Abstract
Hard ticks of the order Ixodidae serve as vectors for numerous human pathogens, including the causative agent of Lyme Disease Borrelia burgdorferi. Tick-associated microbes can influence pathogen colonization, offering the potential to inhibit disease transmission through engineering of the tick microbiota. Here, we investigate whether B. burgdorferi encounters abundant bacteria within the midgut of wild adult Ixodes scapularis, its primary vector. Through the use of controlled sequencing methods and confocal microscopy, we find that the majority of field-collected adult I. scapularis harbor limited internal microbial communities that are dominated by endosymbionts. A minority of I. scapularis ticks harbor abundant midgut bacteria and lack B. burgdorferi. We find that the lack of a stable resident midgut microbiota is not restricted to I. scapularis since extension of our studies to I. pacificus, Amblyomma maculatum, and Dermacentor spp showed similar patterns. Finally, bioinformatic examination of the B. burgdorferi genome revealed the absence of genes encoding known interbacterial interaction pathways, a feature unique to the Borrelia genus within the phylum Spirochaetes. Our results suggest that reduced selective pressure from limited microbial populations within ticks may have facilitated the evolutionary loss of genes encoding interbacterial competition pathways from Borrelia.
Introduction
Vector-borne pathogens infect over one billion people annually and have expanded at an alarming rate in recent years [1–4]. Lyme disease, which is caused by the tick-borne bacterial pathogen Borrelia burgdorferi, is the fifth-most reported infectious disease in the United States, corresponding to over 90% of vector-borne infections in North America [5, 6]. B. burgdorferi transits an enzootic cycle between small mammal reservoir hosts, larger mammals, and birds, vectored by ticks of the genus Ixodes [6]. Transmission of B. burgdorferi to humans via tick bite results in a constellation of inflammatory symptoms requiring an antibiotic treatment regimen for resolution within 2–3 weeks in most cases [7, 8]. Although a vaccine targeting B. burgdorferi had been approved by the FDA, it was subsequently withdrawn and currently there is an urgent need for new strategies to control tick-borne disease transmission [9].
Colonization resistance against pathogens mediated by commensal microorganisms is one such proposed strategy [10, 11]. Studies in mosquitos and tsetse flies, among others, have motivated the characterization of the endogenous microbiota associated with vectors in hopes of identifying means by which pathogen transmission can be interrupted through direct or indirect interactions [12–14]. One mechanism that may govern interactions between pathogens and the microbiota involves direct competition. Many bacteria possess elaborate mechanisms that can mediate interbacterial competition in polymicrobial environments, including specialized pathways such as the type VI secretion system, which delivers toxic effector proteins to target cells [15, 16]. It is thought that the presence and repertoire of interbacterial systems in a given bacterial genome reflects to a certain degree the selective pressures that organism faces in its natural niche [17]. Indeed, these antagonistic systems contribute to bacterial fitness in complex bacterial communities such as the mammalian gut [18–20].
The genus Borrelia comprises a group of pathogenic bacteria that rely upon hematophagous arthropods for infectious transmission to humans, including ticks of the order Ixodida [6]. In hard ticks (Ixodidae), recent surveys of the internal microbiota have to-date identified bacteria that could potentially restrict pathogen transmission [21–35]. However, significant variation in the diversity and identity of tick-associated bacteria was observed in these studies—rendering the potential for bacterial interference within ticks uncertain. In this study, we aimed to determine whether I. scapularis possesses a diverse midgut microbiota that might be encountered by B. burgdorferi. Most previous studies of the microbiome of hard ticks have utilized high-throughput sequencing technologies. We sought to augment sequencing with direct measurements of bacterial load and visualization of bacteria within adult and nymphal I. scapularis ticks by confocal microscopy. With these complementary approaches, we provide evidence that hard ticks lack a stable midgut microbiota.
Methods
Tick collection
Ticks were collected from the following sites: oak woodland habitat in Klickitat River Canyon, Washington (Ixodes pacificus and Dermacentor andersoni); oak-hickory forest in Wolf Creek State Park, Illinois (Ixodes scapularis and Dermacentor variabilis); oak-dominated forest in Gordie Mikkelson Wildlife Management Area and Carlos Avery Wildlife Management Area, Minnesota (abbreviated GM and CA, Ixodes scapularis and Dermacentor variabilis); red pine forest in Kettle Moraine State Forest Southern Unit (KM), mixed hardwood forest in Big Eau Pleine County Park (BEP), and oak-hickory forest in Sandberg Woods Conservancy (SC), Wisconsin (Ixodes scapularis); oak forest near McPherson Preserve, Oklahoma (Amblyomma maculatum). Ticks were collected by the flagging and dragging methods and shipped in 50 mL Falcon tubes with damp paper towels on wet ice to retain moisture. Species, developmental stage, and sex of individual ticks were determined by visual inspection.
DNA isolation
Live ticks were washed three times with sterile water then dried before immobilization on a glass slide using double-sided tape. 21-gauge needles were used to remove the cuticle, and sterile forceps used to dissect and remove the viscera into 500 μL sterile deionized water for subsequent DNA purification. Fresh needles were exchanged, and forceps sterilized by washing three times in sterile water and 70% ethanol between each dissection. The first external wash sample was saved, and pooled across individuals of the same species, sex, and geographical collection location. Samples were frozen at −80 °C prior to DNA isolation. Frozen tissue samples were thawed and subsequently homogenized via glass bead-beating on a MiniBeadBeater (Biospec Products) for two cycles of one minute duration each. Subsequently, phenol-chloroform-isoamyl alcohol extraction of total nucleic acids was performed with a RNase treatment to remove RNA. Following extraction, DNA pellets were resuspended in 25 μL sterile deionized water.
Quantitative PCR
The abundance of bacteria in single tick samples was determined by quantitative PCR using SsoAdvanced Universal SYBR Green Supermix (Biorad) on a BioRad CFX96 Real Time System C1000. Primers used targeted the 16S rRNA gene (331f and 797r [36]) or the B. burgdorferi flaB gene [37]. Standard curves were generated from serial dilutions of purified genomic DNA prepared from monocultures of Escherichia coli DH5a or B. burgdorferi B31-A3. Samples were run in technical triplicates with the mean of each triplicate used for later analysis.
