The bacterium Vibrio cholerae causes severe disease in humans, and strains can persist in the environment in association with a wide diversity of host species. By investigating the molecular mechanisms that underlie these interactions, we can better understand constraints affecting the ecology and evolution of this global pathogen. The Drosophila model of Vibrio cholerae infection has revealed that bacterial regulation of acetate and other small metabolites from within the fly gastrointestinal tract is crucial for its virulence. Here, we demonstrate that genes that may modify the proteome of V. cholerae affect virulence toward Drosophila, most likely by modulating central metabolic pathways that control the consumption of acetate as well as other small molecules. These findings further highlight the many layers of regulation that tune bacterial metabolism to alter the trajectory of interactions between bacteria and their hosts.
KEYWORDS: Drosophila melanogaster, Vibrio cholerae, acetate, acetyl-CoA, acetyl-CoA synthetase, acetylation, carbon metabolism, cholera, posttranslational modification
ABSTRACT
Acetylation is a broadly conserved mechanism of covalently modifying the proteome to precisely control protein activity. In bacteria, central metabolic enzymes and regulatory proteins, including those involved in virulence, can be targeted for acetylation. In this study, we directly link a putative acetylation system to metabolite-dependent virulence in the pathogen Vibrio cholerae. We demonstrate that the cobB and yfiQ genes, which encode homologs of a deacetylase and an acetyltransferase, respectively, modulate V. cholerae metabolism of acetate, a bacterially derived short-chain fatty acid with important physiological roles in a diversity of host organisms. In Drosophila melanogaster, a model arthropod host for V. cholerae infection, the pathogen consumes acetate within the gastrointestinal tract, which contributes to fly mortality. We show that deletion of cobB impairs growth on acetate minimal medium, delays the consumption of acetate from rich medium, and reduces virulence of V. cholerae toward Drosophila. These impacts can be reversed by complementing cobB or by introducing a deletion of yfiQ into the ΔcobB background. We further show that cobB controls the accumulation of triglycerides in the Drosophila midgut, which suggests that cobB directly modulates metabolite levels in vivo. In Escherichia coli K-12, yfiQ is upregulated by cAMP-cAMP receptor protein (CRP), and we identified a similar pattern of regulation in V. cholerae, arguing that the system is activated in response to similar environmental cues. In summary, we demonstrate that proteins likely involved in acetylation can modulate the outcome of infection by regulating metabolite exchange between pathogens and their colonized hosts.
IMPORTANCE The bacterium Vibrio cholerae causes severe disease in humans, and strains can persist in the environment in association with a wide diversity of host species. By investigating the molecular mechanisms that underlie these interactions, we can better understand constraints affecting the ecology and evolution of this global pathogen. The Drosophila model of Vibrio cholerae infection has revealed that bacterial regulation of acetate and other small metabolites from within the fly gastrointestinal tract is crucial for its virulence. Here, we demonstrate that genes that may modify the proteome of V. cholerae affect virulence toward Drosophila, most likely by modulating central metabolic pathways that control the consumption of acetate as well as other small molecules. These findings further highlight the many layers of regulation that tune bacterial metabolism to alter the trajectory of interactions between bacteria and their hosts.
INTRODUCTION
Bacteria finely tune their responses to various environmental conditions by modulating fluxes of nutrients through metabolic pathways, effectively balancing their energy needs with the biosynthetic demands associated with rapid cellular growth. Metabolic fluxes can be regulated most effectively by altering the concentrations of specific enzymes in the cell, but more immediate responses can be achieved by simply switching enzymes “on” and “off” via reversible covalent modifications, such as phosphorylation or acetylation, at key catalytic or allosteric residues. In bacteria, phosphorylation has been studied in the greatest detail because of its central role in mediating signaling via two-component signal transduction systems. However, acetylation similarly controls the activity of a wide variety of proteins in diverse bacterial lineages (1, 2). Acetylation targets specific lysine residues at the ε-amino group and alters one or more residues within a single protein. As bacterial cells progress from exponential- to stationary-phase growth, a surprisingly large proportion of the proteome undergoes acetylation (3–6). Although the impact of this modification on the vast number of proteins is unknown, acetylation affects the activity of specific proteins that, in turn, impact nutrient flux through metabolic pathways. These proteins include acetyl-CoA synthetase (Acs), an enzyme that converts acetate to acetyl-CoA as cells reach stationary phase, during a process termed the “acetate switch” (1). Acetate is first excreted by bacteria during rapid growth. Several theories have been proposed to explain this observation. Recently, a study suggested that growth rate can be maximized by optimally partitioning the proteome between respiratory and fermentative pathways (7). Many other studies have linked acetic acid production to "overflow" metabolism or more specific needs regarding recycling of metabolic intermediates (1). Flipping this switch by inducing Acs allows bacteria to consume the excreted acetate as other carbon sources are depleted (1).
Acs is regulated by acetylation in bacteria and eukaryotes (8–10). In Salmonella, acetylation of a conserved lysine residue, Lys609, regulates the activity of the Acs protein, although other acetylated lysine sites have been identified (5, 10). Lys609 is acetylated and inactivated via a specific acetyltransferase, YfiQ/PatZ/Pka in Salmonella (here referred to as YfiQ), a member of the GCN5-like acetyltransferase (GNAT) family (11). Deacetylation, which reactivates Acs, primarily depends on CobB, a sirtuin-like protein that functions as an NAD+-dependent deacetylase (10). In Bacillus subtilis, a second deacetylase, AcuC, activates Acs as well (12). Acetylation can also occur nonenzymatically; many proteins carry lysine residues that are susceptible to an alternative, direct acetylation mechanism requiring acetyl-phosphate (acetyl-P) (5, 6). Altogether, these acetylation mechanisms target a significant number of proteins in the cell, many of which are situated within central metabolic pathways (2, 5, 6, 13). As a result, acetylation may indirectly mediate the close relationships between bacteria and multicellular organisms that are contingent upon the exchange of metabolic products.
Members of the Vibrio genus are marine bacteria that readily adjust to both free-living and host-associated states. In at least two lineages, regulation of acetate metabolism is critical for the development and stability of their interactions with host organisms. In Vibrio fischeri, the bioluminescent symbiont of the Hawaiian bobtail squid, Euprymna scolopes, Acs promotes colonization of the juvenile squid (14, 15). In Vibrio cholerae, the human pathogen responsible for historical and ongoing global cholera pandemics, Acs is required for virulence in a Drosophila melanogaster model of infection (16). Consumption of acetate via Acs by V. cholerae contributes to the misregulation of triglyceride storage in the fly, resulting in intestinal steatosis, which is necessary for V. cholerae-mediated mortality (16). In V. cholerae, acs transcription is activated by the CrbS/R two-component signal transduction pathway (16, 17). To further define the regulatory network that modulates levels of acetate during colonization and infection, we determined whether genes encoding putative acetyltransferase and deacetylase enzymes in V. cholerae alter acetate metabolism and the infection processes. Here, we demonstrate that CobB and YfiQ, but not AcuC, contribute to the regulation of acetate consumption and virulence in a Drosophila model, suggesting that acetylation is an important regulator of host-microbe interactions in this global pathogen.
RESULTS
Identification and deletion of the V. cholerae CobB and YfiQ genes.
In order to assess whether an acetylation system similar to those of Escherichia coli and Salmonella exists in V. cholerae, we first identified putative yfiQ and cobB genes in the V. cholerae genome via the BLAST algorithm. CobB, an NAD+-dependent protein deacetylase, belongs to the sirtuin family of proteins, which deacetylate a number of different target proteins with wide-ranging biological effects in bacteria, archaea, and eukarya (18). The putative V. cholerae deacetylase protein, encoded by the VC1509 locus, and the E. coli CobB protein share 66% identity and 79% similarity across 88% of the protein, with the catalytic His110 residue being conserved in V. cholerae (19). The Salmonella YfiQ acetyltransferase protein and the putative V. cholerae acetyltransferase protein (VCA0574) share 55% identity and 73% similarity across all but the first four amino acids. The high levels of identity at the amino acid level, including the conservation of key residues required for enzymatic function, strongly suggest that the proteins may share a both functional and evolutionary relationships. Therefore, we refer to these genes as cobB and yfiQ in V. cholerae, respectively.
CobB is required for acetate utilization.
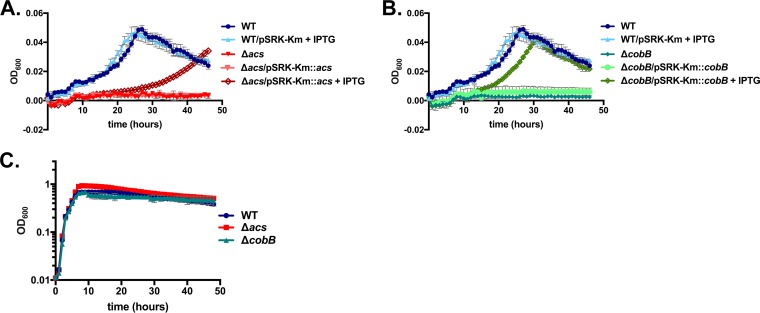
Next, we engineered an in-frame deletion in the putative cobB gene in V. cholerae SIO. Because CobB may function as a deacetylase, we predicted that the deletion may increase the proportion of metabolic enzymes that are acetylated, including those involved in acetate catabolism, such as Acs. As a result, the cells may be unable consume acetate. To test this hypothesis, we inoculated both the acs mutant and the cobB mutant into minimal medium with acetate as the sole carbon source. After ∼48 h, the cobB deletion mutant grew little on acetate in comparison to the wild-type strain, mimicking the phenotype of the acs deletion (Fig. 1A and B). Inducing the expression of cobB from the pSRK-Km plasmid with isopropyl-β-d-thiogalactopyranoside (IPTG) restored growth on acetate, indicating that the lack of cobB is solely responsible for this phenotype (Fig. 1B). Inducing the expression of acs partially complemented the growth of the Δacs mutant (Fig. 1A). Both the acs mutant and the cobB mutant grew similarly to the wild-type strain in LB broth, demonstrating that these mutations do not result in a generalized growth defect in rich medium (Fig. 1C).
FIG 1.
Deletion of V. cholerae SIO cobB prevents growth on acetate minimal medium but not on LB broth. (A and B) V. cholerae SIO Δacs (A) and ΔcobB (B) strains cannot grow on M63 minimal medium supplemented with 10 mM acetate as a sole carbon source, but the addition of the pSRK-Km complementation plasmids carrying the acs and cobB genes, respectively, together with induction with 1 mM IPTG, restores growth. Optical densities were measured once every hour over 46.5 h, and values from triplicate wells of a single experiment are plotted. (C) In LB broth, V. cholerae SIO Δacs and ΔcobB grow at similar rates, and to similar optical densities, relative to the wild-type (WT) strain. Optical densities were measured once every hour over 48 h, and values from duplicate wells of a single experiment in a 96-well plate are plotted. In each panel, error bars depict standard deviations. At some time points, the symbols fully obscure the error bars and therefore are not shown.
