Abstract
Helicobacter (H.) pylori is an important risk factor for gastric malignancies worldwide. Its outer membrane proteome takes an important role in colonization of the human gastric mucosa. However, in zoonotic non-H. pylori helicobacters (NHPHs) also associated with human gastric disease, the composition of the outer membrane (OM) proteome and its relative contribution to disease remain largely unknown. By means of a comprehensive survey of the diversity and distribution of predicted outer membrane proteins (OMPs) identified in all known gastric Helicobacter species with fully annotated genome sequences, we found genus- and species-specific families known or thought to be implicated in virulence. Hop adhesins, part of the Helicobacter-specific family 13 (Hop, Hor and Hom) were restricted to the gastric species H. pylori, H. cetorum and H. acinonychis. Hof proteins (family 33) were putative adhesins with predicted Occ- or MOMP-family like 18-stranded β-barrels. They were found to be widespread amongst all gastric Helicobacter species only sporadically detected in enterohepatic Helicobacter species. These latter are other members within the genus Helicobacter, although ecologically and genetically distinct. LpxR, a lipopolysaccharide remodeling factor, was also detected in all gastric Helicobacter species but lacking as well from the enterohepatic species H. cinaedi, H. equorum and H. hepaticus. In conclusion, our systemic survey of Helicobacter OMPs points to species and infection-site specific members that are interesting candidates for future virulence and colonization studies.
Introduction
H. pylori colonizes the stomach of more than 50% of the human population. Infection with this microbe is considered to be the most important risk factor for gastric malignancies worldwide1,2. In contrast to most other Gram-negative bacteria, the outer membrane of H. pylori is equipped with a remarkably large set of OMPs3. There are approximately 64 well-annotated OMP genes present in H. pylori, encompassing 4% of its entire genome4. This unusually large set of predicted OMPs possibly originates as an adaptation of the gastric bacterium to the hostile environment of the stomach. The H. pylori OMPs can be divided into 6 groups: the Hop (H. pylori OMP), Hor (Hop-related), Hof (Helicobacter OMP), Hom (Helicobacter outer membrane), iron-regulated OMPs and efflux pump OMPs. Most of the H. pylori OMPs belonging to the Hop proteins, are predicted to function as porins or as adhesins that promote binding of the bacterium to the gastric mucosa3,5. More specifically, BabA (HopS), SabA (HopP), OipA (HopH), AlpA (HopC), AlpB (HopB), LabA (HopD), HopQ and HopZ are the best characterized H. pylori Hop adhesins that play a role in its colonization process6–13.
Besides H. pylori, also non-H. pylori Helicobacter (NHPH) species have been associated with gastric disease in humans, including gastritis, gastric and duodenal ulcers and mucosa-associated lymphoid tissue (MALT) lymphoma. Such NHPHs mostly originate from domestic animals, such as H. suis from pigs and H. heilmannii, H. bizzozeronii, H. felis, H. salomonis, H. cynogastricus, H. baculiformis and H. ailurogastricus from cats and dogs14. Recent comparative genomic studies highlighted that gastric NHPHs lack all known H. pylori adhesins described so far15–18. Furthermore, they share only few homologs of the Hor and Hom family. Only genes encoding homologs of the H. pylori Hof protein family are present in these animal-associated helicobacters, but in contrast to H. pylori, their hof genes are located in a ~10-kb locus15. For H. heilmannii, it has recently been shown that HofE and HofF play a key role in adhesion to the gastric mucosa, albeit with a higher affinity for gastric epithelial cells than for mucins19.
Based on the available data, it is clear that gastric NHPH species harbor a different OMP repertoire compared to H. pylori and that the mechanisms by which these NHPHs colonize the gastric mucosa remains largely unknown. Besides the domestic animal-related gastric Helicobacter species, also other gastric helicobacters, including H. acynonichis from wild felines, H. cetorum from marine mamals and H. mustelae from ferrets, with unknown OMP repertoire have been described.
Therefore, in the present study, we screened the genomes in silico from all gastric Helicobacter species known so far, for the presence of genes encoding putative OMPs. The predicted OMPs were then classified into families based on their protein sequence homology, using 3 different protein databases. Finally, phylogenetic analyses of the OMP sequences were applied to study the predicted functional relationships among the OMPs and to unravel the OMPs that could play a role in the colonization and virulence properties of gastric NHPHs. In addition, the genomes of 4 enterohepatic Helicobacter species and 2 closely-related Campylobacter (C.) species were included in the proteomic and phylogenetic analyses for comparison. These species are phylogenetically close to gastric Helicobacter species, yet are ecologically distinct in thriving on the mucosal surfaces of the intestinal tract and/or the liver, so that genomic comparison may point to niche-dependent genetic markers in the OM proteomes20,21. As a reference for functional annotation, we included the genome of Escherichia (E.) coli, for which the biological functions of the majority of its OMPs are well characterized.
Accordingly, this study identified several unique OMP families that are possibly important for colonization and virulence, in gastric Helicobacter species.
Results
Identification of OMP families
Fifty four genomes were analysed for candidate OMPs by screening their open reading frames for proteins with signal peptide and β-strand propensities using the HHomp tool22. A total of 3380 putative OMPs were identified among the different strains of E. coli (500) and the genera Campylobacter (141) and Helicobacter (2739). An overview of the number of OMPs per species and per strain is presented in Table 1. For the Campylobacter and the E. coli strains, the total number of OMPs per genome ranged between 21 and 27, and between 88 and 116, respectively. The mean number of OMPs was lower for the 4 enterohepatic helicobacters (a mean of 33 OMPs) with a minimum of 24 OMPs for H. equorum eqF1 and a maximum of 55 OMPs for H. trogontum R3554), compared to the gastric Helicobacter species (a mean of 66 OMPs) with a minimum of 47 OMPs for H. salomonis M45 and a maximum of 118 OMPs for H. cetorum MIT 00–7128). H. mustelae, which is able to colonize both the gastric and intestinal environment23, contained 49 OMPs (Table 1). Subsequently, these OMPs were classified into families based on protein sequence homology. From the 3380 OMPs, 2794 proteins could be classified into one of the 90 families known from the OMPdb reference database (OMPdb.org). The results are listed in Supplementary Table S1. For each of the 90 OMP families, the name and function is shown as well as the OMPs from the E. coli, Campylobacter and Helicobacter strains that clustered into that family.
Table 1.
Overview of the number of OMPs per species and strain used in this study.
