The results of our study show that C. parvum responded to changes in the intestinal microenvironment induced by a nutritional supplement. This outcome paves the way for research to identify nutritional interventions aimed at limiting the impact of cryptosporidiosis.
KEYWORDS: Cryptosporidium, cryptosporidiosis, probiotics, fecal microbiota, gut microbiota
ABSTRACT
Cryptosporidiosis, a leading cause of diarrhea among infants, is caused by apicomplexan parasites classified in the genus Cryptosporidium. The lack of effective drugs is motivating research to develop alternative treatments. With this aim, the impact of probiotics on the course of cryptosporidiosis was investigated. The native intestinal microbiota of specific pathogen-free immunosuppressed mice was initially depleted with orally administered antibiotics. A commercially available probiotic product intended for human consumption was subsequently added to the drinking water. Mice were infected with Cryptosporidium parvum oocysts. On average, mice treated with the probiotic product developed more severe infections. The probiotics significantly altered the fecal microbiota, but no direct association between ingestion of probiotic bacteria and their abundance in fecal microbiota was observed. These results suggest that probiotics indirectly altered the intestinal microenvironment or the intestinal epithelium in a way that favored proliferation of C. parvum.
IMPORTANCE The results of our study show that C. parvum responded to changes in the intestinal microenvironment induced by a nutritional supplement. This outcome paves the way for research to identify nutritional interventions aimed at limiting the impact of cryptosporidiosis.
INTRODUCTION
In humans, cryptosporidiosis is an enteric infection caused mostly by 2 species of Cryptosporidium parasites, Cryptosporidium parvum and Cryptosporidium hominis. Transmission occurs by ingestion of infectious oocysts in contaminated food or water (1, 2), by fecal-oral contact, and possibly by inhalation (3). Recent surveys have revealed the high prevalence of cryptosporidiosis among infants living in developing nations, where it causes substantial morbidity and death among infants less than 2 years of age (4). The treatment of cryptosporidiosis is limited to supportive care, since no effective drugs are available. Because no vaccines are available either, hygiene and water sanitation to reduce transmission remain the most effective preventative approaches.
Using experimental mouse infections, we showed previously that cryptosporidiosis changes the gut microbiota (5). Given the importance of the microbiota for the physiology of the gut, here we investigated whether a reverse effect, of the microbiota on the parasite, could be demonstrated. We reasoned that the unmet need for anti-Cryptosporidium drugs could be alleviated by probiotics or dietary supplements. This hypothesis does not necessarily imply that the microbial community of the gut directly affects the parasite. Indeed, the transient nature of Cryptosporidium extracellular stages limits the interactions between the resident microbiota and extracellular cryptosporidial life stages, including sporozoites, merozoites, and gametes. It is conceivable that the microbiota affects parasite proliferation by modulating epithelial integrity, affecting the protective mucus layer, or stimulating innate and acquired immune cells.
The literature on the impact of the intestinal microbiota on cryptosporidiosis is sparse. A few studies have investigated the impact of Cryptosporidium parasites on the gut microbiota, but the effect of the gut environment on the course of the infection is not understood and underlying mechanisms are not known. Using germfree severe combined immunodeficiency (SCID) mice, compared to SCID mice colonized with intestinal microbes, Harp et al. showed that normal intestinal microbiota delayed the onset of C. parvum oocyst excretion by several weeks (6). Those authors also showed that resistance of mice to C. parvum could be increased by transferring intestinal mucosa from resistant animals to susceptible infant mice (7). A protective role of the gut microbiota against cryptosporidiosis was also observed in neonatal mice (8, 9). That research found that the gut microbiota synergized with poly(I·C) to elicit protective intestinal immunity against C. parvum. A study on the effect of IMP dehydrogenase inhibitors in C. parvum-infected mice detected an increase in C. parvum virulence in response to the drug; the effect was attributed to alteration of the intestinal microbiota (10).
Research on the effects of probiotics using animal models of other infectious diseases has generated divergent results. A mouse model of rotavirus infection was used to show that administration of Lactobacillus reuteri reduced the duration of diarrhea (11). Similarly, and consistent with findings observed in human trials, probiotics administered to mice had a mitigating effect on colitis induced by Citrobacter rodentium (12). The significant public health impact of nosocomial Clostridium difficile infection has generated a large body of research, including experiments in mice aimed at testing the benefits of fecal transplants (13) and defined probiotics (14–16). Only a few studies reported worse outcomes with probiotic treatment. Working with the cichlid fish tilapia, Liu et al. found that a 14-day treatment with probiotics made fishes more susceptible to infection with Aeromonas hydrophila after the treatment had been discontinued (17). More relevant to the present study, research with mice showed that supplementation of the diet with kefir exacerbated the outcomes of Clostridium difficile infection (18). Indicating that a harmful effect of probiotics is an unusual observation, no other studies in rodent or mammalian models demonstrating increased susceptibility to infection appear to have been published.
We previously reported a significant impact of cryptosporidiosis on the profile of the bacterial intestinal microbiota (5). Replicated experiments with 2 C. parvum isolates in 2 infected groups and 2 control groups of mice revealed that the intestinal microbiota of infected animals differed from that of uninfected animals, regardless of the C. parvum isolate. A taxonomic analysis of bacterial taxa highlighted 2 unclassified Bacteroidetes operational taxonomic units (OTUs), Prevotellaceae, and Porphyromonadaceae as overrepresented in the feces of infected mice, whereas the OTUs most overrepresented in uninfected mice were classified as Porphyromonadaceae and 1 unclassified Bacteroidetes OTU.
Probiotics are typically defined as microorganisms, mostly bacteria, that are consumed with the goal of improving health (19). Examples of gastrointestinal conditions that have been the focus of human clinical trials of probiotic treatment include inflammatory bowel diseases, antibiotic-associated diarrhea following C. difficile infection, peptic ulcer associated with Helicobacter pylori infection, and giardiasis (20). With a goal of developing alternative treatments for cryptosporidiosis, the experiments described here were aimed at assessing whether probiotics could influence the course of cryptosporidiosis. Against expectations, we found that mice consuming probiotics developed more severe infections. Although the goal of this research was to find treatments that mitigate cryptosporidiosis, the results are significant because they show that C. parvum proliferation responds to relatively minor changes in the intestinal microbiota. These observations open the way for targeted editing of the intestinal microbiota (21–23) as a low-cost approach to reducing the impact of these parasites.
RESULTS
Probiotic treatment increases severity of infection.
