Abstract
OmpR/EnvZ is a two-component system that senses osmotic signals and controls downstream gene expression in many species of Enterobacteriaceae. However, the role of OmpR/EnvZ in Klebsiella pneumoniae remains unknown. In this study, we found that production of MrkA, the major subunit of type 3 fimbriae, was decreased under hypertonic conditions. A deletion mutant of ompR and a site-directed mutant with a single amino acid substitution of aspartate 55 to alanine (D55A), which mimics the unphosphorylated form of OmpR, markedly reduced MrkA production under hypertonic conditions. These results indicate that K. pneumoniae type 3 fimbriae expression is activated by the phosphorylated form of OmpR (OmpR∼P). Although no typical OmpR∼P binding site was found in the PmrkA sequence, mrkA mRNA levels and PmrkA activity were decreased in the ΔompR and ompRD55A strains compared with the wild type (WT) strain, indicating that OmpR∼P mediates type 3 fimbriae expression at the transcriptional level. Previous reports have demonstrated that a cyclic-di-GMP (c-di-GMP) related gene cluster, mrkHIJ, regulates the expression of type 3 fimbriae. We found that both the ompR and ompRD55A mutants exhibited decreased mrkHIJ mRNA levels, intracellular c-di-GMP concentration, and bacterial biofilm amount, but increased total intracellular phosphodiesterase activity in response to hypertonic conditions. These results indicate that OmpR∼P regulates type 3 fimbriae expression to influence K. pneumoniae biofilm formation via MrkHIJ and modulation of intracellular c-di-GMP levels. Taken together, we herein provide evidence that OmpR∼P acts as a critical factor in the regulation of the c-di-GMP signaling pathway, type 3 fimbriae expression, and biofilm amount in K. pneumoniae in response to osmotic stresses.
Keywords: Klebsiella pneumoniae, OmpR, type 3 fimbriae, MrkHIJ, c-di-GMP signaling, biofilm
Introduction
Klebsiella pneumoniae is a Gram-negative pathogen, which causes purulent abscess, bacteremia, and urinary as well as respiratory tract infections mostly in patients with underlying diseases (Podschun and Ullmann, 1998). Similar to many enteric bacteria, K. pneumoniae needs to sense various environmental signals, including temperature, nutrition-limitation, pH, oxygen availability, osmolality, as well as other stimuli, in order to achieve successful infection (Carpenter and Payne, 2014; Lustri et al., 2017). Bacterial two-component systems (TCSs), typically consisting of a sensor histidine kinase and a response regulator, play a crucial role in sensing environmental signals and subsequent regulation of gene expression at the transcriptional level (Stock et al., 2000; Laub and Goulian, 2007). In Gram-negative bacteria, the OmpR/EnvZ TCS can sense an osmotic signal to further control downstream gene expression including genes encoding outer membrane proteins and various virulence factors (Mizuno and Mizushima, 1990; Oshima et al., 2002). EnvZ, the sensor kinase, auto-phosphorylates a conserved histidine residue in response to extracellular osmolality and then transfers the phosphate group to aspartate residue 55 on the receiver domain of OmpR to exert DNA binding activity (Taylor et al., 1981; Igo and Silhavy, 1988; Delgado et al., 1993). In uropathogenic Escherichia coli, OmpR promotes bacterial survival in the murine urinary tract and growth in human urine (Schwan, 2009). In addition, deletion of ompR can increase type 1 fimbriae expression via activation of fimB transcription and phase-ON positioning of the fimS element under a high osmolality environment (Rentschler et al., 2013). In E. coli, OmpR increases biofilm amount by negatively regulating the expression of a flagellar regulator (Samanta et al., 2013). Likewise, Yersinia enterocolitica OmpR promotes biofilm formation, but positively regulates flagellar synthesis (Raczkowska et al., 2011). Furthermore, the important role of OmpR in the control of bacterial virulence has been demonstrated in many bacteria including Salmonella Typhimurium, Shigella flexneri, Y. pestis, and Acinetobacter baumannii (Dorman et al., 1989; Bernardini et al., 1990; Reboul et al., 2014; Tipton and Rather, 2016). However, the role of OmpR in K. pneumoniae pathogenesis remains unknown.
Multiple virulence factors have been shown to be involved in K. pneumoniae infection including capsular polysaccharide (), lipopolysaccharides (LPS), type 1 and type 3 fimbriae, iron-acquisition systems, porins, and antibiotic resistance determinants, e.g., carbapenemases, extended-spectrum β-lactamases, efflux systems, DNA gyrase, and topoisomerase IV (Paczosa and Mecsas, 2016; Lee et al., 2017). However, most of these virulence factors are processed or embedded in the cell envelope and enable bacteria to take up nutrients and adhere to diverse surfaces or niches within the human host (Wu and Fives-Taylor, 2001; Kawai et al., 2011). In addition, biofilm formation is considered a key factor in the development of nosocomial infections and increases bacterial resistance to antibiotics, thus hindering medical treatment (Murphy and Clegg, 2012). CPS, LPS, and fimbriae are involved in biofilm formation in K. pneumoniae (Schembri et al., 2005; Balestrino et al., 2008). Interestingly, the thick capsule of K. pneumoniae inhibits fimbriae activity and assembly (Schembri et al., 2005) and cross-regulated expression between fimbriae and CPS for efficient infection has been suggested. Of these factors, type 3 fimbriae are considered the major determinant of biofilm formation in K. pneumoniae (Di Martino et al., 2003; Jagnow and Clegg, 2003; Wu et al., 2012). Although most K. pneumoniae strains possess type 1 and 3 fimbriae, type 3 fimbriae are mainly expressed in heavily encapsulated K. pneumoniae strains, while the production of type 1 fimbriae is poor and phase-variable (Schembri et al., 2005; Struve et al., 2008). Therefore, elucidating the regulation of type 3 fimbriae is required to gain a deeper understanding of K. pneumoniae pathogenesis.
In K. pneumoniae, type 3 fimbriae are encoded by the mrkABCDF operon (Di Martino et al., 2003; Jagnow and Clegg, 2003; Wu et al., 2012). A cyclic di-GMP (c-di-GMP) related gene cluster, mrkHIJ, located downstream of the type 3 fimbrial genes, has been demonstrated to play a central role in regulating type 3 fimbriae expression (Wilksch et al., 2011; Murphy and Clegg, 2012; Wu et al., 2012; Yang et al., 2013). Reverse-transcription PCR analysis revealed that while mrkHIJ is transcribed as a polycistronic mRNA, mrkJ is also independently transcribed (Wu et al., 2012). MrkH possesses that a PilZ-domain that binds c-di-GMP to activate its own promoter activity and type 3 fimbriae expression (Wilksch et al., 2011; Yang et al., 2013; Tan et al., 2015). MrkI is a predicated LuxR-type transcriptional regulator to activate its own operon and type 3 fimbriae expression (Wu et al., 2012). MrkJ contains an EAL domain with functional c-di-GMP phosphodiesterase (PDE) activity for hydrolysis of c-di-GMP and further repression of type 3 fimbriae expression (Johnson and Clegg, 2010). Although MrkHIJ play a critical role in regulating type 3 fimbriae expression, the regulation of mrkHIJ remains unclear. c-di-GMP is a bacterial secondary messenger that modulates biofilm formation and the expression of various virulence factors (Tamayo et al., 2007; Jenal et al., 2017). The intracellular concentration of c-di-GMP in bacteria is modulated by di-guanylate cyclases (DGCs) and PDEs (Simm et al., 2004; Hengge, 2009). K. pneumoniae YfiN harbors DGC domain and has a positive role in the control of type 3 fimbriae expression (Wilksch et al., 2011), while YjcC possess a PDE domain and plays a negative role in type 3 fimbriae expression (Huang et al., 2013). In addition to c-di-GMP-related proteins, several transcriptional regulators have been reported to be involved in the control of type 3 fimbriae expression in K. pneumoniae including histone-like nucleoid-structuring protein (H-NS), CRP, and ferric uptake regulator (Fur) (Wu et al., 2012; Ares et al., 2016; Lin et al., 2016). Thus, the regulation of type 3 fimbriae in K. pneumoniae in response to different environmental stimuli is complicated.
In this study, we found that type 3 fimbriae expression in K. pneumoniae was decreased under hypertonic conditions. In addition, phosphorylated OmpR (OmpR∼P) activated type 3 fimbriae expression through the c-di-GMP signaling pathway and further affected biofilm amount in response to osmolality stresses.
