Abstract
During embryogenesis vertebrates develop a complex craniofacial skeleton associated with sensory organs. These structures are primarily derived from two embryonic cell populations the neural crest and cranial placodes, respectively. Neural crest cells and cranial placodes are specified through the integrated action of several families of signaling molecules, and the subsequent activation of a complex network of transcription factors. Here we describe the expression and function of Anosmin-1 (Anos1), an extracellular matrix protein, during neural crest and cranial placodes development in Xenopus laevis. Anos1 was identified as a target of Pax3 and Zic1, two transcription factors necessary and sufficient to generate neural crest and cranial placodes. Anos1 is expressed in cranial neural crest progenitors at early neurula stage and in cranial placode derivatives later in development. We show that Anos1 function is required for neural crest and sensory organs development in Xenopus, consistent with the defects observed in Kallmann syndrome patients carrying a mutation in ANOS1. These findings indicate that anos1 has a conserved function in the development of craniofacial structures, and indicate that anos1-depleted Xenopus embryos represent a useful model to analyze the pathogenesis of Kallmann syndrome.
Keywords: anosmin-1, cranial placode, Kallmann syndrome, neural crest, sensory organs, Xenopus
1. Introduction
The vertebrate head is characterized by a complex craniofacial skeleton and paired sensory organs. These structures are derived from two embryonic cell populations, neural crest and cranial placodes, that are specified at the anterior border of the neural plate at the end of gastrulation. There is abundant literature indicating that molecules of the fibroblast growth factor (Fgf), Wnt and bone morphogenetic protein (Bmp) families must be precisely modulated to specify neural crest and cranial placodes in the embryo [1–4]. These growth factors in turn activate a unique repertoire of transcription factors that are responsible for initiating the differentiation program of each one of these cell populations [5,6]. Among these transcription factors, Pax3 and Zic1 are especially critical to promote neural crest and cranial placode fates [7–12]. To characterize the molecular events downstream of Pax3 and Zic1, several years ago we performed a microarray screen using Xenopus animal cap explants expressing varying combinations of these factors [13]. Here we describe the expression and function of one of these targets Anosmin-1. Anosmin-1 is a secreted molecule of the extracellular matrix, encoded by the anos1 gene. In human, mutations in ANOS1 cause Kallmann syndrome, a condition characterized by craniofacial defects, anosmia, deafness and hypogonadotropic hypogonadism [14–15]. Using knockdown approaches in Xenopus embryos and animal cap explants we show that Anosmin-1 is required for neural crest specification and for cranial placode-derived sensory organs formation, consistent with defects observed in Kallmann syndrome patients. There is no anos1 ortholog in mouse therefore anos1-depleted Xenopus embryos may represent a unique model to analyze the etiology and pathogenesis of Kallmann syndrome.
2. Materials and methods
2.1. Plasmid constructs and morpholino antisense oligonucleotides
The ORF of Xenopus laevis anos1 was amplified by PCR from neurula stage embryo cDNA using the following primers, F: 5’-ATGTGGCTGAGGGAGCCAGGC-3’ and R: 5’-TCAGTACTTCTCTGGGGATGG-3’. Anos1 (anos1MO; 5’-GAGAACCTCGCTCCCTCAGCCACAT-3’), Pax3 (pax3MO; [7]), and Zic1 (zic1MO; [8]) morpholino antisense oligonucleotides (MOs) were purchased from GeneTools (Philomath, OR). The pax3GR and zic1GR hormone-inducible constructs were generated as previously described [9]. Synthetic mRNAs encoding pax3GR, zic1GR, noggin, wnt8a, anos1, and β-galactosidase [9,16] were synthesized in vitro using the Message Machine kit (Ambion, Grand Island NY).
2.2. Embryos, injections and explants culture
Xenopus laevis embryos were staged as previously described [17] and raised in 0.1X NAM (Normal Amphibian Medium; [18]). The procedures were approved by NYU Institutional Animal Care and Use Committee, under animal protocol # 150201. Embryos were injected in one blastomere at the 2-cell stage with MOs and synthetic mRNA together with 500pg of β-galactosidase mRNA as a lineage tracer. For animal cap explant experiments, pax3GR (100pg), zic1GR (100pg), noggin (200pg), wnt8a (10pg) and anos1 (3ng) mRNAs and anos1MO (40ng) were injected in the animal pole region at the 2-cell stage, explants were dissected at the late blastula stage and cultured for several hours in NAM 0.5X plus 10μM of dexamethasone (Dex; Sigma-Aldrich). The explants were subsequently analyzed by quantitative PCR (qRT-PCR). For whole embryo injections and animal cap explant assays each experiment was performed on at least three independent batches of embryos.
