Abstract
Germ-line and somatic mutations in genes that promote homology-directed repair (HDR), especially BRCA1 and BRCA2, are frequently observed in several cancers, in particular, breast and ovary but also prostate and other cancers. HDR is critical for the error-free repair of DNA double-strand breaks and other lesions, and HDR factors also protect stalled replication forks. As a result, loss of BRCA1 or BRCA2 poses significant risks to genome integrity, leading not only to cancer predisposition but also to sensitivity to DNA-damaging agents, affecting therapeutic approaches. Here we review recent advances in our understanding of BRCA1 and BRCA2, including how they genetically interact with other repair factors, how they protect stalled replication forks, how they affect the response to aldehydes, and how loss of their functions links to mutation signatures. Importantly, given the recent advances with poly(ADP-ribose) polymerase inhibitors (PARPi) for the treatment of HDR-deficient tumors, we discuss mechanisms by which BRCA-deficient tumors acquire resistance to PARPi and other agents.
Keywords: homologous recombination, BRCA1, BRCA2, ATM, double-strand break repair, replication fork protection
1. INTRODUCTION
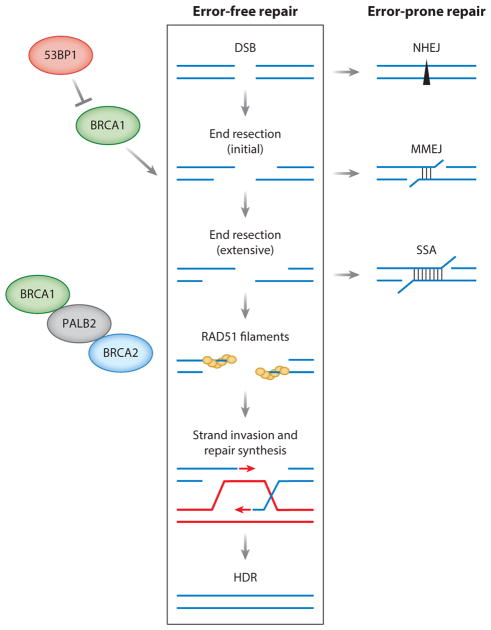
DNA damage poses a significant threat to genome integrity and can lead to tumorigenesis if not properly repaired. A double-strand break (DSB) is considered one of the most cytotoxic types of DNA damage, so it is perhaps not surprising that cells have multiple pathways to repair DSBs (Figure 1). Two major pathways are homologous recombination, also termed homology-directed repair (HDR), and nonhomologous end joining (NHEJ) (Chapman et al. 2012b, Jasin & Rothstein 2013) (Figure 1). Other pathways include microhomology-mediated end joining (MMEJ) and single-strand annealing (SSA), although the physiological roles of these latter two pathways in normal cells are uncertain (Sfeir & Symington 2015). HDR is considered the most precise of the DSB repair pathways because it uses a homologous sequence, usually the sister chromatid, to template repair. Thus, defects in HDR lead to error-prone repair of DSBs, which ultimately leads to mutations, gross chromosomal rearrangements, and genome instability (Moynahan & Jasin 2010). DSBs arise from several exogenous agents and cellular processes, with replication considered to be a major source in every cell cycle (Kass et al. 2016b).
Figure 1.
Homology-directed repair (HDR) and other pathways for the repair of double-strand breaks (DSBs). HDR, also termed homologous recombination, is a major pathway for the error-free repair of DSBs, which occurs primarily during S/G2 phases of the cell cycle. An early determinant of DSB repair pathway choice is DNA end resection—the processing of DNA ends to generate 3′ single strands. Whether end resection occurs is governed by BRCA1 in competition with the antagonistic protein 53BP1, and end resection can occur in two steps, an initial phase to result in short 3′ overhangs (<100 base pairs) and a more extensive phase to result in longer 3′ overhangs (hundreds of base pairs), as demonstrated in yeast. BRCA1 also promotes the later steps of HDR via interaction with the PALB2 protein, which in turn recruits BRCA2. The defining step of HDR, which is controlled by BRCA2, is the formation of RAD51 filaments on the 3′ single strands, followed by strand invasion into a homologous DNA, typically the sister chromatid, priming repair DNA synthesis. Most HDR is completed by a simple gene conversion, although other outcomes are possible (Jasin & Rothstein 2013). DSBs can also be repaired by error-prone DNA repair pathways. Canonical nonhomologous end joining (NHEJ) can occur during any cell cycle phase and is considered the other major DSB repair pathway. In NHEJ, DNA ends are protected from end resection but often undergo some processing before joining, which results in small deletions or insertions. Initial and extensive end resection also provides intermediates for two other DSB repair pathways, alternative NHEJ involving microhomology, also termed microhomology-mediated end joining (MMEJ), and single-strand annealing (SSA), involving longer stretches of homology. Consistent with their involvement in different steps of HDR, BRCA1 promotes HDR and SSA, whereas BRCA2 promotes HDR and suppresses SSA.
Mutations in several HDR genes—in particular, BRCA1 and BRCA2—are associated with predispositions to breast, ovary, and, at lower frequency, prostate, pancreas, and other cancers. More recently, mutations in other HDR genes, including PALB2, RAD51C, RAD51D, and ATM, have also been associated with increased susceptibility to several of the same cancers as BRCA1/2, such that the fraction of some tumor types with HDR gene mutations can be quite substantial (from 10% to >20%) (Lord & Ashworth 2016, Pennington et al. 2014, Robinson et al. 2015). Due to defects in DNA repair, cells with mutations in HDR genes are particularly sensitive to crosslinking agents such as platinum-based chemotherapeutic drugs. The more recent discovery that cells with BRCA1/2 mutations can be selectively targeted by treatment with poly(ADP-ribose) polymerase inhibitors (PARPi) has driven research toward identifying more cancers with impaired HDR (Bryant et al. 2005, Farmer et al. 2005). The term “BRCAness” has been coined to describe tumors that share molecular characteristics with BRCA-mutant cancers and thus may be responsive to treatment with PARPi and platinum-based therapies (Lord & Ashworth 2016).
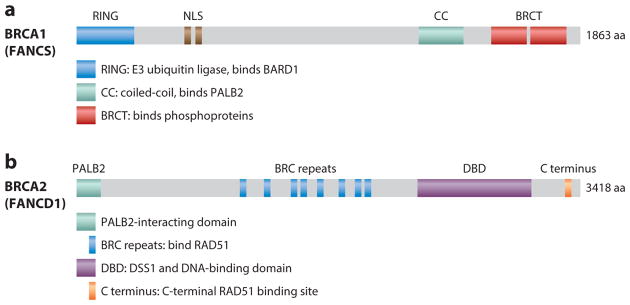
BRCA1 and BRCA2 are both large proteins with multiple functional domains (Prakash et al. 2015) (Figure 2). Although they are not highly conserved in evolution (~50% identity with the mouse proteins), BRCA1/2 are essential for embryonic development in the mouse and likely in humans as well. Germ-line tumor-predisposing mutations, mostly truncating but also missense, occur throughout both genes, with common founder mutations in some populations. Variants of unknown significance have often been identified, leading to the development of functional assays for HDR to evaluate whether the variants may be pathogenic (Anantha et al. 2017, Bouwman et al. 2013, Kuznetsov et al. 2008, Millot et al. 2012). Tumor formation is effectively suppressed in the heterozygous state; thus, a primary requirement for tumorigenesis in heterozygous individuals is loss of the wild-type allele in pretumorigenic cells. The rare inheritance of biallelic mutations requires that at least one allele has partial function. This scenario is associated with subtypes of the rare genetic disorder Fanconi anemia (FA) (Ceccaldi et al. 2016), which is characterized by developmental issues and is typically associated with early childhood onset of malignancies, including medulloblastoma (Meyer et al. 2014). More recently, somatic mutations in these genes have also been identified in tumors, which may impact therapeutic responses (Lord & Ashworth 2016).
Figure 2.
Domain structures of BRCA1 and BRCA2. (a) BRCA1 comprises an N-terminal RING domain with E3 ubiquitin ligase activity that interacts with the RING domain of its binding partner BARD1; a central region encoded by the largest exon (exon 11) of BRCA1 containing nuclear localization signals (NLSs) and other activities; a coiled-coil (CC) domain for PALB2 binding, with an overlapping SQ cluster targeted by ATM kinase; and tandem BRCT repeats at the C terminus for interaction with several phosphoproteins, including Abraxas, BRIP1, and CtIP, leading to the mutually exclusive formation of the BRCA1 A, B, and C complexes, respectively. BRCA1 is also considered a Fanconi anemia gene (FANCS). Readers are referred to Prakash et al. (2015) for more detail. (b) BRCA2 comprises an N-terminal domain that interacts with PALB2, a central region that has eight repeats of a BRC motif that interact with several molecules of RAD51, a DSS1 and DNA-binding domain (DBD), and a distinct RAD51 binding site in the C terminus. BRCA2 is also considered a Fanconi anemia gene (FANCD1). Abbreviation: aa, amino acids.