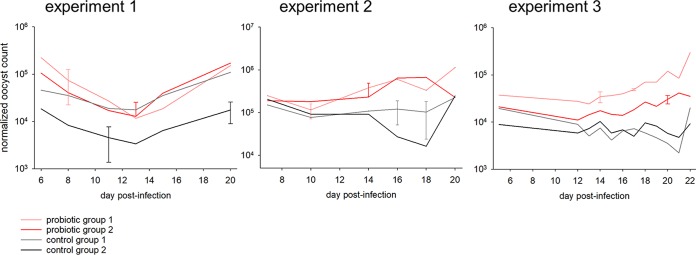
To test whether the probiotic product affects the severity of C. parvum infection, fecal oocyst output was measured by flow cytometry (FCM), as described in Materials and Methods. In experiment 1, a total of 92 oocyst concentration values were acquired from 16 mice at 6 time points over a 15-day period. In experiment 2, 79 data points were obtained from the same number of mice. In experiment 3, 12 time points for 4 groups generated 48 FCM values (Fig. 1 and Table 1). In experiment 1, mice that received probiotics excreted a significantly higher concentration of oocysts. A mean oocyst output of 76,463 oocysts/g feces (n = 44) was measured, compared with a mean of 26,732 oocysts/g for the control mice (n = 48). The difference between treatments was highly significant (Mann-Whitney U = 541, P < 0.001). An analogous significant probiotic effect was obtained for experiment 2 (mean of 378,736 oocysts/g [n = 37] for the probiotic-treated group versus mean of 68,778 oocysts/g [n = 42] for the control group; U = 269.5, P < 0.001). Similarly, the probiotic effect was significant in experiment 3 (Kruskal-Wallis one-way analysis of variance [ANOVA] on ranks, P = 0.001). Figure 1 shows the pattern of normalized oocyst outputs for the 3 experiments, plotted on a logarithmic scale.
FIG 1.
Effect of probiotics on the severity of C. parvum cryptosporidiosis in immunosuppressed mice. The graphs show oocyst counts (on a logarithmic scale), expressed as oocysts per gram of feces, for 3 independent experiments. In experiments 1 and 2, mice were sampled individually. Values represent the means for 4 mice. Error bars show SDs. In experiment 3, each group of 4 mice was sampled together. Error bars for experiment 3 show SDs based on 5 replicate FCM counts for selected samples.
TABLE 1.
Summary of experiments
| Experiment | Isolate | No. of groupsa | Dexamethasone treatment | Oocyst age (days) | Feces collection | Onset of oocyst shedding (days p.i.) |
|---|---|---|---|---|---|---|
| 1 | MD | 4 | In water, at 10 mg/liter and 16 mg/liter | 13 | By mouse | 4 |
| 2 | TU114 | 4 | In water from day −7 to day 1 p.i. subcutaneously from day 2 p.i. onward | 110 | By mouse | 6 |
| 3 | MD | 4 | In water, at 16 mg/liter | 62 | By group | 4 |
Two treated groups and 2 control groups.
Probiotic treatment significantly affects fecal microbiota.
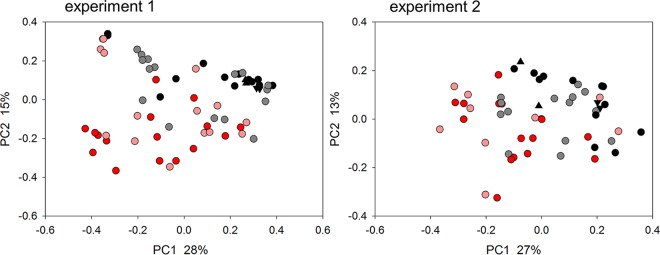
To assess whether probiotic treatment affected the fecal microbiota, pairwise weighted UniFrac distances (24) between sequence data for 64 fecal samples from experiment 1 were visualized on a principal-coordinate analysis (PCoA) plot (Fig. 2). Samples collected from day 5 of probiotic treatment (day 4 postinfection [p.i.]) to day 16 of treatment (day 15 p.i.) were included. The number of samples in this analysis was smaller than that shown in Fig. 1 because not all fecal samples were sequenced. Consistent with an impact of probiotic consumption on the profile of the intestinal microbiota, this analysis revealed a nonoverlapping distribution of data points according to treatment. Analysis of similarity (ANOSIM) (25) was used to check the significance of the treatment effect. The test returned a highly significant R value of 0.305 (P < 10−5). Analogous results were obtained for experiment 2, based on 55 samples collected from day 10 after initiation of probiotic treatment (day 9 p.i.) to day 20 (day 19 p.i.). As for experiment 1, clustering by treatment was significant (R = 0.210, P < 10−5). In experiment 3, microbiota clustering by treatment was also significant (R = 0.09 [n = 34], P = 0.002).
FIG 2.
Impact of probiotics on the fecal microbiome of C. parvum-infected mice. PCoA was used to display weighted UniFrac distances between pairs of fecal microbiome samples. Experiment 1 analysis (left) included data for 64 fecal samples collected from individual mice from day 5 of treatment (day 4 p.i.) to day 16 of treatment (day 15 p.i.). For experiment 2, 55 samples from individual mice were analyzed. Each data point represents 1 sample, color coded according to treatment and group as shown in Fig. 1. Matching triangle symbols indicate replicate analyses of the same fecal samples. Data from experiment 3 are not shown because groupwise sample collection resulted in a small number of data points. PC, principal coordinate.
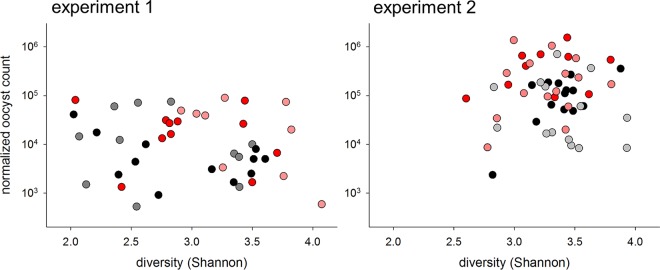
Bacterial α-diversity does not correlate with oocyst output.
Having detected effects of probiotic consumption on oocyst output and the fecal microbiota, the FCM and small-subunit 16S rRNA data were analyzed jointly to identify possible associations between the bacterial microbiota profile and the severity of cryptosporidiosis. These analyses included all samples for which 16S rRNA and FCM data were acquired. A total of 44 samples were included in experiment 1 and 53 in experiment 2. Because dysbiosis typically is characterized by low bacterial diversity and often is associated with increased susceptibility to enteric infections (26), the first global analysis examined the correlation between microbiota α-diversity (within-sample diversity) and total oocyst output (Fig. 3). Regardless of whether data were pooled by experiment or samples from probiotic-treated mice and control mice were analyzed separately, the correlation between α-diversity and oocyst output was very weak, explaining at most 9% of the oocyst count. These analyses indicate that the gut of mice excreting large numbers of oocysts generally is not populated with less diverse microbiota.