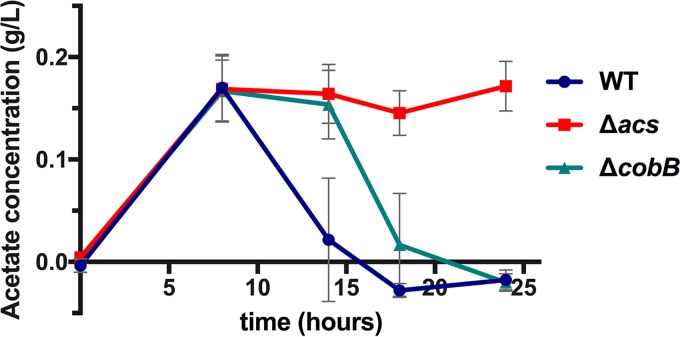
To determine whether the defect in growth on acetate resulted from an inability to remove acetate from the medium, we directly measured acetate concentrations in LB broth over time. We observed that deleting acs or cobB did not impact acetate excretion; by 8 h, acetate concentrations rose to levels similar to those observed in the wild-type strain (Fig. 2). This is consistent with observations in E. coli in which the acs mutant also accumulates acetate to the same extent as the wild type (20). The wild-type V. cholerae strain flipped the acetate switch after 8 h of growth, and all acetate was removed from the medium prior to 20 h postinoculation (Fig. 2). In contrast, acetate consumption by the acs mutant was halted entirely. Deletion of cobB, however, delayed but did not halt the initiation of acetate removal in rich medium (Fig. 2).
FIG 2.
Deletion of V. cholerae SIO cobB delays, but does not halt, acetate consumption. Acetate concentrations relative to those in uninoculated LB broth were measured over time. Here, results from three biological replicates, each performed in duplicate, are depicted. Acetate concentrations in cultures of wild-type SIO and SIO ΔcobB strains are significantly different from one another after 14 h (P = 0.0022) and 18 h (P = 0.0087) of growth (by a Mann-Whitney test). Error bars depict standard deviations.
CobB modulates V. cholerae virulence in Drosophila.
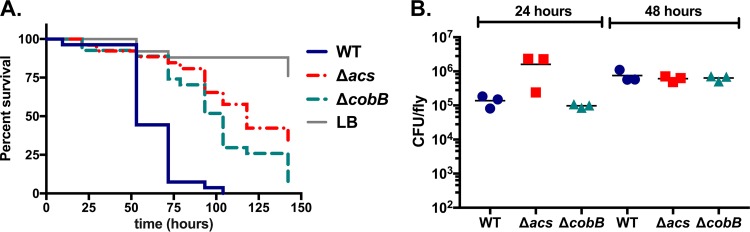
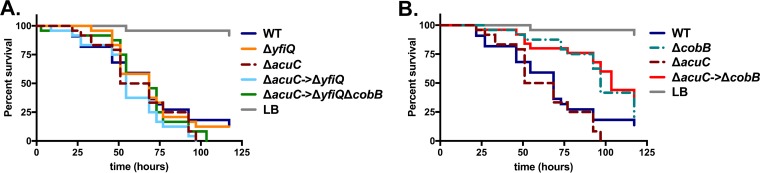
Because cobB alters acetate utilization during growth in culture, we hypothesized that this gene may be necessary for V. cholerae virulence during infection of Drosophila. In this model system, V. cholerae pathogenicity is dependent upon the colonization of specific regions of the gastrointestinal (GI) tract and manipulation of host metabolism (16, 21–24). In order to test whether cobB-dependent alteration of acetate metabolism is relevant in vivo, we fed this mutant to adult Drosophila flies in a standard oral infection assay (25). Ingestion of wild-type V. cholerae is lethal to Drosophila over the course of 48 to 96 h, while a strain carrying a deletion in acs is significantly less lethal to flies despite populating the GI tract to the same level as the wild type (16). This phenotype is observed in strains of both environmental and clinical origins, independent of whether they carry genes that encode cholera toxin and toxin-coregulated pilus (16, 17). Deletion of cobB significantly delayed fly mortality relative to the wild-type strain (P < 0.0001), but it did not render the strain avirulent (Fig. 3A). To ensure that the cobB deletion did not reduce levels of bacteria in the flies, Drosophila flies infected with these bacterial strains were homogenized at 24 and 48 h postinfection, and numbers of CFU per fly were determined by plating. The cobB and acs mutations did not reduce bacterial numbers in the flies, as expected (P > 0.05 by a Mann-Whitney test) (Fig. 3B).
FIG 3.
Deletion of V. cholerae SIO cobB significantly improves survival of Drosophila without appreciably affecting colonization. (A) Survival of flies fed the V. cholerae wild-type SIO, SIO ΔcobB, or SIO Δacs strain, or uninoculated LB broth as a control, was monitored over 143 h. This assay was performed with V. cholerae strains added to triplicate vials, with 8 to 10 flies per vial. This result is representative of data from five separate trials. Flies fed the SIO ΔcobB or SIO Δacs strain survived significantly longer than did flies fed the wild-type strain (P < 0.0001 by a log rank test). (B) Colonization of flies infected with the V. cholerae SIO wild-type, SIO ΔcobB, or SIO Δacs strain. The assay was performed in triplicate vials, and all surviving flies were collected after 24 and 48 h of infection, homogenized, and plated on selective medium. The bacterial loads of flies infected with the mutant strains were not significantly different from bacterial loads of those infected with the wild-type strain (P > 0.05 by a Mann-Whitney test).
YfiQ, a putative acetyltransferase, counteracts the effects of CobB in vitro and in vivo.
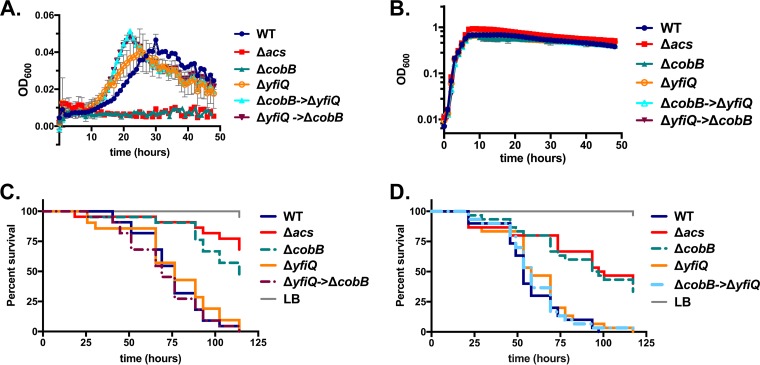
While CobB removes acetyl groups from proteins, the addition of acetyl moieties to specific residues by acetylation can occur both via enzyme-dependent processes mediated by acetyltransferases and via the accumulation of acetyl-P, which can nonenzymatically acetylate target proteins (5, 6). To determine whether YfiQ, the putative acetyltransferase, functions to counteract the effects of CobB, we engineered an in-frame deletion in the yfiQ gene in both the wild-type V. cholerae strain and the ΔcobB strain. To ensure that background mutations did not affect the phenotype, we also created the same strain by introducing the ΔcobB allele into the ΔyfiQ background. We then tested these three strains (ΔyfiQ; ΔcobB into the ΔyfiQ background, designated ΔcobB→ΔyfiQ; and ΔyfiQ→ΔcobB as the reverse construction) for their ability to grow on acetate minimal medium. Relative to the wild-type strain, deletion of yfiQ did not impact growth on acetate (Fig. 4A). Introduction of the yfiQ deletion into the ΔcobB background (and vice versa) restored the ability of the strain to grow on acetate without affecting growth on LB broth (Fig. 4A and B). These results are consistent with the deletion of yfiQ preventing acetylation and subsequent inactivation of one or more key enzymes required for growth on acetate minimal medium, including Acs. In this background, CobB, the putative deacetylase, would have little effect on acetylation levels or enzyme activity, as the deletion of yfiQ would ensure that acetylation is minimal, and targeted enzymes would be maximally active.
FIG 4.
Introduction of the complete deletion of yfiQ restores both virulence in Drosophila and growth on acetate minimal medium to the V. cholerae ΔcobB strain. (A) Deletion of yfiQ does not impact growth on minimal medium supplemented with acetate (10 mM), with growth being similar to that of the wild-type strain. Introduction of the ΔyfiQ mutation restores growth to the strain carrying the deletion in ΔcobB, as both the ΔyfiQ→ΔcobB and the ΔcobB→ΔyfiQ strains grow similarly to the wild-type strain in acetate minimal medium. (B) In LB broth, all strains grow similarly to the wild type. (C) YfiQ does not significantly alter virulence of Drosophila (P > 0.05). In seven independent trials, deletion of yfiQ significantly increased virulence in one assay (P = 0.0003), reduced virulence in a second assay (P = 0.0143), and did not alter virulence in the remaining five assays, including the representative assay presented here. When infected with the ΔyfiQ→ΔcobB strain, fly mortality is increased significantly relative to the strain carrying the ΔcobB deletion alone (P < 0.0001 by a log rank test). This result was reproducible in five independent trials. (D) When flies were infected with the ΔcobB→ΔyfiQ strain, virulence was similarly restored relative to the strain carrying the single cobB deletion (P < 0.0001 by a log rank test).
To determine whether yfiQ function is relevant in vivo, we infected Drosophila flies with the ΔyfiQ, ΔyfiQ→ΔcobB, and ΔcobB→ΔyfiQ strains. Based on our results presented above, we hypothesized that the deletion of yfiQ might reduce the proportion of acetylated Acs proteins (or other metabolic enzymes), which could, in turn, increase acetate consumption and virulence toward Drosophila, resulting in more rapid mortality of flies. However, the ΔyfiQ allele did not consistently alter virulence; in five of seven independent assays, the rate of survival of flies fed this strain was similar to that of flies fed the wild-type strain, while flies died slightly more quickly in one assay, and in the last assay, the flies died more slowly (Fig. 4C and D). A phenotype for YfiQ was observed in the ΔcobB→ΔyfiQ strain, which killed flies more quickly than did the strain carrying the ΔcobB deletion alone (P < 0.0001 in each of five independent assays) (Fig. 4C). In a confirmatory experiment, introduction of the ΔyfiQ allele into the ΔcobB background also restored virulence to the ΔcobB strain (P > 0.05 compared to the wild type and P < 0.0001 compared to the ΔcobB strain in one assay) (Fig. 4D). This is consistent with the hypothesis that the acetylation system is operational in V. cholerae and relevant during in vivo infection.
