole. In particular, some bacteria have been assigned a role as a keystone species, present at a relatively low abundance but exerting a strong stabilizing influence on the community (18). Only a single study reported such a role for Lactobacillus, specifically, Lactobacillus reuteri DSM 17938 (19). For the human isolate Lactobacillus paracasei subsp. paracasei 8700:2, beneficial effects toward other members of the gut microbiota have been observed due to its extracellular breakdown of inulin-type fructan, but the extent of this effect on other strains has not been investigated in detail (17, 20).
In this study, we aimed to identify which factors influence FOS/inulin degradation by pure cultures and defined cocultures of probiotic Lactobacillus strains. The results show that an extracellular family GH32 enzyme employed by Lactobacillus paracasei subsp. paracasei W20 increases the availability of suitable FOS carbon and energy substrates to Lactobacillus salivarius W57 in cocultures. To our knowledge, it has not been shown before that an extracellular GH32 enzyme can mediate cross-feeding propensity to another bacterium. These stimulatory properties of L. paracasei toward another gut bacterium offer possibilities for multispecies synbiotic combinations.
RESULTS
Structural analysis of commercial β-(2-1)-fructans.
An increasing number of β-(2-1)-fructans is commercially available. We first analyzed the structural composition of 9 commercial β-(2-1)-fructans (Table 1). Two types of compounds were found (6): fructan chains with a terminal sucrose unit on one end (GF type) and β-(2-1)-linked fructosyl residues only (FF type). GF-type compounds were present in all products analyzed, while FF-type compounds were found only in 5 of 9 (Table 1). When we compared the intensities of peaks obtained by high-pH anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) analysis (data not shown), the peak responses for FF-type compounds were the highest in Frutalose OFP and FOS P1. These compounds were classified as oligofructoses. For Frutafit HD, Frutafit CLR, and Inulin P2, the signal responses for FF-type compounds were lower, while the numbers of GF-type compounds found in chromatograms increased. These fructans were thus classified as inulin-type β-(2-1)-fructans. The highest diversity in structural composition overall was found in the degree of polymerization (Table 1). Frutafit HD and Inulin P2 offered the broadest range of fructan compounds (DP 2 to DP 60), while Frutalose OFP, FOS P1, and scFOS P6 had a limited range (DP 2 to DP 7). Most preparations still contained some glucose and fructose monosaccharides (relative peak heights ranging between 0.5% and 8.2%) and sucrose (constituting 2% to 15% relative peak heights). The 1-kestose and nystose samples were pure and comprised only these specific DP-3 and DP-4 compounds, respectively. This comparative structural analysis of 9 commercial fructans provided a firm basis for the subsequent study of the utilization of specific compounds by probiotic bacteria.
TABLE 1.
Composition of commercial prebiotic β-(2-1)-fructans obtained by comparative HPAEC-PAD analysis
| Commercial name | Supplier | Description | DPa |
|
|---|---|---|---|---|
| GF type | FF type | |||
| Frutafit HD | Sensus BV | Native inulin | 2–60 | 2–14 |
| Frutafit CLR | Sensus BV | sc-inulin | 2–40 | 2–14 |
| Frutafit TEX | Sensus BV | lc-inulin | 10–60 | |
| Frutalose OFP | Sensus BV | Oligofructose | 2–7 | 2–7 |
| FOS P1 | Winclove BV | Oligofructose | 2–7 | 2–7 |
| Inulin P2 | Winclove BV | Native inulin | 2–60 | 2–14 |
| scFOS P6 | Winclove BV | scFOS | 2–6 | |
| Nystose | CarboSynth | GF3 | 4 | |
| 1-Kestose | CarboSynth | GF2 | 3 | |
DP, degree of polymerization.
Growth of selected probiotic bacteria on short-chain inulin.
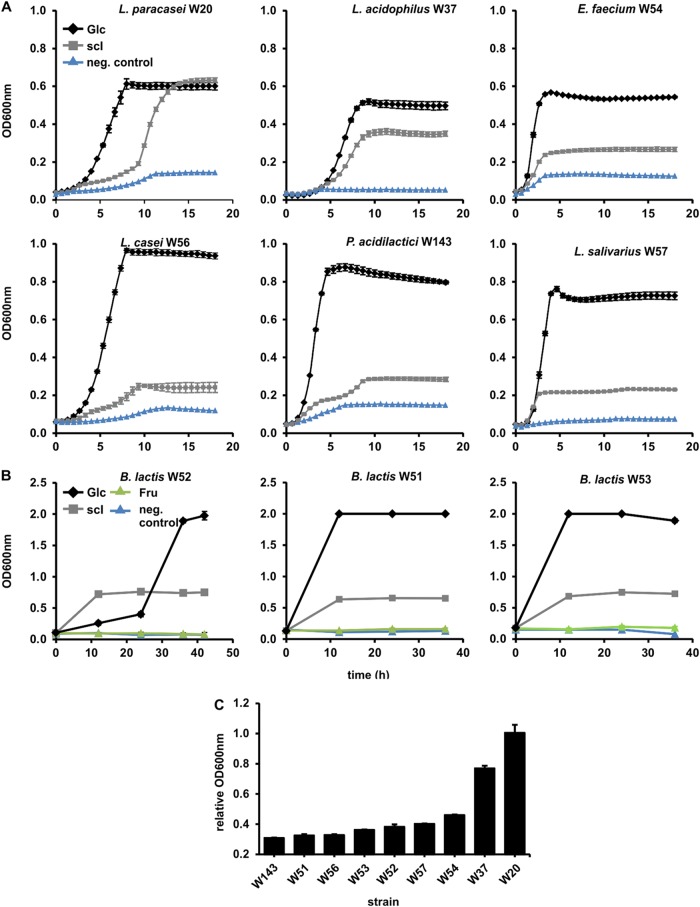
Various probiotic bacteria were tested for their ability to grow on short-chain inulin (sc-inulin; Frutafit CLR). This prebiotic has been derived from inulin and comprises over 50 individual compounds with various DPs (see below), of both GF- and FF-type β-(2-1)-fructans (Table 1). Frutafit CLR thus shows all the structural variations that are unique to prebiotic β-(2-1)-fructans, enabling an analysis of their effects on the growth of individual probiotic bacteria. Various strains of lactobacilli and bifidobacteria, plus Enterococcus faecium and Pediococcus acidilactici, were tested. The growth responses of individual strains with sc-inulin varied greatly, e.g., in the lengths of the lag phase and in the final values for optical density at 600 nm (OD600) reached (Fig. 1A to C). All strains grew well on glucose (positive control), reaching final (100%) OD600 values at stationary phase of approximately 1.0 in microtiter plates and 2.0 in Hungate tubes, set as a relative OD600 value of 1.00. When incubated with sc-inulin, most strains reached a stationary-phase relative OD600 of around 0.4, indicating that sc-inulin compounds were supporting the growth of these strains only partly compared to that of the positive control (Fig. 1C).
FIG 1.
Anaerobic growth of various probiotic bacterial strains with 5 mg/ml prebiotic sc-inulin (scI). (A) Lactic acid bacterial strains were grown in 160- to 200-μl aliquots in microtiter plates under constant N2 flow and growth was followed over time using a microtiter plate reader. (B) Bifidobacterium strains were grown as 3 ml cultures in Hungate tubes under a CO2 atmosphere, and growth was followed by measuring OD600 at 4 or 5 time points. (C) Relative OD600 values at stationary phase (using maximum OD600 of positive controls on glucose to normalize OD600 obtained with sc-inulin). All values shown are means from 3 biological replicates. Some standard deviations are smaller than the size of the symbol and therefore not apparent.
Carbohydrate analysis of sc-inulin consumption.
