Abstract
Involvement of life stress in Late-Onset Alzheimer’s Disease (LOAD) has been evinced in longitudinal cohort epidemiological studies, and endocrinologic evidence suggests involvements of catecholamine and corticosteroid systems in LOAD. Early Life Stress (ELS) rodent models have successfully demonstrated sequelae of maternal separation resulting in LOAD-analogous pathology, thereby supporting a role of insulin receptor signalling pertaining to GSK-3beta facilitated tau hyper-phosphorylation and amyloidogenic processing. Discussed are relevant ELS studies, and findings from three mitogen-activated protein kinase pathways (JNK/SAPK pathway, ERK pathway, p38/MAPK pathway) relevant for mediating environmental stresses. Further considered were the roles of autophagy impairment, neuroinflammation, and brain insulin resistance.
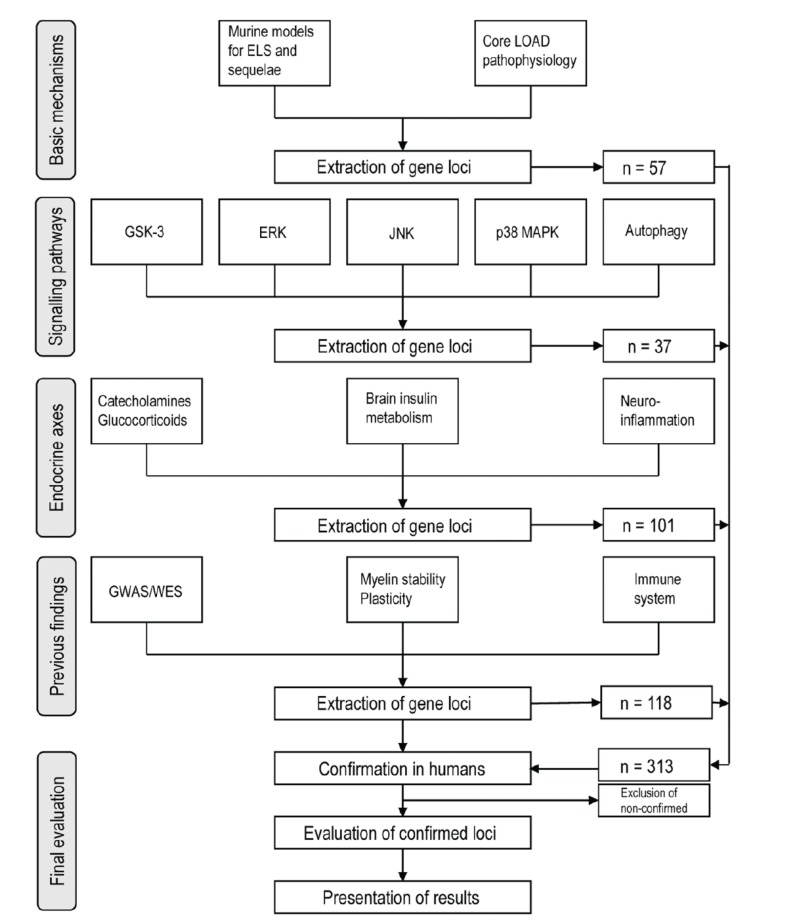
For the meta-analytic evaluation, 224 candidate gene loci were extracted from reviews of animal studies of LOAD pathophysiological mechanisms, of which 60 had no positive results in human LOAD association studies. These loci were combined with 89 gene loci confirmed as LOAD risk genes in previous GWAS and WES. Of the 313 risk gene loci evaluated, there were 35 human reports on epigenomic modifications in terms of methylation or histone acetylation. 64 microRNA gene regulation mechanisms were published for the compiled loci.
Genomic association studies support close relations of both noradrenergic and glucocorticoid systems with LOAD. For HPA involvement, a CRHR1 haplotype with MAPT was described, but further association of only HSD11B1 with LOAD found; however, association of FKBP1 and NC3R1 polymorphisms was documented in support of stress influence to LOAD. In the brain insulin system, IGF2R, INSR, INSRR, and plasticity regulator ARC, were associated with LOAD. Pertaining to compromised myelin stability in LOAD, relevant associations were found for BIN1, RELN, SORL1, SORCS1, CNP, MAG, and MOG. Regarding epigenetic modifications, both methylation variability and de-acetylation were reported for LOAD. The majority of up-to-date epigenomic findings include reported modifications in the well-known LOAD core pathology loci MAPT, BACE1, APP (with FOS, EGR1), PSEN1, PSEN2, and highlight a central role of BDNF. Pertaining to ELS, relevant loci are FKBP5, EGR1, GSK3B; critical roles of inflammation are indicated by CRP, TNFA, NFKB1 modifications; for cholesterol biosynthesis, DHCR24; for myelin stability BIN1, SORL1, CNP; pertaining to (epi)genetic mechanisms, hTERT, MBD2, DNMT1, MTHFR2. Findings on gene regulation were accumulated for BACE1, MAPK signalling, TLR4, BDNF, insulin signalling, with most reports for miR-132 and miR-27. Unclear in epigenomic studies remains the role of noradrenergic signalling, previously demonstrated by neuropathological findings of childhood nucleus caeruleus degeneration for LOAD tauopathy.
Keywords: Late-onset Alzheimer’s disease, Early life stress, Stress neuropsychobiology, Candidate genes, Epigenomics, Epigenetic programming, Gene regulation, MicroRNA, Methylation, Acetylation, Pathophysiology, Ageing biology, Mitogen activated kinases, Neuroinflammation, Catecholamines, Corticosteroids, Insulin signalling
1. INTRODUCTION
1.1. Psychological Stress as a Risk Factor for Late Onset Alzheimer Disease (LOAD)
The last decade has brought about a profound reorganisation of knowledge on the age-related senile dementia of the Alzheimer type (LOAD). It has nowadays become broadly accepted that life stresses must play a mediatory role [1] in the complex genotype-environmental interaction which precedes clinical manifestation of the fatal Alzheimer endo-phenotype [2]. In contrast to all other neurodegenerative disorders, sporadic late-onset dementia of the Alzheimer type is a condition, which is almost probabilistically related to increasing age: it is increasing to almost 50% among those ≥90 years old (2.78% at 65 with almost linear increase to 56.13% at 95 per 1000 person years, slightly peaking at 85) [3].
The demographic change in combination with extending life expectancies is a major sociological risk for increasing incidences of LOAD in developed ageing societies. It has been stipulated that its aetiopathophysiology should accordingly adopt a life-span developmental perspective [4, 5]. The 2014 World Alzheimer Report estimates a worldwide prevalence of 44 million sufferers from this fatal disease, and expects that this number is to triple by the year 2050 [4]. For the US alone, an incidence rate of one million new patients per year is forecasted by the Alzheimer’s Association [6], and the total expected prevalence 2050 is 13.8 million [6]. A proportion of 75% of all dementia cases have LOAD pathology (adding to a co-morbidity with vascular dementia towards 90% of all post-mortem dissected and thus histologically verified cases) [4], with highest prevalence figures in North America and Europe [1], where the life-time prevalence risk is currently 17% for females, and 9% for males [7]. Several European population-based cohort studies have provided evidence in the past five years that the age-specific incidence of dementia has decreased in the past 20 years [8], but incidences increased in China and threshold countries, a fluctuation possibly attributable to life-style factors.
The main difference to familial presenile AD (Morbus Alzheimer proper) is based on autosomal dominant mutations in the highly homologous presenlin 1 (PSEN1 14q24.1), presenilin 2 (PSEN2 1q42.13), and amyloid precursor protein (APP 21q21.3) genes. Up to date, there is still too little knowledge about the exact function and disorders of Amyloid Precursor Protein (APP) [9]. In the mutations linked to early-onset AD, pathogenic presenilin isoforms become part of the enzyme gamma-secretase responsible for the neurotoxic 42-aminoacid isomer of the cleaved APP [10]. In addition, the presenilins interact with Notch1 receptors and are involved in the Notch signalling pathways related to neuronal differentiation and neuritic outgrow. Specifically, in the notch pathway, gamma secretase releases the intracellular domain of the notch receptor protein 1 (NOTCH1 9q34.3), a relative of the epidermal growth factor (EGF), regulating nuclear gene expression, and synaptic stability through synaptic plasticity protein Arc (Section 3.2.). Notch signalling is also involved in oligodendrocyte differentiation and upregulation of myelin-associated glycoprotein MAG [11], thus constituting a direct biochemical link to myelination integrity and late-life myelin breakdown in LOAD. The further main commonality then shared with LOAD is the general pathophysiology (amyloid beta cascade and tau pathologies, in particular), which is the focus of the following sections.
In LOAD, the major genetic risk is the apolipoprotein E (APOE 19q32.13) epsilon4 allele, specifically in heterozygotic genotype with Odds Ratios (ORs) 2.6~3.2 [12, 13]. Apolipoprotein E is crucial for cholesterol transport and metabolism, and in the brain synthesised by astrocytes and microglia. In LOAD, the APOE epsilon4 allele is present in 40% [12, 13] -50% [8] of cases, and therefore constitutes the largest known single genomic risk, however, in 15-fold probability (OR 14.9) [13] for homozygotic carriers. Amongst these APOE epsilon4 carriers, the growth factor receptor-bound protein 2 associated binder protein 2 (GAB2 11q14.1) has been found to have an interaction with tangle bearing neurons leading to an overexpression of GAB2 [14, 15] (however not replicated in all ethnicities). GAB2 addresses PI3K/Akt/mTOR and ERK/MAPK (Section 2.2.) pathways and interacts with APP [16].
Several other rare genetic variants have been encircled by Genome Wide Association Studies (GWAS) [15, 17], Whole Exome Studies (WES) [18, 19], and family/twin studies (adjusted h2=0.32-0.42 for memory) [20], (57-78% for onset time) [21, 22], (h2=0.67-0.74 for liability) [23] (Table 1: confirmed candidate genes, Section 4.1.). However, the lack of single large genomic bases lead to the conclusion that other mechanisms, such as epigenetic modification, transcriptional regulations and/or gene-environment interactions could play a substantial role in pathogenesis. Whereas global genetic expression rates (within age correlations) increase only slightly from 0.089 in the 4th towards 0.23 in the 7th decade [24], the major GWAS risk genes (Section 4.1.) showed marked methylation changes in LOAD over age as compared to a control population [25, 26]. Polygenic risk associations for cerebral substrates of LOAD, as well as amyloid-beta (Abeta) levels, prove significant only in younger ages, but not in higher ages [27, 28], suggesting that ageing per se is an independent process leading to cognitive decline. Hence, these authors led by Mormino conclude that “genetic risk may begin in early life and make an individual more susceptible to cognitive impairment in late life” (p. 481) [27].
Table 1.
Confirmed risk gene loci.
| Locus | Gene | Function | References |
|---|---|---|---|
| 9q31.3 | ABCA1 | ATP-binding cassette transporter subfamily A member 1: regulates cholesterol efflux to forming HDL | [476, 604] |
| 19p13.3 | ABCA7 | ATP binding cassette subfamily A member 7: lipid homeostasis of immune cells, efflux of phospholipids, phagocytosis | [541, 542, 557, 558, 569, 605-608] |
| 15q23.3 | ADAM10 | A-disintegrin and metalloproteinase domain-containing protein 10: excitatory hippocampal synapses | [556, 559] |
| 8p11.21 | ANK1 | Ankyrin 1: integral membrane protein in the spectrin-actin cytoskeleton, neuron motility, activation, proliferation, contact | [476, 569, 570] |
| 19q13.32 | APOE | Apolipoprotein E: neuronal cholesterol and triglyceride transport shuttle | [605, 606] |
| 21q21.2 | APP | Amyloid precursor protein: cell surface receptor, cleavage products binding acetyltransferase complex APBB1/TIP60, promoting transcriptional activation | [124, 211, 545, 546] |
| 9q33.1 | ASTN2 | Neuronal protein astrotactin 2: neuron migration and connectivity | [578] |
| 17q25.1 | ATP5H/KCTD2 | Mitochondrial ATP synthase H+ transporting: catalyses ATP synthesis, involved in OXPHOS, mitochondrial energy and cellular stress | [541] |
| 11q23.3 | BACE1 | Beta-secretase 1: transmembrane protease performing first cleavage step for amyloid beta | [547] |
| 2q14.3 | BIN1 | Myc box-dependent-interacting protein 1: involved in synaptic vesicle endocytosis, endosomal vesicle cycling, clathrin-mediated endocytosis, related to tauopathy | [540, 542, 546, 557, 563, 564, 569, 572, 605-607] |
| 19q13.32 | BLOC1S3 | Biogenesis of lysosomal organelles complex1, subunit 3: autophagy and apoptosis | [572] |
| 22q13.1 | CARD10 | Caspase recruitment domain family, member 10: NFkappaB signalling pathway, hippocampal volume, hippocampal neurodegeneration with PARP1 in APOE epsilon3 carriers | [541, 609] |
| 20q13.31 | CASS4 | Cas scaffolding protein family member 4: cytoskeletal stabilisation, axonal transport, binding to CD2AP, involved in APP pathology, tauopathy | [542, 557, 605, 606] |
| 6p12.3 | CD2AP | CD2 associated protein: endosomal vesicle movement/cycling, cytoskeletal reorganisation, modulating with GRB2 metabolism of APP |
[540, 542, 546, 557, 558, 566, 605-607] |
| 19q13.41 | CD33 | CD33 molecule: microglial/immune/inflammatory response, cell-cell interactions, related to cognitive decline, Abeta clearance | [541, 542, 557, 605-607, 610] |
| 10q23.1 | CDH23 | Cadherin related 23: calcium dependent cell-cell adhesion glycoprotein, neuronal differentiation, neuronal transmission | [543, 569] |
| 11p11.2 | CELF1 | CUGBP Elav-like family member 1: regulating pre-mRNA alternative splicing, editing, translation, including MADD MAPK–activating death domain, affecting long-term neuronal viability | [542, 557, 605, 606, 611] |
| 16q13 | CETP | Cholesteryl ester transfer protein: transfer of cholesteryl ester from HDL to other lipoproteins, related to myelination, white matter integrity, MetS components | [541, 612, 613] |
| 1q31.1 | CFHR1 | Complement factor H related 1: controlling complement, facilitating tissue invasion, neuroinflammation with IL-6, neurodegeneration | [541, 614] |
| 8p21.1 | CLU | Clusterin: molecular chaperone, clearing cellular debris and apoptosis, cholesterol and lipid metabolism, regulation of cell proliferation | [541, 542, 546, 567, 605-608] |
| 1q32.2 | CR1 | Complement receptor 1: membrane glycoprotein binding immune complexes, immune response, regulation of complement activation, neuroinflammation | [540, 541, 542, 546, 557, 562, 605-607] |
| Locus | Gene | Function | References |
| 15q21.1 | CYP19A1 | Enzyme aromatase: oestrogen pathway, involved in tauopathy, Abeta cascade, interaction with IL-10 | [544, 615, 616] |
| 14q32.2 | CYP46A1 | Cholesterol 24S-hydroxylase (CYP46A1) promoter: monooxygenase related to synthesis of cholesterol hydroxylase, steroids and other lipids, cholesterol transport through BBB | [617-619] |
| 1p23.3 | DHCR24 | 24-dehydrocholesterol reductase: oxidoreductase catalysing cholesterol biosynthesis, cholesterol efflux | [476, 620, 621] |
| 21q22.3 | DIP2A | Disco interacting protein 2 homolog A: CNS axon patterning | [569] |
| 18q12.1 | DSG2 | Desmoglein 2: calcium-binding transmembrane glycoprotein component of desmosomes, cell-cell junctions, glycoprotein generation, involved in APP processing | [606, 622] |
| 1p36.12 | ECE-1b promoter | Endothelin converting enzyme 1: related to Abeta degradation | [573, 574] |
| 18q12.2-q21.1 | EPG5 | Ectopic P-granules autophagy protein 5 homologue: autophagy | [551, 623] |
| 7q34-35 | EPHA1 | Ephrin receptor: mediating CNS development, modulating cell migration, axon guidance, synapse development and plasticity, cerebral glucose levels, atrophy, endosomal vesicle cycling, immune system | [542, 546, 557, 565, 567, 605-607] |
| 2q33.3 | FASTKD2 | Fas-associated serine/threonine kinase domains 2: mitochondrial inner compartment protein, COX signalling, memory performance | [541, 624-626] |
| 14q22.1 | FERMT2 | Fermitin family member 2: cytoskeletal function and axonal transport, actin assembly and cell shape modulation, cell matrix adhesion structures, activates integrins, related to tauopathy | [542, 557, 605, 606] |
| 9p24.2 | GLIS3 | GLI-similar family zinc finger 3: repressor and activator of transcription, development of pancreatic beta cells, tauopathy, APP metabolism | [19, 541, 627] |
| 3q28 | GMNC | Geminin coiled-coil domain containing: involved chromosomal DNA replication, preferentially expressed in proliferating neurons, related to tauopathy | [19] |
| 6p21.31 | HLA-DRB5/DRB1 | Major histocompatibility complex class II, DRbeta5 and DRbeta1: immuno- competence, encoding human leukocyte antigen, neuroinflammation | [542, 557, 605, 606] |
| 14q32.33 | IGHV1-67 | Immunoglobulin heavy variable 1-67: neuroinflammation | [628] |
| 2q37.1 | INPP5D | Inositol polyphosphate-5-phosphatase: Immune response, gene regulation, posttranslational modification of proteins, microglial and myeloid function, interacting with CD2AP, neuroinflammation | [542, 557, 605, 606] |
| 17q21.31 | KANSL1 | KAT8 regulatory NSL complex subunit 1: involved with histone acetylation, related to MAPT expression | [629] |
| 12q24.31 | KDM2B | Lysine demethylase 2B: phosphorylation-dependent ubiquitination, posttranslational modification | [630] |
| 9q34.12 | LAMC3 | Laminin subunit gamma 3: extracellular matrix glycoprotein related to cellular morphogenesis, in interaction with LRRK2 and MADD related to age-of-onset | [577] |
| 12q12 | LRRK2 | Leucine rich repeat kinase 2: Ras-Raf signalling, cytoplasm and mitochondrial outer membrane, related to age-of-onset | [577] |
| 11p11.2 | MADD | MAP kinase activating death domain: TNFalpha signalling with the death domain of TNFalpha receptor 1, MAPK apoptotic signal transduction, related to age-of-onset | [577, 631, 632] |
| 17q21.31 | MAPT | Microtubule-associated protein tau: neuron stabilisation, transcript undergoes complex, regulated alternative splicing, producing several mRNA species | [546, 629] |
| Locus | Gene | Function | References |
| 5q14.3 | MEF2C | Myocyte enhancer factor 2C: immune response, neural development, synaptic connectivity and plasticity, limiting excessive activity-dependent synapse formation, facilitating hippocampal-dependent learning and memory, neuroinflammation | [557, 605, 606] |
| 3p25.2 | MME/NEP | Membrane metalloendopeptidase/neprisylin: glycoprotein neutral endopeptidase, related to WM hyperintensities, related to cleavage of peptides, related to Abeta degradation, covariation with WM disintegrity | [573, 633, 634] |
| 11q12.2 | MS4A4/MS4A6E | Membrane spanning 4-domains: involved in signal transduction as a component of a multimeric receptor complex, IgE receptor, immune response, neuroinflammation | [540, 542, 546, 605-608, 635] |
| 20q13.33 | MYT1 | Myelin transcription factor: zinc finger transcription factor expressed on neurons, myelination onset, binding to the significant promoter CYP46A1 polymorphism, in haplotypes with CYP46A1, related to REST, MECP2 | [617] |
| 20p11.21 | NANP | N-acetylneuraminic acid phosphatase: implicated in insulin signalling, glycation, related to brain atrophy in neurodegeneration | [630] |
| 20p11.21 | NINL | Ninein-like protein: implicated in dynein-dynactin-interaction, related to stabilisation of microtubuli, tauopathy, related to brain atrophy in non-APOE epsilon4 carriers in haplotype with NANP | [630] |
| 7p14.1 | NME8 | Thioredoxin domain-containing protein 3: implicated in dynein-related microtubular transport function, related to cognitive decline, neurodegeneration | [542, 557, 605, 606] |
| 10q23.1 | NRG3 | Neuregulin 3: EGF-related ligand to transmembrane receptors, implicated in neuroblast proliferation, migration, differentiation, and survival or apoptosis, related to age-of-onset | [579] |
| 12q22 | NTN4 | Netrin 4: EGF-related, responsible for axon guidance, neurite growth, neuron migration, angiogenesis, in haplotype with common variants | [577] |
| 1p35.3 | OPRD1 | Delta-opioid receptor: small cerebral volume, implicated with APP processing | [541, 636-638] |
| 3q28 | OSTN | Osteocrin: primate-specific regulator of synapse formation, restricting activity-dependent dendritic growth, related to transcription factor MEF2 in the SASP (Section 1.4.) | [19, 541, 639] |
| 1q42.12 | PARP1 | Poly(ADP-ribose) polymerase 1: neuron proliferation, hippocampal volume, hippocampal neurodegeneration in APOE epsilon3 carriers | [541, 609] |
| 11q14.2 | PICALM | Phosphatidylinositol binding clathrin assembly protein: membrane recycling, autophagy, endosomal vesicle cycling, trafficking of synaptic vesicle proteins | [7, 540, 541, 542, 546, 557, 561, 606, 607, 640] |
| 19q13.2 | PLD3 | Enzyme phospholipase D: catalysis of membrane phospholipids, influence to processing of APP | [19, 540, 542, 557, 558, 568] |
| 7q31.32 | POT1 | Protection of telomeres protein 1: hyper-phosphorylated tau, inflammatory response IL-6, ventricular dilation, cognitive decline | [541, 575] |
| 2p14 | PPP3R1 | Protein phosphatase 3 regulatory subunit B alpha: calcineurin gene, implicated in cytokine release, TLR4 signalling, related to tauopathy | [567, 607, 641, 642] |
| 8p21.2 | PTK2B | Protein tyrosine kinase 2 beta: related to MAPK signalling, ionotropic receptors, neuron migration, synaptic function, LTP in hippocampal CA1 neurons, implicated in tauopathy | [542, 543, 557, 569, 605, 606, 632] |
| 4q12 | REST | RE1-silencing transcription factor: non-autonomous Wnt signalling, neuroprotection against OS and Abeta, autophagy, longevity | [541, 643] |
| Locus | Gene | Function | References |
| 17q25.1 | RHBDF2 | Rhomboid 5 homolog 2: intramembrane serine protease, related to EGF receptor signalling, age-of-onset | [569] |
| 19q13.33 | RPL13A | 60S ribosomal protein L13a: protein synthesis, close to BAX and IRF3 loci, component of the IFNgamma-activated inhibitor of translation (GAIT) complex, repression of inflammatory genes | [569] |
| 14q32.12 | SCL24A4 | Solute carrier family 24 member 4: Na+/Ca++ exchanger, involved in neural development, hypertension | [542, 557, 605, 606] |
| 14q32.13 | SERPINA3 | Alpha-1 antichymotrypsin: pro-inflammatory protein, related to amyloid plaques | [545] |
| 17p13.1 | SERPINF1 | Pigment epithelium-derived factor: serpin F1 gene, neurotrophic functions, related to hypoxic stress, increases gamma-secretase activity, apoptotic signaling through p38 MAPK pathway | [569] |
| 17p13.3 | SERPINF2 | Alpha 2-antiplasmin: serine protease inhibitor, involved in protein degradation, anti-inflammatory | [569] |
| 2q36.2 | SLC19A3 | Solute carrier family 19 member A3: biotin-thiamine transporter, related to HIF1A, hypoxic stress | [577] |
| 14q24.2 | SLC8A3 | Solute carrier family 8 member A3: Na+/Ca++ exchanger, based in intracellular organelle membranes, related to oligodendrocyte maturation, myelination, involved in memory and sensory pathways | [577, 644] |
| 15q22.31 | SNX1 | Sorting nexin 1: regulating the cell-surface expression of EGF receptor, lysosome formation, autophagy | [546, 645] |
| 6q21 | SNX3 | Sorting nexin 3: involved in intracellular trafficking, regulating phagocytosis, interacting with cargo-selective retromer complex, involved in APP processing | [546, 645] |
| 11q24.1 | SORL1 | Sortilin related receptor 1: neuronal LDL receptor, VPS10 receptor, endosomal vesicle cycling, vesicle trafficking, APP pathology | [1, 542, 546, 548-551, 605, 606, 646] |
| 11p15.2 | SPON1 | Spondin 1: related to reelin signalling, involved in axon guidance, neural cell adhesion and neurite extension, formation of anatomical connectivity, involved in Abeta cleavage | [541, 647, 648] |
| 5p15.33 | TERT | Telomerase reverse transcriptase: serving as template for the telomere repeat, chromosome repair, age-of-onset | [203, 649, 650] |
| 19q13.32 | TOMM40 | Translocase of outer mitochondrial membrane 40: outer mitochondrial membrane protein, protein import, mitochondrial dysfunction | [541, 555] |
| 8q22.1 | TP53INP1 | Tumor protein p53 inducible nuclear protein 1: autophagy, caspase signalling | [628] |
| 6p21.1 | TREM2 | Triggering receptor expressed on myeloid cells 2: microglial cytokine signalling, related to TYRO protein, autophagy, Abeta clearance, neuroinflammation, interacting with IL4 and TYROBP | [540-542, 557, 560, 571, 606, 651] |
| 15q22.31 | TRIP4 | Thyroid hormone receptor interactor 4: containing tetrameric nuclear activating signal co-integrator 1 (ASC-1) complex, associating with transcriptional coactivators, related to intrinsic histone acetyltransferase activity | [557, 650] |
| 2p22.3 | TTC27 | Tetratricopeptide repeat domain 27: scaffolding protein-protein interactions, related to myelin formation, white-matter integrity | [652] |
| 19q13.12 | TYROBP | TYRO protein tyrosine kinase binding protein: transmembrane signalling polypeptide, tyrosine-based immunoreceptor, related to cerebral myelination, neuroinflammation, interacting with CD33, MS4A4A, MS4A6A, TREM2, IL4 | [540, 542, 571, 651] |
| Locus | Gene | Function | References |
| 8p12 | WRN | Werner syndrome RecQ like helicase: DEAH family of DNA and RNA helicases, involved in DNA metabolism, transcription, replication, recombination, repair, age-of onset |
[577] |
| 7q22.1 | ZCWPW1 | Zinc finger CW-type and PWWP domain containing 1: early embryonic development, regulating chromatin methylation, regulating epigenetic modification | [8, 542, 557, 605, 606, 611] |
| 6q14.3 | ZNF292 | Zinc finger protein 292: transcription factor, binding GH promoter, steroid receptor, posttranscriptional modifications, brain atrophy, neurodegeneration |
[640] |
| 19q13.42 | ZNF628 | Zinc finger protein 628: transcription regulator | [541, 653] |
| 4q31.21-q31.22 | ZNF827 | Zinc finger protein 827: transcription regulator, related to tauopathy, haplotype block with NANP and NINL | [630] |
Note: Tabulated are gene loci from GWAS, WES, case-control studies, and pathway analysis studies.
Ageing is the single largest known risk in LOAD pathophysiology, but LOAD onset is mainly dependent on premorbid intactness of the cerebral neuropil [5]. Age specifically contributes to LOAD progression due to myelin content loss [29] associated with intra-neuronal agglomeration of hyper-phosphorylated tau protein (h-tau181). Clinical manifestation of open LOAD is preceded by 20 years or more [30] latent prodromal phase, with no or few mnestic signs, but onset of Abeta plaques. A phase with subtle subjective memory problems, called Mild Cognitive Impairment (MCI), specifically in declarative and autobiographic memory systems, is a typical transition period, until with executive disturbances the capability for self-governance is lost. According to clinical literature, MCI and manifest LOAD states may switch forth and back before terminal neurodegeneration fully overrides executive functions. Although Abeta plaque precipitation also occurs within normal ageing, preclinical MCI is associated with more Abeta deposition [31], which is in turn related to global cognition, verbal and arithmetic memory, and executive functioning.
Several other risk factors were identified by epidemiological studies, including a meta-analysis of 18 British cohorts [32], once age effects were excluded:
Sociological: social class, educational attainment, gender, family history, occupation, intellectual activities.
Medical: small birth weight [33], concussion and contusion, traumatic encephalopathy, cerebrovascular diseases, Metabolic Syndrome (MetS) components (e.g. insulin resistance, hypertension, total cholesterol, BMI), sedentary lifestyle.
Nutritional: dietary habits (e.g. cholesterol rich nutrition), vitamin D3 depletion, toxic environmental factors (e.g. metals, electromagnetic fields, traffic proximity) [7, 34-37], oestrogen supplementation [2].
Protective (i.e. reducing risks): higher education with high cognitive function at baseline [3], regular physical exercise, non-steroid anti-inflammatory agents (e.g. aspirin, diclofenac), regular coffee and wine consumption [38].
In summary, several of these factors are strongly pointing towards a role of (early) life stress [7, 35] and of MetS (which is at 38% itself cortisol-level related [39]). In fact, available evidence also suggests that (a) major medical risk factor exposure was present in early adulthood, and (b) that present cognitive peculiarities date back to childhood [30, 33].
Looking at a closer connection of cognitive functioning and psychosocial stresses, the key pathogenic protein tau is a plausible target of Early Life Stress (ELS), because interference in its expression in early ontogeny [40] may leave genomic imprinting effects in neuropil structures increasing late-life LOAD susceptibility [41]. Phosphorylation of tau is regulated by protein kinase N1 (PKN1 19p13.12) and other kinases [42], being disruptive of neuronal microtubule organisation: Hyper-phosphorylation of protein tau produces inclusions in microtubules, later resulting in the formation of paired helicoidal filaments, and in a further stage to assembly of neurofibrillary tangles (NFT). Hyper-phosphorylation and formation of paired helicoidal filaments occurs with all six isoforms of the tau protein. Phosphorylation of tau occurs already during embryonic CNS development, where it is developmentally induced [40], thus making it a likely candidate for ELS effects in ontogeny: abnormalities in microtubule associated h-tau may therefore transport early developmental risks.
Further mechanisms related to brain neuropil involve anatomical alterations as an impact of stress psychophysiology. Hippocampal atrophy is a common consequence of exaggerated glucocorticoid action, being the main structural biomarker in LOAD (as well as e.g. in Post-Traumatic Stress Disorder, PTSD). But also in more subtle ways is chronic psychological stress inducing neuronal oxidative stress and thus accelerating biological ageing [43]. Further clinical arguments indicate a role of distress with high sympathetic arousal states in transition to LOAD. Here, late-life depression, which is discussed as one possible precursor [31] of LOAD, is regularly associated with high anxiety levels. The resulting state of so-called ‘pseudo-dementia’ with mnestic disorder in MCI, linking depression to dementia [44] includes vascular disease, alterations in glucocorticoid steroid levels and hippocampal atrophy, increased deposition of Abeta plaques, inflammatory changes, and depletion of neural growth factors [45]. A two-fold to five-fold increased risk of dementia was found to be associated with late-life depression, whilst another study found that even one additional depressive symptom increased dementia risk by 20% (odds ratios, ORs 1.4~4.6) [45]. Thus, the detrimental complex role of stress to LOAD may be present at several developmental time points.
1.2. Epidemiological Evidence for a Role of Stress in LOAD
Although many clinical researchers and the major scholarly societies now acknowledge roles of stresses in its pathophysiology, a direct linkage between stress and LOAD is still not firmly established [46], partly because of a lack of post-mortem autopsy data pertaining to stress biomarkers. In epidemiology, effect sizes for direct concurrent links in humans are modest, and have best supported inflammatory causations, also by inclusion of other diseases. Still, however, a vast majority of prospective epidemiological evidence demonstrates that an active and socially integrated lifestyle is protective against LOAD [47], which suggests in reverse conclusion negative influences of psychological stresses. Nonetheless, there is classical evidence existing in support of life stress to manifestation of LOAD.
The best-known evidence is the Wilson study of catholic clergy utilising retrospective autobiographic data. Nuns with highest levels of distress measure had 2.7 times the risk of being specifically diagnosed with AD [46], but generally with a 10-fold more rapid cognitive decline [48]. In the Johansson study in Swedish women, life stress escalated Hazard Ratios (HRs) specifically for LOAD in over 35 years from 1.6 through 2.51 [49, 50], and in the Deng five-year prospective study of Chinese, a HR 1.5 was found for personal life adversities on the Folstein MMSE (Mini Mental State Examination, a neuropsychological test discriminative for MCI and AD) [51]. In the normative Leng prospective Norfolk study, there was an OR up to 1.24 for each score unit in self-perceived stress and MMSE scores after ten years [52]. In 77.9% of a Greek risk-sample, incident life stresses were found concrete triggers of clinical LOAD manifestation [53]. Findings in the Helsinki birth cohort study [54] support the notion of alteration of physiological functioning due to ELS particularly with cardiometabolic risk including Type 2 Diabetes Mellitus (T2DM), and also intellectual functioning. As meta-analyses show, T2DM has a specific LOAD-Relative Risk (RR) of 1.46 [55]. The classic Helsinki study found that those subjects with ELS had -0.28 SD units lower verbal ability, -0.13 SD units lower visuospatial ability, -0.18 SD units lower arithmetic ability scores as compared to non-separated with strongest relationship between ELS and lower scores on verbal reasoning [56] (these domains being early cognitive markers of LOAD).
Age as a risk factor is based on alterations in myelin content in white matter (evident as WM lesions) as recent MRI research revealed [57]. The longitudinal data from the Lothian birth cohort further indicated: There are also relations of early cognitive capacities and late-life cortical thickness (GM volume) [58]. The linkage of later white matter MR hyperintensities (indicating WM lesions, myelin vulnerability or myelin breakdown) with LOAD [59, 60], are assumed to be caused by neuropil disintegrities originated in early life, specifically related to myelination cycles in early childhood [57]. Findings from several approaches increasingly support the notion that early regional neuropil dysfunctions may be conducive to late life LOAD susceptibilities [57, 61, 62].
The World Alzheimer Report [4] acknowledges direct influence of ELS in terms of early life adversities, orphanage, divorce, foster care, evacuation, poverty, and other hazards to be related to later AD diagnosis. Having experienced socioeconomic hardship in early life increases the risk to developing MCI, the precursor of LOAD (OR 1.68) [63].
Four major prospective cohort studies include reports of ELS and LOAD:
A. Gothenburg Study with 9-year follow-up: RR 6.3 [64]
B. Cache County population study: OR 2.3 [65]
C. Israel Ischaemic Heart Disease study: ORs 2.15~4.22 [66]
D. Scottish Lothian 1932 birth cohort: threefold risk increase [67]
It is furthermore likely that the majority of ELS conditions are conducive to mid-life appearance of other psychopathological problems and/or increased mortality. Specifically, ELS programming predisposed towards later PTSD in a tenfold order [68] (see other contributions on this topic in the Special Section of this journal). And, further, PTSD increased LOAD risk considerably: PTSD had a 7-year cumulative incident dementia rate of 10.6% [46], as compared to 7% in age-matched controls. And, also, the degree of stress has a measurable impact: AD was increased in veterans who were POWs (HR 1.61) compared to those who suffered PTSD only (HR 1.52) [69]. Taken together all findings, available evidence suggests that ELS and/or later severe psychological life stresses can increase incidence risk for LOAD to a degree of major genetic risks.
1.3. Experimental Evidence for ELS Effects on LOAD
The notion of cognitive reserves as decisive for LOAD manifestation dates back to 1960s [70]. From an epidemiological perspective, Borenstein reviewed several lines of evidence pointing towards intrauterine, perinatal or postnatal factors influencing the early childhood cerebral growth spurt at 94% largely determinative for final brain size and, therefore, for such cognitive reserves [71]. The majority of studies indicated that head circumference (reflecting cerebral myelin content) as index of intracranial volume is predictive of MMSE status and later LOAD, but dependent on genetic risk status, vascular health and educational attainment [71, 72]. The summary of these findings let neuroepidemiologists conclude that those factors in neural differentiation, which are determinative for brain volume [33], are also critical for structural abnormalities in memory networks [73], and so could likely be conducive to late-life LOAD.
The prevalence of adversities leading to Early Life Stress (ELS) is estimated between 11-35% [74, 75]. Beyond dispute is the correlation of ELS with psychiatric morbidity in general: ELS is found in 44.6% of all childhood-onset disorders and with 25.9% to 32.0% in later-onset disorders [76]. The study of prenatal adverse effects is based on the well-known Barker hypothesis of “fetal programming” of diseases. There is furthermore accumulating evidence for a transgenerational transmission of fetal stress programming [77]. Besides adverse foetal programming, the most common ELS results from adverse parental care [78, 79], maternal separation [73, 80] or childhood physical or sexual abuse [81], neglect, or maltreatment [75]. Much of the human research is based on the Early Life Stress Questionnaire (ELSQ) [82], which assesses the occurrence of 17 Adverse Childhood Experiences (ACEs) (e.g. physical abuse, sexual abuse, neglect, family conflict, bullying). However, the core postnatal risk for toxic ELS is inconsistent caretaking [79], whereas, in contrast, attachment security resulting from contingently sensitive parenting is the “critical buffer” for the integrity of the HPA axis [78]. Attachment insecurity, the result of inconsistent caretaking, is associated with heightened sympathetic output, and elevated cortisol levels [83, 84].
Adverse Childhood Experiences (ACE) are well established in attenuating global physiological functioning [85], specifically cardiovascular health [33]. Recent studies have substantiated further epigenetic programming between methylation changes in stress and cardiometabolic candidate genes (Sections 3.1. and 3.2.) for lipid transport ABCA1 (9q31.1), read-through insulin-like growth factor INS-IGF2 (11p15.5), leptin LEP (7q32.1), cortisol converter HSD11B2 (16q22.1), and glucocorticoid receptor NR3C1 (5q31.3) due to early life adverse circumstances, independent of confounds [5]. Intrauterine cortisol excess has been shown to result in low birthweight, which, in turn, has been found a dementia risk [86]. Adverse early socioeconomic (SES) conditions were found to exert hypomethylation of the serotonin transporter (SERT/SLC6A4 17q11.2) resulting in amygdalar hyperreactivity in adolescence [87]. Specifically, glucocorticoid resistance, chronic latent inflammation, increased central corticotropin-releasing hormone (CRH) activity and decreased activity of the protective prosocial neuropeptide oxytocin [75] (an endocrine attachment-related counterweight for glucocorticoids) were found related to ELS. Available evidence also supports further interactions of ELS with nutrition, metabolic hormones, and epigenetic mechanisms [88, 89] therein. Animal studies found ELS CpG modification effects in the promoter of cortisol converter enzyme gene HSD11B2, and those greater in the hypothalamus than the cortex [90]. HSD11B2 modification also showed interaction with sugar, fat and protein diets [91].
1.3.1. Long-term ELS Effects
Review of ELS in animal models reveals that its sequelae result in anxiety/depression behaviours, and memory deficits on the cognitive side [79, 92]. Alterations of gene expression patterns were observed in amygdala, hippocampus, hypothalamus, and frontal cortices. These resulted in amygdalar hyperreactivity, as in PTSD, as well as alterations of neurogenesis [79, 80, 93]. Additionally, long-term dynamic methylation may suppress synaptic plasticity in the hippocampus [94]. Specific long-term sequelae of ELS next to amygdalar hyperreactivity and psychopathology were seen in higher order, complex cognitive abilities, with heteromodal brain regions undergoing protracted postnatal development, which are particularly vulnerable to the deleterious effects of ELS [68] (see below).
1.3.2. Cortisol-related Consequences
Human ELS studies investigated fetal programming effects on hypothalamic-pituitary adrenocortical (HPA) axis-related genes in post-partum placenta as index of embryonic environment. In this approach, adversity effects have been documented for the DNA methylation of the cortisol converter enzyme 11-beta-dehydrogenase 2 (HSD11B2 16q22.1) (Section 3.1.) locus [95, 96]. Maternal stress levels indicated by elevated plasma cortisol levels [97] predicted increased activation of the regulatory promoter region exon 1F of the glucocorticoid receptor (1F NR3C1 5q31.3) (Section 3.1.) [98], and decreased brain derived neurotrophic factor exon IV promoter (BDNF IV 11p14.1) methylation. A further consequence of fetal HSD11B2 modification has been found in programming towards obesity, MetS and T2DM, specifically insulin resistance mediated by gluconeogenesis controller phosphoenolpyruvate carboxykinase (PEPCK) (PCK1 20q13.31) (Section 3.2.) mRNA and activity [99]. The short-term ELS model assumes an impact on stress neurobiology, the emotion appraisal system, and emotion regulation [78], producing a specific vulnerability in the early postnatal years. Short-term effects of ELS on HPA functioning consisted in a lack of or in blunted cortisol secretion in rodent pups [74, 100, 101]. Long-term consequences in human infants to prenatal glucocorticoid exposure consisted in HPA-axis alterations with prolonged cortisol secretion [102]. In rats, chronic ELS resulted in specific amygdalar pro-convulsive cortisol releasing hormone (in humans: CRH1 8q13.1) (Section 3.1) secretion and infantile spasms [103]. In mice, HPA-alterations due to maternal separations result in diminished capabilities for coping of stress in adulthood [101]. ELS associated with later CRH hypersecretion was observed leading to six times greater ACTH in response to external stressors [81].
1.3.3. Brain Structural Growth and LOAD
Recent review on early-life predictors identified factors related to brain growth [33, 104]: early life adversity, early body growth and differentiation, socioeconomic status, to which adult risk factors accumulate to pacing abnormal neurodevelopmental trajectories. Low birthweight and small head circumference, indicators of poor myelination, were found related to later LOAD [4, 33]. Experience-expectant and experience-dependent mechanisms determine early brain maturation with simultaneous neuropil growth spurt, neuron differentiation, migration, synaptic pruning, and selective stabilisation of circuitries [105]. Myelination patterns are genetically triggered cycles following exercise-induced utilisation of effective connectivity [106, 107] in the context of experience-dependent development [108]. In this general frame, studies on structural brain growth in 2-25 month-old infants suggested differences in white matter myelin water fraction and grey-matter volume between APOE epsilon4 carriers and non-carriers [109]. This may suggest that differences in lipoprotein metabolism could influence myelin protein expression and storage of lipid content, and thus increase vulnerability for late-life WM lesions by decreased myelin stability.
Effects on cerebral structures established are amygdalar enlargement, hippocampal volume reduction and of adjacent medial temporal cortex, decreases in volume of orbital-frontal cortex [75]. Besides susceptibility for hippocampal atrophy, ELS is characterised by enlargement of the amygdalae, with blunted reactivity, and clinically, with depression, anxiety, and alexithymia [110]. Also, blunting of reward processing in terms of abnormal ventral striatum [111], and differences in corticostriatal circuitry, have been described with ELS, with lower activity after exposure to early social deprivation [112]. Genome-wide association of ELS through maternal care [113] showed highest correlations with clusters of protocadherins alpha, beta, and gamma involved in synaptogenesis [114], and related to gamma-secretase [115], by DNA methylation, histone acetylation, and transcriptional changes in humans and animals [114]. Accelerated age-related cognitive decline and neurodegeneration can thus be primed by ELS [116]. According to threshold theory [117, 118], LOAD onset depends (a) on previous white matter lesions acquired during life span, and (b) whether a basic reserve of normal functioning can be maintained. LOAD entry, according to this theory, then occurs once plaque deposition finally hampers this memory functioning reserve, as classic findings of Roth and colleagues [70] suggest.
Evidence suggests that, whilst some early effects are reversible in enriched environments, specifically structural hippocampal deficits are likely to persist [119], presumably maintained by profound alterations in the hippocampal neurogenesis structure dentate gyrus [100]. Hippocampal neurogenesis itself has been shown dependent on maternal separation/and or care [120, 121]. Experimental manipulations of high vs. low stress prone rodent pups suggest that ELS experiences have lasting effects also for adult neurogenesis [93, 122]. This would provide an explanation, why adult neurogenesis could not compensate for late-life neurodegenerative processes in ELS individuals.
1.3.4. Neurotrophic and Growth Factors
Next to glucocorticoid action, neurotrophin signalling pathways critical for neuronal differentiation, dendrite outgrowth, axon guidance, and synaptic integrity [123], are subject to ELS. Glucocorticoid and neurotrophin pathways converge specifically in the hippocampus, harming BDNF and its receptor, tropomyosin-related kinase receptor B (TrkB) (NTRK2 9q21.33) [124] expression. The majority of ELS paradigms (e.g. maternal separation, restraint, cold shock) report decreases in BDNF levels, and concomitant decreases in p-GSK-3beta, p-ERK1, p-ERK2 in the hippocampus, and overexpression of stress response protein dual specificity phosphatase 1 (DUSP1 5q35.1) [123]. The MAPK pathway is considered the linking mechanism between glucocorticoid and BDNF systems, where over-activation of DUSP1 is inhibitive of neural differentiation and suppressing axonal outgrowth [123]. But there is also a direct effect of BDNF signalling on the GRs, where BDNF via its receptor TrkB enhanced the transcriptional activity of a synthetic GR reporter [124]. BDNF signalling thus modulates the effect of GRs on gene expression in primary neurons by modulating NC3R1 posttranslational modifications through phosphorylation [124]. Other ELS studies showed a down-regulation of Bdnf expression, associated with hypermethylation of CpG sites in the gene regulatory region, whereby epigenetic changes were found within the CREB-binding region (CpG1B) [125] There has also a relation of ELS been observed, in terms of restraint stress, with an enhancement of BDNF promoter activity in hippocampal cornu ammonis subfield CA3, based on transcription co-activator CREBBP (CREBBP 16q13.3) signalling triggered through ERK1/2, dependent on glucocorticoid receptor activation [126]. BDNF depletion is therefore considered a biomarker of LOAD, since progression from MCI towards LOAD is accompanied with a significant decline in peripheral baseline BDNF levels [127], which then persists.
1.3.5. ELS Animal Models for LOAD
Of the three transgenic strains of murine models typically used in Alzheimer research (Tg2576, APPswe/PS1dE9, and 3xTg-AD) [7, 128], the preferred animal model for the isolation stress ELS paradigm is the Tg2576 mouse model of AD. This ELS paradigm resulted in an 59% increase of soluble Abeta40 and Abeta42 [7], accompanied by hippocampal and neocortical neuritic plaques. Moreover, this stress exposure paradigm caused a rise in basal plasma corticosterone levels, paralleled with an increased expression of the Gr and Crh [7]. Later adaptations were, however, reported resulting in reduced gluco- and mineralcorticoid receptor expression [129]. Animal evidence suggests that lacking maternal caregiving behaviours increase Gr expression in the offspring [114] via increased hippocampal serotonergic tone accompanied by increased histone acetylase transferase activity, histone acetylation and DNA demethylation mediated by the early growth response protein EGR-1/NGFI-A (EGR1 5q31.2) [130]. In a murine model of ELS by Crh (CRH1 8q13.1) overexpression, it was observed that MCI-analogous memory problems develop already in midlife [131]: These were caused by hippocampal tau phosphorylation based on activation of the type-1 corticotropin-releasing factor receptor (CRHR1 17q21.31) (Section 3.1.) via increased GSK-3beta activation. Furthermore, the mitogen-activated protein kinases (MAPKs) (Section 2.1.) p38 MAPK, CDK5 activator protein, p35, and ERK1/2 kinases, and also JNK [154], were robustly upregulated.
ELS murine studies pertaining to LOAD were pioneered by work of Nasser Zawia and co-workers, who found in the context of early lead exposure that early toxic impact bears consequences in structural growth in cortex, cerebellum, and hippocampus [132], mediated by EGR-1/NGFI-A and NGF altering synapsin gene expression. With respect to neonatal environmental impact, it was observed that toxic exposure transiently stimulated APP overexpression [133], which later returned in aged animals after a latency period [134]. These early insights were seminal to trigger the assumption of gene expression and regulation abnormalities in LOAD [135]. It was found that oxidative damage introduced by environmental influences during brain development inhibited DNA methyltransferases, thus altering methylation profiles in promoters of LOAD core pathology genes [136, 137]. Stress generally induces tau hyper-phosphorylation [138], for which a structural time course has been observed in the dentate gyrus neurons, wandering from somata to dendrites. In this context, it had been suggested that repeated stresses induce a dysregulation [139] of protein tau through phosphorylation of protein kinase GSK-3beta (Section 2.1) and of calmodulin kinase (CaMKII) (Section 2.1) (CAMK2A 5q32) during memory encoding in hippocampus and amygdala. It is furthermore suggested that, in later life, oxidative stresses in form of protein oxidation and lipid peroxidation then accelerate Abeta deposition, tau phosphorylation, and gliosis [140], leading to insults that cause memory dysfunctions. In addition, metabolic stresses contributed to vascular inflammation, astrocyte reactivity, and cerebral glucose metabolism in APP/PS1 mice [141].
Maternal stress proneness can be transmitted by methylome through gametic programming to the offspring [142], thus providing a stress diathesis phenotype. Furthermore, even mild stresses are conducive to programming of a LOAD phenotype [143]. The type of memory deficits induced by ELS was particularly found in recognition memory [73], as mediated by prefrontal, hippocampal and perirhinal oscillatory electrophysiological coupling. Differential results for Abeta deposition in the hippocampus [144] and amygdalar and MFC dendritic structure, but not for other cortices, were seen for ELS vs. caringly handled animals. ELS animals developed LOAD pathology earlier and fewer life expectancies. In contrast, increased levels of maternal care during the early life period delayed Abeta deposition and cognitive decline in the APPswe LOAD mouse model, involving the hippocampus, but not the amygdala [145]. Maternal separation studies yielded evidence that early and later stresses mainly alter cognitive function in memory tasks, with underlying impairment of neurogenesis in the hippocampal dentate gyrus [120]. This was accompanied by early rises, but long-term depletions, of Brain-Derived Neurotrophic Factor (BDNF), TrkB, Insulin-like Growth Factor-1 (IGF-1), and type 1 IGF receptor (IGF-1R) ligands and receptors [146]. Decreased Bdnf expression was found related to both increased histone acetylation and methylation of H3K9 in cortical neurons [147], as compared to wild-type animal neurons, where epigenomic modifications tend to decrease over age.
Lesion studies in maternal separation rats implied greater impairment in the cholinergic system [148], along with alterations of the HPA in increased CRF mRNA expression and a reduction of GR densities. Decreases in NGF and increases in NGF-p75NTR expression were correlated with memory impairments and learning deficits. The impairments in HPA feedback signalling involved both noradrenergic and glucocorticoids [80, 149], particularly mediated by hypothalamic PVN, and resulting in HPA hypersensitivity. HPA axis is activated in 3xTG-AD mice from early life [150], with exaggerated mRNA levels of Mr and Gr in the hippocampus, Gr and Crh in the PVN, Gr and Crh in the central nucleus of the amygdala, and Crh in the bed nucleus of the stria terminalis. This suggests that LOAD neuropathology is intrinsically connected with central HPA activation and neuroendocrine dysregulation. Related to HPA bias towards exaggerated reactivity reflected in glucocorticoid levels, c-fos expression was found enhanced [151]. This neuronal activity marker interacts with c-jun/AP-1 transcription markers (FOS 14q24.3), associated with the JNK signalling pathway [152]. The HPA bias and glucocorticoid levels interacted with the brain insulin system, inducing phosphorylation of insulin receptors, and so modifying downstream signalling of p-Akt, p-GSK-3beta, p-tau, and p-ERK1 [153], resulting in a shift towards amyloidogenic processing. The activation of JNK signalling pathway, upregulating BACE1 expression with increased Abeta levels, was shown directly related to glucocorticoid increases, and mediated by hypomethylation of the CpG7 site of the BACE promoter [154].
1.3.6. ELS and Tau Pathophysiology
There is a clear direct link of stress response to the LOAD pathophysiological mechanism of tau phosphorylation. Murine models lacking one or both corticoid releasing hormone receptors CRHR1 (17q21.31) and CRHR2 (7p14.3) (Section 3.1.) receptors enabled the demonstration that tau hyper-phosphorylation specifically in the hippocampus, is dependent on CRHR1 and CRHR2 signalling [155]. CRH expression in the paraventricular nucleus [77] of the hypothalamus (PVN) is attenuated by ELS [156], mainly by cAMP response element (CRE), transcription factor AP-1, and Glucocorticoid Response Element (GRE) (Section 3.1.). In addition, methyl CpG-binding protein 2 (MeCP2) [96] and a functional restrictive silencing sequence of the element-1/neuron restrictive silencing element (RE-1/NRSE) (REST 4q12) connect to a sequence on the CRH1 locus, which regulates CRH expression [156]. Other stress hormones than glucocorticoids have also been linked to inducing tau hyper-phosphorylation [86] (Sections 1.4. and 3.1.).
ELS related gene methylation changes have been observed in the hippocampus, but not in other cerebral regions. Szyf has hypothesised that three mechanisms are in specific pertaining to transport of epigenomic changes responsible for life-long ELS effects in animals in induced by ELS-neuronal activation: (a) Neuronal activity leads the calmodulin kinase II (CamKII, above) into changing the affinity of MeCP2 by phosphorylation, and thus induces demethylation of BDNF promoter [157]. (b) Furthermore, the methyl-CpG binding protein 2 MeCP2 (MECP2 Xq28) has been found regulating the expression of arginine vasopressin (AVP) neurons in the hypothalamic PVN [158] following ELS. This process has been identified being responsible for the loss of noradrenergic neurons in the locus caeruleus (Section 3.1.) [159]. (c) The interaction of the AVP (20p13) promoter with MeCP2 and CamKII induces hypomethylation of the AVP gene [157]. EGR-1/NGFI-A is likely to induce histone acetylation triggered by CREB binding protein CREBBP (CREBBP 16p13.3), and the methyl-CpG domain binding protein 2 MBD2 (MBD2 18q21.2) facilitates demethylation of NC3R1 exon 17 promoter [157] (see below). Other loci such as the human GABAA receptor (GABRA1 5q34) promoter within the frontopolar cortex and the tropomyosin-related kinase B receptor TRKB (for BDNF) have, however, not yet been investigated for ELS effects [157]. The hypophysiotropic stress neuropeptide pituitary adenylatecyclase-activating polypeptide (ADCYAP1 18p11.32) and its receptor PAC1 [160, 114] playing a role in PTSD was found to exert neuroprotective effects in murine AD models by supporting BDNF signalling [161].
1.3.7. ELS and Methylation Findings
A meta-analysis of human ELS studies confirmed methylation of five CpG sites preceding promoter region exon 1F NR3C1 coding for GR in children, here resulting from maternal chronic psychosocial stress as experienced during pregnancy [162]. Animal studies have demonstrated that there are causal relations amongst epigenomic states based on DNA methylation, early growth response protein EGR-1 (an ERK transcription factor) binding, GR expression and hypothalamic-pituitary-adrenal (HPA) responses to stress in the offspring [163]. Environmental cues become effective on methylation by means of transcription factors [164], and for maternal care this is early growth response protein EGR-1/ Nerve Growth Factor-Inducible Protein A (NGFI-A) [163, 165]. EGR1 initiates, by means of serotoninergic activation, and as a ligand to the promoter on exon 1F for the GR locus NC3R, the transcription of the latter [74, 165]. Involved in this mechanism are transcription factors TFIID complex (TBP 6q27), and coactivator CREBBP (see above) entailing histone acetyltransferase activity [74, 157]. ELS is generally mediated by the methyltransferases: Activity of DNA methyltransferase 1 (DNMT1 19q13.2), usually responsible for methylation stability, results in DNA demethylation in post-mitotic neurons [113]. De novo methyltransferases DNMT3A (2p23.2) and DNMT3B (20q11.21) induce methylation, also in mature neurons [157]. DNMT3A was found responsible for maintenance of cognitive abilities during ageing [166]. Activity of these methyltransferases and of the MBD2 demethylase [167] was observed in LOAD animal models in methylation of the PSEN1 locus.
1.3.8. ELS and Adult Neurogenesis
It is currently concluded [168] that environmental influences in early neurogenesis are dynamic, and change over time, and also that their relations with adult neurogenesis are still unclear. ELS in terms of maternal neglect has been found to increase the methylation of EGR-1-binding sequences, causing lower expression of EGR-1 target genes later in adult life on CpGs overlapping with the EGR1(NGFI-A)/Zif268/Krox-24-binding sequence that is positioned close to the putative binding sites of neurogenins (NEUROG1 5q23-31, NEUROG2 4q25, NEUROG3 10q21.3) [169, 39], the transcription factors involved in specifying neuronal differentiation. Current views on ageing assume methylation relaxation effects due to losses in chromatin modifiers resulting in increasing genome instability [170]. In the brain, however, this effect is assumed to be dependent of neuronal activity, mediated by Wnt signal transduction. Wnt signalling globally mediates external environmental cues, by means of signal transduction in the GSK-3beta pathway, and is crucial for both embryonal and adult neurogenesis stimulated by astrocytes (with Wnt3, Wnt10b, and Wnt2), involving GSK-3beta and beta-catenin participating in the Wnt signalling pathway. Rats exposed to prenatal stress had accelerated, age-related decline in spatial and working memory [171], however, no primate data are available on this subject. Furthermore, both human and animal studies suggest in summary, that early stress can also improve cognitive functioning. However, if there is a deprivation of early maternal care, effects show impaired juvenile neurogenesis and increased adult apoptosis [172].
Epigenetic changes throughout ageing has been studied in a variety of species [173], indicating either globally decreasing or locally increasing methylation patterns. However, only two studies have yet focused on cerebral structures, and therefore, no universally reliable information is present to date. ELS may have direct effects on ageing as indexed by shorter telomere length in adulthood in leukocytes thus predisposing to generally poorer health outcome [174, 175]. Telomere length shortening has been observed in CVD, T2DM, MetS [176], and all dementias [175], in this context.
1.4. Core Processes in LOAD Pathophysiology
Aetiological mechanisms in LOAD remain unexplained in exact detail, as most experts agree, but the basic neuropathological features in the AD brain currently accepted are (a) widespread axosomatic and neuritic extracellular amyloid plaques leading to dystrophic neurites, (b) intracellular neurofibrillary tangles (NFTs) consisting of previously soluble tau proteins, (c) reduced cytochrome c oxidase (COX) activity in mitochondria, (d) reactive microgliosis, (e) impairment in autophagy related to neurons, and (f) oxidative stress combined with abnormal neuroendocrine signalling (e.g. acetylcholine, BDNF). These hallmarks eventually terminate in synapse atrophy, neuron loss, and widening of pre-existing white matter disintegrities.
1.4.1. APP Cleavage and Abeta Deposition
In contrast to presenile, familial AD, there is typically no genotypic APP pathology present in LOAD [177], and also no PSEN mutations (Section 1.1.). Normally, mature APP (whose exact physiological function remains unknown) becomes cleaved successively by alpha- or beta-, and then gamma-secretases in the late protein secretory pathway and the plasma membrane. The still immature APP is stored in the early secretory pathway such as Endoplasmic Reticulum (ER) or Golgi apparatus, where for imAPP no cleavages take place [178]. Whereas alpha-secretase cleavage does not result in amyloid pathogenic for LOAD, there are two relevant aberrant cleavage steps: (a) beta-site amyloid precursor protein cleaving enzyme 1 (BACE1 11q23.3) [179, 180], (b) gamma secretase (triggered by the gamma secretase activating protein pigeon homolog, PION 7q11.23) [8], and (c) through mediation of delta-opioid receptor [10]: The extracellular APP-cleavage through BACE1 produces the soluble cell membrane-bound fragment C99. Gamma-secretase cuts the transmembrane domain of C99, thus releasing intracellular Abeta. Dependently on three possible cleavage sites, where the enzyme gamma secretase acts on the cleavage products of beta-secretase, are the products either Abeta38 through Abeta43 [181] isoforms, dependent on additional 15 known mutations [35, 180]. The insoluble isoform Abeta42 constitutes the extracellular precipitate on neuron somata and axons. Extracellular Abeta42 neuritic plaque deposition, then, occurs only at late neuropathological Braak amyloid-stage C, once the intracellular Abeta has caused neuron leakage and atrophy [182]. Soluble Abeta40, in contrast, will deposit earlier in arterioles resulting into vascular angiopathy [183]. Oligomeric Abeta40-42 is likely to spread from one neuron to another in similarity to prion diseases [184], which is promoted by impaired autophagy and clearance. Gamma-secretase is specifically activated by pro-inflammatory interferon-gamma, IL-1beta, or TNFalpha, and cytokine-induced gamma-secretase activity was found to be blocked by a JNK (Section 2.4.) inhibitor [185]. Animal evidence suggests that childhood physical exercise normalizes APP physiology, and in reverse, that early sedentary lifestyle may foster early Abeta deposition [186]. Recent prospective PET-imaging in MCI patients has recently supported the assumption that it is the cerebral Abeta deposit in basal and mesial temporal, orbitofrontal, and anterior and posterior cingulate cortices interacting then with total CSF p-tau [187].
1.4.2. Abeta Cascade
LOAD is generally assumed to be the result of a cascading process starting with stronger Abeta deposition (which also occurs in “normal” ageing, but to lesser amounts), with the accession of further pathogenic elements such as tauopathy, mitochondrial dysfunction, and microgliosis (Section 3.3.). Abeta is matured in the Golgi, ER and endosomal/lysosomal system [188], and in the secretory pathway, were beta- and gamma-secretases are to originate neurotoxic Abeta42 and Abeta40 monomers (the amyloidogenic pathway), which subsequently polymerise into oligomers, and finally aggregate into amyloid plaque aggregates. However, the quantity of Abeta42 is not related to cognitive decline [35] per se, as its aggregation also occurs in healthy ageing, and in 20-40% post-mortem dissected cases to a degree also seen in AD, but without respective pathognomonic memory impairment present ante-mortem.
The prevailing theory, that only neurons carry the pathogenic agents Abeta42 and NFT, had to be extended by findings of involvement of astrocytes and oligodendrocytes [189, 190]. Recent studies have shown that Abeta42 oligomers are produced by cooperative activities of both neurons and its associated astrocytes [190]. In this context, are adjacent myelin-producing oligodendrocytes particularly vulnerable to h-tau181 and Abeta42 [29]. Activation of alerted microglia (Section 3.3.) results in production and release of pro-inflammatory cytokines, including IL-1beta, IL-6, TNFalpha, and IFNgamma [191]. In turn, these cytokines stimulate the nearby astrocyte-neuron pairs to produce further amounts of Abeta42 oligomers, thus activating even more Abeta42 production and dispersal [190]. By means of exocytosis of oligomeric Abeta42 (Section 3.3.), associated astrocytes and oligodendrocytes can be infected, and in turn become themselves producer cells of Abeta and tau oligomers [190].
Unclear also remains the actual Abeta42 residue induction in LOAD, but it is believed to be triggered from Reactive Oxidative Species (ROS) (see below). OS may result from several cellular processes such as ageing, hypoxia (mitochondrial dysfunction, HIF-1alpha), hyperglycaemia (advanced glycation endproducts, AGEs; Section 3.2.), NFkappaB), and hypercholesterolemia (oxysterols, sterol regulatory elements, SREBPs activation) [192]. Accordingly, it is the incidence of known risk factors for LOAD (Section 1.1.), which are ultimately inductive of gamma-secretase and BACE1 activation through JNK/DUSP signalling. Evidence of lipid peroxidation and protein peroxidation appears very early in LOAD pathogenesis (corresponding to Braak tau-stages I and II) [192]. It has been shown recently, that cellular cholesterol transduction is associated with APP mis-cleavage by gamma-secretase, so increasing Abeta42 deposition [193]. In specific, cholesterol sequestration is related to an impairment of lyosomal autophagy, with Abeta accumulation leading to oxidative stress and vulnerability for H202 toxicity. Abeta42 reduces neuron-specific endophilin-B1 (SH3GLB1 1q22), a protein involved in beclin-2 mediated apoptosis, autophagy and mitochondrial function, which in turn enhances Abeta42 accumulation and neuronal vulnerability to stress [194].
1.4.3. Tauopathy in LOAD
Protein tau is a physiological stabiliser of microtubuli in neuron cells, and hence present very early in ontogeny. Besides its role in cytoskeleton formation and functioning, a second neurophysiological role of tau has been detected in neuron nuclei [195]: In healthy neurons, tau has a function in posttranslational histone modification (associated with heterochromatin proteins H3K9me and HP1alpha), resulting in dense chromatin packaging, which is lost under stress conditions in LOAD brains, resulting in disordered heterochromatin organisation [195]. Tau is therefore also itself directly involved in epigenetic modification, although a tau gene (MAPT 17q21.31) alternate splicing variant is present in, but not specific to LOAD: The haplotype H1-specific expression of MAPT exon 10 [196] is active also in other tauopathies, and caused no signal in LOAD GWAS [197]; however, supported has been an epigenetic abnormal hypermethylation of MAPT in post-mortem LOAD neurons [198] (Section 4.3.). (Table 2).
Table 2.
