Abstract
The pathogenic bacterium Salmonella enterica serovar Typhimurium utilizes two type III secretion systems (T3SS) to inject effector proteins into target cells upon infection. The T3SS secretion apparatus (the injectisome) is a large macromolecular assembly composed of over twenty proteins, many in highly oligomeric states. A sub‐structure of the injectisome, termed the basal body, spans both membranes and the periplasmic space of the bacterium. It is primarily composed of three integral membranes proteins, InvG, PrgH, and PrgK, that form ring structures through which components are secreted. In particular, PrgK possesses a periplasmic region consisting of two globular domains joined by a linker polypeptide. We showed previously that in isolation, this region adopts two distinct conformations, of with only one is observed in the assembled basal body complex. Here, using NMR spectroscopy, we further characterize these two conformations. In particular, we demonstrate that the interaction of the linker region with the first globular domain, as found in the intact basal body, is dependent upon the cis conformation of the Leu77‐Pro78 peptide. Furthermore, this interaction is pH‐dependent due to coupling with hydrogen bond formation between Tyr75 and His42 in its neutral Nδ1H tautomeric form. This pH‐dependent interaction may play a role in the regulation of the secretion apparatus disassembly in the context of bacterial infection.
Keywords: NMR, protein dynamics, bacterial secretion systems, Salmonella enterica serovar Typhimurium
Introduction
Bacteria use specialized secretion systems, numbered I to IX, to transport proteins across their membrane(s) and cell wall.1 Secreted proteins, termed “effectors,” play numerous functions in bacterial biology, from chemotaxis to infectivity and immune evasion. Of these systems, the type III secretion system (T3SS) has the particularity to be contact‐dependent such that secretion is activated upon contact with a target cell.2, 3, 4 In addition, unlike most secretion systems, T3SS effectors are not just secreted to the extracellular environment, but rather injected directly into the cytosol of the infected cell.
The human pathogen Salmonella enterica serovar Typhimurium encodes two T3SSs, in different genome locations termed the Salmonella Pathogenicity Islands 1 and 2 (SPI‐1 and SPI‐2).5 The SPI‐1 T3SS is active early during infection in the host gut, and its effectors induce actin rearrangements and membrane invagination of the target cell. This leads to the incorporation of bacteria into intracellular vesicles, termed the Salmonella‐Containing Vacuoles (SCVs). Acidification of the SCV in turn activates the SPI‐2 T3SS, whose effectors prevent fusion with lysogenic vesicles and permit bacterium survival.6
The SPI‐1 T3SS is the best‐characterized T3SS at the molecular level.7 Its secretion apparatus, the injectisome, consists of a structure called the basal body, which spans both membranes and the intervening periplasmic space. On the cytosolic side, a set of proteins forming the inner membrane anchored complex of the T3SS system, known as the “export apparatus”, is responsible for the selection and active secretion of effectors. On the extracellular side, an extended needle of species‐specific length projects outwards, toward target cells.5 Genetic studies have demonstrated that the export apparatus assembles first in the inner membrane. Subsequently, the basal body assembles around the export apparatus, forming a secretion‐competent complex. This leads to the secretion of the protein forming the needle, as well as of additional regulatory components, such as the rod and ruler proteins.8, 9, 10, 11
Extensive work, including NMR spectroscopic, X‐ray crystallographic and biochemical studies,12, 13, 14, 15, 16, 17 combined with the recent near‐atomic resolution cryo‐electron microscopy (EM),18 have defined the structure of the SPI‐1 T3SS basal body. In particular, the inner‐membrane proteins PrgH and PrgK, and the outer‐membrane protein InvG, are found to consist largely of small globular domains with a conserved fold. This fold is termed the Ring‐Building Motif (RBM)14 in reference to the involvement of these proteins in assembly of oligomeric ring‐shaped structures.
We previously showed that the periplasmic domain of PrgK consists of two RBMs (termed D1 and D2), joined by a short linker polypeptide [Fig. 1(A,B)]. Furthermore, we found that the periplasmic domain adopts two conformations that are dependent upon the presence of the linker region.12 More specifically, we proposed that these two conformations differ by whether or not the linker is non‐covalently bound to D1.12 In this study, we verified this proposal, using NMR spectroscopy to show that these two conformations indeed correspond to the linker‐bound D1 (denoted as conformer A) and linker‐unbound D1 (conformer B) states of PrgK. We also show that these two conformational states differ in the cis versus trans isomerization of the Leu77‐Pro78 peptide. These conserved linker residues are essential for T3SS function. Furthermore, we report that the equilibrium between the two conformers is dependent upon the protonation state of His42, and hence modulated by hydronium ion activity (sample pH value). This leads us to hypothesize that variations in local pH could contribute to the structural arrangement of PrgK as required for injectisome dynamics in vivo.
Figure 1.
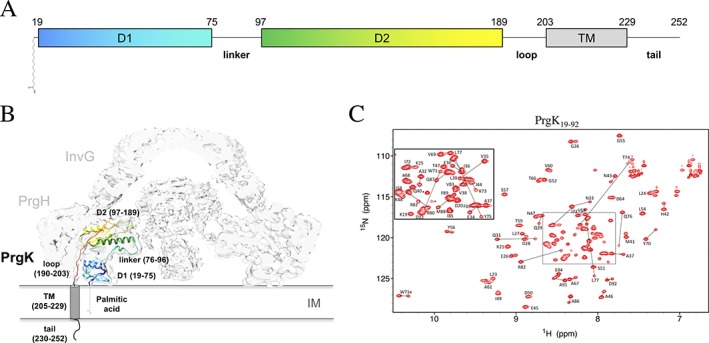
Structure and conformations of the PrgK periplasmic domain. (A) Schematic representation of PrgK, with the boundaries of the D1, linker, D2, loop and TM indicated, along with an N‐terminal cysteine‐linked palmitic acid. (B) Cartoon representation of the PrgK structure (colored; PDB ID 5TCP) in the cryo‐EM map of the basal body complex17 (EMDB ID 8398). (C) Assigned 15N‐HSQC spectrum of the PrgK19–92 construct, reproduced from reference 12. The lines connect the two 1HN‐15N peaks yielded by the residues that differ in the two conformers of this construct
Results
Structures of the two PrgK 19–92 conformers using CS‐Rosetta
We previously showed that a purified PrgK construct encompassing D1, the linker region, and D2, adopts two conformations, as detected by the presence of two sets of resolved amide 1HN‐15N signals from many residues in its 15N‐HSQC spectrum.12 Due to extensive spectral overlap, we were unable to fully assign the backbone resonance frequencies for this construct. However, we also showed that the two conformations, denoted A and B, were recapitulated in a shorter construct encompassing D1 and the linker region (PrgK19–92).12 With a simpler NMR spectrum, and by considering patterns of relative peak intensities, we were able to unambiguously assign the signals from both the major conformer A and minor conformer B of this construct [Fig. 1(C)]. For this reason, we used PrgK19–92 as a proxy to characterize the two conformations adopted by the full PrgK periplasmic domain in isolation.
