Abstract
The expression of miRNAs in cancer has been widely studied and has allowed the definition of oncomirs and oncosuppressors. We note that it is often underestimated that many mRNAs are expressed, but translationally silent. In spite of this, systematic identification of miRNAs in equilibrium with their target mRNAs on polysomes has not been widely exploited. In order to identify biologically active oncomirs, we performed a screen for miRNAs acting on the polysomes of malignant mesothelioma (MPM) cells. Only a small percentage of expressed miRNAs physically associated with polysomes. On polysomes, we identified miRNAs already characterized in MPM, as well as novel ones like miR-24-3p, which acted as a promigratory miRNA in all cancer cells tested. miR-24-3p positively regulated Rho-GTP activity, and inhibition of miR-24-3p reduced growth in MPM cells. Analysis of miR-24-3p common targets, in two mesothelioma cell lines, identified a common subset of down-regulated genes. These same genes were down-regulated during the progression of multiple cancer types. Among the specific targets of miR-24-3p was cingulin, a tight junction protein that inhibits Rho-GTP activity. Overexpression of miR-24-3p only partially abrogated cingulin mRNA, but completely abrogated cingulin protein, confirming its action via translational repression. We suggest that miR-24-3p is an oncomir, and speculate that identification of polysome-associated miRNAs efficiently sorts out biologically active miRNAs from inactive ones.
Introduction
It is well established that miRNAs are powerful regulators of gene expression and induce translational repression (1,2). miRNA-mediated gene silencing, guided by sequence complementarity to their mRNA targets, consists of several steps: mRNA deadenylation, mRNA degradation and translation inhibition (3). The temporal sequence of these steps is not totally clear. Some evidences show that translational repression is an early common event, later followed by mRNA destabilization (1,3–6). Initiation of translation is a rate-limiting event, and most studies suggest that miRNAs repress translation at the initiation phase (3,7). Translation initiation is controlled by eukariotic Initiation Factors, eIFs. Two eIFs exert a predominant role on tumorigenesis, the eIF4F complex (7,8) and eIF6 (9,10). eIF4F complex is composed by the cap binding protein eIF4E, the RNA helicase eIF4A and the scaffold protein eIF4G. eIF4E recruitment in the eIF4F complex is inhibited by dephosphorylated 4E-binding proteins, 4E-BPs. mTORC1 phosphorylates 4E-BPs and relieves their inhibition (7,11,12). Importantly, the availability of eIF4E is required for efficient translational repression by miRNAs (7). As for eIF6, the evidence that it may affect miRNA repression of translation is dubious (13).
Translated mRNAs are in large molecular complexes known as polysomes. Translational repression of a specific mRNA is accompanied by its loss from translating ribosomes. It is evident that a miRNA that targets a specific coding mRNA that is recruited to polysomes for translation must transiently associate with polyribosomes. In spite of this, studies have not taken advantage from the mechanistic feature of miRNA dynamic association with translated mRNAs, in order to isolate functional miRNAs that counteract the translation of their targets. Here, we evaluated whether polysomal association of miRNAs could allow us to identify oncogenic miRNAs relevant for cancer progression.
MPM is an incurable cancer (14), characterized by clinical heterogeneity and variability at the transcriptional level with at least three subtypes that do not share extensive transcriptional signatures (15). This suggests that other mechanisms controlling gene expression, such as translational regulation, may be important in generating oncogenic properties. Previous works have shown that the translational machinery of MPM cells is deregulated, with aberrant initiation checkpoints (16,17). The mTOR pathway, inhibited by rapamycin, drives oncogenic signalling to the translational machinery, and MPM is largely insensitive to rapamycin inhibition (16). Activation of eIF4F complex by mTOR kinase is a signal for cell growth and transformation (18). Because studies on MPM suggests that eIF4E activity is constitutive (19) and its availability is required for miRNAs translation inhibition (7,8), it is likely that in MPM miRNAs hijack the translational machinery for the repression of specific targets.
Previous works on MPM cells have shown a remarkable heterogeneity in miRNAs expression. Different sample sources, variability of investigated histological types or heterogeneity of control groups led to divergent results (20). Alternatively, we hypothesize that miRNAs are oncogenic for their presence on the translational machinery, where they are in equilibrium with their targets, rather than for their simple expression or loss. By using an approach based on the definition of miRNA dynamically associated with polysomes, we discovered miR-24-3p, a new miRNA affecting MPM biology. Strikingly, manipulation of miR-24-3p levels in various cell types and conditions, led to a consistent biological effect on their migration properties. We suggest that miR-24-3p shows oncogenic properties in MPM, and hypothesize that our miRNA screen may unveil relevant connections in cancer studies.
Materials and methods
Mice
All experiments involving mice were performed in accordance with national and EU regulations. Experimental protocols were reviewed and approved by local Institutional Animal Care and Use Committees (IACUC no 688). Adult immunocompromised NOD-SCID male mice (Charles River Laboratories) were used for detecting tumor growth after intraperitoneal (i.p.) injection of REN cells manipulated for miR-24-3p expression level, as indicated. A cohort of these mice were used to identify the overall survival rate. The remaining experimental mice were sacrificed at 30 days post-injection to analyze tumor development and metastasis formation.
Cell lines
In this study, we used cell lines from human Malignant Pleural Mesothelioma: REN (21) and H28 (ATCC® CRL-5820™) for epithelioid subtypes, MM98 (22) for sarcomatoid subtypes and MSTO-211H (ATCC® CRL-2081™) for biphasic subtypes. MeT-5A (ATCC® CRL-9444™) are immortalized mesothelial cells used as non tumorigenic control. MCF-7 (ATCC® HTB22™) are metastatic breast cancer cells. 293T cells (ATCC® CRL-3216™) were used for in vitro luciferase assay. Each cell line was grown in DMEM (Lonza) except for MSTO-211H and MeT-5A cells which were grown in RPMI1640. All media were supplemented with 10% FBS and 1% penicillin, streptomycin, L-glutamine, and cells were maintained at 37°C and 5% CO2. Meso 3T, 7T, 8T, 9T, 12T, 13T, 14T and 17T are human primary mesothelioma cells established in the MRC Toxicology Unit, Leicester, UK. Primary cell lines were authenticated by short tandem repeat (STR) DNA profiling (23). All cell lines were tested by PCR for Mycoplasma detection and were not cultured for more than 2 months.
Antibodies and reagents
The following antibodies were used: rabbit polyclonal anti-cingulin (HPA027587, Sigma), mouse monoclonal antibodies against RhoA (240302, Cell Biolabs) and β-Actin (A5441, Sigma). For apoptosis analysis, staurosporine was bought from Sigma. For mRNA stability experiments, Actinomycin D was bought from ThermoFisher.
