Abstract
Centromeres are the chromosomal domains required to ensure faithful transmission of the genome during cell division. They have a central role in preventing aneuploidy, by orchestrating the assembly of several components required for chromosome separation. However, centromeres also adopt a complex structure that makes them susceptible to being sites of chromosome rearrangements. Therefore, preservation of centromere integrity is a difficult, but important task for the cell. In this review, we discuss how centromeres could potentially be a source of genome instability and how centromere aberrations and rearrangements are linked with human diseases such as cancer.
Centromeres are the chromosomal domains that regulate assembly of the components required for chromosome separation. Here the authors review how centromeres are a potential source of genome instability and link centromere aberrations and rearrangements to human diseases such as cancer.
Introduction
During each cell cycle, cells duplicate their genetic material before dividing it equally among daughter cells, so that the genetic information is faithfully distributed. To achieve this goal all eukaryotes have developed a common mechanism by which chromosomes are attached by microtubule fibers organized in a spindle that physically pull the two chromatids of the same chromosome towards opposite poles. Centromeres are key elements in this process. They are the site for the assembly of the kinetochore, the complex protein structure required for the interaction between spindle fibers and chromosomes, and for recruiting signaling components that ensure proper kinetochore-microtubule attachments (reviewed in ref. 1). They are also essential to maintain sister chromatids together by mediating assembly of the cohesin complex until proper chromosome separation occurs (reviewed in ref. 2).
With few exceptions, such as the point centromeres found in budding yeast, monocentric centromeres from plant to animal species are normally established on highly repetitive DNA arrays such as satellite DNAs and transposable elements, although the sequence itself is not conserved3–5. Human centromeres are built on a series of head-to-tail tandem repeats of 171 basepair (bp) AT-rich DNA6,7 named alpha satellites, that extend for several megabases (Mb) and make up ~3% of the genome (reviewed in ref. 8). Adjacent monomers can form higher-order repeat (HOR) units in which blocks of multiple repeats form a larger domain that can in turn be repeated thousands of times, giving rise to the Mb-sized human centromeres. Individual monomers show 50–70% sequence identity while HORs show up to 95% identity (reviewed in ref. 8). Pericentromeric DNA is also organized in tandemly and short repeated sequences, mainly containing Satellite I (chromosomes 3, 4 and the acrocentrics 13–15, 21, and 22), Satellite II (chromosomes 1, 2, 7, 10, 16, and 22) and Satellite III (chromosomes 1, 9, 10, Y and the acrocentrics 13–15, 21, and 22) (reviewed in ref. 9).
A fraction of centromere repeats contains a 17 bp motif named the CENP-B box. CENP-B boxes are the binding site for CENP-B (Centromere Protein B), the only sequence-specific centromeric DNA-binding protein identified so far10. The importance of CENP-B in centromere biology is currently a paradox. Its binding to DNA has been shown to play an important role in centromere function and chromosome segregation at least in mouse and humans11,12. Nevertheless, CENP-B and CENP-B boxes have not been identified outside vertebrates, with some rare exceptions in yeasts and insects. Furthermore, they are absent from neocentromeres and the male Y chromosome and CENP-B is not essential in mice (reviewed in ref. 13).
In addition, centromeric DNA sequence is not sufficient to maintain centromere position, but rather centromeres are epigenetically identified. From fission yeast to humans the histone H3 variant CENP-A was demonstrated to be the epigenetic mark for centromere identity and function (reviewed in ref. 14) by forming an unique centromeric chromatin in complex with the other canonical histones (H2A, H2B, and H4). CENP-A only marks active centromeres independently from DNA sequence15 and mediates centromere assembly through tightly regulated complex processes (reviewed in ref. 14).
Defects in any of the pathways that regulate centromere assembly and function can lead to chromosome mis-segregation and aneuploidy, common features of cancer cells (reviewed in ref. 14). However, in addition to inducing numerical chromosome alterations, centromere dysfunctions could also destabilize chromosome integrity, leading to structural alterations. In many cases, the centromere region itself is the site of chromosome breakage. This intrinsic fragility is probably due to the high density of repetitive sequences that makes the centromere more vulnerable and prone to rearrangements. In this review, we summarize the current knowledge on the types, causes, and consequences of alterations that lead centromeres to become potential sites of chromosome fragility and how these are linked with human diseases.
Centromere breaks and disease
As mentioned above, failure in chromosome segregation leads to numerical and structural alterations, both common features of tumor cells1. Despite the fact that these alterations are known to participate in early steps of tumorigenesis and in cancer heterogeneity, the mechanisms underlying mitotic errors that lead to structural chromosome alterations still remain elusive. It is interesting to note that chromosome rearrangements and breaks often involve (peri)centromeric regions, with a frequency of up to 40–60% in certain cancer cell lines such as those deriving from colorectal carcinomas and adenocarcinomas (based on the analysis of 345 human carcinoma lines from the NCBI SKY/CGH database16,17). Different methods have allowed the identification of several cases of translocation involving centromeres in various tumors (Table 1)18–33 although the distinction between pericentromeric and centromeric regions is often hindered by the use of low-resolution technology and difficulties in aligning sequencing reads of repetitive DNA. For this reason, when related to breaks, the term “centromere” used in this review includes also its flanking heterochromatin.
Table 1.
List of human tumors with the reported chromosome breaks around the centromere region
| Chromosome involved | Type of tumor | Technique(s) used | Reference |
|---|---|---|---|
| 18, 8, 7 | Breast cancer | FISH, aCGH | Alsop et al.18, Cooke et al.19, Bièche et al.26 |
| 1, 18 | Melanoma | FISH | Alsop et al.18, Smedley et al.27 |
| 18 | Colorectal | FISH | Alsop et al.18 |
| 18, 8 | Pancreatic | FISH, aCGH | Alsop et al.18, Cooke et al.19 |
| Several | Squamous cell carcinoma | FISH, SKY, aCGH | Hermsen et al.28 |
| Several | Adenocarcinoma | FISH, SKY, aCGH | Hermsen et al.28 |
| Several | Oral squamous cell carcinoma | FISH | Hermsen et al.29 |
| Mostly 3, 5 | Head and neck squamous cell carcinoma | FISH, fiber-FISH, aCGH, MLPA | Martinez et al.30, Martins et al. 31 |
| 1 | Non-Hodgkin’s lymphoma | FISH | Baccon et al.32 |
| 1 | Multiple myeloma | FISH | Baccon et al.32 |
| 1 | Ovarian cancer | FISH | Thompson et al.33 |
| 10, 14, 7, 21 | Cervical squamous cell carcinoma | FISH | Backsch et al.20 |
| Mostly 17 | Hematologic malignancies | FISH | Adeyinka et al.21 |
| Several | Prostate cancer | G-banding, SKY | Balachandar et al.22 |
| 17 | Male breast cancer | MLPA, FISH | Lacle et al.23 |
| 1, 8 | Hepatocellular carcinoma | Whole genome sequencing, aCGH | Fernandez-Banet et al.24 |
| 1, 7 | Acute myeloid leukemia | FISH | Ganly et al.25 |
FISH fluorescence in situ hybridization, f-FISH fiber FISH, aCGH array-based comparative genomic hybridization, SKY spectral karyotyping, MLPA multiplex ligation-dependent probe amplification, G-banding Giemsa banding, WGS whole-genome sequencing
A functional genomic analysis on 8000 human cancer genomes from high-resolution array-CGH platforms present in the GEO database revealed that whole-chromosome arm gain, loss, or translocation are in fact common alterations in tumors34. Altogether, this suggests that centromeres are indeed fragile and prone to rearrangements during tumorigenesis.
Structural consequences of centromere breakage
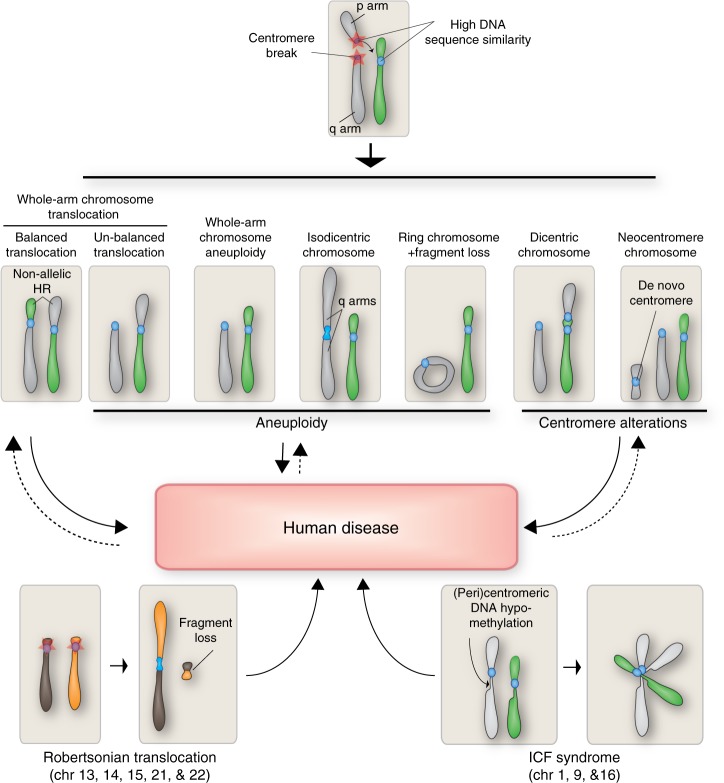
Centromere breaks can generate whole-arm chromosome translocations in which the whole-arm of a chromosome fuses to another chromosome with either no loss of genetic information (balanced translocation) or with loss of genetic material and generation of a derivative chromosome (unbalanced translocation) (Fig. 1). One potential mechanism for the formation of whole-arm chromosome translocations are non-allelic exchanges in non-homologous chromosomes (Non Allelic Homologous Recombination, NAHR) at centromeric repeats during DNA repair of the break. This recombination event between homologous regions located at different loci on different chromosomes might be driven by the high DNA sequence similarity between centromere repeats that favors the occurrence of NAHR. This type of recombination event has been frequently observed in samples from couples with balanced reciprocal translocations during a study for preimplantation genetic diagnosis. Here, it was observed that the centromere itself was the actual breakpoint of the translocation in the majority of analyzed individuals35, suggesting that damage at this site is not so rare. The result of these translocations was the appearance of derivative chromosomes with two centromeres very close to one other often with mixed and different lengths of alpha-satellite repeats. Occasionally, more than one chromosome was involved in this breakage at the same time and, in some cases, the derivative chromosomes were conserved in the family, indicating that they can be meiotically and mitotically stable.
Fig. 1.
Centromere breakage and human diseases. Representation of the genomic alterations deriving from centromere breaks such as aneuploidy, dicentric, and neocentromere chromosomes, Robertsonian translocations together with loss of methylation at (peri)centromere (typical of ICF syndrome), all known to contribute to human diseases
Centromere breaks and translocations can also induce the formation of isochromosomes, chromosomes that carry identical copies of the same arm joined through a single centromere, therefore generating partial trisomy (Fig. 1). It was observed that the smaller the arm, the more likely the survival of the embryo, probably due to the reduced amount of duplicated genetic material. Two main molecular mechanisms have been suggested to generate isochromosomes. The first one described is a phenomenon called “centromere mis-division”: instead of dividing longitudinally, the chromosome would undergo a transverse splitting by separating p- and q- arms36. However, most isochromosomes are rarely monocentric, but isodicentric with two centromeres very close to one another (see “Causes and consequences of dicentric chromosomes” paragraph). The second mechanism known as U-Type exchange, an event that takes place when two sister chromatids undergo a double-strand break and the two broken ends fuse creating a dicentric chromosome37,38. Wolff et al. demonstrated that the majority of breakpoints resulting in isochromosome X formation—the most common isochromosome, responsible for some cases of Turner syndrome—involved the pericentromere and not the centromere itself39. They also observed that it is the proximal short arm that is prone to break and re-join the sister chromatids or the homologous X chromosomes via U-Type exchange. However, more recent characterization of the pericentromeric region of chromosomes highlighted the presence of inverted repeats that could be subject to NAHR giving rise to isochromosomes40–42. Other types of replication-dependent recombination mechanisms such as FoSTeS (Fork Stalling and Template Switching) and MMBIR (microhomology-mediated break-induced replication) could also promote isochromosome formation41.
