Abstract
Similar to other mammalian viruses, the life cycle of hepatitis B virus (HBV) is heavily dependent upon and regulated by cellular (host) functions. These cellular functions can be generally placed in to two categories: (a) intrinsic host restriction factors and innate defenses, which must be evaded or repressed by the virus; and (b) gene products that provide functions necessary for the virus to complete its life cycle. Some of these functions may apply to all viruses, but some may be specific to HBV. In certain cases, the virus may depend upon the host function much more than does the host itself. Knowing which host functions regulate the different steps of a virus’ life cycle, can lead to new antiviral targets and help in developing novel treatment strategies, in addition to improving a fundamental understanding of viral pathogenesis. Therefore, in this review we will discuss known host factors which influence key steps of HBV life cycle, and further elucidate therapeutic interventions targeting host-HBV interactions.
Keywords: Hepatitis B virus (HBV), Hepatitis, Liver, Virus-host interaction, Antiviral agents, Direct acting antiviral agents, Host targeting agents
1. Introduction
There are currently seven medications approved by the United States (US) Food and Drug Administration (FDA) for management of chronic hepatitis B virus (HBV) infection, including two interferons (standard and PEGylated IFN-α) and five nucleos(t)ide analogue polymerase (pol) inhibitors (Belongia et al., 2009; Block et al., 2007, 2015). Although the treatments have provided hope to those affected and have demonstrated the potential of medical intervention, these drugs are all of limited value. They result in sustained benefit in fewer than half of those in whom they are given (Belongia et al., 2009). Moreover, they are limited by toxicity (in the case of the interferons) or development of drug resistance as in the case of the viral DNA polymerase inhibitors (Hoofnagle et al., 2007; Zoulim and Locarnini, 2009). Therefore, development of novel direct acting agents (DAAs) and host targeting agents (HTAs) against HBV infection is warranted to enrich the landscape of HBV antivirals.
This review will discuss about the host functions which are involved in the various steps of HBV life cycle, and further explain possibilities of exploitation of host factors in therapeutic intervention of HBV. We will focus on the constitutive host systems that regulate HBV, and then further sub divide these categories in to their component pathways. The “induced” systems, such as innate host defense systems that negatively influence the viral life cycle also represents an important biology, but will be only considered briefly, in the discussion. The challenges of toxicity and selectivity, associated with targeting host functions for antiviral purposes are especially important, and are also discussed.
2. Hepatitis B virus replication and disease
HBV causes acute and chronic infections. Acute HBV infection can be either asymptomatic or present with symptomatic acute hepatitis. Most adults infected with the virus recover, but ~5% of infected adults and 90% of infected neonates fail to mount an immune response that clears the virus, and they develop a life-long chronic infection that may progress to chronic active hepatitis, cirrhosis, and primary hepatocellular carcinoma (HCC) (Block et al., 2003; El-Serag and Mason, 1999; Hoofnagle, 2004). Despite an effective prophylactic vaccine, it remains a major public health challenge. Between 250 and 290 million people are chronically infected with HBV worldwide which results in 1 million deaths per year (Razavi-Shearer et al., 2018).
HBV is liver tropic virus with a ~3 kb partially double stranded (ds) DNA genome that specifies only 7 viral proteins, which include DNA polymerase (Pol), capsid protein (Core), HBeAg (hepatitis B e antigen), X protein, and three envelope proteins: LHBs (L), MHBs (M) and SHBs (S) (Block et al., 2007). The polymerase gene product is the only enzyme encoded by HBV, thus, virus life cycle is heavily dependent on host or cellular factors. Host functions are involved in almost every step in the HBV life cycle, from entry to secretion (Block et al., 2007; Ganem and Prince, 2004; Watanabe et al., 2007). HBV uses multiple host factors for its replication, and the interplay between host functions and HBV leads to severe disease consequences. Outlining the host factors involved in the life cycle of HBV can provide new insight for discovery of anti-HBV therapies, in which drugs targeting cellular factors can be developed.
3. Viral binding and entry into hepatocytes
3.1. Biology of viral binding and entry in to hepatocytes
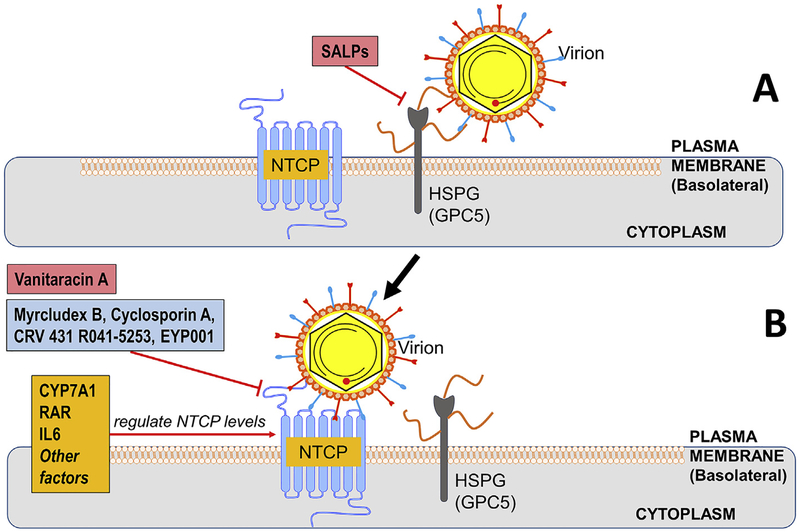
Host functions mediating binding and entry of HBV in to hepatocytes are illustrated in Fig. 1. HBV initiates infection by binding to a low affinity receptor, a heparan sulfate proteoglycan (HSPG) through the infectivity determinant in the surface-exposed antigenic loop (AGL), a polypeptide present in the S domain common to all envelope proteins (Li and Tong, 2015; Schulze et al., 2007; Sureau and Salisse, 2013). Glypican-5 (GPC5), a protein associated with proteoglycans, has recently been reported to be involved in entry of HBV and hepatitis delta virus (HDV, a satellite virus of HBV by using HBV envelope proteins for cell entry), providing further clarity on how HSPG mediate HBV entry (Verrier et al., 2016). Proteoglycan binding is followed by a more specific, and higher affinity, interaction of the N-terminal myristoylated peptide in the pre-S1 domain of L protein with the host cellular plasma membrane bearing Sodium Taurocholate Cotransporting Polypeptide (NTCP) receptor expressed by host SLC10A1 gene (Bruss et al., 1996; Hagenbuch and Meier, 1994; Ni et al., 2014; Yan et al., 2012). The NTCP receptor is on the sinusoidal plasma membrane. The normal physiological function of NTCP is to facilitate hepatocyte uptake of sodium dependent bile acids from the circulation (Stieger, 2011; Takeyama et al., 2010), and its expression is mostly liver specific (Hagenbuch and Meier, 1994). NTCP is itself highly regulated, and its abundance on the sinusoidal membrane changes with pathological and physiological conditions, varying with blood bile salt levels (Roma et al., 2008). HBV infection appears to result in alteration of bile metabolism. For example, the level of cholesterol 7α-hydroxylase (CYP7A1), the rate limiting step in bile acid synthesis from cholesterol, was reported to be significantly elevated in HBV infected murine system with humanized liver and in liver biopsies from infected people (Oehler et al., 2014), suggesting an interplay between HBV infection and cellular genes involved in NTCP and bile acid regulation. One question that remains is the extent to which other host receptor(s) and co-receptor component(s), in addition to NTCP, and t the hepatocyte microenvironment or soluble blood components (as seen in Adenoviruses infecting liver (Morizono and Chen, 2011; Shayakhmetov et al., 2005)) have an effect upon HBV entry into hepatocytes. Understanding of the entry process in depth will help us identify the target(s) for drug intervention against hepatitis B infection.
Fig. 1. Binding to and entry in to hepatocytes.
The hepatitis B virion is shown in (A) making an initial “docking” on to heparin sulfate proteoglycans (HSPG), followed by (B) “rolling” to Sodium Taurocholate Polypeptide (NTCP) receptors, which are believed to be the higher affinity receptor for the virus. Cellular functions mediating these steps are indicated in orange, research phase compounds that interfere with these steps are shown in pinkish red, with compounds that are clinical phase, or approved, in light blue. This scheme was illustrated by using the Biology Bundle of Motifolio Drawing Toolkits (www.motifolio.com) (the same as following figures). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Therapeutic interference with HBV binding and entry by targeting host factors
HBV binding to its receptor on hepatocytes has been successfully used as an antiviral target. Briefly, as indicated in Fig. 1, the synthetic anti-lipopolysaccharide peptides (SALPs) inhibit HBV infection through binding to heparan sulfate moieties on the cell surface (Krepstakies et al., 2012). Myrcludex B is a HBV preS1-derived synthetic lipopoly-peptide that has been shown to specifically bind to NTCP and block de novo HBV infection and prevent intrahepatic viral spreading both in vitro and in vivo (Blank et al., 2016), and clinical trials to manage chronic HBV/HDV infection are well under way, and antiviral efficacy is being demonstrated (Bogomolov et al., 2016). Cyclosporin A (CsA) is an immunosuppressant that can directly bind to NTCP and interrupt the interaction between NTCP and the preS1 region (Watashi et al., 2014). Interestingly, the amino acid motifs of NTCP critical for HBV entry overlap with that for bile salts uptake by NTCP, and the inhibition of HBV entry by NTCP natural ligands has been observed in cell cultures (Yan et al., 2014). Furthermore, a tricyclic polyketide, vanitaracin A, has been shown to inhibit HBV and HDV entry by directly interacting with NTCP and inhibiting its transporter activity (Kaneko et al., 2015). Nonetheless, it is worth noting that certain CsA analogs that abrogate the immunosuppressive activity possess a stronger anti-HBV activity without interfering NTCP transporter function, perhaps through an allosteric mechanism (Shimura et al., 2017). Elucidation of the structure of preS1-NTCP complex will help to explain the exact mode of action of the identified HBV entry inhibitors.
Inhibition of HBV entry can also be achieved by reducing the level of NTCP expression. The retinoic acid receptor (RAR) has been proposed to regulate the promoter activity of the human NTCP (hNTCP) gene and an RAR-selective antagonist, R041–5253 decreases cellular susceptibility to HBV infection by inhibiting hNTCP promoter activity (Tsukuda et al., 2015). The interleukin cytokine, IL-6, has also been found to regulate NTCP expression. Pretreatment of HepaRG cells with IL-6 led to 98% reduction in NTCP mRNA expression, along with an 80% decrease in NTCP-mediated taurocholate uptake and 90% inhibition of HBV entry (Bouezzedine et al., 2015). However, IL-6 is a proinflammatory cytokine and its over-use might lead to development of autoimmune diseases. Thus, proper dosage or endogenous induction level need to be identified that would minimize the side-effects in further development.
In addition, N-Myristoylation of the terminal glycine of the preS1 domain of the LHBs polypeptide is essential for virus infectivity and is a post-translational event, mediated by specific cell acetylases (N-myristoyltransferases), providing another host function that can serve as an antiviral target (Bruss et al., 1996; Gripon et al., 1995).
4. De-envelopment and translocation of the nucleocapsid to the nucleus
4.1. Biology of de-envelopment and antegrade transport of nucleocapsid to the nucleus
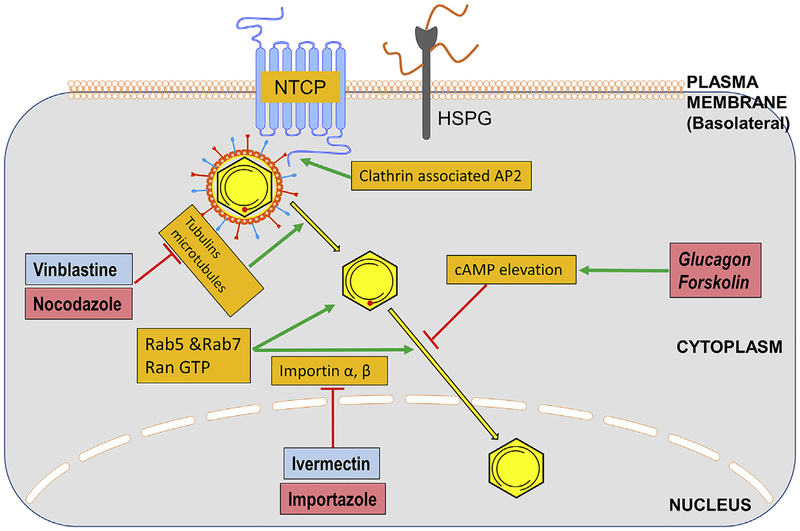
De-envelopment and transport of the virion to the nucleus, and capsid disassembly, are not well characterized for HBV, although host cellular factors and processes are clearly involved. Huang et al. used an immortalized human primary hepatocyte cell line, HuS-E/2, transduced with human telomerase reverse transcriptase (hTERT) and human papillomavirus E6E7 (HPV/E6E7), to show that clathrin-dependent endocytosis is critical for HBV infection through interaction of clathrin associated heavy protein and adaptor (AP-2) with the pre-S1 domain of L protein (Huang et al., 2012). Fig. 2 shows these steps. Following internalization of HBV virions, there is a Rab5-and Rab7-dependent transport through the late endosomal compartment (distinct from recycling endosomes) which is also important to the de-envelopment of capsid (Macovei et al., 2013). Functional caveolin-1 was shown to be necessary for productive HBV infection in HepaRG cells (Macovei et al., 2010). It is speculated that disulfide-reduction within the endosomal compartments is involved in the de-envelopment process as well (Feener et al., 1990; Ohgami et al., 2005). Nonetheless, de-envelopment must occur and the de-enveloped virion cargo of partially double stranded viral DNA is delivered to the nucleus.
