Abstract
AKR1B10 is an aldose reductase (AR) homologue overexpressed in liver cancer and various forms of that enzyme in carcinomas catalyze the reduction of anticancer drugs, potential cytostatic drug, and DL-glyceraldehyde but do not catalyze the reduction of glucose. Kinetic parameters for wild-type and C299S mutant AKR1B10 indicate that substitution of serine for cysteine at position 299 reduces the affinity of this protein for DLglyceraldehyde and enhances its catalytic activity. Fibrates suppress peroxisome proliferation and the development of liver cancer in human. Here we report the potency of fibrate-mediated inhibition of the carbonyl reduction catalyzed by wild-type and C299S mutant AKR1B10 and compare it with known AR inhibitors. Wild-type AKR1B10-catalyzed carbonyl reduction follows pure non-competitive inhibition kinetics using zopolrestat, EBPC or sorbinil, whereas fenofibrate, Wy 14,643, ciprofibrate and fenofibric acid follow mixed non-competitive inhibition kinetics. In contrast, catalysis of reaction by the C299S AKR1B10 mutant is not inhibited by sorbinil and EBPC. Despite these differences, the C299S AKR1B10 mutant still manifests kinetics similar to the wild-type protein with other fibrates including zopolrestat, fenofibrate, Wy 14,346, gemfibrozil and ciprofibrate that show mixed non-competitive inhibition kinetics. The reaction of the mutant AKR1B10 is inhibited by fenofibric acid, but manifests pure non-competitive inhibition kinetics that are different from those demonstrated for the wild-type enzyme.
Keywords: AKR1B10, Fenofibrate, Cancer, Liver, Lung
1. Introduction
Aldo–ketoreductase(AKR)proteinfamilymember,AKR1B10, is overexpressed in most cases of squamous cell carcinoma (SCC) and adenocarcinoma both of which are associated with smoking (Fukumoto et al., 2005). AKR1B10 belongs to AR subfamily (AKR1B) and was discovered as an enzyme overexpressed in human liver cancers (Cao et al., 1998; Hyndman and Flynn, 1998; Jez et al., 1997; Scuric et al., 1998). Furthermore, the anticancer drugs daunorubicin, which is used in the treatment of lung cancer, and oracin, a potential cytostatic drug for oral use, which is, at present, in phase II of clinical trials, are reduced by AKR1B10 (Martin et al., 2006). The carbonyl groups in the daunorubicin and oracin are converted to the corresponding alcohols and inactivated by AKR1B10. The question arises concerning how drugs containing a carbonyl moiety can be used for successful chemotherapy of liver cancers in the presence of overexpressed AKR1B10? To address this issue we explored AR inhibitors, which may provide insight for the development of combination drugs to solve that problem.
Using a novel approach similar to fragment-based, structure-guided inhibitor design we have demonstrated that AR is a target of action for several fibratederivatives(Balendiran and Rajkumar, 2005; Balendiran et al., 2007; Klemin et al., 2006). Fibrates, which are also known as peroxisome proliferators, lead to the development of liver tumors in rats and mice (Hess et al., 1965; Reddyetal.,1980).However,humans appear to beresistant to the induction of peroxisome proliferation and the development of liver cancer by fibrates (Gariot et al., 1983; Gonzalez, 2002). Understanding the role of AKR1B10 in fibrate action is critical since: 1) AKR1B10 is a member of AKR family which shows a high sequence homology with AR. 2) AKR1B10 and AR like molecules are overexpressed in liver and lung cancers as well as hepatocellular and squamous cell carcinomas.
Residue Cys299 in AKR1B10, equivalent to the Cys298 found in the active site vicinity of AR, is conserved in many members of aldo–keto reductase family of proteins. Several lines of evidence distinguish Cys298 as an important regulatory site on AR. In AR, conversion of Cys298 to serine (C298S) resulted in an enzyme form that is resistant to modification with reagents that are known to cause functional changes in enzyme activity (Bohren and Gabbay, 1993; Petrash et al., 1992). Residue Cys298 of AR is also a site for thiolation by oxidized glutathione (Cappiello et al., 1996) and glutath iolated AR is catalytically inactive (Cappiello et al., 1996). The carbonyl containing compound, 4-hydroxy-2-nonenal (HNE) modifies AR predominantly at the Cys298 position, resulting in an enzyme form with reduced sensitivity to AR inhibitors. Therefore, the residue Cys299 in AKR1B10 may play a sensitive role in the inhibition properties of compounds.
Several AR inhibitors, for example sorbinil, have reached human clinical trials,but have been with drawn due to adverse side effects (Group, 1990; Vander Jagt et al., 1996). The adverse side effects are suspected to occur via a closely-related enzyme of the AKR family, aldehyde reductase (AKR1A1, EC 1.1.1.2) (Sato and Kador, 1990; Vander Jagt et al., 1996). Aldehyde reductase and AR are functionally linked through broad and overlapping substrate specificity (Bohrenetal.,1989).Most importantly many AR inhibitors inhibit aldehyde reductase, consequently impeding proper AR. The striking difference between AR and aldehyde reductase is Cys298. Since Cys298 is not conserved in aldehyde reductase, understanding the contribution of the related Cys299 residue in AK1B10 will not only offer an indirect approach to study the side effects caused by aldehyde reductase against the potential inhibitors but will also help evaluate the regulatory role of this residue. Therefore, understanding the contribution of the Cys299 residue is essential in the rational design of a compound that specifically regulates AKR1B10.
In the present study we have characterized the potential of several fibrate derivatives in the inhibition of carbonyl reduction activity of purified recombinant human wild-type and the C299S mutant form of AKR1B10 using DL-glyceraldehyde as the substrate. We have also tested the effectiveness of these compounds to inhibit the AKR1B10-catalyzed reduction of the anticancer drug daunorubicin.
2. Materials and methods
All the reagents used in the study were obtained from SigmaAldrich Chemical Company (St. Louis, USA) with the exception of EBPC, which was purchased from Tocris, USA, and sorbinil and zopolrestat were gift from Pfizer. Fenofibric acid was prepared and characterized following the procedures we reported earlier (Rath et al., 2005).