| Species and strain designation | Isolated from | NCBI accession number* Chromosome |
NCBI accession number* Plasmid |
Number of OMPs |
|---|---|---|---|---|
| Campylobacter coli | ||||
| 15-157360 | human | NC_022660° | NC_022656 | 21 |
| 76339 | human | NC_022132° | 24 | |
| N29710 | chicken | NC_022347° | NC_022355 and NC_02234 | 27 |
| Campylobacter jejuni | ||||
| 00-2425 | human | NC_022362° | 23 | |
| 4031 | contaminated tap water | NC_022529° | 24 | |
| M1 | human | NC_017280° | 22 | |
| Escherichia coli $ | ||||
| 536 | human | NC_008253° | 110 | |
| IAI1 | human | NC_011741° | 89 | |
| IAI39 | human | NC_011750° | 97 | |
| K12-W3110 | human | NC_007779° | 88 | |
| TW14359 | human | NC_013008° | NC_013010 | 116 |
| Helicobacter acinonychis | ||||
| Hacino1 | Cheetah | FZMD00000000°° | 95 | |
| Hacino2 | Sumatran tiger | FZLX00000000°° | 92 | |
| Hacino3 | Lion | NC_008229° | NC_008230 | 106 |
| Hacino4 | Bengal tiger | FZLV00000000°° | 94 | |
| Helicobacter ailurogastricus | ||||
| ASB7 | cat | CDMG00000000°° | 59 | |
| ASB9 | cat | CDMN00000000°° | 60 | |
| ASB11 | cat | CDML00000000°° | 60 | |
| ASB13 | cat | CDMH00000000°° | 60 | |
| Helicobacter baculiformis | ||||
| M50 | cat | FZMF00000000°° | 62 | |
| Helicobacter bizzozeronii | ||||
| 10 | dog | FZEH00000000°° | 69 | |
| 14 | dog | FZMK00000000°° | 65 | |
| CIII | human | NC_015674° | NC_015670 | 68 |
| M7 | dog | FZLK00000000°° | 70 | |
| Helicobacter cetorum | ||||
| MIT 00-7128 | whale | NC_017737° | NC_017738 | 118 |
| MIT 99-5656 | dolphin | NC_017735° | NC_017736 | 101 |
| Helicobacter cinaedi $$ | ||||
| BAA_847 | human | NC_020555° | 29 | |
| Helicobacter cynogastricus | ||||
| JKM4 | dog | FZMQ00000000°° | 62 | |
| Helicobacter equorum | ||||
| eqF1 | horse | FZPO00000000°° | 24 | |
| Helicobacter felis | ||||
| CS1 | cat | NC_014810° | 65 | |
| CS6 | cat | FZKM00000000°° | 66 | |
| CS7 | cat | FZKX00000000°° | 67 | |
| DS1 | dog | FZNI00000000°° | 61 | |
| Helicobacter heilmannii | ||||
| ASB1 | cat | CDMK00000000°° | 53 | |
| ASB2 | cat | CDMP00000000°° | 55 | |
| ASB3 | cat | CDMJ00000000°° | 56 | |
| ASB6 | cat | CDMM00000000°° | 56 | |
| ASB14 | cat | CDMI00000000°° | 55 | |
| Helicobacter hepaticus | ||||
| ATCC 51449 | mouse | NC_004917° | 28 | |
| Helicobacter mustelae | ||||
| ATCC 12198 | ferret | NC_013949° | 49 | |
| Helicobacter pylori | ||||
| Puno120 | human (Amerindian) | NC_017378° | NC_017377 | 69 |
| G27 | human (Europe) | NC_011333° | NC_011334 | 69 |
| F30 | human (East Asia) | NC_017365° | NC_017369 | 72 |
| J99 | human (USA) | NC_000921° | 71 | |
| India7 | human (India) | NC_017372° | 74 | |
| SouthAfrica7 | human (South Africa) | NC_022130° | 69 | |
| Helicobacter salomonis | ||||
| M45 | dog | FZLZ00000000°° | 47 | |
| R1053 | dog | OANQ00000000°° | 48 | |
| KokIII | dog | FZMA00000000°° | 49 | |
| Helicobacter suis | ||||
| HS2 | pig | FZLI00000000°° | 53 | |
| HS4 | pig | FZKI00000000°° | 52 | |
| HS7 | pig | FZKH00000000°° | 53 | |
| HS9 | pig | FZLE00000000°° | 53 | |
| Helicobacter trogontum | ||||
| R3554 | rat | FZNG00000000°° | 55 | |
$$For the gastric and the enterohepatic Helicobacter species, genomes were chosen based on their availability at the moment of analysis in the NCBI database.
For H. pylori, six fully annotated genomes from strains of different regions (one strain per geographical area) were selected (Smet et al. 2018 DOI: 10.1038/s41396-018-0199-5).
Campylobacter and E. coli strains were included as reference strains.
$For E. coli, each strain was chosen out of a different phylogroup based on the paper of Bohlin et al. 2014. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4200225/).
*NCBI accession numbers are available for the species and strains with fully annotated genomes. On the basis of these genomes, OMP extraction was started.
°Complete genome.
°°Draft genome.
The remaining 586 protein sequences were then searched against the Pfam protein database, resulting in the additional classification of 308 proteins into 31 putative OMP families with unknown biological function. These families were assigned a systematic family ID from “X1” to “X31”. The names of these 31 candidate OMP families and the predicted OMPs from the E. coli, Campylobacter and Helicobacter strains that clustered among these families, are shown in Supplementary Table S2.
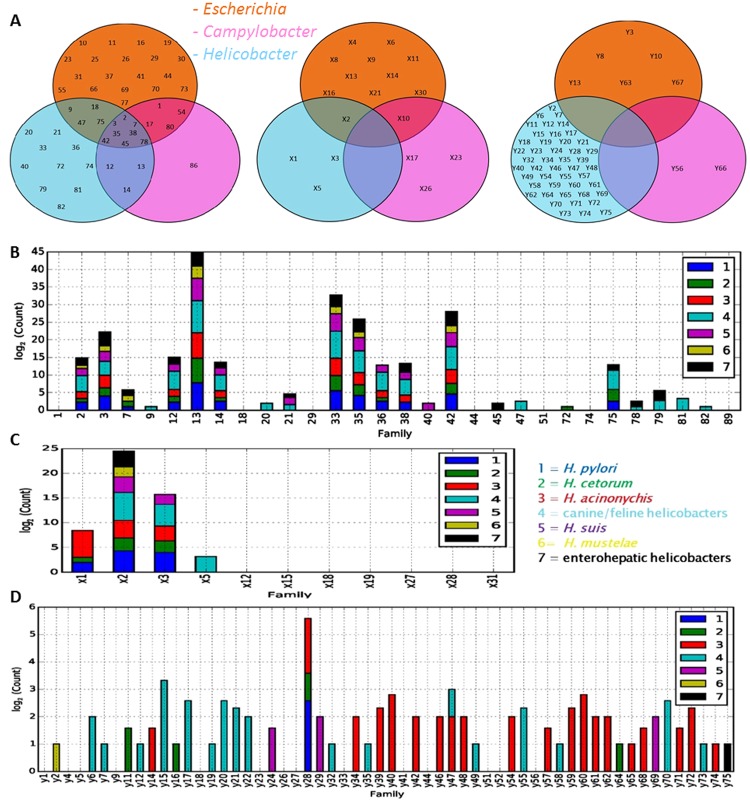
Finally, the remaining 278 unclassified protein sequences were clustered into phylogenetic groups using the CD-HIT program with algorithm settings to produce the fewest possible number of clusters. With this method, 106 clusters were produced. The clusters with single members that had a length of less than 120 amino acids (a total of 52 sequences) were excluded. This final clustering revealed 75 families of unknown annotation or function, which were assigned a systematic family ID of “Y1” to “Y75” (Supplementary Table S3). To visualize the relative distribution of the OMP families amongst E. coli and the Campylobacter and Helicobacter species, the predicted OMPdb families, and X and Y families were presented in Venn diagrams (Fig. 1A). The OMP families from the OMPdb database seemed to be well-conserved among E. coli and Helicobacter, and to a lesser extent in Campylobacter (Fig. 1A). From the 90 OMPdb families, 19 families were unique for E. coli, 10 families for the Helicobacter species, and only 1 family for the Campylobacter species. Several of the X-families from the Pfam database were found to be genus-specific (Fig. 1A). From the 31 X-families, 10 families were unique for E. coli, 3 families for Campylobacter, and 3 for Helicobacter. From the 75 Y-families, 46 families were unique for Helicobacter (especially in the helicobacters from cats and dogs), 2 families for Campylobacter and 6 were unique for E. coli (Fig. 1A).
Figure 1.
(A) Venn diagrams of the 90 OMPdb families, the 31 Pfam-derived “X” families and the 75 “Y” families derived from CD-HIT-generated clusters. Shown are the OMP families for the Escherichia, Campylobacter- and Helicobacter genera. Distribution plots of the Helicobacter OMPs from (B) the OMPdb-, (C) the “X”- and (D) the “Y” families. Families consisting of at least one OMP were included in the plots. Dark blue = H. pylori, green = H. cetorum, red = H. acinonychis, light blue = canine and feline gastric helicobacters, purple = H. suis, yellow = H. mustelae, black = enterohepatic helicobacters. For these 7 groups, the numbers of members (log2 values) in each OMP family are shown. For families with only one OMP member, bars are invisible on the log2 scale.
Furthermore, distribution plots of OMPs were made to search for distinguishing components of the OMP proteomes in gastric canine/feline helicobacters, H. suis, H. pylori, H. cetorum, H. acinonychis, H. mustelae, and enterohepatic helicobacters (Fig. 1B–D). Nine families of the OMPdb database were found well-conserved among gastric and enterohepatic helicobacters (Family 2,3,12,13,14,33,35,38 and 42), and one family (Family 36) was conserved among the gastric species only. The other 20 families were specific to only a few species and strains (Fig. 1B, Supplementary Table S1). From the X-families, only family X2 was well-conserved among the genus Helicobacter and family X3 among the gastric species. OMPs from family X1 were only found in H. pylori, H. cetorum, and H. acinonychis and OMPs from family X5 in H. felis and H. bizzozeronii. OMPs from the other 7×-families were only present in one Helicobacter species (Fig. 1C, Supplementary Table S2). Each Y-family was mainly composed of OMPs from only one Helicobacter species. Most of these Y-families were found in canine and feline gastric helicobacters and in H. acinonychis (Fig. 1D, Supplementary Table S3). In following paragraphs, we elaborate on OMP families of particular interest for Helicobacter infections and disease.