Fig. 2. Transport of the virus nucleocapsid to the nucleus.
Following entry in to the cell, the virus is de-enveloped and the nucleocapsid is transported to the nucleus. Cellular functions mediating these steps are indicated in orange, research phase compounds that interfere with these steps are shown in pinkish red, with compounds that are clinical phase, or approved, in light blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4.2. Therapeutic interference with de-envelopment and translocation to the nucleus
Given that there are many steps involved in the transport of nucleocapsids to the nucleus following entry, numerous host functions can be imagined as targets for antiviral intervention. For example, it has been reported that phosphorylation of the capsid induces exposure of nuclear localization signal (NLS) in the COOH-terminal portion of the core protein that allows core binding to the nuclear pore complex (NPC) by the importin- (karyopherin-) mediated pathway (Kann et al., 1999). Elevation of cell cAMP, and stimulation of protein kinase A (PKA) with glucagon has been reported to interfere with the transport of capsids to the nucleus, at least in duck hepatitis B virus (DHBV) systems, further highlighting the role of phosphorylation in the early steps of the viral life cycle (Hild et al., 1998). That said, the targets of PKA (presumably viral core or polymerase) have not been determined. Another cellular kinase candidate for core proteins is CDK2 (Ludgate et al., 2012).
As is true for most mammalian viruses, a role for microtubule-dependent transfer of the HBV capsid to the nucleus and a subsequent release of the viral genomes into the karyoplasm has also been observed (Schmitz et al., 2010). Thus, HBV nucleocapsids are transported to the nuclear periphery by mechanisms involving microtubules where the capsids bind to cellular importin α and β for nuclear transport (Rabe et al., 2006). In line with this, ivermectin, a specific inhibitor of importin α/β-mediated nuclear import, has been shown to inhibit HBV and other viruses (Longarela et al., 2013; Wagstaff et al., 2012). It has been reported that the nucleocapsid-importin complex arriving at the nuclear pore complex binds to nucleoporin 153, and probably other cell proteins followed by importin α and β dissociation by RanGTP and the release of viral DNA and capsid proteins into the nucleoplasm (Pante and Kann, 2002; Schmitz et al., 2010; Schneider et al., 2014). While various host cellular and nuclear machinery have been identified to be involved in de-envelopment and entry of HBV genome into the nucleus, a detailed molecular mechanism is yet to be determined.
5. Deproteination of viral DNA and synthesis of cccDNA
5.1. Biology of deproteination of viral DNA and synthesis of cccDNA
Intact, enveloped virions, as bound to the NTCP receptor, contain “relaxed circular” (rc) DNA, which are incomplete double strands of the viral genome (Seeger and Mason, 2000). The “minus” strand (complimentary to the RNA) is nearly complete, but is covalently linked to the HBV polymerase (pol) at its 5′ end, and a short 8–9 nt terminal redundant sequence on both termini. Compared with the minus strand, the plus strand is several hundred nucleotides shorter at the 3′ end and contains a covalently attached RNA primer at its 5’ end (Guo et al., 2007; Nassal, 2015). To complete its life cycle, these modifications must be removed from rcDNA, and the gaps need to be filled and ligated to have a proper genomic template for transcription. The resultant completely circular genomes are called covalently circular closed (ccc) DNA. cccDNA is the stable nuclear form of the genome that is the template for all progeny genomes.
The mechanism by which rcDNA is converted into cccDNA is not fully known but, as discussed further below, it is believed that the host DNA repair enzymes as well as other cellular enzymes are used (Guo and Guo, 2015). Removal of pol from the rcDNA minus strand, a process called “deproteination”, is thought to be mediated by host cellular enzymes. These are either proteases, endonucleases and, or phosphodiesterases (Gao and Hu, 2007; Guo et al., 2007, 2010; Kock et al., 2010). Topoisomerases (TOP1 or TOP2) regulate viral DNA super-coiling by incising within one or both DNA strands and forms a 3′ or 5’ tyrosyl-DNA phosphodiester bond at the break sites (Champoux, 2001). Pol bound to minus strand of rcDNA is structurally similar to the TOPDNA adduct in terms of the tyrosyl-DNA phosphodiester bond. TOPDNA adducts are substrates for tyrosyl-DNA-phosphodiesterases (TDPs), which cause a trans-esterification reaction and release of the protein (Pommier et al., 2014). Recent studies have demonstrated that TDP2 can catalytically unlink the tyrosyl-minus strand DNA bond between pol and rcDNA in vitro, but the role of this enzyme in cccDNA formation in virus-replicating cells remains controversial (Cui et al., 2015; Koniger et al., 2014).
As mentioned above, rcDNA contains gaps, one on each strand, which need to be filled in and sealed for cccDNA formation. The gaps on rcDNA can be considered as two single strand DNA breaks and hence, induces host DNA damage and repair response which are then presumably exploited by the virus (Guo and Guo, 2015; Schreiner and Nassal, 2017). HBV pol inhibitors do not apparently prevent cccDNA formation from incoming virus, and thus, host polymerases and ligases are implicated in this process. Consistent with this reasoning, Qi et al. reported that cellular DNA polymerase κ plays a role in cccDNA formation during de novo HBV infection in cell cultures (Qi et al., 2016), presumably through filling in the gap on plus strand of the incoming viral genome. The cellular enzyme, flap-like structure specific endonuclease 1 (FEN 1), which is involved in cellular DNA replication and repair, has been reported to be involved in cccDNA formation, possibly at the step of removing one copy of the terminal redundant sequences from the minus strand of rcDNA (Kitamura et al., 2018). Furthermore, host DNA ligase 1 and 3 have been shown to exert overlapping functions in cccDNA formation, likely at the last step of cccDNA biosynthesis through covalently joining the extremities of rcDNA (Long et al., 2017). However, the detailed molecular mechanism and pathway for cccDNA formation awaits further investigation.
In addition to rcDNA, a minor portion of HBV virions contain double stranded linear (dsl) DNA. Sequence analysis of virus-host DNA joints and cccDNA recombinant joints have shown that dslDNA that can be converted into cccDNA and integrated into host chromosome (Bill and Summers, 2004; Yang and Summers, 1995, 1998, 1999). However, while rcDNA is repaired to form wild-type cccDNA, the dslDNA-derived cccDNA carries deletions or insertions around the site of end-joining (Yang and Summers, 1995), suggesting that rcDNA and dslDNA are converted into cccDNA via different DNA repair pathways. In this regard, it was demonstrated that Ku80 and ligase 4, the components of NHEJ DNA repair pathway, are essential for the formation of DHBV cccDNA from dslDNA, but not rcDNA (Guo et al., 2012; Long et al., 2017). It should be noted that integration of viral dslDNA into the host chromosomes is also dependent upon NHEJ (Bill and Summers, 2004). Integration of HBV DNA into host chromosomes is not required for HBV replication, but it may play a role in viral pathogenesis (Tu et al., 2017). In woodchucks, where viremia is extremely high and liver cirrhosis does not often occur, the integrations appear to be a major mechanism of proto-oncogene activation and liver cancer (Fourel et al., 1992, 1994; Hansen et al., 1993; Wei et al., 1992). In patients, the contribution of integrated HBV DNA to activation of cellular proto-onco-genes as a mechanism of liver cancer is apparently much less common than in woodchucks, but can serve as a marker for clonal expansion (Mason et al., 2016). On the other hand, HBV genomes integrated in to host chromosomes, may be an alternative source of HBsAg (Wooddell et al., 2017).
5.2. Therapeutic interference with HBV cccDNA synthesis
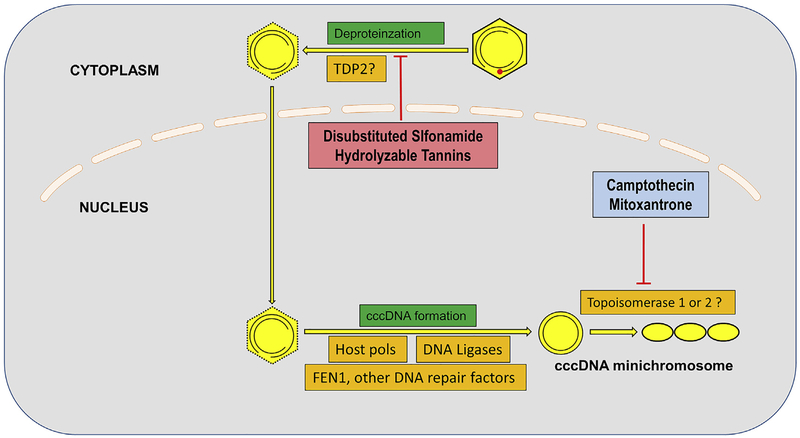
HBV cccDNA is the template for transcription of viral pgRNA (genomic RNA) and is thus essential for production of progeny production. It is stable, and is responsible for long term viral persistence, even after years of Pol inhibitor therapy and suppression of all detectable viral DNA in the circulation (Alweiss and Dandri, 2017). Thus, targeting cccDNA is considered the most important goal in the field of anti-HBV drug design (Alter et al., 2018). The host functions involved in cccDNA formation and the drugs that target them are shown in Fig. 3. Identification of DNA repair enzymes involved in formation of cccDNA may provide a new antiviral drug targets. Disubstituted sulfonamide compounds and hydrolyzable tannins have been identified as inhibitors of cccDNA formation by interfering with rcDNA deproteination (Cai et al., 2012; Liu et al., 2016), though their host or viral target(s) awaits further investigation.
Fig. 3. Deproteination of the viral DNA, and synthesis of viral cccDNA.
The viral polymerase protein within the capsid is covalently attached to the 5′ end of “minus” strand of DNA, and must be removed for the successful production of covalently closed circular DNA (cccDNA), which is the template for all viral transcripts in the normal infection (shown as the 3 interlocked circles). The host functions mediating this process are not definitively known, but TDP2 may be involved. Host polymerase kappa (polK), FEN1, and DNA ligase (LIG) 1 and 3 are needed to complete the formation of cccDNA. Topoisomerases and various DNA repair functions may be involved. Cellular functions mediating these steps are indicated in orange, research phase compounds that interfere with these steps are shown in pinkish red, with compounds that are clinical phase, or approved, in light blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Interferon (IFN)-α and -γ, Tumor Necrosis Factor (TNF)-α, and the activation of lymphotoxin (LT) β (LTβ) receptor have been shown to cause degradation of cccDNA through activation of nuclear APOBEC3 deaminases and subsequently inducing deamination and formation of an Apurinic/Apyrimidinic site in cccDNA for degradation without affecting the host genome (Lucifora et al., 2014; Xia et al., 2016). Thus, use of LTβ receptor agonists or adoptive T cell therapy might lead to receptor-mediated cccDNA degradation and remission of chronic HBV infection. Another cytidine deaminase, the activation-induced cytidine deaminase (AID), has been shown to be upregulated by both IL-1β and TNFα, and induce HBV DNA deamination, however, the AID-mediated anti-HBV activity is not relying on its deaminase activity (Watashi et al., 2013). Such deamination-independent anti-HBV activity has also been observed in study of APOBEC3G (Nguyen et al., 2007).
CRISPR/Cas9 is yet another tool that has emerged to target HBV cccDNA. A series of studies have confirmed the therapeutic effectiveness of this editing tool in targeting HBV DNA both in vitro and in vivo (Kennedy et al., 2015; Lin et al., 2014; Moyo et al., 2017; Ramanan et al., 2015; Seeger and Sohn, 2014, 2016). However, therapeutic use of CRISPR/Cas9 faces significant challenges with respect to delivery, efficiency and safety.
6. Transcription of cccDNA and transport of RNA out of the nucleus
6.1. Biology of transcription of cccDNA and transport of RNA out of the nucleus
As indicated, cccDNA is the major template for all five viral transcripts leading to all progeny viral genomic DNA, and probably most viral mRNA. Recent reports suggested that the integrated viral HBV genome may be a significant contributor of transcripts for mRNA for HBsAg (S), especially in HBeAg-negative patients (Wooddell et al., 2017), although the clinical significance of this is not yet determined. That being said, as with cccDNA, transcription of HBV integrants is mediated by the host RNA pol II (Rall et al., 1983), which recognizes cis acting elements on the viral DNA, and represents a body of prospective host targeting interventions. cccDNA is complexed with cellular histone and non-histone proteins, assembling in to chromatin like structures, as has been called a mini-chromosome (Bock et al., 1994, 2001; Newbold et al., 1995). Enrichment of host RNA pol II on cccDNA has been reported to be associated with active posttranscriptional modifications (PTMs) on ccc-DNA bound histone (Riviere et al., 2015; Zhang et al., 2017).