Lentiviral vectors
REN, MM98 and MSTO-211H cells were stably transduced either with a miR-Zip construct expressing a short hairpin inhibiting miR-24-3p or an empty vector, used as control. MeT-5A cells were permanently infected with a miR construct expressing a miR-24-3p mimic or a scramble vector, used as control. All lentiviral vectors used in this study were provided by System Biosciences.
mRNA extraction and quantitative RT–PCR
Total RNA was prepared using mirVana (Thermo-Fisher Scientific) and treated with RNase-free Dnase I (Promega). cDNA was generated using the SuperScript III First-Strand Synthesis kit (Thermo-Fisher Scientific). Real time PCR amplification was conducted in StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) using Sybr Green technique. The following primers were used to amplify selected genes: CGN (for: CCCGGCTAGGGACTCCTC rev: TGGACTCCATGGTCTACGGG), CXADR (for: GCTGTCGTAAAAAGCGCAGA rev: AGTGGACGTACGGCTCTTTG), BCAM (for: GAGACGCCAGCTTCCACTG rev: GTGGGATAGTGCAGGGTGAG). ACTIN (for: AGAGCTACGAGCTGCCTGAC rev: CGTGGATGCCACAGGACT) was used as endogenous control to normalize samples using the ΔΔCt method. For the analysis of microRNAs expression levels, cDNA was synthesized using the Universal cDNA Synthesis (Exiqon) and qRT-PCR was performed with miRCURY LNA™ Universal RT microRNA PCR (Exiqon) on an ABIPRISM 7900HT Sequence Detection System (Applied Biosystems). U6 snRNA was used as internal control to normalize samples by ΔΔCt method. miR-24-3p expression level in primary mesothelioma cells was established by TaqMan Assay (Thermo Fisher Scientific) following the standard procedure. miR-24-3p assay ID is 000402. The internal control U6B snRNA assay ID is 001093.
Polysomal profiles analysis and RNA purification
Growing cells were lysed in 50 mM Tris HCl pH 7.5, 100 mM NaCl, 30 mM MgCl2, 0.1% Nonidet P-40, 5 mM DTT, 100 μg/ml Cycloheximide and 30 U/ml RNasin, and cytoplasmic lysates were clarified at 15000 g for 15 min at 4°C. In order to distinguish miRNAs that sedimented with heavy polysomes, but were not physically bound to them, we disrupted polysomes by adding 30 mM EDTA after lysate preparation.
Equal amounts of RNA were loaded on 10-50% sucrose gradient dissolved in 25 mM Tris HCl pH 7.4, 25 mM NaCl, 5 mM MgCl2,1 mM DTT, with or w/o 10mM EDTA, and centrifuged at 39000 rpm for 3 h in a SW41Ti Beckman rotor at 4 °C. The gradient was then analyzed by continuous flow absorbance at 254 nm, recorded by BioLogic LP software (BioRad) and fractions were collected and kept on ice to avoid RNA degradation. The following fractions were collected: 1) subpolysomal pool, from the top of the gradient to the 80S peak; 2) light polysomes and 3) heavy polysomes. Samples were incubated with proteinase K and SDS 1% for 1 h at 37°C. RNA was extracted by phenol/chloroform/isoamyilic alcohol method. The same defined amounts of synthetic RNA spike-ins (osa-miR-414, ath-miR-159a, cel-miR-248) are added to all samples during preparation, to normalize RNA measurement.
microRNAs profiling
RNAs were processed with the NanoString nCounter Human v2 miRNA Expression Assay (nanoString, Seattle, Washington, USA) in the Genomics Shared Resource at The Ohio State University. The miRNA v2 panel can detect 699 endogenous miRNAs (with 654 probes), 5 housekeeping transcripts and 5 Spike-in specific for not mammalian small RNA transcript used as controls when processing RNAs. One hundred ng of total RNA were annealed with multiplexed DNA tags (miR-tag) and target specific bridges. Mature microRNAs were bound to specific miR-tags using a ligase enzyme and all the tags in excess were removed by enzymatic clean-up step. The tagged microRNAs product was diluted 1 to 5 and 5 μl were combined with 20 μl of reported probes in hybridization buffer and 5 μl of capture probes. After overnight hybridization (16 to 20 hours) at 65°C allowed to complex probes sequence specific with targets, probe excess was removed using two-step magnetic beads based purification on an automated fluidic handling system (nCounter Prep Station). Target/probe complexes were immobilized on the cartridge for data collection. The nCounter Digital Analyzer collected data by taking images of immobilized fluorescent reporters in the sample cartridge with a CCD camera through a microscope objective lens. For each cartridge, a high-density scan encompassing 600 fields of view was performed.
Cell proliferation, cell cycle and cell death analysis
Proliferation rate of MPM cells was analyzed by CellTiter-Blue assay (G8080, Promega). Briefly, 4x104 cells were plated al 100 μl/well in a 96-well plate, and assayed after 24, 48 and 72 hours. CTB was added and left on cells for 3 hours at 37° C and 5% CO2. The number of viable cells was measured recording fluorescence at 560Ex/590Em nm in a Victor3 multilabel counter (PerkinElmer, Germany). Cell cycle analysis was performed on 48 hours serum-starved MPM cells, restimulated with serum. At the indicated time points, cells were fixed, stained with propidium iodide (PI, P1304MP, ThermoFisher Scientific) and acquired on a BD FACS CANTO II flow cytometer (Becton Dickinson, Bioscences). Cell cycle analysis was performed using the FCS Express software. Cell death detection was performed using APC-Annexin V (BioLegend) and recorded on a BD FACS ARIA flow cytometer (Becton Dickinson, Bioscences).
Wound-healing assay
Cells were plated in a 12-well plate at 6x105 cells/well. Cells were grown to a confluency of 90% and then a thin wound was introduced by scratching with a sterile pipette tip. The wound closure was monitored on a Nikon Eclipse Timelapse every 4 h and analysis was performed using NIS-Elements Viewer.
RNA isolation and RNA sequencing
Total RNA from miR-24-3p knockdown REN and MM98 cells was isolated using mirVana Isolation Kit (ThermoFisher Scientific). RNA quality was controlled with BioAnalyzer (Agilent, Santa Clara, CA, USA). Random primed cDNA libraries were constructed from 100 ng of total RNA. A paired-end (2×125) run was performed on a Genome Sequencer Illumina HiSeq 2500 (GATC Biotech, Germany) with 50 million reads per sample.
RNA-seq data analysis
Read pre-processing and mapping
Three biological replicates were analyzed for REN and MM98 cell lines. Raw reads were checked for quality by FastQC software (version 0.11.2, S., A.FastQC: a quality control tool for high throughput sequence data. 2010; Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc), and filtered to remove low quality calls by Trimmomatic (version 0.32) using default parameters and specifying a minimum length of 50. Processed reads were then aligned to Homo sapiens genome assembly GRCm38 (Ensembl version 83) with STAR software (version 2.4.1c).
Gene expression quantification and differential expression analysis
The number of reads per gene was counted while mapping using the option --quantMode GeneCounts from the STAR software. A read is counted if it overlaps (1nt or more) one and only one gene. Both ends of the paired end read are checked for overlaps. The counts coincide with those produced by HTSeq-count algorithm with default parameters (gene annotation release 83 from Ensembl). To estimate differential expression, the matrix of gene counts produced by HTSeq was analyzed by DESeq2 (version DESeq2_1.12.4). The differential expression analysis by DeSeq2 algorithm was performed on the entire dataset composed by both REN and MM98 samples. The two following comparisons were analyzed: REN miR-Zip24 versus REN miR-empty and MM98 miR-Zip24 versus MM98 miR-empty. Reads counts were normalized by calculating a size factor, as implemented in DESeq2. Independent filtering procedure was then applied, setting the threshold to the 60 percentile; 24178 genes were therefore tested for differential expression. Significantly modulated genes in REN miR-Zip24 and MM98 miR-Zip24 conditions were selected by considering a false discovery rate lower than 10%. Regularized logarithmic (rlog) transformed values were used for heat map representation of gene expression profiles. Analyses were performed in R version 3.3.1 (2016-06-21, Computing, T.R.F.f.S. R: A Language and Environment for Statistical Computing. Available from: http://www.R-project.org/).
