Abstract
Tumors cells reprogram their metabolism to fuel rapid growth. The ability to trace nutrient fluxes in the context of specific alterations has provided new mechanistic insight into the process of oncogenic transformation. A broad array of complementary genetic, epigenetic, transcriptional and translational mechanisms has been identified, revealing a metabolic landscape of cancer. However, cancer metabolism is not a static or uniform process, including within a single tumor. Tumor cells adapt to changing environmental conditions, profoundly shaping the enzymatic dependencies of individual cells. The underlying molecular mechanisms of adaptation, and the specific interactions between tumor genotype, oncogenic signaling, and tissue/biochemical context, remain incompletely understood. In this review, we examine dynamic aspects of how metabolic dependencies develop in cancer, shaped both by genotype and biochemical environment, and review how these interlaced processes generate targetable metabolic vulnerabilities.
Keywords: Cancer metabolism, Metabolic co-dependency, Oncogenic signaling, Tissue context, ecDNA, Heterogeneity
1. Introduction
Altered cellular metabolism is one of the most characteristic phenotypic changes that occurs during the process of tumor formation, progression and drug resistance. Beginning with the pioneering work of Otto Warburg in the 1940’s and culminating in significant and rapidly accelerating progress in the past decade, a picture has begun to emerge of how cancer cells take up and use nutrients to drive cell autonomous growth and rapid adaptation to changing conditions. This line of inquiry has: 1) identified new drug targets; 2) shed light on interplay between mutated genes and altered metabolism; 3) highlighted the diversity of genetic, epigenetic, transcriptional, translational, and post-translational mechanisms that regulate tumor cell metabolism; 4) provided new insight into the flexibility of metabolic pathways that cancer cells use and 5) revealed the heterogeneity of the metabolic pathways used in different parts of a tumor [1-3]. These discoveries have moved the field beyond the initial phase of characterizing the metabolic landscape of tumor cells, into a new and exciting era of trying to understand how it works, with the ultimate goal of using this information to develop more effective cancer treatments.
Clear lessons have begun to emerge, framing the challenge ahead. Cancer cells stop behaving like normal cells in a multicellular organism and start behaving like single celled entities [3, 4]. A molecular basis for this switch to cell autonomous metabolism and proliferation has also become clearer. Gain of function mutation and gene amplification of key components of the growth factor system, the very instructional cues that normal cells require for nutrient uptake and utilization, are frequent events in cancers of almost all histological types, providing a genetic basis for cell autonomous metabolism. How specific genetic alterations interact with the tumor microenvironment remains an open question. How does the biochemical milieu interact with corrupted growth factor signaling pathways to influence metabolic fluxes? How do they enable rapid adaption to changing conditions? What vulnerabilities do these adaptations expose and can they be targeted? The challenge in the field is moving from the critical step of developing a biochemical map of cancer metabolism, towards a dynamic adaptive view of how tumor metabolism changes over the life of a cancer and in response to local cues, and how it can be therapeutically exploited. In this review, we focus on the dynamic aspects of how metabolic dependencies develop in a tumor shaped both by genotype and biochemical environment, and how these interlaced processes generate targetable metabolic vulnerabilities.
2. Gene amplification, deletion and mutation drive metabolic phenotypes
Nearly 90 years ago, Otto Warburg [5] showed that most cancer cells avidly consume glucose and convert it into lactate even in the presence of abundant oxygen, unlike normal cells that metabolize glucose to carbon dioxide via mitochondrial oxidative phosphorylation. The Warburg effect is a biochemical adaptation that benefits cancer cells by enabling them to generate the metabolic ingredients needed for building biomass for cell proliferation, while still yielding sufficient energy to power cellular reactions. Warburg’s observation was the first, and probably the seminal demonstration that cancer cells have an altered metabolic phenotype. However, tumor cells also display major shifts in other metabolic facets, including in amino acid, nucleotide and lipid metabolism. Initially, a molecular basis for these biochemical shifts was not clear, but extensive research, particularly over the past 10 years, a molecular picture has emerged– common genetic alterations in tumor cells drive specific metabolic shifts.
The first major lesson to emerge, is that common genetic alterations in genes that encode core proteins in the growth factor signaling cascade, play a central role in cancer metabolic reprogramming. In multicellular organisms, growth factor signaling instructs cells to take up nutrients and direct them towards biomass generation, coupling nutrient flux with the transcriptional, translational and post-translational programs that regulate cell proliferation. In cancer, amplification and gain of function mutations of genes whose protein products are key components of the growth factor signaling system, including cell surface receptor tyrosine kinases such as EGFR, and downstream effectors PI3K and Akt, is a relatively common event, providing a mechanistic basis for the transition of tumors to cell autonomous nutrition. These activating lesions are often complemented by genetic deletion and/or loss of function mutations of PTEN, a suppressor of PI3K signaling (Fig. 1). This signaling cascade regulates the level and activity of nutrient uptake, such as levels of glucose transporters and helps determine which biosynthetic pathways will be utilized [6-12]. Other genes whose protein products are involved in nutrient uptake and utilization, including the signaling protein Ras and the transcription factor c-Myc, are also commonly amplified and or mutated in cancer [13, 14], and determine other nutrient fluxes including glutamine and other amino acid uptake [15-21] and opportunistic pathways that have recently been described [22-25] (Fig.1).
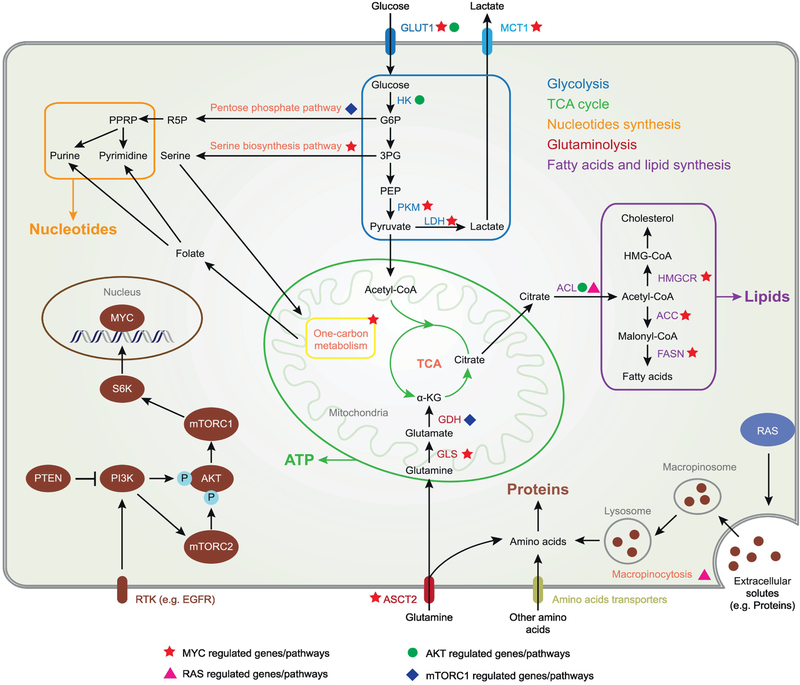
Fig. 1. Oncogenic signaling reprograms metabolism of cancer cells to support rapid cell growth.
PI3K-AKT-mTOR signal promotes glucose uptake, glycolysis and the flux of glucose carbon into fatty acids, lipid and nucleotides. Transcriptional factor MYC further enhances the glycolysis while also facilitates glutamine uptake and utilization, fatty acids, lipid and nucleotides de novo synthesis. Ras induces macropinocytosis as an alternative way of amino acids uptake. Filled green triangle target genes/pathways for PI3K; filed green circle target genes/pathways for AKT; filled green square target genes/ pathways for mTORC1; filled orange star target genes/pathways for MYC; filled pink triangle target process for Ras. GLUT1, Glucose transporter 1; MCT1, Monocarboxylate transporters 1; HK, Hexokinase; PKM, Pyruvate kinase; LDH, Lactate dehydrogenase; PDH, Pyruvate dehydrogenase; GDH, Glutamate dehydrogenase; GLS, Glutaminase; ACL, ATP citrate lyase; HMGCR, Hydroxymethylglutaryl-CoA reductase; ACC, Acetyl-CoA carboxylase; FASN, Fatty acid synthase; ASCT2, ASC amino acid transporter 2; G6P, Glucose 6-phosphate; 3PG, Glycerate 3-phosphate; PEP, Phosphoenolpyruvate; R5P, Ribose 5-phosphate; PPRP, Phosphoribosyl pyrophosphate; α-KG, α-ketoglutarate.
The second major lesson is that common genetic alterations in tumor cells co-opt signaling networks and transcriptional programs that control cellular metabolism through cooperative and tightly coordinated interactions. For example, recent work in glioblastoma, which is highly glycolytic [26], demonstrates how a genetic alteration in EGFR, which occurs in a high fraction of glioblastoma [27, 28], drives glycolysis through three complementary pathways that integrate EGFR signaling through the PI3K pathway, with dysregulation of c-Myc. First, EGFRvIII, through and Akt-mTORC1-dependent pathway, leads to the splicing of the Myc interacting partner Max, generating a gain of function protein, Delta Max, that potently drives glycolysis in a c-Myc dependent fashion [29]. Second, EGFRvIII remodels the enhancer landscape of glioblastoma cells, potently driving SOX9 and FOXG1 to regulate c-Myc-dependent transcription [30]. Lastly, EGFRvIII promotes glycolysis in tumor cells through mTORC2-dependent acetylation of FoxO1 and subsequent regulation of c-Myc protein levels [31].