Centromere breaks in disease
Several tumor types that exhibit chromosomal breakpoints occurring at centromeric regions with high frequency are also associated with high degree of chromosomal instability (CIN) (Table 1). For example, the cytogenetic feature of oral squamous cell carcinoma is the high presence of isochromosomes and unbalanced whole-arm translocations that originate from chromosomal breakpoints. These chromosomal alterations with consequent loss of genetic material mainly occur at centromeric regions (134 out of 218 chromosomal breaks, >60%) and involve several chromosomes including acrocentrics29. Similarly, head and neck squamous cell carcinoma is characterized by the presence of whole-arm translocations which exhibit breakpoints close to the centromere (84%)30. This high percentage suggests that centromeric breaks are directly involved in the origin and evolution of this tumor type although, even in this case, we cannot discriminate whether centromere breakage is a direct cause or simply a consequence of high levels of rearrangements. Indeed, data collected from snapshots of late stage cancer tissues cannot really tell us when such abnormalities occurred during tumorigenesis. Some specific alterations characterize other cancer types such as gain or loss of 1q, 2p, 6p, or 16q arm in retinoblastoma43 and correlate with the degree of malignancy and prognosis21. Hematologic malignancies also show a high frequency of the isochromosome i(17)q, whose breakpoint maps in a complex pericentromeric region characterized by palindromic LCRs separated by a spacer, which would support the hypothesis that the isochromosomes are originated by NAHR40.
Centromere alterations and breaks have also been found in human genetic diseases like the Immunodeficiency, Centromeric region instability, Facial anomalies syndrome (ICF), where mutations of DNA Methyl-Transferase 3 Beta (DNMT3B) (55% of reported cases), ZBTB24, CDCA7, or HELLS44,45 correlate with loss of DNA methylation, pericentromeric breaks, and rearrangements near the centromere with consequent whole-arm deletion (Fig. 1). The exact molecular mechanisms that lead to these defects are still unknown, but previous reports suggest a link between defects in DNA replication and/or chromosome segregation and DNA hypomethylation. Indeed, reduced DNA methylation at centromere/pericentromere has been correlated with aneuploidy46 and with accumulation of micronuclei that contain centromere-positive fragments in human lymphocytes with global hypomethylation47.
Altogether, these evidences suggest that a significant fraction of chromosome instability typically observed in cancer potentially arises from centromeric breakage. Martinez and van Wely described how most of the copy number variations (genetic dosage imbalance) observed in cancer correspond to entire chromosomes or chromosome arms, suggesting that the (peri-)centromere is the preferential break site48. Thus, an initial centromere break could make chromosome segments more prone to several rounds of breakage, therefore directly contributing to cell heterogeneity during cancer transformation. This “centromeric fission model” would explain the high degree of chromosome alterations typically observed in tumors. Given that tumors exhibit both centromere breaks and aneuploidy, it would not be too rash to think that they arise from a common mechanism and that centromere breaks drive a fraction of the CIN observed in cancer.
Causes of centromere breakage
The mechanisms that lead to centromere breaks are still not understood. Below we summarize the current state of knowledge on the possible sources of centromeric ruptures.
Chromosome mis-segregation
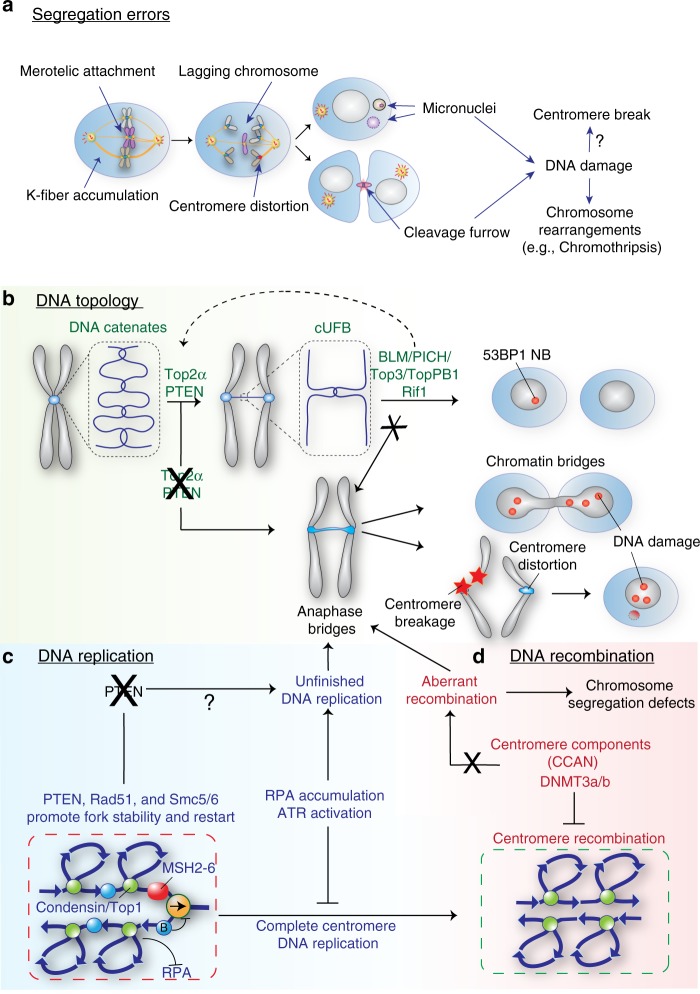
Chromosome segregation errors themselves are one of the proposed potential sources of structural chromosomal alterations and centromere breaks. For example, increased incidence of lagging chromosome formation was linked to the accumulation of DNA damage markers at the cleavage furrow, activation of the ATM/Chk2 response and to an increasing fraction of structural chromosome alterations49 (Fig. 2a). Lagging chromosomes are generally caused by merotelic attachments in which a single kinetochore is attached to microtubules arising from both spindle poles, forcing a single chromatid into a tug-of-war between both poles50. Intriguingly, a more recent study revealed a mitotic role for ATR in preventing lagging chromosomes51. ATR is normally activated by persistent single-strand DNA originated by stalled replication forks or DNA damage. In mitosis ATR localizes at centromeres through Aurora A and CENP-F where it binds RPA-positive-ssDNA generated by R-loops (DNA-RNA hybrids) at centromeres. Here it phosphorylates and activates Chk1 which in turn activates Aurora B, necessary to correct errors in the microtubules–kinetochore attachment1. What generates RNA loops at centromeres and if they contribute to the overall centromere stability together with ATR is unknown, but the presence of ATR and RNA loops reinforces the concept that centromeres are potential fragile regions that need a dedicated surveillance mechanism.
Fig. 2.
Origins of centromeric breaks. Schematic of the possible causes that lead to chromosome breaks at centromere. a Segregation errors: merotelic attachment and k-fiber accumulation are the sources of lagging chromosome formation and centromere distortion in mitosis leading to micronuclei and centromere breaks. b DNA topology: the complex DNA topology enriched in catenanes is normally resolved by topoisomerases prior to completion of sister chromatid separation. Resolution of UFBs is also crucial for maintenance of genome integrity in which a fraction of those results in 53BP1 nuclear bodies (NB) that are repaired in the next cell cycle. Alterations in the centromere decatenation pathway can lead to anaphase bridges and DNA breaks. c, d DNA replication and recombination: due to its highly repetitive sequences a non-canonical pathway of DNA damage and homologous recombination proteins is implicated in the correct completion of centromere replication in order to avoid formation of anaphase bridges, unscheduled recombination and, consequently, segregation errors and breaks
Mis-segregated chromosomes can also be encapsulated in micronuclei, which are a source of genome instability. Indeed lagging or mis-aligned chromosomes generated by microtubule depolymerizing drugs or selective centromere inactivation, respectively, are encapsulated in micronuclei, where they undergo chromosome shattering and re-shuffling, a phenomenon known as chromothripsis52. However, the fate of centromeric regions during the process of chromothripsis was not determined, likely due to the current difficulty of mapping these repetitive regions by conventional DNA sequencing. A direct correlation between centromere breaks and chromosome mis-segregation was proposed to exist upon alteration of the mitotic spindle53. Mouse embryonic fibroblasts (MEFs) depleted of Dido3—structural component of the spindle pole—or human cells carrying a mutation in the tumor suppressor APC that regulates microtubules and kinetochore attachment, accumulate lagging chromosomes, distorted centromeres/kinetochores and eventually micronuclei with γH2AX foci (an early DNA damage marker) in proximity to or at centromeric regions (up to 75%). The authors of these studies propose that the accumulation of kinetochore fibers (K-fibers) and loss of spindle control are the sources of these breaks (Fig. 2a). However, it is still under debate if microtubules can generate enough force to break the double helix.
Further studies are needed to demonstrate a direct causal effect between chromosome mis-segregation and the formation of centromere breaks. Without doubt the centromeres are subjected to high level of mechanical stress during chromosome segregation due to microtubules pulling on the centromere/kinetochore platform. It is tempting to speculate that, to support this stress the centromeres have evolved to accumulate long stretches of repetitive sequences that could form a unique DNA architecture.
Centromere topology
Due to the repetitive nature of their DNA sequences, it is likely that repetitive centromeres have a highly complex DNA topology, which may lead to accumulation of DNA catenanes and formation of DNA loops between their DNA repetitive arrays. Both DNA catenanes and loops represent perfect substrates for the DNA recombination machinery and DNA topoisomerases thus making centromeres potential “fragile” regions in the genome. Pioneering studies in flies54 or using human BACs inserted in Xenopus55 first described the architecture of centromeres. In Drosophila melanogaster, in vitro studies using nuclear magnetic resonance, circular dichroism, and mass spectrometry show complex and stable structures at centromeres called i-motifs54. These are four-stranded structures formed by the association of two duplex DNA strands that are organized in an antiparallel way, reinforced by the occurrence of hemi-protonated C:C+ base pairs. Despite the fact that the specific region analyzed in this study (the G-rich dodeca satellite) was recently found not to be enriched with CID56 (CENP-A homolog in flies), this data suggests that at least regions surrounding functional centromeres (generally marked by CID) contain specific secondary structures. Likewise, using electron microscopy on a BAC containing mainly centromeric DNA from chromosome 17, Aze et al. observed the presence of positive supercoiled DNA loops following incubation with Xenopus egg extract55. These stable structures were proposed to be replication- and condensation-dependent, since full proteinase K treatment led to the disappearance of DNA loops. Indeed, mass spectrometry analysis on reconstituted centromeric BAC revealed the presence of subunits of the condensin complex and topoisomerase II (Top2). The enrichment of Top2 supports previous in vitro data that showed a Top2-mediated DNA cleavage activity on oligonucleotide-forming hairpins of alpha-satellite DNA57.
During mitosis centromeric DNA strands intertwine as a natural consequence of DNA replication, causing an accumulation of catenanes at centromeric regions. At early stages of mitosis, these structures play a role in preventing premature sister chromatid disjunction58, but during anaphase they need to be resolved to preserve centromere stability (Fig. 2b). This catenated DNA also leads to the formation of a particular class of nucleosome-free DNA bridges defined centromeric ultra fine bridges (cUFBs), not detectable with DAPI or other usual DNA staining59. The origins and the role the of this type of centromeric UFBs are still unclear, but it was suggested that they are physiological structures that arise from unresolved topological domains and/or recombination intermediates issuing from potential DNA loops at centromeric regions60.
DNA decatenation is mainly performed by Top2a and its inhibition leads to an increased number of UFBs and chromosomal segregation defects59 (Fig. 2b). In support of its role in DNA decatenation at (but not only) centromeric regions after DNA replication, Top2a accumulates at centromeric regions in budding yeast61 and was shown to be required to resolve topological domains at converging forks also at centromeric regions62. Top2a function at UFBs is supported by two helicases: Bloom’s syndrome protein (BLM) and Plk1-interacting checkpoint helicase (PICH), which both decorate UFBs59,63 (Fig. 2b). PICH is recruited at cUFBs in prometaphase and is necessary for BLM localization64. In addition, PICH has been shown to have a nucleosome remodeling activity required to remove histones from UFBs. This function was proposed to give a sufficient temporal window for Top2a to easily access DNA catenates, and, in turn, to decatenate sister chromatids DNA64.