FIG 3.
Lack of relatedness of oocyst output to microbiome diversity. Oocyst output, normalized for fecal volume, was plotted against Shannon diversity for 44 samples, collected on 3 days, that were analyzed for both properties in experiment 1 and for 53 samples from experiment 2. Color coding indicates the experimental group, as described in Fig. 1. Samples were collected from individual mice. Due to the small number of data points, data for experiment 3 were not analyzed.
Redundancy analysis (RDA) was used to assess whether fecal oocyst concentrations significantly correlated with microbiota profiles and to identify bacterial OTUs correlating in relative abundance with the oocyst output. In experiment 1, a Monte Carlo test with 44 samples collected over the same time period as shown in Fig. 1 and with 1,000 permutations indicated a significant correlation between the oocyst concentration and the OTU profile (pseudo-F = 1.0, P = 0.014). This effect remained significant (F = 2.7, P = 0.024) after accounting for treatment, i.e., defining treatment (with or without probiotic) as a covariate, or accounting for “mouse,” i.e., defining mouse as a covariate (pseudo-F = 3.8, P = 6 × 10−5). The analogous test in experiment 2 also returned a significant pseudo-F ratio of 4.2 (n = 44; P = 0.0016). When the effect of treatment or mouse was removed by defining these variables as covariates, the association remained significant (pseudo-F values of 2.6 [P = 0.015] and 5.2 [P = 3 × 10−4], respectively). These and subsequent analyses were not performed for experiment 3 because samples from mice in the same group were pooled.
Reasoning that the microbiota at the time of infection could be important for subsequent parasite proliferation, we analyzed whether the microbiota in treated and control mice differed on the day of infection, i.e., 1 and 2 days after initiation of microbiota administration in experiments 1 and 2, respectively. In both experiments, a significant correlation between probiotic treatment and the composition of the bacterial fecal microbiota was observed (pseudo-F values of 5.3 [P = 0.028] and 3.5 [P = 0.055] in experiments 1 and 2, respectively). The permutation test is explained in Materials and Methods.
Abundance of facultative anaerobes in severe infections.
With a significant correlation between the fecal oocyst concentration and the OTU profile having been identified, the taxonomic composition of the fecal microbiota was examined in more detail. First, we used the program LEfSe (27) to identify OTUs that significantly defined the difference between samples with high versus low oocyst concentrations. This analysis was based on 20 samples for each experiment, namely, 10 samples with the highest oocyst concentrations and 10 samples with the lowest concentrations collected from the entire experiment. In experiment 1, 7/10 samples in the high-oocyst group originated from mice treated with probiotics and 7/10 samples in the low-oocyst group came from control mice (chi-square = 3.2, P = 0.07). In experiment 2, 9/10 samples in the high-oocyst group originated from treated mice and 8/10 samples in the low-oocyst group originated from control mice (chi-square = 9.9, P = 0.002). Feces from highly infected animals were characterized by a high Proteobacteria abundance, whereas feces from lightly infected animals were significantly enriched for Firmicutes. As observed for the effects of the probiotic on the severity of cryptosporidiosis (Fig. 1) and on the global microbiota profile (Fig. 2), a similar shift toward higher Proteobacteria abundance was observed in the 2 experiments (Fig. 4).
FIG 4.
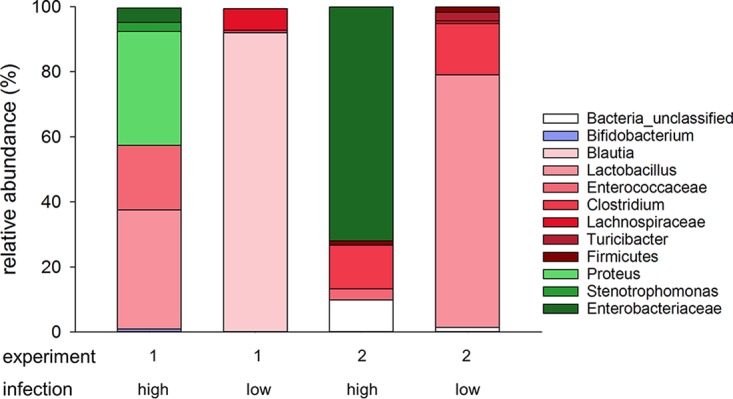
Taxonomy of the fecal microbiome in heavily infected and lightly infected mice, showing that heavy infections were associated with increased abundance of Proteobacteria. For each experiment, bacterial taxa significantly associated with the severity of infection were identified using LEfSe (24), in a comparison of 10 samples with the highest (high) and lowest (low) oocyst concentrations; 20 samples were included for each experiment. The color indicates the phylum and the color intensity the genus or highest taxonomic level (e.g., family or order) identified. Green, Proteobacteria; red, Firmicutes; blue, Actinobacteria.
A second taxonomic analysis aimed at identifying OTUs enriched in highly infected animals was performed with RDA (28). For experiment 1, in the 10 OTUs that best correlated in relative abundance with the oocyst concentration, 61% of the sequences were classified as Lactobacillus, 24% as Proteus, and 14% as Enterococcus. In the 10 OTUs that most negatively correlated with the oocyst concentration, as seen with LEfSe, Blautia was the most abundant classification (61% of reads), followed by Clostridiaceae (27%), Lachnospiraceae (10%), Romboutsia (8%), and unclassified Firmicutes. For experiment 2, the same analysis of the 10 OTUs that best correlated in relative abundance with the oocyst concentration indicated that 92% of sequences originated from Enterobacteriaceae and 8% from Firmicutes. In the 10 OTUs with the lowest oocyst concentrations, 70% of sequences were classified as Lactobacillus, 21% as Turicibacter, 6% as unclassified Lactobacillales, and 1% as Stenotrophomonas (phylum Proteobacteria) (see Table S1 in the supplemental material). The results obtained with RDA are thus is close agreement with the LEfSe results shown in Fig. 4.
Severity of infection correlates with fecal microbiota profiles.
