Abstract
Knowledge about the modulation of gut microbiota improves our understanding of the underlying mechanism by which probiotic treatment benefits the chickens. This study examined the effects of Bacillus subtilis DSM 32315 on intestinal structure and microbial composition in broilers. Broiler chicks were fed basal diets without or with B. subtilis supplementation (1.0 × 109 spores/kg of diet). Supplemental B. subtilis increased average body weight and average daily gain, as well as elevated villus height and villus height to crypt depth ratio of ileum in broilers. Multi-dimension analysis showed a certain degree of separation between the cecal microbiota from treatment and control groups. Increased Firmicutes abundance and reduced Bacteroidetes abundance in cecum were observed responded to B. subtilis addition, which also increased the abundances of Christensenellaceae and Caulobacteraceae, and simultaneously decreased the abundances of potentially harmful bacteria such as Vampirovibrio, Escherichia/Shigella and Parabacteroides. Network analysis signified that B. subtilis addition improved the interaction pattern within cecal microbiota of broilers, however, it exerted little influence on the metabolic pathways of cecal microbiota by comparison of the functional prediction of metagenomes. In conclusion, supplemental B. subtilis DSM 32315 improved growth performance and intestinal structure of broilers, which could be at least partially responsible by the manipulation of cecal microbial composition.
Introduction
The current tendency in animal production is towards a reduction or banning of the use of antibiotics, and an increase in application of non-antibiotic approaches that can provide similar benefits, due to that widespread use of antibiotics has led to the emergence of resistant bacteria and drug residues in animal products. In this context, researchers have been compelled to find alternates to ongoing therapeutic regimes, mainly dependent over antibiotics. There was an interest to characterize the probiotics as a kind of viable alternatives to promote growth and health status of poultry by multiple ways1. Despite a large amount of microorganisms served as probiotics in poultry production, the form of supplemental probiotics through the hostile environment such as low pH value and high concentration of bile salt within gastrointestinal tract are a severe challenge for their survival2. As a result, spore-forming bacteria such as Bacillus subtilis are gaining interest in animal health related functional additive research, due to their high tolerance and survivability under hostile environment in gastrointestinal tract3. An obvious advantage of B. subtilis applied in feed is the stability and extended shelf life without losing viability. Supplemental B. subtilis has earned many benefit claims, including the immune-modulation, promotion of nutrients digestibility, along with improvements of intestinal health and growth performance in animals4,5. However, many properties of probiotic bacteria vary as a function of strain6. With respect to B. subtilis, its probiotic effects are highly strain-specific and the mechanistic details remain largely elusive7. It was reported that the effects of B. subtilis supplementation on growth performance and intestinal physiology in broilers were markedly strain-dependent8,9, providing a need for the continuous studies on the various strains of B. subtilis to understand their respective properties in animals. B. subtilis DSM 32315, a unique probiotic strain, was derived from a multi-parameter selection process, in which more than 500 environmentally sampled Bacillus strains were screened. This strain exhibited several distinct properties that are vital for its beneficial effects in gastrointestinal tract10. The preliminary trial has shown an improvement of growth performance in broilers associated with B. subtilis DSM 32315 addition11. However, far less is known about the effects of this strain on gut health in chickens.
Gut microbiota, an important inhabitant of host, has profound impacts on the bioavailability of dietary components, playing prominent roles in host nutritional and physiological processes12. A full picture of gut microbiota may be crucial for the dietary intervention to promote growth performance and health status of host12. It was thought that the improvements growth performance and intestinal functions of animals by probiotics could be established by the ability of balancing gut microflora which helped to attenuate intestinal inflammation and recover intestinal mucosa from injury13,14. Previous studies in chickens indicated a change in bacterial enumeration in gut after B. subtilis supplementation5,15, however, a comprehensive understanding is lacking about the shift of the composition and functionality of gut microbiota with B. subtilis addition. In keeping with this, the main objective of this study was to investigate the effects of supplemental B. subtilis DSM 32315 on intestinal microbial composition, which may subsequently benefit intestinal structure and growth performance of broilers.
Results
Growth performance
Supplemental B. subtilis DSM 32315 significantly increased (P < 0.05) average body weight (ABW) of broilers at 28 and 42 d of age (Table 1), along with average daily gain (ADG) of birds during the grower period (1−28 d) and overall period (1−42 d). Besides, average daily feed intake (ADFI) during the overall period was significantly higher (P < 0.05) in birds receiving B. subtilis supplementation. However, no significant difference (P > 0.05) in feed conversion ratio (FCR) of birds was observed between the two groups during the grower or overall period.
Table 1.
Effects of Bacillus subtilis DSM 32315 addition on growth performance in broiler chickens (n = 10). ABW, average body weight; ADG, average daily gain; ADFI, average daily feed intake; FCR, feed conversion ratio. a,bDifferent letters within rows indicate differences between treatment groups (P < 0.05).
| Control | B. subtilis | P-value | ||
|---|---|---|---|---|
| 28 d | ABW (g) | 1430 ± 38b | 1469 ± 38a | 0.033 |
| 42 d | ABW (g) | 2670 ± 97b | 2774 ± 89a | 0.022 |
| Grower 1−28 d | ADG (g) | 49.2 ± 1.3b | 50.6 ± 1.4a | 0.028 |
| ADFI (g) | 67.5 ± 1.9 | 68.9 ± 2.1 | 0.135 | |
| FCR | 1.37 ± 0.04 | 1.36 ± 0.02 | 0.381 | |
| Overall 1−42 d | ADG (g) | 60.4 ± 2.5b | 62.7 ± 2.3a | 0.041 |
| ADFI (g) | 89.9 ± 2.7b | 93.1 ± 2.8a | 0.017 | |
| FCR | 1.49 ± 0.04 | 1.49 ± 0.02 | 0.740 |
Intestinal histomorphology
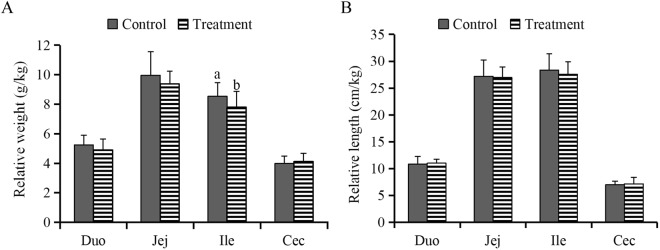
Supplemental B. subtilis DSM 32315 significantly decreased (P < 0.05) the relative weight of ileum of broilers (Fig. 1A), but it had no significant influence (P > 0.05) on the relative weight of duodenum, jejunum, and cecum, or the relative length of these intestinal segments (Fig. 1A,B). The villus height (VH) and VH-to-crypt depth (CD) ratio (VCR) of ileum rather than of duodenum and jejunum were significantly higher (P < 0.05) in treatment group relative to control (Table 2).
Figure 1.
Effects of Bacillus subtilis DSM 32315 addition on the relative weight (A) and relative length (B) of various intestinal segments in broiler chickens (n = 10). Relative weight of intestines was expressed as the ratio of intestinal weight (g) to body weight (kg). Relative length of intestines was expressed as the ratio of intestinal length (cm) to body weight (kg). Duo, duodenum; Jej, Jejunum; Ile, ileum; Cec, cecum. a,bMean values with unlike letters indicate differences between treatment groups (P <0.05).
Table 2.