Epigenetic modification findings in risk genes for late-onset Alzheimer disease.
| Locus | Gene | Findings | References |
|---|---|---|---|
| 12p13.31 | A2M | negative | - |
| 9q31.3 | ABCA1 | negative | - |
| 19p13.3 | ABCA7 | ABCA7 methylation was associated with paired helicoidal filament tau tangle density. ABCA7 methylation was associated with density of neuritic plaques. Index SNP (rs3764650) was associated with neuritic plaque burden, no association with the level of methylation at cg02308560 | [26, 569, 787] |
| 21q22.3 | ABCG1 | negative for LOAD, confirmed for CHD | - |
| 15q23.3 | ADAM10 | negative | - |
| 18p11.32 | ADCYAP1 | negative | - |
| 10q25.2 | ADRA2A | negative | - |
| 10q25.3 | ADRB1 | negative | - |
| 5q31-32 | ADRB2 | negative | - |
| 8p11.23 | ADRB3 | negative | - |
| 6p21.32 | AGER | negative | - |
| 10q11.21 | ALOX5 | Consistent reduction in DNA methylation at 5-LOX gene promoter in LOAD | [788] |
| 8p11.21 | ANK1 | Methylation level at cg11823178 associated with LOAD neuropathology | [569] |
| 4p14-p13 | APBB2 | negative | - |
| 19q13.32 | APOE | APOE CGI is differentially demethylated in LOAD post-mortem probes | [789] |
| 21q21.2 | APP | Aberrant CpG methylation in non APOE epsilon4 carriers | [198] |
| 8q24.3 | ARC | negative | - |
| 12q23.2 | ASCL1/MASH1 | negative | - |
| 9q33.1 | ASTN2 | negative | - |
| 22q13.1 | ATF4 | negative | - |
| 17q25.1 | ATP5H/KCTD2 | negative | - |
| 20p13 | AVP | negative | - |
| 11q23.3 | BACE1 | negative in vivo | - |
| 19q13.33 | BAX | negative | - |
| 3q26.1 | BCHE | negative | - |
| 11p14.1 | BDNF | Hypo- and hypermethylation of CpG islands BDNF promoter region. Methylation in four CpG sites in the promoter of the BDNF were elevated, and correlated negatively with APOE, glucose level, positively with CRP in peripheral blood. Significantly increased hippocampal HDAC2 relates to promoter region of BDNF exon VI; contributing to the histone H3 deacetylation and BDNF suppression in hippocampal CA1. Aberrations in histone acetylation related to ApoE epsilon4 increase nuclear translocation of HDACs in neurons, thereby reducing BDNF exon IV expression, mediated by LRP1, specifically in hippocampus | [790-793] |
| 17q21.31 | BECN1 | negative | - |
| 2q14.3 | BIN1 | Two CpG sites showed associations with LOAD, three with Abeta, and five with NFTs. Association of cg22883290 in the BIN1 with LOAD | [26, 569] |
| 19q13.32 | BLOC1S3 | negative | - |
| Locus | Gene | Findings | References |
| 5q32 | CAMK2A | negative | - |
| 22q13.1 | CARD10 | negative | - |
| 20q13.31 | CASS4 | negative | - |
| 17q12 | CCL2 | negative | - |
| 17q12 | CCL3 | negative | - |
| 17q4 | CCL4 | negative | - |
| 6p12.3 | CD2AP | negative | - |
| 19q13.41 | CD33 | negative | - |
| 10q23.1 | CDH23 | Altered methylation at cg23968456, but unclear if not confounded by enhanced astrocyte activation indicated by GFAD expression | [569] |
| 10q21.2 | CDK1 | negative | - |
| 7q36.1 | CDK5 | negative | - |
| 11p11.2 | CELF1 | Methylation change not directly in loco | [794] |
| 16q13 | CETP | negative | - |
| 1q31.1 | CFHR1 | negative | - |
| 8p21.1 | CLU | negative | - |
| 17q21.2 | CNP | Reduced cortical and allocortical expression of CNP RNA not attributable to DNA methylation at CNP promoter | [672] |
| 22q11.2 | COMT | negative | - |
| 1q32.2 | CR1 | negative | - |
| 2q33.3 | CREB1 | Hypermethylation of DUSP22 inhibiting PKA and CREB1 activity, thereby determining tau phosphorylation status | [795] |
| 9p13.3 | CREB3 | negative | - |
| 11p11.2 | CREB3L1 | negative | - |
| 7q33 | CREB3L2 | negative | - |
| 19p13.3 | CREB3L3 | negative | - |
| 1q21.2 | CREB3L4 | negative | - |
| 7p15.1-p14.3 | CREB5 | negative | - |
| 16p13.3 | CREBBP | Hyper- and hypomethylated CpG islands in promoter regions for CREBBP | [793] |
| 8q13.1 | CRH1 | negative | - |
| 5q13.3 | CRHBP | negative | - |
| 17q21.31 | CRHR1 | negative | - |
| 7p14.3 | CRHR2 | negative | - |
| 1q23.2 | CRP | Methylation in four CpG sites in the promoter of the BDNF were elevated, and correlated negatively with APOE, glucose level, positively with CRP in peripheral blood | [790] |
| 18q21.1 | CTIF | negative | - |
| 16q21 | CX3CL1 | negative | - |
| 3p22.2 | CX3CR1 | negative | - |
| 15q21.1 | CYP19A1 | negative | - |
| Locus | Gene | Findings | References |
| 14q32.2 | CYP46A1 | negative | - |
| 9q34.2 | DBH | negative | - |
| 16p12.1 | DCTN5 | negative | - |
| 1q23.3 | DDR2 | negative | - |
| 1p23.3 | DHCR24 | Study identified GC-rich element in the DHCR24 promoter, which was shown to determine DHCR24 expression levels, also includingnacetylation of histones H3 and H4 to the enhancer region | [796] |
| 21q22.3 | DIP2A | Altered methylation at cg00621289 related to LOAD, interrelation with SORL1 and PLD3 | [569] |
| 17p13.1 | DLG4 | negative | - |
| 19q13.2 | DNMT1 | General DNA hypermethylation in LOAD by higher DNMT1 expression. Interindividual variation in DNMT1 modification | [597, 788] |
| 2p23.2 | DNMT3A | Tested but no LOAD-specific results reported | [797] |
| 20q11.21 | DNMT3B | Tested but no LOAD-specific results reported | [797] |
| 18q12.1 | DSG2 | negative | - |
| 5q35.1 | DUSP1 | negative | - |
| 21q22.13 | DYRK1A | negative | - |
| 11p13 | EAAT2/SLC1A2 | negative | - |
| 1p36.12 | ECE-1b promoter | negative | - |
| 6p24.1 | EDN1 | negative | - |
| 4q31.22-23 | EDNRA | negative | - |
| 13q22.3 | EDNRB | negative | - |
| 5q31.2 | EGR1 | H4 acetylation at EGR1 and FOS promoters regulated by APP | [798] |
| 2p22.2 | EIF2AK2 | negative | - |
| 15q15.1 | EIF2AK4 | negative | - |
| 14q23.3 | EIF2S1 | negative | - |
| 18q12.2-q21.1 | EPG5 | negative | - |
| 7q34-35 | EPHA1 | negative | - |
| 2q33.3 | FASTKD2 | negative | - |
| 14q22.1 | FERMT2 | negative | - |
| 12p13.33 | FKBP4 | negative | - |
| 6p21.31 | FKBP5 | FKBP51 levels increased relative to age and LOAD, corresponding with demethylation of the regulatory regions in the FKBP5 gene. Higher FKBP51 levels were associated with LOAD progression. Age-associated increases in FKBP51 levels show interaction with Hsp90 promoting neurotoxic NFT accumulation | [686] |
| 14q24.3 | FOS | H4 acetylation at EGR1 and FOS promoters | [798] |
| 11q14.1 | GAB2 | negative | - |
| 6p21.3 | GABBR1 | negative | - |
| 9q22.33 | GABBR2 | negative | - |
| 5q34 | GABRA1 | Tested but no LOAD-specific results reported | [797] |
| Locus | Gene | Findings | References |
| 2q31.2 | GAD1 | Tested but no LOAD-specific results reported | [797] |
| 10p12.1 | GAD65 | negative | - |
| 5p13.2 | GDNF/GDNFOS | negative | - |
| 9p24.2 | GLIS3 | negative | - |
| 1p35.3 | GMEB1 | negative | - |
| 20q13.33 | GMEB2 | negative | - |
| 3q28 | GMNC | negative | - |
| 12p13.31 | GNB3 | negative | - |
| 5q33.1 | GRIA1 | negative | - |
| 4q32.1 | GRIA2 | negative | - |
| Xq25 | GRIA3 | negative | - |
| 11q22.3 | GRIA4 | negative | - |
| 12p12 | GRIN2B | negative | - |
| 6q24.3 | GRM1 | negative | - |
| 11q14.2-3 | GRM5 | negative | - |
| 17q21.31 | GRN | negative | - |
| 19q13.2 | GSK3A | negative | - |
| 3q13.33 | GSK3B | Aberrant CpG methylation GSK3B in LOAD non APOE epsilon4 carriers. PI3K/Akt/GSK-3beta inhibits leucine-309 demethylation of protein phosphatase-2A modulating phosphorylation of tau | [198, 799-801] |
| 2q31.1 | HAT1 | negative | - |
| 6q21 | HDAC2 | negative | - |
| 7p21.1 | HDAC9 | negative | - |
| 14q23.2 | HIF1A | negative | - |
| 7q34 | HIPK2 | negative | - |
| 6p21.31 | HLA-DRB5/DRB1 | Methylation in cg17606183 was associated with Abeta load (3 CpG sites) and NFT density (9 CpG sites) | [26] |
| 13q12.3 | HMGB1 | negative | - |
| 5q13.3 | HMGCR | negative | - |
| 20q13.12 | HNF4A | negative | - |
| 1q32-q41 | HSD11B1 | negative | - |
| 16q22.1 | HSD11B2 | negative | - |
| 6q21.31 | HSPA1A | negative | - |
| 12p12.1 | IAPP | negative | - |
| 12q23.2 | IGF1 | negative | - |
| 15q26.3 | IGF1R | negative | - |
| 11p15.5 | IGF2 | negative | - |
| 6q25.3 | IGF2R | negative | - |
| 7p12.3 | IGFBP1 | negative | - |
| Locus | Gene | Findings | References |
| 14q32.33 | IGHV1-67 | negative | - |
| 1q32.1 | IL10 | negative | - |
| 3q25.33 | IL12A | negative | - |
| 11q23.1 | IL18 | negative | - |
| 2q14.1 | IL1A | negative | - |
| 2q14.1 | IL1B | negative | - |
| 9p24.1 | IL33 | negative | - |
| 5q31.1 | IL4 | negative | - |
| 7p15.3 | IL6 | negative | - |
| 2q37.1 | INPP5D | negative | - |
| 11p15.5 | INS | negative | - |
| 11p15.5 | INS-IGF2 | negative | - |
| 19p13.2 | INSR | negative | - |
| 1q23.1 | INSRR | negative | - |
| 19q13.33 | IRF3 | negative | - |
| 2q36.3 | IRS1 | negative | - |
| 13q34 | IRS2 | negative | - |
| 17q21.31 | KANSL1 | negative | - |
| 12q24.31 | KDM2B | negative | - |
| 9q34.12 | LAMC3 | negative | - |
| 7q32.1 | LEP | negative | - |
| 12q13.3 | LRP1/APOER | negative | - |
| 2q31.1 | LRP2 | negative | - |
| 12q12 | LRRK2 | negative | - |
| 11p11.2 | MADD | negative | - |
| 19q13.12 | MAG | negative | - |
| 2q21.1 | MANI | negative | - |
| 5q13.2 | MAP1B | negative | - |
| 16q24.2 | MAP1LC3A/B | negative | - |
| 6q23.3 | MAP3K5 | negative | - |
| 22q11.22 | MAPK1 | negative | - |
| 22q13.11 | MAPK11 | negative | - |
| 22q13.3 | MAPK12 | negative | - |
| 6p21 | MAPK13 | negative | - |
| 6p21.31 | MAPK14 | negative | - |
| 16p11.2 | MAPK3 | negative | - |
| 10q11.22 | MAPK8 | negative | - |
| 11p11.2 | MAPK8IP1 | negative | - |
| Locus | Gene | Findings | References |
| 16p13.3 | MAPK8IP3 | negative | - |
| 5q35.3 | MAPK9 | negative | - |
| 17q21.31 | MAPT | Aberrant CpG methylation in APOE epsilon4 non-carriers. MMSE performance correlated with methylation level. Hypoacetylation of KXGS motifs enables phosphorylation of tau, reversed by HDAC6. Total histone H3 and total histone H4 protein levels significantly increased in post-mortem LOAD temporal cortices | [198, 802-804] |
| 18q21.2 | MBD2 | In entorhinal cortex layer II, DNMT1, MBD2, and p66 with rRNA was nearly absent: loss of methylation markers and methylation stabilisers in LOAD neurons may cause deficits in rRNA gene expression, rRNAs, ribosomes, and ribosomal protein synthesis | [805] |
| 18q23 | MBP | negative | - |
| 18p11.2 | MC2R | negative | - |
| Xq28 | MECP2 | negative | - |
| 5q14.3 | MEF2C | negative | - |
| 22q13.1 | MGAT3 | negative | - |
| 3p25.2 | MME/NEP | negative | - |
| 6p22.1 | MOG | negative | - |
| 7q22 | MPO | negative | - |
| 11q12.2 | MS4A4/MS4A6E | negative | - |
| 8p22 | MSR1 | negative | - |
| 1p36.22 | MTHFR | MTHFR showed methylation levels ranging from 5% to 75% in LOAD, expressing large interindividual variation | [597, 806] |
| 11q12.2 | MYRF | negative | - |
| 20q13.33 | MYT1 | negative | - |
| 20p11.21 | NANP | negative | - |
| 17q21.1 | NBR1 | negative | - |
| 8q13.3 | NCOA2 | negative | - |
| 16q12.2 | NET/SLC6A2 | negative | - |
| 5q23-31 | NEUROG1 | negative | - |
| 4q25 | NEUROG2 | negative | - |
| 10q21.3 | NEUROG3 | negative | - |
| 4q24 | NFKB1 | Hyper- and hypomethylated CpG islands in promoter regions for NFkappaB frontal cortex | [793] |
| 10q24.32 | NFKB2 | negative | - |
| 14q13.3 | NFKBIA | negative | - |
| 2q37.1 | NGEF | negative | - |
| 20p11.21 | NINL | negative | - |
| 17p13.2 | NLRP1 | negative | - |
| 1q44 | NLRP3 | negative | - |
| 7p14.1 | NME8 | negative | - |
| 7q36.1 | NOS3 | negative | - |
| Locus | Gene | Findings | References |
| 9q34.3 | NOTCH1 | negative | - |
| 4q32.2 | NPY1R | negative | - |
| 5q31.3 | NR3C1 | negative | - |
| 9q33.3 | NR5A1 | negative | - |
| 7q32.2 | NRF1 | negative | - |
| 2q31.2 | NRF2 | negative | - |
| 10q23.1 | NRG3 | negative | - |
| 12q22 | NTN4 | negative | - |
| 9q21.33 | NTRK2 | negative | - |
| Xq23 | NXT2 | In promoter region of NXT2, one CpG site located at -432 was differentially unmethylated in APP-sw cells | [675] |
| 1p35.3 | OPRD1 | negative | - |
| 3q28 | OSTN | negative | - |
| 12q24.31 | P2RX7 | negative | - |
| 1q42.12 | PARP1 | negative | - |
| 20q13.31 | PCK1 | negative | - |
| 2p11.2 | PERK | negative | - |
| 11q14.2 | PICALM | negative | - |
| 7q11.23 | PION | negative | - |
| 19p13.12 | PKN1 | negative | - |
| 19q13.2 | PLD3 | negative | - |
| Xq22.2 | PLP1 | negative | - |
| 7q31.32 | POT1 | negative | - |
| 11q13 | PP1 | negative | - |
| 3p25.2 | PPARG | negative | - |
| 4p15.1 | PPARGC1A | negative | - |
| 5q32 | PPARGC1B | negative | - |
| 2p14 | PPP3R1 | negative | - |
| 1p36.33 | PRKCZ | negative | - |
| 17p13.1 | PSD95/DLG4 | negative | - |
| 14q24.1 | PSEN1 | Methylation markers changed. Methylation increased or decreased in peripheral blood cells. Methylation increased in promoter regions in LOAD. Deacetylation of H4 reported from animal models | [167, 807] |
| 1q42.13 | PSEN2 | Methylation increased in promoter regions in LOAD | [807] |
| 14q22.1 | PTGER2 | negative | - |
| 1p31.1 | PTGER3 | negative | - |
| 5p13.1 | PTGER4 | negative | - |
| 9q33.2 | PTGS1 | negative | - |
| 1q31.1 | PTGS2 | Hypo- and hypermethylation of CpG islands in COX-2 promoter region | [793] |
| Locus | Gene | Findings | References |
| 8p21.2 | PTK2B | negative | - |
| 11p15.1 | PTPN5 | negative | - |
| 16p11.2 | PYCARD | negative | - |
| 6p23 | RANBP9 | negative | - |
| 21q22.1 | RCAN1 | negative | - |
| 7q22.1 | RELN | negative | - |
| 4q12 | REST | negative | - |
| 17q25.1 | RHBDF2 | RHBDF2 centered on cg13076843 associated CpG was found in close proximity to two genes, and RNA expression was altered relative to LOAD; RHBDF2 related to PTK2B | [569] |
| 19q13.33 | RPL13A | RPL13A related to cg03169557 found associated to LOAD diagnosis | [569] |
| 12q24.31 | SCARB1 | negative | - |
| 4q21.1 | SCARB2 | negative | - |
| 7q11.2 | SCARB3 | negative | - |
| 14q32.12 | SCL24A4 | negative | - |
| 14q32.13 | SERPINA3 | negative | - |
| 17p13.1 | SERPINF1 | SERPINF1 related to cg19803550 found associated to Abeta cascade network | [569] |
| 17p13.3 | SERPINF2 | SERPINF2 related to cg19803550 found associated to Abeta cascade network | [569] |
| 17q11.2 | SERT/SLC6A4 | negative | - |
| 6q23.2 | SGK1 | negative | - |
| 1q22 | SH3GLB1 | negative | - |
| 1q21.2 | SHC1 | negative | - |
| 2q36.2 | SLC19A3 | negative | - |
| 14q24.2 | SLC8A3 | negative | - |
| 12p12.1 | SLCO1A2 | negative | - |
| 4q22.1 | SNCA | SNCA mRNA expression in AD subjects was significantly higher. mRNA expression and methylation rate of SNCA intron 1 was lower in LOAD at seven CpG sites | [808] |
| 15q22.31 | SNX1 | negative | - |
| 6q21 | SNX354654552331 | negative | - |
| 1q25.2 | SOAT1 | negative | - |
| 21q22.1 | SOD1 | negative | - |
| 10q25.1 | SORCS1 | negative | - |
| 11q24.1 | SORL1 | Methylation at 4 CpG sites (cg15241519, cg08441314, cg11606444, and cg22136098) were associated with both Abeta and tauopathy | [26] |
| 11p15.2 | SPON1 | negative | - |
| 17q21.2 | STAT3 | negative | - |
| Xp11.23 | SYP | negative | - |
| 6q27 | TBP | negative | - |
| 5p15.33 | TERT | hTERT methylation frequency was associated with the aging process in LOAD | [809] |
| Locus | Gene | Findings | References |
| 10q21.1 | TFAM | negative | - |
| 12q13.12-13 | TFCP2 | negative | - |
| 6p21 | TFEB | negative | - |
| 19q13.2 | TGFB1 | negative | - |
| 9q33.1 | TLR4 | negative | - |
| 6p21.33 | TNFA | Hypomethylation of the TNFA promoter was found in LOAD brains | [810] |
| 8p21.2 | TNFRSF10A | negative | - |
| 8p21.2 | TNFRSF10B | negative | - |
| 12p13.31 | TNFRSF1A | negative | - |
| 1p36.22 | TNFRSF1B | negative | - |
| 13q14.11 | TNFSF11 | negative | - |
| 19q13.32 | TOMM40 | negative | - |
| 8q22.1 | TP53INP1 | negative | - |
| 6p21.1 | TREM2 | negative | - |
| 15q22.31 | TRIP4 | negative | - |
| Xq22.3 | TSC22D3 | negative | - |
| 2p22.3 | TTC27 | negative | - |
| 19q13.12 | TYROBP | negative | - |
| 12q24.33 | ULK1 | negative | - |
| 5q31.1 | VDAC1 | negative | - |
| 6p21.1 | VEGFA | negative | - |
| 9p24.2 | VLDLR | negative | - |
| 8p12 | WRN | negative | - |
| 7q22.1 | ZCWPW1 | Methylation change not directly in loco | [794] |
| 6q14.3 | ZNF292 | negative | - |
| 19q13.42 | ZNF628 | negative | - |
| 4q31.21-q31.22 | ZNF827 | negative | - |
Method remarks: Eligible for inclusion were human studies published until May 2017. PubMed was searched using the search terms “Alzheimer human” and gene locus and “epigenomics” or “DNA methylation” or “histone acetylation” or “ubiquitination”. Reported are only positive findings from LOAD patients, “negative” means here that no such results were available for this locus.
Tau kinases are activated already through fetal development and been detected in AD brains: cyclin-dependent kinases (CDK5 7q36.1), MAP kinases (MAPK) and GSK-3 proteins phosphorylate protein tau [199-201]. Tau, once hyper-phosphorylated, is no longer capable of docking to microtubuli, but accumulating in neurons, whilst forming paired helicoidal filaments. These paired helicoidal filaments formed as consequence of the hyper-phosphorylation of immunoreactive microtubule-associated tau destabilise neuronal microtubules [188], instead. H-tau is truncated in its N-terminal and impairs axonal transport of organelles, specifically mitochondria, to synaptic terminals forth and back. The N-terminal fragment of tau induces abnormal mitochondrial dynamics, defective mitochondrial function with increased ROS, decreased cytochrome c oxidase (COX), and decreased ATP production, conducive to synapse deprivation and loss [202]. Hence is it the h-tau contributing through synaptic suffocation to neuron dysfunction and apoptosis [203].
No unequivocal relation between Abeta cascade and tauopathy has yet been registered in biomedical research. CDK5, however, is suspected to be the key link between Abeta and tau pathologies [204], possibly due to a correlation still unknown. Although It is still unexplained how tau pathology could exactly relate to amyloid pathology, however, as has been suggested, tau aggregation is caused by post-translational modifications [35, 198]. However, there is also a clinical phenotype with NFT formation only (Tangle-Only Dementia (TOD) and a Primary Age-Related Tauopathy (PART)) [205]. Recently, the neuropathologist Braak detected primary diffuse tauopathy early in childhood brains from four years onwards into early adolescence, with histologically confirmed post-mortem starting points in locus caeruleus, further subcortical relays of the noradrenergic system, and the transentorhinal region [206]. This discovery may suggest a close relation of tauopathy with the stress-related catecholaminergic system (Section 3.1.), and can support an early developmental origin of LOAD.
1.4.4. Neuropathological Braak Staging, LOAD Latency and Clinical Manifestation
Early in the disease course, diffuse Abeta plaques are seen in the frontal and parietal lobes, including the praecuneus [207], followed by hippocampi, basal ganglia, brainstem, and cerebellum. The synergistic interaction in early Braak stage regions was then related to subsequent overall 24-month metabolic decline. A localisation of phospo-MEK1 preceding ERK (for MAPKs see Section 2.) phosphorylation in neuronal nuclei was detected in Braak tau-stages I-II, indicating abnormal nuclear trafficking of this normally cytosolic kinase [208]. This Braak stage I-II is consistent with an onset of hyper-phosphorlyation of protein tau, however, not yet conglomerated as tangles [209]. The extracellular Abeta42 neuritic plaque deposition occurs only at late Braak amyloid-stage C once the intracellular Abeta40 has caused neuron leakage and atrophy [182]. The Braak stage III corresponds to MCI, termed the “limbic stage” [209] of cerebral neuropathy. In the Braak stages V/VI for tau NFTs and Braak stage C for senile Abeta plaques, there is the extracellular Abeta deposition as neuritic plaques [182], and the intracellular NFT [209] formation. In the clinical manifestation of LOAD, the main risk genes that were found to be associated with the MCI-AD transition (interacting with Abeta deposition) are amylin (IAPP 12p12.1) SNP rs73069071 and the neighbouring hepatic solute carrier organic anion transporter family member 1A2 SLCO1A2 (SLCO1A2 12p12.1) [210], the latter responsible for the uptake of bile acids, bromosulphophthalein, and steroidal compounds. Amylin is a calcitonin family peptide hormone synthesised in pancreatic beta cells together with insulin. It is the main component of amyloid deposition caused by T2DM in Langerhans islets. This may suggest that metabolic processes pertaining to T2DM could also play a role in LOAD manifestation (Section 3.2.).
1.4.5. Oxidative Stress and Cognitive Impairment
Several studies also revealed that, relating to the Abeta cascade, chronic oxidative stress is the conditio sine qua non for the development of tau pathology [211]. The Abeta cascade involves excitotoxity, Abeta aggregation, inflammation, tau hyperphosphorylation, microglial activation [212], increased GSK-3beta activity, and deregulation of the neuronal calcium metabolism [188]. Oxidative stress appears triggered by free ROS such as the hydroxyl radical, the superoxide anion, and hydrogen peroxide, and also as ER stress [185]. Oxidative stress leading to lipid peroxidation precede the appearance of Abeta plaques due to Abeta oligomerisation. Resende [213] also observed increased activity of the antioxidant enzymes glutathione peroxidase (family 1-8) and superoxide dismutase (SOD1 21q22.1) as indicators of ROS. The final steps in oxidation stress consist in in lipid peroxidation, glutamatergic excitotoxicity, inflammation, and activation of the cascade of apoptotic cell death [212]. Neither Abeta nor intracerebral NFT themselves were found clearly related to cognitive decline. Yet in contrast, onset of cognitive decline was associated with increased markers of oxidative stress, caspase-9 activation, an index of neuron death, and decreased hippocampal synaptophysin levels, a synaptic vesicle glycoprotein [214] expressed by the SYP gene (Xp11.23). Cognitive impairments were associated with praecuneus, subcallosal cortex, cingulum, posterior to anterior frontal regions [207], and adjacent to these regions, white matter hyperintensities in periventricular regions and callosal fascicles predicted cognitive impairment at >0.8 [207]. Hence, these associations with cognitive decline point towards underlying structural WM alterations and general oxidative status.
Oxidative stress is thus a common component of the neurodegenerative process. OS has been shown to induce serum glucocorticoid regulated kinase SGK1 (SGK1 6q23.2) involved in neurodifferentiation and neurodegeneration. SGK expression through a p38 MAPK-dependent pathway, inhibits apoptosis [215]. SGK1 expression is mainly regulated by glucocorticoids (Section 3.1.), and glucocorticoids were demonstrated to enhance exercise-related memory consolidation in a range of in animals. Whilst chronic glucocorticoid excess impairs hippocampal neurogenesis by NR3C1 activation, this mechanism is perpetuated by kinase SGK1.
1.4.6. Mitochondrial Dysfunction
Mitochondrial dysfunction is increasingly in focus as associated with tauopathy [203, 211, 216]. Available evidence suggests that tau hyper-phosphorylation is accompanied by multiple factors, including intraneuronal Abeta-oligomers, chronic oxidative stress, reduced insulin-like growth factor 1 (IGF1 12q23.2), and astrocytically mediated Abeta and caspase activation [211]. Pertaining to this ensemble is the observation that LOAD patients carry systemic mitochondrial dysfunction that causes brain pathology [177, 216]. COX reduction is already present at the MCI stage of LOAD pathogenesis, and also shows co-localisation with Abeta42 generation [216]. Abeta42, in turn, and h-tau interacted with the mitochondrial outer membrane protein VDAC1 (VDAC1 5q31.1) [217]. Of the three isoforms, only VDAC1 is expressed in neurons and may thus be relevant for neurodegeneration. VDAC forms the mitochondrion, maintains the mitochondrial permeability transition pore (mPTP, Section 2.6.), binds to apoptosis regulator beclin, and is thus important for apoptotic signalling. Reddy established that Abeta interacts with VDAC1, to the effect of mitochondrial dysfunction, with reduction of ATP and COX activity, lipid peroxidation, free radical production, and mitochondrial fission-linked GTPase activity [217], indicating ROS activation. Dysfunctional mitochondria contribute to malignant APP processing by (a) excess production of ROS, (b) releasing proteins ASK1 (MAP3K5 6q23.3) and glutathione S-transferases (GSTs), (c) thus leading to JNK release, and (d) deacetylation of histones and demethylation of APP and BACE1 gene promoters, which trigger Abeta production [177, 216].
Mitochondrial dysfunctions, however, appear to be a function of normal ageing as well. Evidence indicates age-related mtDNA changes associated with CVD risks also occurring within the human brain basal ganglia. Ageing increases transcription of a 5-kilobase deletion that particularly affects mtDNA COX genes [218]. Overexpression of COX genes is present very early in ontogeny in murine AD models [211]. Heteroplasmic mutations in the COX-1 gene (PTGS1 9q32-q33.2), specifically a COX gene subunit located on the mtDNA, were increased during ageing, but doubled in LOAD subjects [219]. Multiple mutations were detected and the mutation frequency increased with age [216]. The mitochondrial dysfunction cascade assumes that such genomic variability is at the baseline of mitochondrial activity. Age-related perturbed mitochondrial function loss, which influences brain ageing and initiates compensatory responses with Abeta production, tau phosphorylation, and synaptic degeneration [216]. It has been shown that Fus1, a tumor suppressor protein residing in mitochondria, is crucial in regulating inflammatory and stress responses by cytokine and NFkappaB activation [220]. Early signs of LOAD are produced by Fus1 knock-out murine models [220]. Mitochondrial dysfunction and cognitive abilities have been shown improved by blockade of shc1 (in humans SHC1 1q21.2) and its p66shc isoform regulating life-span, ROS production, and apoptosis [221]. Triggered through growth and insulin receptors, sirtuin-1 related p66SHC activates the Ras-ERK pathway but inhibiting ERK1/2 activity. p66SHC promotes stress induced apoptosis, mediating steroid action through the redox signalling pathway [222].
1.4.7. Mitochondrial Oxidative Phosphorylation (OXPHOS)
According to this current insight, mitochondrial damage therefore marks LOAD disease onset. The mitochondrial dysfunction onset early in manifest LOAD seems to be a result of molecular defects in OXPHOS [223], where expression of COX-1 gene PTGS1, for redox coenzyme NADH [224], and for ATPase delta-subunit, were up- or downregulated [224]. Mitochondrial overproduction of ROS energy in the form of ATP is efficiently produced via OXPHOS in the mitochondrial respiratory chain [203], but in LOAD, the mitochondrial surface membrane produces free radicals via H2O2. With Abeta42 deposition, ROS are produced and become virulent, whilst in addition, LOAD patients appear to have a deficient antioxidant defence system, which may allow for excessive oxidative damage in mitochondrial DNA [203].
1.4.8. Reactive Microgliosis
Another hypothesis on LOAD is based on age-dependent alterations in microglial number, their cytotoxic activation, and related TGFbeta-SMAD (TGF ligands, transcription regulators) signalling [225] in the context of neuroinflammation (Section 3.3.). Phosphorylation of MAPKs and the activation of the nuclear factor kappa B (NFkappaB) pathway, which cause the release of inflammatory cytokines, activate astrocyte and microglia hyperfunctionalities. MAPKs comprise three pathways (Section 2.1.), (a) extracellular signal-regulated protein kinases (ERKs), (b) stress activated protein kinases c-Jun-NH2-terminal kinase (JNK), and (c) the p38 pathway. The age-related changes in elevation of pro-inflammatory cytokines, and microglial cell production of ROS, combined with the mitochondrial dysfunction are currently strongly hypothesised as triggering LOAD neurodegeneration. The decline of lysosomal and mitochondrial functions thus would result in an exacerbated generation of ROS by microglia. Ageing effects are further present in the resulting microglial incapability to phagocytise Abeta [225].
1.4.9. Role of APOE Uptake and Removal
Apolipoprotein (APO) E is used by neurons for repair following oxidative damage [201], however, becomes cytotoxic if not cleared by autophagy. APOE epsilon4 genotype is a risk probably due to the APOE epsilon4 isoform being less efficient in Abeta clearance than the APOE epsilon2 and APOE epsilon3 isoforms [201]. The APOE allele epsilon4 isoform utilises the very low-densitiy lipoprotein (VLDL) receptor (VLDLR 9p24.2) [226, 227] instead of LDL receptor related protein LRP1/APOER (12q13.3), as used by epsilon2 and epsilon3 allele isoforms, for removal of Abeta-APOE epsilon4 particles through the BBB, thus slowing Abeta clearance. In addition, the LRP1, which is central in the cholesterol import by apolipoprotein E critical for neuron functioning, is gradually lost in LOAD progression. VLDLR is otherwise used in the cerebral triglyceride metabolism [227], and plays a crucial role for gyrification in early brain development. APOE epsilon4 thus effects on the lipid metabolism in decelerating or precluding Abeta autophagy, in disinhibiting cyclophilin A signalling in the pericytes of the cerebral vasculature, in accelerating neurodegeneration, and in causing leakages of the blood-brain barrier (BBB) [8] (Section 3.3.).
However, the APOE epsilon4 genotype is only present in 40-50% of LOAD cases [12, 13, 109] and, because of its relatively rare allele frequency of 13.7% [13], it not considered a common variant of risk genes. Clinical studies revealed that APOE epsilon4 carrier status did not influence CSF levels of Aβ42, and cortical Aβ accumulation is in fact independent of APOE genotype [228]. However, when in coincidence, their interaction produces most rapid cognitive decline in MCI patients [229, 230]. APOE epsilon4 status revealed malignancy in the Baltimore ageing study when combined with higher cortisol levels, depression, and chronic illnesses, such as diabetes, and hypercholesteinaemia [231]. Normative data from a prospective longitudinal study on CSF Abeta42, p-tau181, gliosis markers including a Pittsburgh Compound B (a radioactive neuroimaging contrast label of Abeta) PET neuroimaging study [232] indicate that, early in midlife, CSF Abeta42 levels decrease, whereas cerebral Abeta42 increase, as part of normal ageing. CSF NFT and gliosis neuroinflammation marker YKL-40 increased then in later adulthood to a stronger degree if a APOE epsilon4 allele genotype was present.
1.4.10. Role of Alpha-synuclein
An alpha-synuclein fragment is known as the non-Abeta component (NAC) of AD amyloid, and synuclein aggregation was confirmed for familial AD. The alpha-synuclein protein (SCN 4q22.1) interacts with tau, inducing its phosphorylation and aggregation while, simultaneously in reverse, tau enhances alpha-synuclein aggregation [233]. Protein alpha-synuclein, which is relevant for presynaptic vesicles and dopamine regulation, is thought to be pivotal to Lewy body pathologies PD, LBD, and TDP-43, thought to be central in non-dementive neurodegeneration, but is likely also present in LOAD [234], and is conceived to possibly play a certain role in its aetiopathogenesis [235]. Specificically, alpha-synuclein aggregation has been observed to be involved in neuroinflammatory processes in LOAD [236] and subsequent autophagy impairment.
2. NEURODEVELOPMENTAL DIFFERENTIATION AND LATE-LIFE NEURODEGENERATION
2.1. Introduction to Stress and Cerebral Plasticity
Enduring stress-provoking early-life experiences may influence childhood and adolescent cognitive and emotional outcomes by disrupting the maturation of the underlying brain networks [79]. Particularly, this leads to alterations of SMA and HPA stress networks with an impairment of attachment learning essential for respective mental representations [84]. Modest empirical support yet exists for disruptions in neurogenesis, myelination, and synaptic pruning as underlying structural changes [237]. Thereby, it is believed that ELS interferes with the critical waves of neurogenesis, synaptic overproduction, and stabilisation of synaptic connections [238]. Specifically, ELS was found leading to 4.2%-6.3% volume reductions in hippocampal cornu ammonis subfields CA2-CA4 and subiculum [239]. Multiple meta-analyses [240-242] have confirmed reductions of hippocampal tissue and reduction of total cerebral white matter volume in adults who experienced ELS.
Another key player (next to protein tau, Section 1.1.) in early neural development are the Glycogen Synthase Kinase (GSK) proteins: GSK-3 proteins play a pivotal role in controlling neuronal progenitor proliferation and neuronal migration [243, 244], and specifically also cytoskeletal organisation. It has been found that upstream and downstream signals modulating neuronal GSK-3 function are critical for neuroplasticity and cognitive functioning across the life-span [243]. In the differentiation of neuropil, GSK-3 proteins are activated downstream to growth factor triggered activation of the tyrosine kinase RTK family of receptors (including e.g. growth factor receptor classes, cytokine and hormone receptors) and the PI3K/Akt/p38 pathway, and are engaged in neurogenesis, neuron orientation, polarisation, axonal and possibly dendritic outgrowth, and synaptogenesis [244]. In the function of stabilisation of microtubule dynamics, GSK-3s interact with the microtubule activated protein 1B (MAP1B 5q13.2) and tau, as well as MEK and ERK. In the coordination of microtubule dynamics, there is interaction with Wnt, mTor, and stress-related MAP kinases [244, 245], with beta-catenin, c-myc, c-jun, and CREB at the transcriptional level.
2.1.1. GSK-3 and MAP Kinases in LOAD
Whilst GSK-3 proteins have been implicated in early neurodevelopmental disorders including neuropsychiatric conditions [244], its late-life roles make it a central focus of LOAD pathology. It has been assumed that, while GSK-3 kinases induce tau hyperphosphorylation [246, 247], the stress kinases SAPK/JNK and p38/MAPK are triggered by Abeta42 in such a way that there is a parallel precipitation of h-tau, Abeta, JNK and p38 in neurites [248] or co-localisation with tau deposits in astrocytes [249, 250]. But in addition, Akt/GSK-3beta, MAPK and NFkappaB activity can be elicited by oxidative stress induction [251]. JNK and p38 MAPK, and NFkappaB are the key mediating links with environmental stresses, ROS production to transcriptional regulation of BACE1, and APP genes [252]. For translational regulation of BACE1, the key players are the eukaryotic translation initiation factor-2 alpha subunit 1 (eIF2alpha, EIF2S1 14q23.3) and protein kinase R PKR (EIF2AK2 2p22.2), a double-stranded RNA dependent protein kinase [252]. The PKR stress signalling pathway was hereby contributing to activation of JNK, and insulin receptor substrate for the metabolic inflammasome in T2DM and LOAD [253] (Sections 3.2. and 3.3.).
2.1.2. The MAPK Family of Mitogen-activated Kinases
Three pathways of MAPK are established [254, 255]: ERK1/2 signalling pathway, c-jun NH3-terminal kinases SAPK, and p38 MAPK. All three are characterised generically as the “three-tiered” MAPK pathways [256, 257], where MAPKKKs are receivers of a variety of inputs and their “gatekeepers”, the mediators MAPKKs for input signals, and the core of kinases MAPKs as functional effectors of phosphorylation. To a certain degree, the three pathways can share phosphorylation activity and be dephosphorylated by dual-specificity phosphatases (DUSPs) (Section 2.3.) [256, 257].
Each of the MAPK pathways have established roles in neuronal differentiation and development, (b) functional neuroplasticity including learning and memory (long-term potentiation, LTP), (c) in neuronal injury and repair, and (d) in apoptosis in the context of neurodegeneration [249, 250, 255]. Each of these enzymes exists in several isoforms: ERK1 to ERK8; p38alpha, -beta, -gamma, and -delta; and JNK1 to JNK3 [185]. All three MAP kinases, ERK1/2, c-Jun NH3-terminal kinases and p38 MAPK are involved in LTP and long-term depression (LTD) in the hippocampus, but by separate mechanisms [258]. In complement to NFkappaB, the three MAPK pathways trigger innate and adaptive immune responses upon activation by MAPKKK [256]. In LOAD, the three MAPK kinases dynamically interact with CREB and calmodulin-dependent protein kinase (CaMKII) in oxidative stress-mediated abnormal hyper-phosphorylation of tau and “exclusively” promote the pathogenesis of LOAD [259]. It has been concluded that conjoint activation of all three MAPK kinase pathways together is essential for LOAD, since (a) ERK and JNK are present in early Braak stages, whilst p38 is not, and (b) because healthy subjects can exhibit activation of each single pathway without signs of dementia pathology [260].
2.2. Glycogen Synthase Kinase 3 (GSK-3)
The glycogen synthase kinase 3 (GSK-3) is involved in the canonical Wnt/beta-catenin signalling pathway [217], where it acts, dependent on Wnt signals, in a proliferative manner; alternatively, in pro-apoptotic decision, it phosphorylates transcription factors. Of the GSK-3 proteins, the major two isoforms exist in GSK-3beta (GSK3B 3q13.33) and GSK-3alpha (GSKA 19q13.2), are encoded by 85% homologous separate genes, which produce additional splice variants [244]. GSK-3beta activity, regulated by alpha-globin transcription factor CP2 (TFCP2 12q13.12-13) [261], is dependent of Akt [262], which plays a key role in switching GSK-3beta between active and passive forms [263]. In neurodifferentiation, GSK-3beta signalling antagonises the PI3-K/Akt/mTOR signalling pathways [264] and CREB signalling [265]. Psychological stress-induced activation of GSK-3beta leads to activation of the NLRP3/IL-1beta pathway [266] involved in inflammasome regulation (Section 3.3.). GSK-3beta has been shown to be the main link to neuroinflammation by targeting inflammasome receptor genes NLRP3 and TLR4 [267, 268] (Section 3.3.), NFkappaB, and inversely, with anti-inflammatory IL-10 activation [266]. In addition, it exerts in parallel immune functions by regulating T-cells and macrophages [266].
GSK-3beta is the key enzyme in the regulation of the cell cycle; it fulfills major roles in early brain development, energy metabolism, neuronal cell development, and body pattern formation. A lacuna of human GSK-3 studies for LOAD has been stated by De Paula [188], but recently it has been shown that both GSK-3alpha and GSK-3beta are also essential in plasticity of dendritic spines, and thus for reliability of neuronal signal transmission [269]. GSK-3alpha is responsible for short-term spine structural plasticity via LTD [269], whereas long-term GSK-3beta depletion (but also its over-activity, see below) generally reduces synaptic spine densities. In LOAD, GSK-3beta plays in neurons a pivotal role in the regulation of tau phosphorylation (i.e., overactive GSK-3beta leads to hyper-phosphorylation of tau - alone [270] and together with CDK5 [200]): GSK-3alpha, GSK-3beta, and MAPK13 were found to be the most active tau kinases in phosphorylating tau at all epitopes tested [271]. There is a direct link between APP, beta-secretase activity, with GSK-3beta and tau phosphorlylation [272]. Abeta42, however, triggers GSK-3beta only by means of interruption of Akt signalling [273], which induces NFT formation by tau hyper-phosphorylation [246, 247]. If tau phosphorylation is reduced by GSK-3beta inhibitors, Abeta42 neurotoxicity is curtailed [234]. Moreover, it has been demonstrated that GSK-3alpha is involved in the preparatory steps of Abeta synthesis [274]: GSK-3alpha inhibitor lithium blocks the production of Abeta42 isomers by interfering with APP cleavage at the gamma-secretase step. GSK-3beta activity itself is negatively regulated by several signal transduction cascades that protect neurons against apoptosis [259]. In response to oxidant stress, GSK-3beta translocates to the mitochondria, increases cytotoxic ROS from mitochondria [259], and GSK-3beta is proposed to activate VDAC1 (Section 1.4.) phosphorylation that ultimately leads to mitochondrial dysfunction and synaptic damage in AD [217]. Activation of GSK-3beta, even in the absence of Abeta produces dendritic spine loss in neurons in vitro, whilst pharmacological inhibition of GSK-3beta prevents spine loss and increases expression of CREB-target genes like BDNF [217, 247].
There is also an epistatic genetic expression interaction between the genes: GSK3B is proposed to activate VDAC1 phosphorylation that ultimately leads to mitochondrial dysfunction and synaptic damage in AD [275, 276]. The interaction between GSK3B (rs334543) and Abeta precursor binding protein APBB2 (4p14-p13) (rs2585590) was confirmed in a post-mortem LOAD autopsy sample [275]. Also, epistatic effects for genetic interactions have been found for GSK3B and the tau MAPT gene in LOAD, which determine beta-catenin levels [277].
2.3. The Extracellular Signal-related Kinases (ERK)
Extracellular signal-related kinases are one of three specific subsets of the MAPK family and have neurodevelopmental functions in meiosis, mitosis and post-mitosis of neurons, activated by growth hormones, cytokines, and G-coupled receptors. The Ras-Raf-MEK-ERK [254] pathway is involved in the regulation of growth and cell differentiation pathways through phosphorylation cascades. Once the signalling cascade is activated by receptors or ion channels, signals are transduced by MAPKK to adaptors which activate Raf, MEK1/2 and ERK, the core components of the pathway [254]. Downstream, ERKs activate nuclear transcription factors NFkappaB, ELK1, c-fos, c-myk, GSK-3 and CREB via phosphorylation. Their main inhibitor is cAMP [254]. As with the GSK-3s, the two major isoforms are overlapping at 85%, are ERK2 (MAPK1=p42 22q11.22) involved in developmental growth, and ERK1 (MAPK3=p41 16p11.2) essential for T-cell development, and roles in enhanced neuron proliferation, and differentiation.
ERK and Akt are two major intracellular signalling pathways activated by BDNF involved in neuron survival [278, 279]. In LOAD, however, Abeta-induced OS was aligned with an increment of the activation of Akt and ERK1/2, and activity of GSK-3 [214]. Abeta42 presence reduces Akt, while increasing GSK-3beta [217]. However, in contrast to GSK-3beta, ERK1/2 are not themselves involved in tau phosphorylation [280], but have been found early in NFT appearance preceding Abeta deposition [209, 281]. It has been shown that a brief signal involving Akt/PTB inhibiting ERK1/2 activation may have neuroprotective survival effects, whereas sustained ERK1/2 signalling with p38 and JNK co-activation could exert pro-apoptotic death signal effects [279]. In LOAD, aberrant expression of ERK and other MAP kinases has been observed [255]. Furthermore, there is growing evidence for the assumption that the neuroprotective effects of ERK are reversed in the context of peroxidation [255]. It is likely that phosphorylated ERKs become inhibited in neurons or are sequestrated in mitophagy (Section 2.6.) of defunct mitochondria [255]. Abeta42 docks to the MAPK/ERK cascade, binds to alpha7 nicotinic acetylcholine receptors (nAChRs) [208, 282], and downregulates both CREB and ERK2. Memory impairments are induced by direct Abeta42 infusion to the hippocampus [283], which affects the NMDA/ERK/CREB pathway. These memory impairments are reversible by MEK inhibitors [284], making clear that ERK over-activation is responsible for memory defects in LOAD.
Memory formation is mediated by acetylcholine release and uptake by ACh-receptors, where LTP in the postsynaptic membrane activates ERK thus enabling neuronal plasticity. Plasticity-related protein synthesis is regulated by mTOR controlling LTM-related translation of synaptic scaffold proteins PSD-95 (DLG4 17p13.1) [257], and insulin receptor substrates IRS1/2 (Section 3.2), specifically in hippocampus and prefrontal cortex [285]. A co-activation of muscarinic ACh and beta-adrenoceptors facilitates the conversion of short term into long term synaptic plasticity through an ERK- and mTOR-dependent translation initiation [286]. In LOAD, mTOR and GSK-3 have been found interrupted by Abeta42, precluding synaptic plasticity [287], which explains anterograde amnesia in LOAD. Microgliosis (Sections 1.4. and 3.3.) in LOAD is also induced by ERK, by involvement of the MAPK pathway through phosphorylation of MARCKS, a kinase involved in phagocytosis, expressed by microglia to Abeta plaques [288]. Phosphorylation of ERK1/2 and p38 in Abeta42-activated microglia triggers production of pro-inflammatory cytokines TNFalpha, IL-6 and IL-1beta, amongst others, from microglia [191] (Section 3.3.).
Kamat and colleagues [259] have proposed that ionotropic glutamate receptor (NMDAR) dysfunction with OS and reduction of free influx of Ca+ is critical for LOAD development, because NMDARs are central to cerebral development. NMDAR are glutamate receptors decisive for strengthening or weakening synaptic responses. Synaptic activations by glutamate input and cell depolarisation changes determine circuit maintenance, which is crucial for mnestic encoding and storage of memory content [289]. However, it was shown that NMDAR is also a receptor for Abeta42 oligomers, and the interaction of NMDAR and Abeta is neurotoxic [290]. Hence, NMDARs contribute to excitotoxicity in LOAD pathology caused by NMDAR dysfunctions under stress conditions. NMDARs induce ERK and CREB activation, and limit Ca++ influx [259, 291], whereby Abeta42 promotes NMDAR dysfunction. In turn, improves and restores the stimulation of the ERK pathway NMDAR functioning and LTP in the hippocampus [292]. Aberrant Ca++-flow in neurons affected by Abeta is associated with loss of dendritic spines and neuritic dystrophy, mediated in part by the Ca++-dependent protein phosphatase calcineurin [259]. RCAN1 (RCAN1 21q22.1), the regulator of calcineurin, is involved in adaptive responses to OS induced by soluble oligomeric Abeta [201]. RCAN1 inhibits calcineurin, a p-tau phosphatase, leading to tau hyperphosphorylation [201], as triggered by GSK-3beta.
2.4. The c-jun N-terminal Kinases (JNK) Pathway
The c-jun N-terminal kinases (JNK, also termed Stress-Activated Protein Kinases, SAPK) consist of 16 isoforms derived from three closely related [257] genes: JNK1/SAPK-gamma (MAPK8 10q11.22) (four splice isoforms), JNK2/ SAPK-alpha (MAPK9 5q35.3) (four splice isoforms) and JNK3/SAPK-beta (MAPK10 4q21.3) (eight isoforms), each with nine or ten exons (6a/b, and alternative translation initiation site for JNK3) [257, 293]. Generally, JNK activity regulates several important cellular functions including cell growth, differentiation, survival and apoptosis, and is triggered by a multiplicity of both biotic and abiotic stressors [257, 294]. JNK activating kinases (the so-called MAPK Kinases (MAPKKs)), MKK4 and MKK7 [293]. JNK serves as a “primer” or “master kinase” for GSK-3beta phosphorylation activity on tau [257], and putatively, a role in chromatin remodelling and transcription effect on many targets [257]. Here, JNK has recently been implicated in the regulation of histone H3 acetylation in trigeminal neurons following their exposure to an environmental neurotoxin and neuroinflammatory agent [293].
During early development, JNK1 is critical for neurogenesis, cell differentiation, proliferation, migration, early brain development, apoptosis and neurodegeneration [257], metabolic homeostasis, but also inflammatory conditions and cytokine production mediated by activation protein 1 AP-1, IL-8 and GM-CSF. JNK2 is critical for regulation of fibroblast regulation, macrophage and T cell activity, other immune functions, and wound repair [257]. JNK3 is later only present in the CNS [257] and related to neurodegeneration [293]. Their central function is revealed by the localisation of JNKs in the mitochondria and microtubule network in addition to their roles in the nucleus to regulate gene transcription [293]. Here, JNK is a link between various divergent functions [257], such as interactions with nuclear transcription factors (e.g. c-myc, ELK1, sirtuin, beta-catenin, histone H3.1, runx2), hormones (glucocorticoid receptor GR, glutamate receptors, lipoprotein receptor LSR, Wnt co-receptor LRP6, lipid sensor SREBP1, nuclear hormone receptor PPAR-gamma), cytoskeleton proteins (e.g. tau, MAP2), vesicular transport adaptors (JIP1, JIP3), cell membrane receptors (BMPR2), and mitochondrial pore proteins (Section 2.6.) (e.g. beclin targets apoptosis regulators Bcl2, Mcl1, Sab).
2.4.1. Pro-apoptotic vs. Pro-survival Functions
JNK activity is responsible for cell cycle arrest and caspase recruitment initiated by Tumor Necrosis Factor (TNF)-related apoptosis [293]. In LOAD, the specific pathogenic role for JNK consists in priming phosphorylation of the Bcl2-related protein family member Mcl-1 to allow subsequent phosphorylation by GSK-3beta. Stress-induced Mcl-1 degradation therefore requires the coordinated activity of JNK and GSK-3beta [293]. The presence of oligomeric Abeta42, then in turn, increased the levels of phospho-JNK [295] in hippocampal neurons. In reverse, the inhibition of p-JNK decreased Abeta-induced ER stress, and increased pro-survival mitochondrial proteins [296], suggesting that JNK is mainly responsible for detrimental effects of Abeta42. This is further supported by the observation that pro-survival mitochondrial proteins including nuclear respiratory factor-1 (NRF-1), peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PPARGalpha), PGC1-alpha, and mitochondrial transcription factor A (TFAM) are suppressed by JNK [296], suggesting a negative correlation of JNK and CREB activation. JNK regulates NC3R1 (Section 3.1.) by means of phosphorylation, thus inhibiting GR-mediated transcription. The biological effect of the GR phosphorylation consists of enhancement of its nuclear export [297].
2.4.2. Axonal Swellings and Vesicle Transport Proteins
Yearly appearing new reports have encircled that c-jun N-terminal kinase-interacting protein 1 (JIP1) (MAPK8IP1 11p11.2) has crucial importance for fast axonal transport of APP and tau in coaction with kinesins [298-301]. Cargo transport proteins exert a strong feedback function for JNK signalling, supported by the notion that JIP1 can bind several MAP3Ks (MLKs and DLK) as well as MKK7 [257]. JIP4, in contrast, is related to and specific for the p38 pathway [257-301]. Recent evidence confirmed a role of MAPK8IP1 and its mutations in pathological APP transport due to affinity for 42-residue APP protein [302, 303]. The inclusion of soluble h-tau into the neuron soma and its aggregation by JIP1 has been identified as one key neurotoxic event in LOAD pathogenesis [304, 305]. Margevicius have in jip1-ko-mice shown that JNK signalling is critical for tau hyper-phosphorylation, loss of synapses and memory decline, even under conditions of Abeta overexpression [306]. In addition, APP is also phosphorylated by JNK [299, 300]. JNK can itself hyper-phosphorylate JIP1 and other JIPs [257-294], however, only part of its binding regions were affected by the post-translational modifications, amongst them, those for JNK, MAP kinase kinase, and RAC-Ser/Thr protein kinase, but not those for kinesin or Abeta [294]. JIP1 has been identified linking APP protein and reelin receptor ApoER2, by docking to AID terminals on APP [294, 307]. This mechanism is, however, not confirmed in human post-mortem brains [307]. Cytokine-induced gamma-secretase activity is mediated by JNK and to be enhanced by the expression of a constitutively active form of MEKK1, implicating the JNK signalling pathway in the regulation of gamma-secretase activity [185]. JIP1-kinesin linkage in the JNK pathway includes MAPKKK and MAPKK [294, 307-309]. Kinesin light-chain subunits of kinesin-I (KLC) related to JIP3 (MAPK8IP3 16p13.3), were shown related to kinesin-1 and choline acetyltransferase accumulation indicative of axonal swellings were present in early Braak stages I-III before Abeta deposition [310].
2.5. P38 Mitogen-activated Protein Kinases (p38 MAPKs)
The p38 mitogen-activated protein kinases characterise the third MAP kinase pathways pertaining neuronal cell differentiation, apoptosis, and autophagy. Four p38 MAP kinase isoforms, p38alpha MAPK (MAPK14 6p21.31), p38beta MAPK (MAPK11 22q13.11), p38gamma MAPK (MAPK12/ERK6 22q13.3), and p38delta MAPK (MAPK13/ SAPK4 6p21), have been identified. Analogous to the JNK/SAPK pathway, p38 MAP kinase is activated by a pleiotropy of cellular and extracellular stresses including excitotoxic shock, inflammatory cytokines, and endotoxins. In neurodifferentiation, p38 MAPK signalling antagonises the PI3K/Akt/mTOR signalling pathway [264] and CREB signalling [265]. p38 is one of the major oxidative stress sensors triggered by its upstream activator, Mitogen-Activated Protein Kinase Kinase 6 (MAPKK6) [201]. New evidence implies that also p38 will induce tau hyper-phosphorylation in presence of oxidative stress [311], and antioxidants will inhibit this mechanism [201]. Specifically, the formation of paired helicoidal filaments with microtubule-associated h-tau, which impairs intracellular traffic, is induced by p38 [312]. In neurons, the upstream activators of p38 consist of MLK and MAPKK3/6 [313]. Once activated, this pathway initiates production of the pro-apoptotic transcription factors c-myc, Pax6, neuronal transcription factor MEF2, p53, Elk1, CREB, NFkappaB, and inflammatory cytokines, termed the Senescence‐Associated Secretory Phenotype (SASP) [314, 315]. The SASP is mainly released from aged astrocytes in the LOAD brain [315], induced by the p38 MAPK pathway causing DNA damage by strand breaks [316]. Via its substrate MSK1/2, p38 MAPK is also related to the histone H3 [317].
2.5.1. Neuroinflammation and Synaptic Impairment
P38alpha MAPK, but not the other isoforms, shape the pro-inflammatory response (Section 3.3.) in the brain, released from microglia [317] and astrocytes [301, 318], including IL-1beta and TNFalpha release [319, 320]. There is evidence that also p38delta MAPK may play roles in inflammatory processes, TD2M and neurodegeneration [321]. Another cytokine, IL-33 released by oligodendrocytes, and its receptor ST2 on microglia, are involved in enhancement of Abeta phagocytosis by microglia [322]. Microglial activation on neurons have been found mediated by activation of p38alpha MAPK, because activated microglia stimulated p38-MAPK phosphorylation in neurons, thus influencing IL-1beta towards tau phosphorylation and increasing synaptophysin levels [323], the presynaptic synaptic vesicle protein [324] encoded by SYP (Xp11.23), as well as presynaptic synapsin I and postsynaptic glutamate receptor I [325]. Thereby, p38 MAPK contributes to microstructural alterations in synapses, increased neurotransmitter release and loss of plasticity [325]. Also in astrocytes, p38 MAPK has functions in glutamate excitotoxicity involved in AD pathophysiology [318]. P38 MAPK activity suppresses BDNF signalling [326], upregulated by IL-1beta [318]. BDNF signal transduction is dependent on phosphorylation of insulin receptor substrate 1 (IRS-1, Section 3.2.), a protein coupling activation of the BDNF receptor TrkB BDNF/NT-3 neurotrophic factor receptor (encoded by NTRK2 9q21.33), to downstream signalling pathways regulating CREB [326]. This mechanism can, however, be prevented by preceding BDNF activation [327].
In the inflammatory response following high-fat diet induced insulin resistance (Section 3.2.) by p38 MAPK, together with JNK pathway, and with the inflammatory NFkappaB pathways (Section 3.3.); this is accompanied by Abeta deposition, NFT formation and decrease in synaptic plasticity [328]. P38 and JNK develop neurotoxic properties under anaesthetic influence of isoflurane, whereas ERK1/2 are more neuroprotective [329].
2.5.2. P38 in LOAD Pathogenesis
As mentioned, p38 is one of the proteins involved in hyper-phosphorylating tau [201] but in combination with MEK3/MAPKK3 [271]. As a sensor for neuroinflammation, p38 MAPK expression reduced the lysosomal degradation of BACE1, enhanced BACE1 protein activity, and also inhibited autophagy [330]. At an early stage of LOAD pathogenesis, p38gamma MAPK located postsynaptically at scaffold protein PSD-95 sites was found to induce tau phosphorylation, which was found protective against Abeta42 action [324]. Although only in vitro, this finding may suggest that, initially, tau phosphorylation may have a self-repair quality at postsynaptic locations. Clinical studies have shown that p38 and JNK increase with LOAD illness duration, which is related to memory performance decline [331].
The receptor for advanced glycosylation specific products (AGER 6p21.32) (Section 3.2.) activates p38 MAPK phosphorylation brought about by Abeta, resulting in LTP impairment [332]. p38 facilitates hippocampal gamma and theta EEG oscillations through ionotropic NMDAR [333], and is therefore pivotal for synaptic functioning. This is related to post-synaptic density protein PSD-95 (DLG4 17p31.1) [320], a marker of synaptic integrity [334, 335]. From observations in murine AD models it was inferred that synaptic impairment in LOAD consists of suppression of LTP and enhancement of LTD [336]. LTD in the dentate gyrus and entorhinal cortex [332] is induced by p38 MAPK under soluble Abeta42, and is facilitated by metabotropic group I receptors GluR1/5 (GRM1 6q24.3, GRM5 11q14.2-3) [313, 336, 337] and ionotropic AMPA-type subunits GluR1-4 (GRIA1 5q33.1, GRIA2 4q32.1, GRIA3 Xq25, GRIA4 11q22.3) [338], striatal enriched protein tyrosine phosphatase STEP (PTPN5 11p15.1), and caspase-3 activation.
Recently it has been found that p38 MAPK and ERK1/2 play a pivotal role in the regional distribution of Abeta42 into hippocampus and parietal cortex [229, 339]. The regional distribution of Abeta42 is accompanied by intensified nAChRalpha7 expression [339, 340]. By interacting of p38 with LDL receptor protein 1 (LRP1/APOER 12q13.3), Abeta42 was internalised into cortical neurons and astrocytes, and distributed into mitochondria, lysosomes, and endoplasmic reticula [339]. This LDL/APOE receptor is crucial for the alpha2-macroglobulin-mediated clearance of Abeta plaques. Expression of this gene decreases with age and has been found to be lower than controls in brain tissue from LOAD patients. However, this clearing mechanism is missing in APOE epsilon4 homozygosity (Section 1.4.).
2.6. Role of (Macro-)autophagy Impairment in LOAD Pathophysiology
Escalation of cytoplasmic protein aggregation is a common feature of neurodegenerative disorders. By means of their lysosomes, cells self-digest protein aggregates and damaged organelles [341], and impairment of this autophagic process accelerates ageing specifically in microglia [342]. Neurons in LOAD suffocate from swellings induced by immature autophagic vacuoles leading to dystrophic and degenerating neurites, caused by physical impairment of retrograde transport by lyosomes to cell somata [343, 344]. The key trigger of autophagic clearance is release of mTOR signalling in response to growth factor sensing indirectly activating PKB/PI3K/Akt activating the mTOR activator Ras homolog enriched in brain (Rheb) [236]. Closely tied to neuroinflammatory processes (Section 3.3.) [236], autophagy is genetically triggered from autophagy genes orchestrated by the identified master regulators NFE2L2/NRF2 (2q31) (nuclear factor, erythroid 2 like 2) [224, 345] and transcription factor EB (TFEB) (6p21) for lysosomal pathways [346, 347], where NFE2L2 is related to both APP/Abeta and MAPT/tau expression in LOAD. Under physiological conditions, stress-responsive transcription factors including p53, NFkappaB and transcription master activator STAT3 (STAT3 17q21.2) have been found regulating the autophagic response [348], and the p53, TNFalpha, mTOR pathway have been described central to LOAD [236, 341]. Specifically, the interplay between p38alpha MAPK14 and the Akt/mTOR pathways is decisive for pro- vs. anti-inflammatory direction in response to environmental stress [349], where a disturbance leads to impairment of plaque macroautophagy. The resulting insufficient clearance of Abeta oligomers of all isoforms causes cell-to-cell transmission of Abeta oligomers [184], which promotes propagation of neurotoxic elements.
The role of autophagy in neurodegeneration has been described [236, 350, 351] in the context of mitochondrial dysfunction [236]: A reduction of autophagic flux leads to a persistence of dysfunctional mitochondria and mitochondrial fissures [352], which generate ROS and are vulnerable to apoptotic and inflammatory agents. One such agent, regulator beclin-2, induces mitochondrial rupture-induced apoptosis [353], where the mitochondrial permeability transition pore (mPTP) results in influx of solutes and water, as well as inner membrane swelling. Destruction of the outer membrane releases cytotoxic proteins into the cytosol and produces necrotic neuron death.
In LOAD, ER stress inducing autophagy is triggered by aggregated Abeta42 [354-356] and decreased solubility of parkin, an E3 ubiquitin ligase involved in autophagy [357] and mitophagy [351]. Expression of parkin, in turn, ubiquitinates both intracellular and extracellular Abeta, stimulating the autophagy process initiating regulator beclin-1 (ATG6) (BECN1 17q21.31), while attenuating caspase activity [358]. GSK-3beta inhibits mTOR by activating the autophagosome enzyme ULK1 (ULK1 12q24.33). GSK-3beta modulates protein aggregation through the phosphorylation of the macroautophagy cargo receptor NBR1 (NBR1 17q21.1) [359]. Activation of the ULK1 phosphorylates beclin-1, which regulates the activity of transcription factor vps34 [360] normally leading to autophagosome formation [351]. Loss of BECN1 hampers the phagocytic function and induces inflammatory responses [361] in microglia and disruption of trophic support from astrocytes [291, 354]. The completion of the autophagosome is marked by the release of LC3beta-II (microtubule-associated protein-light chain 3 beta 2, ATG8, MAP1LC3A/B 16q24.2) from the autophagosome membrane [236], a marker found related to cognitive decline [342]. Abeta42 was found hyperactivating the phosphoinositide3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) axis, which plays a central role in proteostasis, already at MCI/”limbic” Braak stage III [362]. mTOR activation reduces autophagy and induces insulin resistance. Activation and deactivation of mTOR, a kinase implicated in ageing and nutrition [352], is in mutual dependence of autolyosomal substrate digestion [350].
3. ENDOCRINOLOGICAL, METABOLIC, AND NEUROINFLAMMATORY PROCESSES IN LOAD
3.1. Stress Hormones and LOAD Pathology
Amyloid pathology is assumed causative for apoptosis in neurotransmitter producing neuron groups. Their cellular dysfunction is held responsible for disturbances in acetylcholine, noradrenaline, and serotonin transmitter systems [212]. However, the focus is kept here on cortisol, noradrenaline, and neuropeptide Y (NPY), which characterise stress systems; all three have been shown to trigger release of pro-inflammatory cytokines (Section 3.3.), and to bias thyroid hormones and immune reactions [363-365]. It is commonly accepted that at least noradrenaline and glucocorticoid systems are mutually interrelated [366, 367].
