Abstract
Purpose
Inflammatory bowel disease-associated colorectal cancers (IBD-CRCs) are associated with a higher mortality than sporadic colorectal cancers. The poorly defined molecular pathogenesis of IBD-CRCs limits development of effective prevention, detection and treatment strategies. We aimed to identify biomarkers using whole exome sequencing of IBD-CRCs to guide individualised management.
Experimental Design
Whole-exome sequencing was performed on 34 formalin-fixed paraffin-embedded primary IBD-CRCs and 31 matched normal lymph nodes. Computational methods were used to identify somatic point mutations, small insertions and deletions, mutational signatures, and somatic copy number alterations. Mismatch repair status was examined.
Results
Hypermutation was observed in 27% of IBD-CRCs. All hypermutated cancers were from the proximal colon; all but 1 of the cancers with hypermutation had defective mismatch repair or somatic mutations in the proofreading domain of DNA POLE. Hypermutated IBD-CRCs had increased numbers of predicted neo-epitopes, which could be exploited using immunotherapy. We identified 6 distinct mutation signatures in IBD-CRCs, 3 of which corresponded with known mechanisms of mutagenesis. Driver genes were also identified.
Conclusions
IBD-CRCs should be evaluated for hypermutation and defective mismatch repair to identify patients with a higher neo-epitope load who may benefit from immunotherapies. Prospective trials are required to determine whether immunohistochemistry to detect loss of MLH1 expression in dysplastic colonic tissue could identify patients at increased risk of developing IBD-CRC. We identified mutations in genes in IBD-CRCs with hypermutation that might be targeted therapeutically. These approaches would complement and individualise surveillance and treatment programmes.
Keywords: Crohn’s disease, Ulcerative colitis, hypermutation, DNA repair, immunotherapy
Introduction
Inflammatory bowel disease associated colorectal cancer (IBD-CRC) is an aggressive complication of chronic inflammation accounting for 10-15% of deaths from IBD. IBD-CRC patients are younger and have a higher mortality than those with sporadic colorectal cancer (1). IBD-CRC arises diffusely in chronically inflamed epithelium via low-grade dysplasia evolving to high-grade dysplasia and eventually adenocarcinoma, although it may also arise without these preceding changes. Technological advances in endoscopy have improved the ability to discriminate between normal and pre-cancerous mucosa (1), however, a third of cancers arise in the interval between scheduled colonoscopies, suggesting poor efficacy of surveillance programmes (2). Colonoscopy-directed mucosal samples are analysed for the presence of dysplasia, which remains the gold standard for predicting future cancer risk in IBD when reported by an expert gastrointestinal pathologist.
The poorly defined molecular and genetic pathogenesis of IBD-CRC impairs our ability to detect and treat colorectal cancer effectively. There is an urgent need to identify biomarkers to improve early detection of colorectal cancer and guide individualised therapy (2).
We elected to study primary IBD-CRC since molecular changes identified in the resected cancer specimen are most likely to represent biomarkers of cancer development. We characterised the hypermutator status of the cancers, defined the mutational signatures and underlying biological processes, and explored the potential clinical translation of this work with respect to immune therapy, and improving current surveillance programmes.
Materials and Methods
Ethical Approval and Case Identification
The study was approved by the Lothian NHS Research Scotland Human Annotated BioResource, which is an NHS Health Research Authority Ethics Committee-approved Research Tissue Bank (www.hra.nhs.uk; 15/ES/0094) for the use off human tissue surplus to diagnostic requirements. The study was conducted in accordance with the Declaration of Helsinki and the guidance from the Human Tissue Authority on the use of human tissue from diagnostic archives. The Lothian (Edinburgh, UK) pathology database was searched for IBD-CRCs using the terms inflammatory bowel disease, Crohn’s disease, ulcerative colitis, colon cancer, colorectal and rectal cancer between 1990 and 2011 by the Tissue Bank appointed pathologist and provided the anonymised tissue samples with linked clinical data. Two independent expert GI pathologists (MJA and CJB) and a specialist IBD Consultant Physician (SD) verified the IBD-CRCs following careful review of the patient’s anonymised medical records, the histopathological evidence for IBD in previous colonic samples and in the IBD-CRC resection specimen.
Cases were only included from patients with a previous histopathological diagnosis of IBD and where evidence of IBD in the resected colonic specimen was confirmed by both GI specialist pathologists (CJB and MJA) independently. Sporadic adenomas and colorectal cancers which arose in an area of the colon without prior evidence of IBD in that colonic area were excluded. For each case we have clear histopathological evidence of pre-existing IBD at each tumour site and have confirmed that we have selected IBD-associated colorectal cancers.
Formalin fixed paraffin embedded (FFPE) cancer and uninvolved normal lymph node blocks, removed at the time of surgery, were sectioned, H&E stained and used for further analysis. In total 31 cases (15 Crohn’s Disease (CD): 16 Ulcerative Colitis (UC)) with 34 cancers were used in this study.
Clinical Phenotype Data: Statistical Analysis
The ‘survival’ package within the R software environment (3) was used to generate the Kaplan-Meier survival curves and perform statistical analyses. Categorical clinical phenotype data was analyzed using the two-tailed Fisher’s exact test; a P-value of <0.05 was considered significant. No survival data was available for case 2L (hypermutator) which was excluded from the survival analysis. Cases 15G1 and 15G2 were considered as one case for the purpose of statistical comparison, as they appear to originate from the same precursor clone.
Sequencing Data Generation, Processing and Analyses
Nucleic acids were extracted using Qiagen Allprep FFPE DNA extraction kits, and DNA/RNA was quantified using Agilent GeneChips. For WES, DNA was captured using the Agilent SureSelectXT Human All Exon V5 platform following the manufacturer’s protocol. Sequencing was performed using the Illumina HiSeq 2000 platform at the Wellcome Trust Sanger Institute. Raw paired-end sequencing reads (75bp) were aligned and a modified version of the reference genome which includes the GRCh37 primary assembly and additional human contigs and viral sequences that reduce the number of reads erroneously mapped to the primary assembly. Further details on data processing, data quality assessment and sequence alignment are available in the Supplementary Materials and Methods. Somatic point mutations and small insertions and/or deletions (InDels) were identified by comparing cancer and matched normal samples using MuTect (v1.1.7) (4) and Strelka (v1.0.14) (5), respectively. BCFtools (v1.3) (6) was used to identify variants in each patient’s germline (normal lymph nodes) relative to the human reference genome described above. The Ensembl Variant Effect Predictor (7) software was used to predict the effect of each variant on protein sequences, and the clinical significance of the changes was predicted by comparison to variants in the ClinVar database (release date 2016-05-31). For genes with multiple transcripts, we report the effect of the variant on the protein derived from the canonical transcript as annotated in Ensembl release 81 for GRCh37 (Supplementary Table S7; Supplementary Materials and Methods). We also provide the predicted effect on all transcripts in Supplementary Table S8. The Sequenza software package (version 2.1.2) (8) was used to identify somatic copy number alterations. For the non-hypermutator cases, MutSigCV (version 1.4) (9), which identifies significantly mutated genes, and dNdScv (10), which identifies genes under positive selection in cancer, were used to identify driver genes. For the hypermutator cases, microsatellite InDels were excluded from analysis with MutSigCV. The microsatellite InDels were analysed with a modified version of MSMutSig (11) obtained from the author, along with required input files. In dNdScv, which is able to analyze both the SNV and InDel mutations together in both hypermutator and non-hypermutator cohorts, two InDel models are available; one model considers the total number of InDel mutations per gene, and the other model considers unique InDel sites per gene (the “unique-sites model”). For the hypermutator cohort, dNdScv was run using both models, and the results were compared. Only the “unique-sites” model was used for the non-hypermutator cohort. Restricted hypothesis testing was performed on the results from dNdScv, using known cancer genes from the Cancer Gene Census version 81 (12). Further details are available in the Supplementary Materials and Methods. Mutation signatures were identified with the Bioconductor package SomaticSignatures (version 2.6.0) (13) using the non-negative matrix factorization (NMF) algorithm. Details of software parameters, databases used, variant annotation, variant filtering, validation and the identification of driver genes are available in the Supplementary Materials and Methods.