Figure 3 illustrates an important difference between the experiments; the mean fecal oocyst concentration, across all time points, treatments, and mice, in experiment 2 was 2.4 × 105 oocysts/g (standard deviation [SD], 3.4 × 105 oocysts/g [n = 79]), or about 5 times higher than that in experiment 1 (mean, 5.0 × 104 oocysts/g; SD, 8.7 × 104 oocysts/g [n = 92]) and >8 times higher than that in experiment 3 (mean, 2.8 × 104 oocysts/g; SD, 4.6 × 104 oocysts/g [n = 48]). Oocyst outputs were significantly different between experiments (Kruskal-Wallis ANOVA on ranks, H = 34.3 [2 degrees of freedom], P < 0.001). Pairwise comparisons between experiment 1 and experiment 2 and between experiment 3 and experiment 2 were significant, according to Dunn's multiple-comparison test, whereas that between experiment 1 and experiment 3 was not. It is likely that these differences were caused by the dexamethasone administration route (drinking water only in experiments 1 and 3 versus drinking water followed by subcutaneous injection in experiment 2) (Table 1). The more severe infection in experiment 2 represented an unplanned opportunity to further assess the impact of the infection on the gut environment. If heavier infections cause an increase in the relative abundance of Proteobacteria, as suggested by the LEfSe analysis (Fig. 4) and by the RDA described above, then a greater proportion of Proteobacteria in samples originating from severely infected mice in experiment 2 would be expected, which is exactly what was observed (Fig. 4 and RDA results). Furthermore, severe cryptosporidiosis can be expected, based on research involving human patients suffering from other enteric infections (29–31), to lead to changes in bacterial microbiota toward populations enriched for facultative anaerobes, possibly resulting in loss of diversity. To test this hypothesis, mean pairwise weighted UniFrac distances for microbiota from heavily infected samples were compared to distances for lightly infected samples. In experiment 1, the mean β-diversity (between-sample diversity) among the 10 high-oocyst samples was 0.536 (SD, 0.142 [45 pairwise distances]), whereas the mean β-diversity among the low-output samples was 0.590 (SD, 0.121 [45 pairwise distances]; Mann-Whitney T = 1819.0, P = 0.07). In experiment 2, the mean β-diversity values were 0.312 (SD, 0.117 [45 pairwise distances]) and 0.411 (SD, 0.137 [45 pairwise distances]) for the 10 samples with the highest and lowest oocyst concentrations, respectively (Mann-Whitney U = 590, P < 0.001). Although the effect was not significant for experiment 1, together these results are consistent with the model postulated above, i.e., that severe infection leads to a convergence of the microbiota toward a population rich in Proteobacteria and low in Firmicutes.
Loss of microbiota functional diversity in heavily infected mice.
To extend the observed taxonomic differences between severely and lightly infected mice to the metagenome, the program PICRUSt (32) was used to infer microbiota function from OTU profiles. Given the more severe infection in experiment 2, metagenomic analyses are reported only for this experiment. PICRUSt identified 39 KEGG level 2 categories in the combined metagenome. A PCoA based on pairwise distances between KEGG abundance values normalized across KEGG categories revealed a tight clustering of samples with high oocyst concentrations, relative to the samples with lower oocyst concentrations (Fig. S1). This visual assessment was tested by comparing pairwise distances between KEGG profiles. The distances for the 10 samples with the lowest oocyst concentrations averaged 69.9 (SD, 47.6) and those for the same number of samples with the highest concentrations averaged 54.8 (SD, 41.41), which was not statistically significant (Mann-Whitney rank sum test, P = 0.121). If only 8 samples with the highest or lowest oocyst concentrations were tested (28 pairwise distance values for each group), then the distances between the high-concentration samples were significantly smaller (Mann-Whitney rank sum test, P = 0.015). As for the taxonomic analysis described above, we conclude from these results that proliferation of C. parvum leads to a convergence of the inferred bacterial metagenome.
As described above for the taxonomic analysis, LEfSe was used to identify KEGG pathways that differed significantly in abundance between the 10 samples from highly infected mice and the same number of samples from lightly infected mice. Underscoring the difference at the metagenomic level between fecal samples from severely and mildly infected mice, 22 of 39 KEGG level 2 pathways were significantly different between the 2 groups (Table S2). In comparison to the severely infected mice, the microbiota of mice with mild infections was characterized by a high abundance of pathways related to replication, such as carbohydrate, amino acid, and nucleic acid metabolism. These results extend the taxonomy presented in Fig. 4, suggesting that the mouse dysbiotic cryptosporidiosis metagenome is selected for functions other than bacterial replication.
DISCUSSION
In a comprehensive review of the literature, Kristensen et al. (33) found a small number of publications describing randomized controlled trials of probiotics that included the characterization of fecal microbiota. The surprising conclusion of that survey was that no publications reported a significant change in the microbiota based on OTU richness, evenness, or diversity analysis. As no uninfected controls were included in our experiments, the impact of the probiotics on the microbiota could be observed only in the initial phase of the experiment, as described above for days 1 and 2 after initiation of the probiotic treatment. The increase in facultative aerobes later in the infection likely represents the effect of C. parvum multiplication in the intestinal epithelium, rather than a direct impact of probiotics.
Because we did not observe a significant increase in probiotic bacteria in the feces of treated mice, we postulate that some of the bacterial or prebiotic ingredients present in the probiotic product induced changes in the mouse intestinal environment, favoring the proliferation of C. parvum. Proliferation of the parasite then led to extensive secondary modifications of the microbiota, as shown in Fig. 4. An impact of the prebiotics present in the product, i.e., acacia gum, larch gum, oligosaccharides, and l-glutamine, on the microbiota cannot be excluded. Elucidation of the mechanism by which probiotic administration promotes proliferation of C. parvum will require testing of individual probiotic species or defined combinations of species and/or prebiotics (34) and metabolomic analysis to identify mediators of the probiotic effect. This research is of primary importance to enable targeted manipulations of the microbiota aimed at limiting the proliferation of Cryptosporidium parasites. Zhu et al. (21) described methods to “edit” the gut microbiota, in that case by inhibiting the multiplication of facultative anaerobes. An analogous approach could be used to investigate the causal link between parasite proliferation and dysbiosis. Although mice infected with C. parvum do not develop diarrhea, the fecal microbiota from heavily infected animals in our experiments resembles the fecal microbiota of humans suffering from cholera diarrhea (29–31) or diarrhea of other etiologies (31). This observation is significant, because it indicates that neither the actual pathogen nor diarrhea is important for inducing dysbiosis. A characteristic of many intestinal pathologies of infectious or other causes is an increase in the proportion of Gammaproteobacteria (35). Although exceptions to this trend have been reported (36), a shift toward facultative anaerobes, reflecting increased permeability of the gut epithelium, is a hallmark of infectious (29, 30, 37, 38), inflammatory (39–41), and other (42) intestinal pathologies. The abundance of Gammaproteobacteria in the distal gut of mice heavily infected with C. parvum indicates a shift in the luminal oxygen gradient (43), likely a consequence of epithelial erosion and villus atrophy (44–46). These observations raise the question of whether Cryptosporidium proliferation responds to the oxygen concentration in the gut lumen. Selective inhibition or promotion of oxygen-consuming bacteria (21), to increase or to deplete luminal O2 temporarily (22), could potentially be investigated to assess the response of C. parvum and to explore dietary interventions to mitigate the severity and duration of cryptosporidiosis.