Functional analysis by topGO
The Gene Ontology enrichment analysis was performed using topGO R Bioconductor package (version topGO_2.24.0) as described in (24). The annFUN.db function is used to extract the gene-to-GO mappings from the genome wide annotation library org.Hs.eg.db for H. Sapiens.
RhoA activity assay
Pull-down assay to detect RhoA-GTP was performed using RhoA activation assay kit (Cell Biolabs). REN and MeT-5A cells were transduced with lentiviral vectors to inhibit or overexpress miR-24-3p, as before. Cells in growing conditions were lysed 24h after plating and GTP pulldown assay was performed for 1h at 4°C. After washing, samples were resuspended in SDS-PAGE sample buffer and loaded in a polyacrylamide gel.
Luciferase reporter assay
The plasmids containing wild-type (Cat# HmiT088611-MT06) or mutant (Cat# CS-HmiT088611-MT06) Luc-CGN 3’ UTR, wild-type (Cat# HmiT064331-MT06) or mutant (Cat# CS-HmiT064331-MT06) Luc-CXADR 3’ UTR and wild-type (Cat# HmiT010922-MT06-01) or mutant (Cat# CS-HmiT010922--MT06-01) Luc-BCAM 3’ UTR were synthesized (GeneCopoeia) and used in luciferase reporter assays. These plasmids contain firefly luciferase, fused to the 3’ UTR of analyzed genes and Renilla luciferase used as reporter gene. Luciferase activity assays were performed following manufacturer’s protocol. In details, 293T cells were seeded in 12-well plates and co-transfected with miRNA mimic for hsa-miR-24-3p (MC10737, Ambion) and either wild-type or 3’ UTR mutated genes. Cells were harvested after 48h and the firefly and Renilla luciferase activities were measured sequentially using the dual luciferase assay kit (GeneCopoeia). Luciferase activities were normalized with Renilla luciferase. Experiments were run in triplicate.
Statistical analysis
Each experiment was repeated at least three times, as biological replicates; means and standard deviations between different experiments were calculated. Statistical p-values obtained by two-tailed Student t-test were indicated: three asterisks *** for p-values less than 0.001, two asterisks ** for p-values less than 0.01 and one asterisk * for p-values less than 0.05.
Data Availability Statement
The RNA seq data are available at www.ebi.ac.uk/arrayexpress with accession number ID: E-MTAB-7079.
Results
High-throughput analysis of ribosomes-associated microRNAs
We hypothesized that subcellular miRNA localization is essential to mediate an oncogenic effect. Considering the relevance of translational control in cancer biology (8), we aimed at the specific isolation of miRNAs physically associated with the translational machinery. We used as a model system the malignant mesothelioma cell line REN, in which we previously observed dysregulated translation characterized by insensitivity to mTOR inhibition (16). In this cell line, eIF4E, necessary for translational repression by miRNAs at initiation (8), is at higher levels compared to its inhibitors 4E-BPs, explaining the insensitivity to mTOR inhibition. We expected that miRNAs that act by repressing translation were in continuous equilibrium with their target mRNAs and could co-sediment with polysomes. Growing REN cells had well defined polysomes (Supplementary Figure S1A). We used fractionation of polysomes by sucrose density gradient sedimentation, followed by high-throughput analysis of associated miRNAs (Figure 1A). We collected subpolysomal, light polysomal and heavy polysomal fractions into three separate pools (Supplementary Figure S1A). Subpolysomal fractions contain untranslated mRNAs and soluble miRNAs. Heavy polysomal fractions contain highly translated mRNAs; light polysomes may contain a mixture of the two, and we considered them as a cushion phase (25,26). Next, we performed a high-throughput analysis of microRNAs by the nanoString nCounter system (nanoString, Seattle, Washington, USA), and measured the expression levels of 699 microRNAs. Only 66 microRNAs were associated with the translation machinery of REN cells, indicating they are in equilibrium with their targets (Figure 1B). Due to the fact that the presence of miRNAs in the polysomal fraction could be due to interactions with high molecular weight structures, we refined the analysis on EDTA-treated polysomes. EDTA treatment disrupts polysomes, and leads to their dissociation in free 40S and 60S ribosomal subunits (Supplementary Figure S1B). Concomitant with polysomes disaggregation, few miRNAs were EDTA-insensitive, indicating that they were not associated with polysomes, whereas several miRNAs showed an expected shift from polyribosomes to monosomes. These last were further analyzed. To evaluate the preference of a miRNA for polysomal association, we calculated the miRNA polysome occupancy (25). We observe that the bulk of miRNAs are characterized by a medium occupancy, which shrinks after EDTA treatment (Supplementary Figure S1C). Finally, ratio of EDTA-treated/EDTA untreated miRNA polysome occupancy indicates that a restricted set of miRNAs dramatically changes its distribution, like miR-15a-5p, miR-16-5p, miR-24-3p, miR-191-5p and let-7g-5p (Figure 1C). Notably, three of these miRNAs were already linked to MPM through previous analysis, miR-15/16 (27,28) and miR-191-5p (29). These miRNAs represent bona fide highly active miRNAs that act in a dynamic equilibrium with polysomes and their target mRNAs.
Figure 1. Specific microRNAs are associated with translating polysomes.
A) Diagram of the high-throughput analysis of miRNA expression on the translational machinery of REN cells. We performed a sucrose density gradient sedimentation in both EDTA and EDTA-free conditions, followed by microRNA profiling. B) Heat map with microRNAs expressed along the polysomal profile of REN cells, in the absence of EDTA. C) miRNA polysome association following EDTA disruption: a limited set of miRNAs is very sensitive to EDTA condition (light blue), indicating that they associate dynamically with polysomes. Most miRNAs analyzed are less sensitive to EDTA (blue and dark blue), suggesting that they are not active on polysomes, but may rather cosediment with other high molecular weight structures.
To assess whether miRNA expression levels influenced miRNA polysome occupancy we performed a correlation analysis between miRNAs expression level and miRNAs occupancy (Supplementary Figure S1D). Importantly, miRNAs occupancy and expression were not associated (R2= 0.35182), indicating that the presence of miRNAs on the polyribosomes is due to other variables, such as the mRNA target level. If this last hypothesis was correct, we reasoned that in silico analysis of miRNA potential targets (http://mirsystem.cgm.ntu.edu.tw/) may lead to a common signature pathway. The most significant polysome enriched miRNAs (miR-15a-5p, miR-16-5p, miR-24-3p, miR-191-5p and let-7g-5p) were commonly predicted to regulate genes in the cell cycle and in the TCR pathway (Supplementary Figure S1E). We conclude that our screen identifies a minimal set of miRNAs that, by being in active equilibrium with polysomes, may dynamically control at the posttranscriptional level important biological functions in REN cells.