Effects of Bacillus subtilis DSM 32315 addition on intestinal morphology in broiler chickens (n = 10). VH, villus height; CD, crypt depth; VCR, villus height to crypt depth ratio. a,bDifferent letters within rows indicate differences between treatment groups (P < 0.05).
| Control | B. subtilis | P-value | ||
|---|---|---|---|---|
| Duodenum | VH (µm) | 895.1 ± 211.8 | 894.3 ± 138.3 | 0.992 |
| CD (µm) | 174.7 ± 26.1 | 195.6 ± 26.3 | 0.092 | |
| VCR | 5.12 ± 0.95 | 4.63 ± 0.88 | 0.248 | |
| Jejunum | VH (µm) | 688.0 ± 118.9 | 730.6 ± 102.3 | 0.402 |
| CD (µm) | 142.2 ± 24.1 | 163.7 ± 40.9 | 0.169 | |
| VCR | 4.92 ± 0.98 | 4.61 ± 0.84 | 0.464 | |
| Ileum | VH (µm) | 421.9 ± 58.3b | 551.6 ± 110.9a | 0.004 |
| CD (µm) | 101.2 ± 10.0 | 109.0 ± 24.9 | 0.365 | |
| VCR | 4.18 ± 0.47b | 5.12 ± 0.62a | 0.001 |
Relative mRNA expression of ileal genes
As indicated in Fig. 2, there were no significant changes (P > 0.05) in the expression of tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, IL-10, IL-4, interferon (IFN)-γ, toll-like receptor (TLR)-4 and TLR-2 in ileum responded to B. subtilis DSM 32315 addition.
Figure 2.
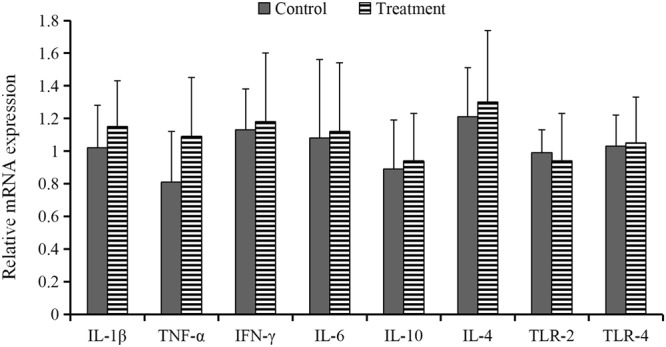
Effects of Bacillus subtilis DSM 32315 addition on intestinal lesion scoring (A) and relative mRNA expression of inflammation-related genes of ileum (B) in broiler chickens (n = 10).
Diversity of cecal microbiota
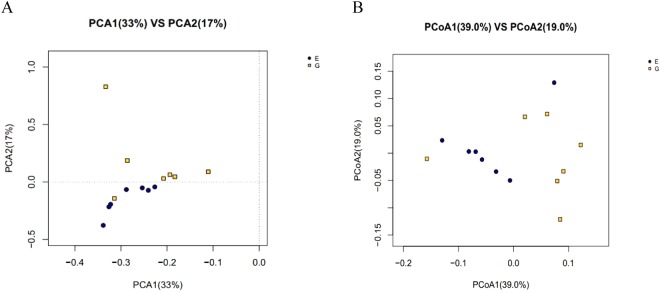
Coverage indice, used to evaluate the percentage of total bacterial Operational Taxonomic Units (OUTs) represented in a sample, were over 99.7% (data not shown), indicating the current 16S rDNA results from each library could represent the complete microbial communities of cecum. However, the α-diversity including Shannon, Simpson, ACE, and Chao1 indexes as well as richness diversity of cecal microbiota were not significantly different (P > 0.05) between treatment and control groups (Supplementary Fig. S1). To estimate the similarity (β-diversity) of microbial community structure between groups, principal component analysis (PCA) based on OUT profile and principal coordinate analysis (PCoA) based on weighted UniFrac distance were performed. As presented in Fig. 3A, PCA plot defined groups where the samples from control and treatment groups occupied distinct positions. The first axis of the PCA explained 33% of the variation in microbial diversity while the second axis explained 17% of it. PCoA plot also showed a trend of separation of microbial communities between the two groups (Fig. 3B).
Figure 3.
Beta-diversity analysis of microbial communities by using principal component analysis (PCA) based on Operational Taxonomic Units (OTUs) and principal co-ordinates analysis (PCoA) based on weighted UniFrac distances (n = 7). Abscissa represents the first principal component, ordinate represents the second principal component, and the percentage represents the contribution of the principal component to the sample difference. E, control group; G, treatment group.
Composition of cecal microbiota
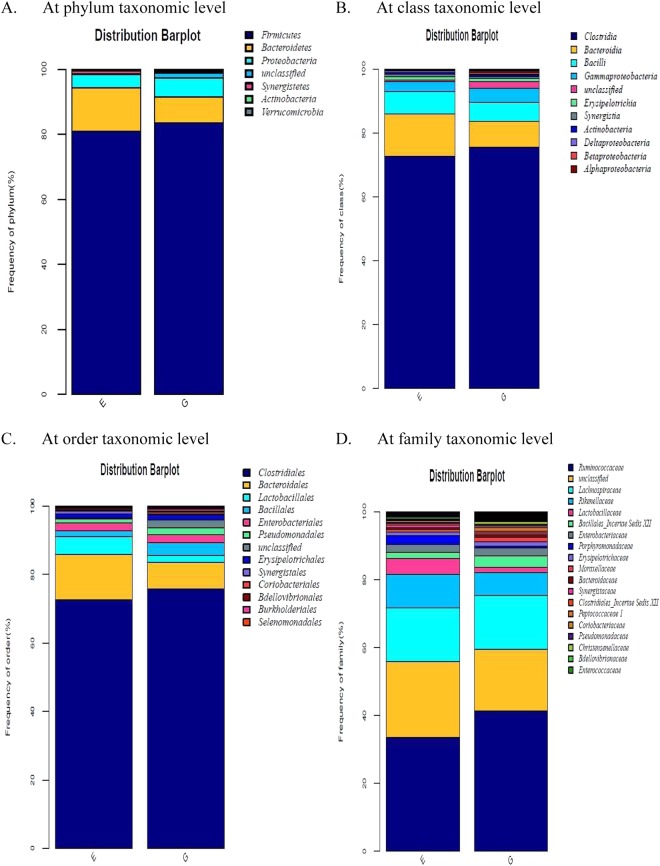
The relative abundance of OTUs of cecal microbiota was analysed at different ranking levels from phylum to genus. The dominant phyla across the two groups were Firmicutes, Bacteroidetes, and Proteobacteria, together contributing greater than 95% of the whole phyla (Fig. 4A). Birds supplemented with B. subtilis had a higher abundance of Firmicutes and a lower abundance of Bacteroidetes. Class level microbiota analysis showed that the cecal microbiota was dominated by Clostridia followed by Bacteroidia and Bacilli, which collectively occupied more than 90% of the total sequences (Fig. 4B). Within phylum Firmicutes, the abundance of Clostridia was higher while Bacilli abundance was lower in treatment group relative to control, which was concomitant with a depletion of Bacteroidetes in treatment group. At order taxonomic level (Fig. 4C), Clostridiales and Bacteroidales accounted for the largest proportion of the community, which were respectively increased and decreased by B. subtilis addition. Within Clostridiales, the majority belonged to the Ruminococcaceae and Lachnospiraceae families (Fig. 4D). An increase of Ruminococcaceae abundance responded to B. subtilis treatment occurred.