AcuC, a second putative deacetylase, does not alter acetate-dependent virulence.
The activity of Acs in Bacillus subtilis, a Gram-positive soil bacterium, is controlled by two deacetylases, SrtN, its sirtuin-like deacetylase, and an additional deacetylase, AcuC (12). In the B. subtilis genome, the acuC gene is adjacent to the acsA gene (12). V. cholerae carries a homolog of AcuC at locus VC2042, but it is not colocalized with acs or other genes involved in altering acetylation. The two proteins are 31% identical and 43% similar across 294 amino acids. To determine whether acuC similarly regulates acetate metabolism in V. cholerae, we introduced an in-frame deletion of this gene into the wild-type, ΔcobB, ΔyfiQ, and ΔyfiQ→ΔcobB backgrounds and monitored its effects on Drosophila survival. We observed that the deletion of acuC alone did not alter the mortality of Drosophila flies (Fig. 5A). When the ΔacuC mutation was introduced into the ΔcobB background, survival was not further decreased, indicating that redundancy with cobB was not masking its phenotype (Fig. 5B). We further observed no impact on survival in the ΔyfiQ or ΔyfiQ→ΔcobB background, as expected (Fig. 5A). Therefore, acuC does not affect Acs activity or acetate metabolism during infection of Drosophila.
FIG 5.
Deletion of the putative deacetylase AcuC does not alter virulence in Drosophila. (A) Survival of flies fed the wild-type, ΔyfiQ, ΔacuC, ΔacuC ΔyfiQ, or ΔacuC ΔyfiQ ΔcobB strain. Deletion of acuC did not alter fly survival relative to the wild type in each of two separate assays, one of which is represented here (P > 0.05 by a log rank test). (B) Survival of flies fed the wild-type, ΔcobB, ΔacuC, or ΔacuC ΔcobB strain. Deletion of acuC in the ΔcobB strain did not alter survival relative to the ΔcobB deletion alone in each of two separate assays (P > 0.05 by a log rank test).
Point mutations in Acs modulate Acs activity.
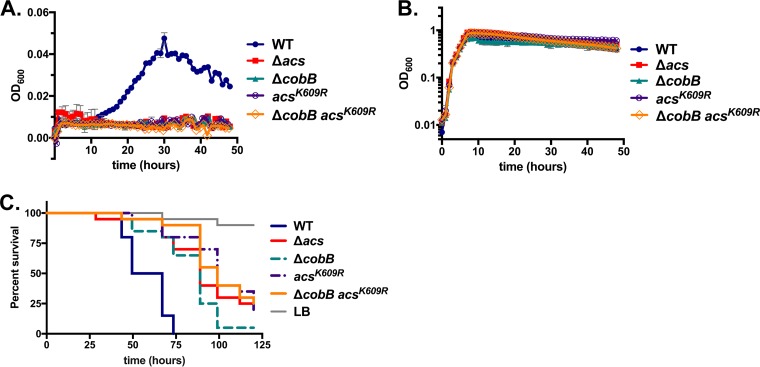
In order to further define the effects of CobB on Acs, we identified point mutations in Acs that might prevent acetylation. We hypothesized that the introduction of these mutations into a cobB deletion strain could support higher rates of acetate consumption, which could then improve growth on acetate minimal medium. We began by engineering specific mutations in conserved Acs residues shown by previous studies in E. coli and Salmonella to modulate acetylation. First, we replaced the conserved catalytic lysine residue K609 in the chromosomal copy of acs with arginine, which is thought to mimic a nonacetylated lysine (26). We reasoned that this mutation could result in one of two outcomes. First, it is possible that this mutation simply reduces the enzymatic activity of Acs, since it may interrupt the catalytic site. Alternatively, this mutation may prevent acetylation while allowing for some level of catalytic activity. We observed that the AcsK609R mutation completely abrogated growth on acetate minimal medium, suggesting that it primarily reduced the protein's catalytic activity (Fig. 6). To test the possibility that the mutation may prevent acetylation while permitting a low level of activity, we introduced this mutation into the ΔcobB strain, reasoning that the K609R mutation may increase Acs activity to a level slightly higher than that observed in the cobB mutant alone. However, growth of the ΔcobB AcsK609R deletion strain on acetate minimal medium was negligible and identical to those of the AcsK609R, Δacs, and ΔcobB strains (Fig. 6A). Thus, introducing the K609R mutation did not increase Acs activity, nor did it overcome the acetate consumption defect observed in the cobB deletion. The K609R mutation did not affect growth on rich LB broth, suggesting that it did not confer a generalized growth defect (Fig. 6B). Next, we tested whether this mutation altered pathogenicity during Drosophila infection. We observed that the K609R mutant mimicked the virulence of the acs mutant (P > 0.05 in three independent assays), further supporting the hypothesis that it abrogates the function of Acs (Fig. 6C). In addition, the introduction of the K609R mutation into the ΔcobB background did not increase virulence but instead further reduced virulence relative to the single mutation in cobB alone (Fig. 6C). These results are each consistent with the conclusion that the mutation simply reduced the ability of Acs to carry out its primary catalytic function, regardless of its effect on the acetylation state. However, confirmation of this conclusion awaits direct biochemical testing with a purified Acs protein carrying this mutation.
FIG 6.
Mutation of lysine 609 abrogates Acs activity. (A) Mutation of the putative catalytic residue of Acs, K609, to arginine entirely halts growth on M63 minimal medium supplemented with 10 mM acetate. The introduction of this mutation has no effect on the growth of the ΔcobB strain. (B) Mutations of acs have no effect on growth on LB broth, consistent with previous findings. (C) Introduction of the acsK609R mutation into the wild-type strain reduces fly mortality (P < 0.0001 by a log rank test). The introduction of the same mutation into the ΔcobB strain does not restore virulence but rather further reduces virulence (P = 0.018 by a log rank test). These results are representative of data from three independent trials.
Next, we introduced a mutation in Acs at residue 641 that changed the Leu residue to Ala. In Salmonella, this mutation restores growth on acetate minimal medium to a strain that also carries deletions in cobB and pta, because the AcsL641A mutant prevents YfiQ from acetylating Acs (27). This permits growth on acetate despite the absence of CobB, which is otherwise needed to deacetylate and activate Acs (27). (In Salmonella, deletion of pta in addition to cobB is necessary to prevent background growth on acetate minimal medium. However, we observed that deletion of cobB alone is sufficient to halt the growth of V. cholerae on acetate [Fig. 1], suggesting that pta does not play a role in the consumption of acetate at these concentrations, as reported previously for E. coli [1].) In Salmonella, the observation that the cobB deletion can be suppressed by the AcsL641A mutation alone, without additional mutations in other enzymes in the acetate assimilation pathway, supports the conclusion that the YfiQ/CobB system may modulate acetate metabolism primarily via its regulation of Acs activity, as opposed to any effects that it may have on other catabolic enzymes.
To determine whether the mutation of a single residue of Acs similarly alters CobB-mediated growth of V. cholerae on acetate, we engineered the same Leu-to-Ala substitution at residue 641 of Acs by replacing the wild-type copy with the acsL641A allele on the chromosome. As with the AcsK609R mutant, the mutation of Leu641 to Ala could affect V. cholerae Acs function in several ways. The mutation could generate an enzyme that is functional but entirely blind to acetylation. In this scenario, we would expect that introducing the AcsL641A mutation into the ΔcobB background would restore the ability of this strain to assimilate acetate, grow on acetate minimal medium, and fatally infect Drosophila flies. This would be most evident if both YfiQ and CobB are primarily acting through Acs to regulate acetate catabolism. If mutation of Leu641 has no effect on the acetylation of Acs, then we would expect the mutation to have no effect on the cobB phenotype. On the other hand, this residue could play an alternative role in Acs function or stability, and its mutation could affect Acs activity through some means other than acetylation.
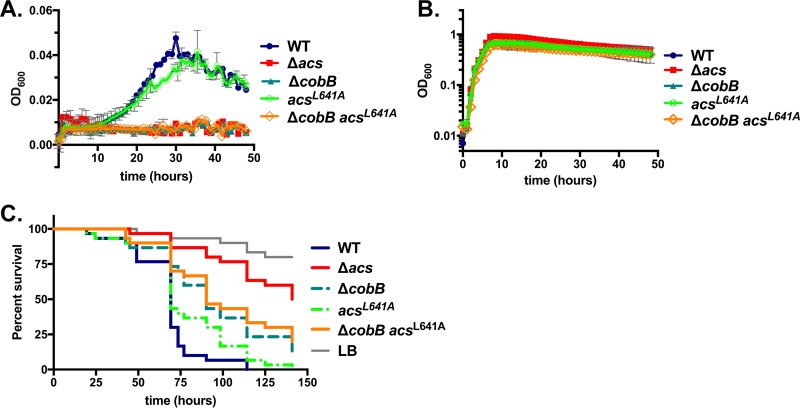
The AcsL641A mutation did not affect growth on LB medium and only minimally affected on growth on acetate, suggesting that the mutation does not significantly impair Acs function (Fig. 7). Next, we tested whether this mutation improves the growth of the ΔcobB strain on acetate minimal medium. We observed that the ΔcobB acsL641A strain grew similarly to the cobB deletion alone; that is, the mutant was unable to grow on acetate minimal medium but grew to wild-type levels on LB medium (Fig. 7). This finding is consistent with each of two possible conclusions: (i) that the introduction of the L641A point mutation has no effect on Acs function or acetylation because its role in YfiQ binding is not conserved or (ii) that conferring resistance to acetylation upon Acs via the introduction of the L641A mutation is insufficient to alter acetate removal, because CobB may be acting multifactorially to control acetate metabolism.
FIG 7.
Mutation of Leu641 to Ala does not restore acetate metabolism in the ΔcobB strain. (A) Mutation of Leu641 does not severely impede growth on M63 minimal medium supplemented with 10 mM acetate. The introduction of this mutation has no effect on the growth of the ΔcobB strain. (B) Mutations of acs have no effect on growth on LB broth, consistent with previous findings. (C) Introduction of the acsL641A mutation into the wild-type strain slightly reduces fly mortality (P = 0.0260 by a log rank test), consistent with findings in four of five independent trials. The introduction of the same mutation into the ΔcobB strain does not affect the virulence of the ΔcobB mutant (P > 0.05 by a log rank test), which was again observed in four of five independent trials.