Few studies have reported on the effects of diet on C. parvum infection. Liu et al. (47) found that protein deficiency increased the concentration of C. parvum DNA in feces; in that study, however, the difference between normal and protein-deficient animals was reported at 20 h p.i. Since C. parvum is not known to complete its life cycle in less than 72 h (48), the results are difficult to interpret. Another study found a positive effect of pomegranate extract on cryptosporidiosis in calves (49). The authors reported that calves fed milk supplemented with extract excreted fewer oocysts. The very limited range of the literature on the effects of diet on cryptosporidiosis illustrates the need for additional research, particularly basic research on mechanistic aspects of parasite-microbiota interactions.
Although the results across experiments were consistent, differences were also noticed. Most notably, the average oocyst output in experiment 2 was higher, indicating more severe infection. Corroborating the model discussed above, more severe infection was associated with greater relative abundance of Gammaproteobacteria (Fig. 4). The reason for the difference in the severity of infection is difficult to determine, but the different route of dexamethasone administration in experiment 2 might have contributed to this outcome. Differences in fecal oocyst outputs between cohoused mice were also observed in experiments 1 and 2 (Fig. 1). As described in Materials and Methods, in those experiments, animals from the same cage were housed individually for 16 h three times a week for collection of feces but otherwise were housed together in 2 cages per treatment (4 cages for each experiment). The differences in oocyst outputs and microbiota profiles between cage mates are difficult to explain, given the close contact between animals. This phenomenon justifies mice being sampled individually, rather than being sampled by group, as is common practice, and is the reason why experiment 3 samples were not subjected to a full set of analyses as for samples from experiments 1 and 2.
In the absence of effective drugs to control cryptosporidiosis, a search for alternative treatments is warranted. In most cases, however, we do not know how perturbation of the microbiota, whether induced by diet, probiotics, antibiotics, or prebiotics, affects enteric pathogens, particularly Cryptosporidium parasites. Identifying specific mechanisms affecting pathogen virulence in response to diet may enable the development of targeted microbiota-editing measures to mitigate the severity of cryptosporidiosis. Methods designed to detect changes in the metabolome (50) will be needed to supplement taxonomic analyses based on 16S amplicon sequencing. Lastly, enhancing the value of the rodent cryptosporidiosis model, the observed shift toward facultative anaerobes in the infected gut indicates common pathogenic changes in the human and rodent intestines in response to enteric infections.
MATERIALS AND METHODS
Parasites.
Oocysts from C. parvum isolate MD (51) were used in experiments 1 and 3, whereas mice in experiment 2 were infected with C. parvum isolate TU114 (52). MD is a zoonotic isolate, and TU114 belongs to the anthroponotic subgroup characterized by an IIc GP60 genotype (53). Oocysts were purified from mouse feces on gradients of Nycodenz (Alere Technologies, Oslo, Norway), as described previously (54). The ages of the oocysts were 13, 110, and 62 days for experiments 1, 2, and 3, respectively.
Mouse experiments.
To test the effect of a commercially available probiotic, 3 experiments were conducted in mice. In each experiment, 16 female CD-1 mice approximately 6 weeks of age were used. Upon delivery, each mouse was individually tagged and randomly assigned to 1 of 4 groups of 4 mice. Mice were immunosuppressed by adding dexamethasone 21-phosphate disodium (catalog no. D1169; Sigma) to the drinking water, at a concentration of 16 mg/liter (55), starting on the day of arrival, defined as day −7 p.i. (i.e., 7 days prior to infection). To deplete the native intestinal microbiota, vancomycin and streptomycin were added to the drinking water at concentrations of 500 mg/liter and 5 g/liter, respectively, starting on day −6 p.i. Metronidazole at a dose of 20 mg/kg was given daily by gavage, starting on day −6 p.i. The antibiotic treatment was discontinued on day −2 p.i. Starting on day −1 p.i., the drinking water was supplemented with 1.3 g of probiotic added to 500 ml of water. The product contained 15 bacterial strains, including strains belonging to the genera Bifidobacterium (4 species) and Lactobacillus (9 species), as well as Streptococcus thermophilus. In addition to bacteria, 1 g of product contained 8 mg acacia gum, 540 mg larch gum, 115 mg galacto-oligosaccharide, 212 mg l-glutamine, and 150 IU vitamin D3; when the product was dissolved in 500 ml of water, the final concentrations of these ingredients were 20 μg/ml, 1.4 mg/ml, 0.3 mg/ml, 0.55 mg/ml, and 0.4 IU/ml, respectively. The product was flavorless. Drinking water with dexamethasone and probiotic was replaced every 3 days. Mice were infected orally with approximately 5 × 104 C. parvum oocysts on day 0 p.i. In experiment 1, to compensate for increased water uptake in the 2 groups receiving probiotic, the concentration of dexamethasone phosphate in the water of the 2 treated groups was reduced to 10 mg/liter starting on day 10 p.i. In experiment 2, to avoid possible differences in dexamethasone uptake with drinking water, dexamethasone (catalog no. D1756; Sigma) was given only subcutaneously every second day starting on day 2 p.i., following 9 days of dexamethasone administration in the water (Table 1). A volume of 100 μl of a 10-mg/ml dexamethasone suspension was injected alternately into the right and left sides of the abdomen. In experiment 3, 16 mg/liter dexamethasone was added to the drinking water of all 4 groups of mice. To obtain fecal pellets for analysis of the microbiota, mice were individually transferred to a 1-liter plastic beaker and pellets were collected upon defecation. In experiment 3, feces were collected by group, not by mouse. Pellets were stored at −20°C. To collect feces for oocyst enumeration, mice were individually transferred overnight to collection cages fitted with a wire bottom. Feces collected from these cages were stored at 4°C. In the morning, the mice were returned to their respective group cages, such that they were housed individually for 14 to 16 h on the days when feces were collected for oocyst enumeration. While in conventional cages, the mice were always housed with the same cage mates. Animal experiments adhered to the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Tufts University Animal Care and Use Committee.
Enumeration of oocysts.