2.1. Expression and purification of recombinant wild-type and C299S mutant AKR1B10
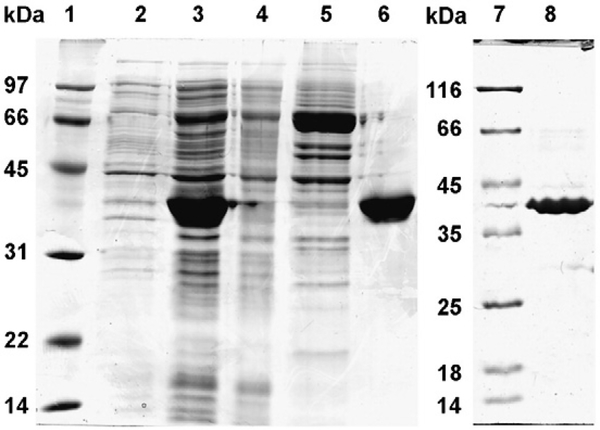
AKR1B10 gene in pQE-70 (QIAGEN) vector was transformed in SG13009 cells (QIAGEN, Valencia, USA). The cells were grown in Luria–Bertani broth containing ampicillin (100 μg/ml) and kanamycin (25 μg/ml) with constant shaking to reach the OD600=0.6–0.8 at 37 °C at 240 rpm. The protein expression was induced by supplementing 1 mM isopropyl-1thio-galactopyranoside (IPTG) in the culture medium. The cells were harvested after 5 h by centrifugation (6000 g, 10 min) and resuspended in 20 mM Tris–HCl buffer (pH 7.4) containing 100 mM NaCl, 10 mM imidazole and 1 mM 2-mercaptoethanol and lysed by sonication. Wild-type AKR1B10 was isolated from the lysate separated by centrifugation at 10,000 g for 45 min at 4 °C. The supernatant containing hexa-HisAKR1B10 was incubated for 1–2 h by constant gentle mixing with Talon metal affinity matrix (Clontech, Mountain View, USA), later matrix slurry was passed through column and washed with 20 mM Tris buffer (pH 7.4) having 100 mM NaCl and 1.0 mM 2-mercaptoethanol. The protein was eluted with 150 mM imidazole in 20 mM Tris buffer (pH 7.4) containing 100 mM NaCl and 1 mM 2-mercaptoethanol and dialyzed in the 20 mM Tris buffer (pH 7.4) containing 1 mM 2-mercaptoethanol. His-AKR1B10 wild-type protein was further purified by anion exchange on DEAE Sephadex A25 column by negative binding with DEAE Sephadex A25 matrix. The concentration of the AKR1B10 proteins was determined by the Bradford assay (Bio-Rad, Hercules, USA) (Bradford, 1976), the purity was assessed by SDS-PAGE (Fig. 1) and the enzyme activity was determined by using 10 mM DL-glyceraldehyde and 0.2 mM NADPH as substrate and cofactor, respectively. Recombinant C299S mutant AKR1B10 was overexpressed and purified (Fig. 1) following the procedures described above for the wild-type AKR1B10.
Fig. 1.
Purification of recombinant wild-type and C298S mutant AKR1B10. Lane assignments are as follows: 1) protein marker, 2) extract of whole cells before induction, 3) soluble proteins after cell lysis, 4) column flow-through fraction, 5) column wash fraction, 6) eluted AKR1B10, wild-type, 7) marker proteins and 8) eluted C299S AKR1B10 mutant.
2.2. Kinetics of DL-glyceraldehyde reduction by AKR1B10
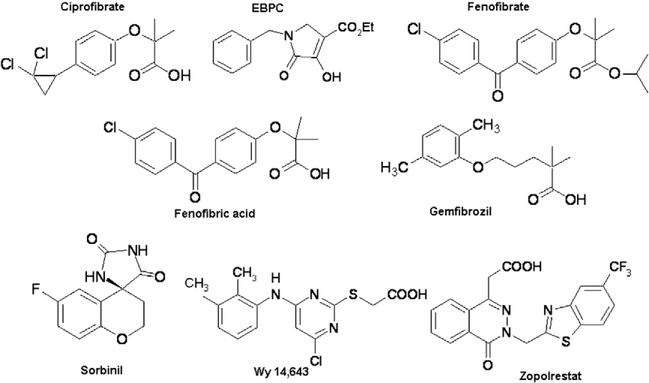
The carbonyl reduction activity of his-tagged AKR1B10 wild-type and C299S mutant proteins were monitored spectrophotometrically, by measuring decrease in the absorbance of the cofactor NADPH at 340 nm (Balendiran and Rajkumar, 2005; Crosas et al., 2003; Nishimura et al., 1991) and 25 °C with time course for 3 min. The assay was carried out in 100 mM sodium phosphate buffer (pH 7.5) using 0.2 mM NADPH, 0.3 μM (wild-type and C299S mutant) AKR1B10, and varied concentrations (0.5–7.5 mM for wild-type and 1.0– 100 mM for C299S) of DL-glyceraldehyde and inhibitors (zopolrestat, fenofibrate, Wy 14,643, sorbinil, ciprofibrate, fenofibric acid, gemfibrozil, EBPC (Fig. 2)). One unit of AKR1B10 enzyme activity is defined as μmoles of NADPH oxidized/min.
Fig. 2.
Chemical names and structures of various inhibitors used for AKR1B10 wild-type and mutant proteins inhibition studies. Ciprofibrate for 2-[p-(2,2dichlorocyclopropyl)-phenoxy]-2-methylpropanoic acid, EBPC for ethyl-1-benzyl-3-hydroxy-2(5H)-oxopyrrole-4-carboxylate, fenofibrate for the isopropyl ester of 2-[4-(4-chlorobenzoyl)-phenoxy]-2-methylpropanoic acid, fenofibric acid for 2-[4-(4-chlorobenzoyl)-phenoxy]-2-methylpropanoic acid, gemfibrozil for 2,2dimethyl-5-[2,5-dimethylphenoxy]-pentanoic acid, sorbinil for 2,4-dihydro-6-fluorospiro[4H-1-benzopyran-4,4′-imidazolidine-]-2′,5′-dione, Wy 14,643 for 4-chloro6-(2,3-xylidino)-2-pyrimidinylthioacetic acid and zopolrestat for 3,4-dihydro-4-oxo-3-{[5-(trifluoromethyl)-2-benzothiazolyl]methyl}−1-phthalazineacetic acid.