Materials and Methods
All chemicals and reagents were purchased from Sigma (St. Louis, MO, United States) unless otherwise stated. All enzymes were purchased from New England Biolabs unless otherwise stated. General molecular techniques, e.g., PCR and eletroporation, were performed according to standard protocols (Sambrook and Russell, 2000).
Bacterial Strains, Plasmids, and Culture Conditions
All bacterial strains and plasmids used in this study are listed in Table 1. Primers used in this study are listed in Table 2. Each strain was grown overnight in Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, and 1% [∼170 mM] sodium chloride [NaCl]) (BD DifcoTM) with respective antibiotics at 37°C for 16 h, and then 1/200 of overnight culture were inoculated into LB broth at concentrations of 0, 50, 200, or 400 mM of NaCl until the bacteria were grown after that to reach exponential phase (OD600nm = 0.6–0.8). However, instead of 1% NaCl, we used LB medium supplemented with 2.32% NaCl (= 400 mM) to analyze the hypertonic effects in K. pneumoniae. The antibiotics used include ampicillin (100 μg/ml), kanamycin (25 μg/ml), streptomycin (500 μg/ml), and tetracycline (12.5 μg/ml).
Table 1.
Bacterial strains and plasmids used in this study.
| Strains or plasmids | Descriptions | Reference or source |
|---|---|---|
| K. pneumoniae | ||
| CG43S3 | CG43 Smr | Lai et al., 2001 |
| ΔompR | CG43S3ΔompR | This study |
| ompRD55A | CG43S3ompRD55A | This study |
| ΔlacZ | CG43S3ΔlacZ | Lin et al., 2006 |
| ΔlacZ-ΔompR | CG43S3ΔlacZΔompR | This study |
| ΔlacZ- ompRD55A | CG43S3ΔlacZompRD55A | This study |
| E. coli | ||
| BL21(DE3) | F- ompT hsdSB[rB-mB-]gal dcm [DE3] | New England Biolabs |
| S17-1 λ pir | hsdR recA pro RP4-2 [Tc::Mu; Km::Tn7] [λpir] | Miller and Mekalanos, 1988 |
| Plasmids | ||
| yT&A | ApR, TA PCR cloning vector | Yeastern |
| pKAS46 | Apr, Kmr, Positive selection suicide vector, rpsL | Skorupski and Taylor, 1996 |
| pACYC184 | Cmr, Tcr, plasmid with p15A origin of replication | Chang and Cohen, 1978 |
| pmrkAZ15 | Cmr, 402 bp fragment containing the region upstream of mrkA cloned into placZ15 | Lin et al., 2016 |
| pompR | Cmr, 1079 bp fragment containing an ompR allele cloned into pACYC184 | This study |
| pET30b-OmpR | Kmr, 716 bp fragment encoding full-length ompR allele cloned into pET30b | This study |
Table 2.
Primers used in this study.
| Primer | Sequence (5′ → 3′) | Enzyme cleaved | |
|---|---|---|---|
| GT254 | GGGATCCAGACCGTCTGGGGTCT | BamHI | |
| GT255 | CTCTAGACAGCGGGTGCATACG | XbaI | |
| GT256 | CGGATCCATCTGTTCGGCGTTC | BamHI | |
| GT257 | CGAGCTCTACCTGTTCGGCTCTGGC | SacI | |
| GT284 | GGGATCCGCTCTCCATCAATGCTAA | BamHI | |
| GT285 | GAGATCTGAAAGACGTAGCGTACAGC | BglII | |
| GT288 | CGGATCCAGACAAAATGGAGGGAACCCTA | BamHI | |
| GT290 | GCAATAGCAACATTCTGATTGG | ||
| GT336 | CCATATGCAAGAGAATTATAAGATTCT | NdeI | |
| GT337 | GAAGCTTGCCTTAGAACCGTCCGGGAC | HindIII | |
| GT356 | ATGCTGCCGGGCGAAGATGGT | ||
| GT357 | TTCTAGATACGAATAAACAGCCA | XbaI | |
| GT363 | CAGAGCCAGCACCATCAGGTGGA | ||
| GT367 | GACATCAATCTGAAAGGCCAG | ||
| GT368 | AGGGATCACCACTGCCAGAA | ||
| CC323 | GGATCCTGCATGCTGTTGCGGTCAC | BamHI | |
| CC324 | GGATCCGCGGTTGCCATTGCTGCAGAG | BamHI | |
| SY01 | GGGATCCGGTAGTCCATCTCCTGCTTCA | BamHI | |
| SY02 | GAAGCTTGACGAACAGCAAGGTGACG | HindIII | |
| For qRT-PCR | Sequence (5′ → 3′) | TaqMan probes | Target |
| RT11 | GGTAGGGGAGCGTTCTGTAA | 67 | 23S rRNA |
| RT12 | TCAGCATTCGCACTTCTGAT | ||
| RT29 | TAAGCAAACTGGGCGTGAA | 20 | mrkA |
| RT30 | TAGCCCTGTTGTTTGCTGGT | ||
| GT46 | GTTTAAGTTCCGCCATCTCG | 120 | mrkH |
| GT47 | TTGCGCTTGGCTTCTAAGAT | ||
| GT42 | AGTTATGCCGATGTCATCCAT | 59 | mrkI |
| GT43 | GATTCTGATGGCAGAAATATCCTT | ||
| GT54 | TTTCGAGGTAACCGAAAACG | 84 | mrkJ |
| GT55 | GAGGTATCCTGTGGGCTCTG | ||
| RT159 | CTGGCTGGATGATTTTGGTC | 67 | D364_06025 |
| RT160 | CCACTTTGACGCAATCGAA | ||
| RT161 | CGTAGGCTGGCTGATGGA | 40 | D364_08130 |
| RT162 | TCTGTCTCGGTGACCTCGAT | ||
| RT164 | GGGTTTGCTGATATCGATGG | 67 | D364_13295 |
| RT165 | AGCCAACGTTTTGACATGCT | ||
| RT169 | ATCGATTTTCGCCATCAGAG | 70 | fimK |
| RT170 | GAAAAGCAGTCGTCCAGCAT | ||
| RT175 | AATGGTCATCCGGGAAGC | 20 | yjcC |
| RT176 | CCGCTGAACACAACTCACC | ||
| RT244 | GCATCGTAACCTGGTCTTCAA | 36 | D364_02175 |
| RT245 | GAGCAGGCTGGTGAAAATG | ||
| RT273 | CCAGGATCCTCGCCACTAT | 22 | D364_04060 |
| RT274 | TCCCGACCTTACCAAAACG | ||
| RT275 | CGCCGGTACTGATCCAAT | 28 | D364_11845 |
| RT276 | ATTCACCACCAGCATCACG | ||
| RT277 | GCTACCTGGTGCGCAATATAA | 68 | D364_16830 |
| RT278 | TCGCCATAAATACAGCGTAGC | ||
Construction of the ompR Deletion Mutant and Its Complementation
A specific ompR gene deletion was introduced into K. pneumoniae CG43S3 using an allelic exchange strategy as previously described (Lai et al., 2003). Briefly, the upstream and downstream regions (approximately 1000 bp DNA fragments) of ompR were cloned into suicide vector pKAS46 containing rpsL, which allows positive selection for the loss of vector with streptomycin (Skorupski and Taylor, 1996). The resulting plasmid was then mobilized from E. coli S17-1λpir (Miller and Mekalanos, 1988) to K. pneumoniae CG43S3 or CG43S3-derived strains by conjugation. Overnight cultures of donor and recipient strains were mixed in a ratio of 2:1, and the mixture was washed in saline (0.9% NaCl). An aliquot of 30 μl of the mixture was spotted on an LB plate incubated at 37°C for 24 h and plated on M9 agar plates (6.78 g/L Na2HPO4, 3 g/L KH2PO4, 1 g/L NH4Cl, 0.5 g/L NaCl, 20% glucose, 1.5% agar) with ampicillin (100 μg/ml) and kanamycin (25 μg/ml) for selection of transconjugants. Several of the ampicillin and kanamycin-resistant transconjugants were picked, grown in LB broth supplemented with 500 μg/ml streptomycin to exponential phase at 37°C, and then spread on LB agar plate containing 500 μg/ml streptomycin. Following the occurrence of a double crossover, streptomycin-resistant and kanamycin-sensitive colonies were selected and the deletion was verified by PCR and Southern hybridization (data not shown).
To obtain the complementation plasmid (pOmpR), a DNA fragment containing the promoter and coding sequence of ompR was PCR amplified using primer pair SY01/SY02 (Table 2) and cloned into the pACYC184 shuttle vector (Chang and Cohen, 1978). Next, pompR and pACYC184 were transformed into ΔompR strain by electroporation.