HLA Typing and Neo-epitope Predictions
Human leukocyte antigen (HLA)-I 4-digit typing was performed using the OptiType 1.0 algorithm and neo-epitopes from missense mutations were predicted by mapping the corresponding protein sequences to the human proteome database (version GRCh37.74). Neo-epitopes with a relative percentile rank ≤ 1% for each HLA-I allele were considered binders (additional details are provided in Supplementary Materials and Methods).
Mismatch Repair Immunohistochemistry
Immunohistochemistry for MLH1, MSH2, MSH6 and PMS2 was undertaken on a cancer tissue microarray and staining was independently scored by two pathologists (MJA and AO). Discordant scores were resolved by staining whole tissue sections with additional MLH1, MSH2, MSH6 and/or PMS2 antibodies.
MLH1 Promoter Methylation Analysis
MLH1 promoter methylation was analysed using the EZ DNA Methylation Kit Gold (Zymo Research). Cases from patient 32N failed methylation analyses.
Further experimental details are described in the Supplementary Materials and Methods.
Results
To identify biomarkers associated with IBD-CRC, WES was performed on 34 IBD-CRCs and matched normal lymph node pairs. Twenty-nine patients had one cancer, one patient had three primary cancers separated anatomically (32N) and one patient had two cancers in close proximity to each other (15G) (Supplementary Table S1). WES yielded between 45- and 90-fold coverage of the cancer samples and 26- and 93-fold coverage of the normal lymph node samples (Supplementary Fig. 1). When considering protein coding and splice site mutations only, the cancers from 32N had only 6 mutations common to at least 2 of the 3 cancers, while the two cancers from 15G had over 2345 common mutations (22% of the total mutant positions in these cancers).
Somatic Mutation Rates in IBD-CRC
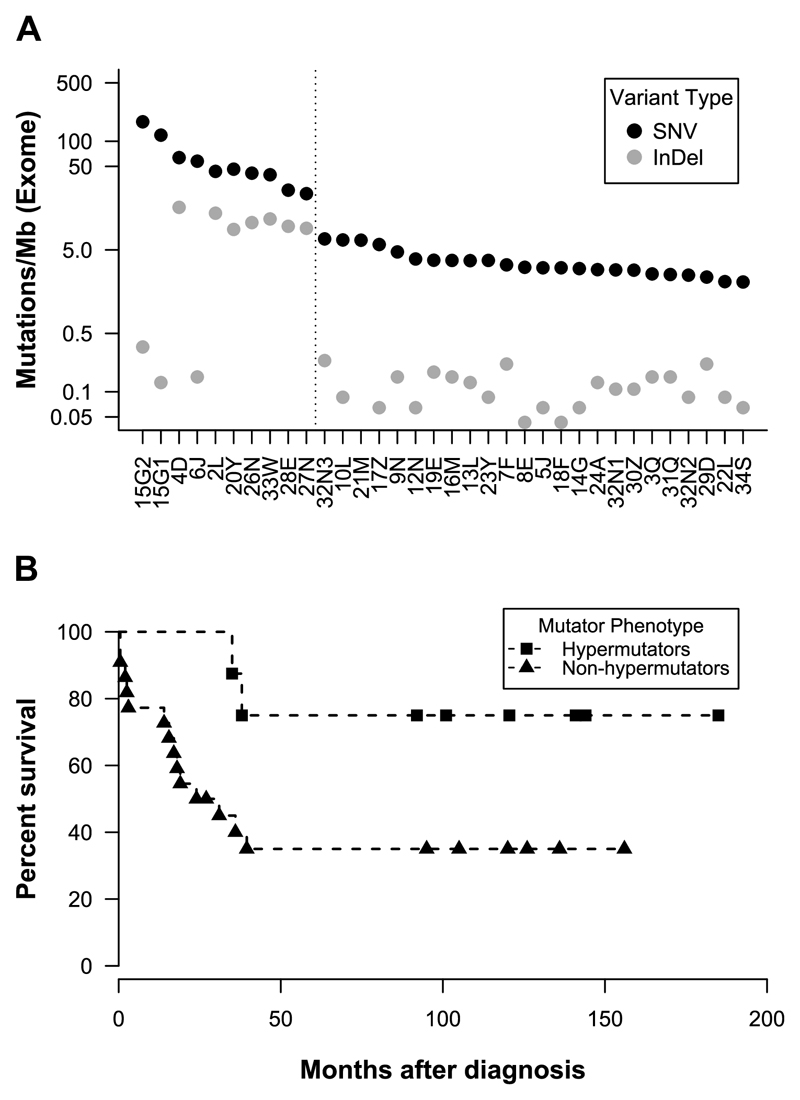
Somatic point mutations and InDels were identified by comparing exome sequences from cancer tissue with those from uninvolved lymph nodes removed at the time of surgery (Supplementary Tables S7 - S8).The 34 cancers were divided into two groups based on distinct somatic mutation rates. There were 24 non-hypermutator cancers with 2.0-7.0 mutations/Mb, and 10 hypermutator cancers with 32.6-171.3 mutations/Mb (Fig. 1A and Table 1). Two of the 10 cases, 15G1 and 15G2, have a somatic mutation burden greater than 100 mutations/Mb and could thus be defined as ultra-hypermutators.
Figure 1. Somatic mutational rates and survival analysis of IBD-CRC.
(A). Mutational frequency in each of the 34 IBD-CRCs ordered by overall mutation rate. There is a clear separation between the 10 hypermutator cancers and the 24 non-hypermutator cancers. With the exception of 15G1, 15G2, and 6J, the hypermutator cancers showed elevated mutation rates of both SNVs and InDels. No InDels were found in the exome of 21M. (B) Kaplan-Meier plot of overall survival stratified by cancer mutator phenotype. Patients with hypermutators cancers had increased survival compared with patients with non-hypermutator cancers (log rank test, P=.04).
Table 1. Mutational rates in IBD-CRC and sporadic CRC.
| Tumour Phenotype | TCGA(25) | DIN | ROBLES(31) |
|---|---|---|---|
| Hypermutators | >12 (728) | 32.6-171.3 (56.2) | 18.19-22.2 (N/A) |
| Non-hypermutators | <8.24 (58); Non-silent mutations only |
2.1-7.0 (3.1) | 0.3-5.1 (1.3) |
Mutational rates (mutations/Mb) from the whole-exome sequence analysis of TCGA sporadic CRC cohort (25) and IBD-CRCs from this study and Robles et al. (31) in the hypermutator and non-hypermutator cancers. Numbers in parentheses are median mutation rates. Two hypermutators were present in the Robles cohort. The rates provided include total mutations with the exception of the median mutation rate of non-hypermutator cancers in the TCGA cohort.
The proportion of hypermutators in our IBD-CRC data set was 27%, or 9/33 (cases 15G1 and 15G2 were counted once, since they appear to originate from the same precursor clone) and was not significantly higher than the 16% present in a cohort of 382 sporadic CRCs downloaded from The Cancer Genome Atlas (TCGA) Data Portal (see Supplementary Materials and Methods) (right sided binomial test, P=.07), nor the 28% observed in a larger cohort of 619 sporadic CRCs from Giannakis et al. (binomial test, P=1) (14). The hypermutator cases had elevated levels of both single nucleotide variants (SNVs) and InDels, with the exception of cases 15G1, 15G2 and 6J, which have similar InDel mutation rates to the non-hypermutator cases (Fig. 1A). The median age at diagnosis was not significantly different between the hypermutators (median 66.2 years; IQR 57.4-78.1 years) and non-hypermutators (median 65.8 years; IQR 51.0-77.0 years).