BLM and PICH are essential for correct chromosome segregation. Indeed, their depletion is associated with changes in centromeric structure, highlighted by a reduced signal of centromeric probes by FISH and by an accumulation of denser centromeric chromatin observed by electron microscopy65. PICH and BLM loss also correlates with non-disjunction of sister chromatids, formation of micronuclei, and lack of recruitment of Top2a65. Finally, BLM is also required to recruit other components to UFBs critical for their resolution such as Topo 3α, that in turn stimulates BLM activity66. Interestingly, Top3 together with Rqh1 (a RecQ helicase) was demonstrated to regulate CENP-ACnp−1 binding at centromere in fission yeast possibly regulating the interplay between topology, recombination, and transcription67. Other factors implicated in the resolution of c-UFBs are TopBP168 and Rif169 that either control Top2 activity, or directly promote c-UFB resolution, respectively.
In summary, a correct regulation of the “centromere decatenation pathway” is critical for the maintenance of genome stability, since any defects in this process lead to segregation errors, chromosome breaks and rearrangements (including at centromeric regions) and hence, potentially, to diseases. One example of disease is Bloom’s syndrome, an inherited disorder characterized by short stature, sun sensitivity and a strong predisposition to any kind of cancer. This syndrome is caused by mutations of BLM that leads to sister chromatids exchanges (SCE), a marker for increased homologous recombination (reviewed in ref. 70).
DNA replication
Unfinished or incorrect DNA replication is another possible source that contributes to centromere breaks. Correct replication of the genome is a prerequisite for the maintenance of genome stability. The licensing of DNA replication origins is restricted to one round per cell cycle by cyclin-dependent kinases, so that the replicative apparatus cannot reinitiate a new synthesis session in the same cell cycle. Along the same line, the presence of only one centromere per sister chromatid is guaranteed to ensure the faithful segregation of chromosomes (see “Causes and consequences of dicentric chromosomes” paragraph). Indeed, induction of centromere re-replication in budding yeast was shown to alter chromosome segregation and induce aneuploidy by destroying the symmetry of normal sister chromatids leading to bipolar attachments of the same chromatid71.
Because of the centromeres’ repetitive nature, their replication might represent a demanding job that generally makes this region unstable, prone to replication errors and to recombination due to the formation of secondary structures (reviewed in ref. 72). How higher Eukaryotes deal with replication problems at centromeres is still unknown. Recent studies on reconstituted chromatin of BACs containing human centromeric DNA in Xenopus laevis showed that centromeric regions display similar replication patterns to other repetitive-free DNA regions. However, centromere DNA was enriched in DNA repair factors such as MRE11/RAD50, Ku80, PARP1, and the MSH2-6 complex (involved in mis-match repair) suggesting that these factors are necessary for correct centromere replication55. Interestingly, following replication stress by aphidicolin treatment, centromeric regions were shown to be depleted of the ssDNA-binding protein RPA-1 (normally enriched under this circumstance) and, consequently, the ATR pathway was not activated. This checkpoint silencing, likely mediated by the positive supercoiling of centromeric DNA structures, together with an increased activity of the MSH2-6 complex, was proposed to be required for successful finalization of DNA replication at centromeric regions55 (Fig. 2c).
Curiously, a direct link between the centromeric component CENP-B and DNA replication has been shown. CENP-B has been shown to take part in centromere replication preserving its integrity during replicative fork stall in both yeast and humans. In fission yeast CENP-B homologs (autonomously replicating sequence-binding protein 1, Abp1, and CENP-B homologs 1 and 2, cbh1/2) were shown to induce the silencing of long terminal repeat (LTR) retrotransposons, recruitment site of Sap1, in order to prevent replication stress by making the forks progress73. In humans, alpha-satellite sequences seem to behave like origins of replication74 and CENP-B, which binds these sequences, is required to regulate the assembly of the replication apparatus on these origins. Consequently, CENP-B depletion results in a change of alpha-satellite chromatin that leads to an increased number of replication origins at centromere74. These evidences would suggest that beside its role in centromere function12,75, CENP-B binds the CENP-B boxes in the alpha-satellite repeats in order to preserve centromere integrity by regulating DNA synthesis and replication fork stall.
The presence of repetitive sequences per se might not be the only problem that replication forks have to deal with at centromeres. For example, during DNA synthesis of the non-repetitive centromere of budding yeast that contains a defined sequence of 125 bp enriched with AT, replication forks pause while approaching centromeric DNA, due to the presence of the centromeric protein complex76. Additionally, at least half of centromeric regions in budding yeast were identified as sites of converging forks, therefore generating even more steric impediment for fork progression62. If not solved correctly, the ensuing replication termination sites can be the source of DNA breaks as observed in the topoisomerase top2 mutant, further demonstrating how centromeres can potentially become fragile sites.
DNA replication stress seems to be one of the main causes of chromosomal instability in some colorectal cancers. Silencing of the so called “CIN suppressor genes” (such as PIGN, MEX3C, and ZNF516) in this type of tumors drives to a variety of chromosomal instability and segregation defects (e.g., lagging chromosomes, acentric fragments, anaphase bridges, chromosome fusion leading to dicentric chromosomes)77. These abnormalities that lead to DNA damage can be rescued by the addition of nucleosides, strongly suggesting the involvement of replication stress in the generation of this chromosomal instability. Replication stress induced by aphidicolin was also shown to lead to enrichment of double-strand breaks (DSBs) at centromeric regions in several cancer cell lines78. This intrinsic fragility might also be enhanced by the fact that, at least in humans, centromeres are late replicated regions79 and therefore, under certain conditions, they may be more prone to stall or incomplete DNA replication, with consequent emergence of UFBs and/or genome instability.
A wide range of other solid tumors which exhibit CIN also have mutations in PTEN, a tumor suppressor gene involved in the repression of phospoinositide 3-kinase (PI3-K) signaling cascade. Interestingly, PTEN was shown to bind centromeres by interacting with CENP-C through its C-terminus80. PTEN centromeric localization is important to prevent centromere breakage and to maintain chromosome stability80 possibly by regulating homologous recombination (HR) during the recovery of replication fork stall, via Rad51 recruitment81. Additionally, PTEN has also been correlated with sister chromatids decatenation, a process occurring after DNA replication and essential for proper chromosome segregation (see section: “Centromere topology”). Indeed, PTEN C-terminus physically binds and regulates DNA topoisomerase IIα (TOP2A) expression in a p53-independent manner by stabilizing TOP2A via its deubiquitination through OTUD382. In summary, by associating with centromeric regions via CENP-C, PTEN is involved in the resolution of replication fork pausing by promoting HR and in regulating DNA decatenation via TOP2A (Fig. 2b, c). This would explain why tumors that suppress PTEN function exhibit chromosome instability.
In conclusion, replicating centromeric DNA is a difficult task for the cells due to the accumulation of large regions of repetitive DNA. Repetitive sequences are generally unstable and might form secondary structures that could induce replication fork stalling and high levels of recombination. This, in turn, could lead to DNA breakage in certain genetic backgrounds. In this view, it is not surprising that centromeres evolved to have a unique replication apparatus rich in DNA repair factors normally required for DSB processing and repair.
Heterochromatin and recombination
In vertebrates, heterochromatic regions surround centromeres in which the chromatin assembly factor CAF-1 deposits H3 and promotes H3K9 mono-methylation. This is in turn recognized by the histone-lysine N-methyl-transferase Suv39h1 which tri-methylates H3 on Lysine 9 (H3K9m3). H3K9m3 creates the docking sites for the heterochromatin protein 1 (HP1) complex, whose recruitment is essential for heterochromatin formation at those peri-centromeric regions (reviewed in ref. 83). Interestingly, in fission yeast pericentromeric heterochromatin is characterized by the constitutive presence of low levels of γH2AX84, typically associated with DNA damage. This intrinsic stress might be caused by continuous stalling of replication forks. In support of this idea, both the SMC5/6 and Brc1, that binds γH2AX, are enriched at the centromere to ensure replication fork restart85,86.
It has been established that stalled replication forks are mainly resolved through homologous recombination. Unresolved HR intermediates during replication fork re-start lead to replication errors, which in turn lead to deleterious consequences during chromosome segregation such as anaphase bridges and centromere breaks. Centromere repeats provide a substrate for HR by promoting break-induced replication (BIR) even in the absence of stress conditions87,88. In C. albicans it was proposed that the main HR regulators Rad51 and Rad52 control centromere fork stall and restart by directly controlling CENP-A deposition since it acts as a physical barrier for fork progression89. It has to be tested if a similar mechanism exists in higher Eukaryotes.
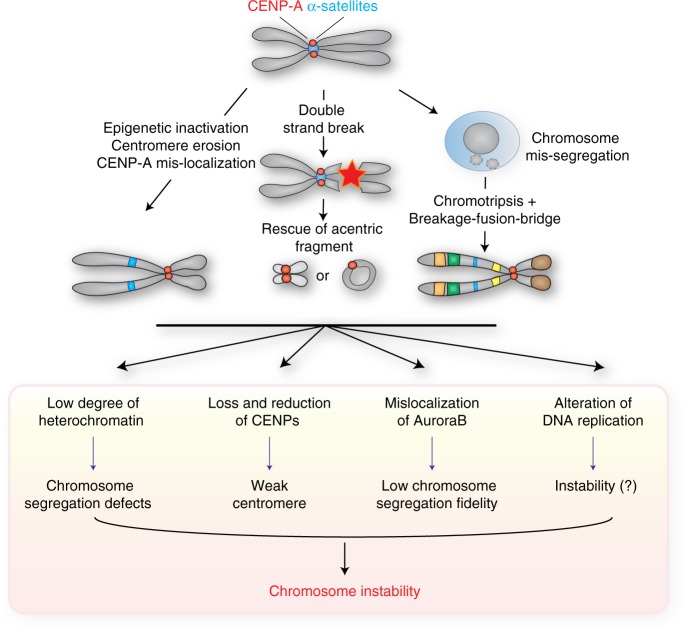
This suggests that recombination is an essential event in centromere maintenance, although it might expose centromeres to potential chromosome rearrangements. The heterochromatic state of the pericentromere region itself is necessary to maintain centromere integrity by protecting it from high levels of recombination. Indeed, defects in heterochromatin formation correlate with gross chromosomal rearrangements and segregation defects as observed in fission yeast90. In mouse it has been shown that an increase in recombination at centromere leads to changes in the length of the repeats, that in turn affects centromere integrity91. Moreover, loss of DNA methylation at mouse minor satellite repeats due to the absence of DNA methyl-transferases 3A/3B, leads to a deregulation of recombination at centromere and shortening of centromere repeats91 (Fig. 2d). Mechanistically, how DNA methylation is involved in centromere organization, function and stability has not been fully explored and remains an open question. We can speculate that either the modified epigenetic status of chromatin at minor satellites following DNA methylation loss or the shortening of centromeric repeats, or both events, may affect the binding of centromeric proteins, thus resulting in genomic instability. Finally, it has been recently shown that gradual removal of CENP-A and some of its associated components such as CENP-C/T/W from human centromeres correlates with a drastic increase in centromere aberrations observed by cen-CO-FISH (centromere chromosome orientation fluorescent in situ hybridization), where more than 90% of centromeres per metaphase displayed abnormal patterns of the centromeric probes92 (Fig. 2d). In addition, CENP-A loss led to excision of centromeric repeats, assessed by the presence of centromere probes outside the centromeres92.
To which extent recombination at centromere/pericentromere is beneficial for centromere is unclear. Centromeres are highly dynamic regions whose size and diversity acquired during evolution could be explained by recombination. However, it is well-known that recombination at centromere is normally repressed during meiosis as demonstrated by the fact that the crossing over between homologous chromosomes is suppressed (so-called “recombination cold spots”) (reviewed in ref. 93). This suppression mechanism is necessary since recombination would affect chromosome cohesion and/or correct kinetochore attachment to fibers radiating from spindle poles, and in turn could generate aneuploid nonviable gametes (reviewed in ref. 94). Nevertheless, it has also been observed that the occurrence of homologous recombination between centromere repetitive sequences leads to covalently closed loops and that these structures can have an important role in the establishment of a functional centromere60. In addition, non-crossover gene conversion has been shown to occur at centromere probably promoting sequence exchanges95. The importance of recombination within centromeric DNA repeats needs to be further demonstrated, in particular in light of the existence of the stably inherited neocentromere that can form at non-repetitive regions96.