Feces collected overnight were weighed, diluted 1:5 in distilled water, and homogenized with a vortex mixer. To remove debris, fecal homogenates were filtered through 100-μm cell strainers (catalog no. 431752; Corning) by centrifugation at 1,300 × g. The filtrates were homogenized, and volumes of 1 ml were centrifuged at 6,700 × g for 5 min. The supernatant was discarded, and the pellet was suspended in 500 μl of phosphate-buffered saline (PBS) supplemented with 10% fetal bovine serum (FBS). A volume of 20 μl of this suspension was transferred to a 1.5-ml microcentrifuge tube with 20 μl of a 1:5 dilution of monoclonal antibody 5F10 cell culture supernatant. This antibody reacts specifically with the Cryptosporidium oocyst wall without binding to other parasite antigens (A. S. Sheoran, unpublished observation). Samples were incubated for 30 min at room temperature. Following incubation with the primary antibody, the samples were centrifuged for 10 min at 6,700 × g, and the pellet was washed once in 500 μl of PBS with 10% FBS and incubated for 30 min at room temperature with 20 μl of secondary antibody (Alexa Fluor 488-conjugated goat anti-mouse IgG). After incubation, 500 μl of PBS was added and the samples was washed once in PBS. For each experiment, 5 samples were randomly selected for replication. Replication involved processing and labeling 5 separate aliquots originating from each strained and washed sample. Labeled samples were analyzed by FCM using a Becton Dickinson Accuri C6 cytometer. Distance matrices were calculated with GenAlEx 6.5 (56), based on the pairwise difference between oocyst concentrations. Specifically, the distance between sample A containing an oocyst concentration of xA per gram of feces and sample B containing a concentration of xB per gram of feces was calculated as (xA − xB)2. The accuracy of the FCM oocyst enumeration method was evaluated by correlating the counts against the Cryptosporidium 18S rRNA gene copy number, as estimated using real-time PCR with published primers (57). This analysis based on 5 randomly chosen samples from experiment 1 generated a correlation coefficient of 0.69.
Microbiota analysis.
DNA was extracted from 10 mg of feces collected individually from each mouse. DNA was extracted in a QIACube instrument using the QIAamp PowerFecal DNA kit (catalog no. 12830-50; Qiagen), according to the manufacturer's protocol. DNA was eluted in 50 μl of elution buffer and stored at −20°C. A previously described PCR protocol to prepare 16S V1V2 amplicons libraries for high-throughput sequencing was used (5). The only deviation from this procedure was the downstream primer; instead of the canonical primer 338R, we used primer Bac R V2 short (GTTCAGACGTGTGCTCTTCCGATCtgctgcctcccgtaggagt, with the lowercase letters being equivalent to the conserved 338R sequence) (58). One microliter of primary PCR mixture was subjected to a secondary PCR to incorporate a 6-nucleotide barcode into each sample. The secondary amplification was as described previously (5), except that the downstream primer CAAGCAGAAGACGGCATACGAGATnnnnnnGTGACTGGAGTTCAGACGTGTGCTCTTCC was used; the italicized nucleotides represent the Illumina adaptor and the lowercase letters the barcode unique to each sample. To assess the quality of the PCRs, a portion of the amplification product was electrophoresed on a 1.5% agarose gel and visualized using GelRed (Biotium). The concentration of each final amplicon was measured in a Qbit spectrophotometer, and up to 80 amplicons were pooled at approximately equal concentrations. The pooled library was size selected with a Pippin HT library size selection system (Sage BioScience). Libraries were sequenced in an Illumina MiSeq sequencer at Tufts University Genomics Core Facility, using a single-end 300-nucleotide strategy. To control for technical variations introduced during library preparation and sequencing, each library included 2 replicates of 2 randomly chosen samples. Replication involved the process of duplicate fecal samples being processed, amplified, and barcoded individually.
Bioinformatic analysis.
FASTQ-formatted sequences were processed using programs found in mothur (59), essentially as described previously (5). Briefly, random subsamples of 5,000 sequences per sample were processed. This procedure is not expected to bias the analysis (60), since the numbers of sequences per barcode were relatively constant. The mean number of sequences per sample in experiment 1 was 1.03 × 105 sequences/sample (SD, 2.44 × 104 sequences/sample) and the average number of reads in experiment 2 was 1.01 × 105 reads/sample (SD, 3.22 × 104 reads/sample), representing coefficients of variation of 0.23 and 0.31, respectively. V1V2 sequences were trimmed to 200 nucleotides to eliminate 3′ sequences with mean Phred quality scores of <30. Sequences were aligned and sequences with the following properties were removed: sequences that did not align, sequences with ambiguous base calls, and sequences with homopolymers of >8 nucleotides. To further reduce the number of putative sequence errors, the program pre.cluster was used to merge unique sequences differing by 1 nucleotide position with the majority sequence (61). This sequence curation protocol removed 15,748 sequences from 400,000 sequences (3.9%) in experiment 1 and 21,546 sequences from 355,000 sequences (6.0%) in experiment 2. Pairwise UniFrac phylogenetic distances (24) between samples were calculated in mothur. ANOSIM (25) was used to test the significance of clustering by treatment. The program anosim was run in mothur using a weighted UniFrac distance matrix as input. OTUs were obtained using the program cluster, also found in mothur, using the OptiClust clustering method (62) and a distance cutoff value of 3%.
Linear discriminant analysis, as implemented in the program LEfSe (27), was used to identify statistically significant differences in OTU abundance profiles between 2 groups of samples, defined, independent of treatment, as heavily infected and lightly infected. Heavy and light infections were defined on the basis of fecal oocyst concentrations determined by FCM as described above. In the experiment-wide LefSe analyses, the 10 samples with the highest oocyst concentrations and the 10 samples with the lowest oocyst concentrations were selected. When subsamples of probiotic-treated and control mice were analyzed separately, the 5 samples with the highest oocyst concentrations and the same number of samples with the lowest oocyst concentrations were tested using LEfSe (27).
RDA was used to test the significance of associations between OTU profiles and oocyst concentrations or between KEGG function profiles and oocyst concentrations. The program was run in CANOCO (28). The pseudo-F statistic was calculated by Monte Carlo permutation of samples between treatment groups. OTU abundance values for the 100 most abundant OTUs or KEGG functions (n = 39) inferred with the metagenome prediction tool PICRUSt (32) served as dependent variables. Oocyst concentrations determined by FCM, as described above, served as the independent variable. Pairwise distances between KEGG profiles were calculated as described above for oocyst concentrations, except that the squared differences were summed over all KEGG categories; this metric is equivalent to the square of the Euclidean distance.
Accession number(s).
Raw sequence data were deposited in the ENA Sequence Read Archive under study accession numbers PRJEB25162 and PRJEB25164.
Supplementary Material
ACKNOWLEDGMENTS
Lucas Vinicius Shigaki de Matos, Olga Douvropoulou, and Kevin Huynh performed preliminary experiments that led to the design of the research presented here.
Support from the National Institute of Allergy and Infectious Diseases (grant 5R21AI125891) is gratefully acknowledged.
B.C.M.O. and G.W. designed the experiments and analyzed the data, B.C.M.O. performed the experiments, and G.W. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01408-18.