EGFRvIII-dependent metabolic reprogramming is not restricted to glycolysis. Cancer cells require not only glucose, but also amino acids, nucleotides and lipids in order to proliferate. Recent work sheds some light on how the same genetic alterations that regulate glycolysis, also coordinately regulate these other metabolic processes. For example, EGFRvIII coordinately regulates fatty acid synthesis through and Akt-SREBP1-dependent mechanism [32], and also controls intratumor cholesterol levels via an LDLR-dependent mechanism [33, 34]. Considering the recent work linking c-Myc and mTORC1 with one carbon metabolism, amino acid regulation and nucleotide biosynthesis [35-38], it is possible that further studies will reveal an even more tightly regulated integration of a diverse set of metabolic events downstream of EGFRvIII in glioblastoma.
3. Metabolic co-dependency shaped by environment
A purely genotype-based view of cancer metabolism is missing half the picture. The metabolic phenotype of tumors is determined not only by the genotype of the cancer cells that it contains, but also by non-cell-autonomous environmental factors, such as nutrient availability, tissue context and biochemical environment. The interactions between these components determines the tumor’s metabolic preferences and range of adaptive possibilities. It also generates metabolic dependencies of cancer cells that may be actionable drug targets.
3.1. Nutrient availability in microenvironment determines metabolic dependencies
Environments supply cells diverse nutrients, such as glucose, amino acids, lipids, O2, and macromolecules, which the cells then use to produce ATP, synthesize macromolecules and modulate redox state for cell survival and proliferation (Fig. 2A). Cancer cells flux abundant nutrients, such as glucose, glutamine and fatty acids, for ATP and Intermediate metabolites[4]. Except for the essential amino acids and fatty acids, cancer cells can synthesis most of required metabolites from the intermediates. Lipids and amino acids of cancer cells can be achieved through synthesis from intermediates or import from environments. Nucleic acids of cancer cells are synthesized from glucose, glutamine and some non-essential amino acids. Under nutrient deprivation condition, cancer cells gain metabolic flexibility by remodeling their metabolic pathways to use alternative nutrient source from environment for cell survival and proliferation, which allows cancer cells to bypass the nutrient limitation and renders them dependent on available nutrients [39]. The tumor cell must balance what is available in the environment vs. what must be synthesized, and at what energetic cost. Thus, nutrient availability in microenvironment is a major factor in determining the metabolic state of tumor cells and its potential metabolic vulnerabilities.
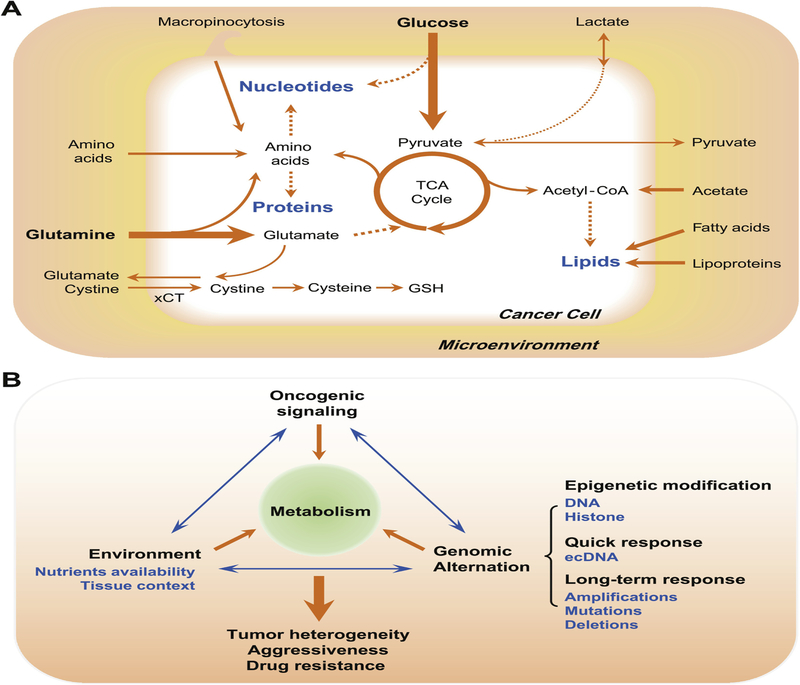
Fig. 2. Metabolic co-dependency of cancer cells shaped by environment and the interactions between environment, oncogenic signaling and genomic alternations.
(A) Nutrient availability in the microenvironment determines metabolic dependencies of cancer cells. Cancer cells can synthesize proteins, lipids and nucleotides from different sources of nutrients, either through directly uptake from the environment or through synthesis from intermediate metabolites. (B) The interactions between environment, oncogenic signaling and genomic alternation, select favorable metabolic profiles and oncogenic signatures for tumor cells, contributing to tumor heterogeneity, tumor aggressiveness and therapeutic resistance. Metabolic stress from environment and oncogenic signaling influents DNA and histone modification of tumor cells. As a quick response, extrachromosomal DNA (ecDNA) profoundly contributes to accelerated tumor evolution and intratumoral genetic heterogeneity. Tumors also accumulate amplifications, mutations and deletions in their genome, which alters the metabolic and signaling profiles of cancer cells.
The richness of potential biochemical interactions, the ability of enzymes to work in two directions, and the potentially wide array of metabolites make it very difficult to predict what a cancer cell will do, without considering its environmental context. For example, acetate and lactate are important alternative carbon sources for cancer cells. Glucose and glutamine, two main nutrients and substrates for metabolism, provides cancer cells major source of carbon. Acetyl-CoA, a precursor for fatty acid and cholesterol de novo synthesis, can be mainly acquired from glucose and glutamine though glycolysis and α-ketoglutarate, and represents a central node of carbon metabolism. Acetate was found as an important bioenergetic substrate for human glioblastoma and brain metastases [40]. Under low-oxygen and lipid-depleted conditions, cancer cells activate their utilization of acetate from microenvironments, providing one alternative source of acetyl-CoA for lipid synthesis and histone acetylation [41]. Depletion of Acetyl-CoA Synthetase 2 (ACSS2), one main enzyme to convert acetate to acetyl-coA and upregulated in a large proportion of tumors, inhibits cancer cells growth and suppresses tumor development [42]. Lactate was also reported as an important carbon source to feed tricarboxylic acid (TCA) cycle for human non-small-cell lung tumors [43]. Consistent with this finding, another systematical study on the fluxes of circulating metabolites in mice reveals that circulating lactate is the primary source of carbon for the TCA cycle and exceeds the contribution of glucose to TCA metabolism in most normal tissues and the genetically engineered lung and pancreatic cancer tumors [44].
Besides glutamate, the availability of other amino acids in the environment also generate the metabolic dependency of cancer cells. For example, many cancers depend on extracellular serine availability, one central precursor for biosynthetic metabolism [45]. Besides directly offering the serine headgroup for cellular metabolism, serine is a precursor of glycine and cysteine, which are required for the synthesis of purine and glutathione (GSH). Serine also supplies carbon source of one-carbon metabolism for thymidine, purine and methionine synthesis. Serine depletion impairs glycolysis and induces oxidative stress in p53-deficient cancer cells, leading to a metabolic vulnerability for p53-deficient cancer cells [46]. Serine contributes to nucleotide synthesis through glycine. A high throughput analysis on metabolic profiles of NCI-60 cancer cell lines found that glycine consumption is strongly correlated with cell proliferating rate and antagonizing glycine uptake impairs cancer cell proliferation[47], indicating a metabolic dependency for rapid proliferating cancer cells on extracellular glycine. Aspartate is required for both purine and pyrimidine de novo synthesis. As Blood aspartate level in human is very low [48] and uptake of aspartate may be not sufficient for cell proliferation, a primary function of respiration in proliferating cell was found to support aspartate biosynthesis, ensuring cancer cell survival and proliferation [49, 50].
3.2. Tissue context shapes metabolic dependencies
Tissue context provides a unique metabolic environment for cancer cells with specific nutrient availability, extracellular matrix and interactions with stromal cells, which creates dependencies on metabolism for cancer cells. Stromal cells and other cells within the tumor microenvironment are genetically stable and thus provides a stable metabolic environment for cancer cells [51]. Kras-driven Non-Small Cell lung tumors use nutrients differently for TCA cycle than cultured cancer cells [52], indicating in vivo tissue environment determines nutrient dependency of cancer cells. Tissue context determines metabolic dependencies of cancer cells. For example, ovarian tumor cells near the adipocytes prefer to directly acquire fatty acids from nearby adipocytes through upregulation of the fatty acid binding protein 4 (FABP4) [53]. A systematically analysis across 20 different cancer types indicates cancers undergo a tissue-specific metabolic rewiring of metabolic genes expression [54].
Although with the same oncogenic drivers, tumors arising from different tissues may have totally different metabolic dependencies. Oncogenic mutations, such as mutations in KRAS, TP53 and EGFR, are common genetic drivers found in many cancers. Interestingly, Kras-driven pancreatic ductal adenocarcinoma (PDAC) and non-small cell lung carcinoma (NSCLC) were found to use branched-chain amino acids (BCAAs) differently in mice model [55]. NSCLC tumors incorporate free BCAAs as nitrogen source whereas PDAC tumors decreases their uptake of BCAAs from microenvironment. Blocking the enzymes for BCAAs use specifically impairs NSCLC tumor formation, suggesting tissue of origin determines the metabolic requirements of cancer cells and targeting metabolic dependencies based on the tissue context may be a potential therapeutic strategy for certain cancers. Myc-induced lung tumors and liver tumors also show tissue-specific difference in metabolism [56]. By using intraoperative 13C-glucose infusions in human Non-small cell lung tumors, Hensley et al. reveals the regional difference of tumor nutrient utilization that less perfuse regions of human NSCLC tumors have elevated glucose oxidation and highly perfused regions use non-glucose alternative nutrients for oxidation[57], suggesting the tumor metabolic heterogeneity between patients and within individual tumors and highlighting importance of microenvironment in metabolic dependency. Furthermore, metastatic cancer cells in bone, lung and liver from the same primary breast cancer cells were found to display unique metabolic signatures on glycolysis and oxidative phosphorylation (OXPHOS) [58], which indicates the metabolic dependency of cancer metastasis may be defined by the tissue of metastasis sites.