2.3. Lineage tracing, in situ hybridization and histology
Xenopus embryos were fixed in MEMFA and processed for Red-Gal (Research Organics, Cleveland OH) staining to visualize β-galactosidase prior to ISH. Whole-mount ISH was performed as previously described [19]. Digoxygenin (DIG)- labeled antisense RNA probes (Roche Diagnostics, Indianapolis, IN) were synthesized using template cDNA encoding Anos1, Snai2 [20], Sox10 [21], Sox2 [22], Six1 [23], Foxi4.1 [24], Dmrta1 [25], Pax8 [26], Emx2 [27], Ebf2 [28] and Foxe1 [29].
2.4. Western blot analysis
For Western blots, embryos were injected at the 2-cell stage with mRNA (500pg) encoding a myc tagged version of anos1 along with increasing amounts of anos1MO and cultured up to stage 15. Pools of 10 embryos were homogenized in lysis buffer (0.5% Triton X-100, 10mM Tris–HCI at pH 7.5, 50mM NaCl, 1mM EDTA), containing Halt™ Protease Inhibitor Cocktail (ThermoFisher Scientific; Waltham, MA). Extracts were resolved on a 10% NuPAGE Bis-Tris gel and transferred onto a PVDF membrane using the iBlot system (Invitrogen, Grand Island NY). Blots were subsequently incubated overnight with anti-Myc polyclonal antibody (Abcam, Cambridge, MA; 1.5μg/ml dilution). The blots were then washed and incubated with donkey anti-goat IgG coupled to horseradish peroxidase (Santa Cruz Biotechnology, Dallas, TX; 1:5000 dilution). Peroxidase activity was detected with the Western Blotting Luminol Reagent (Santa Cruz Biotechnology) and imaged on a ChemiDoc MP Biorad gel documentation system (Hercules, CA).
2.5. RNA preparation and qRT-PCR
Total RNAs from embryos were extracted using the RNeasy Micro Kit (Qiagen, Valencia CA). For mRNA extraction from animal cap explants, pools of 8 explants were homogenized and mRNAs were isolated using Dynabeads® mRNA DIRECT™ Micro Kit (Invitrogen). cDNA synthesis from total RNA and mRNA were performed using Superscript VILO cDNA Synthesis Kit (Invitrogen, Grand Island, NY). qRT-PCR was performed on an Eco Real-Time PCR System (Illumina, San Diego CA) using the primers shown in Table 1, and the Power SYBR Green PCR Master Mix (Invitrogen). The reaction mixture consisted of 10µl of Power SYBR Green PCR Master Mix, 200nM primers, and 2µl of cDNA in a total volume of 10µl. The PCR conditions were as follows: 95°C for 10min; 40 cycles at 95°C for 10sec and at 60°C for 30sec. The ∆∆CT method was used to analyze the qRT-PCR results. Each reaction included a standard curve of serial dilution points (in 10-fold increments) of test cDNA. odc1 or ef1α was used for normalization.
Table 1:
Primers for qRT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| anos1 | 5’- GGAGAAGGTCCAGCAACAATAA −3’ | 5’- GGGATGGTTTGTAGTGATGAGG −3’ |
| snai2 | 5’- CATGGGAATAAGTGCAACCA −3’ | 5’- AGGCACGTGAAGGGTAGAGA −3’ |
| sox2 | 5’-TCACCTCTTCTTCCCATTCG-3’ | 5’-CGACATGTGCAGTCTGCTTT-3’ |
| sox8 | 5’- AAGGTCTCTGGTGGCTGAAA −3’ | 5’- CACCGCCACATTTCAGAGTA −3’ |
| odc1 | 5’-ACATGGCATTCTCCCTGAAG-3’ | 5’-TGGTCCCAAGGCTAAAGTTG-3’ |
| ef1α | 5’-ACCCTCCTCTTGGTCGTTTT-3’ | 5’-TTTGGTTTTCGCTGCTTTCT-3’ |
3. Results
3.1. Anos1 is expressed in the neural crest and cranial placodes
Ansomin-1 (Anos1, formerly Kal1; [30]), an extracellular matrix protein, was independently isolated in two microarray screens designed to identify targets of Pax3 and Zic1 [13,31]. Both transcription factors are necessary and sufficient to specify the neural crest and placodes [7–9]. By ISH anos1 transcripts are first detected at the early neurula stage (NF stage 14) at the lateral edge of the anterior neural plate, in the prospective cranial neural crest region (Fig. 1A). As the neural plate closes, anos1 remains spatially confined to the neural crest territory (Fig. 1B,C). As development proceed, anos1 is expressed in the pharyngeal arches, somites, and anteriorly at the midline in the prospective pituitary gland (Fig. 1D-F). At the tailbud stage (NF stage 27) anos1 transcripts persist in the pharyngeal arches, somites, and anterior pituitary, and appeared to accumulate in the ventral aspect of the otic vesicles (Fig 1G,H). Using qRT-PCR, we compared the temporal expression profile of anos1 to that of snai2, a well-established early neural crest gene [22]. We found that anos1 is maternally expressed (NF stage 8), and towards the end of gastrulation (NF stage 12) the escalation of snai2 and anos1 expression follows a very similar pattern (Fig. 1I).
