Abstract
In this paper, we investigate if the MR prostate segmentation performance could be improved, by only providing one-point labeling information in the prostate region. To achieve this goal, by asking the physician to first click one point inside the prostate region, we present a novel segmentation method by simultaneously integrating the boundary detection results and the patch-based prediction. Particularly, since the clicked point belongs to the prostate, we first generate the location-prior maps, with two basic assumptions: (1) a point closer to the clicked point should be with higher probability to be the prostate voxel, (2) a point separated by more boundaries to the clicked point, will have lower chance to be the prostate voxel. We perform the Canny edge detector and obtain two location-prior maps from horizontal and vertical directions, respectively. Then, the obtained location-prior maps along with the original MR images are fed into a multi-channel fully convolutional network to conduct the patch-based prediction. With the obtained prostate-likelihood map, we employ a level-set method to achieve the final segmentation. We evaluate the performance of our method on 22 MR images collected from 22 different patients, with the manual delineation provided as the ground truth for evaluation. The experimental results not only show the promising performance of our method but also demonstrate the one-point labeling could largely enhance the results when a pure patch-based prediction fails.
1 Introduction
Prostate cancer is one of the most leading cause of male death [1]. Previous clinical studies demonstrate that the radiotherapy can provide effective treatment for prostate cancer. During the radiotherapy process, the prostate cancer tissues should be killed while the normal tissues could not be hurt at the same time. Therefore, an accurate prostate segmentation is significant from the clinical perspective, which indicates that the success of the radiotherapy highly depends on the accuracy of prostate segmentation. In fact, the segmentation of prostate is conventionally done by physician with slice-by-slice delineation, which is very time-consuming.
In recent years, many automatic prostate segmentation methods have been proposed with promising results for different modalities, e.g. CT and Ultrasound [2]. In this paper, we focus on segmentation of MR images. The prostate segmentation is a challenging task due to the low tissue contrast, irregular prostate motion, as well as large shape variation among different patients. Many previous attempts were developed to address the aforementioned challenges. Atlas-based prostate segmentation in MR images is popular in the last decade [3]. Also, prior information is employed to improve the segmentation performance. Gao et al. [4] incorporated both local image statistics and learnt shape prior to guide better segmentation. In [5] the spatial locations of the prostate identified via spectra are used as the initial ROI for a 2D Active Shape Model (ASM). These approaches largely depend on the discriminative ability of the hand-crafted features, with different types of features to choose and also lots of parameters to select. Fortuantely, in recent years, deep learning provides a more feasible way for learning good features. Liao et al. in [6] propose a deep learning framework using independent subspace analysis to extract useful features for segmentation.
However, the performance of these previous automatic segmentation methods largely depends on the consistency of sample distributions between training and testing images. A natural question is whether we could use a simple interaction from physicians to improve the performance. To overcome this issue, several interactive segmentation methods are presented by performing prostate segmentation with a few interactions from physician, although these methods are not specifically designed for segmentation in MR images. In [7], physician’s interactive labeling information is first required to label a small set of prostate and non-prostate voxels, and then transductive Lasso is used to select the most discriminative features. Moreover, interactive methods based on prior knowledge of training data have been proposed: Park et al. [8] used the spatial relationship of adjacent patches to constrain the specific shapes of organs.
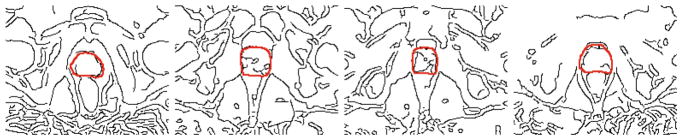
In this paper, a novel interactive method for MR prostate segmentation is proposed, by only requiring one-point interaction from physicians. This is motivated by the following two facts. First, for a new MR image, physician focuses on the region which has a high probability of being prostate, instead of wasting time on other regions. Second, the borders in image provide more information for segmentation than the inside regions. As what Fig. 1 conveys, although the exact boundary of prostate cannot be detected, the edges in image are useful to infer the location of prostate. Specifically, we first ask a physician to provide one point on the MR image, which is probably in the center of prostate. Then, we use Canny operator to obtain the detected edges. By simultaneously combining detected edges and one-point labeling information, we could obtain two prostate location-prior maps from the vertical and horizontal directions, respectively, with two basic assumptions: (1) a point closer to the clicked point should be with higher probability to be prostate voxel, (2) a point separated by more boundaries to the clicked point will have lower chance to be the prostate voxel. The location-prior maps are regarded as new channels and added to the original raw image. We finally train a multi-channel patch-based FCN [9] to perform the patch-based prediction, and employ the refinement for the final segmentation results.