Overall, there was a significant survival difference between the patients with hypermutator cancers and those with non-hypermutator cancers (log rank test, P=.04), with the estimated 10-year survival being 75% in the former group as compared with 36% in the latter (Fig. 1B). However, it is likely that additional factors other than mutator status including stage and age influenced patient survival although we were unable to measure the affects of these variables due to the small sample size.
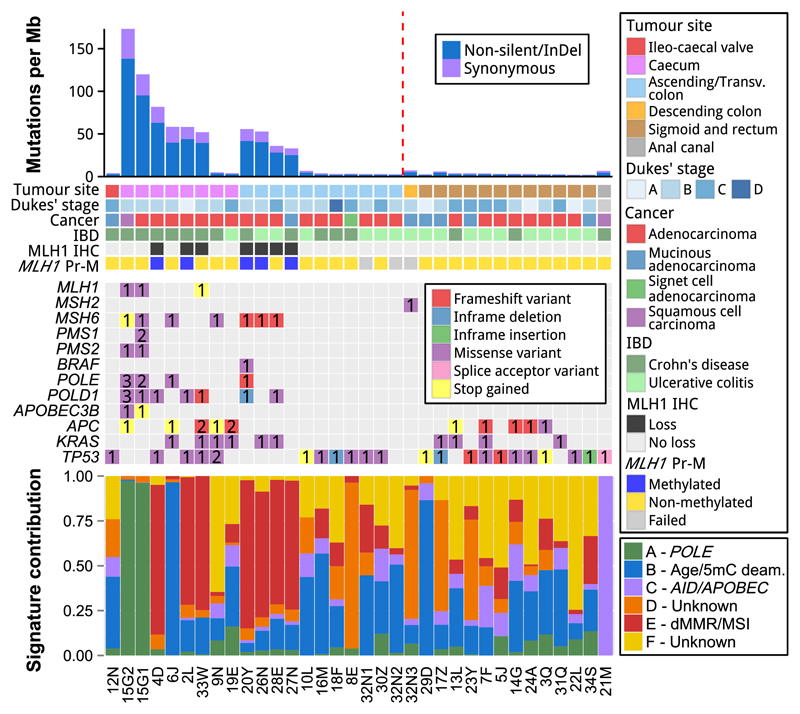
Strikingly, the hypermutator cancers were all located in the proximal colon and none in the distal colon (two-tailed Fisher’s exact test, P=.004) (Fig. 2).
Figure 2. Clinical and genetic characteristics of the IBD-CRC.
Top panel: Mutational rate of each cancer case ordered by tumour site and mutation rate. The red line segregates the proximal and distal bowel cancers. Second panel: Tumour site, Dukes’ stage, cancer type, underlying IBD, immunohistochemical testing of MLH1 (MLH1 IHC) and promoter methylation status of MLH1 (MLH1 Pr-M). Third panel: selected genes and somatic nonsense, non-silent and InDel mutations. The colours indicate the predicted effect of the mutation on the protein sequence, and the number indicates the number of mutations with the same predicted effect. Note that in cases where a gene has multiple mutations with different predicted effects, only the effect type with the highest priority will be shown. Further details are in the Supplementary Methods and Materials. The complete list of mutations and their effects is in Supplementary Tables S2A - S2B. Lower panel: Contribution of signatures of mutational processes (A-F) in each cancer case. Each signature A-F corresponds to a signature identified by Alexandrov et al. (Signatures 10, 1, 13/2, 17, 6 and 5, respectively) with cosine similarities ranging from 0.82 to 0.97 (Supplementary Table S3).
Mismatch Repair Abnormalities
To characterise the difference in mutational rates, the cancers were analysed for genetic aberrations associated with dMMR. Seven out of the ten hypermutator cancers had a high frequency of InDels (Fig. 1A), which is indicative of MSI, and showed loss of expression of MLH1 and its heterodimeric binding partner PMS2 (Fig. 2; Supplementary Fig. 2). Loss of MLH1 protein expression results in dMMR, leading to increased somatic substitution and susceptibility to cancer (15). Loss of MLH1 expression can be explained by MLH1 promoter hypermethylation in 5 of these 7 cancers (Fig. 2; Supplementary Fig. S3). One of the two remaining cancers (33W), had a somatic nonsense mutation within the ATPase domain (R100*) of the MLH1 gene, resulting in a truncated protein with a predicted loss of function. We did not find other coding mutations that would cause MLH1 to be biallelically inactivated in case 33W. We did not detect any point or InDel mutations in MLH1 in the germline or tumour in case 28E, nor epigenetic silencing by promoter hypermethylation that could explain the loss of MHL1 expression in this case. The normal expression of MLH1 and low level of somatic InDels in the remaining 3 hypermutator cancers, 15G1, 15G2, and 6J, point to alternative mechanisms leading to hypermutation. In contrast, loss of expression, promoter hypermethylation and non-silent somatic mutations were not observed in MLH1 in the non-hypermutator cases (Fig. 2; Supplementary Table S2B).
Mutations in DNA Polymerase Proof-reading Domains
During DNA replication, DNA fidelity is maintained by the proof-reading function of DNA polymerases. Germline mutations in the exonuclease proof-reading domains of the DNA polymerases POLD1 (codons 245-571) and POLE (codons 223-517) have been shown to predispose to the development of hypermutated microsatellite stable (MSS) sporadic colorectal cancer (16). Non-silent somatic mutations in POLE and POLD1 were identified in 4 and 7 of the hypermutator cancers, respectively (Fig. 2; Supplementary Table S2A). Two shared somatic POLE mutations (P286R and F348S) affected the exonuclease proof-reading domain in cases 15G1 and 15G2, which were two adjacent cancers (adenocarcinoma and squamous cell carcinoma) in the same patient (Fig. 1A; Fig. 2).
No frameshift or non-silent somatic POLE or POLD1 point mutations were identified in the non-hypermutator cancers.
The mechanism of hypermutation in case 6J remains undefined. Case 6J was an adenocarcinoma from a patient with a 20 year history of colonic Crohn’s disease and autoimmune liver disease for which she received potent immunosuppression (prednisolone, tacrolimus and azathioprine) throughout this period. This patient had 4 basal and 2 squamous cell carcinomas of skin removed but did not have any frameshift or non-silent somatic or germline point mutations in PTCH1, excluding Gorlin syndrome as a contributing factor to the underlying cancer predisposition (17). In addition, germline mutations that could explain the hypermutator phenotype were not found in other DNA repair genes including those involved in nucleotide and base excision. It is possible that the continuous, prolonged immunosuppression in this patient has altered the immune cancer response resulting in both colonic and multiple skin cancers.