Figure 1: Developmental expression of anos1 by whole-mount ISH.
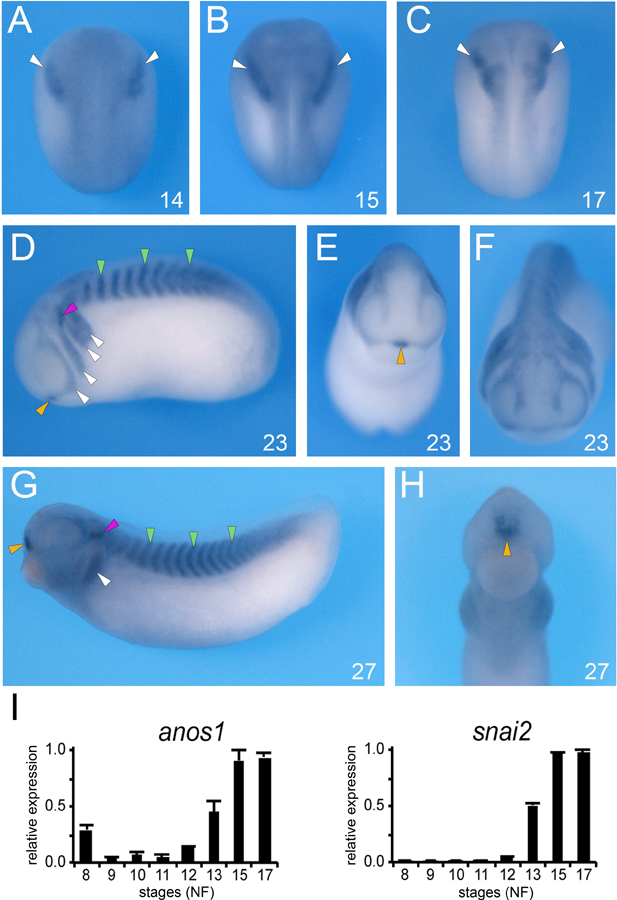
(A-C) At the neurula stage (NF stage 14–17), anos1 is detected in the prospective neural crest territory (white arrowheads). (D-F) At stage 23, anos1 is now more broadly expressed, to include the somites (green arrowheads), otic vesicle (red arrowhead), the anterior pituitary (yellow arrowhead) in addition to the branchial arches (white arrowheads). (G-H) Later in development (NF stage 27) anos1 persists in all these tissues. (A-C) dorsal views, anterior to top. (D, G) lateral views, dorsal to top, anterior to left. (E, F, H) frontal views, dorsal to top. The embryonic stages (NF) are indicated in the lower right corner of each panel. (I) Relative expression levels of anos1 and snail2 analyzed by qRT-PCR at the indicated stages. The values were normalized to odc1.
3.2. Anos1 is a true target of Pax3 and Zic1
To confirm that anos1 is a target of Pax3 and Zic1, we performed perturbation experiments in the embryo using pax3MO and zic1MO [7–9]. Pax3 or Zic1 knockdown resulted in a dramatic reduction or loss of anos1 expression in a vast majority of the embryos, demonstrating that both factors are independently required for anos1 expression (Fig. 2A). In animal cap explants (Fig. 2B), expression of pax3GR and/or zic1GR showed that Pax3 and Zic1 are sufficient to induce anos1 expression and their activity is additive, while Pax3 and Zic1 synergistically activate snai2 (Fig. 2C). Taken together, these results indicate that anos1 is expressed in Xenopus cranial neural crest and its expression is regulated by Pax3 and Zic1.
Figure 2: anos1 is a target of Pax3 and Zic1.