Unfortunately, the added complexity of two conformations prevented us from unambiguously assigning inter‐proton NOEs to one population or the other. This in turn precluded structural determination of the PrgK19–92 conformers by conventional NMR spectroscopic approaches. We therefore used CS‐Rosetta19 to generate structures for both conformers with backbone chemical shift information as the sole experimental constraints. This led to convergent structural models with good energy parameters [Fig. 2(A,B); Supporting Information Fig. S1(A), S1(B)]. As will be justified below, the Leu77‐Pro78 peptide was set to the cis isomer for conformer A, and flexible residues (79–92 for conformer A, 75–92 for conformer B) were trimmed by CS‐Rosetta. We then combined the 20 lowest energy models for each to generate the structural ensembles shown in Figure 2. Both exhibit very good geometry, with overall backbone RMSD values of 0.5 and 0.6 Å for the conformer A and conformer B ensembles, respectively.
Figure 2.
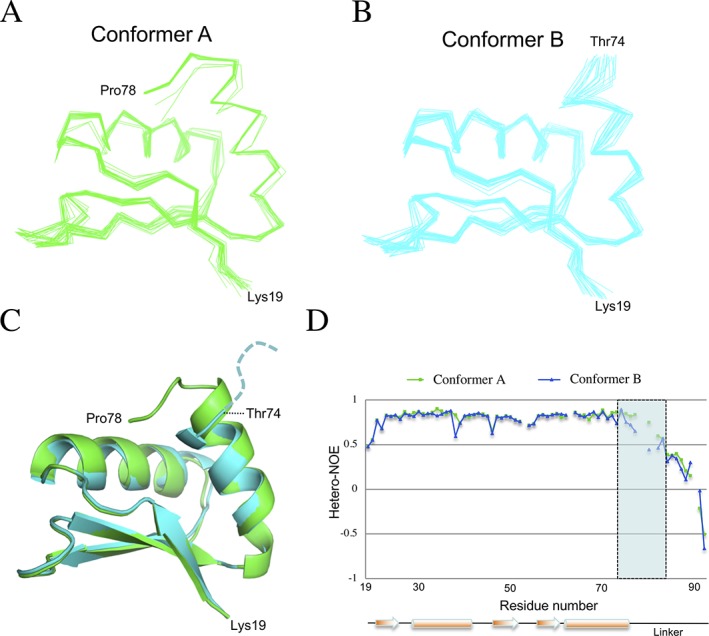
Structural ensembles of the two PrgK19–92 conformers. Line representation of the backbone atoms for the twenty lowest‐energy models for PrgK19–92 in the (A) conformer A and (B) conformer B, obtained with CS‐Rosetta.20 (C) The two averaged structures are overlaid in cartoon representation, with conformer A in green and conformer B in cyan. The only significant difference lies at their C‐termini, where residues 75–78 of the linker are ordered in conformer A but not in conformer B. (D) The heteronuclear NOE values for the two populations of PrgK19–92 plotted versus sequence, with the secondary structure shown at the bottom. Decreasing NOE values indicate increasing mobility of the 1HN‐15N bond on the sub‐nsec timescale. This confirms that residues 75–78 (shaded rectangle) are ordered in conformer A, but flexible in conformer B
The two structural ensembles are very similar, with an average pairwise backbone RMSD of 0.68 Å for residues 19–74. The major differences localize to residues 75–78, which are ordered in the conformer A structural ensemble, but not in that of conformer B [Fig. 2(C)]. This is consistent with the previously reported chemical shift differences between the two conformers of PrgK19–92, and the reduced chemical shift‐derived helical propensity of residues at the C‐terminus of helix 2 in conformer B.12 Importantly, the CS‐Rosetta ensembles are in agreement with the proposed linker‐bound and linker‐free conformations for the two conformers.12 In particular, the structure of conformer A derived from CS‐Rosetta is highly similar to the X‐ray crystallographic structure of PrgK19–92 [PDB ID 4W4M; Supporting Information Fig. S1(D)], as well as to the structure of full sized PrgK in the intact basal body determined by cryo‐EM [PDB ID 5TCP; Supporting Information Fig. S1(C)]. In this later structure, the entire linker is well ordered, and bound to the D1 surface in the manner as observed in the conformer A model. This indicates that within the fully assembled basal body complex, PrgK exists as the conformer A.
To further confirm that the difference between the two conformers lies in the linker region, we measured the heteronuclear 1H‐15N NOE ratios for the PrgK19–92 construct. The heteronuclear NOE is a sensitive indicator of protein dynamics, with decreasing values indicating increasing mobility of the 1HN‐15N bond on the sub‐nanosecond timescale.21 The only significant difference between the two conformers is located at the beginning of the linker region (residues 74–84), with conformer B exhibiting higher flexibility [Fig. 2(D)]. This is consistent with our previous chemical shift‐based analysis of the backbone dynamics of the two PrgK19–92 conformations,12 and the CS‐Rosetta‐derived models described above. Taken together, these data demonstrate that conformers A and B of PrgK observed by NMR spectroscopy correspond to the linker‐bound and linker‐free states of D1, respectively.
The PrgK conformers differ by the cis/trans isomerization of the Leu77‐Pro78 peptide
We next investigated the mechanisms underlying the conformational equilibrium of PrgK19–92. Since several residues yielded two distinct peaks in a 15N‐HSQC spectrum, the conformers must be in the slow exchange regime (i.e., k ex ≪ Δω, where k ex is the interconversion rate constant and Δω is the chemical shift difference between conformers A and B). We therefore collected a 15N‐EXSY experiment22 on this protein. However, we did not observe any exchange cross‐peaks, indicating that k ex ≾ 1 s−1 23 [Supporting Information Fig. S2(A)]. This prompted us to investigate the temperature dependency of the conformational equilibrium. We collected 15N‐HSQC spectra as a function of temperature from 10 to 45°C. As expected, many amide peaks shifted with temperature [Supporting Information Fig. S2(B)]. However, the relative intensity ratios between the peaks assigned to conformer A and conformer B was unchanged. Based on these observations, we concluded that the two conformers do not interconvert on the seconds timescale, even at 45°C.