2.3. Analysis of kinetics data
The glyceraldehyde reduction kinetics was analyzed according to the Michaelis–Menten model. The Michaelis–Menten constant, Km, the maximum velocity, Vmax, for the reactions catalyzed by AKR1B10 were determined by plotting rate of carbonyl reduction, v versus substrate concentration, S using the Eq. (1).
| (1) |
The inhibition of the reduction of DL-glyceraldehyde by various inhibitors was fit to competitive, non-competitive and uncompetitive models of inhibition given by the Eqs. (2), (3) and (4) respectively, and the corresponding inhibition constants were determined by fitting the kinetics data in the following equations.
| (2) |
| (3) |
| (4) |
where v is the initial rate of reaction, Vmax is the maximum initial velocity for the uninhibited reaction at saturated substrate concentration, Km the Michaelis–Menten constant, S and I are the concentrations of substrate and inhibitor respectively. Kis is the slope inhibition constant and Kii is the intercept inhibition constant.
2.4. Determination of IC50 of AKR1B10 inhibitors
The IC50-value of the inhibitors were determined using the assay mixture containing 0.1 M sodium phosphate buffer (pH 7.5), 7.5 mM DL-glyceraldehyde, 0.2 mM NADPH, 0.3 μM AKR1B10 wild-type protein and varying concentrations of inhibitors depending on their inhibition potency. In the case of the C299S mutant IC50 was determined at 50 mM of DL-glyceraldehyde by varying the concentrations of various inhibitors. The IC50-values were determined by nonlinear regression analysis of the percent inhibition plotted versus the log of the inhibitor concentration. Values were expressed as the mean±standard error for three replicate experiments.
2.5. Inhibition kinetics of daunorubicin reduction by AKR1B10
The inhibition kinetics of daunorubicin reduction by histagged AKR1B10 wild-type protein was monitored spectrophotometrically, by measuring decrease in the absorbance of the cofactor NADPH at 340 nm (Balendiran and Rajkumar, 2005, Martin et al., 2006; Crosas et al., 2003; Nishimura et al., 1991) and at 25 °C with a 10 min time course. The assay was carried out in 100 mM sodium phosphate buffer (pH 7.5) using 0.2 mM NADPH, 0.3 μM wild-type AKR1B10 at 1.0 mM daunorubicin, the concentration equal to Km, daunorubicin (Martin et al., 2006), and varied concentrations of various inhibitors (zopolrestat, fenofibrate, Wy 14,643, sorbinil, ciprofibrate, fenofibric acid and EBPC (Fig. 2)). The rate of reduction of daunorubicin was corrected by subtracting the value of rate of auto degradation of NADPH for the time course of 10 min. As for the glyceraldehyde reduction reaction described above one
3. Results
The kinetic parameters, Km, DL-glyceraldehyde, kcat (NADPH, DLglyceraldehyde) and kcat/Km values for DL-glyceraldehyde reduction by wild-type AKR1B10 were 2.2±0.2 mM, 0.71±0.05 s−1, 0.32±0.03 s−1 mM−1, respectively. In the DL-glyceraldehyde reduction catalyzed by the C299S AKR1B10 mutant, the Km, DL-glyceraldehyde was 15.8±1.0 mM, the kcat (NADPH, DL-glyceradehyde) and kcat/Km were 2.8±0.2 s−1, 0.18±0.01 s−1 mM, respectively. The comparison of kinetic parameters for wild-type and C299S mutant AKR1B10 indicates that substitution of serine by cysteine at position 299 reduces the protein affinity for DL-glyceraldehyde and enhances its catalytic activity. Substrate specificity of AKR1B10 is drastically affected by the mutation of the residue 299 from Cys to Ser. Therefore, both the binding and the catalytic rate of DL-glyceraldehyde reduction depend on residue 299 in AKR1B10.