miR-24-3p is up-regulated in Malignant Pleural Mesothelioma and is enriched on polyribosomes of REN cells
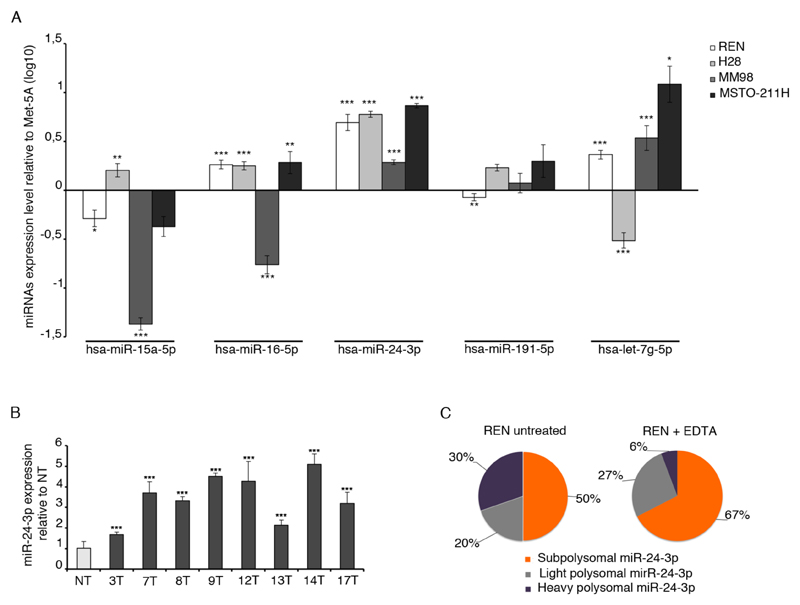
Estabilished that only a restricted number of miRNAs is associated with the polysomal machinery, we evaluated miRNAs expression in multiple MPM samples. We performed qRT-PCR on MPM cell lines, representative of the different MPM histological subtypes: REN and H28 for epithelial type, MM98 for sarcomatoid type and MSTO-211H for biphasic one. Each analyzed miRNA, except for miR-24-3p, displayed a variable expression. miR-24-3p was found up-regulated in all MPM cell lines analyzed, compared to MeT-5A control cells (Figure 2A). miR-24-3p was the only one not previously identified in MPM tumors by other approaches and was further analyzed. Patient derived primary mesothelioma cells well represent the native state of the tumor and reflect the disease heterogeneity observed in patients (23). We used as control, a primary untransformed human mesothelium. We found that miR-24-3p is expressed above the control in all primary mesothelioma cells analyzed (Figure 2B), as well in other tumor cell lines (Supplementary Figure S2). By qRT-PCR, we validated the association of miR-24-3p on polyribosomes of cells. We confirmed that EDTA causes a 5-fold loss of miR-24-3p expression from highly translating ribosomes, from 30% to 6%, and an increase of its expression in monosomes (Figure 2C). In conclusion, we show the up-regulation of miR-24-3p in MPM and its association with actively translated mRNAs, suggesting that a polysome-enrichment analytical approach efficiently identifies biologically active miRNAs that may have escaped previous works.
Figure 2. miR-24-3p is a marker of MPM cells and is associated with actively translating polysomes.
A) qRT-PCR of selected microRNAs found associated with polyribosomes. Analysis performed on MPM cell lines representative of MPM hystological types. miR-24-3p is upregulated in all cell lines, compared to the MeT-5A cells used as control. B) qRT-PCR on primary patient-derived mesothelioma cells shows that miR-24-3p is upregulated compared to primary non tumoral cells (NT). C) miR-24-3p distribution on polysomes (violet) detected in the absence of EDTA (left) or upon EDTA (right) shows a drop from 30% to 6%. The reduction of heavy polysomal area (violet) after EDTA treatment indicates that miR-24-3p is associated with actively translating ribosomes. Histograms represent the mean ± SD of three independent experiments. p-values, are indicated (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001).
Knockdown of miR-24-3p reduces migration in MPM and breast cancer cells
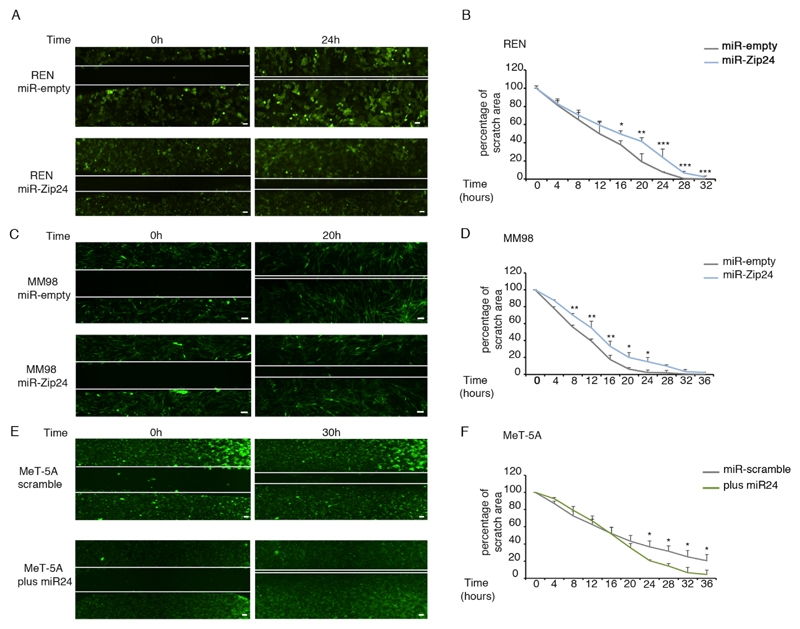
To investigate the biological role of miR-24-3p in MPM, we stably knocked down miR-24-3p in three MPM cell lines, representative of the main histopathological subtypes (REN cells for the epithelioid subtypes, MM98 cells for the sarcomatoid and MSTO-211H cells for mixed type). All cell lines were sorted for GFP co-reporter, and miR-24-3p knockdown was confirmed by quantitative RT-PCR. The efficiency of inhibition was around 70% in all cell lines analyzed (Supplementary Figure S3A). Next, we analyzed MPM cell growth at 72 hours after plating and upon miR-24-3p depletion. The reduction of miR-24-3p slightly reduces cell number relative to control (supplementary Figure S3B), does not affect the apoptotic rate (Supplementary Figure S3C), and causes a mild reduction of cells in S phase (Supplementary Figure S3D). Next, we investigated migratory properties of MPM cells by performing wound-healing assays. All MPM cell lines depleted of miR-24-3p had a lowered migratory capability, as compared to controls (Figure 3 A-D; Supplementary Figure S4A). To further challenge the pro-migratory role of miR-24-3p, we used the mesothelial control cell line MeT-5A that expresses little amounts of miR-24-3p. We transduced MeT-5A cells with a Lenti-miR vector containing miR-24-3p, obtaining 3.5-fold expression, compared to control (Supplementary Figure S4B). Scratch assay analysis showed that MeT-5A cells transduced with miR-24-3p acquired higher migratory capability (Figure 3E-F). Next, we investigated the migratory role of miR-24-3p in MCF-7 cells, a metastatic breast cancer cell line, analyzing both miR-24-3p knockdown and overexpression. We found that in MCF-7 cells the migration capability is positively correlated with miR-24-3p levels (Supplementary Figure S4C-D). In conclusion, our analysis demonstrates that a miRNA isolated by its association with polysomes has a consistent biological effect in all tested cells.
Figure 3. miR-24-3p enhances the migratory properties of all MPM cell lines.