16S rRNA gene sequencing and analysis
Genomic DNA from tick samples, external washes, and water controls was submitted for sequencing and individually barcoded for high-throughput sequencing of V3-V4 16S rRNA amplicons on an Illumina MiSeq, in three separate runs (performed by MrDNA). Sequencing reads were subsequently de-multiplexed and merged into a single fastq file. The UPARSE pipeline was used to process the samples using default settings [38, 39]. Following taxonomy prediction and OTU assignment, the OTU table were filtered to only retain OTUs that appeared at greater than 1% relative abundance in at least one sample. Alpha-diversity and beta-diversity metrics were calculated in QIIME (MacQIIME v1.9.0) [40]. For determination of B. burgdorferi infection status, tick samples in which the sum of the relative abundance of the family Spirochaetaceae exceeded 1% were considered to be infected, while those less than 1% were considered to be uninfected. The sum of the relative abundance of the genera Bacillus and Pseudomonas was calculated for comparison of B. burgdorferi infection status across samples. For LEfSe analysis, the OTU table was converted to relative abundances and, along with associated metadata, was uploaded to the Huttenhower Lab Galaxy web application. Statistical tests and generation of plots were performed in R version 3.3.2 [41].
Histology
Preparing tick sections
Ticks were washed three times in sterile water and immobilized with double-sided tape attached to glass slides. Cuticles were removed with sterile 21-gauge needles. Following removal of the cuticle, dissected ticks were fixed in formalin for at least 5 days at room temperature. Fixed ticks were then embedded in paraffin and four 4-micron sections were prepared from each tick. Slides were de-paraffinized in a series of washes in xylenes, followed by rehydration in washes of decreasing percent ethanol solutions.
Antibody-based microscopy
Deparaffinized slides were first subjected to a 40-min incubation in sodium citrate buffer at 95oC in a vegetable steamer (Black and Decker). Slides were then washed in 2 × SSC + 0.3% Tween 20, blocked in PBS + 5% BSA, and incubated with BacTrace fluorescein isothiocyanate goat anti-Borrelia whole cell antibodies (Kirkegard and Perry Laboratories) for 1 h at room temperature or overnight at 4 °C. Slides were then washed, dried, and mounted in ProLong Diamond plus DAPI (Thermo Fisher Scientific). Coverslips were sealed with nail polish.
In situ hybridization analysis
Deparaffinized slides were washed in 2 × SSC + 0.3% Tween20, then were lightly digested with Proteinase K to allow probe access into tissues, washed and dried. Probes used were EU338 [42] or specific to Rickettsia. For Rickettsia-specific probes, the nucleotide sequence of the Rick_1442 16 S rRNA probe [43] was aligned to the publically available REIS genome [44] and was confirmed to be 100% identical. ISH-probes (5′ biotin-labeled) were diluted 1:100 to 50 μM in ethylene carbonate hybridization buffer (15% ethylene carbonate, 20% dextran sulfate, 600 mM NaCl, 10 mM sodium citrate [45]). Slides were covered with Parafilm, placed in humidified chambers, and allowed to incubate for 1 h at 42 °C. Slides were subsequently washed in 2 × SSC at 37 °C then allowed to air dry. Alexa Fluor 488-coupled tyramide signal amplification (Thermo Fisher Scientific kit T20912) was performed to increase probe signal above background auto-fluorescence from tick midgut digestive products [46, 47]. Slides were mounted in Diamond Prolong mounting medium plus DAPI (Thermo Fisher Scientific) and coverslips sealed with nail polish prior to imaging.
Imaging
Confocal microscopy was performed at the University of Washington Keck Microscopy Center, on a Leica SP8X confocal microscope using Leica Application Suite X. Two-micron z-stacks were imaged at ×63 magnification, using ×6 averaging. Images were extracted from raw.lif files, maximum projected, channels merged, cropped, and pseudocolored using Fiji [48].
Identification of interbacterial effector–immunity gene homologs in bacterial genomes
Multi-alignments for 220 putative and validated effector and immunity proteins were acquired from the supplemental materials of Zhang et al. [17]. Amino acid seed sequences of interbacterial effector and immunity proteins were queried via tBLASTn against a custom database of 65 complete and draft genomes from the phylum Spirochaetes, 17 complete and draft genomes of tick-associated endosymbionts and pathogens from the phylum Proteobacteria (see Table S1 for accession information), and 2 genomes from the phylum Deferribacteres. All genomes were acquired via download from RefSeq. All hits with e-values≤10−3 across the full length of the seed sequence were considered to be homologs of interbacterial effector or immunity proteins. The distribution of effector and immunity genes per genome within each genus was calculated by normalizing the total number of BLAST hit by the number of genomes searched. Turneriella genomes were included within the closely-related Leptospira genus for this analysis. The number of complete genomes analyzed per clade is as follows: Anaplasma [4], Borrelia [18], Brachyspira [7], Coxiella [2], Deferribacteres [2], Erlichia [8], Leptospira [17], Rickettsia [3], Spirochaeta [11], Treponema [10], Turneriella [2].
Results
Borrelia lacks interbacterial effector–immunity genes
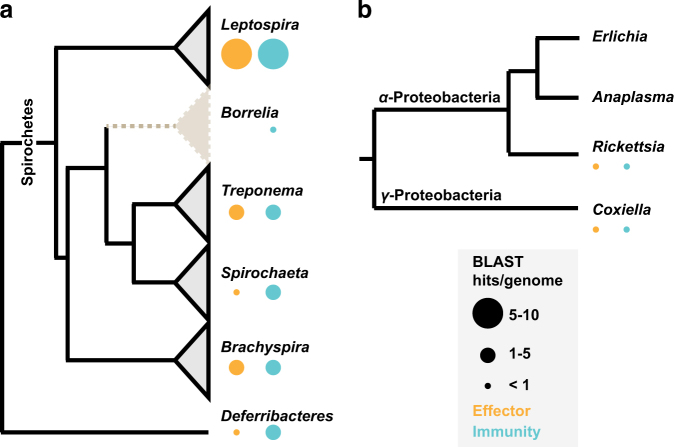
Members of the phylum Spirochaetes inhabit diverse environments, from the dense and competitive oral cavity and gastrointestinal tract of mammals to the midguts of arthropods like termites and ticks. How members of the phylum Spirochaetes engage in interactions with other bacteria is not understood. We therefore sought to characterize the distribution of interbacterial effector and immunity genes in the genomes of Spirochaetes. We first compiled a database of Spirochaetes genomes, encompassing a total of 65 genomes representing all major genera. These genomes were queried for the presence of homologs of 220 interbacterial effector and immunity genes [17], including those with characterized domains found associated with contact-dependent inhibition [49], type VI secretion system [16], and ESX/T7SS antagonistic pathways [50, 51]. This analysis revealed the presence of genes encoding interbacterial effector and immunity domains throughout the Spirochaetes, particularly in species known to inhabit polymicrobial environments such as the mammalian gut microbiome (Treponema succinifaciens and Brachyspira spp) and the oral microbiome (Treponema denticola) (Fig. 1a). In contrast, we failed to detect effector gene homologs and identified only a limited group of immunity gene homologs encoded by any species within the genus Borrelia, including representatives from the sensu stricto and sensu lato genospecies [52]. Since the phylum Spirochaetes is considered to be monophyletic, with extant genera descending from a single common ancestor [53], parsimony supports the conclusion that Borrelia lost interbacterial effector genes early in the evolution of the genus. Parallel investigation of the genomes of tick-associated endosymbionts and tick-transmitted intracellular pathogens including Rickettsia, Coxiella, Anaplasma, and Ehrlichia revealed that these genomes also largely lack interbacterial effector–immunity genes (Fig. 1b).