Figure 4.
Effects of Bacillus subtilis DSM 32315 addition on cecal microbial composition at different taxonomic levels in broiler chickens (n = 7). E, control group; G, treatment group.
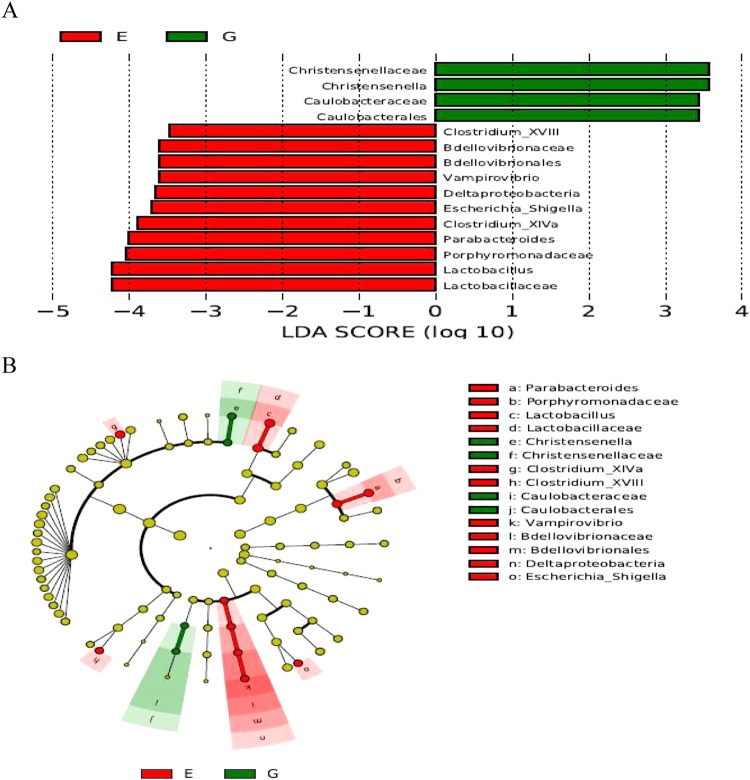
We further applied linear discriminant analysis (LDA) combined effect size measurements (LEfSe) to explore the relative richness (P < 0.05, LDA > 2.0) of bacterial members in control or treatment groups. As presented in Fig. 5, Christensenella (Christensenellaceae) and Caulobacterales (Caulobacteraceae) were enriched in cecum of treatment group, whereas control microbiota was differentially enriched with Clostridium XVIII, Bdellovibrionales (Bdellovibrionaceae), Vampirovibrio, Deltaproteobacteria, Escherichia/Shigella, Clostridium XIVa, Parabacteroides, Porphyromonadaceae and Lactobacillaceae (Lactobacillus).
Figure 5.
Linear discriminant analysis (LDA) combined effect size measurements (LEfSe) analysis of cecal microbiota in broiler chickens (n = 7). (A) Species with significant difference that have an LDA score greater than the estimated value (the default score is 2.0). The length of the histogram represents the LDA score. (B) The cladogram diagram shows the microbial species with significant differences between the two groups. The species classification at the level of phylum, class, order, family, and genus exhibit from the inside to the outside. Red and green nodes in the phylogenetic tree represent microbial species that play an important role in control and treatment groups, respectively. Yellow nodes represent species with no significant difference. E, control group; G, treatment group.
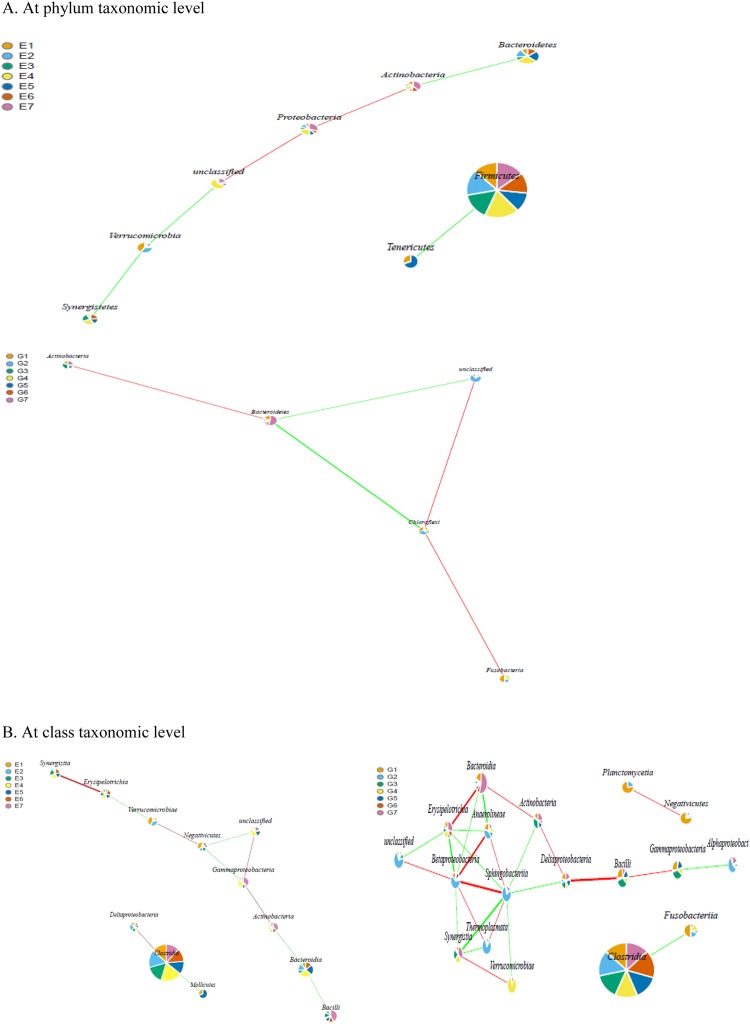
Network pattern of cecal microbiota
We employed correlation network analysis to identify interactions between gut microbiota that changed with administration of B. subtilis DSM 32315. As shown in Fig. 6A, there was a negative correlation between Firmicutes and Tenericutes, as well as between Bacteroidetes and Actinobacteria in control microbiota. However, a destruction to this network was found in treatment group, in which Bacteroidetes was positively and negatively correlated with Actinobacteria and Chloroflexi, respectively, while no correlation was detected between Firmicutes and other members. Besides, a more complicate microbial interaction pattern that evidenced by the presence of more core nodes (such as Sphingobacteria, Betaproteobacteria and Erysipelotrichia that had the highest number of connections with the rest of bacterial members) was found in response to B. subtilis DSM 32315 addition (Fig. 6B). For control group, Clostridia was positively and negatively correlated with Deltaproteobacteria and Mollicutes, respectively, while Bacteroidia was negatively correlated with Bacilli and Actinobacteria. In treatment group, however, there was a negative correlation between Clostridia and Fusobacteriia, as well as a highly positive correlation between Bacilli and Deltaproteobacteria. Remarkably, Bacteroidia was positively correlated with both Actinobacteria and Erysipelotrichia. Moreover, the classes Sphingobacteria, Betaproteobacteria and Erysipelotrichia had the highest number of connections with the rest of the community in treatment group. Network analysis at both order and family taxonomic levels also revealed more complex co-occurrence relationships within cecal microbiota of treatment group in comparison with control, as exhibited by a higher number of edges in the network from treatment group than that from control group (Supplementary Fig. S2).