3.1.1. Noradrenaline Signalling
The sympathetic branch of the ANS is triggered from the amygdala (central and basolateral nuclei) in the presence of external stress signals [368]. Noradrenaline is considered the principle neurotransmitter of the SNS branch. In the CNS brainstem, the main conversion (from dopamine) site in the rostral pons for noradrenaline is the locus caeruleus (formerly coeruleus, LC) besides the adrenal medulla. Their projections to the major midbrain and cortical regions are exerted by noradrenergic neurons, and specifically target transentorhinal memory structures [289]. Abeta42 deposition is increased in these noradrenaline projection areas from locus caeruleus [367]. It is hypothesised that CRH afferents from the central amygdaloid nucleus into the locus caeruleus mediate the conveyance of environmental stresses [367].
Catecholamine release is directly related to SNS hyperreactivity, primarily mediating short-term stress response (also termed sympatho-medullo-adrenal (SMA) axis). SMA mediation of stress is under regulation by the catechol-o-methyltransferase (COMT 22q11.2) gene [369]. During normal ageing, COMT and brain-derived neurotrophic factor (BDNF) showed additive effects on decline in executive functioning in interaction with apolipoprotein E metabolism [370]. The COMT SNP rs4680 has also been found associated with LOAD, with its Met allele interacting with APOE epsilon4 status [371, 372]. Stress reactivity mediated by noradrenaline signalling is also involved in insulin release vs. inhibition from pancreatic beta cells. This mechanism ensures that blood glucose levels rise in stress states, but may also be causative for abnormal cerebral insulin signalling and insulin resistance in LOAD (Section 3.2.). Insulin release is stimulated by beta-adrenoceptors, and inhibited by adrenoceptor alpha2A [373]. These receptor types are also associated with LOAD.
3.1.2. Adrenoceptor Gene Loci and LOAD
There are overall nine subtypes of adrenoceptors, the majority of which have proven functions in spatial and emotional memory, and specifically in STM [374]. The known implications of adrenoceptors for LOAD are currently confined to alpha2A, beta1, beta2, and beta3-adrenoceptor subtypes. Adrenoceptor alpha2A (ADRA2A 10q25.2) has been shown to mediate the interaction between SORL1 (11q24.1) (the neuronal APOE receptor, Section 1.4.) and the sortilin-related vaculuolar protein sorting VPS10 domain containing receptor 1 (SORCS1 10q25.1). Stimulation of the alpha2A adrenoceptor increases APP redirection into endosomes and miscleavage by beta-secretase [375]. Adrenoceptors beta1-3 are more specifically involved in metabolic processes, their binding triggers intracellular concentrations of cAMP as second messenger. In their genomic bases, ADRB1 (10q25.3) releases heterodimers that influence BMI, body weight regulation, blood pressure, and basic metabolic rate. Beta1-adrenoceptor gene ADRB1 interacts with guanine nucleotide-binding protein subunit beta3 (GNB3 12p13.31) to increase LOAD pathophysiology [376]: the GNB3 T allele increases risk for homozygosity of the ADRB1 C allele. The co-expression of GNB3 T and ADRB1 C alleles produced increased cAMP levels and MAPK signalling.
Specifically, adrenoceptors beta2 and beta3 have in murine models been observed to interact with core LOAD pathophysiological processes (Section 1.4.) [374, 377]: Abeta can bind to the beta2-adrenoceptors, and their activation triggers the protein kinase A (PKA)-JNK pathway [378], resulting in tau hyper-phosphorylation. However, a restoring effect for memory functions has, in contrast, also been found for beta2-adrenoceptor [379]. Different polymorphic forms, other variants, and epigenetic modification of the ADRB2 (5q31-32) gene are related to its mediation of hepatic blood flow with glycogenolysis and gluconeogenesis, and insulin secretion from the pancreas. Murine ko-studies indicated that β2-adrenoceptors expressed on astrocytes are essential in LTP and spatial memory [380]. Polymorphisms Gly16, Glu27 and the beta2-adrenoceptor haplotype Gly16 Glu27 have been associated with LOAD, but dependent on APOE epsilon4 status [381]. Specifically, blockade of beta2-adrenoceptors reduced Abeta production induced by acute stress in mice [377]. The haplotype of both the 16Gly allele and the 27Glu allele of ADRB2 exhibited increased risk for LOAD, and a significant interaction with APOE epsilon4 [381]. Activation of beta2-adrenoceptors with presenilin-1 increased gamma-secretase activity [382] in lysosomes, upregulating Abeta42 cleavage, and subsequent plaque deposition. ADRB3 (8p11.23) is mainly expressed in brown and white adipose tissue, and becomes activated in energy expenditure, thermogenesis and lipolysis. Hypermethylation of the ADRB3 gene promoter in blood and visceral tissue is associated with metabolic disturbances [383], such as dyslipidaemia. Reductions of all adrenoceptor subtypes have been observed in LOAD pathology [374].
3.1.3. Noradrenaline Transporter Gene
The noradrenaline transporter gene SLC6A2 (NET 16q12.2) is central to noradrenaline homeostasis and presynaptic reuptake. Its SNP rs2242446 has been correlated to anxious arousal and PTSD [384]. SLC6A2 expression is restricted to noradrenergic neurons that innervate the adrenal medulla. An epigenetic mechanism (hypermethylation of CpG islands in the NET gene promoter region) that results in reduced expression of the noradrenaline has been implicated in stress-related disorders [385]. Oxidoreductase dopamine-beta-hydroxylase (DBH 9q34.2) converts dopamine into noradrenaline, and may therefore influence noradrenaline levels. In combination with cytokines, a SNP (allele rs1611115) of DBH and polymorphisms of the pro-inflammatory cytokine genes, IL1A and IL6 (Section 3.3.), were observed associated with LOAD [386].
3.1.4. Noradrenaline, Tauopathy and Locus Caeruleus Degeneration
Findings from the Lund Longitudinal Dementia Study [387] suggest that pontine locus caeruleus (LC) degeneration is common in and typical for LOAD. Relations of the degree of LC degeneration with white matter lesions, LOAD severity, nor duration have not yet been substantiated [387], however, this could be based on the scoring method used. Whether there are age-related changes in the LC is currently undecided, as small databases prohibit any conclusions [289]. Post-mortem findings in LOAD indicate that the amount of LC neuron loss is about 50% [289], and its level coincides with NFT deposition found in children from 6 years onwards [289], thus possibly providing a direct neuropathological link with ELS. Noradrenaline converted in the 15k LC neurons has additional anti-inflammatory functions on neuron membrane surfaces, glial cells and blood vessels in the neocortex and hippocampus [388, 389]. Noradrenaline induces microglia to suppress Abeta-induced production of cytokines and their phagocytosis of Abeta [389]. It is therefore likely that NFT deposition in LC and loss of its neuronal projections could promote increased Abeta deposition in LOAD brains [389]. In addition, have noradrenaline depletion studies resulted in impairments of working memory and social memory [374]. It is furthermore suspected that noradrenergic hypofunction is an impairment to effective neuron-glia interaction, finally resulting into abnormal glial reaction, and fostering neuron degeneration [388]. Nucleus Basalis of Meynert (NBM) degeneration is also ubiquitous in LOAD, however NBM neuron loss not unique to LOAD, but also present in Parkinson’s, Pick’s disease (FTD), and Lewy body dementia. Acetylcholine depletion is also common amongst those neurodegenerative diseases, and thus not specific to LOAD.
3.1.5. Glucocorticoid System Involvement in LOAD
Cortisol secretion is a physiological response to cope with repeated anxiety triggers resulting from chronic stress [390-392]. Glucocorticoid excess has implications for ageing by reducing life-spans [393]. Cortisol levels in LOAD are directly related to hippocampal sizes, and hippocampus size predicts cognitive performance with increasing age [394]. In addition, CSF cortisol levels increase from MCI progression to LOAD, suggesting an increase during pathogenesis [395, 396], where cortisol increases amyloid neurotoxicity [397]. Cortisol serum levels are elevated in LOAD patients, where they are believed to contribute to a neuroprotective effect, as inverse correlations with p-tau and tau-Abeta relations, and positive association in CSF with Abeta42, suggest [398]. Similar effects were found for non-demented APOE epsilon4 carriers [228]. In the Baltimore Memory Study [231], cortisol levels tended to negatively associate with memory domains language, visuospatial, visuomotor, executive etc.; however, two alleles of epsilon4 showed strong negative predictions in all memory domains.
3.1.6. Corticotropin-releasing Hormone Gene
Long-lasting stress effects are maintained through secretion of the Corticotropin-Releasing Hormone (CRH) resulting in HPA axis bias. In contrast to SMA activation, however, cortisol release in response to repeated stress habituates more quickly [399], but leads eventually into immunodeficiency by impairing CD19-promoted B-cell generation [400]. Synthesised in the hypothalamic paraventricular nucleus (PVN), CRH release is also triggered by TNFalpha and IL-6 resulting from inflammatory states, thus dampening the immune response, and controlling the inflammatory response. Environmental stress activates PVN CRH neurons resulting in a repertoire of stress-related coping behaviours [401]. The expression of CRH1 (8q13.1) is normally inhibited by a negative feedback loop consisting of peripheral glucocorticoid signalling through GRs in hippocampus, hypothalamus and pituitary to the HPA [130, 365]. A second vertebrate corticotropin-releasing hormone gene CRH2 has recently been discovered, but there is still a lacuna in human research [402]. Significant reduction in CRH1 expression has been observed in association with Alzheimer’s disease [403].
The glucocorticoid cascade hypothesis holds that LOAD and co-morbid depression being caused by accumulation of free cortisol responsible for age-related hippocampal tissue damage [397]. Strong co-morbidity of depression and LOAD exists at 50%. Post-mortem studies (although with small N) show that the PVNs of LOAD patients have significantly more CRH mRNA than healthy controls, but less than depressed [397]. Cortisol effects consist in (a) modulating neuroplasticity, circuitry and neurotransmitter systems, (b) regulating neuron death or survival, (c) releasing of structural proteins from glia, and (d) suppressing myelin content [404]. LOAD is associated with profound changes in HPA, with chronic hyperactivation of CRH neurons and plasma hypercortisolaemia [397], which is related to MMSE scores and APOE epsilon4 presence. Correlational studies indicate that, in LOAD, hypercortisolism is associated with disease severity, disease progression and accelerated cognitive decline, as indicated by hippocampal atrophy [404].
3.1.7. Interactions of Glucocorticoid Receptor with MAPK and Cytokines
In humans, the glucocorticoid receptor protein is encoded by the NR3C1 (5q31-32) gene. NR3C1 has a function as nuclear transcription factor [405], whereby glucocorticoids induce mitochondrial DNA transcription, and so bias mitochondrial physiology in the hippocampus. The NR3C1 gene consists of 9 exons, where alternative splicing in exon 9 generates mainly two highly homologous receptor isoforms, labelled alpha and beta [406]. It is bound in inactive state in the cytoplasm until glucocorticoid transgresses the membrane and the ligand-receptor conglomerates dock to Glucocorticoid-Response Elements (GREs) in the promoter regions of glucocorticoid-regulated genes [215]. These GREs are either facilitative or inhibitive, dependent on positive or negative binding, to transcription and transactivation [406], respectively. As floating monomers, the interaction with other transcription factors such as NFkappaB and CREB is repressive for dependent genes, such as those of cytokines IL-1beta, IL-2, TNFalpha and inducible NO-synthase (iNOS) [407, 408].
As mentioned, alternative splicing of NR3C1 generates two main splice variants, the GRalpha and the minor isoform GRbeta [409], with three other isoforms [410]. GRbeta retains the ability to bind DNA and has been observed acting as negative inhibitor of GRalpha [409, 410]. GRalpha has at least 8 different translation initiation sites resulting in isoforms A, B, C1, C2, C3, D1, D2, and D3, which may be restricted to specific cell types, thus resulting in highly individual susceptibility patterns [411] in specific tissues. Loss of GR is observed during both normal ageing and LOAD, and more pronounced in presence of NMDARs [404], and similar processes were observed under stress.
There exists interference of GR with signalling pathways of MAPKs: ERK1/2, p38 MAPK, and JNK, GSK-3/Wnt signalling. Specifically, GR induced the expression of MAP stress protein DUSP1, presumably via the binding of GR to putative GREs in regulatory regions of the mkp-1 promoter [412]. Equivocal results were, however, found on the role of GSK-3beta. There is a reciprocal interaction of GRE with CREB, activator protein AP-1, NFkappaB, and NGFI-B [409, 411, 412]. Transactivated genes through GREs include the insulin-like growth factor binding protein 1 (IGFBP1 7p12.3) FK506 binding protein 5 (FKBP5 6p21.31) (see below), NFkappaB inhibitor alpha (IkBa, NFKBIA 14q13.3), anti-inflammatory interleukin 10 (IL-10) receptor activator of nuclear factor kappa-B ligand (RANKL 13q14.11) [409]. Transrepressed genes through tethering include pro-inflammatory TNFalpha, IL-6, IL-8, IL-11 [409]. Transrepression has been reported also for a growing list of immune-regulating transcription factors, including NFkappaB, AP-1, CREB [410], of transcription coactivators such as interferone regulatory factor IRF3 (IRF3 19q13.33) [410, 412]. Epigenetic modifications have been observed in NC3R1 with interference with histone tail modifications on H3 [412], and for HDAC2 (6q21) in association with NFkappaB [410] activation. The HPA deregulation typically present in manifest LOAD can be targeted by selective synthetic GR regulating agents [413]; their application in murine models resulted in Abeta clearance, restoration of hippocampal vesicle protein synaptotagmine, and reduction of caspases.
3.1.8. Corticotropin Receptors and ACTH Signalling
There are two subtypes of CRH receptors, mainly located in the anterior pituitary, also in amygdala, hippocampus and locus caeruleus. Their activation product, ACTH, is synthesised in basophile neurons of the anterior pituitary under regulation by CRH, and triggers release of glucocorticoids from the adrenal medulla [365]. Like free cortisol, ACTH levels are therefore increased in LOAD. The HPA signalling pathway is mainly dependent on corticotropin-releasing hormone receptor type 1 (CRHR1) polymorphism on exon 6 of CRHR1 (17q21.31) [414]. CRHR1s are most abundant within the hippocampus, but also present in liver tissue. CRHR1 activation has been demonstrated to introduce hippocampal tau hyper-phosphorylation [403]; stress-induced h-tau accumulations were observed specifically in dendritic and axonal processes, with an increase of axonal mitochondrial transport, but with CREB and BDNF downregulation, which was reversible by GSK-3beta inactivation [403]. The CRHR2 (7p14.3) gene has been described involved in cardiovascular homeostasis, PTSD, and susceptibility towards stress [415].
3.1.9. Cortisone-cortisol Interconversion HSD-11β Gene Loci
The protein encoded by the HSD11B1 (1q32-q41) gene is a microsomal enzyme that catalyses the conversion of the stress hormone cortisol to the inactive metabolite cortisone and reverse. Because of its amplificatory action of active glucocorticoids, isoform 1 HSD-11β, has been assumed being a pathogenic factor in MetS, T2DM, and age-related cognitive decline [416]. Isoform 11β-HSD-2 has been implicated in neurodevelopmental susceptibility for the programming of diathesis towards chronic stress [417]. Too much cortisol can lead to central obesity, and several variations (rs10082248, rs2298930, and rs4545339) in the HSD11B1 gene have been associated with obesity and insulin resistance in children [418]. The regulation of both 11β-HSD isoform-genes is dependent on NFkappaB [419] in adipose tissue. HSD11B2 (16q22.1) protects cells from the growth-inhibiting and/or pro-apoptotic effects of cortisol, particularly during embryonic development. Mutations in this latter locus cause the syndrome of apparent mineralocorticoid excess and hypertension. Polymorphisms can regulate maternal cortisol levels in utero and regulate postnatal weight gain [420].
Of eleven genes tested from the glucocorticoid system (corticotropin-releasing hormone CRH (8q13.1), corticotropin-releasing hormone binding protein CRHBP (5q13.3), ACTH receptor 2 MC2R (18p11.2), 11β-hydroxysteroid dehydrogenase type 1 and 2 (HSD11B1 1q32.2, HSD11B2 16q22.1), glucocorticoid receptor NR3C1 (5q31.3), glucocorticoid modulatory element binding protein 1 and 2 GMEB1 (1p35.3), GMEB2 (20q13.33), steroidogenic factor 1 NR5A1 (9q33.3), nuclear receptor coactivator 2 NCOA2 (8q13.3)), only HSD11B1 SNPs rs846911 and rs860185, representing a rare T-A and a frequent C-A haplotype were found directly related to LOAD [12]. However, the significance level of this association was comparable to the significance of the APOE epsilon4 allele, but the effect size with an OR=6.2 was more than doubled [12].
3.1.10. FKBP5 and EGR1 in LOAD
Two other genes have gained considerable interest for their stress programming capabilities through epigenetic mechanisms. The co-chaperone FK506 binding protein 5 (FKBP5 6p21.31) in the cytoplasm is part of the GR-inactivating immunophilin proteins preventing and reducing affinity of GR to its ligand glucocorticoid [406]. GR and HPA [421] co-regulator FKBP5 negatively regulated glucocorticoid function pertaining to PTSD [422]. Four SNPs significantly interacted (rs9296158, rs3800373, rs1360780, rs9470080) with ELS in childhood and later PTSD [423]. The functional FKBP5 risk T-allele (rs1360780), which is related to HPA axis hyperreactivity, showed demethylation at intron 7 of the FKBP5 gene [424]. This activation effect has been found being ageing-related and having impact on brain structure [425-427]: the T-allele carriers were found deficient in cognitive-attentional functioning, based on GM volume in the dorsal anterior cingulate and WM disintegrities. Yet, up to date, no clear associations with LOAD were reported in humans for calcineurin (Section 1.4.) inhibitor FKBP5. However, the binding proteins 51 (FKBP5 6p21.31) and 52 (FKBP4 12p13.33) [74, 428] are initiators of NC3R activation, and these proteins have been found regulative for (a) tau oligomerisation, and (b) Abeta toxicity [429].
Its downstream target, the early growth response 1 EGR-1, is a nerve growth factor-inducible protein (NGFI-A) and member of a family of immediate-early gene-encoded transcription factors. Its binding site located in the hippocampal NR3C1 gene exon 1F (17 in animals) promoter has become of interest due to hypermethylation as a result of ELS [406], related to PTSD and depression. The exon 1F nuclear receptor NR3C1 gene promoter, with at least 14 splice variants [406], has come to focus of neuropsychiatry because of ELS mediation (Section 1.3.). Brains of suicide victims who experienced ELS by childhood abuse, revealed reduced GR mRNA and increased levels of methylation of the NR3C1 exon 1F, which regulates GR expression in the hippocampus [98]. Anacker and colleagues describe that EGR1 also mediates compensatory activation to early stress responses [422] by maternal licking and grooming, which then increases EGR-1 binding in hippocampal NR3C1 exon 17. In parallel, glutamate decarboxylase Gad1 (GAD1 2q31.2) [422], related to insulin signalling, increases. In result of both activations, histone code H3K9 acetylation stabilises GR promoter initiation in the hippocampus. Histone code H3K9ac was found critical for the recruitment of TFIID at the IFNgamma locus, and is hence considered important in eliciting immune responses. Negative ELS long-term effects are further mediated by the neuronal transcriptional repressor REST [169] (Section 1.3.), the expression of which is reduced in LOAD. However, EGR1 itself is not methylated in the hippocampus, yet, is believed relating indirectly to expression of the PSEN2 locus.
3.1.11. NPY and Interactions with Growth Factors
Properties of NPY in CNS consist of neuroprotection, stimulation of neurogenesis, inhibition of neuroinflammation, stimulation of autophagy, and increase of trophic metabolism [103]. In LOAD, severely altered structure of NPY neurons exist in cortex and hippocampus, reduction of NPY binding sites, and lower NPY CSF and plasma concentrations [103]. NPY neurons have been found damaged at early stages of LOAD, suggesting NPY system may have a role in pathogenesis [430], but which is not yet characterised. NPY has a resilience function in ELS through moderation of contextual fear learning, possibly exerted though NPY1R (4q32.2) in the amygdala and prefrontal cortex [431]. A resilience function has also been postulated for NPY in LOAD [430], based on in vitro studies. It is speculated that anxiolysis mediated by NPY2/NPY5 receptors may prevent excitotoxicity by inhibiting glutamate release. However, in humans, have genomic association studies hitherto failed to find respective effects. In rat experiments, pharmacologically induced LOAD-simulations were counteracted in terms of protective effects for spatial memory encoding and retention [431], mediated by NPY1R. NPY administration reduced Abeta40 in neurons, consistent with the observed loss of NPY neurons in LOAD [431]. NPY interacts with ERK1/2 and JNK/SAPK pathways through NPYR1, 2 and 5 [103], mutual upregulation of NPY and BDNF expression. NPY might induce BDNF upregulation through CREB phosphorylation, because BDNF and its receptor TrkB are CREB-target genes.
3.2. Indirect Metabolic Long-term Mediation of Stress
3.2.1. Interaction of Metabolic Factors with Stress and ELS Programming
A considerable part of the GWAS-confirmed risk genes (Section 4.1.) points towards engagement or alterations of metabolic processes as essential for LOAD abnormalities. Such metabolic processes as the moderators of cognitive function include inflammatory mediators, rheological factors, and dysregulation of the HPA axis [432]. Many of the long-term ELS effects induced by e.g. maternal separation and isolation cause programming towards weight gain, glucose intolerance, insulin resistance, and other MetS components [92] (Section 1.3.). There exists direct longitudinal evidence of ELS effects by maternal separation into senescence related to LOAD symptoms demonstrated by Solas and colleagues [153] in a rat model: ELS induced altered HPA axis reactivity evincing persistent cognitive impairments. Ageing effects there consisted in reductions in insulin receptors, phosphorylated insulin receptors, in MAPK-related signalling pathways (p-Akt, p-GSK-3beta, p-tau, and p-ERK1/2 levels), and in the plasticity marker Arc, the regulator of Notch signalling [433] (Section 1.1.). The activity-regulated cytoskeleton protein Arc (ARC 8q24.3) [153] is a CREB binding [434] synaptic protein responsible for stabilisation of neural circuits arising from mnemonic encoding, in the MAPK signalling cascade under regulation of BDNF and other neurotrophins. ARC expression is reduced in animal models of Alzheimer, and subject to posttranslational modification by CpG methylation, or histone ubiquitination [433]. Probably in concert with PSD-95, Arc is the master regulator of de novo experience-dependent plasticity through LTP and LTD, and this function becomes disrupted by Abeta [434]. Directly related to LOAD core pathophysiology (Section 1.4.) were in the Solas study increased C99 ratio, Abeta42 level, and BACE1 level in the hippocampus of ELS rats [153].
In human epidemiology, obesity and nutrition-related ELS programming effects were described in the 1958 British National Child cohort [85], where early life adversities contributed to MetS in 37.5% prevalence for males and 19.8% for females. Co-morbidities with MetS-related diseases (e.g. CVD, T2DM) sum up to 74% [6]-81% [267, 435, 436] of all LOAD cases. Increased risk of insulin resistance for LOAD was first reported in the Rotterdam ageing study [437] with RRs 1.9~4.3. Other epidemiological studies also revealed that T2DM is associated with a 1.5-2.5-fold increased risk of dementia [432]. Elevating fasting glucose levels increased LOAD risk also in nondiabetic elders at HR=1.19 per 0.9 nmol/L difference [438]. Epigenomic modifications, e.g. in IGF2 methylation, are by now particularly well documented for obesity [439] and subsequent conditions like MetS and T2DM. Such investigation also showed concurrent epigenomic modifications in loci relevant to stress processing (NR3C1, HSD11B2, SCL4A6), and immune functions (PI3KCD, TNFA, IL6 promoter) thus connecting ELS and later obesity [439], with late-life LOAD.
3.2.2. Insulin Signalling and Glucose Transport in LOAD as Sequelae of ELS
Deceleration in cerebral glucose transport is characteristic of LOAD. This glucose transportation abnormality and the dysfunction of intracellular glucose catabolism [440] is already present in MCI and predictive to clinical manifestation of LOAD. It is partly responsible for cognitive deficits and neuronal degeneration, and (a) altered functional status of thiamine metabolism, and (b) brain insulin resistance were conjectured as major pathogenic mechanisms in LOAD [440]. Moreover, abnormal glucose metabolism induces oxidative stress and mitochondrial dysfunction [440]. Programming towards glucose intolerance is also a known effect of low birthweight commonly attributed to glucocorticoid excess exposure. This has been found to modify insulin sensitivity of tissues, and altered insulin-secreting capacity of the endocrine pancreas [99]. Modification of HSD11B2 increasing foetal glucocorticoid load in rodents, resulted in permanent hyperglycaemia and hyper-insulinaemia in the offspring with life-long elevations in phosphoenolpyruvate carboxykinase (PEPCK) (Section 1.3.) mRNA and activity [99]. The hepatocyte nuclear factor 4 alpha HNF4A (20q13.12) and GR regulate PEPCK by binding its promoter in the liver [99]. Also, 11beta-HSD type 1 (HSD11B1) methylation, providing reinforcement of local tissue glucocorticoid levels, was identified mediating ELS effects in MetS [39].
Insulin has further to glucose transport essential functions in the CNS, in neural development, dendritic arborisation, synaptic stability, and cognitive functioning [441], which are mediated by insulin receptor expression in diencephalon and telencephalon. However, insulin permeability across the BBB is restricted to hypothalamus, hippocampus and parietal cortex, whereas occipital and other midbrain regions lack permeability. Impairment of the insulin signalling pathway IR/PI3K/Akt is assumed to be responsible for insulin resistance in the brain [441]. There is a strikingly reduced CNS expression of genes encoding insulin (INS 11p15.5), IGF-1 (IGF1 12q23.2), and IGF-2 (IGF2 11p15.5), as well as the insulin and IGF-1 receptor (IGF1R 15q26.3) coinciding with abnormalities in insulin, and IGF-1/IGF-2 signalling diminished in LOAD [442]. Correspondingly, levels of insulin mRNA, IGF, and their receptors are typically reduced in post-mortem LOAD brains [436] (but see [443]), in proportion to progression with neuropathological Braak stageing [436]. This progression coincides with reduced levels of insulin receptor substrate (IRS1 2q36.3) mRNA, tau mRNA, IRS-associated PI3K/p-Akt, GSK-3beta activity [436], and increased amyloid precursor protein mRNA expression. In addition to the PI3K/Akt/GSK-3beta pathway, decreased insulin levels activate also the other MAPK pathways [444] early in LOAD pathogenesis, thus both contributing to NFT aggregation. The IGF-1 in the brain is transported through BBB from the serum by LDL receptors megalin/LRP2 (2q31.1) and LRP1 (Secton 2.5.). IGF-1 binds to megalin/ LRP2 on the endothelial cell surface; IGF-1 import can also be facilitated by LRP1, which is regulated by neuronal activity [443].
There are reduced brain glucose levels reported generally in LOAD brains [441], which have been found accompanied by reduction of the state of O-GlcNAAcylation [445], in turn increasing tau phosphorylation. Hereby, the monosaccharide beta-N-acetylglucosamine (GlcNAc) attaches to serine/threonine residues via an O-linked glycosidic bond in human brain tau. This finding suggests that abnormal hyper-phosphorylation of tau also results from decreased tau O-GlcNAcylation, which is probably induced by deficient brain glucose uptake/metabolism in AD [445]. Reduced glucose metabolism in LOAD is attributable to significant reductions of glucose transporters GLUT-1 and GLUT-2, responsible for neuronal glucose uptake, and to down-regulation of hypoxia-inducible factor 1 alpha subunit (HIF1A 14q23.2), the regulator of GLUT1 and GLUT3, coinciding with decreased O-GlcNAcylation, with hippocampal atrophy, hyper-phosphorylation of tau protein, and density of NFTs [446]. GLUT-4 is only present in hippocampal neurons, but not responsible for glucose transport [447]. All four GLUTs are regulated by IGF1 and IGF1R [443, 447]. The reduction in glucose metabolism, in turn, promotes OXPHOS (Section 1.4.) malfunctions, mitochondrial dysfunctions, and impairment of cyclo-oxygenase COX-2 (PTGS2 1q25.2-q25.3) activity [166, 448].
Neurotoxic Advanced Glycation Endproducts (AGEs) indicate accelerated Maillard reactions due to ageing in T2DM, and are found in LOAD hippocampal neurons [449, 450] and glial cells. AGEs and their receptor RAGE (AGER 6q21.31) resulting from chronic hyperglycaemia provide critical links between diabetes and LOAD [451]. The RAGE gene AGER produces number of splice variants, and resulting protein isoforms of RAGE, including a secreted extracellular form and a N-truncated form [450]. In LOAD, AGEs co-localise with both NFT and Abeta42 plaques [436], thereby increasing oxidative stress through generation of ROS interactions with NFT. Because AGE albumin is synthesised and secreted by microglia, and in turn induced the expression of RAGE, which is also a receptor for Abeta42 [451], and high mobility group protein B1 (HMGB1 13q12.3) (see below), AGEs and RAGEs become critically co-involved in LOAD core pathophysiology [436, 442]. Abeta42 interaction with RAGE, in turn, triggers downstream interaction of the JNK/SAPK pathway [192], in analogy also to MetS [293].
3.2.3. Insulin Receptor-related Receptors, Insulin Receptor Substrates and Insulin Degrading Enzyme
Transport of insulin involves not only insulin (INS 11p15.5), insulin-like growth factors IGF-1 (IGF1 12q23.2), IGF-2 (IGF2 11p15.5), the insulin/IGF-1 receptor (IGF1R 15q26.3), and the myelin-related IGF-2 receptor (IGF2R 6q25.3) [435], but also further regulators such as the insulin receptor-related receptor (INSRR 1q23.1) and the family of insulin receptor substrates, with four members [452, 453], two of which are relevant to LOAD. IRS act as docking sites for adaptor regulators and enzymes [452]. Both insulin receptor substrate 1 (IRS1 2q36.3) [447] and IRS-2 (IRS2 13q34) are decisive for early structural brain growth and functioning, glucose uptake and insulin transport [452]. Specifically, IRS-1 can induce tau phosphorylation through (a) the PI3K/Akt/GSK-3 pathway and (b) through Ras/Mek/ MAPK pathways [452]. Also for the IGF-1R, downstream targets include activation of the MAPK/ERK and PI3K/Akt pathways [443]. Insulin/IGF-1 signalling defects pertain to PI3K/Akt pathway through malignant cascades in glucose metabolism, reduction in their expression correlated with GSK3B activation [440]. This association is covaried with BDNF signalling in charge of maintenance of axonal transport [454]. However, of positive associations with metabolic risk factors [455] for LOAD tested, survived only JNK1 (MAPK8 10q11.22) the analysis in post-mortem study, but not IKK-beta, IRS1, or PKR [455] associations.
Insulin degrading enzyme (IDE 10q23-q25) is correlated to hippocampal Abeta42, with general relations to MAPK signalling in pathophysiology. Insulysin aka insulin-degrading enzyme, which promotes the cleavage catabolism of insulin, is also the major Abeta42 degrading enzyme [436, 456], however only in its monomeric form [457]. Sequestration of insulysin due to hyperinsulinaemia therefore hampers Abeta autophagy [451]. Several of the physiological functions of protease IDE have not yet been elucidated, but are suggested by different locations in cytosol, peroxisomes, endosomes, proteasome complexes. On outer cell surfaces, IDE was found down-regulating the levels of secreted Abeta extracellularly in oligomeric form [458]. PKMzeta, an alternative splice product of the protein kinase Czeta gene (PRKCZ 1q36.33), has been identified important for stabilisation of LTP thus subserving long-term memory engrammes. As one of two atypical isoforms of PKMzeta, it is important in insulin stimulated glucose transport, and its lack consistent with memory loss [459, 460]. Insulin induces phosphorylation of Serine831 GluR1 subunit of AMPAR and induces over-expression of PKMzeta [461]. Insulin-dependent PKMzeta over-expression and MAPK/ERK1/2 phosphorylation is negatively correlated with Abeta42 aggregates, suggesting in conclusion, that Abeta interrupts insulin turnover in LOAD.
3.2.4. Commonalities Between LOAD and T2DM
Glucose hypo-metabolism is an invariant biomarker in LOAD [440], as had become evident from (radioactively labelled glucose) PET neuroimaging, and because it is herein different from all other dementias, LOAD has been termed “T3DM” [435]. Moreover, insulin resistance was found post-mortem also in those LOAD sufferer brains [462], who were not diabetic, specifically in the hippocampi with reduced IGF-1 responsivity in the IGF-1R/IRS-2/PI3K signalling pathway. Brain insulin resistance reduced expression insulin receptor (IRs) and lead to defects in Akt/Foxo3a insulin signalling. Reduced levels of p-Akt and increased levels of Foxo3a were found in the nuclei of neurons with pro-apototic genes activated [463]. Insulin resistance is now generally accepted of being induced by TNFalpha (TNFA 6p21.3) as part of cytokine release. It has been proposed that the common pathogenic link between T2DM/T3DM and LOAD thus consists in the key innate immune receptor TLR4 (TLR4 9q33.1) signalling, activating the JNK/SAPK pathway (Section 2.1.) [268], thereby triggering NFkappaB. Exciting TNFalpha and IL-6 will act into a pro-inflammatory state, whereas chronic TLR4 activation may activate the PI3K/Akt signalling pathway ultimately leading into insulin insensitivity [268, 442]. Further to NFkappaB, TLR4 triggers DUSP1, STAT1, and IRF3, which mediates pro-inflammatory cytokine release [464]. In the cerebrum, TLR4s are expressed mainly in microglia, astrocytes and neurons, where TLR4 contributes to Abeta-induced microglial neurotoxicity, and upregulation of cytokines TNFalpha, IL-1, IL-10 and IL-17 [436, 465]. In the presence of Abeta42, TLR4 converts microglia towards neurotoxicity.
There may be further parallelisms between LOAD and T2DM in amyloid plaque deposition, by the presence of insoluble amyloid protein oligomers in brain and pancreas, respectively [436, 442]. The physiological function of pancreatic amylin appears to be in regulation of insulin secretion from pancreatic beta cells [436]. Amylin (Section 1.4.) (hIAPP) aggregation is associated with pancreatic beta-cell loss, whilst Abeta aggregation is associated with neuronal and synaptic dysfunction [166]. T2DM increases Abeta deposition in hippocampal neurons, and decreases autophagy as indexed by lysosome markers LAMP1 and LAMP2 [466]. Voltage-dependent anion-selective channel 1 (VDAC1 5q31.1) on the outer mitochondrial membrane is responsible for Ca++ transport and ATP efflux into neuron cytosol (Section 1.4.). The interaction of VDAC1 with Abeta42 was found cytotoxic due to conductance increase and resulted in mitochondrially induced apoptosis [467] or mitochondrial dysfunction and neuroinflammation through APP/hIAPP catabolism [468]. Autophagy impairment exacerbates insulin resistance via reductions of insulin secretion in pancreatic beta cells of Langerhans islets [166, 469]. Wang and colleagues observed elevation in the expression of histone deacetylases (HDACs) class IIa in T2DM brains pari passu with altered expression of synaptic proteins, associated with increased susceptibility to oligomeric Abeta42 deposition leading to synaptic dysfunction in the hippocampal formation [470].
It has been proposed that T2DM and LOAD are further characterised by common metabolic factors in the glia-neuron “cholesterol shuttle” constituting one single pathway [261]: (a) lipoproteins APOA1, APOA4, APOC1, APOC2, APOC3, APOD, APOE and LPA, (b) cholesterol transporters ABCA1, ABCA2, CETP, (c) lipoprotein receptors LDLR, LRP1, LRP8 and VLDLR, (d) metabolising enzymes CYP46A1, CH25H, SOAT1, LIPA whose oxysterol products activate APP. APP metabolises cholesterol to 7-betahydroxycholesterol, a substrate of SOAT1 and cortisol converter HSD11B1, tethering to LRP1. BACE1 then cleaves both APP and LRP1. Gamma-secretase cleaves LRP1, LRP8, APP, so regulating transcription factor TFCP2, which controls GSK3B expression responsible for hyper-phosphorylation of microtubular protein tau (MAPT) [261].
A further link between T2DM and LOAD is the heat shock protein Hsp72 (HSPA1A 6p21.33), which is a stress-related chaperone with correction, DNA repair and degradation functions [471]. Being essential to neurodifferentiation during embryonic development, it is also related to involution by regulating caspase activation stimulating the JNK pathway, and inhibiting insulin signalling through NFkappaB and protein kinase C (PKC).
3.2.5. Cholesterol and Lipid Metabolism in LOAD
Lipid traits are important to healthy brain functions equally for neurons and glia. Cholesterol is required by neurons for axonal growth and functioning, and for the synaptic membrane, and there is respective depletion in LOAD post-mortem cases. Cholesterol and triglycerides are necessary ingredients and metabolic precursors of myelin phospholipids. Ko-studies revealed that cholesterol accounts for >70% of myelin lipid bilayer growth [472]. Consequently, cholesterol depletion increases risk for LOAD, as myelin breakdown is accelerated. Lipidomics studies reviewed by Trushina [473] revealed but further group differences between healthy, MCI and LOAD cases: Specifically, in systems phospholipids, phosphatidylinositol, sphingomyelin, ceramide, triacylglycerol, and cholesterol esters. These findings indicate substantial pathological changes in brain myelin content referring to role of myelin breakdown. Related is the deterioration of lipid metabolism in LOAD, and the myelination breakdown tied to apolipoproteins E, C1, J [474], with midlife plasma cholesterol transport. The common cardiovascular and metabolic conditions are therefore at the same time also risk factors for later LOAD manifestation.
Cholesterol transport is mediated by apolipoprotein E (APOE). APOE in the brain maintains BBB integrity and regulates the lipid shuttle between lipoproteins [227] (see above). The circumstance that epsilon4 processed Abeta42 utilises VLDLRs (Section 1.4.) affects healthy BBB functioning in the metabolisms of insulin, triglycerides, free fatty acids (FFAs), and other metabolites involved in cognitive processes [227]: There is initial evidence suggesting that APOE epsilon4 carriers (a) are less effective in insulin utilisation, and (b) are less able to utilise triglycerides in lipoprotein transport [227]. Receptors that can bind APOE, which is also generated by astroglia and neurons, are [475]: the LDLR, very-low-density lipoprotein receptor (VLDLR), LDLR-related protein 8 (LRP8, also known as APOE receptor 2), LRP1 (also known as LRP), and the sortilin-related receptor SORL1 (SORL1 11q24.1). Receptors LRP1, LRP1B, and SORL1 regulate endocytosis of APP, trafficking, and trapping into the Golgi [475] in physiological condition. Other LDL receptor family members, which are not capable binding APOE, include LRP4 (also known as multiple EGF-repeat-containing protein 7 MEGF7), LRP5 and LRP6 [475]. Lipid receptors are essential for embryonic neural development [475], and may therefore be potential candidates for ELS susceptibilities. Astrocytes and neurons participate in cholesterol synthesis through 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR 5q13.3), mobilisation through ATP-binding cassette transporter types A1 and G1 (ABCA1/ABCG1), and transport through BBB by neuron-specific enzyme cholesterol 24S-hydroxylase, encoded by CYP46A1 (14q32.2). The genes related also to cholesterol biosynthesis, DHCR24, and to cholesterol efflux, ATP-binding cassette transporter (ABCA1) [476], have furthermore been related to LOAD.
Signalling through the isoprenoid cascade provides a biochemical chain through which cholesterol synthesis can indirectly modulate production of p-tau in the CNS: from geranylgeranyl diphosphate (GGPP) to cyclin-dependent kinase 5 (CDK5); to GSK-3beta, to protein kinase A (PKA) [474]. As central receptors involved in LOAD [477] were identified: Apolipoprotein E receptor 2 (ApoER2/LRP8) and very-low-density lipoprotein receptor (VLDLR), and low-density lipoprotein receptor-related protein 1 (LRP1) have central roles in lipid clearance of APOE (Section 1.4.) The receptors of the LDL receptor family VLDLR, LRP4, LRP5 and LRP6 are involved in Wnt signalling and are implicated in synaptic plasticity, adult neurogenesis and LOAD pathogenesis [477]. Neuronal stress, e.g. through ER stress or ROS, specifically upregulates VLDLR expression (also known to be preferentially used in APOE epsilon4 allele genotype, Section 1.4.). As memory performance in T2DM is moderated by APOE epsilon4 status [478], this mechanism, together with loss of LRP1s during disease progression, is central to LOAD pathophysiology. Stress impact herein was found accompanied by transiently increased stabilisation and decreased expression of hypoxia-inducible factor 1 alpha subunit (HIF-1alpha, involved in glucose transport, Section 3.2.) and decreased beta-catenin levels affecting the Wnt pathway through GSK-3beta phosphorylation [477]. This mechanism is also promoted by nutritional factors such as excessive LDL intake (see below).
Deficient insulin signalling and glucose metabolism has been widely shown to be influenced by nutritional factors [328]. High-fat plus high sucrose diet triggers expression of insulin receptor, decreasing its tyrosine phosphorylation and, increasing serine phosphorylation of IRS-1. In parallel, inflammatory response by NFkappaB and activations of stress kinase pathways p38 MAPK and JNK/SAPK (Section 2.1.) in whole brain lysate were registered [328]. These changes primed the murine brains towards deposition of Abeta, formation of NFTs, and decreased synaptic plasticity. High fat diet alone increased p-tau levels, microglial activation and induced cognitive impairment, but preferentially in aged animals [479]. Unsaturated fatty acids (PUFAs omega-3 and omega-6) enable in contrast more fluent and effective cholesterol metabolism [183].
3.3. Neuroinflammation in LOAD
3.3.1. Reciprocal Relations of Sympathetic Arousal and Inflammation
Neuroinflammation in LOAD is increasingly discussed as revealed by rising publication numbers. A proportion of GWAS candidate genes pertaining to immune functions (Table 1; Section 4.1.) indicate a strong role for neuroinflammation in LOAD. These findings will be presented as to their specific relevance for the systematic review as tied to ELS: A principle relation consists of the psychophysiological coupling of inflammatory response and noradrenergic signalling. This provides a direct and reciprocal relation of the SNS stress response and cytokines [39].
There is consensus that chronic low-level neuroinflammation in LOAD [480, 481] drives exacerbating neuron degeneration. It is still not conclusively resolved if neuroinflammation is cause or consequence of LOAD [181], but current views [481, 482] increasingly emphasise that neuroinflammation is the driving force for the progression of MCI to clinical manifestation of LOAD. Neuroinflammation alone is sufficient to excite neurodegeneration essential in LOAD pathophysiology [481]. However, neuroinflammation is (such as tauopathy) not specific to LOAD, but exists in a variety of neurodegenerative and autoimmune-related conditions [483]. Suggested by microglial physiology (below) is that early life inflammation is related to late life morbidity, and epidemiology data suggest that LOAD manifestation can be postponed or even prevented by anti-inflammatory agents [484].
3.3.2. Essential Role of Neuroinflammation in LOAD
As inflammatory processes also appear in healthy ageing, their differentiation against LOAD is a matter of dispute [181]. However, neuroinflammatory processes in LOAD are predominant in vulnerable regions with Abeta and NFT deposition [485], where plaques provide stimulation [485] for pro-inflammatory agents such as acute phase proteins and cytokines. Such circumscribed lesion regions are present early in LOAD pathogenesis in dispersed, micro-localised, and enduring manner [485]. When astrocytes and microglia tend to accumulate at such inflammation sites, a process termed ‘reactive gliosis’ [486] appears, which coincides with persistent microglial activation.
Reactive microgliosis (Section 1.4.) is accompanied by processes involving classical complement and alternative complement pathways of the innate immune response [181, 487]. Findings of Abeta42 in and on microglia and astrocytes may suggest that both cell types can be involved in phagocygotic processes [487]. In microglia and astrocytes [488-490], the plaque-associated cytokines interleukin-1(IL-1), IL-6, IL-8, tumor necrosis factor alpha (TNFalpha), interferon gamma (IFNgamma), transforming growth factor beta (TGFbeta), major histocompatibility complex (MHC) class I&II complex proteins [489, 490], and macrophage inflammatory protein-1 alpha and beta (CCL3 17q12 upstream, CCL4 17q4 downstream) [448] and chemokine ligand MCP-1/CCL2 (17q12 cluster) [491], are upregulated in LOAD, a process controlled by the neuroprotective chemokine fractalkine (CX3CL1 16q21) [492]. Free radical generation by activated microglia has been demonstrated by a wide range of in vitro experiments using monocytes/macrophages and microglia, notably ROS, COX-2 [448], increased astrocytic NO [493], and inducible nitric oxide synthase iNOS [488], prostaglandin E2 (PGE-2, the major enzymatic product of COX-2) [448], lipophilic amines [480]. Heneka and colleagues have concluded that Locus Caeruleus (LC) and Nucleus Basalis of Meynert (NBM) (Section 3.1.) degeneration are induced by COX-2 activity [448, 491]. Cytokine release is accompanied directly by CDK5 and JNK/SAPK overexpression increasing with age [494]. In LOAD patients, tumor necrosis factor receptor TNF-R1 (TNFRSF1A 12p13.31) levels are increased, whereas anti-apoptotic TNF-R2 (TNFRSF1B 1p36.22) levels are decreased [495].
3.3.3. Priming and Activation of Microglia in LOAD
Microglia are leukocytes and the brain’s myeloid cells [482], occurring as both free migrating monocytes and tissue bound macrophages [483]. Activated microglia at the inflammation site change their morphology (polarisation), express increased levels of Major Histocompatibility Complex (MHC) antigens, and become phagocytic [236]. Four microglial forms according to stage of activation are known in the literature: (a) the amoeboid stage during early brain development, contributing to cerebral morphology, as freely moving phagocytes; (b) a ramified stage, with provision of stable immune function and as macrophages; (c) a polarised neuroinflammatory stage, with retraction of branches, antigen presenting, production of chemokine and complement component proteins [496]; (d) the dystrophic microglia as a result of the ageing process [483], and related to cerebral neurodegeneration.
Neuroinflammation in LOAD is characterised by innate immune responses and monocyte activation (and in contrast to other neuroinflammatory diseases, with adaptive immune responses), whereby monocytes retain these capabilities also under Abeta burden [497]. Microglia-depleted rodent models revealed that dystrophic microglia are involved in propagation of h-tau via exosomes [498], and thus transregional spreading of NFTs. Tau interacts with APOE epsilon4 allele [499] to boost cytokine release from microglia involved in neuroinflammation [483]: Deficiency of microglial-specific fractalkine receptor (CX3CR1 3p22.2) was observed leading to increased microglial IL-1beta release thus increasing tauopathy through p38 MAPK activation [500]. Progranulin expressed on microglia (GRN 17q21.31) is important for microglial [483] complement expression and lysosome maturation, may in LOAD be protective for synapse functioning [501], and inhibitive of plaque deposition [502]. GRN polymorphisms were found associated with LOAD risk [502].
Inflammatory priming of microglia is a further possible link between ELS and LOAD pathology. It was found that a prenatal pro-inflammatory environment decreases adult neurogenesis by persisting microglial activation and by downregulated expression of Transforming Growth Factor beta-1 (TGFbeta1) [503]. Important for this state is the so-called priming phase, when microglia are first stimulated towards cytokine release [100, 482]. Such microglial priming can occur as an early life event, including stressful events or infections, and be suppressed by epigenetic programming of microglial histone deacetylase inhibitors [100], and also being mediated by nutrition. Ageing or other insult can then simply reactivate the suppressed inclination towards neuroinflammation [100]. Once activated, microglia differentiate in phenotypes M1 and M2 [504]: M1 classically activated, respond to LPS in combination with IFNgamma in pro-inflammatory profiles, whereas M2, or alternatively activated microglia, respond to IL-4 and IL-13 with anti-inflammatory profile. Dependent pro-inflammatory profiles are inhibitive of adult neurogenesis, whereas anti-inflammatory microglia phenotype stimulates hippocampal neurogenesis and oligogendrogenesis through immune modulators such as TGFbeta1 [505].
3.3.4. Role of Microglial Action in LOAD
The microglial involvement consists of a dual contribution [480]: (a) microglia help eliminating Abeta aggregation via phagocytosis; and (b) microglia reinforce Abeta accumulation through the release of neurotoxic proteases and IFN Regulatory Factor 1-dependent (IRF1-dependent) inflammatory response [506]. Control of neuroinflammation is exerted through the anti-inflammatory reflex (see below) cytokines IL-4, IL-10, IL-13 [480]. However, if polarised, microglia present pro-inflammatory cytokines and chemokines, and this will result in self-harm, whereby ‘burnt-out’ microglia become dystrophic [482]. Microglia are furthermore the source of the heme enzyme myeloperoxidase (MPO 7q22) in LOAD brains [507]. Increased expression of MPO mRNA is detected in microglial cells upon treatment with aggregated Abeta peptide, and MPO immunoreactivity has been observed in microglia co-localised with neuritic plaques [507]. High-mobility-group-protein B1 (HMGB1 13q12.3) was found the missing link between neuroinflammation and NFT tauopathy in LOAD [508], where it has a cytokine-like function. Its main receptor is TLR4/2, and its most important signalling pathways are p38 MAPK and NFkappaB. The HMGB1 locus is known for posttranslational modifications by acetylation, methylation, ADP-ribosylation, phosphorylation, and glycosylation.
3.3.5. Inflammasome Participation in LOAD Neuroinflammation
Several studies suggested that inflammasome activation may be linked to disease severity in LOAD by increasing memory deficits. Involvement of inflammasome, a multiprotein oligomer consisting of caspase 1, caspase 5, pattern recognition NOD-like receptor NALP, and apoptosis associated protein PYCARD with its adaptor ASC (PYCARD 16p11.2), is usually a defence against external infection. Abeta is probably acknowledged as such, as it is known to trigger the activation of the cryopyrin gene (NLRP3 1q44), a sensor protein playing a critical role in microglial activation and hence LOAD initiation [509, 510]. Inflammasome activation is triggered by autophagy impairment (Section 1.4.), mediated by beclin-1 and the interacting anti-apoptotic Bcl-2 family members [360]. These are ageing-related, but still overexpressed in LOAD brains [360, 361]. The core complex beclin-1/Atg14L/Vps34/Vps15 is under regulation of CDK5, and inversely related to Abeta42 deposition [361].
The NLR (‘Nod-like receptor’) family NLRP3 gene provides instructions for synthesis of the protein cryopyrin (NALP3), the activation sensor of the inflammasome [511], an intracellular oligomeric organelle consisting of the proteins caspase-1/5, PYCARD, and NALP. Cryopyrin interacts with the apoptosis-associated speck-like protein PYCARD and its inflammasome adaptor ASC, which contains a caspase recruitment domain (CARD), and is a member of the NALP3-inflammasome complex. Psychological stress-induced activation of GSK-3beta leads to activation of the NLRP3/IL-1beta pathway [266], thus enhancing active caspase-1 expression [511, 512]. This NALP-inflammasome complex functions as an upstream activator of NFkappaB signalling thereby triggering the NACHT, LRR and PYD domains-containing protein 3 (NLRP3); the inflammasome induces mitochondrial apoptotic signalling by beclin-2, stimulating the production of interleukin-1beta (IL-1beta) [513]. Oxidised mitochondrial DNA released into the cytosol induces the formation of the NLRP3 inflammasome. Autophagy, in contrast, has been found to be essential for restoring mitochondrial dynamics during NLRP3 inflammation [451]. In addition to NLRP3, also NLRP1 (17p13.2) and its polymorphisms, have been implicated in LOAD [512].
Four prostaglandin E receptors (EP1-EP4) are present on microglia [448], of which EP2 (PTGER2 14q22.1) expression is regulated by methylation, and OS related [506, 514]; also EP3 (PTGER3 1p31.1) with at least eight isoforms [515, 516], and EP4 (PTGER4 5p13.1) [517], were found related to LOAD. Their activation of ERK induces expression of EGR1/NGFI-A (an ERK transcription factor, 5q31.2) (Sections 1.3. and 3.1.), and suppresses the activation of cytokines by inhibiting the NFkappaB pathway. Recent murine studies [506] have demonstrated that inhibition of the COX-2/PGE2/EP2 immune pathway by deletion of the microglia-specific ptger2 restored microglial chemotaxis and Abeta clearance, suppressed toxic inflammation, and increased cytoprotective Insulin-like Growth Factor 1 (IGF1) signalling [506].
3.3.6. Leakage of Blood-brain Barrier and Exacerbation of Neuroinflammation in LOAD
The key process in LOAD neuroinflammation consists probably in RAGE (Section 3.2.) malfunction-caused leakage of the inflammation BBB [100, 480, 518], accompanied by LPR1 expression downregulation (thus pathogenic irrespective of APOE allelic status) (Section 1.4.) [181], and dysregulation of enzymatic degradation (such as zinc matrix metalloprotease MMP-9 involved in learning and memory [MMP9 20q13.12]) [480, 519]. Whereas Abeta clearance by autophagy is more effective in younger age ranges, in higher ages, RAGEs tend to interact with glycated adducts on endothelial cells leading into diabetic vascular injury. Production of pro-inflammatory cytokines and vasoconstrictor endothelin-1 (EDN1 6p24.1) then reduces cerebral blood flow and induces mutually malignant potentiation of Abeta influx and RAGE expression [518]. Of four known isoforms, the endothelin receptor genes EDNRA (4q31.22-23), and EDNRB (13q22.3), were found involved in BBB leakage. The increased BBB permeability gives rise to age-dependent progressive BBB breakdown, by activating the pro-inflammatory cyclophilin A and the NFkappaB-dependent matrix MPP-9 pathway [181], resulting in general cerebral vulnerability to peripheral immune processes [181].
It has been suggested that increased LOAD risk of HR 1.07-1.12 due to proximity to major roads may be associated with disruption of BBB and activation of microglia [37]. The effects of increased NFkappaB activation, mitochondrial dysfunction and cytoskeletal disintegration causing death of oligodendrocytes give rise to myelin lesions and terminal myelination breakdown in LOAD [181]. Likely is a co-action of brain insulin resistance (Section 3.2.), with ROS generation, and with neuroinflammation [447]. Another potential mechanism of neurotoxicity potentiation consists in the blockade of intracellular insulin actions, thus by downstream inhibition of insulin signalling events, through pro-inflammatory cytokine action on p-IRS1. In parallel, were markers of ER stress p-ERK, and eIF2alpha, observed elevated in LOAD brains [447].
3.3.7. Abeta Phagocytosis by Microglia
In manifest LOAD is Abeta phagocytosis mediated by scavenger receptors and modified LDL-clearance performed by polarised microglia [520]. Scavenger receptors, part of innate immunity, are expressed by microglia, related to atherosclerotic lesions, with secreting cytokines [496]. There are currently ten classes (A-J) encompassing 19 receptor types with highly divergent domain structures [496, 520], four of which have been described relevant to LOAD [480]: Class-A scavenger receptor/CD204 gene (MSR1 8p22, 3 isoforms) expresses the SR-A isoform1, which is related to ROS production and microglial interaction with Abeta. SR-A isoform2 promotes phagocytosis by binding fibrillar Abeta and AGEs. SR-A isoform 3 is inhibitive of types 1 and 2. SR-B2/CD36 (SCARB3 7q11.2) is also relevant to LOAD, as activated SR-B2 signal transduction involves the tyrosine kinase Fyn, p38 MAPK and JNK [520]. SR-B1 (SCARB1 12q24.31) is the microglial Abeta receptor and promoting cerebrovascular oxidative dysfunction. SR-B3 (SCARB2 4q21.1) mediates post-Abeta binding activation and lyosomal transport [480]. Activation by ligand Abeta on CD36 triggers p38 MAPK, ERK1/2, JNK, and NFkappaB pathways and stimulates production of pro-inflammatory cytokines and of pro-apoptotic signals [496]. Specifically, the pathogenic interaction of Abeta with engagement of CD36–TLR4-TLR6 receptor complexes on microglia results in neuroinflammation and consequent damage to local tissues at sites of Abeta accumulation [496].
Abeta may itself act as a pro-inflammatory agent [236], and microglia can also be activated by Abeta42 oligomers upon mediation by the scavenger receptor SR-A, and the activated potassium channel KCa3.1, before fibrillary deposits are formed [510]. Several Abeta-degrading enzymes such as neprysilin NEP, insulysin IDE, endothelin converting enzymes ECE-1 and ECE-2, angiotensin converting enzyme ACE, cathepsin B, and plasmin are involved in neuroinflammation, which inhibit their microglial activity to degrade and clear Abeta [480, 519]. Mononuclear cells (PBMC’s) stressed with Abeta upregulated the glycosyltransferase MGAT-III (MGAT3 22q13.1) by a factor of 327. The gene product of MGAT3 may have a critical role in phagocytosis [519]. In addition, microglial-expressed purinergic ionotropic receptor P2X7 (P2RX7 12q24.31) participates in phagocytosis, however, a role in LOAD remains to be clarified [482].
It was shown that neuroinflammation upregulates BACE1 expression, and that knock out of the mapk14 gene encoding p38alpha MAPK enhanced BACE1-induced lyosomal degradation and autophagy [330]. TGFbeta1 decreased Abeta burden in an AD mouse model by promoting microglial Abeta clearance. However, blocking TGFbeta1 and downstream SMAD2-SMAD3 signalling specifically in CD11c-positive myeloid cells also reduced Abeta-like pathology [225]. TGFbeta1 is secreted by astrocytes, and binding of TGFbeta1 to its receptor activates the SMAD3 pathway, as well as MAPKs and PI3K signalling in microglia. Thus, TGFbeta1 regulates the inflammatory activation of microglia, but this capability is lost in ageing or chronic activation, in contrast to MAPK signalling [225].
3.3.8. Proinflammatory Cytokines in LOAD Memory Impairment
Ageing promotes infiltration of immune cells into the brain owing to damage to the Blood Brain Barrier (BBB), thus leading to the exacerbation of central inflammation [451], a process greatly accelerated in LOAD. In vivo, microglia express pro-inflammatory cytokines IL-1, IL-6, granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-12 and IL-23, and TNFalpha [482]. These directly impair hippocampal LTP [521], and for IL-1, IL-6, TNFalpha, and alpha1-antichymotrypsin (SERPINA3 14q32.13), genomic polymorphisms have been documented for LOAD [491]. The ratio of the pro-inflammatory cytokine IL-1 to the anti-inflammatory cytokine IL-10 is greatly elevated in the serum of AD patients, resulting in a chronic neuroinflammation state [236]. Novel evidence supports the notion that tau hyper-phosphorylation can be induced by C-reactive protein (CRP 1q23.2) a pro-inflammatory marker via the Akt/GSK-3beta signalling pathway [522], and thereby priming neuronal apoptosis. Other cytokines such as TNFalpha, IL-1beta, IL-6 were shown involved in a previous study [523], but only increased in the hippocampus, when CRP application in vitro triggered APP expression and Abeta42 cleavage. In contrast, phosphorylation of Akt (Ser473) and GSK-3beta (Ser9) were decreased by CRP treatment, whereas phosphorylation of ERK and p38 were not affected [522]. Taken together, the experiments demonstrate that also the acute-phase protein CRP is capable of triggering key pathogenic mechanisms in AD in vitro.
The arachinodate 5-lipoxygenase (5LO) (ALOX5 10q11.21) is a pro-inflammatory enzymatic pathway widely distributed within the central nervous system and is up-regulated in LOAD [524] in regions of progressed neurodegeneration. Through mediation of CDK5 [524], GSK-3beta, MAPK, and as part of innate immunity, it subserves chronic neuroinflammation, known to modifying tau hyper-phosphory- lation and inhibiting postsynaptic marker PSD-95 (Section 2.1.) [524]. The multiplicity of splice variants from this gene suggest susceptibility for environmental influences.
The enzyme endothelial nitric oxide synthase eNOS (NOS3 7q35-36), an important vasodilator, has been shown to be inhibitive of microglial activation and the pro-inflammatory phenotype [525]. eNOS-deficient mice showed increased expression of APP and BACE1, therefore NOS3 is a modulator of LOAD pathophysiology. NOS3 is extensively documented for multiple epigenetic modifications und has been shown to be related to early uterine longitudinal conditions [526], where 5-hydroxymethylcytosine, H3K9ac and histone 2A (H2A) interact.
3.3.9. Polymorphisms for Cytokines Related to LOAD
TNFalpha [527], which has partly neuroprotective effects [528], when interacting with NFkappaB and IL-1beta [529], and binding tumor necrosis factor receptor 1 TNF-R1 [530]. TNF-R2 signalling mediates most of the protective and regulatory effects of TNFalpha in the CNS [528]. Recent analyses have shown that neuroprotective effects of NFkappaB are exerted by p65 dimers, whereas p50 dimers are pro-apoptotic [531, 532]. Therefore the p65:p50 ratio is decisive for direction of NFkappaB action. Anti-inflammatory and immunosuppressive protein glucocorticoid induced leucine zipper GILZ (TSC22D3 Xq22.3), which is a p65-binding protein that sequesters activated p65, and thereby inhibits transactivation of inflammatory and apoptotic factors. In contrast is Abeta40 synthesis related to p50-preferential signalling [531]. GILZ regulates multiple signal transduction pathways involved in cell growth, cell differentiation, and cell survival. Reviewed for transethnic LOAD association [533] were the following cytokine loci: IL1A (2q14.1), IL1B (2q14.1), IL6 (7p15.3), TNFA (6p21.33), VEGFA (6p21.1), IL18 (11q23.1), IL33 (9p24.1), IL12A (3q25.33), IL12B (5q33.2), IL4 (5q31.1), IL10 (1q32.1), TGFB1 (19q13.2). The cytokine SNPs have often divergent risks, showing decreased vs. increased risk, dependent on the population [533] tested.
3.3.10. Anti-inflammation Reflex
Inflammatory action of NFkappaB and ERK pathways is inhibited by peroxisome proliferator-activated receptor gamma (PPARG 3p25.2) activation, and many intervention address this mechanism [489, 534] or its regulator transcription coactivator PGC-1alpha [535] (PPARGC1A 4p15.2). PPARGC1A, a regulator of metabolic genes, OXPHOS (Section 1.4), and mitochondrial biogenesis, activates CREB and is involved in mitochondrial ROS generation, the reason why it was recently nominated to be a master regulator of neuroinflammation in LOAD [536]. It has been demonstrated that receptor PGC-1alpha action regulates BACE1 expression and is downregulated in LOAD patients [535]. PPARGC1A and MetS-related PPARGC1B (5q32), which stimulates expression of glucocorticoid receptors, is furthermore involved in T2DM; both genes are transcriptionally and post-transcriptionally regulated [536].
3.3.11. Apoptosis Related to Neuroinflammation
It has been shown that neuronal cell death in LOAD is causally induced by the proto-oncogene c-myc [537], whose overexpression is counter-regulated and apoptosis is induced [538]. This occurs when neurons are (a) deprived of NGF and (b) exposed to Abeta42 cytotoxicity. Consequently, myc expressions are preferentially located at diffuse plaques, dystrophic neurites, synaptosomes, and in astrocytes [539], apparently interacting with NFkappaB. The death receptors DR4 (TNFRSF10A 8p21.2) and DR5 (TNFRSF10B 8p21.2) were shown to mediate cerebral microvascular endothelial cell apoptosis that was induced by oligomeric Abeta [236]. RNA translocation protein RanBP9 (RANBP9 6p23) is a scaffold protein that is increased in brains of AD patients [236]. Heightened activation of inflammatory responses produces phosphorylation of p38 MAPK, JNK, and NFkappaB pathways [488, 493] thus inducing apoptosis. Neurotoxic elements are conducive to apoptotic signalling through activator protein AP-1-dependent beclin-associated protein BAX (BAX 19q13.33) and caspase-9 in oligodendrocytes and astrocytes [488], thus accelerating myelination breakdown in LOAD. Abeta42 also directly induces cell death in oligodendrocyte cell cultures [490].
4. EPIGENETICS OF RISK GENES AND GENE REGULATION
4.1. Summary of LOAD Risk Genes in the Reviewed Literature
In sporadic AD, there is no Mendelian heritability, and also familial transmission showed variable concordance rates. There is no single genetic risk for LOAD [540], therefore network analyses of genes are currently considered most promising. A growing number of GWAS in the past two decades have confirmed candidate genes, where common variants, such as ABCA7 [541] or CR1 [542], typically account only for 3-4% variance [8, 543]. It has been suspected that APOE epsilon4 masking effects have hitherto precluded the detection of other genetic risk loci [544]. Studies of gene expression and neuronal activity have, however, been scarcely done [540], a promising combination, which could identify neuroimaging patterns in relation to epigenetics.
Risk genes derived from core pathophysiological processes (Section 1.4.), such as APP [545, 546], MAPT [546], and BACE1 [547] have been found transcriptionally downregulated in LOAD [545], possibly to maintain protein homeostasis. For APP processing, to date >160 mutations in APP have been reported, most of which increase the production of the amyloidogenic Abeta42 isoform of Abeta [475]. APOE epsilon4 allele and APP [211] are, however, classified rare variants in LOAD [546] due to their allele frequencies. The MAPT H1 haplotype, then, is characteristic of several late-onset neurodegenerative disorders [211]. The neuronal receptor of apolipoprotein E (SORL1 11q24.1) is the only firmly established risk gene further to APOE epsilon4 [1, 475, 542, 548-553]. The sortilin-related receptors SORCS1 (10q25.1), SORCS2 (4p16.1) and SORCS3 (10q25.1) (vacuolar protein sorting 10 [VPS10] domain-containing receptors 1-3) are inhibitive of APP processing [554]. The channel-forming subunit of mitochondrial translocase (TOMM40, 19q13.32) [541] is critical for general protein precursor import into mitochondria, and is considered a common variant. The risk status of its SNP rs2075650 was confirmed in a recent meta-analysis although not for all populations [555]. The clusterin gene (CLU 8p21.1) [183, 188, 541, 542] is a molecular Golgi chaperone, but under stress involved in intracellular clearing, lipid transport and apoptosis. The cytochrome P450 enzyme CYP19A1 [92, 544] (15q21.1) aromatase or oestrogen synthase showed association likely due to the neuroprotective effects of oestrogen.
