Summary
Substantial improvements in enzyme activity demand multiple mutations at spatially proximal positions in the active site. Such mutations, however, often exhibit unpredictable epistatic (non-additive) effects on activity. Here, we describe FuncLib - an automated method for designing multipoint mutations at enzyme active sites using phylogenetic analysis and Rosetta design calculations. We applied FuncLib to two unrelated enzymes, a phosphotriesterase and an acetyl-CoA synthetase. All designs were active and most showed activity profiles that significantly differed from the wild type and from one another. Several dozen designs with only 3-6 active-site mutations exhibited 10-4,000-fold higher efficiencies with a range of alternative substrates, including the hydrolysis of the toxic organophosphate nerve agents soman and cyclosarin and the synthesis of butyryl-CoA. FuncLib is implemented as a web-server (http://FuncLib.weizmann.ac.il); it circumvents iterative, high-throughput screens and opens the way to design highly efficient and diverse catalytic repertoires.
Introduction
Mutations that alter enzyme activity profiles are essential for adaptation to an organism’s changing needs, such as metabolizing new substrates. Such mutations are also highly desired in basic research, biotechnology, and biomedicine to enable efficient and environmentally safe solutions, for instance in the synthesis of useful molecules or the degradation of harmful ones. Most mutations, however, are deleterious to protein activity and stability, constraining the emergence of improved variants through natural evolution or protein engineering. Furthermore, due to mutational epistasis, a mutation’s effect on activity depends on whether or not other mutations were previously acquired (de Visser, Cooper, and Elena 2011; Phillips 2008; Starr and Thornton 2016; Miton and Tokuriki 2016). In the extreme case, known as sign epistasis, two mutations that are individually deleterious, enhance activity when combined, or vice versa (Dellus-Gur et al. 2015; Weinreich, Watson, and Chao 2005). In natural evolution, mutations usually occur one at a time, and thus, epistatic combinations of mutations must accumulate in a specific order, since all intermediates must be at least as active as their predecessors or they would be purged by selection. The high prevalence of sign epistasis in improved mutants further reduces the likelihood of obtaining beneficial combinations (Weinreich et al. 2006). Protein evolution is additionally constrained by stability-threshold effects, whereby activity-enhancing mutations may destabilize the protein (Wang, Minasov, and Shoichet 2002; Shoichet et al. 1995) and therefore accumulate only up to a threshold in which additional mutations are no longer tolerated (Bershtein et al. 2006; Bloom et al. 2006). To overcome stability-threshold effects, stabilizing mutations, both in proximity to the active-site pocket and in distant regions, are essential for the accumulation of function-enhancing mutations (Goldsmith et al. 2017).
Due to epistasis and stability-threshold effects, the evolution of variants with significant enhancement in an enzyme activity demands multiple mutations of different type and affecting different regions of the protein. Laboratory-evolution experiments, for instance, may comprise more than a dozen rounds of genetic diversification and selection for improved mutants (Bloom and Arnold 2009; Tracewell and Arnold 2009; Bigley et al. 2013; Goldsmith et al. 2017), and substantial improvements by three orders of magnitude or more require on average ten mutations(Badenhorst and Bornscheuer 2018; Goldsmith and Tawfik 2017). The majority of these mutations occur outside the catalytic pocket and are likely to affect activity only indirectly by enhancing tolerance to function-enhancing mutations. Another complication is that laboratory-evolution experiments are laborious and demand high-throughput or even ultrahigh-throughput screening (>106 variants per round) (Esvelt, Carlson, and Liu 2011). Such screens, however, are only applicable to certain enzyme activities and typically employ synthetic model substrates.
In principle, computational protein design strategies could bypass the need for multiple rounds of experimental optimization, since they are unconstrained by mutational trajectories. Previous applications of protein design computed favorable point mutants or focused libraries for experimental screening (Pavelka, Chovancova, and Damborsky 2009; Reetz, Wang, and Bocola 2006; Damborsky and Brezovsky 2009; Hilvert 2013; Wijma et al. 2015), yielding limited gains in activity (Cherny et al. 2013; Badenhorst and Bornscheuer 2018), and de novo designed enzymes exhibited low catalytic efficiencies (Khersonsky et al. 2012; Blomberg et al. 2013; Baker 2010; Khare et al. 2012). Overall, computational enzyme design remains a specialized expertise and still depends on laboratory evolution to reach comparable efficiencies to those seen in natural enzymes(Khersonsky et al. 2012; Blomberg et al. 2013; Baker 2010; Khare et al. 2012). Thus, substantial gaps remain in our understanding and control of the basic principles of enzyme design.
To address these gaps, we developed a new automated strategy that designs stable networks of interacting residues at the active site and selects a small set of diverse designs amenable to low-throughput screening. Our strategy addresses stability-threshold effects by first designing a stable enzyme scaffold and addresses epistasis by designing dense and preorganized networks of interacting active-site multipoint mutants. It does not a priori target a specific substrate, as this demands accurate models of the enzyme transition-state complex, and such models are rarely attainable. Rather, this strategy results in a repertoire of stable and highly efficient enzymes that can be screened for the activities of interest. Starting from two different enzymes, we used this strategy to design functionally diverse repertoires comprising dozens of enzymes that exhibited 10-4,000 fold improvements in a range of activities. Encouraged by the robustness and effectiveness of the new strategy and by the outcome of our previous publicly available protein-stabilization platform PROSS(Goldenzweig et al. 2016), we implemented the new algorithm, which we called FuncLib, as an automated web-accessible server (http://FuncLib.weizmann.ac.il).
Design
FuncLib’s goal is to design a small set of stable, efficient, and functionally diverse multipoint active-site mutants suitable for low-throughput experimental testing. The design strategy is general and can be applied, in principle, to any natural enzyme using its molecular structure and a diverse set of homologous sequences (Figure 1).
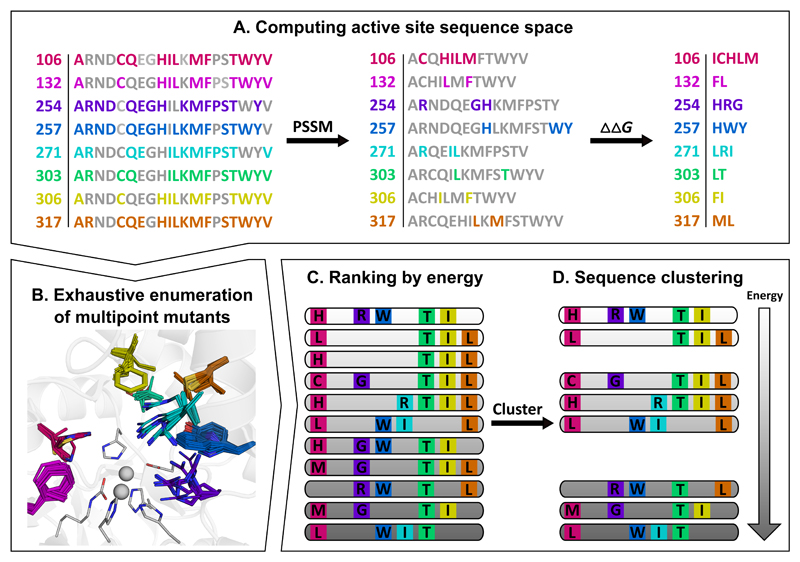
Figure 1. Flow chart illustrating key steps in the design of functional repertoires.
The images illustrate steps in the generation of a repertoire of phosphotriesterase enzymes starting from a bacterial phosphotriesterase (PTE; PDB entry: 1HZY). (A) Active-site positions are selected for design, and at each position, sequence space is constrained by evolutionary-conservation analysis (PSSM) and mutational-scanning calculations (ΔΔG). (B) Multipoint mutants are exhaustively enumerated using Rosetta atomistic design calculations. The PTE active site comprises a bimetal center (gray spheres) of Zn2+ ions that are liganded by six highly conserved residues (gray sticks); eight additional residues (colored sticks) comprise the active-site wall and are less conserved. (C) The designs are ranked by energy, and (D) the sequences are clustered to obtain a repertoire of diverse, low-energy designs for experimental testing. Designed positions are colored consistently in all panels. See also Table S1.