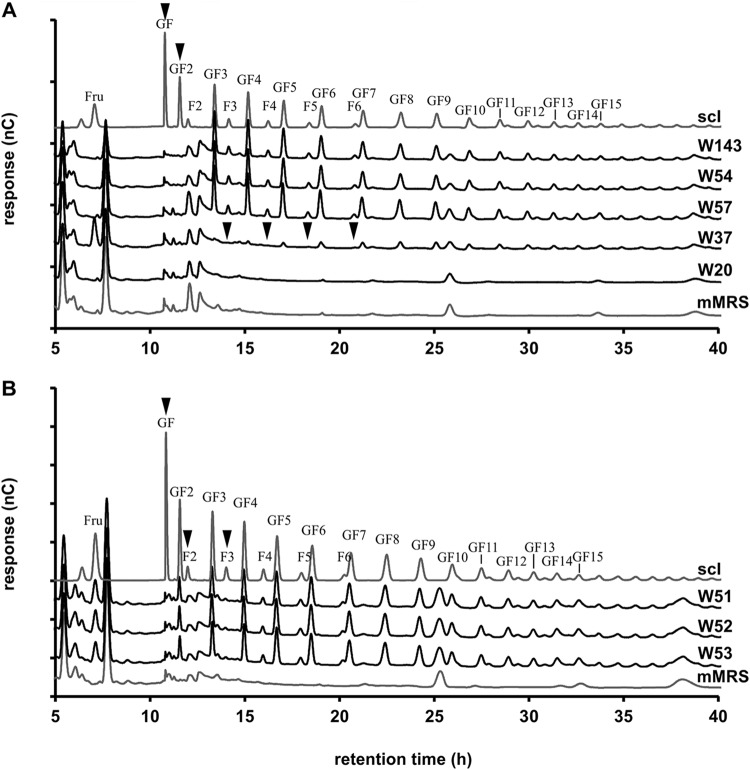
Following bacterial growth, the culture supernatants were analyzed for the presence of any remaining components from the sc-inulin mixture. A carbohydrate analysis revealed four patterns. (i) L. salivarius W57, E. faecium W54, and P. acidilactici W143 specifically used only fructose, GF (sucrose), and GF2 (1-kestose) (Fig. 2A). (ii) Bifidobacteria utilized sc-inulin compounds with the preference F2 > F3 > F4 and GF > GF2 > GF3 > GF4, but only the F2 [β-d-Frup-(2→1)-d-Fru] and F3 [β-d-Frup-(2→1)-β-d-Frup-(2→1)-d-Fru] compounds were completely used, at 24 h for Bifidobacterium animalis subsp. lactis W51 and B. lactis W53 and at 36 h for B. lactis W52 (Fig. 2B). (iii) Lactobacillus acidophilus W37 utilized short-chain compounds of the FF and GF types. (iv) L. paracasei W20 utilized all GF- and FF-type compounds present in sc-inulin. These results clearly highlight that probiotic bacteria have diverse substrate specificities for the utilization of β-(2-1)-fructans.
FIG 2.
HPAEC-PAD analysis of FOS/inulin components (DPs of 2 to 20 are shown) in culture supernatants of probiotic bacterial strains grown on sc-inulin (Frutafit CLR) (see Fig. 1). Peaks were identified using inulin (DP, 2 to 16; polymer) as the standard. (A) Selected probiotic lactic acid bacteria grown for 18 h. (B) B. lactis W51 and B. lactis W53 grown for 24 h and B. lactis W52 grown for 36 h. mMRS, modified MRS medium.
Utilization of β-(2-1)-fructans by L. paracasei W20.
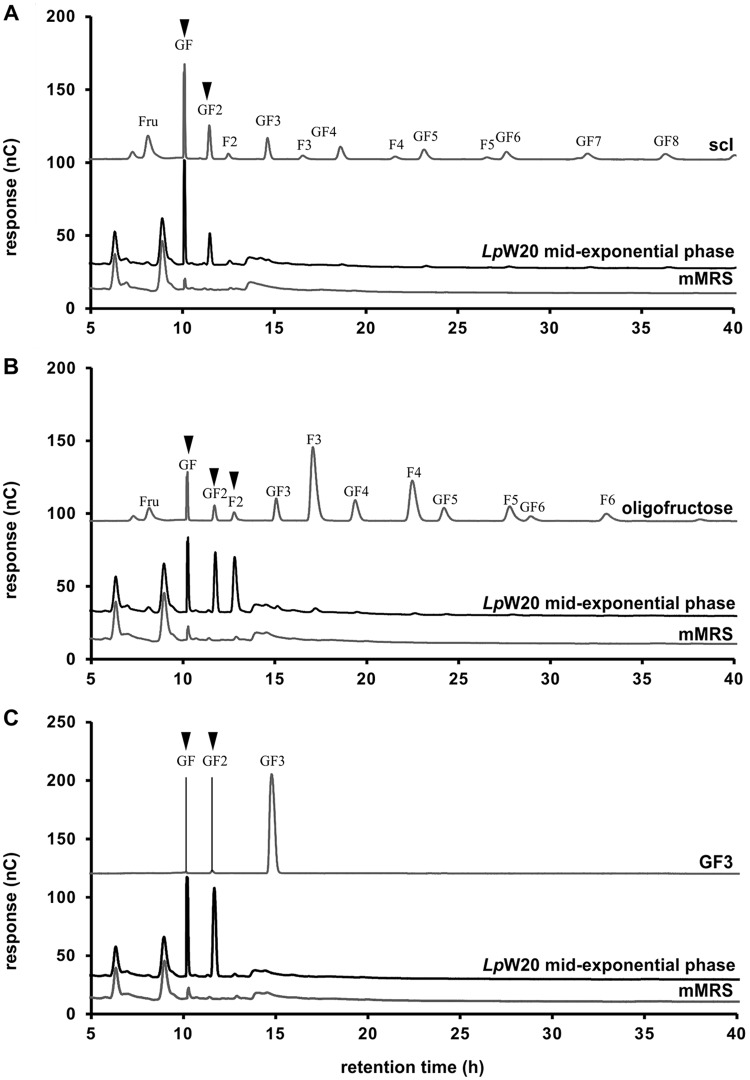
L. paracasei W20 was clearly exceptional by consuming all GF- and FF-type compounds present in sc-inulin. To analyze its fructan metabolism in more detail, we grew L. paracasei W20 on sc-inulin and GF3 (both GF-type fructans) and Frutalose oligofructose (FF-type fructan). An analysis of the changes in fructan compositions over time revealed a preferential degradation of fructose and all sc-inulin compounds larger than a DP of 3 in culture supernatants harvested from L. paracasei W20 at the mid-exponential phase (Fig. 1A) (L. paracasei, t = 12 h), while sucrose (GF) and 1-kestose (GF2) were present in relatively large amounts (Fig. 3A). Also, the degradation of the pure GF3 compound nystose by L. paracasei W20 first resulted in the release of 1-kestose (not shown) in the supernatant, followed by sucrose GF (Fig. 3C). In time, oligofructose degradation resulted in the formation of GF and GF2 and of the disaccharide F2 (Fig. 3B). All compounds had been consumed completely in stationary-phase samples (not shown). These results show that L. paracasei W20 degrades β-(2-1)-fructans in a sequential manner, leading to temporary increases of GF and GF2 in the case of GF-type fructans and F2 in the case of FF-type fructans.
FIG 3.
Temporary accumulation of breakdown products in culture supernatants of L. paracasei strain W20 growing on different prebiotic β-(2-1)-fructans. HPAEC-PAD analysis of sc-inulin Frutafit CLR (A), oligofructose Frutalose OFP (B), and GF3 nystose (C) and compounds derived in culture supernatants of L. paracasei W20 at mid-exponential growth phase (Fig. 1A, L. paracasei, t = 12 h).
Identification and activity of a putative extracellular family GH32 β-fructosidase.