Neocentromere implication in genome instability and disease
The neocentromere is a rare and novel class of centromere, described in humans and other species, that normally lacks repetitive DNA sequences and their binding partner CENP-B (reviewed in ref. 97). Due to the absence of alpha satellites, neocentromere formation was among the strongest evidence in support of the centromere being epigenetically defined. Neocentromeres appear in chromosomal locations different from the original centromere and usually form in euchromatic and non-repetitive regions, with some exceptions located in heterochromatic regions such as the ones in the Y chromosome (reviewed by ref. 98) and the repetitive L1 long interspersed nuclear elements (LINEs)99. Interestingly, evolutionary studies suggest the existence of recurrent hotspots for neocentromere seeding, suggesting that centromeres are often re-used after they have been inactivated in ancestral species (reviewed in ref. 100) and might acquire repetitive sequences over time. Curiously, the majority of neocentromeres artificially formed by centromere depletion in chicken cells and C. albicans arise close to the original centromeres101,102 regardless of their chromatin state102. This suggests that CENP-A accumulates also at pericentromeric regions forming so-called “CENP-A clouds” around the centromeres103. In few other cases, neocentromeres formed at other repetitive sequences such as telomeric regions similar to that described in fission yeast104.
Neocentromere and disease
It is still unclear if there is a selective advantage in forming a neocentromere. Neocentromere formation could play a role in maintaining the correct chromosome content in the case of centromere inactivation or loss. Despite this positive role, neocentromere formation is generally associated with cancer or genetic dysfunction (reviewed in ref. 97). Indeed, since their first observation in 1993 during the routine karyotyping of a boy with learning difficulties96, several different constitutional human neocentromeres have been described in individuals with congenital abnormalities, development delay and intellectual disability97. Up to now little is known about the molecular mechanisms that lead to neocentromere formation (Fig. 3). Certain cancers bear neocentromeres, suggesting that chromosome rearrangements may be one of the driver events for de novo centromere formation. Indeed, neocentromere formation was hypothesized to be a mechanism that cancer cells use to rescue the acentric chromosomal fragments arising from their gross chromosomal rearrangements to allow cell proliferation (reviewed in ref. 105). This hypothesis might also be sustained by the fact that cancer cells continuously undergo genomic rearrangements that could also potentially affect the function of the native centromere. However, the correlation between neocentromeres and cancer may be severely underestimated since cancer cells’ karyotype is rarely analyzed with respect to centromere regions using alpha-satellite FISH.
Fig. 3.
Neocentromeres and chromosome instability. A neocentromere forms at chromosomal locations different from the centromere’s original position. The possible causes of neocentromere formation, depicted here, span from epigenetic inactivation, centromere erosion, CENP-A mislocalization to rescue of acentric DNA fragments following chromosome rearrangements. Neocentromeres display, however, a low degree of heterochromatin, loss and reduction of centromeric proteins (CENPs), mislocalization of Aurora B and alteration of DNA replication that cause a decrease in chromosome segregation fidelity
For example, neocentromere formation was correlated with the generation of small supernumerary marker chromosomes (sSMCs; the presence of a structurally abnormal 47th chromosome in the genotypic makeup) in 3% of published sSMC cases (reviewed in ref. 97). How and when a neocentromere is formed is still a matter of debate and much still needs to be characterized. The most common rearrangements involving centromeres are inverted duplications and interstitial deletions. Both originate from chromosomal breaks that in the first case generate a chromosome fragment without a centromere and with an inverted duplication. In interstitial deletions, chromosomal breaks lead to the formation of a ring chromosome and the presence of an additional acentric or centric fragment depending on the site of deletion (paracentric or pericentric respectively) (reviewed in ref. 97). In contrast to other circular chromosomal rearrangements such as double minutes (chromosome fragments carrying gene amplification), these ring chromosomes resulting from interstitial deletions can acquire a neocentromere. For example, it was observed that a patient with intellectual disability, brachyphalangy and growth hormone deficiency carried a 20 Mb deleted region present, as a mosaicism, in a supernumerary ring chromosome stabilized by the neocentromere at 13q31-q32106. However, in vitro observations showed that ring chromosomes are prone to mis-segregation regardless the presence of the neocentromere106.
When neocentromeres form to rescue inverted duplicated fragments107, the resulting cell will be partially tetrasomic if the fragment segregates together with the intact chromatid; alternatively, if the fragment segregates together with the deleted chromatid, the cell will be partially trisomic. Indeed, neocentromere presence is associated with partial monosomy, trisomy, tetrasomy or ring chromosomes. These aberrations are in turn accompanied by developmental delays, short stature, digital malformations, facial dysmorphisms, renal defects and Turner-like syndrome106,108–111. In addition, some tumors such as liposarcoma, retinoblastoma, non-Hodgkin’s lymphoma, acute myeloid leukemia and lung cancer show complex rearranged chromosomes with the presence of neocentromeres112–116. For example, in some cases of liposarcoma neocentromeres are markers of tumor-like SMC rings and giant rods containing the amplification of the 12q1415 region117, or the alphoid-deprived inverted 10q duplication and the chromosome 8-derived ring chromosome in acute myeloid leukemia118.
Neocentromeres have also been associated with gigantic (up to 600 Mb) supernumerary accessory chromosomes (or neochromosomes) typically observed in well-differentiated/dedifferentiated liposarcoma (WD/DDLPS). Studies that followed the evolution of five of these neochromosomes show evidence of chromothripsis, after several cycles of breakage and re-ligation of chromosomes (breakage fusion bridge, BFB), centromeric corrosion followed by de novo centromere formation and telomere capture119. This process of centromere erosion with loss of up to 78% of α-satellites and surrounding pericentromeric DNA observed in these WD/DDLPS cells could be one of the first events that lead to neocentromere establishment. Indeed, this reduction of α-satellite DNA was also observed in other cases of less rearranged chromosomes containing a neocentromere15,120.
Instability of chromosomes containing neocentromeres
This correlation between chromosome alterations and neocentromeres can be envisaged in either direction, with neocentromeres themselves potentially representing a source of genomic rearrangements (Fig. 3). Indeed, a neocentromere is characterized by a lower degree of heterochromatin around CENP-A binding sites with respect to a constitutive centromere121. This peculiarity seems to be correlated with the high frequency of cohesion defects at neocentromeres that, in turn, affect chromosome segregation. Importantly, the absence of CENP-B, another feature of neocentromeres, has been correlated with reduction of centromeric CENP-C and consequent increase of errors during chromosomal segregation75. Additionally, an example of neocentromere present on chromosome 415 displays mislocalization of Aurora B, an essential protein for the error-correction of kinetochore-microtubule attachment122. A further characterization of neocentromere-containing chromosome is required to determine if other molecular mechanisms contribute to its instability.
Neocentromere formation is also believed to change the timing of DNA replication around its chromosomal area, although the function and consequences of this alteration are still not clear. Studies on the well-characterized neocentromere of chromosome 10-derived marker chromosome [mardel(10)] showed that neocentromere formation correlates with a delay in the replication timing of the area surrounding its location123. Consistently, artificially generated neocentromeres in chicken cells101 or in the yeast C. albicans124 lead to a change in replication timing initiation of the chromosomal domain that hosts the de novo centromeres (from early to late in chicken and from mid/late to early in yeast). These results suggest that the formation of a new centromere affects the initiation of DNA replication for reasons that still need to be investigated. It is possible that late replication timing of centromeres in vertebrates such as the one observed in humans79 is a requisite for centromere function and/or stability.
Artificial neocentromere formation
Many methods have been devised to induce neocentromere formation using several model systems, in order to understand how they arise (for space constraints we will not discuss experiments in plants, but we recommend this review on the topic125). Using Drosophila melanogaster, Williams et al. discovered that acentric mini-chromosomes acquire neocentromere activity following chromosome breaks induced by γ-irradiation, although the presence of centromeric proteins was never tested126. In a follow-up study using a supernumerary minichromosome screening in flies after irradiation, Maggert and Karpen observed neocentromere formation only in fragments juxtaposed to an active centromere127. A further link between DNA damage and neocentromere formation came from experiments done in human cells in which transiently overexpressed CENP-A was shown to bind sites of DSBs128. This report, together with the identification of CENP-A chaperone HJURP at sites of DNA damage129, from that originates its name, Holliday Junction Recognition Protein—suggests that sites of DSBs bound by CENP-A might initiate neocentromere formation as a mechanism to prevent the loss of otherwise acentric chromosomes.
Neocentromeres could also be formed by transient overexpression of CENP-A in flies without evidence of DNA damage130 where telomeres and heterochromatin regions are shown to be hotspots for CENP-A islands and de novo centromere formation131. With the exception of budding yeast that harbors a point centromere, neocentromeres can also be induced in fungi by the deletion of the native centromere using a homologous recombination strategy in C. albicans102,132 or the Cre-Lox system in fission yeast104. In human cells overexpression of CENP-A does not lead to neocentromere formation, with the exception of overexpression of its variant constitutively ubiquitylated on K124R-Ub133, although the function of this CENP-A ubiquitination is controversial134.
In some rare cases, other neocentromeres have been reported in an apparently unarranged chromosome15,120,135. It has to be pointed out that cytogenetic analysis is rarely performed in a healthy population, therefore these “rare cases” of neocentromeres may be underestimated. During neocentromere formation in an apparently unarranged chromosome, the functional centromere has been shifted to a new position along the chromosome arm forming a neocentromere, while the original centromere becomes inactive. This configuration of a neocentromere formed on a chromosome that still harbors alphoid DNA is called a pseudodicentric chromosome. Centromere inactivation is required to prevent the formation of a chromosome with two functional centromeres named dicentric chromosome. The mechanisms via which one of the centromeres is inactivated are mainly uncharacterized.
Causes and consequences of dicentric chromosomes
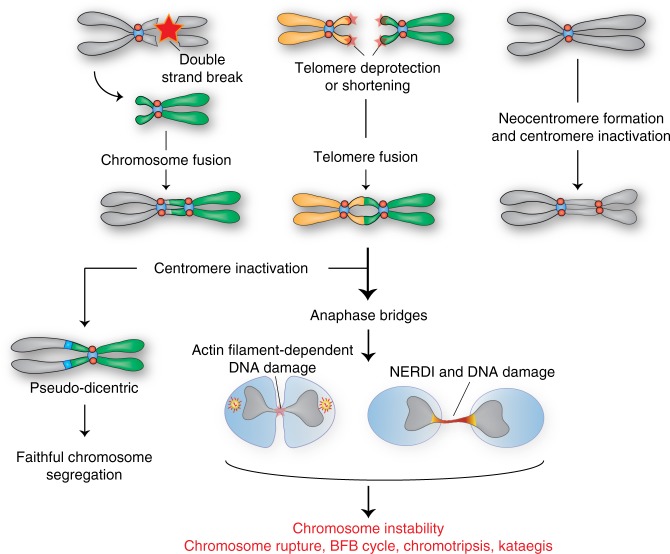
The origin and fate of dicentric chromosomes have been acquiring remarkable importance in the past few years. Dicentric chromosomes were firstly described in maize by Barbara McClintock as the result of a type of crossing over between a broken chromosome and its normal homolog136. Indeed, it is believed that dicentric chromosomes arise after double-strand break formation leading to inversions and translocations or chromosome fusion via telomere deprotection (reviewed in ref. 137) (Fig. 4). In the case of chromosome rearrangements, a physical breakage of two chromosomes, homologs or not, can produce sticky ends that recombine end to end (inversion) generating one dicentric chromosome and two acentric fragments (one in the case of reciprocal translocation). Telomere shortening (e.g. during aging) or dysfunctional telomeres (e.g., loss of the protective Shelterin complex) can also generate telomeric fusions as observed during oncogenesis138 that, in turn, will generate dicentric chromosomes. Dicentric chromosomes can also originate in meiosis from U-Type exchange and NAHR (isodicentric chromosomes; as discussed in the section “Centromere breaks and disease”) and from Robertsonian translocations (see the section “Pseudodicentric chromosomes”)41,139.
Fig. 4.