It is well established that X‐Pro peptide bonds within random coil polypeptides are approximately 10%–20% in the cis isomer, with little temperature dependence and an interconversion rate constant of ∼0.025 s−1.24 We therefore postulated that the two populations of PrgK19–92 differ by the cis and trans isomerization of the Leu77‐Pro78 amide. Indeed, in the previously reported crystal structure of PrgK19–92 in conformer A (PDB ID 4W4M), the Leu77‐Pro78 amide adopts the less common cis geometric arrangement [Fig. 3(A)]. To test this postulate, we assigned the 13C chemical shifts of all prolines in the PrgK19–92 construct. As documented by Schubert et al.,26 the (13Cβ ‐ 13Cγ) chemical shift difference of a proline is highly diagnostic of the X‐Pro amide conformation. This chemical shift difference demonstrates that in conformer A, Leu77‐Pro78 is in the cis isomer, whereas in conformer B, it is in the trans isomer [Fig. 3(B)]. In contrast, all other X‐Pro amides are found as the trans isomer in both conformers (except in the case of Met89‐Pro90, for which only one set of signals could be identified).
Figure 3.
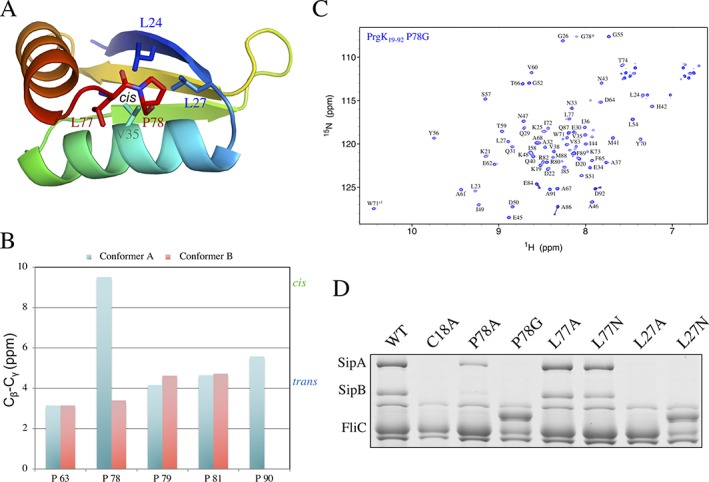
The two conformers correspond to the cis or trans isomers of the Leu77‐Pro78 peptide. (A) Cartoon representation of the ordered residues (19–78) from the lowest‐energy model for the conformer A structure, in rainbow coloring. The residues involved in the linker‐D1 interaction are in stick representation. (B) Difference between the chemical shifts of the 13Cβ and 13Cγ nuclei of each proline residue in the two PrgK19–92 conformers. For most prolines, the difference is ∼4.2 ppm (blue dashed line), corresponding to the average value for a trans X‐Pro isomer.25 In contrast, for Pro78 in conformer A, the difference is 9.6 ppm, corresponding to that of a cis X‐Pro isomer (green dashed line). (C) 15N‐HSQC spectrum of PrgK19–92 with the P78G mutation. The peak for G78, that was introduced instead of a proline in this mutant, is indicated with an asterisk. For most residues, a single 1HN‐15N peak is visible, confirming that this mutation abrogates the linker‐D1 interaction and leads to only conformer B. Residues Glu84, Ala86 and Asp92 still yield two peaks, possibly corresponding to the trans (major) and cis (minor) isomers of Met89‐Pro90. (D) Secretion assay for mutants of Leu27, Leu77 and Pro78. SipA and SipB are effector proteins, secreted when a functional T3SS is formed. FliC is used as a loading control. Mutation of Pro78 and Leu27 abrogates secretion, unlike mutation of Leu77, which has no phenotype. For some mutants (P78G and L27N), FliC runs slower on the gel, likely due to variable glycosylation9, 12
To further confirm the role of Pro78 in the two conformations of PrgK, we engineered the P78G mutation in the PrgK19–92 construct. The trans isomer of the Leu77‐Gly78 peptide should be strongly favored. Indeed, the resulting protein yielded single amide 1HN‐15N peaks in 15N‐HSQC experiments, demonstrating that this mutation disrupts the presence of two predominant conformations [Fig. 3(C)]. Comparison of the spectrum of this protein with that of the wild‐type protein, or that of the protein construct including D1 but not the linker,12 confirms that the peaks yielded by the P78G mutant correspond to the conformer B (Supporting Information Fig. S3). This is also consistent with the conclusion that the Leu77‐Pro78 peptide has cis‐ and trans‐ isomers in conformers A and B, respectively.
In the basal body cryo‐EM map (as well as in the conformer A structural ensemble), Leu77 and Pro78 from the linker region are buried in a hydrophobic pocket formed by Leu24, Leu27, Val35, Val38 and Ile72 [Fig. 3(A)]. This pocket is conserved in PrgK homologs of other T3SS's (Supporting Information Fig. S4), with hydrophobic residues present in all systems, including the highly divergent Chlamydia and Rasltonia Cds and Hrp T3SSs. To verify the role of these residues in T3SS function, we engineered several mutations in a plasmid encoding for the prgK gene, and used it to complement a ΔprgK strain of Salmonella enterica serovar Typhimurium LT2. We then used a secretion assay to monitor T3SS assembly in the context of the corresponding PrgK mutants. As we reported previously, PrgK WT rescued secretion in the ΔprgK strain [repeated as a control here in Fig. 3(D)], but mutation of the normally lipidated N‐terminal Cys18 to Ala, required for stable localization of the basal body to the inner membrane, abrogated secretion.12 Mutating Pro78 to Gly or (to a lesser extent) Ala also impairs secretion [Fig. 3(D)], demonstrating that this residue is important for T3SS function. Similarly, mutating Leu27 to Ala or Asn is detrimental to T3SS function. In contrast, we observed no phenotype for mutations in Leu77 to either Ala or Asn. These results indicate that the interaction between Pro78 and Leu27, observed in the cryo‐EM map of the basal body, and present in conformer A but not in conformer B, is essential for T3SS function.