Helicobacter-specific OMP families with known or putative roles in virulence or colonization
Family 13 - Helicobacter Outer Membrane Protein Family 1 (Hop/Hor/Hom)
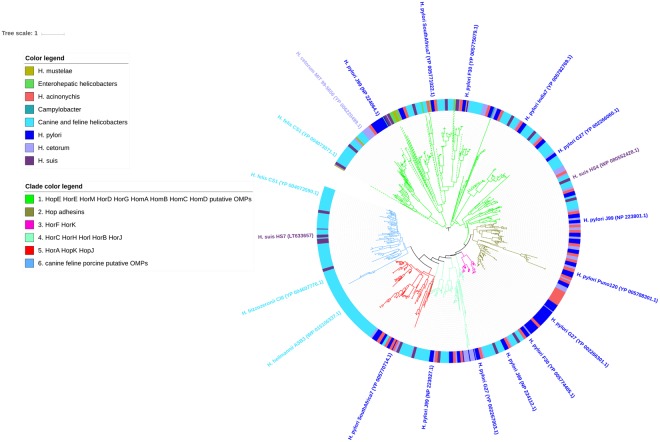
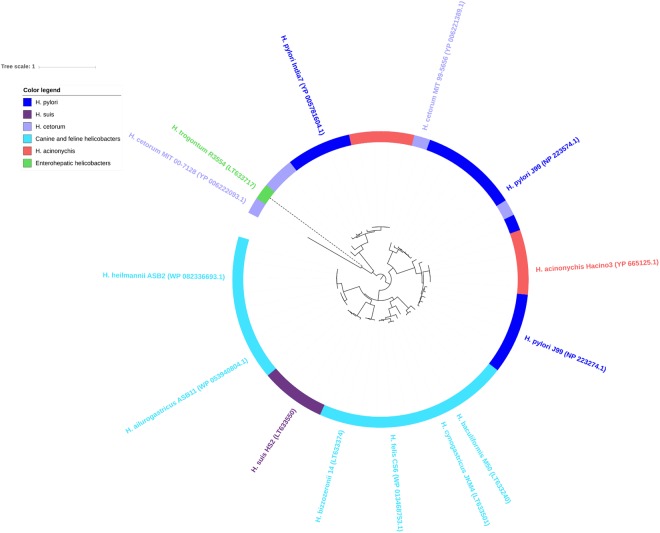
A total of 1,155 orthologous OMPs were clustered among the Helicobacter outer membrane protein family 1 (Family 13 of the OMPdb database, Supplementary Table S1). This family encompasses the H. pylori Hop, Hor and Hom proteins. Orthologs were present in all examined gastric and enterohepatic Helicobacter species. Only one orthologous protein was present in C. coli (76339) and C. jejuni (00–2425) and there were no members of this family detected in E. coli. The phylogenetic tree of this OMP family shows 6 subgroups (clades) could be distinguished (Fig. 2). Strikingly, the known Hop adhesins (2) all cluster into a monophyletic clade (Fig. 2, clade 2) that is specific for H. pylori (dark blue), its closest relative H. acinonychis (red), and H. cetorum (light purple) and which were found to be completely absent in canine, feline or porcine gastric NHPH. In H. pylori family 13, clade 2 comprises several group members, of which AlpA, AlpB, HopA, HopF, HopI, HopG, HopL and OipA were found to be conserved in H. acinonychis and H. cetorum, which additionally held orthologs of BabB and HopD, and of SabA respectively (Fig. 2). BLAST analyses revealed that several putative OMPs from the canine, feline and porcine gastric NHPH species clustered into these subgroups. For the enterohepatic Helicobacter species and H. mustelae, only orthologous putative OMPs of HorD and HorG were present in this family (clade 1 in Fig. 2). The orthologous OMP from C. coli (76339) and C. jejuni (00-2425) also clustered in this latter subgroup. Interestingly, subgroup 6 (Fig. 2) contained putative OMPs that were present in canine, feline and porcine Helicobacter species only.
Figure 2.
Phylogenetic tree of Family 13 – the Helicobacter outer membrane protein family 1. The H. pylori Hop, Hor and Hom adhesins, including BabA (HopS), SabA (HopP), AlpA (HopC) and AlpB (HopB), OipA (HopH), HopZ, HopQ, LabA (HopD), HorB and HomB clustered into this family. Orthologous OMPs are present in all examined gastric and enterohepatic Helicobacter species, only one orthologous protein is present in C. coli (76339) and C. jejuni (00-2425) and there are no orthologous OMPs detected in E. coli. Indicated are 6 different subgroups: putative OMPs specific for canine, feline and porcine helicobacters (6); H. pylori HorA, HopK and HopJ (5) with orthologous putative OMPs in all other gastric NHPH species, except H. mustelae; H. pylori HorC, HorH, HorI, HorB and HorJ (4) with orthologous putative OMPs in all other gastric NHPH species, except H. mustelae; H. pylori HorF and HorK (3) with orthologous putative OMPs in other gastric NHPH species, except H. baculiformis and H. mustelae; H. pylori Hop adhesins (2) that are absent in other gastric NHPH species except for H. acinonychis and H. cetorum; H. pylori HopE, HorE, HorM, HorD, HorG, HomA, HomB, HomC and HomD (1) with orthologous putative OMPs in all other gastric NHPH species. Also, the 1 orthologous OMP from C. coli (76339) and C. jejuni (00-2425) (dark cyan) cluster in the latter subgroup, as well as a few OMPs from H. mustelae (yellow-green) and enterohepatic helicobacters (green). The position of the different H. pylori OMPs in each subgroup are clockwise indicated. OMPs of H. mustelae, enterohepatic helicobacters and Campylobacter species are indicated by dashed clade lines. Per clade, minimum one accession number is added.
Family 33 - Helicobacter Outer Membrane Protein Family 2 (Hof)
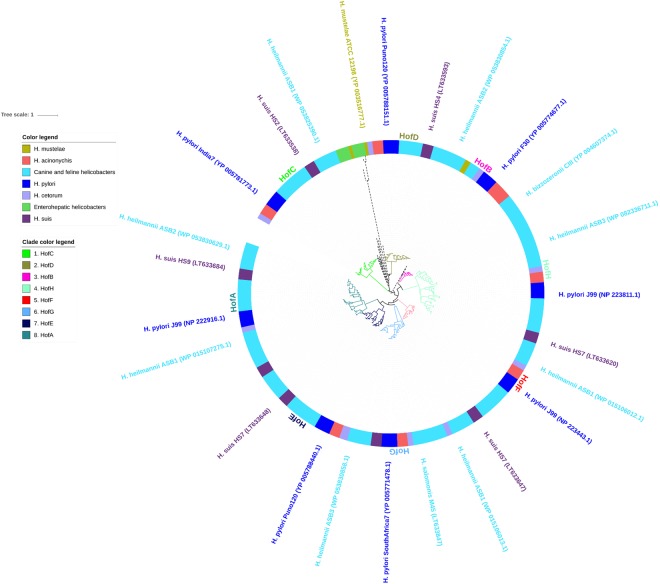
A total of 347 orthologous OMPs clustered in the Helicobacter outer membrane protein family 2 (Family 33 of the OMPdb database, Supplementary Table S1). This family comprises orthologs of the H. pylori Hof proteins. Members of these Hof OMPs were present in all examined Helicobacter species and absent in C. coli, C. jejuni or E. coli. The phylogenetic tree of this OMP family reveals 8 subgroups, corresponding with the H. pylori HofA, HofB, HofC, HofD, HofE, HofF, HofG and HofH proteins (Fig. 3A). These 8 subgroups were highly conserved among H. acinonychis, H. cetorum, as well as canine, feline and porcine NHPH species, except for HofB, which is only present in H. bizzozeronii. On the contrary, the analysed enterohepatic helicobacters and H. mustelae only contained a few putative Hof-like OMPs, which did not cluster with one of the 8 Hof proteins known from H. pylori.
Figure 3.