Mutational Signatures in IBD-CRC
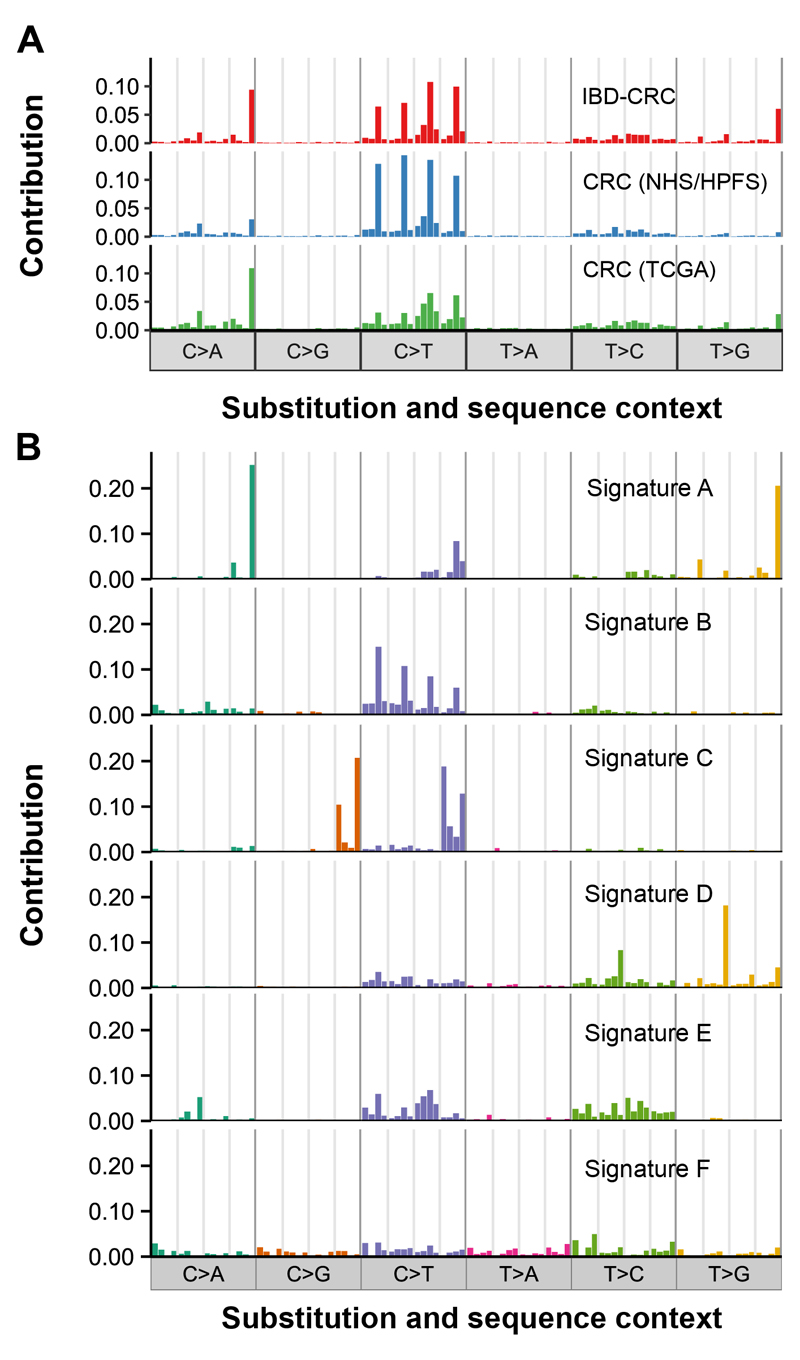
It has been demonstrated that different mutational processes in cancers generate specific patterns of mutation, or ‘signatures’, with 30 distinct signatures identified thus far by Alexandrov et al. (18). The overall mutational spectrum our IBD-CRC cohort was very similar to the spectra derived from cohorts of sporadic CRCs from Giannakis et al. (14) (cosine similarity=.87) and from TCGA Data Portal (cosine similarity=.91) (Fig. 3A; Supplementary Materials and Methods). Six distinct signatures, designated A-F, were extracted from the catalogue of IBD-CRC somatic mutations (Fig. 3B) and corresponded well to Alexandrov Signatures 10, 1, 13/2, 17, 6 and 5, respectively, with cosine similarities ranging from .82 to .97 (Fig. 2 lower panel; Supplementary Table S3). Some of the signatures have been associated with specific mutational mechanisms (18). Of particular note, are IBD-CRC Signature A (Alexandrov Signature 10/POLE) that was predominant in the hypermutator cancers with DNA POLE mutations (15G1 and 15G2); and IBD-CRC Signature E (Alexandrov Signature 6/dMMR and MSI) that was predominant in the 7 hypermutator cancers which had loss of expression of the MLH1 protein (Fig. 2). IBD-CRC Signature C (Alexandrov Signature 13 and 2/AID/APOBEC) is the major contributor in case 21M, however as we our analysis is limited to the exome regions only, we have no evidence of AID/APOBEC activation in this case. Transcriptional upregulation of APOBEC3B is commonly found in bladder, cervical, lung, head/neck and breast cancers with kataegis (18), however, we did not find evidence of kataegis in case 21M (Supplementary Fig. S4). It has been hypothesised that one cause of APOBEC3B upregulation may be infection by viruses, including Human Papillomavirus (HPV) (19). We did not find HPV DNA associated with case 21M, however, since whole-exome sequencing was used in this study, any viral sequence present would be excluded unless it has integrated into the genome in the targeted sequences.
Figure 3. Signatures of mutational processes in IBD-CRC.
(A). The somatic mutation spectra of the IBD-CRC and sporadic CRC datasets are represented by each of the six possible substitution types, C>A, C>G, C>T, T>A, T>C, T>G (including their reverse complements) in the context of their immediate 5’ and 3’ flanking bases, giving 96 possible motif combinations. For each substitution type, the 16 motifs are listed in order from left to right by the 5’ flanking base (A, C, G, then T), then by the 3’ flanking base (A, C, G, then T). The IBD-CRC panel shows the mutation spectrum for 34 IBD-associated colorectal cancers; the CRC (TGCA) panel shows the mutation spectrum for 115 colon adenocarcinomas and 267 rectum adenocarcinomas from downloaded from TCGA Data Portal, and the CRC (NHS/HPFS) panel show the mutation spectrum for a cohort of 619 CRCs from Giannakis et al. (14). (B) Six distinct mutation signatures (A-F) were extracted from IBD-CRCs and on comparison with the Alexandrov Signatures 1 to 30 (33) using cosine similarity (Supplementary Table S3), the correspondence is: Signature A and Signature 10 (POLE mutations); Signature B and Signature 1 (age/spontaneous deamination of 5-methylcyotosine); Signature C and Signature 13/2 (AID/APOBEC activation); Signature D and Signature 17 (unknown aetiology); Signature E and Signature 6 (mismatch repair deficiency and microsatellite instability); Signature F and Signature 5 (unknown aetiology; found in all cancer types).
Although Alexandrov signatures 2 and 13 have not yet been identified in CRC, an APOBEC mutation pattern has been found, using different methods, in a variety of cancer types including whole-genome sequenced sporadic CRCs (20). Signatures 1, 5, 6 and 10 have previously been found in sporadic colorectal cancers (18) and signature 17 has been identified in MSS CRC (21). Taken together, these results indicate that the mutational mechanisms in IBD-CRC and sporadic CRC are similar, although a much larger IBD-CRC cohort and whole-genome sequencing may reveal additional or novel signatures.
Recently, a signature attributed to the persistence of 8-oxoguanine G>T/C>A mismatches due to biallelic inactivation of MUTYH has been identified in MUTYH-associated polyposis (MAP) CRCs (22). Unsurprisingly, we did not find this signature in our IBD-CRCs as our cases did not have a history of MAP nor any germline non-silent or splice site point or InDel mutations in MUTYH other than common SNPs. We did not discover a novel inflammation-associated mutational signature in our cohort. Similarly, others have not found an inflammation-signature common to chronic inflammatory-associated gastrointestinal cancers (such as Barrett’s oesaphagus) (23). Rather, epithelial regeneration and the subsequent cumulative effect of chronic inflammation-associated damage appears to be a major mechanism of promoting carcinogenesis in IBD (24).
Driver Genes in IBD-CRC
To identify driver genes in non-hypermutator IBD-CRC, we used MutSigCV (9) to identify significantly mutated genes, and dNdScv to find genes under positive selection in cancer (10) (Methods and Supplementary Materials and Methods). In our 24 cases, TP53, PIK3CA, APC, and KRAS were identified as driver genes (FDR-adjusted P-value, or q<.10) (Fig. 4A), all of which are also driver genes in sporadic CRC (25). As observed in sporadic CRC (25), significantly more non-hypermutator cancers had non-silent somatic mutations in TP53 than the hypermutator cancers (79% vs. 33%; Two-tailed Fisher’s exact test, P=.03) (Table 2).