Dicentric chromosomes and chromosome instability. Representation of the possible origins of dicentric chromosomes that arise from chromosome/telomere fusions or neocentromere formation in the presence of a functional original centromere. During anaphase onset, the dicentric chromosome that is attached by opposing mitotic spindle fibers is trapped, leading to anaphase and chromosome bridges and, consequently, DNA damage and chromosome instability. Centromere inactivation could restore faithful segregation by preventing incorrect spindle attachment
Consequences of dicentric chromosome formation
Dicentric chromosomes are genetically unstable during cell division because microtubules pull in opposite directions on the two centromeres of the same chromatid. This normally leads to the formation of chromosome bridges during anaphase, causing DNA breakage (see below). This phenomenon could potentially lead to cycles of BFB, as first documented by McClintock140.
In budding yeast the two centromeres of a single dicentric chromosome artificially induced by telomere HR and centromere silencing via active transcription141 were shown to attach to opposite mitotic spindle poles. Simultaneous pulling towards the opposite poles led to the formation of anaphase bridges142. Subsequently, dicentric chromosomes are broken at mitotic exit during cytokinesis in an actin filament-dependent mechanism (Fig. 4). The breakage site normally corresponds to the telomere fusion site in the case of dicentrics formed through telomere deprotection143 or to pericentromeric regions within a 25–30 kb range if the dicentric chromosome formation did not involve telomere fusion142. On the contrary, about 50% of dicentric chromosomes induced in human cells by manipulating a component of the shelterin complex appear to be (unexpectedly) stable after several cell cycles (up to 20 generations)144. Eventually, chromosome fragments containing mainly alpha-satellite DNA appear after several weeks. It is important to note that using this system, dicentric chromosomes were mainly generated between acrocentric chromosomes (up to 80% frequency), although stable dicentrics with an intergenic distance of more than 20 Mbs were observed144. Using a very similar approach in pRB/p21 negative human cells, dicentric chromosomes were shown to produce anaphase bridges that did not break during mitosis and persisted for 3–20 h after anaphase145. However, when extensively stretched, these long DNA bridges underwent transient nuclear envelope rupture during interphase (NERDI) giving access to cytoplasmic exonucleases such as TREX1. Nuclease activity led to ssDNA formation (indeed, 80% of these chromatin bridges accumulated RPA) favoring DNA repair and APOBEC-mediated editing of the fragmented chromatin bridge DNA. This chromatin fragmentation and repair was shown to generate chromothripsis and Kataegis (clusters of closely localized base substitutions)145 (Fig. 4).
Similarly, generation of a dicentric chromosome in humans by ectopic formation of a new centromere/kinetochore complex via CENP-T-LacI tethering to LacO gave rise to chromosome breakages. These breakages can be reduced by cytokinesis inhibition, suggesting that, at least in part, cytokinesis forces are involved in dicentric chromosome rupture146. The fragmented chromosome (of the dicentric chromosome) can in turn undergo chromosome fusions that result in new dicentric chromosomes recapitulating all the steps of the BFB cycle. Altogether, these processes/events will generate gene deletions, genetically unbalanced gametes and acquisition of features of transformed cells, such as invasiveness146.
Altogether, this suggests that the appearance of a dicentric chromosome could directly contribute and further exacerbate cellular transformation and cancer heterogeneity. Accordingly, stable dicentric chromosomes have been observed in some human tumors, such as in myeloid malignancies (myelodysplastic syndrome and acute myeloid leukemia), where they involve intercentromeric or centromere deletion, ring chromosomes and centromere inactivation (reviewed in ref. 137).
Pseudodicentric chromosomes
Dicentric chromosomes are generally unstable (especially when generated experimentally), however there are cases of stable dicentric chromosomes, namely pseudodicentric chromosomes (Fig. 4). Their stability is conferred either by deletion of centromeric DNA sequence144 or (mostly) by epigenetic inactivation of one of the two centromeres, causing loss of centromeric components such as CENP-A and CENP-C147,148. An example of pseudodicentric chromosomes are those formed via Robertsonian translocations (ROBs) (Fig. 1). ROBs are a form of rearrangement involving vertebrates acrocentric chromosomes (13, 14, 15, 21, and 22), that mostly break at the pericentromeric region within satellite 3 DNA and that, through chromosomal fusion, generate dicentrics149. In the majority of cases, one of the two centromeres becomes inactive without deletion of centromeric DNA150. However, ROBs with functional dicentrics have been found139 most likely reflecting differences in the intercentromeric distances (reviewed in ref. 151). Two functional centromeres were also found in the dicentric chromosome X152 and in the isochromosome X in the case of selected somatic cell hybrids153. In some cases such as in ROBs in mice, the resulting fused chromosome originated from telocentric chromosomes gives rise to a metacentric chromosome following deletion of both telomeric and (part of) centromeric sequences (reviewed in ref. 154). Interestingly, the newly generated metacentric chromosome is stable and, in one documented case, it was even shown to be stably transmitted by increasing its centromere strength155. However, in humans carriers of ROBs have an increased risk of infertility since the resulting embryos can be un-viable or have elevated rates of Down Syndrome156. Even individuals that carry the most common ROB (13;14)—which constitutes 75% of all cases of this type of rearrangement and occurs in about 1/1000 new-borns157—have unfavorable pregnancy outcomes (miscarriages, chromosomal aberrations, stillbirth, and malformations)158, though whether they have an increased infertility rate is controversial158,159. Recently, this translocation has been correlated with developmental delay in a boy whose mother, also a ROB (13;14) carrier, was phenotypically healthy, suggesting a role of the (epi-)genomic environment in the outcome of the ROBs160. Carriers of the rare ROB (15; 21) show a 2700-fold increased risk of developing childhood acute lymphoblastic leukemia with intrachromosomal amplification of one copy of chromosome 21 (iAMP21 ALL). This high incidence correlated to the presence of rearrangements compatible with chromothripsis observed on the Roberstonian chromosome161. Other types of pseudodicentric chromosomes are Non-ROBs (in 85% of the cases involving one acrocentric chromosome) and are associated with several syndromes such as Kabuki syndrome, a genetic disorder with multiple congenital anomalies, intellectual disability, and growth deficiency162, Edward syndrome, a trisomy of chromosome 18163 and isodicentric X [i(X)] or Y [i(Y)] chromosomes such as Turner syndrome (as described in the paragraph “Centromere breaks and disease”).
Misregulation of centromeric proteins
As observed for the dicentric chromosomes, the presence of centromeric components regulates centromere functionality. This is particularly true for CENP-A, the epigenetic marker that distinguishes the centromere from the rest of the chromatin. Although CENP-A is not exclusively found at centromeric regions, it is highly enriched at centromeres by ~40-fold compared to other regions in the chromosome and this accumulation is necessary to maintain centromeric identity164. For this reason both its expression and loading occur through a very tightly regulated process165–168. What are the consequences for the cells if CENP-A expression is deregulated?
On the molecular level, CENP-A overexpression (OE) leads to its massive delocalization outside the centromeres, mainly at regulatory elements169,170. Using chromatin immuno-precipitation, the Almouzni group demonstrated that promiscuous CENP-A incorporation is dependent on DAXX and occurs at CTCF (CCCTC-binding factor) locations and at sites with high histone turnover such as at H3.3 or H2AZ-rich regions or with H3K4me1 and H3K27ac post-translational modifications169.
Following precursor studies in yeast170–172 and flies130, Shrestha et al. showed that CENP-A OE and consequent mislocalization affected chromosome segregation in HeLa cells173. Interestingly, CENP-A OE altered CENP-C intensity at centromere and its localization, with CENP-C found at non-centromeric regions too. On the contrary, other centromere and kinetochore proteins, like CENP-T and Nuf2, were reduced following CENP-A OE but did not mis-localize. The authors suggested that this reduction contributes to a reduced inter-kinetochore distance that weakened the kinetochores and chromosome segregation fidelity, but without formation of ectopic kinetochores173. This finding is in agreement with what was previously observed by Van Hooser et al. where CENP-A OE was able to recruit a set of kinetochore proteins at ectopic sites that, however, were insufficient to form a fully functional kinetochore174, suggesting that centromere formation is regulated by multiple components. On the contrary, ectopic kinetochores following CENP-A OE can be observed in flies130. Interestingly, CENP-A overexpression in humans leads to increased tolerance of UV-induced DNA damage169. This suggests that acentric chromosome fragments that arise following DNA damage can be preserved in the cells by forming functional centromeres due to CENP-A enrichment.
CENP-A expression might be regulated by tumor suppressor genes such as pRB (Retinoblastoma protein)175. Interestingly, pRB depletion, frequently found mutated in human cancers, correlates with compromised chromatids cohesion and centromere distortion that negatively compromise mitosis176. It is likely that a fraction of these mitotic defects can be directly attributed to CENP-A overexpression, since restoration of CENP-A levels by siRNA prevents micronuclei formation and aneuploidy175 and prevents hepatocellular carcinoma growth177. Indeed, increased levels of CENP-A and its chaperone HJURP178,179 are found in several tumors like hepatocellular carcinoma, breast cancer and colorectal cancer as described above129,177,180–187. In fact, a comprehensive study on public gene expression data sets of normal and cancer tissues (HBI, expO, and CCLE) revealed that core kinetochore and cell cycle genes were significantly up-regulated in tumors. Interestingly, the authors show that upregulation of these genes correlates with high levels of Forkhead Box M1 (FoxM1), a transcription factor known to bind the promoter of the majority of kinetochore genes and cell cycle players188. Accordingly, a more recent bionformatics analysis on 13 data sets from 12 different types of human cancer in the Gene Expression Omnibus (GEO) database, revealed that there is a progressive increase in centromere and kinetochore gene expression (Centromere and kinetochore gene Expression Score, CES) during disease progression of breast, prostate and liver cancers189. These findings suggest that it is possible to draw a gene expression signature predicting cancer patient outcome based on CES. Interestingly, CENP-A is overexpressed in almost all the analyzed cancers, independently of the progression stage, suggesting that this alteration could contribute to cell transformation. More precisely, Filipescu et al. found that CENP-A is overexpressed in p53-null tumors and that CENP-A (and HJURP) OE is a consequence of p53 inactivation since it occurred in MEFs where p53 function was suppressed190. They also showed that HJURP OE is necessary to sustain growth of p53-null transformed cells due to an increase in proliferation versus normal cells.
On the contrary, CENP-A reduction goes in parallel with cell senescence and aging: CENP-A abundance decreases in cells during aging and it becomes almost undetectable in human islets after 29 years of age191. CENP-A reduction by shRNA in an in vitro tissue culture system blocks cell cycle progression in G1 in a p53-dependent manner (likely in response to aneuploidy) leading to either premature senescence192 or activation of the apoptotic pathway177.
Similarly, reduction of HJURP causes a decrease in cell proliferation that correlates with p53 and p21 stabilization193. This cell cycle block is likely to occur in response to aneuploidy, although if this is the only consequence of p53 activation was never directly demonstrated. For example, Giunta and Funabiki described that CENP-A-depleted cells and senescent cells undergo centromeric aberrations likely due to an imbalance in DNA recombination at centromeric regions92. This increase in recombination could generate centromere erosion due to shortening of repetitive DNA. Intriguingly, centromere deterioration was observed during aging194. Senescent cells are also subject to several chromatin changes such as variations in the degree of compaction of pericentromeric/centromeric regions named senescence-associated distension of satellites (SADS). This change was proposed to be an early event that would mark senescent cells195.
The amount of centromeric components at centromeric regions might also have an impact on the frequency of chromosome mis-segregation, a concept defined as “centromere strength”. A good example of this concept is the Y chromosome, that not only is completely devoid of CENP-B boxes and CENP-B, known to support centromere function, but also contains the shortest alpha-satellite array of all chromosomes196. This CENP-B-free human chromosome has reduced levels of some centromeric components such as CENP-A197 and CENP-C75 and mis-segregates at elevated frequencies12. Intriguingly, the Y chromosome was found to mis-segregate at higher rates in an age- and smoke-dependent manner (2.4 to 4.3 fold increase) and its loss correlates with high risk of blood cancer and shortened survival (50% probability of survival)198–200.
Concluding remarks
Centromeres are the fundamental units of chromosome inheritance. Nevertheless, DNA breaks, rearrangements and structural aberrations at centromeric regions are commonly observed in cancer cells and some genetic syndromes. The causes underlying this intrinsic fragility are still untested.