The focal point of our study was the metalloenzyme phosphotriesterase (PTE) from Pseudomonas diminuta(Holden et al. 2001). In addition to highly efficient hydrolysis of the organophosphate pesticide paraoxon (kcat/KM approximately 108 M-1s-1), PTE promiscuously hydrolyzes esters, lactones, and diverse organophosphates, including toxic nerve agents, such as VX, Russian VX, soman (GD), and cyclosarin (GF), albeit with kcat/KM values that are orders-of-magnitude lower than for paraoxon (Figure 2A)(Jackson et al. 2009). Effective organophosphate detoxification for in vivo protection, however, demands high catalytic efficiency, with a minimal kcat/KM of 107 M-1min-1, thereby motivating several recent enzyme-engineering efforts that targeted PTE (Bigley et al. 2013, 2015; Goldsmith et al. 2017; Cherny et al. 2013). Furthermore, the threat from a new generation of nerve agents (“Novichoks”), similar in structure to VX and GF(Peplow 2018), reinforces the need for broad-spectrum nerve-agent hydrolases.
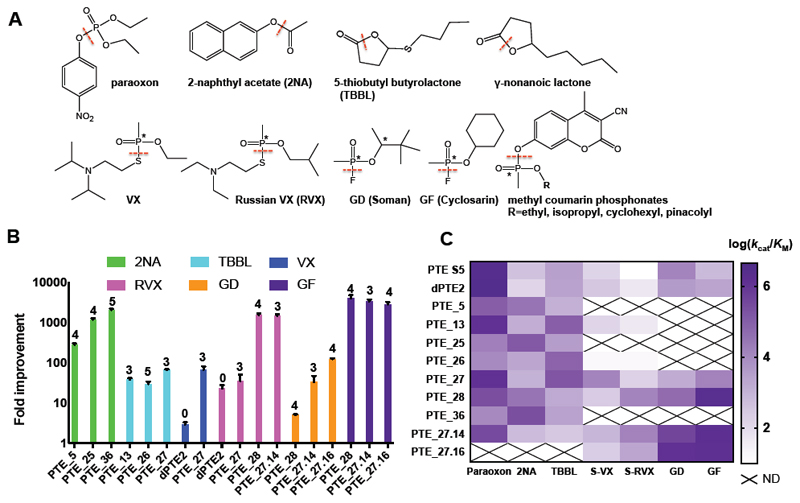
Figure 2. A designed repertoire of phosphotriesterases exhibits orders of magnitude improvement in a range of promiscuous activities, including the hydrolysis of highly toxic nerve agents.
(A) The bacterial PTE is a paraoxonase exhibiting additional promiscuous hydrolase activities. The dashed red lines indicate the bonds that PTE hydrolyses in each of the substrates tested in this study, and the asterisks indicate chiral centers. (B) Fold improvement in catalytic efficiency (kcat/KM) of the top FuncLib designs relative to PTE-S5, showing remarkable >1,000-fold improvement in nerve-agent hydrolysis efficiency in several designs. The number of active-site mutations is indicated above the bars. (C) Activity profiles of the top PTE designs. Several designs, most prominently PTE_27, PTE_28, and PTE_27.14, exhibit substantially broadened substrate selectivity relative to PTE S5. Data for nerve agents are shown for the more toxic Sp stereoisomers. Data are represented as mean ± standard deviations of duplicate measurements. N.D. - not determined. See also Tables S1-S3.
Since active-site mutations often impair protein stability, we started active-site design calculations from dPTE2, a variant of PTE-S5(Roodveldt and Tawfik 2005) with 20 stabilizing mutations outside the active-site pocket that was previously designed in our laboratories using the PROSS stability-design algorithm(Goldenzweig et al. 2016). dPTE2 exhibited higher stability and fivefold higher bacterial-expression yields than PTE-S5, while retaining wild-type levels of activity.
We selected eight active-site positions that comprise the PTE active-site wall (first-shell) for design. FuncLib starts by defining a sequence space comprising active-site point mutations that are predicted to be individually tolerated (Figure 1A). First, it retains only mutations with at least a modest probability of occurrence in the natural diversity according to a multiple-sequence alignment of homologs. Second, it eliminates point mutations that substantially destabilize the wild-type protein according to Rosetta atomistic modeling. Applied to the PTE active-site pocket, for instance, no mutations were allowed in its Zn2+-chelating residues, whereas other first-shell positions were allowed even radical mutations (Figure 1A-B). The two-step filtering drastically reduced the combinatorial space of multipoint mutants at the eight active-site positions from 1010 mutants, if all 20 amino acids were allowed at each position, to <105. From this filtered set, we modeled and refined all the multipoint mutants that comprised 3-5 mutations relative to wild-type PTE in Rosetta, including backbone and sidechain minimization to enable radical mutations such as from small to large amino acid sidechains (Figure 1B). Next, we ranked all multipoint mutants according to their predicted stability (Figure 1C). Thus, the top-ranked designs were predicted to exhibit stable and preorganized active-site pockets — a prerequisite for high catalytic efficiency(Warshel 1998; Lapidoth et al. 2018). Surprisingly, we found that hundreds of unique active-site designs exhibited energies that were as favorable as or better than that of wild-type PTE, suggesting that a very large space of potentially tolerated multipoint mutants at the active site was accessible by computational design. Finally, we clustered the designs (Figure 1D), eliminating ones that differed by fewer than two active-site mutations from one another or from wild-type PTE and selected the top 49 designs for experimental testing (Table S1).
Note that in this implementation, FuncLib does not require a model of the enzyme-transition state complex. Instead, it computes diverse yet stable networks of interacting residues at the active-site pocket, thereby encoding different stereochemical complementarities for alternative substrates that do not need to be defined a priori. We therefore anticipated that the designs would collectively form a functional repertoire, from which individual designs that efficiently hydrolyzed various target substrates could be isolated. In applications that target a specific substrate, by contrast, sequence space can be further constrained by designing the enzyme in the presence of the substrate or transition-state model, and this option is enabled in the FuncLib web-server.
Results
Orders of magnitude efficiency gains in a designed catalytic repertoire
All PTE designs retained detectable levels of paraoxonase activity (Table S1), demonstrating that their active site was intact and functional despite the high sequence diversity. We next measured the designs’ specific activities with alternative, promiscuous substrates including phosphotriesters other than paraoxon, phosphonodiesters, carboxy-esters, and lactones (Figure 2A). Following this initial screen, the catalytic efficiencies of the most active designs were determined. Most designs exhibited efficiency gains relative to the wild type enzyme with respect to at least one substrate: Ten designs exhibited improved efficiencies in hydrolyzing the pesticide malathion by up to 14-fold, 15 showed similar levels of improvement (up to 16-fold) in lactonase efficiency, and 35 exhibited remarkable gains of up to 1,000-fold in esterase efficiency (Figure 2B-C, Table S1). The largest gains in activity were observed for substrates that showed weak activities in the wild-type enzyme, demonstrating that FuncLib could improve a range of weak promiscuous activities.
In addition to exhibiting improved catalytic efficiencies, the designs also showed vast changes in substrate selectivity. For instance, PTE-S5 is selective for paraoxon over the ester 2-naphthyl acetate (2NA) by 3x104-fold. Through only five active-site mutations, selectivity is reversed in design PTE_36 to 0.04 — a nearly million-fold selectivity switch. Similarly, PTE-S5 favors paraoxon over the synthetic lactone tetrabutyl butyrolactone (TBBL) by 103-fold, whereas in design PTE_26 selectivity is switched to 0.1 (Table 1). Remarkably, these designs retained substantial paraoxonase activity (kcat/KM ≥104 M-1s-1), demonstrating that some of the designs broadened substrate recognition rather than only trading off one activity for another (Figure 2C). Consistent with this conclusion, several designs exhibited increased efficiency with respect to the disfavored stereoisomer of methyl coumarin phosphonates relative to the wild type, while retaining high efficiency against the natively favored stereoisomer (Table S1).