Construction of a K. pneumoniae ompRD55A Mutant
A DNA fragment carrying ompR and approximately 1000 bp adjacent regions on either side was amplified by PCR using primer pairs GT257/GT357 (Table 2) and cloned into yT&A vector (Yeastern). The resulting plasmid was used as the template for the inverse-PCR (Ochman et al., 1988) with the primer pair GT356/GT363 (Table 2) to generate a mutant ompR allele encoding the D55A mutation, which was confirmed by Sanger sequencing (Genomics, Taiwan). Subsequently, the mutant allele of ompR was subcloned into pKAS46 (Skorupski and Taylor, 1996) and confirmed by DNA sequencing. Then, the plasmid was mobilized from E. coli S17-1 λpir to the K. pneumoniae ΔompR strain by conjugation, and the subsequent selection was performed as described above.
Western Blotting
The total proteins of exponential phase K. pneumoniae cultures were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (approximately 5 μg per lane) and transferred to polyvinylidene difluoride membrane. Western analysis was followed as previously described (Lin et al., 2016). Rabbit anti-MrkA antibody (customer antibody service from LTK BioLaboratories) and Goat anti-rabbit immunoglobulin G antibody conjugated to horseradish peroxidase (Abcam) were used as the primary antibody and the secondary antibody, respectively. After incubation with the secondary antibody, the signal in the membranes was collected by ImageQuant LAS 4000 mini (GE Health, United States) after the visualization with an enhanced chemiluminescence ECL western blotting luminal reagent (PerkinElmer, Wellesley, MA, United States).
Quantitative Reverse-Transcription PCR (qRT-PCR)
As previous study, total RNA extraction, reverse transcription of isolated mRNA to cDNA, qRT-PCR, and data analysis were performed (Lin et al., 2013). Primers and probes for selected target sequences were designed by using Universal ProbeLibrary Assay Design Center (Roche-applied science) and shown in Table 2. Relative gene expressions were quantified using the comparative threshold cycle 2-ΔΔCT method with 23S rRNA as the endogenous reference.
Measurement of Promoter Activity
To measure the mrkA promoter activity, K. pneumoniae strains was transformed with the promoter-reporter plasmid pmrkAZ15 into by electroporation, respectively. As previously described (Lin et al., 2006), the β-galactosidase activity of logarithmic phase bacteria was measured.
Construction and Purification of OmpR::His6
The coding region of ompR was PCR amplified with primer sets GT336/GT337 (Table 2) and cloned into the NdeI/HindIII site in pET30b (Novagen, 205 Madison, Wis). The resulting plasmid pET30b-OmpR was then transformed into E. coli BL21(DE3) (New England Biolabs), and overproduction of the recombinant protein was induced by the addition of 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 h at 37°C. The recombinant proteins were then purified from the soluble fraction of the total cell lysate by affinity chromatography using His-Bind resin (Novagen, Madison, Wis). Finally, the purified proteins were dialyzed against GMS buffer (50 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, and 10% glycerol) at 4°C overnight, and the purity was determined by SDS-PAGE.
Electrophoretic Mobility Shift Assay (EMSA)
DNA fragments of the putative promoter region of mrkA, mrkHI, mrkJ, and ompF were individually PCR-amplified with primer pairs CC323/CC324, GT288/GT290, GT284/GT285, and GT367/GT368 sets by Pfu-polymerase to generate DNA probes for EMSA. Briefly, the purified OmpR::His6 was incubated with 10 ng DNA in a 10 μl solution containing 10 mM Tris-HCl, pH 7.4, 50 mM KCl, 1 mM DTT, 10 mM MgCl2, 10% glycerol, and 25 mM acetyl-phosphate at room temperature for 20 min. The samples were then loaded onto a native gel of 5% non-denaturing polyacrylamide in 0.5X TB buffer (45 mM Tris-HCl, pH 8.0, 45 mM boric acid). Gels were electrophoresed with a 20-mA current at 4°C and then stained with SYBR Green I dye (Invitrogen). The assay was repeated in at least 3 independent experiments.
Intracellular Concentration of c-di-GMP
The intracellular c-di-GMP in late exponential phase K. pneumoniae cultures (OD600nm = 1.0–1.2) was extracted according to the previous study (Morgan et al., 2006). The dried extracts were solubilized in distill water and further were measured the c-di-GMP level by a ELISA kit (Wuhan EIAab Science Co., Ltd). The c-di-GMP concentration was normalized by total protein concentration. Relative percentage of c-di-GMP content was calculated by the c-di-GMP concentration of extracts is relative to that of wild type (WT) strain.
PDE Activity
The PDE activity of the crude extracts in exponential phase K. pneumoniae strains was performed as previous study by using bis(p-nitrophenyl) phosphate (bis-pNPP) (Johnson and Clegg, 2010). Briefly, 10 μg of total protein in assay buffer (50 mM Tris-HCl, 1 mM MnCl2 [pH 8.5]) supplemented with 5 mM bis-pNPP at 37°C for 5 min. The PDE activity was determined by measuring the release of p-nitrophenol at OD410nm. Relative percentage of PDE activity was calculated by the OD410nm of crude extracts is relative to that of WT strain.
Quantification of Biofilm Amount
Biofilm amount was assessed by the ability of the cells to adhere to the walls of 96-well microtitre dishes made of PVC (TPP 96 flat) with some modification of the reported protocol (Lembke et al., 2006). The plate contained an aliquot of 1:10 diluted overnight bacteria culture and then was incubated at 37°C statically for 24 h for biofilm formation. The biofilms were washed triply with 200 μl PBS to remove non-adherent bacteria and then adherent bacteria was stained with 200 μl of 0.1% safranin solution at room temperature for 30 min. The plates was rinsed twice with deionizer water to remove excess stain. Finally, the safranin stained biofilm was solubilized in 200 μl of 95% ethanol and the absorbance determined at OD492nm.
Statistical Method
The results of qRT-PCR analysis, promoter activity, c-di-GMP concentration, PDE activity, and biofilm amount were performed by biological replicates at least triplicate. The results are showed as the mean and standard deviation. Differences between groups were evaluated by an unpaired t-test. Values of P < 0.05 and P < 0.01 were considered statistically significant differences.
Results
The Role of OmpR in Regulating MrkA Production Under Hypertonic Conditions
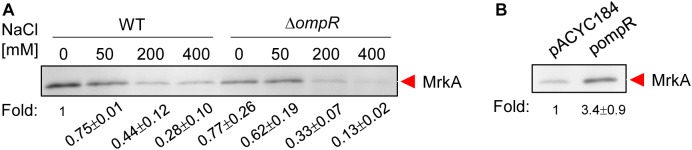
To investigate whether the expression of type 3 fimbriae in K. pneumoniae is affected by hypertonic conditions, K. pneumoniae CG43S3 was grown in LB broth with increasing concentrations of NaCl and the MrkA production, the major subunit of type 3 fimbriae, was determined by western blotting analysis. As shown in Figure 1A, MrkA production decreased when K. pneumoniae was grown in LB broth with 50, 200, and 400 mM NaCl, compared to LB broth without NaCl. This indicates that MrkA production was repressed in response to increasing hypertonic stress. To determine whether OmpR is involved in this regulation, a deletion mutant of ompR was generated in K. pneumoniae CG43S3. MrkA production similarity decreased in the ΔompR strain in response to NaCl. However, MrkA production was lower in the ΔompR strain than in the WT strain in LB broth without NaCl; in LB broth with 400 mM NaCl, MrkA production was markedly reduced in the ΔompR strain compared to the WT strain. To confirm the effect of ompR deletion on MrkA production, the complementation plasmid, pompR, and the empty vector, pACYC184, were introduced into the ΔompR strain and MrkA production was evaluated in LB broth with 400 mM NaCl. As shown in Figure 1B, MrkA production was higher in the ΔompR[pompR] strain than in the ΔompR[pACYC184] strain; this confirms that OmpR can increase MrkA production in K. pneumoniae CG43S3 in response to hypertonic stresses.
FIGURE 1.
Hypertonic stress and OmpR affect MrkA production ount in K. pneumoniae CG43S3. (A) K. pneumoniae CG43S3 WT and ΔompR strains were grown overnight at 37°C with agitation in LB broth with or without NaCl as indicated to observe the MrkA production by western blot analysis against MrkA. Likewise, the strains, ΔompR (pACYC184) and ΔompR (pOmpR), were grown to mid-log phase at 37°C in LB broth with 400 mM NaCl to observe the MrkA production (B). The fold change of MrkA production calculated by Image J software is also shown.