Fig. 1.
Distribution of interbacterial effector–immunity gene homologs across the phylum Spirochaetes. a The distribution of interbacterial effector and immunity gene homologs in genomes from across a phylogenetic tree of genera within the phylum Spirochaetes. Deferribacteres is shown as the outgroup. BLAST hits are normalized to the number of genomes analyzed for each genus. The genus Borrelia shows a significant deviation from the expected frequency relative to other genera (p < 0.05). b Interbacterial effector and immunity gene homologs in genomes of bacterial endosymbionts and pathogens associated with Ixodidae ticks from the phylum Proteobacteria.
Assessment of bacterial abundance and diversity in I. scapularis
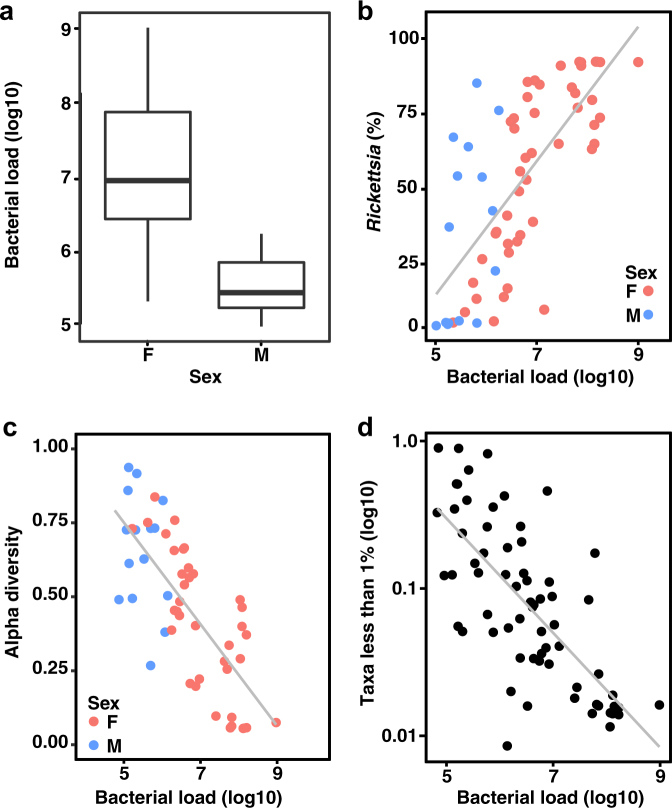
The evolutionary loss of effector–immunity genes from the genus Borrelia led us to hypothesize that interbacterial interactions might be limited within I. scapularis. We therefore sought to quantify the microbial communities associated with wild I. scapularis. We first isolated DNA from the dissected viscera (a combination of internal tissues that included midgut, reproductive tissues, and salivary glands) of 61 adult ticks collected from 5 distinct geographic sites in the Midwestern US. We then performed quantitative polymerase chain reaction (qPCR) targeting conserved regions of the 16 S rRNA gene. Our analysis demonstrated that the mean internal bacterial load of unfed adult I. scapularis ticks varies over several orders of magnitude (Fig. 2a). Male ticks analyzed harbored less bacterial load than did females (4.5 × 105 vs 5.3 × 107, t-test p-value < 0.001, N = 61). qPCR with primers specific to the B. burgdorferi flaB gene revealed infection frequencies similar to that previously reported from the Midwestern geographic regions (Fig. S1A) [54]. To investigate the differences in bacterial community underlying the sex-specific differences that we observed, we performed 16 S rRNA gene sequencing on the same samples used for qPCR analysis. In addition, we sequenced the external washes from a subset of ticks, and a water-only control for each group. In total 178 OTUs were detected in the water-only controls, including taxa commonly implicated in reagent contamination and some previously reported to be associated with ticks, such as Sphingomonas and Comamonas [26, 27, 35]. These OTUs were subsequently eliminated from all other samples in downstream analyses. To describe the taxa that comprise I. scapularis-associated bacterial communities following removal of water-borne contamination, we calculated the taxonomic relative abundance averaged across all internal and external samples. This revealed that 258 (95%) of 270 OTUs fell below 1% relative abundance when averaged. In contrast, other taxa exceeded 1% average relative abundance in both internal and external samples, including Bacillus, Pseudomonas, and Enterobacteriaceae (Fig. S2). Importantly, the taxon that was most abundant in internal samples but entirely absent in external samples was the genus Rickettsia (average relative abundance of 52%), which includes the dominant Rickettsia endosymbiont of I. scapularis [44]. The relative abundance of Rickettsia exhibited a positive correlation with total bacterial load (Fig. 2b), indicating that Rickettsia is the primary driver of bacterial abundance in most I. scapularis ticks. The family Spirochaetaceae also was often abundant in viscera samples but absent in washes (Fig. S2).
Fig. 2.
Inflation of bacterial diversity in Ixodes scapularis. a The bacterial load of male and female I. scapularis adult ticks, determined by qPCR amplifying the 16 S rRNA gene (N = 61). Male ticks harbor significantly less bacteria than do females, (t-test p < 0.001). b Correlation between the relative abundance of Rickettsia and the total bacterial load across all adult I. scapularis ticks in unfiltered samples (Spearman’s rho = 0.75, p < 0.001). c Alpha diversity measured by the Simpson’s value (log2) negatively correlates with bacterial load (log10) across all adult I. scapularis samples without filtering of low abundance taxa (Spearman’s rho = −0.74, p < 0.001). Male (blue) and female (pink) samples appear stratified according to both load and diversity. d Relative abundance of the sum of all taxa whose average relative abundance was <1% correlates negatively with bacterial load in adult I. scapularis samples (log10), Spearman’s rho = −0.73, p < 0.001
The number of taxa with low relative abundance could indicate that the internal environment of I. scapularis is particularly well suited to fostering diverse microbial communities. However, an alternate possibility is that low abundance OTUs represent signal from contamination. Indeed, several studies have examined the effect of low biomass input on diversity metrics in 16 S rRNA gene sequencing surveys, with the notable result that alpha diversity (within-sample diversity) correlated negatively with the abundance of input DNA [55–57]. We therefore examined the relationship between alpha diversity and bacterial load as quantified by qPCR in our I. scapularis samples. When examined across all 61 I. scapularis samples, we found a strong negative correlation between total bacterial load and alpha diversity that was independent of geographic origin (Spearman’s rho = −0.74, p < 0.001) (Fig. 2c and S3). These data imply that the low bacterial biomass associated with adult I. scapularis ticks can impact the interpretation of 16 S rRNA gene sequencing surveys by artificially inflating alpha diversity. We further observed a strong negative correlation between bacterial load and the relative abundance of the group of taxa contributing to 1% or less of the total across all I. scapularis samples (Fig. 2d). We found only six taxa to be present in all I. scapularis viscera samples, represented by eight OTUs, including the genera Rickettsia, Borrelia, Pseudomonas, Francisella, and Escherichia, and the family Enterobacteriaceae (Fig. S2). Linear discriminant analysis effect size (LEfSe) revealed the taxa most likely to explain differences between external wash samples and internal viscera samples. External samples are characterized by taxa from the phyla Proteobacteria and Actinobacteria, while viscera samples are distinguished by the orders Spirochaetales and Rickettsiales (Fig. S4) [58].