2. BRCA1 REGULATES END RESECTION–DEPENDENT DOUBLE-STRAND BREAK REPAIR AND LATER STEPS IN HOMOLOGY-DIRECTED REPAIR
BRCA1 was first conclusively shown to be required for HDR when researchers examined the repair of a DSB introduced into a defined site in the genome (Moynahan et al. 1999, 2001a). BRCA1 participates in both early and later steps of HDR: Following DSB formation, BRCA1 is involved in counteracting the NHEJ factor 53BP1 to facilitate DNA end resection (see below), an early step in HDR (Figure 1). BRCA1 also interacts with the PALB2 protein (Sy et al. 2009, Zhang et al. 2009) (Figure 2), which in turn binds BRCA2 to facilitate RAD51 filament formation, a later step in HDR. Patient-derived mutations of BRCA1 that impair HDR typically also reduce SSA, another DSB repair pathway that requires end resection (Anantha et al. 2017, Stark et al. 2004). The exceptions are missense mutations that specifically disrupt BRCA1 interaction with PALB2, which result in reduced HDR but increased SSA, similar to BRCA2 deficiency (Anantha et al. 2017, Stark et al. 2004).
The mutual antagonism between BRCA1 and 53BP1 controls levels of end-resected DNA in a cell cycle–dependent manner (Panier & Boulton 2014, Zimmermann & de Lange 2014). In G1 phase, 53BP1 recruits several factors in an ATM-dependent manner, thus allowing for efficient NHEJ. Deficiency of 53BP1 abrogates the protection of DNA ends and predisposes mice to lymphoma due to the increased levels of end processing and oncogenic translocations (Bothmer et al. 2010, Difilippantonio et al. 2008, Jankovic et al. 2013). On the other hand, in S/G2 phases, BRCA1 is required to counteract 53BP1 to promote the availability of end-resected intermediates. This ensures that HDR occurs when sister chromatids are available to template HDR. Importantly, loss of 53BP1 rescues the HDR defects and lethality of BRCA1- but not BRCA2-deficient cells (Bouwman et al. 2010, Bunting et al. 2010, Cao et al. 2009). Thus, the role of BRCA1 in antagonizing 53BP1 is likely more pivotal than a direct role for BRCA1 in end resection per se.
Given the additional, later role for BRCA1 in HDR in promoting PALB2-BRCA2 functions, it seems surprising that 53BP1 loss can fully restore HDR in BRCA1-deficient settings. Possibly, 53BP1 loss creates more resected DNA that attenuates the requirement for BRCA1 in recruiting PALB2 and BRCA2 for subsequent RAD51 filament formation (Buisson et al. 2010, Dray et al. 2010). Alternatively, additional factors such as ATM/ATR-mediated phosphorylation may contribute to the regulation of PALB2 in this context (Ahlskog et al. 2016). Importantly, some aspects of BRCA1 function cannot be rescued by 53BP1 loss, such as interstrand crosslink repair (Bunting et al. 2012) and replication fork stabilization (Ray Chaudhuri et al. 2016) (see below), suggesting a distinct requirement for BRCA1 in these processes, for example, crosstalk with the FA pathway (D’Andrea 2013).
ATM-mediated phosphorylation of the N terminus of 53BP1 is required for the interaction of several 53BP1-binding proteins, including RIF1 and PTIP, to suppress end resection in G1 phase (Callen et al. 2013, Chapman et al. 2013, Di Virgilio et al. 2013, Escribano-Diaz et al. 2013, Feng et al. 2013, Zimmermann et al. 2013). Loss of RIF1 in mice leads to many of the same phenotypes as 53BP1-deficient mice. However, RIF1 loss does not rescue the HDR defect of BRCA1-deficient cells to the same extent as 53BP1 loss. By contrast, although PTIP deficiency does not affect NHEJ in the same way as 53BP1 deficiency, loss of PTIP in BRCA1-deficient cells almost completely rescues HDR and suppresses chromosome aberrations following PARPi treatment. These observations illustrate the complexities of 53BP1 and interacting proteins in affecting end resection.
The mechanism by which BRCA1 counteracts 53BP1 is not well understood and is likely to be complex. CtIP, which binds BRCA1, is a critical resection factor (Sartori et al. 2007), such that 53BP1 loss fails to rescue the lethality of CtIP-deficient mice (Polato et al. 2014). It is possible that BRCA1 stimulates the end resection machinery, including CtIP, which in turn displaces NHEJ components. For example, activities of the resection factors MRE11 and CtIP limit the accumulation of the NHEJ factor Ku at DSB ends (Chanut et al. 2016, Mimitou & Symington 2010). However, depletion of Ku cannot rescue cellular lethality associated with BRCA1 deficiency, despite a reduction in chromosome aberrations (Bunting et al. 2012). Given that Ku binds only to DNA ends (Walker et al. 2001), whereas 53BP1 binds methylated and ubiquitylated histones surrounding the DSB site (Botuyan et al. 2006, Fradet-Turcotte et al. 2013, Huyen et al. 2004), the presence of single-stranded DNA (ssDNA) alone may not be sufficient to abolish 53BP1 action. Consistent with the notion, the transition from a 53BP1 block of HDR to BRCA1-mediated HDR can be facilitated by several chromatin-modifying proteins (e.g., histone acetyltransferases TIP60/KAT5 and MOF/KAT8) or chromatin remodelers (e.g., SMARCAD1) (Densham et al. 2016, Gupta et al. 2014, Tang et al. 2013).
BRCA1 may also actively disrupt the function of the 53BP1 complex to abrogate the blockade of HDR. For example, BRCA1 disrupts the interaction of 53BP1 and RIF1 in S/G2 phases by promoting dephosphorylation of 53BP1 by the PP4C phosphatase and targeting RIF1 by an E3 ubiquitin ligase UHRF1 (Isono et al. 2017, Zhang et al. 2016). BRCA1 also induces the exclusion of 53BP1 from the core of DNA damage foci in S/G2 phases (Chapman et al. 2012a, Kakarougkas et al. 2013).
3. BRCA2 PROMOTES RAD51 FILAMENT FORMATION AND STABILITY
Like BRCA1, the requirement for BRCA2 in HDR was directly demonstrated using a reporter for DSB-induced HDR (Moynahan et al. 2001b). BRCA2 is clearly involved at a distinct step from BRCA1 during HDR because the end resection–dependent SSA pathway increases with BRCA2 deficiency, unlike with BRCA1 but similar to RAD51 deficiency, apparently due to the increase in end resected intermediates that cannot be channeled into HDR (Stark et al. 2004). Given this later role in HDR, it is not surprising that 53BP1 loss does not rescue the survival of BRCA2-deficient cells as it does with BRCA1-deficient cells (Bouwman et al. 2010). More recent work has demonstrated the requirement for both proteins in replication-induced interstrand crosslink HDR (Nakanishi et al. 2011) and in suppressing error-prone long-tract gene conversion induced by either a DSB or replication fork stalling (Willis et al. 2014).
The critical biochemical function of BRCA2 in HDR is to promote RAD51 filament assembly onto ssDNA that arises from end resection (Prakash et al. 2015). BRCA2 directly interacts with RAD51 at multiple sites (Figure 2) to facilitate RAD51 filament assembly at two levels. First, BRCA2 helps RAD51 overcome the inhibitory effect of the high-affinity ssDNA-binding protein RPA, which normally coats ssDNA and prevents RAD51 loading. Second, by preferentially binding to ssDNA over double-stranded DNA (dsDNA), BRCA2 specifically promotes the productive assembly of RAD51 filaments onto ssDNA, which is critical for strand invasion of a homologous DNA, while preventing the nonproductive formation of RAD51 filaments onto dsDNA (Jensen et al. 2010, Liu et al. 2010, Thorslund et al. 2010). Full-length BRCA2 protein may function as an oligomer, as indicated by structural and imaging studies (Reuter et al. 2014, Sanchez et al. 2017, Shahid et al. 2014). In cells, BRCA2 is responsible for recruiting RAD51 to DNA damage sites (Tarsounas et al. 2003, Yuan et al. 1999). A recent study using super-resolution microscopy revealed that BRCA2 forms local clusters that progressively overlap with the more elongated RAD51 foci, suggesting dynamic interactions during the repair process (Sanchez et al. 2017).
When HDR activity is compromised through BRCA1/2 loss, cells become reliant on other factors or alternative pathways to repair DSBs. Thus, BRCA1/2-deficient tumor cells show synergistic genome instability and synthetic lethality with additional loss of the MMEJ pathway factor POLθ (Ceccaldi et al. 2015, Mateos-Gomez et al. 2015) or the minor HDR factor RAD52 (Feng et al. 2011, Lok et al. 2013), although conversely disabling canonical NHEJ through inhibition of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) has been reported to diminish PARPi sensitivity and genome instability (Patel et al. 2011).
RAD52 may have an additional role besides being a backup HDR factor. It facilitates DNA synthesis during mitosis, which is thought to relieve the sequelae of replication stress arising in the previous S phase (Bhowmick et al. 2016). Replication stress is clearly linked to BRCA2-deficiency (Feng & Jasin 2017), such that mitotic DNA synthesis may contribute to the survival of BRCA-deficient cells (Bhowmick et al. 2016). These observations provide a rationale for developing RAD52 (or POLθ) inhibitors to target BRCA-deficient tumors.
In addition to HDR, BRCA2 has also been implicated in G2/M checkpoint maintenance (Menzel et al. 2011), R-loop processing (Bhatia et al. 2014), the spindle assembly checkpoint (Choi et al. 2012), and cytokinesis (Daniels et al. 2004, Mondal et al. 2012), all having an impact on genome or chromosomal stability. Some of these processes are distinguishable from HDR (Bhatia et al. 2014, Menzel et al. 2011, Mondal et al. 2012), but additional studies are required to fully understand the role of BRCA2 in these processes.