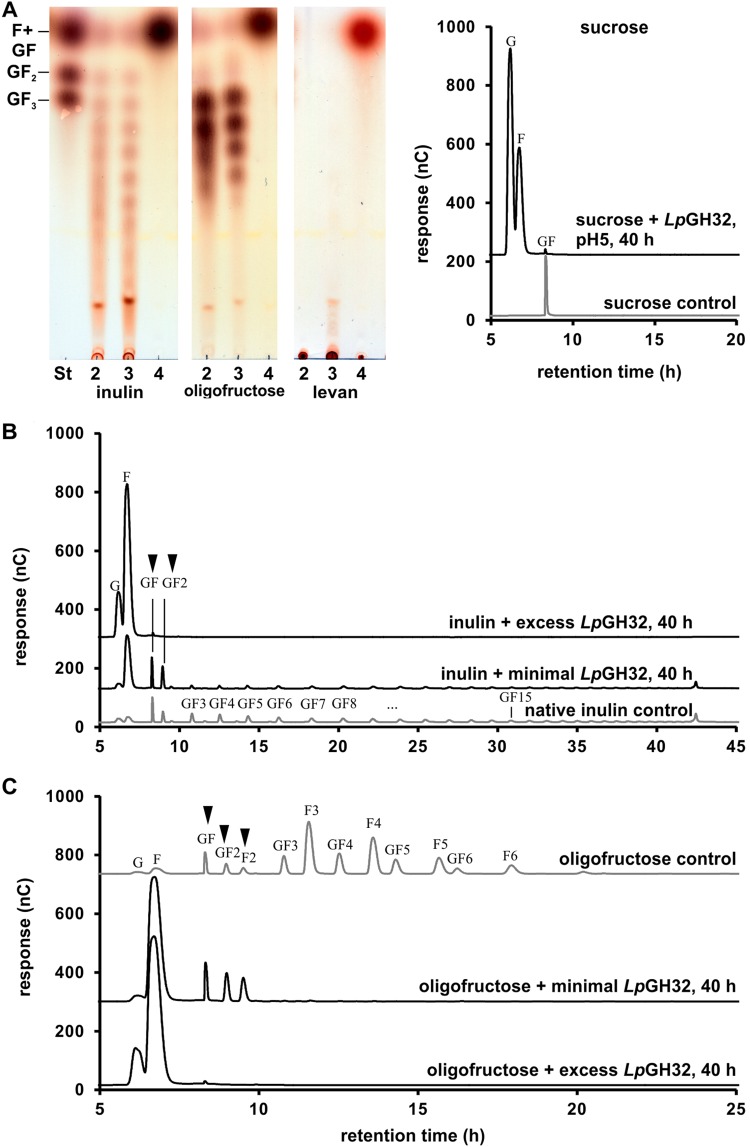
Following growth on several β-(2-1)-fructans, we detected β-fructosidase activity in L. paracasei W20 culture supernatants. This activity degraded sc-inulin into sucrose, fructose, and glucose (see Fig. S1 in the supplemental material). The supernatants obtained from the growth of L. paracasei W20 on glucose or fructose clearly had much lower activities. Via a search of the Carbohydrate-Active enZYmes Database (www.cazy.org) (dbCAN [21]), the genome of L. paracasei W20 was found to encode one putative β-fructosidase protein of 1,031 amino acids (family GH32) including a cell wall anchor motif for Gram-positive bacteria (LPQAG) (GenBank accession no. MH047828). The subcellular localization of this protein is predicted to be >99% extracellular. The protein sequence showed 97% identity to β-fructosidases and sucrose-6-phosphorylases present in various strains of L. paracasei and L. casei and around 30% identity to exo-inulinases from Bacteroides species and Bacillus subtilis (22, 23). To characterize the activity of this putative GH32 enzyme, we cloned the predicted catalytic domain (residues 188 to 739) into the pET15bLIC vector which added an N-terminal 6-His tag (called LpGH32). The overexpression of LpGH32 in Escherichia coli BL21 yielded a dominant band by SDS-PAGE at the expected molecular mass of 61 kDa; this protein was purified using His-trap column affinity chromatography. The incubation of 0.9 μg/ml (excess amount) of the purified LpGH32 with β-(2-1)-fructans revealed activity on inulin and oligofructose (only at pH 5.0), which was observed before with culture supernatants. The final products of enzymatic conversion after 40 h were identified as monosaccharides (Fig. 4A, inulin and oligofructose) and subsequently separated and identified by HPAEC-PAD as glucose and fructose. This activity was similar to what was observed before within the culture supernatants of L. paracasei W20 (Fig. S1). We further tested the activity of the enzyme with sucrose and the β-(2-6)-fructan levan (Fig. 4A, levan and sucrose). While all sucrose was converted into fructose and glucose after 40 h, levan was not a substrate at pH 4.0 and only partly converted into fructose at pH 5.0 (indicated by a remaining spot at the bottom of the thin-layer chromatography [TLC] plate) (Fig. 4A, levan, lanes 3 and 4). Product formation at 40 h was also analyzed by adding a minimal amount (0.15 μg/ml) of LpGH32 to incubations with inulin. Besides the monosaccharides glucose and fructose, GF sucrose and GF2 1-kestose accumulated from inulin conversion (Fig. 4B). With oligofructose, we observed an increase of DP-2 F2 plus DP-2 sucrose and DP-3 1-kestose (derived from GF impurities present in the oligofructose) (Fig. 4C). These results show that larger β-(2-1)-fructan chains are preferably converted by the LpGH32 enzyme, leading to an intermediate accumulation of shorter DP-2 to DP-3 β-(2-1)-fructan chains.
FIG 4.
Substrate specificity of LpGH32 and release of breakdown products during conversion of different β-(2-1)-fructans. (A) Products formed by purified LpGH32 enzyme (0.9 μg/ml) incubated with fructan [β-(2-1) and β-(2-6)] substrates: native inulin [from chicory, β-(2-1)], oligofructose [β-(2-1)], levan [β-(2-6), bacterial origin], and sucrose [α-(1-2)]. St, standard containing fructose (Fruc), sucrose (Suc), 1-kestose (GF2), and nystose (GF3). Incubation was at pH 4.0 (lane 3) and pH 5.0 (lane 4) for 40 h; lane 2, substrate control. Accumulation of intermediate breakdown products liberated by LpGH32 (excess, 0.9 μg/ml, and minimal, 0.15 μg/ml, amounts) during incubations with GF-type fructan native inulin (B) and FF-type fructan oligofructose (also contains GF-type fructans) (C) and stopping reactions after 40 h. Main product released was fructose; accumulation of intermediate breakdown products is highlighted with arrowheads.
Genome analysis explains different fructan utilization patterns in probiotic bacterial strains.
Our results show that L. paracasei W20 employs an extracellular exo-inulinase (EC 3.2.1.80, family GH32) to degrade prebiotic β-(2-1)-fructans (LpGH32). Among the bacteria tested in this study, this enzyme accounts for the ability of L. paracasei W20 to degrade β-(2-1)-fructans independent of their structural parameters (GF and FF types; DP 2 to DP 60). Due to the preference of LpGH32 to degrade fructan chains larger than a DP of 3 first, seen both with the purified enzyme and in supernatants of L. paracasei W20, the intermediate breakdown products GF and GF2 for GF-type fructans and GF, GF2, and F2 for FF-type fructans accumulated (Fig. 3 and 4).