Figure 6.
The co-network pattern of cecal microbiota at phylum (A) and class (B) levels in broiler chickens (n = 7). The significance of correlation was set as P <0.05 and expressed by the dotted line, while the high significance of correlation was set as P <0.01 and expressed by the full line. The red and green lines among nodes stand for the positive and negative correlations, respectively. The weight of the lines correspond to the correlation coefficients, whose values were higher and lower than 0.8 were represented by the thick and thin lines, respectively. The diameter of the nodes was proportional to the relative abundance. Taxa in brackets are based on annotations suggested by the Greengenes database. E, control group; G, treatment group.
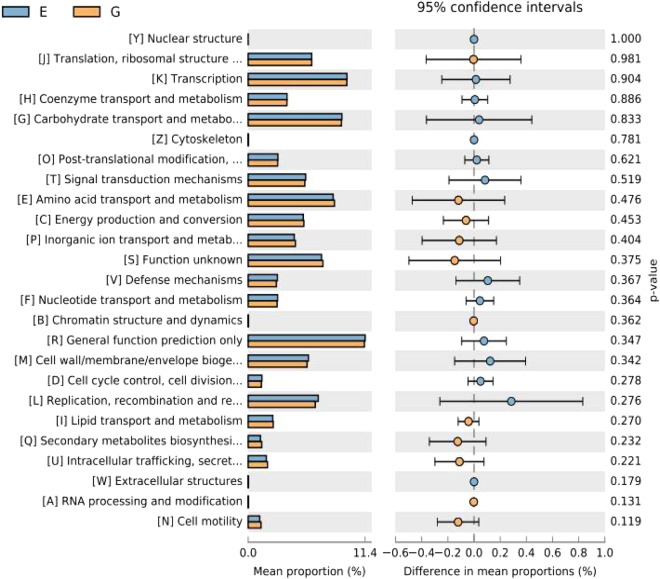
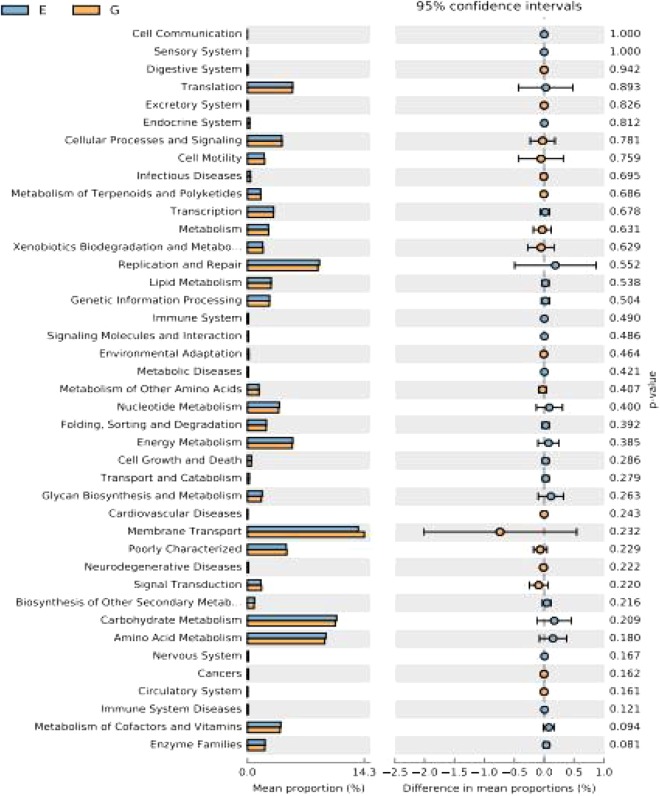
Functional prediction of cecal microbiota
The changes in the presumptive functions of cecal microbiota were examined by predicting the metagenomes using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt). Before the examination, the functional accumulation curve was employed to evaluate the sufficiency of sampling number and the richness of functional components, as it can reflect the rate of emergence of new COG (Clusters of Orthologous Groups) categories or KEGG (Kyoto Encyclopedia of Genes and Genome) orthologs responded to the continuous sampling. As exhibited in Supplementary Fig. S3, the functional accumulation curve rise rapidly in the case of small sample and become smooth while the sampling number growing, indicating a plenitude of sampling in the present study. Metagenomic prediction based on COG categories revealed that the functional pathways enriched within cecal microbiota of broilers were general function prediction only, transcription, carbohydrate transport and metabolism, amino acid transport and metabolism, replication, recombination and repair, cell wall/membrane/envelope biogenesis (Supplementary Fig. S4). In terms of the prediction based on KEGG orthologs, the abundant functional annotations of cecal microbiota were those corresponding to membrane transport, carbohydrate metabolism, amino acid metabolism, replication and repair, translation, energy metabolism (Supplementary Fig. S5). However, there was little difference in the predicted pathways between the microbiota from treatment and control groups based on COG categories or KEGG orthologs through Welch’s t-test (Figs 7 and 8) and heatmap analysis (Supplementary Figs S6 and S7).
Figure 7.
Functional comparison of cecal metagenomic sequence based on Clusters of Orthologous Groups (COG) categories in broiler chickens (n = 7). The differences between the levels of the predicted functions were tested using a two-sided Welch’s t test. E, control group; G, treatment group.
Figure 8.
Functional comparison of cecal metagenomic sequence based on Kyoto Encyclopedia of Genes and Genome (KEGG) orthologs in broiler chickens (n = 7). The differences between the levels of the predicted functions were tested using a two-sided Welch’s t test. E, control group; G, treatment group.
Discussion
Growing evidences indicated that administration of B. subtilis into diets improved the growth performance of animals4,5,8. Contradictorily, some other studies found no relation of B. subtilis to growth performance of chickens16,17. It could be speculated that the effects of B. subtilis treatment are uncertain depending largely on various factors such as the bacterial strains4, since there was a distinction between the effects of different strains of B. subtilis on chicken performance4. Herein, we found an improvement of growth performance, as characterized by the increased ADG of broilers in the presence of B. subtilis DSM 32315, whose efficacy may derive from a modulation on the intestinal environment4,18. To verify this, we next investigated the shift of intestinal structure and microbial composition of birds with B. subtilis DSM 32315 addition.
Improvement of intestinal morphology including elevated VH and VCR suggest an increased absorptive surface area and expression of brush border enzymes that capable of greater digestion and absorption19, conducing to the improved growth performance in animals. In concert with a previous study5, we found increases in ileal VH and VCR responded to B. subtilis DSM 32315 addition, which might be partially responsible for the elevated ADG and ABW of broilers. The relative weight and relative length of intestinal segments are important estimators of gut histology. Herein, supplemental B. subtilis DSM 32315 decreased the relative weight of ileum in broilers, which was similar to the study of Reis et al.20 who reported an improved growth performance concomitant with a lessened relative weight of duodenum of broilers after B. subtilis (DSM 17299) addition. Increased weight of intestine could be an adaptive response to an increased need for digestion and absorption21. It was confirmed that supplementing some functional additives such as enzymes to diets could promote digestion and absorption, but which potentially attenuate the secretory function of intestine, leading to a reduction of the organ weight in animals22. B. subtilis was proposed to secrete a wide range of digestive enzymes within intestinal tract aid in digestion and absorption23. Thus, the present reduction of the relative weight of ileum may result from an enhanced absorption capacity, as supported by the improvement of ileal morphological structure with B. subtilis DSM 32315 addition.