As shown in Fig. 4, cccDNA is transcribed into 5 major transcripts: (a) 3.5 kb preCore mRNA (specifies preCore polypeptide and HBeAg), (b) 3.5 kb pre-genomic(pg) RNA (the template for reverse transcription of viral DNA), (c) 2.4 kb specifying LHBs (L), (d) 2.1 kb specifying MHBs (M) and SHBs (S), and (e) 0.7 kB transcript specifying X. These 5 mRNA are from 4 promoters, Core, preS, S and X, with M/S coming from the S promoter.
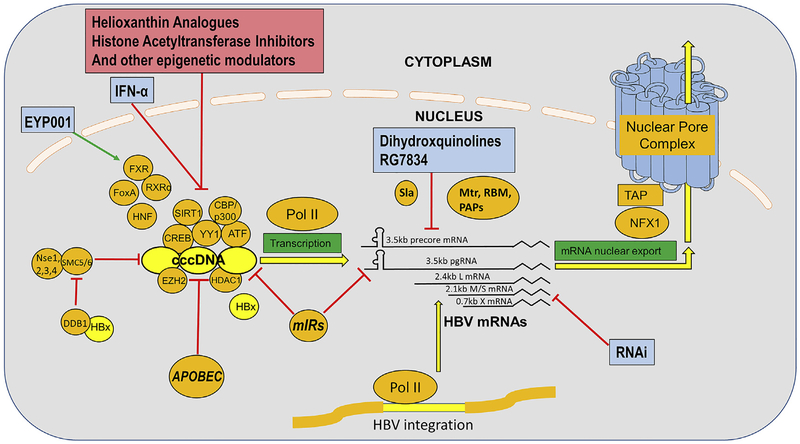
Fig. 4. Transcription of cccDNA and transport of viral RNA out of the nucleus.
HBV cccDNA is associated with, and presumably regulated by numerous host transcriptional factors, and unmodified and modified histones. Host SMC5/6 complex is shown, in this illustration, “attacking” (and transcriptionally repressing) HBV cccDNA, with HBx binding host protein DBB1 which, causes the degradation of SMC5/6, and thus promotes HBV cccDNA transcription. Host pol II transcribes HBV cccDNA as well as HBV DNA integrated in to the host chromosomes, and these transcripts are processed and transported out of the nucleus using numerous host functions, including polyadenylation, although the precise pathways of processing are not well established. Cellular functions mediating these steps are indicated in orange, research phase compounds that interfere with these steps are shown in pinkish red, with compounds that are clinical phase, or approved, in light blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
There is evidence that splicing of HBV RNA occurs, and could lead to transcripts with unappreciated functions and even viral polypeptides not previously recognized (Bayliss et al., 2013; Su et al., 1989). Splice variants have been found in high amounts in many chronically infected patients. Polypeptides encoded by HBV RNA spliced variant transcripts have been hypothesized to play role in viral persistence (Lee GH, 2008). The significance of these splice variants in the normal viral life cycle is unclear, although elevations in circulating levels of them have been associated with increased risk of liver cancer (Bayliss et al., 2013). Thus, understanding the role and importance of the host-regulated splicing event can be another possible point of intervention for drug targets.
Since HBV cccDNA exists as a mini-chromosome, its transcription is regulated by host factors and subject to host epigenetic modification(s) (Hong et al., 2017). Using explants of human livers, a direct correlation was shown between HBV replication activity and the acetylation status of HBV cccDNA-bound H3 and H4 histones (Pollicino et al., 2006). Transcription factors ATF, YY1, p300/CBP and CREB, and proteins that modify chromatin such as STAT1, and EZH2 have also been implicated in regulation of HBV gene expression. There is active co-recruitment of cellular histone acetyltransferases-p300/CBP and PCAF/GCN5- and the histone deacetylases (HDAC1 and hSirt1) and polycomb repressor complex (EZH2 and YY1) in vivo and in vitro onto cccDNA that correlate with high and low viral replication, respectively (Belloni et al., 2009, 2012). In other attempts to study the histone methylation status of cccDNA and regulation of its transcription, PRMT5 (protein methyltransferase 5) has been shown to induce a symmetric di-methylation of H4R3 on cccDNA and regulate the binding of host pol II and host Brg1-based hSW1/SNF chromatin remodeler contributing to the transcriptional repression RNA of cccDNA (Zhang et al., 2017); in addition, hSirt3 was found to restrict HBV cccDNA transcription by acting cooperatively with histone methyltransferase SUV39H1 to decrease the recruitment of SETD1A, leading to a marked increase of H3K9me3 and a decrease of H3K4me3 on cccDNA (Ren et al., 2018).
Consistent with the liver tropism of HBV, liver “specific” transcription factors regulate pgRNA synthesis and activate replication. For example, HNF4 and RXRα/PPARα. HNF3β antagonizes the viral replication by inhibiting pgRNA synthesis in mouse NIH 3T3 fibroblasts (Tang and McLachlan, 2001). Epigenetic changes in the cccDNA alter the binding of various liver-enriched transcription factors to its enhancer region. While HNF4α can bind to various HBV enhancer regions and promote viral transcription (Moolla et al., 2002; Quasdorff et al., 2008; Quasdorff and Protzer, 2010), higher HNF4α expression levels were seen in CHB patients whereas HNF3β levels were observed to be downregulated (Long et al., 2009).
Other, general transcription factors that bind to HBV promoters and enhancers include nuclear factor (NF1), specificity protein 1 (SP1), activator protein 1 (AP1), TATA-binding protein (TBP), prosperous-related homeobox protein 1(PROX1), c-AMP-response element-binding protein (CREB), nuclear factor-kappa B (NF-kB), octane transcription factor 1 (OCT1), and nuclear respiratory factor 1 (NRF-1). HNF1α has been shown to control HBV regulatory elements, such as, preS1 promoter by binding directly to it (Raney et al., 1991) and enhancer II (EnhII) by interacting with it through hB1F1 or B element in the enhancer region in cccDNA (Cai et al., 2003) and thus, regulating cccDNA transcription. HNF3α and HNF3β binding motifs were observed in the EnhII region and competitive binding of HNF3β and HNF4 was also noticed in HBV EnhII region (Li et al., 1995). HNF6 is involved in liver homeostasis and liver cell proliferation and has been revealed to inhibit HBV gene expression in HepG2 cells by suppressing preS2/S promoter activity (Hao et al., 2015). A liver-enriched transcription factor, C/EBP has also been demonstrated to bind to HBV promoters and enhancers and promote HBV transcription by transactivation of EnhII and synergistically activate EnhII through interaction with HBx (Lopez-Cabrera et al., 1991). C/EBP has also been proven to suppress core promoter (Lopez-Cabrera et al., 1990) and bind to enhancer I (EnhI) to repress HBV transcription (Pei and Shih, 1990). The Fox A transcription factors, which are critical in mediating development and tissue specific expression, appear to be important to the regulation of the expression of the HBV genome, which contains multiple FoxA binding sites. Briefly, the HBV transgene in the hepatocytes of FoxA deficient mice was highly methylated, and was not efficiently transcribed (McFadden et al., 2017). It is worth noting that the above studies are largely performed with transfected HBV plasmid in cell culture or transgenic HBV DNA in mice. In addition, many transcription factors that influence HBV cccDNA expression are also associated with host cell metabolism and nutrition. In this regard, CREB, HNFs and Farnesoid X receptor (FXR) fall in to this category, although the FXR may regulate HBV via direct affects upon viral functions, and by indirect affects upon bile salts (Ramiere et al., 2008; Wang et al., 1999). Nonetheless, the FXR deserves special attention, since a FXR agonist (EYP001) is now in clinical trials (Erken et al., 2018).
Methylation of CpG islands within integrated HBV DNA occurs (Miller and Robinson, 1983) and has been correlated with HCC progression (Chen et al., 1988). CpG methylation within the HBV cccDNA may also contribute to viral gene regulation during viral infection, highlighting another system of host factors affecting the HBV genome of cccDNA, has been reported (Jain et al., 2015; Mogul and Schwarz, 2011; Zhang et al., 2014). Interaction of the HBV core protein with cccDNA has been observed to be correlated with association of host CREB binding protein (CBP), hypomethylation and decreased HDAC1 binding on CpG island of cccDNA (Guo et al., 2009).
Taken together, host epigenetic readers, writers and erasers and host cytokines play an important role in regulating HBV cccDNA transcription the detailed proteomics and epigenomics of cccDNA mini-chromosome and may provide opportunities for drug intervention targeting the most important HBV player, cccDNA.
The HBV “X” (HBx) polypeptide has been reported to affect HBV cccDNA transcription activity by its interaction with cellular polypep-tides. Initially it was observed that p300 recruitment to cccDNA was impaired while there was increased recruitment of HDACs, hSirt1 and HDAC1, in HBx mutant (cells replicating with HBx defective virus). Accordingly, while the HBx mutant cells didn’t show any difference in the cccDNA pool compared to the wild type, there was impairment of cccDNA transcription that lead to reduced pgRNA formation suggesting HBx’s role in epigenetic regulation of cccDNA transcription. Also, the kinetics of HBx recruitment was concurrent with HBV replication (Belloni et al., 2009; Lucifora et al., 2011).
Recently, HBx has been reported to function by recruiting the DDB1 ubiquitin ligase to degrade SMC5/6, a putative negative regulator of HBV transcription (Decorsiere et al., 2016; Murphy et al., 2016). HBx has been reported to also work via AP-2, CREB, TFIIB, TFIH and has also been thought to oppose the cccDNA repressive activities of host protein Spindlin 1 and possibly to “overcome” SETDB1 mediated H3Kme3 and HP1 condensation of cccDNA (Riviere et al., 2015). See Fig. 4.
Cellular microRNAs (miRNAs) are another host system that may influence HBV gene expression and hence its life cycle. Although a direct cause and relationship between specific miRNAs and HBV gene expression has not been unambiguously shown, there is a growing body of evidence associating levels of specific miRNAs and HBV levels in people, as well as evidence of an interplay between HBx and a number of microRNAs (Novellino et al., 2012; Wei et al., 2013a, 2013b; Zhang et al., 2013). miRNA-18a prevents the expression of ER-α and thus inhibiting the repressive effect of ER-α on HBV replication through its interaction with HNF4α (Liu et al., 2009). Other miRNAs, such as, miRNA-1, 148a, 152, 210 and 449a, have been speculated to regulate HBV replication by targeting host epigenetic factors-DNMT and HDAC (Braconi et al., 2010; Huang et al., 2010; Zhang et al., 2009, 2011). A number miRNAs are known to target HBV transcripts, such as, miRNA-122, which is abundant in hepatocytes and targets and binds a highly conserved site on pgRNA, reducing the level of viral expression (Chen et al., 2011b; Qiu et al., 2010; Wang et al., 2012). miRNA125a-5p, miRNA199–3p, miRNA210, miR-224 have all been reported to specifically bind HBV mRNA. miRNA-199a-3p and miRNA-125a-5p bind to the 2.1 kb RNA species, and miRNA-210 bind to 2.4 kb RNA species, and directly and suppress translation (Potenza et al., 2011; Zhang et al., 2010). Moreover, miRNA-1231 targets pgRNA and suppresses replication (Kohno et al., 2014). Bioinformatics analyses have also identified miRNA-199a-3p, miRNA-210 and miRNA-345 binding sites on 3.5 kb RNA; miRNA-let7, −196b and −511 binding sites on 2.4 kb RNA and miRNA-433 binding on 2.1 kb RNA and miRNA-205 on 0.7kbRNA (Liu et al., 2009; Potenza et al., 2011; Wu et al., 2015; Zhang et al., 2010, 2013). However, the biological significance of these binding sites is not firmly established.
HBV transcripts contain significant, and unusual, secondary structures, at their 5′ and near the 3’ ends, such as “epsilon” and PRE (post transcriptional element). Both of these elements may have cis functions to facilitate either transport, translation, stability and, or packaging in to capsids (in the case of epsilon). They are likely to interact with host functions that mediate these processes, and given how unusual are their structures (found more often in noncoding than in coding RNAs), they may present opportunities for antiviral targeting. Curiously, there is evidence that HBV pregenomic RNA may use the host TRAMP pathway normally used for quality control and degradation of defective host transcripts, for its maturation (Javanbakht et al., 2017; Mueller et al., 2018; Zhou et al., 2018).
It has been reported that the host zinc finger antiviral protein (ZAP) and ribonuclease ISG20 target the epsilon structure of HBV RNA for degradation in nucleus (Liu et al., 2017; Mao et al., 2013). In addition, AID has been shown to interact with HBV pol and RNA processing exosome to degrade HBV RNA (Liang et al., 2015a). Lastly, the nuclear export of HBV RNA is regulated by PRE and involves a cellular factor TAP/NXF1 (Tip-associated protein/nuclear export factor-1) (Li et al., 2010; Lim and Brown, 2016; Yang et al., 2014).