Tissue environment can dictate metabolic dependencies across tumor types with distinct oncogenic signatures. An example comes from brain tumor and brain metastasis. Brain contains over 20 % of unesterified cholesterol of whole human body [59]. The Blood-brain barrier (BBB) generates a separated cholesterol pool for brain, in which astrocytes are response for synthesizing most of cholesterol in brain from glucose, glutamine and acetate [59]. In contrast to normal astrocytes, glioblastoma cells suppress cholesterol and liver X receptor (LXR) ligand synthesis, and are largely reliant on LDL-derived cholesterol uptake for survival, suggesting a metabolic dependency on cholesterol [33, 34]. Targeting this dependency by activating LXR with LXR-623, a highly brain-penetrant synthetic LXR ligand, selectively kills brain cancer cells and suppresses tumor growth in a patient-derived glioblastoma (GBM) xenograft model [34]. More exciting, the brain metastatic breast cancer cell lines were found also highly sensitive to LXR-623, indicating tissue microenvironments can shape the metabolism of tumors with different genetic backgrounds, offering a broad targeting strategy for tumors from the same tissue origin. The metabolic similarities identified between human KRAS-driven lung tumors and EGFR-driven lung tumors [52, 57], all point to the same direction that tissue context plays an important role in metabolic rewiring and dependency.
3.3. Oncogene and environment interactions select favorable metabolic co-dependencies
The interactions between oncogenic signaling and environment, select favorable metabolic pathways and oncogenic signatures for tumor cells, contributing to tumor heterogeneity, tumor aggressiveness and therapeutic resistance (Fig. 2B).
Environment coordinates with oncogenic signaling to regulate the reversible posttranslational protein modification. Acetyl-CoA links environmental nutrients to oncogenic signaling through protein acetylation [60]. Acetyl-CoA also functions as a central metabolite in carbon metabolism of cancer cells, glucose or acetate derived acetyl-CoA can directly modify the growth factor signaling in cancers by providing acetyl group for protein acetylation. Glucose-dependent Rictor acetylation promotes mTORC2 signaling, ensuring that growth factor receptor signaling leads to cellular proliferation in tumor cells only when there are sufficient levels of glucose to support it, although acetate can provide an alternative route for Acetyl-CoA generation to drive growth [61]. This adaptation also potentially renders tumor cells relatively resistant to EGFR-PI3K-Akt axis based targeted therapies [61]. Although the acetylation of Skp2 and MDM2 dictates their oncogenic function [62, 63], it is unclear whether environmental nutrients also affect these comportments in oncogenic signaling pathways through acetyl-CoA mediated acetylation. O-GlcNAcylation is another reversible post-translational modification that can dynamically response to environmental glucose concentration [64]. Increased glucose flux of cancer cells elevates cellular UDP-GlcNAc level and cellular O-GlcNAcylation in oncogenic transcriptional factors [65]., including c-Myc [66], p53 [67], β-catenin [68], and NF-kB [69], which promotes tumor development and progression in broad types of cancers. Elevated glucose import of cancer cells can also activate SREBP activity and downstream lipogenesis through SCAP N-glycosylation in tumors [70].
Nutrient sensors mediate the interactions between intrinsic and extrinsic factors of cancer cells. The cystine-glutamate antiporter system xc− (xCT) imports cystine and secrets glutamate from extracellular environment [71]. Disruption of the cystine-glutamate antiporter xCT improves cell viability under glucose-deficient conditions [72, 73]. Growth factor signaling regulates metabolism of cancer cells in response to rapid changing microenvironment. mTORC2 was found as an important regulator of glutamate and glutathione metabolism in cancer through directly phosphorylating the cystine-glutamate antiporter xCT on serine 26 [74]. Interestingly, xCT was also shown to be phosphorylated by AKT at the same site serine 26 to promotes the methionine dependency in breast cancer cells [75], which could be a shared mechanism with mTORC2 from growth factor signaling in cancer cells. Inhibition of mTORC2 enhances xCT-mediated cystine uptake and glutathione synthesis, which offers an adaptation mechanism to metabolic stress [74]. Hypoxia-inducible factor 1 and 2 (HIF1/2) mediate the hypoxic response of cancer cells [76, 77]. Under normoxic conditions, large amounts of glutamate secreted by triple-negative breast cancers, in turn ensures oncogenic HIF1 stability through inhibition of xCT antiporter and HIF prolyl-hydroxylases [78], providing another link between environment metabolites and oncogenic signaling. mTORC1 signaling is highly activated in many cancers [79, 80]. As a group of intracellular amino acid sensors has been identified to activate mTORC1 signaling [81], it is interesting to understand whether extracellular amino acids will affect tumor progression by directly modulating mTORC1 activity of tumor cells in the in vivo conditions.
Environmental factors also contributes to epigenetic reprograming of cancer cells through metabolism, which is exploited by cancer cells to modulate the expression levels of oncogenes and tumor suppressors. Cellular metabolism directly provides methyl group and acetyl group for cancer cell epigenetic modification. Aberrant DNA methylation has been found as an important epigenetic events occurring in different types of cancers [82]. S-adenosyl methionine (SAM), the primary methyl group donor for DNA or histone methylation, can be synthesized from methionine in one-carbon metabolism [83]. In a pan-cancer analysis of The Cancer Genome Atlas (TCGA) data, the expression of methylation cycle and related serine, glycine, one-carbon (SGOC) network genes was found to highly associate with the tumor DNA methylation and patient survival [84]. As serine metabolism supports the methionine cycle, serine starvation decreases the DNA/RNA methylation of cancer cells [85]. Methionine restriction diet alters methionine metabolism and histone methylation in human liver [86], highlighting the determination of nutrient availability in epigenetic modification in physiological condition. ATP citrate lyase (ACL), the enzyme that converts glucose-derived citrate in to acetyl-CoA, promotes histone acetylation and confirms the transcriptional regulation from growth factor signaling [87]. Tumor acidosis induces metabolic rewiring toward fatty acid oxidation in cancer cells through mitochondrial hyperacetylation and histone deacetylation [88]. Together, these results suggest environmental nutrients influence epigenetic reprograming through metabolism.
In response to metabolic stress from environment and oncogenic signaling, tumor cells bearing the genomic alternations with the greatest fitness for the environment will be selected (Fig. 2B). As a consequence, it is possible that cells within different regions of a tumor may be exposed to distinct microenvironments, influencing their genetic complement, potentially contributing to metabolic heterogeneity within a tumor. This diversity confers a considerable advantage to tumors cells, as phenotypic heterogeneity is the fuel for natural selection and enhanced diversity increases the likelihood of finding a tumor cell that is optimally fit for its environment no matter what those conditions might be or how frequently they may change. Further studies to elucidate the transcriptional and epigenetic mechanisms will be required. In addition, the link between tumor genetic heterogeneity and metabolic heterogeneity needs to be further examined. Tumors accumulate amplifications, mutations and deletions in their genome [89, 90], which alters the metabolic and signaling profiles of cancer cells. The recent finding that nearly 40% of cancers contain oncogenes amplified on extrachromosomal DNA (ecDNA); that it profoundly contributes to accelerated tumor evolution and intratumoral genetic heterogeneity, and that some of the most commonly amplified oncogenes on ecDNA are known drivers of altered cellular metabolism, including EGFR and c-Myc, raises the possibility that ecDNA oncogene amplification contributes to metabolic heterogeneity in cancer [91]. It may also help explain how tumor cells might change oncogene copy number, and metabolic pathway activation relatively quickly. GBM cells harbor high levels of oncogenic EGFRvIII on ecDNA [91, 92]. In response to tyrosine kinase inhibitors treatment and withdrawal, GBM cells change their expression of EGFRvIII protein by directly altering EGFRvIII positive ecDNA elements, which promotes the cancer drug resistance [92]. It is also noted that PDX tumor samples have much higher ecDNA counts than cultured tumor cell lines [91], and that copy number of oncogene amplified on ecDNA can change relatively quickly in tumors in culture, raising the possibility that the local environment influences oncogene copy number and possibly metabolic phenotype.
4. Targeting metabolic co-dependency in cancer
4.1. Targeting mutated metabolic gene
To date, eight genes (FH, SDHA, SDHB, SDHC, SDHD, SDHAF2, IDH1 and IDH2) encoding three types of enzymes in TCA cycle, are found to be mutated in a portion of cancer [93]. The first reported mutated metabolic gene is SDHD, encoding succinate dehydrogenase (SDH) in familial paraganglioma [94]. Subsequently, all SDH subunit genes SDHA, SDHB, SDHC and the assembly factor SDHAF2, were all found to be mutated in a subset of tumors including paragangliomas, gastric stromal sarcoma and pheochromocytoma, as documented in Leiden Open Variation Database [95], and in renal cell carcinoma [96] and papillary thyroid carcinoma [97]. Mutations of FH gene (encoding fumarate hydratase) are found in inherited uterine fibroids, skin leiomyomata, papillary renal cell cancer [98], and leydig cell tumors [99].
Mutations of IDH1 and IDH2, encoding isocitrate dehydrogenases, are the most well characterized mutated metabolic genes in cancer. Recurrent heterozygous IDH1 mutation was first reported in glioblastoma (GBM), affecting its amino residue R132, majorly in the form of R132H [100]. It was then characterized as an early mutation in lower grade astrocytoma and secondary GBM, but rare in primary GBM. Together with IDH2 heterozygous mutation, mainly affecting R172 residue, IDH status is now used as an important diagnostic marker for brain tumors [101]. Heterozygous mutations of R132 in IDH1 and R172 in IDH2 have also been found in acute myeloid leukemia, with the majorly in the cytogenetically normal subtype [102].