Cytokines such as IL-1β and TNF-α are small immunoregulatory peptides that produced mainly by the activated macrophages or T lymphocytes through the activation of TLRs signaling pathways, aiding cell-to-cell communication during cellular immune responses24. B. subtilis was documented to trigger production of several kinds of cytokines, acting as the response of mononuclear cells to the stimulus contained in probiotic bacteria23. However, we found that B. subtilis DSM 32315 exerted little effect on the expression of ileal cytokines and TLRs, which was not in accordance with a previous study regarding the immunoregulation of B. subtilis B10 addition in broilers25. This discrepancy may be attributed to differences in the bacterial strains.
Gut microbiota conduce to the maintenance of normal physiological structure and function of intestinal tract, providing a variety of benefits for their host12–14. It is generally recognized that the improved growth performance and intestinal structure by probiotics are largely ascribed to a healthy modulation of gut microbiota13,26. In chickens, it was observed a change in bacterial enumeration in gut responded to B. subtilis addition5,15, however, to what extent it can influence the composition and functionality of gut microbiota is poorly understood. In the current study, though B. subtilis DSM 32315 addition failed to modify the α-diversity of cecal microbiota of broilers, it conferred several changes to the microbial composition, which were evidenced by a higher abundance of Firmicutes and a lower abundance of Bacteroidetes in treatment group, accompanied by the increased ABW and ADG of broilers. Firmicutes plays influential roles in polysaccharide decomposition and energy utilization in gut because of genes coding for the secretion of glycan-degrading enzymes27. Moreover, growth performance of animals was positively correlated with the abundance of Firmicutes especially the ratio of it to Bacteroidetes abundance in gut13,28. Hence, the elevation in Firmicutes to Bacteroidetes ratio in treatment group, reflected by the relevant increase in Firmicutes and reduction of Bacteroidetes, could at least partially explain the observed increases in ABW and ADG of broilers fed with B. subtilis DSM 32315. Clostridia comprises mostly non-pathogenic commensal bacteria including the members that capable of degrading complex glycans and proteins, connecting with the improved growth performance of chickens29,30. Ruminococcaceae, one of the most abundant family from the order Clostridiales in gut, has a high copy of genes equipped to degrade a serious of glycans such as cellulose31. Therefore, the increased abundances of Clostridia and Ruminococcaceae were likely associated with the increased ADG and ABW due to B. subtilis 32315 addition. LEfSe analysis identified more representative species as biomarkers to distinguish the microbiota of these two groups, including Christensenellaceae and Caulobacteraceae, along with Clostridium XVIII, Vampirovibrio, Deltaproteobacteria, Escherichia/Shigella, Parabacteroides, Porphyromonadaceae and Lactobacillus. Christensenellaceae was proposed to be associated with feed intake in animals32, whose reduction in gut was observed in reaction to gut inflammation33. Caulobacteraceae was characterized as the microorganism responsible for decomposing cellulose in ecosystems34. Clostridium XVIII was proved to be increased in the patients with bowel movement disorder35, highlighting a negative role of this bacterium in the regulation of gut health. The genus Vampirovibrio and class Deltaproteobacteria were highly enriched in gut of infected chickens, and associated with intestinal inflammation and injury36,37. Escherichia/Shigella, as opportunistic pathogenic bacteria, were described to destroy intestinal structure and exert pro-inflammatory activities through multiple ways such as the production of virulence factors38, resulting in an increased risk to the infection and diarrhea of host39. Parabacteroides also represents an opportunistic pathogen in gut, due to its frequent involvement in infectious diseases together with its ability to develop resistance to antimicrobial drugs40. Besides, Parabacteroides was considered to be linked with the reduction of body weight of host animals41. The increased Porphyromonadaceae was identified as a response to gut inflammation and tumorigenesis42, leading to an elevated risk of the occurrence of several intestinal diseases43. Taken together, the increased abundances of Christensenellaceae and Caulobacteraceae, along with the decreased abundances of Clostridium XVIII, Deltaproteobacteria, Escherichia/Shigella, Parabacteroides, Porphyromonadaceae in gut of treatment group could contribute to the improved growth performance and intestinal structure of broilers fed with B. subtilis DSM 32315. Strikingly, there was a reduction of Lactobacillus abundance of broilers with B. subtilis DSM 32315 addition. Lactobacillus is generally used as a kind of probiotics in animal production, as some strains of it can exert several benefits for host1,2. However, the negative correlation was also noted between certain strains of Lactobacillus and chicken growth performance13. There might be a positive relationship between Lactobacillus abundance and pathogen-induced gut dysfunction44, based on an enrichment of Lactobacillus in gut after enteric infection by pathogenic bacteria such as Salmonella44,45. The reasons for the reduced abundance of Lactobacillus in treatment group are presumably involved with the complex microbial-host milieu. One possibility is that the reduced Lactobacillus members in this study are functionally different from the strains used as probiotics. Alternatively, the reduction of Lactobacillus may be a feedback of the lessened potential pathogens and alleviated gut inflammation after B. subtilis DSM 32315 addition46.
Dynamic balance in microbial ecosystem depends on the interactions between bacteria and host environment as well as among bacterial members47. To our knowledge, no published study was available regarding the alteration of microbe-microbe interactions of gut microbiota induced by dietary intervention in broilers. Network analysis of taxon co-occurrence patterns provides new insights into the internal structure of complex microbial communities48. The positive correlations indicate cooperative interactions or the presence of common biological functions between taxa, while negative correlations could be indicative of competitive interactions between taxa48. A previous study confirmed an improved interaction pattern of gut microbiota in pigs fed with yeast probiotic49. In the present study, a diet-dependent co-occurrence network of cecal microbiota in broilers was observed. For example, there was a negative correlation between Tenericutes and Firmicutes in control group, while this microbial linkage observed in treatment group was absent. Besides, B. subtilis DSM 32315 addition increased relative degree of connectance of Bacteroidetes within the microbiota, and reversed the negative correlation between Actinobacteria and Bacteroidia, as well as promoted the positive correlation of Bacilli and Actinobacteria with some other bacteria, suggesting that B. subtilis DSM 32315 addition could fortify the interaction between Actinobacteria and other members, who can benefit from the metabolic activity of each other, namely mutualistic relationship. Such communities can thereby effectively utilize nutrients within intestinal tract and suppress the colonization by opportunistic bacteria50. In addition, Actinobacteria was regarded as “keystone taxa” that exert essential roles in modulating the functionality of gut microbiota and as a key source of novel bacteriocines and other metabolites, which were helpful for the maintenance of overall microbial structure, along with host growth and intestinal health51,52. Thereby, the potential inhibition of Actinobacteria by Bacteroidetes (as the second abundant phylum) in control group might be adverse to the performance and gut health of broilers, but which could be attenuated by B. subtilis DSM 32315 addition. With regard to the network analysis at order and family levels, higher degree of connectance of bacterial members within the community also occurred in treatment group, which agreed with the analysis results at phylum and class levels. To sum up, supplemental B. subtilis DSM 32315 resulted in a more intricate interaction pattern of cecal microbiota, which might be translated into improvements of growth performance and gut health of broilers49.