6.2. Therapeutic interference with HBV cccDNA transcription and transcripts
Drug discovery approaches aiming to target the epigenetic enzymes are well established in anti-cancer therapy. The presence of upregulated acetylated H3 and H4 on cccDNA offers the possibility of incorporating HAT inhibitors as treatment options for chronic patients, although toxicity inherent in these approaches presents a challenge. Use of PRMT5 agonists might be another approach to repress HBV cccDNA transcription. Also, as illustrated in Fig. 4, HBV cccDNA transcription is regulated positively and negatively by host transcription factors, and as the contributions of these factors become better understood, it is possible to imagine drugs targeting the factors upon which the virus is most, and perhaps selective dependent, that could have useful antiviral activity. Helioxanthin analogues, for example, have been shown to have a potent antiviral effect in hepatoma cells stably producing HBV by down-regulation of critical transcription factors (Ying et al., 2007). Again, of course, the major concern with histone and transcriptional modifiers is their selectivity, or possible lack thereof, for viral functions.
IFN-α treatment, in vitro, using HBV producing hepatoma cells and, in vivo, using mouse models, repressed HBV cccDNA transcription (Belloni et al., 2012). In cell-based systems, IFN-α, has been shown to suppress HBV cccDNA transcription by promoting recruitment of HDAC1 to HBV mini-chromosome and reducing the acetylation of cccDNA-associated histone H3 lysine 9 (H3K9) and 27 (H3K27) (Belloni et al., 2009; Liu et al., 2013).
Exploiting the ability of SMC5/6 to repress HBV cccDNA, is certainly worth exploring, possibly by way of inhibiting the HBx poly-peptide, since HBx was reported to oppose the SMC5/6-mediated repression of HBV cccDNA (Decorsiere et al., 2016; Murphy et al., 2016).
Recently, we and others have reported that a small molecule, a dihydroxyquinoline, can cause the rapid degradation of HBV transcripts (Mueller et al., 2018; Zhou et al., 2018). The degradation occurs in the nucleus, and is selective, to the extent that most other host transcripts and proteins tested are not significantly affected, over the period of study. The drug’s action requires that the viral transcript contain the highly structured PRE region, which could be the basis for a viral selectivity. However, the PRE hair pin structures are similar to sub sets of noncoding cellular RNAs, suggesting that host functions may also be targeted by this compound. The ultimate clinical usefulness of this approach remains to be seen, but these data certainly establish the principle of targeting viral RNA stability and transport for selective antiviral therapy.
7. HBV RNA translation, capsid formation, pgRNA encapsidation and reverse transcription
7.1. HBV RNA translation, capsid formation, pgRNA encapsidation and reverse transcription
There are at least 5 HBV mRNA species that translate 7 HBV proteins (see Introduction) in cytoplasm by using cellular translation machinery. Production of HBV e antigen (HBeAg) is mediated by a host furin endopeptidase that cleaves the C-terminus of HBeAg precursor, preCore protein, and the product (HBeAg) is secreted into the serum (Ito et al., 2009).
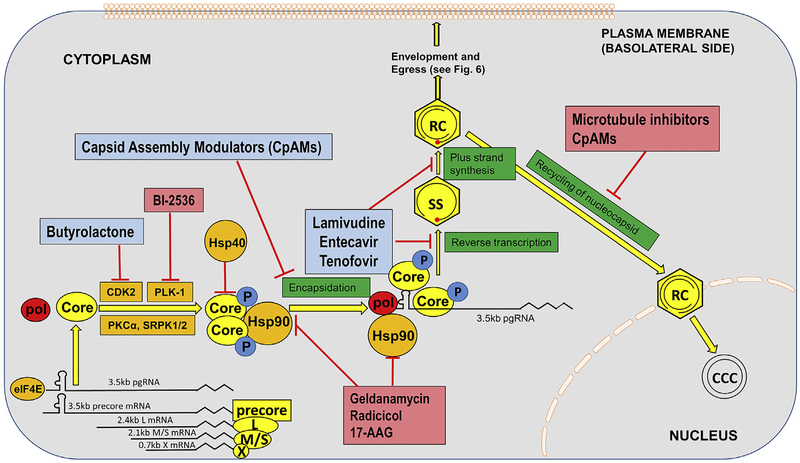
As indicated in Fig. 5, Core and pol are translated from the same mRNA, the pgRNA (Ou et al., 1990). There are 240 core proteins in every capsid, and only one pol. Thus, it is likely that the translation efficiencies of Core and pol are regulated by host, as well as viral, factors. Due to the bicistronic nature of Core and pol ORFs within pgRNA, translation of pol occurs by more than one mechanism. The majority of pol is translated by re-initiation at the first AUG after translation termination of a mini-cistron that is translated from the second AUG between the Core and Pol ORFs (Hwang and Su, 1998). Pol translation is also initiated via at least two mechanisms: (i) leaky scanning through the four AUGs between core and pol ORFs and (ii) backward scanning to the pol AUG after translation termination of the core ORF (Sen et al., 2004).
Fig. 5. HBV RNA translation, capsid formation, encapsidation, reverse transcription and recycling of the nucleocapsid to the nucleus.
Five main species of HBV mRNA are translated in the cytoplasm in to the viral polypeptides precore (precursor of HBeAg, from the 3.5 kb precore mRNA), core and pol (from the 3.5 kb pgRNA), envelope polypeptides L (from the 2.4 kb mRNA), envelope polypeptide, M and S (from the 2.1 kb mRNA) and X from the 0.7 kb mRNA. Core rapidly dimerizes, is modified by phosphorylations, and cooperates with pol and pgRNA to form the nucleocapsid. Host factors modulate translation, phosphorylation, and even folding through chaperons of the viral functions. Encapsidated viral pgRNA is reverse transcribed, and nucleocapsids are either enveloped and secreted (see Fig. 6), or not enveloped and recycled to the nucleus, where they may then begin the cycle of cccDNA production, and transcription of new viral gene products. Cellular functions mediating these steps are indicated in orange, research phase compounds that interfere with these steps are shown in pinkish red, with compounds that are clinical phase, or approved, in light blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
HBV core protein is ~183–185 amino acid (aa) long, depending on the genotype, and forms dimers and multimers which are assembled into the capsid (Gallina et al., 1989; Nassal, 1992; Zlotnick et al., 1997). The C-terminal domain (CTD) of core has serine-phosphorylation sites, which are substrates for host kinases. This phosphorylation correlate with pgRNA encapsidation and the subsequent dephosphorylation has been concurrent with capsid maturation, envelopment and egress from the cell (Lan et al., 1999; Melegari et al., 2005; Perlman and Hu, 2003). The host cellular kinases involved include cdc2-like kinase, CDK2, Polo-like-kinase 1 (PLK-1), Protein kinase C-α (PKC-α), SRPK1 and SRPK2 (Barrasa et al., 2001; Daub et al., 2002; Diab et al., 2017; Kann and Gerlich, 1994; Kau and Ting, 1998; Ludgate et al., 2012). However, the detailed roles of the above-mentioned kinases need to be further characterized. Wittkop et al. showed that inhibition of PKC-α doesn’t affect genome maturation but hinders virion formation and leads to capsid accumulation due to defects in the latter’s development (Wittkop et al., 2010). SRPK1 is also considered to gate and help in capsid assembly (Chen et al., 2011a). NIRF (Np95/ICBP90-like ring finger protein) is an ubiquitin ligase that interacts with the core protein to regulate its half-life, and causes its proteosomal degradation and destabilizes the capsid (Qian et al., 2012). Along the same lines, while Hsp90 interacts with the core to stabilize the capsid, Hsp40 is yet another chaperone protein that antagonizes this protein-protein interaction to destabilize capsid and also degrades HBx protein (Shim et al., 2011; Sohn et al., 2006). Whether HBV capsid assembly occurs in a specific cell compartment is largely unknown, but a recent study indicates that an intact cellular microtubule network is required for capsid formation (Iwamoto et al., 2017).
After translation, in-situ binding of pol to epsilon (ε) stem-loop structure of pgRNA inhibits viral translation and triggers nucleocapsid assembly by specific incorporation of both pgRNA and pol into viral capsids, which is called pgRNA encapsidation (Ryu et al., 2008, 2010; Seeger and Mason, 2000). The proper folding of pol and its binding to pgRNA are assisted by host chaperons including HSP90 and its cofactors (Hu et al., 2002). HBV DNA replication exclusively takes place in viral nucleocapsid, where the pgRNA is reverse transcribed into rcDNA by pol (Nassal, 2008). Several host proteins have been shown to be packaged into HBV nucleocapsid via interaction with pol and potentially regulate the reverse transcription, including eIF4E, DDX3, APOBEC3G, etc (Kim et al., 2010; Nguyen and Hu, 2008; Wang et al., 2009).
7.2. Therapeutic interference with HBV RNA translation, capsid formation and pgRNA encapsidation and reverse transcription
At present, there are 5 Direct Acting Antiviral (DAA) polymerase inhibitors (Liang et al., 2015b). As indicated in this review, it is possible, in theory, to inhibit the viral reverse transcriptase by targeting host functions, and drugs that affect cell chaperons involved in reverse transcription have shown to be antiviral. Indeed, interference with any of the host proteins involved in capsid and virion formation, could have antiviral activity. Hsp90 inhibitors have been gaining importance in cancer treatment for its potential for combinatorial targeting of multiple oncogenic protein pathways. Since Hsp90 is involved in pol-ε complex formation and Hsp90-Core interaction stabilizes the capsid, it is possible that Hsp90 small molecule inhibitors, such as geldanamycin and radicicol (natural products) as well as the semisynthetic derivatives 17-N-Allylamino-17-demethoxygeldanamycin (17AAG), could have anti-HBV potential. Hsp40 analogues or agonists might also be employed to reduce the stability of the capsid, by causing degradation of HBc and HBx proteins (Sohn et al., 2006). Moreover, since HBx has been shown to regulate the transcription of cccDNA, Hsp40 analogues may also inhibit cccDNA transcription.
Various kinase inhibitors are presently employed in the field of cancer treatment, and thus kinase inhibition is well established. For HBV intervention, since phosphorylation of core plays a role in regulating capsid formation and nucleocapsid maturation, inhibition of the host kinases involved is a possible antiviral strategy. Butyrolactone I is a selective inhibitor of CDK2 and cdc2 kinase (Kitagawa et al., 1993), and has been used in management of cancer. Several other small molecule kinase inhibitors specific for PKC-α and cdc-2, which mediate capsid formation, are now commercially available. However, as with any host targeting strategy, toxicity is always a concern. Encouragingly, administration of PLK-1 inhibitor BI-2536 to HBV-infected humanized liver FRG mice strongly inhibited HBV infection, and the compound was safely tolerated by the animals (Diab et al., 2017).
8. Envelope protein synthesis, viral particle assembly and egress
8.1. Biology of envelope protein synthesis, viral particle assembly and egress
Hepatitis B surface polypeptides (the envelope proteins) are translated from 2 species of mRNA (2.1 and 2.4 kb). There are 3 envelope polypeptides, the “large”, 42 kD LHBs (L) middle sized 33 kD MHBs (M) and the most abundant, small 26 kD, SHBs (S) polypeptide. They are expressed from a single open reading frame, with alternate start codons. L is translated from the 2.4 kb mRNA, and S and M from the 2.1 kb mRNA. The S polypeptide is secreted from the cell in far greater amounts than are L and M. The envelope polypeptides are N-glycosylated. Glycosylation and possibly other post-translational modifications play key roles in morphogenesis and secretion of the virions and subviral particles (Lu et al., 1995; Mehta et al., 1997; Werr and Prange, 1998).
In addition to a common N glycosylation site on all 3 polypeptides, M possesses a second N-glycan and an O-glycan site near its N-terminus, and this plays a role in its folding. Following synthesis and N-glycosylation, envelope polypeptides “oligomerize” at the ER/ERGIC membranes, with spherical sub viral particles apparently returning to the Golgi for possibly further processing and secretion. Folding of M and L envelope proteins are dependent upon N-glycan processing, host chaperone proteins, such as heat shock protein (Hsp) 70 and ER resident proteins BiP and calnexin (Fig. 6). Maturation of S may be independent of at least N-glycan processing and calnexin, but still involves the ER luminal protein, protein disulfide isomerase (PDI) that seems to monitor the disulfide linkages for cross-linking of S chains during envelope assembly (Prange, 2012).
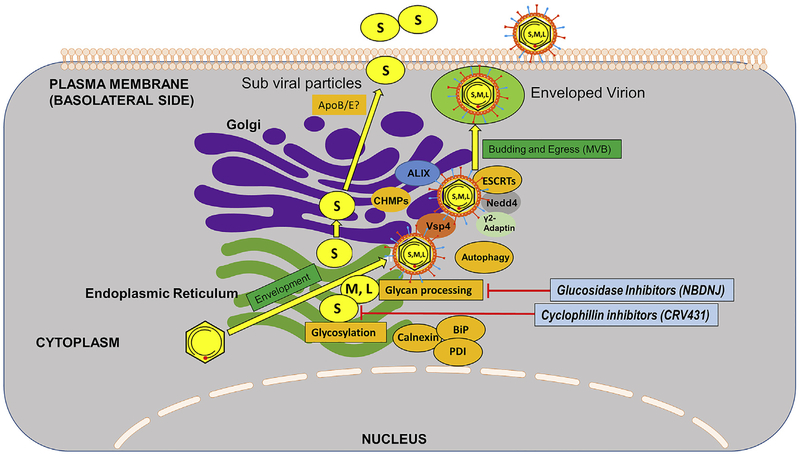
Fig. 6. Morphogenesis, envelopment and secretion of HBV.