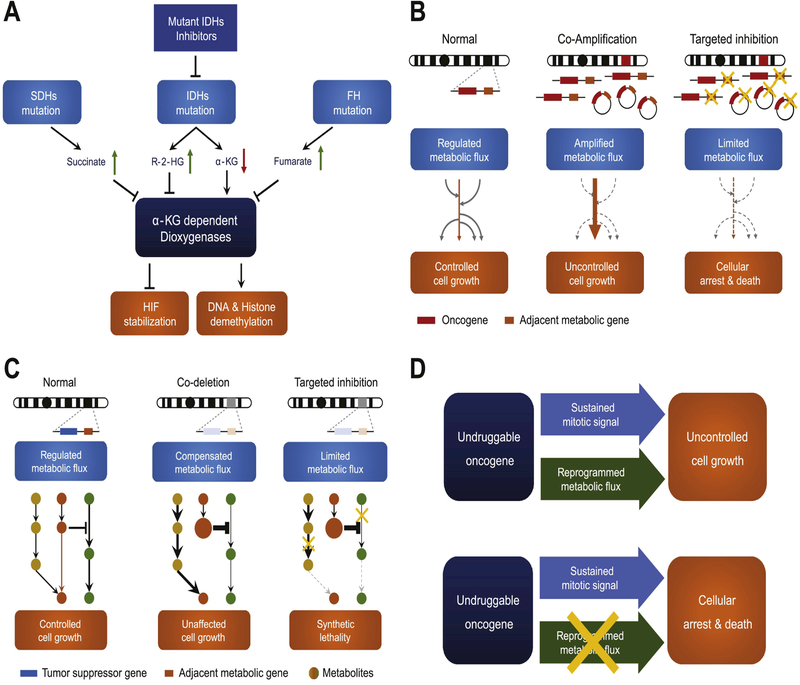
Germline or somatic mutations of SDHs and FH often result in loss or reduction of enzyme function, leading to the accumulation of their substrates, while mutated IDH1 and IDH2 acquire a new function to convert α-ketoglutarate (α-KG) into oncogenic metabolite R(−)-2-hydroxyglutarate (R-2-HG). Mutations of these eight metabolic genes seem to have a convergent consequence that disrupts α-KG-dependent dioxygenases process (Fig. 3A). Produced in TCA cycle, α-KG is not only a metabolite participating in energy and substance production, but also a key player of regulating signaling pathway and epigenetic modification. HIF-α prolyl hydroxylase (PHD) utilizes α-KG as a co-substrate to destabilize HIF, in the presence of oxygen, iron and ascorbate. In SDHs mutated cancer, accumulated succinate impedes PHD activity, leading to stabilization of HIF and promote oncogenesis [103]. Excessive fumarate resulted from FH mutation can also achieve the same effect on stabilizing HIF by directly inhibiting PHD [104]. R-2-HG is structurally similar to α-KG and play as a competitive inhibitor of α-KG-dependent enzymes, including histone lysine demethylases, and ten-eleven translocation DNA hydroxylases involved in DNA demethylation [105], and therefore reprograms the epigenome [106]. But surprisingly, R-2-HG acts as an agonist of PHD to maintain the low level of HIF, and benefits leukemia and low grade glioma development [107, 108].
Fig. 3. Targeting metabolic co-dependency in cancer.
(A) Mutations of SDHs and FH genes lead to accumulation of their individual substrates succinate and fumarate. Mutated IDHs acquire new function to convert α-KG into R-2-HG. These metabolites can inhibit α-KG dependent dioxygenases, and subsequently abolish HIF de-stabilization and DNA, histone demethylation. (B) When a metabolic gene recurrently co-amplified with a nearby oncogene, either in the form of chromosomal or extrachromosomal DNA, can drive the metabolic flux to a specific direction to aid malignant transformation, creating a metabolic co-dependency. Hitting co-amplified metabolic gene is a potential strategy to halt cancer cells. (C) Metabolic flux in normal cells comprises parallel or redundant pathways with feedback regulation mechanism, allowing metabolic plasticity. Metabolic gene co-deletion with a tumor suppressor impairs the plasticity, creating the potential to exploit synthetic lethality. (D) Oncogenes confer uncontrolled cell growth by providing sustained mitotic signal and reprogramming metabolism. Targeting reprogrammed metabolism becomes an alternative strategy when oncogene is not ideally druggable.
Preclinical studies of mutant IDH1/2 inhibitors have shown promising results. IDH1-R132H mutant homodimer inhibitor AGI-5198 can delay glioma cells growth and promote differentiation [109]. Targeting IDH2-R140Q suppresses the growth of patient-derived leukemia cells and also promote differentiation [110]. Currently, a series of compounds targeting mutant IDHs for cancer treatment are under Phase I/II clinical trials [111].
4.2. Targeting co-amplified metabolic genes
More than 60% of all human genes are annotated to involve in “metabolic process” according to gene ontology, but so far, only 8 major metabolic genes exclusively in TCA cycle were found to be recurrently mutated in a small fraction of tumors. In fact, most metabolic genes in cancer are affected by somatic copy number alteration (SCNA). One amplified genomic region usually carries multiple genes. Except for the canonical oncogenic drivers, the nearby co-amplified genes have so far been less evaluated, and usually defined as passenger genes -- the term to describe a gene that does not have any effect on the tumor fitness. However, this notion needs to be reconsidered.
How frequently does a metabolic gene co-amplify with an oncogenic driver? In an SCNA pattern analysis of The Cancer Genome Atlas (TCGA) Pan-Cancer data set, 70 peak regions of recurrent amplifications have been identified. Of the 37 peak regions, the most associated functional features are “histone”, “cytochrome”, “mitochondrial” and “acetyltransferase” [112]. In another pan-cancer analysis of >6,500 tumor and normal samples, 44 metabolic genes were identified to exhibit frequent somatic copy number gains or amplifications [113]. These findings strongly suggest that, in cancer, metabolic genes amplification is also a hotspot event, and usually accompany with the canonical oncogenic drivers. The non-stochastic pattern also suggests that, those co-amplified metabolic genes could potentially aid oncogenic drivers by reprogramming the metabolic pathways to confer survival and growth advantage, thus having positively selected during tumor development. In fact, gene set enrichment analysis on the SCNA of metabolic genes has revealed a hypoxia responding signature [113], and a glycolytic phenotype which strongly associating with glucose uptake and de novo nucleotide synthesis [90], enabling rapid cellular growth under selection the pressure of a relatively hypoxic microenvironment.
Given that co-amplified metabolic genes are shifting the metabolic flux in cancer, metabolic dependency is thus created, opening a therapeutic window (Fig. 3B). A recently research highlight the significance of targeting co-amplified metabolic driver. Ubiquitin-specific peptidase belongs to deubiquitinase family that deubiquitinates target proteins to regulate protein turnover, trafficking and cell signaling. USP13 was reported to be co-amplified with PIK3CA in 3q26 locus in 29.3% of high-grade serous ovarian cancers. It functions as a master metabolic regulator of glutaminolysis and mitochondrial function by directly deubiquitinates ACL and oxoglutarate dehydrogenase (OGDH), thus promoting ovarian cancer progression. Targeting USP13 by RNAi suppresses cancer growth in vitro and in vivo [114].
Another example comes from the discovery of metabolic genes co-amplified with MYC, comprising SQLE, PYCRL, TSTA3, CYC1 and SLC39A4. SQLE gene encodes squalene monooxygenase in sterol biosynthesis pathway. Inhibition of SQLE by compound NB-598 was shown to suppress cancer cell survival in vitro [113]. The other 4 co-amplified metabolic genes have not yet been deeply studied, especially on whether they could cooperate with MYC to promote oncogenic function, and whether targeted inhibition could have selective therapeutic effect in MYC-driven cancer. Nonetheless, evidence has shown that SLC39A4 can enhance cell migration, confer cisplatin resistance, and associate with poor survival in non-small cell lung cancer [115]. The mechanism of MYC-promoted proline biosynthesis has also been reported by which up-regulating PYCRL and its isozymes [116].
4.3. Exploiting synthetic lethality in cancer with co-deleted metabolic gene
Deep deletion of tumor suppressors such as CDKN2A/2B, PTEN, and RB1, is wide-spread across cancer types. Like oncogene amplification, deletion of a genomic locus containing tumor suppressor may involve adjacent metabolic genes. Loss-of-function of a metabolic gene, either by mutation or deletion, sometimes results in metabolic deficiency and reduced fitness. But how do cancers develop a mechanism to avoid negative selection?
The most possible mechanism is that complementary metabolic pathways are adopted to compensate the deficiency (Fig. 3C). In the cases of SDH mutant, which could lead to completely loss of SDH activity to convert succinate to fumarate, and to reduce ubiquinone into ubiquinol, apparently it is not critically lethal to the cell, but instead tumor-prone. It is possible that anaplerotic reactions can be employed to supplement necessary ingredients to the TCA cycle. And as long as the mitochondrial complex I is still fully functional, ubiquinol can still be generated and passed to complex III for the following electron transportation.
In line with this inference, it also strongly suggests that, tumors with a certain metabolic deficiency will have higher dependency on the complementary metabolism gene or pathway, therefore creating a rational targeting strategy (Fig. 3C). One representative example comes from the studies of ME2 co-deletion with SMAD4. Tumor suppressor SMAD4 is frequently inactivated in PDAC tumors by mutation or deletion, facilitating KRAS-driven tumorigenesis [117]. In the cases of SMAD4 homozygous deletion, its neighboring gene ME2 (encoding malic enzyme 2) is co-deleted in more than a half of the samples. ME2 is a critical enzyme in TCA cycle, converting malate to pyruvate and generating NADH from NAD+ in mitochondria. A previous study has suggested that ME2 is highly expressed across cancer types, and knocking down ME2 can suppress tumor growth [118]. But how do PDACs manage to survive and expand under the context of ME2 deficiency? Malic enzymes have three isozymes, with ME1 located in cytosol and ME2/3 in mitochondria. In the ME2-null PDAC, NADP+− dependent isozyme gene ME3 is compensatorily up-regulated to maintain the metabolic flux in the mitochondria. Therefore, depletion ME3 is capable of inducing mitochondrial defect and reactive oxygen species (ROS) overload in ME2-null cancer, leading to synthetic lethality [119].