REFERENCES
- 1.Rochelle PA, Di Giovanni GD. 2014. Cryptosporidium oocysts in drinking water and recreational water, p 489–513. In Caccìo SM, Widmer G (ed), Cryptosporidium: parasite and disease. Springer, New York, NY. [Google Scholar]
- 2.Cacciò SM, Putignani L. 2014. Epidemiology of human cryptosporidiosis, p 43–79. In Caccìo SM, Widmer G (ed), Cryptosporidium: parasite and disease. Springer, New York, NY. [Google Scholar]
- 3.Mor SM, Tumwine JK, Ndeezi G, Srinivasan MG, Kaddu-Mulindwa DH, Tzipori S, Griffiths JK. 2010. Respiratory cryptosporidiosis in HIV-seronegative children in Uganda: potential for respiratory transmission. Clin Infect Dis 50:1366–1372. doi: 10.1086/652140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 5.Ras R, Huynh K, Desoky E, Badawy A, Widmer G. 2015. Perturbation of the intestinal microbiota of mice infected with Cryptosporidium parvum. Int J Parasitol 45:567–573. doi: 10.1016/j.ijpara.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Harp JA, Chen W, Harmsen AG. 1992. Resistance of severe combined immunodeficient mice to infection with Cryptosporidium parvum: the importance of intestinal microflora. Infect Immun 60:3509–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harp JA. 2003. Cryptosporidium and host resistance: historical perspective and some novel approaches. Anim Health Res Rev 4:53–62. doi: 10.1079/AHRR200352. [DOI] [PubMed] [Google Scholar]
- 8.Lacroix-Lamande S, Guesdon W, Drouet F, Potiron L, Lantier L, Laurent F. 2014. The gut flora is required for the control of intestinal infection by poly(I:C) administration in neonates. Gut Microbes 5:533–540. doi: 10.4161/gmic.29154. [DOI] [PubMed] [Google Scholar]
- 9.Lantier L, Drouet F, Guesdon W, Mancassola R, Metton C, Lo-Man R, Werts C, Laurent F, Lacroix-Lamande S. 2014. Poly(I:C)-induced protection of neonatal mice against intestinal Cryptosporidium parvum infection requires an additional TLR5 signal provided by the gut flora. J Infect Dis 209:457–467. doi: 10.1093/infdis/jit432. [DOI] [PubMed] [Google Scholar]
- 10.Gorla SK, McNair NN, Yang G, Gao S, Hu M, Jala VR, Haribabu B, Striepen B, Cuny GD, Mead JR, Hedstrom L. 2014. Validation of IMP dehydrogenase inhibitors in a mouse model of cryptosporidiosis. Antimicrob Agents Chemother 58:1603–1614. doi: 10.1128/AAC.02075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preidis GA, Saulnier DM, Blutt SE, Mistretta TA, Riehle KP, Major AM, Venable SF, Finegold MJ, Petrosino JF, Conner ME, Versalovic J. 2012. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. FASEB J 26:1960–1969. doi: 10.1096/fj.10-177980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues DM, Sousa AJ, Johnson-Henry KC, Sherman PM, Gareau MG. 2012. Probiotics are effective for the prevention and treatment of Citrobacter rodentium-induced colitis in mice. J Infect Dis 206:99–109. doi: 10.1093/infdis/jis177. [DOI] [PubMed] [Google Scholar]
- 13.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondepudi KK, Ambalam P, Karagin PH, Nilsson I, Wadstrom T, Ljungh A. 2014. A novel multi-strain probiotic and synbiotic supplement for prevention of Clostridium difficile infection in a murine model. Microbiol Immunol 58:552–558. doi: 10.1111/1348-0421.12184. [DOI] [PubMed] [Google Scholar]
- 15.Colenutt C, Cutting SM. 2014. Use of Bacillus subtilis PXN21 spores for suppression of Clostridium difficile infection symptoms in a murine model. FEMS Microbiol Lett 358:154–161. doi: 10.1111/1574-6968.12468. [DOI] [PubMed] [Google Scholar]
- 16.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Liu W, Ran C, Hu J, Zhou Z. 2016. Abrupt suspension of probiotics administration may increase host pathogen susceptibility by inducing gut dysbiosis. Sci Rep 6:23214. doi: 10.1038/srep23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spinler JK, Brown A, Ross CL, Boonma P, Conner ME, Savidge TC. 2016. Administration of probiotic kefir to mice with Clostridium difficile infection exacerbates disease. Anaerobe 40:54–57. doi: 10.1016/j.anaerobe.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlundt J. 2006. Report of a joint FAO/WHO expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Food and Agriculture Organization of the United Nations, World Health Organization, Rome, Italy: http://www.fao.org/3/a-a0512e.pdf. [Google Scholar]
- 20.Koretz RL. 2018. Probiotics in gastroenterology: how pro is the evidence in adults? Am J Gastroenterol 113:1125–1136. doi: 10.1038/s41395-018-0138-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Buttner L, de Lima Romao E, Behrendt CL, Lopez CA, Sifuentes-Dominguez L, Huff-Hardy K, Wilson RP, Gillis CC, Tukel C, Koh AY, Burstein E, Hooper LV, Baumler AJ, Winter SE. 2018. Precision editing of the gut microbiota ameliorates colitis. Nature 553:208–211. doi: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao MS, Huang S, Chang WL, Hsieh MF, Huang CJ, Gallo RL, Huang CM. 2017. Microbiome precision editing: using PEG as a selective fermentation initiator against methicillin-resistant Staphylococcus aureus. Biotechnol J 12. doi: 10.1002/biot.201600399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theriot CM. 2018. Beyond structure: defining the function of the gut using omic approaches for rational design of personalized therapeutics. mSystems 3:e00173-. doi: 10.1128/mSystems.00173-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone C, Hamady M, Knight R. 2006. UniFrac: an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371–385. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 26.Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. 2014. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 27.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ter Braak CJF, Smilauer P. 2002. CANOCO reference manual and CanoDraw for Windows user's guide: software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca, NY. [Google Scholar]