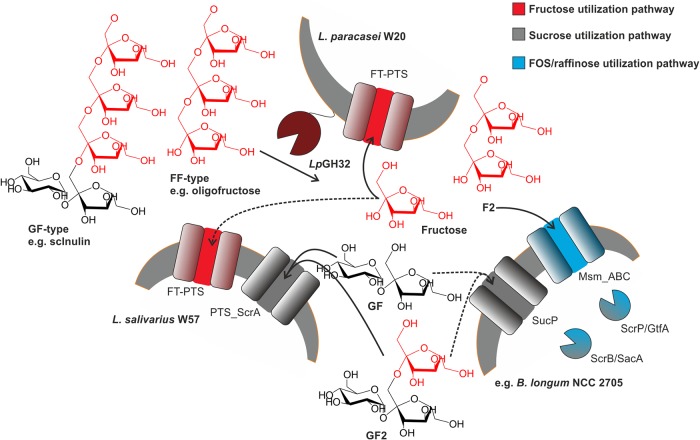
We searched bacterial genomes for genes involved in known pathways of fructose utilization, sucrose utilization, and fructooligosaccharide/raffinose utilization (22, 24–31) to explain the strain differences in utilization of GF-type and FF-type components. Specifically, we identified key glycoside hydrolases and transporters involved in fructose metabolism. We found that these strains differed highly when comparing genes from all three pathways mentioned above (Fig. 5 and Table S1). None of these strains combined all three pathways; rather, one or two pathways were predicted by the genomic annotation. The genomes of L. paracasei W20 and L. salivarius W57 both encode a fructose utilization pathway with a PTS transport system, which may account for the uptake of the free fructose released by LpGH32. The L. salivarius W57 genome contains a predicted sucrose utilization system with a sucrose PTS transport system, which was not found in the L. paracasei W20 genome. The Bifidobacterium strains studied lacked a fructose utilization pathway (also observed experimentally, bifidobacterial strains tested could not grow on pure fructose) (Fig. 1B), but sucrose utilization was predicted in the bifidobacterial representative strain B. longum NCC 2705, with a sucrose permease and a FOS/raffinose utilization pathway with an ABC transporter and an intracellular β-fructosidase (Fig. 5 and Table S1). On the basis of the results from the genome analyses of these probiotic strains, we hypothesize that the diverse distribution of these pathways may enable interspecies interactions resulting in cross-feeding during the degradation of inulin. If interspecies interactions occur, these may generate interesting biotechnological opportunities in the application of combinations of probiotic strains with the prebiotic inulin as novel synbiotic mixtures on the basis of interspecies cross-feeding properties.
FIG 5.
Metabolism of prebiotic β-(2-1)-fructans by L. paracasei W20, L. salivarius W57, and B. longum NCC 2705. Fructose uptake via a fructose-PTS transport system (FT-PTS), sucrose uptake via a sucrose-PTS system (PTS_ScrA) or a sucrose permease (SucP), and a FOS/raffinose utilization pathway with a FOS-ABC transport system (Msm_ABC) followed by intracellular β-fructosidases ScrP/GtfA and ScrB/SacA.
Interspecies interactions between L. salivarius W57 and L. paracasei W20 involving LpGH32 degradation products.
To test the interspecies cross-feeding hypothesis and to identify possible involvement of particular enzymes and transporters harbored by probiotic bacteria in the degradation of prebiotic fructan components, we studied the interspecies interactions of the two probiotic strains L. paracasei W20 and L. salivarius W57. We chose to use these strains on the basis of the results of their genome analysis and their different carbohydrate-utilization profiles: while L. salivarius incubated with sc-inulin showed very specific utilization of fructan compounds with a DP of <4 (Fig. 2A), L. paracasei was able to utilize all components of β-(2-1)-fructans, employing an extracellular exo-inulinase. Both strains differ at the transporter level, with a single sucrose transporter present in L. salivarius W57 (PTS_ScrA) (Fig. 5) but lacking in L. paracasei W20.
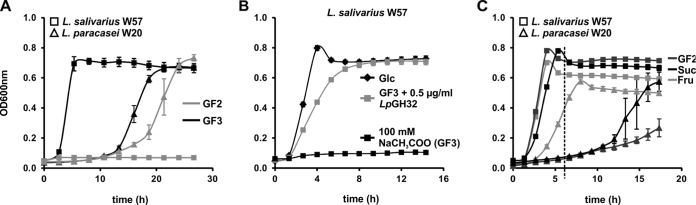
To identify any preference of L. salivarius W57 for GF-type fructans with distinct sizes, the strain was grown on pure 1-kestose (GF2, DP-3 FOS) and nystose (GF3, DP-4 FOS). The results show that L. salivarius only grew on 1-kestose, while L. paracasei grew on both 1-kestose and nystose compounds (Fig. 6A). Nystose thus is the smallest DP fructooligosaccharide that demonstrates selectivity between the two Lactobacillus strains and therefore requires the activity of LpGH32 in order to be metabolized. We then looked for the possible beneficial cross-feeding effects of LpGH32 by adding the purified enzyme to L. salivarius W57 cultures incubated with nystose. L. salivarius W57 was unable to grow on nystose (Fig. 6A), while the addition of LpGH32 enabled the strain to completely metabolize this DP-4 fructan (Fig. 6B). To identify the products of LpGH32 produced by L. paracasei W20 that are potentially available for cross-feeding by L. salivarius W57, we grew L. paracasei W20 and L. salivarius W57 on several LpGH32 inulin products. While L. salivarius W57 grew equally well on fructose, sucrose, and 1-kestose, reaching stationary phase after 5 h, L. paracasei W20 used fructose rapidly (stationary after 9 h) but grew quite poorly on 1-kestose (after a long lag phase of 12 h) and on sucrose (Fig. 6C). These results suggest that L. paracasei W20 overall has a high affinity for fructose, while the GF-type compounds sucrose and GF2 1-kestose are used at a lower preference. Therefore, sucrose and 1-kestose liberated by LpGH32 activity may be available as cross-feeding products from sc-inulin for metabolism by other bacteria that are in close proximity to L. paracasei.
FIG 6.
Beneficial effects of LpGH32 on growth of L. salivarius W57 and abilities of L. salivarius W57 and L. paracasei W20 to grow on LpGH32 breakdown products liberated from prebiotic inulin conversion. (A) Anaerobic growth of L. paracasei W20 and L. salivarius W57 individually on 5 mg/ml pure 1-kestose (GF2) and nystose (GF3). (B) Growth of L. salivarius W57 on 5 mg/ml glucose (positive control) or nystose (DP-4 FOS, GF3) with (0.5 μg/ml) or without added LpGH32 enzyme activity. mMRS, modified MRS medium, 100 mM NaCH3COO enzyme buffer. (C) Growth of L. salivarius W57 and L. paracasei W20 on 5 mg/ml of individual LpGH32 (intermediate) inulin degradation products: fructose (Fruc), sucrose (Suc), and 1-kestose (GF2). All values shown are means from 3 biological replicates. Some standard deviations are smaller than the size of the symbol and therefore not apparent.
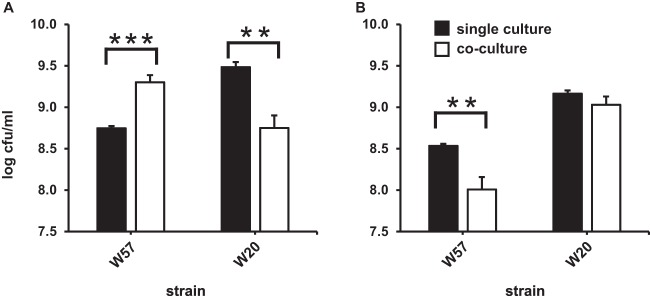
To test for in vivo cross-feeding, we cocultured L. salivarius W57 and L. paracasei W20 on two types of β-(2-1)-fructans, sc-inulin containing mainly GF-type fructan chains and oligofructose (Frutalose OFP) containing mostly pure fructose-composed β-(2-1)-fructan chains. In the case of sc-inulin, coculturing resulted in a clear increase in CFU/ml for L. salivarius W57 in the cocultures compared to that in the single cultures (Fig. 7A). Cross-feeding thus occurred between the two strains, stimulating the growth of L. salivarius W57, which benefited from the presence of L. paracasei W20, employing the extracellular GH32 enzyme for the degradation of the GF-type fructan sc-inulin (resulting in an accumulation of short-chain FOS DP-2 to DP-3 compounds). The degradation of oligofructose yields free fructose, which does not result in cross-feeding of L. salivarius W57 (Fig. 7B).