The conformational equilibrium of PrgK is pH‐dependent due to His42
We then sought to determine if the propensity of the protein to adopt the two conformers is dependent upon buffer composition. We observed that ionic strength (monitored by varying NaCl concentration from 0 to 1 M) does not affect the ratio between the peaks formed by conformer A and conformer B (not shown). In contrast, varying the pH of the sample clearly altered the population ratio [Fig. 4(A)]. To quantify this observation, we measured relative peak intensities for selected residues with identifiable peak pairs at pH 5, 7 and 9. As shown on Figure 4(B), at pH 5 the peaks for both conformers have comparable intensities (average I A/(I A + I B) peak intensity ratio = 0.58), suggesting that the two conformations are approximately equally populated (i.e, [A]/[B] ∼ 1.4). At high pH, we observed that the relative intensities of the conformer A peaks increased, with those of conformer B decreasing correspondingly. Based on an average of peak intensity ratios for amides within the ordered core of PrgK19–92, the relative population of conformer A increases to 0.74 at pH 7 and 0.85 at pH 9 [Fig. 4(A)]. The latter corresponds to [A]/[B] ∼ 5.7. Importantly, this series of measurements also demonstrates that conformers A and B are in a conformational equilibrium, and do not arise simply from some form of irreversible chemical heterogeneity of PrgK19–92.
Figure 4.
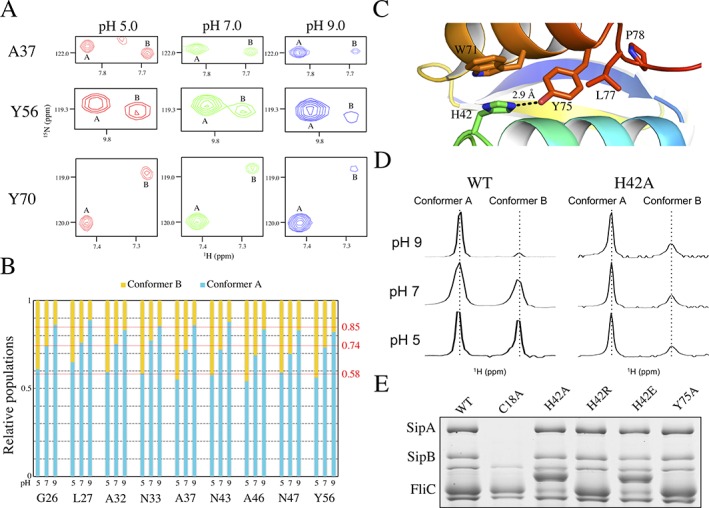
The PrgK population ratio is affected by pH. (A) Selected regions from the overlaid 15N‐HSQC spectra of PrgK19–92 at pH 5 (red), pH 7 (green) and pH 9 (blue). With increasing pH, the corresponding signals from conformer B become weaker relative to those from conformer A. (B) The relative populations of the A and B conformers, IA/(IA + IB) and IB/(IA + IB), determined from corresponding 15N‐HSQC peak intensity ratios, are plotted for selected residues as a function of sample pH value. The relative population of conformer A increases from ∼0.58 at pH 5 to 0.74 at pH 7 and 0.85 at pH 9. Thus, PrgK19–92 preferentially adopts conformer A under more alkaline conditions. Note that peak intensities are also dependent upon amide hydrogen exchange and the relaxation behavior of the 1HH and 15N nuclei, which may differ between conformers A and B. Hence, these intensity ratios are only an approximation of population ratios. (C) Cartoon representation of the PrgK D1 structure in conformer A (PDB ID 4W4M), centered around the salt bridge formed by residues His42 and Tyr75. (D) Expanded, aligned regions of the 1D 1H‐NMR spectra corresponding to the resolved downfield indole 1Hɛ1 signal (∼ 10.5 ppm) of Trp71, for PrgK19–92 WT or with the H42A mutation, at various pH conditions. In the WT protein, two peaks corresponding to the two populations are clearly visible, and their relative intensity varies with sample pH value, as also seen for amides in the spectra of panel A. In contrast, with the H42A mutant, the relative peak intensities are pH‐independent. Although conformer B appears minor in these 1D spectra, in 15N‐HSQC spectra, most amides show approximately equal 1HN‐15N peak intensities for conformers A and B (Fig. S5). (E) Secretion assay for mutants of His42 and Tyr75, performed as in Figure 3(B). None of the mutations affects secretion in this assay
Based on the conformer A structures (PDB ID 4W4M, 5TCP and this study), we observed that the imidazole Nɛ2 of His42 is within hydrogen bonding distance (∼3 Å) of the phenolic Oη of Tyr75 [Fig. 4(C)], which is part of the linker region. It is likely that the cis isomer of the Leu77‐Pro78 peptide is needed for the sidechain of Pro78 to pack against Leu27 [Fig. 3(A)], which in turn stabilizes the His42‐Tyr75 interaction. In contrast, in the NMR‐derived structural ensemble of PrgK19–76 (PDB ID 2MKY) and the CS‐Rosetta model of conformer B structure reported herein [Supporting Information Fig. S1(E)], Tyr75 is disordered and likely does not interact with His42. Since histidine is the amino acid most commonly titratable around neutral pH values,27 we postulated that it plays a role in the equilibrium between the two populations.
To verify this, we mutated His42 to an alanine. The resulting protein still yields two populations, but with approximately equal intensity ratios [Fig. 4(D), Supporting Information Fig. S5]. We note that the significant difference in the 15N‐HSQC spectrum of the H42A mutant versus that of the WT species prevented us from unambiguously assigning all amide signals. However, regardless of assignments, the relative peak intensities of conformers A and B are approximately equal in the H42A mutant (Supporting Information Fig. S5), and most importantly, did not change between pH 5 and pH 9 [Fig. 4(D)].
We then sought to verify the role of this pH‐dependent phenomenon within the cellular context. To that end, we engineered mutations in His42 for a secretion assay as described above. Surprisingly, we observed that mutating this residue to a neutral (alanine), negatively charged (glutamic acid) or positively charged (arginine) residue has no significant effect on secretion in this assay, as shown on Figure 4(E). Similarly, mutating Tyr75 to an Ala shows no phenotype change [Fig. 4(E)]. Taken together, these results demonstrate that while Pro78, which mediates the interaction between the linker and D1, is essential for secretion [Fig. 3(B)], neither His42 nor Tyr75 are required for secretion in this assay. This indicates that the pH‐dependency of the interconversion between the two conformers is not required for assembling a functional T3SS.