In the ME2-null context, ME3 is the only complementary gene to support cancer metabolism. Thus, targeting ME3 can achieve substantial effect. However, when complementary routes are ample, hitting these pathways may not show satisfying outcome. Nonetheless, in particular genetic background, other metabolic vulnerability can still be exploited.
Homozygous deletion of MTAP, encoding methylthioadenosine phosphorylase, is frequently seen in human tumors, as the gene body is only ~32 kb away from CDKN2A in human genome. MTAP catalyzes substrate S-methyl-5'-thioadenosine (MTA) to S-methyl-5-thio-alpha-D-ribose 1-phosphate, playing a major role in adenine and methionine salvage pathway. MTAP deletion seems to be silent, as two other two-step reactions can compensate the deficiency according to the KEGG pathway database, though with slower metabolic rate. By RNAi screening, two groups have independently found that, MTAP-null tumors are highly vulnerable to protein arginine methyltransferase 5 (PRMT5) depletion [120, 121]. In MTAP-deficient tumor, its substrate MTA accumulates. In line with the previous findings suggesting that MTA can act as a protein methyltransferase inhibitor, elevated intracellular MTA levels correlated with lower activity of PRMT5 in MTAP-null cell lines. In fact, PRMT5 is highly sensitive to inhibition by MTA among a panel of methyltransferases [122]. Therefore, these cancers are sensitive to further PRMT5 inhibition. Another study further demonstrates that the synthetic lethality in MTAP-null cancer can extend to both upstream and downstream depletion of PRMT5 [122].
The study of how TP53 regulates metabolism has also shed new light on the role of tumor suppressors in cancer metabolism, and potentially suggests a path for developing a new cancer therapy. TP53 is among the most frequently mutated genes in cancer. Besides acting as the guardian of the genome, TP53 extensively participates in regulating cellular metabolism, including glycolysis, oxidative phosphorylation and autophagy [123]. One of the interesting aspects of TP53 is the ability to resolve oxidative stress, though it could be a pro-oxidant gene as well when persistently or highly activated. TP53 exert its antioxidative function majorly through two mechanisms: 1) Up-regulates antioxidative enzymes such as glutathione peroxidase and peroxiredoxins [124, 125]; 2) Promote synthesis of antioxidant coenzyme NADPH [126]. Targeting ROS has long been a controversial topic in cancer therapy, given the fact that ROS can be both cancer-prone and inhibitory. We believe it is context dependent: Is TP53-deficient in germline or somatic fashion? Are we dealing with tumor prevention or therapy? Germline Trp53-null mice exhibit increased levels of ROS in normal tissue, showing aneuploidy and develop malignant tumors in a ROS-dependent manner. Therefore, feeding parental mice with antioxidant N-Acetyl-Cysteine before Trp53-null zygogenesis and continuously through pregnancy and through the lifetime of the progeny, can abrogate the tumor onset [127]. In this case, systematically high ROS is promoting cancer by creating genomic instability. And as other normal tissues also lose part of the ability to resolve ROS, pro-oxidant treatment could have severe consequence. What about treating an established cancer with somatic TP53-deficency? Would pro-oxidant therapy work, as the tumor has compromised ROS-resolving ability whereas normal tissues are intact? This hypothesis needs to be further examined.
4.4. Targeting reprogrammed metabolic pathway
As discussed above, during malignant transformation and tumor progression, metabolic pathways are usually reprogrammed by oncogenic signaling, through altering metabolic gene expression, and shaped by specific microenvironments and biochemical milieus, to accommodate energy and nutrient demand. This creates metabolic co-dependencies, generates potentially targetable vulnerabilities, and opens a new therapeutic window to cancer, including opening a pipeline of drugs that may not come from the oncology pipeline (Fig. 3D).
For many types of cancer, such as glioblastoma, the genomic landscape of protein coding genes has been largely revealed [27, 128], identifying a discrete set of targetable genomic alterations led primarily by EGFR amplification and mutation, which occur in close to 60% of patients. However, targeting EGFR has proven to be quite difficult because of pharmacokinetic barriers imposed by the blood brain barrier. Poor brain/plasma ratios for many targeted compounds including many EGFR tyrosine kinase inhibitors make it extremely difficult to achieve sufficient intratumoral concentrations in the brain to engage and inhibit their targets, without causing serious and dose-limiting toxicities [129] [130]. mTOR kinase inhibitors have also, to date, had difficulty fully accessing their targets in the brain, especially mTORC2, greatly limiting efficacy. Therefore, broadening the pharmacopeia for patients with brain tumors, whether they have glioblastomas or tumors that metastasize to the brain, is a compelling priority. Some of the drugs that have been shown to target metabolic vulnerabilities generated by tumor genotype and brain microenvironment, may have far better ability to access their targets in the brain, suggesting a potentially fruitful new approach for developing new treatments for patients with brain cancers. Recent work on altered cholesterol metabolism in glioblastoma provides a compelling example.
Over 20% of total cholesterol locates in the brain. However, BBB prevents peripheral cholesterol from transporting into the brain. Thus, almost all cholesterol in the brain requires de novo synthesis. As discussed above, the primary oncogenic driver EGFR suppresses energy-demanding cholesterol synthesis in GBM, rendering GBM’s dependency of uptake of exogenous cholesterol for survival. To prevent excessive cellular cholesterol accumulation, neurons and astrocytes utilize LXR pathway to govern the efflux and reduce the uptake of cholesterol [131]. Therefore, activating LXR pathway by a highly brain permeable agonist LXR-623 has shown selective lethal effect in a PDX GBM orthotopic model [34]. LXR-623 is a relatively unique compound that preferentially accumulated in the brain [132], raising the possibility that this drug or similar members of this class of drugs could be repurposed for GBM therapy.
5. Conclusions and Future Perspectives
Cancer metabolism is a field in transition. We are moving from a static view towards a dynamic consideration that embraces the complexity inherent in the process of tumor development and progression. The early geneticists understood well that genotype is not phenotype, a lesson well-worth remembering. We have a more well-developed genomic landscape of cancer, with clear metabolic correlates, making it tempting to view caner metabolism largely through a genetic lens. However, the evidence is becoming overwhelming – metabolic phenotypes are flexible and heterogeneous, and deeply influenced by environment. This tumor metabolic flexibility underlies the ability of tumors to adapt and thrive in rapidly changing conditions. What are the specific molecular interactions that drive metabolic dependencies, including its ability to rapidly adapt? Can they be anticipated and therapeutically exploited? If so, must we consider a gamut of ways to influence tumor microenvironments in addition to targeting tumor cell autonomous processes? Is there a role for diet, lifestyle or pharmacotherapy in modulating tumor microenvironments in a way that may impede tumor progression and improve anti-cancer treatments, including metabolically targeted agents? With the collective goal of better understanding cancer metabolism in order to develop more effective treatments for patients, a more dynamic, interactive and nuanced understanding of cancer metabolism is required, including examining the molecular mechanisms that tumor cells use to adapt, diversify, resist and thrive in changing environments. This line of research is exciting, as it promises to reveal context-dependent enzymatic vulnerabilities that may not traditionally be thought of as cancer targets. These insights promise to pave the way for new cancer treatments, including by developing and/or repurposing drugs that may not come from traditional oncology pipelines.
Acknowledgements
This work was supported by the Ludwig Institute for Cancer Research and grants from National Institute for Neurological Diseases and Stroke (NS73831), the Defeat GBM Program of the National Brain Tumor Society, the Ben and Catherine Ivy Foundation, an award from the Sharpe/National Brain Tumor Society Research Program, and a generous donations from the Ziering Family Foundation in memory of Sigi Ziering (P.S.M.). We regret that due to space limitations, we have not been able to include many important studies that have shaped, and continue to shape our understanding of cancer metabolism.