4. BRCA1 AND BRCA2 PROTECT NASCENT DNA STRANDS AT STALLED REPLICATION FORKS
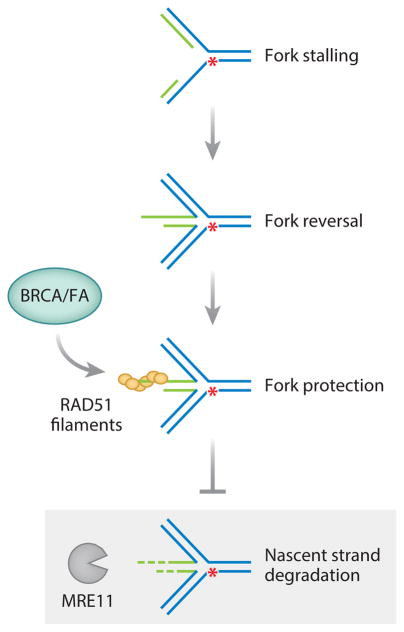
More recently, BRCA1 and BRCA2, as well as FA proteins, were discovered to be required for replication fork protection, a pathway that can be genetically separable from HDR (Kolinjivadi et al. 2017; Schlacher et al. 2011, 2012) (Figure 3). Under replication stress conditions leading to fork stalling, BRCA/FA proteins prevent the degradation of newly synthesized DNA strands by the MRE11 nuclease (Schlacher et al. 2011, 2012; Ying et al. 2012). Despite this degradation, once the replication stress is removed, forks can restart seemingly normally, although it is likely that some will collapse leading to an increase in chromosome aberrations. Thus, these proteins both repair DNA damage and prevent DNA damage from occurring.
Figure 3.
Protection of nascent DNA strands from degradation at stalled replication forks. Replication fork stalling can occur when a fork encounters a lesion or nucleotides become limiting. Nascent strands (green) can pair with each other such that the fork regresses (fork reversal). BRCA1, BRCA2, and other homology-directed repair and Fanconi anemia (FA) proteins protect nascent strands from degradation by stabilizing RAD51 filaments at stalled forks. In the absence of these proteins (shaded gray box), nascent strands can become a substrate for degradation by MRE11 and other nucleases, resulting in genome instability. The red asterisk indicates a lesion that prevents fork progression.
The molecular mechanism by which the BRCA/FA proteins function in fork protection is still unclear. One model is that the stalled replication fork reverses and RAD51 forms a nucleoprotein filament on the extruded nascent strand to prevent nuclease access (Figure 3). A separation-of-function mutant in BRCA2 has been identified, which is predicted to lead to less stable RAD51 filaments; this mutant is defective in fork protection but proficient at HDR and resistant to PARPi and crosslinking agents (Schlacher et al. 2011). Thus, in this model, both forming and stabilizing RAD51 filaments are critical for fork protection (Schlacher et al. 2011, 2012). While FA-deficient cells are profoundly deficient in protecting stalled replication forks (Schlacher et al. 2012), they show only a mild defect in HDR of DSBs (Nakanishi et al. 2005, 2011). Therefore, despite involving some overlapping biochemical activities such as RAD51 filament formation, replication fork protection and HDR are functionally separable processes.
A recent report has complicated this simple model by showing that RAD51 depletion does not affect nascent strand stability upon fork stalling (Thangavel et al. 2015). Moreover, another report suggested that RAD51 itself promotes replication fork reversal (Zellweger et al. 2015). One way to reconcile these findings is to theorize that preventing RAD51 from reversing stalled replication forks prevents their degradation, whereas impairing RAD51 filament formation on nascent strands of reversed forks leads to their degradation. The role of RAD51, BRCA2, and other proteins in this process is an active area of investigation.
5. THE TOXIC EFFECTS OF ENVIRONMENTAL AND METABOLIC ALDEHYDES ON BRCA1/2-DEFICIENT CELLS
Aldehydes, including acetaldehyde and formaldehyde, are natural byproducts of cellular metabolism and a source of endogenous DNA damage in cells. They are also found exogenously: Formaldehyde is abundant in tobacco smoke, while acetaldehyde is a product of alcohol metabolism. The carcinogenic effects of these aldehydes stem from overexposure and saturation of cellular processes involved in aldehyde detoxification, which leads to DNA-DNA and DNA-protein crosslinks and ultimately to mutagenesis (Ross & Shipley 1980, Yu et al. 2010).
Genes in the HDR and FA pathways play a critical role in counteracting DNA damage toxicity from aldehydes (Langevin et al. 2011, Ridpath et al. 2007). Mice doubly deficient in FANCD2 and an enzyme required for formaldehyde [alcohol dehydrogenase 5 (ADH5)] or acetaldehyde [aldehyde dehydrogenase 2 (ALDH2)] catabolism show evidence of aldehyde-induced genotoxicity, including increased DNA damage, bone marrow failure, and leukemia (Garaycoechea et al. 2012, Langevin et al. 2011, Pontel et al. 2015). Notably, Adh5−/−;Fancd2−/− mice also develop kidney dysfunction and liver failure (Pontel et al. 2015), suggesting tissue tropism in the accumulation or catabolism of endogenous formaldehyde. Incidentally, Aldh2−/−;Fancd2−/− mouse embryos require maternal ALDH2 function to survive, highlighting the importance of acetaldehyde detoxification during embryonic development (Langevin et al. 2011).
Although the role of BRCA1/2 in counteracting the toxic effects of aldehydes has not been studied in the same capacity using mouse models, it has been investigated extensively in human cancer cell lines and mouse embryonic fibroblasts (MEFs) (Tacconi et al. 2017). BRCA1/2-deficient human cancer cells are sensitive to treatment with acetaldehyde or disulfiram, an inhibitor of ALDH2, and Aldh2-null MEFs show synthetic lethality with BRCA1/2 deficiency. The effects of acetaldehyde on these cells are manifested by elevated replication stress, DSBs, chromosomal aberrations, and checkpoint activation. Notably, acetaldehyde treatment reduced the survival of both Brca1-and Brca2-deleted mouse mammary tumor–derived cell lines and even an olaparib-resistant line (Brca1−/−, 53bp1 deficient). These findings suggest the potential clinical use of disulfiram and other ALDH2 inhibitors, which have been approved for treating chronic alcoholism (Koppaka et al. 2012), as antitumor agents for treating BRCA1/2-deficient tumors, including those that have acquired resistance to PARPi.
However, an unexpected effect of exogenous aldehyde exposure was recently reported that is cautionary with regard to BRCA2 carriers. Exposure of cells to either formaldehyde or acetaldehyde was found to lead to proteasomal degradation of BRCA2 (Tan et al. 2017). As a result of reduced protein levels, cells heterozygous for cancer-associated BRCA2 mutations, which normally behave like wild-type cells due to the presence of wild-type BRCA2 from one allele, show evidence of BRCA2 loss of function, including loss of replication fork protection and increased chromosomal aberrations. The mechanism leading to proteasomal degradation is unclear but is relatively selective, as only a small number of other cellular proteins appeared degraded upon formaldehyde exposure. The effects of aldehyde exposure on BRCA1 heterozygous cells are still unknown, although BRCA1 protein levels seem at most modestly affected by formaldehyde-triggered degradation. The lability of BRCA2 observed upon aldehyde exposure is interesting given that the protein is also susceptible to heat-induced degradation (Krawczyk et al. 2011).
6. MUTATIONAL SIGNATURES FROM BRCA1/2 LOSS
Patterns of DNA sequence changes—mutational signatures—have recently been identified by examining >1,000 genomes from many different cancer types (see http://cancer.sanger.ac.uk/cosmic/signatures). Mutational signatures are typically classified according to the mutated base and the two flanking bases (for 96 possible mutated trinucleotides) and are associated with intrinsic processes involving DNA metabolism, mutagen exposure, and defects in DNA repair (Helleday et al. 2014, Kass et al. 2016b). An unusually broad distribution of base substitution mutations has been associated with BRCA1 or BRCA2 loss (Nik-Zainal et al. 2012), termed mutational signature 3, although why loss of these proteins causes base substitutions is unclear. This signature is also associated with elevated numbers of insertions and deletions (indels) of <50 base pairs (bp) with microhomology at the breakpoint junctions (typically 1–5 bp). The increase in indels likely results from reliance on more error-prone DSB repair pathways, such as MMEJ (Figure 1).
In addition to the base and indel mutational signatures, rearrangement signatures have been characterized depending on whether they involve deletions, tandem duplications, or inversions and on the size and clustering of the events (Nik-Zainal et al. 2016). BRCA1/2 loss is associated with rearrangement signature 5, characterized by nonclustered deletions of <100 kb. Further, cancers with BRCA1 loss, but not BRCA2 loss, are specifically associated with rearrangement signature 3, characterized by small (<10 kb) tandem duplications.
The presence of these distinct mutational signatures—including indels associated with micro-homology, base substitution signature 3, and rearrangement signatures 3 and 5—is now being used among other parameters to accurately predict not only BRCA1/2-deficient tumors but also tumors with other HDR pathway defects, potentially identifying more cancers that may respond to treatment with PARPi or platinum-based drugs (Davies et al. 2017).
7. MECHANISMS OF RESISTANCE TO POLY(ADP-RIBOSE) POLYMERASE INHIBITORS AND PLATINUM THERAPIES
Although tumors with BRCA1/2 or other HDR deficiencies can be targeted by PARPi and platinum drugs, the development of therapy resistance is a critical problem in the clinic. Several resistance mechanisms have been proposed, the best validated of which are secondary mutations within the gene that restore the reading frame of the primary truncating mutations.