The repetitive nature of centromeric DNA sequences might provide a favorable environment that helps maintain centromere position and assembly. This in turn sustains faithful chromosome segregation, ultimately contributing to preserving genome integrity. Given the recurrence of repetitive DNA in centromeres throughout evolution it has been suggested that the centromeric sequences were naturally selected more for the ability to form a particular structure than for the DNA sequence itself. The resulting highly organized region could play a role in supporting the mechanical stress generated by the spindle microtubules pulling on the centromere/kinetochore platform during chromosome segregation. However, maintaining repetitive DNA also has major drawbacks. Repetitive sequences are generally unstable and might form secondary structures that could potentially induce replication fork stalling, topological problems, and high levels of recombination. Altogether, these anomalies could lead to DNA breakage in certain genetic backgrounds. Indeed, due to the repetitive nature of their DNA sequences, human centromeres are proposed to be enriched with DNA catenates and loops between their DNA repetitive arrays. A well-defined and organized machinery would then be required to solve those structures prior to segregation in order to avoid centromere breaks. In addition, centromeres might also be difficult to replicate leading to an accumulation of replication fork stalling and collapse (especially since centromeres are commonly late replicating regions), which could result in double-strand breaks. It seems that the centromeric structures themselves are necessary to activate unique pathways essential to complete DNA replication prior to chromosome segregation. Curiously, it was proposed that some components normally present during DNA replication such as RPA-1 and ATR are excluded from centromeric regions but become essential during mitosis for correct chromosome segregation. Altogether, this highlights the uniqueness of centromeric regions and how changes in these intricate mechanisms can lead to errors and breakage.
Centromere alterations such as the appearance of neocentromeres and dicentric chromosomes generated by chromosome rearrangements are also found in disease and most of the time, once formed, are remarkably stable during both mitosis and meiosis. While more is known about how dicentric chromosomes form and contribute to genome instability, the generation and the role of neocentromere as a source of instability is still not fully explored. This is why models of de novo centromere formation coupled with centromere inactivation are extremely valuable to understand neocentromeres genesis and its pathological outcome. In addition, varying the expression levels of centromeric components, first and foremost CENP-A and its chaperone HJURP, has been shown to have opposite effects on cells: while downregulation promotes both cell proliferation arrest and senescence, upregulation leads to chromosome instability and cancer transformation, although via molecular mechanisms that need to be clarified; indeed, CENP expression levels could now be used as biomarkers for some types of cancers.
In conclusion, centromeres can be the subject of alterations that contribute to chromosome instability and that are found in human diseases. To what extent and at which stage centromere dysfunctions directly participate to these pathological events is still unclear. For this reason, models and technologies to induce and assess centromere alterations are needed. Until now, centromere analysis was largely limited by the lack of appropriate DNA sequencing technologies for such large repetitive sequences, making centromere sequences the black hole of the human genome. Because of these technical limitations, it is still unclear if the chromosomal breakpoints observed at centromeric regions are actually at the centromeres per se or at the surrounding pericentromeric regions rich in heterochromatin. The advent of more sophisticated technologies will be essential to reveal instability within these regions in human disease to finally shed light on the dark side of the centromeres.
Acknowledgements
We would like to thank all the members of the D. Fachinetti laboratory, Sarah McClelland (Barts Cancer Institute, UK), C. Bartocci (I. Curie) for helpful suggestions. D.F. receives salary support from the CNRS. D.F. has received support by Labex “CelTisPhyBio”, the Institut Curie and the ATIP-Avenir 2015 program. V.B. received salary support from the ATIP-Avenir 2015 program. This work has also received support under the program “Investissements d’Avenir” launched by the French Government and implemented by ANR with the references ANR-10-LABX-0038 and ANR-10-IDEX-0001-02 PSL. We apologize to our colleagues whose important contributions to centromere specification, function, and stability could not be cited due to space constraints.
Author contributions
V.B. and D.F. conceived and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka TU, Clayton L, Natsume T. Three wise centromere functions: see no error, hear no break, speak no delay. EMBO Rep. 2013;14:1073–1083. doi: 10.1038/embor.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willard HF, Waye JS. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J. Mol. Evol. 1987;25:207–214. doi: 10.1007/BF02100014. [DOI] [PubMed] [Google Scholar]
- 4.Grady DL, et al. Highly conserved repetitive DNA sequences are present at human centromeres. Proc. Natl Acad. Sci. USA. 1992;89:1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 6.Waye JS, Willard HF. Chromosome-specific alpha satellite DNA: nucleotide sequence analysis of the 2.0 kilobasepair repeat from the human X chromosome. Nucleic Acids Res. 1985;13:2731–2743. doi: 10.1093/nar/13.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vissel B, Choo KH. Human alpha satellite DNA--consensus sequence and conserved regions. Nucleic Acids Res. 1987;15:6751–6752. doi: 10.1093/nar/15.16.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miga KH. The promises and challenges of genomic studies of human centromeres. Prog. Mol. Subcell. Biol. 2017;56:285–304. doi: 10.1007/978-3-319-58592-5_12. [DOI] [PubMed] [Google Scholar]
- 9.Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC. Human centromeric DNAs. Hum. Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- 10.Muro Y, et al. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J. Cell Biol. 1992;116:585–596. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okada T, et al. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann S, et al. CENP-A is dispensable for mitotic centromere function after initial centromere/kinetochore assembly. Cell Rep. 2016;17:2394–2404. doi: 10.1016/j.celrep.2016.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumont, M. & Fachinetti, D. DNA sequences in centromere formation and function. Prog. Mol. Subcell. Biol.56, 305–336 (2017). [DOI] [PubMed]
- 14.McKinley KL, Cheeseman IM. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016;17:16–29. doi: 10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amor DJ, et al. Human centromere repositioning "in progress". Proc. Natl Acad. Sci. USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knutsen T, et al. The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosom. Cancer. 2005;44:52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knutsen T, et al. Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines. Genes Chromosom. Cancer. 2010;49:204–223. doi: 10.1002/gcc.20730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsop AE, Teschendorff AE, Edwards PAW. Distribution of breakpoints on chromosome 18 in breast, colorectal, and pancreatic carcinoma cell lines. Cancer Genet. Cytogenet. 2006;164:97–109. doi: 10.1016/j.cancergencyto.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Cooke SL, et al. High-resolution array CGH clarifies events occurring on 8p in carcinogenesis. BMC Cancer. 2008;8:288. doi: 10.1186/1471-2407-8-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Backsch C, et al. Two novel unbalanced whole arm translocations are frequently detected in cervical squamous cell carcinoma. Cancer Genet. 2011;204:646–653. doi: 10.1016/j.cancergen.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Adeyinka A, Wei S, Sanchez J. Loss of 17p is a major consequence of whole-arm chromosome translocations in hematologic malignancies. Cancer Genet. Cytogenet. 2007;173:136–143. doi: 10.1016/j.cancergencyto.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Balachandar V, et al. Identification of a high frequency of chromosomal rearrangements in the centromeric regions of prostate cancer patients. J. Zhejiang. Univ. Sci. B. 2007;8:638–646. doi: 10.1631/jzus.2007.B0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacle MM, et al. Chromosome 17 copy number changes in male breast cancer. Cell. Oncol. 2015;38:237–245. doi: 10.1007/s13402-015-0227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Banet J, et al. Decoding complex patterns of genomic rearrangement in hepatocellular carcinoma. Genomics. 2014;103:189–203. doi: 10.1016/j.ygeno.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Ganly P, McDonald M, Spearing R, Morris CM. Constitutional t(5;7)(q11;p15) rearranged to acquire monosomy7q and trisomy 1q in a patient with myelodysplastic syndrome transforming to acute myelocytic leukemia. Cancer Genet. Cytogenet. 2004;149:125–130. doi: 10.1016/j.cancergencyto.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Bièche I, Khodja A, Driouch K, Lidereau R. Genetic alteration mapping on chromosome 7 in primary breast cancer. Clin. Cancer Res. 1997;3:1009–1016. [PubMed] [Google Scholar]
- 27.Smedley D, et al. Characterization of chromosome 1 abnormalities in malignant melanomas. Genes Chromosomes Cancer. 2000;28:121–125. doi: 10.1002/(SICI)1098-2264(200005)28:1<121::AID-GCC14>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 28.Hermsen M, et al. Centromeric chromosomal translocations show tissue-specific differences between squamous cell carcinomas and adenocarcinomas. Oncogene. 2005;24:1571–1579. doi: 10.1038/sj.onc.1208294. [DOI] [PubMed] [Google Scholar]
- 29.Hermsen MA, et al. Centromeric breakage as a major cause of cytogenetic abnormalities in oral squamous cell carcinoma. Genes Chromosomes Cancer. 1996;15:1–9. doi: 10.1002/(SICI)1098-2264(199601)15:1<1::AID-GCC1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 30.Martínez JG, Pérez-Escuredo J, Llorente JL, Suárez C, Hermsen MA. Localization of centromeric breaks in head and neck squamous cell carcinoma. Cancer Genet. 2012;205:622–629. doi: 10.1016/j.cancergen.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Martins C, et al. Fluorescent in situ hybridisation (FISH) characterisation of pericentromeric breakpoints on chromosome 5 in head and neck squamous cell carcinomas. Eur. J. Cancer. 1999;35:498–501. doi: 10.1016/S0959-8049(98)00368-2. [DOI] [PubMed] [Google Scholar]
- 32.Baccon PLe, et al. Novel evidence of a role for chromosome 1 pericentric heterochromatin in the pathogenesis of B-cell lymphoma and multiple myeloma. Genes Chromosom. Cancer. 2001;32:250–264. doi: 10.1002/gcc.1189. [DOI] [PubMed] [Google Scholar]
- 33.Thompson FH, et al. Band 1p36 abnormalities and t(1;17) in ovarian carcinoma. Cancer Genet. Cytogenet. 1997;96:106–110. doi: 10.1016/S0165-4608(96)00307-X. [DOI] [PubMed] [Google Scholar]
- 34.Kim, T. et al. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res.23, 217–227 (2013). The screening of more than 8000 tumors reveals a high recurrence of entire chromosome arms alterations and allows the detection of occurred chromothripsis. [DOI] [PMC free article] [PubMed]
- 35.Wang JC, Hajianpour A, Habibian R. Centromeric α-satellite DNA break in reciprocal translocations. Cytogenet. Genome Res. 2009;125:329–333. doi: 10.1159/000235939. [DOI] [PubMed] [Google Scholar]
- 36.Darlington CD. Misdivision and the genetics of the centromere. J. Genet. 1939;37:341–364. doi: 10.1007/BF02982733. [DOI] [Google Scholar]
- 37.Rowe LR, et al. U-type exchange is the most frequent mechanism for inverted duplication with terminal deletion rearrangements. J. Med. Genet. 2009;46:694–702. doi: 10.1136/jmg.2008.065052. [DOI] [PubMed] [Google Scholar]
- 38.Shaffer LG, Jackson-Cook CK, Meyer JM, Brown JA, Spence JE. A molecular genetic approach to the identification of isochromosomes of chromosome 21. Hum. Genet. 1991;86:375–382. doi: 10.1007/BF00201838. [DOI] [PubMed] [Google Scholar]
- 39.Wolff DJ, Miller AP, Van Dyke DL, Schwartz S, Willard HF. Molecular definition of breakpoints associated with human Xq isochromosomes: implications for mechanisms of formation. Am. J. Hum. Genet. 1996;58:154–160. [PMC free article] [PubMed] [Google Scholar]
- 40.Barbouti A, et al. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am. J. Hum. Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koumbaris G, et al. FoSTeS, MMBIR and NAHR at the human proximal Xp region and the mechanisms of human Xq isochromosome formation. Hum. Mol. Genet. 2011;20:1925–1936. doi: 10.1093/hmg/ddr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott SA, Cohen N, Brandt T, Warburton PE, Edelmann L. Large inverted repeats within Xp11.2 are present at the breakpoints of isodicentric X chromosomes in Turner syndrome. Hum. Mol. Genet. 2010;19:3383–3393. doi: 10.1093/hmg/ddq250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D, Gallie BL, Squire JA. Minimal regions of chromosomal imbalance in retinoblastoma detected by comparative genomic hybridization. Cancer Genet. Cytogenet. 2001;129:57–63. doi: 10.1016/S0165-4608(01)00427-7. [DOI] [PubMed] [Google Scholar]