FIG 7.
Growth (CFU/ml) of L. paracasei W20 plus L. salivarius W57 in single cultures and cocultures on 5 mg/ml sc-inulin (Frutafit CLR) (A) and oligofructose (Frutalose OFP) (B). Cocultures and single cultures of L. paracasei W20 were grown for 20 h; single cultures of L. salivarius W57 were grown for 6 h. Values shown are the means from 3 biological replicates. **, P < 0.01; ***, P < 0.001 (Student's t test).
DISCUSSION
The selective stimulation of the growth of beneficial bacteria in the human gut may yield in vivo health effects. This study reports the structural characterization of commercial prebiotic β-(2-1)-fructans and their utilization for growth by selected probiotic bacteria.
The carbohydrate utilization properties of probiotic strains from the genera Lactobacillus and Bifidobacterium were investigated previously. The ability of Lactobacillus strains to utilize prebiotic carbohydrates appeared much more limited than that of Bifidobacterium strains (32). The transporters and GH enzymes encoded by the genomes of these probiotic bacteria varied even among strains from the same species (13). For instance, Kaplan and Hutkins investigated L. paracasei 1195 for its ability to degrade FOS, attributing its FOS utilization to an intracellular β-fructosidase enzyme (30). Later, Hutkins and colleagues identified by whole-genome microarray analysis the fos utilization operon in the genome of L. paracasei 1195 encoding an extracellular β-fructosidase enzyme and a fructose/mannose transporter (22). An extracellularly located β-fructosidase was also found in L. paracasei DSM 23505, used to effectively convert FOS/inulin into lactic acid (33). Both studies characterized the extracellular enzyme activity on FOS/inulin of L. paracasei 1195 and L. paracasei DSM 23505 as exo-inulinase, EC 3.2.1.80. To characterize the extracellular β-fructosidase of L. paracasei W20, we purified the LpGH32 enzyme after overexpression in E. coli and identified it as an exo-inulinase (EC 3.2.1.80). Hutkins and colleagues observed for L. paracasei 1195 that the degradation of FOS DP-4 and DP-5 occurred first, resulting in a temporary increase of glucose, sucrose, and FOS DP-3. A temporary increase of short-chain fructooligosaccharides was also mentioned in the study using L. paracasei 8700:2, a human isolate releasing fructose, GF2, and F2 upon oligofructose degradation or fructose, glucose, and sucrose upon inulin degradation (20). Cultures of L. paracasei W20 showed preferential utilization of FOS components of a DP of >3 and thus temporal accumulation of sucrose and DP-3 FOS. This degradation pattern was also observed using purified LpGH32. These data show that the preferential degradation of FOS/inulin components with a DP of >3 is due to the activity of LpGH32 synthesized by L. paracasei W20 and not to selective uptake systems in the host (Fig. 3 and 4).
Unlike L. paracasei W20, L. salivarius W57 displayed a highly specific fructan utilization profile which enabled the strain to exclusively utilize GF-type β-(2-1)-fructan with a DP of 2 and a DP of 3. The preferential utilization of FOS components with a low DP has not been described for L. salivarius strains before; however, Saulnier et al. reported that Lactobacillus plantarum WCFS1 consumes GF2 and GF3 compounds. Using whole-genome microarrays, three genes were identified as participating in scFOS metabolism of L. plantarum, i.e., genes encoding a sucrose PTS uptake system, a β-fructofuranosidase, and a fructokinase (34). Our annotation of the L. salivarius W57 genome revealed the presence of a PTS_ScrA gene, explaining the preference of L. salivarius W57 for scFOS compounds, which, in comparison to L. plantarum WCFS1, shows a higher specificity for GF-type FOS components with DPs of only 2 to 3.
To specifically stimulate the growth of human gut microorganisms, possibly even at the species/strains level, the generation of a given metabolic activity which may then lead to a given health effect is required. Where bacterial cells are limited in their ability to utilize prebiotic FOS/inulin compounds, the presence of extracellular β-fructosidase enzymes may stimulate degradation and thus result in an improved growth of a probiotic bacterium on a given prebiotic (17, 35). For instance, bifidobacterial growth is very well known to be stimulated by prebiotic FOS/inulin utilization. The enrichment of this genus through FOS/inulin utilization is so dominant that this was labeled as the bifidogenic effect. However, Bifidobacterium strains in pure culture are only able to use FOS/inulin components with a DP of <20 (12). When growing bifidobacteria in cocultures with (commensal) gut species expressing extracellular enzymes from family GH32, cross-feeding occurs and results in increased bifidobacterial growth (35). As shown in the present study, L. paracasei W20 exhibits the ability to degrade FOS and inulin extracellularly using a family GH32 enzyme, an exo-inulinase (LpGH32) that yields intermediate degradation products of GF- and FF-type β-(2-1)-fructans, respectively (Fig. 4). GF-type fructan use results in an accumulation of the short-chain FOS DP-2 to -3 compounds, whereas FF-type fructan use only yields fructose as a product from LpGH32 activity. This accumulation of intermediate FOS DP-2 to -3 products leads to the stimulation of growth of another Lactobacillus strain, L. salivarius W57, that specifically takes up GF (sucrose) and GF2 (1-kestose) using a sucrose transporter. In contrast, any released fructose (e.g., from FF-type fructans) is more likely to be directly taken up by L. paracasei W20 using a fructose/mannose transporter (Fig. 5). On the basis of the results of our study, we can conclude that the ability of a Lactobacillus strain to benefit from cross-feeding during growth on prebiotic β-(2-1)-fructans is highly dependent on the use of GF-type (cross-feeding stimulating) or FF-type (relatively low cross-feeding) fructans. In comparison, cross-feeding mediated by bifidobacteria toward butyrate-producing members of the gut microbiome was mediated by short fractions of oligofructose (FF-type fructan) and thus not the GF-type fructan as observed in our study (15, 16). Moreover, we conclude that the extent of cross-feeding among lactobacilli also depends on fructan chain length: β-(2-1)-fructans with a relatively low DP contain at the molar level a relatively high content of GF-type molecules, thus resulting in higher concentrations of the breakdown products of the GF type with DPs of 2 to 3. Our results therefore show that the extent of bacterial cross-feeding among probiotic lactobacilli is clearly controlled by the structure of the prebiotic fructan substrate used.
These results provide insights into the mechanism and role of cross-feeding by β-(2-1)-fructan degradation products, stimulating the growth of bacteria living in the same environment. To stimulate the growth of probiotic bacteria at the strain level, the prebiotic β-(2-1)-fructan should be carefully chosen, taking into account its structural composition (Table 1). In synbiotic mixtures using L. paracasei W20 employing an extracellular GH32 enzyme, FF-type FOS is preferred when only L. paracasei W20 is supposed to benefit from prebiotic fructan supplementation (Table 2). When aiming to stimulate the growth of another strain, preferably GF-type fructan of a relatively low chain length should be considered. Our results further highlight the advantageous properties of L. paracasei and its extracellular GH32 enzyme mediated through the release of intermediate degradation products from large-chain inulin, demonstrating the potential ability of L. paracasei to act as a keystone species among probiotic lactobacilli.
TABLE 2.