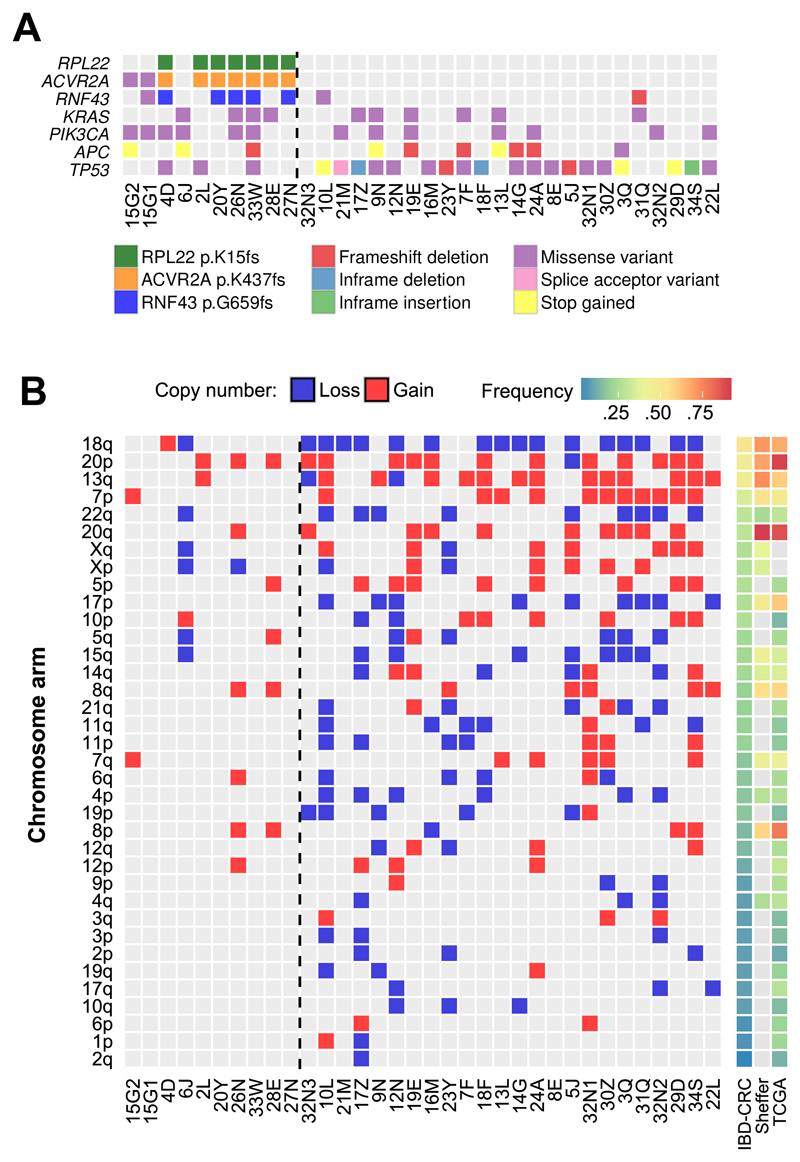
Figure 4. Hotspot microsatellite InDels, driver genes and recurrent somatic copy number alterations in IBD-CRC genomes.
(A). Hotspot InDels and driver genes in IBD-CRC. Somatic SNVs and InDels were used to identify hotspot InDels and driver genes in hypermutator (n=9) and non-hypermutator cancers (n=24). Because hypermutator case 15G1 and 15G2 were related, SNV and InDels were merged for this analysis. (B) Chromosome arm-level somatic copy number alterations. Shown are predicted SCNAs that cover at least 90% of a chromosome arm (left panel), and the frequency of the SCNAs in IBD-CRC (this study), and in sporadic CRC studies by Sheffer et al. (33) and TCGA (25) (right panel). Chromosome arms are listed in descending order of frequency in IBD-CRC. Grey boxes in the frequency columns indicate unknown values. IBD-CRC cases are ordered left to right by overall SNV and InDel mutation rate. Cases to the left of the dashed line are hypermutator cancers.
Table 2. Frequency of TP53, KRAS and APC mutations in IBD-CRC and sporadic CRC.
In the hypermutator cases, no driver genes were identified using MutSigCV (9) or MSMutsig (11), due to small sample size. When using dNdScv (10), followed by restricted hypothesis testing with a list of known cancer genes from the Cancer Gene Census (version 81) (12), KRAS (q=.03) was identified as a driver gene in the hypermutator cohort, in which 3 of the 4 cases with KRAS mutations are p.G12D (Fig. 4A). Importantly, this analysis also identified hotspot InDel mutations in RPL22 (p.K15fs; q=.001), ACVR2A (p.K437fs; q=.03) and RNF43 (p.G659fs; q=.1) (Fig. 4A; Supplementary Materials and Methods). The same ACVR2A and RNF43 mutations have previously been identified as driver events in a much larger cohort of sporadic CRC (11), and while they may represent driver events in IBD-CRC, they could also be genomic sites that are particularly prone to mutation. A much larger cohort of MSI IBC-CRCs would be required to confirm these as driver events. As reported for sporadic CRCs (26), we find enrichment of RNF43 mutations in MSI cases, and mutual exclusivity of RNF43 truncating mutations (which includes frameshift InDels, nonsense mutations and splice site mutations) with APC truncating mutations in our IBD-CRC cohort (Fig. 4A).
Several of the genes we identified are clinically actionable. For example, tumours with RNF43 mutations are sensitive to porcupine inhibitors (27). In the same way RPL22 mutant cancers may be susceptible to MDM2-p53 pathway compounds (28). Notably, the same RPL22 p.K15fs hotspot mutation we identified in our IBD-CRC cohort has been frequently found in endometrial, gastric and colorectal cancers (28).
Somatic IDH1 R132 Mutations
A potentially targetable IDH1 hotspot mutation at R132, which was mutated in 11% (5/47) of IBD-CRCs and only in CD-associated CRCs, was previously reported by Yaeger et al. (29). In our cohort, one IDH1 R132C mutation was present in UC-associated CRC case 32N3. The observed frequency here is not significantly different than that of Yaeger et al. (29) (Chi squared test, P=.21). We note, however, that the median 17-fold coverage (range 9-36-fold in the tumour samples) at this site was lower than the whole-exome median of 70-fold, as this mutation is only 20bp from an exon boundary where coverage tends to be lower in whole-exome sequencing studies.
Somatic Copy Number Alterations in IBD-CRC
As previously observed with sporadic CRC (30), somatic copy number alterations (SCNAs) in IBD-CRCs were significantly more prevalent (one-tailed Student’s t-test P=.001) in the non-hypermutator cases compared with hypermutator cases (Supplementary Fig. S5; Supplementary Table S6).
Robles et al. (31) reported 15 focal copy number gains, 8 of which were observed in more than one tumour and Shivakumar et al. (32) reported 26 focal SCNAs in pooled IBD-associated dysplastic and carcinoma biopsy samples. Focal SCNAs common to our IBD-CRC cohort and these studies are gains in 12p13.33-12p13.31, 22q11.21, 4p16.3, 8q24.3, 10q26, 13q12, 20q11.23 and 20q13; however, none were common to all three.
We compared the frequency of predicted chromosome arm-level SCNAs to recurrent SCNAs found by Sheffer et al. (33), and to SCNAs identified by TCGA (25) (Fig. 4B; Supplementary Table S6). Our novel data demonstrate that the frequencies of specific arm-level SCNAs in IBD-CRC broadly concurs with those found in sporadic CRC (25, 33).