Fig. 1.
Typical boundary image of our training data. The black curves denote the boundary detected by Canny edge detector. The red curves denote the ground-truth manual delineation. (Color figure online)
2 Our Method
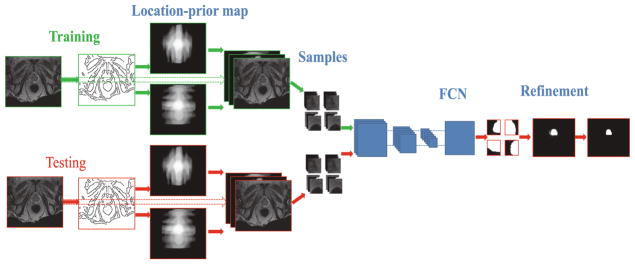
We now first present how to generate the location-prior maps and then illustrate details of network structure. Finally, we will introduce the result refinement for generating final segmentations. The pipeline of the whole framework is shown in Fig. 2.
Fig. 2.
Illustration of the pipeline of our method.
Generating location-prior maps
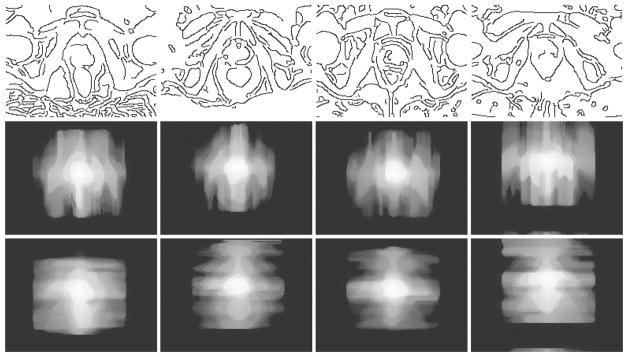
For a new MR image, we ask the physician to roughly find the center of prostate for generating the location-prior maps. Please note that, we do not mean that the physician must find the true center of prostate, and the later evaluation also shows the robustness of our method to different initializations. Then more attention is paid to the regions extending from the center of prostate, especially those with border information. As what Fig. 1 conveys, partial edges of prostate can be detected by common edge detector (i.e., Canny edge detector), which are close to the ground-truth manual delineation. Also, the borders of rectum and bladder can be detected at the same time, which are in fact surrounding the prostate region. Although it is infeasible that all the images could provide relative clear and complete edge of prostate region, the border of other organs surrounding prostate is generally clear and can thus provide effective heuristic information for generating the location-prior maps. Figure 3 shows several typical examples.
Fig. 3.
Typical location-prior maps generated from our dataset. Images in first row are the border images obtained by Canny edge detector, while the second and the third are the location-prior maps obtained from vertical and horizontal directions, respectively.
For each testing image, we ask the physician to first provide one point to indicate the location of prostate, which is generally in the central area of prostate. For the sake of convenience, we take the point as a voxel in the image with exact coordinates. Please note that, the one point information for training images is given by calculating the mass center, since the manual delineation of training images are available during the training stage. Normally, to generate the prior-location maps, we follow the two basic assumptions: (1) a point closer to the clicked point should be with higher probability to be the prostate voxel, (2) a point separated by more boundaries to the clicked point will have lower chance to be the prostate voxel. After using Canny edge detector to detect all edges in the MR image, two horizontal and vertical rays are extended out from one-point voxel (we set the intensity value as 255) with the intensity reducing strategy as follows for each move: in the location-prior map, the intensity value is reduced by 1, and in particular, when the move crosses through a detected edge by Canny edge detector, the intensity value is reduced by 10. From each voxel in all lines, rays are extended with the same intensity reducing strategy as above. Then we use a median filter to smooth these two calculated images as the location-prior maps from two different directions, respectively. Finally, the original MR image along with the two location-prior maps are combined as a multi-channel image in the subsequent learning process.