A-D) Representative images of wound-healing assay on miR-24-3p knockdown REN (A) and MM98 cells (C) show that depletion of miR-24-3p reduces the speed of wound healing. The rate of wound healing was analyzed within indicated times after the scratch in REN (B) and MM98 cells plates (D). E-F) Representative images of wound-healing assay on MeT-5A cells overexpressing miR-24-3p show that miR-24-3p enhances the rapidity of wound healing (E). Scratch area was measured at indicated time after the cut (F). Experiments are performed as biological replicates. Data are expressed as percentage of control. p-values, are indicated (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001). Scale bar corresponds to 10 μm.
miR-24-3p down-regulation in vivo reduces migration and is associated with modulation of RhoA activity
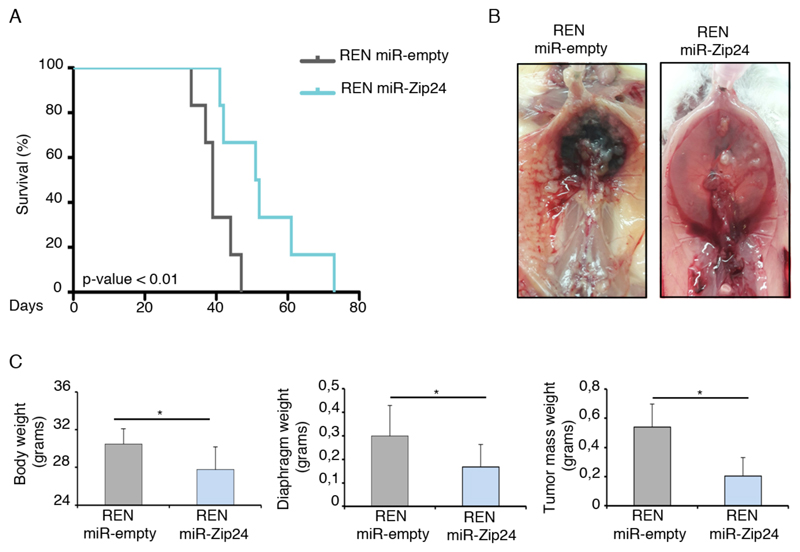
The effect of miR-24-3p on MPM cells proliferation and migration led us to investigate the role of miR-24-3p on tumor growth, in vivo. We injected i.p. REN miR-Zip24 and REN miR-empty control cells into immunocompromised NOD-SCID mice and evaluated overall survival, tumor volumes and metastasis formation. As reported in Kaplan-Meier curve, a negative correlation between miR-24-3p expression and mice overall survival was found (Figure 4A). A subset of mice were sacrificed at 30 days post-injection and analysed. At autopsy, we observed a consistent reduction of metastasis formation and intraperitoneal hemorrhage in mice injected with REN cells depleted of miR-24-3p (Figure 4B). We measured total body, tumor mass and diaphragm weight and found that knockdown of miR-24-3p causes a significant reduction in tumor volume in vivo (Figure 4C and Supplementary Table S1). These results confirm that miR-24-3p may function as a proncogenic miRNA also in vivo.
Figure 4. miR-24-3p has a pro-tumorigenic role in vivo, in MPM.
REN transduced with miR-empty and miR-Zip24 vectors were injected i.p. in NOD-SCID mice. Experimental groups: mice with miR-empty n=12; mice with miR-Zip24 n=12. A) Mice overall survival. KM indicates that knockdown of miR-24-3p increases mice survival. B) Representative images of control and knockdown mice: depletion of miR-24-3p causes a reduction of metastasis and less bleeding. C) miR-24-3p knockdown significantly reduces body, diaphragm and tumor mass weight. Data are represented as mean ± SD. Statistical p-values are indicated (*p ≤ 0.05).
Several properties of malignant progression and migration have been attributed to aberrant signaling through RHO-GTPase proteins. The Rho-GTPases play crucial role in cell migration, tumor growth and metastasis (30). We investigated RhoA activity in cells depleted for miR-24-3p or overexpressing it. RhoA-GTP pulldown assays showed that miR-24-3p knockdown causes a reduction of RhoA activity, in REN cells (Figure 5A), whereas miR-24-3p overexpression in MeT-5A cells caused an increment of RhoA activity (Figure 5B). We noted that the modulation of RhoA activity is independent from RhoA mRNA and protein levels (Figure 5C-D). These data suggest that miR-24-3p indirectly regulates RhoA activity in MPM.
Figure 5. miR-24-3p increases RhoA activity in MPM cells.
A) Representative WB analysis of GTP pulldown in REN miR-empty and miR-Zip-24 cells shows that miR-24-3p partial depletion reduces RhoA activity, related to control. B) Conversely, RhoA pulldown of miR-24-3p overexpressing Met-5A cells indicates that miR-24-3p enhances RhoA activity, related to control. C) qRT-PCR and representative WB analysis on miR-24-3p knockdown REN cells: RhoA mRNA and protein remain unchanged in the analyzed conditions. D) qRT-PCR and representative WB analysis on miR-24-3p overexpressing MeT-5A cells indicates that RhoA mRNA and protein remain similar at the indicated conditions. Experiments were performed as biological triplicates.
miR-24-3p knockdown causes a widespread multitarget suppression consistent with its biological effects and acting at the translation level
Next, we searched for miR-24-3p direct targets that explain the pro-migratory phenotype. We aimed at defining in an unbiased way genes regulated at the translation level by miR-24-3p. Technical issues complicate a direct analysis of translation targets. We exploited the well-known observation that miRNA translational repression of mRNA targets is followed by their degradation (4,28). We reasoned that if translational control is involved in a dynamic fashion (equilibrium between a miRNA on polysomes and continuous transcription of the targeted mRNA), we may unveil situations in which a reduction in the target mRNA is present, but weaker than the reduction in the encoded protein. Thus, we performed RNAseq analysis of REN and MM98 in which miR-24-3p was knocked down. We observed that the genes up and downregulated in the two cell lines analyzed were highly dissimilar at the steady-state mRNA level. Neverthless, most miR-24-3p modulated genes were protein-coding genes (Supplementary Figure S5A).
To bring order in this scenario we clustered, by Gene Ontology analysis, differentially expressed genes in functional groups and we observed that miR-24-3p modulates several mRNAs involved in cell adhesion and migration in REN and MM98 cells (Figure 6A). Thus, in spite of an apparent heterogeneous signature, the biological landscape evoked by miR-24-3p is similar, with a contribution of targets involved in migration. Next, we overlapped all genes upregulated by mir-24-3p depletion in REN and MM98 cells and found that the two MPM cell lines share 194 genes (Supplementary Table S2) which we arbitrarily considered biologically relevant for explaining the consistent biological results obtained by miR-24-3p manipulation in multiple cell lines. To further restrict targets, we performed a miRNA target prediction analysis using 5 different miRNA prediction software (PITA, MiRWalk, RNAHybrid, Targtescan, miRanda) and we matched the obtained list with the 194 common genes derived from RNA-seq analysis. Thus, we shortlisted 33 genes that are bona fide direct targets of miR-24-3p (Supplementary Figure S5B). We then asked whether these genes are involved in cancer progression and are shared among different tumor types. We interrogated PRECOG (PREdiction of Clinical Outcomes from Genomic profiles) system to analyze the association between the 33 miR-24-3p regulated genes and cancer outcomes. Many genes identified as miR-24-3p direct targets correlate with favorable survival across different cancer types (Supplementary Figure S5C).
Figure 6. miR-24-3p is able to repress multiple target mRNAs and translationally regulates cingulin.