Germline Variants in IBD Patients
Colorectal cancer affects up to 2.5% of the patients who suffer from IBD (1), and germline variations conferring cancer susceptibility have been poorly described in this population of patients. We examined the germlines of our cohort for variants in genes implicated in susceptibility to colorectal cancer (Supplementary Tables S4 - S5). The CHEK2 frameshift mutation at T367 (c.1100delC; rs555607708), in the germline of case 15N, is known to predispose to breast cancer, but association with colon cancer has not been resolved (34). The R242H mutation in SDHB (rs74315368) in case 3Q has been characterized in paragangliomas (35) but not, to our knowledge, in colon cancer. The remaining germline variants we have reported in Supplementary Table S5 are in genes known to be associated with increased risk of CRC, however, they have unknown or mixed reported clinical significance and currently we are unable to clarify as to whether these variants confer a predisposition to colorectal cancer in the setting of chronic inflammation.
Neo-epitope and Immune Infiltrate Analysis
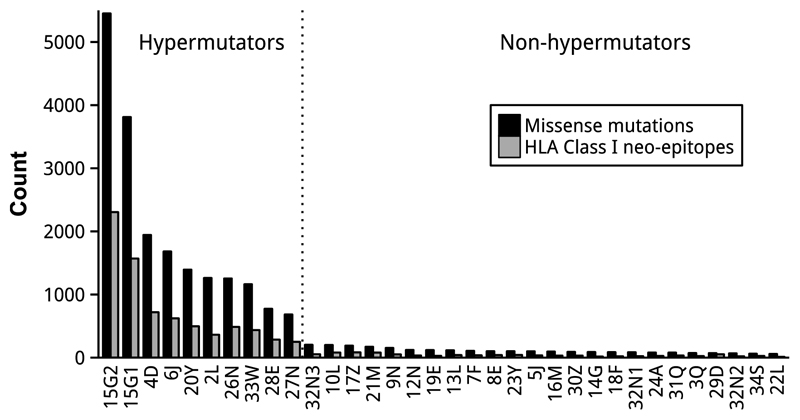
Non-synonymous somatic mutations in cancer can generate novel antigens, (neo-epitopes) that can be exploited in cancer immunotherapy. The number of predicted neo-epitopes is expected to be directly proportional to the mutational load of cancers (36). As expected, the IBD-CRC hyermutator cancers generated the largest number of HLA Class I neo-epitopes, and were significantly higher than the number found in non-hypermutators (one-tailed Student’s t-test, P=.004) (Fig. 5). Similar to our study the neo-epitope load was higher in sporadic CRC with dMMR/MSI-high status, than in pMMR/MSS tumours, and notably, they responded favourably to PD1 blockade (14). IBD is characterized by a dysregulated immune response and many therapeutic modalities targeting inflammation are directed at cytokines. Analysis of the cytokine gene immune expression profile demonstrated that overall the cytokine gene expression profile was similar in the hypermutator IBD-CRCs and the non-hypermutator IBD-CRCs (Supplementary Fig. S6), although this analysis was limited by the very small size of tumour epithelium (with very little immune cell-containing stroma) sampled.
Figure 5. Predicted HLA Class I neo-epitopes in IBD-CRCs.
The cancers with the highest mutational rates (hypermutators) generated the largest number of predicted HLA Class I neo-epitopes.
Discussion
We have undertaken a comprehensive analysis of 34 IBD-CRCs from 31 patients using WES. Hypermutator cancers were observed in both our study and Robles et al. (31) and the mutational rates were within range of the larger TCGA sporadic CRC cohort (25).
In sporadic and hereditary colorectal cancers, the hypermutator phenotype is most frequent in cancers of the right (proximal) colon (25). In this study, we observed a strong association between CD-associated CRCs occurring in the proximal colon (two-tailed Fisher’s exact test, P=.03) and the hypermutator phenotype (two-tailed Fisher’s exact test, P=.05. Long standing, extensive colonic CD has been shown to be associated with proximal cancers (37) and we have clear histopathological evidence of pre-existing IBD at each tumour site, more than half of which were in the proximal colon. Similar to previous studies of IBD-CRC (38) a higher proportion of UC-associated CRCs occurred in the left colon (Fig. 2) when compared with CD-associated CRCs (two-tailed Fisher’s exact test, P=.03).
Previous studies have reported a variable frequency of MLH1 promoter hypermethylation and loss of expression of MLH1 in IBD-CRCs (39). The frequency of MLH1 promoter methylation in our series may be associated with the more advanced age at time of IBD-CRC diagnosis, although it remains lower than that of sporadic MSI-high CRC with CpG (MLH1) hypermethylation (40). Lennerz et al. (39) have exclusively studied CRC complicating Crohn’s colitis and described the median age of cancer diagnosis to be 58 years (range 34–77) which is also slightly higher than that conventionally reported for IBD-CRCs. In addition to our strict inclusion criteria, and similar to previous studies (39), the mutation analysis identified a single BRAF V600E mutation in one hypermutator IBD-CRC (Fig. 2) confirming that these cancers are unlikely to be sporadic in nature.
BRAF V600E mutations occur in the majority (>85%) of hypermethylated sporadic MSI high CRCs (25) and are therefore established in diagnostic algorithms to differentiate between sporadic and familial Lynch Syndrome cases of colorectal cancer (41). Notably, we did not identify germline mutations in genes such as MLH1, MSH2, APC, and MUTYH that predispose to the development of CRC in our IBD-CRC cohort. Although the proportion of hypermutator cancers in our cohort was not significantly different than that in sporadic CRC, these differential mutations distinguish our hypermutated IBD-CRC cohort from hypermutated sporadic and familial forms of CRC.
The effect of dMMR/MSI on survival outcomes in IBD-CRC has not been reported. A recent meta analysis of 20 studies including 571,278 patients by Reynolds and colleagues (42) has reported that IBD-CRC does not affect the overall 5 year survival compared with sporadic CRC without any adjustment for molecular subtypes. In our series, the hypermutator IBD-CRCs (which included 2 MSS hypermutators 15G and 6J) had a significantly better survival compared with non-hypermutator IBD-CRCs. The increased survival of sporadic MSI cancers (43) is comparable to the data presented here for IBD-CRCs. The improved survival of early stage sporadic dMMR/MSI cancers is postulated to be mediated by (anti-) tumour infiltrating lymphocytes (TILs) in response to the neo-epitopes generated by the high mutational rate. Immune blockade therapies targeting immune checkpoints and enhancing the anti-TIL response are currently being used in various cancers including CRC (44). In our series, the hypermutator IBD-CRCs had a higher predicted neo-epitope load but we were unable to identify any obvious differences in the immune related gene expression profile. This is unsurprising as our samples had been enriched for cancer-containing cells and not the adjacent immune rich stromal compartment. Molecular phenotyping of all colorectal cancers for targeted therapy is a compelling reason for universal screening for dMMR. Of note, the United States Food and Drug Administration has approved Pembrolizumab (targeting the programmed cell death 1 receptor) for the treatment of all unresectable or metastatic MSI-H/dMMR tumours that have progressed after initial treatment (45). Immune checkpoint blockade has been associated with immune-mediated colitis and this is particularly relevant for patients with co-existing inflammatory bowel disease (46). Although our results require validation in a randomized prospective cohort, it is exciting to postulate that immune blockade therapies may be useful adjuncts in treating patients with dMMR (and/or hypermutator phenotype) associated IBD-CRC that have undergone a total colectomy as part of their treatment programme, negating the risk of immunotherapy mediated colitis. Importantly, immune therapies should be used with caution or avoided in IBD-CRC patients who have undergone a partial colectomy or those with Stage IV disease with an intact colon as it can aggravate the underlying colitis (47).