Nucleocapsids associated with newly synthesized, envelope polypeptides, S, M, and L, which would have been co- and post-translationally modified by N-glycosylations and glycan processing (L, M, S) and O-glycosylation (M), and myristolyation (L). M, and possibly L, are “folded” by Calnexin, and BiP. L, M and S are all heavily disulfide bonded, are use Protein Disulphide Isomerase (PDI) is their maturation. Numerous cell functions mediate vesicle transport. Subviral particles of oligomerized S bud in to the lumen of the ER-Golgi and are secreted out of the cell via constitutive secretory mechanisms. Envelopment of nucleocapsids, to form virions, probably occurs at the ER and then a Multi Vesicular Body (MVP). MVP derived exosomes containing enveloped virions are thought to fuse with plasma membranes and result in virion release in to the blood. Note that this occurs in a “polarized” hepatocyte, in which secretion of virions is through the basolateral side of the cell. Cellular functions mediating these steps are indicated in orange, research phase compounds that interfere with these steps are shown in pinkish red, with compounds that are clinical phase, or approved, in light blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
HBV is thought to exploit the Multi Vesicular Body (MVB) machinery for virion maturation and egress (Rost et al., 2008). HBV envelope proteins have been found to co-localize with Class E MVB proteins- AIP1/ALIX and VPS4B- in hepatoma cells through immunofluorescence and a dominant negative (DN) AIP1 mutant inhibited production and/or release of enveloped virions without significant effects on intracellular nucleocapsid formation, whereas DN VPS4B inhibited both nucleocapsid production and budding. By contrast, DN AIP1 and VPS4 had no effect on the efficiency of release of HBV subviral particles, which are secreted via the ER-Golgi constitutive secretory pathway (Watanabe et al., 2007). Lambert et al. used DN ESCRT III mutant proteins (CHMPs) and ATPase defective Vps4A and B to show that the expression of these defective mutant players of MVB inhibited virus assembly and release (Lambert et al., 2007). The E3 ubiquitin ligase- γ−2 adaptin of the AP-I complex have been identified as important players in MVB-mediated HBV assembly and egress. Indeed, HBV core and L protein have been found to co-localize with γ−2 adaptin in the late endosomal compartments and disruption of HBV/γ2-adaptin interactions inhibits virus production. While depletion of γ−2 adaptin inhibited HBV release (Rost et al., 2006), overexpression of the adapter produced similar results, as observed with mutant MVB proteins-entrapment of HBV core and L protein in detergent-insoluble compartments (Lambert et al., 2007). NEDD4 has been shown to interact with HBV core protein in vivo through PPAY motif (thought to be exposed on the capsid as seen by cryo-EM) through co-immunoprecipitation and generating PPAY sequence mutant (Rost et al., 2008). HBV has also been found to utilize the ESCRT-II complex, comprised of EAP20, EAP30, and EAP45, for secretion (Prange, 2012). Thus, HBV exploits various ubiquitin receptors together with endosomal sorting pathway functions for egress from hepatocytes.
Autophagy may be involved in the HBV life cycle. Both the viral X protein and the S envelope proteins have been reported to be inducers of autophagy and found in close association with LC3, an autophagosomal membrane protein (Li et al., 2011; Sir et al., 2010). Thus, it seems possible that the HBV envelope proteins may use autophagic pathways in their transport to the endosomal system. In line with this notion, GTPase Rab7 has been identified as a central regulator of HBV transport and secretion. HBV precore protein has been shown to enhance Rab7 activity causing tubulations of MVB and autophagic compartments and interaction between the former mentioned two compartments with lysosomes and thus regulating its own secretion. Depletion of Rab7 or inhibition of lysosome function caused an increase in HBV secretion (Inoue et al., 2015). It is worthwhile to note that cellular lipids and sterols are also implication in virion secretion, and inhibitors of HMG CoA reductase have been reported to inhibit virion secretion. S has been reported to require cholesterol (synthesized by HMG CoA reductase) for its secretion without affecting the release for Dane particles (Lin et al., 2003). The budding and egress of HBV respects cell polarity, favoring egress in to the basal membranes (in to the circulation) and not cell to cell and not into the bile (Bhat et al., 2011).
Taken together, host factors are clearly critical for morphogenesis and exit of subviral particles and enveloped infectious virions from the cell. However, the pathway used for infectious virion and sub viral particle secretion are distinct, at least functionally. While γ−2 Adaptin, Nedd4, ESCRT II, III and vsp4 are essential for virion secretion, they are dispensable for subviral secretion. It is therefore conceivable that drugs can be found that selectively repress one, but not the other, pathway. Identifying the exact route and the factors involved in the secretion pathway of different kinds of viral particles can help us better understand the egress of the virus and will prove a better outlook towards designing drugs to inhibit the egress.
8.2. Therapeutic interference with HBV morphogenesis
Cyclophilin A (Cyp A) has been identified as a binding partner for S-protein and is speculated to help in envelope assembly and release through interaction with the proline residues on the first cytosolic loop of S (Tian et al., 2010). Therefore, cyclophilin inhibitors, such as CRV431 and alisporivir have been proposed as antiviral agents (Phillips et al., 2015; Trepanier et al., 2017).
S protein has also been shown to associate with LC3 autophagosomal protein, indicating use of the host autophagy machinery in viral envelopment (Li et al., 2011). Autophagy was also reported to be required for HBV replication (Sir et al., 2010). Autophagy inhibitors may be useful against HBV.
Glucosidase inhibitors have been shown to have anti-HBV effect by inhibiting envelope protein glycosylation processing (the removal of terminal glucose residues from the N-glycan on the polypeptide) in the ER and thereby, inhibiting viral morphogenesis and infectivity in vitro and in the woodchuck model (Block et al., 1998).
9. Conclusion and perspective
Viruses interact with many host cellular pathways to achieve their replication cycle. Entry into the host cell, transport to the viral replication sites or viral exit from the host cell are all steps that require specific interactions between the virus and its host. Additionally, the evasion from the host immune response requires a lot of viral proteins to associate with and inhibit cellular proteins with antiviral functions. Viruses display remarkable specificity in both the host species and the cell types that they infect. Understanding this specificity reveals insight into the basic host components that are required for the viral life cycle and host restriction factors that limit the virus. HBV is a small virus, encoding only four open reading frames. Because it is so compact it must be adapted to its host to take full advantage of host systems, something commonly seen in viruses. Consequently, HBV proteins must be multi-functional. In this review we tried to identify the different host factors that interact and effect HBV as it proceeds along its life cycle starting from its entry to a cell-specific receptor to egress of new viral particles. While there are host restriction and immune factors that work towards fighting against the infection, the virus utilizes the basic host machinery to sustain within the host cell. We emphasize the toxicity concerns inherent in an intervention that targets host functions, and have taken care to distinguish between compounds that target host functions but serve only as research tools, and compounds that have promise as therapeutic leads. This review aims at identifying feasible druggable targets that helps us take a step forward towards understanding HBV-host interaction and working towards fighting the infection especially in cases of chronic patients. Combination therapy might be the best way to move forward considering the very limited treatment options and development of viral resistance to existing drugs. Thus, using different approaches to target the virus directly or proviral host factors at more than one step along the viral life cycle might be the future of HBV treatment. While the path to understanding and targeting host factors as antivirals have been less traveled and significant challenges remain, delivering the most effective antiviral regimen, including both host and viral targets, should be well worth the effort.
Acknowledgments
Ms. Judith Marchand is thanked for her help in the manuscript preparation. We apologize to numerous investigators whose original contributions could not be cited due to space limitations.
This study is supported by the US National Institutes of Health (NIH) grants (AI094474, AI104636, AI110762, AI123271, AI134818), the Commonwealth of Pennsylvania, and the Carol and Edmund Blake Foundation.
References
- Allweiss L, Dandri M, 2017. The role of cccDNA in HBV maintenance. Viruses 9, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter H, Block T, Brown N, Brownstein A, Brosgart C, Chang KM, Chen PJ, Chisari FV, Cohen C, El-Serag H, Feld J, Gish R, Glenn J, Greten T, Guo H, Guo JT, Hoshida Y, Hu J, Kowdley KV, Li W, Liang J, Locarnini S, Lok AS, Mason W, McMahon B, Mehta A, Perrillo R, Revill P, Rice CM, Rinaudo J, Schinazi R, Seeger C, Shetty K, Tavis J, Zoulim F, 2018. A research agenda for curing chronic hepatitis B virus infection. Hepatology 67, 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrasa MI, Guo JT, Saputelli J, Mason WS, Seeger C, 2001. Does a cdc2 kinase-like recognition motif on the core protein of hepadnaviruses regulate assembly and disintegration of capsids? J. Virol 75, 2024–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss J, Lim L, Thompson AJ, Desmond P, Angus P, Locarnini S, Revill PA, 2013. Hepatitis B virus splicing is enhanced prior to development of hepatocellular carcinoma. J. Hepatol 59, 1022–1028. [DOI] [PubMed] [Google Scholar]
- Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M, 2009. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc. Natl. Acad. Sci. U.S.A 106, 19975–19979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M, 2012. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. JCI (J. Clin. Investig.) 122, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belongia EA, Consta J, Gareen IF, Grem JL, Inadomi JM, Kern ER, McHugh JA, Peterson GM, Rein MF, Sorrell MF, Strader DB, Trotter HT, 2009. National instittutes of health consensus development conference statement: management of hepatitis B. Ann. Intern. Med 150, 104–110. [DOI] [PubMed] [Google Scholar]
- Bhat P, Snooks MJ, Anderson DA, 2011. Hepatocytes traffic and export hepatitis B virus basolaterally by polarity-dependent mechanisms. J. Virol 85, 12474–12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill CA, Summers J, 2004. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc. Natl. Acad. Sci. U.S.A 101, 11135–11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank A, Markert C, Hohmann N, Carls A, Mikus G, Lehr T, Alexandrov A, Haag M, Schwab M, Urban S, Haefeli WE, 2016. First-in-human application of the novel hepatitis B and hepatitis D virus entry inhibitor myrcludex B. J. Hepatol 65, 483–489. [DOI] [PubMed] [Google Scholar]
- Block TM, Lu X, Mehta A, Park J, Blumberg BS, Dwek R, 1998. Role of glycan processing in hepatitis B virus envelope protein trafficking. Adv. Exp. Med. Biol 435, 207–216. [DOI] [PubMed] [Google Scholar]
- Block TM, Mehta AS, Fimmel CJ, Jordan R, 2003. Molecular viral oncology of hepatocellular carcinoma. Oncogene 22, 5093–5107. [DOI] [PubMed] [Google Scholar]
- Block TM, Guo H, Guo JT, 2007. Molecular virology of hepatitis B virus for clinicians. Clin. Liver Dis 11, 685–706 vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block TM, Rawat S, Brosgart CL, 2015. Chronic hepatitis B: a wave of new therapies on the horizon. Antivir. Res 121, 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock CT, Schranz P, Schroder CH, Zentgraf H, 1994. Hepatitis B virus genome is organized into nucleosomes in the nucleus of the infected cell. Virus Gene. 8, 215–229. [DOI] [PubMed] [Google Scholar]
- Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H, 2001. Structural organization of the hepatitis B virus minichromosome. JMB (J. Mol. Biol.) 307, 183–196. [DOI] [PubMed] [Google Scholar]
- Bogomolov P, Alexandrov A, Voronkova N, Macievich M, Kokina K, Petrachenkova M, Lehr T, Lempp FA, Wedemeyer H, Haag M, Schwab M, Haefeli WE, Blank A, Urban S, 2016. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: first results of a phase Ib/IIa study. J. Hepatol 65, 490–498. [DOI] [PubMed] [Google Scholar]
- Bouezzedine F, Fardel O, Gripon P, 2015. Interleukin 6 inhibits HBV entry through NTCP down regulation. Virology 481, 34–42. [DOI] [PubMed] [Google Scholar]
- Braconi C, Huang N, Patel T, 2010. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 51, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss V, Hagelstein J, Gerhardt E, Galle PR, 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218, 396–399. [DOI] [PubMed] [Google Scholar]