Table 1. Specificity changes (as ratios of catalytic efficiency, kcat/KM) in PTE variants.
| PTE variant | Paraoxon/2-naphthyl acetate | Specificity switch relative to dPTE2 | Paraoxon/TBBL | Specificity switch relative to dPTE2 |
|---|---|---|---|---|
| dPTE2 | 31048.6 | 1 | 1406.5 | 1 |
| PTE_5 | 3.41 | 9104 | 98.7 | 14 |
| PTE_13 | 1149.3 | 27 | 15.7 | 90 |
| PTE_24 | 25.65 | 1210 | 7.6 | 186 |
| PTE_25 | 0.13 | 246732 | 5.2 | 272 |
| PTE_26 | 4.61 | 6737 | 0.1 | 11219 |
| PTE_27 | 1454.3 | 21 | 8.8 | 161 |
| PTE_28 | 7.60 | 4086 | 148.0 | 10 |
| PTE_36 | 0.04 | 741664 | 4.1 | 347 |
| PTE_27.14 | 591 | 53 | 1206.5 | 1 |
We next measured catalytic efficiency of the designs that retained high phosphotriesterase activity with the toxic nerve agents VX, Russian VX (RVX), soman (GD), and cyclosarin (GF) (Table S2). PTE_27 exhibited 66-fold increase in VX hydrolysis efficiency relative to wild-type PTE, and PTE_28 exhibited remarkable gains in efficiency of 1,550 and 3,980-fold in hydrolyzing RVX and GF, respectively. Starting from PTE_27, we tested a second round of design, this time directing FuncLib to model all combinations of 3-5 mutations that occurred in the best nerve-agent hydrolases tested in the first round and eliminating designs that were predicted to be unstable (>8 Rosetta energy units relative to PTE_27). We experimentally tested the 14 resulting designs, finding that designs PTE_27.14 and PTE_27.16 exhibited increased activities towards GD (32-fold and 122-fold, respectively), and both designs exhibited a 3,000-fold increase in hydrolyzing GF. These designs, with kcat/KM ≥ 107 M-1min-1 for the highly toxic nerve agents RVX, GD, and GF, may be suitable for in vivo detoxification(Wille et al. 2016).
Note that the efficiency gains observed by testing 63 designs were comparable to the best variants from the application of more than a dozen rounds of diversification and experimental testing of thousands of variants using conventional laboratory-evolution strategies (Badenhorst and Bornscheuer 2018; Bigley et al. 2013; Goldsmith et al. 2017) (Table S3). Furthermore, laboratory-evolution experiments demand separate selection campaigns for each substrate, whereas the designed repertoire comprised dozens of enzymes that exhibited improved efficiency towards each of the substrates we tested. Additionally, all of the designs showed bacterial-expression levels comparable to the highly expressed dPTE2 design (>300 mg protein per liter culture, Figure S1)(Goldenzweig et al. 2016). These results demonstrate that the combination of PROSS and FuncLib may not exhibit the stability-threshold bottlenecks that have constrained the laboratory evolution of many enzymes, including PTE(Bershtein et al. 2006; Bloom et al. 2006; Goldsmith et al. 2017; Goldenzweig and Fleishman 2018). Thus, FuncLib results in a small but functionally highly diverse repertoire of stable and efficient enzymes and may in some cases bypass the requirement for high-throughput screens.
Structural bases of high catalytic efficiency and selectivity
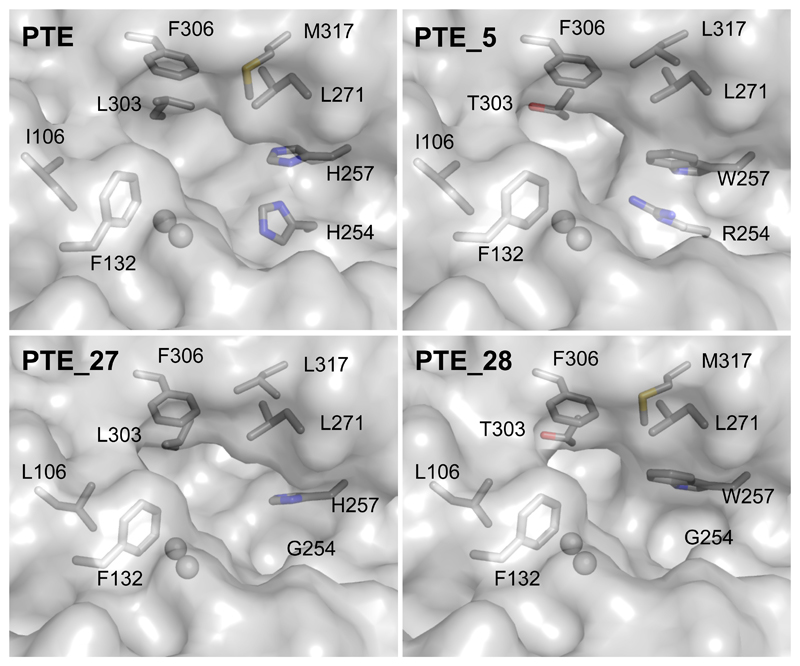
To understand what molecular factors underlie the high gains in catalytic efficiency in some designs, we used X-ray crystallography and determined the molecular structures of PTE_5 (280-fold improved activity with 2NA), PTE_27 (65-fold improved activity with TBBL and 103-fold improved activity with S-VX), and PTE_28 (3,980-fold improved activity with GF) (Figure 3; Table S4). All three structures showed high accuracy relative to their respective models (root mean square deviation [rmsd] <0.5 Å over the backbone and 0.3 Å all-atom rmsd in mutated active-site residues)(Figure S2), confirming that the design process resulted in precise and preorganized active-site pockets as required for high-efficiency catalysis(Lapidoth et al. 2018).
Figure 3. The stereochemical properties of the designed active-site pockets underlie selectivity changes in PTE designs.
PTE_27 and PTE_28 exhibit a larger active-site pocket than PTE and high catalytic efficiency against bulky V- and G-type nerve agents. In clockwise order from top-left, molecular renderings are based on PDB entries: 1HZY, 6GBJ, 6GBK, and 6GBL (see Table S4). Spheres indicate ions of the bimetal center. Figure S2 shows models of the designed enzymes with docked substrates. An animated Interactive 3D Complement (I3DC) is available in Proteopedia at http://proteopedia.org/w/Journal:Molecular_Cell:2
We used molecular docking simulations to model the toxic Sp stereoisomers of VX, RVX, and GD in the active-site pockets of PTE_27, PTE_28, and PTE_27.14, respectively. The resulting models indicated that the designed active-site pockets were large enough to accommodate the bulky nerve agents and form direct contacts with them, mostly due to two large-to-small substitutions, His254Gly and Leu303Thr (Figure 3 and Figure S2). These direct contacts may also underlie the high enantioselectivity observed in some designs (>104 for design PTE_28; Table S2). Furthermore, several improved esterases and lactonases (PTE 13-15, 30-34, and 36) encoded the His254Arg mutation, which changed the sterics and electrostatics of the active-site pocket, as also reported in laboratory-evolution studies that enhanced these activities(Afriat-Jurnou, Jackson, and Tawfik 2012; Tokuriki et al. 2012) (Figure 3). We therefore concluded that the FuncLib mutations only affected the structure of the active-site pocket, that the designed repertoire encoded substantial stereochemical diversity in the active site leading to large selectivity changes, and that a handful of active-site mutations was sufficient to effect orders-of-magnitude improvements in catalytic efficiency and selectivity for several substrates.
Sign epistasis among designed mutations
In each design, the mutations are spatially clustered. We therefore anticipated that some designs would show complex epistatic relationships, whereby the effects of multipoint mutants could not be simply predicted based on the effects of the single-point mutants. We therefore measured the specific activities of all single- and double-point mutants comprising three of the best designs: PTE_5, PTE_27, and PTE_32 with four, three, and four active-site mutations relative to PTE, respectively (Figure 4, Figure S3). In PTE_5 and PTE_32, the point mutations improved catalytic efficiency relative to the wild type, but some double mutants exhibited efficiencies that were substantially lower than those of the wild type.
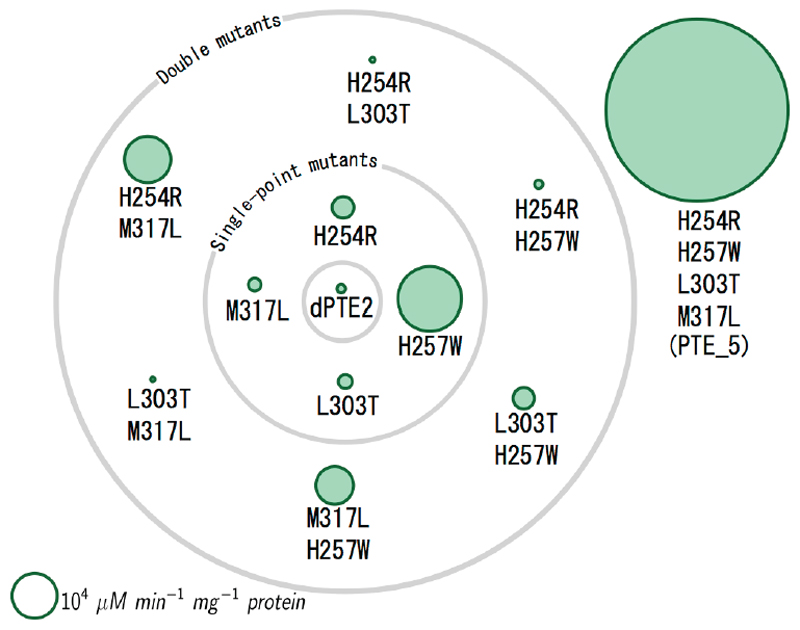
Figure 4. Designed mutations exhibit sign-epistatic relationships.