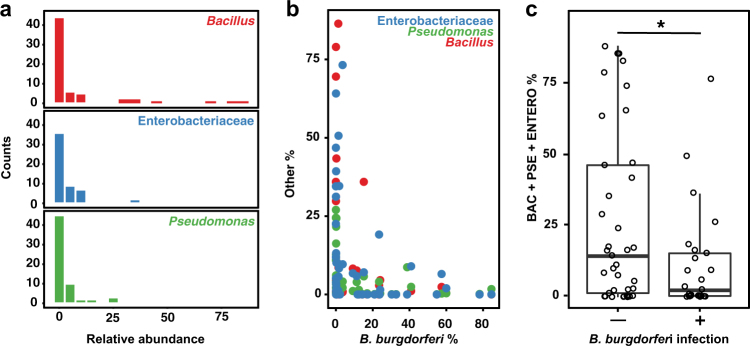
While the internal bacteria associated with most wild adult I. scapularis are dominated by Rickettsia and B. burgdorferi, a minority of samples exhibit high relative abundances of three putatively environmental taxa, including the genera Bacillus and Pseudomonas, and the family Enterobacteriaceae (Fig. 3a) all of which have been previously described as associated with I. scapularis [35, 59, 60]. Colonization of ticks appeared to be independent for each taxon since, although co-occurrence was detected in a few samples, most ticks harbored only one dominant environmental taxon. While total bacterial load was not correlated with colonization by environmental bacteria, samples with high abundance of these bacteria—deriving from each of the five geographically isolated collection sites—were less frequently infected by B. burgdorferi than expected (Fig. 3b, c). This observation suggests that colonization by Bacillus, Pseudomonas, and Enterobacteriaceae may limit B. burgdorferi infection.
Fig. 3.
Transient environmental bacteria limit B. burgdorferi infection. a Histograms depict the relative abundance of bacteria (Bacillus, red; Enterobacteriaceae, blue; Pseudomonas, green) detected in I. scapularis viscera samples, in bins of 5%. Counts indicate number of samples. b Scatter plot of the relationship between the relative abundance of environmental bacteria detected in I. scapularis viscera and the relative abundance of B. burgdorferi. Color scheme as in a. c Barplots indicate the sum of the relative abundance for each of three abundant taxa between samples in which B. burgdorferi relative abundance exceeds 1% (infected) and those samples for which abundance is <1% (uninfected). The difference is statistically significant (Mann Whitney U test, p < 0.001)
Immuno-staining and confocal microscopy reveals I. scapularis midgut biogeography
To orthogonally and directly validate our findings of a limited internal microbiome in ticks, we turned to confocal microscopy in order to visualize bacteria distribution and localization within I. scapularis. Our sequencing results suggest that Rickettsia comprise the dominant internal microbial inhabitants of most wild adult I. scapularis ticks. However, we questioned (i) if our analyses failed to detect low-abundance yet highly diverse communities of midgut-associated bacteria masked by the high abundance of Rickettsia and (ii) if the limited midgut colonization by environmental bacteria that we observed could be visualized and therefore validated. Ticks present a number of challenges that hinder standard microscopy techniques. These challenges include the physical barrier of the thick outer cuticle, a high degree of internal auto-fluorescence derived from the cuticle, and remnants of previous blood-meals including heme crystals [46]. In order to preserve tissue integrity and organization, we utilized formalin-fixation and paraffin embedding of dissected ticks, followed by thin sectioning and staining for visualization. Use of biotin-labeled ISH-probes allowed us to perform tyramide-signal amplification (TSA), dramatically increasing the signal to noise ratio within tick sections.
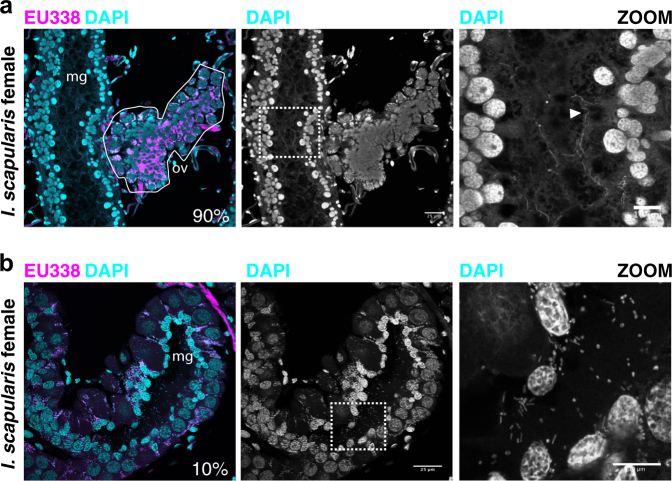
With these methods, we characterized the distribution of bacteria within tissues of individual adult I. scapularis ticks (N = 41). Abundant and DAPI-intense cocci that co-stained with probes targeting universally conserved 16 S rRNA sequences (EU338) were observed in the cytoplasm of ovarian cells of all female I. scapularis ticks (Fig. S5). These are presumed to be vertically transmitted Rickettsia [61]. In striking contrast to ovaries, the midgut of most unfed adult I. scapularis lacked bacilli or cocci as visualized by DAPI signal or ISH signal localized to the lumen or luminal epithelium (Fig. 5a, 90%, N = 41 total ticks). This differed for B. burgdorferi-infected I. scapularis ticks, in which spirochete cells constituted the only detected midgut bacteria (Fig. 4a, S5). However, a minority of I. scapularis ticks from a single site (CA) exhibited midgut colonization by bacterial cells with distinct cocci and bacilli cellular morphologies (10% of total I. scapularis examined, N = 41) (Fig. 4b). Since I. scapularis nymphs have been reported to possess a diverse midgut biofilm-forming microbiome, with very low relative abundance of Rickettsia [26, 62], we considered the possibility that while adult I. scapularis ticks may lack a diverse internal microbiome, earlier life stages could be colonized to a greater extent. To test this possibility, we imaged sections prepared from whole-mounted I. scapularis nymphs, using similar formalin-fixation and staining methodology as before. We found that I. scapularis nymphs also exhibit low DAPI and ISH probe staining within midgut tissues (Fig. S6). Since nymphs are considerably smaller than adults, they may be more sensitive to artefactual noise associated with low-biomass high-throughput sequencing. In support of this notion, the alpha diversity of the I. pacificus microbiome negatively correlates with life stage progression from larvae to adults as body size increases [31].