To determine whether Drosophila infection assays might more sensitively detect slight alterations in virulence caused by the L641A mutation, we fed these strains to flies and monitored survival. The L641A mutation alone reduced virulence in 4 of 5 independent assays compared to the wild-type strain (P < 0.05). Introducing this L641A mutation into the ΔcobB strain background increased virulence significantly in only one of five assays, although a nonsignificant trend was observed in a second assay (P = 0.0687). Therefore, we cannot conclude that the Leu641 residue modulates virulence or acetylation in any way (Fig. 7). Interestingly, the amino acids directly adjacent to Leu641 are not highly conserved between Salmonella enterica and V. cholerae, suggesting that this residue may not function similarly in the two homologous proteins, and the mutation may have no effect on YfiQ binding or interactions.
Effect of the CobB deletion on triglyceride storage in the fly.
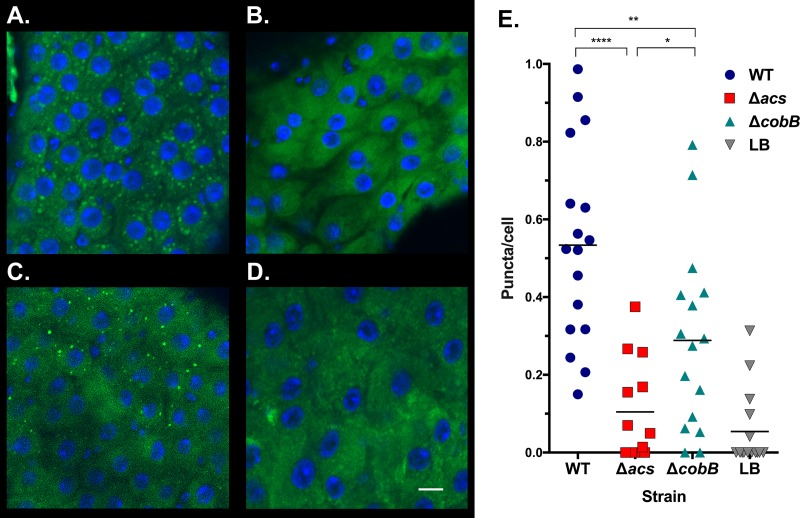
In order to confirm that the cobB gene is affecting fly survival via its manipulation of acetate or other metabolites, we examined the GI tracts of flies for effects on the storage of lipids. When fed LB broth in the absence of infection, flies accumulate and store triglycerides in the fat body (16). However, consumption of acetate by V. cholerae causes triglyceride droplets to amass in cells lining the GI tract (16). This is directly related to V. cholerae-dependent processes that remove acetate and other metabolites during infection (16, 21). In order to determine whether cobB similarly affects fat storage, we fed flies wild-type V. cholerae, LB medium alone, or the strains carrying mutations in cobB or acs for 48 h. We then removed the GI tracts, performed staining with BODIPY dye, and quantified puncta of triglycerides in cells (21). Consistent with previous findings, infection with the wild-type strain drastically increased the number of puncta of accumulating triglycerides relative to those in uninfected flies, while the acs mutant maintained reduced triglyceride storage (16) (Fig. 8). The cobB strain exhibited an intermediate phenotype. In two of three experimental replicates, the cobB mutant significantly reduced triglyceride storage relative to the wild type, and the trend was similar but did not reach significance in a third trial (Fig. 8). In comparison to the acs mutant, infection with the ΔcobB strain increased triglyceride storage in the enterocytes significantly (Fig. 8). These results suggest that the cobB gene alters bacterial metabolism, which in turn affects the accumulation of lipids in the fly GI tract in a manner that hastens mortality.
FIG 8.
CobB alters accumulation of triglycerides in the Drosophila gastrointestinal tract. (A to D) Representative images of Drosophila GI tracts infected with the V. cholerae wild-type (A), Δacs (B), or ΔcobB (D) strain or provided sterile LB broth (D); visualized by confocal microscopy; and stained with DAPI and BODIPY. Bar, 10 μm. (E) Quantification of triglyceride puncta stained with BODIPY 493/503 in midguts of Drosophila flies infected with the V. cholerae wild-type, Δacs, or ΔcobB strain or sterile LB broth, with the levels of significance indicated (****, P < 0.0001; **, P = 0.0060; *, P = 0.0127). These results were representative of data from three trials comparing the wild-type and ΔcobB strains, two of which included the Δacs strain.
Transcriptional regulatory mechanisms of yfiQ are conserved in V. cholerae.
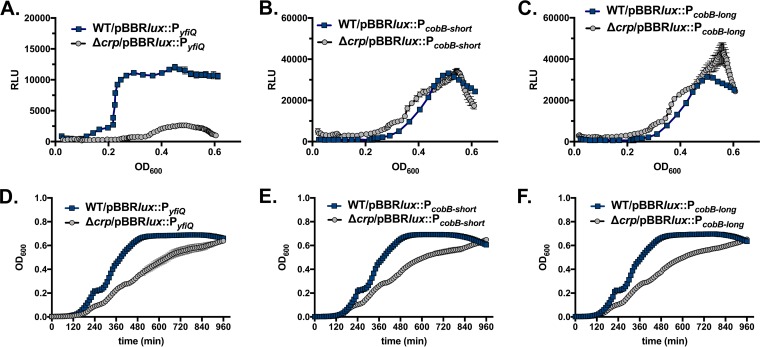
In order to place the acetylation system in a broader regulatory context in V. cholerae, we tested whether this system is subject to conserved regulatory controls. In E. coli and Salmonella, the yfiQ gene is transcriptionally regulated by the cAMP-cAMP receptor protein (CRP) system, but cobB expression levels change little across different phases of growth (28). To test whether the cAMP-CRP system similarly activates yfiQ but not cobB in V. cholerae, we cloned the putative promoters of yfiQ and cobB into the pBBRlux plasmid and conjugated the plasmids into both wild-type V. cholerae and V. cholerae carrying a deletion in crp. We tested two regions of the cobB promoter because of the presence of a poorly conserved putative crp binding site. The site was included in the longer fragment but was excluded from the small promoter fragment. We then monitored both growth and luminescence continuously in a 96-well multimode plate reader. Deletion of crp significantly reduced luminescence driven by the promoter of yfiQ, but crp had no effect on expression driven by the cobB promoters (Fig. 9). Therefore, CRP activates the yfiQ promoter, but cobB is controlled independently of CRP. The latter result also serves as a control to confirm that the CRP-cAMP system is not required for regulating other metabolic pathways that feed into the luminescence reaction. We identified a putative CRP binding site [TGCGA(N6)TCATA] within the yfiQ promoter region that differs from the consensus sequences of E. coli CRP [TGTGA(N6)TCACA] and V. cholerae CRP [GTGA(N6)TCAC] at two nucleotides (29–32). This binding site is centered 225 bp upstream of the translational start site of the yfiQ gene. A putative σ70 bacterial promoter, with −35 and −10 regions, was predicted using BPROM (Softberry, Inc.), which places the putative transcriptional start site roughly 120 bp upstream of the translational start and ∼105 bp downstream of the putative CRP binding site. Altogether, these findings support the hypothesis that the CRP-cAMP system activates the transcription of yfiQ but not cobB. This is consistent with a previous whole-genome microarray transcriptome analysis of V. cholerae O1 El Tor A1552, which identified yfiQ as being downregulated in strains lacking CRP (33), although a second microarray approach with another V. cholerae El Tor strain carrying a crp deletion did not identify yfiQ as a target (34).
FIG 9.
CRP regulates transcription of yfiQ but not cobB. (A to C) The promoter of yfiQ (A) and both a short region (B) and a longer region (C) of the cobB promoter were cloned into the pBBRlux plasmid and conjugated into V. cholerae wild-type and V. cholerae Δcrp bacteria. Bioluminescence was monitored in a multimode plate reader over 1,000 min and is depicted as relative light units (RLU), defined as luminescence normalized to the OD600. Due to the growth defect of the crp mutants, the y axis depicts the OD600, and RLU are shown over the time period during which the cultures were at an OD600 of 0.02 to 0.60. The yfiQ promoter is strongly induced, but the deletion of crp prevents the expression of yfiQ. In contrast, the deletion of crp does not affect expression from the cobB promoter fragments. (D to F) The Δcrp strain exhibits a growth defect as cells approach stationary phase. Growth of the WT and Δcrp strains carrying the designated promoters was monitored in the multimode plate reader by measurement of OD600.
DISCUSSION
Acetylation, one means of posttranslational regulation in bacteria, alters the structures of a wide variety of proteins across bacterial proteomes. Studies in E. coli and Salmonella, together with findings from other diverse bacteria, are revealing that acetylation mechanisms, alongside the enzymes targeted by this modification, are well conserved across evolutionary time scales (2, 5, 6). However, the biological and physiological impacts of acetylation-dependent regulation on protein function, particularly in the context of host-microbe interactions, remain unclear. The Vibrio cholerae-Drosophila model system is uniquely suited for the study of the effects of this posttranslational modification in vivo. In this system, V. cholerae virulence is not toxin mediated, but rather, pathogenesis is dependent upon the regulation of metabolites in the GI cavity by the pathogen (16, 21). In this work, we have shown that Vibrio cholerae genes likely involved in protein acetylation are critical for the proper regulation of acetate metabolism and pathogenesis in this arthropod model of infection.
The gene encoding CobB in V. cholerae is remarkably well conserved with its orthologs in E. coli and Salmonella, strongly suggesting that a functional relationship also exists. Although this conclusion awaits confirmation via biochemical and proteomic approaches, our findings are consistent with this hypothesis. Deletion of cobB prevents the growth of V. cholerae on acetate as a sole carbon source and slows the removal of acetate from rich media. This result is consistent with the idea that the cobB deletion increases the acetylation stoichiometry of key enzymes, such as Acs, that are involved in acetate catabolism. However, deleting cobB does not entirely halt acetate consumption, while deleting acs itself completely prevents both acetate consumption as well as growth on acetate minimal medium. This discrepancy is consistent with several conclusions. As one possibility, cobB may acetylate multiple enzymes within the acetate assimilation pathway. In this case, acetate assimilation may be halted in the ΔcobB strain because multiple enzymes are highly acetylated and locked in inactive states, reducing flux through this catabolic pathway such that growth cannot be supported. It is also possible that the ΔcobB strain may continue to remove acetate from the medium because (i) a second deacetylase can restore Acs activity or (ii) acetylation may not fully inactivate Acs. To examine a role for a second deacetylase, we introduced a deletion in acuC, a putative deacetylase that acts on Acs in B. subtilis (12). However, we observed no effect on Drosophila survival in any strain background, suggesting that the second deacetylase, if present in V. cholerae, is not AcuC. Our results, explained below, lead us to favor the conclusion that CobB targets multiple proteins controlling acetate assimilation, although these scenarios are not mutually exclusive.