A) RNA-seq analysis of miR-24-3p depleted REN and MM98 cells: GO analysis identifies migration and adhesion genes reprogramming in both cell lines. The scale represents statistical significance obtained by using the Fisher’s exact test. B-D) Left: predicted miR-24-3p binding sites within 3’ UTR of CGN (B), BCAM (C) and CXADR mRNAs (D). miR-24-3p seed region is showed in bold. Mutations to disrupt miRNA-mRNA binding were performed within the seed region; for each gene the mutated binding site is evidenced in grey. Right: validation of miR-24-3p binding sites within 3’ UTR of interested genes. Luciferase activity measurement shows that the presence of miR-24-3p binding sites within 3’UTR of CGN (B), BCAM (C) and CXADR (D) impairs expression of luciferase mRNA in miR-24-3p expressing cells. The effect was abolished considering a mutant form of 3 ‘UTR or lower levels of miR-24-3p expression. E) qRT-PCR analysis of cingulin mRNA validates CGN gene upregulation after miR-24-3p inhibition both in REN, MM98 and MSTO-211H cells. F) WB analysis shows that inhibition of miR-24-3p results in an increase of cingulin protein levels in REN, MM98 and MSTO-211H. G) qRT-PCR on MeT-5A cells transduced with miR-24-3p overexpressing vector underlines that CGN mRNA is partially reduced related to scramble control. H) In the same conditions cingulin protein is completely absent. Values are represented as mean ± SD of three independent experiments. All experiments are performed as biological replicates. p-values are indicated (*p ≤ 0.05; ***p ≤ 0.001).
Genes involved in cell migration are translationally regulated by miR-24-3p
Our RNAseq data indicate that miR-24-3p hits relevant biological pathways in cancer, including cell motility and migration. In order to validate NGS results and identify miR-24-3p direct target mRNAs, we analyzed genes with specific involvement in cell migration and tight junctions structures. We scanned the 3’ UTRs of three selected genes, CGN, BCAM and CXADR, for potential miR-24-3p binding sites, and identified in their 3’ UTR sequence elements complementary to the seed sequence of human miR-24-3p (Figure 6B-D left). We hypothesized that miR-24-3p binding to CGN, BCAM and CXADR mRNAs may inhibit their expression. In support of this idea, miR-24-3p expression in 293T cells is able to specifically decrease the activity of a luciferase reporter gene fused to the wild-type 3’ UTR of CGN, whereas the presence of a mutant form of CGN 3’ UTR has no effect on the luciferase activity (Figure 6B right). We conclude that the effect of miR-24-3p on the luciferase mRNA translation is dependent on the presence of miR-24-3p binding site within the 3’ UTR of the gene. Similar results were obtained for BCAM and CXADR genes (Figure 6C-D).
Next, we validated the changes of cingulin, BCAM and CXADR mRNAs in REN, MM98 and MSTO-211H cells, after miR-24-3p partial depletion (Fig. 6E). In summary, we observed an increment of the expression levels of these gene transcripts in miR-24-3p knockdown MPM cell lines (Figure 6E; Supplementary Figure S5D). We tested in parallel the protein levels of cingulin, BCAM and CXADR. Unfortunately, commercial antibodies against BCAM and CXADR did not perform well and were discarded. Data are therefore shown only for cingulin target. As expected, we found that miR-24-3p reduction induces an increase in cingulin protein levels in REN, MM98 and MSTO-211H (Figure 6F). Next, we explored cingulin, BCAM and CXADR mRNAs and cingulin protein levels in miR-24-3p overexpressing MeT-5A cells and found a partial reduction of target mRNAs expression (Figure 6G; Supplementary Figure S5E) accompanied by a total loss of cingulin protein (Figure 6H). The total loss of cingulin protein in miR-24-3p overexpression conditions (Figure 6H), combined with a partial reduction of cingulin mRNA transcript levels, suggests that, as hypothesized, miR-24-3p is able to regulate cingulin expression first at the translation level, and subsequently at the mRNA stability level (Supplementary Figure S5F).
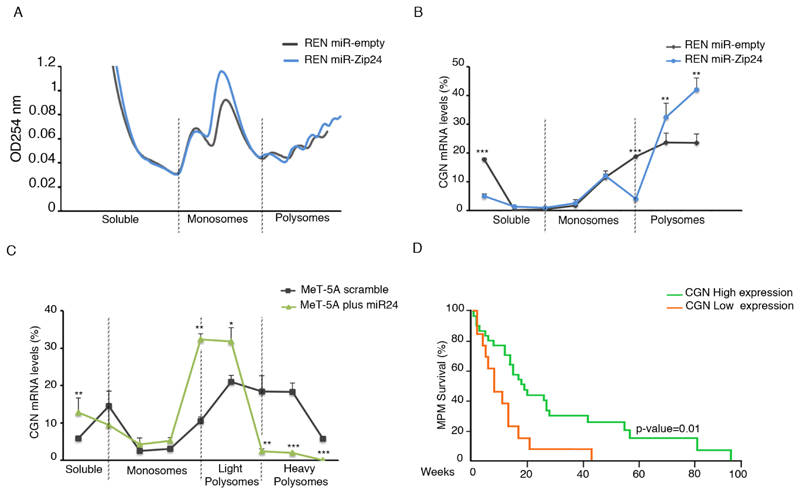
Next, we addressed the relative contributions of translation and mRNA stability. We first evaluated the association of cingulin mRNA with polysomes, when miR-24-3p is expressed at either low or high levels. REN cells with normal or reduced miR-24-3p expression levels were characterized by similar polysomal profiles, indicating that miR-24-3p has no effect on global translation (Figure 7A). We analyzed cingulin mRNA distribution along the polysomal profile of normal or miR-24-3p knockdown REN cells and found that reduced levels of miR-24-3p cause an accumulation of cingulin mRNA in highly translating polysomes (Figure 7B). The effect was consistently accompanied by a reduction of cingulin mRNA in the soluble fraction (Figure 7B). Identical results were obtained for BCAM and CXADR mRNAs (Supplementary Figure 6A-B). Next, we investigated the distribution of cingulin mRNA in MeT-5A cells that expressed miR-24-3p, compared to control. Strikingly, when miR-24-3p is overexpressed, the residual transcript moves from heavy polysomes to light ribosomes, confirming that its translation is inhibited (Figure 7C). We then evaluated whether the loss of polysomal association of cingulin mRNA with polysomes in REN, MM98 and MSTO-211H cells, results also in its destabilization. Data show that cingulin mRNA has a shorter half-life in cells with increased miR-24-3p. In summary, our data demonstrate that miR-24-3p strongly acts at the translational level by its association with target mRNAs on polysomes, followed by mRNA destabilization.
Figure 7. Cingulin is regulated translationally by miR-24-3p.
A) Polysomal profile analysis of REN miR-empty and miR-Zip24 cells suggests that global translation is similar. Soluble, monosomes and polysomal fractions are indicated. B) Distribution of cingulin mRNA along the translation machinery evidences that cingulin mRNA is more abundant on polysomes of REN cells with depleted miR-24-3p. C) Conversely, when miR-24-3p is overexpressed, the distribution of cingulin mRNA on heavy polysomes is lost and cingulin mRNA shifts to poorly translating ribosomes (light polysomes). D) SynTarget tool analysis shows that CGN expression is positively correlated with survival of MPM patients (high expression of CGN n=31; low expression of CGN n=13). Histograms represent the mean ± SD of three independent experiments. All experiments are performed as biological replicates. p-values are indicated (*p ≤ 0.05; ***p ≤ 0.001).
Finally, the striking regulation of cingulin driven by miR-24-3p led us to address whether cingulin expression is related to the survival of MPM patients. Analysis of CGN expression in MPM tissues, according to the SynTarget tool (27,29), predicts that high levels of cingulin are a predictor for survival (Figure 7D).