3.1. Inhibition kinetics of wild-type AKR1B10
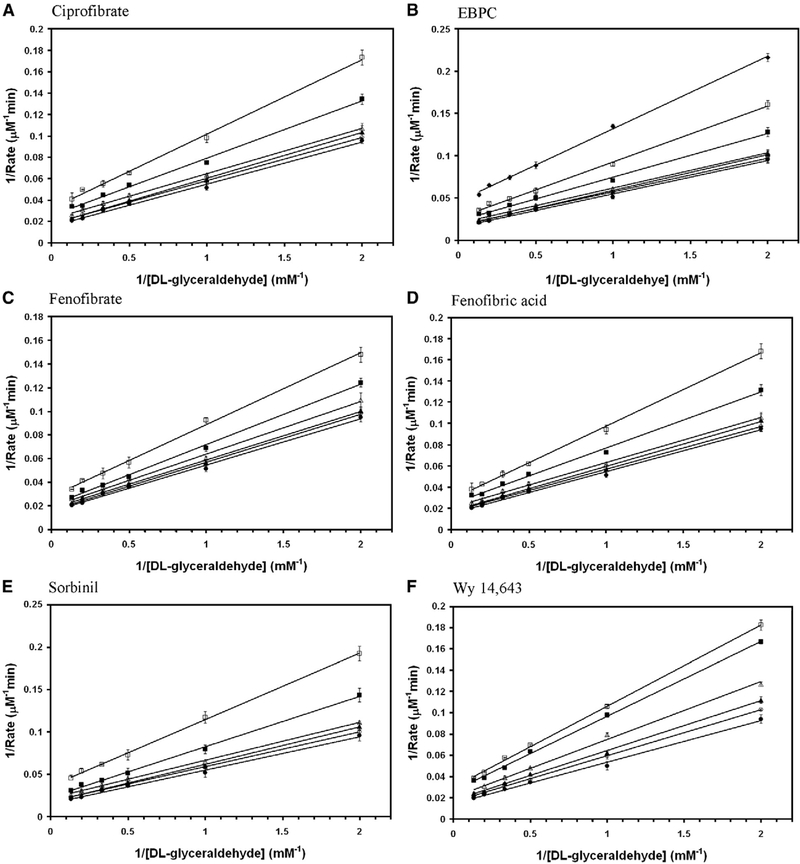
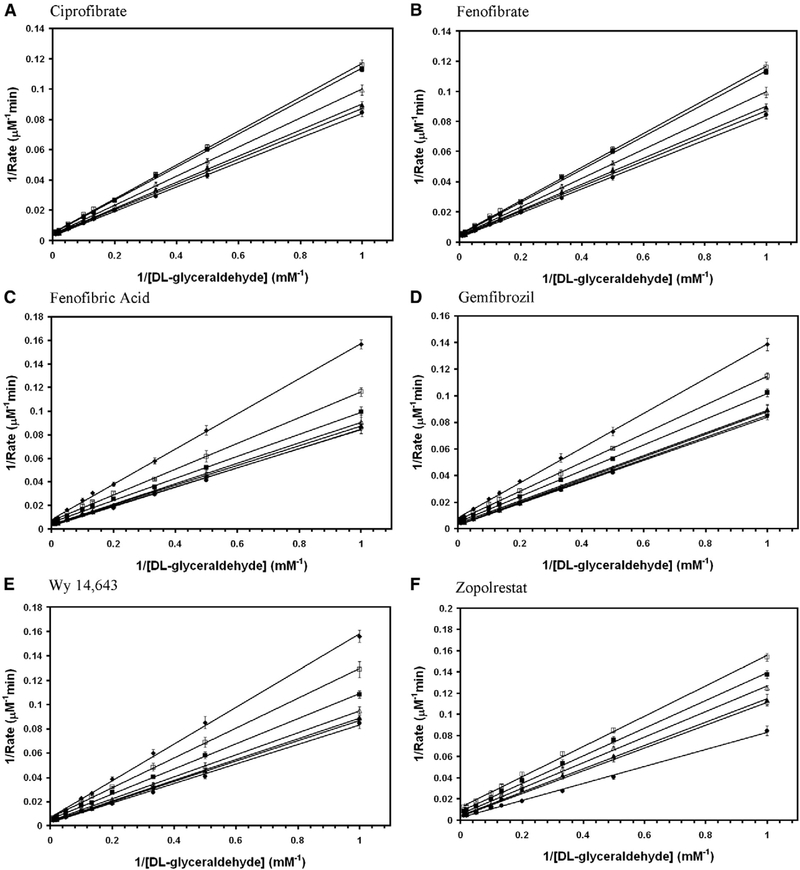
Aldose reductase inhibitors were tested for the inhibition of DL-glyceraldehyde reduction activity of wild-type AKR1B10. Among them zopolrestat, EBPC and sorbinil were noncompetitive whereas, fenofibrate, Wy 14,643, ciprofibrate and fenofibric acid were mixed non-competitive (Fig. 3). The inhibition kinetics constants for the glyceraldehyde reduction activity of wild-type AKR1B10 are reported in Table 1. Several fibrate derivatives with diverged chemical structures are capable of inhibiting the reduction of DL-glyceraldehyde by wild-type AKR1B0 in the presence of NADPH.
Fig. 3.
Double reciprocal plot of the rate of reduction of glyceraldehyde by wild-type AKR1B10. Lineweaver–Burk plots of rate of reduction of DL-glyceraldehyde in the presence of various concentrations of (A) ciprofibrate (● — 0 μM; ○ — 10 μM; ▲ — 20 μM; Δ — 50 μM; ■ — 100 μM; □ — 200 μM), (B) EBPC (● — 0 μM; ○ — 0.5 μM; ▲ — 1 μM; Δ — 2 μM; ■ — 5 μM; □ — 10 μM; ♦ — 20 μM), (C) fenofibrate (● — 0 μM; ○ — 1 μM; ▲ — 2 μM; Δ — 5 μM; ■ — 10 μM; □ —20 μM), (D) fenofibric acid (● — 0 μM; ○ — 10 μM; ▲ — 20 μM; Δ — 50 μM; ■ — 100 μM; □ — 200 μM), (E) sorbinil (● — 0 μM; ○ — 5 μM; ▲ — 10 μM; Δ — 20 μM; ■ — 50 μM; □ — 100 μM), (F) Wy 14,643 (● — 0 μM; ○ — 5 μM; ▲ — 10 μM; Δ — 20 μM; ■ — 40 μM; □ — 50 μM) and (G) zopolrestat (● — 0 μM; ○ — 1 μM; ▲ — 2 μM; Δ — 5 μM; ■ — 10 μM; □ — 20 μM). Each individual rate measurement was evaluated in triplicate.unit of AKR1B0 enzyme activity is defined as μmoles of NADPH oxidized/min..
Table 1.
Inhibition kinetics parameters for DL-glyceraldehyde reduction activity of wildtype AKR1B10 by various inhibitors
| Compound | Inhibition constant (μM) |
Mode of inhibition | |||
|---|---|---|---|---|---|
| IC50 | Kii | Kis | Comparison | ||
| Zopolrestat | 8.0±1.0 | 10.0±1.0 | 10.5±1.0 | Kii≈Kis | pNC |
| EBPC | 12.3±0.7 | 13.5±0.5 | 13.0±0.5 | Kii≈Kis | pNC |
| Fenofibrate | 25.0±5.0 | 30.2±3.0 | 27.5±2.0 | Kii≠Kis | mNC |
| Wy 14,643 | 60.0±10.0 | 60.0±5.0 | 45.0±5.0 | Kii≠Kis | mNC |
| Sorbinil | 100.0±15 | 80±10.0 | 90±10.0 | Kii≈Kis | pNC |
| Ciprofibrate | 210.0±15.0 | 190.0±10.0 | 240.0±15.0 | Kii≠Kis | mNC |
| Fenofibric acid | 300.0±30.0 | 220.0±20.0 | 250.0±15.0 | Kii≠Kis | mNC |
mNC — mixed non-competitive; pNC — pure non-competitive.
3.2. Inhibition kinetics of C299S mutant AKR1B10
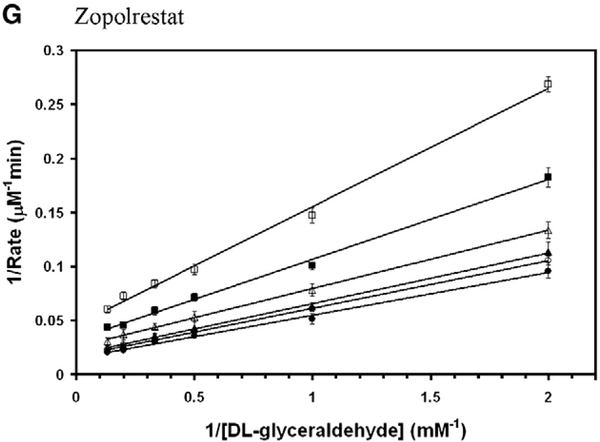
All the above compounds that are shown in Fig. 2 were tested for inhibition potency of the C299S mutant AKR1B10 activity. Gemfibrozil did not inhibit wild-type AKR1B10 activity. Gemfibrozil, which is a member of fibrate family of compounds but with slightly elongated skeleton than the others containing three methylene groups inserted between phenoxy moiety and dimethyl group, was also tested. Among them zopolrestat, fenofibrate, Wy 14,643, ciprofibrate and gemfibrozil were found mixed non-competitive, whereas fenofibric acid demonstrated purely non-competitive mode of inhibition (Fig. 4). The inhibition kinetics constants for the DL-glyceraldehyde reduction activity of C299S mutant AKR1B10 are shown in Table 2. Although several fibrate derivatives were able to inhibit both the wild-type and the C299S mutant AKR1B10, gemfibrozil is only effective in inhibiting the C299S mutant AKR1B10.