Phylogenetic tree of Family 33 – the Helicobacter outer membrane protein family 2. Orthologous OMPs are present in all examined Helicobacter species, but are absent in Campylobacter and E. coli. Indicated are 8 different subgroups, corresponding to the different H. pylori Hof proteins: HofA (8) with orthologous putative OMPs in gastric NHPH except H. acinonychis, H. salomonis and H. mustelae; HofB (3) with orthologous putative OMPs in H. acinonychis, H. bizzozeronii and H. cetorum; HofC (1), HofD (2), HofE (7), HofF (5), HofG (6) and HofH (4) with orthologous putative OMPs in all gastric Helicobacter species except H. mustelae. H. mustelae (yellow-green) and enterohepatic helicobacters (green) contain a few uncharacterized OMPs that cluster separately. OMPs of H. mustelae and enterohepatic helicobacters are indicated by dashed clade lines. Per clade, minimum one accession number is added.
Hof proteins lack close homologs outside the genus Helicobacter. They are of unknown function except for reports in H. heilmannii that implicate HofE and HofF in adherence to human gastric mucins19. To gain insight into the putative Hof function, representative sequences of the 8 Hof subgroups were subjected to remote homology and fold recognition searches using the Protein Homology/analogy Recognition Engine 2 (PHYRE2)24 and RaptorX25. Both algorithms picked up high confidence structural homology to 18-stranded β-barrels of the Outer membrane Carboxylate Channel (Occ) family proteins (formerly Opd/Opr) found in Pseudomonas and Acinetobacter26,27, as well as to the 18-stranded Major Outer Membrane Protein (MOMP) of C. jejuni28. In support of this homology assignment, the transmembrane elements in the 3D threaded structure are also found by the transmembrane β-barrel protein predictor BOCTOPUS29, have plausible hydrophobic residue distributions, and show the presence of 2 aromatic “belts” as is frequently observed in OM β-barrels (Supplementary Fig. S1). The Occ family OMPs are monomeric 18-stranded porins found in species lacking large channel trimeric porins, and are implicated in non-specific diffusion of small carboxylate-containing solutes across the OM26,27. C. jejuni MOMP can be found both as a monomeric or trimeric porin of 18-stranded β-barrels involved in cation selective passive diffusion over the OM30, though has also been implicated in bacterial adherence31.
Family 36 - Systemic factor protein A (SfpA/LpxR)
The prototype of this family is the systemic factor protein A (SfpA) of Yersinia enterocolitica. Lipid A deacylase (LpxR), a SfpA homologue in Salmonella Typhimurium, has been shown to be important for immune evasion through lipopolysaccharide modification32. Also in H. pylori, LpxR homologs play a key role in the establishment of long-term colonization33.
Except for H. cinaedi, H. equorum, and H. hepaticus, the examined Helicobacter strains contained between 1 and 2 orthologs of SfpA/LpxR per genome (Family 36 of the OMPdb database, Supplementary Table S1), suggesting that LPS remodeling by lipid A deacylation is a common characteristic in most Helicobacter species. In contrast, only one orthologous protein was present in E. coli strain TW14359 and there were no members of the SfpA/LpxR family detected in Campylobacter and the other E. coli strains (Supplementary Table S1). As shown in the phylogenetic tree (Fig. 4), the H. mustelae ortologous OMP clustered closer to enterohepatic helicobacters than to gastric NHPH species.
Figure 4.
Phylogenetic tree of Family 36 – Systemic factor protein A (SfpA/LpxR). Orthologous OMPs are present in the examined strains of Helicobacter except for H. cinaedi, H. equorum and H. hepaticus, and only one orthologous OMP is present in E. coli strain TW14359 (orange). In the other E. coli strains and in Campylobacter, no orthologous OMPs are detected. H. mustelae (yellow-green) clusters closer to the enterohepatic species H. trogontum (green) than to gastric NHPH species. OMPs of H. mustelae and enterohepatic helicobacters are indicated by dashed clade lines. Per clade, minimum one accession number is added.
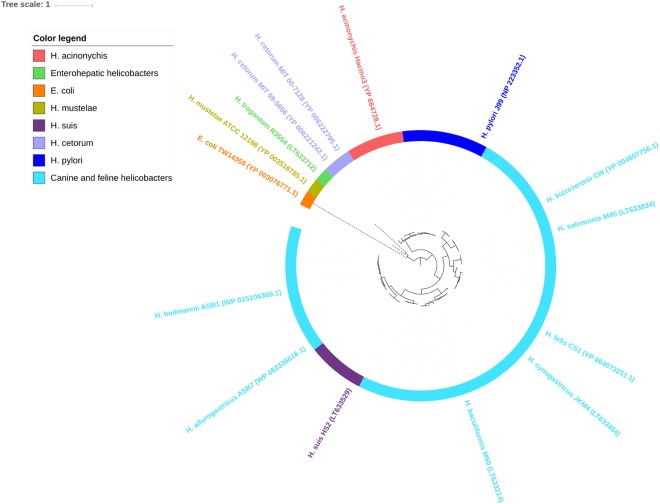
Family X1 and X3 – (Putative) vacuolating cytotoxin family
The secreted vacuolating cytotoxin A (VacA), belonging to the autotransporter OMP family, is an important virulence factor of H. pylori. After binding to and internalization into host epithelial cells, VacA induces cellular vacuolation and various other responses34,35. Strikingly, most other Helicobacter species lack VacA homologs, except for H. cetorum and H. acinonychis.
In the present study, all analysed H. pylori and H. cetorum strains each harbored one vacA copy (Family X1 of the Pfam database, Supplementary Table S2), whilst H. acinonychis contained more than one vacA copy, though these were inactivated by insertion sequences compared to H. pylori and H. cetorum as described before15,36. Besides VacA, H. pylori also contains 3 VacA-like autotransporters that each enhance its capacity to colonize the stomach35. Previously, we reported the presence of such vacA-like autotransporter gene in canine, feline and porcine gastric Helicobacter species37. At the protein level, these VacA-like autotransporters showed to be highly divergent amongst the different Helicobacter species15. In the present study, 55 homologs of the VacA-like protein were classified into the putative vacuolating cytotoxin family (Family X3 of the Pfam database, Supplementary Table S2). Orthologs were present in the examined strains of Helicobacter except for H. cinaedi, H. equorum, H. hepaticus, H. mustelae and H. bizzozeronii 10. There were no orthologs of this autotransporter present in Campylobacter and E. coli (Fig. 5).
Figure 5.
Phylogenetic tree of Family X3 – putative vacuolating cytotoxin. Orthologous OMPs are present in the examined strains of Helicobacter except for H. cinaedi, H. equorum, H. hepaticus, H. mustelae and H. bizzozeronii 10. Orthologous OMPs are absent in E. coli and Campylobacter species. Most Helicobacter strains harbor one VacA-like protein, whereas H. acinonychis, H. cetorum MIT 00-7128 and H. pylori Puno120 and F30 harbor two copies, and H. cetorum MIT 99-5656 and H. pylori G27, J99, India7 and SouthAfrica harbor tree copies of the VacA-like OMP. OMPs of enterohepatic helicobacters are indicated by dashed clade lines. Per clade, minimum one accession number is added.
Well-conserved OMP families present among Helicobacter, Campylobacter and/or E. coli with a role in virulence or colonization
Family 14 - Imp/OstA
Imp (increased membrane permeability) or OstA (organic solvent tolerance) is an organic solvent tolerance protein in Gram-negative bacteria that participates in outer membrane biogenesis and integrity38,39. The Imp/OstA protein is implicated in the translocation and insertion of LPS into the outer leaflet of the OM bilayer. It has also been associated with membrane permeability, organic solvent tolerance and resistance to antibiotics in H. pylori40,41. We identified 48 orthologous outer membrane proteins in the Imp/OstA family (Family 14 of the OMPdb database, Supplementary Table S1). In each of the examined strains of Campylobacter and Helicobacter, except for H. trogontum, one orthologous Imp/OstA protein was found. However, the latter species harbored an additional putative OstA paralog (Family X18, Supplementary Table S2). The phylogenetic tree of OMP family 14 is shown in Supplementary Fig. S2. In accordance to the other families, H. mustelae clustered closer to enterohepatic helicobacters than to gastric NHPH species.