OmpR Increases mrkA Expression at the Transcriptional Level
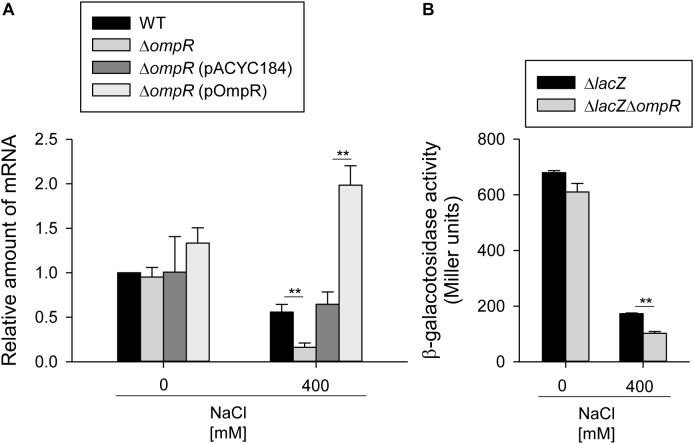
To analyze the regulatory role of OmpR in type 3 fimbriae expression in response to hypertonic stimulus, the effect of ompR deletion on mrkA mRNA levels was measured in the WT and ΔompR strains grown in LB broth with or without 400 mM NaCl using qRT-PCR. As shown in Figure 2A, mrkA mRNA levels were lower in the ΔtextitompR strain than in the WT strain in LB broth with 400 mM NaCl, while no apparent effect was observed in LB without NaCl. In addition, introduction of the complementation plasmid pompR into ΔompR reversed the effect of the deletion in response to 400 mM NaCl. To further investigate whether OmpR affects the promoter activity of mrkA, a plasmid carrying PmrkA fused to a lacZ reporter gene was introduced into the ΔlacZ and ΔlacZΔompR strains. As shown in Figure 2B, PmrkA activity was significantly lower in ΔlacZΔompR than in ΔlacZ, suggesting that OmpR increases type 3 fimbriae expression at the transcriptional level in response to hypertonic stresses.
FIGURE 2.
OmpR activates mrkA expression. (A) qRT-PCR analyses of the mrkA expression for WT, ΔompR, ΔompR (pACYC184), and ΔompR (pOmpR) strains in LB broth with or without 400 mM NaCl. (B) β-galactosidase activities of K. pneumoniae CG43S3ΔlacZ and the isogenic strain (ΔlacZΔompR) carrying the reporter plasmid pmrkAZ15 (PmrkA::lacZ) were determined using log-phase cultures grown in LB broth with or without 400 mM NaCl. The results are representative of three independent experiments. Error bars indicate standard deviations. ∗∗P < 0.01 compared to the indicated group.
Phosphorylated OmpR Is Required for Increasing MrkA Expression
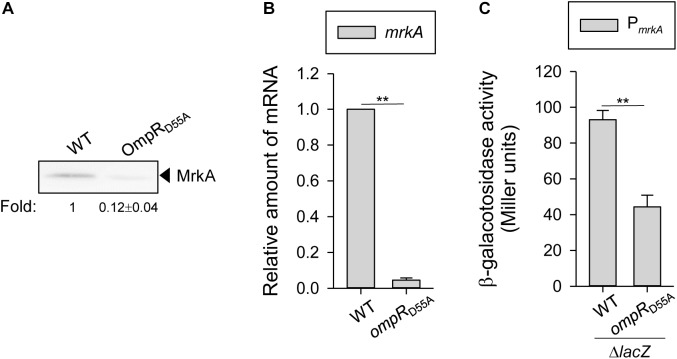
To test whether the effect of OmpR on mrkA transcription and protein production might require phosphorylation of OmpR at aspartate 55, a site-directed mutant with a single amino acid substitution of aspartate 55 to alanine (D55A), which prevents phosphorylation, was generated. As shown in Figure 3A, MrkA production was decreased in the ompRD55A mutant than in the WT strain when grown in LB broth with 400 mM NaCl. Furthermore, mrkA mRNA levels were lower in the ompRD55A mutant compared to the WT strain when grown in LB broth with 400 mM NaCl (Figure 3B). Similarly, lower mrkA promoter activity was observed in the OmpRD55A mutant (Figure 3C). These results suggest that the phosphorylated form of OmpR is required for increasing type 3 fimbriae expression.
FIGURE 3.
Effect of phosphorylated OmpR on regulating MrkA expression in K. pneumoniae. K. pneumoniae CG43S3 WT and ompRD55A strains was grown to mid-log phase at 37°C in LB broth with 400 mM NaCl to observe the type 3 fimbriae expression by (A) western blot analysis against MrkA, and (B) qRT-PCR analyses of mrkA gene expression. (C) β-galactosidase activities of K. pneumoniae CG43S3 ΔlacZ and the isogenic strains (ΔlacZ_OmpRD55A) carrying the reporter plasmid pmrkAZ15 (PmrkA::lacZ) were determined using log-phase cultures grown in LB broth with 400 mM NaCl. The results are representative of three independent experiments. Error bars indicate standard deviations. ∗∗P < 0.01 compared to the indicated group. The fold change of MrkA production calculated by Image J software is also shown.
OmpR-Mediated Regulation of Type 3 Fimbriae Expression Is Probably Indirect
To investigate the mechanism underlying OmpR regulation of mrkA transcription, the consensus sequence of the E. coli OmpR∼P binding site was used as a reference for analyzing the mrkA promoter sequence (Yoshida et al., 2006). However, no typical OmpR binding site was found upstream of mrkA. To further confirm the absence of OmpR∼P binding activity in PmrkA, the interaction between a recombinant OmpR::His6 protein phosphorylated by acetyl-phosphate and DNA fragments containing PmrkA and PompF (positive control) were examined by EMSA. As shown in Figure 4, a DNA-protein-binding complex was observed following incubation of 1.3 μM phosphorylated OmpR::His6 and 10 ng PompF. However, no binding activity was observed for the phosphorylated OmpR::His6 protein with PmrkA (Figure 4). These results indicate that OmpR activation of type 3 fimbriae expression may be indirect.
FIGURE 4.
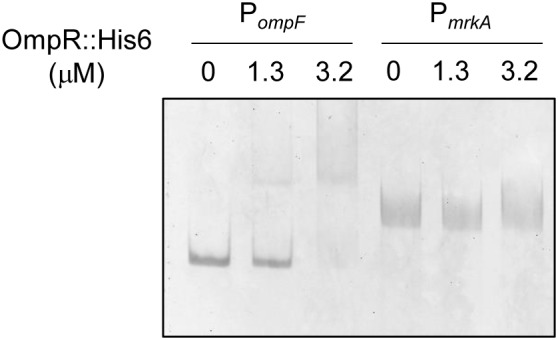
Binding activity of phosphorylated OmpR::His6 with PompF and PmrkA. Different concentrations of purified OmpR::His6 were incubated with 25 mM acetyl phosphate and 10 ng of the upstream regions of ompF and mrkA. Following incubation at room temperature for 20 min, the mixtures were analyzed on a 5% non-denaturing polyacrylamide gel. The gel was stained with SYBR Green I dye and photographed.
Regulatory Effect of OmpR on mrkHIJ Expression
Previous studies have demonstrated that MrkHIJ play an important role in regulating type 3 fimbriae expression (Murphy and Clegg, 2012; Wu et al., 2012). To further examine whether MrkHIJ are involved in OmpR regulon, the mRNA levels of these genes were measured in the WT and ΔompR strains grown in LB broth with or without 400 mM NaCl by qRT-PCR. In contrast to the lack of an apparent effect in the WT and ΔompR strains in LB without NaCl, mrkH, mrkI, and mrkJ mRNA levels were markedly decreased in the ΔompR strain in LB broth with 400 mM NaCl (Figure 5). In addition, a similar reduction was also observed in the ompRD55A mutant. This indicates that the phosphorylated form of OmpR is required for increasing mrkH, mrkI, and mrkJ mRNA expression. However, no typical OmpR∼P binding site was found upstream of mrkHI and mrkJ and no binding activity was observed for the phosphorylated OmpR::His6 protein with PmrkHI and PmrkJ (data not shown), suggesting that OmpR∼P positive regulation of mrkH, mrkI, and mrkJ expression is indirect.
FIGURE 5.