Fig. 4.
Double reciprocal plot of the rate of reduction of glyceraldehyde by C299S mutant AKR1B10. Lineweaver–Burk plots of rate of reduction of DLglyceraldehyde in the presence of various concentrations of (A) ciprofibrate (● — 0 μM; ○ — 20 μM; ▲ — 50 μM; Δ — 100 μM; ■ — 200 μM; □ — 250 μM), (B) fenofibrate (● — 0 μM; ○ — 0.5 μM; ▲ — 1 μM; Δ — 2 μM; ■ — 5 μM; □ — 10 μM; □ — 20 μM), (C) fenofibric acid (● — 0 μM; ○ — 5 μM; ▲ — 10 μM; Δ — 20 μM; ■ — 50 μM; □ — 100 μM; ♦ — 200 μM), (D) gemfibrozil. (● — 0 μM; ○ — 10 μM; ▲ — 20 μM; Δ — 50 μM; ■ — 100 μM; □ — 200 μM; ♦ — 400 μM), (E) Wy 14,643 (● — 0 μM; ○ — 10 μM; ▲ — 20 μM; Δ — 50 μM; ■ — 100 μM; □ — 200 μM; ♦ — 300 μM) and (F) zopolrestat (● — 0 μM; ○ — 0.5 μM; ▲ — 2 μM; Δ — 5 μM; ■ — 10 μM; □ — 20 μM). Each individual rate measurement was evaluated in triplicate.
Table 2.
Inhibition kinetic parameters for DL-glyceraldehyde reduction activity of C299S mutant AKR1B10 by various inhibitors
| Compound | Inhibition constant (μM) |
Mode of inhibition | |||
|---|---|---|---|---|---|
| IC50 | Kii | Kis | Comparison | ||
| Zopolrestat | 9.0±0.6 | 11.0±0.5 | 12.5±0.5 | Kii≠Kis | mNC |
| Fenofibrate | 10.0±0.5 | 11.5±0.5 | 16.5±0.6 | Kii≠Kis | mNC |
| Wy 14,346 | 350.0±20.0 | 360.0±20.0 | 240.0±10.0 | Kii≠Kis | mNC |
| Fenofibric acid | 220.0±15.0 | 250.0±10.0 | 240.0±10.0 | Kii≈Kis | pNC |
| Gemfibrozil | 380.0±20.0 | 415.0±12.0 | 370.0±30.0 | Kii≠Kis | mNC |
| Ciprofibrate | ND | 815.0±45.0 | 705.0±60.0 | Kii≠Kis | mNC |
mNC — mixed non-competitive; pNC — pure non-competitive; ND — not detectable; EBPC and sorbinil did not show measurable level of inhibiting C299S mutantAKR1B10.
3.3. Inhibiting the carbonyl reduction of daunorubicin catalyzed by AKR1B10
The same collection of compounds (zopolrestat, fenofibrate, Wy 14,643, sorbinil, ciprofibrate, fenofibric acid, gemfibrozil, EBPC (Fig. 2)) that were tested for their inhibition potential of the DL-glyceraldehyde reduction above were tested for the reduction of daunorubicin, an anticancer drug. About 50% inhibition was achieved by zopolrestat and EBPC, 35–40% was observed by other compounds and their inhibition potency could be ranked in the following order, fenofibrate, Wy 14,643 and sorbinil are comparable followed by ciprofibrate and fenofibric acid (Table 3). Noticeably the prodrug, fenofibrate is 25-fold effective than its hydrolyzed fibrate derivative, fenofibric acid in inhibiting the AKR1B10 catalyzed reduction of daunorubicin.
Table 3.
Inhibition potency (percentage inhibition) of various inhibitors for the reduction of daunorubicin by AKR1B10 wild-type protein
| Inhibitors | Concentration (μM) | Inhibition (%) |
|---|---|---|
| Zopolrestat | 20 | 43 |
| 50 | 47 | |
| EBPC | 100 | 54 |
| 250 | 57 | |
| Fenofibrate | 20 | 35 |
| Wy 14,643 | 250 | 37 |
| 500 | 50 | |
| Sorbinil | 100 | 31 |
| 250 | 37 | |
| Ciprofibrate | 250 | 33 |
| 500 | 39 | |
| Fenofibric acid | 250 | 25 |
| 500 | 35 |
4. Discussion
AKR1B10 shares 70% sequence identity with human aldose reductase (AR) and contains 316 amino acid residues. The kinetics studies reveal that AKR1B10 is able to catalyze the reduction of DL-glyceraldehyde efficiently similar to AR but unable to utilize glucose as a substrate. AR is known to use glucose in the open-form which is around 0.003% of the total in solution (Grimshaw, 1986; Inagaki et al., 1982) with the kcat/Km of about 106 s−1 M−1 range. Furthermore, residue Cys299 in AKR1B10 is analogous to AR Cys298, which is located in the AR active site pocket. Although this residue is considered noncatalytic, the oxidation and reduction state of this residue plays an important role in AR inhibition kinetics. The C299S mutation in AKR1B10 causes about 7-fold increase in Km and 4-fold increase in the kcat. This results in an approximate 2-fold decrease in the pseudo second order rate constant kcat/Km for DL-glyceraldehyde reduction. These observations suggest that although AKR1B10 shares high sequence similarity with AR, the natural substrate (glucose) specificity of AKR1B10 is different from that of AR. This difference in substrate selectivity may reflect the diversity in the functional role of AKR1B10.