Each green circle represents a mutant of dPTE2, and the gray orbits represent the number of mutations from dPTE2. The area of each circle is proportional to the design’s specific activity in hydrolyzing the aryl ester 2-naphthyl acetate (2NA). PROSS-stabilized design dPTE2, which was used as the starting point for FuncLib design, exhibits low specific activity. Each of the point mutants exhibits improved specific activity, but specific activity declines in the double mutants. Last, the quad-mutant, design PTE_5, substantially improves specific activity relative to all single or double mutants. See also Figure S3.
PTE_5 provided a compelling case of sign epistasis (Figure 4)(Weinreich, Watson, and Chao 2005). All point mutations improved specific activity with the ester 2NA. All double mutants, however, were worse than the single-point His257Trp, and three of the double mutants were even worse than the starting point dPTE2. Most revealing, the combination of two double mutants that exhibited lower specific activities than dPTE2, His254Arg/His257Trp and Leu303Thr/Met317Leu, resulted in the most active design PTE_5, which improved specific activity by two orders of magnitude relative to dPTE2 and by three orders of magnitude relative to the Leu303Thr/Met317Leu double mutant. Furthermore, at the level of DNA, the point mutations His→Trp and Leu→Thr require three and two nucleotide exchanges, respectively, drastically reducing the odds for the emergence of PTE_5 through stepwise accumulation of mutations. A previous analysis of mutational trajectories leading to enhanced fitness in clinically isolated β-lactamase mutants noted the pervasiveness of sign epistasis among function-enhancing mutations; and yet, a fraction of the trajectories in that case showed monotonous, and therefore evolutionarily selectable, improvement in activity(Weinreich et al. 2006). For PTE_5, by contrast, our analysis suggested not even a single mutational trajectory of monotonously increasing activity. Hence, FuncLib may access mutants that cannot be obtained through the stepwise accumulation of beneficial mutations that is a prerequisite for natural or laboratory evolution(Tracewell and Arnold 2009).
Broadened substrate selectivity in a repertoire of acyl-CoA synthetases
To test FuncLib’s generality, we next applied it to the Salmonella enterica acetyl CoA synthetase (ACS), which ligates CoA and acetate in a two-step reaction (Figure 5A). Acyl-CoA formation is a first and critical step leading to a variety of useful modifications, including the reduction of carboxylic acids to aldehydes that in turn can be reduced to alcohols or alkanes, which can be used as biofuels; aldehydes can also be used to form new carbon-carbon bonds via aldol condensation. ACS variants that efficiently ligate different aliphatic acids may therefore open the way to new and efficient synthetic pathways.
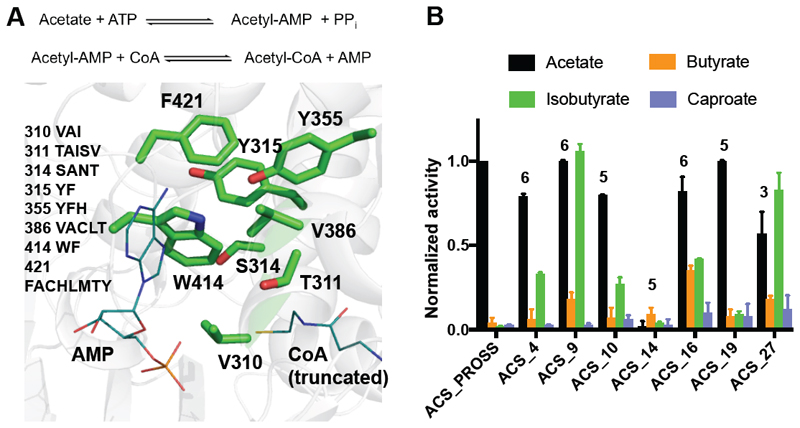
Figure 5. A designed repertoire of acyl-CoA-synthetases and substantially broadened substrate selectivity.
(A) The ACS active site. Positions where design was enabled are shown as green sticks, and AMP and CoA substrates are shown as teal lines. Amino acid positions and identities allowed during design are indicated. (B) Specific activity of ACS variants, normalized to that of ACS_PROSS with acetate, demonstrates substantial broadening of substrate selectivity in designs ACS_9, ACS_16, and ACS_27. See also Tables S5-S6 and Figures S4-S5.
Effective protein engineering of non-hydrolytic enzymes, such as ACS, is a challenge(Kaltenbach and Tokuriki 2014). Indeed, ACS binds three substrates (an acid and two cofactors, AMP and CoA) and performs two reactions in a ping-pong mechanism. Similar to many other non-hydrolytic enzymes, ACS exhibits fairly limited catalytic efficiency even towards its native substrate acetate (kcat/kM=4.5 x 104 M-1s-1)(Reger, Carney, and Gulick 2007), is not amenable to high-throughput screening and has not been subjected to in vitro evolution so far and therefore presented an intriguing test case for FuncLib. In addition to acetate-CoA ligation, ACS accepts other aliphatic acids, foremost propionate(Reger, Carney, and Gulick 2007); however, the substrate diversity observed in natural ACS enzymes is much more restricted than that observed in PTE.
The ACS active site presented a challenge to our design strategy, since it was much more evolutionarily conserved than that of PTE, and mutations at several active-site positions were prohibited by the evolutionary-conservation filter used by FuncLib. We therefore extended design to second-shell positions Ser314 and Tyr315 (Figure 5A), anticipating that such mutations might cooperate with first-shell mutations to further alter the shape of the active site and allow access to larger acids than acetate. With the intent of obtaining a stable starting enzyme for FuncLib designs, we first tested two automatically generated PROSS-stabilized variants(Goldenzweig et al. 2016) of the S. enterica ACS, and chose the one that showed the highest level of acetate-CoA synthesis activity (ACS_PROSS). This variant comprised 47 mutations outside the active-site pocket and exhibited 100-fold higher bacterial expression than S. enterica ACS. We then selected 29 FuncLib designs with 3-6 mutations at eight active-site positions for experimental testing (Table S5).
As with PTE, all ACS designs exhibited extremely high bacterial-expression yields (>300 mg protein per liter culture, Figure S1). The designs’ specific activity was determined with different acids (acetate, butyrate, isobutyrate, glycine, valerate, isovalerate, caproate, formate and phenyl acetate) using endpoint-thiol detection of CoA with DTNB (Table S5). All the designs retained detectable levels of acetyl-CoA-ligase activity, despite the complexity of the active-site pocket and the catalytic mechanism. Seven designs demonstrated increased activity with both butyrate and isobutyrate (up to tenfold and 47-fold, respectively), and four designs exhibited up to fourfold increased activity with caproate (Figure 5B, Figure S4).
The specificity profiles of the best designs changed substantially relative to ACS_PROSS. ACS_9 and ACS_27 exhibited comparable specific activity with isobutyrate and acetate. In fact, these two designs reached specific activities with butyrate and isobutyrate that ACS_PROSS exhibited with its native substrate acetate, whereas ACS_PROSS exhibited at least 50-fold lower specific activity with isobutyrate (Table S6). The increased catalytic efficiency of ACS designs toward larger acids was confirmed using a NADH-consumption assay, which couples CoA synthesis with lactate dehydrogenase activity (Reger, Carney, and Gulick 2007) (Figure S5).
Collectively, the results indicate that FuncLib can reliably improve low starting activities, whether in enzymes that exhibit high sequence diversity and substrate promiscuity, such as PTE, or high conservation, restricted substrate specificity, and a complex multistep catalytic mechanism, such as ACS.
Discussion
Natural and laboratory evolution of altered activities depend on the stepwise accumulation of mutations, each of which must be at least neutral in fitness (Weinreich, Watson, and Chao 2005; Tracewell and Arnold 2009). Following a few mutations, however, improvements in activity often plateau due to epistasis or stability-threshold effects(Bershtein et al. 2006; Bloom et al. 2006; Goldsmith et al. 2017). Typical evolutionary trajectories leading from one highly efficient enzyme to another are therefore time-consuming and often comprise dozens of enabling mutations outside the active site, most of which only contribute to the activity indirectly, for instance by stabilizing the enzyme(Badenhorst and Bornscheuer 2018). The strategy presented in this manuscript rationalizes and accelerates the generation of stable enzymes exhibiting altered activities: It starts by designing stable and highly expressed enzyme variants(Goldenzweig et al. 2016) and then designs dozens of variants that encode preorganized networks of active-site mutants exhibiting different physicochemical features. FuncLib requires a single molecular structure of the target enzyme and a set of diverse sequence homologs (in the two reported test cases, hundreds of unique sequences were available). The combination of evolutionary-conservation analysis and Rosetta atomistic modeling focuses design calculations on stable, preorganized, and functional active-site constellations. Although FuncLib uses evolutionary-conservation analysis, the resulting designs show high-efficiency catalysis even towards nerve agents — human-made compounds for which efficient enzymes are unlikely to exist in nature. FuncLib can therefore expand on the functional diversity that can be accessed through genome mining and, in combination with PROSS stability design, yields stable, highly expressed, and functionally diverse enzymes.