Commensal bacteria might increase or decrease certain specific metabolic pathways to react to the change in host intestinal environment that can be induced by dietary intervention in broilers53. In order to infer the alterations of metabolic pathways of cecal microbiota in response to B. subtilis addition, the PICRUSt analysis was employed in the present study. Unexpectedly, broilers shared a core set of predicted metagenomes of cecal microbiota regardless of B. subtilis addition, suggesting a stability of the functional capabilities of cecal microbiota in broilers. The metagenomic prediction based on COG categories revealed that cecal microbiota of broiler was enriched with the functional pathways associated with general function prediction only, transcription, carbohydrate transport and metabolism. In terms of the prediction based on KEGG orthologs, the abundant functional annotations of cecal microbiota were those corresponding to membrane transport, carbohydrate metabolism, amino acid metabolism. Nevertheless, B. subtilis DSM 32315 addition exerted little impact on the predicted pathways of cecal microbiota in broilers based on COG or KEGG database.
In conclusion, supplemental B. subtilis DSM 32315 improved the growth performance and intestinal structure of broilers, the possible mechanism for which was its role in shaping gut microbial composition. Specifically, B. subtilis DSM 32315 supplementation modified cecal microbiota of broilers towards a healthy balance by increasing the beneficial bacteria and decreasing the potentially harmful bacteria, as well as improving the interaction pattern within the community. This study can expand our fundamental knowledge concerning the roles of B. subtilis in the maintenance of intestinal microecological balance in animals.
Materials and Methods
Ethics Statement
All experimental protocols involving animals in this study were carried out in accordance with the institutional and national guidelines and approved by the Animal Care and Use Committee of Chinese Academy of Agricultural Sciences. Every effort was made to minimize animal pain, suffering, and distress.
Animals and experimental design
A total of 240 1-day-old male Arbor Acre broiler chicks were randomly allocated into 2 groups with 10 replicates of each. Each replicate pen involving 12 birds. Initial body weights of birds were similar across all the replicates. Birds were received basal diets without or with 0.5 g/kg B. subtilis (DSM 32315, 2.0 × 109 spores/g, Evonik Nutrition & Care GmbH, Germany) throughout the trial period. The basal diets were formulated based on the feeding standards of China (China′s Ministry of Agriculture, 2004) for broilers. The composition and nutrient levels are shown in Supplementary Table S1. All birds were raised in battery cages in an environmentally controlled room with continuous incandescent white light throughout the experiment. The room temperature was maintained at 33 °C for the first week, and then reduced by 3 °C per week until it reached 24 °C. Feed and fresh water were available ad libitum.
Sample collection
At 42 d of age, birds were randomly selected from each replicate pen and slaughtered rapidly for sample collection. Intestinal segments (duodenum, jejunum, ileum and cecum) from these broilers (10 birds per group) were excised and weighed to determine the relative weight and length of them, which were expressed as the ratio of intestinal weight (g) and intestinal length (cm) to body weight (kg), respectively. The mid regions of duodenum, jejunum and ileum of these broilers (10 birds per group) were then cut off and fixed in 4% paraformaldehyde solution for morphology measurement. In addition, a little patch of ileum from each bird (10 birds per group) was harvested and rinsed, which was then quick-frozen in liquid nitrogen and kept at −80 °C for the quantification of gene expression. Besides, cecal content of broilers (7 birds per group) was collected and quick-frozen using liquid nitrogen, followed by storage at −80 °C for DNA extraction.
Performance measurement
Body weight and feed intake were recorded for each replicate at 28 and 42 d of age. Average body weight at 28 and 42 d of age, along with average daily gain, average daily feed intake, and feed conversion ratio during the grower period (1−28 d) and overall period (1−42 d) were calculated.
Intestinal morphological analysis
The cross-sections of fixed duodenal, jejunal and ileal tissues were obtained after staining with toluidine blue using standard paraffin-embedding procedures. For each sample, ten intact and straight villi in the section were chosen for morphology examination using a light microscope. Villus height was determined from the tip of villus to the junction of villus and crypt, while crypt depth was defined as the depth of invagination between adjacent villi, villus height to crypt depth ratio was calculated. For the purpose of statistical analysis, the average of these measured values was used.
Quantitative real-time PCR for ileal genes
Total RNA was extracted from the ileum using Trizol Reagent (Invitrogen, Carlsbad, USA). Extracted RNA was dissolved in RNase-free water and quantified using an UV/Visible spectrophotometer (Amersham Bioscience, Sweden) at an absorbance of 260 nm. The quality of RNA was estimated from the absorbance ratio at 260 to 280 nm and by determination of the 18S and 28S bands after electrophoresis in 1% agarose gels stained with ethidium bromide. The relative mRNA expression of immune-related genes was determined using quantitative real-time PCR according to the procedures of Zhang et al.54. Gene specific primer sequences are shown in Supplementary Table S2. The results of relative mRNA expression of intestinal genes were calculated using the 2−ΔΔCt method55.
16S rDNA sequencing of cecal microbiota
Total bacteria DNA was extracted from 200 mg of digesta from cecum using the E.Z.N.A.TM Mag-Bind DNA Kit (Omega Bio-Tek, Norcross, USA) following the manufacturer′s instructions. The quality and concentration of extracted DNA were measured using gel electrophoresis and Qubit 2.0 fluorometer (Life Technology, Carlsbad, USA). Bacterial 16S rDNA sequences spanning the variable regions V3–V4 were amplified using primer 341 F (5′-CCCTACACGACGCTCTTCCGATCTG-3′) and 805 R (5′-GACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′)56. The PCR mixture contained 15 μl of Taq Master Mix (Vazyme, Nanjing, China), 1 μL of forward and reverse primers, and 10 ng template DNA in a total volume of 30 μL. The amplification profile consisted of the following procedure: initial denaturation at 94 °C for 3 min, 5 cycles of three steps (94 °C for 30 s, 45 °C for 20 s, and 65 °C for 30 s), 20 cycles of three steps (94 °C for 20 s, 55 °C for 20 s, and 72 °C for 30 s), followed by a final step at 72 °C for 5 min. The PCR products were detected by agarose gel electrophoresis and purified using a PCR Purification kit (SAGON SK8131 kit) (Sangon, Shanghai, China) to remove excess primer dimers and dNTPs. 16S rDNA sequencing was carried out on the Illumina MiSeq 2 × 250 platform (Illumina, San Diego, USA) at Sangon Bioengineering Co. Ltd. (Shanghai, China). Raw tags were produced by merging paired-end reads using FLASH software (v1.2.7)57 and were demultiplexed and quality-filtered using QIIME (v1.7.0)58 to obtain the high quality clean tags. Besides, the chimera sequences were removed to obtain effective tags by using the UCHIME algorithm59. Clustering of filtered sequences into OTUs was achieved using UPARSE (v7.0.1001)60 at 97% sequence identity. Taxonomic classification at different taxonomic levels of OTU sequences were performed by comparing sequences to the GreenGene database61. Shannon and Simpson diversity indices, Chao1 and ACE richness estimators, as well as diversity coverage were included in α-diversity analysis by using the MOTHUR program62. The PCA and PCoA plots were used to estimate pairwise distances between samples and to establish β-diversity. In addition, LEfSe was employed to identify the biological differences between groups (P value less than 0.05 and LDA score higher than 2.0 were considered as significant). Correlation network analysis was used to uncover the internal interaction within the microbial community. Metagenome functional content from 16S rDNA was predicted using PICRUSt63 based on KEGG and COG databases. The results of pathway analysis of each sample were visualized in a heatmap generated by using R package (http://www.r-project.org).