Family 42 - Outer membrane factor (OMF)
Gram-negative bacteria possess energy-dependent transport systems to export proteins, carbohydrates, drugs and heavy metals across the two membranes of the cell envelope42. Type 1 protein secretion systems (e.g. HlyBD-TolC) and RND multidrug efflux systems (e.g. AcrA/B-TolC or MexAB-OprM) consist of a cytoplasmic or inner membrane (IM) export system, a membrane fusion protein (MFP) and an outer membrane factor (OMF). TolC, the prototype OMF found in E. coli forms a trimeric 12-stranded β-barrel (3 × 4 strands) with an extended coiled coil domain reaching into the periplasm and contacting the IM-localized export system via the MFP43. These transport systems have been shown to play a role in protein export and multidrug efflux, the latter producing both intrinsic and elevated multidrug resistance42,44,45. In total, 219 orthologous proteins were identified to belong to the OMF family (Family 42 of the OMPdb database, Supplementary Table S1). Different members of the OMF family were present in all the analysed E. coli, Campylobacter and Helicobacter strains. With protein BLAST, several different OMPs subgroups could be distinguished in the OMF family (Supplementary Table S4). These different subgroups are indicated in the phylogenetic tree of the OMF family (Supplementary Fig. S3). Also, here, orthologs of the OMP family of H. mustelae clustered with enterohepatic helicobacters rather than with gastric Helicobacter species.
Family 3 - Outer Membrane Receptor (OMR-TonB Dependent Receptor) and Family X2 - TonB-dependent Receptor Plug Domain
TonB-dependent transporters are bacterial outer membrane proteins that bind and transport ferric chelates called siderophores, as well as vitamin B12, nickel complexes, and carbohydrates into the periplasm. For which they use energy from the proton motive force of the cytoplasmic membrane via the TonB-ExhB-ExbD membrane proteins. The TonB-dependent outer membrane receptors have also been shown to be required for bacterial virulence46. In the present study, two TonB-dependent outer membrane receptor families were identified.
Family 3 of the OMPdb database comprised the TonB-dependent outer membrane receptors, including Btub, CfrA, FhuA, FhuE, Fiu, FecA, FepA, and FerA, for the uptake of iron (siderophores), nickel and vitamin B12. A total of 123 orthologous OMP proteins clustered in this family. Members of this family are present in all examined strains of E. coli, Campylobacter and Helicobacter except for H. bizzozeronii and H. salomonis. Helicobacter genomes contained an average of 2 paralogs per strain. The phylogenetic tree of OMP family 3 is shown in Supplementary Fig. S4. The TonB-dependent receptors of H. mustelae clustered together with those of enterohepatic helicobacters which on their turn clustered together with the Campylobacter TonB-dependent receptors.
Family X2 of TonB-dependent receptors comprised 116 orthologous proteins (Family X2 of the Pfam database, Supplementary Table S2). Members of this family were present in all examined strains of E. coli and Helicobacter apart from H. salomonis. OMPs belonging to this TonB-dependent receptor family were absent in C. coli and C. jejuni. The phylogenetic tree of OMP family X2 is shown in Supplementary Fig. S5.
Family 38 - Outer Membrane Phospholipase (OMPLA)
The outer membrane phospholipase A (OMPLA), encoded by the pldA gene which is widespread among Gram-negative bacteria, hydrolyses acyl ester bonds in phospholipids and lysophospholipids47,48. OMPLA has been described as a virulence factor. For instance, in C. coli, OMPLA was identified as a major hemolytic factor and H. pylori OMPLA has been shown to be involved in the colonization and invasion of the human gastric mucosa47,49–52. Moreover, H. pylori isolates with high OMPLA activity have been associated with peptic ulcer disease in human patients53,54. In the present study, 53 proteins were classified in the OMPLA family (Family 38 of the OMPdb database, Supplementary Table S1). All E. coli, Campylobacter and Helicobacter strains, apart from H. mustelae, H. equorum, H. cetorum MIT 00-7128, and H. pylori G27, harbored 1 OMPLA ortholog, whereas 4 different paralogs were present in H. trogontum. Phylogenetically, the OMPLA orthologs of all strains clustered separately per genera and per species (Supplementary Fig. S6).
Discussion
In this study, we have analysed the genome sequences from a total of 54 different strains of the genera Helicobacter, Campylobacter and E. coli for their presence of OMPs. Considering the genome length and the total protein count, gastric Helicobacter species harbor proportionally more OMPs than other helicobacters or the Campylobacter or Escherichia reference genomes. Gastric helicobacters have small genomes and proteomes compared to the average E. coli genome, which holds a median genome length and protein count of, respectively, 5.17 Mb and 4931 proteins according to the NCBI database. Instead, H. pylori has a median genome length of only 1.63 Mb with a median protein count of 1451. However, H. pylori’s surface-localized proteome did not shrink proportionally, and ~4% of its total protein count constitutes of OMPs, compared to just ~2% for E. coli. This large collection of OMPs in an otherwise reductionist genome suggests they form important fitness factors in the survival and adaptation to the harsh gastric environment4. Among the gastric Helicobacter species, the total OMP number was the highest for the H. acinonychis (a mean of 97) and H. cetorum strains (a mean of 110), although it should be noted that these species contain multiple fragmented OMPs (Table 1).
In general, we found that the clustering of a strain or species’ OMPs in the phylogenetic trees is similar to the phylogenetic clustering of their full genomes. The phylogenetic reconstructions of the different families revealed a clear and evident division between enterohepatic and gastric Helicobacter species. The gastric helicobacters could be further divided into H. pylori and its two closest relatives, H. acinonychis and H. cetorum and NHPH species including the canine, feline, and porcine helicobacters clades. Interestingly, H. mustelae, which has been associated with gastritis, peptic ulcers, MALT lymphoma, and adenocarcinoma in domestic ferrets55,56, clustered within the clade of the enterohepatic Helicobacter species. This supports previous hypotheses, which emphasize the capability of H. mustelae to colonize both the stomach and the intestinal tract15.
The primary function of the outer membrane of Gram-negative bacteria is to form a barrier against hazardous substances from the environment such as enzymes, detergents, and antimicrobials. The permeability of the outer membrane is determined by the presence of OMPs that function as porins. They contain transmembrane diffusion channels through which small hydrophilic molecules, nutrients, and small antibiotics can be transported across the outer membrane4,57. In our study, two Helicobacter-specific porin families were found, namely Family 13 and 33 (Supplementary Table S1, Figs 2 and 3). Both families mainly contain OMP orthologs from the genus Helicobacter albeit with a greater extent in the gastric helicobacters than in the enterohepatic ones. C. coli and C. jejuni only harbor one such OMP and members of these families are even lacking in E. coli. The families 13 and 33 were thus probably acquired by Helicobacter after splitting-off from a last common ancestor. Moreover, most gastric NHPH species lack all H. pylori Hop adhesins, suggesting that these OMPs were acquired after H. pylori speciation15–18. The H. pylori-specific Hop proteins function as adhesins for gastric epithelial cells4. Interestingly, the adhesive properties of the blood group antigen binding adhesin BabA was found to be pH responsive and to provide the bacteria with a reversible adherence profile that is fine-tuned to the pH-gradients in the stomach mucosa58. Also the canine, feline and porcine gastric NHPH species have been shown to attach to the gastric mucosa15,19. The absence of the H. pylori Hop adhesins in these NHPHs suggests that other OMPs function as adhesins in these organisms. Indeed, genes encoding orthologs of H. pylori Hof proteins seem to be well conserved in the canine, feline and porcine gastric NHPH species of which HofE and HofF have recently been identified as adhesins in H. heilmannii19. HofF has also been shown to be important for H. pylori colonization, but the function of the other H. pylori Hof OMPs remains largely unknown19,59. Furthermore, the exact role of the other NHPH OMPs from Families 13 and 33 in NHPH colonization remains to be further elucidated. Remote homology recognition and 3D threading of family 33 (Hof) members indicates structural similarity with 18-stranded porins of the Occ family in Pseudomonas and Acinetobacter, and the C. jejuni major outer membrane protein – MOMP, which are implicated in cation-selective solute diffusion across the OM, as well as in adherence in case of MOMP28,30,31.
The explicit OMP variation among species might also be favorable for evasion of the host’s immune response. H. pylori has developed a very large repertoire of mechanisms to evade both innate and adaptive immune recognition60. One way of H. pylori to evade the immune response is the avoidance of recognition by Toll-like receptors of its bacterial surface molecules such as LPS and flagellin. Here, we identified the SfpA/LpxR OMP family (Family 36, Supplementary Table S1, Fig. 4) present in all gastric Helicobacter species, which might be involved in immune evasion by removing the 3′-acyloxyacyl group of lipid A61,62. For the enterohepatic Helicobacter species tested here, SfpA/LpxR was only detected in H. trogontum. This OMP family might thus be more specific for gastric species than their enterohepatic members within the genus Helicobacter.