- 29.David LA, Weil A, Ryan ET, Calderwood SB, Harris JB, Chowdhury F, Begum Y, Qadri F, LaRocque RC, Turnbaugh PJ. 2015. Gut microbial succession follows acute secretory diarrhea in humans. mBio 6:e00381-. doi: 10.1128/mBio.00381-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monira S, Nakamura S, Gotoh K, Izutsu K, Watanabe H, Alam NH, Nakaya T, Horii T, Ali SI, Iida T, Alam M. 2013. Metagenomic profile of gut microbiota in children during cholera and recovery. Gut Pathog 5:1. doi: 10.1186/1757-4749-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain MA, Oundo J, Tamboura B, Mai V, Astrovskaya I, Corrada Bravo H, Rance R, Stares M, Levine MM, Panchalingam S, Kotloff K, Ikumapayi UN, Ebruke C, Adeyemi M, Ahmed D, Ahmed F, Alam MT, Amin R, Siddiqui S, Ochieng JB, Ouma E, Juma J, Mailu E, Omore R, Morris JG, Breiman RF, Saha D, Parkhill J, Nataro JP, Stine OC. 2014. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol 15:R76. doi: 10.1186/gb-2014-15-6-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristensen NB, Bryrup T, Allin KH, Nielsen T, Hansen TH, Pedersen O. 2016. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med 8:52. doi: 10.1186/s13073-016-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. 2014. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med 6:220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter SE, Lopez CA, Baumler AJ. 2013. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker-Dreps S, Allali I, Monteagudo A, Vilchez S, Hudgens MG, Rogawski ET, Carroll IM, Zambrana LE, Espinoza F, Azcarate-Peril MA. 2015. Gut microbiome composition in young Nicaraguan children during diarrhea episodes and recovery. Am J Trop Med Hyg 93:1187–1193. doi: 10.4269/ajtmh.15-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Petri WA Jr, Haque R, Ahmed T, Gordon JI. 2014. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barash NR, Maloney JG, Singer SM, Dawson SC. 2017. Giardia alters commensal microbial diversity throughout the murine gut. Infect Immun 85:e00948-. doi: 10.1128/IAI.00948-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. 2012. IBD: what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol 9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- 40.Munyaka PM, Rabbi MF, Khafipour E, Ghia JE. 2016. Acute dextran sulfate sodium (DSS)-induced colitis promotes gut microbial dysbiosis in mice. J Basic Microbiol 56:986–998. doi: 10.1002/jobm.201500726. [DOI] [PubMed] [Google Scholar]
- 41.Gillis CC, Hughes ER, Spiga L, Winter MG, Zhu W, Furtado de Carvalho T, Chanin RB, Behrendt CL, Hooper LV, Santos RL, Winter SE. 2018. Dysbiosis-associated change in host metabolism generates lactate to support Salmonella growth. Cell Host Microbe 23:54–64.e6. doi: 10.1016/j.chom.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manor O, Levy R, Pope CE, Hayden HS, Brittnacher MJ, Carr R, Radey MC, Hager KR, Heltshe SL, Ramsey BW, Miller SI, Hoffman LR, Borenstein E. 2016. Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Sci Rep 6:22493. doi: 10.1038/srep22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Espey MG. 2013. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med 55:130–140. doi: 10.1016/j.freeradbiomed.2012.10.554. [DOI] [PubMed] [Google Scholar]
- 44.Tzipori S, Rand W, Griffiths J, Widmer G, Crabb J. 1994. Evaluation of an animal model system for cryptosporidiosis: therapeutic efficacy of paromomycin and hyperimmune bovine colostrum-immunoglobulin. Clin Diagn Lab Immunol 1:450–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Argenzio RA, Liacos JA, Levy ML, Meuten DJ, Lecce JG, Powell DW. 1990. Villous atrophy, crypt hyperplasia, cellular infiltration, and impaired glucose-Na absorption in enteric cryptosporidiosis of pigs. Gastroenterology 98:1129–1140. doi: 10.1016/0016-5085(90)90325-U. [DOI] [PubMed] [Google Scholar]
- 46.Vitovec J, Koudela B. 1992. Pathogenesis of intestinal cryptosporidiosis in conventional and gnotobiotic piglets. Vet Parasitol 43:25–36. doi: 10.1016/0304-4017(92)90045-B. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Bolick DT, Kolling GL, Fu Z, Guerrant RL. 2016. Protein malnutrition impairs intestinal epithelial cell turnover, a potential mechanism of increased cryptosporidiosis in a murine model. Infect Immun 84:3542–3549. doi: 10.1128/IAI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Current WL, Haynes TB. 1984. Complete development of Cryptosporidium in cell culture. Science 224:603–605. doi: 10.1126/science.6710159. [DOI] [PubMed] [Google Scholar]
- 49.Weyl-Feinstein S, Markovics A, Eitam H, Orlov A, Yishay M, Agmon R, Miron J, Izhaki I, Shabtay A. 2014. Short communication: effect of pomegranate-residue supplement on Cryptosporidium parvum oocyst shedding in neonatal calves. J Dairy Sci 97:5800–5805. doi: 10.3168/jds.2013-7136. [DOI] [PubMed] [Google Scholar]
- 50.Jansson JK, Baker ES. 2016. A multi-omic future for microbiome studies. Nat Microbiol 1:16049. doi: 10.1038/nmicrobiol.2016.49. [DOI] [PubMed] [Google Scholar]
- 51.Okhuysen PC, Rich SM, Chappell CL, Grimes KA, Widmer G, Feng XC, Tzipori S. 2002. Infectivity of a Cryptosporidium parvum isolate of cervine origin for healthy adults and interferon-γ knockout mice. J Infect Dis 185:1320–1325. doi: 10.1086/340132. [DOI] [PubMed] [Google Scholar]
- 52.Widmer G, Lee Y, Hunt P, Martinelli A, Tolkoff M, Bodi K. 2012. Comparative genome analysis of two Cryptosporidium parvum isolates with different host range. Infect Genet Evol 12:1213–1221. doi: 10.1016/j.meegid.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mallon ME, MacLeod A, Wastling JM, Smith H, Tait A. 2003. Multilocus genotyping of Cryptosporidium parvum type 2: population genetics and sub-structuring. Infect Genet Evol 3:207–218. doi: 10.1016/S1567-1348(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 54.Widmer G, Feng X, Tanriverdi S. 2004. Genotyping of Cryptosporidium parvum with microsatellite markers. Methods Mol Biol 268:177–187. [DOI] [PubMed] [Google Scholar]
- 55.Yang S, Healey MC. 1993. The immunosuppressive effects of dexamethasone administered in drinking water to C57BL/6N mice infected with Cryptosporidium parvum. J Parasitol 79:626–630. doi: 10.2307/3283395. [DOI] [PubMed] [Google Scholar]
- 56.Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel: population genetic software for teaching and research: an update. Bioinformatics 28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stroup SE, Roy S, McHele J, Maro V, Ntabaguzi S, Siddique A, Kang G, Guerrant RL, Kirkpatrick BD, Fayer R, Herbein J, Ward H, Haque R, Houpt ER. 2006. Real-time PCR detection and speciation of Cryptosporidium infection using Scorpion probes. J Med Microbiol 55:1217–1222. doi: 10.1099/jmm.0.46678-0. [DOI] [PubMed] [Google Scholar]
- 58.Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westcott SL, Schloss PD. 2017. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere 2:e00073-17. doi: 10.1128/mSphereDirect.00073-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.