7.1. Secondary Mutations in BRCA1 and BRCA2 and Other Homology-Directed Repair Genes
A source of cross resistance to platinum-based chemotherapies and PARPi emerged in 2008 when two papers described secondary mutations of BRCA2 6174delT that restored the reading frame in breast, pancreatic, and ovarian cancers and cell lines (Edwards et al. 2008, Sakai et al. 2008). These reversion mutations often involved small indels near the truncating mutation and even direct reversion to the wild-type sequence, but large deletions reaching nearly 60 kb were also observed that encompassed the truncating mutation and several exons, demonstrating the plasticity of BRCA2 protein for maintaining function (Siaud et al. 2011). These mutations were found to restore HDR and, when checked, fork protection (Schlacher et al. 2011). Secondary mutations were also identified that same year in platinum-resistant BRCA1-mutated ovarian cancers (185delAG and 2594delC) (Swisher et al. 2008). BRCA1 and BRCA2 secondary mutations continue to be reported in multiple cancer types (e.g., Afghahi et al. 2017, Pishvaian et al. 2017) and, more recently, in other HDR pathway genes, in particular, the RAD51 paralog genes RAD51C and RAD51D (Kondrashova et al. 2017).
Multiple secondary mutations in individual patients with either a BRCA1 or BRCA2 mutation have also been reported. Sequencing of ascites DNA from patients with platinum-resistant high-grade serous ovarian cancers identified an extreme case involving a patient carrying BRCA2 c.767_771delTCAAA, in which five frame-restoring mutations were identified (Patch et al. 2015). Another seven independent mutations were identified at several metastatic sites, often in abdominal tissue. These 12 secondary mutations were primarily deletions between 1 and 40 bp that restored the reading frame. This patient did not have debulking surgery, leading to the proposal that the large tumor mass provided a pool from which multiple resistance mutations could occur.
Patients’ cell-free DNA (cfDNA) has also been used to identify secondary mutations that are likely to mediate therapy resistance, suggesting a significant clinical utility of liquid biopsies for predicting treatment response. In both high-grade serous ovarian cancer and metastatic prostate cancer, multiple secondary mutations in both germ-line and somatically mutated BRCA2 and PALB2 were identified in cfDNA from patients with recurrent disease (Christie et al. 2017, Goodall et al. 2017, Quigley et al. 2017). Interestingly, in the case of prostate cancers treated with PARPi (either olaparib or talazoparib), analysis of mutations in cfDNA revealed evidence of multiclonal heterogeneity of BRCA2 reversion mutations, information that would be difficult to glean from a single tumor biopsy sample (Quigley et al. 2017). In the most extreme case in this study, 34 secondary mutations for BRCA2 c.775_776delAG were identified involving deletions of 1 to 42 bp. As every one of these mutations restores the reading frame, the confidence that these mutations confer therapy resistance is extremely high. cfDNA analysis was also suggested to have prognostic value, as responders were more likely to have reductions in cfDNA with the primary mutation compared with nonresponders (Goodall et al. 2017).
Given the HDR defect found in cells with BRCA1/2 mutation, it seems likely that cells attempt to repair DNA damage arising from chemotherapeutics by alternative pathways, in particular, error-prone NHEJ, that then result in the occurrence of resistance mutations. This scenario has implications for therapy regimens, as multiple rounds of therapy and prolonged treatment likely accelerate the development of resistance mutations (Norquist et al. 2011) such that dose levels and scheduling may be an important consideration in treating these tumors.
7.2. Restoration of Homology-Directed Repair by Modulating Levels of Resected DNA
One mechanism identified for PARPi resistance in BRCA1-deficient mouse tumors is the restoration of resected DNA for HDR. Constitutional loss of 53bp1 reduces mammary tumor occurrence in BRCA1-deficient mice (Bunting et al. 2010, Cao et al. 2009), consistent with the notion that HDR proficiency suppresses tumors. However, BRCA1-deficient mammary tumors arising in 53BP1 wild-type mice can acquire PARPi resistance through somatic loss of 53BP1 (Jaspers et al. 2013). Evidence for the relevance of this finding in human tumors is that loss of 53BP1 expression is more prevalent in BRCA1-mutated tumors, although it is surprisingly also associated with BRCA2-mutated tumors (Bouwman et al. 2010). Furthermore, selection of PARPi-resistant BRCA1-mutated tumor cells is also associated with reduced 53BP1 (Jaspers et al. 2013, Johnson et al. 2013). Notably, however, because BRCA1 has a distinct role in interstrand crosslink repair (Long et al. 2014, Sawyer et al. 2015), 53BP1 loss does not lead to cross-resistance to platinum drugs (Bunting et al. 2012).
Loss of other proteins that affect levels of end-resected DNA, such as REV7/MAD2L2 and HELB, similarly confers PARPi resistance to BRCA1-deficient cells (Boersma et al. 2015, Patel et al. 2011, Tkac et al. 2016, Wang et al. 2014, Xu et al. 2015, Zimmermann & de Lange 2014). These proteins could operate in the same step as 53BP1 or in distinct pathways to control levels of resected DNA. It will be important to determine whether loss of these or other players controlling end resection intermediates is an important resistance mechanism in BRCA1-deficient patient tumors.
Notably, given that ATM contributes independently to HDR in BRCA1-53BP1 double mutant cells (Bunting et al. 2010, Chen et al. 2017), evidence suggests that HDR-restored BRCA1-deficient tumor cells can be resensitized to PARPi by ATM kinase inhibition (Tkac et al. 2016, Xu et al. 2015). ATR kinase also has a role in bypassing the requirement for BRCA1 for HDR in PARPi-resistant cells by promoting RAD51 recruitment to damage sites (Yazinski et al. 2017). Given the use of bioavailable inhibitors of ATM or ATR in combination with genotoxic therapies in mouse tumor models (Degorce et al. 2016; Fokas et al. 2012; Vendetti et al. 2015, 2017), we envision the potential of combining PARPi with ATM/ATR kinase inhibitors in the treatment of BRCA1-deficient tumors.
7.3. Restoration of Replication Fork Stability Without Homology-Directed Repair
Restoration of replication fork protection has also been proposed as a mechanism leading to PARPi and cisplatin resistance in BRCA1/2-deficient cells. In this scenario, nascent strand degradation is prevented by precluding the recruitment of MRE11 nuclease to stalled replication forks, which can be achieved by ablation of PTIP, MLL3/4, CHD4, and even PARP1 itself (Ding et al. 2016, Guillemette et al. 2015, Ray Chaudhuri et al. 2016). Interestingly, HDR activity is not restored in these cases, indicating in the cell types tested that fork protection alone is sufficient to confer chemoresistance. While it remains to be determined whether restoration of fork protection is an important resistance mechanism in patients, high PTIP expression in BRCA2-deficient ovarian tumors treated with platinum drugs has been associated with longer progression-free survival (Ray Chaudhuri et al. 2016).
7.4. Hypomorphic BRCA1 Alleles and Resistance Mechanisms
In the following sections, we discuss several studies that have pointed to additional mechanisms of therapy resistance in tumors with specific BRCA1 mutations.
7.4.1. RING-less and RING mutations of BRCA1
At the N terminus of BRCA1 is the highly conserved RING domain (Figure 1a), which interacts with the RING domain of the BARD1 protein to form a BRCA1-BARD1 heterodimer with E3 ubiquitin ligase activity (Hashizume et al. 2001). The interaction is considered to be crucial, as loss of either protein destabilizes the other in early mouse embryos (McCarthy et al. 2003), and loss of either protein or both together leads to mammary tumors in mice with similar latency and characteristics (Shakya et al. 2008). However, specific loss of E3 ligase activity does not appear to be essential in the mouse during development or for cancer suppression (Shakya et al. 2011).
Cancer-predisposing mutations in the BRCA1 RING domain have recently been modeled in mice. BRCA1 C61G is a frequently reported BRCA1 missense mutation, which encodes a protein that impairs BRCA1-BARD1 heterodimerization and ubiquitin ligase activity (Hashizume et al. 2001). Similar to nullizygous mice, mice with this mutation die during embryogenesis, and those with conditional expression develop mammary tumors, which display genomic instability (Drost et al. 2011). However, unlike mammary tumors with null alleles, the Brca1 C61G mouse mammary tumors express a mutant protein and respond poorly to PARPi (Drost et al. 2011). Whereas nullizygous tumors never develop resistance to cisplatin, BRCA1 C61G mammary tumors frequently develop resistance. Notably, resistant tumors displayed some level of RAD51 focus formation, indicating that the BRCA1 C61G protein has partial HDR activity. Whether therapy resistance involves increased expression of the mutant protein or some other cellular change is as yet unclear.
Building on these observations, recent studies have shed additional light on the RING domain. Human breast cancer cells carrying the common truncating mutation BRCA1 185delAG and tumors from mice with a mimicking mutation or deletion of the RING coding sequence were shown to express a RING-less BRCA1 protein that uses an alternative translation start site downstream of the RING domain (Drost et al. 2016, Li et al. 2016, Wang et al. 2016b). As with the Brca1 C61G mutation, mammary tumors from Brca1 185stop mice respond much worse to PARPi therapy than mice with a null allele, indicating resistance, even though the mutation also results in embryonic lethality (Drost et al. 2016). The RING-less BRCA1 demonstrates some HDR activity, which has been proposed to be insufficient to prevent tumor formation but proficient enough to mediate a level of PARPi resistance. Alternatively, the RING-less BRCA1 may be deficient in maintaining genomic integrity independent from its role in HDR (Li et al. 2016). In human BRCA1 185delAG tumor cells, PARPi resistance is associated with increased expression of the RING-less protein, although increased protein expression is not evident in the mouse tumors (Drost et al. 2016, Wang et al. 2016b). However, the RING-less human protein is significantly smaller than the mouse protein due to use of a different downstream translation start site, which could differentially affect protein stability. Thus, BRCA1 185delAG provides two potential routes for therapy resistance: through expression of a hypomorphic protein and as a secondary mutation, as discussed above.