A very well-studied outer membrane virulence factor of H. pylori is the secreted vacuolating cytotoxin A (VacA; Family X1, Supplementary Table S2) that causes uncontrolled cellular vacuolation34,35. In agreement with previous research15,36, we identified the VacA OMP only in H. pylori and H. cetorum and short fragments of this protein in H. acinonychis, but not in other NHPH species. We additionally found a Helicobacter-specific putative VacA-like cytotoxin family (Family X3, Supplementary Table S2, Fig. 5). The VacA-like autotransporters belonging to this family enhance the bacterium’s colonization capacity of the stomach63. The VacA-like OMP is well conserved among the different gastric Helicobacter species, although their protein sequences exhibit much variation15. Besides, we also showed variation in the number of VacA-like autotransporters, not only between species but also at the species level. For instance, in H. bizzozeronii strain 10, no VacA-like autotransporter could be identified, whereas the other examined strains of H. bizzozeronii each contained one copy of this OMP. This underlines the genetic diversity among strains within a species. However, it should be noted that the genomes of most NHPH species that were analysed in this study, including that of H. bizzozeronii strain 10, are draft genomes that lack approximately 5% of the full genome sequence. Therefore, it cannot be excluded that the gene encoding the VacA-like autotransporter of this H. bizzozeronii strain is part of the lacking 5% of its genome sequence. The genomes of enterohepatic Helicobacter species, except for H. trogontum, lack a VacA-like autotransporter as well. This OMP family may therefore be more specific for gastric helicobacters. However, in the H. mustelae strain included in our study, the VacA-like OMP is absent as well. Also for all other families that were analysed, the OMPs of this H. mustelae strain clustered closer together with enterohepatic helicobacters, or separately between enterohepatic and gastric species. Thus, although H. mustelae has been described as a gastric Helicobacter species associated with gastric malignancies, recent studies suggest that this species is an enterohepatic species by origin, but adapted its colonization niche from the intestinal to the gastric environment23,56,64,65.
In addition, several OMP families were found to be well conserved among E. coli and the Campylobacter, and Helicobacter genera. The Imp/OstA family (Family 14, Supplementary Table S1, Fig. S2), an organic solvent tolerance protein, and OMF family (Family 42, Supplementary Table S1, Fig. S3), part of type 1 protein secretion systems and RND multidrug efflux systems, maintain a barrier for antimicrobial agents and play a role in the resistance to drugs. In contrast to the Helicobacter-specific outer membrane porins which utilize passive diffusion for solute uptake, outer membrane receptor proteins, such as TonB-dependent receptors, carry out high-affinity binding and energy-dependent uptake of specific substrates including iron. In this study, two TonB-dependent receptor families (Family 3 and Family X2, Supplementary Tables S1 and S2, Figs S4 and S5) were identified that contribute to bacterial virulence46. Remarkably, OMPs from both Family 3 and Family X2 were lacking in H. salomonis. This may suggest that this species has other iron uptake mechanisms at their disposal to maintain iron homeostasis.
Finally, a virulence factor that has been shown to influence the colonization capacity and pathogenicity of Gram-negative bacteria, is the outer membrane phospholipase A (OMPLA) (Family 38, Supplementary Table S1, Fig. S6). Orthologs of this family could not be found in the genomes of H. cetorum strain MIT 00-7128, H. pylori strain G27, H. equorum, and H. mustelae. Whether the absence of OMPLA in these strains influences their virulence and colonization capacity remains to be further investigated.
In conclusion, several important OMP families, mainly from gastric Helicobacter species, were determined by using comparative genomic and phylogenetic analyses. To our knowledge, this is the first report analysing OMP occurrence and diversity in NHPH species, since previous studies on the OMP repertoire in the genus Helicobacter have mostly concentrated on the human pathogen H. pylori. Two Helicobacter-specific outer membrane protein families with possible functions in adhesion (Family 13, i.e. Hop, Hor and Hom; and Family 33, Hof), the Helicobacter-specific SfpA/LpxR OMP (Family 36) that functions in immune evasion, and a Helicobacter-specific VacA-like cytotoxin family with a role in colonization capacity (Family X3), were identified primarily in gastric species. Furthermore, we showed that most Helicobacter species contain an outer membrane factor (OMF; Family 42) and Imp/OstA (Family 14), both involved in antimicrobial resistance, TonB-dependent OMPs with a function in metal and vitamin-uptake (Family 3 and Family X2), and an outer membrane phospholipase (OMPLA; Family 38) that plays a role in colonization capacity.
In summary, our systemic survey of Helicobacter OMPs points to species and infection-site specific members that are interesting candidates for future virulence and colonization studies.
Methods
Escherichia coli, Campylobacter and Helicobacter species included
For the gastric and the enterohepatic Helicobacter species, genomes were chosen based on their availability at the moment of analysis. Therefore, the genomes from 12 gastric Helicobacter species, namely H. acinonychis (4 strains), H. ailurogastricus (4 strains), H. baculiformis (1 strain), H. bizzozeronii (4 strains), H. cetorum (2 strains), H. cynogastricus (1 strain), H. felis (4 strains), H. heilmannii (5 strains), H. mustelae (1 strain), H. pylori (6 strains), H. salomonis (3 strains), and H. suis (4 strains) and from 4 enterohepatic Helicobacter species, namely H. cinaedi (1 strain), H. equorum (1 strain), H. hepaticus (1 strain), and H. trogontum (1 strain) were analysed in this study. Since several genomes are described for H. pylori, we selected six fully annotated ones from strains of different geographical regions as described by Smet et al. (2018, DOI: 10.1038/s41396-018-0199-5). For comparison, the genomes from the Campylobacter species C. coli (3 strains) and C. jejuni (3 strains), which are closely related to Helicobacter, were included as well. Also, the genomes from different E. coli strains (5 strains) were analysed, since the biological functions of most E. coli OMPs are well known. Each strain was chosen out of a different phylogroup based on the paper of Bohlin et al. (2014, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4200225/). An overview of the analysed species and strains, their origin and their accession numbers is shown in Table 1.
Data management and integration
Genomic data was modeled using the Django object relational mapper (ORM) (http://www.djangoproject.com) and database tables were automatically created using the management command ‘syncdb’. The FASTA protein sequences of putative outer membrane proteins (OMP) of the selected strains of E. coli, Campylobacter and Helicobacter were extracted from the genomes by using the HHomp tool22 as described before15. The strains of which their corresponding genome sequences are fully known were deposited in the EMBL databases and the accession numbers are shown in the tables. From the other strains, with unknown genome sequences, their OMPs were uploaded in the database. Subsequently, all OMPs were combined to a single ‘combined.fasta’ file using the Unix ‘cat’ command. Next, a Phython script was written using IPhython Notebook (http://ipython.org/notebook.html), to load all sequences to the database.
Classification into OMP families
The seed alignments from the 90 OMP families that are defined in OMPdb.org were downloaded. For each of these families, the seed alignments were converted to HMM profiles using the HMMER3 suite of programs (http://hmmer.janelia.org). Specifically, the ‘hmmbuild’ program was executed for each seed alignment as follows: ‘hmmbuild <hmmfile_out> <seed_alignment>’. The generated HMM profiles were then stored to the database. Next, the HMM profiles for all of the OMP families were collected and “pressed” for faster searches using the program ‘hmmpress’ in HMMER3, resulting in an HMM database of OMP families. Then, each OMP protein sequence was searched against this HMM database in order to classify them into any of those OMP families. The remaining unclassified proteins were searched against the Pfam database using ‘hmmscan’ in the HMMER3 web server.
Finally, the last remaining 278 unclassified proteins were clustered into groups with CD-HIT using the settings ‘word length = 2, identity cutoff = 40%’ in order to produce the fewest possible number of clusters. The clusters with single members with an amino acid length of <120 (52 sequences in total) were dropped. The distribution of the OMP families from the Escherichia-, Campylobacter- and Helicobacter genera was determined with ‘pandas’, a data processing package in Python (http://pandas.pydata.org). For plotting and visualization, the ‘matplotlib’ plotting package was used (http://matplotlib.org).