In the two case studies presented here, small-scale testing of FuncLib designs was sufficient to identify variants that exhibited orders-of-magnitude changes in enzyme activity profiles without loss in apparent protein stability. FuncLib can therefore be used to rapidly optimize specific activities or generate functional repertoires from enzymes that are not amenable to high-throughput screening. Indeed, the emergence of a new generation of nerve agents(Peplow 2018) adds urgency for rapid design of highly efficient functional repertoires as demonstrated here. Furthermore, whereas conventional active-site design strategies rely on transition-state modeling, FuncLib computes diverse and stable networks of interacting active-site mutations, enabling design even in the cases discussed here, for which enzyme transition-state models are uncertain. Although the designed mutations conserve the wild type backbone structure, some designs exhibit sign-epistatic relationships, which render these designs all but inaccessible to stepwise mutational trajectories. Thus, the sequence space of an enzyme active site provides a vast resource of functional diversity that defies exploration by natural and laboratory evolution but can now be accessed through computational protein design.
Limitations
FuncLib requires a molecular structure and a diverse set of sequence homologues as input. Although in some cases comparative modeling may provide a starting point for design, we note the high sensitivity of computational design to atomic details, particularly in dense regions such as an enzyme active site. In the absence of sufficient diversity of sequence homologues, FuncLib may productively combine mutations that were observed in experimental screens, and the FuncLib web-server provides an option to manually define the allowed sequence space based on such observations.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sarel J. Fleishman (sarel.fleishman@weizmann.ac.il).
Experimental Model and Subject Details
PTE, ACS and CobB proteins were expressed in Escherichia coli BL21 DE3 cells. The bacteria were grown at 37°C prior to induction and at 20°C after induction with IPTG.
Method Details
Computational methods
PROSS stability design calculations. In case of PTE, the previously described design dPTE2 (Goldenzweig et al. 2016) was used as the starting point for FuncLib calculations. ACS from S. enterica (PDB entry: 1PG4) was stabilized using the PROSS web-server (http://pross.weizmann.ac.il), which automatically collected 478 unique homologs. Two designs were experimentally tested, and the design with higher acetyl-CoA synthetase activity (ACS_PROSS) was chosen as the starting point for FuncLib calculations.
FuncLib calculations. We defined the active-site residues to be designed by visual examination of the enzyme molecular structures (PDB entries 1HZY and 1PG4 for PTE and ACS, respectively). Evolutionary conservation scores were computed from Position-Specific Scoring Matrices (PSSMs) and ΔΔG values were computed essentially as described previously(Goldenzweig et al. 2016). Tolerated amino acid identities at the active site were filtered according to the following thresholds: PSSM ≥ -2 and ΔΔG ≤ +6 R.e.u. and PSSM ≥ -1 and ΔΔG ≤ +5 R.e.u. for PTE and ACS, respectively. In the case of ACS, we also carried out the same calculations with cutoffs PSSM ≥ -2 and ΔΔG ≤ +4 R.e.u.
We next exhaustively enumerated all possible combinations of at least three and as many as five and six mutations in PTE and ACS, respectively. Each mutant was modeled in Rosetta, including combinatorial sidechain packing, and the backbone and sidechains of all residues were minimized, subject to harmonic restraints on the Cα coordinates of the entire protein. All designs were ranked according to all-atom energy, and the top-ranked designs were chosen for experimental analysis after removing designs with fewer than two mutations relative to one another.
FuncLib web-server. The web-server implements several important improvements relative to the method used to design the reported PTE and ACS variants. In designing the PTE and ACS variants, a multiple-sequence alignment (MSA) was computed for the entire protein sequence, and wherever loops were observed in the query structure, any aligned sequence that exhibited gaps relative to the query was eliminated to reduce alignment ambiguity (see ref. (Goldenzweig et al. 2016) for details). In the FuncLib web-server, by contrast, all secondary-structure elements are subjected to this filtering, resulting in improved PSSM accuracy, particularly in the active-site pocket. Furthermore, the web-server implements more accurate atomistic modeling and scoring: it uses the recent Rosetta energy function ref15(Park et al. 2016) with improved electrostatics and solvation potentials relative to previous Rosetta energy functions; implements harmonic coordinate restraints on sidechain atoms of essential amino acid residues in the catalytic pocket to guarantee their preorganization; restricts refinement to amino acids within 8 Å of designed positions instead of refining the entire protein; allows the user to modify the tolerated sequence space (for instance, based on prior experimental and structural analysis); and enables modeling of small-molecule ligands or transition-state complexes.
Substrate docking. The Adaptive-PELE method(Lecina, Gilabert, and Guallar 2017) with an ε-greedy reward phase based on the protein-ligand interaction energy was used to dock the Sp stereoisomers of nerve agent substrates VX, RVX and GD into PTE_27, PTE_28, and PTE_27.14 designs, respectively. Catalytic poses were obtained using 50 PELE trajectories, each running for six hours in a computing core. Substrates were initially positioned in the vicinity of the active-site pocket and were diffused by Adaptive-PELE to their binding catalytic poses.
Quantification and Statistical Analysis
Statistical parameters are stated in the figure legends and under the appropriate Methods detail section. All the reported values represent the averages ±standard deviations based on at least two independent measurements.
Data and Software Availability
The Rosetta software suite for biomolecular design is available for download at http://www.rosettacommons.org. Rosetta git version 627f7dd22223c3074594934b789abb4f4e2e3b10 was used for all simulations. All Rosetta modeling and design was done using RosettaScripts(Fleishman et al. 2011), which are available with their command lines and flag files in Supplemental Information. All design calculations used the Rosetta talaris14 all-atom energy function, which is dominated by van der Waals packing, hydrogen bonding, solvation, and electrostatics(O’Meara et al. 2015).
The coordinates of PTE_5, PTE_27 and PTE_28 were deposited in the RCSB Protein Data Bank with accession codes 6GBJ, 6GBK and 6GBL respectively. The structures will be released upon publication.
The plasmids with genes of active designs are available from AddGene (deposit number 75507).
Additional Resources
PROSS web-server (http://pross.weizmann.ac.il) was used for protein stabilization.
FuncLib web-server is available online (http://FuncLib.weizmann.ac.il).
Detailed protocol
Cloning. Synthetic genes for the parental enzymes and the designed variants were codon optimized for efficient E. coli expression, and custom synthesized as linear fragments by Twist Bioscience. The genes of PTE designs were amplified and cloned into the pMal C2 vector with N-terminal MBP fusion tag through the EcoRI and PstI restriction sites. The variable segment of ACS gene was introduced into pET21d plasmid containing the PROSS-stabilized ACS gene by RF cloning(van den Ent and Löwe 2006). The plasmids were transformed into E. coli BL21 DE3 cells, and DNA was extracted for Sanger sequencing to validate accuracy.
Protein expression. 2 ml of 2YT medium supplemented with 100 μg/ml ampicillin (and 0.1 mM ZnCl2 in case of PTE) were inoculated with a single colony and grown at 37°C for ~15h. 10ml 2YT medium supplemented with 100 μg/ml ampicillin (and 0.1 mM ZnCl2 in case of PTE) were inoculated with 0.2 ml overnight culture and grown at 37°C to an OD600 of ~0.6. Overexpression was induced with 0.2 mM IPTG, and the cultures were grown for ~24h at 20°C. After centrifugation and storage at -20°C, the pellets were resuspended in lysis buffer and lysed by sonication.
PTE purification. PTE lysis buffer: 50 mM Tris (pH 8.0), 100 mM NaCl, 10 mM NaHCO3, 0.1 mM ZnCl2, benzonase and 0.1 mg/ml lysozyme. The protein was bound to amylose resin (NEB), washed with 50 mM Tris with 100 mM NaCl and 0.1 mM ZnCl2, and the proteins were eluted with wash buffer containing 10 mM maltose. The elution fraction was used for SDS-PAGE gel and before activity assays the proteins were dialyzed in wash buffer. For crystallization, the PTE variants were recloned into pETMBPH vector containing an N-terminal 6xHis tag and MBP fusion(Peleg and Unger 2008) and the expression was performed with 500 ml culture. After purification, the protein was digested with TEV protease to remove the MBP fusion tag (1:20 TEV, 1 mM DTT, 24-48h/RT). The MBP fusion was removed by binding to Ni2+-NTA resin, and the protein was purified by gel filtration (HiLoad 26/600 Superdex75 preparative grade column, GE).