Discussion
Several works have identified miRNAs involved in cancer biology by their differential expression in tumor versus control, and correlation between miRNA expression and prognosis. In some cases, miRNAs have been identified for their cellular localization, as in exosomes. One prominent effect of miRNAs is their effect on translational repression. Starting from this observation, we reasoned that miRNAs that dynamically modify gene expression counteract the translation of continuosly transcribed genes and associate with the polysomal machinery. In this work, we suggest a screening approach for the identification of biologically active miRNAs in cancer through a rapid analysis of polysome-associated miRNAs. In this study, we confirm miRNAs previously associated with MPM. In addition, we identified miR-24-3p, a polysome-associated miRNA, which is expressed in MPM and in other solid tumors. Strikingly, our data establish that miR-24-3p expression consistently affects similar cellular properties in multiple cell lines. We conclude that our approach is suitable for the identification of biologically relevant miRNAs.
The observed localization of miRNAs with actively translating ribosomes has not been systematically exploited for the identification of active miRNAs. It is often not fully appreciated that many mRNAs are transcribed, but not translated, and that translational control exerts a fundamental role in gene expression (8). Indeed, initiation factors like eIF4E or eIF6 are rate limiting in translation (31), can regulate the translation of specific mRNAs (9) and control tumor progression (17). In summary, a miRNA in order to modulate gene expression at the protein level must target a mRNA that is continuously transcribed, and brought to ribosomes for translation. miRNAs in D. melanogaster and in C. elegans sediment with polysomes (32,33), where miRNA-mRNA target interactions occurs (34). We have found that the association of a miRNA with polysomes is independent from miRNA abundance, in line with a model in which the affinity and abundance of target mRNA sequences regulate miRNA association with ribosomes (25). Our experiments identified, beside miR-24-3p, let-7 (35) and miR-191-5p (36), which are well established in connection to cancer. Furthermore, we observed that miR-15 and miR-16 cosediment with polysomes. TargomiR targeting miR-16 (37) are in phase I clinical trial, in MPM patients (clinicaltrials.gov NCT02369198). It is therefore evident that the identification of miRNAs bound to ribosomes is predictive of physiological relevance.
We decided to characterize miR-24-3p, among the miRNAs associated with the polysomal machinery for several reasons. First, it was not previously identified in MPM studies; second, it was expressed in both sarcomatoid and epithelial tumors; third, its putative function was puzzling. According to the literature, miR-24-3p has been described either as a pro-oncogenic (38–40) or as a tumor suppressor (41,42). The two-faced role of a miRNA is not uncommon in miRNA biology. Whether the two-faced role reflects a biological role or the product of different testing conditions is often not clear. Many models claim a miRNA context-dependent function, stressing the fact that evolutionary pressure acts on the 3’ UTR of the target gene, rather than to the miRNA itself. Alternatively, methodological approaches may often be the cause. Most studies, for instance, rely on the overexpression of miRNA in pathological conditions, without addressing its localization. By our approach, we first pinpointed the potential biological relevance of miR-24-3p by its association with polysomes, independently from its expression levels. Next, we confirmed that miR-24-3p acts as a promigratory miRNA in multiple cell types. In the past, reports of a tumor suppressor role for miR-24-3p relied on the use of a mimic miRNA in cells normally devoid of it, without parallel down-regulation experiments (43). We conclude that in physiological conditions, miR-24-3p positively regulates migratory properties, rather than suppressing cancer. Supporting the conclusion of miR-24-3p acting as an oncogenic facilitator, PRECOG analysis showed that the expression of the vast majority of the common targets of miR-24-3p in MPM, is down-regulated in the progression of most tumors. Among genes identified, ZNF185 may function as tumor suppressor gene (44) whilst SALL3 governs methyltransferase activity in cancer and its silencing predicts poor survival (45). One drawback of our approach is that it is not fully suitable for antiassociation studies between target mRNAs and microRNAs. As we have shown in Figure 7, cingulin protein is annihilated by miR-24-3p, but its mRNA is not. This can be mechanistically explained by the fact that ongoing transcription may compensate for the loss of mRNA, but as soon as the mRNA reaches polysomes, its translation is first blocked and then it is degraded.
Mechanistically, the activity of RhoA was significantly reduced in a miR-24-3p dependent manner. RhoA signaling pathways normally correlate with the ability of tumor cells to invade and establish metastases (46). Interfering with the Rho-kinase pathway may inhibit migration and invasion of NSCLC (47). Hyperactivation of the Rho pathway is generally associated with more aggressive tumour properties (30) and RhoA expression is associated with a significant reduction of survival of MPM patients (48). miR-24-3p controls RhoA activity through at least one target, cingulin. Indeed, it has been reported that the binding of GEF-H1 to the tight junction-associated adaptor cingulin results in inhibition of Rho signaling (49), consistently with our data. Besides cingulin, discussed above, also CXADR, a transmembrane protein involved in TJ structure and function, is regulated by miR-24-3p and is essential for TJ integrity. Unfortunately, we could not validate more targets due to the lack of good antibodies.
Even if miR-24-3p affects migration in all tested cells, including the MCF-7 metastatic breast cancer cells, its primary identification in mesothelioma is intriguing. In MPM most patients succumb to the effect of the primary tumor mass rather than to metastatic burden. At the time of diagnosis most patients are described as late staging, presenting a highly infiltrative disease. However, the landscape of cancer diagnosis and treatment confirm the presence of unusual metastases for MPM patients (50).
In summary, with our study we propose an original approach for identifying novel miRNAs with relevant function in cancer biology and we suggest that miR-24-3p expression is relevant for cancer progression and metastasization.
Supplementary Material
Acknowledgements
This work was supported by Gruppo Italiano Mesothelioma (GIMe), AIRC IG 2014 number 19793 and European Research Council (ERC) Translate number 338999 to SB. We thank Dr. Raphael Bueno and his collaborator David Severson for helpful insights. We also thank Riccardo Rossi for bioinformatic support.
Footnotes
Conflict-of-interest statement:
Authors declare no conflict of interests for this article.