Statistical analysis
Data were presented as mean ± standard deviation. Pens were used as the experimental unit for growth performance parameters, whereas an individual bird served as the experimental unit for other parameters. The t-test (SPSS 18.0 software) was used to measure the effects of dietary treatment. Significance was defined as P < 0.05.
Electronic supplementary material
Acknowledgements
The authors appreciate the financial support provided by Beijing Innovation Consortium of Agriculture Research System (BAIC04-2018) and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ZDXT2018008).
Author Contributions
Y.M. performed the experiments. W.W. analysed the data and drafted the manuscript. H.Z. and J.W. contributed to the writing of the manuscript. S.W and G.Q. conceived and designed the experiments. W.Z. and J.G. contributed to the experimental design. All authors approved the submitted versions.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Youbiao Ma and Weiwei Wang contributed equally.
Contributor Information
Shugeng Wu, Email: wushugeng@caas.cn.
Guanghai Qi, Email: qiguanghai@caas.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33762-8.
References
- 1.Alagawany M. The use of probiotics as eco-friendly alternatives for antibiotics in poultry nutrition. Environ. Sci. Pollut. R. 2018;25:10611–10618. doi: 10.1007/s11356-018-1687-x. [DOI] [PubMed] [Google Scholar]
- 2.Sarao LK, Arora M. Probiotics, prebiotics, and microencapsulation: A review. Crit. Rev. Food Sci. Nutr. 2017;57:344–371. doi: 10.1080/10408398.2014.887055. [DOI] [PubMed] [Google Scholar]
- 3.Hong HA, Duc LH, Cutting SM. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 2005;29:813–835. doi: 10.1016/j.femsre.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Gadde UD, et al. Dietary Bacillus subtilis-based direct-fed microbials alleviate LPS-induced intestinal immunological stress and improve intestinal barrier gene expression in commercial broiler chickens. Res. Vet. Sci. 2017;114:236–243. doi: 10.1016/j.rvsc.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Al-Fataftah AR, Abdelqader A. Effects of dietary Bacillus subtilis on heat-stressed broilers performance, intestinal morphology and microflora composition. Anim. Feed Sci. Tech. 2014;198:279–285. doi: 10.1016/j.anifeedsci.2014.10.012. [DOI] [Google Scholar]
- 6.Dowarah R, Verma AK, Agarwal N, Singh P, Singh BR. Selection and characterization of probiotic lactic acid bacteria and its impact on growth, nutrient digestibility, health and antioxidant status in weaned piglets. PLoS ONE. 2018;13:e0192978. doi: 10.1371/journal.pone.0192978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen N, et al. Characterization of Bacillus spp. strains for use as probiotic additives in pig feed. Appl. Microbiol. Biot. 2014;98:1105–1118. doi: 10.1007/s00253-013-5343-6. [DOI] [PubMed] [Google Scholar]
- 8.Gadde U, et al. The Effects of direct-fed microbial supplementation, as an alternative to antibiotics, on growth performance, intestinal immune status, and epithelial barrier gene expression in broiler chickens. Probiotics Antimicro. 2017;9:397–405. doi: 10.1007/s12602-017-9275-9. [DOI] [PubMed] [Google Scholar]
- 9.Rhayat L, et al. Bacillus subtilis strain specificity affects performance improvement in broilers. Poult. Sci. 2017;96:2274–2280. doi: 10.3382/ps/pex018. [DOI] [PubMed] [Google Scholar]
- 10.Petri, D. et al. New Bacillus subtilis strain comprising B. subtilis and mutant of B. subtilis, useful for e.g. degrading water-insoluble cellulose and protein under anaerobic conditions, and inhibiting growth of Clostridium perfringens bacteria. US Patent, US2017340683-A1 (2017).
- 11.Doranalli, K., Whelan, R., Petri, D., Srinongkote, S. & Rao, S. R. Feeding Bacillus subtilis DSM 32315 consistently improves performance of broilers reared in various feeding conditions. Poult. Sci. (Abstr. #T125) (2017).
- 12.Susan EP, Paul WO, Catherine S, Ross RP, Fitzgerald GF. Intestinal microbiota, diet and health. Br. J. Nutr. 2014;111:387–402. doi: 10.1017/S0007114513002560. [DOI] [PubMed] [Google Scholar]
- 13.Crisol-Martínez E, Stanley D, Geier MS, Hughes RJ, Moore RJ. Understanding the mechanisms of zinc bacitracin and avilamycin on animal production: linking gut microbiota and growth. Appl. Microbiol. Biotechnol. 2017;101:4547–4559. doi: 10.1007/s00253-017-8193-9. [DOI] [PubMed] [Google Scholar]
- 14.Wang WW, et al. Dietary live yeast and mannan oligosaccharide supplementation attenuate intestinal inflammation and barrier dysfunction induced by Escherichia coli in broilers. Br. J. Nutr. 2016;116:1878–1888. doi: 10.1017/S0007114516004116. [DOI] [PubMed] [Google Scholar]
- 15.Molnar AK, et al. Effect of different concentrations of Bacillus subtilis on growth performance, carcase quality, gut microflora and immune response of broiler chickens. Br. Poult. Sci. 2011;52:658–665. doi: 10.1080/00071668.2011.636029. [DOI] [PubMed] [Google Scholar]
- 16.Lee KW, Lillehoj HS, Jang SI, Lee SH. Effects of salinomycin and Bacillus subtilis on growth performance and immune responses in broiler chickens. Res. Vet. Sci. 2014;97:304–308. doi: 10.1016/j.rvsc.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 17.Zhang LL, et al. Dietary effects of Bacillus subtilis fmbj on the antioxidant capacity of broilers at an early age. Poult. Sci. 2017;96:3564–3573. doi: 10.3382/ps/pex172. [DOI] [PubMed] [Google Scholar]
- 18.Mountzouris KC, et al. Effects of probiotic inclusion levels in broiler nutrition on growth performance, nutrient digestibility, plasma immunoglobulins, and cecal microflora composition. Poult. Sci. 2010;89:58–67. doi: 10.3382/ps.2009-00308. [DOI] [PubMed] [Google Scholar]
- 19.Pluske JR, et al. Maintenance of villus height and crypt depth, and enhancement of disaccharide digestion and monosaccharide absorption, in piglets fed on cows’ whole milk after weaning. Br. J. Nutr. 1996;76:409–422. doi: 10.1079/BJN19960046. [DOI] [PubMed] [Google Scholar]
- 20.Reis MP, et al. Effect of Bacillus subtilis (DSM 17299) on performance, digestibility, intestine morphology, and pH in broiler chickens. J. Appl. Poult. Res. 2017;26:573–583. doi: 10.3382/japr/pfx032. [DOI] [Google Scholar]
- 21.Brenes A, Smith M, Guenter W, Marquardt RR. Effect of enzyme supplementation on the performance and digestive tract size of broiler chickens fed wheat- and barleybased diets. Poult. Sci. 1993;72:1731–1739. doi: 10.3382/ps.0721731. [DOI] [PubMed] [Google Scholar]
- 22.Hashemipour H, Khaksar V, Rubio LA, Veldkamp T, van Krimpen MM. Effect of feed supplementation with a thymol plus carvacrol mixture, in combination or not with an NSP-degrading enzyme, on productive and physiological parameters of broilers fed on wheat-based diets. Anim. Feed Sci. Tech. 2016;211:117–131. doi: 10.1016/j.anifeedsci.2015.09.023. [DOI] [Google Scholar]