4.1. AKR1B10 modulators
Zopolrestat, EBPC and sorbinil show pure non-competitive inhibition kinetics for wild-type AKR1B10 while fenofibrate, Wy 14,643, ciprofibrate and fenofibric acid follow mixed noncompetitive inhibition kinetics in the reduction of glyceraldehyde. Several of the compounds have an acidic moiety similar to that found in zopolrestat and may bind to AKR1B10 in a mode similar to AR. However, fenofibrate and EBPC which are isopropyl and ethyl esters of the carboxylic acids may bind differently due to steric effects caused by the ester groups. All of these compounds inhibit the reduction of DL-glyceraldehyde catalyzed by AR non-competitively (Balendiran and Rajkumar, 2005). Reduction of DL-glyceraldehyde by C299S mutant AKR1B10 is inhibited by zopolrestat, fenofibrate, Wy 14,643, gemfibrozil and ciprofibrate following mixed non-competitive mode of inhibition whereas fenofibric acid follows a pure noncompetitive pattern of inhibition. Mutation of Cys299 to Ser resulted in changes in the kinetic parameters as well as in the mode of inhibition of the reduction that is catalyzed by AKR1B10.
Zopolrestat has the lowest IC50 value (8.0±1.0 μM) inhibiting the glyceraldehyde reduction activity of wild-type AKR1B10 and it is not noticeably affected by C299S mutation. Though sorbinil has an IC50 value of 100.0+15 μM, which is 12.5-fold higher than zopolrestat for the wild-type AKR1B10, sorbinil does not show any detectable inhibition for the C299S mutant AKR1B10. The IC50 value of EBPC is about 1.5-fold higher than zopolrestat but EBPC and ciprofibrate do not inhibit the C299S mutant activity.
In contrast to the other compounds, fenofibrate inhibits AKR1B10 the IC50 value of fenofibrate is 25.0±5.0 μM for the wild-type AKR1B10 and it is reduced 2.5-fold for the C299S mutant enzyme to a value close to that of zopolrestat. Moreover the IC50 value offenofibric acid is decreased 1.4-fold upon Cys to Ser mutation of residue 299. While fenofibric acid is 12-fold less potent than fenofibrate in inhibiting wild-type enzyme, it is 22fold less effective in inhibiting the C299S mutant AKR1B10. Similar to fenofibrate and fenofibric acid another interesting trend is manifested by gemfibrozil that inhibits the C299S mutant AKR1B10, whereas gemfibrozil does not inhibit wild-type AKR1B10. The inhibition kinetic parameters Kii, Kis and mode of inhibition also reflect similar trends revealed by the IC50 values upon mutation. These inhibition kinetic studies reflect the inhibitor selectivity over a group of compounds capable of modulating the reaction which catalyses the common substrate by two different enzymes with high sequence homology and also reveals the role of a conserved residue in both the enzymatic reaction and inhibition.
Both fenofibrate and fenofibric acid demonstrated 35% inhibition of ARK1B10 catalyzed reduction of daunorubicin but 20 and 500 μM concentrations of respective compounds were required to achieve the same level of inhibition. This indicates that the prodrug, fenofibrate which is the isopropyl ester of fenofibric acid has higher potency than the hydrolyzed acid form. The reduced percentage inhibition potential demonstrated by cleavage of the isopropyl ester bond when daunorubicin was used as the substrate is parallel to the trend seen with IC50 values when DL-glyceraldehyde was utilized as the substrate. Furthermore, in the reduction of DL-glyceraldehyde C299S mutation did not alter this phenomenon.
The compounds used in this study such as ciprofibrate, fenofibric acid, fenofibrate and gemfibrozil are structurally divergent class of fibrates. Fibrates are antilipidemic drugs that are in wide clinical use for the treatment of hyperglycemia (Forcheron et al., 2002; Miller and Spence, 1998) due to their known pharmacological properties (Fruchart et al., 1998; Jones et al., 1990; Rubins et al., 1999). Therefore the current study implies that AKR1B10 plays a central role in mediating the mechanism of ciprofibrate, fenofibrate, fenofibric acid, gemfibrozil and Wy 14,643 action. Hence, AKR1B10 may be an added target of action for these classes of compounds beyond PPAR or the action mediated by other targets.
4.2. Inhibitor selectivity
Although the modulators of AKR1B10 overlap that of AR, their specificities vary. Structural studies reveal that carboxylic acid head group of zopolrestat forms ionic interactions with residues Tyr48, His110, Trp111 and nicotinamide ring of NADP+ in AR (PDB:2HVO) (Steuber et al., 2006; Wilson et al., 1993). Conservation of these residues implies similar interactions are anticipated to be present in AKR1B10 with zopolrestat and hence same catalytic role played in the reduction of aldehydes. Sorbinil is a spirohydantoin class of compound that inhibits AR potently like the carboxylic acid class of AR inhibitors. The catalytic residues Tyr48, His110 and Trp111 of pig AR form polar interactions with the carbonyl and amino (NH) groups of cyclic imide moiety of sorbinil (PDB:1AH4) (Urzhumtsev et al., 1997). Side chain atoms of Cys298 are within van der Waals (vdW) distance from carbon atom C6 of sorbinil in pig AR (PDB:1AH4) and nitrogen atom N2 of zopolrestat in human AR (PDB:2HVO) structures. Also the side chain atoms of Cys298 make contacts with the carbon atoms C4 and C5 of nicotinamide ring in pig and human AR complex with sorbinil (PDB:1AH4) and zopolrestat (PDB:2HVO) structures. The molecular interactions from the corresponding residue Cys299 of AKR1B10 are anticipated to be similar to those described above for AR. Overall sorbinil is a weaker inhibitor for AKR1B10 than AR and this may be due to the combined outcome of interactions caused by the residue that are lining the inhibitor-binding pocket.