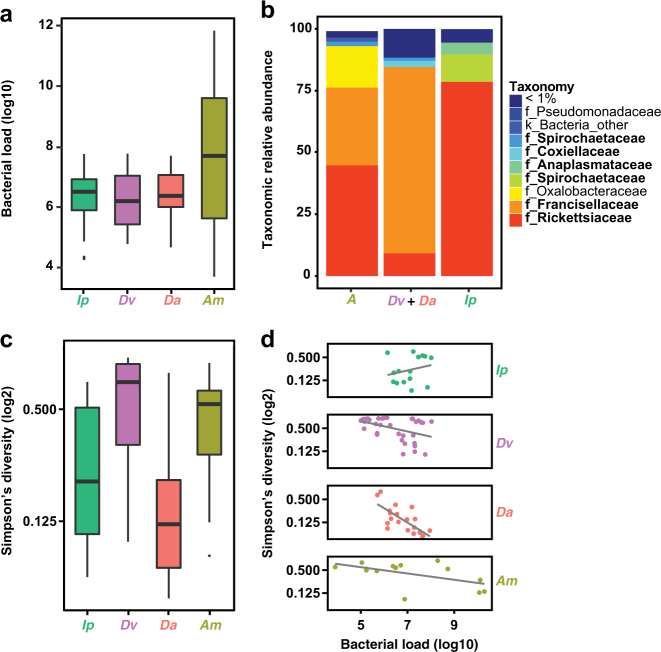
Fig. 5.
Impact of limited bacterial load on bacterial diversity estimates in hard ticks. a Box and whisker plots describing the internal bacterial load (log10) across all hard ticks sampled (N = 139), as determined by qPCR. Species names abbreviated to Ip (Ixodes pacificus), Dv (Dermacentor variabilis), Da (Dermacentor andersoni), and Am (Amblyomma maculatum). b Relative taxonomic abundance for bacteria detected by 16 S rRNA gene sequencing averaged across all samples for A. maculatum, D. variabilis and D. andersoni, and I. pacificus. Taxa previously known to be internally associated with these species are bolded in legend. c Box and whisker plots describing Simpson’s Diversity (log2) of all hard tick internal samples without filtering, with same color scheme as in a. d Bacterial load plotted against Simpson’s Diversity (log2). Spearman’s rank correlation: Ip rho = 0.07, p > 0.1; Dv rho = −0.88, p < 0.001; Da rho = −0.73, p < 0.001; Am rho = −0.62, p < 0.05
Fig. 4.
Midgut-associated bacteria in I. scapularis. a Cross-section of the ovary and midgut of a B. burgdorferi-infected adult female I. scapularis tick. Region highlighted by zoom indicated by white box. “mg” indicates midgut lumen. Ovary indicated by white outline labeled with “ov”. Percent indicates the proportion of analyzed ticks with similar internal bacterial biogeography (N = 41). Arrowhead in zoom indicates B. burgdorferi cells. b Staining of an I. scapularis female tick collected from CA site with midgut-associated bacteria. White dashed box indicates the region highlighted by zoom. Percent indicates the percent of total ticks analyzed with similar internal bacterial content. Scale bars indicate 25 microns for wide view and 10 microns for zoomed panels
16S rRNA gene sequencing of diverse wild adult ticks
We next sought to test whether our findings of a limited internal bacterial load and restricted diversity in I. scapularis were generalizable across Ixodidae. Vector competence for Borrelia spp. varies across genera within the Ixodidae. Ixodid ticks also vector pathogenic Anaplasma and Ehrlichia spp which must transit the midgut during their enzootic cycles [63]. We collected six species of hard tick from multiple geographic locations, including species from the genera Amblyomma, Dermacentor, and Ixodes. qPCR-based assessment of internal bacterial load revealed a variability similar to that of I. scapularis, with Amblyomma maculatum exhibiting the highest load and broadest variation across individual ticks (Fig. 5a). As observed for I. scapularis, the dominant bacterial taxa associated with each tick species were species-specific endosymbionts (Fig. 5b). These include Francisella and Rickettsia in Amblyomma maculatum, Francisella in Dermacentor spp, and Rickettsia in Ixodes spp, as has been previously reported [25, 28]. Although alpha diversity varies broadly across species (Fig. 5c), negative correlations with total bacterial load across hard tick samples (an exception being I. pacificus) supported a similar pattern as seen for I. scapularis (Fig. 5d).
Beta diversity (between-samples) analysis using the weighted UniFrac metric (which accounts for differences in taxon abundance between samples) revealed significant clustering of samples by tick genus (ADONIS, p = 0.001) (Fig. S7A), as expected for stable microbiomes or those largely composed of endosymbionts. In contrast, external wash samples cluster together regardless of which tick they were associated with. This suggests that external microbiomes share common features (e.g., the presence of diverse low-abundance taxa and lack of highly abundant endosymbionts) that differ from internal microbiomes of ticks. This interpretation is supported by beta diversity analyses using the unweighted UniFrac metric (in which each OTU is given equal weight regardless of relative abundance in the sample) which revealed no such internal-external (Fig. S7B) or genus-level clustering, suggesting that the low-abundance taxa in samples do not sufficiently drive differences between tick genera.
Finally, we sought to extend our microscopy findings across hard tick species. Similar to I. scapularis, we found that A. maculatum, D. variabilis, and I. pacificus ticks lack abundant midgut-associated bacteria, while females belonging to these species contained abundant DAPI-intense cocci within ovaries (Fig. S8). We also observed that midgut sections near ovaries sometimes contained localized micro-colonies, similar to those shown to be Rickettsia in I. scapularis. These may be species-specific endosymbionts, as previously reported [23, 64].
Discussion
Here we have provided multiple lines of evidence showing that unfed wild hard ticks possess a limited internal microbiome. Our findings extend upon early studies which found little evidence for culturable bacteria within I. scapularis and add to the growing literature suggesting that not all animals are intimately associated with a complex and abundant internal microbiome [65–68]. This is in contrast to recent reports that suggest a diverse microbiome is a characteristic feature of hard ticks across species and life stages [21, 26–29, 31, 33–35, 62]. We find in our samples that measurements of elevated diversity derived from high-throughput sequencing of tick samples are largely a function of low bacterial biomass. This is a widespread problem inherent in high-throughput sequencing studies that rely upon PCR-amplification during the sample preparation process [69, 70]. We propose that pooling multiple samples to increase input biomass is one method that might lower noise from reagent-based or external contamination [71].