ACS purification. ACS lysis buffer: 25 mM Hepes (pH 7.5), 100 mM NaCl, 1mM TCEP, benzonase and 0.1 mg/ml lysozyme. The protein was bound to Ni-NTA resin (Merck) supplemented with 10mM imidazole, washed with 20 mM imidazole in 25 mM Hepes (pH 7.5), 100 mM NaCl, and the proteins were eluted with wash buffer containing 250 mM imidazole. The elution fraction was analyzed by SDS-PAGE gel and the proteins were dialyzed in 25 mM Hepes (pH 7.5) with 100 mM NaCl. Prior to activity assays ACS variants had to be deacetylated with CobB for activation, due to in vivo acetylation of the catalytic lysine.
CobB production and ACS deacetylation. E. coli cells over-expressing CobB were taken from ASKA library(Kitagawa, Ara, and Arifuzzaman 2005), 10 ml 2YT supplemented with 35 μg/ml chloramphenicol and 1% glucose were grown at 37°C for ~15h. 1.5L 2YT medium supplemented with 35 μg/ml chloramphenicol were inoculated with 10 ml overnight culture and grown at 37°C to an OD600 of ~1. Overexpression was induced with 0.5 mM IPTG, and the cultures were grown for ~20h at 16°C. After centrifugation and storage at -20°C, the pellets were resuspended in lysis buffer and lysed by sonication. CobB lysis buffer: 20mM Tris (pH 8). 100mM NaCl, 1mM DTT, benzonase, and 0.1mg/ml lysozyme. The protein was bound to Ni-NTA resin (Merck) supplemented with 10mM imidazole, washed with 30 mM imidazole in 20 mM Tris (pH 8) with 100 mM NaCl, and eluted with wash buffer containing 250 mM imidazole. The elution fraction was used for SDS-PAGE gel and the proteins was dialyzed in 50mM Tris (pH 8), 100mM NaCl, 10% glycerol, and 0.5mM DTT. Deacetylation was performed overnight at 37°C in a mix containing 3 μM CobB, 20-40 μM ACS designs and 1mM NAD+.
Kinetic measurements. The kinetic measurements of PTE designs were performed with purified proteins in activity buffer (50 mM Tris pH 8.0 with 100 mM NaCl, and 0.1 mM ZnCl2). A range of enzyme concentrations was used, depending on the activity. The activity of PTE designs was tested colorimetrically with phosphotriesters (paraoxon (0.5mM), malathion (0.25mM), EMP, IMP, CMP, PMP (0.1mM)), esters (p-nitrophenyl acetate (0.5mM), p-nitrophenyl octanoate (0.1mM), 2-naphthyl acetate (0.3mM)), and lactones (TBBL (0.5mM)(Khersonsky and Tawfik 2006), γ-nonanoic lactone (0.5mM, pH-sensitive assay, by monitoring the absorbance of m-cresol indicator at 577 nm). The kinetic measurements were performed in 96-well plates (optical length – 0.5cm), and background hydrolysis rates were subtracted.
The rate of hydrolysis of the V-type nerve agents in presence of OP hydrolases was performed as described (Cherny et al. 2013). The in situ conversion of the coumarin surrogates to the corresponding G nerve agents in diluted aqueous solutions and the monitoring of the rate of detoxification of the G agents by OP hydrolases were performed as previously described by (Gupta et al. 2011; Yacov Ashani et al. 2011). Note that the concentration of the in situ generated G-and V-agents is non-hazardous foremost because the in situ synthesis was performed on a small (mg) scale in diluted aqueous solutions. Nonetheless, due to their high potency as inhibitors of AChE, all safety requirements were strictly observed.
Catalytic efficiencies (kcat/KM) were determined for the most active PTE designs by measuring the activity at several low substrate concentrations in the approximated first-order kinetics region of the Michaelis-Menten equation.
The activity of ACS designs was determined by detection of remaining CoA with DTNB. Reaction mixture (100 μl) contained 5 μM purified deacetylated ACS variant, 25mM Hepes pH 7.5, 1mM MgCl2, 0.25mM coenzyme A, 5mM ATP and 1mM of the substrates (formate, acetate, propionate, butyrate, isobutyrate, valerate, isovalerate, glycine, caproate, and L-lactate). After 15 minutes incubation at 37°C, 100 μl of 1mM DTNB were added to the reaction mixture, and OD412 was measured in BioTEK H1 plate reader. Subtraction of the obtained OD from blank OD (no enzyme in reaction mixture) is the OD of reacted CoA.
A NADH consumption assay (Reger, Carney, and Gulick 2007)was also used to assess the activity of ACS variants. Reaction mixture contained: 50mM Hepes pH 7.5, 5mM MgCl2, 1mM TCEP, 3mM phosphoenol pyruvate, 5U of myokinase, 1U of pyruvate kinase, 5.5U of lactate dehydrogenase, 0.5mM NADH, 2.5mM ATP, 1.5mM CoA, 0-10mM substrate (acetate, glycolate, butyrate, isobutyrate and caproate), and 0.5-1μM purified deacetylated ACS, and the loss of NADH was monitored at 340nm. Unfortunately, it was not possible to obtain the kinetic parameters from the coupled assay, since it gave unrealistically low KM values, probably due to inhibition of the coupled assay enzymes by substrates other than acetate.
Structure determination and refinement. Crystals of PTE_5, PTE_27 and PTE_28 were obtained using the hanging-drop vapor-diffusion method with a Mosquito robot (TTP LabTech). All data sets were collected at 100 K on a single crystal on in-house RIGAKU RU-H3R X-ray. The crystals of PTE_5 were grown from 0.85M Lithium sulfate and 0.05M HEPES pH=7.0. The crystals formed in the space group P43212, with one dimer per asymmetric unit and diffracted to 1.63 Å resolution. Crystals of PTE_27 were grown from 0.1M MgCl2*6H2O, 10% PEG 4000 and 0.05M Tris pH=7.5. The crystals formed in the space group C2, with one dimer per asymmetric unit and diffracted to 1.9 Å resolution. Crystals of PTE_28 were grown from 0.1M Mg(OAC)2*4H2O, 8% PEG 8000 and 0.05M Na cacodylate pH=6.4. The crystals formed in the space group C2, with one dimer per asymmetric unit and diffracted to 1.95 Å resolution.
Diffraction images of PTE_5, PTE_27 and PTE_28 crystals were indexed and integrated using the Mosflm program(Randy J. Read, Joel L Sussman 2007), and the integrated reflections were scaled using the SCALA program(Evans 2006). Structure factor amplitudes were calculated using TRUNCATE(French and Wilson 1978) from the CCP4 program suite. The PTE_5, PTE_27 and PTE_28 structures were solved by molecular replacement with the program PHASER(McCoy 2007). The model used to solve the PTE_5, PTE_27 and PTE_28 structures was the engineered organophosphorous hydrolase (PDB entry: 1QW7).
All steps of atomic refinement were carried out with the CCP4/REFMAC5 program(Murshudov, Vagin, and Dodson 1997) and by Phenix refine (Afonine et al. 2012). The models were built into 2mFobs - DFcalc, and mFobs - DFcalc maps by using the COOT program(Emsley and Cowtan 2004). Details of the refinement statistics of the PTE_5, PTE_27 and PTE_28 structures are described in Table S4.
Supplementary Material
Acknowledgements
We thank Jonathan Weinstein for code development, Shira Warszawski for help in developing FuncLib, and S. W., Adi Goldenzweig, and Nir London for critical reading. The research was supported by a Starting Grant from the European Research Council (335439), the Israel Science Foundation through its Center of Excellence in Structural Cell Biology (1775/12), a research grant from Sheri and David E. Stone and the Yeda-Sela Center for Basic Research, and by a charitable donation from Sam Switzer and family (to S.J.F.), and by a Defense Threat Reduction Agency (DTRA) Grant (CB10265/HDTRA1724528) to D.S.T.