His42 has different pKa values in the two conformers of PrgK 19–92
To further investigate the equilibrium between the two conformers of PrgK, we used NMR spectroscopy to determine the apparent pK a values of His42 in each conformer. Specifically 13C/15N‐histidine‐labeled PrgK19–92 was monitored from approximately pH 5 to 9 using 13C‐HSQC (Fig. 5) and Hβ(Cβ)Cγ spectra (not shown).28 Although the signals from conformer B weakened with increasing sample pH values, it was still possible to obtain titration curves. Fitting these pH‐dependent chemical shifts yielded average apparent pK a values of 7.0 ± 0.1 for His 42 in conformer A and 7.7 ± 0.1 for conformer B (Fig. 6).
Figure 5.
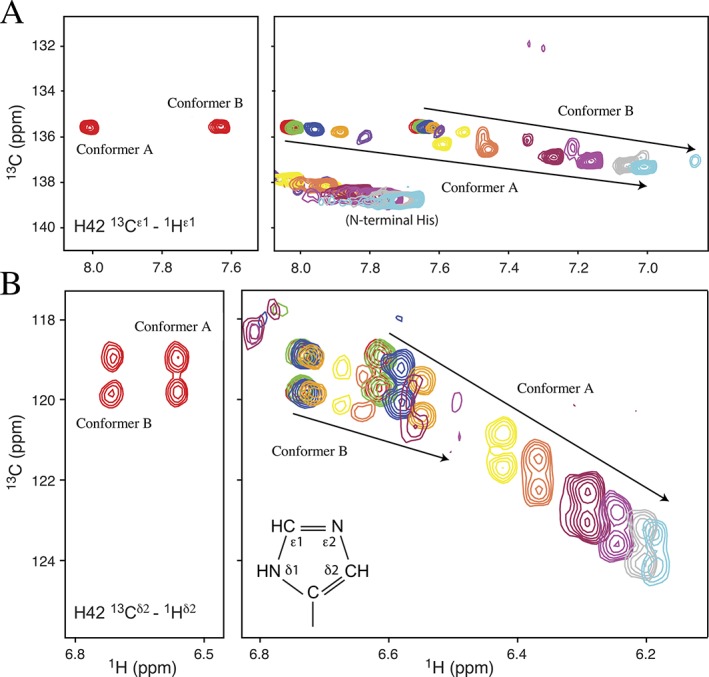
pH‐dependent chemical shifts of His42. The pK a values of His42 were determined from 15N‐decoupled 13C‐HSQC spectra of PrgK19–92 selectively‐labeled with 13C/15N‐histidine. (Left) Assigned spectra recorded at pH 5.2 showing the (A) 13Cɛ1‐1Hɛ1 and (B) 13Cδ2‐1Hδ2 signals. Each 13Cδ2 signal is a doublet due to scalar coupling with adjacent the 13Cγ. (Right) Eleven superimposed spectra recorded as the sample was titrated in small steps from pH 5.2 (red) to 8.7 (light blue). Signals from conformer B diminish in intensity as conformer A is increasingly favored under alkaline conditions. The nomenclature is illustrated for the neutral Nδ1H tautomer of a histidine sidechain
Figure 6.
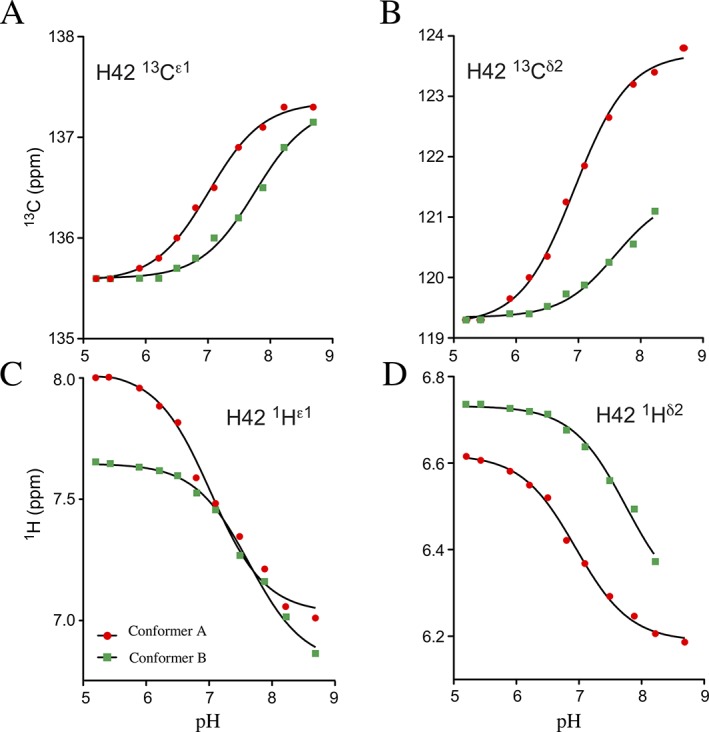
pK a determination for His42. Averaging the individual fits of the pH‐dependent chemical shifts of four His42 nuclei (from Fig. 5) to the equation for a single acid‐base equilibrium yields apparent pK a values of 7.0 ± 0.1 for conformer A (red circles) and 7.7 ± 0.1 for conformer B (green squares)
It is also noteworthy that upon deprotonation, the 13Cδ2 signal of His42 in conformer A shifts downfield by 4.5 ppm to 123.7 ppm [Fig. 5(B)]. Furthermore, its 13Cγ signal shifts up‐field by > 1 ppm (not shown). These highly diagnostic patterns of pH‐dependent chemical shift changes demonstrate that His42 of conformer A predominantly adopts the neutral Nδ1H tautomer under alkaline conditions.27, 29 In contrast, the 13Cδ2 signal of His42 in conformer B shifts downfield by only ∼ 2 ppm to an extrapolated chemical shift of 121.4 ppm. These spectral changes indicate that, at high pH, His42 in this conformer exists as an approximately equimolar mixture of both neutral tautomers. For comparison, the non‐native N‐terminal histidine in the PrgK19–92 construct has a pK a value of 6.5 ± 0.2 and, upon deprotonation, is predominantly in the more common Nɛ2H tautomeric state (Supporting Information Fig. S6). This closely matches the properties expected for an unperturbed histidine in a disordered polypeptide.27
Discussion
In this study, we further characterize the two conformations adopted by the periplasmic domain of PrgK. Specifically, we report the CS‐Rosetta structures of the two conformers, showing that they correspond to the linker‐free and linker‐bound states of PrgK D1. We also demonstrate that the conformers differ in the cis/trans isomerization of the Leu77‐Pro78 peptide. In addition, we demonstrate that pH alters the population ratio between the two conformers, and that this coincides with the fact that the interaction between D1 and the linker is favored upon deprotonation of His42.