Since different regions of prostate and non-prostate regions do not distribute uniformly, we sample patches from both prostate and non-prostate regions from training images as previous methods [10,11]. Typically, we sample patches in the following ways: (1) densely sampling the patches which locate inside the prostate region or are centered close to prostate boundary, and (2) sparsely sampling patches which are far from prostate boundary.
Neural network structure
To predict the prostate-likelihood for the patches from testing images, we design a multi-channel fully convolutional network inspired by FCN [9]. The traditional FCN [9] is modified to fit the small size input patches in our task. Specifically, the whole structure is similar to traditional FCN [9], but with fewer layers. Please refer to Table 1 for details. Training a deep neural network will be a challenging task when internal covariance shift problem happens [12]: the distribution of internal nodes changes when the parameter of previous layer is updated. Fortunately, batch normalization [12] provides an efficient way to tackle this issue. To avoid the above issue, we perform batch normalization towards reducing internal covariance shift. Typically, we add one batch normalization layer to each convolutional layer. We observe that, with batch normalization [12], the neural network will end up with a poor local optimum. The Adam [13] optimizer is employed for optimization, where initial learning rate is set 0.001 and other parameters keep default.
Table 1.
Network structure.
| Layer | Kernel | Pooling | Activation |
|---|---|---|---|
| Conv | 5 × 5 × 3 × 32 | N.A. | Relu |
| Conv | 5 × 5 × 32 × 32 | N.A. | Relu |
| Conv | 5 × 5 × 32 × 64 | Max | Relu |
| Conv | 4 × 4 × 64 × 64 | N.A. | Relu |
| Conv | 4 × 4 × 64 × 2 | Upsample | Softmax |
We implement our method with TensorFlow toolbox [14], on a PC with 3.7 GHz CPU, 128 GB RAM, and Nvidia Tesla K80 GPU. The number of epoch is set to 30. The running time for training requires 2 to 3 h. Currently, our method only takes 1–5 s to segment a new coming image.
Refinement
It is noteworthy that, in the training stage, for each image we randomly sample patches from the whole image to train our model, while, in the testing stage, we use a different sample pattern to choose patches fed into the neural network: a number of rays in equal degree intervals are extended from the one-point pixel labeled by physicians, we choose testing patches whose center point locates at these rays. Their responding segmentation will be integrated to forming the prostate likelihood map. Experiments show that, in this way, we can capture more precise boundaries of prostate. Since the patient-specific shape prior is not available, on the obtained prostate-likelihood maps, we perform the level-set method [15] to generate the final segmentations.
3 Experimental Results
Setting
Our dataset consists of 22 MR images, scanned from 22 different patients. The resolution of each MR image after image preprocessing is 193 × 152 × 60: the in-plane voxel size is 1 × 1 mm2 and the inter-slice thickness is 1 mm. The manual delineation results are available for each image that can be used as ground truth for evaluation. Meanwhile, for segmenting each testing image, we will first ask the physician to provide an additional one-point coordinate. We perform 2-fold cross validation on these 22 MR images. For evaluation metrics, we employ the Dice ratio and centroid distance (CD) along 3 directions (i.e., lateral x-axis, anterior-posterior y-axis, and superior-inferior z-axis), which are widely used in previous studies [10,11].
Qualitative results
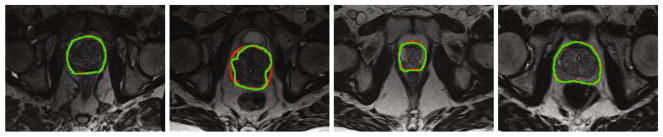
Figure 4 shows the typical MR images in our dataset along with the detected prostate boundaries by our segmentation method (green) and manual rater (red). It is obvious that our method can achieve promising result.
Fig. 4.
