Molecular targeted therapy against driver gene aberrations has a significantly high positive response rate compared with traditional cytotoxic chemotherapies, and has thus dramatically changed the treatment strategies for malignant tumors, including non-small cell lung cancer (NSCLC) (1). Lung adenocarcinoma (LADC) can be treated with targeted therapy against driver gene aberrations using tyrosine kinase inhibitors (TKIs). A large genomic sequencing study showed that mutations of EGFR, KRAS, BRAF, and HER2, and gene rearrangements involving ALK, RET, ROS1, and MET exon 14 skipping are mutually exclusively identified as driver gene alterations in LADC (2). The National Comprehensive Cancer Network (NCCN) Guidelines Insights focus on recent updates in targeted therapy against driver genes for patients with advanced NSCLC (3). According to the NCCN guidelines, testing for EGFR mutations, BRAF mutations, ALK fusions, and ROS1 fusions, and assessing PD-L1 expression levels are recommended in patients with advanced NSCLC before the initial treatment. The guidelines recommend first-line specific TKIs for driver-positive patients and immunotherapy and/or chemotherapy for driver-negative patients. Efforts to confirm the therapeutic utility of TKIs and to identify resistance mutations leading to the development of novel TKIs for overcoming acquired resistance are continuous. For example, oncogenic RET fusions were discovered in LADC (4-6) and confirmed as a promising target (4,7). The efficacy of vandetanib and cabozantinib in patients with RET fusions (8,9) promoted the design of clinical trials to evaluate the efficacy of a novel RET-specific inhibitor that was developed to overcome the weak points of multikinase inhibitors (10,11). A recent report describes the mechanism underlying acquired resistance to vandetanib (12), and a novel RET-inhibitor was developed to suppress acquired resistance associated with mutations (13).
We recently demonstrated that the frequency of driver gene aberrations in LADC differs according to ethnicity, sex, and smoking, which leads to differences in treatment efficacy (14). In this report, comparison of Asian and European/US cohorts showed that EGFR mutations are more prevalent in Asian patients, whereas KRAS mutations, which are not druggable, are more prevalent in European/US patients. When stratified by sex and smoking status, EGFR mutations are frequently detected in women and never-smokers in both Asian and European/US patients. EGFR mutations are more likely to be detected in Asian patients than in European/US patients, whereas KRAS and BRAF mutations occur preferentially in men and ever smokers, especially among European/US patients. On the other hand, oncogenic fusions, such as ALK, RET, and ROS1 fusions, are frequently detected in women never smokers in both cohorts. Lastly, driver-negative cases are more prevalent among smokers in both cohorts. There are currently no targeted therapies against driver-negative cases, underscoring the need to identify new druggable targets.
Recently, de Semir et al. reported that pleckstrin homology domain-interacting protein (PHIP) could be a novel therapeutic target for driver-negative malignancies including NSCLC (15). These authors demonstrated that PHIP plays a role in promoting the progression of driver-negative NSCLC. PHIP knockdown and overexpression in vitro revealed that increased expression of PHIP promotes tumor cell proliferation and invasion. In clinical specimens, PHIP is upregulated in the bronchioid subtype of LADC without EGFR mutations, KRAS mutations, and ALK fusions. This study demonstrated the possible therapeutic utility of anti-PHIP bromodomain inhibitors for the treatment of patients with elevated PHIP expression.
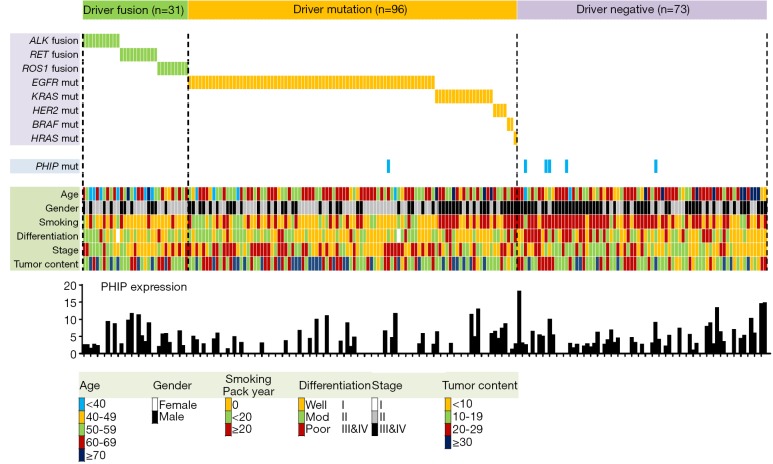
To explore the therapeutic potential of PHIP inhibitors, we extracted data on PHIP expression from 200 LADC cases that were previously included in a comprehensive genetic analysis performed by our group (16). This study included activating mutations in the EGFR, KRAS, HER2, BRAF, and HRAS oncogenes (n=96), oncogenic ALK, RET, or ROS1 fusions (n=31), and driver-negative cases (n=73). As shown in Figure 1, LADC cases with significant PHIP upregulation were preferentially detected among driver negative cases, suggesting the therapeutic utility of PHIP targeting in LADC, especially in driver-negative cases. Analysis of PHIP mutations showed that they were more frequent in the driver-negative group (5/73, 6.8%) than in the other two groups (1/127, 0.8%). These results suggest that PHIP gene aberrations preferentially occur in driver-negative patients with LADC with accumulated non-synonymous mutations. As indicated by de Semir et al., identifying new targetable mutations and adapting immunotherapies are critical issues in driver-negative tumors. In driver-negative cases, MET amplification, RIT1 mutations, MEK1 (MAP2K1) mutations, and loss of functional NF1 mutations were identified and considered therapeutic targets (2,14). In addition, truncating mutations of SMARCA4/BRG1 and ARID1A are prevalent in driver-negative tumors and targetable using a synthetic lethal approach (14). Currently, the most promising additional personalized therapy for driver-negative cases is immunotherapy (18-20). Because mutation burden is associated with anti- programmed death (PD)-ligand 1 (PD-L1) drug sensitivity (21), immune checkpoint blockade of PD-1 and PD-L1 is expected to be effective in driver-negative LADC cases. PHIP or other targeted therapy and immunotherapy will hopefully lead to the development of effective targeted therapies providing a survival benefit in patients with NSCLC.