References
- 1.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 2.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 3.Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World journal of biological chemistry. 2017;8:45–56. doi: 10.4331/wjbc.v8.i1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–7. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–33. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 6.Nishimura T, Fabian MR. Scanning for a unified model for translational repression by microRNAs. Embo J. 2016;35:1158–9. doi: 10.15252/embj.201694324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–7. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 8.Loreni F, Mancino M, Biffo S. Translation factors and ribosomal proteins control tumor onset and progression: how? Oncogene. 2014;33:2145–56. doi: 10.1038/onc.2013.153. [DOI] [PubMed] [Google Scholar]
- 9.Brina D, Miluzio A, Ricciardi S, Biffo S. eIF6 anti-association activity is required for ribosome biogenesis, translational control and tumor progression. Biochim Biophys Acta. 2015;1849:830–5. doi: 10.1016/j.bbagrm.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Miluzio A, Beugnet A, Grosso S, Brina D, Mancino M, Campaner S, et al. Impairment of cytoplasmic eIF6 activity restricts lymphomagenesis and tumor progression without affecting normal growth. Cancer Cell. 2011;19:765–75. doi: 10.1016/j.ccr.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Chapat C, Jafarnejad SM, Matta-Camacho E, Hesketh GG, Gelbart IA, Attig J, et al. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc Natl Acad Sci U S A. 2017;114:5425–30. doi: 10.1073/pnas.1701488114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–5. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 13.Galicia-Vazquez G, Chu J, Pelletier J. eIF4AII is dispensable for miRNA-mediated gene silencing. Rna. 2015;21:1826–33. doi: 10.1261/rna.052225.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbone M, Ly BH, Dodson RF, Pagano I, Morris PT, Dogan UA, et al. Malignant mesothelioma: facts, myths, and hypotheses. J Cell Physiol. 2012;227:44–58. doi: 10.1002/jcp.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48:407–16. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 16.Grosso S, Pesce E, Brina D, Beugnet A, Loreni F, Biffo S. Sensitivity of global translation to mTOR inhibition in REN cells depends on the equilibrium between eIF4E and 4E-BP1. PLoS One. 2011;6:e29136. doi: 10.1371/journal.pone.0029136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miluzio A, Oliveto S, Pesce E, Mutti L, Murer B, Grosso S, et al. Expression and activity of eIF6 trigger malignant pleural mesothelioma growth in vivo. Oncotarget. 2015;6:37471–85. doi: 10.18632/oncotarget.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–61. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson BA, Thumma SC, Jay-Dixon J, Patel MR, Dubear Kroening K, Kratzke MG, et al. Targeting eukaryotic translation in mesothelioma cells with an eIF4E-specific antisense oligonucleotide. PLoS One. 2013;8:e81669. doi: 10.1371/journal.pone.0081669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truini A, Coco S, Alama A, Genova C, Sini C, Dal Bello MG, et al. Role of microRNAs in malignant mesothelioma. Cell Mol Life Sci. 2014;71:2865–78. doi: 10.1007/s00018-014-1584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smythe WR, Kaiser LR, Hwang HC, Amin KM, Pilewski JM, Eck SJ, et al. Successful adenovirus-mediated gene transfer in an in vivo model of human malignant mesothelioma. The Annals of thoracic surgery. 1994;57:1395–401. doi: 10.1016/0003-4975(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 22.Orecchia S, Schillaci F, Salvio M, Libener R, Betta PG. Aberrant E-cadherin and gamma-catenin expression in malignant mesothelioma and its diagnostic and biological relevance. Lung Cancer. 2004;45(Suppl 1):S37–43. doi: 10.1016/j.lungcan.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 23.Chernova T, Sun XM, Powley IR, Galavotti S, Grosso S, Murphy FA, et al. Molecular profiling reveals primary mesothelioma cell lines recapitulate human disease. Cell Death Differ. 2016;23:1152–64. doi: 10.1038/cdd.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calamita P, Miluzio A, Russo A, Pesce E, Ricciardi S, Khanim F, et al. SBDS-Deficient Cells Have an Altered Homeostatic Equilibrium due to Translational Inefficiency Which Explains their Reduced Fitness and Provides a Logical Framework for Intervention. PLoS genetics. 2017;13:e1006552. doi: 10.1371/journal.pgen.1006552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molotski N, Soen Y. Differential association of microRNAs with polysomes reflects distinct strengths of interactions with their mRNA targets. Rna. 2012;18:1612–23. doi: 10.1261/rna.033142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nature chemical biology. 2010;6:209–17. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amelio I, Tsvetkov PO, Knight RA, Lisitsa A, Melino G, Antonov AV. SynTarget: an online tool to test the synergetic effect of genes on survival outcome in cancer. Cell Death Differ. 2016;23:912. doi: 10.1038/cdd.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–40. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonov AV. BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Res. 2011;39:W323–7. doi: 10.1093/nar/gkr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridley AJ. Rho GTPase signalling in cell migration. Current opinion in cell biology. 2015;36:103–12. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, et al. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature. 2008;455:684–8. doi: 10.1038/nature07267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, et al. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–4. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 33.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 34.Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. Rna. 2004;10:387–94. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A. 2007;104:11400–5. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma S, Rajendran V, Kulshreshtha R, Ghosh PC. Enhanced efficacy of anti-miR-191 delivery through stearylamine liposome formulation for the treatment of breast cancer cells. International journal of pharmaceutics. 2017;530:387–400. doi: 10.1016/j.ijpharm.2017.07.079. [DOI] [PubMed] [Google Scholar]
- 37.Reid G, Pel ME, Kirschner MB, Cheng YY, Mugridge N, Weiss J, et al. Restoring expression of miR-16: a novel approach to therapy for malignant pleural mesothelioma. Ann Oncol. 2013;24:3128–35. doi: 10.1093/annonc/mdt412. [DOI] [PubMed] [Google Scholar]
- 38.Du WW, Fang L, Li M, Yang X, Liang Y, Peng C, et al. MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. Journal of cell science. 2013;126:1440–53. doi: 10.1242/jcs.118299. [DOI] [PubMed] [Google Scholar]
- 39.Liu R, Zhang H, Wang X, Zhou L, Li H, Deng T, et al. The miR-24-Bim pathway promotes tumor growth and angiogenesis in pancreatic carcinoma. Oncotarget. 2015;6:43831–42. doi: 10.18632/oncotarget.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Duan J, Qu Y, Deng T, Liu R, Zhang L, et al. Onco-miR-24 regulates cell growth and apoptosis by targeting BCL2L11 in gastric cancer. Protein & cell. 2016;7:141–51. doi: 10.1007/s13238-015-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y, Liu Y, Du L, Li J, Qu A, Zhang X, et al. Down-regulation of miR-24-3p in colorectal cancer is associated with malignant behavior. Medical oncology. 2015;32:362. doi: 10.1007/s12032-014-0362-4. [DOI] [PubMed] [Google Scholar]
- 42.Yin Y, Zhong J, Li SW, Li JZ, Zhou M, Chen Y, et al. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget. 2016;7:86755–65. doi: 10.18632/oncotarget.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang H, Rho JG, Kim C, Tak H, Lee H, Ji E, et al. The miR-24-3p/p130Cas: a novel axis regulating the migration and invasion of cancer cells. Sci Rep. 2017;7:44847. doi: 10.1038/srep44847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang JS, Gong A, Young CY. ZNF185, an actin-cytoskeleton-associated growth inhibitory LIM protein in prostate cancer. Oncogene. 2007;26:111–22. doi: 10.1038/sj.onc.1209769. [DOI] [PubMed] [Google Scholar]
- 45.Shikauchi Y, Saiura A, Kubo T, Niwa Y, Yamamoto J, Murase Y, et al. SALL3 interacts with DNMT3A and shows the ability to inhibit CpG island methylation in hepatocellular carcinoma. Mol Cell Biol. 2009;29:1944–58. doi: 10.1128/MCB.00840-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol. 2010;12:457–67. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asnaghi L, Vass WC, Quadri R, Day PM, Qian X, Braverman R, et al. E-cadherin negatively regulates neoplastic growth in non-small cell lung cancer: role of Rho GTPases. Oncogene. 2010;29:2760–71. doi: 10.1038/onc.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Rienzo A, Archer MA, Yeap BY, Dao N, Sciaranghella D, Sideris AC, et al. Gender-Specific Molecular and Clinical Features Underlie Malignant Pleural Mesothelioma. Cancer Res. 2016;76:319–28. doi: 10.1158/0008-5472.CAN-15-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aijaz S, D'Atri F, Citi S, Balda MS, Matter K. Binding of GEF-H1 to the tight junction-associated adaptor cingulin results in inhibition of Rho signaling and G1/S phase transition. Developmental cell. 2005;8:777–86. doi: 10.1016/j.devcel.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Sibio S, Sammartino P, Accarpio F, Biacchi D, Cornali T, Cardi M, et al. Metastasis of pleural mesothelioma presenting as bleeding colonic polyp. The Annals of thoracic surgery. 2011;92:1898–901. doi: 10.1016/j.athoracsur.2011.04.117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA seq data are available at www.ebi.ac.uk/arrayexpress with accession number ID: E-MTAB-7079.