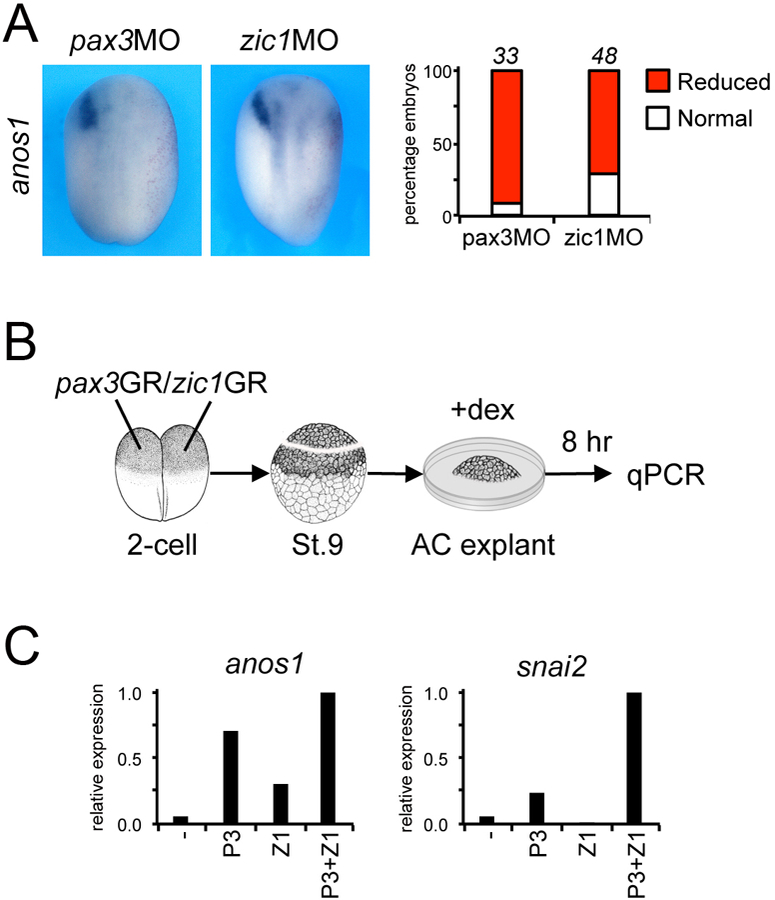
(A) Injection of pax3MO (40ng) or zic1MO (40ng) reduces anos1 expression at the neurula stage. Dorsal views, anterior to top. The injected side is to the right as indicated by the presence of the lineage tracer (Red-Gal). The graph indicates the percentage of embryos with normal (white) or reduced/lost (red) anos1 expression. The number of embryos analyzed is indicated on top of each bar. (B) mRNA encoding pax3GR and zic1GR (100pg each), alone or in combination were injected into both blastomeres in the animal pole at the 2-cell stage. At the blastula stage (stage 9), animal cap (AC) explants were dissected and cultured for 8 hours in the presence of dexamethasone (+dex). (C) anos1 and snai2 expression in pax3GR and zic1GR injected AC explants analyzed by qRT-PCR.
3.3. Anos1 is necessary but not sufficient for neural crest development
To determine whether anos1 function is implicated in neural crest development we performed anos1 knockdown using MOs. anos1MO was designed to specifically interfere with translation of anos1 mRNA. The activity of the MO was confirmed by Western blot. In embryos injected with mRNA encoding a myc-tagged version of Xenopus anos1, increasing doses of anos1MO blocks Anos1 protein accumulation (Fig. 3A). Unilateral injection of anos1MO at the 2-cell stage led to a reduction of expression of several neural crest genes including snai2 and sox10 at stage 14 (Fig. 3B,C). The loss of neural crest genes correlated with an expansion of sox2 expression domain in morphant embryos (Fig. 3B). We also tested the function of Anos1 in an animal cap explant assay. Activation of the Wnt/β-catenin pathway in conjunction with attenuation of BMP signaling induces neural crest genes in animal cap explants [32]. We found that the induction of the neural crest genes, snai2 and sox8, by wnt8a and noggin was significantly repressed in Anos1-depleted (MO-injected) animal cap explants (Fig. 3D). The reduction in snai2 and sox8 expression was associated with an increase in sox2 expression, consistent with a loss of neural crest fate. Therefore, in the embryos and in animal cap explants Anos1 function is required to specify the neural crest.
Figure 3: Anos1 is required for neural crest formation.