Prebiotic β-(2-1)-fructans used in this study, ordered according to their potential to stimulate cross-feeding between L. paracasei W20 and L. salivarius W57a
| Commercial name | Description |
|---|---|
| 1-Kestose | GF2 |
| Nystose | GF3 |
| scFOS P6 | scFOS |
| Frutafit CLR | sc-inulin |
| Inulin P2 | Native inulin |
| Frutafit HD | Native inulin |
| Frutafit TEX | lc-inulin |
| FOS P1 | Oligofructose |
| Frutalose OFP | Oligofructose |
From top (starting with 1-kestose; high cross-feeding) to bottom (ending with oligofructose; low cross-feeding), the fructans are ranked on the basis of the activity of the extracellular L. paracasei W20 LpGH32 enzyme for releasing more of the GF (sucrose) and 1-kestose (GF2) molecules from the shorter β-(2-1)-fructans and from fructans comprising more GF-type molecules (and not from oligofructans).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The commercial probiotic bacterial strains Lactobacillus paracasei subsp. paracasei W20 (L. paracasei W20), Lactobacillus acidophilus W37, Lactobacillus salivarius W57, Lactobacillus casei W56, Enterococcus faecium W54, Pediococcus acidilactici W143, Bifidobacterium animalis subsp. lactis W51 (B. lactis W51), Bifidobacterium animalis subsp. lactis W52 (B. lactis W52), and Bifidobacterium animalis subsp. lactis W53 (B. lactis W53) were provided by Winclove Probiotics B.V. (Amsterdam, The Netherlands). All strains belonging to lactic acid bacteria (LAB) were maintained in de Man-Rogosa-Sharpe (MRS) broth (Oxoid, Basingstoke, UK) containing 2% glucose as the carbon source. Liquid cultures (5 ml) were inoculated in anaerobic culture glass tubes (Boom, Meppel, The Netherlands) under anaerobic conditions provided by the Hungate system with nitrogen (36) and incubated at 37°C. For permanent storage, fresh 200-μl aliquots of the cultures were diluted with sterile glycerol to 15% (vol/vol) in 1.5-ml tubes, and the tubes were maintained at −80°C. The purity of the cultures was frequently checked by spreading aliquots on agar plates prepared with MRS and Luria-Bertani (LB) medium and by light microscopy. For growth experiments with β-(2-1)-fructans, modified MRS medium (mMRS) was used with LAB strains prepared according to reference 32. In brief, 1 liter mMRS contained 10 g peptone, 2.5 g granulated yeast extract, 3 g tryptose, 1 ml Tween 80, 3 g K2HPO4, 3 g KH2PO4, 2 g ammonium citrate, 0.2 g pyruvic acid sodium salt, 0.575 g MgSO4·7H2O, 0.12 g MnSO4·H2O, and 0.034 g FeSO4·7H2O. The components were dissolved in double-distilled H2O by bringing the medium to a boil and then adding 0.5 g/liter cysteine-HCl and adjusting the pH to 6.8. The medium was sterilized by autoclaving (15 min at 121°C).
Bifidobacterial strains were maintained in Bifidobacterium medium (BM) that was prepared according to reference 37. One liter BM contained 10 g Trypticase peptone, 2.5 g yeast extract, 3 g tryptose, 3 g K2HPO4, 3 g KH2PO4, 2 g triammonium citrate, 0.3 g pyruvic acid, 1 ml Tween 80, 0.574 g MgSO4·H2O, 0.12 g MnSO4·H2O, and 5 g NaCl in 1 liter of distilled water. After autoclaving, the BM was supplemented with 0.05% (wt/vol) filter-sterilized cysteine-HCl, and strains were grown at 37°C under anaerobic conditions maintained by 100% CO2.
Carbohydrates.
Native inulin (Frutafit HD; inulin of DP of 2 to 60, ≥90% [wt/wt]; average chain length, 11), long-chain inulin (lc-inulin, Frutafit TEX; inulin ≥99.5% [wt/wt]; average chain length, 22), short-chain inulin (sc-inulin, Frutafit CLR; inulin/oligofructose, ≥85% [wt/wt]; average chain length, 7), and oligofructose (Frutalose OFP; oligofructose, ≥92% ± 2% [wt/wt]; average chain length, 7) were provided by SENSUS (Roosendaal, The Netherlands). 1-Kestose (≥98%) and nystose (≥98%) were purchased from CarboSynth (Compton, UK). FOS P1 (oligofructose of DP of 2 to 8, ≥93% [wt/wt]), Inulin P2 (inulin of DP of 2 to 60, ≥90% [wt/wt]; average chain length, 10), and scFOS P6 (oligofructose of DP of 3 to 5, ≥95% [wt/wt]) were obtained from Winclove Probiotics B.V. Levan was produced by sucrose conversion with B. subtilis as described previously (38). Carbohydrates were dissolved to 1% (wt/vol) in Milli-Q water and filter sterilized using 0.2-μm cellulose acetate membrane filters (VWR International, Radnor, PA); lc-inulin was dissolved and sterilized by autoclaving.
Growth experiments.
Growth experiments with LAB strains were carried out using 96-well microtiter plates (Greiner Bio-one, Frickenhausen, Germany). Prior to the inoculation of the plates, cultures of bacterial strains grown overnight were harvested by centrifugation (2,300 × g for 2 min) and diluted 25-fold in 2× mMRS. Separately, carbohydrate solutions were added in triplicates to wells of plates, while glucose and double-distilled water served as the positive and negative controls, respectively. Subsequently, diluted bacterial suspensions and plates containing carbohydrate solutions were transferred to an anaerobic glove box (Bohlender, Grünsfeld, Germany) with constant nitrogen flow, and cultures were inoculated with 2% (vol/vol) of bacteria (total volume of cultures per well, 160 to 200 μl) to provide anaerobic conditions in microtiter plates. Then, the plate was sealed with an airtight transparent seal (Simport, Beloeil, Canada) and placed in a plate reader (at 37°C) for the monitoring of the OD600 at 5-min intervals with continuous shaking at medium speed. The plate reader was installed inside an AtmosBag (Sigma-Aldrich, Schnelldorf, Germany) and constantly flushed with nitrogen throughout the OD600 measurements. For growing L. salivarius W57 in the presence of the GH32 enzyme from L. paracasei W20 (LpGH32), 10 μl of the recombinantly produced and purified enzyme was added per well, yielding a final concentration of 0.5 μg/ml. For coculture experiments, cultures were inoculated with 1% (vol/vol) of overnight cultures of L. salivarius W57 and L. paracasei W20. To determine the CFU/ml, bacterial cultures were harvested at 18 h (L. paracasei W20 single cultures and L. paracasei plus L. salivarius cocultures) and 8 h (L. salivarius W57 single cultures). Harvested cultures were diluted to appropriate levels and spread on MRS agar (MRS medium supplemented with 1.5% [wt/vol] agar). Subsequently, the plates were incubated anaerobically in jars using the GasPak EZ container system (BD, Sparks, MD). The plates incubated with both strains were kept at 18°C (growth of L. paracasei W20 only) and 37°C (growth of both strains).
Bifidobacterial strains were grown in 3-ml cultures in BM supplemented with 0.5% (wt/vol) carbohydrates using anaerobic glass tubes and maintained under anaerobic conditions with 100% CO2 at 37°C. BM with and without 5 mg/ml glucose was used as the positive and negative controls, respectively. Growth was followed over time by measuring the OD600 manually directly in anaerobic glass tubes using the cell density meter WPA CO 8000 (Biochrom, Cambridge, UK).
Structural analysis of fructooligosaccharides.
FOS composition was analyzed using thin-layer chromatography (TLC) as reported previously (39). In brief, samples of 1 μl were spotted up to 3 times on TLC sheets (silica gel 60 F254; Merck, Darmstadt, Germany) that were developed with n-butanol-ethanol-water (5:5:3 [vol/vol]). The bands were visualized by urea staining. FOS composition was determined by high-pH anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) by diluting the samples to an appropriate concentration in 80% (vol/vol) dimethyl sulfoxide. The samples were analyzed on a CarboPac PA1 column (4 mm by 250 mm; Dionex, Sunnyvale, CA) using a linear gradient (buffer A, 0.1 M NaOH; buffer B, 0.6 M NaOAc in 0.1 M NaOH): 0 min, 0% B; 10 min, 22% B; 25 min, 40% B; 25.1 min, 100% B. Glucose, fructose, 1-kestose, and nystose were used as the standards to determine monosaccharides and short oligosaccharides (40). Inulin synthesized by the InuJ enzyme (DP range, 2 to 16; inulin polymer) was used to annotate any larger oligosaccharides (39).