Footnotes
Authors contributions
O.K., R.L., Z.A., D.S.T., and S.J.F. designed the research; O.K., R.L., Z.A., and S.J.F. designed proteins; R.L. and S.J.F. developed design methods; O.K., Z.A., M.G., Y.A., H.L., D.L.T. performed biochemical experiments; O.D. and S.R. determined crystallographic structures; R.L. and J.P. designed the web-server; P.A.R. and V.G. performed nerve-agent docking simulations; O.K., D.S.T., and S.J.F. wrote the manuscript with contributions from all the authors; S.J.F. supervised the research.
Declaration of interests
The authors declare no competing interests.
References
- Afonine Pavel V, Grosse-Kunstleve Ralf W, Echols Nathaniel, Headd Jeffrey J, Moriarty Nigel W, Mustyakimov Marat, Terwilliger Thomas C, Urzhumtsev Alexandre, Zwart Peter H, Adams Paul D. Towards Automated Crystallographic Structure Refinement with Phenix.refine. Acta Crystallographica. Section D, Biological Crystallography. 2012;68(Pt 4):352–67. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afriat-Jurnou Livnat, Jackson Colin J, Tawfik Dan S. Reconstructing a Missing Link in the Evolution of a Recently Diverged Phosphotriesterase by Active-Site Loop Remodeling. Biochemistry. 2012;51(31):6047–55. doi: 10.1021/bi300694t. [DOI] [PubMed] [Google Scholar]
- Ashani Yacov, Goldsmith Moshe, Leader Haim, Silman Israel, Sussman Joel L, Tawfik Dan S. In Vitro Detoxification of Cyclosarin in Human Blood Pre-Incubated Ex Vivo with Recombinant Serum Paraoxonases. Toxicology Letters. 2011;206(1):24–28. doi: 10.1016/j.toxlet.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashani Y, Gupta RD, Goldsmith M, Silman I, Sussman JL, Tawfik DS, Leader H. Stereo-Specific Synthesis of Analogs of Nerve Agents and Their Utilization for Selection and Characterization of Paraoxonase (PON1) Catalytic Scavengers. Chemico-Biological Interactions. 2010;187(1–3):362–69. doi: 10.1016/j.cbi.2010.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst Christoffel PS, Bornscheuer Uwe T. Getting Momentum: From Biocatalysis to Advanced Synthetic Biology. Trends in Biochemical Sciences. 2018;43(3):180–98. doi: 10.1016/j.tibs.2018.01.003. [DOI] [PubMed] [Google Scholar]
- Baker David. An Exciting but Challenging Road Ahead for Computational Enzyme Design. Protein Science: A Publication of the Protein Society. 2010;19(10):1817–19. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HA, Leonard K. Chiral Reactions of Acetylcholinesterase Probed with Enantiomeric Methylphosphonothioates. Noncovalent Determinants of Enzyme Chirality. The Journal of Biological Chemistry. 1989;264(7):3942–50. [PubMed] [Google Scholar]
- Bershtein Shimon, Segal Michal, Bekerman Roy, Tokuriki Nobuhiko, Tawfik Dan S. Robustness-Epistasis Link Shapes the Fitness Landscape of a Randomly Drifting Protein. Nature. 2006;444(7121):929–32. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- Bigley Andrew N, Mabanglo Mark F, Harvey Steven P, Raushel Frank M. Variants of Phosphotriesterase for the Enhanced Detoxification of the Chemical Warfare Agent VR. Biochemistry. 2015;54(35):5502–12. doi: 10.1021/acs.biochem.5b00629. [DOI] [PubMed] [Google Scholar]
- Bigley Andrew N, Xu Chengfu, Henderson Terry J, Harvey Steven P, Raushel Frank M. Enzymatic Neutralization of the Chemical Warfare Agent VX: Evolution of Phosphotriesterase for Phosphorothiolate Hydrolysis. Journal of the American Chemical Society. 2013;135(28):10426–32. doi: 10.1021/ja402832z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg Rebecca, Kries Hajo, Pinkas Daniel M, Mittl Peer RE, Grütter Markus G, Privett Heidi K, Mayo Stephen L, Hilvert Donald. Precision Is Essential for Efficient Catalysis in an Evolved Kemp Eliminase. Nature. 2013;503(7476):418–21. doi: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- Bloom Jesse D, Arnold Frances H. In the Light of Directed Evolution: Pathways of Adaptive Protein Evolution. Proceedings of the National Academy of Sciences of the United States of America. 2009 Jun;106(Suppl 1):9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom Jesse D, Labthavikul Sy T, Otey Christopher R, Arnold Frances H. Protein Stability Promotes Evolvability. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(15):5869–74. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny Izhack, Greisen Per, Jr, Ashani Yacov, Khare Sagar D, Oberdorfer Gustav, Leader Haim, Baker David, Tawfik Dan S. Engineering V-Type Nerve Agents Detoxifying Enzymes Using Computationally Focused Libraries. ACS Chemical Biology. 2013;8(11):2394–2403. doi: 10.1021/cb4004892. [DOI] [PubMed] [Google Scholar]
- Damborsky Jiri, Brezovsky Jan. Computational Tools for Designing and Engineering Biocatalysts. Current Opinion in Chemical Biology. 2009;13(1):26–34. doi: 10.1016/j.cbpa.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Dellus-Gur Eynat, Elias Mikael, Caselli Emilia, Prati Fabio, Salverda Merijn LM, de Visser J Arjan GM, Fraser James S, Tawfik Dan S. Negative Epistasis and Evolvability in TEM-1 β-Lactamase--The Thin Line between an Enzyme’s Conformational Freedom and Disorder. Journal of Molecular Biology. 2015;427(14):2396–2409. doi: 10.1016/j.jmb.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley Paul, Cowtan Kevin. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallographica. Section D, Biological Crystallography. 2004;60(Pt 12 Pt 1):2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- van den Ent Fusinita, Löwe Jan. RF Cloning: A Restriction-Free Method for Inserting Target Genes into Plasmids. Journal of Biochemical and Biophysical Methods. 2006;67(1):67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Esvelt Kevin M, Carlson Jacob C, Liu David R. A System for the Continuous Directed Evolution of Biomolecules. Nature. 2011;472(7344):499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans Philip. Scaling and Assessment of Data Quality. Acta Crystallographica. Section D, Biological Crystallography. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Fleishman Sarel J, Leaver-Fay Andrew, Corn Jacob E, Strauch Eva-Maria, Khare Sagar D, Koga Nobuyasu, Ashworth Justin, et al. RosettaScripts: A Scripting Language Interface to the Rosetta Macromolecular Modeling Suite. PloS One. 2011;6(6):e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S, Wilson K. On the Treatment of Negative Intensity Observations. Acta Crystallographica. Section A, Foundations of Crystallography. 1978;34(4):517–25. [Google Scholar]
- Goldenzweig Adi, Fleishman Sarel. Principles of Protein Stability and Their Application in Computational Design. Annual Review of Biochemistry. 2018;87:105–129. doi: 10.1146/annurev-biochem-062917-012102. [DOI] [PubMed] [Google Scholar]
- Goldenzweig Adi, Goldsmith Moshe, Hill Shannon E, Gertman Or, Laurino Paola, Ashani Yacov, Dym Orly, et al. Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability. Molecular Cell. 2016;63(2):337–46. doi: 10.1016/j.molcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith Moshe, Aggarwal Nidhi, Ashani Yacov, Jubran Halim, Greisen Per, Jr, Ovchinnikov Sergey, Leader Haim, et al. Overcoming an Optimization Plateau in the Directed Evolution of Highly Efficient Nerve Agent Bioscavengers. Protein Engineering, Design & Selection: PEDS. 2017;30(4):333–45. doi: 10.1093/protein/gzx003. [DOI] [PubMed] [Google Scholar]
- Goldsmith Moshe, Tawfik Dan S. Enzyme Engineering: Reaching the Maximal Catalytic Efficiency Peak. Current Opinion in Structural Biology. 2017 Dec;47:140–50. doi: 10.1016/j.sbi.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Gupta Rinkoo D, Goldsmith Moshe, Ashani Yacov, Simo Yair, Mullokandov Gavriel, Bar Hagit, Ben-David Moshe, et al. Directed Evolution of Hydrolases for Prevention of G-Type Nerve Agent Intoxication. Nature Chemical Biology. 2011;7(2):120–25. doi: 10.1038/nchembio.510. [DOI] [PubMed] [Google Scholar]
- Hilvert Donald. Design of Protein Catalysts. Annual Review of Biochemistry. 2013;82:447–70. doi: 10.1146/annurev-biochem-072611-101825. [DOI] [PubMed] [Google Scholar]
- Holden HM, Benning MM, Raushel FM, Shim H. High resolution structure of the Zn-containing phosphotriesterase from Pseudomonas diminuta. Biochemistry. 2001;40(9):2712–2722. doi: 10.1021/bi002661e. [DOI] [PubMed] [Google Scholar]