Abbreviations
- BBB
Blood-brain barrier
- BCAAs
Branched-chain amino acids
- ecDNA
extrachromosomal DNA
- FH
Fumarate hydratase
- GBM
Glioblastoma multiforme
- HIF1/2
Hypoxia-inducible factor 1/2
- IDH1/2
Isocitrate dehydrogenases 1/2
- LXR
Liver X receptor
- MTA
S-methyl-5'-thioadenosine
- MTAP
Methylthioadenosine phosphorylase
- mTORC1/2
Mammalian target of rapamycin complex 1/2
- NSCLC
Non-small cell lung carcinoma
- PDAC
Pancreatic ductal adenocarcinoma
- PHD
Prolyl hydroxylase
- PRMT5
Protein arginine methyltransferase 5
- R-2-HG
R(−)-2-hydroxyglutarate
- ROS
Reactive oxygen species
- SCNA
Somatic copy number alteration
- SDH
Succinate dehydrogenase
- TCA
Tricarboxylic acid
- xCT
Cystine-glutamate antiporter system xc-
- α-KG
α-ketoglutarate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
PSM, JB, SW and WZ wrote the manuscript and prepared the figures. JB, SW and WZ contributed equally to this work. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.DeBerardinis RJ, Chandel NS, Fundamentals of cancer metabolism, Science advances, 2 (2016) e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP, Cancer metabolism: a therapeutic perspective, Nature reviews. Clinical oncology, 14 (2017) 11-31. [DOI] [PubMed] [Google Scholar]
- 3.Pavlova NN, Thompson CB, The Emerging Hallmarks of Cancer Metabolism, Cell Metab, 23 (2016) 27-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palm W, Thompson CB, Nutrient acquisition strategies of mammalian cells, Nature, 546 (2017) 234-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warburg O, Wind F, Negelein E, The Metabolism of Tumors in the Body, J Gen Physiol, 8 (1927) 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD, Activation of a metabolic gene regulatory network downstream of mTOR complex 1, Molecular cell, 39 (2010) 171-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barthel A, Okino ST, Liao J, Nakatani K, Li J, Whitlock JP, Jr., Roth RA, Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1, J Biol Chem, 274 (1999) 20281-20286. [DOI] [PubMed] [Google Scholar]
- 8.Majewski N, Nogueira V, Bhaskar P, Coy PE, Skeen JE, Gottlob K, Chandel NS, Thompson CB, Robey RB, Hay N, Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak, Mol Cell, 16 (2004) 819-830. [DOI] [PubMed] [Google Scholar]
- 9.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, Blenis J, The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4, Cell, 153 (2013) 840-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, Yu Y, Liu X, He L, Cavaliere PM, Chavez A, Zhang E, Isik M, Couvillon A, Dephoure NE, Blackwell TK, Yu JJ, Rabinowitz JD, Cantley LC, Blenis J, Post-transcriptional Regulation of De Novo Lipogenesis by mTORC1-S6K1-SRPK2 Signaling, Cell, 171 (2017) 1545-1558 e1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB, ATP citrate lyase is an important component of cell growth and transformation, Oncogene, 24 (2005) 6314-6322. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Fridman A, Blackledge W, Connelly S, Wilson IA, Pilz RB, Boss GR, The phosphatidylinositol 3-kinase/akt cassette regulates purine nucleotide synthesis, J Biol Chem, 284 (2009) 3521-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang CV, MYC on the path to cancer, Cell, 149 (2012) 22-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D, RAS oncogenes: weaving a tumorigenic web, Nat Rev Cancer, 11 (2011) 761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Slebos RJ, Lorkiewicz PK, Higashi RM, Fan TW, Dang CV, Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells, Cell metabolism, 15 (2012) 110-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV, c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism, Nature, 458 (2009) 762-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doherty JR, Yang C, Scott KE, Cameron MD, Fallahi M, Li W, Hall MA, Amelio AL, Mishra JK, Li F, Tortosa M, Genau HM, Rounbehler RJ, Lu Y, Dang CV, Kumar KG, Butler AA, Bannister TD, Hooper AT, Unsal-Kacmaz K, Roush WR, Cleveland JL, Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis, Cancer Res, 74 (2014) 908-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM, c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry, Oncogene, 28 (2009) 2485-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David CJ, Chen M, Assanah M, Canoll P, Manley JL, HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer, Nature, 463 (2010) 364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV, c-Myc transactivation of LDH-A: implications for tumor metabolism and growth, Proc Natl Acad Sci U S A, 94 (1997) 6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL, Global mapping of c-Myc binding sites and target gene networks in human B cells, Proceedings of the National Academy of Sciences of the United States of America, 103 (2006) 17834-17839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim JP, Gleeson PA, Macropinocytosis: an endocytic pathway for internalising large gulps, Immunology and cell biology, 89 (2011) 836-843. [DOI] [PubMed] [Google Scholar]
- 23.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, Bar-Sagi D, Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells, Nature, 497 (2013) 633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, Thompson CB, Rabinowitz JD, Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein, Cancer Res, 75 (2015) 544-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson SM, Jonas O, Keibler MA, Hou HW, Luengo A, Mayers JR, Wyckoff J, Del Rosario AM, Whitman M, Chin CR, Condon KJ, Lammers A, Kellersberger KA, Stall BK, Stephanopoulos G, Bar-Sagi D, Han J, Rabinowitz JD, Cima MJ, Langer R, Vander Heiden MG, Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors, Nat Med, 23 (2017) 235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priolo C, Pyne S, Rose J, Regan ER, Zadra G, Photopoulos C, Cacciatore S, Schultz D, Scaglia N, McDunn J, De Marzo AM, Loda M, AKT1 and MYC induce distinctive metabolic fingerprints in human prostate cancer, Cancer research, 74 (2014) 7198-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, Manyam G, Zoppoli P, Ling S, Rao AA, Grifford M, Cherniack AD, Zhang H, Poisson L, Carlotti CG, Jr., Tirapelli DP, Rao A, Mikkelsen T, Lau CC, Yung WK, Rabadan R, Huse J, Brat DJ, Lehman NL, Barnholtz-Sloan JS, Zheng S, Hess K, Rao G, Meyerson M, Beroukhim R, Cooper L, Akbani R, Wrensch M, Haussler D, Aldape KD, Laird PW, Gutmann DH, Network TR, Noushmehr H, Iavarone A, Verhaak RG, Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma, Cell, 164 (2016) 550-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS, Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma, Nat Rev Cancer, 15 (2015) 302-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B, Zhu S, Gu Y, Villa GR, Akhavan D, Nathanson D, Gini B, Mareninov S, Li R, Camacho CE, Kurdistani SK, Eskin A, Nelson SF, Yong WH, Cavenee WK, Cloughesy TF, Christofk HR, Black DL, Mischel PS, EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer, Cell Metab, 17 (2013) 1000-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Hon GC, Villa GR, Turner KM, Ikegami S, Yang H, Ye Z, Li B, Kuan S, Lee AY, Zanca C, Wei B, Lucey G, Jenkins D, Zhang W, Barr CL, Furnari FB, Cloughesy TF, Yong WH, Gahman TC, Shiau AK, Cavenee WK, Ren B, Mischel PS, EGFR Mutation Promotes Glioblastoma through Epigenome and Transcription Factor Network Remodeling, Mol Cell, 60 (2015) 307-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masui K, Tanaka K, Akhavan D, Babic I, Gini B, Matsutani T, Iwanami A, Liu F, Villa GR, Gu Y, Campos C, Zhu S, Yang H, Yong WH, Cloughesy TF, Mellinghoff IK, Cavenee WK, Shaw RJ, Mischel PS, mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc, Cell Metab, 18 (2013) 726-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo D, Prins RM, Dang J, Kuga D, Iwanami A, Soto H, Lin KY, Huang TT, Akhavan D, Hock MB, Zhu S, Kofman AA, Bensinger SJ, Yong WH, Vinters HV, Horvath S, Watson AD, Kuhn JG, Robins HI, Mehta MP, Wen PY, DeAngelis LM, Prados MD, Mellinghoff IK, Cloughesy TF, Mischel PS, EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy, Sci Signal, 2 (2009) ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo D, Reinitz F, Youssef M, Hong C, Nathanson D, Akhavan D, Kuga D, Amzajerdi AN, Soto H, Zhu S, Babic I, Tanaka K, Dang J, Iwanami A, Gini B, Dejesus J, Lisiero DD, Huang TT, Prins RM, Wen PY, Robins HI, Prados MD, Deangelis LM, Mellinghoff IK, Mehta MP, James CD, Chakravarti A, Cloughesy TF, Tontonoz P, Mischel PS, An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway, Cancer Discov, 1 (2011) 442-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villa GR, Hulce JJ, Zanca C, Bi J, Ikegami S, Cahill GL, Gu Y, Lum KM, Masui K, Yang H, Rong X, Hong C, Turner KM, Liu F, Hon GC, Jenkins D, Martini M, Armando AM, Quehenberger O, Cloughesy TF, Furnari FB, Cavenee WK, Tontonoz P, Gahman TC, Shiau AK, Cravatt BF, Mischel PS, An LXR-Cholesterol Axis Creates a Metabolic Co-Dependency for Brain Cancers, Cancer Cell, 30 (2016) 683-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun L, Song L, Wan Q, Wu G, Li X, Wang Y, Wang J, Liu Z, Zhong X, He X, Shen S, Pan X, Li A, Wang Y, Gao P, Tang H, Zhang H, cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions, Cell Res, 25 (2015) 429-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikiforov MA, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD, A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism, Molecular and cellular biology, 22 (2002) 5793-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Sahra I, Howell JJ, Asara JM, Manning BD, Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1, Science, 339 (2013) 1323-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN, Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis, Science, 339 (2013) 1320-1323. [DOI] [PubMed] [Google Scholar]