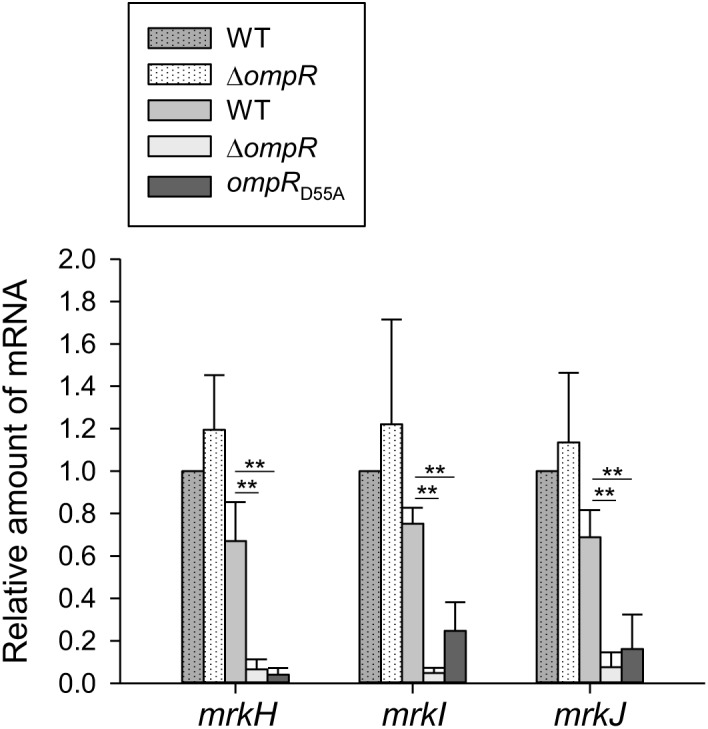
Effect of OmpR on regulating mrkH, mrkI, and mrkJ expression. qRT-PCR analyses of the mrkH, mrkI, and mrkJ expressions for WT, ΔompR, and ompRD55A strains in LB broth with (solid bars) or without 400 mM NaCl (dot bars). The results are representative of three independent experiments. Error bars indicate standard deviations. ∗P < 0.05 and ∗∗P < 0.01 compared to the indicated group.
Effect of OmpR on Intracellular c-di-GMP Concentration, PDE Activity, and PDE-Related Genes Expression
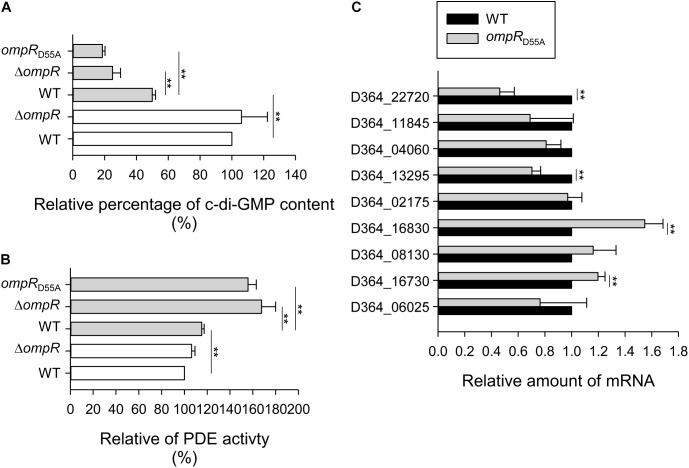
Based on the previous study, the expression of type 3 fimbriae and MrkHI is c-di-GMP dependent (Tan et al., 2015). To further investigate whether OmpR affects the c-di-GMP production, thus increasing type 3 fimbriae and MrkHI expression, the intracellular concentration of c-di-GMP was measured in the WT and ΔompR strains grown in LB broth with or without 400 mM NaCl. As shown in Figure 6A, the c-di-GMP concentration of K. pneumoniae decreased in LB with 400 mM NaCl. Furthermore, the c-di-GMP concentration was lower in the ΔompR strain than in the WT strain in LB broth with 400 mM NaCl, while this effect was not observed in LB without NaCl. An apparent lower intracellular c-di-GMP concentration was also found in the ompRD55A strain relative to the WT strain when grown in LB with 400 mM NaCl. These results indicate that OmpR∼P is required for increasing intracellular c-di-GMP production in K. pneumoniae under hypertonic conditions. In bacteria, intracellular c-di-GMP is degraded by PDEs (Simm et al., 2004; Hengge, 2009). Therefore, we evaluated the in vitro PDE activity of crude extracts from the WT and ΔompR strains grown in LB with or without 400 mM NaCl using a colorimetric assay. As shown in Figure 6B, the WT strain demonstrated a slightly higher PDE activity in LB with 400 mM NaCl. Furthermore, the PDE activity in the ΔompR extract was higher than in the WT extract in LB with 400 mM NaCl, but not in LB without NaCl. In addition, the ompRD55A mutant exhibited higher PDE activity in response to 400 mM NaCl compared to the WT strain. Therefore, we hypothesized that OmpR∼P can decrease PDE activity to further increase intracellular c-di-GMP concentrations, resulting in elevated type 3 fimbriae and mrkHI expression. Analysis of the upstream sequences of the c-di-GMP PDE related open reading frames (ORFs) in K. pneumoniae CG43, including mrkJ (D364_16630), D364_06025, fimK (D364_16730), D364_04060, D364_08130, D364_16830, ylaB (D364_02175), yoaD (D364_11845), rtn (D364_13295), and yjcC (D364_22720), did not reveal any typical OmpR∼P binding sites (data not shown). To further investigate the effect of OmpR∼P on the expression of these c-di-GMP PDE related ORFs, the mRNA expression of these ORFs was measured in the WT and ompRD55A strains in response to 400 mM NaCl. As shown in Figure 6C, the mRNA levels of fimK and D364_16830 were increased, while the mRNA levels of D364_13295 and yjcC were decreased in the ompRD55A strain. Therefore, fimK and D364_16830 may be involved in repression of PDE activity by OmpR∼P.
FIGURE 6.
Effect of phosphorylated OmpR on c-di-GMP concentration, PDE activity, and PDE-related genes expression. (A) c-di-GMP concentration of WT, ΔompR, and ompRD55A strains in LB broth with 400 mM NaCl (solid gray bars) or without (open white bars) was quantified by ELISA according to the manual (Wuhan EIAab Science). Relative percentage of c-di-GMP content was calculated by the c-di-GMP concentration of extracts is relative to that of WT strain. (B) The PDE activity of crude extracts of WT, ΔompR, and ompRD55A strains in LB broth with 400 mM NaCl (solid gray bars) or without (open white bars) was determined by using bis-pNPP as substrate and measuring the absorbance at OD410. Relative percentage of PDE activity was calculated by the OD410 of crude extracts is relative to that of WT strain. (C) qRT-PCR analyses of the PDE-related genes expression for WT and ompRD55A strains in LB broth with 400 mM NaCl. The results are representative of three independent experiments. Error bars indicate standard deviations. ∗∗P < 0.01 compared to the indicated group.
OmpR Increases Biofilm Amount Under Hypertonic Conditions
Type 3 fimbriae are a major determinant modulating K. pneumoniae biofilm formation (Di Martino et al., 2003; Jagnow and Clegg, 2003; Wu et al., 2012). To further investigate the role of OmpR in K. pneumoniae biofilm formation, the biofilm amount of WT and ΔompR grown in LB broth with and without 400 mM NaCl was measured. As shown in Figure 7, biofilm amount was lower in ΔompR than in the WT strain in LB broth with 400 mM NaCl, while no apparent effect was observed in LB broth without NaCl. Furthermore, introduction of the complementation plasmid, pompR, into the ΔompR strain increased biofilm amount, compared to ΔompR carrying the empty vector in LB broth supplemented with 400 mM NaCl. These results indicate that OmpR can increase biofilm amount in K. pneumoniae under hypertonic conditions. In addition, we evaluated biofilm amount in the ompRD55A mutant grown LB broth supplemented with 400 mM NaCl; biofilm amount was decreased in the ompRD55A strain compared to the WT strain, indicating that the phosphorylated form of OmpR increased biofilm amount in K. pneumoniae CG43S3 under hypertonic conditions.
FIGURE 7.