Alignment and phylogenetic analysis
The OMP families with at least 3 identified members were subjected to phylogenetic analysis. The OMP protein sequences of these families were written into multi-sequence FASTA files (one for each family). Multiple sequence alignment was performed using the Clustal Omega program (http://www.clustal.org/omega/). The alignments were trimmed with ‘TrimAl’ program (http://trimal.cgenomics.org) in order to remove sequence regions that potentially blur the phylogenetic signal such as highly variable loop regions. The trimmed alignments were then fed into the ‘FastTree’ phylogenetic tree-building program (http://www.microbesonline.org/fasttree). The phylogenetic trees were visualized and edited by the online tool ‘Interactive Tree Of Life’ (iTol)66. The best-fitting root was selected with TempEst v1.567, formerly known as ‘Path-O-Gen’. The OMP protein sequences of all species and strains were subjected to protein BLAST (ncbi). In this way, the individual OMPs belonging to each family as well as possible subgroups could be distinguished. The relative positions of all species’ OMPs in the phylogenetic trees were studied and evaluated, and the possible roles in virulence and colonization are presented.
Accession codes
The NCBI or EMBL accession numbers of the species and strains with fully annotated genomes are provided in Table 1.
Electronic supplementary material
Acknowledgements
We thank Nathalie Van Rysselberghe and Sofie De Bruyckere for their skilled technical assistance. This work was supported by the Flemish Agency for Innovation by Science and Technology (IWT), Grants No. SB-121092 and G033717N, the Research Foundation Flanders (FWO Vlaanderen), the Research Fund of Ghent University, Belgium (code GOA 01G00408), the FWO Vlaanderen Odysseus program (Grant No. G.0902.09), the Special Research Fund (BOF) for a Visiting Foreign Researcher (VBO), the Vice Chancellor of the University of Western Australia, the NHMRC Sir Macfarlane Burnet Fellowship grant (572723), the German Science Foundation (project B10 in CRC-796 and project A04 in CRC-1181), the US National Institute of Environmental Health Sciences (NIH T32- OD010978, R01-OD011141 and P30-ES002109) and the Flanders Institute for Biotechnology (VIB; project grant PRJ9).
Author Contributions
E.B. and M.J. participated in the design of the study, performed the experiments, analysed the data and drafted the manuscript. J.T., M.R., A.D., A.T. and F.P. participated in the design of the study and in the experiments, helped to interpret the results and edited the manuscript. S.B., J.F. and R.D. participated in the design of the study and edited the manuscript. H.R., F.H. and A.S. coordinated the study, participated in the design of the study, helped to interpret the results and edited the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Eva Bauwens, Myrthe Joosten, Joemar Taganna and Mirko Rossi contributed equally
Han Remaut, Freddy Haesebrouck and Annemieke Smet jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32476-1.
References
- 1.Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014;345:196–202. doi: 10.1016/j.canlet.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 2.De Falco M, et al. Molecular Mechanisms of Helicobacter pylori Pathogenesis. J. Cell. Physiol. 2015;230:1702–1707. doi: 10.1002/jcp.24933. [DOI] [PubMed] [Google Scholar]
- 3.Oleastro M, Ménard A. The Role of Helicobacter pylori Outer Membrane Proteins in Adherence and Pathogenesis. Biology (Basel) 2013;2:1110–34. doi: 10.3390/biology2031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm RA, et al. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 2000;68:4155–68. doi: 10.1128/IAI.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Exner MM, Doig P, Trust TJ, Hancock RE. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 1995;63:1567–72. doi: 10.1128/iai.63.4.1567-1572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilver D, et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–7. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 7.Mahdavi J, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–8. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaoka Y, Kwon DH, Graham DY. A Mr 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 1999;31:1537–48. doi: 10.1046/j.1365-2958.1999.01300.x. [DOI] [PubMed] [Google Scholar]
- 10.Rossez Yannick, Gosset Pierre, Boneca Ivo G., Magalhães Ana, Ecobichon Chantal, Reis Celso A., Cieniewski-Bernard Caroline, Joncquel Chevalier Curt Marie, Léonard Renaud, Maes Emmanuel, Sperandio Brice, Slomianny Christian, Sansonetti Philippe J., Michalski Jean-Claude, Robbe-Masselot Catherine. The LacdiNAc-Specific Adhesin LabA Mediates Adhesion of Helicobacter pylori to Human Gastric Mucosa. The Journal of Infectious Diseases. 2014;210(8):1286–1295. doi: 10.1093/infdis/jiu239. [DOI] [PubMed] [Google Scholar]
- 11.Backert S, Clyne M, Tegtmeyer N. Molecular mechanisms of gastric epithelial cell adhesion and injection of CagA by Helicobacter pylori. Cell Commun. Signal. 2011;9:28. doi: 10.1186/1478-811X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Königer V, et al. Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat. Microbiol. 2016;2:16188. doi: 10.1038/nmicrobiol.2016.188. [DOI] [PubMed] [Google Scholar]
- 13.Javaheri A, et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat. Microbiol. 2016;2:16189. doi: 10.1038/nmicrobiol.2016.189. [DOI] [PubMed] [Google Scholar]
- 14.Flahou Bram, Haesebrouck Freddy, Smet Annemieke. Helicobacter pylori Research. Tokyo: Springer Japan; 2016. Non-Helicobacter pylori Helicobacter Infections in Humans and Animals; pp. 233–269. [Google Scholar]
- 15.Joosten M, et al. Divergence between the highly virulent zoonotic pathogen Helicobacter heilmannii and its closest relative, the low virulent Helicobacter ailurogastricus sp. nov. Infect. Immun. 2015;84:IAI.01300–15. doi: 10.1128/IAI.01300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermoote M, et al. Genome sequence of Helicobacter suis supports its role in gastric pathology. Vet. Res. 2011;42:51. doi: 10.1186/1297-9716-42-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold IC, et al. Comparative Whole Genome Sequence Analysis of the Carcinogenic Bacterial Model Pathogen Helicobacter felis. Genome Biol. Evol. 2011;3:302–308. doi: 10.1093/gbe/evr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schott T, Kondadi PK, Hänninen M-L, Rossi M. Comparative genomics of Helicobacter pylori and the human-derived Helicobacter bizzozeronii CIII-1 strain reveal the molecular basis of the zoonotic nature of non-pylori gastric Helicobacter infections in humans. BMC Genomics. 2011;12:534. doi: 10.1186/1471-2164-12-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, et al. The Helicobacter heilmannii hofE and hofF Genes are Essential for Colonization of the Gastric Mucosa and Play a Role in IL-1β-Induced Gastric MUC13 Expression. Helicobacter. 2016;21:504–522. doi: 10.1111/hel.12307. [DOI] [PubMed] [Google Scholar]
- 20.Schauer, D. B. EnterohepaticHelicobacterSpecies. Helicobacter pylori: Physiology and Genetics (2001).