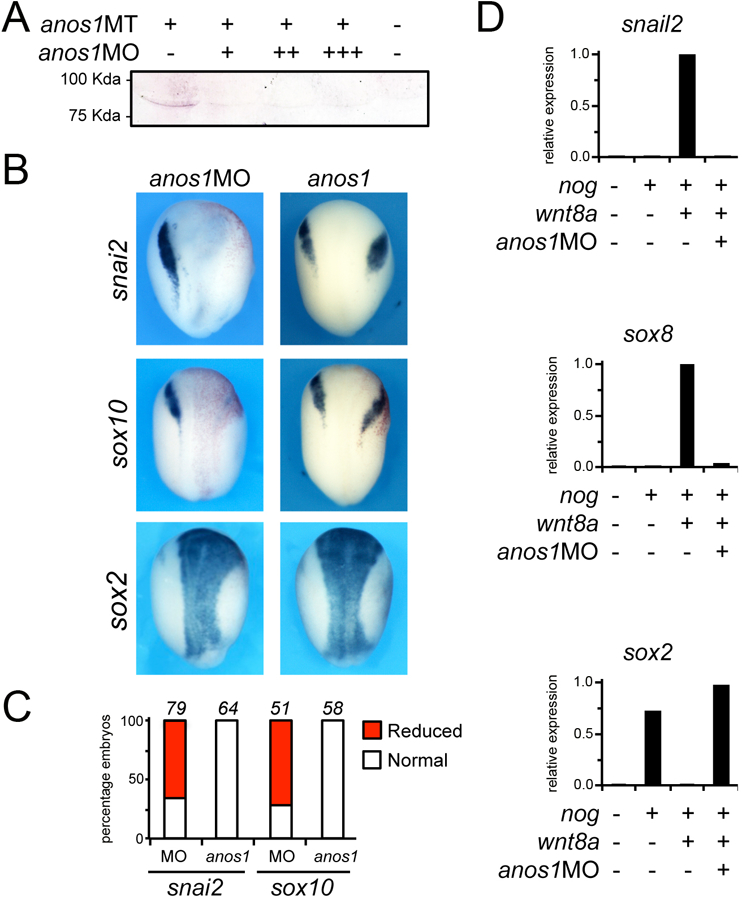
(A) Western blot of embryos injected with mRNA encoding a myc-tagged version of Xenopus anos1, alone or in combination with increasing doses of anos1MO, 10ng (+), 20ng (++), and 40ng (+++), showed that anos1MO blocks Anos1 protein accumulation. (B) Unilateral injection of anos1MO (40ng) at the 2-cell stage causes a reduction/loss of expression of snail2 and sox10, and a lateral expansion of sox2 expression domain. Injection of Xenopus anos1 mRNAs (3ng) did not significantly affect snail2 and sox10 expression levels, although their expression domain was shifted laterally. The expression of sox2 was only marginally expanded in these embryos. Dorsal views, anterior to top. The injected side is to the right (Red-Gal). (C) The graphs indicate the percentage of embryos with normal (white), reduced/lost (red) expression. The number of embryos analyzed is indicated on top of each bar. (D) In explants, the induction of snail2 and sox8 by co-injection of noggin (200pg) and wnt8a (10pg) mRNA is dramatically reduced in the context of embryos injected with anos1MO (40ng). This reduction in neural crest genes expression is associated by an increase in sox2 expression. The values were normalized to ef1α. A representative experiments is shown from three independent experiments.
We also performed gain-of-function experiments. Unilateral injection of anos1 mRNA at the 2-cell stage did not significantly affect snail2 and sox10 expression levels, although their expression appeared shifted laterally (Fig. 3B,C). Moreover, in animal cap explant anos1 expression was unable to activate snail2 or sox8 expression (not shown). These results support the view that anos1 participates in neural crest formation downstream of Pax3 and Zic1, but is not sufficient to activate a neural crest development program.
3.4. Anos1 is essential for sensory placodes development
In human, mutations in ANOS1 have been associated with sensory deficits, including anosmia and hearing loss, and hypogonadotropic hypogonadism due to malfunction of the gonadotropin releasing hormone (GnRH) system [15]. To determine whether Anosmin-1 is also critical for sensory organs development in Xenopus we performed unilateral injection of anos1MO at the 2-cell stage. These injections resulted in the reduction or loss of expression of two pan-placodal genes, six1 and foxi4.1, as well as genes restricted to individual placodal domains such as dmrta1 for the adenohypophyseal and olfactory placodes, and pax8 for the otic and lateral line placodes (Fig. 4A-D). Later in development, these embryos exhibited reduced dmrta1, emx2 and ebf2 expression in the presumptive olfactory epithelium (Fig. 4E-G), and a marked decrease of foxe1 expression in the prospective pituitary (Fig. 4I). These observations are consistent with a broad function of anos1 in the regulation of cranial placode-derived sensory organs development.
Figure 4: Anos1 is essential for the formation of cranial placodes and their derivatives.
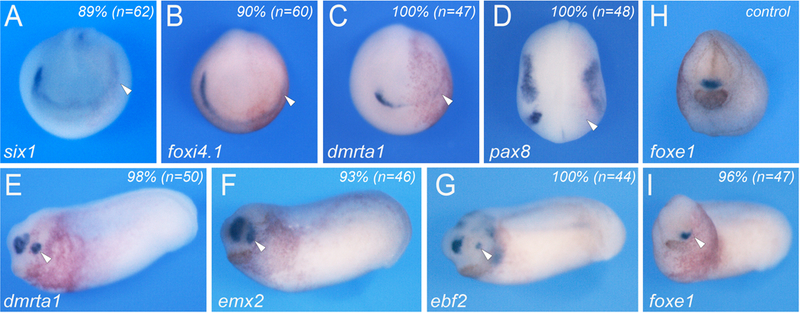
(A-D) Unilateral injection of anos1MO (40ng) at the 2-cell stage causes a reduction/loss of expression of two pan-placodal genes, six1 (A) and foxi4.1 (B), as well as genes restricted to individual placodal domains such as dmrta1 (C) and pax8 (D). (E-G) Later in development, the injected embryos exhibited reduce dmrta1 (E), emx2 (F) and ebf2 (G) expression in the olfactory epithelium. (H-I) The expression of foxe1 in the developing pituitary was also reduced (I) as compared to sibling control (H). (A-I) Anterior views, dorsal to top. The injected side (arrowheads) is to the right. The percentage of affected embryos is indicated in the upper right corner of each panel.