- Cai YN, Zhou Q, Kong YY, Li M, Viollet B, Xie YH, Wang Y, 2003. LRH-1/hB1F and HNF1 synergistically up-regulate hepatitis B virus gene transcription and DNA replication. Cell Res. 13, 451–458. [DOI] [PubMed] [Google Scholar]
- Cai D, Mills C, Yu W, Yan R, Aldrich CE, Saputelli JR, Mason WS, Xu X, Guo JT, Block TM, Cuconati A, Guo H, 2012. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob. Agents Chemother. 56, 4277–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ, 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem 70, 369–413. [DOI] [PubMed] [Google Scholar]
- Chen JY, Hsu HC, Lee CS, Chen DS, Zuckerman AJ, Harrison TJ, 1988. Detection of hepatitis B virus DNA in hepatocellular carcinoma: methylation of integrated viral DNA. J. Virol Meth 19, 257–263. [DOI] [PubMed] [Google Scholar]
- Chen C, Wang JC, Zlotnick A, 2011a. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog. 7, e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shen A, Rider PJ, Yu Y, Wu K, Mu Y, Hao Q, Liu Y, Gong H, Zhu Y, Liu F, Wu J, 2011b. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. Faseb. J 25, 4511–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Guo JT, Hu J, 2015. Hepatitis B virus covalently closed circular DNA formation in immortalized mouse hepatocytes associated with nucleocapsid destabilization. J. Virol 89, 9021–9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Blencke S, Habenberger P, Kurtenbach A, Dennenmoser J, Wissing J, Ullrich A, Cotten M, 2002. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J. Virol 76, 8124–8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decorsiere A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, Livingston CM, Niu C, Fletcher SP, Hantz O, Strubin M, 2016. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 531, 386–389. [DOI] [PubMed] [Google Scholar]
- Diab A, Foca A, Fusil F, Lahlali T, Jalaguier P, Amirache F, N’Guyen L, Isorce N, Cosset FL, Zoulim F, Andrisani O, Durantel D, 2017. Polo-like-kinase 1 is a proviral host factor for hepatitis B virus replication. Hepatology 66, 1750–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag HB, Mason AC, 1999. Rising incidence of hepatocellular carcinoma in the United States. N. Engl. J. Med 340, 745–750. [DOI] [PubMed] [Google Scholar]
- Erken R, Stelma F, Roy E, Diane S, Andre P, Vonderscher J, Eric M, Tim S, Philippe P, Christian L, Scalfaro P, Reesink H, Sousa CM, Jacob E, 2018. First clinical evaluation in chronic hepatitis B patients of the synthetic farnesoid X receptor agonist EYP001. J. Hepatol 68, S488–S489. [Google Scholar]
- Feener EP, Shen WC, Ryser HJ, 1990. Cleavage of disulfide bonds in endocytosed macromolecules. A processing not associated with lysosomes or endosomes. J. Biol. Chem 265, 18780–18785. [PubMed] [Google Scholar]
- Fourel G, Transy C, Tennant BC, Buendia MA, 1992. Expression of the woodchuck N-myc2 retroposon in brain and in liver tumors is driven by a cryptic N-myc promoter. Mol. Cell Biol 12, 5336–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Couturier J, Wei Y, Apiou F, Tiollais P, Buendia MA, 1994. Evidence for long-range oncogene activation by hepadnavirus insertion. EMBO J. 13, 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina A, Bonelli F, Zentilin L, Rindi G, Muttini M, Milanesi G, 1989. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J. Virol 63, 4645–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D, Prince AM, 2004. Hepatitis B virus infection–natural history and clinical consequences. N. Engl. J. Med 350, 1118–1129. [DOI] [PubMed] [Google Scholar]
- Gao W, Hu J, 2007. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J. Virol 81, 6164–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripon P, Le Seyec J, Rumin S, Guguen-Guillouzo C, 1995. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 213, 292–299. [DOI] [PubMed] [Google Scholar]
- Guo JT, Guo H, 2015. Metabolism and function of hepatitis B virus cccDNA: implications for the development of cccDNA-targeting antiviral therapeutics. Antivir. Res 122, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT, 2007. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J. Virol 81, 12472–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li Y, Mu S, Zhang J, Yan Z, 2009. Evidence that methylation of hepatitis B virus covalently closed circular DNA in liver tissues of patients with chronic hepatitis B modulates HBV replication. J. Med. Virol 81, 1177–1183. [DOI] [PubMed] [Google Scholar]
- Guo H, Mao R, Block TM, Guo JT, 2010. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J. Virol 84, 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Xu C, Zhou T, Block TM, Guo JT, 2012. Characterization of the host factors required for hepadnavirus covalently closed circular (ccc) DNA formation. PLoS One 7, e43270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch B, Meier PJ, 1994. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. JCI (J. Clin. Investig.) 93, 1326–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LJ, Tennant BC, Seeger C, Ganem D, 1993. Differential activation of myc gene family members in hepatic carcinogenesis by closely related hepatitis B viruses. Mol. Cell Biol 13, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao R, He J, Liu X, Gao G, Liu D, Cui L, Yu G, Yu W, Chen Y, Guo D, 2015. Inhibition of hepatitis B virus gene expression and replication by hepatocyte nuclear factor 6. J. Virol 89, 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hild M, Weber O, Schaller H, 1998. Glucagon treatment interferes with an early step of duck hepatitis B virus infection. J. Virol 72, 2600–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Kim ES, Guo H, 2017. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: implications for epigenetic therapy against chronic hepatitis B. Hepatology 66, 2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle JH, 2004. Hepatocellular carcinoma: summary and recommendations. Gastroenterology 127, S319–S323. [DOI] [PubMed] [Google Scholar]
- Hoofnagle JH, D. E, Liang TJ, Fleischer R, Lok A, 2007. Management of hepatitis B: summary of a clinical research workshop. Hepatology 45, 1056–1075. [DOI] [PubMed] [Google Scholar]
- Hu J, Toft D, Anselmo D, Wang X, 2002. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol 76, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang Y, Guo Y, Sun S, 2010. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology 52, 60–70. [DOI] [PubMed] [Google Scholar]
- Huang HC, Chen CC, Chang WC, Tao MH, Huang C, 2012. Entry of hepatitis B virus into immortalized human primary hepatocytes by clathrin-dependent endocytosis. J. Virol 86, 9443–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WL, Su TS, 1998. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J. Gen. Virol 79 (Pt 9), 2181–2189. [DOI] [PubMed] [Google Scholar]
- Inoue J, Krueger EW, Chen J, Cao H, Ninomiya M, McNiven MA, 2015. HBV secretion is regulated through the activation of endocytic and autophagic compartments mediated by Rab7 stimulation. J. Cell Sci 128, 1696–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Kim KH, Lok AS, Tong S, 2009. Characterization of genotype-specific carboxyl-terminal cleavage sites of hepatitis B virus e antigen precursor and identi-fication of furin as the candidate enzyme. J. Virol 83, 3507–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Cai D, Sugiyama M, Suzuki R, Aizaki H, Ryo A, Ohtani N, Tanaka Y, Mizokami M, Wakita T, Guo H, Watashi K, 2017. Functional association of cellular microtubules with viral capsid assembly supports efficient hepatitis B virus replication. Sci. Rep 7, 10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Chang TT, Chen S, Boldbaatar B, Clemens A, Lin SY, Yan R, Hu CT, Guo H, Block TM, Song W, Su YH, 2015. Comprehensive DNA methylation analysis of hepatitis B virus genome in infected liver tissues. Sci. Rep 5, 10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht H, Mueller H, Ottosen O, Pedersen L, 2017. In: Treaty PC (Ed.), Nucleic Acid Molecules for Reduction of PAPD5 or PAPD7 mRNA for Treating Hepatitis B Infection. [Google Scholar]
- Kaneko M, Watashi K, Kamisuki S, Matsunaga H, Iwamoto M, Kawai F, Ohashi H, Tsukuda S, Shimura S, Suzuki R, Aizaki H, Sugiyama M, Park SY, Ito T, Ohtani N, Sugawara F, Tanaka Y, Mizokami M, Sureau C, Wakita T, 2015. A novel tricyclic polyketide, vanitaracin A, specifically inhibits the entry of hepatitis B and D Viruses by targeting sodium taurocholate cotransporting polypeptide. J. Virol 89, 11945–11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M, Gerlich WH, 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J. Virol 68, 7993–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A, 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol 145, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau JH, Ting LP, 1998. Phosphorylation of the core protein of hepatitis B virus by a 46-kilodalton serine kinase. J. Virol 72, 3796–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Kornepati AV, Cullen BR, 2015. Targeting hepatitis B virus cccDNA using CRISPR/Cas9. Antivir. Res 123, 188–192. [DOI] [PubMed] [Google Scholar]
- Kim S, Wang H, Ryu WS, 2010. Incorporation of eukaryotic translation initiation factor eIF4E into viral nucleocapsids via interaction with hepatitis B virus polymerase. J. Virol 84, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Okabe T, Ogino H, Matsumoto H, Suzuki-Takahashi I, Kokubo T, Higashi H, Saitoh S, Taya Y, Yasuda H, et al. , 1993. Butyrolactone I, a selective inhibitor of cdk2 and cdc2 kinase. Oncogene 8, 2425–2432. [PubMed] [Google Scholar]
- Kitamura K, Que L, Shimadu M, Koura M, Ishihara Y, Wakae K, Nakamura T, Watashi K, Wakita T, Muramatsu M, 2018. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 14, e1007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock J, Rosler C, Zhang JJ, Blum HE, Nassal M, Thoma C, 2010. Generation of covalently closed circular DNA of hepatitis B viruses via intracellular recycling is regulated in a virus specific manner. PLoS Pathog. 6, e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno T, Tsuge M, Murakami E, Hiraga N, Abe H, Miki D, Imamura M, Ochi H, Hayes CN, Chayama K, 2014. Human microRNA hsa-miR-1231 suppresses hepatitis B virus replication by targeting core mRNA. J. Viral Hepat 21, e89–97. [DOI] [PubMed] [Google Scholar]
- Koniger C, Wingert I, Marsmann M, Rosler C, Beck J, Nassal M, 2014. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc. Natl. Acad. Sci. U.S.A 111, E4244–E4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepstakies M, Lucifora J, Nagel CH, Zeisel MB, Holstermann B, Hohenberg H, Kowalski I, Gutsmann T, Baumert TF, Brandenburg K, Hauber J, Protzer U, 2012. A new class of synthetic peptide inhibitors blocks attachment and entry of human pathogenic viruses. J. Infect. Dis 205, 1654–1664. [DOI] [PubMed] [Google Scholar]
- Lambert C, Doring T, Prange R, 2007. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J. Virol 81, 9050–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan YT, Li J, Liao W, Ou J, 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259, 342–348. [DOI] [PubMed] [Google Scholar]
- Lee GH, W. S.L. S, 2008. Hepatitis B pregenomic RNA splicing–the products, the regulatory mechanisms and its biological significance. Virus Res 136, 1–7. [DOI] [PubMed] [Google Scholar]
- Li J, Tong S, 2015. From DCPD to NTCP: the long journey towards identifying a functional hepatitis B virus receptor. Clin. Mol. Hepatol 21, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xie Y, Wu X, Kong Y, Wang Y, 1995. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology 214, 371–378. [DOI] [PubMed] [Google Scholar]
- Li HC, Huang EY, Su PY, Wu SY, Yang CC, Lin YS, Chang WC, Shih C, 2010. Nuclear export and import of human hepatitis B virus capsid protein and particles. PLoS Pathog. 6, e1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z, 2011. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J. Virol 85, 6319–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Liu G, Kitamura K, Wang Z, Chowdhury S, Monjurul AM, Wakae K, Koura M, Shimadu M, Kinoshita K, Muramatsu M, 2015a. TGF-beta suppression of HBV RNA through AID-dependent recruitment of an RNA exosome complex. PLoS Pathog. 11, e1004780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang TJ, Block TM, McMahon BJ, Ghany MG, Urban S, Guo JT, Locarnini S, Zoulim F, Chang KM, Lok AS, 2015b. Present and future therapies of hepatitis B: from discovery to cure. Hepatology 62, 1893–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CS, Brown CM, 2016. Hepatitis B virus nuclear export elements: RNA stem-loop alpha and beta, key parts of the HBV post-transcriptional regulatory element. RNA Biol. 13, 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-L, Shiao M-S, Mettling C, Chou C-K, 2003. Cholesterol requirement of hepatitis B surface antigen (HBsAg) secretion. Virology 314, 253–260. [DOI] [PubMed] [Google Scholar]
- Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, Wu FY, Kao JH, Chen DS, Chen PJ, 2014. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Molecular therapy. Nucleic acids 3, e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY, Chen DS, Chen PJ, 2009. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology 136, 683–693. [DOI] [PubMed] [Google Scholar]