- 44.Weemaes CM, et al. Heterogeneous clinical presentation in ICF syndrome: correlation with underlying gene defects. Eur. J. Hum. Genet. 2013;21:1219–1225. doi: 10.1038/ejhg.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thijssen PE, et al. Mutations in CDCA7 and HELLS cause immunodeficiency-centromeric instability-facial anomalies syndrome. Nat. Commun. 2015;6:7870. doi: 10.1038/ncomms8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costa G, Barra V, Lentini L, Cilluffo D, Leonardo ADI. DNA demethylation caused by 5-Aza-2’-deoxycytidine induces mitotic alterations and aneuploidy. Oncotarget. 2016;7:3726–3739. doi: 10.18632/oncotarget.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho YH, et al. The association of LINE-1 hypomethylation with age and centromere positive micronuclei in human lymphocytes. PLoS ONE. 2015;10:1–10. doi: 10.1371/journal.pone.0133909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martínez-A C, van Wely KHM. Centromere fission, not telomere erosion, triggers chromosomal instability in human carcinomas. Carcinogenesis. 2011;32:796–803. doi: 10.1093/carcin/bgr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssen A, Van Der Burg M, Szuhai K, Kops GJPL, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 50.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 2001;152:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabeche L, Nguyen HD, Buisson R, Zou L. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science. 2018;359:108–114. doi: 10.1126/science.aan6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guerrero AA, et al. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proc. Natl Acad. Sci. USA. 2010;107:4159–4164. doi: 10.1073/pnas.0912143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garavís M, et al. The structure of an endogenous Drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat. Cell Biol. 2016;18:684–691. doi: 10.1038/ncb3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talbert PB, Kasinathan S, Henikoff S. Simple and complex centromeric satellites in Drosophila sibling species. Genetics. 2018;208:977–990. doi: 10.1534/genetics.117.300620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonstrup AT, et al. Hairpin structures formed by alpha satellite DNA of human centromeres are cleaved by human topoisomerase IIα. Nucleic Acids Res. 2008;36:6165–6174. doi: 10.1093/nar/gkn640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang LHC, Mayer B, Stemmann O, Nigg EA. Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J. Cell Sci. 2010;123:806–813. doi: 10.1242/jcs.058255. [DOI] [PubMed] [Google Scholar]
- 59.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McFarlane RJ, Humphrey TC. A role for recombination in centromere function. Trends Genet. 2010;26:209–213. doi: 10.1016/j.tig.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. doi: 10.1016/S1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 62.Fachinetti D, et al. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol. Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baumann C, Körner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 64.Ke Y, et al. PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. EMBO J. 2011;30:3309–3321. doi: 10.1038/emboj.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rouzeau, S. et al. Bloom’s syndrome and PICH helicases cooperate with topoisomerase IIα in centromere disjunction before anaphase. PLoS ONE7, e33905 (2012). BLM and PICH helicases localize to centromeres from G2-phase to mitosis and are necessary to solve ultrafine anaphase bridges arising from centromeres by recruiting Top2a. [DOI] [PMC free article] [PubMed]
- 66.Yang J, Bachrati CZ, Ou J, Hickson ID, Brown GW. Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J. Biol. Chem. 2010;285:21426–21436. doi: 10.1074/jbc.M110.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Norman-Axelsson Ulrika, Durand-Dubief Mickaël, Prasad Punit, Ekwall Karl. DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast. PLoS Genetics. 2013;9(3):e1003371. doi: 10.1371/journal.pgen.1003371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Broderick R, Nieminuszczy J, Blackford AN, Winczura A, Niedzwiedz W. TOPBP1 recruits TOP2A to ultra-fine anaphase bridges to aid in their resolution. Nat. Commun. 2015;6:6572. doi: 10.1038/ncomms7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hengeveld RCC, et al. Rif1 is required for resolution of ultrafine DNA bridges in anaphase to ensure genomic stability. Dev. Cell. 2015;34:466–474. doi: 10.1016/j.devcel.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 70.Amor-Guéret M. Bloom syndrome, genomic instability and cancer: the SOS-like hypothesis. Cancer Lett. 2006;236:1–12. doi: 10.1016/j.canlet.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 71.Hanlon SL, Li JJ. Re-replication of a centromere induces chromosomal instability and aneuploidy. PLoS Genet. 2015;11:1–30. doi: 10.1371/journal.pgen.1005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carvalho CMB, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zaratiegui M, et al. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469:112–115. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erliandri I, et al. Replication of alpha-satellite DNA arrays in endogenous human centromeric regions and in human artificial chromosome. Nucleic Acids Res. 2014;42:11502–11516. doi: 10.1093/nar/gku835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fachinetti D, et al. DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Greenfeder SA, Newlon CS. Replication forks pause at yeast centromeres. Mol. Cell. Biol. 1992;12:4056–4066. doi: 10.1128/MCB.12.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ten Hagen KG, Gilbert DM, Willard HF, Cohen SN. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol. Cell. Biol. 1990;10:6348–6355. doi: 10.1128/MCB.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 81.He J, Kang X, Yin Y, Clifford Chao KS, Shen WH. PTEN regulates DNA replication progression and stalled fork recovery. Nat. Commun. 2015;6:1–11. doi: 10.1038/ncomms8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang X, et al. PTEN stabilizes TOP2A and regulates the DNA decatenation. Sci. Rep. 2015;5:1–12. doi: 10.1038/srep17873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nikolov I, Taddei A. Linking replication stress with heterochromatin formation. Chromosoma. 2016;125:523–533. doi: 10.1007/s00412-015-0545-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rozenzhak S, et al. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet. 2010;6:e1001032. doi: 10.1371/journal.pgen.1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Irmisch A, Ampatzidou E, Mizuno K, O’Connell MJ, Murray JM. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 2009;28:144–155. doi: 10.1038/emboj.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee SY, Rozenzhak S, Russell P. γH2A-binding protein Brc1 affects centromere function in fission yeast. Mol. Cell. Biol. 2013;33:1410–1416. doi: 10.1128/MCB.01654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakamura K, et al. Rad51 suppresses gross chromosomal rearrangement at centromere in Schizosaccharomyces pombe. EMBO J. 2008;27:3036–3046. doi: 10.1038/emboj.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tinline-Purvis H, et al. Failed gene conversion leads to extensive end processing and chromosomal rearrangements in fission yeast. EMBO J. 2009;28:3400–3412. doi: 10.1038/emboj.2009.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mitra Sreyoshi, Gómez-Raja Jonathan, Larriba Germán, Dubey Dharani Dhar, Sanyal Kaustuv. Rad51–Rad52 Mediated Maintenance of Centromeric Chromatin in Candida albicans. PLoS Genetics. 2014;10(4):e1004344. doi: 10.1371/journal.pgen.1004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li PC, et al. Replication fork stability is essential for the maintenance of centromere integrity in the absence of heterochromatin. Cell Rep. 2013;3:638–645. doi: 10.1016/j.celrep.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jaco I, Canela A, Vera E, Blasco MA. Centromere mitotic recombination in mammalian cells. J. Cell Biol. 2008;181:885–892. doi: 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giunta S, Funabiki H. Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc. Natl Acad. Sci. USA. 2017;114:1928–1933. doi: 10.1073/pnas.1615133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choo KH. Why is the centromere so cold? Genome Res. 1998;8:81–82. doi: 10.1101/gr.8.2.81. [DOI] [PubMed] [Google Scholar]
- 94.Nambiar M, Smith GR. Repression of harmful meiotic recombination in centromeric regions. Semin. Cell. Dev. Biol. 2016;54:188–197. doi: 10.1016/j.semcdb.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shi J, et al. Widespread gene conversion in centromere cores. PLoS Biol. 2010;8:e1000327. doi: 10.1371/journal.pbio.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Voullaire LE, Slater HR, Petrovic V, Choo KH. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am. J. Hum. Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 97.Marshall OJ, Chueh AC, Wong LH, Choo KHA. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tyler-Smith, C., Corish, P. & Burns, E. Neocentromeres, the Y chromosome and centromere evolution. Chromosome Res.6, 65–71 (1998). [DOI] [PubMed]
- 99.Chueh Anderly C., Northrop Emma L., Brettingham-Moore Kate H., Choo K. H. Andy, Wong Lee H. LINE Retrotransposon RNA Is an Essential Structural and Functional Epigenetic Component of a Core Neocentromeric Chromatin. PLoS Genetics. 2009;5(1):e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rocchi M, Archidiacono N, Schempp W, Capozzi O, Stanyon R. Centromere repositioning in mammals. Heredity. 2012;108:59–67. doi: 10.1038/hdy.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shang, W. H. et al. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell24, 635–648 (2013). ChIP analyses show that CENP-A localizes at neocentromeres without sequence-specificity and in the absence of the usual histone marks. [DOI] [PMC free article] [PubMed]
- 102.Thakur, J. & Sanyal, K. Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function. Genome Res.23, 638–652 (2013). [DOI] [PMC free article] [PubMed]
- 103.Shang WH, et al. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010;20:1219–1228. doi: 10.1101/gr.106245.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ishii K, et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 105.Burrack LS, Berman J. Neocentromeres and epigenetically inherited features of centromeres. Chromosom. Res. 2012;20:607–619. doi: 10.1007/s10577-012-9296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Amor DJ, Voullaire L, Bentley K, Savarirayan R, Choo KHA. Mosaic monosomy of a neocentric ring chromosome maps brachyphalangy and growth hormone deficiency to 13q31.1-13q32.3. Am. J. Med. Genet. 2005;133A:151–157. doi: 10.1002/ajmg.a.30527. [DOI] [PubMed] [Google Scholar]
- 107.Murmann AE, et al. Inverted duplications on acentric markers: mechanism of formation. Hum. Mol. Genet. 2009;18:2241–2256. doi: 10.1093/hmg/ddp160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burnside RD, et al. Interstitial deletion of proximal 8q including part of the centromere from unbalanced segregation of a paternal deletion/marker Karyotype with neocentromere formation at 8p22. Cytogenet. Genome Res. 2011;132:227–232. doi: 10.1159/000322815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hemmat M, et al. Neocentric X-chromosome in a girl with Turner-like syndrome. Mol. Cytogenet. 2012;5:1. doi: 10.1186/1755-8166-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma R, et al. Partial trisomy 2q33.3-q37.3 in a patient with an inverted duplicated neocentric marker chromosome. Mol. Cytogenet. 2015;8:1–7. doi: 10.1186/s13039-014-0104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mascarenhas A, et al. First prenatally detected small supernumerary neocentromeric derivative chromosome 13 resulting in a non-mosaic partial tetrasomy 13q. Cytogenet. Genome Res. 2008;121:293–297. doi: 10.1159/000138901. [DOI] [PubMed] [Google Scholar]
- 112.Blom E, Heyning FH, Kroes WGM. A case of angioimmunoblastic T-cell non-Hodgkin lymphoma with a neocentric inv dup(1) Cancer Genet. Cytogenet. 2010;202:38–42. doi: 10.1016/j.cancergencyto.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 113.de Figueiredo AF, et al. A case of childhood acute myeloid leukemia AML (M5) with a neocentric chromosome neo(1)(qter→q23∼24::q23∼24→q43→neo→q43→qter) and tetrasomy of chromosomes 8 and 21. Cancer Genet. Cytogenet. 2009;193:123–126. doi: 10.1016/j.cancergencyto.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 114.Italiano A, et al. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23∼p24 amplification including JAK2 and JMJD2C. Cancer Genet. Cytogenet. 2006;167:122–130. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 115.Italiano A, et al. Variability of origin for the neocentromeric sequences in analphoid supernumerary marker chromosomes of well-differentiated liposarcomas. Cancer Lett. 2009;273:323–330. doi: 10.1016/j.canlet.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 116.Morrissette JJD, et al. Boy with bilateral retinoblastoma due to an unusual ring chromosome 13 with activation of a latent centromere. Am. J. Med. Genet. 2001;99:21–28. doi: 10.1002/1096-8628(20010215)99:1<21::AID-AJMG1122>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 117.Pedeutour F, et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosom. Cancer. 1999;24:30–41. doi: 10.1002/(SICI)1098-2264(199901)24:1<30::AID-GCC5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 118.Abeliovich D, Yehuda O, Ben-Neriah S, Kapelushnik Y, Ben-Yehuda D. dup(10q) lacking alpha-satellite DNA in bone marrow cells of a patient with acute myeloid leukemia. Cancer Genet. Cytogenet. 1996;89:1–6. doi: 10.1016/0165-4608(95)00300-2. [DOI] [PubMed] [Google Scholar]