4. Discussion
Here we describe the expression and function of Anosmin-1 (Anos1) during early Xenopus laevis development. Anos1 was identified as a target of Pax3 and Zic1 [13,31], two important regulators of neural crest and cranial placode fates in the embryo [7–12]. anos1 is first detected at the early neurula stage at the lateral edges of the anterior neural plate, in the cranial neural crest forming region. Later in development, anos1 expression persists in neural crest cells as they populate the branchial arches, and is activated in the prospective nasal and otic placodes, anterior pituitary, and in the somites. MO-mediated knockdown of Anosmin-1 resulted in a reduction of neural crest and cranial placode genes expression suggesting that Anosmin-1 is required for the specification of both cell populations.
ANOS1 encodes a 100-kDa glycoprotein of the extracellular matrix with great affinity for cell membrane heparin sulfate proteoglycans. It is highly conserved across species, with anos1 orthologs present in Drosophila, C. elegans, zebrafish, and most mammals. Interestingly, asnos1 has not been identified in rat and mouse [30]. Anos1 is composed of several functional domains including a cysteine rich region, a whey acidic protein-like domain (typically present in serine protease inhibitors), four consecutive domains Fibronectin-like type III domains (implicated in cell-cell adhesion) and an histidine-rich C-terminal region [30,33]. Anos1 has been implicated in cell-cell adhesion, cell migration and differentiation of multiple cell types in the nervous system. Anos1 regulates migration of immortalized GnRH producing neurons as well as neuronal and oligodendrocyte precursors. It has also a role in axon guidance, neurite outgrowth and the genesis of axon collaterals from neurons in the olfactory system [34–36].
Molecules of the Bmp, Fgf and Wnt families have been implicated in neural crest and cranial placode induction [1–4], however little is known how the activity of these growth factors is controlled in the extracellular space. Functional studies in vivo and in vitro have shown that Anos1 physically interacts with fibroblast growth factor receptor 1 (FgfR1) and its ligand Fgf2 to activate signaling [37–40]. More recently, the role of Anos1 was evaluated during cranial neural crest formation in chicken embryos. This study showed that Anos1 upregulates Fgf8 and Bmp5 gene expression, and binds directly to Fgf8, Bmp5 and Wnt3a to modulate their activity locally and promote cranial neural crest formation [41]. While anos1 is also expressed in the anterior neural fold region in chicken embryos, it is unclear whether its activity is also involved in the regulation of cranial placode development. Our observations indicate that in addition to the cranial neural crest, Anos1 plays a critical role in cranial placodes formation, as Anos1 knockdown affects the development of adenohyphyseal, olfactory and otic placode derivatives. While we did not detect anos1 transcripts in the placodal region at the early neurula stage, Anos1 knockdown resulted in the reduction of two pan-placodal genes, six1 and foxi4.1. Since Anos1 is a secreted protein it is likely that knocking down its function in the cranial neural crest will also affect adjacent cell populations, including the formation of cranial placode progenitors, which depends on similar signaling events for their specification.
ANOS1 is the gene responsible for X-linked Kallmann’s syndrome [42,43]. This condition is the result of defects in the development of GnRH and olfactory neurons, both cell types originating from the nasal placode. As a consequence these patients suffer from hypogonadotropic hyogonadism and anosmia. Associated defects include renal agenesis, cleft lip with or without cleft palate, hearing loss and abnormal tooth development. The genetics and cellular pathogenesis of Kallmann syndrome are still incompletely understood [15,44], therefore it is essential to develop suitable experimental model systems to gain insights into the molecular basis underlying this condition. We propose that anos1-depleted Xenopus embryos may represent an excellent model to investigate the pathogenesis of Kallmann syndrome and the mechanisms of action of Anos1.
Acknowledgements
We are grateful to Drs. Brandli, El-Hodiri, Knochel, Vetter and Vignali for reagents.
Funding sources
This work was supported by grants from the National Institutes of Health to J-P S-J (R01-DE014212 and R01-DE025468).