- Liu F, Campagna M, Qi Y, Zhao X, Guo F, Xu C, Li S, Li W, Block TM, Chang J, Guo JT, 2013. Alpha-interferon suppresses hepadnavirus transcription by altering epigenetic modification of cccDNA minichromosomes. PLoS Pathog. 9, e1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Cai D, Zhang L, Tang W, Yan R, Guo H, Chen X, 2016. Identification of hydrolyzable tannins (punicalagin, punicalin and geraniin) as novel inhibitors of hepatitis B virus covalently closed circular DNA. Antivir. Res 134, 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nie H, Mao R, Mitra B, Cai D, Yan R, Guo JT, Block TM, Mechti N, Guo H, 2017. Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog. 13, e1006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Chen E, Liu C, Huang F, Zhou T, He F, Liu L, Liu F, Tang H, 2009. The correlation of hepatocyte nuclear factor 4 alpha and 3 beta with hepatitis B virus replication in the liver of chronic hepatitis B patients. J. Viral Hepat 16, 537–546. [DOI] [PubMed] [Google Scholar]
- Long Q, Yan R, Hu J, Cai D, Mitra B, Kim ES, Marchetti A, Zhang H, Wang S, Liu Y, Huang A, Guo H, 2017. The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog. 13, e1006784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longarela OL, Schmidt TT, Schoeneweis K, Romeo R, Wedemeyer H, Urban S, Schulze A, 2013. Proteoglycans act as cellular hepatitis delta virus attachment receptors. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Cabrera M, Letovsky J, Hu KQ, Siddiqui A, 1990. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc. Natl. Acad. Sci. U.S.A 87, 5069–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Cabrera M, Letovsky J, Hu KQ, Siddiqui A, 1991. Transcriptional factor C/EBP binds to and transactivates the enhancer element II of the hepatitis B virus. Virology 183, 825–829. [DOI] [PubMed] [Google Scholar]
- Lu X, Mehta A, Dwek R, Butters T, Block T, 1995. Evidence that N-linked glycosylation is necessary for hepatitis B virus secretion. Virology 213, 660–665. [DOI] [PubMed] [Google Scholar]
- Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, Zoulim F, Hantz O, Protzer U, 2011. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J. Hepatol 55, 996–1003. [DOI] [PubMed] [Google Scholar]
- Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Huser N, Durantel D, Liang TJ, Munk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U, 2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343, 1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludgate L, Ning X, Nguyen DH, Adams C, Mentzer L, Hu J, 2012. Cyclin-dependent kinase 2 phosphorylates s/t-p sites in the hepadnavirus core protein C-terminal domain and is incorporated into viral capsids. J. Virol 86, 12237–12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei A, Radulescu C, Lazar C, Petrescu S, Durantel D, Dwek RA, Zitzmann N, Nichita NB, 2010. Hepatitis B virus requires intact caveolin-1 function for productive infection in HepaRG cells. J. Virol 84, 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei A, Petrareanu C, Lazar C, Florian P, Branza-Nichita N, 2013. Regulation of hepatitis B virus infection by Rab5, Rab7, and the endolysosomal compartment. J. Virol 87, 6415–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, Cuconati A, Block TM, Guo JT, Guo H, 2013. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog. 9, e1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason WS, Gill US, Litwin S, Zhou Y, Peri S, Pop O, Hong ML, Naik S, Quaglia A, Bertoletti A, Kennedy PT, 2016. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology 151 986–998 e984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden VC, Shalaby RE, Iram S, Oropeza CE, Landolfi JA, Lyubimov AV, Maienschein-Cline M, Green SJ, Kaestner KH, McLachlan A, 2017. Hepatic deficiency of the pioneer transcription factor FoxA restricts hepatitis B virus bio-synthesis by the developmental regulation of viral DNA methylation. PLoS Pathog. 13, e1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Lu X, Block TM, Blumberg BS, Dwek RA, 1997. Hepatitis B virus (HBV) envelope glycoproteins vary drastically in their sensitivity to glycan processing: evidence that alteration of a single N-linked glycosylation site can regulate HBV secretion. Proc. Natl. Acad. Sci. U.S.A 94, 1822–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melegari M, Wolf SK, Schneider RJ, 2005. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J. Virol 79, 9810–9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RH, Robinson WS, 1983. Integrated hepatitis B virus DNA sequences specifying the major viral core polypeptide are methylated in PLC/PRF/5 cells. Proc. Natl. Acad. Sci. U.S.A 80, 2534–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogul D,T. M, Schwarz KB, 2011. Epigenetic regulation of hepatitis B virus infection. Curr. Hepat. Rep 10, 277–284. [Google Scholar]
- Moolla N, Kew M, Arbuthnot P, 2002. Regulatory elements of hepatitis B virus transcription. J. Viral Hepat 9, 323–331. [DOI] [PubMed] [Google Scholar]
- Morizono K, Chen IS, 2011. Receptors and tropisms of envelope viruses. Curr. Opin. Virol 1, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyo B, Bloom K, Scott T, Ely A, Arbuthnot P, 2017. Advances with using CRISPR/Cas-mediated gene editing to treat infections with hepatitis B virus and hepatitis C virus. Virus Res. 244, 311–320. [DOI] [PubMed] [Google Scholar]
- Mueller H, Wildum S, Luangsay S, Walther J, Lopez A, Tropberger P, Ottaviani G, Lu W, Parrott NJ, Zhang JD, Schmucki R, Racek T, Hoflack JC, Kueng E, Point F, Zhou X, Steiner G, Lutgehetmann M, Rapp G, Volz T, Dandri M, Yang S, Young JAT, Javanbakht H, 2018. A novel orally available small molecule that inhibits hepatitis B virus expression. J. Hepatol 68, 412–420. [DOI] [PubMed] [Google Scholar]
- Murphy CM, Xu Y, Li F, Nio K, Reszka-Blanco N, Li X, Wu Y, Yu Y, Xiong Y, Su L, 2016. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Rep. 16, 2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M, 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol 66, 4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M, 2008. Hepatitis B viruses: reverse transcription a different way. Virus Res 134, 235–249. [DOI] [PubMed] [Google Scholar]
- Nassal M, 2015. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64, 1972–1984. [DOI] [PubMed] [Google Scholar]
- Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S, 1995. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J. Virol 69, 3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Hu J, 2008. Reverse transcriptase-and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J. Virol 82, 6852–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Gummuluru S, Hu J, 2007. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J. Virol 81, 4465–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, Sultmann H, Urban S, 2014. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146, 1070–1083. [DOI] [PubMed] [Google Scholar]
- Novellino L, Rossi RL, Bonino F, Cavallone D, Abrignani S, Pagani M, Brunetto MR, 2012. Circulating hepatitis B surface antigen particles carry hepatocellular microRNAs. PLoS One 7, e31952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler N, Volz T, Bhadra OD, Kah J, Allweiss L, Giersch K, Bierwolf J, Riecken K, Pollok JM, Lohse AW, Fehse B, Petersen J, Urban S, Lutgehetmann M, Heeren J, Dandri M, 2014. Binding of hepatitis B virus to its cellular receptor alters the expression profile of genes of bile acid metabolism. Hepatology 60, 1483–1493. [DOI] [PubMed] [Google Scholar]
- Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD, 2005. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat. Genet 37, 1264–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou JH, Bao H, Shih C, Tahara SM, 1990. Preferred translation of human hepatitis B virus polymerase from core protein-but not from precore protein-specific transcript. J. Virol 64, 4578–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N, Kann M, 2002. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 13, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei DQ, Shih CH, 1990. Transcriptional activation and repression by cellular DNA-binding protein C/EBP. J. Virol 64, 1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D, Hu J, 2003. Duck hepatitis B virus virion secretion requires a double-stranded DNA genome. J. Virol 77, 2287–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Chokshi S, Chatterji U, Riva A, Bobardt M, Williams R, Gallay P, Naoumov NV, 2015. Alisporivir inhibition of hepatocyte cyclophilins reduces HBV replication and hepatitis B surface antigen production. Gastroenterology 148, 403–414 e407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M, 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130, 823–837. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Huang SY, Gao R, Das BB, Murai J, Marchand C, 2014. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair 19, 114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A, 2011. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 39, 5157–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prange R, 2012. Host factors involved in hepatitis B virus maturation, assembly, and egress. Med. Microbiol. Immunol 201, 449–461. [DOI] [PubMed] [Google Scholar]
- Qi Y, Gao Z, Xu G, Peng B, Liu C, Yan H, Yao Q, Sun G, Liu Y, Tang D, Song Z, He W, Sun Y, Guo JT, Li W, 2016. DNA polymerase kappa is a key cellular factor for the formation of covalently closed circular DNA of hepatitis B virus. PLoS Pathog. 12, e1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian G, Jin F, Chang L, Yang Y, Peng H, Duan C, 2012. NIRF, a novel ubiquitin ligase, interacts with hepatitis B virus core protein and promotes its degradation. Biotechnol. Lett 34, 29–36. [DOI] [PubMed] [Google Scholar]
- Qiu L, Fan H, Jin W, Zhao B, Wang Y, Ju Y, Chen L, Chen Y, Duan Z, Meng S, 2010. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem. Biophys. Res. Commun 398, 771–777. [DOI] [PubMed] [Google Scholar]
- Quasdorff M, Protzer U, 2010. Control of hepatitis B virus at the level of transcription. J. Viral Hepat 17, 527–536. [DOI] [PubMed] [Google Scholar]
- Quasdorff M, Hosel M, Odenthal M, Zedler U, Bohne F, Gripon P, Dienes HP, Drebber U, Stippel D, Goeser T, Protzer U, 2008. A concerted action of HNF4alpha and HNF1alpha links hepatitis B virus replication to hepatocyte differentiation. Cell Microbiol. 10, 1478–1490. [DOI] [PubMed] [Google Scholar]
- Rabe B, Glebe D, Kann M, 2006. Lipid-mediated Introduction of Hepatitis B Virus Capsids into Nonsusceptible Cells Allows Highly Efficient Replication and Facilitates the Study of Early Infection Events. [DOI] [PMC free article] [PubMed]
- Rall LB, Standring DN, Laub O, Rutter WJ, 1983. Transcription of hepatitis B virus by RNA polymerase II. Mol. Cell Biol 3, 1766–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN, 2015. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci. Rep 5, 10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiere C, Scholtes C, Diaz O, Icard V, Perrin-Cocon L, Trabaud MA, Lotteau V, Andre P, 2008. Transactivation of the hepatitis B virus core promoter by the nuclear receptor FXRalpha. J. Virol 82, 10832–10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney AK, Easton AJ, Milich DR, McLachlan A, 1991. Promoter-specific transactivation of hepatitis B virus transcription by a glutamine- and proline-rich domain of hepatocyte nuclear factor 1. J. Virol 65, 5774–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi-Shearer D, Gamkrelidze I, Nguyen MH, Chen D-S, Van Damme P, Abbas Z, Abdulla M, Abou Rached A, Adda D, Aho I, Akarca U, Hasan F, Al Lawati F, Al Naamani K, Al-Ashgar HI, Alavian SM, Alawadhi S, Albillos A, Al-Busafi SA, Aleman S, Alfaleh FZ, Aljumah AA, Anand AC, Anh NT, Arends JE, Arkkila P, Athanasakis K, Bane A, Ben-Ari Z, Berg T, Bizri AR, Blach S, Brandão Mello CE, Brandon SM, Bright B, Bruggmann P, Brunetto M, Buti M, Chan HLY, Chaudhry A, Chien R-N, Choi MS, Christensen PB, Chuang W-L, Chulanov V, Clausen MR, Colombo M, Cornberg M, Cowie B, Craxi A, Croes EA, Cuellar DA, Cunningham C, Desalegn H, Drazilova S, Duberg A-S, Egeonu SS, El-Sayed MH, Estes C, Falconer K, Ferraz MLG, Ferreira PR, Flisiak R, Frankova S, Gaeta GB, García-Samaniego J, Genov J, Gerstoft J, Goldis A, Gountas I, Gray R, Guimarães Pessôa M, Hajarizadeh B, Hatzakis A, Hézode C, Himatt SM, Hoepelman A, Hrstic I, Hui Y-TT, Husa P, Jahis R, Janjua NZ, Jarčuška P, Jaroszewicz J, Kaymakoglu S, Kershenobich D, Kondili LA, Konysbekova A, Krajden M, Kristian P, Laleman W, Lao W.-c.C., Layden J, Lazarus JV, Lee M-H, Liakina V, Lim Y-SS, Loo C.-k.K., Lukšić B, Malekzadeh R, Malu AO, Mamatkulov A, Manns M, Marinho RT, Maticic M, Mauss S, Memon MS, Mendes Correa MC, Mendez-Sanchez N, Merat S, Metwally AM, Mohamed R, Mokhbat JE, Moreno C, Mossong J, Mourad FH, Müllhaupt B, Murphy K, Musabaev E, Nawaz A, Nde HM, Negro F, Nersesov A, Nguyen VTT, Njouom R, Ntagirabiri R, Nurmatov Z, Obekpa S, Ocama P, Oguche S, Omede O, Omuemu C, Opare-Sem O, Opio CK, Örmeci N, Papatheodoridis G, Pasini K, Pimenov N, Poustchi H, Quang TD, Qureshi H, Ramji A, Razavi-Shearer K, Redae B, Reesink HW, Rios CY, Rjaskova G, Robbins S, Roberts LR, Roberts SK, Ryder SD, Safadi R, Sagalova O, Salupere R, Sanai FM, Sanchez-Avila JF, Saraswat V, Sarrazin C, Schmelzer JD, Schréter I, Scott J, Seguin-Devaux C, Shah SR, Sharara AI, Sharma M, Shiha GE, Shin T, Sievert W, Sperl J, Stärkel P, Stedman C, Sypsa V, Tacke F, Tan SS, Tanaka J, Tomasiewicz K, Urbanek P, van der Meer AJ, Van Vlierberghe H, Vella S, Vince A, Waheed Y, Waked I, Walsh N, Weis N, Wong VW, Woodring J, Yaghi C, Yang H-I, Yang C-L, Yesmembetov K, Yosry A, Yuen M-F, Yusuf MAM, Zeuzem S, Razavi H, 2018. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol. Hepatol 3, 383–403. [DOI] [PubMed] [Google Scholar]