To date, we have not been able to use genetic or molecular markers to improve the detection or treatment of IBD-CRC. In 2009, a specialist committee in the United States of America recommended universal screening for Lynch syndrome in all newly diagnosed colorectal cancers (48) and this has not been adopted for several reasons including financial, technical, ethical and health economical limitations. In the United Kingdom, the National Institute for Health and Care Excellence (NICE) is advocating use of mismatch repair immunohistochemistry on all colorectal cancers (including IBD cases) to detect Lynch syndrome (41). A cost effective analysis limited to early onset colorectal cancers suggests that this would be economically viable assuming that all of the necessary subsequent health interventions are fully implemented to reduce cancer mortality and morbidity (49). Our data supports this recommendation to detect dMMR in IBD-CRC, but not necessarily to detect Lynch syndrome as we did not identify any known clinically significant pathogenic germline mutations in MMR genes in this IBD-CRC cohort. Whilst these important issues are resolved our data can be used to support the rational for universal testing in IBD-CRCs to profile tumours for the use of targeted therapies, which is not routinely undertaken at all institutions.
The immunohistochemical analysis of TP53 protein has not been widely accepted to aid in the discrimination between dysplastic and inflamed colonic epithelium. Prospective studies are required to determine whether analyzing for TP53 abnormalities and loss of expression of MLH1 together, in colonic biopsies with potentially dysplastic epithelium, could aid in the evaluation of those at highest risk of developing CRC, similar to the molecular profiling used in gastric cancer (50).
Our retrospective study has inherent limitations and the small sample size may result in a reporting bias. Notwithstanding these issues, the power of next-generation sequencing technology has provided detailed information allowing clinically relevant statistical analyses to be undertaken. Future similar studies may discover many more similarities and differences between IBD-CRC and other types of CRC.
Conclusion
The categorisation of disease guides individualised therapeutic strategies and can predict the response to therapy and prognosis. Our analysis demonstrates that proximal IBD-CRCs have high mutational rates associated with defects in MMR and DNA POLE proof-reading function. This results in a predicted higher neo-epitope load, which could be exploited using immunotherapies. The hypermutator phenotype of this cohort is mostly associated with loss of MLH1 expression and this could be evaluated in colonic dysplastic lesions detected in IBD patients. The identification of driver genes in hypermutated IBD-CRCs can be used to develop therapeutic agents targeting the corresponding molecular pathways. In our series up to 90% of the hypermutated IBD-CRCs have actionable mutations. These approaches would complement and individualise current surveillance and treatment programmes for IBD-CRC.
Supplementary Material
Statement of Translational Relevance.
There is an urgent need to identify predictive biomarkers in inflammatory bowel disease-associated colorectal cancers (IBD-CRCs) to individualise the current prevention, surveillance and treatment programmes. This analysis demonstrates that proximal IBD-CRCs have high mutational rates associated with defects in MMR (MLH1 loss) and DNA POLE proofreading function. Hypermutation is associated with a predicted higher neo-epitope load, which could be exploited using immunotherapies in selected patients with hypermutated IBD-CRCs. Prospective studies are required to determine whether analysis for loss of MLH1 in surveillance colonic biopsies could discriminate those at increased risk of developing CRC, permitting a more targeted approach for cancer treatment and surveillance. The identification of driver genes in hypermutated IBD-CRCs could also be used to develop therapeutic agents targeting the corresponding molecular pathways. Our comprehensive analysis of the mutational landscape of IBD-CRCs has revealed several novel approaches that may complement and personalise surveillance and treatment programmes.
Acknowledgements
CpG Methylation primer sequences were generously provided by RH Masalmeh and D Sproul (University of Edinburgh).
Financial Support
This work was supported by the European Research Council Combat Cancer Program, Cancer Research UK and the Wellcome Trust.
SD acknowledges the financial support of NHS Research Scotland (NRS), through NHS Lothian.
AJS was supported by a doctoral fellowship from the Cancer Research UK Cambridge Institute and the Mexican National Council of Science and Technology (CONACyT).
Abbreviations
- CD
Crohn’s disease
- dMMR
defective mismatch repair
- FFPE
formalin-fixed paraffin-embedded
- IBD-CRC
inflammatory bowel disease associated colorectal cancer
- InDels
insertions and/or deletions
- IQR
inter-quartile range
- Mb
megabase
- MMR
mismatch repair
- MSI
micro-satellite instability
- MSS
micro-satellite stable
- SNVs
single nucleotide variants
- TCGA
The Cancer Genome Atlas
- TILs
tumor infiltrating lymphocytes
- UC
ulcerative colitis
Footnotes
Exome Sequencing:
All exome sequencing data is available from the European Genome-phenome Archive under accession number EGAS00001001129.
Author Contributions
SD acquired the ethical approval, provided the study samples, clinical data and analysis, created the cancer tissue micro-array, co-coordinated the study (with DJA and MJA), provided intellectual input on the project and interpretation of data, and drafted the manuscript.
KW analysed the sequencing data (including variant calling and validation, somatic copy number alterations, recurrently mutated genes and signatures of mutational processes), generated figures and tables, and co-wrote the manuscript.
MFM conducted the MLHI promoter methylation analysis.
AO conducted the mismatch repair immunohistochemistry and associated analysis.
JH performed the DNA/RNA extractions and WES.
CJB reviewed and approved the IBD-CRC cases.
MLM and AJS conducted the HLA Neo-epitope predictions and critically reviewed the manuscript.
MR and RR performed the Nanostring analysis.
JS provided intellectual input and critically reviewed the manuscript.
DJA provided expert guidance, intellectual input on the project, coordinated and designed experiments with KW, JH, MM, AJS, MR and, RR and critically reviewed the manuscript.
MJA provided expert guidance, intellectual input on the project, reviewed the histology for the IBD-CRC cases, assessed the mismatch repair immunohistochemistry, designed experiments with MM and critically reviewed the manuscript.
Conflict of Interest disclosures:
The authors declare no potential conflicts of interest.
Contributor Information
Shahida Din, NHS Lothian, Gastrointestinal Unit, Western General Hospital, Edinburgh, Scotland, UK.
Kim Wong, Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK.
Mike F Mueller, Division of Pathology, Centre for Comparative Pathology, Edinburgh Cancer Research Centre, Institute of Genetics & Molecular Medicine, Western General Hospital, University of Edinburgh, Edinburgh, Scotland, UK.
Anca Oniscu, NHS Lothian, Department of Molecular Pathology, Laboratory Medicine, Royal Infirmary of Edinburgh, Edinburgh, Scotland, UK.
James Hewinson, Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK.
Catherine J Black, NHS Lothian, Department of Pathology, Western General Hospital, Edinburgh, Scotland, UK.
Martin L Miller, Cancer Research UK Cambridge Institute, University of Cambridge, Li Ka Shing Centre, Robinson Way, Cambridge UK.
Alejandro Jiménez-Sánchez, Cancer Research UK Cambridge Institute, University of Cambridge, Li Ka Shing Centre, Robinson Way, Cambridge UK.
Roy Rabbie, Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK.
Mamunar Rashid, Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK.
Jack Satsangi, Centre for Genomic and Experimental Medicine, University of Edinburgh, Edinburgh, Scotland, UK.
David J Adams, Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK.
Mark J Arends, Division of Pathology, Centre for Comparative Pathology, Cancer Research UK Edinburgh Centre, Institute of Genetics & Molecular Medicine, Western General Hospital, University of Edinburgh, Edinburgh, Scotland, UK.