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- [1].Stuhlmiller TS, Garcia-Castro MI, Current perspectives of the signaling pathways directing neural crest induction, Cell. Mol. Life Sci 69 (2012) 3715–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bae CJ, Saint-Jeannet JP, Induction and Specification of Neural Crest Cells: Extracellular Signals and Transcriptional Switches, in: Trainor P (Ed.), Neural Crest Cells: Evolution, Development and Disease, Elsevier Academic Press, San Diego, 2014, pp. 27–49. [Google Scholar]
- [3].Saint-Jeannet JP, Moody SA, Establishing the pre-placodal region and breaking it into placodes with distinct identities, Dev. Biol 389 (2014) 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schlosser G, Making senses development of vertebrate cranial placodes, Int. Rev. Cell Mol. Biol 283 (2010) 129–234. [DOI] [PubMed] [Google Scholar]
- [5].Sauka-Spengler T, Bronner-Fraser M, A gene regulatory network orchestrates neural crest formation, Nat. Rev. Mol. Cell Biol 97 (2008) 557–568. [DOI] [PubMed] [Google Scholar]
- [6].Grocott T, Tambalo M, Streit A, The peripheral sensory nervous system in the vertebrate head: A gene regulatory perspective, Dev. Biol 370 (2012) 3–23. [DOI] [PubMed] [Google Scholar]
- [7].Monsoro-Burq AH, Wang E, Harland R, Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction, Dev. Cell 8 (2005) 167–178. [DOI] [PubMed] [Google Scholar]
- [8].Sato T, Sasai N, Sasai Y, Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm, Development 132 (2005) 2355–2363. [DOI] [PubMed] [Google Scholar]
- [9].Hong CS, Saint-Jeannet JP, The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border, Mol. Biol. Cell 18 (2007) 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cornish EJ, Hassan SM, Martin JD, Li S, Merzdorf CS, A microarray screen for direct targets of Zic1 identifies an aquaporin gene, aqp-3b, expressed in the neural folds, Dev. Dyn 238 (2009) 1179–1194. [DOI] [PubMed] [Google Scholar]
- [11].Garnett AT, Square TA, Medeiros DM, BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border, Development 139 (2012) 4220–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jaurena MB, Juraver-Geslin H, Devotta A, Saint-Jeannet JP, Zic1 controls placode progenitor formation non-cell autonomously by regulating retinoic acid production and transport, Nat. Commun 6 (2015) 7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bae CJ, Park BY, Lee YH, Tobias JW, Hong CS, Saint-Jeannet JP,. Identification of Pax3 and Zic1 targets in the developing neural crest, Dev. Biol 386 (2014) 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kallmann F, Schoenfeld W, Barrera S, The genetic aspects of primary eunuchoidism, Am. J. Ment. Defic 48 (1944) 33. [Google Scholar]
- [15].Dode C, Hardelin JP, Kallmann syndrome, Eur. J. Hum. Genet 17 (2009) 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hong CS, Park BY, Saint-Jeannet JP, Fgf8a induces neural crest indirectly through the activation of Wnt8 in the paraxial mesoderm, Development 135 (2008) 3903–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nieuwkoop PD, Faber J, Normal table of Xenopus laevis (Daudin) North Holland Publishing Company, Amsterdam: 1967. [Google Scholar]
- [18].M Slack J, Forman D, An interaction between dorsal and ventral regions of the marginal zone in early amphibian embryos, J. Embryol. Exp. Morpho.l 56 (1980) 283–299. [PubMed] [Google Scholar]
- [19].Saint-Jeannet JP, Whole-mount in situ hybridization of Xenopus embryos, Cold Spring Harb. Protoc 2017 (2017) pdb.prot0972876. [DOI] [PubMed] [Google Scholar]
- [20].Mayor R, Morgan R, Sargent MG, Induction of the prospective neural crest of Xenopus, Development 121 (1995) 767–777. [DOI] [PubMed] [Google Scholar]
- [21].Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet JP, Sox10 regulates the development of neural crest-derived melanocytes in Xenopus, Dev. Biol 259 (2003) 19–33. [DOI] [PubMed] [Google Scholar]
- [22].Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y, Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction, Development 125 (1998) 579–587. [DOI] [PubMed] [Google Scholar]
- [23].Pandur PD, Moody SA, Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines, Mech. Dev 96 (2000) 253–257. [DOI] [PubMed] [Google Scholar]
- [24].Pohl BS, Knochel S, Dillinger K, Knochel W, Sequence and expression of FoxB2 (XFD-5) and FoxI1c (XFD-10) in Xenopus embryogenesis, Mech. Dev 117 (2002) 283–287. [DOI] [PubMed] [Google Scholar]