7.4.2. BRCA1-Δ11q isoform
Exon 11 is the largest exon in BRCA1, encoding more than half of the protein, although the key BRCA1 protein domains (i.e., the RING, CC, and BRCT domains; see Figure 1a) are encoded by upstream and downstream exons. Mice with a deletion of exon 11 survive longer during embryogenesis than those with null alleles, and mouse cells expressing a Brca1 exon 11 splice isoform, fully skipping exon 11, have some level of HDR activity (Moynahan et al. 1999, Westermark et al. 2003, Xu et al. 2001), demonstrating that the resulting peptide has partial function.
Human tumors and cell lines with truncating mutations in exon 11, as well as normal cells, express a related splice variant, BRCA1-Δ11q, resulting from partial skipping of exon 11 (Wang et al. 2016a). However, this variant is not in cells with truncating mutations downstream of exon 11. Evidence suggests that the BRCA1-Δ11q isoform affects therapy response. Ovarian cancer patients with BRCA1 truncating mutations in exon 11 show reduced survival. Moreover, human cell lines containing these mutations, which have been selected for PARPi and cisplatin resistance, express higher levels of the BRCA1-Δ11q isoform than sensitive cells, as do PARPi-resistant tumors, and PARPi resistance can be suppressed with spliceosome inhibitors. Thus, alternative splicing can affect therapy response.
7.4.3. BRCT domain of BRCA1
The BRCT domain of BRCA1 interacts with several phosphoproteins involved in various aspects of the DNA damage response and HDR, including CtIP, BRIP1, and Abraxas (Wu et al. 2015). Through these interactions, BRCA1 may have a role in fine-tuning the length of resected DNA through distinct BRCT complexes for optimal HDR function. Several missense and truncating mutations in the BRCT domain have been identified in patients, compromising multiple aspects of BRCA1 function, including damage foci localization, protein stability, and resection-dependent HDR and SSA (Anantha et al. 2017, Chen et al. 2017, Lee et al. 2010, Li & Yu 2013, Nelson & Holt 2010). Mice carrying a Brca1 S1598F point mutation in the BRCT domain, which is analogous to the patient-derived mutation S1655F, show accelerated tumorigenesis similar to complete inactivation of Brca1 (Shakya et al. 2011).
Mutations in the BRCA1 BRCT domain frequently result in protein products that are unstable and subject to proteasome-mediated degradation (Williams et al. 2004). Stabilization of these mutant polypeptides has been proposed as a mechanism of acquired therapy resistance in tumors. One proposed mechanism of stabilization is via interaction with HSP90, as tumor cells can be resensitized to PARPi using HSP90 inhibitors (Johnson et al. 2013, Stecklein et al. 2012), providing a new potential avenue of treatment of tumors with BRCT domain mutations. Interestingly, stabilization of the BRCA1-mutant protein may be combined with another resistance mechanism, since lower 53BP1 protein levels can also be observed (Johnson et al. 2013).
8. TISSUE TROPISM OF BRCA-MUTATED CANCERS AND CELL OF ORIGIN OF BRCA1 BREAST TUMORS
One mystery in the field is the tissue tropism of BRCA-deficient cancers: Whereas HDR is generally considered a critical DSB repair pathway for all proliferative cells, germ-line mutations in BRCA1/2 predominantly predispose to cancers of the breast and ovary. One possibility is that breast and ovarian cells are more reliant on HDR than other tissues. This hypothesis was supported by recent in vivo observations of particularly robust HDR activity in proliferative mammary cells compared to other proliferative tissues (Kass et al. 2016a). Despite more robust HDR overall, all tissues examined in this study show a similar reliance on BRCA2 for repair of a particular lesion, consistent with BRCA2 having a general role in HDR. In addition to a higher reliance on HDR, another non–mutually exclusive possibility is that breast and ovarian tissues may provide a supportive microenvironment to prevent the otherwise lethal effects of BRCA loss, for example, through growth-stimulating hormonal signaling. Other hypotheses include increased DNA damage from estrogen exposure (Caldon 2014, Stork et al. 2016), which may lead to enhanced LOH at the BRCA1/2 loci in breast and ovarian tissue.
Besides the tissue preference, breast cancers arising in BRCA1 versus BRCA2 mutation carriers are distinct. BRCA2 mutation is mainly associated with the luminal subtype of breast cancer, which tends to be estrogen receptor positive (Foulkes et al. 2003, Jonsson et al. 2010, Sorlie et al. 2003, Waddell et al. 2010). By contrast, BRCA1 breast cancers are predominantly basal-like and triple negative, showing a lack of human epidermal growth factor receptor 2 amplification and estrogen and progesterone receptor expression.
Despite their basal-like properties, studies in both mouse and human tissues have provided evidence that BRCA1-associated breast cancers likely arise from luminal epithelial progenitor cells (Lim et al. 2009, Molyneux et al. 2010, Proia et al. 2011). Mice in which Brca1 is deleted in mammary epithelial luminal progenitors develop tumors similar to human BRCA1-associated breast cancers, whereas mice with Brca1 deleted in basal cells develop tumors that are distinct (Molyneux et al. 2010). Similarly, luminal cells from human BRCA1 mutation carriers form tumors with increased basal differentiation in a humanized mouse system (Proia et al. 2011). Notably, precancerous tissue samples from patients carrying heterozygous BRCA1 mutations (BRCA1mut/+) contain an aberrant population of luminal progenitor cells, suggesting that defects may exist in progenitor cell lineage commitment even in normal tissue of mutation carriers (Lim et al. 2009).
In support of the idea that there are preexisting differences in the DNA damage response even in mammary epithelium from women carrying cancer-predisposing mutations, increased genomic instability has been observed in primary cultures of BRCA1mut/+ mammary epithelial cells from carriers (Sedic et al. 2015). BRCA1mut/+ cells have also shown haploinsufficiency for BRCA1’s role in stalled replication fork repair (Pathania et al. 2014), further suggesting that functional defects associated with haploinsufficiency for BRCA1 in breast epithelial cells may at least partially explain the tissue specificity of BRCA1-associated cancers.
Linking BRCA1’s roles in proper mammary cell differentiation and DSB repair, recent studies have shed light on why luminal progenitor cells may be more sensitive to defects in BRCA1’s DSB repair function (Nolan et al. 2016, Sau et al. 2016). Luminal progenitors in preneoplastic breast tissue from BRCA1 mutation carriers separate into two distinct populations, RANK+ and RANK−, based on expression of the receptor for RANK ligand, a paracrine effector of progesterone signaling. RANK+ luminal progenitors are highly proliferative, show higher levels of baseline DNA damage, and are particularly sensitive to ionizing radiation, suggesting aberrant DNA repair, even in normal BRCA1mut/+ tissue (Nolan et al. 2016). Combined with the observation that the RANK+ cells have a molecular signature more similar to basal-like breast cancer than any other subtype, these findings suggest that the RANK+ luminal progenitor cells may represent the cell-of-origin population for the basal-like breast cancers arising in BRCA1 mutation carriers and that RANK ligand (RANKL) may be a valuable therapeutic target in these patients (Nolan et al. 2016). Similarly, the NF-κB pathway is persistently activated in a subset of luminal progenitor cells from preneoplastic BRCA1mut/+ breast tissue (Sau et al. 2016). Sau et al. (2016) found that progesterone-stimulated proliferation increases NF-κB signaling and the accumulation of DNA damage. Thus, it has been hypothesized that overly active progesterone-induced signaling via the NF-κB pathway and paracrine mediator RANKL may account for the tissue specificity and cell of origin for BRCA1-associated breast cancers.
9. ATM KINASE–ASSOCIATED TUMORIGENESIS
The ATM kinase has critical and diverse roles in the DNA damage response and checkpoint regulation following DSBs (Shiloh & Ziv 2013). Biallelic ATM mutations result in the syndrome ataxiatelangiectasia (A-T), which is highly associated with tumor predisposition, primarily lymphoid tumors during childhood whose incidence correlates with the absence of any residual ATM kinase activity. Adult A-T patients are also prone to solid tumors, in particular of the breast (~30-fold increased risk) (Reiman et al. 2011, Verhagen et al. 2012). Monoallelic ATM mutation in carriers also leads to a significant increase in breast cancer risk (~2-fold) (Renwick et al. 2006, Thompson et al. 2005). Most ATM mutations associated with A-T are truncations; however, studies suggest that those associated with cancers tend to be missense mutations, frequently in the kinase domain of ATM (Scott et al. 2002, Yamamoto et al. 2016). Inhibition of ATM kinase activity promotes the transformation of normal human mammary epithelial cells, consistent with its association with breast cancer suppression (Mandriota et al. 2010).