There is great interest in exploiting natural microbial communities to benefit human health and combat disease. One such avenue of active research involves leveraging the native microbial communities associated with medically relevant arthropod vectors (like mosquitos and ticks) to prevent pathogen transmission [14, 72]. We note that B. burgdorferi may be particularly susceptible to inhibition by competitor bacteria if encountered due to a paucity of genetic mechanisms for direct interbacterial interactions. This is supported by our findings that the increased abundance of Bacillus, Enterobacteriaceae, and Pseudomonas within the midgut is associated with decreased B. burgdorferi infection. Notably, these bacteria were also detected in external wash samples. This intriguing pattern suggests that some tick-associated external bacteria can colonize the I. scapularis midgut and, in doing so, might competitively exclude B. burgdorferi, either by displacement or by inhibition of infection during a subsequent blood meal. Members of these taxa encode an arsenal of mechanisms with which to compete with other bacterial cells, including type IV, VI, and VII secretion systems, contact-dependent inhibition (CDI) mechanisms, and the ability to produce bacteriocins [16, 49, 50, 73, 74]. It is unknown if environmental bacteria can persist within the midgut during transstadial molts, but it is worth noting that detection of Bacillus and other bacteria within ticks long preceded the 16 S rRNA gene sequencing era [59]. Direct inhibition of B. burgdorferi by tick-associated bacteria remains an active area of investigation by our laboratories.
Despite the detection of environmental bacteria within ticks, our findings suggest that the midgut of hard ticks may largely be an environment that is ill-suited for bacterial growth, perhaps due in part to the exceedingly low levels of the vital nutrient thiamin in I. scapularis [75]. While B. burgdorferi has evolved unique metabolic strategies in order to persist in this thiamin-limited environment, other less-specialized bacteria may not be able to do so. Additional factors that might limit the overall bacterial load of hard ticks are the possession of conserved or unique innate immunity factors that could target bacteria [76, 77], heme toxicity [78], and the effects of nutrient limitation and desiccation during the extended period between blood meals [6, 79].
Borrelia burgdorferi may encounter a limited diversity of bacteria within the I. scapularis midgut, yet co-infections with other pathogens such as Anaplasma are common [54]. The mechanisms by which B. burgdorferi co-exists with other microbial pathogens within the tick remain unexplored. While Anaplasma, Ehrlichia, and Rickettsia spp are intracellular and may not engage in direct physical interactions with Borrelia spp, these bacteria may influence colonization through other means [80, 81]. B. burgdorferi may not frequently encounter diverse bacteria within the midgut of the tick, however, multi-strain infections may be common within single ticks [82–86]. Furthermore, it is as yet unclear the extent to which different B. burgdorferi strains engage in interactions or how multi-strain infections might influence transmission to humans and pathogenicity.
Our bioinformatic analyses suggests that the genus Borrelia lost genes for mediating interbacterial interactions during the course of its evolution. While Ixodes species are the only vectors for B. burgdorferi in North America, ticks of the genus Ornithodoros (soft ticks) vector the relapsing fever spirochete B. hermsii [87]. We did not detect interbacterial effector–immunity gene homologs encoded by B. hermsii. Although soft ticks were not included in our 16 S rRNA gene sequencing or microscopy, we speculate that B. hermsii encounters a limited internal microbiome within Ornithodoros ticks. Our results further imply that lice of the order Phthiraptera (class Insecta), which vectors B. recurrentis, may also possess a similarly limited internal microbiome [88]. Borrelia spp may have therefore evolved to exploit evolutionarily divergent hematophagous arthropods that share the common lack of a stable internal microbiota.
Data accessibility
Sequencing data were deposited at the NCBI Sequence Read Archive, BioProject accession PRJNA414923.
Electronic supplementary material
Acknowledgements
We thank the Fred Hutch Experimental Histopathology Core facility and Dan Long at Rocky Mountain Labs for tick sectioning and slide preparation. We thank Barbara Simon, Jim Ruppa, David Simon, and June Reznikoff for their indefatigable efforts in collection of ticks from the Klickitat River “Ant Ranch”, and Susan Little for collection of Amblyomma ticks. We thank the UW Keck Imaging Center for providing equipment and assistance in confocal microscopy and acknowledge its support from the NIH (S10 OD016240). We thank Mr. DNA for sequencing support. We are grateful to our colleagues for careful review of the manuscript, and members of the Mougous lab for helpful discussions. This work was supported by the National Institutes of Health grant R21AI114923 (JDM). JDM holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (BWF 1010010) and is an HHMI investigator. BDR was supported by a Simons Foundation-sponsored Life Sciences Research Foundation postdoctoral fellowship. SC was supported by the Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation.
Author contributions
B.D.R., S.C. and J.D.M. conceived the study. B.D.R., S.C. and J.D.M. designed the study. B.D.R., S.C., B.H. and M.C.R. conducted experimental work. X.L., T.J., D.N. and J.B. collected ticks. B.D.R., S.C. and J.D.M. wrote the paper. All authors read and approved the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Benjamin D. Ross, Email: bdross@uw.edu
Seemay Chou, Email: seemay.chou@ucsf.edu.
Joseph D. Mougous, Email: mougous@u.washington.edu
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0161-6) contains supplementary material, which is available to authorized users.