Differencesin the inhibitor selectivity displayed by ARK1B10 compared to AR may be hidden in the subtle disparity in the primary sequence between these protein molecules. This phenomenon could also be exhibited in their substrate specificity. Amino acid residues that are involved in the cofactor binding are highlyconservedbetweenAKR1B10andARindicatingthattheir cofactor dependency may be similar. However the intriguing differences seen in some of the residues like Ser304, Val301, Asn300 and Pro81 that are located in the substrate or inhibitorbinding region of AKR1B10 may play a combined role in the inhibitorbindingandselectivity.TheresidueCys303inARforms arene–sulfur interactions with zopolrestat (PDB:2HVO) but this residue is Ser304 in AKR1B10. Moreover replacement of the residues Val301 and Asn300 in AKR1B10 to Leu300 and Ala299 in AR may cause differences in their substrate as well as the inhibitor-binding properties due to alteration in the ionic nature, the size and shape of the cavity. The main chain amino (NH) group of Leu300 in AR is at the hydrogen bonding distance from atom nitrogenatom(N3)of zopolrestat(PDB:2HVO).Also in AR beta carbon atom (CB) of Leu300 is located 3.3 Å away from the nitrogen atom (N3) of zopolrestat (PDB:2HVO) but AKR1B10 has Val301 which is a residue with larger side chain instead at this location. In addition, there are changes in residues Pro81 and Phe84 of AKR1B10 in other locations than the cofactor and ligand binding regions. The closest distance between the thiol group of Cys80 in AR and any atoms of zopolrestat is about 4.3 Å (PDB:2HVO). Due to restricted flexibility as well as the absence of arene–sulfur interactions of residue Pro81 which replaces Cys in AKR1B10 may contribute differently than AR. This nonconservative replacement may also reflect in the substrate specificity and inhibitor selectivity as well. Furthermore, the distance between gamma oxygen atom (OG1) of Thr113 and fluorine atom (F2) of zopolrestat is 3.48 Å (PDB:2HVO) and that between gamma oxygen atom (OG2) of Thr113 and fluorine atoms (F1 and F2) of zopolrestat (PDB:2HVO) are 3.3 Å and 3.2 Å in AR indicating existence of van der Waals (vdW) interaction but this residue is replaced by Gln114 in AKR1B10. Therefore, substrate specificity and inhibitor selectivity is governed by minor changes in the amino acid sequence, which defines the shape, size and the electronic potential of the substrate and inhibitor-binding pocket.
4.3. AKR1B10 inhibitors as antineoplastic agents
Identification of structurally diverged set of lead compounds that are able to regulate the activity of AKR1B10 will have multiple benefits in several forms of cancer because of the following reasons. Recent studies on the effect of small interfering RNA (siRNA)-mediated downregulation of AKR1B10 on proliferation of colorectal cancer cells (HCT-8) estimated its potential as a tumor intervention target (Yan et al., 2007). Moreover, AKR1B10 was detected in 20.0% cervical cancer casesand15.8%endometrialcancercasesinsamplesfromuterine cancer patients (Yoshitake et al., 2007). In addition, statistical analysis indicated that AKR1B10 expression was associated with tumor recurrence after surgery and keratinization of squamous cell carcinoma in cervical cancer (Yoshitake et al., 2007). Also bioinformatics analysis of the public gene expression data and validation of clinical specimens reveal AKR1B10 as a potential diagnostic marker for non-small-cell lung cancer (Kim et al., 2007).Severalfibrateshavebeenprescribedtohumanpatientsfor the treatment of hyperlipidemias for more than 30 years (Fruchart et al., 1998; Peters et al., 2005). Therefore, the compounds especially fibrates and their derivatives described in the current study may have the potential in the regulation of AKR1B10 and similar proteins that are various cancer drug targets.
Acknowledgments
We thank Dr. Peter Oates for reagents. This work was supported by funding from the American Diabetes Association.
References
- Balendiran GK, Rajkumar B, 2005. Fibrates inhibit aldose reductase activity in the forward and reverse reactions. Biochem. Pharmacol 70, 1653–1663. [DOI] [PubMed] [Google Scholar]
- Balendiran GK, Verma M, Perry E, 2007. Chemistory of fibrates. Curr. Chem. Biol 1, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohren KM, Bullock B, Wermuth B, Gabbay KH, 1989. The aldo–keto reductase superfamily. cDNAs and deduced amino acid sequences of human aldehyde and aldose reductases. J. Biol. Chem 264, 9547–9551. [PubMed] [Google Scholar]
- Bohren KM, Gabbay KH, 1993. Cys298 is responsible for reversible thiolinduced variation in aldose reductase activity. Adv. Exp. Med. Biol 328, 267–277. [DOI] [PubMed] [Google Scholar]
- Bradford MM, 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Cao D, Fan ST, Chung SS, 1998. Identification and characterization of a novel human aldose reductase-like gene. J. Biol. Chem 273, 11429–11435. [DOI] [PubMed] [Google Scholar]
- Cappiello M, Voltarelli M, Cecconi I, Vilardo PG, Dal Monte M, Marini I, Del Corso A, Wilson DK, Quiocho FA, Petrash JM, Mura U, 1996. Specifically targeted modification of human aldose reductase by physiological disulfides. J. Biol. Chem 271, 33539–33544. [DOI] [PubMed] [Google Scholar]
- Crosas B, Hyndman DJ, Gallego O, Martras S, Pares X, Flynn TG, Farres J, 2003. Human aldose reductase and human small intestine aldose reductase are efficient retinal reductases: consequences for retinoid metabolism. Biochem. J 373, 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcheron F, Cachefo A, Thevenon S, Pinteur C, Beylot M, 2002. Mechanisms of the triglyceride- and cholesterol-lowering effect of fenofibrate in hyperlipidemic type 2 diabetic patients. Diabetes 51, 3486–3491. [DOI] [PubMed] [Google Scholar]
- Fruchart JC, Brewer HB Jr., Leitersdorf E, 1998. Consensus for the use of fibrates in the treatment of dyslipoproteinemia and coronary heart disease. Fibrate Consensus Group. Am. J. Cardiol 81, 912–917. [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Yamauchi N, Moriguchi H, Hippo Y, Watanabe A, Shibahara J, Taniguchi H, Ishikawa S, Ito H, Yamamoto S, Iwanari H, Hironaka M, Ishikawa Y, Niki T, Sohara Y, Kodama T, Nishimura M, Fukayama M, Dosaka-Akita H, Aburatani H, 2005. Overexpression of the aldo–keto reductase family protein AKR1B10 is highly correlated with smokers’ non-small cell lung carcinomas. Clin. Cancer Res 11, 1776–1785. [DOI] [PubMed] [Google Scholar]