Based on the data reported here, we propose a model for the conformational equilibria of PrgK19–92 that is summarized schematically in Figure 7. The linker, including residues Leu77 and Pro78, interacts with a hydrophobic surface of D1 [Fig. 3(A)]. This interaction is also coupled with the ordering of the C‐terminal end of the second helix of D1, which includes Tyr75 [Figs. 2(F) and 4(C)]. However, favorable packing of the linker along the hydrophobic surface only occurs with the less favorable cis isomer of the Leu77‐Pro78 peptide bond. As a result, under acidic conditions, conformers A and B are approximately equally populated and interconverting with kex ≾ 1 s−1. Mutation of Pro78 to glycine restricts the resulting Leu77‐Gly78 peptide bond to the trans isomer and thus PrgK19‐92 P78G only adopts conformer B [Fig. 3(C)]. Functionally, this impairs PrgK‐dependent secretion of effectors SipA and SipB via the T3SS [Fig. 3(D)].
Figure 7.

Model of the PrgK conformational equilibria. Schematic summary of the conformational equilibria of PrgK involving coupling of linker binding and the ordering of the C‐terminal helix of protein with the cis/trans isomerization of Leu77/Pro78 and the pH‐dependent interaction of Tyr75 with His42. See text for further discussion
The equilibrium between conformers A and B is also dependent upon pH. This is attributed to a favorable interaction for conformer A in which Tyr75 donates a hydrogen bond via its OηH to the Nɛ2 of neutral His42. Evidence for this interaction includes the lower pK a value of His42 in conformer A (7.0 ± 0.1) versus conformer B (7.7 ± 0.1), and its deprotonation to the less common neutral Nδ1H tautomer to accept the hydrogen bond (Figs. 5 and 6). However, increasing the sample pH from 5 to 9 increases the [A]/[B] population ratio by ∼ 4‐fold, which corresponds to a rather small ΔΔG ∼ 0.8 kcal/mol. Thus the interaction of Tyr75 with neutral His42 only modestly stabilizes conformer A. It is also noteworthy that mutation of His42 to Ala does not abrogate conformer A. Rather, similar to the WT protein under acidic conditions, conformers A and B are approximately equally populated in the H42A mutant. This suggests that any interaction of Tyr75 with a positively‐charged His42 (i.e., which cannot occur in the H42A mutant) does not markedly impact the conformational equilibrium of PrgK.
Parenthetically, we note that His42 has an elevated pK a value in conformer A, and even more so in conformer B, relative to that of an unperturbed histidine in a disordered peptide (∼ 6.5). This is somewhat unexpected as His42 lacks any neighboring negatively charged groups that would otherwise promote its positively charged form. Also, based on the X‐ray crystallographic structure of PrgK19–92 (conformer A), its pK a is predicted to be ∼ 6 using empirical and computational approaches.30, 31 However, the stacking of His42 over the sidechain of Trp71 [Fig. 4(C)] may lead to its elevated pK a value due to favorable π‐cation interactions.32 Regardless, the NMR‐monitored pH titrations support the model of Figure 7 in which the pH‐dependent interaction of Tyr75 and His42 are coupled with the cis/trans isomerization of the Leu77‐Pro78 peptide and the interaction of the linker residues with the D1 domain of this protein.
Importantly, the interaction between the linker and D1 observed in conformer A, is also found in the cryo‐EM map of the intact basal body18 (although the limited resolution does not allow determination of the physicochemical details). The in vivo assays reported here [Fig. 3(D)] demonstrate that Pro78 and Leu27 are necessary for T3SS secretion, suggesting that this interaction is critical for assembly or function of the complex. In contrast, the residues involved in the pH‐dependence of this interaction (His42, Tyr75) can be mutated with no phenotype in our secretion assay [Fig. 4(E)], and we do not have any evidence that conformer B plays any role in T3SS assembly or function. Based on this, we cannot exclude the possibility that conformer B, and the destabilization of conformer A at lower pH, pertains only to the constructs used for the in vitro biochemical and structural studies.
Nevertheless, we note that His42 is conserved in the closely related Shigella, Burkholderia T3SSs and others of the Inv/Mxi/Spa family (Supporting Information Fig. S4), where T3SS function triggers endocytosis of the bacteria into endosome vacuoles, by non‐phagocytic cell types. In Burkholderia and Shigella, this is followed by endosomal membrane lysis, whereas in Salmonella, decreasing pH activates the SPI‐2 T3SS, preventing lysosome fusion. In all cases, the phagocytosis‐inducing T3SS is no longer required after vesicle formation, but the fate of assembled complexes has not been investigated to our knowledge. We speculate that upon endosome acidification, the drop in pH may alter the linker‐driven interactions, contributing to T3SS disassembly. Unfortunately the latter cannot be tested by our secretion assay, thus potentially explaining why no phenotype was observed for the His42 mutants. Alternatively, it is also possible that the pH‐dependency actually mimics the interaction with other components of the full complex, and therefore reflects a coordinated assembly/disassembly of the complex. Specifically, His42 is in close proximity of the PrgH oligomeric ring in the intact basal body,17 and it is conceivable that the proximity of a charged residue from PrgH alters the protonation state of His42 in the full complex, corresponding to the pH‐mediated events observed in isolation. Further in vivo studies of T3SS complexes, potentially using the pH‐dependency mutants reported here, will be required to further test these hypotheses.
Materials and Methods
Cloning, protein expression, and purification
Protocols for the cloning, mutagenesis, expression and purification of PrgK19–92 has been described previously.12 Briefly, the pET28a expression plasmids encoding wild‐type PrgK19–92 or variants with site‐directed mutations were transformed into E. coli BL21(λDE3) strains. Transformants were grown to log phase, at which point expression was induced with 1 mM IPTG. After 16 h at 20°C, cells were collected by centrifugation and lysed by sonication. The protein was purified by Ni+2‐affinity chromatography, and the 6xHis tag was cleaved with thrombin (Roche). The protein was further purified by gel filtration using a Superdex75 column (GE Healthcare). For NMR spectroscopy, proteins were expressed in M9 minimal media supplemented with 99% 15N NH4Cl (0.5 g/L) and/or 99% 13C6 D‐glucose (2.5 g/L), and purified as above. For pKa measurements, PrgK19–92 was expressed in the histidine auxotroph E. coli strain BL21 hisG::Tn10 (λDE3)33 grown in a synthetic medium34 containing 50 mg/L 98% 13C6‐15N3‐L‐histidine (Sigma‐Aldrich).