Further GWAS-confirmed candidate genes (Table 1) (additional suspected genes extracted from preceding review sections listed in Table 3) are the ABC1 transporter gene ABCA7 [542] (19p13.3), the A-disintegrin and metalloproteinase domain-containing protein gene ADAM10 [556] (15q23.3), the bridging integrator protein gene, involved in axonal ensheathment by oligodendrocytes and white matter tracts, BIN1 [546] (2q14.3), the CD2-associated protein gene CD2AP [542, 557] (6p12.3), the complement C3b/C4b receptor type 1 gene CR1 [542, 546] (1q32.2), the ephrin type-A receptor 1 EPHA1 [542, 546, 557] (7q34-35), the membrane-spanning 4-domains subfamily A member 4 MS4A4A/ MS4A6A [542] (11q12.2), the phosphatidylinositol binding clathrin assembly protein PICALM [542] (11q14.2), the phospholipase D family member 3 gene PLD3 [542, 557] (19q13.2), and triggering receptor expressed on myeloid cells 2 TREM2 [542] (6p21.1), which have been found associated specifically with LOAD [540]. The ABCA7 transporter is known affecting the production and clearance of Abeta [557, 558]. The ADAM10 encodes a proteolytic enzyme at the alpha secretase cleavage stage, accumulating in hippocampal synapses [556, 559]. The TREM2 encodes microglial cytokine signalling elements related to chronic inflammation, autophagy and Abeta clearance [541, 542, 557, 560]. The PICALM [7, 541, 542, 546, 557, 561] gene produces a clathrin protein involved in membrane recycling and autophagy. The CR1 complement receptor 1 gene [8, 541, 542, 546, 557, 562], a membrane glycoprotein binding to immune complexes. BIN1 [542, 546, 557] produces a nucleocytoplasmic adaptor protein important for synaptic functioning and myelination [563, 564]. The EPHA1 gene product was found regulating cerebral glucose levels, and brain atrophy [565]. The CD2AP encodes a cytoplasmic protein implicated in membrane trafficking, related to Abeta40/42 levels [566]. The MS4A4A/MS4A6A was found significantly related to CSF Abeta42 [567]. The PLD3 enzyme phospholipase D is described catalysing membrane phospholipids, thereby exerting influence upon processing of APP [568]. ABCA7 and BIN1, ANK1, CDH23, DIP2A, RHBDF2, RPL13, SERPINF1 and SERPINF2 [569], ANK1, a well-known susceptibility gene for T2DM [476], RPL13 and RHBDF2, are biologically linked to PTK2B, a known LOAD-associated gene. The fourth, the cadherin gene CDH23, is involved in neuronal differentiation [543]. The ANK1 gene was associated with neuropathology in the entorhinal cortex, and was confirmed as being substantially hypermethylated in superior temporal gyrus and prefrontal cortex [570].
Table 3.
Extracted risk genes for late-onset Alzheimer disease.
| Locus | Gene | Function | References |
|---|---|---|---|
| 12p13.31 | A2M | Alpha2-macroglobulin: cytokine transporter, Abeta degradation with LRP1, interaction with MPO | [654] |
| 21q22.3 | ABCG1 | ATP binding cassette subfamily G member 1 | [275, 276, 655, 656] |
| 18p11.32 | ADCYAP1 | Adenylate cyclase activating polypeptide 1: proprotein increasing cyclic adenosine monophosphate (cAMP) levels, stress response | [656] |
| 10q25.2 | ADRA2A | Adrenoceptor alpha 2A: sympathetic transmission | [657, 658] |
| 10q25.3 | ADRB1 | Adrenoceptor beta 1: sympathetic transmission | [376] |
| 5q31-32 | ADRB2 | Adrenoceptor beta 2: sympathetic transmission | [381] |
| 8p11.23 | ADRB3 | Adrenoceptor beta 3: sympathetic transmission | [659] |
| 6p21.32 | AGER | Advanced glycosylation endproduct (AGE) receptor: neuroinflammation | [660] |
| 10q11.21 | ALOX5 | Arachidonate 5-lipoxygenase: regulator of gamma-secretase | [661] |
| 4p14-p13 | APBB2 | Amyloid beta precursor protein binding family B member 2: Abeta cascade | [275, 276, 662] |
| 21q21.3 | APP | Amyloid beta precursor protein: Abeta cascade | [663] |
| 8q24.3 | ARC | Activity-regulated cytoskeleton-associated protein: synaptic plasticity, regulator of Notch | [664] |
| 12q23.2 | ASCL1/MASH1 | Achaete-scute family bHLH transcription factor 1: ANS neurogenesis, noradrenergic neurons | [665] |
| 22q13.1 | ATF4 | Activating transcription factor 4: neurodegenerative signal | Not tested |
| 20p13 | AVP | Arginine vasopressin: neuropeptide implicated in stress | Not tested |
| 11q23.3 | BACE1 | Beta-secretase 1: Abeta cascade | [666, 667] |
| 19q13.33 | BAX | BCL2 associated X apoptosis regulator: interacting with mitochondrial voltage-dependent anion channel (VDAC) | [668] |
| 3q26.1 | BCHE | Butyrylcholinesterase: cholinergic transmission, acetylcholine catabolism | [275, 276] |
| 11p14.1 | BDNF | Brain-derived neurotrophic factor: age of onset | [669, 670] |
| 17q21.31 | BECN1 | Beclin 1: autophagy | Not tested |
| 5q32 | CAMK2A | Calcium/calmodulin dependent protein kinase II alpha: synaptic plasticity, LTP | [671] |
| 17q12 | CCL2 | C-C motif chemokine ligand 2: immune | Negative finding |
| 17q12 | CCL3 | C-C motif chemokine ligand 3: immune | Negative finding |
| 17q4 | CCL4 | C-C motif chemokine ligand 4: immune | Negative finding |
| 10q21.2 | CDK1 | Cyclin-dependent kinase: cell cycle control, haplotype with p38α, JNK1-3, MEK2, and ERK2 loci | [671] |
| 7q36.1 | CDK5 | Cyclin-dependent kinase: brain development | [275, 276, 612] |
| 17q21.2 | CNP | 2',3'-Cyclic-nucleotide 3'-phosphodiesterase: oligodendrocyte differentiation, microtubule formation | [672] |
| 22q11.2 | COMT | Catechol-O-methyltransferase: catecholamine regulation | Negative finding |
| 2q33.3 | CREB1 | cAMP response element binding protein 1: transcription induction | Not tested |
| 9p13.3 | CREB3 | cAMP response element binding protein 3: transcription induction | Not tested |
| 11p11.2 | CREB3L1 | cAMP responsive element binding protein 3 like 1: ER stress | Not tested |
| 7q33 | CREB3L2 | cAMP responsive element binding protein 3 like 1: ER stress | Not tested |
| Locus | Gene | Function | References |
| 19p13.3 | CREB3L3 | cAMP responsive element binding protein 3 like 1: ER stress | Not tested |
| 1q21.2 | CREB3L4 | cAMP responsive element binding protein 3 like 1: ER stress | Not tested |
| 7p15.1-p14.3 | CREB5 | cAMP response element binding protein 5: trans-activator | Not tested |
| 16p13.3 | CREBBP | CREB binding protein: chromatin remodelling to transcription factor recognition | Not tested |
| 8q13.1 | CRH1 | Corticotropin releasing hormone: HPA | Not tested |
| 5q13.3 | CRHBP | Corticotropin releasing hormone binding protein: CRH inactivation | Not tested |
| 17q21.31 | CRHR1 | Corticotropin releasing hormone receptor 1: HPA regulation, association with MAPT H1 haplotype | [673] |
| 7p14.3 | CRHR2 | Corticotropin releasing hormone receptor 2: HPA regulation | Not tested |
| 1q23.2 | CRP | C-reactive protein: inflammation | [674] |
| 18q21.1 | CTIF | Cap binding complex dependent translation initiation factor: translation | [675] |
| 16q21 | CX3CL1 | C-X3-C motif chemokine ligand 1: fractalkine | Not tested |
| 3p22.2 | CX3CR1 | C-X3-C motif chemokine receptor 1 | Not tested |
| 9q34.2 | DBH | Dopamine beta-hydroxylase: noradrenalin converter, interaction with IL1A and IL6 | [386] |
| 16p12.1 | DCTN5 | Dynactin subunit 5: vesicle transport | Not tested |
| 1q23.3 | DDR2 | Discoidin domain receptor tyrosine kinase 2: cell-environment communication | [675] |
| 1p32.3 | DHCR24 | 24-dehydrocholesterol reductase: cholesterol biosynthesis | [275, 276, 620] |
| 17p13.1 | DLG4 | Discs large MAGUK scaffold protein 4: postsynaptic density protein 95 | [676] |
| 19q13.2 | DNMT1 | DNA methyltransferase 1: maintaining methylation patterns | [92, 677] |
| 2p23.2 | DNMT3A | DNA methyltransferase 3A: de novo methylation | [678] |
| 20q11.21 | DNMT3B | DNA methyltransferase 3B: de novo methylation | [679] |
| 5q35.1 | DUSP1 | Dual specificity phosphatase 1: cellular response to environmental stress | Not tested |
| 21q22.13 | DYRK1A | Dual specificity tyrosine phosphorylation regulated kinase 1A: brain maturation | [275, 276, 680] |
| 11p13 | EAAT2/SLC1A2 | Excitatory amino acid transporter 2/glial high affinity glutamate transporter: synaptic glutamate clearance | [583, 681] |
| 6p24.1 | EDN1 | Endothelin 1: vasoconstrictor | [682] |
| 4q31.22-23 | EDNRA | Endothelin receptor type A: vasoconstriction | Not tested |
| 13q22.3 | EDNRB | Endothelin receptor type B: vasoconstriction | Not tested |
| 5q31.2 | EGR1 | Early growth response 1: transcriptional regulator | [683] |
| 2p22.2 | EIF2AK2 | Eukaryotic translation initiation factor 2 alpha kinase 2: translation initiation | [253, 684] |
| 15q15.1 | EIF2AK4 | Eukaryotic translation initiation factor 2 alpha kinase 4: protein synthesis | [602] |
| 14q23.3 | EIF2S1 | Eukaryotic translation initiation factor 2 subunit alpha: protein synthesis initiation | Not tested |
| 12p13.33 | FKBP4 | FK506 binding protein 4: steroid receptor signalling, immunoregulatory gene expression, tauopathy | [685] |
| 6p21.31 | FKBP5 | FK506 binding protein 5: immunosuppression, calcineurin inhibition, tauopathy, LOAD progression | [686] |
| 14q24.3 | FOS | Fos proto-oncogene AP-1 transcription factor subunit: proliferation, apoptosis | [687] |
| 11q14.1 | GAB2 | Growth factor receptor-bound protein 2 associated binding protein 2: interaction receptor tyrosine kinases, PI3K/ERK/Akt pathway, brain growth | [688-690] |
| Locus | Gene | Function | References |
| 6p21.3 | GABBR1 | Gamma-aminobutyric acid type B receptor subunit 1: inhibitory neurotrans-mission | Not tested |
| 9q22.33 | GABBR2 | Gamma-aminobutyric acid type B receptor subunit 2: inhibitory neurotrans-mission | Not tested |
| 5q34 | GABRA1 | Gamma-aminobutyric acid type A receptor alpha1 subunit: inhibitory neurotrans- mission | Not tested |
| 2q31.2 | GAD1 | Glutamate decarboxylase 1: GABA synthesis | Negative |
| 10p12.1 | GAD65 | Glutamate decarboxylase 2: GABA synthesis | [691] |
| 5p13.2 | GDNF/GDNFOS | Glial cell derived neurotrophic factor: activation of SMAD family transcription factors, neuroprotection | [692] |
| 1p35.3 | GMEB1 | Glucocorticoid modulatory element binding protein 1: transactivation of GRs bound to GREs | [12] |
| 20q13.33 | GMEB2 | Glucocorticoid modulatory element binding protein 2: transactivation of GRs bound to GREs | [12] |
| 12p13.31 | GNB3 | Guanine nucleotide-binding protein subunit beta 3: regulators of signal transduction receptors, interacting with ADRB1/MAPK signalling in LOAD | [376] |
| 5q33.1 | GRIA1 | Glutamate ionotropic receptor AMPA type subunit 1: excitatory neurotransmission | Not tested |
| 4q32.1 | GRIA2 | Glutamate ionotropic receptor AMPA type subunit 2: excitatory neurotransmission | [693] |
| Xq25 | GRIA3 | Glutamate ionotropic receptor AMPA type subunit 3: excitatory neurotransmission | Not tested |
| 11q22.3 | GRIA4 | Glutamate ionotropic receptor AMPA type subunit 4: excitatory neurotransmission | [694] |
| 12p12 | GRIN2B | Glutamate ionotropic receptor NMDA type subunit 2B: ROS, excitotoxicity, excessive Ca++ influx, interacting with ADRA1A | [695] |
| 6q24.3 | GRM1 | Glutamate metabotropic receptor 1: glutamatergic neurotransmission, activation of ERK1/2, LTD/LTP | Not tested |
| 11q14.2-3 | GRM5 | Glutamate metabotropic receptor 5: glutamatergic neurotransmission, receptor for cellular prion protein PrPC bound soluble Abeta42 oligomers | [696, 697] |
| 17q21.31 | GRN | Granulin precursor progranulin: regulating cell growth | [698, 699] |
| 19q13.2 | GSK3A | Glycogen synthase kinase 3 alpha: regulating transcription factor JUN, Wnt/PI3K/signalling, Abeta cascade | Not tested |
| 3q13.33 | GSK3B | Glycogen synthase kinase 3 beta: regulating transcription factor JUN, Wnt/PI3K/signalling, energy metabolism, neuronal cell development, Abeta cascade | [275, 276, 700, 701] |
| 2q31.1 | HAT1 | Histone acetyltransferase 1: rapid acetylation of newly synthesised cytoplasmic histones | [586, 589] |
| 6q21 | HDAC2 | Histone deacetylase 2: deacetylation of lysine residues at histones, transcriptional repression, transcription factor activation | [702] |
| 7p21.1 | HDAC9 | Histone deacetylase 9: recruitment of multi-component co-repressor complexes | [586, 589] |
| 14q23.2 | HIF1A | Hypoxia inducible factor 1 alpha subunit: regulating cellular response to hypoxia, Abeta cascade | [233, 703] |
| 7q34 | HIPK2 | Homeodomain-interacting protein kinase 2: transcription factor, interaction with HIF1A, Abeta cascade | [704] |
| Locus | Gene | Function | References |
| 13q12.3 | HMGB1 | High mobility group box 1: regulating transcription, neuroinflammation, interaction with AGER, Abeta cascade | [705] |
| 5q13.3 | HMGCR | 3-hydroxy-3-methylglutaryl-CoA reductase: rate-limiting enzyme for cholesterol synthesis | [541, 706-708] |
| 20q13.12 | HNF4A | Hepatocyte nuclear factor 4 alpha: nuclear transcription factor, insulin signalling | Negative finding |
| 1q32-q41 | HSD11B1 | Hydroxysteroid 11-beta dehydrogenase 1: cortisol converter enzyme | [12] |
| 16q22.1 | HSD11B2 | Hydroxysteroid 11-beta dehydrogenase 2: cortisol converter enzyme | Negative finding |
| 6q21.31 | HSPA1A | Heat shock protein family A (Hsp70) member 1A: chaperone heat shock protein, trigger of neuronal apoptosis, caspase activation | [346] |
| 12p12.1 | IAPP | Islet amyloid polypeptide amylin: regulating blood glucose levels, brain insulin resistance, rs73069071 related to SLCO1A2 | [210] |
| 12q23.2 | IGF1 | Insulin like growth factor 1: mediating growth and development | [709] |
| 15q26.3 | IGF1R | Insulin like growth factor 1 receptor: anti-apoptotic agent, enhancing cell survival signaling neuroprotection, healthy ageing | [710, 711] |
| 11p15.5 | IGF2 | Insulin like growth factor 2: mediating growth and development, overlap with INS-IGF2 | [712] |
| 6q25.3 | IGF2R | Insulin like growth factor 2 receptor: activation of TGFbeta, degrading IGF2, covariation with WM disintegrity | [634] |
| 7p12.3 | IGFBP1 | Insulin like growth factor binding protein 1: binding both IGFs, mediating interaction with cell surface receptors | [712] |
| 1q32.1 | IL10 | Interleukin 10: anti-inflammatory cytokine produced by monocytes, anti-inflamma- tory reflex, interaction with TNFA and CYP19A1 | [713, 714] |
| 3q25.33 | IL12A | Interleukin 12A: cytokine receptor, induction of IFNgamma | [715, 716] |
| 11q23.1 | IL18 | Interleukin 18: pro-inflammatory cytokine, augmenting killer cell, stimulation IFNgamma | [716] |
| 2q14.1 | IL1A | Interleukin 1A: pro-apoptotic proprotein, interaction with DBH and IL6 attributed to LC neuron loss | [386] |
| 2q14.1 | IL1B | Interleukin 1B: pro-inflammatory response cytokine, mediated by caspase-1, induction of COX-2 | [716] |
| 9p24.1 | IL33 | Interleukin 33: induction of TH2-cells, activation of NFkappaB and MAPK pathways, histone binding | [717] |
| 5q31.1 | IL4 | Interleukin 4: adaptive immunity, chronic inflammation, mediating neurodegeneration, regeneration cascade through effector STAT6, interacting with TREM2 and TYROBP | [651, 718] |
| 7p15.3 | IL6 | Interleukin 6: cytokine both pro- and anti-inflammatory, activating PI3K pathway, targeting PKB, activating DNMT1/DNMT3A/DNMT3B/HDAC1 | [716, 718] |
| 11p15.5 | INS | Insulin: binding to the insulin receptor stimulates glucose uptake, overlap with INS-IGF2 | [719, 720] |
| 11p15.5 | INS-IGF2 | INS-IGF2 read-through: activating ERK and MAPK pathways | [712] |
| 19p13.2 | INSR | Insulin receptor: activating insulin signalling pathway, covariation with WM disintegrity | [634] |
| 1q23.1 | INSRR | Insulin receptor related receptor: activating insulin signalling pathway, covariation with WM disintegrity | [634] |
| 19q13.33 | IRF3 | Interferon regulatory factor 3: transcription factor, forming complex with CREBBP | [721] |
| Locus | Gene | Function | References |
| 2q36.3 | IRS1 | Insulin receptor substrate 1: signal transduction from INSR and IGF1R to PI3K/Akt and ERK/MAPK pathways, related to Beclin-1 | [462] |
| 13q34 | IRS2 | Insulin receptor substrate 1: mediating effects of insulin, insulin-like growth factor 1, cytokines, interacting with IL4 | Not tested in humans |
| 7q32.1 | LEP | Leptin: secreted by white adipocytes, regulation of immune functions and inflammatory responses, interaction with IAPP |
Not tested |
| 12q13.3 | LRP1/APOER | LDL receptor related protein 1: alpha 2-macroglobulin-mediated clearance of Abeta, reducing LOAD risk in APOE epsilon4 carriers | [722, 723] |
| 2q31.1 | LRP2 | LDL receptor related protein 2 megalin: reuptake of lipoproteins and hormones, MAPK signalling and JNK interacting proteins | [724] |
| 19q13.12 | MAG | Myelin-associated glycoprotein: mediating certain myelin-neuron interactions, myelination process | [682, 725, 726] |
| 2q21.1 | MANI | Family with sequence similarity 168 member B: myelin-associated neurite-outgrowth inhibitor, differentiation into catecholaminergic neurons | [726] |
| 5q13.2 | MAP1B | Microtubule associated protein 1B: microtubule assembly, neurogenesis, tauopathy | [727] |
| 16q24.2 | MAP1LC3A/B | Microtubule associated proteins 1 light chain 3 alpha/beta: cytoskeleton interaction, neurogenesis, tauopathy | [728] |
| 6q23.3 | MAP3K5 | Mitogen-activated protein kinase kinase kinase 5: activation of JNK/SAPK | [729] |
| 22q11.22 | MAPK1 | Mitogen-activated protein kinase 1 ERK2: neuronal proliferation, differentiation, transcription regulation | [671] |
| 22q13.11 | MAPK11 | Mitogen-activated protein kinase 11 p38beta: neuronal development | Not tested |
| 22q13.3 | MAPK12 | Mitogen-activated protein kinase 12 p38gamma: signal transducer | Not tested |
| 6p21 | MAPK13 | Mitogen-activated protein kinase 12 p38delta: microtubule dynamics regulator | [271] |
| 6p21.31 | MAPK14 | Mitogen-activated protein kinase 12 p38alpha: stress related transcription, genotoxic stress response | [349, 671] |
| 16p11.2 | MAPK3 | Mitogen-activated protein kinase 3 ERK1: neuronal proliferation | Not tested |
| 10q11.22 | MAPK8 | Mitogen-activated protein kinase 8 JNK1: mediating immediate-early gene expression, co-induction of apoptosis with TNFalpha | [671, 730] |
| 11p11.2 | MAPK8IP1 | Mitogen-activated protein kinase 8 interacting protein 1 JIP1: regulator of JNK1 mediated activation of transcription factors | [302, 303, 731] |
| 16p13.3 | MAPK8IP3 | Mitogen-activated protein kinase 8 interacting protein 2 JIP2: regulator of JNK1 mediated activation of transcription factors | Not tested |
| 5q35.3 | MAPK9 | Mitogen-activated protein kinase 2 JNK2: mediating immediate-early gene expression | [730] |
| 18q21.2 | MBD2 | Methyl-CpG binding domain protein 2: mediator of the biological consequences of the methylation signal | Not tested |
| 18q23 | MBP | Myelin basic protein: major myelin constituent | [725, 732] |
| 18p11.2 | MC2R | Melanocortin 2 receptor: ACTH signalling | [12] |
| Xq28 | MECP2 | Methyl-CpG binding protein 2: binding to methylated DNA | Not tested |
| 22q13.1 | MGAT3 | Mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase: glycosyltransferase, Abeta phagocytosis | [733] |
| 6p22.1 | MOG | Myelin oligodendrocyte glycoprotein: oligodendrocyte surface and outermost of myelin sheath surface, maintenance | [725, 726] |
| Locus | Gene | Function | References |
| 7q22 | MPO | Myeloperoxidase: myeloid differentiation, interacting with A2M | [654] |
| 8p22 | MSR1 | Macrophage scavenger receptor 1: mediating endocytosis of LDLs | [734] |
| 1p36.22 | MTHFR | Methylenetetrahydrofolate reductase: rate-limiting enzyme in the methyl cycle, methylation homeostasis | [92, 735] |
| 11q12.2 | MYRF | Myelin regulatory factor: directly promoting myelin gene expression | Not tested |
| 17q21.1 | NBR1 | Autophagy cargo receptor: autophagic degradation of peroxisomes | Not tested |
| 8q13.3 | NCOA2 | Nuclear receptor coactivator 2: transcriptional coactivator for nuclear hormone receptors | [736] |
| 16q12.2 | NET/SLC6A2 | Solute carrier family 6 member 2: noradrenaline reuptake | [737, 738] |
| 5q23-31 | NEUROG1 | Neurogenin 1: regulating neuronal differentiation, interacting with CREBBP | Not tested |
| 4q25 | NEUROG2 | Neurogenin 2: controlling cortical neuron migration | [739] |
| 10q21.3 | NEUROG3 | Neurogenin 3: transcription factor involved in neurogenesis, determinative for pancreatic islet cell phenotype | Negative finding |
| 4q24 | NFKB1 | Nuclear factor kappa B subunit 1 p50: transcription regulator, neuroinflammation | [740, 741] |
| 10q24.32 | NFKB2 | Nuclear factor kappa B subunit 2: transcription regulator, neuroinflammation | [741] |
| 14q13.3 | NFKBIA | NFKB inhibitor alpha: anti-inflammatory reflex protein | [740, 741] |
| 2q37.1 | NGEF | Neuronal guanine nucleotide exchange factor ephexin: Abeta cascade | [742] |
| 17p13.2 | NLRP1 | NLR family pyrin domain containing 1: neuronal inflammasome, mediating apoptosis, interaction with caspase-2 and caspase-9 | [509, 743, 744] |
| 1q44 | NLRP3 | NLR family pyrin domain containing 3: neuronal inflammasome, mediating apoptosis, upstream activator of NFkappaB signalling | [266, 509, 744] |
| 7q36.1 | NOS3 | Nitric oxide synthase 3: reactive free radical, neurotransmission | [745] |
| 9q34.3 | NOTCH1 | Notch 1: EGF-related, neuropil development | [711] |
| 4q32.2 | NPY1R | Neuropeptide Y receptor Y1: mobilisation of intracellular calcium, neuroprotection | Not tested |
| 5q31.3 | NR3C1 | Nuclear receptor subfamily 3 group C member 1 glucocorticoid receptor: transcription factor, binding to glucocorticoid response elements | [746] |
| 9q33.3 | NR5A1 | Nuclear receptor subfamily 5 group A member 1: transcription factor | Not tested |
| 7q32.2 | NRF1 | Nuclear respiratory factor 1: mitochondrial biogenesis pathway, transcription factor, neurite outgrowth, correlation with PGC-1a | [296, 747] |
| 2q31.2 | NRF2 | Nuclear respiratory factor 2: transcription factor, oxidative stress | [748] |
| 9q21.33 | NTRK2 | Neurotrophic receptor tyrosine kinase 2: binding neurotrophins, signalling in MAPK pathway, neuronal differentiation | [749] |
| Xq23 | NXT2 | Nuclear transport factor 2 like export factor 2: mRNA nuclear export | [675] |
| 12q24.31 | P2RX7 | Purinergic receptor P2X 7: formation of membrane pores | [750] |
| 20q13.31 | PCK1 | Phosphoenolpyruvate carboxykinase 1: regulation of gluconeogenesis through insulin, glucocorticoids | [751] |
| 2p11.2 | PERK | Eukaryotic translation initiation factor 2 alpha kinase 3: modulating mitochondrial function, ER stress | [602] |
| 7q11.23 | PION | Gamma-secretase activating protein: increasing Abeta production, interaction with gamma-secretase and APP-CTF | [752] |
| Locus | Gene | Function | References |
| 19p13.12 | PKN1 | Protein kinase N1: mediating insulin signalling, tauopathy | [753] |
| Xq22.2 | PLP1 | Proteolipid protein: transmembrane protein, myelin compaction, maintenance, interacting with EDN1 | [682, 725] |
| 11q13 | PP1 | Protein phosphatase 1: hepatic blood-glucose levels, glycogen metabolism | [754] |
| 3p25.2 | PPARG | Peroxisome proliferator activated receptor gamma: adipocyte differentiation, T2DM | Negative |
| 4p15.1 | PPARGC1A | PPARG coactivator 1 alpha: regulation of CREB and NRFs, neuronal cholesterol homeostasis | [296, 747] |
| 5q32 | PPARGC1B | PPARG coactivator 1 beta: non-oxidative glucose metabolism, regulating transcription factors and glucocorticoid receptor | [755] |
| 1p36.33 | PRKCZ | Protein kinase zeta: stabilisation of LTP, atypical isoform: insulin stimulated glucose transport, consistent with memory loss | [459, 460] |
| 17p13.1 | PSD95/DLG4 | Postsynaptic density protein 95, LTP | [756] |
| 14q24.1 | PSEN1 | Presenilin 1: regulating APP processing, cleavage of Notch receptor | Negative |
| 1q42.13 | PSEN2 | Presenilin 2: regulating APP processing, cleavage of Notch receptor | Negative |
| 14q22.1 | PTGER2 | Prostaglandin E receptor 2: glial cells, neuroinflammation, GSK-3beta, beta-catenin signalling, COX-2 induction | [757] |
| 1p31.1 | PTGER3 | Prostaglandin E receptor 3: reducing cAMP-dependent signalling, COX-2 induction | [515] |
| 5p13.1 | PTGER4 | Prostaglandin E receptor 4: activating transcription factor CREP signalling, EGR-1 expression through ERK, p38, PI3K/Akt/mTOR pathways | [758] |
| 9q33.2 | PTGS1 | Prostaglandin-endoperoxide synthase 1 COX-1: catalysing prostaglandin biosynthesis | [759] |
| 1q31.1 | PTGS2 | Prostaglandin-endoperoxide synthase 1 COX-2: inducible isozyme, prostanoid biosynthesis, neuroinflammation | [760, 761] |
| 11p15.1 | PTPN5 | Protein tyrosine phosphatase, non-receptor type 5 STEP: dephosphorylation of ERK1/2, p38, internalisation of NMDARs, synaptic strength | [762] |
| 16p11.2 | PYCARD | PYD and CARD domain containing: apoptotic signalling pathway, activation of caspase | [763] |
| 6p23 | RANBP9 | RAN binding protein 9: translocation of RNA, Abeta cascade | [764] |
| 21q22.1 | RCAN1 | Regulator of calcineurin 1: inhibiting calcineurin-dependent signalling pathways, CNS development, activation of caspase-3, linking tau and Abeta | [765] |
| 7q22.1 | RELN | Reelin: neurogenesis, synaptic positioning, LTP, interaction with CDK5, GSK-3b, interaction with APP | [766] |
| 4q12 | REST | RE1 silencing transcription factor: neuron-restrictive silencer element, master negative regulator of neurogenesis | Not tested |
| 12q24.31 | SCARB1 | Scavenger receptor class B member 1: HDL receptor | Negative |
| 4q21.1 | SCARB2 | Scavenger receptor class B member 2: lyosomal membranes | Not tested |
| 7q11.2 | SCARB3 | Scavenger receptor class B member 3 CD36: fatty acid transport | [767] |
| 14q32.13 | SERPINA3 | Serpin family A member 3: plasma protease inhibitor | [768] |
| 17q11.2 | SERT/SLC6A4 | Solute carrier family 6 member 4: serotonin transporter | Negative |
| 6q23.2 | SGK1 | Serum/glucocorticoid regulated kinase 1: anti-apoptotic cellular stress protector, Akt homologue | [769] |
| Locus | Gene | Function | References |
| 1q22 | SH3GLB1 | Endophilin-B1: Bax-interacting factor 1 Bif-1 | [194] |
| 1q21.2 | SHC1 | SHC adaptor protein 1: mitochondrial matrix, ROS regulator | [770] |
| 12p12.1 | SLCO1A2 | Hepatic solute carrier organic anion transporter family member 1A2: uptake bile acids, steroidal compounds, interacting with IAPP | [210] |
| 4q22.1 | SNCA | Alpha synuclein: presynaptic terminals | [771] |
| 1q25.2 | SOAT1 | Sterol O-acyltransferase 1: cholesterol acyltransferase, endoplasmic reticulum, cholesterol equilibrium, Abeta cascade | [275, 276] |
| 21q22.1 | SOD1 | Superoxide dismutase 1: ROS converter | [772] |
| 10q25.1 | SORCS1 | Sortilin related VPS10 domain containing receptor 1: covariation with WM disintegrity | [634, 773] |
| 11q24.1 | SORL1 | Sortilin related receptor 1: neuronal LDL receptor, covariation with WM disintegrity | [634, 773] |
| 17q21.2 | STAT3 | Signal transducer and activator of transcription 3: respondent to cytokines and growth factors | [741] |
| Xp11.23 | SYP | Synaptophysin: binding cholesterol, targeting of vesicle-associated synaptobrevin | Not tested |
| 6q27 | TBP | TATA-box binding protein: element of transcription factor IID (TFIID) | [774] |
| 10q21.1 | TFAM | Mitochondrial transcription factor A: mitochondrial DNA replication and repair | [296, 775] |
| 12q13.12-13 | TFCP2 | Transcription factor CP2: binding the alpha-globin promoter, inflammatory response | [776] |
| 6p21 | TFEB | Transcription factor EB: lyosomal biogenesis, autophagy, activated by PGC-1alpha | Not tested |
| 19q13.2 | TGFB1 | Transforming growth factor beta 1: regulating cell proliferation, and growth, covariation with WM disintegrity | [634, 777] |
| 9q33.1 | TLR4 | Toll like receptor 4: activation of innate immunity | [778] |
| 6p21.33 | TNFA | Tumor necrosis factor alpha: cytokine, cell proliferation, apoptosis, neuroinflammation | [779] |
| 8p21.2 | TNFRSF10A | TNF receptor superfamily member 10A: inducing cell apoptosis | [780] |
| 8p21.2 | TNFRSF10B | TNF receptor superfamily member 10B: transducing apoptosis signal | [780] |
| 12p13.31 | TNFRSF1A | TNF receptor superfamily member 1A: inhibiting inflammation | [781] |
| 1p36.22 | TNFRSF1B | TNF receptor superfamily member 1B: mediating anti-apoptotic signals, anti-oxidative pathways, neuroprotection | Negative finding |
| 13q14.11 | TNFSF11 | TNF superfamily member 11 RANKL: regulation of cell apoptosis | [782] |
| Xq22.3 | TSC22D3 | TSC22 domain family member 3: anti-inflammatory protein glucocorticoid (GC)-induced leucine zipper, immunosuppression through NFkappaB | Not tested |
| 12q24.33 | ULK1 | Unc-51 like autophagy activating kinase 1: autophagy regulator | Not tested |
| 5q31.1 | VDAC1 | Voltage dependent anion channel 1: metabolite exchange in mitochondria | [217, 783] |
| 6p21.1 | VEGFA | Vascular endothelial growth factor A: embryonic blood vessel formation, metabolic abnormalities | [784] |
| 9p24.2 | VLDLR | Very low density lipoprotein receptor: VLDL-triglyceride metabolism, reelin signalling pathway | [785, 786] |
Method remarks: This table contains suspected gene loci extracted from preceding text sections, based on animal research, and suspected risk genes from additional literature. These loci were tested for associations in human LOAD genetic studies in PubMed and/or AlzGene databases. If not available, human pathophysiology studies are reported instead.
Additional rare variants were also detected in APP, TREM2, PLD3 [542, 571]. The SNP rs597668 near BLOC1S3 (19q13.32) biogenesis of lysosomal organelles complex1 subunit 3 is responsible for functioning of the lyosomal system [572]. The highly polymorphic ECE-1b (1p36.12) promoter [573] was found relevant for APOE epsilon4 non-carriers: endothelin converting enzyme 1 is the key enzyme in endothelin biosynthesis and crucial for receptor recycling and Abeta degradation [574]. The protection of telomeres protein 1 gene POT1 (7q31.32, rs4728029) was detected as related to h-tau, inflammatory response cytokine IL-6 [575], ventricular enlargement and cognition. The TYRO protein tyrosine kinase-binding protein gene DAP12/TYROBP (19q13.12) encodes a transmembrane signalling polypeptide containing an immunoreceptor, and is crucial for brain myelination, inflammation and causal regulation of microglial genes [576]. In LOAD, genetic variants in sortilin related receptor 1, clusterin, complement component receptor 1, CD2AP, CD33 [541], EPHA1, and MS4A4/MS4A6E [542, 546] genes contribute to late-onset age. Further common variants identified for age-of-onset are: WRN, NTN4, LAMC3 [577], and HMGCR [541]; rare variants for age-of-onset are SLC8A3, SLC19A3, MADD and LRRK2 [577].
4.1.1. Myelin Structural and Neuronal Plasticity Genes in LOAD
Stability of myelin and integrity of WM tracts are critical for the functioning cognitive reserve during ageing, and disruptions are structural biomarkers of neurodegeneration. GWAS have supported a central role of myelin maintenance by revealing indirect myelination markers such as SORL1, SORCS1, BIN1, TYROBP. However, traditional myelin protein genes, such as myelin basic protein MBP, myelin oligodendrocyte protein MOG, myelin associated glycoprotein MAG, have rarely been studied in the context of LOAD. Genes crucial for onset of white matter myelination and myelin transcription factor MYT1, myelin regulatory factor MYRF, 2',3'-cyclic-nucleotide 3'-phosphodiesterase CNP, and cholesteryl ester transfer protein CETP [541] have not been studied systematically in LOAD. More interest have, in contrast, raised synaptic plasticity and anatomical connectivity genes, such as neuronal protein astrotactin 2 ASTN2 [578], neuregulin 3 NRG3 [579], spondin SPON1 [541] and reelin RELN. The mammal-specific factors osteocrin OSTN and the related MEF2C are highly expressed in human cortices, here contributing to cognition. For these genes, it has been shown that epigenetic mechanisms exist, where HDACs deacetylate targets indicated by lysine histone codes H3K9 and H4K12, to regulate downstream genes important for neuroplasticity (BDNF, EGR1/NGFI-A, CDK5, SYT1, SYP, and GRN).
The plasticity marker activity-regulated cytoskeleton-associated protein Arc (ARC 8q24.3) (Section 3.1.) belongs to a class of genes transcribed in presence of protein synthesis inhibitors, and localised to NMDARs [580] in the synaptic cleft, there stabilising learning-related plastic changes as a Synaptic Activity Response Element (SARE) essential for associative learning and conditioning. Arc is also related to glutamatergic signalling via AMPA receptors [581], dopamine receptor 1, and LTP, responsive to neurotrophic signals including epidermal growth factor EGF, nerve growth factor NGF, and brain-derived neurotrophic factor BDNF [581]. Its gene is regulated by CREB, dependent of the MAPK [580] pathway following ERK1/2 phosphorylation [580], and the microtubular mRNA transport locally translated at dendritic spines and activated synapses [582].
Overall, brain growth is a LOAD risk-factor, and the delta-opioid receptor OPRD1 was related to small cerebral volume [541] (and to amyloidogenic processing, Section 1.4.). Glutamate transporter EAAT2 is overactive in LOAD leading to excess glutamate in synaptic functioning [583]. The fas-associated serine/threonine kinase domains 2 FASTKD2 was associated with memory performance [541]. Poly(ADP-ribose) polymerase 1 PARP1, and caspase recruitment domain family, member 10 CARD10 genes correlated with hippocampal volume [541], whereas astrotactin 2 ASTN2, and ionotropic glutamate receptor, N-methyl D-aspartate 2B, GRIN2B correlated with hippocampal and temporal atrophy [541]. The biological ageing-related gene POT1 (protection of telomeres 1) has been observed in correlation with ventricular dilation [541], whereas neuroprotection was promoted by expression of RE1-silencing transcription factor REST [541].
4.1.2. Immune System Genes in LOAD
Under physiological conditions, triggering receptor expressed on myeloid cells 2 (TREM2) pairs with the adaptor protein DNAX-activating protein (DAP12/TYROBP) to induce phagocytosis of apoptotic neurons without inflammatory responses, and to regulate toll-like receptor (TLR4/2) mediated inflammatory responses, and microglial activation [584]. It has been detected recently that its natural ligands consist of stress-related mitochondrial heat shock proteins [584]. Further GWAS-genes associated with neuroinflammation [482, 504] were summarised with CR1 complement receptor, CFHR1 [541] complement receptor, immunoreceptor CD33, membrane-spanning proteins MS4A6A/MS4A4E, interleukin-6 receptor IL6R [541], geminin coiled-coil domain containing GMNC, transcription repressor GLIS family zinc finger GLIS3 [541], and mitochondrial energy and cellular stress genes ATP5H/KCTD2 [541]. Genes associated with microglial function [498] were summarised as TREM2, CD33, CR1, CLU, CD2AP, EPHA1, ABCA7, and INPP5D. Genes relating to neuroinflammation [510] were summarised in MRP14, MS4A6A, MS4A4AE. Autophagy genes identified: (a) ULK1, ULK2, BECN1, (b) ATG13, ATG2A, ATG2B, ATG4C, ATG4D (c) MAPLC3A, MAPLC3B2 (d) LAMP1 [341, 345, 352]. Mitophagy genes identified are: PINK, PARK1, MAPLC3A, MAPLC3B2, HDAC6 [351, 352], and ectopic P-granules autophagy protein 5 homologue EPG5 [551].
Table 3 lists the candidate genes extracted from the previous sections of pathophysiological mechanisms in LOAD, mainly from animal studies, with their loci and brief functional descriptions. The references then pertain to human association, case-control, or pathophysiology studies in LOAD patients, where the genes have been reported involved. For 60 of 224 gene loci, no positively confirming results were reported in humans.
4.2. Meta-analytic Evaluation of Genes Nominated from the Collection of Studies
Because known LOAD risk genes are not sufficient in explaining the heritability estimates (Section 1.1.), epigenetic mechanisms are increasingly assumed to playing a key role in LOAD aetiopathology [1]. It is herein supposed that epigenetic modification mechanisms are typically associated to SNPs [1] occurring in risk loci. It has been proposed for LOAD pathogenesis, that Lahiri’s “Latent Early Associated Regulation” (LEARn) theory, which was formulated for all neurodegenerative disorders, provides a sufficient theoretical framework for the contribution of epigenomics towards clinical manifestation. Consistent with the notion of ELS priming of stress for brain functioning, the LEARn model posits that early environmental exposures can change gene expression for long-lasting developmental time periods. Alterations of gene expression would hence be conducive to neuropathology that only becomes apparent later in life. As an example, the concept of a fetal basis of amyloidogenesis [133] assumes that the regulatory regions of the genes involved in amyloid processing, become biased specifically through changes in methylation and oxidation status within the promoter of specific genes [585], and therefore epigenetically programmed towards late-life neurodegeneration.
Whilst a variety of currently known post-translational modifications can occur: acetylation, methylation, ubiquitylation, phosphorylation, sumoylation, crotonlylation and numerous less well-studied modifications [586, 587], the probes yet examined for LOAD have included only the two main mechanisms DNA methylation and histone acetylation.
4.2.1. Previous Epigenomic Findings in LOAD
Best cues to epigenetic modification currently stem from histone deacetylases (HDAC) in model systems of AD and in off-label treatments of LOAD patients with HDAC inhibitors [569]. HDAC (isoforms 1, 2 and 3) can also be PET-neuroimaged in humans, and in normals showed greatest variability in cortical grey matter [588]. Epigenetics is specifically relevant in cognitive disturbances [586, 589], and enzymes responsible for histone acetylation (HAT) and deacetylation (HDAC) here act as the regulators of synaptic plasticity and memory. Disrupted epigenetic regulation is suspected in LOAD [586, 589] with respect to memory-related genes such as CREB, CREBBP, FOS, ARC, PSD95, CDK5, GLUR, GABBR, PP1, NFkappaB, MECP2, alpha-synuclein, Ephexin5, ERK/MAP, reelin RELN [589]. But HDACs furthermore epigenetically control Abeta expression, beta/gamma secretases, and the Abeta degradation enzyme Mmp2 [590]. The parallel downregulation of HDACs and upregulation of histone acetyltransferases at the p300/CREB-binding protein locus (CREBBP) was observed with concurrent DNA methyltransferase reduction [591]. Epigenetic regulations of Abeta42 processing are therefore exerted jointly by DNA methyltransferase DNMT, histone acetyltransferase HAT, and histone deacetylase complex HDAC genes. In vivo and post-mortem findings suggest also that soluble polymeric Abeta can itself trigger epigenetic regulation of transcriptional activation and inhibition [592], specifically of histone H3 homeostasis [591-593].
Histone deacetylase 2 (HDAC2) associates with and reduces the histone acetylation of genes important for learning and memory. Overexpression of HDAC2, but not that of HDAC1, decreased dendritic spine density, synapse number, synaptic plasticity and memory formation. However, this was found reversible, which suggests that cognitive capacities following neurodegeneration are not entirely lost, but merely impaired by this epigenetic blockade [594, 595]. The HDAC2 exhibited a relation with chronic stress experience and reward related plasticity in the ventral striatum [596]. Chronic stress resulted there in significant increase in the level of HDAC2 and in its binding to the promoter region of glial cell-derived neurotrophic factor (GDNF), thereby causing down-regulation of Gdnf expression in the mesolimbic nucleus accumbens (NAcc).
Methylation studies in twelve LOAD patients revealed evidence for an “epigenetic drift” with interindividual variabilities in APOE promoter; DNMT1, MTHFR and TFAM were hypermethylated compared to the norm [92]. Controls did not exhibit notable epigenetic distances from the norm in cerebral methylation for LOAD candidate genes. Similar results pertaining to large interindividual variation in candidate genes are reported by a number of studies (Table 2). Epigenetic drifts were substantiated in LOAD for genes involved in amyloid processing (PSEN1, APOE promoter) and methylation homeostasis (MTHFR, DNMT1) [597]. Furthermore, regulations of Abeta42 are exerted by DNA methyltransferase DNMT genes. While it remains difficult to measure impact of early life adversity over the entire life-span, available evidence suggests that environmental influences tend to inhibit DNA methyltransferases during brain development, which results e.g. in hypomethylation of APP gene [136]. It is therefore plausible that analogous alterations are leading to genomic imprints for late-life susceptibility of LOAD (corresponding to the Lahiri LEARn model):
Both hypermethylation and hypomethylation were found to be involved in oxidative stress damage induced by Abeta miscleavage [136]. In hypermethylation of AD genes, methylcytosines restrict repair of adjacent hydroxyguanosines, thus increasing susceptibility to oxidative stress [136]. Recent studies isolated methylation and hydroxymethylation as putative key epigenetic mechanisms in LOAD [598]. The respective markers 5-methylcytidine (5-mC) and 5-hydroxy- methylcytidine (5-hmC) were decreased in a magnitude of 20% in LOAD-hippocampi and negatively correlated with Abeta plaques [598]. One further mechanism related to ER stress is the homocysteine metabolism known to be altered during ageing. Hepatic S-adenosylmethionine (SAM) metabolism [599, 600] is also reduced in AD and found to cause hypomethylation in APP and PSEN1 promoter genes [600], thus leading to increased Abeta expression [600].
In vitro results demonstrate that environmental stress MAPK pathways p38 and JNK are activators of Abeta by means of an ensemble of epigenetic modulations: demethylation in three gene promoters responsible for reduction of methyltransferases (DNMTs) enhanced expression of APP, BACE1, and PSEN1 [591]. Histone H3 hyperacetylation in concord with DNMT-dependent hypomethylation inducing Abeta overexpression are thus the putative consequence of MAPK stress signalling [591]. Another putative mechanism for JNK activation in stress signalling is the pro-apoptotic kinase PKR (double-stranded RNA dependent protein kinase) phosphorylating eIF2alpha (eukaryotic translation initiation factor-2 alpha) [601] in translational regulation of BACE1. The two kinases related to elF2alpha, PERK and GCN2, were found responsible for impairments in spatial memory and synaptic plasticity [602], and are dependent of phosphorylation of eIF2alpha. Phosphorylated eIF2alpha generally stops protein translation in response to cellular stress. AD genes, in contrast, including BACE1, are activated by eIF2alpha [252, 544].
For the evaluation presented in Table 2, the 313 candidate risk genes of Tables 1 and 3 were combined. It was searched for results from human studies. 35 studies were identified containing human epigenomics findings. In addition, gene regulation studies pertaining to candidate genes were compiled. In this literature, 64 separate gene regulation mechanisms were described. Fig. (1) illustrates the extraction of genes and analysis steps.
Fig. (1).
PRISMA flowchart depicting levels of data extraction and meta-analysis.
4.2.2. Summary of Present Findings on Epigenomics in LOAD
The meta-analysis on epigenetic modification of risk genes (Table 2) is mainly extending the lines of the literature detailed above. Special care in generalisation of findings should be at hand, as the relative small amount of studies tend to cluster to relative few well-known gene loci. Previous reports of findings tend to attract further studies in the same loci, whereas the less well-known risk genes remain unresearched. Given the fact that epigenomics is assumed to be crucial in LOAD pathophysiology for more than a decade, little research effort has been invested up to date in comparison. An explanatory reason for this slow progression is that epigenomic studies of post-mortem tissue are difficult to accomplish. For these reasons, the compiled findings are likely to reflect a certain sampling bias. In addition, the studies have mostly small sample sizes. Epigenomic findings encompass seven groups:
Genes relevant to familial EOAD APP, PSEN1, APOE, MAPT;
Central role of BDNF;
Myelin-related genes BIN1, CNP, SORL1;
ELS-related genes: FKBP5, EGR1/NFI-A, GSK3B;
Lipid metabolism: ABCA7, ALOX5, DHCR24;
Gene modification itself: CREB, CREBBP, FOS, MBD2, MTHFR2, DNMT1;
Neuroinflammation: NFKB1, TNFA, ANK1, CRP.
Given the large number of candidate genes, it is likely that the research field in epigenomics of LOAD is only in its initial stage.
4.2.3. Summary of Present Findings on Gene Regulation by MicroRNA
The compilation of microRNA resulting from candidate genes is listed in Table 4. Search for microRNA biomarkers has been conducted since 2007, but without reaching consensus yet [560]. Special areas were gene regulation during neurogenesis [116] and microRNAs related to memory impairments as implicated in LOAD [603]. It is likely that only a small proportion of LOAD-related microRNA has been identified.
Table 4.
Gene regulation.
| Number of micro-RNA | Target/Function | References |
|---|---|---|
| miR-106b | ABCA1 | [476, 546] |
| miR-107 | BACE1 | [811] |
| miR-124 | CREB, APP, BACE1 | [546] |
| miR-124 | Neurogenesis (e.g. SOX9, Jag1 and Dlx2) | [799] |
| miR-125b | Tau hyperphosphorylation, p35, p44/42, CDK5, MAPK signalling | [812-814] |
| miR-128b | CREB1, ROS, Abeta | [546, 815] |
| miR-132 | Morphological rearrangements of neurons, memory formation | [546, 813] |
| miR-132 | MeCP | [546] |
| miR-132 | TLR4 | [268] |
| miR-132 | NBM degeneration, cholinergic system, related to EGR1 | [683] |
| miR-132/107 | - | [560] |
| Number of micro-RNA | Target/Function | References |
| miR-132/128 | - | [560] |
| miR-132/132 | - | [560] |
| miR-132/138 | - | [560] |
| miR-132/191 | - | [560] |
| miR-132/206 | - | [560] |
| miR-132/212 | Tau expression, tau hyperphosphorylation, tau aggregation, GSK-3beta, PP2B | [816] |
| miR-132/384 | - | [560] |
| miR-134 | BDNF, MCI | [546] |
| miR-135a/-193b/-200b/-384 | APP/BACE1 | [817-819] |
| mir-135b | Hippocampal cell proliferation, neuroprotection, memory capacities | [820] |
| miR-138 | RARA/GSK-3b pathway | [551] |
| miR-144 | Negatively related to ADAM10, regulated by AP-1 | [821] |
| miR-146a | TLR4, microglia, NRF2 | [268, 822] |
| miR-153 | APP | [823] |
| miR-155 | JNK pathway, B-cell maturation and immunoglobulin production | [293] |
| miR-155 | TLR4 | [268] |
| miR-16/101 | APP | [546] |
| miR-181c | SIRT1, stress, inflammation | [813] |
| miR-182 | Cortactin | [546] |
| miR-186 | BACE1 | [824] |
| miR-188-3p | PPARgamma, MAGL | [825] |
| miR-193b | APP | [819] |
| miR-195 | BACE1 | [826] |
| miR-200 | APP, PSEN1 | [817] |
| miR-206 | IGF1 | [827] |
| miR-206 | BDNF | [828] |
| miR-20a | APP | [546] |
| miR-26b | IGF1 | [829] |
| miR-26b | Aberrant cell cycle entry (CCE), CDK5 kinase activity, tau-phosphorylation | [814, 830] |
| miR-27-3p | Adipocyte differentiation, extracellular signal related kinase 5 (ERK5), peroxisome proliferator-activated receptor gamma (PPARgamma), CCAAT/enhancer binding protein (C/EBP) | [476] |
| miR-27a | - | [560] |
| miR-27b | TNFalpha, IL-6, PPARgamma | [831] |
| miR-298/328/195 | BACE1 | [546] |
| miR-299-5p | Suppression of autophagy, neuroprotection, attenuation of caspase-mediated apoptosis | [832] |
| miR-29a/20b-1/9b | BACE1, SPTLC2 | [546] |
| miR-29b | BACE1 | [813, 833] |
| miR-29c | Positively to DNMT3, negatively to BDNF | [834] |
| Number of micro-RNA | Target/Function | References |
| miR-29c | BACE1, increasing CREBBP | [835, 836] |
| miR-33 | Cholesterol homeostasis | [476] |
| miR-33 | ABCA1 expression, Abeta reduction, APOE lipidation | [837] |
| miR-339-5p | BACE1 | [838] |
| miR-34 | Tau aggregation | [170, 546, 814] |
| miR-34a | TREM2, NFkappaB | [839] |
| miR-511 | FKBP5 interaction | [840] |
| miR-512 | cFLIP, MCL1, h-tau | [841] |
| miR-574 | Neuritin | [842] |
| miR-603 | LRP1 | [843] |
| miR-613 | BDNF | [844] |
| miR-758 | ABCA1 | [476] |
| miR-9 | Abeta, orphan receptor TLX, expressed in neural progenitors, neuronal differentiation | [546, 813] |
| miR-922 | Ubiquitin carboxyterminal hydrolase L1 (UCHL1), tau hyperphosphorylation | [845] |
| miR-98 | IGF-1, autophagy, GSK-3beta, tau hyperphosphorylation | [546, 846] |
Index of gene loci in the text
With respect to frequency of the target, 10 microRNAs relate to BACE1, 7 to MAPK signalling, 5 to APP, 5 to TLR4, 4 to BDNF, and 3 to insulin signalling. Most mechanisms are related to microRNA-132 and microRNA-27. It must be stated that in this research domain no tentative conclusions can be reached yet.
4.3. Evaluation with Respect to Possible Impact of Early Life Stress
Returning to the initial question of ELS effects in LOAD, the present state of research permits the conclusion that rodent studies have indeed demonstrated such effects. Gaining direct evidence from human studies remains difficult for the larger life-spans, and the greater genomic complexity of the organism homo sapiens.
The summary of findings from the genome level supports an involvement of SMA and HPA in LOAD. The accumulated evidence from association studies includes support for the assumption of strong role of structural brain growth factors. This relates to the microtubule associated protein tau gene MAPT and further neuronal cell stabilisers. Association studies also suggest a strong role of genes related to myelin
ABCA1 9q31.1, 11; ADCYAP1 18p11.32, 16; ADRA2A 10q25.2, 35; ADRB1 10q25.3, 35; ADRB2 5q31-32, 36; ADRB3 8p11.23, 36; AGER 6p21.32, 32; ALOX5 10q11.21, 52; APBB2 4p14-p13, 26; APOE 19q32.13, 6; APP 21q21.3, 6; ARC 8q24.3, 42; AVP 20p13, 16; BACE1 11q23.3, 18; BAX 19q13.33, 54; BDNF 11p14.1, 12; BECN1 17q21.31, 33; CAMK2A 5q32, 14; CCL2 17q12, 48; CCL3 17q12, 48; CCL4 17q4, 48; CDK5 7q36.1, 20; COMT 22q11.2, 35; CREBBP 16p13.3, 16; CREBBP 16q13.3, 13; CRH1 8q13.1, 12, 37; CRHBP 5q13.3, 39; CRHR1 17q21.31, 15, 39; CRHR2 7p14.3, 15, 39; CRP 1q23.2, 52; CX3CL1 16q21, 48; CX3CR1 3p22.2, 49; CYP46A1 14q32.2, 46; DBH 9q34.2, 36; DLG4 17p13.1, 27; DLG4 17p31.1, 32; DNMT1 19q13.2, 16; DNMT3A 2p23.2, 16; DNMT3B 20q11.21, 16; DUSP1 5q35.1, 13; EDN1 6p24.1, 51; EDNRA 4q31.22-23, 51; EDNRB 13q22.3, 51; EGR1 5q31.2, 14; EIF2AK2 2p22.2, 24; EIF2S1 14q23.3, 24; FKBP4 12p13.33, 40; FKBP5 6p21.31, 38; FOS 14q24.3, 15; GAB2 11q14.1, 7; GABRA1 5q34, 16; GAD1 2q31.2, 40; GMEB1 1p35.3, 39; GMEB2 20q13.33, 39; GNB3 12p13.31, 36; GRIA1 5q33.1, 32; GRIA2 4q32.1, 32; GRIA3 Xq25, 32; GRIA4 11q22.3, 32; GRM1 6q24.3, 32; GRM5 11q14.2-3, 32; GRN 17q21.31, 49; GSK3A 19q13.2, 26; GSK3B 3q13.33, 26; HDAC2 6q21, 38; HIF1A 14q23.2, 43; HMGB1 13q12.3, 44; HMGCR 5q13.3, 46; HNF4A 20q13.12, 43; HSD11B1 1q32-q41, 39; HSD11B2 16q22.1, 11; HSPA1A 6p21.33, 45; IAPP 12p12.1, 20; IDE 10q23-q25, 44; IGF1 12q23.2, 21, 43; IGF1R 15q26.3, 43, 44; IGF2 11p15.5, 43; IGF2R 6q25.3, 44; IGFBP1 7p12.3, 38; IL10 1q32.1, 53; IL12A 3q25.33, 53; IL12B 5q33.2, 53; IL18 11q23.1, 53; IL1A 2q14.1, 53; IL1B 2q14.1, 53; IL33 9p24.1, 53; IL4 5q31.1, 53; IL6 7p15.3, 53; INS 11p15.5, 43; INS-IGF2 11p15.5, 11; INSRR 1q23.1, 44; IRF3 19q13.33, 38; IRS1 2q36.3, 43; IRS2 13q34, 44; LEP 7q32.1, 11; LRP1/APOER 12q13.3, 23; LRP2 2q31.1, 43; MAP1B 5q13.2, 24; MAP1LC3A/B 16q24.2, 33; MAP3K5 6q23.3, 22; MAPK1 22q11.22, 27; MAPK10 4q21.3, 29; MAPK11 22q13.11, 31; MAPK12 22q13.3, 31; MAPK13 6p21, 31; MAPK14 6p21.31, 31; MAPK3 16p11.2, 27; MAPK8 10q11.22, 29; MAPK8IP1 11p11.2, 30; MAPK8IP3 16p13.3, 30; MAPK9 5q35.3, 29; MAPT 17q21.31, 19; MBD2 18q21.2, 16; MC2R 18p11.2, 39; MECP2 Xq28, 16; MGAT3 22q13.1, 52; MPO 7q22, 50; MSR1 8p22, 51; NBR1 17q21.1, 33; NCOA2 8q13.3, 39; NET 16q12.2, 36; NEUROG1 5q23-31, 17; NEUROG2 4q25, 17; NEUROG3 10q21.3, 17; NFKBIA 14q13.3, 38; NLRP1 17p13.2, 50; NLRP3 1q44, 50; NOS3 7q35-36, 53; NOTCH1 9q34.3, 6; NPY1R 4q32.2, 40; NR3C1 5q31.3, 11; NR3C1 5q31-32, 38; NR5A1 9q33.3, 39; NRF2 2q31, 33; NTRK2 9q21.33, 13; P2RX7 12q24.31, 52; PCK1 20q13.31, 12; PION 7q11.23, 18; PKN1 19p13.12, 8; PPARG 3p25.2, 53; PPARGC1A 4p15.2, 53; PPARGC1B 5q32, 53; PSEN1 14q24.1, 6; PSEN2 1q42.13, 6; PTGER2 14q22.1, 50; PTGER3 1p31.1, 50; PTGER4 5p13.1, 50; PTGS1 9q32-q33.2, 22; PTGS2 1q25.2-q25.3, 43; PTPN5 11p15.1, 32; PYCARD 16p11.2, 50; RANBP9 6p23, 54; RANKL 13q14.11, 38; RCAN1 21q22.1, 28; REST 4q12, 15; SCARB1 12q24.31, 51; SCARB2 4q21.1, 51; SCARB3 7q11.2, 51; SCN 4q22.1, 23; SERPINA3 14q32.13, 52; SERT/SLC6A4 17q11.2, 11; SGK1 6q23.2, 21; SH3GLB1 1q22, 19; SHC1 1q21.2, 22; SLCO1A2 12p12.1, 20; SOD1 21q22.1, 21; SORCS1 10q25.1, 35; SORL1 11q24.1, 35; STAT3 17q21.2, 33; SYP Xp11.23, 21; TBP 6q27, 16; TFCP2 12q13.12-13, 26; TFEB 6p21, 33; TGFB1 19q13.2, 53; TLR4 9q33.1, 45; TNFA 6p21.33, 53; TNFRSF10A 8p21.2, 54; TNFRSF10B 8p21.2, 54; TNFRSF1A 12p13.31, 49; TNFRSF1B 1p36.22, 49; TSC22D3 Xq22.3, 53; ULK1 12q24.33, 33; VDAC1 5q31.1, 21, 45; VEGFA 6p21.1, 53; VLDLR 9p24.2, 23
stability BIN1, CNP, MOG, MAG, RELN, SORL1, SORCS1. Structural MRI studies have previously demonstrated WM loss in LOAD.
SNPs in genes of adrenoceptors alpha and beta providing noradrenergic signalling were found associated with LOAD. However, genomic research have no clues yet for the connection between noradrenaline systems and LC degeneration in LOAD. Unclear also remains the physiological regulation between noradrenaline and disrupted cerebral insulin signalling present in LOAD. Association studies for genes related to HPA functioning and LOAD have ruled out most loci for steps of glucocorticoid processing. The sole surviving significant gene HSD11B1 for association with LOAD is, however, a strong argument for ELS effects in LOAD. Further support for the assumption of ELS effects in LOAD are also associations of the trauma-related genes FKBP5 and NR3C1 in glucocorticoid signalling. Because rat models implicated insulin signalling abnormalities, and altered levels of the protein Arc related to active neurites and synapses, and activated by MAPK signalling, insulin metabolism was specifically included. The finding of genomic associations for genes involved in cerebral insulin signalling IGF2R, INSR, INSRR, and the related plasticity marker ARC, is therefore also supportive for ELS in LOAD. Generally, were metabolic biases as consequence of ELS, as conducive to LOAD, well supported animal and human studies.
The summary of findings on the epigenomic level is also able to extend these conclusions. On the epigenomic level, the present state of the literature suggests that the assumption of priming influences of environment in terms of epigenetic mechanism may be supported for core pathology genes of the Abeta cascade APP, BACE1, PSEN1, PSEN2, and of tauopathy MAPT. Mutations of these genes were found relevant for EOAD, but not LOAD. Findings of epigenetic modifications can now provide explanations as to their analogous pathophysiologies. As LOAD is dependent on ageing effects, epigenetic mechanisms in lipid metabolism genes ABCA7, ALOX5, DHCR24, APOE promoter are explanative. Modification in major neuroinflammation markers NFKB1, TNFA, ANK1, CRP extend recent analogous findings from GWAS. Metabolic and inflammatory processes are likely related to modifications in myelin-related genes BIN1, CNP, SORL1. ELS effects are assumable through modifications in ELS-related genes FKBP5, EGR1/NGFI-A, GSK3B. Finally, modifications in CREB, CREBBP, FOS, MBD2, MTHFR2, DNMT1 suggest alterations of synaptic functioning, transcription, and epigenetic gene modification itself in LOAD.
CONCLUSION AND FURTHER DIRECTIONS
In conclusion, it is noteworthy to say that the present analysis supports plausibility for an assumption of trauma-related ELS effects in LOAD, specifically FKBP5 and EGR1 mediated, and early life stress effects through glucocorticoid converter HSD11B1. The state of research suggests a coupling of the glucocorticoid receptor to the MAPT gene. Therefore, it is plausible that early tau neuronal mechanisms could be affected. This is also suggested by findings of early childhood LC degeneration and tauopathy. It is also possible, and also paediatric findings of a relation between reduced early cerebral growth and LOAD support this assumption, that early myelin formation could be compromised. Lesser myelin stability could be conducive to WM disintegrities as a structural biomarker of LOAD in old age. Similarly, early microglial priming towards an inflammatory phenotype, and early metabolic priming towards insulin resistance are plausible consequences of ELS effects. Unclear remains the functioning of the noradrenaline system in the context of locus caeruleus degeneration-related neuropathology.