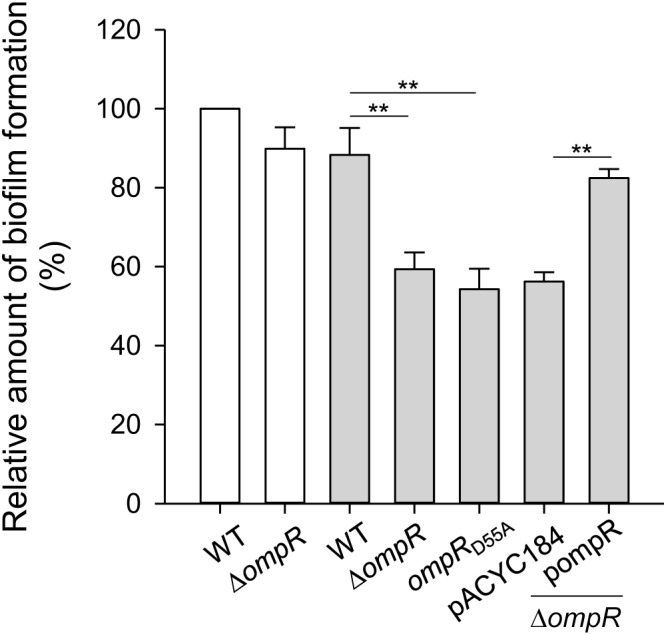
Effect of OmpR on biofilm formation. K. pneumoniae strains CG43S3 WT, ΔompR, ompRD55A, ΔompR (pACYC184), and ΔompR (pOmpR) were grown at 37°C for 24 h in LB broth with 400 mM NaCl (solid gray bars) or without (open white bars), and bacterial biofilm formation was quantified as described in Materials and Methods. The results are representative of three independent experiments. Error bars indicate standard deviations. ∗∗P < 0.01 compared to the indicated group.
Discussion
OmpR/EnvZ is a well-known TCS in many species of Enterobacteriaceae, which regulates the expression of various virulence genes in response to osmotic stresses (Mizuno and Mizushima, 1990; Oshima et al., 2002). However, the role of OmpR/EnvZ in K. pneumoniae pathogenesis remains largely unknown. In this study, we found that OmpR∼P activated type 3 fimbriae expression and biofilm formation under hypertonic conditions. Additionally, OmpR regulation of type 3 fimbriae expression appeared to involve the c-di-GMP signaling pathway. Our previous studies have also shown that transcription factors, such as Fur, cAMP-CRP, and IscR, coordinate c-di-GMP related proteins, MrkH, MrkI, and MrkJ, to regulate type 3 fimbriae expression in response to iron availability and exogenous glucose stimuli in K. pneumoniae (Wu et al., 2012; Lin et al., 2016, 2017). These findings indicate that K. pneumoniae needs to orchestrate various signaling pathways in response to dynamic environmental cues in order to regulate the expression of virulence factors for successful infection.
In this study, we found that osmotic stress repressed type 3 fimbriae expression in K. pneumoniae CG43S3 (Figure 1A). In E. coli, transcriptional regulators, including CpxAR, RpoS, Lrp, and H-NS, have been demonstrated to play important roles in bacterial responses to osmotic stress (Levinthal and Pownder, 1996; De Wulf et al., 1999; Giuliodori et al., 2007; Hengge, 2008; Pruss, 2017). Of these regulators, H-NS has been shown to directly repress the expression of mrkHIJ, while activating the expression of mrkA (Ares et al., 2016, 2017). It has also been demonstrated that increased osmolality can reduce the polymerization of H-NS tetramers and subsequently decrease the regulatory activity of H-NS (Stella et al., 2006). Thus, whether polymerization of H-NS in response to osmotic stress affects type 3 fimbriae expression in K. pneumoniae should be investigated in future studies. Furthermore, we noted that OmpR activated type 3 fimbriae expression under hypertonic conditions and that the phosphorylation status of OmpR played a crucial role in this regulation (Figures 1, 3). Intracellular OmpR∼P levels are tightly controlled by EnvZ, which exhibits high phosphatase activity and low kinase activity at low osmolality (Mattison and Kenney, 2002). Taken together, these findings suggest that the relatively high level of OmpR∼P present in response to hypertonic conditions can increase type 3 fimbriae expression in K. pneumoniae.
In K. pneumoniae, the type 1 and type 3 fimbrial gene clusters are physically linked and deletion of mrkA increases the production of FimA, the major subunit of type 1 fimbriae (Wang et al., 2013), implying that expression of these fimbriae is coordinately regulated. In E. coli, deletion of ompR increased type 1 fimbriae expression via the activation of fimB transcription (Rentschler et al., 2013). We also found that the mRNA levels of fimA and fimB were increased in the K. pneumoniae ΔompR strain compared to the WT strain when grown in LB broth with 400 mM NaCl (Supplementary Figure S1). This result shows that OmpR also decreases type 1 fimbriae expression in K. pneumoniae; further investigation is required to elucidate the underlying mechanism(s). To our knowledge, the gene clusters, mrkABCDEF and mrkHIJ, are highly conserved among K. pneumniae strains with about 99% amino acid identities, but are not found in other bacteria. Since we focused on the study of the OmpR regulation on the expression of mrk genes, which are not found in the genomes of S. Typhimurium, S. flexneri, Y. pestis, and A. baumanni, it is difficult to make a comparison.
High concentrations of c-di-GMP are linked to bacterial biofilm formation (Caly et al., 2015; Jenal et al., 2017). In K. pneumoniae, intracellular c-di-GMP concentrations influence the regulatory activity of MrkH on type 3 fimbriae expression and bacterial biofilm amount (Wilksch et al., 2011; Yang et al., 2013). Our results might indicate that OmpR indirectly regulates the expression of mrkHIJ (Figure 5). Multiple genes encoding GGDEF- and EAL-domain containing proteins have been identified in the K. pneumoniae genome and their expression, in response to various environmental stimuli, may influence intracellular c-di-GMP concentrations (Ferreira et al., 2008; Kalia et al., 2012). Although OmpR∼P has an apparent effect on modulating c-di-GMP concentration and the intracellular PDE activity, only a slight increase in the mRNA levels of fimK and D364_16830 was observed (Figure 6). Whether OmpR∼P affects the expression of fimK and D364_16830 to increase PDE activity, needs to be clarified. In addition, hybrid proteins with both GGDEF and EAL domains may exert dual activities or only single activity depending on various cellular conditions (Ferreira et al., 2008; Kalia et al., 2012). Therefore, OmpR may also affect the expression of ORFs encoding hybrid proteins with both GGDEF and EAL domains, to modulate PDE activity; this possibility needs to be further investigated. In E. coli, OmpR∼P represses the expression of bolA, which is responsible for cell growth and division (Yamamoto et al., 2000). Deletion of bolA increases intracellular c-di-GMP concentrations and biofilm formation in E. coli (Moreira et al., 2017), suggesting that OmpR∼P represses bolA expression to modulate intracellular c-di-GMP concentrations. We identified a bolA homolog (D364_02025) in the genome of K. pneumoniae CG43, which shared 91.3% amino acid sequence identity with the E. coli protein. However, determining whether OmpR regulates the expression of bolA to affect intracellular c-di-GMP concentrations, type 3 fimbriae production, and biofilm formation in K. pneumoniae will require further investigation.
Type 3 fimbriae expression is thought to play a crucial role in K. pneumoniae biofilm formation; thus, we hypothesized that bacterial biofilm formation is affected by osmotic stimuli. However, no apparent effect on biofilm formation was observed when K. pneumoniae was grown in LB broth with or without 400 mM NaCl (Figure 7). In addition to type 3 fimbriae, multiple factors have been reported to affect K. pneumoniae biofilm formation such as CPS, LPS, quorum-sensing systems, and antibiotic resistance (Balestrino et al., 2005, 2008; Boddicker et al., 2006; Alcantar-Curiel et al., 2013; Khodadadian et al., 2017; Vuotto et al., 2017). We also found that CPS levels were increased under hypertonic conditions (Supplementary Figure S2). As the thick capsule of K. pneumoniae impedes the assembly of type 3 fimbriae (Schembri et al., 2005), the decrease in type 3 fimbriae under hypertonic conditions may also be due to an increase in CPS levels. However, no apparent difference in CPS amount was observed between WT and ΔompR strains grown under hypertonic conditions (data not shown), implying the involvement of other regulator(s). In E. coli, OmpR regulates the expression of the porins OmpF and OmpC in response to osmotic stress (Mizuno and Mizushima, 1990). Furthermore, the deletion of ompF can increase bacterial biofilm formation (Yang et al., 2008). K. pneumoniae possess two classical porins, OmpK35 and OmpK36, which are homologs of E. coli OmpF and OmpC, respectively (Sugawara et al., 2016). In K. pneumoniae, biofilm formation is highly related to antibiotic resistance (Vuotto et al., 2017), which is affected by the expression of porins and efflux pumps (e.g., AcrAB) (Bialek et al., 2010; Pitout et al., 2015). Taken together, these findings suggest that OmpR∼P coordinately regulates the expression of OmpK35, OmpK36, and type 3 fimbriae to mediate K. pneumoniae biofilm formation. This possibility needs to be further investigated.