- Gariot P, Barrat E, Mejean L, Pointel JP, Drouin P, Debry G, 1983. Fenofibrate and human liver. Lack of proliferation of peroxisomes. Arch. Toxicol 53, 151–163. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, 2002. The peroxisome proliferator-activated receptor alpha (PPARalpha): role in hepatocarcinogenesis. Mol. Cell. Endocrinol 193, 71–79. [DOI] [PubMed] [Google Scholar]
- Grimshaw CE,1986Directmeasurementofthe rateofringopeningofD-glucose by enzyme-catalyzed reduction. Carbohydr. Res 148, 345–348. [DOI] [PubMed] [Google Scholar]
- Group SRTR, 1990. A randomized trial of sorbinil, an aldose reductase inhibitor, in diabetic retinopathy. Sorbinil Retinopathy Trial Research Group. Arch. Ophthalmol 108, 1234–1244. [DOI] [PubMed] [Google Scholar]
- Hess R, Staubli W, Reiss W, 1965. Nature of the hepatomegalic effect produced by ethyl-chlorphenoxyisobutyrate in the rat. Nature 208, 856–858. [DOI] [PubMed] [Google Scholar]
- Hyndman DJ, Flynn TG, 1998. Sequence and expression levels in human tissues of a new member of the aldo–keto reductase family. Biochim. Biophys. Acta 1399, 198–202. [DOI] [PubMed] [Google Scholar]
- Inagaki K, Miwa I, Okuda J, 1982. Affinity purification and glucose specificity of aldose reductase from bovine lens. Arch. Biochem. Biophys 216, 337–344. [DOI] [PubMed] [Google Scholar]
- Jez JM, Flynn TG, Penning TM, 1997. A new nomenclature for the aldo– keto reductase superfamily. Biochem. Pharmacol 54, 639–647. [DOI] [PubMed] [Google Scholar]
- Jones IR, Swai A, Taylor R, Miller M, Laker MF, Alberti KG, 1990. Lowering of plasma glucose concentrations with bezafibrate in patients with moderately controlled NIDDM. Diabetes Care 13, 855–863. [DOI] [PubMed] [Google Scholar]
- Kim B, Lee HJ, Choi HY, Shin Y, Nam S, Seo G, Son DS, Jo J, Kim J, Lee J, Kim J, Kim K, Lee S, 2007. Clinical validity of the lung cancer biomarkers identified by bioinformatics analysis of public expression data. Cancer Res. 67, 7431–7438. [DOI] [PubMed] [Google Scholar]
- Klemin S, Calvo RY, Bond S, Dingess H, Rajkumar B, Perez R, Chow L, Balendiran GK, 2006. WY 14,643 inhibits human aldose reductase activity. J. Enzyme Inhib. Med. Chem 21, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HJ, Breyer-Pfaff U, Wsol V, Venz S, Block S, Maser E, 2006. Purification and characterization of AKR1B10 from human liver: role in carbonyl reduction of xenobiotics. Drug Metab. Dispos 34, 464–470. [DOI] [PubMed] [Google Scholar]
- Miller DB, Spence JD, 1998. Clinical pharmacokinetics of fibric acid derivatives (fibrates). Clin. Pharmacokinet 34, 155–162. [DOI] [PubMed] [Google Scholar]
- Nishimura C, Yamaoka T, Mizutani M, Yamashita K, Akera T, Tanimoto T, 1991. Purification and characterization of the recombinant human aldose reductase expressed in baculovirus system. Biochim. Biophys. Acta 1078, 171–178. [DOI] [PubMed] [Google Scholar]
- Peters JM, Cheung C, Gonzalez FJ, 2005. Peroxisome proliferatoractivated receptor-alpha and liver cancer: where do we stand? J. Mol. Med 83, 774–785. [DOI] [PubMed] [Google Scholar]
- Petrash JM, Harter TM, Devine CS, Olins PO, Bhatnagar A, Liu S, Srivastava SK, 1992. Involvement of cysteine residues in catalysis and inhibition of human aldose reductase. Site-directed mutagenesis of Cys-80, −298, and −303. J. Biol. Chem 267, 24833–24840. [PubMed] [Google Scholar]
- Rath NP, Haq W, Balendiran GK, 2005. Fenofibric acid. Acta Crystallogr C. 61, 81–84. [DOI] [PubMed] [Google Scholar]
- Reddy JK, Azarnoff DL, Hignite CE, 1980. Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature 283, 397–398. [DOI] [PubMed] [Google Scholar]
- Rubins HB, Robins SJ, Collins D, Fye CL, Anderson JW, Elam MB, Faas FH, Linares E, Schaefer EJ, Schectman G, Wilt TJ, Wittes J, 1999. Gemfibrozil for the secondary prevention of coronary heart disease in men with low levels of high-density lipoprotein cholesterol. Veterans Affairs High-density Lipoprotein Cholesterol Intervention Trial Study Group. N. Engl. J. Med 341, 410–418. [DOI] [PubMed] [Google Scholar]
- Sato S, Kador PF, 1990. Inhibition of aldehyde reductase by aldose reductase inhibitors. Biochem. Pharmacol 40, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Scuric Z, Stain SC, Anderson WF, Hwang JJ, 1998. New member of aldose reductase family proteins overexpressed in human hepatocellular carcinoma. Hepatology 27, 943–950. [DOI] [PubMed] [Google Scholar]
- Steuber H, Zentgraf M, Gerlach C, Sotriffer CA, Heine A, Klebe G, 2006. Expect the unexpected or caveat for drug designers: multiple structure determinations using aldose reductase crystals treated under varying soaking and co-crystallisation conditions. J. Mol. Biol 363, 174–187. [DOI] [PubMed] [Google Scholar]
- Urzhumtsev A,Tete-Favier F,Mitschler A,Barbanton J,Barth P,Urzhumtseva L,Biellmann JF,Podjarny A,Moras D,1997A’specificity’pocketinferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil. Structure 5, 601–612. [DOI] [PubMed] [Google Scholar]
- Vander Jagt DL, Torres JE, Hunsaker LA, Deck LM, Royer RE, 1996. Physiological substrates of human aldose and aldehyde reductases. Adv. Exp. Med. Biol 414, 491–497. [DOI] [PubMed] [Google Scholar]
- Wilson DK, Tarle I, Petrash JM, Quiocho FA, 1993. Refined 1.8 Å structure of human aldose reductase complexed with the potent inhibitor zopolrestat. Proc. Natl. Acad. Sci. U. S. A 90, 9847–9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zu X, Ma J, Liu Z, Adeyanju M, Cao D, 2007. Aldo–keto reductase family 1 B10 gene silencing results in growth inhibition of colorectal cancer cells: implication for cancer intervention. Int. J. Cancer 121, 2301–2306. [DOI] [PubMed] [Google Scholar]
- Yoshitake H, Takahashi M, Ishikawa H, Nojima M, Iwanari H, Watanabe A, Aburatani H, Yoshida K, Ishi K, Takamori K, Ogawa H, Hamakubo T, Kodama T, Araki Y, 2007. Aldo–keto reductase family 1, member B10 in uterine carcinomas: a potential risk factor of recurrence after surgical therapy in cervical cancer. Int. J. Gynecol. Cancer 17, 1300–1306. [DOI] [PubMed] [Google Scholar]