Detection of extracellular enzyme activity in bacterial culture supernatants.
After 24 h of growth on FOS, cultures of probiotic bacteria were centrifuged (10,000 × g for 15 min). Subsequently, the supernatants were filtered using 0.45-μm cellulose acetate filters (VWR International) and mixed 1:1 with 10 mg/ml sc-inulin in 1.5-ml reaction tubes. After incubating for 72 h at 37°C, the (changes in the) sc-inulin compositions were analyzed by TLC and HPAEC-PAD. The culture supernatants from growth on glucose served as the negative controls.
Cloning, overexpression, and purification of truncated GH32 enzyme from L. paracasei W20.
Genomic DNA isolation from L. paracasei W20 was performed using a GenElute bacterial genomic DNA kit (Sigma-Aldrich). Genomic DNA (10 ng) was used as the template in PCRs to amplify the gene encoding the GH32 enzyme from L. paracasei W20. The primers used in PCRs were LPGH32-Fwd (5′-CAGGGACCCGGTCCATACCGAAACCAGTATCACTACTCAAGTAGC-3′) and LPGH32-Rev (5′-CGAGGAGAAGCCCGGTTAGTTCCAAATTGAAGTAATTGGATTGATAGTTAAGTCGC-3′).
The underlined complementary overhangs were used for joining inserts with vector pET15b, which was modified to enable ligation independent cloning (LIC) of the amplified DNA (41). Phire Hot Start II DNA polymerase (Thermo Scientific, Waltham, MA) was used for amplification in an MJ Mini personal thermal cycler (Bio-Rad, Hercules, CA). The reaction mixtures contained 0.05% (vol/vol) dimethyl sulfoxide. PCR mixtures yielding sufficient amplification of the desired gene were cleaned up using an Illustra GFX PCR DNA and Gel Band purification kit (GE Healthcare, Buckinghamshire, UK). Purified PCR products were then cloned into pET15b according to LIC procedures. In brief, the insert and vector were treated with T4 DNA polymerase for 60 min at room temperature (RT), followed by inactivation of the enzyme (20 min at 75°C). T4 DNA polymerase-treated vector and insert were ligated for 15 min at RT, EDTA was added to a 10 mM final concentration, and the ligation mixture was incubated for another 5 min at RT. Subsequently, the ligation mixture was transformed into E. coli TOP10 competent cells, and positive clones were checked by isolating plasmid DNA followed by digestions with NcoI/XhoI. For the overexpression of the recombinant protein, plasmid DNA containing the desired gene was transformed into E. coli BL21 Star (DE3), and precultures of transformed E. coli BL21 Star (DE3) were grown overnight in 5 ml LB medium containing 50 μg/ml ampicillin. Subsequently, expression cultures of 250 ml LB containing 100 μg/ml ampicillin were inoculated with 1% (vol/vol) of the precultures. The flasks were put into incubator shakers with agitation at 210 rpm at 37°C, and growth was allowed until the OD600 reached 0.5. Then, the cultures were put on ice and expression was started by adding 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Expression was continued in incubator shakers at 18°C and 160 rpm for 16 h. Bacterial pellets were collected by centrifugation (10 min at 3,500 × g), resuspended in 20 ml 20 mM Tris HCl (pH 8.0) supplemented with 1 mM CaCl2, 5 mM β-mercaptoethanol, and 4 mM imidazole, and sonicated 10 rounds for 15 s using a Soniprep 150 Plus (MSE, Lower Sydenham, UK). Between every sonication step, the suspension was put on ice for 30 s. Afterwards, the cell extract was centrifuged (15,000 × g for 20 min) and added to a HisTrap HP 1-ml column (GE Healthcare) connected to a peristaltic pump for protein purification. The column was equilibrated with 1.5 ml buffer A (20 mM Tris HCl [pH 8.0] supplemented with 500 mM NaCl, 3 mM CaCl2, and 3 mM MgCl2), eluted with 1.5 ml of elution buffer (containing 500 mM imidazole), and again equilibrated with 1.5 ml buffer A. Then, the protein sample was applied to the column, washed with 5 column volumes of buffer A, and eluted with 1.5 ml each of imidazole of 10 to 500 mM. Protein content and purity of collected fractions were analyzed by running samples by SDS-PAGE using 12% (vol/vol) polyacrylamide gels. Subsequently, the elution fractions with the highest content of purified protein were combined and dialyzed overnight against 100 mM sodium acetate buffer (pH 5.4) as described previously (39).
Analyzing substrate specificity of GH32 enzyme.
Substrates (10 mg/ml) were incubated with excess and minimal amounts of LpGH32 using 100 mM citric acid buffer at pH 4.0 and pH 5.0. Reaction volumes (100 μl) were supplemented with 0.01% (wt/vol) NaN3 to prevent microbial growth, and incubations were carried out at 37°C for 40 h. Product formation through GH32 enzyme activity was checked by TLC and HPAEC-PAD.
Statistical analysis.
All growth experiments are biological triplicates; the OD600 values are expressed as averages. Medium without bacterial inoculation was used to obtain blank values during OD600 measurements. Differences in the growth results between L. salivarius W57 and L. paracasei W20 in single and coculture were assessed by running nonparametric unpaired t tests. Tests yielding P values of <0.05 were considered significantly different.
Accession number(s).
The putative fructose and fructan utilization gene clusters of L. paracasei W20 were assigned the following accession numbers: fructose utilization, MH035726; fructan utilization, MH047828. The putative fructose and sucrose utilization gene clusters of L. salivarius W57 were assigned the following accession numbers: fructose utilization, MH047826; sucrose utilization, MH047827.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Pieter Grijpstra for assisting with implementing anaerobic culture techniques in our laboratory.
A.L.V.B. is the recipient of a VENI grant from the Netherlands Organization for Scientific Research (NWO). The research of M.C.L.B. was performed in the public-private partnership CarboHealth coordinated by the Carbohydrate Competence Center (CCC) and financed by participating partners and allowances of the TKI Agri&Food program, Ministry of Economic Affairs.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01539-18.