Typical results. Red curves denote the manual delineation. Green curves denote the results obtained by our method. (Color figure online)
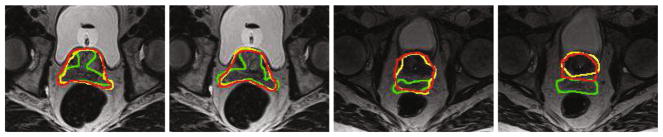
Figure 5 shows that, with the help of additional one-point information, compared to the traditional fully convolutional network, our method can locate prostate more precisely with a significant improvement.
Fig. 5.
Typical results of FCN and our method. Red, green, yellow curves denote the manual delineation, traditional FCN and our method, respectively. (Color figure online)
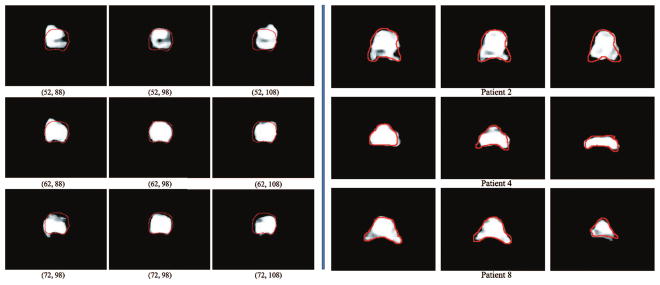
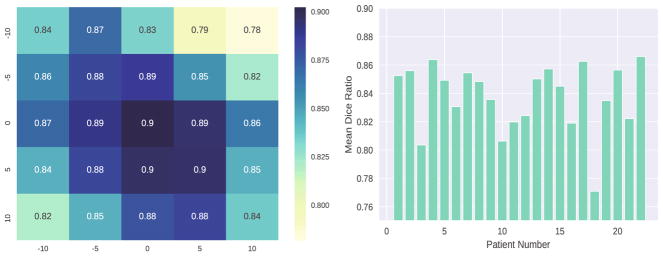
Since the point provided from physician is important in our framework, it is necessary to evaluate how the point coordinates affect the segmentation result. As what Fig. 6 conveys, the coordinates of point truly have effect on the final segmentation result: the farther away from the center area of prostate, the worse the result will be. Besides, we plot a heatmap in Fig. 7 to illustrate the impact. It is noteworthy that, for this image, the Dice ratio by performing traditional FCN [9] is 0.85.
Fig. 6.
Illustration of prostate likelihood maps. Left part demonstrates prostate likelihood maps when different point coordinates are given for the same image (i.e., initial coordinates of point are (62,98)). Right part illustrates the likelihood maps obtained by our method on prostates with irregular shapes. The red curves denote the manual delineation. (Color figure online)
Fig. 7.
(Left) Illustration of the Dice ratio values with changes of coordinates. (Right) The Dice ratio values for 22 individual patients in our dataset.
Quantative results
Our method can obtain mean dice ratio of 0.84 ± 0.02 which is higher than 0.71 ± 0.15 achieved by traditional FCN [9]. Please refer to Fig. 7 for the Dice ratio of each patient. Table 2 shows evaluation compared with traditional FCN [9].
Table 2.
Evaluation comparison with FCN.
| CD-x (mm) | CD-y (mm) | CD-z (mm) | Dice ratio | |
|---|---|---|---|---|
| FCN | 1.91 ± 2.63 | 1.31 ± 0.99 | 0.57 ± 0.37 | 0.71 ± 0.15 |
| Our method | 0.74 ± 0.48 | 0.49 ± 0.44 | 0.27 ± 0.19 | 0.84 ± 0.02 |
Due to the fact that neither executables nor the datasets of other works are publicly available, it is difficult for us to directly compare our method with other MR prostate segmentation method [6,16,17]. Thus, we only cite the results reported in their publications in Table 3 for reference. It is worth noting that our method can obtain competitive results compared with these state-of-art methods.
Table 3.
Comparison with other state-of-the-art methods.