Figure 1.
PHIP expression and gene mutation profile in lung adenocarcinoma according to driver aberrations with clinicopathological characteristics. PHIP expression was expressed as the FPKM (fragments per kilobase of exon per million fragments mapped) value, as previously described (17). PHIP, pleckstrin homology domain-interacting protein.
Acknowledgements
None.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer--is it becoming a reality? Nat Rev Clin Oncol 2010;7:401-14. 10.1038/nrclinonc.2010.64 [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettinger DS, Aisner DL, Wood DE, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 5.2018. J Natl Compr Canc Netw 2018;16:807-21. 10.6004/jnccn.2018.0062 [DOI] [PubMed] [Google Scholar]
- 4.Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375-7. 10.1038/nm.2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382-4. 10.1038/nm.2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. 10.1038/nm.2658 [DOI] [PubMed] [Google Scholar]
- 7.Saito M, Ishigame T, Tsuta K, et al. A mouse model of KIF5B-RET fusion-dependent lung tumorigenesis. Carcinogenesis 2014;35:2452-6. 10.1093/carcin/bgu158 [DOI] [PubMed] [Google Scholar]
- 8.Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013;3:630-5. 10.1158/2159-8290.CD-13-0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 2017;5:42-50. 10.1016/S2213-2600(16)30322-8 [DOI] [PubMed] [Google Scholar]
- 10.Subbiah V, Gainor JF, Rahal R, et al. Precision Targeted Therapy with BLU-667 for RET-Driven Cancers. Cancer Discov 2018;8:836-49. 10.1158/2159-8290.CD-18-0338 [DOI] [PubMed] [Google Scholar]
- 11.Drilon A, Hu ZI, Lai GGY, et al. Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nat Rev Clin Oncol 2018;15:151-67. 10.1038/nrclinonc.2017.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakaoku T, Kohno T, Araki M, et al. A secondary RET mutation in the activation loop conferring resistance to vandetanib. Nat Commun 2018;9:625. 10.1038/s41467-018-02994-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subbiah V, Velcheti V, Tuch BB, et al. Selective RET Kinase Inhibition for Patients with RET-Altered Cancers. Ann Oncol 2018;29:1869-76. 10.1093/annonc/mdy137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito M, Shiraishi K, Kunitoh H, et al. Gene aberrations for precision medicine against lung adenocarcinoma. Cancer Sci 2016;107:713-20. 10.1111/cas.12941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Semir D, Bezrookove V, Nosrati M, et al. PHIP as a therapeutic target for driver-negative subtypes of melanoma, breast, and lung cancer. Proc Natl Acad Sci U S A 2018;115:E5766-E75. 10.1073/pnas.1804779115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito M, Shimada Y, Shiraishi K, et al. Development of lung adenocarcinomas with exclusive dependence on oncogene fusions. Cancer Res 2015;75:2264-71. 10.1158/0008-5472.CAN-14-3282 [DOI] [PubMed] [Google Scholar]
- 17.Saito M, Fujiwara Y, Asao T, et al. The genomic and epigenomic landscape in thymic carcinoma. Carcinogenesis 2017;38:1084-91. 10.1093/carcin/bgx094 [DOI] [PubMed] [Google Scholar]
- 18.Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N Engl J Med 2018;378:2288-301. 10.1056/NEJMoa1716948 [DOI] [PubMed] [Google Scholar]
- 19.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandhi L, Rodriguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:2078-92. 10.1056/NEJMoa1801005 [DOI] [PubMed] [Google Scholar]
- 21.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]