- [25].Huang X, Hong CS, O’Donnell M, Saint-Jeannet JP, The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system, Proc. Nat. Acad. Sci. (USA) 102 (2005) 11349–11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Heller N, Brandli AW, Xenopus Pax-2/5/8 orthologues: novel insights into Pax gene evolution and identification of Pax-8 as the earliest marker for otic and pronephric cell lineages, Dev. Genet 24 (1999) 208–219. [DOI] [PubMed] [Google Scholar]
- [27].Pannese M, Lupo G, Kablar B, Boncinelli E, Barsacchi G, Vignali R, The Xenopus Emx genes identify presumptive dorsal telencephalon and are induced by head organizer signals, Mech. Dev 73 (1998) 73–83. [DOI] [PubMed] [Google Scholar]
- [28].Dubois L, Bally-Cuif L, Crozatier M, Moreau J, Paquereau L, Vincent A, XCoe2, a transcription factor of the Col/Olf-1/EBF family involved in the specification of primary neurons in Xenopus, Curr. Biol 8 (1998) 199–209. [DOI] [PubMed] [Google Scholar]
- [29].El-Hodiri HM, Seufert DW, Nekkalapudi S, Prescott NL, Kelly LE, Jamrich M, Xenopus laevis FoxE1 is primarily expressed in the developing pituitary and thyroid, Int. J. Dev. Biol 49 (2005) 881–884. [DOI] [PubMed] [Google Scholar]
- [30].De Castro F, Seal R, Maggi R, ANOS1: a unified nomenclature for Kallmann syndrome 1 gene (KAL1) and anosmin-1, Brief. Funct. Genomics 16 (2017) 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Plouhinec JL, Roche DD, Pegoraro C, Figueiredo AL, Maczkowiak F, Brunet LJ, Milet C, Vert JP, Pollet N, Harland RM, Monsoro-Burq AH, Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers, Dev. Biol 386 (2014) 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Saint-Jeannet JP, He X, Varmus HE, Dawid IB, Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a, Proc. Natl. Acad. Sci. (USA) 94 (1997) 13713–13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].del Castillo I, Cohen-Salmon M, Blanchard S, Lutfalla G, Petit C, Structure of the X-linked Kallmann syndrome gene and its homologous pseudogene on the Y chromosome, Nat. Genet 2 (1992) 305–310. [DOI] [PubMed] [Google Scholar]
- [34].Lutz B B, Rugarli EI, Eichele G, Ballabio A, X-linked Kallmann syndrome. A neuronal targeting defect in the olfactory system?, FEBS Lett 325 (1993) 128–134. [DOI] [PubMed] [Google Scholar]
- [35].Rugarli EI, Ghezzi C, Valsecchi V, Ballabio A, The Kallmann syndrome gene product expressed in COS cells is cleaved on the cell surface to yield a diffusible component, Hum. Mol. Genet 5 (1996) 1109–1115. [DOI] [PubMed] [Google Scholar]
- [36].Soussi-Yanicostas N, Hardelin JP, Arroyo-Jimenez MM, Ardouin O, Legouis R, Levilliers J, Traincard F, Betton JM, Cabanié L, Petit C, Initial characterization of anosmin-1, a putative extracellular matrix protein synthesized by definite neuronal cell populations in the central nervous system, J. Cell Sci 109 (1996) 1749–1757. [DOI] [PubMed] [Google Scholar]
- [37].Gonzalez-Martinez D, Kim SH, Hu Y, Guimond S, Schofield J, Winyard P, Vannelli GB, Turnbull J, Bouloux PM, Anosmin-1 modulates fibroblast growth factor receptor 1 signaling in human gonadotropin releasing hormone olfactory neuroblasts through a heparan sulfate-dependent mechanism, J. Neurosci 24 (2004) 10384–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bribian A, Barallobre MJ, Soussi-Yanicostas N, de Castro F, Anosmin-1 modulates the FGF-2-dependent migration of oligodendrocyte precursors in the developing optic nerve, Mol. Cell. Neurosci 33 (2006) 2–14. [DOI] [PubMed] [Google Scholar]
- [39].Ayari B, Soussi-Yanicostas N, FGFR1 and anosmin-1 underlying genetically distinct forms of Kallmann syndrome are co-expressed and interact in olfactory bulbs, Dev. Genes Evol 217 (2007) 169–175. [DOI] [PubMed] [Google Scholar]
- [40].Hu Y, Guimond SE, Travers P, Cadman S, Hohenester E, Turnbull JE, Kim SH, Bouloux PM, Novel mechanisms of fibroblast growth factor receptor 1 regulation by extracellular matrix protein anosmin-1, J. Biol. Chem 284 (2009) 29905–29920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Endo Y, Ishiwata-Endo H, Yamada KM, Extracellular matrix protein anosmin promotes neural crest formation and regulates FGF, BMP, and WNT activities, Dev. Cell 23 (2012) 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Legouis R, Hardelin JP, Levilliers J, Claverie JM, Compain S, Wunderle V, Millasseau P, Le Paslier D, Cohen D, Caterina D, et al. , The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules, Cell 67 (1991) 423–435. [DOI] [PubMed] [Google Scholar]
- [43].Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, et al. , A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal pathfinding molecules, Nature 353 (1991) 529–536. [DOI] [PubMed] [Google Scholar]
- [44].Hu Y, Tanriverdi F, MacColl GS, Bouloux PM, Kallmann’s syndrome: molecular pathogenesis, Int. J. Biochem. Cell Biol 35 (2003) 1157–1162. [DOI] [PubMed] [Google Scholar]