4 Conclusion
In this paper, we propose a novel interactive segmentation method for MR prostate, which can achieve the promising results with just one-point labeling from the physician. Specifically, we can make full use of provided interaction information by simultaneously incorporating the additional point labeling and detected edges, in order to generate two location-prior maps, from vertical and horizontal directions, respectively. Upon the obtained location-prior maps along with the original patches from MR images, a multi-channel patch-based fully convolutional network is then used for patch-based prediction. Finally, a level-set based refinement is performed for final segmentation results. The experimental results demonstrate the effectiveness of our method.
Acknowledgments
This work was supported by NSFC (61673203, 61432008), Nan-jing Science and Technique Development Foundation (201503041), Young Elite Scientists Sponsorship Program by CAST (YESS 20160035) and NIH Grant (CA206100).
References
- 1.Cokkinides V, Albano J, Samuels A, et al. American Cancer Society. Cancer Facts and Figures. American Cancer Society; Atlanta: 2005. [Google Scholar]
- 2.Ghose S, Oliver A, Mart R, et al. A survey of prostate segmentation methodologies in ultrasound, magnetic resonance and computed tomography images. Comput Methods Programs Biomed. 2012;108(1):262–287. doi: 10.1016/j.cmpb.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Klein S, van der Heide UA, Lips IM, et al. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med Phys. 2008;35(4):1407–1417. doi: 10.1118/1.2842076. [DOI] [PubMed] [Google Scholar]
- 4.Gao Y, Sandhu R, Fichtinger G, et al. A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery. IEEE Trans Med Imaging. 2010;29(10):1781–1794. doi: 10.1109/TMI.2010.2052065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toth R, Tiwari P, Rosen M, et al. A magnetic resonance spectroscopy driven initialization scheme for active shape model based prostate segmentation. Med Image Anal. 2011;15(2):214–225. doi: 10.1016/j.media.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao S, Gao Y, Oto A, Shen D. Representation learning: a unified deep learning framework for automatic prostate MR segmentation. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. MICCAI 2013. LNCS. Vol. 8150. Springer; Heidelberg: 2013. pp. 254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, Liao S, Gao Y, Zhang D, Gao Y, Shen D. Transductive prostate segmentation for CT image guided radiotherapy. In: Wang F, Shen D, Yan P, Suzuki K, editors. MLMI 2012. LNCS. Vol. 7588. Springer; Heidelberg: 2012. pp. 1–9. [DOI] [Google Scholar]
- 8.Park SH, Yun ID, Lee SU. Data-driven interactive 3D medical image segmentation based on structured patch model. In: Gee JC, Joshi S, Pohl KM, Wells WM, Zöllei L, editors. IPMI 2013. LNCS. Vol. 7917. Springer; Heidelberg: 2013. pp. 196–207. [DOI] [PubMed] [Google Scholar]
- 9.Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015. pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y, Liao S, Gao Y, et al. Prostate segmentation in CT images via spatial-constrained transductive lasso. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2013. pp. 2227–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, Gao Y, Liao S, et al. Semi-automatic segmentation of prostate in CT images via coupled feature representation and spatial-constrained transductive lasso. IEEE Trans Pattern Anal Mach Intell. 2015;37(11):2286–2303. doi: 10.1109/TPAMI.2015.2424869. [DOI] [PubMed] [Google Scholar]
- 12.Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. 2015 arXiv: 1502.03167. [Google Scholar]
- 13.Kingma D, Ba J. Adam: A Method for Stochastic Optimization. 2014 arXiv: 1412.6980. [Google Scholar]
- 14.Abadi M, Barham P, Chen J, et al. TensorFlow: a system for large-scale machine learning. Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI); Savannah, Georgia, USA. 2016. [Google Scholar]
- 15.Li C, Xu C, Gui C, et al. Distance regularized level set evolution and its application to image segmentation. IEEE Trans Image Process. 2010;19(12):3243–3254. doi: 10.1109/TIP.2010.2069690. [DOI] [PubMed] [Google Scholar]
- 16.Coupé P, Manjnó JV, Fonov V, et al. Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. NeuroImage. 2011;54(2):940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Klein S, van der Heide UA, Lips IM, et al. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med Phys. 2008;35(4):1407–1417. doi: 10.1118/1.2842076. [DOI] [PubMed] [Google Scholar]