- 39.Vander Heiden MG, DeBerardinis RJ, Understanding the Intersections between Metabolism and Cancer Biology, Cell, 168 (2017) 657-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, Huang Z, Barnett S, Mickey BE, DeBerardinis RJ, Tu BP, Maher EA, Bachoo RM, Acetate is a bioenergetic substrate for human glioblastoma and brain metastases, Cell, 159 (2014) 1603-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, Horton JD, Hammer RE, McKnight SL, Tu BP, Acetate dependence of tumors, Cell, 159 (2014) 1591-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, Goodwin LM, Smethurst E, Mason S, Blyth K, McGarry L, James D, Shanks E, Kalna G, Saunders RE, Jiang M, Howell M, Lassailly F, Thin MZ, Spencer-Dene B, Stamp G, van den Broek NJ, Mackay G, Bulusu V, Kamphorst JJ, Tardito S, Strachan D, Harris AL, Aboagye EO, Critchlow SE, Wakelam MJ, Schulze A, Gottlieb E, Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress, Cancer Cell, 27 (2015) 57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, Li H, Huet G, Yuan Q, Wigal T, Butt Y, Ni M, Torrealba J, Oliver D, Lenkinski RE, Malloy CR, Wachsmann JW, Young JD, Kernstine K, DeBerardinis RJ, Lactate Metabolism in Human Lung Tumors, Cell, 171 (2017) 358-371 e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Le Z, Yanxiang Guo J, White E, Rabinowitz JD, Glucose feeds the TCA cycle via circulating lactate, Nature, 551 (2017) 115-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattaini KR, Sullivan MR, Vander Heiden MG, The importance of serine metabolism in cancer, J Cell Biol, 214 (2016) 249-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH, Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells, Nature, 493 (2013) 542-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK, Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation, Science, 336 (2012) 1040-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wuu JA, Wen LY, Chuang TY, Chang GG, Amino acid concentrations in serum and aqueous humor from subjects with extreme myopia or senile cataract, Clin Chem, 34 (1988) 1610-1613. [PubMed] [Google Scholar]
- 49.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM, An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis, Cell, 162 (2015) 540-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG, Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells, Cell, 162 (2015) 552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quail DF, Joyce JA, Microenvironmental regulation of tumor progression and metastasis, Nat Med, 19 (2013) 1423-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O'Brien JP, Pierce KA, Gui DY, Sullivan LB, Wasylenko TM, Subbaraj L, Chin CR, Stephanopolous G, Mott BT, Jacks T, Clish CB, Vander Heiden MG, Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer, Cell Metab, 23 (2016) 517-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, Yamada SD, Peter ME, Gwin K, Lengyel E, Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth, Nat Med, 17 (2011) 1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaude E, Frezza C, Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival, Nat Commun, 7 (2016) 13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM, Muir A, Chin CR, Freinkman E, Jacks T, Wolpin BM, Vitkup D, Vander Heiden MG, Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers, Science, 353 (2016) 1161-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, Chen X, Bishop JM, The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type, Cell Metab, 15 (2012) 157-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, Wodzak M, Klimko C, McMillan E, Butt Y, Ni M, Oliver D, Torrealba J, Malloy CR, Kernstine K, Lenkinski RE, DeBerardinis RJ, Metabolic Heterogeneity in Human Lung Tumors, Cell, 164 (2016) 681-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dupuy F, Tabaries S, Andrzejewski S, Dong Z, Blagih J, Annis MG, Omeroglu A, Gao D, Leung S, Amir E, Clemons M, Aguilar-Mahecha A, Basik M, Vincent EE, St-Pierre J, Jones RG, Siegel PM, PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer, Cell Metab, 22 (2015) 577-589. [DOI] [PubMed] [Google Scholar]
- 59.Dietschy JM, Turley SD, Cholesterol metabolism in the brain, Curr Opin Lipidol, 12 (2001) 105-112. [DOI] [PubMed] [Google Scholar]
- 60.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M, The growing landscape of lysine acetylation links metabolism and cell signalling, Nat Rev Mol Cell Biol, 15 (2014) 536-550. [DOI] [PubMed] [Google Scholar]
- 61.Masui K, Tanaka K, Ikegami S, Villa GR, Yang H, Yong WH, Cloughesy TF, Yamagata K, Arai N, Cavenee WK, Mischel PS, Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance, Proc Natl Acad Sci U S A, 112 (2015) 9406-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nihira NT, Ogura K, Shimizu K, North BJ, Zhang J, Gao D, Inuzuka H, Wei W, Acetylation-dependent regulation of MDM2 E3 ligase activity dictates its oncogenic function, Sci Signal, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inuzuka H, Gao D, Finley LW, Yang W, Wan L, Fukushima H, Chin YR, Zhai B, Shaik S, Lau AW, Wang Z, Gygi SP, Nakayama K, Teruya-Feldstein J, Toker A, Haigis MC, Pandolfi PP, Wei W, Acetylation-dependent regulation of Skp2 function, Cell, 150 (2012) 179-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fardini Y, Dehennaut V, Lefebvre T, Issad T, O-GlcNAcylation: A New Cancer Hallmark?, Front Endocrinol (Lausanne), 4 (2013) 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma Z, Vosseller K, Cancer metabolism and elevated O-GlcNAc in oncogenic signaling, J Biol Chem, 289 (2014) 34457-34465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chou TY, Hart GW, Dang CV, c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas, J Biol Chem, 270 (1995) 18961-18965. [DOI] [PubMed] [Google Scholar]
- 67.Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW, Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability, Nat Cell Biol, 8 (2006) 1074-1083. [DOI] [PubMed] [Google Scholar]
- 68.Olivier-Van Stichelen S, Guinez C, Mir AM, Perez-Cervera Y, Liu C, Michalski JC, Lefebvre T, The hexosamine biosynthetic pathway and O-GlcNAcylation drive the expression of beta-catenin and cell proliferation, Am J Physiol Endocrinol Metab, 302 (2012) E417-424. [DOI] [PubMed] [Google Scholar]
- 69.Yang WH, Park SY, Nam HW, Kim DH, Kang JG, Kang ES, Kim YS, Lee HC, Kim KS, Cho JW, NFkappaB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions, Proc Natl Acad Sci U S A, 105 (2008) 17345-17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng C, Ru P, Geng F, Liu J, Yoo JY, Wu X, Cheng X, Euthine V, Hu P, Guo JY, Lefai E, Kaur B, Nohturfft A, Ma J, Chakravarti A, Guo D, Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth, Cancer Cell, 28 (2015) 569-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.D'Angelo JA, Mattox ML, Fiebiger E, Dickinson BL, The cystine/glutamate antiporter regulates the functional expression of indoleamine 2,3-dioxygenase in human dendritic cells, Scand J Immunol, 76 (2012) 448-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shin CS, Mishra P, Watrous JD, Carelli V, D'Aurelio M, Jain M, Chan DC, The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility, Nat Commun, 8 (2017) 15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koppula P, Zhang Y, Shi J, Li W, Gan B, The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate, J Biol Chem, 292 (2017) 14240-14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gu Y, Albuquerque CP, Braas D, Zhang W, Villa GR, Bi J, Ikegami S, Masui K, Gini B, Yang H, Gahman TC, Shiau AK, Cloughesy TF, Christofk HR, Zhou H, Guan KL, Mischel PS, mTORC2 Regulates Amino Acid Metabolism in Cancer by Phosphorylation of the Cystine-Glutamate Antiporter xCT, Molecular cell, 67 (2017) 128-138 e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lien EC, Ghisolfi L, Geck RC, Asara JM, Toker A, Oncogenic PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT, Sci Signal, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giaccia AJ, Simon MC, Johnson R, The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease, Genes & development, 18 (2004) 2183-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wigerup C, Pahlman S, Bexell D, Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer, Pharmacology & therapeutics, 164 (2016) 152-169. [DOI] [PubMed] [Google Scholar]
- 78.Briggs KJ, Koivunen P, Cao S, Backus KM, Olenchock BA, Patel H, Zhang Q, Signoretti S, Gerfen GJ, Richardson AL, Witkiewicz AK, Cravatt BF, Clardy J, Kaelin WG, Jr., Paracrine Induction of HIF by Glutamate in Breast Cancer: EglN1 Senses Cysteine, Cell, 166 (2016) 126-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guertin DA, Sabatini DM, Defining the role of mTOR in cancer, Cancer Cell, 12 (2007) 9-22. [DOI] [PubMed] [Google Scholar]
- 80.Populo H, Lopes JM, Soares P, The mTOR signalling pathway in human cancer, Int J Mol Sci, 13 (2012) 1886-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goberdhan DC, Wilson C, Harris AL, Amino Acid Sensing by mTORC1: Intracellular Transporters Mark the Spot, Cell Metab, 23 (2016) 580-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sandoval J, Esteller M, Cancer epigenomics: beyond genomics, Curr Opin Genet Dev, 22 (2012) 50-55. [DOI] [PubMed] [Google Scholar]
- 83.Hoffman RM, Altered methionine metabolism, DNA methylation and oncogene expression in carcinogenesis. A review and synthesis, Biochim Biophys Acta, 738 (1984) 49-87. [DOI] [PubMed] [Google Scholar]
- 84.Mehrmohamadi M, Mentch LK, Clark AG, Locasale JW, Integrative modelling of tumour DNA methylation quantifies the contribution of metabolism, Nat Commun, 7 (2016) 13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maddocks OD, Labuschagne CF, Adams PD, Vousden KH, Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells, Mol Cell, 61 (2016) 210-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gomez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, Nichenametla SN, Locasale JW, Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism, Cell Metab, 22 (2015) 861-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB, ATP-citrate lyase links cellular metabolism to histone acetylation, Science, 324 (2009) 1076-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Corbet C, Pinto A, Martherus R, Santiago de Jesus JP, Polet F, Feron O, Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation, Cell Metab, 24 (2016) 311-323. [DOI] [PubMed] [Google Scholar]
- 89.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW, Cancer genome landscapes, Science, 339 (2013) 1546-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Graham NA, Minasyan A, Lomova A, Cass A, Balanis NG, Friedman M, Chan SN, Zhao S, Delgado A, Go J, Beck L, Hurtz C, Ng C, Qiao R, ten Hoeve J, Palaskas N, Wu H, Muschen M, Multani AS, Port E, Larson SM, Schultz N, Braas D, Christofk HR, Mellinghoff IK, Graeber TG, Recurrent patterns of DNA copy number alterations in tumors reflect metabolic selection pressures, Molecular Systems Biology, 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, Kornblum HI, Taylor MD, Kaushal S, Cavenee WK, Wechsler-Reya R, Furnari FB, Vandenberg SR, Rao PN, Wahl GM, Bafna V, Mischel PS, Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity, Nature, 543 (2017) 122-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, Eskin A, Hwang K, Wang J, Masui K, Paucar A, Yang H, Ohashi M, Zhu S, Wykosky J, Reed R, Nelson SF, Cloughesy TF, James CD, Rao PN, Kornblum HI, Heath JR, Cavenee WK, Furnari FB, Mischel PS, Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA, Science, 343 (2014) 72-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oermann EK, Wu J, Guan KL, Xiong Y, Alterations of metabolic genes and metabolites in cancer, Semin Cell Dev Biol, 23 (2012) 370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PEM, Rubinstein WS, Myers EN, Richard CW, Cornelisse CJ, Devilee P, Devlin B, Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma, Science, 287 (2000) 848-851. [DOI] [PubMed] [Google Scholar]