References
- 1.Sebastian S, Hernandez V, Myrelid P, Kariv R, Tsianos E, Toruner M, et al. Colorectal cancer in inflammatory bowel disease: results of the 3rd ECCO pathogenesis scientific workshop (I) J Crohns Colitis. 2013;8:5–18. doi: 10.1016/j.crohns.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Sanduleanu S, Rutter MD. Interval colorectal cancers in inflammatory bowel disease: the grim statistics and true stories. Gastrointest Endosc Clin N Am. 2014;24:337–348. doi: 10.1016/j.giec.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 3.RCoreTeam. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. [Google Scholar]
- 4.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 6.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biology. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martincorena Ii, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171:1029–1041.e1021. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maruvka YE, Mouw KW, Karlic R, Parasuraman P, Kamburov A, Polak P, et al. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nature Biotechnology. 2017;35:951. doi: 10.1038/nbt.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nature Reviews Cancer. 2004;4:177. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31:3673–3675. doi: 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giannakis M, Mu Xinmeng J, Shukla Sachet A, Qian Zhi R, Cohen O, Nishihara R, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Reports. 2016;15:857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitzer E, Tomlinson I. Replicative DNA polymerase mutations in cancer. Curr Opin Genet Dev. 2014;24:107–113. doi: 10.1016/j.gde.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gailani MR, Bale SJ, Leffell DJ, DiGiovanna JJ, Peck GL, Poliak S, et al. Developmental defects in Gorlin syndrome related to a putative tumor suppressor gene on chromosome 9. Cell. 1992;69:111–117. doi: 10.1016/0092-8674(92)90122-s. [DOI] [PubMed] [Google Scholar]
- 18.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vieira VC, Leonard B, White EA, Starrett GJ, Temiz NA, Lorenz LD, et al. Human Papillomavirus E6 Triggers Upregulation of the Antiviral and Cancer Genomic DNA Deaminase APOBEC3B. mBio. 2014;5:e02234–02214. doi: 10.1128/mBio.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katainen R, Dave K, Pitkanen E, Palin K, Kivioja T, Valimaki N, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 22.Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RKS, et al. Multiple Colorectal Adenomas, Classic Adenomatous Polyposis, and Germ-Line Mutations in MYH. New England Journal of Medicine. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto T, Shimizu T, Takai A, Marusawa H. Exploring the Mechanisms of Gastrointestinal Cancer Development Using Deep Sequencing Analysis. Cancers. 2015;7:1037–1051. doi: 10.3390/cancers7020823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi CR, Bakir IA, Hart AL, Graham TA. Clonal evolution of colorectal cancer in IBD. Nat Rev Gastroenterol Hepatol. 2017 doi: 10.1038/nrgastro.2017.1. [DOI] [PubMed] [Google Scholar]
- 25.The Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46:1264–1266. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koo B-K, van Es JH, van den Born M, Clevers H. Porcupine inhibitor suppresses paracrine Wnt-driven growth of mutant neoplasia. Proceedings of the National Academy of Sciences. 2015;112:7548–7550. doi: 10.1073/pnas.1508113112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira AM, Tuominen I, van Dijk-Bos K, Sanjabi B, van der Sluis T, van der Zee AG, et al. High frequency of RPL22 mutations in microsatellite-unstable colorectal and endometrial tumors. Hum Mutat. 2014;35:1442–1445. doi: 10.1002/humu.22686. [DOI] [PubMed] [Google Scholar]
- 29.Yaeger R, Shah MA, Miller VA, Kelsen JR, Wang K, Heins ZJ, et al. Genomic Alterations Observed in Colitis-Associated Cancers Are Distinct From Those Found in Sporadic Colorectal Cancers and Vary by Type of Inflammatory Bowel Disease. Gastroenterology. 2016;151:278–287 e276. doi: 10.1053/j.gastro.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Liang L, Fang JY, Xu J. Somatic gene copy number alterations in colorectal cancer: new quest for cancer drivers and biomarkers. Oncogene. 2016;35:2011–2019. doi: 10.1038/onc.2015.304. [DOI] [PubMed] [Google Scholar]
- 31.Robles AI, Traverso G, Zhang M, Roberts NJ, Khan MA, Joseph C, et al. Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology. 2016;150:931–943. doi: 10.1053/j.gastro.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shivakumar BM, Chakrabarty S, Rotti H, Seenappa V, Rao L, Geetha V, et al. Comparative analysis of copy number variations in ulcerative colitis associated and sporadic colorectal neoplasia. BMC Cancer. 2016;16:271. doi: 10.1186/s12885-016-2303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cybulski C, Wokolorczyk D, Kladny J, Kurzwaski G, Suchy J, Grabowska E, et al. Germline CHEK2 mutations and colorectal cancer risk: different effects of a missense and truncating mutations? European Journal of Human Genetics. 2007;15:237–241. doi: 10.1038/sj.ejhg.5201734. [DOI] [PubMed] [Google Scholar]
- 35.Young AL, Baysal BE, Deb A, Young WF., Jr Familial malignant catecholamine-secreting paraganglioma with prolonged survival associated with mutation in the succinate dehydrogenase B gene. J Clin Endocrinol Metab. 2002;87:4101–4105. doi: 10.1210/jc.2002-020312. [DOI] [PubMed] [Google Scholar]
- 36.Platten M, Offringa R. Cancer immunotherapy: exploiting neoepitopes. Cell Res. 2015;25:887–888. doi: 10.1038/cr.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bansal P, Sonnenberg A. Risk factors of colorectal cancer in inflammatory bowel disease. Am J Gastroenterol. 1996;91:44–48. [PubMed] [Google Scholar]
- 38.Choi PM, Zelig MP. Similarity of colorectal cancer in Crohn's disease and ulcerative colitis: implications for carcinogenesis and prevention. Gut. 1994;35:950–954. doi: 10.1136/gut.35.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lennerz JK, van der Sloot KWJ, Le LP, Batten JM, Han JY, Fan KC, et al. Colorectal cancer in Crohn’s colitis is comparable to sporadic colorectal cancer. International Journal of Colorectal Disease. 2016;31:973–982. doi: 10.1007/s00384-016-2574-x. [DOI] [PubMed] [Google Scholar]
- 40.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 41.Molecular testing strategies for Lynch syndrome in people with colorectal cancer. 2017 https://www.nice.org.uk/guidance/dg27/documents/diagnostics-consultation-document.
- 42.Reynolds IS, O’Toole A, Deasy J, McNamara DA, Burke JP. A meta-analysis of the clinicopathological characteristics and survival outcomes of inflammatory bowel disease associated colorectal cancer. International Journal of Colorectal Disease. 2017;32:443–451. doi: 10.1007/s00384-017-2754-3. [DOI] [PubMed] [Google Scholar]
- 43.Sinicrope FA, Sargent DJ. Molecular Pathways: Microsatellite Instability in Colorectal Cancer: Prognostic, Predictive, and Therapeutic Implications. Clinical Cancer Research. 2012;18:1506–1512. doi: 10.1158/1078-0432.CCR-11-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.FDA approves first cancer treatment for any solid tumor with a specific genetic feature. 2017 https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm.
- 46.Michot JM, Bigenwald C, Champiat S, Collins M, Carbonnel F, Postel-Vinay S, et al. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. European Journal of Cancer. 2016;54:139–148. doi: 10.1016/j.ejca.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 47.Johnson DB, Sullivan RJ, Ott PA, et al. Ipilimumab therapy in patients with advanced melanoma and preexisting autoimmune disorders. JAMA Oncology. 2016;2:234–240. doi: 10.1001/jamaoncol.2015.4368. [DOI] [PubMed] [Google Scholar]
- 48.Evaluation of Genomic Applications in Practice and Prevention Working, G. Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genetics in Medicine. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snowsill T, Huxley N, Hoyle M, Jones-Hughes T, Coelho H, Cooper C, et al. A model-based assessment of the cost-utility of strategies to identify Lynch syndrome in early-onset colorectal cancer patients. BMC Cancer. 2015;15:313. doi: 10.1186/s12885-015-1254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cristescu R, Lee J, Nebozhyn M, Kim K, T J. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nature Medicine. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.