Directions for further research must point to the interaction of neuroinflammation, cerebral lipid metabolism, brain insulin resistance, and myelin disintegration, which is suggested by the genomic and epigenomic findings accumulated. Currently, there is not enough knowledge in each of these domains available to answer the questions raised by abnormal cerebral insulin metabolism, myelin genes and maintenance of white matter, under the impact of chronic neuroinflammation.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
REFERENCES
- 1.Reitz C., Brayne C., Mayeux R. Epidemiology of Alzheimer disease. Nat. Rev. Neurol. 2011;7(3):137–152. doi: 10.1038/nrneurol.2011.2. https://www.nature.com/articles/nrneurol.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reitz C., Mayeux R. Endophenotypes in normal brain morphology and Alzheimer’s disease. Rev. Neurosci. 2009;164(1):174–190. doi: 10.1016/j.neuroscience.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kukull W.A., Higdon R., Bowen J.D., McCormick W.C., Teri L., Schellenberg G.D., van Belle G., Jolley L., Larson E.B. Dementia and Alzheimer disease incidence: A prospective cohort study. Arch. Neurol. 2002;59(11):1737–1746. doi: 10.1001/archneur.59.11.1737. [DOI] [PubMed] [Google Scholar]
- 4.Prince M., Albanese E., Guerchet M., Prina M., Pender R., Ferri C., Mazzotti D.R., Piovezan R.D., Padilla I., Luchsinger J.A. World Alzheimer Report. 2016 http://www.alz.co.uk/WAM
- 5.Huang D., Cai Z., Xiao M. Alzheimer’s disease and prenatal maternal stress. Aging Neurodegener. 2014;2(1):1–5. [Google Scholar]
- 6.Alzheimer’s Association 2015 Alzheimer’s disease facts and figures. Alzheimers Dement. 2015;11(3):332–384. doi: 10.1016/j.jalz.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Chouliaras L., Sierksma A.S., Kenis G., Prickaerts J., Lemmens M.A., Brasnjevic I., van Donkelaar E.L., Martinez-Martinez P., Losen M., De Baets M.H., Kholod N., van Leeuwen F., Hof P.R., van Os J., Steinbusch H.W., van den Hove D.L., Rutten B.P. Gene-environment interaction research and transgenic mouse models of Alzheimer’s disease. Int. J. Alzheimers Dis. 2010;2010:859101. doi: 10.4061/2010/859101. https://www.hindawi.com/journals/ ijad/2010/859101/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheltens P., Blennow K., Breteler M.M.B., de Strooper B., Frisoni G.B., Salloway S., Van der Flier W.M. Alzheimer’s disease. Lancet. 2016;388(10043):505–517. doi: 10.1016/S0140-6736(15)01124-1. https://www.thelancet.com/journals/lancet/article/PIIS0140-6736 (15)01124-1/fulltext [DOI] [PubMed] [Google Scholar]
- 9.Muller U.C., Deller T., Korte M. Not just amyloid: Physiological functions of the amyloid precursor protein family. Nat. Rev. Neurosci. 2017;18(5):281–298. doi: 10.1038/nrn.2017.29. [DOI] [PubMed] [Google Scholar]
- 10.Teng L., Zhao J., Wang F., Ma L., Pei G.A. GPCR/secretase complex regulates beta- and gamma-secretase specificity for Abeta production and contributes to AD pathogenesis. Cell Res. 2010;20(2):138–153. doi: 10.1038/cr.2010.3. [DOI] [PubMed] [Google Scholar]
- 11.Kandasamy K., Mohan S.S., Raju R., Keerthikumar S., Kumar G.S., Venugopal A.K., Telikicherla D., Navarro J.D., Mathivanan S., Pecquet C., Gollapudi S.K., Tattikota S.G., Mohan S., Padhukasahasram H., Subbannayya Y., Goel R., Jacob H.K., Zhong J., Sekhar R., Nanjappa V., Balakrishnan L., Subbaiah R., Ramachandra Y.L., Rahiman B.A., Prasad T.S., Lin J.X., Houtman J.C., Desiderio S., Renauld J.C., Constantinescu S.N., Ohara O., Hirano T., Kubo M., Singh S., Khatri P., Draghici S., Bader G.D., Sander C., Leonard W.J., Pandey A. NetPath: A public resource of curated signal transduction pathways. Genome Biol. 2010;11(1):R3. doi: 10.1186/gb-2010-11-1-r3. https://genomebiology. biomedcentral.com/articles/10.1186/gb-2010-11-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Quervain D.J., Poirier R., Wollmer M.A., Grimaldi L.M., Tsolaki M., Streffer J.R., Hock C., Nitsch R.M., Mohajeri M.H., Papassotiropoulos A. Glucocorticoid-related genetic susceptibility for Alzheimer’s disease. Hum. Mol. Genet. 2004;13(1):47–52. doi: 10.1093/hmg/ddg361. [DOI] [PubMed] [Google Scholar]
- 13.Liu C.C., Liu C.C., Kanekiyo T., Xu H., Bu G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013;9(2):106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiman E.M., Webster J.A., Myers A.J., Hardy J., Dunckley T., Zismann V.L., Joshipura K.D., Pearson J.V., Hu-Lince D., Huentelman M.J., Craig D.W., Coon K.D., Liang W.S., Herbert R.H., Beach T., Rohrer K.C., Zhao A.S., Leung D., Bryden L., Marlowe L., Kaleem M., Mastroeni D., Grover A., Heward C.B., Ravid R., Rogers J., Hutton M.L., Melquist S., Petersen R.C., Alexander G.E., Caselli R.J., Kukull W., Papassotiropoulos A., Stephan D.A. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007;54(5):713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertram L., Tanzi R.E. Genome-wide association studies in Alzheimer’s disease. 2009 doi: 10.1093/hmg/ddp406. https://academic.oup.com/hmg/article/18/R2/R137/ [DOI] [PMC free article] [PubMed]
- 16.Pan X.L., Ren R.J., Wang G., Tang H.D., Chen S.D. The Gab2 in signal transduction and its potential role in the pathogenesis of Alzheimer’s disease. Neurosci. Bull. 2010;26(3):241–246. doi: 10.1007/s12264-010-1109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J., Rujescu D., Hampel H., Giegling I., Andreassen O.A., Engedal K., Ulstein I., Djurovic S., Ibrahim-Verbaas C., Hofman A., Ikram M.A., van Duijn C.M., Thorsteinsdottir U., Kong A., Stefansson K. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013;368(2):107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakobsdottir J., van der Lee S.J., Bis J.C., Chouraki V., Li-Kroeger D., Yamamoto S., Grove M.L., Naj A., Vronskaya M., Salazar J.L., DeStefano A.L., Brody J.A., Smith A.V., Amin N., Sims R., Ibrahim-Verbaas C.A., Choi S.H., Satizabal C.L., Lopez O.L., Beiser A., Ikram M.A., Garcia M.E., Hayward C., Varga T.V., Ripatti S., Franks P.W., Hallmans G., Rolandsson O., Jansson J.H., Porteous D.J., Salomaa V., Eiriksdottir G., Rice K.M., Bellen H.J., Levy D., Uitterlinden A.G., Emilsson V., Rotter J.I., Aspelund T., Cohorts for Heart and Aging Research in Genomic Epidemiology consortium Alzheimer’s Disease Genetic Consortium; Genetic and Environmental Risk in Alzheimer’s Disease consortium; O’Donnell, C.J.; Fitzpatrick, A.L.; Launer, L.J.; Hofman, A.; Wang, L.S.; Williams, J.; Schellenberg, G.D.; Boerwinkle, E.; Psaty, B.M.; Seshadri, S.; Shulman, J.M.; Gudnason, V.; van Duijn, C.M. Rare functional variant in TM2D3 is associated with late-onset Alzheimer’s disease. PLoS Genet. 2016;12(10):e1006327. doi: 10.1371/journal.pgen.1006327. http://journals.plos.org/ plosgenetics/article?id=10.1371/journal.pgen.1006327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruchaga C., Karch C.M., Jin S.C., Benitez B.A., Cai Y., Guerreiro R., Harari O., Norton J., Budde J., Bertelsen S., Jeng A.T., Cooper B., Skorupa T., Carrell D., Levitch D., Hsu S., Choi J., Ryten M., UK British Expression Consortium Hardy, J.; Ryten, M.; Trabzuni, D.; Weale, M.E.; Ramasamy, A.; Smith, C.; Sassi, C.; Bras, J.; Gibbs, J.R.; Hernandez, D.G.; Lupton, M.K.; Powell, J.; Forabosco, P.; Ridge, P.G.; Corcoran, C.D.; Tschanz, J.T.; Norton, M.C.; Munger, R.G.; Schmutz, C.; Leary, M.; Demirci, F.Y.; Bamne, M.N.; Wang, X.; Lopez, O.L.; Ganguli, M.; Medway, C.; Turton, J.; Lord, J.; Braae, A.; Barber, I.; Brown, K.; Alzheimer’s Research UK (ARUK) Consortium; Passmore, P.; Craig, D.; Johnston, J.; McGuinness, B.; Todd, S.; Heun, R.; Kolsch, H.; Kehoe, P.G.; Hooper, N.M.; Vardy, E.R.; Mann, D.M.; Pickering-Brown, S.; Brown, K.; Kalsheker, N.; Lowe, J.; Morgan, K.; David Smith, A.; Wilcock, G.; Warden, D.; Holmes, C.; Pastor, P.; Lorenzo-Betancor, O.; Brkanac, Z.; Scott, E.; Topol, E.; Morgan, K.; Rogaeva, E.; Singleton, A.B.; Hardy, J.; Kamboh, M.I.; St. George-Hyslop, P.; Cairns, N.; Morris, J.C.; Kauwe, J.S.; Goate, A.M. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505(7484):550–554. doi: 10.1038/nature12825. https://www.nature.com/articles/nature12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson R.S., Barral S., Lee J.H., Leurgans S.E., Foroud T.M., Sweet R.A., Graff-Radford N., Bird T.D., Mayeux R., Bennett D.A. Heritability of different forms of memory in the late onset Alzheimer’s disease family study. J. Alzheimers Dis. 2011;23(2):249–255. doi: 10.3233/JAD-2010-101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer J.M., Breitner J.C. Multiple threshold model for the onset of Alzheimer’s disease in the NAS-NRC twin panel. Am. J. Med. Genet. 1998;81(1):92–97. doi: 10.1002/(sici)1096-8628(19980207)81:1<92::aid-ajmg16>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen N.L., Posner S.F., Gatz M. Multiple-threshold models for genetic influences on age of onset for Alzheimer disease: Findings in Swedish twins. Am. J. Med. Genet. 2001;105(8):724–728. doi: 10.1002/ajmg.1608. [DOI] [PubMed] [Google Scholar]
- 23.Gatz M., Pedersen N.L., Berg S., Johansson B., Johansson K., Mortimer J.A., Posner S.F., Viitanen M., Winblad B., Ahlbom A. Heritability for Alzheimer’s disease: The study of dementia in Swedish twins. J. Gerontol. A Biol. Sci. Med. Sci. 1997;52A(2):M117–M1125. doi: 10.1093/gerona/52a.2.m117. https://academic.oup.com/biomed gerontology/article/52A/2/M117/651528 [DOI] [PubMed] [Google Scholar]
- 24.Colantuoni C., Lipska B.K., Ye T., Hyde T.M., Tao R., Leek J.T., Colantuoni E.A., Elkahloun A.G., Herman M.M., Weinberger D.R., Kleinman J.E. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–523. doi: 10.1038/nature10524. https://www.nature.com/ articles/nature10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J., Yu L., Gaiteri C., Srivastava G.P., Chibnik L.B., Leurgans S.E., Schneider J.A., Meissner A., De Jager P.L., Bennett D.A. Association of DNA methylation in the brain with age in older persons is confounded by common neuropathologies. Int. J. Biochem. Cell Biol. 2015;67:58–64. doi: 10.1016/j.biocel.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu L., Chibnik L.B., Srivastava G.P., Pochet N., Yang J., Xu J., Kozubek J., Obholzer N., Leurgans S.E., Schneider J.A., Meissner A., De Jager P.L., Bennett D.A. Association of brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol. 2015;72(1):15–24. doi: 10.1001/jamaneurol.2014.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mormino E.C., Sperling R.A., Holmes A.J., Buckner R.L., Jager P.L.D., Smoller J.W., Sabuncu M.R. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology. 2016;87(5):481–488. doi: 10.1212/WNL.0000000000002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson M., Lee B.Y., Hane F.T. Recent progress in Alzheimer’s disease research, part 2: Genetics and epidemiology. J. Alzheimers Dis. 2017;57(2):317–330. doi: 10.3233/JAD-161149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dean D.C., III, Hurley S.A., Kecskemeti S.R., O’Grady J.P., Canda C., Davenport-Sis N.J., Carlsson C.M., Zetterberg H., Blennow K., Asthana S., Sager M.A., Johnson S.C., Alexander A.L., Bendlin B.B. Association of amyloid pathology with myelin alteration in preclinical Alzheimer disease. JAMA Neurol. 2016;74(1):41–49. doi: 10.1001/jamaneurol.2016.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie K., Ritchie C.W., Yaffe K., Skoog I., Scarmeas N. Is late-onset Alzheimer’s disease really a disease of midlife? Alzheimer Dem.: Transl. Res. Clin. Interv. 2015;1(2):122–130. doi: 10.1016/j.trci.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietrzak R.H., Lim Y.Y., Neumeister A., Ames D., Ellis K.A., Harrington K., Lautenschlager N.T., Restrepo C., Martins R.N., Masters C.L., Villemagne V.L., Rowe C.C., Maruff P., Australian Imaging, Biomarkers, and Lifestyle Research Group Amyloid-beta, anxiety, and cognitive decline in preclinical Alzheimer disease: A multicenter, prospective cohort study. JAMA Psychiatry. 2015;72(3):284–291. doi: 10.1001/jamapsychiatry.2014.2476. [DOI] [PubMed] [Google Scholar]
- 32.Russ T.C., Kivimaki M., Starr J.M., Stamatakis E., Batty G.D. Height in relation to dementia death: Individual participant meta-analysis of 18 UK prospective cohort studies. Br. J. Psychiatry. 2014;205(5):348–354. doi: 10.1192/bjp.bp.113.142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller M., Sigurdsson S., Kjartansson O., Jonsson P.V., Garcia M., von Bonsdorff M.B., Gunnarsdottir I., Thorsdottir I., Harris T.B., van Buchem M., Gudnason V., Launer L.J. Birth size and brain function 75 years later. Pediatrics. 2014;134(4):761–770. doi: 10.1542/peds.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Killin L.O., Starr J.M., Shiue I.J., Russ T.C. Environmental risk factors for dementia: A systematic review. BMC Geriatr. 2016;16(1):175. doi: 10.1186/s12877-016-0342-y. https://bmcgeriatr.biomedcentral.com/ articles/10.1186/s12877-016-0342-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kepp K.P. Alzheimer’s disease due to loss of function: A new synthesis of the available data. Prog. Neurobiol. 2016;143:36–60. doi: 10.1016/j.pneurobio.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Lehmann D.J., Schuur M., Warden D.R., Hammond N., Belbin O., Kolsch H., Lehmann M.G., Wilcock G.K., Brown K., Kehoe P.G., Morris C.M., Barker R., Coto E., Alvarez V., Deloukas P., Mateo I., Gwilliam R., Combarros O., Arias-Vasquez A., Aulchenko Y.S., Ikram M.A., Breteler M.M., van Duijn C.M., Oulhaj A., Heun R., Cortina-Borja M., Morgan K., Robson K., Smith A.D. Transferrin and HFE genes interact in Alzheimer’s disease risk: The Epistasis Project. Neurobiol. Aging. 2012;33(1):202.e1–202.e13. doi: 10.1016/j.neurobiolaging.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Chen H., Kwong J.C., Copes R., Tu K., Villeneuve P.J., van Donkelaar A., Hystad P., Martin R.V., Murray B.J., Jessiman B., Wilton A.S., Kopp A., Burnett R.T. Living near major roads and the incidence of dementia, Parkinson’s disease, and multiple sclerosis: A population-based cohort study. Lancet. 2017;389(10070):718–726. doi: 10.1016/S0140-6736(16)32399-6. https://www.thelancet.com/ journals/lancet/article/PIIS0140-6736(16)32399-6/fulltext [DOI] [PubMed] [Google Scholar]
- 38.Lindsay J. Risk factors for Alzheimer’s disease: A prospective analysis from the Canadian study of health and aging. Am. J. Epidemiol. 2002;156(5):445–453. doi: 10.1093/aje/kwf074. [DOI] [PubMed] [Google Scholar]
- 39.Lemche E., Chaban O.S., Lemche A.V. Neuroendorine and epigentic mechanisms subserving autonomic imbalance and HPA dysfunction in the metabolic syndrome. Front. Neurosci. 2016;10:142. doi: 10.3389/fnins.2016.00142. https://www.frontiersin.org/articles/10.3389/ fnins.2016.00142/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanemaru K., Takio K., Miura R., Titani K., Ihara Y. Fetal-type phosphorylation of the tau in paired helical filaments. J. Neurochem. 1992;58(5):1667–1675. doi: 10.1111/j.1471-4159.1992.tb10039.x. [DOI] [PubMed] [Google Scholar]
- 41.Matsuo E.S., Shin R.W., Billingsley M.L., Van deVoorde A., O’Connor M., Trojanowski J.Q., Lee V.M. Biopsy-derived adult human brain tau is phosphorylated at many of the same sites as Alzheimer’s disease paired helical filament tau. Neuron. 1994;13(4):989–1002. doi: 10.1016/0896-6273(94)90264-x. [DOI] [PubMed] [Google Scholar]
- 42.Taniguchi T., Kawamata T., Mukai H., Hasegawa H., Isagawa T., Yasuda M., Hashimoto T., Terashima A., Nakai M., Mori H., Ono Y., Tanaka C. Phosphorylation of tau is regulated by PKN. J. Biol. Chem. 2001;276(13):10025–10031. doi: 10.1074/jbc.M007427200. [DOI] [PubMed] [Google Scholar]
- 43.Miller M.W., Sadeh N. Traumatic stress, oxidative stress and post-traumatic stress disorder: Neurodegeneration and the accelerated-aging hypothesis. Mol. Psychiatry. 2014;19(11):1156–1162. doi: 10.1038/mp.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richard E., Reitz C., Honig L.H., Schupf N., Tang M.X., Manly J.J., Mayeux R., Devanand D., Luchsinger J.A. Late-life depression, mild cognitive impairment, and dementia. JAMA Neurol. 2013;70(3):374–382. doi: 10.1001/jamaneurol.2013.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byers A.L., Yaffe K. Depression and risk of developing dementia. Nat. Rev. Neurol. 2011;7(6):323–331. doi: 10.1038/nrneurol.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greenberg M.S., Tanev K., Marin M.F., Pitman R.K. Stress, PTSD, and dementia. Alzheimers Dement. 2014;10(3) Suppl.:S155–S165. doi: 10.1016/j.jalz.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Fratiglioni L., Paillard-Borg S., Winblad B. An active and socially integrated lifestyle in late life might protect against dementia. Lancet Neurol. 2004;3(6):343–353. doi: 10.1016/S1474-4422(04)00767-7. [DOI] [PubMed] [Google Scholar]
- 48.Wilson R.S., Evans D.A., Bienias J.L., Mendes de Leon C.F., Schneider J.A., Bennett D.A. Proneness to psychological distress is associated with risk of Alzheimer’s disease. Neurology. 2003;61(11):1479–1485. doi: 10.1212/01.wnl.0000096167.56734.59. [DOI] [PubMed] [Google Scholar]
- 49.Johansson L., Guo X., Hallstrom T., Norton M.C., Waern M., Ostling S., Bengtsson C., Skoog I. Common psychosocial stressors in middle-aged women related to longstanding distress and increased risk of Alzheimer’s disease: A 38-year longitudinal population study. BMJ Open. 2013;3(9):e003142. doi: 10.1136/bmjopen-2013-003142. http://bmjopen.bmj.com/content/3/9/e003142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansson L., Guo X., Waern M., Ostling S., Gustafson D., Bengtsson C., Skoog I. Midlife psychological stress and risk of dementia: A 35-year longitudinal population study. Brain. 2010;133(Pt 8):2217–2224. doi: 10.1093/brain/awq116. [DOI] [PubMed] [Google Scholar]
- 51.Deng J., Lian Y., Shen C., Chen Y., Zhang M., Wang Y.J., Zhou H.D. Adverse life event and risk of cognitive impairment: A 5-year prospective longitudinal study in Chongqing, China. Eur. J. Neurol. 2012;19(4):631–637. doi: 10.1111/j.1468-1331.2011.03599.x. [DOI] [PubMed] [Google Scholar]
- 52.Leng Y., Wainwright N.W., Hayat S., Stephan B.C., Matthews F.E., Luben R., Surtees P.G., Khaw K.T., Brayne C. The association between social stress and global cognitive function in a population-based study: The European Prospective Investigation into Cancer (EPIC)-Norfolk study. Psychol. Med. 2013;43(3):655–666. doi: 10.1017/S0033291712001316. [DOI] [PubMed] [Google Scholar]
- 53.Tsolaki M., Papaliagkas V., Kounti F., Messini C., Boziki M., Anogianakis G., Vlaikidis N. Severely stressful events and dementia: A study of an elderly Greek demented population. Psychiatry Res. 2010;176(1):51–54. doi: 10.1016/j.psychres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Eriksson M., Raikkonen K., Eriksson J.G. Early life stress and later health outcomes--findings from the Helsinki Birth Cohort Study. Am. J. Hum. Biol. 2014;26(2):111–116. doi: 10.1002/ajhb.22502. [DOI] [PubMed] [Google Scholar]
- 55.Cheng G., Huang C., Deng H., Wang H. Diabetes as a risk factor for dementia and mild cognitive impairment: A meta-analysis of longitudinal studies. Intern. Med. J. 2012;42(5):484–491. doi: 10.1111/j.1445-5994.2012.02758.x. [DOI] [PubMed] [Google Scholar]
- 56.Pesönen A.K., Räikkönen K. The lifespan consequences of early life stress. Physiol. Behav. 2012;106(5):722–727. doi: 10.1016/j.physbeh.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 57.Alexander G.E. An emerging role for imaging white matter in the preclinical risk for Alzheimer Disease: Linking beta-Amyloid to myelin. JAMA Neurol. 2016;74(1):17–19. doi: 10.1001/jamaneurol.2016.4123. [DOI] [PubMed] [Google Scholar]
- 58.Karama S., Bastin M.E., Murray C., Royle N.A., Penke L., Munoz Maniega S., Gow A.J., Corley J., Mdel C.V.H., Lewis J.D., Rousseau M.E., Lepage C., Fonov V., Collins D.L., Booth T., Rioux P., Sherif T., Adalat R., Starr J.M., Evans A.C., Wardlaw J.M., Deary I.J. Childhood cognitive ability accounts for associations between cognitive ability and brain cortical thickness in old age. Mol. Psychiatry. 2014;19(5):555–559. doi: 10.1038/mp.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bartzokis G., Lu P.H., Geschwind D.H., Tingus K., Huang D., Mendez M.F., Edwards N., Mintz J. Apolipoprotein E affects both myelin breakdown and cognition: Implications for age-related trajectories of decline into dementia. Biol. Psychiatry. 2007;62(12):1380–1387. doi: 10.1016/j.biopsych.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 60.Bartzokis G. Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiol. Aging. 2011;32(8):1341–1371. doi: 10.1016/j.neurobiolaging.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Small S.A. Isolating pathogenic mechanisms embedded within the hippocampal circuit through regional vulnerability. Neuron. 2014;84(1):32–39. doi: 10.1016/j.neuron.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iturria-Medina Y., Sotero R.C., Toussaint P.J., Mateos-Perez J.M., Evans A.C., Alzheimer’s Disease Neuroimaging Initiative Early role of vascular dysregulation on late-onset Alzheimer’s disease based on multifactorial data-driven analysis. 2016 doi: 10.1038/ncomms11934. https://www.nature.com/articles/ [DOI] [PMC free article] [PubMed]
- 63.Hale J.M. Alzheimer’s and the Great Depression: Do early-life deprivation and social inequalities impact late-life cognitive decline?; American Sociological Association Annual Meeting; 2014. [Google Scholar]
- 64.Persson G., Skoog I. A prospective population study of psychosocial risk factors for late onset dementia. Int. J. Geriatr. Psychiatry. 1996;11(1):15–22. [Google Scholar]
- 65.Norton M.C., Smith K.R., Ostbye T., Tschanz J.T., Schwartz S., Corcoran C., Breitner J.C., Steffens D.C., Skoog I., Rabins P.V., Welsh-Bohmer K.A., Cache County Investigators Early parental death and remarriage of widowed parents as risk factors for Alzheimer disease: The Cache County study. Am. J. Geriatr. Psychiatry. 2011;19(9):814–824. doi: 10.1097/JGP.0b013e3182011b38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ravona-Springer R., Beeri M.S., Goldbourt U. Younger age at crisis following parental death in male children and adolescents is associated with higher risk for dementia at old age. Alzheimer Dis. Assoc. Disord. 2012;26(1):68–73. doi: 10.1097/WAD.0b013e3182191f86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whalley L.J., Staff R.T., Murray A.D., Deary I.J., Starr J.M. Genetic and environmental factors in late onset dementia: Possible role for early parental death. Int. J. Geriatr. Psychiatry. 2013;28(1):75–81. doi: 10.1002/gps.3792. [DOI] [PubMed] [Google Scholar]
- 68.Pechtel P., Pizzagalli D.A. Effects of early life stress on cognitive and affective function: An integrated review of human literature. Psychopharmacology (Berl.) 2011;214(1):55–70. doi: 10.1007/s00213-010-2009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meziab O., Kirby K.A., Williams B., Yaffe K., Byers A.L., Barnes D.E. Prisoner of war status, posttraumatic stress disorder, and dementia in older veterans. Alzheimers Dement. 2014;10(3) Suppl.:S236–S241. doi: 10.1016/j.jalz.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 70.Roth M., Tomlinson B.E., Blessed G. Correlation between scores for dementia and counts of ‘senile plaques’ in cerebral grey matter of elderly subjects. Nature. 1966;209(5018):109–110. doi: 10.1038/209109a0. https://www.nature.com/articles/209109a0 [DOI] [PubMed] [Google Scholar]
- 71.Borenstein A.R., Copenhaver C.I., Mortimer J.A. Early-life risk factors for Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2006;20(1):63–72. doi: 10.1097/01.wad.0000201854.62116.d7. [DOI] [PubMed] [Google Scholar]
- 72.Borenstein A.R., Wu Y., Mortimer J.A., Schellenberg G.D., McCormick W.C., Bowen J.D., McCurry S., Larson E.B. Developmental and vascular risk factors for Alzheimer’s disease. Neurobiol. Aging. 2005;26(3):325–334. doi: 10.1016/j.neurobiolaging.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 73.Reincke S.A., Hanganu-Opatz I.L. Early-life stress impairs recognition memory and perturbs the functional maturation of prefrontal-hippocampal-perirhinal networks. Sci. Rep. 2017;7:42042. doi: 10.1038/srep42042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoffmann A., Spengler D. The lasting legacy of social stress on the epigenome of the hypothalamic-pituitary-adrenal axis. Epigenomics. 2012;4(4):431–444. doi: 10.2217/epi.12.34. [DOI] [PubMed] [Google Scholar]
- 75.Heim C., Binder E.B. Current research trends in early life stress and depression: Review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp. Neurol. 2012;233(1):102–111. doi: 10.1016/j.expneurol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 76.Green J.G., McLaughlin K.A., Berglund P.A., Gruber M.J., Sampson N.A., Zaslavsky A.M., Kessler R.C. Childhood adversities and adult psychiatric disorders in the national comorbidity survey replication I: Associations with first onset of DSM-IV disorders. Arch. Gen. Psychiatry. 2010;67(2):113–123. doi: 10.1001/archgenpsychiatry.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grundwald N.J., Brunton P.J. Prenatal stress programs neuroendocrine stress responses and affective behaviors in second generation rats in a sex-dependent manner. Psychoneuroendocrinology. 2015;62:204–216. doi: 10.1016/j.psyneuen.2015.08.010. https://www.psyneuen-journal. com/article/S0306-4530(15)00883-5/fulltext [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loman M.M., Gunnar M.R., Early Experience, Stress, and Neurobehavioral Development Center Early experience and the development of stress reactivity and regulation in children. Neurosci. Biobehav. Rev. 2010;34(6):867–876. doi: 10.1016/j.neubiorev.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y., Baram T.Z. Toward understanding how early-life stress reprograms cognitive and emotional brain networks. Neuropsychopharmacology. 2016;41(1):197–206. doi: 10.1038/npp.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aisa B., Tordera R., Lasheras B., Del Rio J., Ramirez M.J. Cognitive impairment associated to HPA axis hyperactivity after maternal separation in rats. Psychoneuroendocrinology. 2007;32(3):256–266. doi: 10.1016/j.psyneuen.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 81.Heim C., Newport D.J., Heit S., Graham Y.P., Wilcox M., Bonsall R., Miller A.H., Nemeroff C.B. Pituitary-adrenal and autonomic responses to stress in women after sexual and physical abuse in childhood. JAMA. 2000;284(5):592–597. doi: 10.1001/jama.284.5.592. [DOI] [PubMed] [Google Scholar]
- 82.Cohen R.A., Hitsman B.L., Paul R.H., McCaffery J., Stroud L., Sweet L., Gunstad J., Niaura R., MacFarlane A., Bryant R.A., Gordon E. Early life stress and adult emotional experience: An international perspective. Int. J. Psychiatry Med. 2006;36(1):35–52. doi: 10.2190/5R62-9PQY-0NEL-TLPA. [DOI] [PubMed] [Google Scholar]
- 83.Lemche E., Giampietro V.P., Surguladze S.A., Amaro E.J., Andrew C.M., Williams S.C., Brammer M.J., Lawrence N., Maier M.A., Russell T.A., Simmons A., Ecker C., Joraschky P., Phillips M.L. Human attachment security is mediated by the amygdala: Evidence from combined fMRI and psychophysiological measures. Hum. Brain Mapp. 2005;27(8):623–635. doi: 10.1002/hbm.20206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lemche E., Klann-Delius G., Koch R., Joraschky P. Mentalizing language development in a longitudinal attachment sample: Implications for alexithymia. Psychother. Psychosom. 2004;73(6):366–374. doi: 10.1159/000080390. [DOI] [PubMed] [Google Scholar]
- 85.Delpierre C., Fantin R., Barboza-Solis C., Lepage B., Darnaudery M., Kelly-Irving M. 2016 doi: 10.1186/s12889-016-3484-0. https://bmcpublichealth.biomedcentral.com/articles/10.1186/ [DOI] [PMC free article] [PubMed]
- 86.Reynolds R.M. Glucocorticoid excess and the developmental origins of disease: Two decades of testing the hypothesis--2012 Curt Richter Award Winner. Psychoneuroendocrinology. 2013;38(1):1–11. doi: 10.1016/j.psyneuen.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 87.Swartz J.R., Hariri A.R., Williamson D.E. An epigenetic mechanism links socioeconomic status to changes in depression-related brain function in high-risk adolescents. Mol. Psychiatry. 2017;22(2):209–214. doi: 10.1038/mp.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yam K.Y., Naninck E.F., Schmidt M.V., Lucassen P.J., Korosi A. Early-life adversity programs emotional functions and the neuroendocrine stress system: The contribution of nutrition, metabolic hormones and epigenetic mechanisms. Stress. 2015;18(3):328–342. doi: 10.3109/10253890.2015.1064890. [DOI] [PubMed] [Google Scholar]
- 89.Meaney M.J., Szyf M., Seckl J.R. Epigenetic mechanisms of perinatal programming of hypothalamic-pituitary-adrenal function and health. Trends Mol. Med. 2007;13(7):269–277. doi: 10.1016/j.molmed.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 90.Pena C.J., Monk C., Champagne F.A. 2012 http://journals.plos.org/plosone/article?id=10.1371/journal
- 91.Togher K.L., O’Keeffe M.M., Khashan A.S., Gutierrez H., Kenny L.C., O’Keeffe G.W. Epigenetic regulation of the placental HSD11B2 barrier and its role as a critical regulator of fetal development. Epigenetics. 2014;9(6):816–822. doi: 10.4161/epi.28703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vargas J., Junco M., Gomez C., Lajud N. Early life stress increases metabolic risk, HPA axis reactivity, and depressive-like behavior when combined with postweaning social isolation in rats. PLoS One. 2016;11(9):e0162665. doi: 10.1371/journal.pone.0162665. http://journals. plos.org/plosone/article?id=10.1371/journal.pone.0162665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oomen C.A., Soeters H., Audureau N., Vermunt L., van Hasselt F.N., Manders E.M., Joels M., Lucassen P.J., Krugers H. Severe early life stress hampers spatial learning and neurogenesis, but improves hippocampal synaptic plasticity and emotional learning under high-stress conditions in adulthood. J. Neurosci. 2010;30(19):6635–6645. doi: 10.1523/JNEUROSCI.0247-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meadows J.P., Guzman-Karlsson M.C., Phillips S., Brown J.A., Strange S.K., Sweatt J.D., Hablitz J.J. Dynamic DNA methylation regulates neuronal intrinsic membrane excitability. 2016 doi: 10.1126/scisignal.aaf5642. mag.org/content/9/442/ra83 [DOI] [PMC free article] [PubMed]
- 95.Appleton A.A., Armstrong D.A., Lesseur C., Lee J., Padbury J.F., Lester B.M., Marsit C.J. 2013 http://journals.plos.org/plosone/article?id=10.1371/journal
- 96.Harris A., Seckl J. Glucocorticoids, prenatal stress and the programming of disease. Horm. Behav. 2011;59(3):279–289. doi: 10.1016/j.yhbeh.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 97.Braithwaite E.C., Kundakovic M., Ramchandani P.G., Murphy S.E., Champagne F.A. Maternal prenatal depressive symptoms predict infant NR3C1 1F and BDNF IV DNA methylation. Epigenetics. 2015;10(5):408–417. doi: 10.1080/15592294.2015.1039221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McGowan P.O., Sasaki A., D’Alessio A.C., Dymov S., Labonte B., Szyf M., Turecki G., Meaney M.J. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reynolds R.M. Corticosteroid-mediated programming and the pathogenesis of obesity and diabetes. J. Steroid Biochem. Mol. Biol. 2010;122(1-3):3–9. doi: 10.1016/j.jsbmb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 100.Hoeijmakers L., Heinen Y., van Dam A.M., Lucassen P.J., Korosi A. Microglial priming and Alzheimer’s disease: A possible role for (early) immune challenges and epigenetics? Front. Hum. Neurosci. 2016;10:398. doi: 10.3389/fnhum.2016.00398. https://www.frontiersin.org/ articles/10.3389/fnhum.2016.00398/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lajud N., Roque A., Cajero M., Gutierrez-Ospina G., Torner L. Periodic maternal separation decreases hippocampal neurogenesis without affecting basal corticosterone during the stress hyporesponsive period, but alters HPA axis and coping behavior in adulthood. Psychoneuroendocrinology. 2012;37(3):410–420. doi: 10.1016/j.psyneuen.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 102.Waffarn F., Davis E.P. Effects of antenatal corticosteroids on the hypothalamic-pituitary-adrenocortical axis of the fetus and newborn: Experimental findings and clinical considerations. Am. J. Obstet. Gynecol. 2012;207(6):446–454. doi: 10.1016/j.ajog.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duarte-Neves J., Pereira de Almeida L., Cavadas C., Neuropeptide Y. NPY) as a therapeutic target for neurodegenerative diseases. Neurobiol. Dis. 2016;95:210–224. doi: 10.1016/j.nbd.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 104.Seifan A., Schelke M., Obeng-Aduasare Y., Isaacson R. Early life epidemiology of Alzheimer’s disease--A critical review. Neuroepidemiology. 2015;45(4):237–254. doi: 10.1159/000439568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lemche E. Body image development in the first three years of life: On the developmental psychology of the mental representation of one’s own body. Ann Arbor, MI: University Microfilms International; 1996. [Google Scholar]
- 106.Lemche E. The development of the body image in the first three years of life. Psychoanal. Contemp. Thought. 1998;21(2):155–275. [Google Scholar]
- 107.Lemche E. Myelination patterns in the cerebral cortex as indicators of brain-behavior relationships in childhood concept formation: A comparison of staining, immunohistochemical and magnetic resonance imaging studies. Dev. Psychobiol. 2001;41(1):84–85. [Google Scholar]
- 108.Greenough W.T., Black J.E., Wallace C.S. Experience and brain development. Child Dev. 1987;58(3):539–559. [PubMed] [Google Scholar]
- 109.Dean D.C., Jerskey B.A., Chen K., Protas H., Thiyyagura P., Roontiva A., O’Muircheartaigh J., Dirks H., Waskiewicz N., Lehman K., Siniard A.L., Turk M.N., Hua X., Madsen S.K., Thompson P.M., Fleisher A.S., Huentelman M.J., Deoni S.C.L., Reiman E.M. Brain differences in infants at differential genetic risk for late-onset Alzheimer Disease. JAMA Neurol. 2014;71(1):11–22. doi: 10.1001/jamaneurol.2013.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clark U.S., Sweet L.H., Morgello S., Philip N.S., Cohen R.A. High early life stress and aberrant amygdala activity: Risk factors for elevated neuropsychiatric symptoms in HIV+ adults. Brain Imaging Behav. 2016;11(3):649–665. doi: 10.1007/s11682-016-9542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hanson J.L., Albert D., Iselin A.M., Carre J.M., Dodge K.A., Hariri A.R. Cumulative stress in childhood is associated with blunted reward-related brain activity in adulthood. Soc. Cogn. Affect. Neurosci. 2016;11(3):405–412. doi: 10.1093/scan/nsv124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mehta M.A., Gore-Langton E., Golembo N., Colvert E., Williams S.C., Sonuga-Barke E. Hyporesponsive reward anticipation in the basal ganglia following severe institutional deprivation early in life. J. Cogn. Neurosci. 2010;22(10):2316–2325. doi: 10.1162/jocn.2009.21394. [DOI] [PubMed] [Google Scholar]
- 113.Szyf M. DNA methylation, behavior and early life adversity. J. Genet. Genomics. 2013;40(7):331–338. doi: 10.1016/j.jgg.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 114.Szyf M. The genome- and system-wide response of DNA methylation to early life adversity and its implication on mental health. Can. J. Psychiatry. 2013;58(12):697–704. doi: 10.1177/070674371305801208. [DOI] [PubMed] [Google Scholar]
- 115.Bonn S., Seeburg P.H., Schwarz M.K. Combinatorial expression of alpha- and gamma-protocadherins alters their presenilin-dependent processing. Mol. Cell. Biol. 2007;27(11):4121–4132. doi: 10.1128/MCB.01708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Korosi A., Naninck E.F., Oomen C.A., Schouten M., Krugers H., Fitzsimons C., Lucassen P.J. Early-life stress mediated modulation of adult neurogenesis and behavior. Behav. Brain Res. 2012;227(2):400–409. doi: 10.1016/j.bbr.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 117.Satz P. Brain reserve capacity on symptom onset after brain injury: A formulation and review of evidence for threshold theory. Neuropsychology. 1993;7(3):273–295. [Google Scholar]
- 118.Mortimer J. The continuum hypothesis of Alzheimer’s disease and normal aging: The role of brain reserve. Alzheimer Res. 1995;1(1):67–70. [Google Scholar]
- 119.Hui J.J., Zhang Z.J., Liu S.S., Xi G.J., Zhang X.R., Teng G.J., Chan K.C., Wu E.X., Nie B.B., Shan B.C., Li L.J., Reynolds G.P. Hippocampal neurochemistry is involved in the behavioural effects of neonatal maternal separation and their reversal by post-weaning environmental enrichment: A magnetic resonance study. Behav. Brain Res. 2011;217(1):122–127. doi: 10.1016/j.bbr.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 120.Hulshof H.J., Novati A., Sgoifo A., Luiten P.G., den Boer J.A., Meerlo P. Maternal separation decreases adult hippocampal cell proliferation and impairs cognitive performance but has little effect on stress sensitivity and anxiety in adult Wistar rats. Behav. Brain Res. 2011;216(2):552–560. doi: 10.1016/j.bbr.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 121.Koehl M., van der Veen R., Gonzales D., Piazza P.V., Abrous D.N. Interplay of maternal care and genetic influences in programming adult hippocampal neurogenesis. Biol. Psychiatry. 2012;72(4):282–289. doi: 10.1016/j.biopsych.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 122.Koehl M. Gene-environment interaction in programming hippocampal plasticity: Focus on adult neurogenesis. Front. Mol. Neurosci. 2015;8:41. doi: 10.3389/fnmol.2015.00041. https://www.frontiersin.org/articles/10.3389/fnmol.2015.00041/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Daskalakis N.P., De Kloet E.R., Yehuda R., Malaspina D., Kranz T.M. Early life stress effects on glucocorticoid-BDNF interplay in the hippocampus. 2015 doi: 10.3389/fnmol.2015.00068. https://www.frontiersin.org/articles/10.3389/fnmol [DOI] [PMC free article] [PubMed]
- 124.Lambert W.M., Xu C-F., Neubert T.A., Chao M.V., Garabedian M.J., Jeanneteau F.D. Brain-derived neurotrophic factor signaling rewrites the glucocorticoid transcriptome via glucocorticoid receptor phosphorylation. Mol. Cell. Biol. 2013;33(18):3700–3714. doi: 10.1128/MCB.00150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kundakovic M., Gudsnuka K., Herbstman J.B., Tang D., Perera F.P., Champagne F.A. DNA methylation of BDNF as a biomarker of early-life adversity. Proc. Natl. Acad. Sci. USA. 2015;112(22):6807–6813. doi: 10.1073/pnas.1408355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alboni S., Tascedda F., Corsini D., Benatti C., Caggia F., Capone G., Barden N., Blom J.M., Brunello N. Stress induces altered CRE/CREB pathway activity and BDNF expression in the hippocampus of glucocorticoid receptor-impaired mice. Neuropharmacology. 2011;60(7-8):1337–1346. doi: 10.1016/j.neuropharm.2011.01.050. [DOI] [PubMed] [Google Scholar]
- 127.Qin X.Y., Cao C., Cawley N.X., Liu T.T., Yuan J., Loh Y.P., Cheng Y. Decreased peripheral brain-derived neurotrophic factor levels in Alzheimer’s disease: A meta-analysis study (N=7277). Mol. Psychiatry. 2017;22(2):312–320. doi: 10.1038/mp.2016.62. [DOI] [PubMed] [Google Scholar]
- 128.Gimenez-Llort L., Blazquez G., Canete T., Johansson B., Oddo S., Tobena A., LaFerla F.M., Fernandez-Teruel A. Modeling behavioral and neuronal symptoms of Alzheimer’s disease in mice: A role for intraneuronal amyloid. Neurosci. Biobehav. Rev. 2007;31(1):125–147. doi: 10.1016/j.neubiorev.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 129.Zimmer C., Spencer K.A. Modifications of glucocorticoid receptors mRNA expression in the hypothalamic-pituitary-adrenal axis in response to early-life stress in female Japanese quail. J. Neuroendocrinol. 2014;26(12):853–860. doi: 10.1111/jne.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weaver I.C. Epigenetic effects of glucocorticoids. Semin. Fetal Neonatal Med. 2009;14(3):143–150. doi: 10.1016/j.siny.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 131.Campbell S.N., Zhang C., Monte L., Roe A.D., Rice K.C., Tache Y., Masliah E., Rissman R.A. Increased tau phosphorylation and aggregation in the hippocampus of mice overexpressing corticotropin-releasing factor. J. Alzheimers Dis. 2015;43(3):967–976. doi: 10.3233/JAD-141281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Reddy G.R., Zawia N.H. Lead exposure alters Egr-1 DNA-binding in the neonatal rat brain. Int. J. Dev. Neurosci. 2000;18(8):791–795. doi: 10.1016/s0736-5748(00)00048-4. [DOI] [PubMed] [Google Scholar]
- 133.Basha M.R., Wei W., Bakheet S.A., Benitez N., Siddiqi H.K., Ge Y.W., Lahiri D.K., Zawia N.H. The fetal basis of amyloidogenesis: Exposure to lead and latent overexpression of amyloid precursor protein and beta-amyloid in the aging brain. J. Neurosci. 2005;25(4):823–829. doi: 10.1523/JNEUROSCI.4335-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zawia N.H., Basha M.R. Environmental risk factors and the developmental basis for Alzheimer’s disease. Rev. Neurosci. 2005;16(4):325–337. doi: 10.1515/revneuro.2005.16.4.325. [DOI] [PubMed] [Google Scholar]
- 135.Wu J., Basha M.R., Zawia N.H. The environment, epigenetics and amyloidogenesis. J. Mol. Neurosci. 2008;34(1):1–7. doi: 10.1007/s12031-007-0009-4. [DOI] [PubMed] [Google Scholar]
- 136.Zawia N.H., Lahiri D.K., Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic. Biol. Med. 2009;46(9):1241–1249. doi: 10.1016/j.freeradbiomed.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bihaqi S.W., Schumacher A., Maloney B., Lahiri D.K., Zawia N.H. Do epigenetic pathways initiate late onset Alzheimer disease (LOAD): Towards a new paradigm. Curr. Alzheimer Res. 2012;9(5):574–588. doi: 10.2174/156720512800617982. [DOI] [PubMed] [Google Scholar]
- 138.Feng Q., Cheng B., Yang R., Sun F.Y., Zhu C.Q. Dynamic changes of phosphorylated tau in mouse hippocampus after cold water stress. Neurosci. Lett. 2005;388(1):13–16. doi: 10.1016/j.neulet.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 139.Fujio J., Hosono H., Ishiguro K., Ikegami S., Fujita S.C. Tau phosphorylation in the mouse brain during aversive conditioning. Neurochem. Int. 2007;51(2-4):200–208. doi: 10.1016/j.neuint.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 140.Kanamaru T., Kamimura N., Yokota T., Iuchi K., Nishimaki K., Takami S., Akashiba H., Shitaka Y., Katsura K., Kimura K., Ohta S. Oxidative stress accelerates amyloid deposition and memory impairment in a double-transgenic mouse model of Alzheimer’s disease. Neurosci. Lett. 2015;587:126–131. doi: 10.1016/j.neulet.2014.12.033. [DOI] [PubMed] [Google Scholar]
- 141.Yeh C.W., Yeh S.H., Shie F.S., Lai W.S., Liu H.K., Tzeng T.T., Tsay H.J., Shiao Y.J. Impaired cognition and cerebral glucose regulation are associated with astrocyte activation in the parenchyma of metabolically stressed APPswe/PS1dE9 mice. Neurobiol. Aging. 2015;36(11):2984–2994. doi: 10.1016/j.neurobiolaging.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 142.Mitchell E., Klein S.L., Argyropoulos K.V., Sharma A., Chan R.B., Toth J.G., Barboza L., Bavley C., Bortolozzi A., Chen Q., Liu B., Ingenito J., Mark W., Dudakov J., Gross S., Di Paolo G., Artigas F., van den Brink M., Toth M. Behavioural traits propagate across generations via segregated iterative-somatic and gametic epigenetic mechanisms. Nat. Commun. 2016;7:11492. doi: 10.1038/ncomms11492. https://www.nature.com/articles/ncomms11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cuadrado-Tejedor M., Garcia-Osta A. Chronic mild stress assay leading to early onset and propagation of Alzheimer’s Disease phenotype in mouse models. Methods Mol. Biol. 2016;1303:241–246. doi: 10.1007/978-1-4939-2627-5_14. [DOI] [PubMed] [Google Scholar]
- 144.Lesuis S.L., Maurin H., Borghgraef P., Lucassen P.J., Van F. Leuven; Krugers, H.J. Positive and negative early life experiences differentially modulate long term survival and amyloid protein levels in a mouse model of Alzheimer’s disease. Oncotarget. 2016;7(26):39118–39135. doi: 10.18632/oncotarget.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lesuis S.L., van Hoek B., Lucassen P.J., Krugers H.J. Early postnatal handling reduces hippocampal amyloid plaque formation and enhances cognitive performance in APPswe/PS1dE9 mice at middle age. Neurobiol. Learn. Mem. 2017;144:27–35. doi: 10.1016/j.nlm.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 146.Lee K.Y., Miki T., Yokoyama T., Ueki M., Warita K., Suzuki S., Ohta K., Wang Z.Y., Jamal M., Yakura T., Liu J.Q., Hosomi N., Takeuchi Y. Neonatal repetitive maternal separation causes long-lasting alterations in various neurotrophic factor expression in the cerebral cortex of rats. Life Sci. 2012;90(15-16):578–584. doi: 10.1016/j.lfs.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 147.Walker M.P., LaFerla F.M., Oddo S.S., Brewer G.J. Reversible epigenetic histone modifications and Bdnf expression in neurons with aging and from a mouse model of Alzheimer’s disease. Age (Dordr.) 2013;35(3):519–531. doi: 10.1007/s11357-011-9375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Aisa B., Gil-Bea F.J., Marcos B., Tordera R., Lasheras B., Del Rio J., Ramirez M.J. Neonatal stress affects vulnerability of cholinergic neurons and cognition in the rat: Involvement of the HPA axis. Psychoneuroendocrinology. 2009;34(10):1495–1505. doi: 10.1016/j.psyneuen.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 149.Aisa B., Tordera R., Lasheras B., Del Rio J., Ramirez M.J. Effects of maternal separation on hypothalamic-pituitary-adrenal responses, cognition and vulnerability to stress in adult female rats. Neuroscience. 2008;154(4):1218–1226. doi: 10.1016/j.neuroscience.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 150.Hebda-Bauer E.K., Simmons T.A., Sugg A., Ural E., Stewart J.A., Beals J.L., Wei Q., Watson S.J., Akil H. 3xTg-AD mice exhibit an activated central stress axis during early-stage pathology. J. Alzheimers Dis. 2013;33(2):407–422. doi: 10.3233/JAD-2012-121438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nishi M., Horii-Hayashi N., Sasagawa T., Matsunaga W. Effects of early life stress on brain activity: Implications from maternal separation model in rodents. Gen. Comp. Endocrinol. 2013;181:306–309. doi: 10.1016/j.ygcen.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 152.Chiu R., Boyle W.J., Meek J., Smeal T., Hunter T., Karin M. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell. 1988;54(4):541–552. doi: 10.1016/0092-8674(88)90076-1. [DOI] [PubMed] [Google Scholar]
- 153.Solas M., Aisa B., Mugueta M.C., Del Rio J., Tordera R.M., Ramirez M.J. Interactions between age, stress and insulin on cognition: Implications for Alzheimer’s disease. Neuropsychopharmacology. 2010;35(8):1664–1673. doi: 10.1038/npp.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Martisova E., Solas M., Gerenu G., Milagro F.I., Campion J., Ramirez M.J. Mechanisms involved in BACE upregulation associated to stress. Curr. Alzheimer Res. 2012;9(7):822–829. doi: 10.2174/156720512802455368. [DOI] [PubMed] [Google Scholar]
- 155.Rissman R.A., Staup M.A., Lee A.R., Justice N.J., Rice K.C., Vale W., Sawchenko P.E. Corticotropin-releasing factor receptor-dependent effects of repeated stress on tau phosphorylation, solubility, and aggregation. Proc. Natl. Acad. Sci. USA. 2012;109(16):6277–6282. doi: 10.1073/pnas.1203140109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McClelland S., Korosi A., Cope J., Ivy A., Baram T.Z. Emerging roles of epigenetic mechanisms in the enduring effects of early-life stress and experience on learning and memory. Neurobiol. Learn. Mem. 2011;96(1):79–88. doi: 10.1016/j.nlm.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Szyf M. The early life social environment and DNA methylation: DNA methylation mediating the long-term impact of social environments early in life. Epigenetics. 2011;6(8):971–978. doi: 10.4161/epi.6.8.16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Murgatroyd C., Patchev A.V., Wu Y., Micale V., Bockmuhl Y., Fischer D., Holsboer F., Wotjak C.T., Almeida O.F., Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 2009;12(12):1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 159.Taneja P., Ogier M., Brooks-Harris G., Schmid D.A., Katz D.M., Nelson S.B. Pathophysiology of locus ceruleus neurons in a mouse model of Rett syndrome. J. Neurosci. 2009;29(39):12187–12195. doi: 10.1523/JNEUROSCI.3156-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ressler K.J., Mercer K.B., Bradley B., Jovanovic T., Mahan A., Kerley K., Norrholm S.D., Kilaru V., Smith A.K., Myers A.J., Ramirez M., Engel A., Hammack S.E., Toufexis D., Braas K.M., Binder E.B., May V. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470(7335):492–497. doi: 10.1038/nature09856. https://www.nature.com/articles/nature09856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rat D., Schmitt U., Tippmann F., Dewachter I., Theunis C., Wieczerzak E., Postina R., van Leuven F., Fahrenholz F., Kojro E. Neuropeptide pituitary adenylate cyclase-activating polypeptide (PACAP) slows down Alzheimer’s disease-like pathology in amyloid precursor protein-transgenic mice. FASEB J. 2011;25(9):3208–3218. doi: 10.1096/fj.10-180133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Palma-Gudiel H., Cordova-Palomera A., Eixarch E., Deuschle M., Fananas L. Maternal psychosocial stress during pregnancy alters the epigenetic signature of the glucocorticoid receptor gene promoter in their offspring: A meta-analysis. Epigenetics. 2015;10(10):893–902. doi: 10.1080/15592294.2015.1088630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weaver I.C., Cervoni N., Champagne F.A., D’Alessio A.C., Sharma S., Seckl J.R., Dymov S., Szyf M., Meaney M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 164.McGowan P.O., Meaney M.J., Szyf M. Diet and the epigenetic (re)programming of phenotypic differences in behavior. Brain Res. 2008;1237:12–24. doi: 10.1016/j.brainres.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Szyf M., Weaver I.C., Champagne F.A., Diorio J., Meaney M.J. Maternal programming of steroid receptor expression and phenotype through DNA methylation in the rat. Front. Neuroendocrinol. 2005;26(3-4):139–162. doi: 10.1016/j.yfrne.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 166.Correia S.C., Santos R.X., Carvalho C., Cardoso S., Candeias E., Santos M.S., Oliveira C.R., Moreira P.I. Insulin signaling, glucose metabolism and mitochondria: Major players in Alzheimer’s disease and diabetes interrelation. Brain Res. 2012;1441:64–78. doi: 10.1016/j.brainres.2011.12.063. [DOI] [PubMed] [Google Scholar]
- 167.Fuso A., Nicolia V., Cavallaro R.A., Scarpa S. DNA methylase and demethylase activities are modulated by one-carbon metabolism in Alzheimer’s disease models. J. Nutr. Biochem. 2011;22(3):242–251. doi: 10.1016/j.jnutbio.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 168.Covic M., Karaca E., Lie D.C. Epigenetic regulation of neurogenesis in the adult hippocampus. Heredity. 2010;105(1):122–134. doi: 10.1038/hdy.2010.27. [DOI] [PubMed] [Google Scholar]
- 169.Weaver I.C., D’Alessio A.C., Brown S.E., Hellstrom I.C., Dymov S., Sharma S., Szyf M., Meaney M.J. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: Altering epigenetic marks by immediate-early genes. J. Neurosci. 2007;27(7):1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pal S., Tyler J.K. Epigenetics and aging. Sci. Adv. 2016;2(7):e1600584. doi: 10.1126/sciadv.1600584. http://advances.sciencemag.org/content/2/7/e1600584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hedges D.W., Woon F.L. Early-life stress and cognitive outcome. Psychopharmacology (Berl.) 2011;214(1):121–130. doi: 10.1007/s00213-010-2090-6. [DOI] [PubMed] [Google Scholar]
- 172.Banasr M., Duman R.S. Adult neurogenesis: Nature versus nurture. Biol. Psychiatry. 2012;72(4):256–257. doi: 10.1016/j.biopsych.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 173.Benayoun B.A., Pollina E.A., Brunet A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015;16(10):593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Entringer S., Epel E.S., Kumsta R., Lin J., Hellhammer D.H., Blackburn E.H., Wust S., Wadhwa P.D. Stress exposure in intrauterine life is associated with shorter telomere length in young adulthood. Proc. Natl. Acad. Sci. USA. 2011;108(33):E513–E518. doi: 10.1073/pnas.1107759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Entringer S., Buss C., Wadhwa P.D. Prenatal stress, development, health and disease risk: A psychobiological perspective-2015 Curt Richter Award Paper. Psychoneuroendocrinology. 2015;62:366–375. doi: 10.1016/j.psyneuen.2015.08.019. https://www.psyneuen-journal.com/ article/S0306-4530(15)00892-6/fulltext [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lemche A.V., Chaban O.S., Lemche E. Depression contributing to dyslipidemic cardiovascular risk in the metabolic syndrome. J. Endocrinol. Invest. 2017;40(5):539–546. doi: 10.1007/s40618-016-0601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Selfridge J.E. E, L.; Lu, J.; Swerdlow, R.H. Role of mitochondrial homeostasis and dynamics in Alzheimer’s disease. Neurobiol. Dis. 2013;51:3–12. doi: 10.1016/j.nbd.2011.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Saito Y., Akiyama M., Araki Y., Sumioka A., Shiono M., Taru H., Nakaya T., Yamamoto T., Suzuki T. 2011 doi: 10.1371/journal.pone.0022108. http://journals.plos.org/plosone/article?id=10.1371/journal [DOI] [PMC free article] [PubMed]
- 179.Vassar R. BACE1: The beta-secretase enzyme in Alzheimer’s disease. J. Mol. Neurosci. 2004;23(1-2):105–114. doi: 10.1385/JMN:23:1-2:105. [DOI] [PubMed] [Google Scholar]
- 180.Jonsson T., Atwal J.K., Steinberg S., Snaedal J., Jonsson P.V., Bjornsson S., Stefansson H., Sulem P., Gudbjartsson D., Maloney J., Hoyte K., Gustafson A., Liu Y., Lu Y., Bhangale T., Graham R.R., Huttenlocher J., Bjornsdottir G., Andreassen O.A., Jonsson E.G., Palotie A., Behrens T.W., Magnusson O.T., Kong A., Thorsteinsdottir U., Watts R.J., Stefansson K. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488(7409):96–99. doi: 10.1038/nature11283. https://www.nature.com/articles/nature11283 [DOI] [PubMed] [Google Scholar]
- 181.Da Mesquita S., Ferreira A.C., Sousa J.C., Correia-Neves M., Sousa N., Marques F. Insights on the pathophysiology of Alzheimer’s disease: The crosstalk between amyloid pathology, neuroinflammation and the peripheral immune system. Neurosci. Biobehav. Rev. 2016;68:547–562. doi: 10.1016/j.neubiorev.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 182.Pensalfini A., Albay R., III, Rasool S., Wu J.W., Hatami A., Arai H., Margol L., Milton S., Poon W.W., Corrada M.M., Kawas C.H., Glabe C.G. Intracellular amyloid and the neuronal origin of Alzheimer neuritic plaques. Neurobiol. Dis. 2014;71:53–61. doi: 10.1016/j.nbd.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jicha G.A., Markesbery W.R. Omega-3 fatty acids: potential role in the management of early Alzheimer’s disease. Clin. Interv. Aging. 2010;5:45–61. doi: 10.2147/cia.s5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Domert J., Rao S.B., Agholme L., Brorsson A.C., Marcusson J., Hallbeck M., Nath S. Spreading of amyloid-beta peptides via neuritic cell-to-cell transfer is dependent on insufficient cellular clearance. Neurobiol. Dis. 2014;65:82–92. doi: 10.1016/j.nbd.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 185.Kim E.K., Choi E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta. 2010;1802(4):396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 186.Herring A., Munster Y., Metzdorf J., Bolczek B., Krussel S., Krieter D., Yavuz I., Karim F., Roggendorf C., Stang A., Wang Y., Hermann D.M., Teuber-Hanselmann S., Keyvani K. Late running is not too late against Alzheimer’s pathology. Neurobiol. Dis. 2016;94:44–54. doi: 10.1016/j.nbd.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 187.Pascoal T.A., Mathotaarachchi S., Mohades S., Benedet A.L., Chung C.O., Shin M., Wang S., Beaudry T., Kang M.S., Soucy J.P., Labbe A., Gauthier S., Rosa-Neto P. Amyloid-beta and hyperphosphorylated tau synergy drives metabolic decline in preclinical Alzheimer’s disease. Mol. Psychiatry. 2017;22(2):306–311. doi: 10.1038/mp.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.De-Paula V.J., Radanovic M., Diniz B.S., Forlenza O.V. Alzheimer’s disease. Subcell. Biochem. 2012;65:329–352. doi: 10.1007/978-94-007-5416-4_14. [DOI] [PubMed] [Google Scholar]
- 189.Dal Pra I., Chiarini A., Gui L., Chakravarthy B., Pacchiana R., Gardenal E., Whitfield J.F., Armato U. Do astrocytes collaborate with neurons in spreading the “infectious” abeta and Tau drivers of Alzheimer’s disease? Neuroscientist. 2015;21(1):9–29. doi: 10.1177/1073858414529828. [DOI] [PubMed] [Google Scholar]
- 190.Kumar A., Singh A., Ekavali A. A review on Alzheimer’s disease pathophysiology and its management: An update. Pharmacol. Rep. 2015;67(2):195–203. doi: 10.1016/j.pharep.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 191.Shi X., Zheng Z., Li J., Xiao Z., Qi W., Zhang A., Wu Q., Fang Y. Curcumin inhibits Abeta-induced microglial inflammatory responses in vitro: Involvement of ERK1/2 and p38 signaling pathways. Neurosci. Lett. 2015;594:105–110. doi: 10.1016/j.neulet.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 192.Tamagno E., Guglielmotto M., Monteleone D., Tabaton M. Amyloid-beta production: Major link between oxidative stress and BACE1. Neurotox. Res. 2012;22(3):208–219. doi: 10.1007/s12640-011-9283-6. [DOI] [PubMed] [Google Scholar]
- 193.Maulik M., Peake K., Chung J., Wang Y., Vance J.E., Kar S. APP overexpression in the absence of NPC1 exacerbates metabolism of amyloidogenic proteins of Alzheimer’s disease. Hum. Mol. Genet. 2015;24(24):7132–7150. doi: 10.1093/hmg/ddv413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang D.B., Kinoshita Y., Kinoshita C., Uo T., Sopher B.L., Cudaback E., Keene C.D., Bilousova T., Gylys K., Case A., Jayadev S., Wang H.G., Garden G.A., Morrison R.S. Loss of endophilin-B1 exacerbates Alzheimer’s disease pathology. Brain. 2015;138(Pt 7):2005–2019. doi: 10.1093/brain/awv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mansuroglu Z., Benhelli-Mokrani H., Marcato V., Sultan A., Violet M., Chauderlier A., Delattre L., Loyens A., Talahari S., Begard S., Nesslany F., Colin M., Soues S., Lefebvre B., Buee L., Galas M.C., Bonnefoy E. Loss of Tau protein affects the structure, transcription and repair of neuronal pericentromeric heterochromatin. Sci. Rep. 2016;6:33047. doi: 10.1038/srep33047. https://www.nature.com/articles/srep33047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Caffrey T.M., Joachim C., Paracchini S., Esiri M.M., Wade-Martins R. Haplotype-specific expression of exon 10 at the human MAPT locus. Hum. Mol. Genet. 2006;15(24):3529–3537. doi: 10.1093/hmg/ddl429. [DOI] [PubMed] [Google Scholar]