ATM functions in multiple pathways in the DNA damage response, but the linkage of ATM deficiency and breast cancer risk, as well as the sensitivity of ATM-deficient cells to PARPi (Patel et al. 2011, Rass et al. 2013, Weston et al. 2010, Williamson et al. 2010), raises the question of the importance of ATM in HDR. Loss of ATM reduces HDR in some contexts but not in others (Alvarez-Quilon et al. 2014, Beucher et al. 2009, Chen et al. 2017, Kass et al. 2013, Morrison et al. 2000, Rass et al. 2013, White et al. 2010), possibly due in part to redundancy of substrate phosphorylation by other PI3K-related kinase family members, namely ATR and DNA-PKcs (Tomimatsu et al. 2009). However, several ATM targets have been clearly implicated in HDR. For example, ATM-mediated phosphorylation regulates several proteins involved in end resection, including CtIP, MRE11, and EXO1 (Bolderson et al. 2010, Jazayeri et al. 2006, Kijas et al. 2015, Wang et al. 2013, You et al. 2009). ATM also phosphorylates other proteins involved in later steps of HDR (Ahlskog et al. 2016, Bakr et al. 2015). Another aspect of ATM function is to promote heterochromatin relaxation to allow for the general access of the repair machinery (Goodarzi & Jeggo 2012).
Recent studies in mice have shown that the expression of kinase-dead ATM leads to embryonic lethality and a more severe HDR defect than loss of the ATM protein itself, such that the kinase-dead ATM mutations likely pose greater risk for tumorigenesis (Daniel et al. 2012; Yamamoto et al. 2012, 2016). In particular, kinase-dead Atm-mutant mouse cells are more sensitive to a topoisomerase I inhibitor than Atm-null cells, with implications for targeting cancers carrying kinase-inactivating mutations of ATM (Yamamoto et al. 2016). The severity of the mutations may be due to the presence of kinase-dead ATM protein physically blocking DNA ends and preventing repair by other pathways.
The epistatic relationships between ATM, BRCA1, and 53BP1 in the early steps of DSB repair are clearly complex. Although the interaction of 53BP1 with RIF1 or PTIP in G1 phase depends on ATM, inhibition of ATM activity in 53BP1-deficient lymphocytes reduces hyper-resection and partially rescues class switching defects (Bothmer et al. 2010, Tubbs et al. 2014, Yamane et al. 2013), suggesting that ATM-mediated end resection occurs in G1 following 53BP1 loss. On the other hand, ATM also phosphorylates BRCA1 following DNA damage at multiple sites, which facilitates BRCA1 function, at least in checkpoint activation (Cortez et al. 1999, Gatei et al. 2000, Xu et al. 2002). Although the significance of phosphorylation on BRCA1-mediated HDR is not clear, mutation of three phosphorylation sites (S1423, S1387, and S1524) does not support cellular viability (Chang et al. 2009). ATM has recently been shown to play a crucial role in supporting the viability and residual HDR in Brca1 S1598F BRCT mutant mice (Chen et al. 2017). This suggests that although ATM is normally not essential for HDR, its HDR functions could become critical in certain BRCA1 mutant conditions. As ATM is a master regulator at DSB sites, it would be important to determine if ATM has active roles in regulating the choice of DSB repair pathways in response to therapeutic agents.
10. GENOME INSTABILITY AND IMMUNE CHECKPOINT THERAPY
Immunotherapy that enhances the antitumor response from the host immune system has demonstrated promising efficacy against a variety of cancers. For example, antibodies that block the immune-suppressive checkpoints by targeting cytotoxic T-lymphocyte-associated antigen 4 (CTLA4) or programmed cell death-1 (PD-1) receptors can significantly boost endogenous T cell activities to eliminate cancer cells (Sharma & Allison 2015). A key to immunotherapy efficiency is tumor-specific antigens, i.e., neoantigens, which distinguish tumor cells from normal cells and thus can elicit a tumor-specific immune response. Neoantigens can be generated as a result of the intrinsic genome instability of cancer cells that produces mutant polypeptides, a part of which is presented to the cell surface and recognized by the immune system (Schumacher & Schreiber 2015). Thus, a higher mutation load in tumors is thought to lead to more neoantigens and greater immunogenicity (Brown et al. 2014, Rooney et al. 2015). Supporting this, the clinical benefit of immune checkpoint blockade therapies has been observed in cancers with a high mutational burden resulting from environmental exposure, including melanoma and non-small-cell lung cancer (Hugo et al. 2016, Rizvi et al. 2015, Snyder et al. 2014, Van Allen et al. 2015).
Defective DNA repair that increases mutation frequency has also been recognized as an important factor in the antitumor immune response (Mouw et al. 2017), for example, in DNA mismatch repair (Bouffet et al. 2016; Le et al. 2015, 2017). HDR deficiency has also been examined in this context. BRCA1 mutations are shown to be associated with a higher mutation burden in triple-negative breast cancers (Nolan et al. 2017). In Brca1-deficient mouse mammary tumors, combined treatment of cisplatin with anti-PD-1/anti-CTLA4 therapies suppresses the growth of tumors more effectively than cisplatin alone (Nolan et al. 2017). BRCA2 mutations have also been identified in melanomas that show a better response to anti-PD-1 therapy (Hugo et al. 2016). These findings demonstrate the potential of using the mutation status of DNA repair genes, including HDR genes, to predict the immunotherapy response independent of the tumor origins. Moreover, immune checkpoint blockade could be combined with chemotherapies to boost the efficacy in these repair-deficient tumors.
11. CONCLUDING REMARKS
Our knowledge of BRCA1 and BRCA2 has continued to expand since their discovery more than two decades ago. Much progress has already been made using the knowledge gleaned at the bench to develop therapy strategies that benefit clinical practice. However, significant challenges exist to overcoming the drug resistance that almost inevitably arises. Multiple pathways have been proposed as resistance mechanisms based on experimental models. So far, secondary mutations remain the only resistance mechanism clearly validated from patient-derived tumors (Lord & Ashworth 2017). How often these secondary mutations are driven by the therapies themselves needs to be investigated. Future efforts are also needed to validate the clinical relevance of the other proposed resistance mechanisms and to explore additional Achilles’ heels to treating cancers with BRCAness.
Acknowledgments
We thank members of the Jasin lab for helpful discussions. This work was supported by the Breast Cancer Research Foundation (17-078); the Dream Team Translational Research Grant (SU2C-AACR-DT16-15) supported by Stand Up To Cancer, the Ovarian Cancer Research Fund, the Ovarian Cancer National Alliance, the National Ovarian Cancer Coalition, and the American Association for Cancer Research; the Ludwig Center at Memorial Sloan Kettering (MSK) Cancer Center; MSK Cancer Center Support Grant/Core Grant (P30 CA008748); and National Institutes of Health grants K99 CA184133 (E.M.K.), R01 CA185660 (M.J.), and R35 GM118175 (M.J.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Afghahi A, Timms KM, Vinayak S, Jensen KC, Kurian AW, et al. Tumor BRCA1 reversion mutation arising during neoadjuvant platinum-based chemotherapy in triple-negative breast cancer is associated with therapy resistance. Clin Cancer Res. 2017;23:3365–70. doi: 10.1158/1078-0432.CCR-16-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlskog JK, Larsen BD, Achanta K, Sorensen CS. ATM/ATR-mediated phosphorylation of PALB2 promotes RAD51 function. EMBO Rep. 2016;17:671–81. doi: 10.15252/embr.201541455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Quilon A, Serrano-Benitez A, Lieberman JA, Quintero C, Sanchez-Gutierrez D, et al. ATM specifically mediates repair of double-strand breaks with blocked DNA ends. Nat Commun. 2014;5:3347. doi: 10.1038/ncomms4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantha RW, Simhadri S, Foo TK, Miao S, Liu J, et al. Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. eLife. 2017;6:e21350. doi: 10.7554/eLife.21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakr A, Oing C, Kocher S, Borgmann K, Dornreiter I, et al. Involvement of ATM in homologous recombination after end resection and RAD51 nucleofilament formation. Nucleic Acids Res. 2015;43:3154–66. doi: 10.1093/nar/gkv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, et al. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–27. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–65. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- Bhowmick R, Minocherhomji S, Hickson ID. RAD52 facilitates mitotic DNA synthesis following replication stress. Mol Cell. 2016;64:1117–26. doi: 10.1016/j.molcel.2016.10.037. [DOI] [PubMed] [Google Scholar]
- Boersma V, Moatti N, Segura-Bayona S, Peuscher MH, van der Torre J, et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature. 2015;521:537–40. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38:1821–31. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–65. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34:2206–11. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–95. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, van der Gulden H, van der Heijden I, Drost R, Klijn CN, et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013;3:1142–55. doi: 10.1158/2159-8290.CD-13-0094. [DOI] [PubMed] [Google Scholar]
- Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–50. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–17. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Buisson R, Dion-Côté AM, Coulombe Y, Launay H, Cai H, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–54. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46:125–35. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldon CE. Estrogen signaling and the DNA damage response in hormone dependent breast cancers. Front Oncol. 2014;4:106. doi: 10.3389/fonc.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–80. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–41. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature. 2015;518:258–62. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–49. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- Chang S, Biswas K, Martin BK, Stauffer S, Sharan SK. Expression of human BRCA1 variants in mouse ES cells allows functional analysis of BRCA1 mutations. J Clin Investig. 2009;119:3160–71. doi: 10.1172/JCI39836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanut P, Britton S, Coates J, Jackson SP, Calsou P. Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat Commun. 2016;7:12889. doi: 10.1038/ncomms12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–71. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Sossick AJ, Boulton SJ, Jackson SP. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci. 2012a;125:3529–34. doi: 10.1242/jcs.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012b;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chen CC, Kass EM, Yen WF, Ludwig T, Moynahan ME, et al. ATM loss leads to synthetic lethality in BRCA1 BRCT mutant mice associated with exacerbated defects in homology-directed repair. PNAS. 2017;114:7665–70. doi: 10.1073/pnas.1706392114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Park PG, Lee HO, Lee YK, Kang GH, et al. BRCA2 fine-tunes the spindle assembly checkpoint through reinforcement of BubR1 acetylation. Dev Cell. 2012;22:295–308. doi: 10.1016/j.devcel.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Christie EL, Fereday S, Doig K, Pattnaik S, Dawson SJ, Bowtell DDL. Reversion of BRCA1/2 germline mutations detected in circulating tumor DNA from patients with high-grade serous ovarian cancer. J Clin Oncol. 2017;35:1274–80. doi: 10.1200/JCO.2016.70.4627. [DOI] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–66. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- D’Andrea AD. BRCA1: a missing link in the Fanconi anemia/BRCA pathway. Cancer Discov. 2013;3:376–78. doi: 10.1158/2159-8290.CD-13-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Pellegrini M, Lee BS, Guo Z, Filsuf D, et al. Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol. 2012;198:295–304. doi: 10.1083/jcb.201204035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–79. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–25. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degorce SL, Barlaam B, Cadogan E, Dishington A, Ducray R, et al. Discovery of novel 3-quinoline carboxamides as potent, selective, and orally bioavailable inhibitors of ataxia telangiectasia mutated (ATM) kinase. J Med Chem. 2016;59:6281–92. doi: 10.1021/acs.jmedchem.6b00519. [DOI] [PubMed] [Google Scholar]