NMR spectroscopy
Data were collected at 25°C (unless specified), on Bruker Avance III 500 and 600 MHz spectrometers equipped with xyz‐gradient TCI cryoprobes and processed using Topspin3.1, NMRPipe,35 and Sparky.30 Side‐chain chemical shifts for prolines were assigned using HCCH‐TOCSY,36 HC(CO)NH‐TOCSY,37 HACAN,38 and HACA(CO)N22 spectra. Histidine chemical shifts were assigned using 13C‐HSQC, CβHδ and Hβ(Cβ)Cγ spectra.25 The chemical shift values for PrgK19–92 have been deposited in the BMRB database (BMRB ID: 27424). Heteronuclear 1H‐15N NOE and 15N‐EXSY spectra were measured using published procedures.21, 39 Except for the pH titration experiments, all spectra were collected at pH 6.8.
NMR‐monitored pH titrations of His42 in PrgK19–92 were acquired using a Bruker Avance III 600 MHz spectrometer. The sample was titrated in 11 steps from pH 5.2 to 8.7 by the addition of small aliquots of 0.1 to 1 M NaOH or HCl, as appropriate, and monitored via 13C‐HSQC spectra.25 Sample pH values were recorded at room temperature (∼20°C). The pH‐dependent chemical shifts for the individual 13C and 1H nuclei of His42 were fit with GraphPad Prism to the Henderson‐Hasselbalch equation for a single ionizable group to obtain the corresponding pKa values, which were then averaged to yield those reported for the two conformers of PrgK19–92.25, 40
Structure modeling
For structure determination, the chemical shift values for the two populations of PrgK19–92 were separated and provided as input for the CS‐Rosetta server.20 Of 10,000 models generated, the 20 lowest‐energy were combined as the final structural ensemble. Flexible residues (residues 79–92 for conformer A, residues 75–92 for conformer B) were truncated, following the Lange method.20 In the CS‐Rosetta derived model of the conformer A structure, Leu77‐Pro78 is in the trans conformation, whereas its chemical shift clearly indicates that it is in fact in the cis conformation (see Results section). This is likely due to the difficulty of CS‐Rosetta to identify the correct X‐Pro isomer.41 We therefore introduced the cis isomer for the Leu77‐Pro78 peptide in the structural model manually, using Coot.42 Geometry and structure statistics were checked with the Protein Structure Validation Suite server.43
Secretion assay
The assay for T3SS effector secretion has been described previously.8 Briefly, mutations in a plasmid encoding for the prgK gene were engineered by site‐directed mutagenesis, and transformed into a prgK‐deletion strain of Salmonella enterica serovar Typhimurium LT2. Transformants were grown in LB for 5 h, and cells were subsequently harvested. Proteins were precipitated from the supernatant with acetone and run on 12% SDS‐PAGE gels.
Supporting information
Supporting Information
Acknowledgments
We acknowledge instrument support provided by the Canada Foundation for Innovation, the British Columbia Knowledge Development Fund, the UBC Blusson Fund, and the Michael Smith Foundation for Health Research. J.R.C.B. was a MSFHR postdoctoral fellow. N.C.J.S. is a Canada Research Chair Tier 1 in Antibiotic Discovery. This work was supported by operating grants from the Canadian Institute of Health Research (to L.P.M. and N.C.J.S), and the HHMI International Scholar Program (to N.C.J.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Lawrence P. McIntosh, Email: mcintosh@chem.ubc.ca
Natalie C. J. Strynadka, Email: ncjs@mail.ubc.ca
References
- 1. Costa TR, Felisberto‐Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G (2015) Secretion systems in Gram‐negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. [DOI] [PubMed] [Google Scholar]
- 2. Buttner D (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant‐ and animal‐pathogenic bacteria. Microbiol Mol Biol Rev 76:262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burkinshaw BJ, Strynadka NC (2014) Assembly and structure of the T3SS. Biochim Biophys Acta 1843:1649–1663. [DOI] [PubMed] [Google Scholar]
- 4. Galan JE, Lara‐Tejero M, Marlovits TC, Wagner S (2014) Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Ann Rev Microbiol 68:415–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moest TP, Meresse S (2013) Salmonella T3SSs: successful mission of the secret(ion) agents. Curr Opin Microbiol 16:38–44. [DOI] [PubMed] [Google Scholar]
- 6. Haraga A, Ohlson MB, Miller SI (2008) Salmonellae interplay with host cells. Nat Rev Microbiol 6:53–66. [DOI] [PubMed] [Google Scholar]
- 7. Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara‐Tejero M, Sukhan A, Galán JE, Aizawa SI (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605. [DOI] [PubMed] [Google Scholar]
- 8. Kimbrough TG, Miller SI (2002) Assembly of the type III secretion needle complex of Salmonella typhimurium . Microbes Infect 4:75–82. [DOI] [PubMed] [Google Scholar]
- 9. Kimbrough TG, Miller SI (2000) Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci USA 97:11008–11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kubori T, Sukhan A, Aizawa SI, Galan JE (2000) Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA 97:10225–10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, Strynadka NCJ, Finlay BB (2017) Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol 15:323–337. [DOI] [PubMed] [Google Scholar]
- 12. Bergeron JR, Worrall LJ, De S, Sgourakis NG, Cheung AH, Lameignere E, Okon M, Wasney GA, Baker D, McIntosh LP, Strynadka NC (2015) The modular structure of the inner‐membrane ring component PrgK facilitates assembly of the type III secretion system basal body. Structure 23:161–172. [DOI] [PubMed] [Google Scholar]