- 197.Niblock M., Gallo J.M. Tau alternative splicing in familial and sporadic tauopathies. Biochem. Soc. Trans. 2012;40(4):677–680. doi: 10.1042/BST20120091. [DOI] [PubMed] [Google Scholar]
- 198.Iwata A., Nagata K., Hatsuta H., Takuma H., Bundo M., Iwamoto K., Tamaoka A., Murayama S., Saido T., Tsuji S. Altered CpG methylation in sporadic Alzheimer’s disease is associated with APP and MAPT dysregulation. Hum. Mol. Genet. 2014;23(3):648–656. doi: 10.1093/hmg/ddt451. [DOI] [PubMed] [Google Scholar]
- 199.Lovestone S., Reynolds C.H. The phosphorylation of tau: A critical stage in neurodevelopment and neurodegenerative processes. Neuroscience. 1997;78(2):309–324. doi: 10.1016/s0306-4522(96)00577-5. [DOI] [PubMed] [Google Scholar]
- 200.Froelich-Fabre S., Bhat R.V. Mechanisms of tauopathies. Drug Discov. Today Dis. Mech. 2004;1(4):391–398. [Google Scholar]
- 201.Lloret A., Fuchsberger T., Giraldo E., Vina J. Molecular mechanisms linking amyloid beta toxicity and Tau hyperphosphorylation in Alzheimer’s disease. Free Radic. Biol. Med. 2015;83:186–191. doi: 10.1016/j.freeradbiomed.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 202.Ittner L.M., Gotz J. Amyloid-beta and tau--a toxic pas de deux in Alzheimer’s disease. Nat. Rev. Neurosci. 2011;12(2):65–72. doi: 10.1038/nrn2967. [DOI] [PubMed] [Google Scholar]
- 203.Mao P., Reddy P.H. Aging and amyloid beta-induced oxidative DNA damage and mitochondrial dysfunction in Alzheimer’s disease: Implications for early intervention and therapeutics. Biochim. Biophys. Acta. 2011;1812(11):1359–1370. doi: 10.1016/j.bbadis.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tenreiro S., Eckermann K., Outeiro T.F. Protein phosphorylation in neurodegeneration: Friend or foe? Front. Mol. Neurosci. 2014;7:42. doi: 10.3389/fnmol.2014.00042. https://www.frontiersin.org/articles/10.3389/ fnmol.2014.00042/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Crary J.F. Primary age-related tauopathy and the amyloid cascade hypothesis: The exception that proves the rule? J. Neurol. Neuromed. 2016;1(6):53–57. doi: 10.29245/2572.942x/2016/6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Braak H., Del Tredici K. The pathological process underlying Alzheimer’s disease in individuals under thirty. Acta Neuropathol. 2011;121(2):171–181. doi: 10.1007/s00401-010-0789-4. [DOI] [PubMed] [Google Scholar]
- 207.Gordon B.A., Friedrichsen K., Brier M., Blazey T., Su Y., Christensen J., Aldea P., McConathy J., Holtzman D.M., Cairns N.J., Morris J.C., Fagan A.M., Ances B.M., Benzinger T.L. The relationship between cerebrospinal fluid markers of Alzheimer pathology and positron emission tomography tau imaging. Brain. 2016;139(Pt 8):2249–2260. doi: 10.1093/brain/aww139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Young K.F., Pasternak S.H., Rylett R.J. Oligomeric aggregates of amyloid beta peptide 1-42 activate ERK/MAPK in SH-SY5Y cells via the alpha7 nicotinic receptor. Neurochem. Int. 2009;55(8):796–801. doi: 10.1016/j.neuint.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 209.Pei J-J., Braak H., An W-L., Winblad B., Cowburn R.F., Iqbal K., Grundke I. Up-regulation of mitogen-activated protein kinases ERK1/2 and MEK1/2 is associated with the progression of neurofibrillary degeneration in Alzheimer’s disease. Brain Res. Mol. Brain Res. 2002;109(1-2):45–55. doi: 10.1016/s0169-328x(02)00488-6. [DOI] [PubMed] [Google Scholar]
- 210.Roostaei T., Nazeri A., Felsky D., De Jager P.L., Schneider J.A., Pollock B.G., Bennett D.A., Voineskos A.N., Alzheimer’s Disease Neuroimaging Initiative (ADNI) Genome-wide interaction study of brain beta-amyloid burden and cognitive impairment in Alzheimer’s disease. Mol. Psychiatry. 2017;22(2):287–295. doi: 10.1038/mp.2016.35. https://www.nature.com/articles/mp201635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Reddy P.H. Abnormal tau, mitochondrial dysfunction, impaired axonal transport of mitochondria, and synaptic deprivation in Alzheimer’s disease. Brain Res. 2011;1415:136–148. doi: 10.1016/j.brainres.2011.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cummings J.L. Alzheimer’s disease. N. Engl. J. Med. 2004;351(1):56–67. doi: 10.1056/NEJMra040223. [DOI] [PubMed] [Google Scholar]
- 213.Resende R., Moreira P.I., Proenca T., Deshpande A., Busciglio J., Pereira C., Oliveira C.R. Brain oxidative stress in a triple-transgenic mouse model of Alzheimer disease. Free Radic. Biol. Med. 2008;44(12):2051–2057. doi: 10.1016/j.freeradbiomed.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 214.Morroni F., Sita G., Tarozzi A., Rimondini R., Hrelia P. Early effects of Abeta1-42 oligomers injection in mice: Involvement of PI3K/Akt/GSK3 and MAPK/ERK1/2 pathways. Behav. Brain Res. 2016;314:106–115. doi: 10.1016/j.bbr.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 215.Anacker C., Cattaneo A., Musaelyan K., Zunszain P.A., Horowitz M., Molteni R., Luoni A., Calabrese F., Tansey K., Gennarelli M., Thuret S., Price J., Uher R., Riva M.A., Pariante C.M. Role for the kinase SGK1 in stress, depression, and glucocorticoid effects on hippocampal neurogenesis. Proc. Natl. Acad. Sci. USA. 2013;110(21):8708–8713. doi: 10.1073/pnas.1300886110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Swerdlow R.H. Brain aging, Alzheimer’s disease, and mitochondria. Biochim. Biophys. Acta. 2011;1812(12):1630–1639. doi: 10.1016/j.bbadis.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Reddy P.H. Amyloid beta-induced glycogen synthase kinase 3beta phosphorylated VDAC1 in Alzheimer’s disease: Implications for synaptic dysfunction and neuronal damage. Biochim. Biophys. Acta. 2013;1832(12):1913–1921. doi: 10.1016/j.bbadis.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Corral-Debrinski M., Horton T., Lott M.T., Shoffner J.M., Beal M.F., Wallace D.C. Mitochondrial DNA deletions in human brain: Regional variability and increase with advanced age. Nat. Genet. 1992;2(4):324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 219.Lin M.T., Simon D.K., Ahn C.H., Kim L.M., Beal M.F. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum. Mol. Genet. 2002;11(2):133–145. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- 220.Coronas-Samano G., Baker K.L., Tan W.J., Ivanova A.V., Verhagen J.V. Fus1 KO mouse as a model of oxidative stress-mediated sporadic Alzheimer’s disease: Circadian disruption and long-term spatial and olfactory memory impairments. Front. Aging Neurosci. 2016;8:268. doi: 10.3389/fnagi.2016.00268. https://www.frontiersin.org/ articles/10.3389/fnagi.2016.00268/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Derungs R., Camici G.G., Spescha R.D., Welt T., Tackenberg C., Spani C., Wirth F., Grimm A., Eckert A., Nitsch R.M., Kulic L. Genetic ablation of the p66Shc adaptor protein reverses cognitive deficits and improves mitochondrial function in an APP transgenic mouse model of Alzheimer’s disease. Mol. Psychiatry. 2017;22(4):605–614. doi: 10.1038/mp.2016.112. [DOI] [PubMed] [Google Scholar]
- 222.Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–247. doi: 10.1038/35041687. https://www.nature.com/articles/35041687 [DOI] [PubMed] [Google Scholar]
- 223.Manczak M., Park B.S., Jung Y., Reddy P.H. Differential expression of oxidative phosphorylation genes in patients with Alzheimer’s disease: Implications for early mitochondrial dysfunction and oxidative damage. Neuromol. Med. 2004;5(2):147–162. doi: 10.1385/NMM:5:2:147. [DOI] [PubMed] [Google Scholar]
- 224.Shi Q., Gibson G.E. Oxidative stress and transcriptional regulationin Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2007;21(4):276–291. doi: 10.1097/WAD.0b013e31815721c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.von Bernhardi R., Cornejo F., Parada G.E., Eugenin J. Role of TGF β signaling in the pathogenesis of Alzheimer’s disease. Front. Cell. Neurosci. 2015;9:426. doi: 10.3389/fncel.2015.00426. https://www.frontiersin.org/articles/10.3389/fncel.2015.00426/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Deane R., Sagare A., Hamm K., Parisi M., Lane S., Finn M.B., Holtzman D.M., Zlokovic B.V. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J. Clin. Invest. 2008;118(12):4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Salameh T.S., Rhea E.M., Banks W.A., Hanson A.J. Insulin resistance, dyslipidemia, and apolipoprotein E interactions as mechanisms in cognitive impairment and Alzheimer’s disease. Exp. Biol. Med. (Maywood) 2016;241(15):1676–1683. doi: 10.1177/1535370216660770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lautner R., Palmqvist S., Mattsson N., Andreasson U., Wallin A., Palsson E., Jakobsson J., Herukka S.K., Owenius R., Olsson B., Hampel H., Rujescu D., Ewers M., Landen M., Minthon L., Blennow K., Zetterberg H., Hansson O., Alzheimer’s Disease Neuroimaging Initiative Apolipoprotein E genotype and the diagnostic accuracy of cerebrospinal fluid biomarkers for Alzheimer disease. JAMA Psychiatry. 2014;71(10):1183–1191. doi: 10.1001/jamapsychiatry.2014.1060. [DOI] [PubMed] [Google Scholar]
- 229.Ma K.G., Lv J., Hu X.D., Shi L.L., Chang K.W., Chen X.L., Qian Y.H., Yang W.N., Qu Q.M. The p38 mitogen-activated protein kinase signaling pathway is involved in regulating low-density lipoprotein receptor-related protein 1-mediated beta-amyloid protein internalization in mouse brain. Int. J. Biochem. Cell Biol. 2016;76:75–86. doi: 10.1016/j.biocel.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 230.Mormino E.C., Betensky R.A., Hedden T., Schultz A.P., Ward A., Huijbers W., Rentz D.M., Johnson K.A., Sperling R.A. Amyloid and APOE e4 interact to influence short-term decline in preclinical Alzheimer disease. Neurology. 2014;82(20):1760. doi: 10.1212/WNL.0000000000000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lee B.K., Glass T.A., Wand G.S., McAtee M.J., Bandeen-Roche K., Bolla K.I., Schwartz B.S. Apolipoprotein e genotype, cortisol, and cognitive function in community-dwelling older adults. Am. J. Psychiatry. 2008;165(11):1456–1464. doi: 10.1176/appi.ajp.2008.07091532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sutphen C.L., Jasielec M.S., Shah A.R., Macy E.M., Xiong C., Vlassenko A.G., Benzinger T.L., Stoops E.E., Vanderstichele H.M., Brix B., Darby H.D., Vandijck M.L., Ladenson J.H., Morris J.C., Holtzman D.M., Fagan A.M. Longitudinal cerebrospinal fluid biomarker changes in preclinical Alzheimer disease during middle age. JAMA Neurol. 2015;72(9):1029–1042. doi: 10.1001/jamaneurol.2015.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sanchez-Mut J.V., Heyn H., Vidal E., Moran S., Sayols S., Delgado-Morales R., Schultz M.D., Ansoleaga B., Garcia-Esparcia P., Pons-Espinal M., de Lagran M.M., Dopazo J., Rabano A., Avila J., Dierssen M., Lott I., Ferrer I., Ecker J.R., Esteller M. Human DNA methylomes of neurodegenerative diseases show common epigenomic patterns. 2016 doi: 10.1038/tp.2015.214. https://www.nature.com/articles/ [DOI] [PMC free article] [PubMed]
- 234.Yokota O., Terada S., Ishizu H., Ujike H., Ishihara T., Nakashima H., Yasuda M., Kitamura Y., Ueda K., Checler F., Kuroda S. NACP/alpha-synuclein, NAC, and beta-amyloid pathology of familial Alzheimer’s disease with the E184D presenilin-1 mutation: A clinicopathological study of two autopsy cases. Acta Neuropathol. 2002;104(6):637–648. doi: 10.1007/s00401-002-0596-7. [DOI] [PubMed] [Google Scholar]
- 235.Huang Y., Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148(6):1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ghavami S., Shojaei S., Yeganeh B., Ande S.R., Jangamreddy J.R., Mehrpour M., Christoffersson J., Chaabane W., Moghadam A.R., Kashani H.H., Hashemi M., Owji A.A., Los M.J. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog. Neurobiol. 2014;112:24–49. doi: 10.1016/j.pneurobio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 237.Gogtay N., Thompson P.M. Mapping gray matter development: Implications for typical development and vulnerability to psychopathology. Brain Cogn. 2010;72(1):6–15. doi: 10.1016/j.bandc.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Teicher M.H., Tomoda A., Andersen S.L. Neurobiological consequences of early stress and childhood maltreatment: Are results from human and animal studies comparable? Ann. N. Y. Acad. Sci. 2006;1071:313–323. doi: 10.1196/annals.1364.024. [DOI] [PubMed] [Google Scholar]
- 239.Teicher M.H., Anderson C.M., Polcari A. Childhood maltreatment is associated with reduced volume in the hippocampal subfields CA3, dentate gyrus, and subiculum. Proc. Natl. Acad. Sci. USA. 2012;109(9):E563–E572. doi: 10.1073/pnas.1115396109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Villareal G. Reduced hippocampal volume and total white matter volume in posttraumatic stress disorder. Biol. Psychiatry. 2002;52(2):119–125. doi: 10.1016/s0006-3223(02)01359-8. [DOI] [PubMed] [Google Scholar]
- 241.Karl A., Schaefer M., Malta L.S., Dorfel D., Rohleder N., Werner A. A meta-analysis of structural brain abnormalities in PTSD. Neurosci. Biobehav. Rev. 2006;30(7):1004–1031. doi: 10.1016/j.neubiorev.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 242.Hedges D.W., Woon F.L. Alcohol use and hippocampal volume deficits in adults with posttraumatic stress disorder: A meta-analysis. Biol. Psychol. 2010;84(2):163–168. doi: 10.1016/j.biopsycho.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 243.Salcedo-Tello P., Ortiz-Matamoros A., Arias C. GSK3 function in the brain during development, neuronal plasticity, and neurodegeneration. Int. J. Alzheimers Dis. 2011;2011:189728. doi: 10.4061/2011/189728. https://www.hindawi.com/journals/ijad/2011/189728/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hur E.M., Zhou F.Q. GSK3 signalling in neural development. Nat. Rev. Neurosci. 2010;11(8):539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ka M., Condorelli G., Woodgett J.R., Kim W.Y. mTOR regulates brain morphogenesis by mediating GSK3 signaling. Development. 2014;141(21):4076–4086. doi: 10.1242/dev.108282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D’Avanzo C., Chen H., Hooli B., Asselin C., Muffat J., Klee J.B., Zhang C., Wainger B.J., Peitz M., Kovacs D.M., Woolf C.J., Wagner S.L., Tanzi R.E., Kim D.Y. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515(7526):274–278. doi: 10.1038/nature13800. https://www.nature.com/ articles/nature13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.DaRocha-Souto B., Coma M., Perez-Nievas B.G., Scotton T.C., Siao M., Sanchez-Ferrer P., Hashimoto T., Fan Z., Hudry E., Barroeta I., Sereno L., Rodriguez M., Sanchez M.B., Hyman B.T., Gomez-Isla T. Activation of glycogen synthase kinase-3 beta mediates beta-amyloid induced neuritic damage in Alzheimer’s disease. Neurobiol. Dis. 2012;45(1):425–437. doi: 10.1016/j.nbd.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ferrer I., Gomez-Isla T., Puig B., Freixes M., Ribe E., Dalfo E., Avila J. Current advances on different kinases involved in tau phosphorylation, and implications in Alzheimer’s disease and tauopathies. Curr. Alzheimer Res. 2005;2(1):3–18. doi: 10.2174/1567205052772713. [DOI] [PubMed] [Google Scholar]
- 249.Ferrer I., Barrachina M., Tolnay M., Rey M.J., Vidal N., Carmona M., Blanco R., Puig B. Phosphorylated protein kinases associated with neuronal and glial tau deposits. Brain Pathol. 2003;13(1):62–78. doi: 10.1111/j.1750-3639.2003.tb00007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ferrer I., Blanco R., Carmona M., Puig B. Phosphorylated mitogen-activated protein kinase (MAPK/ERK-P), protein kinase of 38kDa (p38-P), stress-activated protein kinase (SAPK/JNK-P), and calcium/calmodulin dependent kinase II (CaM kinase II) are differentially expressed in tau deposits in neurons and glial cells in tauopathies. J. Neural Trans. 2001;108(12):1397–1415. doi: 10.1007/s007020100016. [DOI] [PubMed] [Google Scholar]
- 251.Jiang W., Luo T., Li S., Zhou Y., Shen X.Y., He F., Xu J., Wang H.Q. 2016 http://journals.plos.org/plosone/article?id=10.1371/journal.pone
- 252.Zuo L., Hemmelgarn B.T., Chuang C.C., Best T.M. The role of oxidative stress-induced epigenetic alterations in amyloid-beta production in Alzheimer’s disease. Oxid. Med. Cell. Longev. 2015;2015:604658. doi: 10.1155/2015/604658. https://www.hindawi.com/journals/omcl/2015/604658/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Taga M., Minett T., Classey J., Matthews F.E., Brayne C., Ince P.G., Nicoll J.A., Hugon J., Boche D., Mrc C. Metaflammasome components in the human brain: A role in dementia with Alzheimer’s pathology? Brain Pathol. 2017;27(3):266–275. doi: 10.1111/bpa.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhang W., Liu H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12(1):9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 255.Chu C.T., Levinthal D.J., Kulich S.M., Chalovich E.M., DeFranco D.B. Oxidative neuronal injury. The dark side of ERK1/2. Eur. J. Biochem. 2004;271(11):2060–2066. doi: 10.1111/j.1432-1033.2004.04132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Huang G., Shi L.Z., Chi H. Regulation of JNK and p38 MAPK in the immune system: Signal integration, propagation and termination. Cytokine. 2009;48(3):161–169. doi: 10.1016/j.cyto.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Zeke A., Misheva M., Remenyi A., Bogoyevitch M.A. JNK signaling: Regulation and functions based on complex protein-protein partnerships. Microbiol. Mol. Biol. Rev. 2016;80(3):793–835. doi: 10.1128/MMBR.00043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ito-Ishida A., Kakegawa W., Yuzaki M. ERK1/2 but not p38 MAP kinase is essential for the long-term depression in mouse cerebellar slices. Eur. J. Neurosci. 2006;24(6):1617–1622. doi: 10.1111/j.1460-9568.2006.05055.x. [DOI] [PubMed] [Google Scholar]
- 259.Kamat P.K., Kalani A., Rai S., Swarnkar S., Tota S., Nath C., Tyagi N. Mechanism of oxidative stress and synapse dysfunction in the pathogenesis of Alzheimer’s disease: Understanding the therapeutics strategies. Mol. Neurobiol. 2016;53(1):648–661. doi: 10.1007/s12035-014-9053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhu X., Castellani R.J., Takeda A., Nunomura A., Atwood C.S., Perry G., Smith M.A. Differential activation of neuronal-ERK JNK SAPK and p38 in Alzheimer disease the two hit hypothesis. Mech. Ageing Dev. 2001;123(1):39–46. doi: 10.1016/s0047-6374(01)00342-6. [DOI] [PubMed] [Google Scholar]
- 261.Carter C.J. Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem. Int. 2007;50(1):12–38. doi: 10.1016/j.neuint.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 262.Beurel E., Grieco S.F., Jope R.S. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 2015;148:114–131. doi: 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Jope R.S., Johnson G.V. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 2004;29(2):95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 264.Xu W., Yang L., Li J. Protection against beta-amyloid-induced neurotoxicity by naturally occurring Z-ligustilide through the concurrent regulation of p38 and PI3-K/Akt pathways. Neurochem. Int. 2016;100:44–51. doi: 10.1016/j.neuint.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 265.Peltier J., O’Neill A., Schaffer D.V. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev. Neurobiol. 2007;67(10):1348–1361. doi: 10.1002/dneu.20506. [DOI] [PubMed] [Google Scholar]
- 266.Jope R.S., Cheng Y., Lowell J.A., Worthen R.J., Sitbon Y.H., Beurel E. Stressed and inflamed, Can GSK3 be blamed? Trends Biochem. Sci. 2017;42(3):180–192. doi: 10.1016/j.tibs.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Trotta T., Porro C., Calvello R., Panaro M.A. Biological role of Toll-like receptor-4 in the brain. J. Neuroimmunol. 2014;268(1-2):1–12. doi: 10.1016/j.jneuroim.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 268.Huang N.Q., Jin H., Zhou S.Y., Shi J.S., Jin F. TLR4 is a link between diabetes and Alzheimer’s disease. Behav. Brain Res. 2017;316:234–244. doi: 10.1016/j.bbr.2016.08.047. [DOI] [PubMed] [Google Scholar]
- 269.Cymerman I.A., Gozdz A., Urbanska M., Milek J., Dziembowska M., Jaworski J. Structural plasticity of dendritic spines requires GSK3alpha and GSK3beta. PLoS One. 2015;10(7):e0134018. doi: 10.1371/journal.pone.0134018. http://journals.plos.org/plosone/article? id=10.1371/journal.pone.0134018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Houck A.L., Hernandez F., Avila J. A simple model to study tau pathology. J. Exp. Neurosci. 2016;10:31–38. doi: 10.4137/JEN.S25100. http://journals.sagepub.com/doi/10.4137/JEN.S25100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Cavallini A., Brewerton S., Bell A., Sargent S., Glover S., Hardy C., Moore R., Calley J., Ramachandran D., Poidinger M., Karran E., Davies P., Hutton M., Szekeres P., Bose S. An unbiased approach to identifying tau kinases that phosphorylate tau at sites associated with Alzheimer disease. J. Biol. Chem. 2013;288(32):23331–23347. doi: 10.1074/jbc.M113.463984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Israel M.A., Yuan S.H., Bardy C., Reyna S.M., Mu Y., Herrera C., Hefferan M.P., Van Gorp S., Nazor K.L., Boscolo F.S., Carson C.T., Laurent L.C., Marsala M., Gage F.H., Remes A.M., Koo E.H., Goldstein L.S. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–220. doi: 10.1038/nature10821. https://www.nature.com/ articles/nature10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Takashima A. GSK-3 is essential in the pathogenesis of Alzheimer’s disease. J. Alzheimers Dis. 2006;9(3) Suppl.:309–317. doi: 10.3233/jad-2006-9s335. [DOI] [PubMed] [Google Scholar]
- 274.Phiel C.J., Wilson C.A., Lee V.M., Klein P.S. 2003 https://www.nature.com/articles/
- 275.Hohman T.J., Chibnik L., Bush W.S., Jefferson A.L., De Jaeger P.L., Thornton-Wells T.A., Bennett D.A., Schneider J.A. GSK3beta interactions with amyloid genes: An autopsy verification and extension. Neurotox. Res. 2015;28(3):232–238. doi: 10.1007/s12640-015-9541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hohman T.J., Koran M.E., Thornton-Wells T.A., Alzheimer’s Neuroimaging Initiative Interactions between GSK3beta and amyloid genes explain variance in amyloid burden. Neurobiol. Aging. 2014;35(3):460–465. doi: 10.1016/j.neurobiolaging.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Kwok J.B., Loy C.T., Hamilton G., Lau E., Hallupp M., Williams J., Owen M.J., Broe G.A., Tang N., Lam L., Powell J.F., Lovestone S., Schofield P.R. Glycogen synthase kinase-3beta and tau genes interact in Alzheimer’s disease. Ann. Neurol. 2008;64(4):446–454. doi: 10.1002/ana.21476. [DOI] [PubMed] [Google Scholar]
- 278.Li J., Ding X., Zhang R., Jiang W., Sun X., Xia Z., Wang X., Wu E., Zhang Y., Hu Y. Harpagoside ameliorates the amyloid-beta-induced cognitive impairment in rats via up-regulating BDNF expression and MAPK/PI3K pathways. Neuroscience. 2015;303:103–114. doi: 10.1016/j.neuroscience.2015.06.042. [DOI] [PubMed] [Google Scholar]
- 279.Subramaniam S., Unsicker K. ERK and cell death: ERK1/2 in neuronal death. FEBS J. 2010;277(1):22–29. doi: 10.1111/j.1742-4658.2009.07367.x. [DOI] [PubMed] [Google Scholar]
- 280.Noel A., Poitras I., Julien J., Petry F.R., Morin F., Charron J., Planel E. ERK (MAPK) does not phosphorylate tau under physiological conditions in vivo or in vitro. Neurobiol. Aging. 2015;36(2):901–902. doi: 10.1016/j.neurobiolaging.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 281.Veeranna; Kaji, T.; Boland, B.; Odrljin, T.; Mohan, P.; Basavarajappa, B.S.; Peterhoff, C.; Cataldo, A.; Rudnicki, A.; Amin, N.; Li, B.S.; Pant, H.C.; Hungund, B.L.; Arancio, O.; Nixon, R.A. Calpain mediates calcium-Induced activation of the ERK1/2 MAPK pathway and cytoskeletal phosphorylation in neurons. Am. J. Pathol. 2004;165(3):795–805. doi: 10.1016/S0002-9440(10)63342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Dineley K.T., Westerman M., Bui D., Bell K., Ashe K.H., Sweatt J.D. Beta-amyloid activates the mitogen-activated protein kinase cascade via hippocampal a7 nicotinic acetylcholine receptors: In vitro and in vivo mechanisms related to Alzheimer’s disease. J. Neurosci. 2001;21(12):4125–4133. doi: 10.1523/JNEUROSCI.21-12-04125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Faucher P., Mons N., Micheau J., Louis C., Beracochea D.J. Hippocampal injections of oligomeric amyloid beta-peptide (1-42) Induce selective working memory deficits and long-lasting alterations of ERK signaling pathway. Front. Aging Neurosci. 2015;7:245. doi: 10.3389/fnagi.2015.00245. https://www.frontiersin.org/articles/10.3389/ fnagi.2015.00245/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Feld M., Krawczyk M.C., Sol Fustinana M., Blake M.G., Baratti C.M., Romano A., Boccia M.M. Decrease of ERK/MAPK overactivation in prefrontal cortex reverses early memory deficit in a mouse model of Alzheimer’s disease. J. Alzheimers Dis. 2014;40(1):69–82. doi: 10.3233/JAD-131076. [DOI] [PubMed] [Google Scholar]
- 285.Giovannini M.G. The role of the extracellular signal-regulated kinase pathway in memory encoding. Rev. Neurosci. 2006;17(6):619–634. doi: 10.1515/revneuro.2006.17.6.619. [DOI] [PubMed] [Google Scholar]
- 286.Giovannini M.G., Lana D., Pepeu G. The integrated role of ACh, ERK and mTOR in the mechanisms of hippocampal inhibitory avoidance memory. Neurobiol. Learn. Mem. 2015;119:18–33. doi: 10.1016/j.nlm.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 287.Ma T., Hoeffer C.A., Capetillo-Zarate E., Yu F., Wong H., Lin M.T., Tampellini D., Klann E., Blitzer R.D., Gouras G.K. Dysregulation of the mTOR pathway mediates impairment of synaptic plasticity in a mouse model of Alzheimer’s disease. PLoS One. 2010;5(9):e12845. doi: 10.1371/journal.pone.0012845. http://journals.plos.org/plosone/article? id=10.1371/journal.pone.0012845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hasegawa H., Nakai M., Tanimukai S., Taniguchi T. Akira Terashima; Kawamata, T.; Fukunaga, K.; Miyamoto, E.; Misaki, K.; Mukai, H.; Tanaka, C. Microglial signaling by amyloid beta protein through mitogen-activated protein kinase mediating phosphorylation of MARCKS. Neuroreport. 2001;12(11):2567–2571. doi: 10.1097/00001756-200108080-00055. [DOI] [PubMed] [Google Scholar]
- 289.Mather M., Harley C.W. The locus caeruleus: Essential for maintaining cognitive function and the aging brain. Trends Cogn. Sci. 2016;20(3):214–226. doi: 10.1016/j.tics.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Tominaga-Yoshino K., Uetsuki T., Yoshikawa K., Ogura A. Neurotoxic and neuroprotective effects of glutamate are enhanced by introduction of amyloid precursor protein cDNA. Brain Res. 2001;918(1-2):121–130. doi: 10.1016/s0006-8993(01)02983-3. [DOI] [PubMed] [Google Scholar]
- 291.Tsai S.F., Chen P.C., Calkins M.J., Wu S.Y., Kuo Y.M. Exercise counteracts aging-related memory impairment: A potential role for the astrocytic metabolic shuttle. Front. Aging Neurosci. 2016;8:57. doi: 10.3389/fnagi.2016.00057. https://www.frontiersin.org/articles/10.3389/ fnagi.2016.00057/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Bi R., Broutman G., Foy M.R., Thompson R.F., Baudry M. The tyrosine kinase and mitogen-activated protein kinase pathways mediate multiple effects of estrogen in hippocampus. Proc. Natl. Acad. Sci. USA. 2000;97(7):3602–3607. doi: 10.1073/pnas.060034497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Bogoyevitch M.A., Ngoei K.R., Zhao T.T., Yeap Y.Y., Ng D.C. c-Jun N-terminal kinase (JNK) signaling: Recent advances and challenges. Biochim. Biophys. Acta. 2010;1804(3):463–475. doi: 10.1016/j.bbapap.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 294.D’Ambrosio C., Arena S., Fulcoli G., Scheinfeld M.H., Zhou D., D’Adamio L., Scaloni A. Hyperphosphorylation of JNK-interacting protein 1, a protein associated with Alzheimer disease. Mol. Cell. Proteomics. 2006;5(1):97–113. doi: 10.1074/mcp.M500226-MCP200. [DOI] [PubMed] [Google Scholar]
- 295.Chen B., Teng Y., Zhang X., Lv X., Yin Y. Metformin alleviated abeta-induced apoptosis via the suppression of JNK MAPK signaling pathway in cultured hippocampal neurons. BioMed Res. Int. 2016;2016:1421430. doi: 10.1155/2016/1421430. https://www.hindawi.com/ journals/bmri/2016/1421430/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Yenki P., Khodagholi F., Shaerzadeh F. Inhibition of phosphorylation of JNK suppresses Abeta-induced ER stress and upregulates prosurvival mitochondrial proteins in rat hippocampus. J. Mol. Neurosci. 2013;49(2):262–269. doi: 10.1007/s12031-012-9837-y. [DOI] [PubMed] [Google Scholar]
- 297.Itoh M., Adachi M., Yasui H., Takekawa M., Tanaka H., Imai K. Nuclear export of glucocorticoid receptor is enhanced by c-Jun N-terminal kinase-mediated phosphorylation. Mol. Endocrinol. 2002;16(10):2382–2392. doi: 10.1210/me.2002-0144. [DOI] [PubMed] [Google Scholar]
- 298.Matsuda S., Matsuda Y., D’Adamio L. Amyloid beta protein precursor (AbetaPP), but not AbetaPP-like protein 2, is bridged to the kinesin light chain by the scaffold protein JNK-interacting protein 1. J. Biol. Chem. 2003;278(40):38601–38606. doi: 10.1074/jbc.M304379200. [DOI] [PubMed] [Google Scholar]
- 299.Chiba K., Araseki M., Nozawa K., Furukori K., Araki Y., Matsushima T., Nakaya T., Hata S., Saito Y., Uchida S., Okada Y., Nairn A.C., Davis R.J., Yamamoto T., Kinjo M., Taru H., Suzuki T. Quantitative analysis of APP axonal transport in neurons: Role of JIP1 in enhanced APP anterograde transport. Mol. Biol. Cell. 2014;25(22):3569–3580. doi: 10.1091/mbc.E14-06-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Tamayev R., Zhou D., D’Adamio L. The interactome of the amyloid beta precursor protein family members is shaped by phosphorylation of their intracellular domains. Mol. Neurodegener. 2009;4:28. doi: 10.1186/1750-1326-4-28. https://molecularneurodegeneration. biomedcentral.com/articles/10.1186/1750-1326-4-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Kim E.K., Choi E.J. Compromised MAPK signaling in human diseases: An update. Arch. Toxicol. 2015;89(6):867–882. doi: 10.1007/s00204-015-1472-2. [DOI] [PubMed] [Google Scholar]
- 302.Taylor C.A., Miller B.R., III, Shah S.S., Parish C.A. A molecular dynamics study of the binary complexes of APP, JIP1, and the cargo binding domain of KLC. Proteins. 2017;85(2):221–234. doi: 10.1002/prot.25208. [DOI] [PubMed] [Google Scholar]
- 303.Fu M.M., Holzbaur E.L. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J. Cell Biol. 2013;202(3):495–508. doi: 10.1083/jcb.201302078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Götz J., Lim Y.A., Ke Y.D., Eckert A., Ittner L.M. Dissecting toxicity of tau and beta-amyloid. Neurodegener. Dis. 2010;7(1-3):10–12. doi: 10.1159/000283475. [DOI] [PubMed] [Google Scholar]
- 305.Ittner L.M., Ke Y.D., Gotz J. Phosphorylated tau interacts with c-Jun N-terminal kinase-interacting protein 1 (JIP1) in Alzheimer disease. J. Biol. Chem. 2009;284(31):20909–20916. doi: 10.1074/jbc.M109.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Margevicius D.R., Bastian C., Fan Q., Davis R.J., Pimplikar S.W. JNK-interacting protein 1 mediates Alzheimer’s-like pathological features in AICD-transgenic mice. Neurobiol. Aging. 2015;36(8):2370–2379. doi: 10.1016/j.neurobiolaging.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 307.Scheinfeld M.H., Roncarati R., Vito P., Lopez P.A., Abdallah M., D’Adamio L. Jun NH2-terminal kinase (JNK) interacting protein 1 (JIP1) binds the cytoplasmic domain of the Alzheimer’s beta-amyloid precursor protein (APP). J. Biol. Chem. 2002;277(5):3767–3775. doi: 10.1074/jbc.M108357200. [DOI] [PubMed] [Google Scholar]
- 308.Vagnoni A., Glennon E.B., Perkinton M.S., Gray E.H., Noble W., Miller C.C. Loss of c-Jun N-terminal kinase-interacting protein-1 does not affect axonal transport of the amyloid precursor protein or Abeta production. Hum. Mol. Genet. 2013;22(22):4646–4652. doi: 10.1093/hmg/ddt313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Horiuchi D., Collins C.A., Bhat P., Barkus R.V., Diantonio A., Saxton W.M. Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr. Biol. 2007;17(15):1313–1317. doi: 10.1016/j.cub.2007.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Stokin G.B., Lillo C., Falzone T.L., Brusch R.G., Rockenstein E., Mount S.L., Raman R., Davies P., Masliah E., Williams D.S., Goldstein L.S. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science. 2005;307(5713):1282–1288. doi: 10.1126/science.1105681. http://science.sciencemag.org/content/ 307/5713/1282 [DOI] [PubMed] [Google Scholar]
- 311.Giraldo E., Lloret A., Fuchsberger T., Vina J. Abeta and tau toxicities in Alzheimer’s are linked via oxidative stress-induced p38 activation: Protective role of vitamin E. Redox Biol. 2014;2:873–877. doi: 10.1016/j.redox.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Pinsetta F.R., Taft C.A., de Paula da Silva C.H. Structure- and ligand-based drug design of novel p38-alpha MAPK inhibitors in the fight against the Alzheimer’s disease. J. Biomol. Struct. Dyn. 2014;32(7):1047–1063. doi: 10.1080/07391102.2013.803441. [DOI] [PubMed] [Google Scholar]
- 313.Sanderson T.M., Hogg E.L., Collingridge G.L., Correa S.A. Hippocampal metabotropic glutamate receptor long-term depression in health and disease: Focus on mitogen-activated protein kinase pathways. J. Neurochem. 2016;139(Suppl. 2):200–214. doi: 10.1111/jnc.13592. [DOI] [PubMed] [Google Scholar]
- 314.Freund A., Patil C.K., Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011;30(8):1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Salminen A., Ojala J., Kaarniranta K., Haapasalo A., Hiltunen M., Soininen H. Astrocytes in the aging brain express characteristics of senescence-associated secretory phenotype. Eur. J. Neurosci. 2011;34(1):3–11. doi: 10.1111/j.1460-9568.2011.07738.x. [DOI] [PubMed] [Google Scholar]
- 316.Mombach J.C., Vendrusculo B., Bugs C.A. A model for p38MAPK-induced astrocyte senescence. PLoS One. 2015;10(5):e0125217. doi: 10.1371/journal.pone.0125217. http://journals.plos.org/plosone/article? id=10.1371/journal.pone.0125217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Bachstetter A.D., Xing B., de Almeida L., Dimayuga E.R., Watterson D.M., Van Eldik L.J. 2011 doi: 10.1186/1742-2094-8-79. central.com/articles/10.1186/1742-2094-8-79 [DOI] [PMC free article] [PubMed]
- 318.Munoz L., Ammit A.J. Targeting p38 MAPK pathway for the treatment of Alzheimer’s disease. Neuropharmacology. 2010;58(3):561–568. doi: 10.1016/j.neuropharm.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 319.Munoz L., Ranaivo H.R., Roy S.M., Hu W., Craft J.M., McNamara L.K., Chico L.W., Van Eldik L.J., Watterson D.M. A novel p38 alpha MAPK inhibitor suppresses brain proinflammatory cytokine up-regulation and attenuates synaptic dysfunction and behavioral deficits in an Alzheimer’s disease mouse model. 2007 doi: 10.1186/1742-2094-4-21. central.com/articles/10.1186/1742-2094-4-21 [DOI] [PMC free article] [PubMed]
- 320.Alam J.J. Selective brain-targeted antagonism of p38 MAPKalpha reduces hippocampal IL-1beta levels and improves morris water maze performance in aged rats. J. Alzheimers Dis. 2015;48(1):219–227. doi: 10.3233/JAD-150277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.O’Callaghan C., Fanning L.J., Barry O.P. p38delta MAPK: Emerging roles of a neglected isoform. Int. J. Cell Biol. 2014;2014:272689. doi: 10.1155/2014/272689. https://www.hindawi.com/journals/ijcb/2014/272689/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Fu A.K., Hung K.W., Yuen M.Y., Zhou X., Mak D.S., Chan I.C., Cheung T.H., Zhang B., Fu W.Y., Liew F.Y., Ip N.Y. IL-33 ameliorates Alzheimer’s disease-like pathology and cognitive decline. Proc. Natl. Acad. Sci. USA. 2016;113(19):E2705–E2713. doi: 10.1073/pnas.1604032113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Li Y., Liu L., Barger S.W., Griffin W.S. Interleukin-1 mediates pathological effects of microglia on tau phosphorylation and on synaptophysin synthesis in cortical neurons through a p38-MAPK pathway. J. Neurosci. 2003;23(5):1605–1611. doi: 10.1523/JNEUROSCI.23-05-01605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Ittner A., Chua S.W., Bertz J., Volkerling A., van der Hoven J., Gladbach A., Przybyla M., Bi M., van Hummel A., Stevens C.H., Ippati S., Suh L.S., Macmillan A., Sutherland G., Kril J.J., Silva A.P., Mackay J., Poljak A., Delerue F., Ke Y.D., Ittner L.M. Site-specific phosphorylation of tau inhibits amyloid-beta toxicity in Alzheimer’s mice. 2016 doi: 10.1126/science.aah6205. http://science.sciencemag.org/content/354/6314/ [DOI] [PubMed]
- 325.Koppensteiner P., Trinchese F., Fa M., Puzzo D., Gulisano W., Yan S., Poussin A., Liu S., Orozco I., Dale E., Teich A.F., Palmeri A., Ninan I., Boehm S., Arancio O. Time-dependent reversal of synaptic plasticity induced by physiological concentrations of oligomeric Abeta42: An early index of Alzheimer’s disease. Sci. Rep. 2016;6:32553. doi: 10.1038/srep32553. https:// www.nature.com/articles/srep32553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Tong L., Prieto G.A., Kramar E.A., Smith E.D., Cribbs D.H., Lynch G., Cotman C.W. Brain-derived neurotrophic factor-dependent synaptic plasticity is suppressed by interleukin-1beta via p38 mitogen-activated protein kinase. J. Neurosci. 2012;32(49):17714–17724. doi: 10.1523/JNEUROSCI.1253-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Criscuolo C., Fabiani C., Bonadonna C., Origlia N., Domenici L. BDNF prevents amyloid-dependent impairment of LTP in the entorhinal cortex by attenuating p38 MAPK phosphorylation. Neurobiol. Aging. 2015;36(3):1303–1309. doi: 10.1016/j.neurobiolaging.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 328.Kothari V., Luo Y., Tornabene T., O’Neill A.M., Greene M.W., Geetha T., Babu J.R. High fat diet induces brain insulin resistance and cognitive impairment in mice. Biochim. Biophys. Acta. 2017;1863(2):499–508. doi: 10.1016/j.bbadis.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 329.Liao Z., Cao D., Han X., Liu C., Peng J., Zuo Z., Wang F., Li Y. Both JNK and P38 MAPK pathways participate in the protection by dexmedetomidine against isoflurane-induced neuroapoptosis in the hippocampus of neonatal rats. Brain Res. Bull. 2014;107:69–78. doi: 10.1016/j.brainresbull.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 330.Schnöder L., Hao W., Qin Y., Liu S., Tomic I., Liu X., Fassbender K., Liu Y. Deficiency of neuronal p38alpha MAPK attenuates amyloid pathology in Alzheimer disease mouse and cell models through facilitating lysosomal degradation of BACE1. J. Biol. Chem. 2016;291(5):2067–2079. doi: 10.1074/jbc.M115.695916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Wang S., Zhang C., Sheng X., Zhang X., Wang B., Zhang G. Peripheral expression of MAPK pathways in Alzheimer’s and Parkinson’s diseases. J. Clin. Neurosci. 2014;21(5):810–814. doi: 10.1016/j.jocn.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 332.Origlia N., Righi M., Capsoni S., Cattaneo A., Fang F., Stern D.M., Chen J.X., Schmidt A.M., Arancio O., Yan S.D., Domenici L. Receptor for advanced glycation end product-dependent activation of p38 mitogen-activated protein kinase contributes to amyloid-beta-mediated cortical synaptic dysfunction. J. Neurosci. 2008;28(13):3521–3530. doi: 10.1523/JNEUROSCI.0204-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Ittner A.A., Gladbach A., Bertz J., Suh L.S., Ittner L.M. 2014 https://actaneurocomms.biomedcentral.com/articles/10.1186/
- 334.Meyer D., Bonhoeffer T., Scheuss V. Balance and stability of synaptic structures during synaptic plasticity. Neuron. 2014;82(2):430–443. doi: 10.1016/j.neuron.2014.02.031. [DOI] [PubMed] [Google Scholar]
- 335.Straub C., Sabatini B.L. How to grow a synapse. Neuron. 2014;82(2):256–257. doi: 10.1016/j.neuron.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 336.Chen X., Lin R., Chang L., Xu S., Wei X., Zhang J., Wang C., Anwyl R., Wang Q. Enhancement of long-term depression by soluble amyloid beta protein in rat hippocampus is mediated by metabotropic glutamate receptor and involves activation of p38MAPK, STEP and caspase-3. Neuroscience. 2013;253:435–443. doi: 10.1016/j.neuroscience.2013.08.054. [DOI] [PubMed] [Google Scholar]
- 337.Moult P.R., Correa S.A., Collingridge G.L., Fitzjohn S.M., Bashir Z.I. Co-activation of p38 mitogen-activated protein kinase and protein tyrosine phosphatase underlies metabotropic glutamate receptor-dependent long-term depression. J. Physiol. 2008;586(10):2499–2510. doi: 10.1113/jphysiol.2008.153122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Xiong W., Kojic L.Z., Zhang L., Prasad S.S., Douglas R., Wang Y., Cynader M.S. Anisomycin activates p38 MAP kinase to induce LTD in mouse primary visual cortex. Brain Res. 2006;1085(1):68–76. doi: 10.1016/j.brainres.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 339.Yang W.N., Ma K.G., Qian Y.H., Zhang J.S., Feng G.F., Shi L.L., Zhang Z.C., Liu Z.H. Mitogen-activated protein kinase signaling pathways promote low-density lipoprotein receptor-related protein 1-mediated internalization of beta-amyloid protein in primary cortical neurons. Int. J. Biochem. Cell Biol. 2015;64:252–264. doi: 10.1016/j.biocel.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 340.de Oliveira A.S., Santiago F.E., Balioni L.F., Ferrari Mde F., Almeida M.C., Carrettiero D.C. BAG2 expression dictates a functional intracellular switch between the p38-dependent effects of nicotine on tau phosphorylation levels via the alpha7 nicotinic receptor. Exp. Neurol. 2016;275(Pt 1):69–77. doi: 10.1016/j.expneurol.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 341.He L.Q., Lu J.H., Yue Z.Y. Autophagy in ageing and ageing-associated diseases. Acta Pharmacol. Sin. 2013;34(5):605–611. doi: 10.1038/aps.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Francois A., Julian A., Ragot S., Dugast E., Blanchard L., Brishoual S., Terro F., Chassaing D., Page G., Paccalin M. Inflammatory stress on autophagy in peripheral blood mononuclear cells from patients with Alzheimer’s disease during 24 months of follow-up. PLoS One. 2015;10(9):e0138326. doi: 10.1371/journal.pone.0138326. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0138326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Nixon R.A. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 2007;120(Pt 23):4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 344.Ulamek-Koziol M., Furmaga-Jablonska W., Januszewski S., Brzozowska J., Scislewska M., Jablonski M., Pluta R. Neuronal autophagy: Self-eating or self-cannibalism in Alzheimer’s disease. Neurochem. Res. 2013;38(9):1769–1773. doi: 10.1007/s11064-013-1082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Pajares M., Jimenez-Moreno N., Garcia-Yague A.J., Escoll M., de Ceballos M.L., Van Leuven F., Rabano A., Yamamoto M., Rojo A.I., Cuadrado A. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12(10):1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Xiao Q., Yan P., Ma X., Liu H., Perez R., Zhu A., Gonzales E., Tripoli D.L., Czerniewski L., Ballabio A., Cirrito J.R., Diwan A., Lee J.M. Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing abeta generation and amyloid plaque pathogenesis. J. Neurosci. 2015;35(35):12137–12151. doi: 10.1523/JNEUROSCI.0705-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Zhang Y.D., Zhao J.J. TFEB participates in the abeta-induced pathogenesis of Alzheimer’s disease by regulating the autophagy-lysosome pathway. DNA Cell Biol. 2015;34(11):661–668. doi: 10.1089/dna.2014.2738. [DOI] [PubMed] [Google Scholar]
- 348.Pietrocola F., Izzo V., Niso-Santano M., Vacchelli E., Galluzzi L., Maiuri M.C., Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 2013;23(5):310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 349.Alam J., Scheper W. Targeting neuronal MAPK14/p38alpha activity to modulate autophagy in the Alzheimer disease brain. Autophagy. 2016;12(12):2516–2520. doi: 10.1080/15548627.2016.1238555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Platt F.M., Boland B., van der Spoel A.C. The cell biology of disease: Lysosomal storage disorders: The cellular impact of lysosomal dysfunction. J. Cell Biol. 2012;199(5):723–734. doi: 10.1083/jcb.201208152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Edens B.M., Miller N., Ma Y.C. 2016 https://www.frontiersin.org/articles/10.3389/fncel.2016
- 352.Guebel D.V., Torres N.V. Sexual dimorphism and aging in the human hyppocampus: Identification, validation, and impact of differentially expressed genes by factorial microarray and network analysis. Front. Aging Neurosci. 2016;8:229. doi: 10.3389/fnagi.2016.00229. https://www.frontiersin.org/articles/10.3389/fnagi.2016.00229/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Kubli D.A., Gustafsson A.B. Mitochondria and mitophagy: The yin and yang of cell death control. Circ. Res. 2012;111(9):1208–1221. doi: 10.1161/CIRCRESAHA.112.265819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Cai Y., Arikkath J., Yang L., Guo M.L., Periyasamy P., Buch S. Interplay of endoplasmic reticulum stress and autophagy in neurodegenerative disorders. Autophagy. 2016;12(2):225–244. doi: 10.1080/15548627.2015.1121360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Fonseca A.C., Ferreiro E., Oliveira C.R., Cardoso S.M., Pereira C.F. Activation of the endoplasmic reticulum stress response by the amyloid-beta 1-40 peptide in brain endothelial cells. Biochim. Biophys. Acta. 2013;1832(12):2191–2203. doi: 10.1016/j.bbadis.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 356.Fonseca A.C., Oliveira C.R., Pereira C.F., Cardoso S.M. Loss of proteostasis induced by amyloid beta peptide in brain endothelial cells. Biochim. Biophys. Acta. 2014;1843(6):1150–1161. doi: 10.1016/j.bbamcr.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 357.Lonskaya I., Shekoyan A.R., Hebron M.L., Desforges N., Algarzae N.K., Moussa C.E. Diminished parkin solubility and co-localization with intraneuronal amyloid-beta are associated with autophagic defects in Alzheimer’s disease. J. Alzheimers Dis. 2013;33(1):231–247. doi: 10.3233/JAD-2012-121141. [DOI] [PubMed] [Google Scholar]
- 358.Khandelwal P.J., Herman A.M., Hoe H.S., Rebeck G.W., Moussa C.E. Parkin mediates beclin-dependent autophagic clearance of defective mitochondria and ubiquitinated Abeta in AD models. Hum. Mol. Genet. 2011;20(11):2091–2102. doi: 10.1093/hmg/ddr091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A., Zughaier S.M. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Salminen A., Kaarniranta K., Kauppinen A., Ojala J., Haapasalo A., Soininen H., Hiltunen M. Impaired autophagy and APP processing in Alzheimer’s disease: The potential role of Beclin 1 interactome. Prog. Neurobiol. 2013;106-107:33–54. doi: 10.1016/j.pneurobio.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 361.Salminen A., Kaarniranta K., Kauppinen A. Beclin 1 interactome controls the crosstalk between apoptosis, autophagy and inflammasome activation: Impact on the aging process. Ageing Res. Rev. 2013;12(2):520–534. doi: 10.1016/j.arr.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 362.Tramutola A., Triplett J.C., Di Domenico F., Niedowicz D.M., Murphy M.P., Coccia R., Perluigi M., Butterfield D.A. Alteration of mTOR signaling occurs early in the progression of Alzheimer disease (AD): Analysis of brain from subjects with pre-clinical AD, amnestic mild cognitive impairment and late-stage AD. J. Neurochem. 2015;133(5):739–749. doi: 10.1111/jnc.13037. [DOI] [PubMed] [Google Scholar]
- 363.Ricci S., Fuso A., Ippoliti F., Businaro R. Stress-induced cytokines and neuronal dysfunction in Alzheimer’s disease. J. Alzheimers Dis. 2012;28(1):11–24. doi: 10.3233/JAD-2011-110821. [DOI] [PubMed] [Google Scholar]
- 364.Yu J.T., Wang N.D., Ma T., Jiang H., Guan J., Tan L. Roles of beta-adrenergic receptors in Alzheimer’s disease: Implications for novel therapeutics. Brain Res. Bull. 2011;84(2):111–117. doi: 10.1016/j.brainresbull.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 365.Swaab D.F., Bao A.M., Lucassen P.J. The stress system in the human brain in depression and neurodegeneration. Ageing Res. Rev. 2005;4(2):141–194. doi: 10.1016/j.arr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 366.Schechter D.S., Moser D.A., Paoloni-Giacobino A., Stenz L., Gex-Fabry M., Aue T., Adouan W., Cordero M.I., Suardi F., Manini A., Sancho Rossignol A., Merminod G., Ansermet F., Dayer A.G., Rusconi Serpa S. Methylation of NR3C1 is related to maternal PTSD, parenting stress and maternal medial prefrontal cortical activity in response to child separation among mothers with histories of violence exposure. 2015 doi: 10.3389/fpsyg.2015.00690. https://www.frontiersin.org/articles/10.3389/fpsyg [DOI] [PMC free article] [PubMed]
- 367.Ross J.A., McGonigle P., Van Bockstaele E.J. Locus coeruleus, norepinephrine and Abeta peptides in Alzheimer’s disease. Neurobiol. Stress. 2015;2:73–84. doi: 10.1016/j.ynstr.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Lemche E., Giampietro V.P., Surguladze S.A., Amaro E.J., Andrew C.M., Williams S.C., Brammer M.J., Lawrence N., Maier M.A., Russell T.A., Simmons A., Ecker C., Joraschky P., Phillips M.L. Human attachment security is mediated by the amygdala: Evidence from combined fMRI and psychophysiological measures. Hum. Brain Mapp. 2006;27(8):623–635. doi: 10.1002/hbm.20206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Kopin I.J., Lake R.C., Ziegler M. Plasma levels of norepinephrine. Ann. Intern. Med. 1978;88(5):671–680. doi: 10.7326/0003-4819-88-5-671. [DOI] [PubMed] [Google Scholar]
- 370.Sapkota S., Vergote D., Westaway D., Jhamandas J., Dixon R.A. Synergistic associations of catechol-O-methyltransferase and brain-derived neurotrophic factor with executive function in aging are selective and modified by apolipoprotein E. Neurobiol. Aging. 2015;36(1):249–256. doi: 10.1016/j.neurobiolaging.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Ji Y., Shi Z., Liu M., Liu S., Liu S., Wang J. Association between the COMTVal158Met genotype and Alzheimer’s disease in the han chinese population. Dement. Geriatr. Cogn. Disord. Extra. 2014;4(1):14–21. doi: 10.1159/000357161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Martinez M.F., Martin X.E., Alcelay L.G., Flores J.C., Valiente J.M., Juanbeltz B.I., Beldarrain M.A., Lopez J.M., Gonzalez-Fernandez M.C., Salazar A.M., Gandarias R.B., Borda S.I., Marques N.O., Amillano M.B., Zabaleta M.C., de Pancorbo M.M. The COMT Val158 Met polymorphism as an associated risk factor for Alzheimer disease and mild cognitive impairment in APOE 4 carriers. BMC Neurosci. 2009;10:125. doi: 10.1186/1471-2202-10-125. https://bmcneurosci.biomedcentral.com/articles/10.1186/1471-2202-10-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Layden B.T., Durai V., Lowe J.W.L. 2010 http://art.daneshlink.ir/Handler.ashx?server=1&id =21/scitable/topicpage/g-protein-coupled-receptors-pancreatic-islets-and-14257267
- 374.Gannon M., Che P., Chen Y., Jiao K., Roberson E.D., Wang Q. Noradrenergic dysfunction in Alzheimer’s disease. Front. Neurosci. 2015;9:220. doi: 10.3389/fnins.2015.00220. https://www.frontiersin.org/ articles/10.3389/fnins.2015.00220/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Chen Y., Peng Y., Che P., Gannon M., Liu Y., Li L., Bu G., van Groen T., Jiao K., Wang Q. Alpha(2A) adrenergic receptor promotes amyloidogenesis through disrupting APP-SorLA interaction. Proc. Natl. Acad. Sci. USA. 2014;111(48):17296–172301. doi: 10.1073/pnas.1409513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Bullido M.J., Ramos M.C., Ruiz-Gomez A., Tutor A.S., Sastre I., Frank A., Coria F., Gil P., Mayor F., Jr, Valdivieso F. Polymorphism in genes involved in adrenergic signaling associated with Alzheimer’s. Neurobiol. Aging. 2004;25(7):853–859. doi: 10.1016/j.neurobiolaging.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 377.Yu N.N., Wang X.X., Yu J.T., Wang N.D., Lu R.C., Miao D., Tian Y., Tan L. Blocking beta2-adrenergic receptor attenuates acute stress-induced amyloid beta peptides production. Brain Res. 2010;1317:305–310. doi: 10.1016/j.brainres.2009.12.087. [DOI] [PubMed] [Google Scholar]
- 378.Wang D., Fu Q., Zhou Y., Xu B., Shi Q., Igwe B., Matt L., Hell J.W., Wisely E.V., Oddo S., Xiang Y.K. beta2 adrenergic receptor, protein kinase A (PKA) and c-Jun N-terminal kinase (JNK) signaling pathways mediate tau pathology in Alzheimer disease models. J. Biol. Chem. 2013;288(15):10298–102307. doi: 10.1074/jbc.M112.415141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Gibbs M.E., Maksel D., Gibbs Z., Hou X., Summers R.J., Small D.H. Memory loss caused by beta-amyloid protein is rescued by a beta(3)-adrenoceptor agonist. Neurobiol. Aging. 2010;31(4):614–624. doi: 10.1016/j.neurobiolaging.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 380.Jensen C.J., Demol F., Bauwens R., Kooijman R., Massie A., Villers A., Ris L., De Keyser J. Astrocytic beta2 adrenergic receptor gene deletion affects memory in aged mice. PLoS One. 2016;11(10):e0164721. doi: 10.1371/journal.pone.0164721. http://journals.plos.org/plosone/article? id=10.1371/journal.pone.0164721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Yu J.T., Tan L., Ou J.R., Zhu J.X., Liu K., Song J.H., Sun Y.P. Polymorphisms at the beta2-adrenergic receptor gene influence Alzheimer’s disease susceptibility. Brain Res. 2008;1210:216–222. doi: 10.1016/j.brainres.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 382.Ni Y., Zhao X., Bao G., Zou L., Teng L., Wang Z., Song M., Xiong J., Bai Y., Pei G. Activation of beta2-adrenergic receptor stimulates gamma-secretase activity and accelerates amyloid plaque formation. Nat. Med. 2006;12(12):1390–1396. doi: 10.1038/nm1485. [DOI] [PubMed] [Google Scholar]
- 383.Guay S.P., Brisson D., Lamarche B., Biron S., Lescelleur O., Biertho L., Marceau S., Vohl M.C., Gaudet D., Bouchard L. ADRB3 gene promoter DNA methylation in blood and visceral adipose tissue is associated with metabolic disturbances in men. Epigenomics. 2014;6(1):33–43. doi: 10.2217/epi.13.82. [DOI] [PubMed] [Google Scholar]
- 384.Pietrzak R.H., Sumner J.A., Aiello A.E., Uddin M., Neumeister A., Guffanti G., Koenen K.C. Association of the rs2242446 polymorphism in the norepinephrine transporter gene SLC6A2 and anxious arousal symptoms of posttraumatic stress disorder. J. Clin. Psychiatry. 2015;76(4):e537–e538. doi: 10.4088/JCP.14l09346. http:// www.psychiatrist.com/jcp/article/Pages/2015/v76n04/v76n0426.aspx [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Bayles R., Baker E.K., Jowett J.B., Barton D., Esler M., El-Osta A., Lambert G. Methylation of the SLC6a2 gene promoter in major depression and panic disorder. PLoS One. 2013;8(12):e83223. doi: 10.1371/journal.pone.0083223. http://journals.plos.org/plosone/article?id= 10.1371/journal.pone.0083223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Combarros O., Warden D.R., Hammond N., Cortina-Borja M., Belbin O., Lehmann M.G., Wilcock G.K., Brown K., Kehoe P.G., Barber R., Coto E., Alvarez V., Deloukas P., Gwilliam R., Heun R., Kolsch H., Mateo I., Oulhaj A., Arias-Vasquez A., Schuur M., Aulchenko Y.S., Ikram M.A., Breteler M.M., van Duijn C.M., Morgan K., Smith A.D., Lehmann D.J. The dopamine beta-hydroxylase -1021C/T polymorphism is associated with the risk of Alzheimer’s disease in the Epistasis Project. BMC Med. Genet. 2010;11:162. doi: 10.1186/1471-2350-11-162. https://bmcmedgenet.biomedcentral.com/ articles/10.1186/1471-2350-11-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Haglund M., Sjöbeck M., Englund E. Locus ceruleus degeneration is ubiquitous in Alzheimer’s disease: Possible implications for diagnosis and treatment. Neuropathology. 2006;26(6):528–532. doi: 10.1111/j.1440-1789.2006.00725.x. [DOI] [PubMed] [Google Scholar]
- 388.Carnevale D., De Simone R., Minghetti L. Microglia-neuron interaction in inflammatory and degenerative diseases: Role of cholinergic and noradrenergic systems. CNS Neurol. Disord. Drug Targets. 2007;6(6):388–397. doi: 10.2174/187152707783399193. [DOI] [PubMed] [Google Scholar]
- 389.Heneka M.T., Nadrigny F., Regen T., Martinez-Hernandez A., Dumitrescu-Ozimek L., Terwel D., Jardanhazi-Kurutz D., Walter J., Kirchhoff F., Hanisch U.K., Kummer M.P. Locus ceruleus controls Alzheimer’s disease pathology by modulating microglial functions through norepinephrine. Proc. Natl. Acad. Sci. USA. 2010;107(13):6058–6063. doi: 10.1073/pnas.0909586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Brown M.R., Fisher L.A., Spiess J., Rivier C., Rivier J., Vale W. Corticotropin-releasing factor: actions on the sympathetic nervous system and metabolism. Endocrinology. 1982;111(3):928–931. doi: 10.1210/endo-111-3-928. [DOI] [PubMed] [Google Scholar]
- 391.Fisher L.A., Rivier J., Rivier C., Spiess J., Vale W., Brown M.R. Corticotropin-releasing factor (CRF): Central effects on mean arterial pressure and heart rate in rats. Endocrinology. 1982;110(6):2222–2224. doi: 10.1210/endo-110-6-2222. [DOI] [PubMed] [Google Scholar]
- 392.Rivier C., Brownstein M., Spiess J., Rivier J., Vale W. In vivo corticotropin-releasing factor-induced secretion of adrenocorticotropin, beta-endorphin, and corticosterone. Endocrinology. 1982;110(1):272–278. doi: 10.1210/endo-110-1-272. [DOI] [PubMed] [Google Scholar]
- 393.Zambrano E., Reyes-Castro L.A., Nathanielsz P.W. Aging, glucocorticoids and developmental programming. Age (Dordr.) 2015;37(3):9774. doi: 10.1007/s11357-015-9774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Huang C.W., Lui C.C., Chang W.N., Lu C.H., Wang Y.L., Chang C.C. Elevated basal cortisol level predicts lower hippocampal volume and cognitive decline in Alzheimer’s disease. J. Clin. Neurosci. 2009;16(10):1283–1286. doi: 10.1016/j.jocn.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 395.Popp J., Schaper K., Kolsch H., Cvetanovska G., Rommel F., Klingmuller D., Dodel R., Wullner U., Jessen F. CSF cortisol in Alzheimer’s disease and mild cognitive impairment. Neurobiol. Aging. 2009;30(3):498–500. doi: 10.1016/j.neurobiolaging.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 396.Arsenault-Lapierre G., Chertkow H., Lupien S. Seasonal effects on cortisol secretion in normal aging, mild cognitive impairment and Alzheimer’s disease. Neurobiol. Aging. 2010;31(6):1051–1054. doi: 10.1016/j.neurobiolaging.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 397.Bao A.M., Meynen G., Swaab D.F. The stress system in depression and neurodegeneration: Focus on the human hypothalamus. Brain Res. Brain Res. Rev. 2008;57(2):531–553. doi: 10.1016/j.brainresrev.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 398.Laske C., Stransky E., Fritsche A., Eschweiler G.W., Leyhe T. Inverse association of cortisol serum levels with T-tau, P-tau 181 and P-tau 231 peptide levels and T-tau/Abeta 1-42 ratios in CSF in patients with mild Alzheimer’s disease dementia. Eur. Arch. Psychiatry Clin. Neurosci. 2009;259(2):80–85. doi: 10.1007/s00406-008-0838-3. [DOI] [PubMed] [Google Scholar]