REFERENCES
- 1.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. 2014. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 2.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. 2017. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 3.Mussatto SI, Mancilha IM. 2007. Non-digestible oligosaccharides: a review. Carbohydr Polym 68:587–597. doi: 10.1016/j.carbpol.2006.12.011. [DOI] [Google Scholar]
- 4.Al-Sheraji SH, Ismail A, Manap MY, Mustafa S, Yusof RM, Hassan FA. 2013. Prebiotics as functional foods: a review. J Funct Foods 5:1542–1553. doi: 10.1016/j.jff.2013.08.009. [DOI] [Google Scholar]
- 5.Dominguez AL, Rodrigues LR, Lima NM, Teixeira JA. 2014. An overview of the recent developments on fructooligosaccharide production and applications. Food Bioproc Tech 7:324–337. doi: 10.1007/s11947-013-1221-6. [DOI] [Google Scholar]
- 6.Roberfroid M, Delzenne N. 1998. Dietary fructans. Annu Rev Nutr 18:117–143. doi: 10.1146/annurev.nutr.18.1.117. [DOI] [PubMed] [Google Scholar]
- 7.Puertollano E, Kolida S, Yaqoob P. 2014. Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr Opin Clin Nutr Metab Care 17:139–144. doi: 10.1097/MCO.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 8.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantarel BL, Lombard V, Henrissat B. 2012. Complex carbohydrate utilization by the healthy human microbiome. PLoS One 7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung WSF, Walker AW, Louis P, Parkhill J, Vermeiren J, Bosscher D, Duncan SH, Flint HJ. 2016. Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol 14:3. doi: 10.1186/s12915-015-0224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valdés-Varela L, Ruas-Madiedo P, Gueimonde M. 2017. In vitro fermentation of different fructo-oligosaccharides by Bifidobacterium strains for the selection of synbiotic combinations. Int J Food Microbiol 242:19–23. doi: 10.1016/j.ijfoodmicro.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Falony G, Lazidou K, Verschaeren A, Weckx S, Maes D, De Vuyst L. 2009. In vitro kinetic analysis of fermentation of prebiotic inulin-type fructans by Bifidobacterium species reveals four different phenotypes. Appl Environ Microbiol 75:454–461. doi: 10.1128/AEM.01488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaughlin HP, Motherway MO, Lakshminarayanan B, Stanton C, Ross RP, Brulc J, Menon R, O'Toole PW, van Sinderen D. 2015. Carbohydrate catabolic diversity of bifidobacteria and lactobacilli of human origin. Int J Food Microbiol 203:109–121. doi: 10.1016/j.ijfoodmicro.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Selak M, Riviere A, Moens F, Van den Abbeele P, Geirnaert A, Rogelj I, Leroy F, De Vuyst L. 2016. Inulin-type fructan fermentation by bifidobacteria depends on the strain rather than the species and region in the human intestine. Appl Microbiol Biotechnol 100:4097–4107. doi: 10.1007/s00253-016-7351-9. [DOI] [PubMed] [Google Scholar]
- 15.Belenguer A, Duncan S, Calder A, Holtrop G, Louis P, Lobley G, Flint H. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falony G, Vlachou A, Verbrugghe K, De Vuyst L. 2006. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol 72:7835–7841. doi: 10.1128/AEM.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moens F, Verce M, De Vuyst L. 2017. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int J Food Microbiol 241:225–236. doi: 10.1016/j.ijfoodmicro.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 18.Ze X, Le Mougen F, Duncan SH, Louis P, Flint HJ. 2013. Some are more equal than others: the role of “keystone” species in the degradation of recalcitrant substrates. Gut Microbes 4:236–240. doi: 10.4161/gmic.23998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia Rodenas CL, Lepage M, Ngom-Bru C, Fotiou A, Papagaroufalis K, Berger B. 2016. Effect of formula containing Lactobacillus reuteri DSM 17938 on fecal microbiota of infants born by cesarean-section. J Pediatr Gastroenterol Nutr 63:681–687. doi: 10.1097/MPG.0000000000001198. [DOI] [PubMed] [Google Scholar]
- 20.Makras L, Van Acker G, De Vuyst L. 2005. Lactobacillus paracasei subsp paracasei 8700:2 degrades inulin-type fructans exhibiting different degrees of polymerization. Appl Environ Microbiol 71:6531–6537. doi: 10.1128/AEM.71.11.6531-6537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goh YJ, Lee J, Hutkins RW. 2007. Functional analysis of the fructooligosaccharide utilization operon in Lactobacillus paracasei 1195. Appl Environ Microbiol 73:5716–5724. doi: 10.1128/AEM.00805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrova P, Velikova P, Popova L, Petrov K. 2015. Direct conversion of chicory flour into l(+)-lactic acid by the highly effective inulinase producer Lactobacillus paracasei DSM 23505. Bioresour Technol 186:329–333. doi: 10.1016/j.biortech.2015.03.077. [DOI] [PubMed] [Google Scholar]
- 24.Prior T, Kornberg H. 1988. Nucleotide sequence of fruA, the gene specifying enzyme IIfru of the phosphoenolpyruvate-dependent sugar phosphotransferase system in Escherichia coli K-12. J Gen Microbiol 134:2757–2768. [DOI] [PubMed] [Google Scholar]
- 25.Robillard G, Broos J. 1999. Structure/function studies on the bacterial carbohydrate transporters, enzymes II, of the phosphoenolpyruvate-dependent phosphotransferase system. Biochim Biophys Acta 1422:73–104. doi: 10.1016/S0304-4157(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 26.Saier M, Fagan M, Hoischen C, Reizer J. 1993. Transport mechanisms, p 133–156. In Sonenshein A, Hoch J, Losick R (ed), Bacillus subtilis and other Gram-positive bacteria: biochemistry, physiology and molecular genetics. ASM Press, Washington, DC. [Google Scholar]
- 27.Lengeler J, Jahreis K, Wehmeier U. 1994. Enzymes II of the phospho enol pyruvate-dependent phosphotransferase systems: their structure and function in carbohydrate transport. Biochim Biophys Acta 1188:1–28. doi: 10.1016/0005-2728(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 28.Saier MH., Jr 1998. Molecular phylogeny as a basis for the classification of transport proteins from bacteria, archaea and eukarya. Adv Microb Physiol 40:81–136. doi: 10.1016/S0065-2911(08)60130-7. [DOI] [PubMed] [Google Scholar]
- 29.Barrangou R, Azcarate-Peril M, Duong T, Conners S, Kelly R, Klaenhammer T. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc Natl Acad Sci U S A 103:3816–3821. doi: 10.1073/pnas.0511287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan H, Hutkins R. 2003. Metabolism of fructooligosaccharides by Lactobacillus paracasei 1195. Appl Environ Microbiol 69:2217–2222. doi: 10.1128/AEM.69.4.2217-2222.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parche S, Amon J, Jankovic I, Rezzonico E, Beleut M, Barutcu H, Schendel I, Eddy MP, Burkovski A, Arigoni F, Titgemeyer F. 2007. Sugar transport systems of Bifidobacterium longum NCC2705. J Mol Microbiol Biotechnol 12:9–19. doi: 10.1159/000096455. [DOI] [PubMed] [Google Scholar]
- 32.Watson D, Motherway MO, Schoterman MHC, van Neerven RJJ, Nauta A, van Sinderen D. 2013. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J Appl Microbiol 114:1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 33.Velikova P, Petrov K, Petrova P. 2017. The cell wall anchored beta-fructosidases of Lactobacillus paracasei: overproduction, purification, and gene expression control. Process Biochem 52:53–62. doi: 10.1016/j.procbio.2016.10.010. [DOI] [Google Scholar]
- 34.Saulnier DAA, Molenaar D, de Vos WA, Gibson GR, Kolida S. 2007. Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Appl Environ Microbiol 73:1753–1765. doi: 10.1128/AEM.01151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falony G, Calmeyn T, Leroy F, De Vuyst L. 2009. Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the prebiotic effect of inulin-type fructans. Appl Environ Microbiol 75:2312–2319. doi: 10.1128/AEM.02649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daniels L, Zeikus JG. 1975. Improved culture flask for obligate anaerobes. Appl Microbiol 29:710–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan SM, Fitzgerald GF, van Sinderen D. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl Environ Microbiol 72:5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng G, Futterer K. 2003. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat Struct Biol 10:935–941. doi: 10.1038/nsb974. [DOI] [PubMed] [Google Scholar]
- 39.Anwar MA, Kralj S, van der Maarel MJEC, Dijkhuizen L. 2008. The probiotic Lactobacillus johnsonii NCC 533 produces high-molecular-mass inulin from sucrose by using an inulosucrase enzyme. Appl Environ Microbiol 74:3426–3433. doi: 10.1128/AEM.00377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pijning T, Anwar MA, Boger M, Dobruchowska JM, Leemhuis H, Kralj S, Dijkhuizen L, Dijkstra BW. 2011. Crystal structure of inulosucrase from Lactobacillus: insights into the substrate specificity and product specificity of GH68 fructansucrases. J Mol Biol 412:80–93. doi: 10.1016/j.jmb.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 41.Li MZ, Elledge SJ. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.