To provide additional evidence for a role for CobB in acetylation, we tested whether the effects of its mutation could be rescued by the introduction of a deletion of yfiQ, the putative acetyltransferase, in the ΔcobB background. We hypothesized that the removal of YfiQ would prevent acetylation, essentially locking the targeted enzymes in their most highly active state. In this background, deletion of cobB, the deacetylase, would have no effect on enzyme activity. Indeed, the yfiQ deletion alone does not affect growth on acetate, but introducing the yfiQ deletion into the ΔcobB background restores the ability of this strain to grow on acetate minimal medium, suggesting that the two genes regulate an identical target(s). We observed a similar effect during infection of Drosophila: deletion of CobB reduces mortality, while the introduction of the YfiQ mutation restores virulence to this strain. Interestingly, the yfiQ deletion alone does not increase fly mortality. We have found few mutations that increase lethality in strongly virulent strains, suggesting that there is a maximum rate at which V. cholerae can infect Drosophila, perhaps as a result of an infection bottleneck (16, 22).
Next, we performed a series of experiments with the goal of linking the effects of CobB and YfiQ on acetate metabolism to their impacts on Acs. We first introduced the K609R mutation into Acs at the putative catalytic site. Not surprisingly, this mutation appears to have inactivated the enzyme; none of our results were consistent with an effect on acetylation. As an alternative possibility, this mutation may have generated an Acs protein that cannot be acetylated by CobB but could halt growth via a different mechanism. In Salmonella, overexpression of Acs alleles that are highly active and resistant to acetylation reduces the energy charge of the cell by overproducing AMP, a by-product of the reactions that convert acetate to acetyl-CoA (35). The reduction in energy charge, in turn, impairs growth (35). If this is the case in V. cholerae, then we can expect the acsK609R mutation, as well as the same mutation introduced into the ΔcobB background, to phenocopy the ΔyfiQ deletion, because acetylation cannot reduce Acs activity in these strains. Instead, we found that the phenotypes of the ΔyfiQ mutation resemble those of the wild-type strain in fly survival assays and growth assays, consistent with the idea that reduced levels of acetylation in the ΔyfiQ strain do not impair growth on acetate in V. cholerae. However, strains carrying the acsK609R or the ΔcobB acsK609R mutations cannot grow on acetate, and fly survival is improved, indicating that acetate consumption is reduced. Therefore, it is more likely that this mutation halts enzymatic activity, rather than impairing acetylation and reducing energy charge, to impact growth.
Next, we introduced the L641A mutation, which prevents acetylation in Salmonella Acs, into V. cholerae Acs. This mutation was unable to rescue acetate catabolism in a ΔcobB strain, suggesting that (i) the role of this site in YfiQ binding may not be conserved or (ii) acetylated sites in other enzymes important for acetate assimilation are also controlled by CobB. In the latter case, it would be impossible to isolate mutations in Acs alone that confer growth on acetate because they prevent acetylation. Because the results from these targeted mutation studies yielded multiple interpretations, we attempted to search for alleles of Acs that are capable of resisting acetylation. We created a pool of random mutations in acs, introduced these mutations into a ΔcobB Δacs strain, and selected for those mutations capable of suppressing the growth defect of the ΔcobB strain on acetate minimal medium (27). In Salmonella, acetylation-resistant alleles of Acs were sufficient to complement the growth of a ΔcobB Δacs Δpta strain (27). Ultimately, we did not identify Acs mutants capable of conferring growth to a ΔcobB Δacs strain on acetate minimal medium (data not shown). One of several possible explanations for this result is that the V. cholerae CobB enzyme deacetylates multiple enzymes, as it does in E. coli (3, 5, 19, 36), and the activity of these enzymes may be required for acetate assimilation. This is also consistent with our previous finding that the ΔcobB strain delays acetate consumption but cannot grow on acetate as a sole carbon source. Altogether, we hypothesize that CobB deacetylates Acs to promote acetate consumption, but it is CobB's ability to modify other enzymes, together with its alteration of Acs, that may be important for growth on acetate minimal medium. However, further biochemical and mass spectrometry analyses are needed to confirm this conclusion, as other evidence suggests that the enzymatic acetylation system may in fact target a smaller number of enzymes than proposed previously (37).
The molecule acetyl-P also directly acetylates lysine residues nonenzymatically, and we questioned whether this system could be functioning similarly in V. cholerae. In E. coli, pools of acetyl-P can be modulated via mutations in the pta and ackA genes (5, 6). The Pta protein removes the CoA group from acetyl-CoA to generate acetyl-P, which AckA then converts to acetate. In V. cholerae, the pta and ackA genes are located in an operon. However, V. cholerae also encodes a second ackA gene, on chromosome II. Deletion of pta was sufficient to prevent acetate accumulation in the medium, as expected, but deletion of the ackA gene directly adjacent to pta did not affect acetate excretion (61). This result argues that the two ackA genes encode functionally redundant proteins, and both ackA genes must be deleted from the V. cholerae genome in order to genetically probe the role of acetyl-P in the acetylation-dependent control of metabolism. However, our results suggest that the CobB-YfiQ system is the primary mechanism through which acetylation modifies enzymes involved in acetate metabolism, as the deletion of yfiQ entirely suppressed the deletion of cobB. Thus, YfiQ is likely responsible for acetylating enzymes within this pathway, which may include Acs.
Because the CobB and YfiQ enzymes modulate metabolism in vivo, the regulatory mechanisms controlling their expression could, in turn, play an important role in metabolite-dependent virulence. In Salmonella, the cobB and yfiQ genes are expressed to a greater extent during exponential phase than during stationary phase, and different carbon sources affected their transcription (3). In E. coli, however, yfiQ is upregulated by CRP, a global regulator that controls gene transcription in response to cAMP levels, which rise in the cell as other preferred carbon sources are depleted (28). Our evidence confirms that yfiQ is similarly regulated by CRP in V. cholerae, despite contradictory findings in previous microarray studies (33, 34). We identified a putative CRP binding site in the yfiQ promoter, but a global chromatin immunoprecipitation sequencing (ChIP-Seq) analysis did not detect significant CRP binding in that location (32). Therefore, it is possible that CRP indirectly regulates yfiQ. Nevertheless, our results support a model in which depletion of preferred carbon sources and entry into stationary phase result in two countervailing regulatory mechanisms: (i) acs expression is increased, which flips the acetate switch and increases the flux of acetate through catabolic pathways, and (ii) CRP is activated, which subsequently increases yfiQ expression. High levels of YfiQ may then acetylate and inactivate the newly expressed enzymes required for acetate catabolism, including Acs. This multilayered regulatory strategy may precisely control flux through these metabolic pathways in response to nutrient availability and the energy state of the cell (28). In V. cholerae, acs is upregulated as cells enter stationary phase, but its expression is controlled predominantly via CrbS/R, a two-component signal transduction system that is absent from the E. coli and Salmonella genomes (16, 17). Our evidence suggests that CRP further activates the transcription of crbS and crbR, as well as acs (61). Thus, CRP activates the expression of multiple mechanisms that increase acs transcription but reduce Acs activity.
Acetylation modifies a sizeable proportion of the proteome in a number of bacterial pathogens, including Vibrio parahaemolyticus, Mycobacterium tuberculosis, Staphylococcus aureus, and V. cholerae, but these modifications have been directly linked to virulence only in Salmonella and E. coli (38, 39). Acetylation regulates the expression of Salmonella type III secretion system 1 by controlling the stability and DNA binding capability of a transcriptional regulator (40, 41). A second transcriptional regulator contributing to Salmonella virulence, PhoP of the PhoP/Q two-component system, is also subject to acetylation, which specifically reduces its DNA binding affinity (42). The E. coli transcriptional regulator RscB, which is involved in flagellar biosynthesis, cell division, and capsule biosynthesis, is susceptible to acetylation, which reduces its transcriptional capability (19, 43, 44). Acetylation may mediate the acid tolerance response in Salmonella, during which highly acidic conditions downregulate yfiQ to alter the tricarboxylic acid (TCA) cycle consumption of acetyl-CoA, stabilizing intracellular pH and improving survival (45). Because Salmonella must persist through the acidic environment of the stomach to reach the GI tract and spread to other organs such as the spleen, this mechanism may contribute to virulence in vivo, although this hypothesis has not been tested directly (45). Salmonella persistence specifically in macrophages is aided by a toxin-antitoxin system that alters the acetylation state of aminoacyl-tRNAs, which, in turn, lowers rates of protein synthesis to induce the “persister” phenotype (46, 47). In addition to targeting the aminoacyl-tRNAs, the acetylase toxin also directly modifies its cognate antitoxin protein, which then increases the activity of the toxin (46). In E. coli, chemotaxis may be regulated by acetylation of the CheY response regulator (48). These mechanisms highlight the myriad ways in which acetylation can affect pathogenesis in E. coli and Salmonella. We hypothesize that the V. cholerae cobB and yfiQ genes mediate virulence by directly regulating metabolite levels in the fly gut, based upon the observation that infection with the cobB deletion strain significantly reduced the storage of triglycerides in the enterocytes lining the fly GI tract. Previous studies have linked these changes in fat storage to reductions of both acetate and succinate levels (16, 21). Therefore, these genes may be modulating acetate alone, or both acetate and succinate as well as other active metabolites, in the context of the Drosophila GI tract. However, these examples highlight the potential for acetylation to affect a broad range of processes that could alter virulence in many ways.
A very recent study reported that the proteome of V. cholerae V52, a strain pathogenic to humans, is subject to acetylation (39). A number of global regulators important for both virulence and metabolic regulation are acetylated. These regulators include the AphB and TcpP transcription factors that control the expression of cholera toxin and toxin-coregulated pilus, via ToxT. However, V. cholerae SIO lacks the ctxAB and tcp gene clusters, and virulence in this strain appears to be entirely contingent upon the regulation of metabolite consumption. Intriguingly, Acs in V. cholerae strain V52 is acetylated, but not at the conserved lysine, as in Salmonella and E. coli (39). This observation is consistent with our own results in which mutation of AcsK609 or AcsL641, when introduced into a ΔcobB background, did not improve acetate metabolism or virulence in vivo, suggesting that these mutations could not prevent acetylation. The identification of alternative acetylation sites, however, implies that acetylation may be modifying Acs activity in V. cholerae by a novel mechanism. The physiological effects of these novel Acs acetylation sites on acetate metabolism and virulence should be confirmed and investigated via genetic, biochemical, and in vivo approaches.