- Jackson CJ, Foo J-L, Tokuriki N, Afriat L, Carr PD, Kim H-K, Schenk G, Tawfik DS, Ollis DL. Conformational Sampling, Catalysis, and Evolution of the Bacterial Phosphotriesterase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(51):21631–36. doi: 10.1073/pnas.0907548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach Miriam, Tokuriki Nobuhiko. Dynamics and Constraints of Enzyme Evolution. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution. 2014;322(7):468–87. doi: 10.1002/jez.b.22562. [DOI] [PubMed] [Google Scholar]
- Khare Sagar D, Kipnis Yakov, Greisen Per, Jr, Takeuchi Ryo, Ashani Yacov, Goldsmith Moshe, Song Yifan, et al. Computational Redesign of a Mononuclear Zinc Metalloenzyme for Organophosphate Hydrolysis. Nature Chemical Biology. 2012;8(3):294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky Olga, Kiss Gert, Röthlisberger Daniela, Dym Orly, Albeck Shira, Houk Kendall N, Baker David, Tawfik Dan S. Bridging the Gaps in Design Methodologies by Evolutionary Optimization of the Stability and Proficiency of Designed Kemp Eliminase KE59. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(26):10358–63. doi: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky Olga, Tawfik Dan S. Chromogenic and Fluorogenic Assays for the Lactonase Activity of Serum Paraoxonases. Chembiochem: A European Journal of Chemical Biology. 2006;7(1):49–53. doi: 10.1002/cbic.200500334. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M. Complete Set of ORF Clones of Escherichia Coli ASKA Library (A Complete S et of E. Coli K-12 ORF A Rchive): Unique Resources for Biological Research. DNA Research. 2005;12(5):191–199. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Lapidoth Gideon, Khersonsky Olga, Lipsh Rosalie, Dym Orly, Albeck Shira, Rogotner Shelly, Fleishman Sarel J. Highly Active Enzymes by Automated Combinatorial Backbone Assembly and Sequence Design. Nature Communications. 2018;9(1) doi: 10.1038/s41467-018-05205-5. 2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecina Daniel, Gilabert Joan F, Guallar Victor. Adaptive Simulations, towards Interactive Protein-Ligand Modeling. Scientific Reports. 2017;7(1) doi: 10.1038/s41598-017-08445-5. 8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy Airlie J. Solving Structures of Protein Complexes by Molecular Replacement with Phaser. Acta Crystallographica. Section D, Biological Crystallography. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miton Charlotte M, Tokuriki Nobuhiko. How Mutational Epistasis Impairs Predictability in Protein Evolution and Design. Protein Science: A Publication of the Protein Society. 2016;25(7):1260–72. doi: 10.1002/pro.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica. Section D, Biological Crystallography. 1997;53(Pt 3):240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- O’Meara Matthew J, Leaver-Fay Andrew, Tyka Michael D Mike, Stein Amelie, Houlihan Kevin, DiMaio Frank, Bradley Philip, et al. A Combined Covalent-Electrostatic Model of Hydrogen Bonding Improves Structure Prediction with Rosetta. Journal of Chemical Theory and Computation. 2015;11(2):609–22. doi: 10.1021/ct500864r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Hahnbeom, Bradley Philip, Greisen Per, Jr, Liu Yuan, Mulligan Vikram Khipple, Kim David E, Baker David, DiMaio Frank. Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. Journal of Chemical Theory and Computation. 2016;12(12):6201–12. doi: 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka Antonin, Chovancova Eva, Damborsky Jiri. HotSpot Wizard: A Web Server for Identification of Hot Spots in Protein Engineering. Nucleic Acids Research. 2009;37(Web Server issue):W376–83. doi: 10.1093/nar/gkp410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Yoav, Unger Tamar. Application of High-Throughput Methodologies to the Expression of Recombinant Proteins in E. Coli. Methods in Molecular Biology. 2008;426:197–208. doi: 10.1007/978-1-60327-058-8_12. [DOI] [PubMed] [Google Scholar]
- Peplow Mark. Nerve Agent Attack on Spy Used ‘Novichok’ Poison. Chemical & Engineering News. 2018 Mar 19;96(12) Chemical & Engineering News, 2018 March 19, 2018. [Google Scholar]
- Phillips Patrick C. Epistasis--the Essential Role of Gene Interactions in the Structure and Evolution of Genetic Systems. Nature Reviews. Genetics. 2008;9(11):855–67. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read Randy J, Sussman Joel L., editors. Evolving Methods for Macromolecular Crystallography. Springer; Netherlands, Dordrecht: 2007. [Google Scholar]
- Reetz MT, Wang LW, Bocola M. Directed Evolution of Enantioselective Enzymes: Iterative Cycles of CASTing for Probing Protein-sequence Space. Angewandte Chemie. 2006;45(8):1236–1241. doi: 10.1002/anie.200502746. [DOI] [PubMed] [Google Scholar]
- Reger Albert S, Carney Jill M, Gulick Andrew M. Biochemical and Crystallographic Analysis of Substrate Binding and Conformational Changes in Acetyl-CoA Synthetase. Biochemistry. 2007;46(22):6536–46. doi: 10.1021/bi6026506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodveldt C, Tawfik DS. Directed Evolution of Phosphotriesterase from Pseudomonas Diminuta for Heterologous Expression in Escherichia Coli Results in Stabilization of the Metal-Free State. Protein Engineering, Design & Selection: PEDS. 2005;18(1):51–58. doi: 10.1093/protein/gzi005. [DOI] [PubMed] [Google Scholar]
- Shoichet BK, Baase Wa, Kuroki R, Matthews BW. A Relationship between Protein Stability and Protein Function. Proceedings of the National Academy of Sciences. 1995;92(2):452–56. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr Tyler N, Thornton Joseph W. Epistasis in Protein Evolution. Protein Science: A Publication of the Protein Society. 2016;25(7):1204–18. doi: 10.1002/pro.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki Nobuhiko, Jackson Colin J, Afriat-Jurnou Livnat, Wyganowski Kirsten T, Tang Renmei, Tawfik Dan S. Diminishing Returns and Tradeoffs Constrain the Laboratory Optimization of an Enzyme. Nature Communications. 2012;3:1257. doi: 10.1038/ncomms2246. [DOI] [PubMed] [Google Scholar]
- Tracewell Cara A, Arnold Frances H. Directed Enzyme Evolution: Climbing Fitness Peaks One Amino Acid at a Time. Current Opinion in Chemical Biology. 2009;13(1):3–9. doi: 10.1016/j.cbpa.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J Arjan GM, Cooper Tim F, Elena Santiago F. The Causes of Epistasis. Proceedings Biological Sciences / The Royal Society. 2011;278(1725):3617–24. doi: 10.1098/rspb.2011.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Minasov G, Shoichet BK. Evolution of an Antibiotic Resistance Enzyme Constrained by Stability and Activity Trade-Offs. Journal of Molecular Biology. 2002;320(1):85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- Warshel A. Electrostatic Origin of the Catalytic Power of Enzymes and the Role of Preorganized Active Sites. The Journal of Biological Chemistry. 1998;273(42):27035–38. doi: 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- Weinreich Daniel M, Delaney Nigel F, Depristo Mark A, Hartl Daniel L. Darwinian Evolution Can Follow Only Very Few Mutational Paths to Fitter Proteins. Science. 2006;312(5770):111–14. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Weinreich Daniel M, Watson Richard A, Chao Lin. Perspective: Sign Epistasis and Genetic Constraint on Evolutionary Trajectories. Evolution; International Journal of Organic Evolution. 2005;59(6):1165–74. [PubMed] [Google Scholar]
- Wijma Hein J, Floor Robert J, Bjelic Sinisa, Marrink Siewert J, Baker David, Janssen Dick B. Enantioselective Enzymes by Computational Design and in Silico Screening. Angewandte Chemie. 2015;54(12):3726–30. doi: 10.1002/anie.201411415. [DOI] [PubMed] [Google Scholar]
- Wille Timo, Neumaier Katharina, Koller Marianne, Ehinger Christina, Aggarwal Nidhi, Ashani Yacov, Goldsmith Moshe, et al. Single Treatment of VX Poisoned Guinea Pigs with the Phosphotriesterase Mutant C23AL: Intraosseous versus Intravenous Injection. Toxicology Letters. 2016 Sep;258:198–206. doi: 10.1016/j.toxlet.2016.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.