In this study, we provide evidence that OmpR∼P participates in the regulation of type 3 fimbriae and mrkHIJ expression, c-di-GMP concentration, and biofilm formation in response to osmotic stress, which may play an important role during successful infection.
Author Contributions
T-HL, J-TK, YC, and C-TL conceived and designed the experiments. YC, J-TK, and C-TL performed the experiments. YC, J-TK, Y-CL, and C-TL analyzed the data. T-HL, Y-CL, C-CW, C-FH, and C-TL contributed reagents, materials, and analysis tools. T-HL and C-TL wrote the paper. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Miss Yi-Min Hong for her technical assistance during the study.
Footnotes
Funding. This work was supported by the grant from Ministry of Science and Technology (MOST 106-2320-B-039-031-), China Medical University (CMU106-S-35), China Medical University Hospital (DMR-106-114), and Taichung Tzuchi Hospital, The Buddhist Tzu Chi Medical Foundation (TTCRD104-11).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02405/full#supplementary-material
References
- Alcantar-Curiel M. D., Blackburn D., Saldana Z., Gayosso-Vazquez C., Iovine N. M., De La Cruz M. A., et al. (2013). Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence 4 129–138. 10.4161/viru.22974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M. A., Fernandez-Vazquez J. L., Pacheco S., Martinez-Santos V. I., Jarillo-Quijada M. D., Torres J., et al. (2017). Additional regulatory activities of MrkH for the transcriptional expression of the Klebsiella pneumoniae mrk genes: antagonist of H-NS and repressor. PLoS One 12:e0173285. 10.1371/journal.pone.0173285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M. A., Fernandez-Vazquez J. L., Rosales-Reyes R., Jarillo-Quijada M. D., Von Bargen K., Torres J., et al. (2016). H-NS Nucleoid Protein Controls Virulence Features of Klebsiella pneumoniae by regulating the expression of type 3 Pili and the capsule polysaccharide. Front. Cell. Infect. Microbiol. 6:13. 10.3389/fcimb.2016.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrino D., Ghigo J. M., Charbonnel N., Haagensen J. A., Forestier C. (2008). The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ. Microbiol. 10 685–701. 10.1111/j.1462-2920.2007.01491.x [DOI] [PubMed] [Google Scholar]
- Balestrino D., Haagensen J. A., Rich C., Forestier C. (2005). Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 187 2870–2880. 10.1128/JB.187.8.2870-2880.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini M. L., Fontaine A., Sansonetti P. J. (1990). The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J. Bacteriol. 172 6274–6281. 10.1128/jb.172.11.6274-6281.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek S., Lavigne J. P., Chevalier J., Marcon E., Leflon-Guibout V., Davin A., et al. (2010). Membrane efflux and influx modulate both multidrug resistance and virulence of Klebsiella pneumoniae in a Caenorhabditis elegans model. Antimicrob. Agents Chemother. 54 4373–4378. 10.1128/AAC.01607-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker J. D., Anderson R. A., Jagnow J., Clegg S. (2006). Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect. Immun. 74 4590–4597. 10.1128/IAI.00129-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly D. L., Bellini D., Walsh M. A., Dow J. M., Ryan R. P. (2015). Targeting cyclic di-GMP signalling: a strategy to control biofilm formation? Curr. Pharm. Des. 21 12–24. [DOI] [PubMed] [Google Scholar]
- Carpenter C., Payne S. M. (2014). Regulation of iron transport systems in Enterobacteriaceae in response to oxygen and iron availability. J. Inorg. Biochem. 133 110–117. 10.1016/j.jinorgbio.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P., Kwon O., Lin E. C. (1999). The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181 6772–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado J., Forst S., Harlocker S., Inouye M. (1993). Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol. Microbiol. 10 1037–1047. 10.1111/j.1365-2958.1993.tb00974.x [DOI] [PubMed] [Google Scholar]
- Di Martino P., Cafferini N., Joly B., Darfeuille-Michaud A. (2003). Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 154 9–16. 10.1016/S0923-2508(02)00004-9 [DOI] [PubMed] [Google Scholar]
- Dorman C. J., Chatfield S., Higgins C. F., Hayward C., Dougan G. (1989). Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: ompR mutants are attenuated in vivo. Infect. Immun. 57 2136–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R. B., Antunes L. C., Greenberg E. P., Mccarter L. L. (2008). Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 190 851–860. 10.1128/JB.01462-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliodori A. M., Gualerzi C. O., Soto S., Vila J., Tavio M. M. (2007). Review on bacterial stress topics. Ann. N. Y. Acad. Sci. 1113 95–104. 10.1196/annals.1391.008 [DOI] [PubMed] [Google Scholar]
- Hengge R. (2008). The two-component network and the general stress sigma factor RpoS (sigma S) in Escherichia coli. Adv. Exp. Med. Biol. 631 40–53. 10.1007/978-0-387-78885-2_4 [DOI] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Huang C. J., Wang Z. C., Huang H. Y., Huang H. D., Peng H. L. (2013). YjcC, a c-di-GMP phosphodiesterase protein, regulates the oxidative stress response and virulence of Klebsiella pneumoniae CG43. PLoS One 8:e66740. 10.1371/journal.pone.0066740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M. M., Silhavy T. J. (1988). EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J. Bacteriol. 170 5971–5973. 10.1128/jb.170.12.5971-5973.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagnow J., Clegg S. (2003). Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 149 2397–2405. 10.1099/mic.0.26434-0 [DOI] [PubMed] [Google Scholar]
- Jenal U., Reinders A., Lori C. (2017). Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15 271–284. 10.1038/nrmicro.2016.190 [DOI] [PubMed] [Google Scholar]
- Johnson J. G., Clegg S. (2010). Role of MrkJ, a phosphodiesterase, in type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J. Bacteriol. 192 3944–3950. 10.1128/JB.00304-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia D., Merey G., Nakayama S., Zheng Y., Zhou J., Luo Y., et al. (2012). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42 305–341. 10.1039/c2cs35206k [DOI] [PubMed] [Google Scholar]
- Kawai Y., Marles-Wright J., Cleverley R. M., Emmins R., Ishikawa S., Kuwano M., et al. (2011). A widespread family of bacterial cell wall assembly proteins. EMBO J. 30 4931–4941. 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodadadian R., Rahdar H. A., Javadi A., Safari M., Khorshidi A. (2017). Detection of VIM-1 and IMP-1 genes in Klebsiella pneumoniae and relationship with biofilm formation. Microb. Pathog. 115 25–30. 10.1016/j.micpath.2017.12.036 [DOI] [PubMed] [Google Scholar]
- Lai Y. C., Peng H. L., Chang H. Y. (2001). Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun. 69 7140–7145. 10.1128/IAI.69.11.7140-7145.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y. C., Peng H. L., Chang H. Y. (2003). RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J. Bacteriol. 185 788–800. 10.1128/JB.185.3.788-800.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub M. T., Goulian M. (2007). Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41 121–145. 10.1146/annurev.genet.41.042007.170548 [DOI] [PubMed] [Google Scholar]
- Lee C. R., Lee J. H., Park K. S., Jeon J. H., Kim Y. B., Cha C. J., et al. (2017). Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front. Cell. Infect. Microbiol. 7:483. 10.3389/fcimb.2017.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembke C., Podbielski A., Hidalgo-Grass C., Jonas L., Hanski E., Kreikemeyer B. (2006). Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl. Environ. Microbiol. 72 2864–2875. 10.1128/AEM.72.4.2864-2875.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinthal M., Pownder T. (1996). hns, rpoS and lrp mutations affect stationary-phase survival at high osmolarity. Res. Microbiol. 147 333–342. 10.1016/0923-2508(96)84708-5 [DOI] [PubMed] [Google Scholar]
- Lin C. T., Chen Y. C., Jinn T. R., Wu C. C., Hong Y. M., Wu W. H. (2013). Role of the cAMP-dependent carbon catabolite repression in capsular polysaccharide biosynthesis in Klebsiella pneumoniae. PLoS One 8:e54430. 10.1371/journal.pone.0054430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. T., Huang T. Y., Liang W. C., Peng H. L. (2006). Homologous response regulators KvgA, KvhA and KvhR regulate the synthesis of capsular polysaccharide in Klebsiella pneumoniae CG43 in a coordinated manner. J. Biochem. 140 429–438. 10.1093/jb/mvj168 [DOI] [PubMed] [Google Scholar]