- 21.Backert S, Boehm M, Wessler S, Tegtmeyer N. Transmigration route of Campylobacter jejuni across polarized intestinal epithelial cells: paracellular, transcellular or both? Cell Commun. Signal. 2013;11:72. doi: 10.1186/1478-811X-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Remmert M, Linke D, Lupas AN, Söding J. HHomp—prediction and classification of outer membrane proteins. Nucleic Acids Res. 2009;37:W446–W451. doi: 10.1093/nar/gkp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikkonen TP, Kärenlampi RI, Hänninen M-L. Phylogenetic analysis of gastric and enterohepatic Helicobacter species based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 2004;54:753–758. doi: 10.1099/ijs.0.02839-0. [DOI] [PubMed] [Google Scholar]
- 24.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J, Wang S, Zhao F, Xu J. Protein threading using context-specific alignment potential. Bioinformatics. 2013;29:i257–65. doi: 10.1093/bioinformatics/btt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eren E, et al. Substrate Specificity within a Family of Outer Membrane Carboxylate Channels. PLoS Biol. 2012;10:e1001242. doi: 10.1371/journal.pbio.1001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zahn M, et al. Structural Insights into Outer Membrane Permeability of Acinetobacter baumannii. Struct. Des. 2016;24:221–231. doi: 10.1016/j.str.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Ferrara LGM, et al. MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded β-Barrel Monomers with a Ca 2+ Ion Bound at the Constriction Zone. J. Mol. Biol. 2016;428:4528–4543. doi: 10.1016/j.jmb.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayat S, Peters C, Shu N, Tsirigos KD, Elofsson A. Inclusion of dyad-repeat pattern improves topology prediction of transmembrane β-barrel proteins. Bioinformatics. 2016;32:1571–1573. doi: 10.1093/bioinformatics/btw025. [DOI] [PubMed] [Google Scholar]
- 30.Dé E, et al. MOMP (major outer membrane protein) of Campylobacter jejuni; a versatile pore-forming protein. FEBS Lett. 2000;469:93–7. doi: 10.1016/S0014-5793(00)01244-8. [DOI] [PubMed] [Google Scholar]
- 31.Moser I, Schroeder W, Salnikow J. Campylobacter jejuni major outer membrane protein and a 59-kDa protein are involved in binding to fibronectin and INT 407 cell membranes. FEMS Microbiol. Lett. 1997;157:233–8. doi: 10.1111/j.1574-6968.1997.tb12778.x. [DOI] [PubMed] [Google Scholar]
- 32.Petrone BL, Stringer AM, Wade JT. Identification of HilD-Regulated Genes in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2014;196:1094–1101. doi: 10.1128/JB.01449-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cullen TW, et al. Helicobacter pylori versus the Host: Remodeling of the Bacterial Outer Membrane Is Required for Survival in the Gastric Mucosa. PLoS Pathog. 2011;7:e1002454. doi: 10.1371/journal.ppat.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palframan SL, Kwok T, Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front. Cell. Infect. Microbiol. 2012;2:92. doi: 10.3389/fcimb.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sause WE, Castillo AR, Ottemann KM. The Helicobacter pylori autotransporter ImaA (HP0289) modulates the immune response and contributes to host colonization. Infect. Immun. 2012;80:2286–96. doi: 10.1128/IAI.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kersulyte D, Rossi M, Berg DE. Sequence Divergence and Conservation in Genomes of Helicobacter cetorum Strains from a Dolphin and a Whale. PLoS One. 2013;8:e83177. doi: 10.1371/journal.pone.0083177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joosten M, et al. Divergence between the Highly Virulent Zoonotic Pathogen Helicobacter heilmannii and Its Closest Relative, the Low-Virulence ‘Helicobacter ailurogastricus’ sp. nov. Infect. Immun. 2016;84:293–306. doi: 10.1128/IAI.01300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aono R, Negishi T, Nakajima H. Cloning of organic solvent tolerance gene ostA that determines n-hexane tolerance level in Escherichia coli. Appl. Environ. Microbiol. 1994;60:4624–6. doi: 10.1128/aem.60.12.4624-4626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu H-C, Lin T-L, Wang J-T. Identification and Characterization of an Organic Solvent Tolerance Gene in Helicobacter pylori. Helicobacter. 2007;12:74–81. doi: 10.1111/j.1523-5378.2007.00473.x. [DOI] [PubMed] [Google Scholar]
- 42.Paulsen IT, Park JH, Choi PS, Saier MH. A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol. Lett. 1997;156:1–8. doi: 10.1016/S0378-1097(97)00379-0. [DOI] [PubMed] [Google Scholar]
- 43.Koronakis V, Eswaran J, Hughes C. Structure and Function of TolC: The Bacterial Exit Duct for Proteins and Drugs. Annu. Rev. Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 44.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta - Proteins Proteomics. 2009;1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Symmons MF, Marshall RL, Bavro VN. Architecture and roles of periplasmic adaptor proteins in tripartite efflux assemblies. Front. Microbiol. 2015;6:513. doi: 10.3389/fmicb.2015.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Y, Dang W, Sun L. A TonB-dependent outer membrane receptor of Pseudomonas fluorescens: virulence and vaccine potential. Arch. Microbiol. 2012;194:795–802. doi: 10.1007/s00203-012-0812-3. [DOI] [PubMed] [Google Scholar]
- 47.Kingma RL, Snijder HJ, Dijkstra BW, Dekker N, Egmond MR. Functional importance of calcium binding sites in outer membrane phospholipase. A. Biochim. Biophys. Acta - Biomembr. 2002;1561:230–237. doi: 10.1016/S0005-2736(02)00351-6. [DOI] [PubMed] [Google Scholar]
- 48.Nishijima M, Nakaike S, Tamori Y, Nojima S. Detergent-resistant phospholipase A of Escherichia coli K-12. Purification and properties. Eur. J. Biochem. 1977;73:115–24. doi: 10.1111/j.1432-1033.1977.tb11297.x. [DOI] [PubMed] [Google Scholar]
- 49.Grant KA, Belandia IU, Dekker N, Richardson PT, Park SF. Molecular characterization of pldA, the structural gene for a phospholipase A from Campylobacter coli, and its contribution to cell-associated hemolysis. Infect. Immun. 1997;65:1172–80. doi: 10.1128/iai.65.4.1172-1180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorrell N, et al. Characterization of Helicobacter pylori PldA, a phospholipase with a role in colonization of the gastric mucosa. Gastroenterology. 1999;117:1098–104. doi: 10.1016/S0016-5085(99)70394-X. [DOI] [PubMed] [Google Scholar]
- 51.Vollan HS, Tannæs T, Yamaoka Y, Bukholm G. In silico evolutionary analysis of Helicobacter pylori outer membrane phospholipase A (OMPLA) BMC Microbiol. 2012;12:206. doi: 10.1186/1471-2180-12-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ziprin Richard L., Young Colin R., Byrd J. Allen, Stanker Larry H., Hume Michael E., Gray Sean A., Kim Bong J., Konkel Michael E. Role of Campylobacter jejuni Potential Virulence Genes in Cecal Colonization. Avian Diseases. 2001;45(3):549. doi: 10.2307/1592894. [DOI] [PubMed] [Google Scholar]
- 53.Istivan TS, Coloe PJ. Phospholipase A in Gram-negative bacteria and its role in pathogenesis. Microbiology. 2006;152:1263–1274. doi: 10.1099/mic.0.28609-0. [DOI] [PubMed] [Google Scholar]
- 54.Tannaes T, Bukholm IK, Bukholm G. High relative content of lysophospholipids of Helicobacter pylori mediates increased risk for ulcer disease. FEMS Immunol. Med. Microbiol. 2005;44:17–23. doi: 10.1016/j.femsim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 55.Fox JG, et al. Helicobacter mustelae isolation from feces of ferrets: evidence to support fecal-oral transmission of a gastric Helicobacter. Infect. Immun. 1992;60:606–11. doi: 10.1128/iai.60.2.606-611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fox JG, Marini RP. Helicobacter mustelae infection in ferrets: Pathogenesis, epizootiology, diagnosis, and treatment. Semin. Avian Exot. Pet Med. 2001;10:36–44. doi: 10.1053/saep.2001.19544. [DOI] [Google Scholar]
- 57.Deisenhofer J, et al. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 58.Bugaytsova JA, et al. Helicobacter pylori Adapts to Chronic Infection and Gastric Disease via pH-Responsive BabA-Mediated Adherence. Cell Host Microbe. 2017;21:376–389. doi: 10.1016/j.chom.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kavermann H, et al. Identification and Characterization of Helicobacter pylori Genes Essential for Gastric Colonization. J. Exp. Med. 2003;197:813–822. doi: 10.1084/jem.20021531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lina TT, et al. Immune evasion strategies used by Helicobacter pylori. World J. Gastroenterol. 2014;20:12753–66. doi: 10.3748/wjg.v20.i36.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reynolds CM, et al. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J. Biol. Chem. 2006;281:21974–87. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawasaki K, Teramoto M, Tatsui R, Amamoto S. Lipid A 3′-O-deacylation by Salmonella outer membrane enzyme LpxR modulates the ability of lipid A to stimulate Toll-like receptor 4. Biochem. Biophys. Res. Commun. 2012;428:343–347. doi: 10.1016/j.bbrc.2012.10.054. [DOI] [PubMed] [Google Scholar]
- 63.Radin JN, et al. Flagellar localization of a Helicobacter pylori autotransporter protein. MBio. 2013;4:e00613–12. doi: 10.1128/mBio.00613-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gueneau P, Loiseaux-De Goër S. Helicobacter: molecular phylogeny and the origin of gastric colonization in the genus. Infect. Genet. Evol. 2002;1:215–23. doi: 10.1016/S1567-1348(02)00025-4. [DOI] [PubMed] [Google Scholar]
- 65.Fox JG, et al. Helicobacter mustelae-associated gastritis in ferrets. An animal model of Helicobacter pylori gastritis in humans. Gastroenterology. 1990;99:352–61. doi: 10.1016/0016-5085(90)91016-Y. [DOI] [PubMed] [Google Scholar]
- 66.Letunic I, Bork P. Interactive tree of life (iTOL)v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.