Finally, the global metabolic regulator CRP was also subject to acetylation in V. cholerae V52 (39). We have confirmed that CRP is important for the regulation of yfiQ transcription. If CRP is acetylated by YfiQ to dampen its activity, this could enact a negative-feedback loop in which CRP activates expression of yfiQ, which then suppresses CRP function. However, the functional consequences of these specific acetylation sites in V. cholerae pathogenesis, survival, metabolism, and ecology await further investigation. Together, these studies on acetylation in V. cholerae reveal that this posttranslational modification is widespread within the proteome, has broad functional consequences for metabolic regulation, and is likely controlled, at least in part, by the cobB and yfiQ genes that we have identified here.
Bacterial modulation of metabolites can alter host health, physiology, and development. Alteration of succinate levels by the GI microbiota affects glucose homeostasis in mice (49), and microbiota-dependent alteration of acetate levels can have broad effects on host health (50, 51). A positive correlation between short-chain fatty acid levels and recovery from cholera has been observed for children (52), suggesting a possible link between metabolism and disease. Therefore, mechanisms of global regulation that modulate bacterial metabolic fluxes can drastically alter the effects of microbial colonization on the host. Because acetylation-dependent control of metabolism is widely conserved, the genes involved in regulating acetylation could impact host-microbe interactions across a diversity of systems.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. All experiments were performed in V. cholerae strain SIO, a nontoxigenic non-O1/non-O139 strain isolated just off the coast of San Diego, CA. Vibrio cholerae bacteria were incubated with shaking at 37°C with appropriate antibiotics in LB-Miller broth (Fisher), unless otherwise noted. V. cholerae bacteria were plated onto LB-Miller agar and stored at −80°C in 15% glycerol. Kanamycin was added to a final concentration of 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain | Description | Reference(s) |
|---|---|---|
| E. coli | ||
| MFDpir | MG1655 RP4-2-Tc::[ΔMu1::aac(3)IV-ΔaphA-Δnic35-ΔMu2::zeo] ΔdapA::(erm-pir) ΔrecA Aprar Zeor Ermr | 54 |
| DH5α λpir | F− Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 λ::pir | 58 |
| S17-1 λpir | RP4-2(Km::Tn7 Tc::Mu1) pro-82 λpir recA1 endA1 thiE1 hsdR17 creC510 | 59 |
| AP09 | E. coli WM5406/pHC001B | 53 |
| AP207 | E. coli MFDpir/pHC001B::Δacs | This study |
| AP1083 | E. coli MFDpir/pHC001B::ΔcobB | This study |
| AP1392 | E. coli MFDpir/pHC001B::ΔyfiQ | This study |
| AP302 | E. coli DH5α/pSRK-Km | 55, 56 |
| AP1870 | E. coli MFDpir/pSRK-Km::cobB | This study |
| AP1350 | E. coli MFDpir/pHC001B::acsK609R | This study |
| AP1351 | E. coli MFDpir/pHC001B::acsL641A | This study |
| AP1782 | E. coli S17-1 λpir/pDN001 (pBBRlux::PyfiQ) | This study |
| AP2027 | E. coli MFDpir/pCP001 (pBBRlux::PcobB-long) | This study |
| AP2023 | E. coli MFDpir/pCP002 (pBBRlux::PcobB-short) | This study |
| AP547 | E. coli MFDpir/pHC001B::Δcrp | This study |
| AP2006 | E. coli MFDpir/pHC001B::ΔacuC | This study |
| V. cholerae | ||
| AP95 | V. cholerae strain SIO; wild type | 60 |
| AP218 | V. cholerae strain SIO Δacs | This study |
| AP1150/1151 | V. cholerae strain SIO ΔcobB | This study |
| AP1401a | V. cholerae strain SIO ΔcobB/pSRK-Km | This study |
| AP1882 | V. cholerae strain SIO ΔcobB/pSRK-cobB | This study |
| AP1415 | V. cholerae strain SIO ΔyfiQ | This study |
| AP1427 | V. cholerae strain SIO ΔcobB→ΔyfiQ (cobB into yfiQ) | This study |
| AP1862 | V. cholerae strain SIO ΔyfiQ→ΔcobB (yfiQ into cobB) | This study |
| AP1346 | V. cholerae strain SIO acsL641A | This study |
| AP1348 | V. cholerae strain SIO ΔcobB acsL641A | This study |
| AP1344 | V. cholerae strain SIO acsK609R | This study |
| AP1345 | V. cholerae strain SIO ΔcobB acsK609R | This study |
| AP431 | V. cholerae strain SIO/pPT002 (pBBRlux::Pacs) | 17 |
| AP462 | V. cholerae strain SIO/pBBRlux | 17 |
| AP1787 | V. cholerae strain SIO Δcrp/pDN001 (pBBRlux::PyfiQ) | This study |
| AP1797 | V. cholerae strain SIO/pDN001 (pBBRlux::PyfiQ) | This study |
| AP2033 | V. cholerae strain SIO/pCP001 (pBBRlux::PcobB-long) | This study |
| AP2029 | V. cholerae strain SIO/pCP002 (pBBRlux::PcobB-short) | This study |
| AP2039 | V. cholerae strain SIO Δcrp/pCP001 (pBBRlux::PcobB-long) | This study |
| AP2035 | V. cholerae strain SIO Δcrp/pCP002 (pBBRlux::PcobB-short) | This study |
| AP2041 | V. cholerae strain SIO ΔacuC | This study |
| AP2049 | V. cholerae strain SIO ΔacuC ΔcobB | This study |
| AP2048 | V. cholerae strain SIO ΔacuC ΔyfiQ | This study |
| AP2061 | V. cholerae strain SIO ΔacuC ΔyfiQ ΔcobB | This study |
Bacterial mutant construction and complementation.
The deletion of the acs gene was constructed by using Gibson assembly (New England BioLabs) according the manufacturer's instructions. All other deletion and mutation constructs were generated by using a standard splicing by overlap extension (SOE) PCR protocol, as described previously for the environmental V. cholerae strain SIO (17). All primers are listed in Table 2. Altogether, deletions were constructed in acs (VC0298), cobB (VC1509), yfiQ (VCA0574), and crp (VC2614) (locus tags according to V. cholerae O1 El Tor N16961 assignments). To perform the SOE protocol, ∼750 to 1,100 bp on either side of the gene deletion or modification were amplified with Q5 high-fidelity DNA polymerase (New England BioLabs). The inner primers included an ∼18- to 21-bp region of overlap. For each of the gene deletions, the overlap sequence was 5′-TGCGGCCGCTCGTTA-3′, while primers for specific gene mutations included an overlapping region encompassing the point mutation being constructed. Primers outside the construct incorporated restriction enzyme sites, which are designated in Table 2. The PCR products were digested and ligated into the pHC001B plasmid (53). The resulting plasmids carrying the deletion constructs were transformed into E. coli DH5-α λpir cells or MFDpir cells (54), and the construct was sequenced. The correct plasmid was then conjugated from MFDpir cells into V. cholerae. Selection for double recombinants was accomplished by kanamycin and sucrose selection, as described previously (17). Integration of the correct construct was verified by PCR or sequencing. To generate the complementation plasmids in pSRK-Km carrying the cobB gene or the acs gene (55, 56), the gene was amplified by PCR using a forward primer that incorporated an NdeI site into the start codon. The PCR fragment was digested, ligated into the pSRK-Km vector, and transformed into E. coli SM10 λpir. The plasmid was then conjugated into V. cholerae via standard procedures.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Description |
|---|---|---|
| KL10_SIOdelcobB_P1 | GACAACTAGTTGGCGGCTTTAAGTGGTCTA | SOE deletion of cobB with SpeI |
| KL11_SIOdelcobB_P2 | TAACGAGCGGCCGCAGCCTGCTCCAGTCAGTATCA | SOE deletion of cobB with tag |
| KL12_SIOdelcobB_P3 | TGCGGCCGCTCGTTAGCGAAAGAGAGTGCAGC | SOE deletion of cobB with tag |
| KL13_SIOdelcobB_P4 | CATTGGATCCCAAAATTGCGGGCATCATGG | SOE deletion of cobB with BamHI |
| KL48_SIOdelyfiQ.3_P1 | GACAACTAGTCCACCGCACAAAAGTAGGAT | SOE deletion of yfiQ with SpeI |
| KL49_SIOdelyfiQ.3_P2 | TAACGAGCGGCCGCAGGGTCTAAGCAGTTGATTCAA | SOE deletion of yfiQ with tag |
| KL50_SIOdelyfiQ.3_P3 | TGCGGCCGCTCGTTATTTGCCGTCGATATTCATT | SOE deletion of yfiQ with tag |
| KL51_SIOdelyfiQ.3_P4 | CATTGAGCTCAGGAGAGAGCATTGTTAAGTCCA | SOE deletion of yfiQ with SacI |
| KL44_AcsL641A_P1 | GACAACTAGTTGTATGGACCACTGGCGAAT | SOE mutation of AcsL641A with SpeI |
| KL45_AcsL641A_P2 | TTGTGCTTTTTCGGCAATCGCGCGGTCAACCACGCTTGG | SOE mutation of AcsL641A complete overlap |
| KL46_AcsL641A_P3 | CCAAGCGTGGTTGACCGCGCGATTGCCGAAAAAGCACAA | SOE mutation of AcsL641A complete overlap |
| KL47_AcsL641A_P4 | CATTGGATCCATTTCCAGTTCTGCGATGCC | SOE mutation of AcsL641A with BamHI |
| KL40_AcsK609R_P1 | GACAACTAGTTGTATGGACCACTGGCGAAT | SOE mutation of AcsK609R with SpeI |
| KL41_AcsK609R_P2 | CAAAATACGGCGCATAATTCTACCTGAACGGGTTTTCGG | SOE mutation of AcsK609R complete overlap |
| KL42_AcsK609R_P3 | CCGAAAACCCGTTCAGGTAGAATTATGCGCCGTATTTTG | SOE mutation of AcsK609R complete overlap |
| KL43_acsK609R_P4 | CATTGGATCCGGCACCTAAACCGCAAATCA | SOE mutation of AcsK609R with BamHI |
| MS37_delVC2614_P1 | GACAACTAGTCCAGATGCCCGGTACGTTTAC | SOE mutation of crp with SpeI |
| MS33_delVC2614_P2 | GGTTTCGCGAGAACAGCCGTGAAAGAAACCACTCTAGTG | SOE mutation of crp complete overlap |
| MS34_delVC2614_P3 | CACTAGAGTGGTTTCTTTCACGGCTGTTCTCGCGAAACC | SOE mutation of crp complete overlap |
| MS39_delVC2614_P4 | CATTCTCGAGTGCATCAACTCCTACAAGAAG | SOE mutation of crp with XhoI |
| DN05_pYfiQ_F | GACAACTAGTCACTGTCTGGAGCTTTGACAAC | Cloning of yfiQ promoter with SpeI |
| DN06_pYfiQ_R | CATTGGATCCCAAACGGCAGGATAATAGGG | Cloning of yfiQ promoter with BamHI |
| MMS13_acs_GP1 | actcactatagggcccccccCGGCTACCACATTCGTTACG | Gibson deletion of acs, primer 1 |
| MMS14_acs_GP2 | GGCAATCAGGCGGTCAACCGGATAAATATGGGCTTCAC | Gibson deletion of acs, primer 2 |
| MMS15_acs_GP3 | GTGAAGCCCATATTTATCCGGTTGACCGCCTGATTGCC | Gibson deletion of acs, primer 3 |
| MMS16_acs_GP4 | ggcggccgctctagaaCGCGGTTAGATTGCAGATGT | Gibson deletion of acs, primer 4 |
| DN01_pCobB_F | GACAACTAGTCCAAGATGGTGAGATAGCGAAT | Cloning of cobB long promoter with SpeI |
| DN02_pCobB_R | CATTGGATCCGTGTCGCCACATCTTCGATT | Cloning of cobB promoters with BamHI |
| CP37_pCobB_shortF | GACAACTAGTGGAGCCATTTCAATTTGGAT | Cloning of cobB short promoter with SpeI |
| GR09_VC2042_delSOE_P1 | CATGGATCCGCCAGTCTCTTTATGAAAAGC | SOE deletion of acuC homolog, with BamHI |
| GR10_VC2042_delSOE_P2 | TAACGAGCGGCCGCAGATCATTTTGCTCTGCTCTGG | SOE deletion of acuC homolog, with tag |
| GR11_VC2042_delSOE_P3 | TGCGGCCGCTCGTTATGAGGCGAAATTATGACCGTA | SOE deletion of acuC homolog, with tag |
| GR12_VC2042_delSOE_P4 | GACAACTAGTTTACGCAGTGCATACAACG | SOE deletion of acuC homolog, with SpeI |
Restriction sites are underlined, the tag (additional base pairs) that enables overlapping PCR for gene deletions is in boldface italic type, and primers that create point mutations include the specific mutation are in boldface type. Primers that overlap each other completely are indicated. For Gibson mutation, overlap with the plasmid is indicated in lowercase type.