- Lin C. T., Lin T. H., Wu C. C., Wan L., Huang C. F., Peng H. L. (2016). CRP-Cyclic AMP regulates the expression of type 3 fimbriae via cyclic di-GMP in Klebsiella pneumoniae. PLoS One 11:e0162884. 10.1371/journal.pone.0162884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. H., Tseng C. Y., Lai Y. C., Wu C. C., Huang C. F., Lin C. T. (2017). IscR regulation of type 3 Fimbriae expression in Klebsiella pneumoniae CG43. Front. Microbiol. 8:1984. 10.3389/fmicb.2017.01984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustri B. C., Sperandio V., Moreira C. G. (2017). Bacterial chat: intestinal metabolites and signals in host-microbiota-pathogen interactions. Infect. Immun. 85:e00476-17.. 10.1128/IAI.00476-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K., Kenney L. J. (2002). Phosphorylation alters the interaction of the response regulator OmpR with its sensor kinase EnvZ. J. Biol. Chem. 277 11143–11148. 10.1074/jbc.M111128200 [DOI] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170 2575–2583. 10.1128/jb.170.6.2575-2583.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Mizushima S. (1990). Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol. Microbiol. 4 1077–1082. 10.1111/j.1365-2958.1990.tb00681.x [DOI] [PubMed] [Google Scholar]
- Moreira R. N., Dressaire C., Barahona S., Galego L., Kaever V., Jenal U., et al. (2017). BolA Is Required for the Accurate Regulation of c-di-GMP, a Central Player in Biofilm Formation. mBio 8:e00443-17. 10.1128/mBio.00443-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R., Kohn S., Hwang S. H., Hassett D. J., Sauer K. (2006). BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 188 7335–7343. 10.1128/JB.00599-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. N., Clegg S. (2012). Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future Microbiol. 7 991–1002. 10.2217/fmb.12.74 [DOI] [PubMed] [Google Scholar]
- Ochman H., Gerber A. S., Hartl D. L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics 120 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T., Aiba H., Masuda Y., Kanaya S., Sugiura M., Wanner B. L., et al. (2002). Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46 281–291. 10.1046/j.1365-2958.2002.03170.x [DOI] [PubMed] [Google Scholar]
- Paczosa M. K., Mecsas J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80 629–661. 10.1128/MMBR.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitout J. D., Nordmann P., Poirel L. (2015). Carbapenemase-Producing Klebsiella pneumoniae, a Key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59 5873–5884. 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podschun R., Ullmann U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss B. M. (2017). Involvement of two-component signaling on bacterial motility and biofilm development. J. Bacteriol. 199:e00259-17. 10.1128/JB.00259-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczkowska A., Skorek K., Brzostkowska M., Lasinska A., Brzostek K. (2011). Pleiotropic effects of a Yersinia enterocolitica ompR mutation on adherent-invasive abilities and biofilm formation. FEMS Microbiol. Lett. 321 43–49. 10.1111/j.1574-6968.2011.02308.x [DOI] [PubMed] [Google Scholar]
- Reboul A., Lemaitre N., Titecat M., Merchez M., Deloison G., Ricard I., et al. (2014). Yersinia pestis requires the 2-component regulatory system OmpR-EnvZ to resist innate immunity during the early and late stages of plague. J. Infect. Dis. 210 1367–1375. 10.1093/infdis/jiu274 [DOI] [PubMed] [Google Scholar]
- Rentschler A. E., Lovrich S. D., Fitton R., Enos-Berlage J., Schwan W. R. (2013). OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology 159 316–327. 10.1099/mic.0.059386-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta P., Clark E. R., Knutson K., Horne S. M., Pruss B. M. (2013). OmpR and RcsB abolish temporal and spatial changes in expression of flhD in Escherichia coli biofilm. BMC Microbiol. 13:182. 10.1186/1471-2180-13-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2000). Molecular Cloning: A Laboratory Manual Third Edition. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schembri M. A., Blom J., Krogfelt K. A., Klemm P. (2005). Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 73 4626–4633. 10.1128/IAI.73.8.4626-4633.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan W. R. (2009). Survival of uropathogenic Escherichia coli in the murine urinary tract is dependent on OmpR. Microbiology 155 1832–1839. 10.1099/mic.0.026187-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R., Morr M., Kader A., Nimtz M., Romling U. (2004). GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53 1123–1134. 10.1111/j.1365-2958.2004.04206.x [DOI] [PubMed] [Google Scholar]
- Skorupski K., Taylor R. K. (1996). Positive selection vectors for allelic exchange. Gene 169 47–52. 10.1016/0378-1119(95)00793-8 [DOI] [PubMed] [Google Scholar]
- Stella S., Falconi M., Lammi M., Gualerzi C. O., Pon C. L. (2006). Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J. Mol. Biol. 355 169–174. 10.1016/j.jmb.2005.10.034 [DOI] [PubMed] [Google Scholar]
- Stock A. M., Robinson V. L., Goudreau P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69 183–215. 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- Struve C., Bojer M., Krogfelt K. A. (2008). Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 76 4055–4065. 10.1128/IAI.00494-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara E., Kojima S., Nikaido H. (2016). Klebsiella pneumoniae major porins OmpK35 and OmpK36 allow more efficient diffusion of beta-lactams than their Escherichia coli homologs OmpF and OmpC. J. Bacteriol. 198 3200–3208. 10.1128/JB.00590-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R., Pratt J. T., Camilli A. (2007). Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61 131–148. 10.1146/annurev.micro.61.080706.093426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. W., Wilksch J. J., Hocking D. M., Wang N., Srikhanta Y. N., Tauschek M., et al. (2015). Positive auto-regulation of mrkHI by the c-di-GMP-dependent MrkH protein in the biofilm regulatory circuit of Klebsiella pneumoniae. J. Bacteriol. 197 1659–1667. 10.1128/JB.02615-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. K., Hall M. N., Enquist L., Silhavy T. J. (1981). Identification of OmpR: a positive regulatory protein controlling expression of the major outer membrane matrix porin proteins of Escherichia coli K-12. J. Bacteriol. 147 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton K. A., Rather P. N. (2016). An ompR/envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075. J. Bacteriol. 199:e00705-16. 10.1128/JB.00705-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuotto C., Longo F., Pascolini C., Donelli G., Balice M. P., Libori M. F., et al. (2017). Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J. Appl. Microbiol. 123 1003–1018. 10.1111/jam.13533 [DOI] [PubMed] [Google Scholar]
- Wang Z. C., Huang C. J., Huang Y. J., Wu C. C., Peng H. L. (2013). FimK regulation on the expression of type 1 fimbriae in Klebsiella pneumoniae CG43S3. Microbiology 159 1402–1415. 10.1099/mic.0.067793-0 [DOI] [PubMed] [Google Scholar]
- Wilksch J. J., Yang J., Clements A., Gabbe J. L., Short K. R., Cao H., et al. (2011). MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7:e1002204. 10.1371/journal.ppat.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. C., Lin C. T., Cheng W. Y., Huang C. J., Wang Z. C., Peng H. L. (2012). Fur-dependent MrkHI regulation of type 3 fimbriae in Klebsiella pneumoniae CG43. Microbiology 158 1045–1056. 10.1099/mic.0.053801-0 [DOI] [PubMed] [Google Scholar]
- Wu H., Fives-Taylor P. M. (2001). Molecular strategies for fimbrial expression and assembly. Crit. Rev. Oral Biol. Med. 12 101–115. 10.1177/10454411010120020101 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Nagura R., Tanabe H., Fujita N., Ishihama A., Utsumi R. (2000). Negative regulation of the bolA1p of Escherichia coli K-12 by the transcription factor OmpR for osmolarity response genes. FEMS Microbiol. Lett. 186 257–262. 10.1111/j.1574-6968.2000.tb09114.x [DOI] [PubMed] [Google Scholar]
- Yang J., Wilksch J. J., Tan J. W., Hocking D. M., Webb C. T., Lithgow T., et al. (2013). Transcriptional activation of the mrkA promoter of the Klebsiella pneumoniae type 3 fimbrial operon by the c-di-GMP-dependent MrkH protein. PLoS One 8:e79038. 10.1371/journal.pone.0079038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Ma Q., Wood T. K. (2008). The R1 conjugative plasmid increases Escherichia coli biofilm formation through an envelope stress response. Appl. Environ. Microbiol. 74 2690–2699. 10.1128/AEM.02809-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Qin L., Egger L. A., Inouye M. (2006). Transcription regulation of ompF and ompC by a single transcription factor. OmpR. J. Biol. Chem. 281 17114–17123. 10.1074/jbc.M602112200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.