- 95.Fokkema IFAC, Taschner PEM, Schaafsma GCP, Celli J, Laros JFJ, den Dunnen JT, LOVD v.2.0: The Next Generation in Gene Variant Databases, Human Mutation, 32 (2011) 557-563. [DOI] [PubMed] [Google Scholar]
- 96.Ricketts CJ, Forman JR, Rattenberry E, Bradshaw N, Lalloo F, Izatt L, Cole TR, Armstrong R, Kumar VKA, Morrison PJ, Atkinson AB, Douglas F, Ball SG, Cook J, Srirangalingam U, Killick P, Kirby G, Aylwin S, Woodward ER, Evans DGR, Hodgson SV, Murday V, Chew SL, Connell JM, Blunde TL, MacDonald F, Maher ER, Tumor Risks and Genotype-Phenotype-Proteotype Analysis in 358 Patients With Germline Mutations in SDHB and SDHD, Human Mutation, 31 (2010) 41-51. [DOI] [PubMed] [Google Scholar]
- 97.Ni Y, Zbuk KM, Sadler T, Patocs A, Lobo G, Edelman E, Platzer P, Orloff MS, Waite KA, Eng C, Germline mutations and variants in the succinate dehydrogenase genes in Cowden and Cowden-like syndromes, American Journal of Human Genetics, 83 (2008) 261-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tomlinson IPM, Alam NA, Rowan AJ, Barclay E, Jaeger EEM, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA, Consortium ML, Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer, Nat Genet, 30 (2002) 406-410. [DOI] [PubMed] [Google Scholar]
- 99.Carvajal-Carmona LG, Alam NA, Pollard PJ, Jones AM, Barclay E, Wortham N, Pignatelli M, Freeman A, Pomplun S, Ellis I, Poulsom R, El-Bahrawy MA, Berney DM, Tomlinson IPM, Adult Leydig cell tumors of the testis caused by germline fumarate hydratase mutations, Journal of Clinical Endocrinology & Metabolism, 91 (2006) 3071-3075. [DOI] [PubMed] [Google Scholar]
- 100.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr., Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW, An integrated genomic analysis of human glioblastoma multiforme, Science, 321 (2008) 1807-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW, The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary, Acta Neuropathol, 131 (2016) 803-820. [DOI] [PubMed] [Google Scholar]
- 102.Patel KP, Ravandi F, Ma D, Paladugu A, Barkoh BA, Medeiros LJ, Luthra R, Acute myeloid leukemia with IDH1 or IDH2 mutation: frequency and clinicopathologic features, Am J Clin Pathol, 135 (2011) 35-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E, Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase, Cancer cell, 7 (2005) 77-85. [DOI] [PubMed] [Google Scholar]
- 104.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, Ratcliffe PJ, Linehan WM, Neckers L, HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability, Cancer Cell, 8 (2005) 143-153. [DOI] [PubMed] [Google Scholar]
- 105.Ye D, Ma S, Xiong Y, Guan KL, R-2-hydroxyglutarate as the key effector of IDH mutations promoting oncogenesis, Cancer Cell, 23 (2013) 274-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Turcan S, Makarov V, Taranda J, Wang Y, Fabius AWM, Wu W, Zheng Y, El-Amine N, Haddock S, Nanjangud G, LeKaye HC, Brennan C, Cross J, Huse JT, Kelleher NL, Osten P, Thompson CB, Chan TA, Mutant-IDH1-dependent chromatin state reprogramming, reversibility, and persistence, Nat Genet, 50 (2018) 62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, Travins J, Weiss S, Looper R, Ligon KL, Verhaak RG, Yan H, Kaelin WG, Jr., Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation, Nature, 483 (2012) 484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG, Jr., (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible, Science, 339 (2013) 1621-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, Kunii K, Pedraza A, Schalm S, Silverman L, Miller A, Wang F, Yang H, Chen Y, Kernytsky A, Rosenblum MK, Liu W, Biller SA, Su SM, Brennan CW, Chan TA, Graeber TG, Yen KE, Mellinghoff IK, An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells, Science, 340 (2013) 626-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, Straley K, Kernytsky A, Liu W, Gliser C, Yang H, Gross S, Artin E, Saada V, Mylonas E, Quivoron C, Popovici-Muller J, Saunders JO, Salituro FG, Yan S, Murray S, Wei W, Gao Y, Dang L, Dorsch M, Agresta S, Schenkein DP, Biller SA, Su SM, de Botton S, Yen KE, Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation, Science, 340 (2013) 622-626. [DOI] [PubMed] [Google Scholar]
- 111.Dang L, Yen K, Attar EC, IDH mutations in cancer and progress toward development of targeted therapeutics, Annals of Oncology, 27 (2016) 599-608. [DOI] [PubMed] [Google Scholar]
- 112.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, Sougnez C, Gabriel SB, Hernandez B, Shen H, Laird PW, Getz G, Meyerson M, Beroukhim R, Pan-cancer patterns of somatic copy number alteration, Nat Genet, 45 (2013) 1134-U1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haider S, McIntyre A, van Stiphout RGPM, Winchester LM, Wigfield S, Harris AL, Buffa FM, Genomic alterations underlie a pan-cancer metabolic shift associated with tumour hypoxia, Genome Biol, 17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han C, Yang LF, Choi HH, Baddour J, Achreja A, Liu YH, Li YJ, Li JD, Wan GH, Huang C, Ji G, Zhang XN, Nagrath D, Lu XB, Amplification of USP13 drives ovarian cancer metabolism, Nature Communications, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu DM, Liu T, Deng SH, Han R, Xu Y, SLC39A4 expression is associated with enhanced cell migration, cisplatin resistance, and poor survival in non-small cell lung cancer, Sci Rep, 7 (2017) 7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu W, Hancock CN, Fischer JW, Harman M, Phang JM, Proline biosynthesis augments tumor cell growth and aerobic glycolysis: involvement of pyridine nucleotides, Sci Rep, 5 (2015) 17206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, DePinho RA, Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer, Genes Dev, 20 (2006) 3130-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ren JG, Seth P, Clish CB, Lorkiewicz PK, Higashi RM, Lane AN, Fan TW, Sukhatme VP, Knockdown of malic enzyme 2 suppresses lung tumor growth, induces differentiation and impacts PI3K/AKT signaling, Sci Rep, 4 (2014) 5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dey P, Baddour J, Muller F, Wu CC, Wang H, Liao WT, Lan Z, Chen A, Gutschner T, Kang Y, Fleming J, Satani N, Zhao D, Achreja A, Yang L, Lee J, Chang E, Genovese G, Viale A, Ying H, Draetta G, Maitra A, Wang YA, Nagrath D, DePinho RA, Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer, Nature, 542 (2017) 119-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, Vazquez F, Weir BA, Fitzgerald ME, Tanaka M, Bielski CM, Scott JM, Dennis C, Cowley GS, Boehm JS, Root DE, Golub TR, Clish CB, Bradner JE, Hahn WC, Garraway LA, MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells, Science, 351 (2016) 1214-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mavrakis KJ, McDonald ER, 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, Ruddy DA, Venkatesan K, Yu J, McAllister G, Stump M, deBeaumont R, Ho S, Yue Y, Liu Y, Yan-Neale Y, Yang G, Lin F, Yin H, Gao H, Kipp DR, Zhao S, McNamara JT, Sprague ER, Zheng B, Lin Y, Cho YS, Gu J, Crawford K, Ciccone D, Vitari AC, Lai A, Capka V, Hurov K, Porter JA, Tallarico J, Mickanin C, Lees E, Pagliarini R, Keen N, Schmelzle T, Hofmann F, Stegmeier F, Sellers WR, Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5, Science, 351 (2016) 1208-1213. [DOI] [PubMed] [Google Scholar]
- 122.Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, McVay M, Choe S, Kernytsky A, Gross S, Konteatis Z, Murtie J, Blake ML, Travins J, Dorsch M, Biller SA, Marks KM, MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis, Cell Rep, 15 (2016) 574-587. [DOI] [PubMed] [Google Scholar]
- 123.Vousden KH, Ryan KM, p53 and metabolism, Nat Rev Cancer, 9 (2009) 691-700. [DOI] [PubMed] [Google Scholar]
- 124.Tan M, Li S, Swaroop M, Guan K, Oberley LW, Sun Y, Transcriptional activation of the human glutathione peroxidase promoter by p53, J Biol Chem, 274 (1999) 12061-12066. [DOI] [PubMed] [Google Scholar]
- 125.[] Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM, Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD, Science, 304 (2004) 596-600. [DOI] [PubMed] [Google Scholar]
- 126.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH, TIGAR, a p53-inducible regulator of glycolysis and apoptosis, Cell, 126 (2006) 107-120. [DOI] [PubMed] [Google Scholar]
- 127.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM, The antioxidant function of the p53 tumor suppressor, Nat Med, 11 (2005) 1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G, Discovery and saturation analysis of cancer genes across 21 tumour types, Nature, 505 (2014) 495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, Kubek S, Oldrini B, Chheda MG, Yannuzzi N, Tao H, Zhu S, Iwanami A, Kuga D, Dang J, Pedraza A, Brennan CW, Heguy A, Liau LM, Lieberman F, Yung WK, Gilbert MR, Reardon DA, Drappatz J, Wen PY, Lamborn KR, Chang SM, Prados MD, Fine HA, Horvath S, Wu N, Lassman AB, DeAngelis LM, Yong WH, Kuhn JG, Mischel PS, Mehta MP, Cloughesy TF, Mellinghoff IK, Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors, Cancer Discov, 2 (2012) 458-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Deng Y, Feng W, Wu J, Chen Z, Tang Y, Zhang H, Liang J, Xian H, Zhang S, The concentration of erlotinib in the cerebrospinal fluid of patients with brain metastasis from non-small-cell lung cancer, Mol Clin Oncol, 2 (2014) 116-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Courtney R, Landreth GE, LXR Regulation of Brain Cholesterol: From Development to Disease, Trends Endocrinol Metab, 27 (2016) 404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Katz A, Udata C, Ott E, Hickey L, Burczynski ME, Burghart P, Vesterqvist O, Meng X, Safety, pharmacokinetics, and pharmacodynamics of single doses of LXR-623, a novel liver X-receptor agonist, in healthy participants, J Clin Pharmacol, 49 (2009) 643-649.</References> [DOI] [PubMed] [Google Scholar]