- Ren JH, Hu JL, Cheng ST, Yu HB, Wong VKW, Law BYK, Yang YF, Huang Y, Liu Y, Chen WX, Cai XF, Tang H, Hu Y, Zhang WL, Liu X, Long QX, Zhou L, Tao NN, Zhou HZ, Yang QX, Ren F, He L, Gong R, Huang AL, Chen J, 2018. SIRT3 restricts HBV transcription and replication via epigenetic regulation of cccDNA involving SUV39H1 and SETD1A histone methyltransferases. Hepatology [epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Riviere L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel ML, Buendia MA, Hantz O, Neuveut C, 2015. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J. Hepatol 63, 1093–1102. [DOI] [PubMed] [Google Scholar]
- Roma MG, Crocenzi FA, Mottino AD, 2008. Dynamic localization of hepatocellular transporters in health and disease. World J. Gastroenterol 14, 6786–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost M, Mann S, Lambert C, Doring T, Thome N, Prange R, 2006. Gamma-adaptin, a novel ubiquitin-interacting adaptor, and Nedd4 ubiquitin ligase control hepatitis B virus maturation. J. Biol. Chem 281, 29297–29308. [DOI] [PubMed] [Google Scholar]
- Rost M, Doring T, Prange R, 2008. gamma2-Adaptin, a ubiquitin-interacting adaptor, is a substrate to coupled ubiquitination by the ubiquitin ligase Nedd4 and functions in the endosomal pathway. J. Biol. Chem 283, 32119–32130. [DOI] [PubMed] [Google Scholar]
- Ryu DK, Kim S, Ryu WS, 2008. Hepatitis B virus polymerase suppresses translation of pregenomic RNA via a mechanism involving its interaction with 5’ stem-loop structure. Virology 373, 112–123. [DOI] [PubMed] [Google Scholar]
- Ryu DK, Ahn BY, Ryu WS, 2010. Proximity between the cap and 5’ epsilon stem-loop structure is critical for the suppression of pgRNA translation by the hepatitis B viral polymerase. Virology 406, 56–64. [DOI] [PubMed] [Google Scholar]
- Schmitz A, Schwarz A, Foss M, Zhou L, Rabe B, Hoellenriegel J, Stoeber M, Pante N, Kann M, 2010. Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog. 6, e1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM, 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol 32, 513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner S, Nassal M, 2017. A role for the host DNA damage response in hepatitis B virus cccDNA formation-and beyond? Viruses 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Gripon P, Urban S, 2007. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 46, 1759–1768. [DOI] [PubMed] [Google Scholar]
- Seeger C, Mason WS, 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. : MMBR (Microbiol. Mol. Biol. Rev.) 64, 51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Sohn JA, 2014. Targeting hepatitis B virus with CRISPR/Cas9. Molecular therapy. Nucleic acids 3, e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Sohn JA, 2016. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Mol. Ther. J. Am. Soc. Gene Ther 24, 1258–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N, Cao F, Tavis JE, 2004. Translation of duck hepatitis B virus reverse transcriptase by ribosomal shunting. J. Virol 78, 11751–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayakhmetov DM, Gaggar A, Ni S, Li ZY, Lieber A, 2005. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J. Virol 79, 7478–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim HY, Quan X, Yi YS, Jung G, 2011. Heat shock protein 90 facilitates formation of the HBV capsid via interacting with the HBV core protein dimers. Virology 410, 161–169. [DOI] [PubMed] [Google Scholar]
- Shimura S, Watashi K, Fukano K, Peel M, Sluder A, Kawai F, Iwamoto M, Tsukuda S, Takeuchi JS, Miyake T, Sugiyama M, Ogasawara Y, Park SY, Tanaka Y, Kusuhara H, Mizokami M, Sureau C, Wakita T, 2017. Cyclosporin derivatives inhibit hepatitis B virus entry without interfering with NTCP transporter activity. J. Hepatol 66, 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH, 2010. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl. Acad. Sci. U.S.A 107, 4383–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn SY, Kim SB, Kim J, Ahn BY, 2006. Negative regulation of hepatitis B virus replication by cellular Hsp40/DnaJ proteins through destabilization of viral core and X proteins. J. Gen. Virol 87, 1883–1891. [DOI] [PubMed] [Google Scholar]
- Stieger B, 2011. The role of the sodium-taurocholate cotransporting polypeptide (NTCP) and of the bile salt export pump (BSEP) in physiology and pathophysiology of bile formation In: Fromm MF, Kim RB (Eds.), Drug Transporters. Springer; Berlin Heidelberg, Berlin, Heidelberg, pp. 205–259. [DOI] [PubMed] [Google Scholar]
- Su TS, Lai CJ, Huang JL, Lin LH, Yauk YK, Chang CM, Lo SJ, Han SH, 1989. Hepatitis B virus transcript produced by RNA splicing. J. Virol 63, 4011–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau C, Salisse J, 2013. A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved hepatitis B virus a-determinant. Hepatology 57, 985–994. [DOI] [PubMed] [Google Scholar]
- Takeyama Y, Kanegae K, Inomata S, Takata K, Tanaka T, Ueda S, Yokoyama K, Morihara D, Nishizawa S, Anan A, Irie M, Iwata K, Shakado S, Sohda T, Sakisaka S, 2010. Sustained upregulation of sodium taurocholate cotransporting polypeptide and bile salt export pump and downregulation of cholesterol 7alpha-hydroxylase in the liver of patients with end-stage primary biliary cirrhosis. Med. Mol. Morphol 43, 134–138. [DOI] [PubMed] [Google Scholar]
- Tang H, McLachlan A, 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. U.S.A 98, 1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Zhao C, Zhu H, She W, Zhang J, Liu J, Li L, Zheng S, Wen YM, Xie Y, 2010. Hepatitis B virus (HBV) surface antigen interacts with and promotes cyclophilin a secretion: possible link to pathogenesis of HBV infection. J. Virol 84, 3373–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepanier DJ, Ure DR, Foster RT, 2017. In vitro phase I metabolism of CRV431, a novel oral drug candidate for chronic hepatitis B. Pharmaceutics 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda S, W. K, Iwamoto M, Suzuki R, Aizaki H, Okada M, Sugiyama M, Kojima S, Tanaka Y, Mizokami M, 2015. Dysregulation of retinoic acid receptor diminishes hepatocyte permissiveness to hepatitis b virus infection through modulation of sodium taurocholate cotransporting polypeptide (NTCP) expression. J. Biol. Chem 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu T, Budzinska MA, Shackel NA, Urban S, 2017. HBV DNA integration: molecular mechanisms and clinical implications. Viruses 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier ER, Colpitts CC, Bach C, Heydmann L, Weiss A, Renaud M, Durand SC, Habersetzer F, Durantel D, Abou-Jaoude G, Lopez Ledesma MM, Felmlee DJ, Soumillon M, Croonenborghs T, Pochet N, Nassal M, Schuster C, Brino L, Sureau C, Zeisel MB, Baumert TF, 2016. A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology 63, 35–48. [DOI] [PubMed] [Google Scholar]
- Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA, 2012. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J 443, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM, 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3, 543–553. [DOI] [PubMed] [Google Scholar]
- Wang H, Kim S, Ryu WS, 2009. DDX3 DEAD-Box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J. Virol 83, 5815–5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, Wu E, Ye X, Gao GF, Wang F, Chen Y, Duan Z, Meng S, 2012. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1)-modulated P53 activity. Hepatology 55, 730–741. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P, 2007. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. U.S.A 104, 10205–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watashi K, Liang G, Iwamoto M, Marusawa H, Uchida N, Daito T, Kitamura K, Muramatsu M, Ohashi H, Kiyohara T, Suzuki R, Li J, Tong S, Tanaka Y, Murata K, Aizaki H, Wakita T, 2013. Interleukin-1 and tumor necrosis factor-alpha trigger restriction of hepatitis B virus infection via a cytidine deaminase activation-induced cytidine deaminase (AID). J. Biol. Chem 288, 31715–31727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watashi K, Sluder A, Daito T, Matsunaga S, Ryo A, Nagamori S, Iwamoto M, Nakajima S, Tsukuda S, Borroto-Esoda K, Sugiyama M, Tanaka Y, Kanai Y, Kusuhara H, Mizokami M, Wakita T, 2014. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP). Hepatology 59, 1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Fourel G, Ponzetto A, Silvestro M, Tiollais P, Buendia MA, 1992. Hepadnavirus integration: mechanisms of activation of the N-myc2 retrotransposon in woodchuck liver tumors. J. Virol 66, 5265–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z, Liu R, Wu Z, 2013a. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell. Signal. 25, 1037–1043. [DOI] [PubMed] [Google Scholar]
- Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X, Wu Z, 2013b. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell. Signal. 25, 439–446. [DOI] [PubMed] [Google Scholar]
- Werr M, Prange R, 1998. Role for calnexin and N-linked glycosylation in the assembly and secretion of hepatitis B virus middle envelope protein particles. J. Virol 72, 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkop L, Schwarz A, Cassany A, Grun-Bernhard S, Delaleau M, Rabe B, Cazenave C, Gerlich W, Glebe D, Kann M, 2010. Inhibition of protein kinase C phosphorylation of hepatitis B virus capsids inhibits virion formation and causes intracellular capsid accumulation. Cell Microbiol. 12, 962–975. [DOI] [PubMed] [Google Scholar]
- Wooddell CI, Yuen MF, Chan HL, Gish RG, Locarnini SA, Chavez D, Ferrari C, Given BD, Hamilton J, Kanner SB, Lai CL, Lau JYN, Schluep T, Xu Z, Lanford RE, Lewis DL, 2017. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci. Transl. Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang WJ, Yang L, Fu Q, Xu JJ, Gu JX, 2015. Decreased expression of hepatocyte nuclear factor 4alpha (Hnf4alpha)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J. Biol. Chem 290, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Stadler D, Lucifora J, Reisinger F, Webb D, Hosel M, Michler T, Wisskirchen K, Cheng X, Zhang K, Chou WM, Wettengel JM, Malo A, Bohne F, Hoffmann D, Eyer F, Thimme R, Falk CS, Thasler WE, Heikenwalder M, Protzer U, 2016. Interferon-gamma and tumor necrosis factor-alpha Produced by T Cells reduce the HBV persistence form, cccDNA, without cytolysis. Gastroenterology 150, 194–205. [DOI] [PubMed] [Google Scholar]
- Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W, 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1, e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Peng B, Liu Y, Xu G, He W, Ren B, Jing Z, Sui J, Li W, 2014. Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J. Virol 88, 3273–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Summers J, 1995. Illegitimate replication of linear hepadnavirus DNA through nonhomologous recombination. J. Virol 69, 4029–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Summers J, 1998. Infection of ducklings with virus particles containing linear double-stranded duck hepatitis B virus DNA: illegitimate replication and reversion. J. Virol 72, 8710–8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Summers J, 1999. Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J. Virol 73, 9710–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CC, Huang EY, Li HC, Su PY, Shih C, 2014. Nuclear export of human hepatitis B virus core protein and pregenomic RNA depends on the cellular NXF1-p15 machinery. PLoS One 9, e106683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying C, Li Y, Leung CH, Robek MD, Cheng YC, 2007. Unique antiviral mechanism discovered in anti-hepatitis B virus research with a natural product analogue. Proc. Natl. Acad. Sci. U.S.A 104, 8526–8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu S, Hu T, Liu S, He Y, Sun S, 2009. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 50, 490–499. [DOI] [PubMed] [Google Scholar]
- Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H, 2010. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antivir. Res 88, 169–175. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M, 2011. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology 53, 1476–1485. [DOI] [PubMed] [Google Scholar]
- Zhang X, Hou J, Lu M, 2013. Regulation of hepatitis B virus replication by epigenetic mechanisms and microRNAs. Front. Genet 4, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mao R, Yan R, Cai D, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Huang Y, Guo H, Zhang J, 2014. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One 9, e110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chen J, Wu M, Zhang X, Zhang M, Yue L, Li Y, Liu J, Li B, Shen F, Wang Y, Bai L, Protzer U, Levrero M, Yuan Z, 2017. PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation. Hepatology 66, 398–415. [DOI] [PubMed] [Google Scholar]
- Zhou T, Block T, Liu F, Kondratowicz AS, Sun L, Rawat S, Branson J, Guo F, Steuer HM, Liang H, Bailey L, Moore C, Wang X, Cuconatti A, Gao M, Lee ACH, Harasym T, Chiu T, Gotchev D, Dorsey B, Rijnbrand R, Sofia MJ, 2018. HBsAg mRNA degradation induced by a dihydroquinolizinone compound depends on the HBV posttranscriptional regulatory element. Antivir. Res 149, 191–201. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Cheng N, Stahl SJ, Conway JF, Steven AC, Wingfield PT, 1997. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc. Natl. Acad. Sci. U.S.A 94, 9556–9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Locarnini S, 2009. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 137 1593–1608 e1591–1592. [DOI] [PubMed] [Google Scholar]