- 399.Schommer N.C., Hellhammer D.H., Kirschbaum C. Dissociation between reactivity of the hypothalamus-pituitary-adrenal axis and the sympathetic-adrenal-medullary system to repeated psychosocial stress. Psychosom. Med. 2003;65(3):450–460. doi: 10.1097/01.psy.0000035721.12441.17. [DOI] [PubMed] [Google Scholar]
- 400.McGregor B.A., Murphy K.M., Albano D.L., Ceballos R.M. Stress, cortisol, and B-lymphocytes: A novel approach to understanding academic stress and immune function. Stress. 2016;19(2):185–191. doi: 10.3109/10253890.2015.1127913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Füzesi T., Daviu N., Wamsteeker Cusulin J.I., Bonin R.P., Bains J.S. Hypothalamic CRH neurons orchestrate complex behaviours after stress. Nat. Commun. 2016;7:11937. doi: 10.1038/ncomms11937. https://www.nature.com/articles/ncomms11937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Grone B.P., Maruska K.P. A second corticotropin-releasing hormone gene (CRH2) is conserved across vertebrate classes and expressed in the hindbrain of a basal neopterygian fish, the spotted gar (Lepisosteus oculatus). J. Comp. Neurol. 2015;523(7):1125–1143. doi: 10.1002/cne.23729. [DOI] [PubMed] [Google Scholar]
- 403.Le M.H., Weissmiller A.M., Monte L., Lin P.H., Hexom T.C., Natera O., Wu C., Rissman R.A. Functional impact of corticotropin-releasing factor exposure on tau phosphorylation and axon transport. PLoS One. 2016;11(1):e0147250. doi: 10.1371/journal.pone.0147250. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0147250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Belanoff J.K., Gross K., Yager A., Schatzberg A.F. Corticosteroids and cognition. J. Psychiatr. Res. 2001;35(3):127–145. doi: 10.1016/s0022-3956(01)00018-8. [DOI] [PubMed] [Google Scholar]
- 405.Hunter R.G., Seligsohn M., Rubin T.G., Griffiths B.B., Ozdemir Y., Pfaff D.W., Datson N.A., McEwen B.S. Stress and corticosteroids regulate rat hippocampal mitochondrial DNA gene expression via the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA. 2016;113(32):9099–9104. doi: 10.1073/pnas.1602185113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Farrell C., O’Keane V. Epigenetics and the glucocorticoid receptor: A review of the implications in depression. Psychiatry Res. 2016;242:349–356. doi: 10.1016/j.psychres.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 407.Anacker C., Zunszain P.A., Cattaneo A., Carvalho L.A., Garabedian M.J., Thuret S., Price J., Pariante C.M. Antidepressants increase human hippocampal neurogenesis by activating the glucocorticoid receptor. Mol. Psychiatry. 2011;16(7):738–750. doi: 10.1038/mp.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Anacker C., Zunszain P.A., Carvalho L.A., Pariante C.M. The glucocorticoid receptor: Pivot of depression and of antidepressant treatment? Psychoneuroendocrinology. 2011;36(3):415–425. doi: 10.1016/j.psyneuen.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Nixon M., Andrew R., Chapman K.E. It takes two to tango: Dimerisation of glucocorticoid receptor and its anti-inflammatory functions. Steroids. 2013;78(1):59–68. doi: 10.1016/j.steroids.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 410.Ratman D., Vanden Berghe W., Dejager L., Libert C., Tavernier J., Beck I.M., De Bosscher K. How glucocorticoid receptors modulate the activity of other transcription factors: A scope beyond tethering. Mol. Cell. Endocrinol. 2013;380(1-2):41–54. doi: 10.1016/j.mce.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 411.De Bosscher K., Haegeman G., Elewaut D. Targeting inflammation using selective glucocorticoid receptor modulators. Curr. Opin. Pharmacol. 2010;10(4):497–504. doi: 10.1016/j.coph.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 412.Kassel O., Herrlich P. Crosstalk between the glucocorticoid receptor and other transcription factors: Molecular aspects. Mol. Cell. Endocrinol. 2007;275(1-2):13–29. doi: 10.1016/j.mce.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 413.Pineau F., Canet G., Desrumaux C., Hunt H., Chevallier N., Ollivier M., Belanoff J.K., Givalois L. New selective glucocorticoid receptor modulators reverse amyloid-beta peptide-induced hippocampus toxicity. Neurobiol. Aging. 2016;45:109–122. doi: 10.1016/j.neurobiolaging.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 414.Rogers J., Raveendran M., Fawcett G.L., Fox A.S., Shelton S.E., Oler J.A., Cheverud J., Muzny D.M., Gibbs R.A., Davidson R.J., Kalin N.H. CRHR1 genotypes, neural circuits and the diathesis for anxiety and depression. Mol. Psychiatry. 2013;18(6):700–707. doi: 10.1038/mp.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Wolf E.J., Mitchell K.S., Logue M.W., Baldwin C.T., Reardon A.F., Humphries D.E., Miller M.W. Corticotropin releasing hormone receptor 2 (CRHR-2) gene is associated with decreased risk and severity of posttraumatic stress disorder in women. Depress. Anxiety. 2013;30(12):1161–1169. doi: 10.1002/da.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Seckl J.R., Walker B.R. Minireview: 11beta-hydroxysteroid dehydrogenase type 1- a tissue-specific amplifier of glucocorticoid action. Endocrinology. 2001;142(4):1371–1376. doi: 10.1210/endo.142.4.8114. [DOI] [PubMed] [Google Scholar]
- 417.Sousa N. The dynamics of the stress neuromatrix. Mol. Psychiatry. 2016;21(3):302–312. doi: 10.1038/mp.2015.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Ruan L.L., Xu J., Wang C.L., Zou C.C. Variants of 11beta-hydroxysteroid dehydrogenase (HSD11B) gene type 1 and 2 in Chinese obese adolescents. J. Endocrinol. Invest. 2014;37(6):565–573. doi: 10.1007/s40618-014-0075-8. [DOI] [PubMed] [Google Scholar]
- 419.Lee J.H., Gao Z., Ye J. Regulation of 11beta-HSD1 expression during adipose tissue expansion by hypoxia through different activities of NF-kappaB and HIF-1alpha. Am. J. Physiol. Endocrinol. Metab. 2013;304(10):E1035–E1041. doi: 10.1152/ajpendo.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Rogers S.L., Hughes B.A., Jones C.A., Freedman L., Smart K., Taylor N., Stewart P.M., Shackleton C.H., Krone N.P., Blissett J., Tomlinson J.W. Diminished 11beta-hydroxysteroid dehydrogenase type 2 activity is associated with decreased weight and weight gain across the first year of life. J. Clin. Endocrinol. Metab. 2014;99(5):E821–E831. doi: 10.1210/jc.2013-3254. [DOI] [PubMed] [Google Scholar]
- 421.Binder E.B., Salyakina D., Lichtner P., Wochnik G.M., Ising M., Putz B., Papiol S., Seaman S., Lucae S., Kohli M.A., Nickel T., Kunzel H.E., Fuchs B., Majer M., Pfennig A., Kern N., Brunner J., Modell S., Baghai T., Deiml T., Zill P., Bondy B., Rupprecht R., Messer T., Kohnlein O., Dabitz H., Bruckl T., Muller N., Pfister H., Lieb R., Mueller J.C., Lohmussaar E., Strom T.M., Bettecken T., Meitinger T., Uhr M., Rein T., Holsboer F., Muller-Myhsok B. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat. Genet. 2004;36(12):1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 422.Anacker C., O’Donnell K.J., Meaney M.J. Early life adversity and the epigenetic programming of hypothalamic pituitary adrenal function. Dialogues Clin. Neurosci. 2014;16(3):321–333. doi: 10.31887/DCNS.2014.16.3/canacker. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Binder E.B., Bradley R.G., Liu W., Epstein M.P., Deveau T.C., Mercer K.B., Tang Y., Gillespie C.F., Heim C.M., Nemeroff C.B., Schwartz A.C., Cubells J.F., Ressler K.J. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299(11):1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Klengel T., Mehta D., Anacker C., Rex-Haffner M., Pruessner J.C., Pariante C.M., Pace T.W., Mercer K.B., Mayberg H.S., Bradley B., Nemeroff C.B., Holsboer F., Heim C.M., Ressler K.J., Rein T., Binder E.B. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 2013;16(1):33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Fujii T., Ota M., Hori H., Hattori K., Teraishi T., Matsuo J., Kinoshita Y., Ishida I., Nagashima A., Kunugi H. The common functional FKBP5 variant rs1360780 is associated with altered cognitive function in aged individuals. Sci. Rep. 2014;4:6696. doi: 10.1038/srep06696. https://www.nature.com/articles/srep06696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Fujii T., Ota M., Hori H., Hattori K., Teraishi T., Sasayama D., Higuchi T., Kunugi H. Association between the common functional FKBP5 variant (rs1360780) and brain structure in a non-clinical population. J. Psychiatr. Res. 2014;58:96–101. doi: 10.1016/j.jpsychires.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 427.Fujii T., Hori H., Ota M., Hattori K., Teraishi T., Sasayama D., Yamamoto N., Higuchi T., Kunugi H. Effect of the common functional FKBP5 variant (rs1360780) on the hypothalamic-pituitary-adrenal axis and peripheral blood gene expression. Psychoneuroendocrinology. 2014;42:89–97. doi: 10.1016/j.psyneuen.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 428.Gassen N., Han Y., Rein T. Functional impact of FKBP5 on the phosphatase calcineurin. Pharmacopsychiatry. 2009;42(5):217–218. [Google Scholar]
- 429.Blair L.J., Baker J.D., Sabbagh J.J., Dickey C.A. The emerging role of peptidyl-prolyl isomerase chaperones in tau oligomerization, amyloid processing, and Alzheimer’s disease. J. Neurochem. 2015;133(1):1–13. doi: 10.1111/jnc.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Reichmann F., Holzer P., Neuropeptide Y. A stressful review. Neuropeptides. 2016;55:99–109. doi: 10.1016/j.npep.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Gøtzsche C.R., Woldbye D.P. The role of NPY in learning and memory. Neuropeptides. 2016;55:79–89. doi: 10.1016/j.npep.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 432.Strachan M.W., Reynolds R.M., Marioni R.E., Price J.F. Cognitive function, dementia and type 2 diabetes mellitus in the elderly. Nat. Rev. Endocrinol. 2011;7(2):108–114. doi: 10.1038/nrendo.2010.228. [DOI] [PubMed] [Google Scholar]
- 433.Korb E., Finkbeiner S. Arc in synaptic plasticity: From gene to behavior. Trends Neurosci. 2011;34(11):591–598. doi: 10.1016/j.tins.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Shepherd J.D., Bear M.F. New views of Arc, a master regulator of synaptic plasticity. Nat. Neurosci. 2011;14(3):279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Steen E., Terry B.M., Rivera E.J., Cannon J.L., Neely T.R., Tavares R., Xu X.J., Wands J.R., de la Monte S.M. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease--is this type 3 diabetes? J. Alzheimers Dis. 2005;7(1):63–80. doi: 10.3233/jad-2005-7107. [DOI] [PubMed] [Google Scholar]
- 436.Yang Y., Song W. Molecular links between Alzheimer’s disease and diabetes mellitus. Neuroscience. 2013;250:140–150. doi: 10.1016/j.neuroscience.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 437.Ott A., Stolk R.P., van Harskamp F., Pols H.A., Hofman A., Breteler M.M. Diabetes mellitus and the risk of dementia: The Rotterdam Study. Neurology. 1999;53(9):1937–1942. doi: 10.1212/wnl.53.9.1937. [DOI] [PubMed] [Google Scholar]
- 438.Crane P.K., Walker R., Hubbard R.A., Li G., Nathan D.M., Zheng H., Haneuse S., Craft S., Montine T.J., Kahn S.E., McCormick W., McCurry S.M., Bowen J.D., Larson E.B. Glucose levels and risk of dementia. N. Engl. J. Med. 2013;369(6):540–548. doi: 10.1056/NEJMoa1215740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.van Dijk S.J., Molloy P.L., Varinli H., Morrison J.L., Muhlhausler B.S. EpiScope, M.O. Epigenetics and human obesity. Int. J. Obes. 2015;39(1):85–97. doi: 10.1038/ijo.2014.34. [DOI] [PubMed] [Google Scholar]
- 440.Chen Z., Zhong C. Decoding Alzheimer’s disease from perturbed cerebral glucose metabolism: Implications for diagnostic and therapeutic strategies. Prog. Neurobiol. 2013;108:21–43. doi: 10.1016/j.pneurobio.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 441.Lee S.H., Zabolotny J.M., Huang H., Lee H., Kim Y.B. Insulin in the nervous system and the mind: Functions in metabolism, memory, and mood. Mol. Metab. 2016;5(8):589–601. doi: 10.1016/j.molmet.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Mushtaq G., Khan J.A., Kamal M.A. Biological mechanisms linking Alzheimer’s disease and type-2 diabetes mellitus. CNS Neurol. Disord. Drug Targets. 2014;13(7):1192–1201. doi: 10.2174/1871527313666140917114537. [DOI] [PubMed] [Google Scholar]
- 443.Ostrowski P.P., Barszczyk A., Forstenpointner J., Zheng W., Feng Z.P. Meta-analysis of serum insulin-like growth factor 1 in Alzheimer’s disease. 2016 doi: 10.1371/journal.pone.0155733. http://journals.plos.org/plosone/article?id=10.1371/journal.pone [DOI] [PMC free article] [PubMed]
- 444.Cardoso S., Correia S., Santos R.X., Carvalho C., Santos M.S., Oliveira C.R., Perry G., Smith M.A., Zhu X., Moreira P.I. Insulin is a two-edged knife on the brain. J. Alzheimers Dis. 2009;18(3):483–507. doi: 10.3233/JAD-2009-1155. [DOI] [PubMed] [Google Scholar]
- 445.Liu F., Iqbal K., Grundke-Iqbal I., Hart G.W., Gong C.X. O-GlcNAcylation regulates phosphorylation of tau: A mechanism involved in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2004;101(29):10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Liu Y., Liu F., Iqbal K., Grundke-Iqbal I., Gong C.X. Decreased glucose transporters correlate to abnormal hyperphosphorylation of tau in Alzheimer disease. FEBS Lett. 2008;582(2):359–364. doi: 10.1016/j.febslet.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Verdile G., Keane K.N., Cruzat V.F., Medic S., Sabale M., Rowles J., Wijesekara N., Martins R.N., Fraser P.E., Newsholme P. Inflammation and oxidative stress: The molecular connectivity between insulin resistance, obesity, and Alzheimer’s disease. Mediators Inflamm. 2015;2015:105828. doi: 10.1155/2015/105828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Heneka M.T., O’Banion M.K., Terwel D., Kummer M.P. Neuroinflammatory processes in Alzheimer’s disease. J. Neural Transm. (Vienna) 2010;117(8):919–947. doi: 10.1007/s00702-010-0438-z. [DOI] [PubMed] [Google Scholar]
- 449.Krautwald M., Munch G. Advanced glycation end products as biomarkers and gerontotoxins - A basis to explore methylglyoxal-lowering agents for Alzheimer’s disease? Exp. Gerontol. 2010;45(10):744–751. doi: 10.1016/j.exger.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 450.Srikanth V., Maczurek A., Phan T., Steele M., Westcott B., Juskiw D., Munch G. Advanced glycation endproducts and their receptor RAGE in Alzheimer’s disease. Neurobiol. Aging. 2011;32(5):763–777. doi: 10.1016/j.neurobiolaging.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 451.Pugazhenthi S., Qin L., Reddy P.H. Common neurodegenerative pathways in obesity, diabetes, and Alzheimer’s disease. Biochim. Biophys. Acta. 2017;1863(5):1037–1045. doi: 10.1016/j.bbadis.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Saltiel A.R., Kahn C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. doi: 10.1038/414799a. https://www.nature.com/articles/414799a [DOI] [PubMed] [Google Scholar]
- 453.Chang L., Chiang S.H., Saltiel A.R. Insulin signaling and the regulation of glucose transport. Mol. Med. 2004;10(7-12):65–71. doi: 10.2119/2005-00029.Saltiel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Takach O., Gill T.B., Silverman M.A. Modulation of insulin signaling rescues BDNF transport defects independent of tau in amyloid-beta oligomer-treated hippocampal neurons. Neurobiol. Aging. 2015;36(3):1378–1382. doi: 10.1016/j.neurobiolaging.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 455.Taga M., Minett T., Classey J., Matthews F.E., Brayne C., Ince P.G., Nicoll J.A., Hugon J., Boche D., Mrc C. Metaflammasome components in the human brain: A role in dementia with Alzheimer’s pathology? Brain Pathol. 2017;27(3):266–275. doi: 10.1111/bpa.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Yamamoto N., Tanida M., Ono Y., Kasahara R., Fujii Y., Ohora K., Suzuki K., Sobue K. Leptin inhibits amyloid beta-protein degradation through decrease of neprilysin expression in primary cultured astrocytes. Biochem. Biophys. Res. Commun. 2014;445(1):214–217. doi: 10.1016/j.bbrc.2014.01.168. [DOI] [PubMed] [Google Scholar]
- 457.Yamamoto N., Fujii Y., Kasahara R., Tanida M., Ohora K., Ono Y., Suzuki K., Sobue K. Simvastatin and atorvastatin facilitates amyloid beta-protein degradation in extracellular spaces by increasing neprilysin secretion from astrocytes through activation of MAPK/Erk1/2 pathways. Glia. 2016;64(6):952–962. doi: 10.1002/glia.22974. [DOI] [PubMed] [Google Scholar]
- 458.Qiu W.Q., Walsh D.M., Ye Z., Vekrellis K., Zhang J., Podlisny M.B., Rosner M.R., Safavi A., Hersh L.B., Selkoe D.J. Insulin-degrading enzyme regulates extracellular levels of amyloid beta-protein by degradation. J. Biol. Chem. 1998;273(49):32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- 459.Shao C.Y., Crary J.F., Rao C., Sacktor T.C., Mirra S.S. Atypical protein kinase C in neurodegenerative disease II: PKCiota/lambda in tauopathies and alpha-synucleinopathies. J. Neuropathol. Exp. Neurol. 2006;65(4):327–335. doi: 10.1097/01.jnen.0000218441.00040.82. [DOI] [PubMed] [Google Scholar]
- 460.Crary J.F., Shao C.Y., Mirra S.S., Hernandez A.I., Sacktor T.C. Atypical protein kinase C in neurodegenerative disease I: PKMzeta aggregates with limbic neurofibrillary tangles and AMPA receptors in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2006;65(4):319–326. doi: 10.1097/01.jnen.0000218442.07664.04. [DOI] [PubMed] [Google Scholar]
- 461.Adzovic L., Domenici L. Insulin induces phosphorylation of the AMPA receptor subunit GluR1, reversed by ZIP, and over-expression of Protein Kinase M zeta, reversed by amyloid beta. J. Neurochem. 2014;131(5):582–587. doi: 10.1111/jnc.12947. [DOI] [PubMed] [Google Scholar]
- 462.Talbot K., Wang H.Y., Kazi H., Han L.Y., Bakshi K.P., Stucky A., Fuino R.L., Kawaguchi K.R., Samoyedny A.J., Wilson R.S., Arvanitakis Z., Schneider J.A., Wolf B.A., Bennett D.A., Trojanowski J.Q., Arnold S.E. Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J. Clin. Invest. 2012;122(4):1316–1338. doi: 10.1172/JCI59903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Nuzzo D., Picone P., Baldassano S., Caruana L., Messina E., Marino Gammazza A., Cappello F., Mule F., Di Carlo M. Insulin resistance as common molecular denominator linking obesity to Alzheimer’s disease. Curr. Alzheimer Res. 2015;12(8):723–735. doi: 10.2174/1567205012666150710115506. [DOI] [PubMed] [Google Scholar]
- 464.Roy A., Srivastava M., Saqib U., Liu D., Faisal S.M., Sugathan S., Bishnoi S., Baig M.S. Potential therapeutic targets for inflammation in toll-like receptor 4 (TLR4)-mediated signaling pathways. Int. Immunopharmacol. 2016;40:79–89. doi: 10.1016/j.intimp.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 465.Walter S., Letiembre M., Liu Y., Heine H., Penke B., Hao W., Bode B., Manietta N., Walter J., Schulz-Schuffer W., Fassbender K. Role of the toll-like receptor 4 in neuroinflammation in Alzheimer’s disease. Cell. Physiol. Biochem. 2007;20(6):947–956. doi: 10.1159/000110455. [DOI] [PubMed] [Google Scholar]
- 466.Ma L.Y., Lv Y.L., Huo K., Liu J., Shang S.H., Fei Y.L., Li Y.B., Zhao B.Y., Wei M., Deng Y.N., Qu Q.M. Autophagy-lysosome dysfunction is involved in Abeta deposition in STZ-induced diabetic rats. Behav. Brain Res. 2016;320:484–493. doi: 10.1016/j.bbr.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 467.Smilansky A., Dangoor L., Nakdimon I., Ben-Hail D., Mizrachi D., Shoshan-Barmatz V. The voltage-dependent anion channel 1 mediates amyloid beta toxicity and represents a potential target for Alzheimer disease therapy. J. Biol. Chem. 2015;290(52):30670–30683. doi: 10.1074/jbc.M115.691493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Kandimalla R., Thirumala V., Reddy P.H. Is Alzheimer’s disease a type 3 diabetes? A critical appraisal. Biochim. Biophys. Acta. 2017;1863(5):1078–1089. doi: 10.1016/j.bbadis.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Wang S., Sun Q.Q., Xiang B., Li X.J. Pancreatic islet cell autophagy during aging in rats. Clin. Invest. Med. 2013;36(2):E72–E80. doi: 10.25011/cim.v36i2.19569. [DOI] [PubMed] [Google Scholar]
- 470.Wang J., Gong B., Zhao W., Tang C., Varghese M., Nguyen T., Bi W., Bilski A., Begum S., Vempati P., Knable L., Ho L., Pasinetti G.M. Epigenetic mechanisms linking diabetes and synaptic impairments. Diabetes. 2014;63(2):645–654. doi: 10.2337/db13-1063. [DOI] [PubMed] [Google Scholar]
- 471.Henstridge D.C., Whitham M., Febbraio M.A. Chaperoning to the metabolic party: The emerging therapeutic role of heat-shock proteins in obesity and type 2 diabetes. Mol. Metab. 2014;3(8):781–793. doi: 10.1016/j.molmet.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Saher G., Brugger B., Lappe-Siefke C., Mobius W., Tozawa R., Wehr M.C., Wieland F., Ishibashi S., Nave K.A. High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 2005;8(4):468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- 473.Trushina E., Mielke M.M. Recent advances in the application of metabolomics to Alzheimer’s Disease. Biochim. Biophys. Acta. 2014;1842(8):1232–1239. doi: 10.1016/j.bbadis.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Leduc V., Jasmin-Belanger S., Poirier J. APOE and cholesterol homeostasis in Alzheimer’s disease. Trends Mol. Med. 2010;16(10):469–477. doi: 10.1016/j.molmed.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 475.Andersen O.M., Willnow T.E. Lipoprotein receptors in Alzheimer’s disease. Trends Neurosci. 2006;29(12):687–694. doi: 10.1016/j.tins.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 476.Goedeke L., Fernandez-Hernando C. MicroRNAs: A connection between cholesterol metabolism and neurodegeneration. 2014. [DOI] [PMC free article] [PubMed]
- 477.Kysenius K., Huttunen H.J. Stress-induced upregulation of VLDL receptor alters Wnt-signaling in neurons. Exp. Cell Res. 2016;340(2):238–247. doi: 10.1016/j.yexcr.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 478.Dore G.A., Elias M.F., Robbins M.A., Elias P.K., Nagy Z. Presence of the APOE epsilon4 allele modifies the relationship between type 2 diabetes and cognitive performance: The Maine-Syracuse Study. Diabetologia. 2009;52(12):2551–2560. doi: 10.1007/s00125-009-1497-2. [DOI] [PubMed] [Google Scholar]
- 479.Ledreux A., Wang X., Schultzberg M., Granholm A.C., Freeman L.R. Detrimental effects of a high fat/high cholesterol diet on memory and hippocampal markers in aged rats. Behav. Brain Res. 2016;312:294–304. doi: 10.1016/j.bbr.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Cai Z., Hussain M.D., Yan L.J. Microglia, neuroinflammation, and beta-amyloid protein in Alzheimer’s disease. Int. J. Neurosci. 2014;124(5):307–321. doi: 10.3109/00207454.2013.833510. [DOI] [PubMed] [Google Scholar]
- 481.Rogers J., Webster S., Lue L.F., Brachova L., Civin W.H., Emmerling M., Shivers B., Walker D., McGeer P. Inflammation and Alzheimer’s disease pathogenesis. Neurobiol. Aging. 1996;17(5):681–686. doi: 10.1016/0197-4580(96)00115-7. [DOI] [PubMed] [Google Scholar]
- 482.Heppner F.L., Ransohoff R.M., Becher B. Immune attack: The role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 2015;16(6):358–372. doi: 10.1038/nrn3880. [DOI] [PubMed] [Google Scholar]
- 483.Ransohoff R.M. How neuroinflammation contributes to neurodegeneration. Science. 2016;353(6301):777–783. doi: 10.1126/science.aag2590. http://science.sciencemag.org/content/353/6301/777 [DOI] [PubMed] [Google Scholar]
- 484.Finch C.E., Crimmins E.M. Inflammatory exposure and historical changes in human life-spans. 2004 doi: 10.1126/science.1092556. http://science.sciencemag.org/content/305/5691/ [DOI] [PubMed]
- 485.Akiyama H., Barger S., Barnum S., Bradt B., Bauer J., Cole G.M., Cooper N.R., Eikelenboom P., Emmerling M., Fiebich B.L., Finch C.E., Frautschy S., Griffin W.S., Hampel H., Hull M., Landreth G., Lue L., Mrak R., Mackenzie I.R., McGeer P.L., O’Banion M.K., Pachter J., Pasinetti G., Plata-Salaman C., Rogers J., Rydel R., Shen Y., Streit W., Strohmeyer R., Tooyoma I., Van Muiswinkel F.L., Veerhuis R., Walker D., Webster S., Wegrzyniak B., Wenk G., Wyss-Coray T. Inflammation and Alzheimer’s disease. Neurobiol. Aging. 2000;21(3):383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Bronzuoli M.R., Iacomino A., Steardo L., Scuderi C. Targeting neuroinflammation in Alzheimer’s disease. J. Inflamm. Res. 2016;9:199–208. doi: 10.2147/JIR.S86958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Tuppo E.E., Arias H.R. The role of inflammation in Alzheimer’s disease. Int. J. Biochem. Cell Biol. 2005;37(2):289–305. doi: 10.1016/j.biocel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 488.Badshah H., Ali T., Shafiq-ur R., Faiz-ul A., Ullah F., Kim T.H., Kim M.O. Protective effect of lupeol against lipopolysaccharide-induced neuroinflammation via the p38/c-Jun N-terminal kinase pathway in the adult mouse brain. J. Neuroimmune Pharmacol. 2016;11(1):48–60. doi: 10.1007/s11481-015-9623-z. [DOI] [PubMed] [Google Scholar]
- 489.Lee Y.J., Han S.B., Nam S.Y., Oh K.W., Hong J.T. Inflammation and Alzheimer’s disease. Arch. Pharm. Res. 2010;33(10):1539–1556. doi: 10.1007/s12272-010-1006-7. [DOI] [PubMed] [Google Scholar]
- 490.Meraz-Rios M.A., Toral-Rios D., Franco-Bocanegra D., Villeda-Hernandez J., Campos-Pena V. 2013 doi: 10.3389/fnint.2013.00059. https://www.frontiersin.org/articles/10.3389/fnint.2013.00059/ [DOI] [PMC free article] [PubMed]
- 491.Heneka M.T., O’Banion M.K. Inflammatory processes in Alzheimer’s disease. J. Neuroimmunol. 2007;184(1-2):69–91. doi: 10.1016/j.jneuroim.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 492.Chen P., Zhao W., Guo Y., Xu J., Yin M. CX3CL1/CX3CR1 in Alzheimer’s Disease: A target for neuroprotection. BioMed Res. Int. 2016;2016:8090918. doi: 10.1155/2016/8090918. https://www.hindawi.com/ journals/bmri/2016/8090918/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Hong Z.Y., Shi X.R., Zhu K., Wu T.T., Zhu Y.Z. SCM-198 inhibits microglial overactivation and attenuates Abeta(1-40)-induced cognitive impairments in rats via JNK and NF-kappaB pathways. J. Neuroinflammation. 2014;11:147. doi: 10.1186/s12974-014-0147-x. https://jneuroinflammation.biomedcentral.com/articles/10.1186/s12974-014-0147-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Ontiveros-Torres M.A., Labra-Barrios M.L., Diaz-Cintra S., Aguilar-Vazquez A.R., Moreno-Campuzano S., Flores-Rodriguez P., Luna-Herrera C., Mena R., Perry G., Floran-Garduno B., Luna-Munoz J., Luna-Arias J.P. Fibrillar amyloid-beta accumulation triggers an inflammatory mechanism leading to hyperphosphorylation of the carboxyl-terminal end of tau polypeptide in the hippocampal formation of the 3xTg-AD transgenic mouse. J. Alzheimers Dis. 2016;52(1):243–269. doi: 10.3233/JAD-150837. [DOI] [PubMed] [Google Scholar]
- 495.Granic I., Dolga A.M., Nijholt I.M., van Dijk G., Eisel U.L. Inflammation and NF-kappaB in Alzheimer’s disease and diabetes. J. Alzheimers Dis. 2009;16(4):809–821. doi: 10.3233/JAD-2009-0976. [DOI] [PubMed] [Google Scholar]
- 496.Canton J., Neculai D., Grinstein S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013;13(9):621–634. doi: 10.1038/nri3515. [DOI] [PubMed] [Google Scholar]
- 497.Savchenko E., Malm T., Konttinen H., Hämäläinen R.H., Guerrero-Toro C., Wojciechowski S., Giniatullin R., Koistinaho J., Magga J. 2016 doi: 10.3389/fncel.2016.00279. https://www.frontiersin.org/articles/10.3389/fncel [DOI] [PMC free article] [PubMed]
- 498.Spangenberg E.E., Green K.N. Inflammation in Alzheimer’s disease: Lessons learned from microglia-depletion models. Brain Behav. Immun. 2016;61:1–11. doi: 10.1016/j.bbi.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Shi Y., Yamada K., Liddelow S.A., Smith S.T., Zhao L., Luo W., Tsai R.M., Spina S., Grinberg L.T., Rojas J.C., Gallardo G., Wang K., Roh J., Robinson G., Finn M.B., Jiang H., Sullivan P.M., Baufeld C., Wood M.W., Sutphen C., McCue L., Xiong C., Del-Aguila J.L., Morris J.C., Cruchaga C., Alzheimer’s Disease Neuroimaging Initiative Fagan, A.M.; Miller, B.L.; Boxer, A.L.; Seeley, W.W.; Butovsky, O.; Barres, B.A.; Paul, S.M.; Holtzman, D.M. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017;549(7673):523–527. doi: 10.1038/nature24016. https://www.nature.com/articles/nature24016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Bhaskar K., Konerth M., Kokiko-Cochran O.N., Cardona A., Ransohoff R.M., Lamb B.T. Regulation of tau pathology by the microglial fractalkine receptor. Neuron. 2010;68(1):19–31. doi: 10.1016/j.neuron.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A., Weissman I.L., Chang E.F., Li G., Grant G.A., Gephart M.G.H., Barres B.A. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA. 2016;113(12):E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Minami S.S., Min S.W., Krabbe G., Wang C., Zhou Y., Asgarov R., Li Y., Martens L.H., Elia L.P., Ward M.E., Mucke L., Farese R.V., Jr, Gan L. Progranulin protects against amyloid beta deposition and toxicity in Alzheimer’s disease mouse models. Nat. Med. 2014;20(10):1157–1164. doi: 10.1038/nm.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Graciarena M., Depino A.M., Pitossi F.J. Prenatal inflammation impairs adult neurogenesis and memory related behavior through persistent hippocampal TGFbeta1 downregulation. Brain Behav. Immun. 2010;24(8):1301–1309. doi: 10.1016/j.bbi.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 504.Calsolaro V., Edison P. Neuroinflammation in Alzheimer’s disease: Current evidence and future directions. Alzheimers Dement. 2016;12(6):719–732. doi: 10.1016/j.jalz.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 505.Lucassen P.J., Meerlo P., Naylor A.S., van Dam A.M., Dayer A.G., Fuchs E., Oomen C.A., Czeh B. Regulation of adult neurogenesis by stress, sleep disruption, exercise and inflammation: Implications for depression and antidepressant action. Eur. Neuropsychopharmacol. 2010;20(1):1–17. doi: 10.1016/j.euroneuro.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 506.Johansson J.U., Woodling N.S., Wang Q., Panchal M., Liang X., Trueba-Saiz A., Brown H.D., Mhatre S.D., Loui T., Andreasson K.I. Prostaglandin signaling suppresses beneficial microglial function in Alzheimer’s disease models. J. Clin. Invest. 2015;125(1):350–364. doi: 10.1172/JCI77487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Reynolds W.F., Rhees J., Maciejewski D., Paladino T., Sieburg H., Maki R.A., Masliah E. Myeloperoxidase polymorphism is associated with gender specific risk for Alzheimer’s disease. Exp. Neurol. 1999;155(1):31–41. doi: 10.1006/exnr.1998.6977. [DOI] [PubMed] [Google Scholar]
- 508.Nilson A.N., English K.C., Gerson J.E., Whittle T.B., Crain C.N., Xue J., Sengupta U., Castillo-Carranza D.L., Zhang W., Gupta P., Kayed R. Tau oligomers associate with inflammation in the brain and retina of tauopathy mice and in neurodegenerative diseases. J. Alzheimers Dis. 2017;55(3):1083–1099. doi: 10.3233/JAD-160912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Freeman L.C., Ting J.P. The pathogenic role of the inflammasome in neurodegenerative diseases. J. Neurochem. 2016;136(Suppl. 1):29–38. doi: 10.1111/jnc.13217. [DOI] [PubMed] [Google Scholar]
- 510.Heneka M.T., Golenbock D.T., Latz E. Innate immunity in Alzheimer’s disease. Nat. Immunol. 2015;16(3):229–236. doi: 10.1038/ni.3102. [DOI] [PubMed] [Google Scholar]
- 511.Heneka M.T., Kummer M.P., Stutz A., Delekate A., Schwartz S., Vieira-Saecker A., Griep A., Axt D., Remus A., Tzeng T.C., Gelpi E., Halle A., Korte M., Latz E., Golenbock D.T. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493(7434):674–678. doi: 10.1038/nature11729. https://www.nature.com/articles/nature11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Lénárt N., Brough D., Denes A. Inflammasomes link vascular disease with neuroinflammation and brain disorders. J. Cereb. Blood Flow Metab. 2016;36(10):1668–1685. doi: 10.1177/0271678X16662043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Shimada K., Crother T.R., Karlin J., Dagvadorj J., Chiba N., Chen S., Ramanujan V.K., Wolf A.J., Vergnes L., Ojcius D.M., Rentsendorj A., Vargas M., Guerrero C., Wang Y., Fitzgerald K.A., Underhill D.M., Town T., Arditi M. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Liang X., Wang Q., Hand T., Wu L., Breyer R.M., Montine T.J., Andreasson K. Deletion of the prostaglandin E2 EP2 receptor reduces oxidative damage and amyloid burden in a model of Alzheimer’s disease. J. Neurosci. 2005;25(44):10180–10187. doi: 10.1523/JNEUROSCI.3591-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Shi J., Wang Q., Johansson J.U., Liang X., Woodling N.S., Priyam P., Loui T.M., Merchant M., Breyer R.M., Montine T.J., Andreasson K. Inflammatory prostaglandin E2 signaling in a mouse model of Alzheimer disease. Ann. Neurol. 2012;72(5):788–798. doi: 10.1002/ana.23677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Wood H. Alzheimer disease: Prostaglandin E(2) signalling is implicated in inflammation early in the Alzheimer disease course. Nat. Rev. Neurol. 2012;8(8):411. doi: 10.1038/nrneurol.2012.145. [DOI] [PubMed] [Google Scholar]
- 517.Woodling N.S., Wang Q., Priyam P.G., Larkin P., Shi J., Johansson J.U., Zagol-Ikapitte I., Boutaud O., Andreasson K.I. Suppression of Alzheimer-associated inflammation by microglial prostaglandin-E2 EP4 receptor signaling. J. Neurosci. 2014;34(17):5882–5894. doi: 10.1523/JNEUROSCI.0410-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Bates K.A., Verdile G., Li Q.X., Ames D., Hudson P., Masters C.L., Martins R.N. Clearance mechanisms of Alzheimer’s amyloid-beta peptide: Implications for therapeutic design and diagnostic tests. Mol. Psychiatry. 2009;14(5):469–486. doi: 10.1038/mp.2008.96. [DOI] [PubMed] [Google Scholar]
- 519.Fiala M., Veerhuis R. Biomarkers of inflammation and amyloid-beta phagocytosis in patients at risk of Alzheimer disease. Exp. Gerontol. 2010;45(1):57–63. doi: 10.1016/j.exger.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Zani I.A., Stephen S.L., Mughal N.A., Russell D., Homer-Vanniasinkam S., Wheatcroft S.B., Ponnambalam S. Scavenger receptor structure and function in health and disease. Cells. 2015;4(2):178–201. doi: 10.3390/cells4020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Heneka M.T. Inflammation in Alzheimer’s disease. Clin. Neurosci. Res. 2006;6(5):247–260. [Google Scholar]
- 522.Guo H., Wang H., Wang C., Cheng Y., Zou Z., Li Y., Wu J., Xu J. C-reactive protein induces tau hyperphosphorylation via GSK3beta signaling pathway in SH-SY5Y cells. J. Mol. Neurosci. 2015;56(2):519–527. doi: 10.1007/s12031-015-0572-z. [DOI] [PubMed] [Google Scholar]
- 523.Lin H.B., Yang X.M., Li T.J., Cheng Y.F., Zhang H.T., Xu J.P. Memory deficits and neurochemical changes induced by C-reactive protein in rats: Implication in Alzheimer’s disease. Psychopharmacology (Berl.) 2009;204(4):705–714. doi: 10.1007/s00213-009-1499-2. [DOI] [PubMed] [Google Scholar]
- 524.Giannopoulos P.F., Joshi Y.B., Pratico D. Novel lipid signaling pathways in Alzheimer’s disease pathogenesis. Biochem. Pharmacol. 2014;88(4):560–564. doi: 10.1016/j.bcp.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Katusic Z.S., Austin S.A. Endothelial nitric oxide: Protector of a healthy mind. Eur. Heart J. 2014;35(14):888–894. doi: 10.1093/eurheartj/eht544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Postberg J., Kanders M., Forcob S., Willems R., Orth V., Hensel K.O., Weil P.P., Wirth S., Jenke A.C. CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression. Clin. Epigenetics. 2015;7:9. doi: 10.1186/s13148-014-0042-4. https://clinicalepigeneticsjournal.biomedcentral.com/ articles/10.1186/s13148-014-0042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Di Bona D., Candore G., Franceschi C., Licastro F., Colonna-Romano G., Camma C., Lio D., Caruso C. Systematic review by meta-analyses on the possible role of TNF-alpha polymorphisms in association with Alzheimer’s disease. Brain Res. Brain Res. Rev. 2009;61(2):60–68. doi: 10.1016/j.brainresrev.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 528.Smith J.A., Das A., Ray S.K., Banik N.L. Role of pro-inflammatory cytokines released from microglia in neurodegenerative diseases. Brain Res. Bull. 2012;87(1):10–20. doi: 10.1016/j.brainresbull.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Di Bona D., Plaia A., Vasto S., Cavallone L., Lescai F., Franceschi C., Licastro F., Colonna-Romano G., Lio D., Candore G., Caruso C. Association between the interleukin-1beta polymorphisms and Alzheimer’s disease: A systematic review and meta-analysis. Brain Res. Brain Res. Rev. 2008;59(1):155–163. doi: 10.1016/j.brainresrev.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 530.He P., Zhong Z., Lindholm K., Berning L., Lee W., Lemere C., Staufenbiel M., Li R., Shen Y. Deletion of tumor necrosis factor death receptor inhibits amyloid beta generation and prevents learning and memory deficits in Alzheimer’s mice. J. Cell Biol. 2007;178(5):829–841. doi: 10.1083/jcb.200705042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Valerio A., Boroni F., Benarese M., Sarnico I., Ghisi V., Bresciani L.G., Ferrario M., Borsani G., Spano P., Pizzi M. NF-kappaB pathway: A target for preventing beta-amyloid (Abeta)-induced neuronal damage and Abeta42 production. Eur. J. Neurosci. 2006;23(7):1711–1720. doi: 10.1111/j.1460-9568.2006.04722.x. [DOI] [PubMed] [Google Scholar]
- 532.Srinivasan M., Bayon B., Chopra N., Lahiri D.K. Novel nuclear factor-KappaB targeting peptide suppresses beta-amyloid induced inflammatory and apoptotic responses in neuronal cells. PLoS One. 2016;11(10):e0160314. doi: 10.1371/journal.pone.0160314. http://journals.plos.org/ plosone/article?id=10.1371/journal.pone.0160314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Su F., Bai F., Zhang Z. Inflammatory cytokines and Alzheimer’s disease: A review from the perspective of genetic polymorphisms. Neurosci. Bull. 2016;32(5):469–480. doi: 10.1007/s12264-016-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Liu Z.J., Li Z.H., Liu L., Tang W.X., Wang Y., Dong M.R., Xiao C. Curcumin attenuates beta-amyloid-induced neuroinflammation via activation of peroxisome proliferator-activated receptor-gamma function in a rat model of Alzheimer’s disease. Front. Pharmacol. 2016;7:261. doi: 10.3389/fphar.2016.00261. https://www.frontiersin.org/articles/10.3389/fphar.2016.00261/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Katsouri L., Lim Y.M., Blondrath K., Eleftheriadou I., Lombardero L., Birch A.M., Mirzaei N., Irvine E.E., Mazarakis N.D., Sastre M. PPARgamma-coactivator-1alpha gene transfer reduces neuronal loss and amyloid-beta generation by reducing beta-secretase in an Alzheimer’s disease model. Proc. Natl. Acad. Sci. USA. 2016;113(43):12292–12297. doi: 10.1073/pnas.1606171113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Onyango I.G., Khan S.M., Bennett J.P., Jr Mitochondria in the pathophysiology of Alzheimer’s and Parkinson’s diseases. Front. Biosci. 2017;22:854–872. doi: 10.2741/4521. [DOI] [PubMed] [Google Scholar]
- 537.McShea A., Lee H.G., Petersen R.B., Casadesus G., Vincent I., Linford N.J., Funk J.O., Shapiro R.A., Smith M.A. Neuronal cell cycle re-entry mediates Alzheimer disease-type changes. Biochim. Biophys. Acta. 2007;1772(4):467–472. doi: 10.1016/j.bbadis.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 538.Lee H.P., Kudo W., Zhu X., Smith M.A., Lee H.G. Early induction of c-Myc is associated with neuronal cell death. Neurosci. Lett. 2011;505(2):124–127. doi: 10.1016/j.neulet.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Ferrer I., Blanco R. N-myc and c-myc expression in Alzheimer disease, Huntington disease and Parkinson disease. Brain Res. Mol. Brain Res. 2000;77(2):270–276. doi: 10.1016/s0169-328x(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 540.Gaiteri C., Mostafavi S., Honey C.J., De Jager P.L., Bennett D.A. Genetic variants in Alzheimer disease - molecular and brain network approaches. Nat. Rev. Neurol. 2016;12(7):413–427. doi: 10.1038/nrneurol.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Saykin A.J., Shen L., Yao X., Kim S., Nho K., Risacher S.L., Ramanan V.K., Foroud T.M., Faber K.M., Sarwar N., Munsie L.M., Hu X., Soares H.D., Potkin S.G., Thompson P.M., Kauwe J.S., Kaddurah-Daouk R., Green R.C., Toga A.W., Weiner M.W., Alzheimer’s Disease Neuroimaging Initiative Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 2015;11(7):792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Rosenthal S.L., Kamboh M.I. Late-onset Alzheimer’s disease genes and the potentially implicated pathways. 2014 doi: 10.1007/s40142-014-0034-x. central.com/articles/10.1186/s13024-017-0184-x [DOI] [PMC free article] [PubMed]
- 543.Lord J., Cruchaga C. The epigenetic landscape of Alzheimer’s disease. Nat. Neurosci. 2014;17(9):1138–1140. doi: 10.1038/nn.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Zheng J., Yan H., Shi L., Kong Y., Zhao Y., Xie L., Li J., Huang M., Li J., Zhao S. The CYP19A1 rs3751592 variant confers susceptibility to Alzheimer disease in the Chinese Han population. Medicine (Baltimore) 2016;95(35):e4742. doi: 10.1097/MD.0000000000004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Ciryam P., Kundra R., Freer R., Morimoto R.I., Dobson C.M., Vendruscolo M. A transcriptional signature of Alzheimer’s disease is associated with a metastable subproteome at risk for aggregation. Proc. Natl. Acad. Sci. USA. 2016;113(17):4753–4758. doi: 10.1073/pnas.1516604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Kim D.H., Yeo S.H., Park J.M., Choi J.Y., Lee T.H., Park S.Y., Ock M.S., Eo J., Kim H.S., Cha H.J. Genetic markers for diagnosis and pathogenesis of Alzheimer’s disease. Gene. 2014;545(2):185–193. doi: 10.1016/j.gene.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 547.Deng Y., Wang Z., Wang R., Zhang X., Zhang S., Wu Y., Staufenbiel M., Cai F., Song W. Amyloid-beta protein (Abeta) Glu11 is the major beta-secretase site of beta-site amyloid-beta precursor protein-cleaving enzyme 1(BACE1), and shifting the cleavage site to Abeta Asp1 contributes to Alzheimer pathogenesis. Eur. J. Neurosci. 2013;37(12):1962–1969. doi: 10.1111/ejn.12235. [DOI] [PubMed] [Google Scholar]
- 548.Kölsch H., Jessen F., Wiltfang J., Lewczuk P., Dichgans M., Teipel S.J., Kornhuber J., Frolich L., Heuser I., Peters O., Wiese B., Kaduszkiewicz H., van den Bussche H., Hull M., Kurz A., Ruther E., Henn F.A., Maier W. Association of SORL1 gene variants with Alzheimer’s disease. Brain Res. 2009;1264:1–6. doi: 10.1016/j.brainres.2009.01.044. [DOI] [PubMed] [Google Scholar]
- 549.Kimura R., Yamamoto M., Morihara T., Akatsu H., Kudo T., Kamino K., Takeda M. SORL1 is genetically associated with Alzheimer disease in a Japanese population. Neurosci. Lett. 2009;461(2):177–180. doi: 10.1016/j.neulet.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 550.Li Y., Rowland C., Catanese J., Morris J., Lovestone S., O’Donovan M.C., Goate A., Owen M., Williams J., Grupe A. SORL1 variants and risk of late-onset Alzheimer’s disease. Neurobiol. Dis. 2008;29(2):293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Tan E.K., Lee J., Chen C.P., Teo Y.Y., Zhao Y., Lee W.L. SORL1 haplotypes modulate risk of Alzheimer’s disease in Chinese. Neurobiol. Aging. 2009;30(7):1048–1051. doi: 10.1016/j.neurobiolaging.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 552.Andersen O.M., Rudolph I.M., Willnow T.E. Risk factor SORL1: From genetic association to functional validation in Alzheimer’’s disease. Acta Neuropathol. 2016;132(5):653–665. doi: 10.1007/s00401-016-1615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Rogaeva E., Meng Y., Lee J.H., Gu Y., Kawarai T., Zou F., Katayama T., Baldwin C.T., Cheng R., Hasegawa H., Chen F., Shibata N., Lunetta K.L., Pardossi-Piquard R., Bohm C., Wakutani Y., Cupples L.A., Cuenco K.T., Green R.C., Pinessi L., Rainero I., Sorbi S., Bruni A., Duara R., Friedland R.P., Inzelberg R., Hampe W., Bujo H., Song Y.Q., Andersen O.M., Willnow T.E., Graff-Radford N., Petersen R.C., Dickson D., Der S.D., Fraser P.E., Schmitt-Ulms G., Younkin S., Mayeux R., Farrer L.A., St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Reitz C., Tokuhiro S., Clark L.N., Conrad C., Vonsattel J.P., Hazrati L.N., Palotas A., Lantigua R., Medrano M., Jiménez-Velázquez I.Z., Vardarajan B., Simkin I., Haines J.L., Pericak-Vance M.A., Farrer L.A., Lee J.H., Rogaeva E., George-Hyslop P.S., Mayeux R. SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer’s disease risk. Ann. Neurol. 2011;69(1):47–64. doi: 10.1002/ana.22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.He Y., Li C., Yang Y., Li Y., Wang Y., Yang H., Jin T., Chen S. Meta-analysis of the rs2075650 polymorphism and risk of Alzheimer disease. Aging Clin. Exp. Res. 2016;28(5):805–811. doi: 10.1007/s40520-015-0489-y. [DOI] [PubMed] [Google Scholar]
- 556.Haass C., Kaether C., Thinakaran G., Sisodia S. Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2012;2(5):a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Chouraki V., Seshadri S. Genetics of Alzheimer’s disease. Adv. Genet. 2014;87:245–294. doi: 10.1016/B978-0-12-800149-3.00005-6. [DOI] [PubMed] [Google Scholar]
- 558.Bohm C., Chen F., Sevalle J., Qamar S., Dodd R., Li Y., Schmitt-Ulms G., Fraser P.E., St George-Hyslop P.H. Current and future implications of basic and translational research on amyloid-beta peptide production and removal pathways. 2015. [DOI] [PMC free article] [PubMed]
- 559.Marcello E., Saraceno C., Musardo S., Vara H., de la Fuente A.G., Pelucchi S., Di Marino D., Borroni B., Tramontano A., Perez-Otano I., Padovani A., Giustetto M., Gardoni F., Di Luca M. Endocytosis of synaptic ADAM10 in neuronal plasticity and Alzheimer’s disease. J. Clin. Invest. 2013;123(6):2523–2538. doi: 10.1172/JCI65401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Zhao Y., Bhattacharjee S., Dua P., Alexandrov P.N., Lukiw W.J. MicroRNA-based biomarkers and the diagnosis of Alzheimer’s disease. Front. Neurol. 2015;6:162. doi: 10.3389/fneur.2015.00162. https://www.frontiersin.org/articles/10.3389/fneur.2015.00162/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Gharesouran J., Rezazadeh M., Khorrami A., Ghojazadeh M., Talebi M. Genetic evidence for the involvement of variants at APOE, BIN1, CR1, and PICALM loci in risk of late-onset Alzheimer’s disease and evaluation for interactions with APOE genotypes. J. Mol. Neurosci. 2014;54(4):780–786. doi: 10.1007/s12031-014-0377-5. [DOI] [PubMed] [Google Scholar]
- 562.Li Y., Song D., Jiang Y., Wang J., Feng R., Zhang L., Wang G., Chen Z., Wang R., Jiang Q., Liu G. CR1 rs3818361 polymorphism contributes to Alzheimer’s disease susceptibility in Chinese population. Mol. Neurobiol. 2016;53(6):4054–4059. doi: 10.1007/s12035-015-9343-7. [DOI] [PubMed] [Google Scholar]
- 563.Holler C.J., Davis P.R., Beckett T.L., Platt T.L., Webb R.L., Head E., Murphy M.P. Bridging integrator 1 (BIN1) protein expression increases in the Alzheimer’s disease brain and correlates with neurofibrillary tangle pathology. J. Alzheimers Dis. 2014;42(4):1221–1227. doi: 10.3233/JAD-132450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.De Rossi P., Buggia-Prevot V., Clayton B.L., Vasquez J.B., van Sanford C., Andrew R.J., Lesnick R., Botte A., Deyts C., Salem S., Rao E., Rice R.C., Parent A., Kar S., Popko B., Pytel P., Estus S., Thinakaran G. Predominant expression of Alzheimer’s disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts. Mol. Neurodegener. 2016;11(1):59. doi: 10.1186/s13024-016-0124-1. https://molecularneurodegeneration. biomedcentral.com/articles/10.1186/s13024-016-0124-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Wang H.F., Tan L., Hao X.K., Jiang T., Tan M.S., Liu Y., Zhang D.Q., Yu J.T., Alzheimer’s Disease Neuroimaging Initiative Effect of EPHA1 genetic variation on cerebrospinal fluid and neuroimaging biomarkers in healthy, mild cognitive impairment and Alzheimer’s disease cohorts. J. Alzheimers Dis. 2015;44(1):115–123. doi: 10.3233/JAD-141488. [DOI] [PubMed] [Google Scholar]
- 566.Chen H., Wu G., Jiang Y., Feng R., Liao M., Zhang L., Ma G., Chen Z., Zhao B., Li K., Yu C., Liu G. Analyzing 54,936 samples supports the association between CD2AP rs9349407 polymorphism and Alzheimer’s disease susceptibility. Mol. Neurobiol. 2015;52(1):1–7. doi: 10.1007/s12035-014-8834-2. [DOI] [PubMed] [Google Scholar]
- 567.Elias-Sonnenschein L.S., Helisalmi S., Natunen T., Hall A., Paajanen T., Herukka S.K., Laitinen M., Remes A.M., Koivisto A.M., Mattila K.M., Lehtimaki T., Verhey F.R., Visser P.J., Soininen H., Hiltunen M. Genetic loci associated with Alzheimer’s disease and cerebrospinal fluid biomarkers in a Finnish case-control cohort. 2013 doi: 10.1371/journal.pone.0059676. http://journals.plos.org/plosone/article?id=10.1371/journal [DOI] [PMC free article] [PubMed]
- 568.Wang C., Tan L., Wang H.F., Yu W.J., Liu Y., Jiang T., Tan M.S., Hao X.K., Zhang D.Q., Yu J.T., Alzheimer’s Disease Neuroimaging Initiative Common variants in PLD3 and correlation to amyloid-related phenotypes in alzheimer’s disease. J. Alzheimers Dis. 2015;46(2):491–495. doi: 10.3233/JAD-150110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.De Jager P.L., Srivastava G., Lunnon K., Burgess J., Schalkwyk L.C., Yu L., Eaton M.L., Keenan B.T., Ernst J., McCabe C., Tang A., Raj T., Replogle J., Brodeur W., Gabriel S., Chai H.S., Younkin C., Younkin S.G., Zou F., Szyf M., Epstein C.B., Schneider J.A., Bernstein B.E., Meissner A., Ertekin-Taner N., Chibnik L.B., Kellis M., Mill J., Bennett D.A. Alzheimer’s disease: Early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci. 2014;17(9):1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Lunnon K., Smith R., Hannon E., De Jager P.L., Srivastava G., Volta M., Troakes C., Al-Sarraj S., Burrage J., Macdonald R., Condliffe D., Harries L.W., Katsel P., Haroutunian V., Kaminsky Z., Joachim C., Powell J., Lovestone S., Bennett D.A., Schalkwyk L.C., Mill J. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat. Neurosci. 2014;17(9):1164–1170. doi: 10.1038/nn.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Colonna M., Wang Y. TREM2 variants: New keys to decipher Alzheimer disease pathogenesis. Nat. Rev. Neurosci. 2016;17(4):201–207. doi: 10.1038/nrn.2016.7. [DOI] [PubMed] [Google Scholar]
- 572.Seshadri S., Fitzpatrick A.L., Ikram M.A., DeStefano A.L., Gudnason V., Boada M., Bis J.C., Smith A.V., Carassquillo M.M., Lambert J.C., Harold D., Schrijvers E.M., Ramirez-Lorca R., Debette S., Longstreth W.T., Jr, Janssens A.C., Pankratz V.S., Dartigues J.F., Hollingworth P., Aspelund T., Hernandez I., Beiser A., Kuller L.H., Koudstaal P.J., Dickson D.W., Tzourio C., Abraham R., Antunez C., Du Y., Rotter J.I., Aulchenko Y.S., Harris T.B., Petersen R.C., Berr C., Owen M.J., Lopez-Arrieta J., Varadarajan B.N., Becker J.T., Rivadeneira F., Nalls M.A., Graff-Radford N.R., Campion D., Auerbach S., Rice K., Hofman A., Jonsson P.V., Schmidt H., Lathrop M., Mosley T.H., Au R., Psaty B.M., Uitterlinden A.G., Farrer L.A., Lumley T., Ruiz A., Williams J., Amouyel P., Younkin S.G., Wolf P.A., Launer L.J., Lopez O.L., van Duijn C.M., Breteler M.M. CHARGE Consortium; GERAD1 Consortium; EADI1 Consortium. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Hamilton G., Harris S.E., Davies G., Liewald D.C., Tenesa A., Payton A., Horan M.A., Ollier W.E., Pendleton N., Genetic and Environmental Risk for Alzheimer’s Disease (GERAD1) Consortium Starr, J.M.; Porteous, D.; Deary, I.J. The role of ECE1 variants in cognitive ability in old age and Alzheimer’s disease risk. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2012;159B(6):696–709. doi: 10.1002/ajmg.b.32073. [DOI] [PubMed] [Google Scholar]
- 574.Li Y., Seidel K., Marschall P., Klein M., Hope A., Schacherl J., Schmitz J., Menk M., Schefe J.H., Reinemund J., Hugel R., Walden P., Schlosser A., Volkmer R., Schimkus J., Kolsch H., Maier W., Kornhuber J., Frolich L., Klare S., Kirsch S., Schmerbach K., Scheele S., Grittner U., Zollmann F., Goldin-Lang P., Peters O., Kintscher U., Unger T., Funke-Kaiser H. A polymorphic microsatellite repeat within the ECE-1c promoter is involved in transcriptional start site determination, human evolution, and Alzheimer’s disease. J. Neurosci. 2012;32(47):16807–16820. doi: 10.1523/JNEUROSCI.2636-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Hohman T.J., Koran M.E., Thornton-Wells T.A., The Alzheimer’s Disease Neuroimaging Initiative Genetic modification of the relationship between phosphorylated tau and neurodegeneration. Alzheimers Dement. 2014;10(6):637–645. doi: 10.1016/j.jalz.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., Fluder E., Clurman B., Melquist S., Narayanan M., Suver C., Shah H., Mahajan M., Gillis T., Mysore J., MacDonald M.E., Lamb J.R., Bennett D.A., Molony C., Stone D.J., Gudnason V., Myers A.J., Schadt E.E., Neumann H., Zhu J., Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153(3):707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Saad M., Brkanac Z., Wijsman E.M. Family-based genome scan for age at onset of late-onset Alzheimer’s disease in whole exome sequencing data. Genes Brain Behav. 2015;14(8):607–617. doi: 10.1111/gbb.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Wang K.S., Tonarelli S., Luo X., Wang L., Su B., Zuo L., Mao C., Rubin L., Briones D., Xu C. Polymorphisms within ASTN2 gene are associated with age at onset of Alzheimer’s disease. J. Neural Transm. (Vienna) 2015;122(5):701–708. doi: 10.1007/s00702-014-1306-z. [DOI] [PubMed] [Google Scholar]
- 579.Wang K.S., Xu N., Wang L., Aragon L., Ciubuc R., Arana T.B., Mao C., Petty L., Briones D., Su B.B., Luo X., Camarillo C., Escamilla M.A., Xu C. NRG3 gene is associated with the risk and age at onset of Alzheimer disease. J. Neural Transm. (Vienna) 2014;121(2):183–192. doi: 10.1007/s00702-013-1091-0. [DOI] [PubMed] [Google Scholar]
- 580.Huang F., Chotiner J.K., Steward O. Actin polymerization and ERK phosphorylation are required for Arc/Arg3.1 mRNA targeting to activated synaptic sites on dendrites. J. Neurosci. 2007;27(34):9054–9067. doi: 10.1523/JNEUROSCI.2410-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Rao V.R., Pintchovski S.A., Chin J., Peebles C.L., Mitra S., Finkbeiner S. AMPA receptors regulate transcription of the plasticity-related immediate-early gene Arc. Nat. Neurosci. 2006;9(7):887–895. doi: 10.1038/nn1708. [DOI] [PubMed] [Google Scholar]
- 582.Kawashima T., Okuno H., Nonaka M., Adachi-Morishima A., Kyo N., Okamura M., Takemoto-Kimura S., Worley P.F., Bito H. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc. Natl. Acad. Sci. USA. 2009;106(1):316–321. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Pereira A.C., Gray J.D., Kogan J.F., Davidson R.L., Rubin T.G., Okamoto M., Morrison J.H., McEwen B.S. Age and Alzheimer’s disease gene expression profiles reversed by the glutamate modulator riluzole. Mol. Psychiatry. 2017;22(2):296–305. doi: 10.1038/mp.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Lue L.F., Schmitz C., Walker D.G. What happens to microglial TREM2 in Alzheimer’s disease: Immunoregulatory turned into immunopathogenic? Neuroscience. 2015;302:138–150. doi: 10.1016/j.neuroscience.2014.09.050. [DOI] [PubMed] [Google Scholar]
- 585.Lahiri D.K., Maloney B., Zawia N.H. The LEARN model: An epigenetic explanation for idiopathic neurobiological diseases. Mol. Psychiatry. 2009;14(11):992–1003. doi: 10.1038/mp.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Kleefstra T., Schenck A., Kramer J.M., van Bokhoven H. The genetics of cognitive epigenetics. Neuropharmacology. 2014;80:83–94. doi: 10.1016/j.neuropharm.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 587.Tan M., Luo H., Lee S., Jin F., Yang J.S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Wey H.Y., Gilbert T.M., Zurcher N.R., She A., Bhanot A., Taillon B.D., Schroeder F.A., Wang C., Haggarty S.J., Hooker J.M. Insights into neuroepigenetics through human histone deacetylase PET imaging. Sci. Transl. Med. 2016;8(351):351ra106. doi: 10.1126/scitranslmed.aaf7551. http://stm.sciencemag.org/content/8/351/351ra106 [full]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Rudenko A., Tsai L.H. Epigenetic regulation in memory and cognitive disorders. Neuroscience. 2014;264:51–63. doi: 10.1016/j.neuroscience.2012.12.034. [DOI] [PubMed] [Google Scholar]
- 590.Sung Y.M., Lee T., Yoon H., DiBattista A.M., Song J.M., Sohn Y., Moffat E.I., Turner R.S., Jung M., Kim J., Hoe H.S. Mercaptoacetamide-based class II HDAC inhibitor lowers Abeta levels and improves learning and memory in a mouse model of Alzheimer’s disease. Exp. Neurol. 2013;239:192–201. doi: 10.1016/j.expneurol.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Guo X., Wu X., Ren L., Liu G., Li L. Epigenetic mechanisms of amyloid-beta production in anisomycin-treated SH-SY5Y cells. Neuroscience. 2011;194:272–281. doi: 10.1016/j.neuroscience.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 592.Lithner C.U., Lacor P.N., Zhao W.Q., Mustafiz T., Klein W.L., Sweatt J.D., Hernandez C.M. Disruption of neocortical histone H3 homeostasis by soluble Abeta: Implications for Alzheimer’s disease. Neurobiol. Aging. 2013;34(9):2081–2090. doi: 10.1016/j.neurobiolaging.2012.12.028. [DOI] [PubMed] [Google Scholar]
- 593.Marques S.C., Lemos R., Ferreiro E., Martins M., de Mendonca A., Santana I., Outeiro T.F., Pereira C.M. Epigenetic regulation of BACE1 in Alzheimer’s disease patients and in transgenic mice. Neuroscience. 2012;220:256–266. doi: 10.1016/j.neuroscience.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 594.Gräff J., Rei D., Guan J.S., Wang W.Y., Seo J., Hennig K.M., Nieland T.J., Fass D.M., Kao P.F., Kahn M., Su S.C., Samiei A., Joseph N., Haggarty S.J., Delalle I., Tsai L.H. An epigenetic blockade of cognitive functions in the neurodegenerating brain. 2012 doi: 10.1038/nature10849. https://www.nature.com/articles/ [DOI] [PMC free article] [PubMed]
- 595.Guan J.S., Haggarty S.J., Giacometti E., Dannenberg J.H., Joseph N., Gao J., Nieland T.J., Zhou Y., Wang X., Mazitschek R., Bradner J.E., DePinho R.A., Jaenisch R., Tsai L.H. HDAC2 negatively regulates memory formation and synaptic plasticity. 2009 doi: 10.1038/nature07925. https://www.nature.com/articles/ [DOI] [PMC free article] [PubMed]
- 596.Uchida S., Hara K., Kobayashi A., Otsuki K., Yamagata H., Hobara T., Suzuki T., Miyata N., Watanabe Y. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69(2):359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 597.Wang S.C., Oelze B., Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008;3(7):e2698. doi: 10.1371/journal.pone.0002698. http://journals.plos.org/plosone/article?id= 10.1371/journal.pone.0002698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Chouliaras L., Mastroeni D., Delvaux E., Grover A., Kenis G., Hof P.R., Steinbusch H.W., Coleman P.D., Rutten B.P., van den Hove D.L. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol. Aging. 2013;34(9):2091–2099. doi: 10.1016/j.neurobiolaging.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Scarpa S., Cavallaro R.A., D’Anselmi F., Fuso A. Gene silencing through methylation: An epigenetic intervention on Alzheimer disease. J. Alzheimers Dis. 2006;9(4):407–414. doi: 10.3233/jad-2006-9406. [DOI] [PubMed] [Google Scholar]