- 119.Garsed DW, et al. The architecture and evolution of cancer neochromosomes. Cancer Cell. 2014;26:653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 120.Tyler-Smith C, et al. Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 1999;64:1440–1444. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bassett EA, et al. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J. Cell Biol. 2010;190:177–185. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lo AWI, et al. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 2001;20:2087–2096. doi: 10.1093/emboj/20.8.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koren A, et al. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in s-phase. PLoS Genet. 2010;6:15–17. doi: 10.1371/journal.pgen.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cuacos M, H. Franklin FC, Heckmann S. Atypical centromeres in plants—what they can tell us. Front. Plant Sci. 2015;6:1–15. doi: 10.3389/fpls.2015.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat. Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 127.Maggert KA, Karpen GH. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 2001;158:1615–1628. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zeitlin SG, et al. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc. Natl Acad. Sci. USA. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kato T, et al. Activation of Holliday junction-recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- 130.Heun P, et al. Mislocalization of the drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Olszak AM, et al. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat. Cell Biol. 2011;13:799–808. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- 132.Ketel Carrie, Wang Helen S. W., McClellan Mark, Bouchonville Kelly, Selmecki Anna, Lahav Tamar, Gerami-Nejad Maryam, Berman Judith. Neocentromeres Form Efficiently at Multiple Possible Loci in Candida albicans. PLoS Genetics. 2009;5(3):e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Niikura Y, Kitagawa R, Kitagawa K. CENP-A ubiquitylation is inherited through dimerization between cell divisions. Cell Rep. 2016;15:61–76. doi: 10.1016/j.celrep.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fachinetti D, et al. CENP-A modifications on Ser68 and Lys124 are dispensable for establishment, maintenance, and long-term function of human centromeres. Dev. Cell. 2017;40:104–113. doi: 10.1016/j.devcel.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liehr T, Kosyakova N, Weise A, Ziegler M, Raabe-Meyer G. First case of a neocentromere formation in an otherwise normal chromosome 7. Cytogenet. Genome Res. 2010;128:189–191. doi: 10.1159/000271471. [DOI] [PubMed] [Google Scholar]
- 136.McClintock B. The stability of broken ends of chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.MacKinnon RN, Campbell LJ. The role of dicentric chromosome formation and secondary centromere deletion in the evolution of myeloid malignancy. Genet. Res. Int. 2011;2011:1–11. doi: 10.4061/2011/643628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jones RE, et al. Escape from telomere-driven crisis is DNA ligase III dependent. Cell Rep. 2014;8:1063–1076. doi: 10.1016/j.celrep.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 139.Page SL, Shaffer LG. Chromosome stability is maintained by short intercentromeric distance in functionally dicentric human Robertsonian translocations. Chromosom. Res. 1998;6:115–122. doi: 10.1023/A:1009286929145. [DOI] [PubMed] [Google Scholar]
- 140.McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 142.Lopez V, et al. Cytokinesis breaks dicentric chromosomes preferentially at pericentromeric regions and telomere fusions. Genes Dev. 2015;29:322–336. doi: 10.1101/gad.254664.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pennaneach V, Kolodner RD. Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS ONE. 2009;4:17–26. doi: 10.1371/journal.pone.0006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stimpson Kaitlin M., Song Ihn Young, Jauch Anna, Holtgreve-Grez Heidi, Hayden Karen E., Bridger Joanna M., Sullivan Beth A. Telomere Disruption Results in Non-Random Formation of De Novo Dicentric Chromosomes Involving Acrocentric Human Chromosomes. PLoS Genetics. 2010;6(8):e1001061. doi: 10.1371/journal.pgen.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Maciejowski J, Li Y, Bosco N, Campbell PJ, De Lange T. Chromothripsis and kataegis induced by telomere crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gascoigne KE, Cheeseman IM. Induced dicentric chromosome formation promotes genomic rearrangements and tumorigenesis. Chromosom. Res. 2013;21:407–418. doi: 10.1007/s10577-013-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Earnshaw WC, Ratrie H, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- 148.Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- 149.Gravholt CH, Friedrich U, Caprani M, Jørgensen AL. Breakpoints in Robertsonian translocations are localized to satellite III DNA by fluorescence in situ hybridization. Genomics. 1992;14:924–930. doi: 10.1016/S0888-7543(05)80113-2. [DOI] [PubMed] [Google Scholar]
- 150.Higgins AW, Gustashaw KM, Willard HF. Engineered human dicentric chromosomes show centromere plasticity. Chromosom. Res. 2005;13:745–762. doi: 10.1007/s10577-005-1009-2. [DOI] [PubMed] [Google Scholar]
- 151.Stimpson KM, Matheny JE, Sullivan BA. Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosom. Res. 2012;20:595–605. doi: 10.1007/s10577-012-9302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sullivan BA, Willard HF. Stable dicentric X chromosomes with two functional centromeres. Nat. Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- 153.Higgins AW, Schueler MG, Willard HF. Chromosome engineering: generation of mono- and dicentric isochromosomes in a somatic cell hybrid system. Chromosoma. 1999;108:256–265. doi: 10.1007/s004120050376. [DOI] [PubMed] [Google Scholar]
- 154.Garagna S, Page J, Fernandez-Donoso R, Zuccotti M, Searle JB. The robertsonian phenomenon in the house mouse: mutation, meiosis and speciation. Chromosoma. 2014;123:529–544. doi: 10.1007/s00412-014-0477-6. [DOI] [PubMed] [Google Scholar]
- 155.Chmátal L, et al. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yilmaz A, et al. Chromosome segregation analysis in human embryos obtained from couples involving male carriers of reciprocal or Robertsonian translocation. PLoS ONE. 2012;7:7–13. doi: 10.1371/journal.pone.0046046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nielsen J, Wohlert M. Chromosome abnormalities found among 34910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Hum. Genet. 1991;87:81–83. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- 158.Engels H, et al. Genetic counseling in Robertsonian translocations der(13;14): frequencies of reproductive outcomes and infertility in 101 pedigrees. Am. J. Med. Genet. A. 2008;146A:2611–2616. doi: 10.1002/ajmg.a.32500. [DOI] [PubMed] [Google Scholar]
- 159.Scriven PN, Flinter FA, Braude PR, Ogilvie CM. Robertsonian translocations--reproductive risks and indications for preimplantation genetic diagnosis. Hum. Reprod. 2001;16:2267–2273. doi: 10.1093/humrep/16.11.2267. [DOI] [PubMed] [Google Scholar]
- 160.Dolskiy AA, et al. Robertsonian translocation 13/14 associated with rRNA genes overexpression and intellectual disability. Egypt. J. Med. Hum. Genet. 2018;19:141–145. doi: 10.1016/j.ejmhg.2017.11.002. [DOI] [Google Scholar]
- 161.Li Y, et al. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature. 2014;508:98–102. doi: 10.1038/nature13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lynch SA, Ashcroft KA, Zwolinski S, Clarke C, Burn J. Kabuki syndrome-like features in monozygotic twin boys with a pseudodicentric chromosome 13. J. Med. Genet. 1995;32:227–230. doi: 10.1136/jmg.32.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gravholt CH, et al. A patient with Edwards syndrome caused by a rare pseudodicentric chromosome 18 of paternal origin. Clin. Genet. 1997;52:56–60. doi: 10.1111/j.1399-0004.1997.tb02515.x. [DOI] [PubMed] [Google Scholar]
- 164.Bodor DL, et al. The quantitative architecture of centromeric chromatin. eLife. 2014;2014:1–26. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fachinetti D, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Silva MCC, et al. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 169.Lacoste, N. et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell53, 631–644 (2014). Overexpression of CENP-A leads to DAXX-dependent CENP-A ectopic loading along chromosome arms on functional DNA elements conveying survival advantages to human cells. [DOI] [PubMed]
- 170.Gonzalez M, He H, Dong Q, Sun S, Li F. Ectopic centromere nucleation by CENP-A in fission yeast. Genetics. 2014;198:1433–1446. doi: 10.1534/genetics.114.171173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Choi ES, et al. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-ACnp1 in fission yeast. PLoS Genet. 2012;8:e1002985. doi: 10.1371/journal.pgen.1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics. 2008;179:263–275. doi: 10.1534/genetics.108.088518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shrestha RL, et al. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget. 2017;8:46781–46800. doi: 10.18632/oncotarget.18108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Van Hooser AA, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 175.Amato A, Schillaci T, Lentini L, Di Leonardo A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer. 2009;8:1–14. doi: 10.1186/1476-4598-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Li Y, et al. Shrna-targeted centromere protein a inhibits hepatocellular carcinoma growth. PLoS ONE. 2011;6:19–23. doi: 10.1371/journal.pone.0017794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 179.Foltz DR, et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gu XM, et al. Expression and prognostic relevance of centromere protein A in primary osteosarcoma. Pathol. Res. Pract. 2014;210:228–233. doi: 10.1016/j.prp.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 181.Hu Z, et al. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12:1–15. doi: 10.1186/bcr2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Montes de Oca R, et al. The histone chaperone HJURP is a new independent prognostic marker for luminal A breast carcinoma. Mol. Oncol. 2015;9:657–674. doi: 10.1016/j.molonc.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.de Tayrac M, et al. Prognostic significance of EDN/RB, HJURP, p60/CAF-1 and PDLI4, four new markers in high-grade gliomas. PLoS ONE. 2013;8:e73332. doi: 10.1371/journal.pone.0073332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.McGovern SL, Qi Y, Pusztai L, Symmans WF, Buchholz TA. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012;14:R72. doi: 10.1186/bcr3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sun X, et al. Elevated expression of the centromere protein-A(CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int. J. Cancer. 2016;139:899–907. doi: 10.1002/ijc.30133. [DOI] [PubMed] [Google Scholar]
- 186.Tomonaga T, et al. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- 187.Wu Q, et al. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012;77:407–414. doi: 10.1016/j.lungcan.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 188.Thiru P, et al. Kinetochore genes are coordinately up-regulated in human tumors as part of a FoxM1-related cell division program. Mol. Biol. Cell. 2014;25:1983–1994. doi: 10.1091/mbc.e14-03-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang W, et al. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun. 2016;7:12619. doi: 10.1038/ncomms12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Filipescu D, et al. Essential role for centromeric factors following p53 loss and oncogenic transformation. Genes Dev. 2017;31:463–480. doi: 10.1101/gad.290924.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee JK, Huberman JA, Hurwitz J. Purification and characterization of a CENP-B homologue protein that binds to the centromeric K-type repeat DNA of Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA. 1997;94:8427–8432. doi: 10.1073/pnas.94.16.8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Maehara K, Takahashi K, Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol. Cell Biol. 2010;30:2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Heo JI, Cho JH, Kim JR. HJURP regulates cellular senescence in human fibroblasts and endothelial cells via a p53-dependent pathway. J. Gerontol. A Biol. Sci. Med. Sci. 2013;68:914–925. doi: 10.1093/gerona/gls257. [DOI] [PubMed] [Google Scholar]
- 194.Nakagome Y, Abe T, Misawa S, Takeshita T, Iinuma K. The "loss" of centromeres from chromosomes of aged women. Am. J. Hum. Genet. 1984;36:398–404. [PMC free article] [PubMed] [Google Scholar]
- 195.Swanson EC, Manning B, Zhang H, Lawrence JB. Higher-order unfolding of satellite heterochromatin is a consistent and early event in cell senescence. J. Cell Biol. 2013;203:929–942. doi: 10.1083/jcb.201306073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Miga KH, et al. Centromere reference models for human chromosomes X and y satellite arrays. Genome Res. 2014;24:697–707. doi: 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Irvine DV, et al. Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosom. Res. 2004;12:805–815. doi: 10.1007/s10577-005-5377-4. [DOI] [PubMed] [Google Scholar]
- 198.Dumanski JP, et al. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–83. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Forsberg LA, et al. Mosaic loss of chromosome y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat. Genet. 2014;46:624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nath Joginder, Tucker James D., Hando John C. Y Chromosome aneuploidy, micronuclei, kinetochores and aging in men. Chromosoma. 1995;103(10):725–731. doi: 10.1007/BF00344234. [DOI] [PubMed] [Google Scholar]