Growth assays.
To monitor growth on acetate minimal medium, V. cholerae was streaked for single colonies, inoculated into LB medium supplemented with appropriate antibiotics (if necessary), and incubated with shaking overnight at 37°C. The following day, cells were centrifuged at maximum speed for 3 min, the supernatant was removed, and the cells were resuspended in an equal volume of M63 minimal medium (Amresco). The resuspended cells were then inoculated into M63 medium supplemented with 10 mM acetate to an initial optical density at 600 nm (OD600) of 0.01. From this dilution, 120 μl was added to each of at least three wells in a sterile, flat-bottomed, 96-well plate and incubated for 48 h at 37°C with shaking in a multimode plate reader (Molecular Devices), while the OD600 was monitored every hour. Individual measurements were averaged, and results of assays representative of data from at least two independent replicates are shown.
Acetate concentration in medium.
To monitor acetate concentrations in culture, V. cholerae was streaked for single colonies and grown overnight for 16 h. From this culture, 500 μl was inoculated into 50 ml of LB broth in a 125-ml Erlenmeyer flask in duplicate. At the appropriate times, the OD600 of the culture was measured using a spectrophotometer (catalog number 6320D; Jenway), and a 1-ml sample was removed, centrifuged, filter sterilized, and heat treated at 80°C for 20 min, according to the manufacturer's instructions. Samples were stored at −20°C until they were analyzed with an acetic acid assay kit (catalog no. K-ACETRM; Megazyme), and acetate was measured according to the manufacturer's instructions.
Fly survival and colonization.
V. cholerae strains were streaked for single colonies from glycerol stocks, grown overnight in LB broth, and then diluted 1:10 and added to a cellulose acetate Drosophila vial closure (Genesee Scientific) at the bottom of a standard fly vial. Between 6 and 10 flies were added to each vial, with each assay including at least two vials, and most often three vials, per strain. All survival assays were repeated on at least two separate occasions, unless otherwise indicated. Statistical significance of fly survival curves was measured with the log rank assay implemented in GraphPad Prism (GraphPad, La Jolla, CA). To assess colonization, live flies were removed from vials after 24 and 48 h, homogenized in 300 μl of phosphate-buffered saline (PBS), serially diluted, and plated onto LB agar. After overnight incubation at 37°C, plates with colonies numbering between 30 and 300 were counted to assess CFU per milliliter. To ensure that only V. cholerae colonies were counted, flies infected with sterile LB medium were similarly homogenized and plated, and no colonies were apparent on these plates after 24 h of incubation.
Triglyceride storage determination.
To measure triglycerides, flies were infected with V. cholerae for 48 h. The midgut was then dissected in PBS and fixed in 4% paraformaldehyde in PBS with 0.2% Triton X-100 (PBS-T) for 1 h at room temperature or overnight at 4°C. Midguts were then washed three times in PBS-T, followed by incubation in 1 μg/ml BODIPY 493/503 dye (ThermoFisher/Molecular Probes) and 1 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) in PBS-T for 45 min (21). Midguts were then washed three times in PBS-T and mounted in Vectashield. Slides were stored at 4°C prior to visualization on a Nikon C2 confocal microscope. Puncta were quantified with Fiji, using images taken with a 60× objective, and images were coded so that the researcher performing the quantification was blind as to the bacterial strain with which the fly had been infected. In each of three independent trials, two to four images from each of at least five to six midguts harvested from flies infected with either wild-type V. cholerae SIO or V. cholerae ΔcobB or uninfected flies were quantified. Images were similarly quantified for the V. cholerae Δacs mutant in two of these three independent trials. To assess puncta per cell, the number of puncta was divided by the number of nuclei (as assessed by DAPI) in the image.
Measurement of yfiQ and cobB promoter activities.
In order to assess whether the yfiQ promoter is controlled by crp, a 581-bp segment of the yfiQ promoter was cloned into the pBBRlux plasmid (57). Because the transcriptional start site was unknown, the segment was selected to extend 423 bp upstream and 158 bp downstream of the putative start codon. The yfiQ gene is not predicted to fall within an operon, as the gene upstream of yfiQ, VCA0573, is transcribed in the opposite direction. The resulting plasmid, pDN001, was introduced into both the wild-type V. cholerae SIO strain and a strain carrying a deletion in crp by conjugation from E. coli.
Similarly, in order to assess whether the cobB promoter is controlled by crp, two segments of the cobB promoter were each cloned into the pBBRlux plasmid. The longer of the two was selected to extend 442 bp upstream and 106 bp downstream of the putative start codon. However, a poorly conserved crp binding site was detected within this region. To create a promoter lacking this site, a second promoter segment was designed to reach 235 bp upstream and 106 bp downstream of the putative start codon, covering a total of 341 bp. The resulting plasmids, pCP001 and pCP002, respectively, were introduced into both the wild-type V. cholerae SIO strain and a strain carrying a deletion in crp by conjugation.
To detect bioluminescence, single colonies were inoculated into LB broth with chloramphenicol (5 μg/ml), which were grown for not more than 14 h at 37°C with shaking. The cultures were then diluted 1:500 into fresh LB broth with chloramphenicol, and 120 μl was added to sterile, black, flat, clear-bottomed, 96-well plates that isolate luminescence from adjacent wells (BRAND GMBH + CO KG, Germany). Strains were incubated overnight with shaking, while luminescence and absorption were measured in succession with a 96-well multimode plate reader (Molecular Devices).
ACKNOWLEDGMENTS
We thank Mariame Sylla (Department of Biology, Amherst College [currently at Harvard Medical School]) for construction of the V. cholerae strain carrying the crp deletion. We also thank Maureen Manning, Lori Nichols, and Valerie Hale, all from Amherst College, for additional support and assistance with this project. We are grateful to the three anonymous reviewers, whose comments significantly strengthened the manuscript.