- Densham RM, Garvin AJ, Stone HR, Strachan J, Baldock RA, et al. Human BRCA1–BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat Struct Mol Biol. 2016;23:647–55. doi: 10.1038/nsmb.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–15. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–33. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Ray Chaudhuri A, Callen E, Pang Y, Biswas K, et al. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat Commun. 2016;7:12425. doi: 10.1038/ncomms12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray E, Etchin J, Wiese C, Saro D, Williams GJ, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17:1255–59. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost R, Bouwman P, Rottenberg S, Boon U, Schut E, et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Drost R, Dhillon KK, van der Gulden H, van der Heijden I, Brandsma I, et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J Clin Investig. 2016;126:2903–18. doi: 10.1172/JCI70196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–15. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–83. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem. 2013;288:11135–43. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Jasin M. BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat Commun. 2017;8:525. doi: 10.1038/s41467-017-00634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. PNAS. 2011;108:686–91. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokas E, Prevo R, Pollard JR, Reaper PM, Charlton PA, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–85. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–75. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- Gatei M, Scott SP, Filippovitch I, Soronika N, Lavin MF, et al. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res. 2000;60:3299–304. [PubMed] [Google Scholar]
- Goodall J, Mateo J, Yuan W, Mossop H, Porta N, et al. Circulating free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7:1–12. doi: 10.1158/2159-8290.CD-17-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Jeggo PA. The heterochromatic barrier to DNA double strand break repair: how to get the entry visa. Int J Mol Sci. 2012;13:11844–60. doi: 10.3390/ijms130911844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette S, Serra RW, Peng M, Hayes JA, Konstantinopoulos PA, et al. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 2015;29:489–94. doi: 10.1101/gad.256214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Hunt CR, Hegde ML, Chakraborty S, Chakraborty S, et al. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell Rep. 2014;8:177–89. doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–40. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–98. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–11. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Isono M, Niimi A, Oike T, Hagiwara Y, Sato H, et al. BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep. 2017;18:520–32. doi: 10.1016/j.celrep.2016.12.042. [DOI] [PubMed] [Google Scholar]
- Jankovic M, Feldhahn N, Oliveira TY, Silva IT, Kieffer-Kwon KR, et al. 53BP1 alters the landscape of DNA rearrangements and suppresses AID-induced B cell lymphoma. Mol Cell. 2013;49:623–31. doi: 10.1016/j.molcel.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 2013;3:68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–83. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N, Johnson SF, Yao W, Li YC, Choi YE, et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. PNAS. 2013;110:17041–46. doi: 10.1073/pnas.1305170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson G, Staaf J, Vallon-Christersson J, Ringner M, Holm K, et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res. 2010;12:R42. doi: 10.1186/bcr2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Ismail A, Katsuki Y, Freire R, Shibata A, Jeggo PA. Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res. 2013;41:10298–311. doi: 10.1093/nar/gkt802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Helgadottir HR, Chen CC, Barbera M, Wang R, et al. Double-strand break repair by homologous recombination in primary mouse somatic cells requires BRCA1 but not the ATM kinase. PNAS. 2013;110:5564–69. doi: 10.1073/pnas.1216824110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Lim PX, Helgadottir HR, Moynahan ME, Jasin M. Robust homology-directed repair within mouse mammary tissue is not specifically affected by Brca2 mutation. Nat Commun. 2016a;7:13241. doi: 10.1038/ncomms13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Moynahan ME, Jasin M. When genome maintenance goes badly awry. Mol Cell. 2016b;62:777–87. doi: 10.1016/j.molcel.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijas AW, Lim YC, Bolderson E, Cerosaletti K, Gatei M, et al. ATM-dependent phosphorylation of MRE11 controls extent of resection during homology directed repair by signalling through Exonuclease 1. Nucleic Acids Res. 2015;43:8352–67. doi: 10.1093/nar/gkv754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinjivadi AM, Sannino V, de Antoni A, Techer H, Baldi G, Costanzo V. Moonlighting at replication forks—a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 2017;591:1083–100. doi: 10.1002/1873-3468.12556. [DOI] [PubMed] [Google Scholar]
- Kondrashova O, Nguyen M, Shield-Artin K, Tinker AV, Teng N, et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2017;7:984–98. doi: 10.1158/2159-8290.CD-17-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, et al. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64:520–39. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk PM, Eppink B, Essers J, Stap J, Rodermond H, et al. Mild hypertermia inhibits homologous recombination, induces BRCA2 degradation and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition. PNAS. 2011;108:9851–56. doi: 10.1073/pnas.1101053108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SG, Liu P, Sharan SK. Mouse embryonic stem cell–based functional assay to evaluate mutations in BRCA2. Nat Med. 2008;14:875–81. doi: 10.1038/nm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Green R, Marsillac SM, Coquelle N, Williams RS, et al. Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res. 2010;70:4880–90. doi: 10.1158/0008-5472.CAN-09-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cole F, Patel DS, Misenko SM, Her J, et al. 53BP1 ablation rescues genomic instability in mice expressing ‘RING-less’ BRCA1. EMBO Rep. 2016;17:1532–41. doi: 10.15252/embr.201642497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–62. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32:3552–58. doi: 10.1038/onc.2012.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol Cell. 2014;56:174–85. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–20. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–58. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandriota SJ, Buser R, Lesne L, Stouder C, Favaudon V, et al. Ataxia telangiectasia mutated (ATM) inhibition transforms human mammary gland epithelial cells. J Biol Chem. 2010;285:13092–106. doi: 10.1074/jbc.M109.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–57. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EE, Celebi JT, Baer R, Ludwig T. Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol Cell Biol. 2003;23:5056–63. doi: 10.1128/MCB.23.14.5056-5063.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel T, Nahse-Kumpf V, Kousholt AN, Klein DK, Lund-Andersen C, et al. A genetic screen identifies BRCA2 and PALB2 as key regulators of G2 checkpoint maintenance. EMBO Rep. 2011;12:705–12. doi: 10.1038/embor.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Tischkowitz M, Chandler K, Gillespie A, Birch JM, Evans DG. Fanconi anaemia, BRCA2 mutations and childhood cancer: a developmental perspective from clinical and epidemiological observations with implications for genetic counselling. J Med Genet. 2014;51:71–75. doi: 10.1136/jmedgenet-2013-101642. [DOI] [PubMed] [Google Scholar]
- Millot GA, Carvalho MA, Caputo SM, Vreeswijk MP, Brown MA, et al. A guide for functional analysis of BRCA1 variants of uncertain significance. Hum Mutat. 2012;33:1526–37. doi: 10.1002/humu.22150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–69. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–17. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Mondal G, Rowley M, Guidugli L, Wu J, Pankratz VS, Couch FJ. BRCA2 localization to the midbody by Filamin A regulates CEP55 signaling and completion of cytokinesis. Dev Cell. 2012;23:137–52. doi: 10.1016/j.devcel.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 2000;19:463–71. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw KW, Goldberg MS, Konstantinopoulos PA, D’Andrea AD. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017;7:675–93. doi: 10.1158/2159-8290.CD-17-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–18. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Cui TY, Jasin M. Homology-directed DNA repair, mitomycin-C resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001a;61:4842–50. [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001b;7:263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, et al. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol. 2011;18:500–3. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. PNAS. 2005;102:1110–15. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AC, Holt JT. Impact of RING and BRCT domain mutations on BRCA1 protein stability, localization and recruitment to DNA damage. Radiat Res. 2010;174:1–13. doi: 10.1667/RR1290.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan E, Savas P, Policheni AN, Darcy PK, Vaillant F, et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med. 2017;9:eaal4992. doi: 10.1126/scitranslmed.aal4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan E, Vaillant F, Branstetter D, Pal B, Giner G, et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat Med. 2016;22:933–39. doi: 10.1038/nm.4118. [DOI] [PubMed] [Google Scholar]
- Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29:3008–15. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–94. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. PNAS. 2011;108:3406–11. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Bade S, Le Guillou M, Burke K, Reed R, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20:764–75. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishvaian MJ, Biankin AV, Bailey P, Chang DK, Laheru D, et al. BRCA2 secondary mutation-mediated resistance to platinum and PARP inhibitor-based therapy in pancreatic cancer. Br J Cancer. 2017;116:1021–26. doi: 10.1038/bjc.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polato F, Callen E, Wong N, Faryabi R, Bunting S, et al. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J Exp Med. 2014;211:1027–36. doi: 10.1084/jem.20131939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontel LB, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, et al. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol Cell. 2015;60:177–88. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–63. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley D, Alumkal JJ, Wyatt AW, Kothari V, Foye A, et al. Analysis of circulating cell-free DNA identifies multi-clonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 2017;7:999–1005. doi: 10.1158/2159-8290.CD-17-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E, Chandramouly G, Zha S, Alt FW, Xie A. Ataxia telangiectasia mutated (ATM) is dispensable for endonuclease I-SceI-induced homologous recombination in mouse embryonic stem cells. J Biol Chem. 2013;288:7086–95. doi: 10.1074/jbc.M112.445825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–87. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman A, Srinivasan V, Barone G, Last JI, Wootton LL, et al. Lymphoid tumours and breast cancer in ataxia telangiectasia; substantial protective effect of residual ATM kinase activity against childhood tumours. Br J Cancer. 2011;105:586–91. doi: 10.1038/bjc.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–75. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- Reuter M, Zelensky A, Smal I, Meijering E, van Cappellen WA, et al. BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells. J Cell Biol. 2014;207:599–613. doi: 10.1083/jcb.201405014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–22. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348:124–28. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross WE, Shipley N. Relationship between DNA damage and survival in formaldehyde-treated mouse cells. Mutat Res. 1980;79:277–83. doi: 10.1016/0165-1218(80)90075-0. [DOI] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–20. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez H, Paul MW, Grosbart M, van Rossum-Fikkert SE, Lebbink JHG, et al. Architectural plasticity of human BRCA2-RAD51 complexes in DNA break repair. Nucleic Acids Res. 2017;45:4507–18. doi: 10.1093/nar/gkx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sau A, Lau R, Cabrita MA, Nolan E, Crooks PA, et al. Persistent activation of NF-κB in BRCA1-deficient mammary progenitors drives aberrant proliferation and accumulation of DNA damage. Cell Stem Cell. 2016;19:52–65. doi: 10.1016/j.stem.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–42. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–42. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–16. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- Scott SP, Bendix R, Chen P, Clark R, Dork T, Lavin MF. Missense mutations but not allelic variants alter the function of ATM by dominant interference in patients with breast cancer. PNAS. 2002;99:925–30. doi: 10.1073/pnas.012329699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedic M, Skibinski A, Brown N, Gallardo M, Mulligan P, et al. Haploinsufficiency for BRCA1 leads to cell-type-specific genomic instability and premature senescence. Nat Commun. 2015;6:7505. doi: 10.1038/ncomms8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Symington LS. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci. 2015;40:701–14. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid T, Soroka J, Kong EH, Malivert L, McIlwraith MJ, et al. Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor. Nat Struct Mol Biol. 2014;21:962–68. doi: 10.1038/nsmb.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya R, Reid LJ, Reczek CR, Cole F, Egli D, et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science. 2011;334:525–28. doi: 10.1126/science.1209909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya R, Szabolcs M, McCarthy E, Ospina E, Basso K, et al. The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression. PNAS. 2008;105:7040–45. doi: 10.1073/pnas.0711032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- Siaud N, Barbera MA, Egashira A, Lam I, Christ N, et al. Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLOS Genet. 2011;7:e1002409. doi: 10.1371/journal.pgen.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. PNAS. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–16. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecklein SR, Kumaraswamy E, Behbod F, Wang W, Chaguturu V, et al. BRCA1 and HSP90 cooperate in homologous and non-homologous DNA double-strand-break repair and G2/M checkpoint activation. PNAS. 2012;109:13650–55. doi: 10.1073/pnas.1203326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. eLife. 2016;5:e17548. doi: 10.7554/eLife.17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–86. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. PNAS. 2009;106:7155–60. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconi EM, Lai X, Folio C, Porru M, Zonderland G, et al. BRCA1 and BRCA2 tumor suppressors protect against endogenous acetaldehyde toxicity. EMBO Mol Med. 2017;9:1398–414. doi: 10.15252/emmm.201607446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SLW, Chadha S, Liu Y, Gabasova E, Perera D, et al. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell. 2017;169:1105–18. e15. doi: 10.1016/j.cell.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–25. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–23. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, et al. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol. 2015;208:545–62. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Duedal S, Kirner J, McGuffog L, Last J, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst. 2005;97:813–22. doi: 10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17:1263–65. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkac J, Xu G, Adhikary H, Young JT, Gallo D, et al. HELB is a feedback inhibitor of DNA end resection. Mol Cell. 2016;61:405–18. doi: 10.1016/j.molcel.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Tomimatsu N, Mukherjee B, Burma S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 2009;10:629–35. doi: 10.1038/embor.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs AT, Dorsett Y, Chan E, Helmink B, Lee BS, et al. KAP-1 promotes resection of broken DNA ends not protected by gamma-H2AX and 53BP1 in G1-phase lymphocytes. Mol Cell Biol. 2014;34:2811–21. doi: 10.1128/MCB.00441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendetti FP, Lau A, Schamus S, Conrads TP, O’Connor MJ, Bakkenist CJ. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget. 2015;6:44289–305. doi: 10.18632/oncotarget.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendetti FP, Leibowitz BJ, Barnes J, Schamus S, Kiesel BF, et al. Pharmacologic ATM but not ATR kinase inhibition abrogates p21-dependent G1 arrest and promotes gastrointestinal syndrome after total body irradiation. Sci Rep. 2017;7:41892. doi: 10.1038/srep41892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen MM, Last JI, Hogervorst FB, Smeets DF, Roeleveld N, et al. Presence of ATM protein and residual kinase activity correlates with the phenotype in ataxia-telangiectasia: a genotype–phenotype study. Hum Mutat. 2012;33:561–71. doi: 10.1002/humu.22016. [DOI] [PubMed] [Google Scholar]
- Waddell N, Arnold J, Cocciardi S, da Silva L, Marsh A, et al. Subtypes of familial breast tumours revealed by expression and copy number profiling. Breast Cancer Res Treat. 2010;123:661–77. doi: 10.1007/s10549-009-0653-1. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wang H, Shi LZ, Wong CC, Han X, Hwang PY, et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR–mediated double-strand break repair. PLOS Genet. 2013;9:e1003277. doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Aroumougame A, Lobrich M, Li Y, Chen D, et al. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev. 2014;28:2693–98. doi: 10.1101/gad.252478.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bernhardy AJ, Cruz C, Krais JJ, Nacson J, et al. The BRCA1-Δ11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 2016a;76:2778–90. doi: 10.1158/0008-5472.CAN-16-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krais JJ, Bernhardy AJ, Nicolas E, Cai KQ, et al. RING domain–deficient BRCA1 promotes PARP inhibitor and platinum resistance. J Clin Investig. 2016b;126:3145–57. doi: 10.1172/JCI87033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark UK, Reyngold M, Olshen AB, Baer R, Jasin M, Moynahan ME. BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol Cell Biol. 2003;23:7926–36. doi: 10.1128/MCB.23.21.7926-7936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–87. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- White JS, Choi S, Bakkenist CJ. Transient ATM kinase inhibition disrupts DNA damage–induced sister chromatid exchange. Sci Signal. 2010;3:ra44. doi: 10.1126/scisignal.2000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–25. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- Williamson CT, Muzik H, Turhan AG, Zamó A, O’Connor MJ, et al. ATM deficiency sensitizes Mantle Cell Lymphoma cells to PARP-1 inhibitors. Mol Cancer Ther. 2010;9:347–57. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis NA, Chandramouly G, Huang B, Kwok A, Follonier C, et al. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 2014;510:556–59. doi: 10.1038/nature13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Jubb H, Blundell TL. Phosphopeptide interactions with BRCA1 BRCT domains: more than just a motif. Prog Biophys Mol Biol. 2015;117:143–48. doi: 10.1016/j.pbiomolbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, O’Donnell AH, Kim ST, Kastan MB. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res. 2002;62:4588–91. [PubMed] [Google Scholar]
- Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–44. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Qiao W, Linke SP, Cao L, Li WM, et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–71. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Wang J, Sprinzen L, Xu J, Haddock CJ, et al. Kinase-dead ATM protein is highly oncogenic and can be preferentially targeted by Topo-isomerase I inhibitors. eLife. 2016;5:e14709. doi: 10.7554/eLife.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Wang Y, Jiang W, Liu X, Dubois RL, et al. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J Cell Biol. 2012;198:305–13. doi: 10.1083/jcb.201204098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Robbiani DF, Resch W, Bothmer A, Nakahashi H, et al. RPA accumulation during class switch recombination represents 5′–3′ DNA-end resection during the S–G2/M phase of the cell cycle. Cell Rep. 2013;3:138–47. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazinski SA, Comaills V, Buisson R, Genois MM, Nguyen HD, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31:318–32. doi: 10.1101/gad.290957.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72:2814–21. doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, et al. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–69. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HS, Oyama T, Isse T, Kitagawa K, Pham TT, et al. Formation of acetaldehyde-derived DNA adducts due to alcohol exposure. Chem Biol Interact. 2010;188:367–75. doi: 10.1016/j.cbi.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–51. [PubMed] [Google Scholar]
- Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–79. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–29. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu H, Chen Y, Yang X, Wang P, et al. A cell cycle-dependent BRCA1–UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nat Commun. 2016;7:10201. doi: 10.1038/ncomms10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24:108–17. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–4. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]