References
- 1.Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, et al. Climate warming and disease risks for terrestrial and marine biota. Science. 2002;296:2158–62. doi: 10.1126/science.1063699. [DOI] [PubMed] [Google Scholar]
- 2.Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, Holt RD, et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–52. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vora N. Impact of anthropogenic environmental alterations on vector-borne diseases. Medscape J Med. 2008;10:238. [PMC free article] [PubMed] [Google Scholar]
- 4.Jones BA, Grace D, Kock R, Alonso S, Rushton J, Said MY, et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc Natl Acad Sci USA. 2013;110:8399–404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kugeler KJ, Farley GM, Forrester JD, Mead PS. Geographic distribution and expansion of human lyme disease, United States. Emerg Infect Dis. 2015;21:1455–7. doi: 10.3201/eid2108.141878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wormser GP, Dattwyler RJ, Shapiro ED, Halperin JJ, Steere AC, Klempner MS, et al. The clinical assessment, treatment, and prevention of lyme disease, human granulocytic anaplasmosis, and babesiosis: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis. 2006;43:1089–134. doi: 10.1086/508667. [DOI] [PubMed] [Google Scholar]
- 8.Berende A, ter Hofstede HJ, Vos FJ, van Middendorp H, Vogelaar ML, Tromp M, et al. Randomized trial of longer-term therapy for symptoms attributed to lyme disease. N Engl J Med. 2016;374:1209–20. doi: 10.1056/NEJMoa1505425. [DOI] [PubMed] [Google Scholar]
- 9.Nigrovic LE, Thompson KM. The Lyme vaccine: a cautionary tale. Epidemiol Infect. 2007;135:1–8. doi: 10.1017/S0950268806007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finney CA, Kamhawi S, Wasmuth JD. Does the arthropod microbiota impact the establishment of vector-borne diseases in mammalian hosts? PLoS Pathog. 2015;11:e1004646. doi: 10.1371/journal.ppat.1004646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, et al. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–8. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Dos-Santos ALA, Huang W, Liu KC, Oshaghi MA, Wei G, et al. Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science. 2017;357:1399. doi: 10.1126/science.aan5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirimotich CM, Ramirez JL, Dimopoulos G. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 2011;10:307–10. doi: 10.1016/j.chom.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell AB, Peterson SB, Mougous J. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12:137–48. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wexler AG, Bao Y, Whitney JC, Bobay LM, Xavier JB, Schofield WB, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113:3639–44. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, et al. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe. 2017;22:411–9 e4. doi: 10.1016/j.chom.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–22. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreotti R, Perez de Leon AA, Dowd SE, Guerrero FD, Bendele KG, Scoles GA. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 2011;11:6. doi: 10.1186/1471-2180-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clayton KA, Gall CA, Mason KL, Scoles GA, Brayton KA. The characterization and manipulation of the bacterial microbiome of the Rocky Mountain wood tick, Dermacentor andersoni. Parasit Vectors. 2015;8:632. doi: 10.1186/s13071-015-1245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clay K, Klyachko O, Grindle N, Civitello D, Oleske D, Fuqua C. Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol Ecol. 2008;17:4371–81. doi: 10.1111/j.1365-294X.2008.03914.x. [DOI] [PubMed] [Google Scholar]
- 24.Hawlena H, Rynkiewicz E, Toh E, Alfred A, Durden LA, Hastriter MW, et al. The arthropod, but not the vertebrate host or its environment, dictates bacterial community composition of fleas and ticks. ISME J. 2012;7:221–3. doi: 10.1038/ismej.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rynkiewicz EC, Hemmerich C, Rusch DB, Fuqua C, Clay K. Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol Ecol. 2015;24:2566–79. doi: 10.1111/mec.13187. [DOI] [PubMed] [Google Scholar]
- 26.Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, et al. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe. 2014;15:58–71. doi: 10.1016/j.chom.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakao R, Abe T, Nijhof AM, Yamamoto S, Jongejan F, Ikemura T, et al. A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks. ISME J. 2013;7:1003–15. doi: 10.1038/ismej.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budachetri K, Browning RE, Adamson SW, Dowd E, Chao CC, Ching WM, et al. An Insight Into the Microbiome of the Amblyomma maculatum (Acari: Ixodidae) J Med Entomol. 2014;5:119–29. doi: 10.1603/ME12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budachetri K, Williams J, Mukherjee N, Sellers M, Moore F, Karim S. The microbiome of neotropical ticks parasitizing on passerine migratory birds. Ticks Tick Borne Dis. 2016;8:170–3. doi: 10.1016/j.ttbdis.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams-Newkirk Amanda Jo, Rowe Lori A., Mixson-Hayden Tonya R., Dasch Gregory A. Characterization of the Bacterial Communities of Life Stages of Free Living Lone Star Ticks (Amblyomma americanum) PLoS ONE. 2014;9(7):e102130. doi: 10.1371/journal.pone.0102130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swei A, Kwan JY. Tick microbiome and pathogen acquisition altered by host blood meal. ISME J. 2017;11:813–6. doi: 10.1038/ismej.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khoo JJ, Chen F, Kho KL, Ahmad Shanizza AI, Lim FS, Tan KK, et al. Bacterial community in Haemaphysalis ticks of domesticated animals from the Orang Asli communities in Malaysia. Ticks Tick Borne Dis. 2016;7:929–37. doi: 10.1016/j.ttbdis.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 33.Trout Fryxell RT, DeBruyn JM. The Microbiome of Ehrlichia-Infected and Uninfected Lone Star Ticks (Amblyomma americanum) PLoS ONE. 2016;11:e0146651. doi: 10.1371/journal.pone.0146651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zolnik Christine P., Prill Robert J., Falco Richard C., Daniels Thomas J., Kolokotronis Sergios-Orestis. Microbiome changes through ontogeny of a tick pathogen vector. Molecular Ecology. 2016;25(19):4963–4977. doi: 10.1111/mec.13832. [DOI] [PubMed] [Google Scholar]
- 35.van Treuren W, Ponnusamy L, Brinkerhoff RJ, Gonzalez A, Parobek CM, Juliano JJ, et al. Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl Environ Microbiol. 2015;81:6200–9. doi: 10.1128/AEM.01562-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–66. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 37.Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, et al. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol Microbiol. 2007;64:1358–74. doi: 10.1111/j.1365-2958.2007.05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 39.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 40.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Development Core Team. R: A Language and Environment for Statistical Computing. 2013.
- 42.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–69. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vannini C, Petroni G, Verni F, Rosati G. A bacterium belonging to the Rickettsiaceae family inhabits the cytoplasm of the marine ciliate Diophrys appendiculata(Ciliophora, Hypotrichia) Microb Ecol. 2005;49:434–42. doi: 10.1007/s00248-004-0055-1. [DOI] [PubMed] [Google Scholar]
- 44.Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, et al. A rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol. 2012;194:376–94. doi: 10.1128/JB.06244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthiesen SH, Hansen CM. Fast and non-toxic in situ hybridization without blocking of repetitive sequences. PLoS One. 2012;7:e40675. doi: 10.1371/journal.pone.0040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sojka D, Franta Z, Horn M, Caffrey CR, Mareš M, Kopáček P. New insights into the machinery of blood digestion by ticks. Trends Parasitol. 2013;29:276–85. doi: 10.1016/j.pt.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Biegala IC, Kennaway G, Alverca E, Lennon JF, Vaulot D, Simon N. Identification of bacteria associated with Dinoflagellates (Dinophyceae) Alexandrium Spp. using tyramide signal amplification–fluorescent In situ hybridization and confocal microscopy1. J Phycol. 2002;38:404–11. doi: 10.1046/j.1529-8817.2002.01045.x. [DOI] [Google Scholar]