- 23.Elshaghabee FMF, Rokana N, Gulhane RD, Sharma C, Panwar H. Bacillus as potential probiotics: status, concerns, and future perspectives. Front. Microbiol. 2017;8:1490. doi: 10.3389/fmicb.2017.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jimenez-Dalmaroni MJ, Gerswhin ME, Adamopoulos IE. The critical role of toll-like receptors-from microbial recognition to autoimmunity: a comprehensive review. Autoimmun. Rev. 2016;15:1–8. doi: 10.1016/j.autrev.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajput IR, et al. Effect of Saccharomyces boulardii and Bacillus subtilis B10 on intestinal ultrastructure modulation and mucosal immunity development mechanism in broiler chickens. Poult. Sci. 2013;92:956–965. doi: 10.3382/ps.2012-02845. [DOI] [PubMed] [Google Scholar]
- 26.Wang WW, et al. Effects of Kluyveromyces marxianus supplementation on immune responses, intestinal structure and microbiota in broiler chickens. PLoS ONE. 2017;12:e0180884. doi: 10.1371/journal.pone.0180884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ransom-Jones E, McCarthy AJ, Haldenby S, Doonan J, McDonald JE. Lignocellulose-degrading microbial communities in landfill sites represent a repository of unexplored biomass-degrading diversity. Msphere. 2017;2:e00300–17. doi: 10.1128/mSphere.00300-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han GG, et al. Evaluating the association between body weight and the intestinal microbiota of weaned piglets via 16S rRNA sequencing. Appl. Microbiol. Biotechnol. 2017;101:5903–5911. doi: 10.1007/s00253-017-8304-7. [DOI] [PubMed] [Google Scholar]
- 29.Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanley D, Hughes RJ, Geier MS, Moore RJ. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identifcation of performance enhancing probiotic bacteria. Front. Microbiol. 2016;7:187. doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fbrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. 2013;5:627–640. doi: 10.3390/d5030627. [DOI] [Google Scholar]
- 32.McCormack UM, et al. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl. Environ. Microb. 2017;83:e00380–17. doi: 10.1128/AEM.00380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy NA, et al. The impact of NOD2 variants on fecal microbiota in crohn’s disease and controls without gastrointestinal disease. Inflamm. Bowel Dis. 2018;24:583–592. doi: 10.1093/ibd/izx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinnell LJ, Dunford E, Ronan P, Hausner M, Neufeld JD. Recovering glycoside hydrolase genes from active tundra cellulolytic bacteria. Can. J. Microbiol. 2014;60:469–476. doi: 10.1139/cjm-2014-0193. [DOI] [PubMed] [Google Scholar]
- 35.Shimozato A, et al. Transglucosidase improves the bowel movements in type 2 diabetes mellitus patients: a preliminary randomized double-blind, placebo-controlled study. United Eur. Gastroent. 2017;5:898–907. doi: 10.1177/2050640617692268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yitbarek A, Weese JS, Alkie TN, Parkinson J, Sharif S. Influenza A virus subtype H9N2 infection disrupts the composition of intestinal microbiota of chickens. FEMS Microbiol. Ecol. 2018;94:165. doi: 10.1093/femsec/fix165. [DOI] [PubMed] [Google Scholar]
- 37.Oakley BB, Kogut MH. Spatial and temporal changes in the broiler chicken cecal and fecal microbiomes and correlations of bacterial taxa with cytokine gene expression. Front. Vet. Sci. 2016;3:11. doi: 10.3389/fvets.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaur T, Ganguly NK. Modulation of gut physiology through enteric toxins. Mol. Cell. Biochem. 2003;253:15–19. doi: 10.1023/A:1026088915240. [DOI] [PubMed] [Google Scholar]
- 39.Sousa MAB, Mendes EN, Apolonio ACM, Farias LD, Magalhaes P. Bacteriocin production by Shigella sonnei isolated from faeces of children with acute diarrhea. APMIS. 2010;118:125–135. doi: 10.1111/j.1600-0463.2009.02570.x. [DOI] [PubMed] [Google Scholar]
- 40.Nakano V, et al. Antimicrobial resistance and prevalence of resistance genes in intestinal Bacteroidales strains. Clinics. 2011;66:543–547. doi: 10.1590/S1807-59322011000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng HL, et al. Lotus seed resistant starch regulates gut microbiota and increases short-chain fatty acids production and mineral absorption in mice. J. Agric. Food Chem. 2017;65:9217–9225. doi: 10.1021/acs.jafc.7b02860. [DOI] [PubMed] [Google Scholar]
- 43.Roselli M, et al. Immunomodulating effects of probiotics for microbiota modulation, gut health and disease resistance in pigs. Anim. Feed Sci. Tech. 2017;233:104–119. doi: 10.1016/j.anifeedsci.2017.07.011. [DOI] [Google Scholar]
- 44.Borton MA, et al. Chemical and pathogen-induced inflammation disrupt the murine intestinal microbiome. Microbiome. 2017;5:e47. doi: 10.1186/s40168-017-0264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh P, et al. Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome. 2015;3:2049–2618. doi: 10.1186/s40168-015-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Endt K, et al. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 2010;6:e1001097. doi: 10.1371/journal.ppat.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sam MZ, et al. Network analysis suggests a potentially ‘evil’ alliance of opportunistic pathogens inhibited by a cooperative network in human milk bacterial communities. Sci. Rep. 2015;5:8275. doi: 10.1038/srep08275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiros TG, et al. Effect of live yeast Saccharomyces cerevisiae (Actisaf Sc 47) supplementation on the performance and hindgut microbiota composition of weanling pigs. Sci. Rep. 2018;8:5315. doi: 10.1038/s41598-018-23373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemergut DR, et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013;77:342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trosvik P, Muinck EJ. Ecology of bacteria in the human gastrointestinal tract-identifcation of keystone and foundation taxa. Microbiome. 2015;3:1. doi: 10.1186/s40168-015-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barka EA, et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016;80:1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin DF, et al. Supplemental thymol and carvacrol increases ileum Lactobacillus population and reduces effect of necrotic enteritis caused by Clostridium perfringes in chickens. Sci. Rep. 2017;7:7334. doi: 10.1038/s41598-017-07420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, et al. Stimulation with monochromatic green light during incubation alters satellite cell mitotic activity and gene expression in relation to embryonic and posthatch muscle growth of broiler chickens. Animal. 2014;8:86–93. doi: 10.1017/S1751731113001882. [DOI] [PubMed] [Google Scholar]
- 55.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Roos S, et al. 454 Pyrosequencing analysis on faecal samples from a randomized dbpc trial of colicky infants treated with Lactobacillus reuteri DSM 17938. PLoS ONE. 2013;8:e56710. doi: 10.1371/journal.pone.0056710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 61.DeSantis TZ, et al. Greengenes, a chimera-checked 16SrRNA gene database and work bench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.