This work was funded by Amherst College, the Amherst College Summer Undergraduate Research Fellowship Program to K.L. and E.F., and the National Science Foundation (MCB RUI 1715956) to A.E.P. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Wolfe AJ. 2005. The acetate switch. Microbiol Mol Biol Rev 69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakayasu ES, Burnet MC, Walukiewicz HE, Wilkins CS, Shukla AK, Brooks S, Plutz MJ, Lee BD, Schilling B, Wolfe AJ, Müller S, Kirby JR, Rao CV, Cort JR, Payne SH. 2017. Ancient regulatory role of lysine acetylation in central metabolism. mBio 8:e01894-. doi: 10.1128/mBio.01894-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning Z-B, Zeng R, Xiong Y, Guan K-L, Zhao S, Zhao G-P. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie L, Wang X, Zeng J, Zhou M, Duan X, Li Q, Zhang Z, Luo H, Pang L, Li W, Liao G, Yu X, Li Y, Huang H, Xie J. 2015. Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int J Biochem Cell Biol 59:193–202. doi: 10.1016/j.biocel.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Weinert BT, Iesmantavicius V, Wagner SA, Schölz C, Gummesson B, Beli P, Nyström T, Choudhary C. 2013. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell 51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn ML, Zemaitaitis B, Hu LI, Sahu A, Sorensen D, Minasov G, Lima BP, Scholle M, Mrksich M, Anderson WF, Gibson BW, Schilling B, Wolfe AJ. 2014. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One 9:e94816. doi: 10.1371/journal.pone.0094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, Hwa T. 2015. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallows WC, Lee S, Denu JM. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A 103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. 2006. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A 103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 11.Starai VJ, Escalante-Semerena JC. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol 340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Gardner JG, Escalante-Semerena JC. 2009. In Bacillus subtilis, the sirtuin protein deacetylase, encoded by the srtN gene (formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl coenzyme A synthetase. J Bacteriol 191:1749–1755. doi: 10.1128/JB.01674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baeza J, Dowell JA, Smallegan MJ, Fan J, Amador-Noguez D, Khan Z, Denu JM. 2014. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem 289:21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Studer SV, Mandel MJ, Ruby EG. 2008. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J Bacteriol 190:5915–5923. doi: 10.1128/JB.00148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan M, Schwartzman JA, Dunn AK, Lu Z, Ruby EG. 2015. A single host-derived glycan impacts key regulatory nodes of symbiont metabolism in a coevolved mutualism. mBio 6:e00811-. doi: 10.1128/mBio.00811-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hang S, Purdy AE, Robins WP, Wang Z, Mandal M, Chang S, Mekalanos JJ, Watnick PI. 2014. The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 16:592–604. doi: 10.1016/j.chom.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob K, Rasmussen A, Tyler P, Servos MM, Sylla M, Prado C, Daniele E, Sharp JS, Purdy AE. 2017. Regulation of acetyl-CoA synthetase transcription by the CrbS/R two-component system is conserved in genetically diverse environmental pathogens. PLoS One 12:e0177825. doi: 10.1371/journal.pone.0177825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao K, Chai X, Marmorstein R. 2004. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J Mol Biol 337:731–741. doi: 10.1016/j.jmb.2004.01.060. [DOI] [PubMed] [Google Scholar]
- 19.Castaño-Cerezo S, Bernal V, Post H, Fuhrer T, Cappadona S, Sánchez-Díaz NC, Sauer U, Heck AJR, Altelaar AFM, Cánovas M. 2014. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol Syst Biol 10:762. doi: 10.15252/msb.20145227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumari S, Tishel R, Eisenbach M, Wolfe AJ. 1995. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol 177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamareddine L, Wong ACN, Vanhove AS, Hang S, Purdy AE, Kierek-Pearson K, Asara JM, Ali A, Morris JG Jr, Watnick PI. 2018. Activation of Vibrio cholerae quorum sensing promotes survival of an arthropod host. Nat Microbiol 3:243–252. doi: 10.1038/s41564-017-0065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purdy AE, Watnick PI. 2011. Spatially selective colonization of the arthropod intestine through activation of Vibrio cholerae biofilm formation. Proc Natl Acad Sci U S A 108:19737–19742. doi: 10.1073/pnas.1111530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanhove AS, Hang S, Vijayakumar V, Wong AC, Asara JM, Watnick PI. 2017. Vibrio cholerae ensures function of host proteins required for virulence through consumption of luminal methionine sulfoxide. PLoS Pathog 13:e1006428. doi: 10.1371/journal.ppat.1006428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamareddine L, Robins WP, Berkey CD, Mekalanos JJ, Watnick PI. 2018. The Drosophila immune deficiency pathway modulates enteroendocrine function and host metabolism. Cell Metab 28:449.E5–462.E5. doi: 10.1016/j.cmet.2018.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blow NS, Salomon RN, Garrity K, Reveillaud I, Kopin A, Jackson FR, Watnick PI. 2005. Vibrio cholerae infection of Drosophila melanogaster mimics the human disease cholera. PLoS Pathog 1:e8. doi: 10.1371/journal.ppat.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto H, Higuchi M, Koike M, Ode H, Pinak M, Bunta JK, Nemoto T, Sakudoh T, Honda N, Maekawa H, Saito K, Tsuchida K. 2012. A possible overestimation of the effect of acetylation on lysine residues in KQ mutant analysis. J Comput Chem 33:239–246. doi: 10.1002/jcc.21956. [DOI] [PubMed] [Google Scholar]
- 27.Starai VJ, Gardner JG, Escalante-Semerena JC. 2005. Residue Leu-641 of acetyl-CoA synthetase is critical for the acetylation of residue Lys-609 by the protein acetyltransferase enzyme of Salmonella enterica. J Biol Chem 280:26200–26205. doi: 10.1074/jbc.M504863200. [DOI] [PubMed] [Google Scholar]
- 28.Castaño-Cerezo S, Bernal V, Blanco-Catalá J, Iborra JL, Cánovas M. 2011. cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol Microbiol 82:1110–1128. doi: 10.1111/j.1365-2958.2011.07873.x. [DOI] [PubMed] [Google Scholar]
- 29.Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 30.Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J Mol Biol 293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 31.Ebright RH, Ebright YW, Gunasekera A. 1989. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res 17:10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manneh-Roussel J, Haycocks JRJ, Magán A, Perez-Soto N, Voelz K, Camilli A, Krachler A-M, Grainger DC. 2018. cAMP receptor protein controls Vibrio cholerae gene expression in response to host colonization. mBio 9:e00966-. doi: 10.1128/mBio.00966-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fong JCN, Yildiz FH. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J Bacteriol 190:6646–6659. doi: 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang W, Pascual-Montano A, Silva AJ, Benitez JA. 2007. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 35.Chan CH, Garrity J, Crosby HA, Escalante-Semerena JC. 2011. In Salmonella enterica, the sirtuin-dependent protein acylation/deacylation system (SDPADS) maintains energy homeostasis during growth on low concentrations of acetate. Mol Microbiol 80:168–183. doi: 10.1111/j.1365-2958.2011.07566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinert BT, Satpathy S, Hansen BK, Lyon D, Jensen LJ, Choudhary C. 2017. Accurate quantification of site-specific acetylation stoichiometry reveals the impact of sirtuin deacetylase CobB on the E. coli acetylome. Mol Cell Proteomics 16:759–769. doi: 10.1074/mcp.M117.067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crosby HA, Pelletier DA, Hurst GB, Escalante-Semerena JC. 2012. System-wide studies of N-lysine acetylation in Rhodopseudomonas palustris reveal substrate specificity of protein acetyltransferases. J Biol Chem 287:15590–15601. doi: 10.1074/jbc.M112.352104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren J, Sang Y, Lu J, Yao Y-F. 2017. Protein acetylation and its role in bacterial virulence. Trends Microbiol 25:768–779. doi: 10.1016/j.tim.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Jers C, Ravikumar V, Lezyk M, Sultan A, Sjöling Å, Wai SN, Mijakovic I. 2018. The global acetylome of the human pathogen Vibrio cholerae V52 reveals lysine acetylation of major transcriptional regulators. Front Cell Infect Microbiol 7:537. doi: 10.3389/fcimb.2017.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sang Y, Ren J, Ni J, Tao J, Lu J, Yao Y-F. 2016. Protein acetylation is involved in Salmonella enterica serovar Typhimurium virulence. J Infect Dis 213:1836–1845. doi: 10.1093/infdis/jiw028. [DOI] [PubMed] [Google Scholar]
- 41.Sang Y, Ren J, Qin R, Liu S, Cui Z, Cheng S, Liu X, Lu J, Tao J, Yao Y-F. 2017. Acetylation regulating protein stability and DNA-binding ability of HilD, thus modulating Salmonella Typhimurium virulence. J Infect Dis 216:1018–1026. doi: 10.1093/infdis/jix102. [DOI] [PubMed] [Google Scholar]
- 42.Ren J, Sang Y, Tan Y, Tao J, Ni J, Liu S, Fan X, Zhao W, Lu J, Wu W, Yao Y-F. 2016. Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog 12:e1005458. doi: 10.1371/journal.ppat.1005458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thao S, Chen C-S, Zhu H, Escalante-Semerena JC. 2010. Nε-Lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS One 5:e15123. doi: 10.1371/journal.pone.0015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu LI, Chi BK, Kuhn ML, Filippova EV, Walker-Peddakotla AJ, Bäsell K, Becher D, Anderson WF, Antelmann H, Wolfe AJ. 2013. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J Bacteriol 195:4174–4186. doi: 10.1128/JB.00383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren J, Sang Y, Ni J, Tao J, Lu J, Zhao M, Yao Y-F. 2015. Acetylation regulates survival of Salmonella enterica serovar Typhimurium under acid stress. Appl Environ Microbiol 81:5675–5682. doi: 10.1128/AEM.01009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.VanDrisse CM, Parks AR, Escalante-Semerena JC. 2017. A toxin involved in Salmonella persistence regulates its activity by acetylating its cognate antitoxin, a modification reversed by CobB sirtuin deacetylase. mBio 8:e00708-17. doi: 10.1128/mBio.00708-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheverton AM, Gollan B, Przydacz M, Wong CT, Mylona A, Hare SA, Helaine S. 2016. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol Cell 63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barak R, Prasad K, Shainskaya A, Wolfe AJ, Eisenbach M. 2004. Acetylation of the chemotaxis response regulator CheY by acetyl-CoA synthetase purified from Escherichia coli. J Mol Biol 342:383–401. doi: 10.1016/j.jmb.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 49.De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. 2016. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab 24:151–157. doi: 10.1016/j.cmet.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Marques FZ, Nelson E, Chu P-Y, Horlock D, Fiedler A, Ziemann M, Tan JK, Kuruppu S, Rajapakse NW, El-Osta A, Mackay CR, Kaye DM. 2017. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice: clinical perspective. Circulation 135:964–977. doi: 10.1161/CIRCULATIONAHA.116.024545. [DOI] [PubMed] [Google Scholar]
- 51.Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. 2016. Acetate mediates a microbiome–brain–β-cell axis to promote metabolic syndrome. Nature 534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monira S, Hoq MM, Chowdhury AKA, Suau A, Magne F, Endtz HP, Alam M, Rahman M, Pochart P, Desjeux J-F, Alam NH. 2010. Short-chain fatty acids and commensal microbiota in the faeces of severely malnourished children with cholera rehydrated with three different carbohydrates. Eur J Clin Nutr 64:1116–1124. doi: 10.1038/ejcn.2010.123. [DOI] [PubMed] [Google Scholar]
- 53.Borisova SA, Christman HD, Metcalf MEM, Zulkepli NA, Zhang JK, van der Donk WA, Metcalf WW. 2011. Genetic and biochemical characterization of a pathway for the degradation of 2-aminoethylphosphonate in Sinorhizobium meliloti 1021. J Biol Chem 286:22283–22290. doi: 10.1074/jbc.M111.237735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrieres L, Hemery G, Nham T, Guerout A-M, Mazel D, Beloin C, Ghigo J-M. 2010. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan SR, Gaines J, Roop RM, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan F, Liu Z, Jabeen N, Birdwell LD, Zhu J, Kan B. 2014. Enhanced interaction of Vibrio cholerae virulence regulators TcpP and ToxR under oxygen-limiting conditions. Infect Immun 82:1676–1682. doi: 10.1128/IAI.01377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 58.Merrell DS, Camilli A. 2000. Regulation of Vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J Bacteriol 182:5342–5350. doi: 10.1128/JB.182.19.5342-5350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 60.Purdy A, Rohwer F, Edwards R, Azam F, Bartlett DH. 2005. A glimpse into the expanded genome content of Vibrio cholerae through identification of genes present in environmental strains. J Bacteriol 187:2992–3001. doi: 10.1128/JB.187.9.2992-3001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muzhingi I, Prado C, Sylla M, Diehl FF, Nguyen DK, Servos MM, Ramos SF, Purdy AE. 17 September 2018. Modulation of CrbS-dependent activation of the acetate switch in Vibrio cholerae. J Bacteriol doi: 10.1128/JB.00380-18. [DOI] [PMC free article] [PubMed] [Google Scholar]