- 13. Bergeron JR, Worrall LJ, Sgourakis NG, DiMaio F, Pfuetzner RA, Felise HB, Vuckovic M, Yu AC, Miller SI, Baker D, Strynadka NC (2013) A refined model of the prototypical Salmonella SPI‐1 T3SS basal body reveals the molecular basis for its assembly. PLoS Pathog 9:e1003307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spreter T, Yip CK, Sanowar S, Andre I, Kimbrough TG, Vuckovic M, Pfuetzner RA, Deng W, Yu AC, Finlay BB, Baker D, Miller SI, Strynadka NC (2009) A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat Struct Mol Biol 16:468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanowar S, Singh P, Pfuetzner RA, Andre I, Zheng H, Spreter T, Strynadka NC, Gonen T, Baker D, Goodlett DR, Miller SI (2010) Interactions of the transmembrane polymeric rings of the Salmonella enterica serovar Typhimurium type III secretion system. MBio 1:e00158‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schraidt O, Marlovits TC (2011) Three‐dimensional model of Salmonella's needle complex at subnanometer resolution. Science 331:1192–1195. [DOI] [PubMed] [Google Scholar]
- 17. Schraidt O, Lefebre MD, Brunner MJ, Schmied WH, Schmidt A, Radics J, Mechtler K, Galan JE, Marlovits TC (2010) Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog 6:e1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Worrall LJ, Hong C, Vuckovic M, Deng W, Bergeron JR, Majewski DD, Huang RK, Spreter T, Finlay BB, Yu Z, Strynadka NC (2016) Near‐atomic‐resolution cryo‐EM analysis of the Salmonella T3S injectisome basal body. Nature 540:597–601. [DOI] [PubMed] [Google Scholar]
- 19. Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA 105:4685–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D (2012) Determination of solution structures of proteins up to 40 kDa using CS‐Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci USA 109:10873–10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman‐Kay JD, Kay LE (1994) Backbone dynamics of a free and phosphopeptide‐complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33:5984–6003. [DOI] [PubMed] [Google Scholar]
- 22. Wang AC, Grzesiek S, Tschudin R, Lodi PJ, Bax A (1995) Sequential backbone assignment of isotopically enriched proteins in D2O by deuterium‐decoupled HA(CA)N and HA(CACO)N. J Biomol NMR 5:376–382. [DOI] [PubMed] [Google Scholar]
- 23. Ernst RR, Bodenhausen G, Wokaun A (1988) Principles of nuclear magnetic resonance in one and two dimensions. Berichte Bunsengesell physikal Chem 92:1563–1565. [Google Scholar]
- 24. Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G (1998) Side‐chain effects on peptidyl‐prolyl cis/trans isomerisation. J Mol Biol 279:449–460. [DOI] [PubMed] [Google Scholar]
- 25. Yamazaki T, Forman‐Kay JD, Kay LE (1993) Two‐dimensional NMR experiments for correlating carbon‐13.beta. and proton.delta./.epsilon. chemical shifts of aromatic residues in 13C‐labeled proteins via scalar couplings. J Am Chem Soc 115:11054–11055. [Google Scholar]
- 26. Schubert M, Labudde D, Oschkinat H, Schmieder P (2002) A software tool for the prediction of Xaa‐Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J Biomol NMR 24:149–154. [DOI] [PubMed] [Google Scholar]
- 27. Platzer G, Okon M, McIntosh LP (2014) pH‐dependent random coil 1H, 13C, and 15N chemical shifts of the ionizable amino acids: a guide for protein pK a measurements. J Biomol NMR 60:109–129. [DOI] [PubMed] [Google Scholar]
- 28. Ludwiczek ML, D'Angelo I, Yalloway GN, Brockerman JA, Okon M, Nielsen JE, Strynadka NCJ, Withers SG, McIntosh LP (2013) Strategies for modulating the pH‐dependent activity of a family 11 glycoside hydrolase. Biochemistry 52:3138–3156. [DOI] [PubMed] [Google Scholar]
- 29. Sudmeier JL, Bradshaw EM, Haddad KE, Day RM, Thalhauser CJ, Bullock PA, Bachovchin WW (2003) Identification of histidine tautomers in proteins by 2D 1H/13C(delta2) one‐bond correlated NMR. J Am Chem Soc 125:8430–8431. [DOI] [PubMed] [Google Scholar]
- 30. SPARKY 3 (University of California, San Francisco: ). [Google Scholar]
- 31. Li H, Robertson AD, Jensen JH (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins 61:704–721. [DOI] [PubMed] [Google Scholar]
- 32. Loewenthal R, Sancho J, Fersht AR (1992) Histidine‐aromatic interactions in barnase. Elevation of histidine pKa and contribution to protein stability. J Mol Biol 224:759–770. [DOI] [PubMed] [Google Scholar]
- 33. Waugh DS (1996) Genetic tools for selective labeling of proteins with alpha‐15N‐amino acids. J Biomol NMR 8:184–192. [DOI] [PubMed] [Google Scholar]
- 34. Muchmore DC, McIntosh LP, Russell CB, Anderson DE, Dahlquist FW (1989) Expression and nitrogen‐15 labeling of proteins for proton and nitrogen‐15 nuclear magnetic resonance. Methods Enzymol 177:44–73. [DOI] [PubMed] [Google Scholar]
- 35. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 36. Kay LE, Xu GY, Singer AU, Muhandiram DR, Forman‐Kay JD (1993) A gradient‐enhanced HCCH‐TOCSY experiment for recording side‐chain 1H and 13C correlations in H2O samples of proteins. J Magnet Reson B 101:333–337. [Google Scholar]
- 37. Logan TM, Olejniczak ET, Xu RX, Fesik SW (1993) A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH‐TOCSY triple resonance experiments. J Biomol NMR 3:225–231. [DOI] [PubMed] [Google Scholar]
- 38. Kanelis V, Donaldson L, Muhandiram DR, Rotin D, Forman‐Kay JD, Kay LE (2000) Sequential assignment of proline‐rich regions in proteins: application to modular binding domain complexes. J Biomol NMR 16:253–259. [DOI] [PubMed] [Google Scholar]
- 39. Farrow NA, Zhang O, Forman‐Kay JD, Kay LE (1994) A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR 4:727–734. [DOI] [PubMed] [Google Scholar]
- 40. McIntosh LP, Naito D, Baturin SJ, Okon M, Joshi MD, Nielsen JE (2011) Dissecting electrostatic interactions in Bacillus circulans xylanase through NMR‐monitored pH titrations. J Biomol NMR 51:5–19. [DOI] [PubMed] [Google Scholar]
- 41. Shen Y, Bax A (2010) Prediction of Xaa‐Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR 46:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Cryst D 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhattacharya A, Tejero R, Montelione GT (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66:778–795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