- 48.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- 50.Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T. The type VII secretion system of Staphylococcus aureussecretes a nuclease toxin that targets competitor bacteria. Nat Microbiol. 2016;2:16183. doi: 10.1038/nmicrobiol.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitney JC, Peterson SB, Kim J, Pazos M, Verster AJ, Radey MC, et al. A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife. 2017;6:e26938. 10.7554/eLife.26938. [DOI] [PMC free article] [PubMed]
- 52.Becker NS, Margos G, Blum H, Krebs S, Graf A, Lane RS, et al. Recurrent evolution of host and vector association in bacteria of the Borrelia burgdorferisensu lato species complex. BMC Genom. 2016;17:734. doi: 10.1186/s12864-016-3016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paster BJ, Dewhirst FE, Weisburg WG, Tordoff LA, Fraser GJ, Hespell RB, et al. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991;173:6101–9. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamer SA, Hickling GJ, Walker ED, Tsao JI. Increased diversity of zoonotic pathogens and Borrelia burgdorferistrains in established versus incipient Ixodes scapularispopulations across the Midwestern United States. Infect Genet Evol. 2014;27:531–42. doi: 10.1016/j.meegid.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 55.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016;4:29. doi: 10.1186/s40168-016-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jervis-Bardy J, Leong LE, Marri S, Smith RJ, Choo JM, Smith-Vaughan HC, et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 2015;3:19. doi: 10.1186/s40168-015-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steinhaus EA. Insect microbiology: an account of the microbes associated with insects and ticks, with special reference to the biologic relationships involved. Ithaca, N.Y.: Comstock publishing company, Inc.; 1946. [Google Scholar]
- 60.Moreno CX, Moy F, Daniels TJ, Godfrey HP, Cabello FC. Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ Microbiol. 2006;8:761–72. doi: 10.1111/j.1462-2920.2005.00955.x. [DOI] [PubMed] [Google Scholar]
- 61.Noda H, Munderloh UG, Kurtti TJ. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol. 1997;63:3926–32. doi: 10.1128/aem.63.10.3926-3932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abraham NM, Liu L, Jutras BL, Yadav AK, Narasimhan S, Gopalakrishnan V, et al. Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc Natl Acad Sci USA. 2017;114:E781–E90. doi: 10.1073/pnas.1613422114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ismail N, Bloch KC, McBride JW. Human ehrlichiosis and anaplasmosis. Clin Lab Med. 2010;30:261–92. doi: 10.1016/j.cll.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klyachko O, Stein BD, Grindle N, Clay K, Fuqua C. Localization and visualization of a Coxiella-type symbiont within the lone star tick, Amblyomma americanum. Appl Environ Microbiol. 2007;73:6584–94. doi: 10.1128/AEM.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jing X, Wong AC, Chaston JM, Colvin J, McKenzie CL, Douglas AE. The bacterial communities in plant phloem-sap-feeding insects. Mol Ecol. 2014;23:1433–44. doi: 10.1111/mec.12637. [DOI] [PubMed] [Google Scholar]
- 66.Anderson JF, Magnarelli LA, Burgdorfer W, Barbour AG. Spirochetes in Ixodes Dammini and Mammals from Connecticut. Am J Trop Med Hyg. 1983;32:818–24. doi: 10.4269/ajtmh.1983.32.818. [DOI] [PubMed] [Google Scholar]
- 67.Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease-a tick-borne spirochetosis? Science. 1982;216:1317–9. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 68.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N. Caterpillars lack a resident gut microbiome. Proc Natl Acad Sci Usa. 2017;114:9641–6. doi: 10.1073/pnas.1707186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glassing A, Dowd SE, Galandiuk S, Davis B, Chiodini RJ. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016;8:24. doi: 10.1186/s13099-016-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gall CA, Reif KE, Scoles GA, Mason KL, Mousel M, Noh SM, et al. The bacterial microbiome of Dermacentor andersoni ticks influences pathogen susceptibility. ISME J. 2016; 1-10. [DOI] [PMC free article] [PubMed]
- 72.Narasimhan S, Fikrig E. Tick microbiome: the force within. Trends Parasitol. 2015;31:315–23. doi: 10.1016/j.pt.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, et al. Bacterial killing via a type IV secretion system. Nat Commun. 2015;6:6453. doi: 10.1038/ncomms7453. [DOI] [PubMed] [Google Scholar]
- 74.Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol. 2002;56:117–37. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 75.Zhang K, Bian J, Deng Y, Smith A, Nunez RE, Li MB, et al. Lyme disease spirochaete Borrelia burgdorferi does not require thiamin. Nat Microbiol. 2016;2:16213. doi: 10.1038/nmicrobiol.2016.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chou S, Daugherty MD, Peterson SB, Biboy J, Yang Y, Jutras BL, et al. Transferred interbacterial antagonism genes augment eukaryotic innate immune function. Nature. 2014. [DOI] [PMC free article] [PubMed]
- 77.Palmer WJ, Jiggins FM. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol Biol Evol. 2015;32:2111–29. doi: 10.1093/molbev/msv093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anzaldi LL, Skaar EP. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun. 2010;78:4977–89. doi: 10.1128/IAI.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sonenshine DE, Roe RM. Biology of Ticks. 2nd ed. New York: Oxford University Press; 2014. [Google Scholar]
- 80.Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol. 2010;167:95–107. doi: 10.1016/j.vetpar.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 81.Simhadri RK, Fast EM, Guo R, Schultz MJ, Vaisman N, Ortiz L, et al. The gut commensal microbiome of Drosophila melanogaster is modified by the Endosymbiont Wolbachia. mSphere. 2017;2. [DOI] [PMC free article] [PubMed]
- 82.Strandh M, Råberg L. Within-host competition between Borrelia afzelii ospC strains in wild hosts as revealed by massively parallel amplicon sequencing. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140293. doi: 10.1098/rstb.2014.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Herrmann C, Gern L, Voordouw MJ. Species co-occurrence patterns among Lyme borreliosis pathogens in the tick vector Ixodes ricinus. Appl Environ Microbiol. 2013;79:7273–80. doi: 10.1128/AEM.02158-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Voordouw MJ. Co-feeding transmission in Lyme disease pathogens. Parasitology. 2015;142:290–302. doi: 10.1017/S0031182014001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Durand Jonas, Jacquet Maxime, Paillard Lye, Rais Olivier, Gern Lise, Voordouw Maarten J. Cross-Immunity and Community Structure of a Multiple-Strain Pathogen in the Tick Vector. Applied and Environmental Microbiology. 2015;81(22):7740–7752. doi: 10.1128/AEM.02296-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Durand J, Herrmann C, Genne D, Sarr A, Gern L, Voordouw MJ. Multistrain infections with lyme borreliosis pathogens in the tick vector. Appl Environ Microbiol. 2017;83:e02552–16. [DOI] [PMC free article] [PubMed]
- 87.Dworkin MS, Schwan TG, Anderson DE., Jr Tick-borne relapsing fever in North America. Med Clin North Am. 2002;86:417–33. doi: 10.1016/S0025-7125(03)00095-6. [DOI] [PubMed] [Google Scholar]
- 88.Lescot M, Audic S, Robert C, Nguyen TT, Blanc G, Cutler SJ, et al. The genome of Borrelia recurrentis, the agent of deadly louse-borne relapsing fever, is a degraded subset of tick-borne Borrelia duttonii. PLoS Genet. 2008;4:e1000185. doi: 10.1371/journal.pgen.1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data were deposited at the NCBI Sequence Read Archive, BioProject accession PRJNA414923.