- 600.Lin H.C., Hsieh H.M., Chen Y.H., Hu M.L. S-Adenosylhomocys- teine increases beta-amyloid formation in BV-2 microglial cells by increased expressions of beta-amyloid precursor protein and presenilin 1 and by hypomethylation of these gene promoters. Neurotoxicology. 2009;30(4):622–627. doi: 10.1016/j.neuro.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 601.Mouton-Liger F., Paquet C., Dumurgier J., Bouras C., Pradier L., Gray F., Hugon J. Oxidative stress increases BACE1 protein levels through activation of the PKR-eIF2alpha pathway. Biochim. Biophys. Acta. 2012;1822(6):885–896. doi: 10.1016/j.bbadis.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 602.Ma T., Trinh M.A., Wexler A.J., Bourbon C., Gatti E., Pierre P., Cavener D.R., Klann E. Suppression of eIF2alpha kinases alleviates Alzheimer’s disease-related plasticity and memory deficits. Nat. Neurosci. 2013;16(9):1299–1305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Saab B.J., Mansuy I.M. Neuroepigenetics of memory formation and impairment: The role of microRNAs. Neuropharmacology. 2014;80:61–69. doi: 10.1016/j.neuropharm.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 604.Yassine H.N., Feng Q., Chiang J., Petrosspour L.M., Fonteh A.N., Chui H.C., Harrington M.G. ABCA1-mediated cholesterol efflux capacity to cerebrospinal fluid is reduced in patients with mild cognitive impairment and Alzheimer’s disease. J. Am. Heart Assoc. 2016;5(2):e002886. doi: 10.1161/JAHA.115.002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Van Cauwenberghe C., Van Broeckhoven C., Sleegers K. The genetic landscape of Alzheimer disease: Clinical implications and perspectives. Genet. Med. 2016;18(5):421–430. doi: 10.1038/gim.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., Russo G., Thorton-Wells T.A., Jones N., Smith A.V., Chouraki V., Thomas C., Ikram M.A., Zelenika D., Vardarajan B.N., Kamatani Y., Lin C.F., Gerrish A., Schmidt H., Kunkle B., Dunstan M.L., Ruiz A., Bihoreau M.T., Choi S.H., Reitz C., Pasquier F., Cruchaga C., Craig D., Amin N., Berr C., Lopez O.L., De Jager P.L., Deramecourt V., Johnston J.A., Evans D., Lovestone S., Letenneur L., Moron F.J., Rubinsztein D.C., Eiriksdottir G., Sleegers K., Goate A.M., Fievet N., Huentelman M.W., Gill M., Brown K., Kamboh M.I., Keller L., Barberger-Gateau P., McGuiness B., Larson E.B., Green R., Myers A.J., Dufouil C., Todd S., Wallon D., Love S., Rogaeva E., Gallacher J., St George-Hyslop P., Clarimon J., Lleo A., Bayer A., Tsuang D.W., Yu L., Tsolaki M., Bossu P., Spalletta G., Proitsi P., Collinge J., Sorbi S., Sanchez-Garcia F., Fox N.C., Hardy J., Deniz Naranjo M.C., Bosco P., Clarke R., Brayne C., Galimberti D., Mancuso M., Matthews F., European Alzheimer’s Disease Initiative Genetic and Environmental Risk in Alzheimer’s Disease; Alzheimer’s Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology; Moebus, S.; Mecocci, P.; Del Zompo, M.; Maier, W.; Hampel, H.; Pilotto, A.; Bullido, M.; Panza, F.; Caffarra, P.; Nacmias, B.; Gilbert, J.R.; Mayhaus, M.; Lannefelt, L.; Hakonarson, H.; Pichler, S.; Carrasquillo, M.M.; Ingelsson, M.; Beekly, D.; Alvarez, V.; Zou, F.; Valladares, O.; Younkin, S.G.; Coto, E.; Hamilton-Nelson, K.L.; Gu, W.; Razquin, C.; Pastor, P.; Mateo, I.; Owen, M.J.; Faber, K.M.; Jonsson, P.V.; Combarros, O.; O’Donovan, M.C.; Cantwell, L.B.; Soininen, H.; Blacker, D.; Mead, S.; Mosley, T.H., Jr.; Bennett, D.A.; Harris, T.B.; Fratiglioni, L.; Holmes, C.; de Bruijn, R.F.; Passmore, P.; Montine, T.J.; Bettens, K.; Rotter, J.I.; Brice, A.; Morgan, K.; Foroud, T.M.; Kukull, W.A.; Hannequin, D.; Powell, J.F.; Nalls, M.A.; Ritchie, K.; Lunetta, K.L.; Kauwe, J.S.; Boerwinkle, E.; Riemenschneider, M.; Boada, M.; Hiltuenen, M.; Martin, E.R.; Schmidt, R.; Rujescu, D.; Wang, L.S.; Dartigues, J.F.; Mayeux, R.; Tzourio, C.; Hofman, A.; Nothen, M.M.; Graff, C.; Psaty, B.M.; Jones, L.; Haines, J.L.; Holmans, P.A.; Lathrop, M.; Pericak-Vance, M.A.; Launer, L.J.; Farrer, L.A.; van Duijn, C.M.; Van Broeckhoven, C.; Moskvina, V.; Seshadri, S.; Williams, J.; Schellenberg, G.D.; Amouyel, P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Kamboh M.I., Demirci F.Y., Wang X., Minster R.L., Carrasquillo M.M., Pankratz V.S., Younkin S.G., Saykin A.J., Alzheimer’s Disease Neuroimaging Initiative Jun, G.; Baldwin, C.; Logue, M.W.; Buros, J.; Farrer, L.; Pericak-Vance, M.A.; Haines, J.L.; Sweet, R.A.; Ganguli, M.; Feingold, E.; Dekosky, S.T.; Lopez, O.L.; Barmada, M.M. Genome-wide association study of Alzheimer’s disease. Transl. Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. https://www.nature.com/articles/tp201245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Allen M., Zou F., Chai H.S., Younkin C.S., Crook J., Pankratz V.S., Carrasquillo M.M., Rowley C.N., Nair A.A., Middha S., Maharjan S., Nguyen T., Ma L., Malphrus K.G., Palusak R., Lincoln S., Bisceglio G., Georgescu C., Schultz D., Rakhshan F., Kolbert C.P., Jen J., Haines J.L., Mayeux R., Pericak-Vance M.A., Farrer L.A., Schellenberg G.D., Petersen R.C., Graff-Radford N.R., Dickson D.W., Younkin S.G., Ertekin-Taner N., Alzheimer’s Disease Genetics Consortium (ADGC) Apostolova, L.G.; Arnold, S.E.; Baldwin, C.T.; Barber, R.; Barmada, M.M.; Beach, T.; Beecham, G.W.; Beekly, D.; Bennett, D.A.; Bigio, E.H.; Bird, T.D.; Blacker, D.; Boeve, B.F.; Bowen, J.D.; Boxer, A.; Burke, J.R.; Buros, J.; Buxbaum, J.D.; Cairns, N.J.; Cantwell, L.B.; Cao, C.; Carlson, C.S.; Carney, R.M.; Carroll, S.L.; Chui, H.C.; Clark, D.G.; Corneveaux, J.; Cotman, C.W.; Crane, P.K.; Cruchaga, C.; Cummings, J.L.; De Jager, P.L.; DeCarli, C.; DeKosky, S.T.; Demirci, F.Y.; Diaz-Arrastia, R.; Dick, M.; Dombroski, B.A.; Duara, R.; Ellis, W.D.; Evans, D.; Faber, K.M.; Fallon, K.B.; Farlow, M.R.; Ferris, S.; Foroud, T.M.; Frosch, M.; Galasko, D.R.; Gallins, P.J.; Ganguli, M.; Gearing, M.; Geschwind, D.H.; Ghetti, B.; Gilbert, J.R.; Gilman, S.; Giordani, B.; Glass, J.D.; Goate, A.M.; Green, R.C.; Growdon, J.H.; Hakonarson, H.; Hamilton, R.L.; Hardy, J.; Harrell, L.E.; Head, E.; Honig, L.S.; Huentelman, M.J.; Hulette, C.M.; Hyman, B.T.; Jarvik, G.P.; Jicha, G.A.; Jin, L.W.; Jun, G.; Kamboh, M.I.; Karlawish, J.; Karydas, A.; Kauwe, J.S.; Kaye, J.A.; Kennedy, N.; Kim, R.; Koo, E.H.; Kowall, N.W.; Kramer, P.; Kukull, W.A.; Lah, J.J.; Larson, E.B.; Levey, A.I.; Lieberman, A.P.; Lopez, O.L.; Lunetta, K.L.; Mack, W.J.; Marson, D.C.; Martin, E.R.; Martiniuk, F.; Mash, D.C.; Masliah, E.; McCormick, W.C.; McCurry, S.M.; McDavid, A.N.; McKee, A.C.; Mesulam, M.; Miller, B.L.; Miller, C.A.; Miller, J.W.; Montine, T.J.; Morris, J.C.; Myers, A.J.; Naj, A.C.; Nowotny, P.; Parisi, J.E.; Perl, D.P.; Peskind, E.; Poon, W.W.; Potter, H.; Quinn, J.F.; Raj, A.; Rajbhandary, R.A.; Raskind, M.; Reiman, E.M.; Reisberg, B.; Reitz, C.; Ringman, J.M.; Roberson, E.D.; Rogaeva, E.; Rosenberg, R.N.; Sano, M.; Saykin, A.J.; Schneider, J.A.; Schneider, L.S.; Seeley, W.; Shelanski, M.L.; Slifer, M.A.; Smith, C.D.; Sonnen, J.A.; Spina, S.; St George-Hyslop, P.; Stern, R.A.; Tanzi, R.E.; Trojanowski, J.Q.; Troncoso, J.C.; Tsuang, D.W.; Van Deerlin, V.M.; Vardarajan, B.N.; Vinters, H.V.; Vonsattel, J.P.; Wang, L.S.; Weintraub, S.; Welsh-Bohmer, K.A.; Williamson, J.; Woltjer, R.L. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology. 2012;79(3):221–228. doi: 10.1212/WNL.0b013e3182605801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Nho K., Corneveaux J.J., Kim S., Lin H., Risacher S.L., Shen L., Swaminathan S., Ramanan V.K., Liu Y., Foroud T., Inlow M.H., Siniard A.L., Reiman R.A., Aisen P.S., Petersen R.C., Green R.C., Jack C.R., Weiner M.W., Baldwin C.T., Lunetta K., Farrer L.A., Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) Study Furney, S.J.; Lovestone, S.; Simmons, A.; Mecocci, P.; Vellas, B.; Tsolaki, M.; Kloszewska, I.; Soininen, H.; AddNeuroMed Consortium; McDonald, B.C.; Farlow, M.R.; Ghetti, B.; Indiana Memory and Aging Study; Huentelman, M.J.; Saykin, A.J.; Alzheimer’s Disease Neuroimaging Initiative (ADNI). Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol. Psychiatry. 2013;18(7):781–787. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Li X., Shen N., Zhang S., Liu J., Jiang Q., Liao M., Feng R., Zhang L., Wang G., Ma G., Zhou H., Chen Z., Jiang Y., Zhao B., Li K., Liu G. CD33 rs3865444 polymorphism contributes to Alzheimer’s disease susceptibility in Chinese, European, and North American populations. Mol. Neurobiol. 2015;52(1):414–421. doi: 10.1007/s12035-014-8880-9. [DOI] [PubMed] [Google Scholar]
- 611.Karch C.M., Goate A.M. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry. 2015;77(1):43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Arias-Vasquez A., Isaacs A., Aulchenko Y.S., Hofman A., Oostra B.A., Breteler M., van Duijn C.M. The cholesteryl ester transfer protein (CETP) gene and the risk of Alzheimer’s disease. Neurogenetics. 2007;8(3):189–193. doi: 10.1007/s10048-007-0089-x. [DOI] [PubMed] [Google Scholar]
- 613.Li Q., Huang P., He Q.C., Lin Q.Z., Wu J., Yin R.X. Association between the CETP polymorphisms and the risk of Alzheimer’s disease, carotid atherosclerosis, longevity, and the efficacy of statin therapy. 2014. [DOI] [PubMed]
- 614.Kim S., Swaminathan S., Inlow M., Risacher S.L., Nho K., Shen L., Foroud T.M., Petersen R.C., Aisen P.S., Soares H., Toledo J.B., Shaw L.M., Trojanowski J.Q., Weiner M.W., McDonald B.C., Farlow M.R., Ghetti B., Saykin A.J., Alzheimer’s Disease Neuroimaging Initiative Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel. PLoS One. 2013;8(7):e70269. doi: 10.1371/journal.pone.0070269. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0070269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Chen L.H., Fan Y.H., Kao P.Y., Ho D.T., Ha J.C., Chu L.W., Song Y.Q. Genetic polymorphisms in estrogen metabolic pathway associated with risks of Alzheimer’s disease: Evidence from a Southern Chinese population. J. Am. Geriatr. Soc. 2017;65(2):332–339. doi: 10.1111/jgs.14537. [DOI] [PubMed] [Google Scholar]
- 616.Han M.R., Schellenberg G.D., Wang L.S., Alzheimer’s Disease Neuroimaging Initiative Genome-wide association reveals genetic effects on human Abeta42 and tau protein levels in cerebrospinal fluids: A case control study. BMC Neurol. 2010;10:90. doi: 10.1186/1471-2377-10-90. https://bmcneurol.biomedcentral.com/articles/10.1186/1471-2377-10-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Li Y., Chu L.W., Wang B., Yik P.Y. Huriletemuer; Jin, D.Y.; Ma, X.; Song, Y.Q. CYP46A1 functional promoter haplotypes decipher genetic susceptibility to Alzheimer’s disease. J. Alzheimers Dis. 2010;21(4):1311–1323. doi: 10.3233/JAD-2010-100765. [DOI] [PubMed] [Google Scholar]
- 618.Kölsch H., Lutjohann D., Jessen F., Popp J., Hentschel F., Kelemen P., Schmitz S., Maier W., Heun R. CYP46A1 variants influence Alzheimer’s disease risk and brain cholesterol metabolism. Eur. Psychiatry. 2009;24(3):183–190. doi: 10.1016/j.eurpsy.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 619.He X.M., Zhang Z.X., Zhang J.W., Zhou Y.T., Wu C.B., Tang M.N., Hong Z. An intronic CYP46A1 polymorphism is associated with Alzheimer disease in a Chinese Han population. J. Mol. Neurosci. 2012;47(3):514–518. doi: 10.1007/s12031-012-9778-5. [DOI] [PubMed] [Google Scholar]
- 620.Simpson J.E., Ince P.G., Minett T., Matthews F.E., Heath P.R., Shaw P.J., Goodall E., Garwood C.J., Ratcliffe L.E., Brayne C., Rattray M., Wharton S.B. MRCC Role and Function; Ageing Neuropathology Study Group. Neuronal DNA damage response-associated dysregulation of signalling pathways and cholesterol metabolism at the earliest stages of Alzheimer-type pathology. Neuropathol. Appl. Neurobiol. 2016;42(2):167–179. doi: 10.1111/nan.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Feher A., Juhasz A., Pakaski M., Kalman J., Janka Z. Gender dependent effect of DHCR24 polymorphism on the risk for Alzheimer’s disease. Neurosci. Lett. 2012;526(1):20–23. doi: 10.1016/j.neulet.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 622.Camargo L.M., Zhang X.D., Loerch P., Caceres R.M., Marine S.D., Uva P., Ferrer M., de Rinaldis E., Stone D.J., Majercak J., Ray W.J., Yi-An C., Shearman M.S., Mizuguchi K. Pathway-based analysis of genome-wide siRNA screens reveals the regulatory landscape of APP processing. PLoS One. 2015;10(2):e0115369. doi: 10.1371/journal.pone.0115369. http://journals.plos.org/plosone/article? id=10.1371/journal.pone.0115369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Wang K.S., Liu X., Xie C., Liu Y., Xu C. Non-parametric survival analysis of EPG5 gene with age at onset of Alzheimer’s Disease. J. Mol. Neurosci. 2016;60(4):436–444. doi: 10.1007/s12031-016-0821-9. [DOI] [PubMed] [Google Scholar]
- 624.Ramanan V.K., Saykin A.J. FASTKD2 and human memory: Functional pathways and prospects for novel therapeutic target development for Alzheimer’s disease and age-associated memory decline. Pharmacogenomics. 2015;16(5):429–432. doi: 10.2217/pgs.15.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Ramanan V.K., Nho K., Shen L., Risacher S.L., Kim S., McDonald B.C., Farlow M.R., Foroud T.M., Gao S., Soininen H., Kloszewska I., Mecocci P., Tsolaki M., Vellas B., Lovestone S., Aisen P.S., Petersen R.C., Jack C.R., Jr, Shaw L.M., Trojanowski J.Q., Weiner M.W., Green R.C., Toga A.W., De Jager P.L., Yu L., Bennett D.A., Saykin A.J. FASTKD2 is associated with memory and hippocampal structure in older adults. Mol. Psychiatry. 2015;20(10):1197–1204. doi: 10.1038/mp.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Sekar S., McDonald J., Cuyugan L., Aldrich J., Kurdoglu A., Adkins J., Serrano G., Beach T.G., Craig D.W., Valla J., Reiman E.M., Liang W.S. Alzheimer’s disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol. Aging. 2015;36(2):583–591. doi: 10.1016/j.neurobiolaging.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Chung S.J., Kim M.J., Kim J., Ryu H.S., Kim Y.J., Kim S.Y., Lee J.H. Association of type 2 diabetes GWAS loci and the risk of Parkinson’s and Alzheimer’s diseases. Parkinsonism Relat. Disord. 2015;21(12):1435–1440. doi: 10.1016/j.parkreldis.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 628.Escott-Price V., Bellenguez C., Wang L.S., Choi S.H., Harold D., Jones L., Holmans P., Gerrish A., Vedernikov A., Richards A., DeStefano A.L., Lambert J.C., Ibrahim-Verbaas C.A., Naj A.C., Sims R., Jun G., Bis J.C., Beecham G.W., Grenier-Boley B., Russo G., Thornton-Wells T.A., Denning N., Smith A.V., Chouraki V., Thomas C., Ikram M.A., Zelenika D., Vardarajan B.N., Kamatani Y., Lin C.F., Schmidt H., Kunkle B., Dunstan M.L., Vronskaya M., United Kingdom Brain Expression Consortium (UKBEC) Johnson, A.D.; Ruiz, A.; Bihoreau, M.T.; Reitz, C.; Pasquier, F.; Hollingworth, P.; Hanon, O.; Fitzpatrick, A.L.; Buxbaum, J.D.; Campion, D.; Crane, P.K.; Baldwin, C.; Becker, T.; Gudnason, V.; Cruchaga, C.; Craig, D.; Amin, N.; Berr, C.; Lopez, O.L.; De Jager, P.L.; Deramecourt, V.; Johnston, J.A.; Evans, D.; Lovestone, S.; Letenneur, L.; Hernandez, I.; Rubinsztein, D.C.; Eiriksdottir, G.; Sleegers, K.; Goate, A.M.; Fievet, N.; Huentelman, M.J.; Gill, M.; Brown, K.; Kamboh, M.I.; Keller, L.; Barberger-Gateau, P.; McGuinness, B.; Larson, E.B.; Myers, A.J.; Dufouil, C.; Todd, S.; Wallon, D.; Love, S.; Rogaeva, E.; Gallacher, J.; George-Hyslop, P.S.; Clarimon, J.; Lleo, A.; Bayer, A.; Tsuang, D.W.; Yu, L.; Tsolaki, M.; Bossu, P.; Spalletta, G.; Proitsi, P.; Collinge, J.; Sorbi, S.; Garcia, F.S.; Fox, N.C.; Hardy, J.; Naranjo, M.C.; Bosco, P.; Clarke, R.; Brayne, C.; Galimberti, D.; Scarpini, E.; Bonuccelli, U.; Mancuso, M.; Siciliano, G.; Moebus, S.; Mecocci, P.; Zompo, M.D.; Maier, W.; Hampel, H.; Pilotto, A.; Frank-Garcia, A.; Panza, F.; Solfrizzi, V.; Caffarra, P.; Nacmias, B.; Perry, W.; Mayhaus, M.; Lannfelt, L.; Hakonarson, H.; Pichler, S.; Carrasquillo, M.M.; Ingelsson, M.; Beekly, D.; Alvarez, V.; Zou, F.; Valladares, O.; Younkin, S.G.; Coto, E.; Hamilton-Nelson, K.L.; Gu, W.; Razquin, C.; Pastor, P.; Mateo, I.; Owen, M.J.; Faber, K.M.; Jonsson, P.V.; Combarros, O.; O’Donovan, M.C.; Cantwell, L.B.; Soininen, H.; Blacker, D.; Mead, S.; Mosley, T.H., Jr.; Bennett, D.A.; Harris, T.B.; Fratiglioni, L.; Holmes, C.; de Bruijn, R.F.; Passmore, P.; Montine, T.J.; Bettens, K.; Rotter, J.I.; Brice, A.; Morgan, K.; Foroud, T.M.; Kukull, W.A.; Hannequin, D.; Powell, J.F.; Nalls, M.A.; Ritchie, K.; Lunetta, K.L.; Kauwe, J.S.; Boerwinkle, E.; Riemenschneider, M.; Boada, M.; Hiltunen, M.; Martin, E.R.; Schmidt, R.; Rujescu, D.; Dartigues, J.F.; Mayeux, R.; Tzourio, C.; Hofman, A.; Nothen, M.M.; Graff, C.; Psaty, B.M.; Haines, J.L.; Lathrop, M.; Pericak-Vance, M.A.; Launer, L.J.; Van Broeckhoven, C.; Farrer, L.A.; van Duijn, C.M.; Ramirez, A.; Seshadri, S.; Schellenberg, G.D.; Amouyel, P.; Williams, J.; Cardiovascular Health Study. Gene-wide analysis detects two new susceptibility genes for Alzheimer’s disease. PLoS One. 2014;9(6):e94661. doi: 10.1371/journal.pone.0094661. http://journals.plos.org/plosone/article? id=10.1371/journal.pone.0094661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Jun G., Ibrahim-Verbaas C.A., Vronskaya M., Lambert J.C., Chung J., Naj A.C., Kunkle B.W., Wang L.S., Bis J.C., Bellenguez C., Harold D., Lunetta K.L., Destefano A.L., Grenier-Boley B., Sims R., Beecham G.W., Smith A.V., Chouraki V., Hamilton-Nelson K.L., Ikram M.A., Fievet N., Denning N., Martin E.R., Schmidt H., Kamatani Y., Dunstan M.L., Valladares O., Laza A.R., Zelenika D., Ramirez A., Foroud T.M., Choi S.H., Boland A., Becker T., Kukull W.A., van der Lee S.J., Pasquier F., Cruchaga C., Beekly D., Fitzpatrick A.L., Hanon O., Gill M., Barber R., Gudnason V., Campion D., Love S., Bennett D.A., Amin N., Berr C., Tsolaki M., Buxbaum J.D., Lopez O.L., Deramecourt V., Fox N.C., Cantwell L.B., Tarraga L., Dufouil C., Hardy J., Crane P.K., Eiriksdottir G., Hannequin D., Clarke R., Evans D., Mosley T.H., Jr, Letenneur L., Brayne C., Maier W., De Jager P., Emilsson V., Dartigues J.F., Hampel H., Kamboh M.I., de Bruijn R.F., Tzourio C., Pastor P., Larson E.B., Rotter J.I., O’Donovan M.C., Montine T.J., Nalls M.A., Mead S., Reiman E.M., Jonsson P.V., Holmes C., St George-Hyslop P.H., Boada M., Passmore P., Wendland J.R., Schmidt R., Morgan K., Winslow A.R., Powell J.F., Carasquillo M., Younkin S.G., Jakobsdottir J., Kauwe J.S., Wilhelmsen K.C., Rujescu D., Nothen M.M., Hofman A., Jones L. Indus Consortium; Haines, J.L.; Psaty, B.M.; Van Broeckhoven, C.; Holmans, P.; Launer, L.J.; Mayeux, R.; Lathrop, M.; Goate, A.M.; Escott-Price, V.; Seshadri, S.; Pericak-Vance, M.A.; Amouyel, P.; Williams, J.; van Duijn, C.M.; Schellenberg, G.D.; Farrer, L.A. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol. Psychiatry. 2016;21(1):108–117. doi: 10.1038/mp.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Jiang S., Yang W., Qiu Y., Chen H.Z., Alzheimer’s Disease Neuroimaging Initiative Identification of novel quantitative traits-associated susceptibility loci for APOE epsilon 4 non-carriers of Alzheimer’s disease. Curr. Alzheimer Res. 2015;12(3):218–227. doi: 10.2174/1567205012666150302160145. [DOI] [PubMed] [Google Scholar]
- 631.Del Villar K., Miller C.A. Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer’s disease brain and hippocampal neurons. Proc. Natl. Acad. Sci. USA. 2004;101(12):4210–4215. doi: 10.1073/pnas.0307349101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Dourlen P., Fernandez-Gomez F.J., Dupont C., Grenier-Boley B., Bellenguez C., Obriot H., Caillierez R., Sottejeau Y., Chapuis J., Bretteville A., Abdelfettah F., Delay C., Malmanche N., Soininen H., Hiltunen M., Galas M.C., Amouyel P., Sergeant N., Buee L., Lambert J.C., Dermaut B. Functional screening of Alzheimer risk loci identifies PTK2B as an in vivo modulator and early marker of Tau pathology. Mol. Psychiatry. 2017;22(6):874–883. doi: 10.1038/mp.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Wang D.S., Iwata N., Hama E., Saido T.C., Dickson D.W. Oxidized neprilysin in aging and Alzheimer’s disease brains. Biochem. Biophys. Res. Commun. 2003;310(1):236–241. doi: 10.1016/j.bbrc.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 634.Bai Z., Stamova B., Xu H., Ander B.P., Wang J., Jickling G.C., Zhan X., Liu D., Han G., Jin L.W., DeCarli C., Lei H., Sharp F.R. Distinctive RNA expression profiles in blood associated with Alzheimer disease after accounting for white matter hyperintensities. Alzheimer Dis. Assoc. Disord. 2014;28(3):226–233. doi: 10.1097/WAD.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., Larson E.B., Bird T.D., Boeve B.F., Graff-Radford N.R., De Jager P.L., Evans D., Schneider J.A., Carrasquillo M.M., Ertekin-Taner N., Younkin S.G., Cruchaga C., Kauwe J.S., Nowotny P., Kramer P., Hardy J., Huentelman M.J., Myers A.J., Barmada M.M., Demirci F.Y., Baldwin C.T., Green R.C., Rogaeva E., St George-Hyslop P., Arnold S.E., Barber R., Beach T., Bigio E.H., Bowen J.D., Boxer A., Burke J.R., Cairns N.J., Carlson C.S., Carney R.M., Carroll S.L., Chui H.C., Clark D.G., Corneveaux J., Cotman C.W., Cummings J.L., DeCarli C., DeKosky S.T., Diaz-Arrastia R., Dick M., Dickson D.W., Ellis W.G., Faber K.M., Fallon K.B., Farlow M.R., Ferris S., Frosch M.P., Galasko D.R., Ganguli M., Gearing M., Geschwind D.H., Ghetti B., Gilbert J.R., Gilman S., Giordani B., Glass J.D., Growdon J.H., Hamilton R.L., Harrell L.E., Head E., Honig L.S., Hulette C.M., Hyman B.T., Jicha G.A., Jin L.W., Johnson N., Karlawish J., Karydas A., Kaye J.A., Kim R., Koo E.H., Kowall N.W., Lah J.J., Levey A.I., Lieberman A.P., Lopez O.L., Mack W.J., Marson D.C., Martiniuk F., Mash D.C., Masliah E., McCormick W.C., McCurry S.M., McDavid A.N., McKee A.C., Mesulam M., Miller B.L., Miller C.A., Miller J.W., Parisi J.E., Perl D.P., Peskind E., Petersen R.C., Poon W.W., Quinn J.F., Rajbhandary R.A., Raskind M., Reisberg B., Ringman J.M., Roberson E.D., Rosenberg R.N., Sano M., Schneider L.S., Seeley W., Shelanski M.L., Slifer M.A., Smith C.D., Sonnen J.A., Spina S., Stern R.A., Tanzi R.E., Trojanowski J.Q., Troncoso J.C., Van Deerlin V.M., Vinters H.V., Vonsattel J.P., Weintraub S., Welsh-Bohmer K.A., Williamson J., Woltjer R.L., Cantwell L.B., Dombroski B.A., Beekly D., Lunetta K.L., Martin E.R., Kamboh M.I., Saykin A.J., Reiman E.M., Bennett D.A., Morris J.C., Montine T.J., Goate A.M., Blacker D., Tsuang D.W., Hakonarson H., Kukull W.A., Foroud T.M., Haines J.L., Mayeux R., Pericak-Vance M.A., Farrer L.A., Schellenberg G.D. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 2011;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Sarajarvi T., Marttinen M., Natunen T., Kauppinen T., Makinen P., Helisalmi S., Laitinen M., Rauramaa T., Leinonen V., Petaja-Repo U., Soininen H., Haapasalo A., Hiltunen M. Genetic variation in delta-opioid receptor associates with increased beta- and gamma-secretase activity in the late stages of Alzheimer’s disease. J. Alzheimers Dis. 2015;48(2):507–516. doi: 10.3233/JAD-150221. [DOI] [PubMed] [Google Scholar]
- 637.Sarajarvi T., Tuusa J.T., Haapasalo A., Lackman J.J., Sormunen R., Helisalmi S., Roehr J.T., Parrado A.R., Makinen P., Bertram L., Soininen H., Tanzi R.E., Petaja-Repo U.E., Hiltunen M. Cysteine 27 variant of the delta-opioid receptor affects amyloid precursor protein processing through altered endocytic trafficking. Mol. Cell. Biol. 2011;31(11):2326–2340. doi: 10.1128/MCB.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Roussotte F.F., Jahanshad N., Hibar D.P., Sowell E.R., Kohannim O., Barysheva M., Hansell N.K., McMahon K.L., de Zubicaray G.I., Montgomery G.W., Martin N.G., Wright M.J., Toga A.W., Jack C.R., Jr, Weiner M.W., Thompson P.M., Alzheimer’s Disease Neuroimaging Initiative A commonly carried genetic variant in the delta opioid receptor gene, OPRD1, is associated with smaller regional brain volumes: Replication in elderly and young populations. Hum. Brain Mapp. 2014;35(4):1226–1236. doi: 10.1002/hbm.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Ataman B., Boulting G.L., Harmin D.A., Yang M.G., Baker-Salisbury M., Yap E.L., Malik A.N., Mei K., Rubin A.A., Spiegel I., Durresi E., Sharma N., Hu L.S., Pletikos M., Griffith E.C., Partlow J.N., Stevens C.R., Adli M., Chahrour M., Sestan N., Walsh C.A., Berezovskii V.K., Livingstone M.S., Greenberg M.E. Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature. 2016;539(7628):242–247. doi: 10.1038/nature20111. https://www.nature.com/articles/nature20111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Furney S.J., Simmons A., Breen G., Pedroso I., Lunnon K., Proitsi P., Hodges A., Powell J., Wahlund L.O., Kloszewska I., Mecocci P., Soininen H., Tsolaki M., Vellas B., Spenger C., Lathrop M., Shen L., Kim S., Saykin A.J., Weiner M.W., Lovestone S., Alzheimer’s Disease Neuroimaging Initiative AddNeuroMed Consortium. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol. Psychiatry. 2011;16(11):1130–1138. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Cruchaga C., Kauwe J.S., Mayo K., Spiegel N., Bertelsen S., Nowotny P., Shah A.R., Abraham R., Hollingworth P., Harold D., Owen M.M., Williams J., Lovestone S., Peskind E.R., Li G., Leverenz J.B., Galasko D., Alzheimer’s Disease Neuroimaging Initiative 2010 http://journals.plos.org/plosgenetics/article?id=10
- 642.Peterson D., Munger C., Crowley J., Corcoran C., Cruchaga C., Goate A.M., Norton M.C., Green R.C., Munger R.G., Breitner J.C., Welsh-Bohmer K.A., Lyketsos C., Tschanz J., Kauwe J.S., Alzheimer’s Disease Neuroimaging Initiative Variants in PPP3R1 and MAPT are associated with more rapid functional decline in Alzheimer’s disease: The Cache County dementia progression study. Alzheimers Dement. 2014;10(3):366–371. doi: 10.1016/j.jalz.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Lu T., Aron L., Zullo J., Pan Y., Kim H., Chen Y., Yang T.H., Kim H.M., Drake D., Liu X.S., Bennett D.A., Colaiacovo M.P., Yankner B.A. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507(7493):448–454. doi: 10.1038/nature13163. https://www.nature.com/articles/nature13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Atherton J., Kurbatskaya K., Bondulich M., Croft C.L., Garwood C.J., Chhabra R., Wray S., Jeromin A., Hanger D.P., Noble W. Calpain cleavage and inactivation of the sodium calcium exchanger-3 occur downstream of Abeta in Alzheimer’s disease. Aging Cell. 2014;13(1):49–59. doi: 10.1111/acel.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Vardarajan B.N., Bruesegem S.Y., Harbour M.E., Inzelberg R., Friedland R., St George-Hyslop P., Seaman M.N., Farrer L.A. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol. Aging. 2012;33(9):2231.e15–2231.e30. doi: 10.1016/j.neurobiolaging.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.Vardarajan B.N., Zhang Y., Lee J.H., Cheng R., Bohm C., Ghani M., Reitz C., Reyes-Dumeyer D., Shen Y., Rogaeva E., St George-Hyslop P., Mayeux R. Coding mutations in SORL1 and Alzheimer disease. Ann. Neurol. 2015;77(2):215–227. doi: 10.1002/ana.24305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Jahanshad N., Rajagopalan P., Hua X., Hibar D.P., Nir T.M., Toga A.W., Jack C.R., Jr, Saykin A.J., Green R.C., Weiner M.W., Medland S.E., Montgomery G.W., Hansell N.K., McMahon K.L., de Zubicaray G.I., Martin N.G., Wright M.J., Thompson P.M., Alzheimer’s Disease Neuroimaging Initiative Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proc. Natl. Acad. Sci. USA. 2013;110(12):4768–4773. doi: 10.1073/pnas.1216206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Sherva R., Tripodis Y., Bennett D.A., Chibnik L.B., Crane P.K., de Jager P.L., Farrer L.A., Saykin A.J., Shulman J.M., Naj A., Green R.C. Genetics Consortium; Alzheimer’s Disease Neuroimaging Initiative; Alzheimer’s Disease Genetics Consortium. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimers Dement. 2014;10(1):45–52. doi: 10.1016/j.jalz.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Scarabino D., Broggio E., Gambina G., Pelliccia F., Corbo R.M. Common variants of human TERT and TERC genes and susceptibility to sporadic Alzheimers disease. Exp. Gerontol. 2017;88:19–24. doi: 10.1016/j.exger.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 650.Kalpouzos G., Rizzuto D., Keller L., Fastbom J., Santoni G., Angleman S., Graff C., Backman L., Fratiglioni L. Telomerase gene (hTERT) and survival: Results from two Swedish cohorts of older adults. J. Gerontol. A Biol. Sci. Med. Sci. 2016;71(2):188–195. doi: 10.1093/gerona/glu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651.Roussos P., Katsel P., Fam P., Tan W., Purohit D.P., Haroutunian V. The triggering receptor expressed on myeloid cells 2 (TREM2) is associated with enhanced inflammation, neuropathological lesions and increased risk for Alzheimer’s dementia. Alzheimers Dement. 2015;11(10):1163–1170. doi: 10.1016/j.jalz.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Melville S.A., Buros J., Parrado A.R., Vardarajan B., Logue M.W., Shen L., Risacher S.L., Kim S., Jun G., DeCarli C., Lunetta K.L., Baldwin C.T., Saykin A.J., Farrer L.A., Alzheimer’s Disease Neuroimaging Initiative Multiple loci influencing hippocampal degeneration identified by genome scan. Ann. Neurol. 2012;72(1):65–75. doi: 10.1002/ana.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Kim J.H., Song P., Lim H., Lee J.H., Lee J.H., Park S.A., Alzheimer’s Disease Neuroimaging Initiative Gene-based rare allele analysis identified a risk gene of Alzheimer’s disease. PLoS One. 2014;9(10):e107983. doi: 10.1371/journal.pone.0107983. http://journals.plos.org/plosone/article? id=10.1371/journal.pone.0107983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Zappia M., Manna I., Serra P., Cittadella R., Andreoli V., La Russa A., Annesi F., Spadafora P., Romeo N., Nicoletti G., Messina D., Gambardella A., Quattrone A. Increased risk for Alzheimer disease with the interaction of MPO and A2M polymorphisms. Arch. Neurol. 2004;61(3):341–344. doi: 10.1001/archneur.61.3.341. [DOI] [PubMed] [Google Scholar]
- 655.Beecham G.W., Hamilton K., Naj A.C., Martin E.R., Huentelman M., Myers A.J., Corneveaux J.J., Hardy J., Vonsattel J.P., Younkin S.G., Bennett D.A., De Jager P.L., Larson E.B., Crane P.K., Kamboh M.I., Kofler J.K., Mash D.C., Duque L., Gilbert J.R., Gwirtsman H., Buxbaum J.D., Kramer P., Dickson D.W., Farrer L.A., Frosch M.P., Ghetti B., Haines J.L., Hyman B.T., Kukull W.A., Mayeux R.P., Pericak-Vance M.A., Schneider J.A., Trojanowski J.Q., Reiman E.M., Alzheimer’s Disease Genetics Consortium Schellenberg, G.D.; Montine, T.J. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10(9):e1004606. doi: 10.1371/journal.pgen.1004606. http://journals.plos.org/plosgenetics/article? id=10.1371/journal.pgen.1004606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Davies G., Armstrong N., Bis J.C., Bressler J., Chouraki V., Giddaluru S., Hofer E., Ibrahim-Verbaas C.A., Kirin M., Lahti J., van der Lee S.J., Le Hellard S., Liu T., Marioni R.E., Oldmeadow C., Postmus I., Smith A.V., Smith J.A., Thalamuthu A., Thomson R., Vitart V., Wang J., Yu L., Zgaga L., Zhao W., Boxall R., Harris S.E., Hill W.D., Liewald D.C., Luciano M., Adams H., Ames D., Amin N., Amouyel P., Assareh A.A., Au R., Becker J.T., Beiser A., Berr C., Bertram L., Boerwinkle E., Buckley B.M., Campbell H., Corley J., De Jager P.L., Dufouil C., Eriksson J.G., Espeseth T., Faul J.D., Ford I., Generation S., Gottesman R.F., Griswold M.E., Gudnason V., Harris T.B., Heiss G., Hofman A., Holliday E.G., Huffman J., Kardia S.L., Kochan N., Knopman D.S., Kwok J.B., Lambert J.C., Lee T., Li G., Li S.C., Loitfelder M., Lopez O.L., Lundervold A.J., Lundqvist A., Mather K.A., Mirza S.S., Nyberg L., Oostra B.A., Palotie A., Papenberg G., Pattie A., Petrovic K., Polasek O., Psaty B.M., Redmond P., Reppermund S., Rotter J.I., Schmidt H., Schuur M., Schofield P.W., Scott R.J., Steen V.M., Stott D.J., van Swieten J.C., Taylor K.D., Trollor J., Trompet S., Uitterlinden A.G., Weinstein G., Widen E., Windham B.G., Jukema J.W., Wright A.F., Wright M.J., Yang Q., Amieva H., Attia J.R., Bennett D.A., Brodaty H., de Craen A.J., Hayward C., Ikram M.A., Lindenberger U., Nilsson L.G., Porteous D.J., Raikkonen K., Reinvang I., Rudan I., Sachdev P.S., Schmidt R., Schofield P.R., Srikanth V., Starr J.M., Turner S.T., Weir D.R., Wilson J.F., van Duijn C., Launer L., Fitzpatrick A.L., Seshadri S., Mosley T.H., Jr, Deary I.J. Genetic contributions to variation in general cognitive function: A meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Mol. Psychiatry. 2015;20(2):183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Koutroumani M., Daniilidou M., Giannakouros T., Proitsi P., Liapi D., Germanou A., Nikolakaki E., Tsolaki M. The deletion variant of alpha2b-adrenergic receptor is associated with decreased risk in Alzheimer’s disease and mild cognitive impairment. J. Neurol. Sci. 2013;328(1-2):19–23. doi: 10.1016/j.jns.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 658.Hong C.J., Wang Y.C., Liu T.Y., Liu H.C., Tsai S.J. A study of alpha-adrenoceptor gene polymorphisms and Alzheimer disease. J. Neural Transm. (Vienna) 2001;108(4):445–450. doi: 10.1007/s007020170065. [DOI] [PubMed] [Google Scholar]
- 659.Nalls M.A., Guerreiro R.J., Simon-Sanchez J., Bras J.T., Traynor B.J., Gibbs J.R., Launer L., Hardy J., Singleton A.B. Extended tracts of homozygosity identify novel candidate genes associated with late-onset Alzheimer’s disease. Neurogenetics. 2009;10(3):183–190. doi: 10.1007/s10048-009-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Bennet A.M., Reynolds C.A., Eriksson U.K., Hong M.G., Blennow K., Gatz M., Alexeyenko A., Pedersen N.L., Prince J.A. Genetic association of sequence variants near AGER/NOTCH4 and dementia. J. Alzheimers Dis. 2011;24(3):475–484. doi: 10.3233/JAD-2011-101848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Sery O., Hlinecka L., Povova J., Bonczek O., Zeman T., Janout V., Ambroz P., Khan N.A., Balcar V.J. Arachidonate 5-lipoxygenase (ALOX5) gene polymorphism is associated with Alzheimer’s disease and body mass index. J. Neurol. Sci. 2016;362:27–32. doi: 10.1016/j.jns.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 662.Li Y., Hollingworth P., Moore P., Foy C., Archer N., Powell J., Nowotny P., Holmans P., O’Donovan M., Tacey K., Doil L., van Luchene R., Garcia V., Rowland C., Lau K., Cantanese J., Sninsky J., Hardy J., Thal L., Morris J.C., Goate A., Lovestone S., Owen M., Williams J., Grupe A. Genetic association of the APP binding protein 2 gene (APBB2) with late onset Alzheimer disease. Hum. Mutat. 2005;25(3):270–277. doi: 10.1002/humu.20138. [DOI] [PubMed] [Google Scholar]
- 663.Cruchaga C., Haller G., Chakraverty S., Mayo K., Vallania F.L., Mitra R.D., Faber K., Williamson J., Bird T., Diaz-Arrastia R., Foroud T.M., Boeve B.F., Graff-Radford N.R., St Jean P., Lawson M., Ehm M.G., Mayeux R., Goate A.M., NIA-LOAD/NCRAD Family Study Consortium Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PLoS One. 2012;7(2):e31039. doi: 10.1371/journal.pone.0031039. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0031039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Landgren S., von Otter M., Palmer M.S., Zetterstrom C., Nilsson S., Skoog I., Gustafson D.R., Minthon L., Wallin A., Andreasen N., Bogdanovic N., Marcusson J., Blennow K., Zetterberg H., Kettunen P. A novel ARC gene polymorphism is associated with reduced risk of Alzheimer’s disease. J. Neural Transm. (Vienna) 2012;119(7):833–842. doi: 10.1007/s00702-012-0823-x. [DOI] [PubMed] [Google Scholar]
- 665.Uchida Y. Molecular mechanisms of regeneration in Alzheimer’s disease brain. Geriatr. Gerontol. Int. 2010;10(Suppl. 1):S158–S168. doi: 10.1111/j.1447-0594.2010.00607.x. [DOI] [PubMed] [Google Scholar]
- 666.Gold G., Blouin J.L., Herrmann F.R., Michon A., Mulligan R., Duriaux Sail G., Bouras C., Giannakopoulos P., Antonarakis S.E. Specific BACE1 genotypes provide additional risk for late-onset Alzheimer disease in APOE epsilon 4 carriers. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2003;119B(1):44–47. doi: 10.1002/ajmg.b.10010. [DOI] [PubMed] [Google Scholar]
- 667.Jo S.A., Ahn K., Kim E., Kim H.S., Jo I., Kim D.K., Han C., Park M.H. Association of BACE1 gene polymorphism with Alzheimer’s disease in Asian populations: Meta-analysis including Korean samples. Dement. Geriatr. Cogn. Disord. 2008;25(2):165–169. doi: 10.1159/000112918. [DOI] [PubMed] [Google Scholar]
- 668.Sajan F.D., Martiniuk F., Marcus D.L., Frey W.H., II, Hite R., Bordayo E.Z., Freedman M.L. Apoptotic gene expression in Alzheimer’s disease hippocampal tissue. Am. J. Alzheimers Dis. Other Demen. 2007;22(4):319–328. doi: 10.1177/1533317507302447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 669.Ghisletta P., Backman L., Bertram L., Brandmaier A.M., Gerstorf D., Liu T., Lindenberger U. The Val/Met polymorphism of the brain-derived neurotrophic factor (BDNF) gene predicts decline in perceptual speed in older adults. Psychol. Aging. 2014;29(2):384–392. doi: 10.1037/a0035201. [DOI] [PubMed] [Google Scholar]
- 670.Lin Y., Cheng S., Xie Z., Zhang D. Association of rs6265 and rs2030324 polymorphisms in brain-derived neurotrophic factor gene with Alzheimer’s disease: A meta-analysis. 2014 doi: 10.1371/journal.pone.0094961. http://journals.plos.org/plosone/article? id=10 [DOI] [PMC free article] [PubMed]
- 671.Vazquez-Higuera J.L., Mateo I., Sanchez-Juan P., Rodriguez E., Pozueta A., Calero M., Dobato J.L., Frank-Garcia A., Valdivieso F., Berciano J., Bullido M.J., Combarros O. Genetic variation in the tau kinases pathway may modify the risk and age at onset of Alzheimer’s disease. J. Alzheimers Dis. 2011;27(2):291–297. doi: 10.3233/JAD-2011-110794. [DOI] [PubMed] [Google Scholar]
- 672.Silva P.N., Furuya T.K., Sampaio Braga I., Rasmussen L.T., de Labio R.W., Bertolucci P.H., Chen E.S., Turecki G., Mechawar N., Payao S.L., Mill J., Smith M.C. CNP and DPYSL2 mRNA expression and promoter methylation levels in brain of Alzheimer’s disease patients. J. Alzheimers Dis. 2013;33(2):349–355. doi: 10.3233/JAD-2012-121192. [DOI] [PubMed] [Google Scholar]
- 673.de Jong S., Chepelev I., Janson E., Strengman E., van den Berg L.H., Veldink J.H., Ophoff R.A. Common inversion polymorphism at 17q21.31 affects expression of multiple genes in tissue-specific manner. BMC Genomics. 2012;13:458. doi: 10.1186/1471-2164-13-458. https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-13-458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Kok E.H., Alanne-Kinnunen M., Isotalo K., Luoto T., Haikonen S., Goebeler S., Perola M., Hurme M.A., Haapasalo H., Karhunen P.J. CRP gene variation affects early development of Alzheimer’s disease-related plaques. J. Neuroinflammation. 2011;8:96. doi: 10.1186/1742-2094-8-96. https://jneuroinflammation.biomedcentral.com/ articles/10.1186/1742-2094-8-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.Sung H.Y., Choi E.N., Jo S.A., Oh S., Ahn J.H. Amyloid protein-mediated differential DNA methylation status regulates gene expression in Alzheimer’s disease model cell line. Biochem. Biophys. Res. Commun. 2011;414(4):700–705. doi: 10.1016/j.bbrc.2011.09.136. [DOI] [PubMed] [Google Scholar]
- 676.Yuki D., Sugiura Y., Zaima N., Akatsu H., Takei S., Yao I., Maesako M., Kinoshita A., Yamamoto T., Kon R., Sugiyama K., Setou M. DHA-PC and PSD-95 decrease after loss of synaptophysin and before neuronal loss in patients with Alzheimer’s disease. Sci. Rep. 2014;4:7130. doi: 10.1038/srep07130. https:// www.nature.com/articles/srep07130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677.Klein C.J., Bird T., Ertekin-Taner N., Lincoln S., Hjorth R., Wu Y., Kwok J., Mer G., Dyck P.J., Nicholson G.A. DNMT1 mutation hot spot causes varied phenotypes of HSAN1 with dementia and hearing loss. Neurology. 2013;80(9):824–828. doi: 10.1212/WNL.0b013e318284076d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Ling C., Fangyu D., Wanhua H., Kelong C., Zhimin W., Yuting Z., Rong Z. DNMT3A rs1550117 polymorphism is associated with late-onset Alzheimer’s disease in a Chinese population. Am. J. Alzheimers Dis. Other Demen. 2016;31(3):278–281. doi: 10.1177/1533317515603688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 679.Pezzi J.C., Ens C.M., Borba E.M., Schumacher-Schuh A.F., de Andrade F.M., Chaves M.L., Fiegenbaum M., Camozzato A.L. DNA methyltransferase haplotype is associated with Alzheimer’s disease. Neurosci. Lett. 2014;579:70–74. doi: 10.1016/j.neulet.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 680.Kimura R., Kamino K., Yamamoto M., Nuripa A., Kida T., Kazui H., Hashimoto R., Tanaka T., Kudo T., Yamagata H., Tabara Y., Miki T., Akatsu H., Kosaka K., Funakoshi E., Nishitomi K., Sakaguchi G., Kato A., Hattori H., Uema T., Takeda M. The DYRK1A gene, encoded in chromosome 21 Down syndrome critical region, bridges between beta-amyloid production and tau phosphorylation in Alzheimer disease. Hum. Mol. Genet. 2007;16(1):15–23. doi: 10.1093/hmg/ddl437. [DOI] [PubMed] [Google Scholar]
- 681.Woltjer R.L., Duerson K., Fullmer J.M., Mookherjee P., Ryan A.M., Montine T.J., Kaye J.A., Quinn J.F., Silbert L., Erten-Lyons D., Leverenz J.B., Bird T.D., Pow D.V., Tanaka K., Watson G.S., Cook D.G. Aberrant detergent-insoluble excitatory amino acid transporter 2 accumulates in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2010;69(7):667–676. doi: 10.1097/NEN.0b013e3181e24adb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Thomas T., Miners S., Love S. Post-mortem assessment of hypoperfusion of cerebral cortex in Alzheimer’s disease and vascular dementia. Brain. 2015;138(Pt 4):1059–1069. doi: 10.1093/brain/awv025. [DOI] [PubMed] [Google Scholar]
- 683.Zhu Q.B., Unmehopa U., Bossers K., Hu Y.T., Verwer R., Balesar R., Zhao J., Bao A.M., Swaab D. MicroRNA-132 and early growth response-1 in nucleus basalis of Meynert during the course of Alzheimer’s disease. Brain. 2016;139(Pt 3):908–921. doi: 10.1093/brain/awv383. [DOI] [PubMed] [Google Scholar]
- 684.Bullido M.J., Martinez-Garcia A., Tenorio R., Sastre I., Munoz D.G., Frank A., Valdivieso F. Double stranded RNA activated EIF2 alpha kinase (EIF2AK2; PKR) is associated with Alzheimer’s disease. Neurobiol. Aging. 2008;29(8):1160–1166. doi: 10.1016/j.neurobiolaging.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 685.Giustiniani J., Sineus M., Sardin E., Dounane O., Panchal M., Sazdovitch V., Duyckaerts C., Chambraud B., Baulieu E.E. Decrease of the immunophilin FKBP52 accumulation in human brains of Alzheimer’s disease and FTDP-17. J. Alzheimers Dis. 2012;29(2):471–483. doi: 10.3233/JAD-2011-111895. [DOI] [PubMed] [Google Scholar]
- 686.Blair L.J., Nordhues B.A., Hill S.E., Scaglione K.M., O’Leary J.C., III, Fontaine S.N., Breydo L., Zhang B., Li P., Wang L., Cotman C., Paulson H.L., Muschol M., Uversky V.N., Klengel T., Binder E.B., Kayed R., Golde T.E., Berchtold N., Dickey C.A. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Invest. 2013;123(10):4158–4169. doi: 10.1172/JCI69003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Ghanim H., Monte S.V., Sia C.L., Abuaysheh S., Green K., Caruana J.A., Dandona P. Reduction in inflammation and the expression of amyloid precursor protein and other proteins related to Alzheimer’s disease following gastric bypass surgery. J. Clin. Endocrinol. Metab. 2012;97(7):E1197–E1201. doi: 10.1210/jc.2011-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 688.Schjeide B.M., Hooli B., Parkinson M., Hogan M.F., DiVito J., Mullin K., Blacker D., Tanzi R.E., Bertram L. GAB2 as an Alzheimer disease susceptibility gene: Follow-up of genomewide association results. Arch. Neurol. 2009;66(2):250–254. doi: 10.1001/archneurol.2008.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.Wang G., Pan X.L., Cui P.J., Wang Y., Ma J.F., Ren R.J., Deng Y.L., Xu W., Tang H.D., Chen S.D. Association study of the GAB2 gene with the risk of Alzheimer disease in the Chinese population. Alzheimer Dis. Assoc. Disord. 2011;25(3):283–285. doi: 10.1097/WAD.0b013e318209e63a. [DOI] [PubMed] [Google Scholar]
- 690.Jin C., Wu C.Z., Liu X., Zhang F., Tian L., Yuan J., Wang G., Cheng Z. GAB2 polymorphism rs2373115 confers susceptibility to sporadic Alzheimer’s disease. Neurosci. Lett. 2013;556:216–220. doi: 10.1016/j.neulet.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 691.Schwab C., Yu S., Wong W., McGeer E.G., McGeer P.L. GAD65, GAD67, and GABAT immunostaining in human brain and apparent GAD65 loss in Alzheimer’s disease. J. Alzheimers Dis. 2013;33(4):1073–1088. doi: 10.3233/JAD-2012-121330. [DOI] [PubMed] [Google Scholar]
- 692.Airavaara M., Pletnikova O., Doyle M.E., Zhang Y.E., Troncoso J.C., Liu Q.R. Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J. Biol. Chem. 2011;286(52):45093–45102. doi: 10.1074/jbc.M111.310250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693.Conejero-Goldberg C., Hyde T.M., Chen S., Dreses-Werringloer U., Herman M.M., Kleinman J.E., Davies P., Goldberg T.E. Molecular signatures in post-mortem brain tissue of younger individuals at high risk for Alzheimer’s disease as based on APOE genotype. Mol. Psychiatry. 2011;16(8):836–847. doi: 10.1038/mp.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Jacob C.P., Koutsilieri E., Bartl J., Neuen-Jacob E., Arzberger T., Zander N., Ravid R., Roggendorf W., Riederer P., Grunblatt E. Alterations in expression of glutamatergic transporters and receptors in sporadic Alzheimer’s disease. J. Alzheimers Dis. 2007;11(1):97–116. doi: 10.3233/jad-2007-11113. [DOI] [PubMed] [Google Scholar]
- 695.Hohman T.J., Bush W.S., Jiang L., Brown-Gentry K.D., Torstenson E.S., Dudek S.M., Mukherjee S., Naj A., Kunkle B.W., Ritchie M.D., Martin E.R., Schellenberg G.D., Mayeux R., Farrer L.A., Pericak-Vance M.A., Haines J.L., Thornton-Wells T.A., Alzheimer’s Disease Genetics Consortium Discovery of gene-gene interactions across multiple independent data sets of late onset Alzheimer disease from the Alzheimer Disease Genetics Consortium. Neurobiol. Aging. 2016;38:141–150. doi: 10.1016/j.neurobiolaging.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 696.Um J.W., Kaufman A.C., Kostylev M., Heiss J.K., Stagi M., Takahashi H., Kerrisk M.E., Vortmeyer A., Wisniewski T., Koleske A.J., Gunther E.C., Nygaard H.B., Strittmatter S.M. Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer abeta oligomer bound to cellular prion protein. Neuron. 2013;79(5):887–902. doi: 10.1016/j.neuron.2013.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Lauren J., Gimbel D.A., Nygaard H.B., Gilbert J.W., Strittmatter S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. 2009 doi: 10.1038/nature07761. https://www.nature.com/articles/nature [DOI] [PMC free article] [PubMed]
- 698.Nelson P.T., Wang W.X., Partch A.B., Monsell S.E., Valladares O., Ellingson S.R., Wilfred B.R., Naj A.C., Wang L.S., Kukull W.A., Fardo D.W. Reassessment of risk genotypes (GRN, TMEM106B, and ABCC9 variants) associated with hippocampal sclerosis of aging pathology. J. Neuropathol. Exp. Neurol. 2015;74(1):75–84. doi: 10.1097/NEN.0000000000000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 699.Sassi C., Guerreiro R., Gibbs R., Ding J., Lupton M.K., Troakes C., Al-Sarraj S., Niblock M., Gallo J.M., Adnan J., Killick R., Brown K.S., Medway C., Lord J., Turton J., Bras J., Alzheimer’s Research UK Consortium Morgan, K.; Powell, J.F.; Singleton, A.; Hardy, J. Investigating the role of rare coding variability in Mendelian dementia genes (APP, PSEN1, PSEN2, GRN, MAPT, and PRNP) in late-onset Alzheimer’s disease. Neurobiol. Aging. 2014;35(12):2881.e1–2881.e6. doi: 10.1016/j.neurobiolaging.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700.Lin Q., Cao Y.P., Gao J. Common polymorphisms in the GSK3beta gene may contribute to the pathogenesis of Alzheimer disease: A meta-analysis. J. Geriatr. Psychiatry Neurol. 2015;28(2):83–93. doi: 10.1177/0891988714554712. [DOI] [PubMed] [Google Scholar]
- 701.Kettunen P., Larsson S., Holmgren S., Olsson S., Minthon L., Zetterberg H., Blennow K., Nilsson S., Sjolander A. Genetic variants of GSK3B are associated with biomarkers for Alzheimer’s disease and cognitive function. J. Alzheimers Dis. 2015;44(4):1313–1322. doi: 10.3233/JAD-142025. [DOI] [PubMed] [Google Scholar]
- 702.Gonzalez-Zuniga M., Contreras P.S., Estrada L.D., Chamorro D., Villagra A., Zanlungo S., Seto E., Alvarez A.R. c-Abl stabilizes HDAC2 levels by tyrosine phosphorylation repressing neuronal gene expression in Alzheimer’s disease. Mol. Cell. 2014;56(1):163–173. doi: 10.1016/j.molcel.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 703.Jeong J.H., Yum K.S., Chang J.Y., Kim M., Ahn J.Y., Kim S., Lapchak P.A., Han M.K. Dose-specific effect of simvastatin on hypoxia-induced HIF-1alpha and BACE expression in Alzheimer’s disease cybrid cells. BMC Neurol. 2015;15:127. doi: 10.1186/s12883-015-0390-5. https://bmcneurol.biomedcentral.com/articles/10.1186/s12883-015-0390-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Lanni C., Nardinocchi L., Puca R., Stanga S., Uberti D., Memo M., Govoni S., D’Orazi G., Racchi M. Homeodomain interacting protein kinase 2: A target for Alzheimer’s beta amyloid leading to misfolded p53 and inappropriate cell survival. PLoS One. 2010;5(4):e10171. doi: 10.1371/journal.pone.0010171. http://journals.plos.org/plosone/ article?id=10.1371/journal.pone.0010171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Piras S., Furfaro A.L., Piccini A., Passalacqua M., Borghi R., Carminati E., Parodi A., Colombo L., Salmona M., Pronzato M.A., Marinari U.M., Tabaton M., Nitti M. Monomeric Abeta1-42 and RAGE: Key players in neuronal differentiation. Neurobiol. Aging. 2014;35(6):1301–1308. doi: 10.1016/j.neurobiolaging.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 706.Keller L., Murphy C., Wang H.X., Fratiglioni L., Olin M., Gafvels M., Bjorkhem I., Graff C., Meaney S. A functional polymorphism in the HMGCR promoter affects transcriptional activity but not the risk for Alzheimer disease in Swedish populations. Brain Res. 2010;1344:185–191. doi: 10.1016/j.brainres.2010.04.073. [DOI] [PubMed] [Google Scholar]
- 707.Leduc V., Theroux L., Dea D., Dufour R., Poirier J. Effects of rs3846662 variants on HMGCR mRNA and protein levels and on markers of Alzheimer’s disease pathology. J. Mol. Neurosci. 2016;58(1):109–119. doi: 10.1007/s12031-015-0666-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Leduc V., De Beaumont L., Theroux L., Dea D., Aisen P., Petersen R.C., Alzheimer’s Disease Neuroimaging Initiative Dufour, R.; Poirier, J. HMGCR is a genetic modifier for risk, age of onset and MCI conversion to Alzheimer’s disease in a three cohorts study. Mol. Psychiatry. 2015;20(7):867–873. doi: 10.1038/mp.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709.Wang W., Yu J.T., Tan L., Liu Q.Y., Wang H.F., Ma X.Y. Insulin-like growth factor 1 (IGF1) polymorphism is associated with Alzheimer’s disease in Han Chinese. Neurosci. Lett. 2012;531(1):20–23. doi: 10.1016/j.neulet.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 710.Garcia J., Ahmadi A., Wonnacott A., Sutcliffe W., Nagga K., Soderkvist P., Marcusson J. Association of insulin-like growth factor-1 receptor polymorphism in dementia. Dement. Geriatr. Cogn. Disord. 2006;22(5-6):439–444. doi: 10.1159/000095803. [DOI] [PubMed] [Google Scholar]
- 711.Halaschek-Wiener J., Amirabbasi-Beik M., Monfared N., Pieczyk M., Sailer C., Kollar A., Thomas R., Agalaridis G., Yamada S., Oliveira L., Collins J.A., Meneilly G., Marra M.A., Madden K.M., Le N.D., Connors J.M., Brooks-Wilson A.R. Genetic variation in healthy oldest-old. 2009 doi: 10.1371/journal.pone.0006641. http://journals.plos.org/plosone/article?id= 10.1371/ [DOI] [PMC free article] [PubMed]
- 712.Aberg D., Johansson P., Isgaard J., Wallin A., Johansson J.O., Andreasson U., Blennow K., Zetterberg H., Aberg N.D., Svensson J. Increased cerebrospinal fluid level of insulin-like growth factor-II in male patients with Alzheimer’s Disease. J. Alzheimers Dis. 2015;48(3):637–646. doi: 10.3233/JAD-150351. [DOI] [PubMed] [Google Scholar]
- 713.Ramos E.M., Lin M.T., Larson E.B., Maezawa I., Tseng L.H., Edwards K.L., Schellenberg G.D., Hansen J.A., Kukull W.A., Jin L.W. Tumor necrosis factor alpha and interleukin 10 promoter region polymorphisms and risk of late-onset Alzheimer disease. Arch. Neurol. 2006;63(8):1165–1169. doi: 10.1001/archneur.63.8.1165. [DOI] [PubMed] [Google Scholar]
- 714.Medway C., Combarros O., Cortina-Borja M., Butler H.T., Ibrahim-Verbaas C.A., de Bruijn R.F., Koudstaal P.J., van Duijn C.M., Ikram M.A., Mateo I., Sanchez-Juan P., Lehmann M.G., Heun R., Kolsch H., Deloukas P., Hammond N., Coto E., Alvarez V., Kehoe P.G., Barber R., Wilcock G.K., Brown K., Belbin O., Warden D.R., Smith A.D., Morgan K., Lehmann D.J. The sex-specific associations of the aromatase gene with Alzheimer’s disease and its interaction with IL10 in the epistasis project. Eur. J. Hum. Genet. 2014;22(2):216–220. doi: 10.1038/ejhg.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715.Zhu X.C., Tan L., Jiang T., Tan M.S., Zhang W., Yu J.T. Association of IL-12A and IL-12B polymorphisms with Alzheimer’s disease susceptibility in a Han Chinese population. J. Neuroimmunol. 2014;274(1-2):180–184. doi: 10.1016/j.jneuroim.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 716.Swardfager W., Lanctot K., Rothenburg L., Wong A., Cappell J., Herrmann N. A meta-analysis of cytokines in Alzheimer’s disease. Biol. Psychiatry. 2010;68(10):930–941. doi: 10.1016/j.biopsych.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 717.Tian M., Deng Y.Y., Hou D.R., Li W., Feng X.L., Yu Z.L. Association of IL-1, IL-18, and IL-33 gene polymorphisms with late-onset Alzheimers disease in a Hunan Han Chinese population. Brain Res. 2015;1596:136–145. doi: 10.1016/j.brainres.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 718.Benedet A.L., Labbe A., Lemay P., Zimmer E.R., Pascoal T.A., Leuzy A., Mathotaarachchi S., Mohades S., Shin M., Dionne-Laporte A., Beaudry T., Picard C., Gauthier S., Poirier J., Rouleau G., Rosa-Neto P., Alzheimer’s Disease Neuroimaging Initiative Epistasis analysis links immune cascades and cerebral amyloidosis. J. Neuroinflammation. 2015;12:227. doi: 10.1186/s12974-015-0436-z. https://jneuroinflammation.biomedcentral.com/articles/10.1186/s12974-015-0436-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 719.Kölsch H., Lehmann D.J., Ibrahim-Verbaas C.A., Combarros O., van Duijn C.M., Hammond N., Belbin O., Cortina-Borja M., Lehmann M.G., Aulchenko Y.S., Schuur M., Breteler M., Wilcock G.K., Brown K., Kehoe P.G., Barber R., Coto E., Alvarez V., Deloukas P., Mateo I., Maier W., Morgan K., Warden D.R., Smith A.D., Heun R. Interaction of insulin and PPAR-alpha genes in Alzheimer’s disease: The Epistasis Project. J. Neural Transm. (Vienna) 2012;119(4):473–479. doi: 10.1007/s00702-011-0732-4. [DOI] [PubMed] [Google Scholar]
- 720.Vos S.J., Verhey F., Frolich L., Kornhuber J., Wiltfang J., Maier W., Peters O., Ruther E., Nobili F., Morbelli S., Frisoni G.B., Drzezga A., Didic M., van Berckel B.N., Simmons A., Soininen H., Kloszewska I., Mecocci P., Tsolaki M., Vellas B., Lovestone S., Muscio C., Herukka S.K., Salmon E., Bastin C., Wallin A., Nordlund A., de Mendonca A., Silva D., Santana I., Lemos R., Engelborghs S., Van der Mussele S., Alzheimer’s Disease Neuroimaging Initiative Freund-Levi, Y.; Wallin, A.K.; Hampel, H.; van der Flier, W.; Scheltens, P.; Visser, P.J. Prevalence and prognosis of Alzheimer’s disease at the mild cognitive impairment stage. Brain. 2015;138(Pt 5):1327–1338. doi: 10.1093/brain/awv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 721.de Rivero Vaccari J.P., Brand F.J., III, Sedaghat C., Mash D.C., Dietrich W.D., Keane R.W. RIG-1 receptor expression in the pathology of Alzheimer’s disease. J. Neuroinflammation. 2014;11:67. doi: 10.1186/1742-2094-11-67. https://jneuroinflammation.biomedcentral.com/ articles/10.1186/1742-2094-11-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Grimmer T., Goldhardt O., Guo L.H., Yousefi B.H., Forster S., Drzezga A., Sorg C., Alexopoulos P., Forstl H., Kurz A., Perneczky R. LRP-1 polymorphism is associated with global and regional amyloid load in Alzheimer’s Disease in humans in-vivo. Neuroimage Clin. 2014;4:411–416. doi: 10.1016/j.nicl.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723.Yuan Q., Wang F., Xue S., Jia J. Association of polymorphisms in the LRP1 and A2M genes with Alzheimer’s disease in the Northern Chinese Han population. J. Clin. Neurosci. 2013;20(2):253–256. doi: 10.1016/j.jocn.2012.01.052. [DOI] [PubMed] [Google Scholar]
- 724.Parcerisas A., Rubio S.E., Muhaisen A., Gomez-Ramos A., Pujadas L., Puiggros M., Rossi D., Urena J., Burgaya F., Pascual M., Torrents D., Rabano A., Avila J., Soriano E. Somatic signature of brain-specific single nucleotide variations in sporadic Alzheimer’s disease. J. Alzheimers Dis. 2014;42(4):1357–1382. doi: 10.3233/JAD-140891. [DOI] [PubMed] [Google Scholar]
- 725.Papuc E., Kurys-Denis E., Krupski W., Tatara M., Rejdak K. Can antibodies against glial derived antigens be early biomarkers of hippocampal demyelination and memory loss in Alzheimer’s disease? J. Alzheimers Dis. 2015;48(1):115–121. doi: 10.3233/JAD-150309. [DOI] [PubMed] [Google Scholar]
- 726.Mishra M., Akatsu H., Heese K. The novel protein MANI modulates neurogenesis and neurite-cone growth. J. Cell. Mol. Med. 2011;15(8):1713–1725. doi: 10.1111/j.1582-4934.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 727.Gevorkian G., Gonzalez-Noriega A., Acero G., Ordonez J., Michalak C., Munguia M.E., Govezensky T., Cribbs D.H., Manoutcharian K. Amyloid-beta peptide binds to microtubule-associated protein 1B (MAP1B). Neurochem. Int. 2008;52(6):1030–1036. doi: 10.1016/j.neuint.2007.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Di Domenico F., Coccia R., Cocciolo A., Murphy M.P., Cenini G., Head E., Butterfield D.A., Giorgi A., Schinina M.E., Mancuso C., Cini C., Perluigi M. Impairment of proteostasis network in Down syndrome prior to the development of Alzheimer’s disease neuropathology: Redox proteomics analysis of human brain. Biochim. Biophys. Acta. 2013;1832(8):1249–1259. doi: 10.1016/j.bbadis.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Hashimoto Y., Toyama Y., Kusakari S., Nawa M., Matsuoka M. An Alzheimer disease-linked rare mutation potentiates netrin receptor uncoordinated-5c-induced signaling that merges with amyloid beta precursor protein signaling. J. Biol. Chem. 2016;291(23):12282–12293. doi: 10.1074/jbc.M115.698092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Zhu X., Raina A.K., Rottkamp C.A., Aliev G., Perry G., Boux H., Smith M.A. Activation and redistribution of c-jun N-terminal kinase/stress activated protein kinase in degenerating neurons in Alzheimer’s disease. J. Neurochem. 2001;76(2):435–441. doi: 10.1046/j.1471-4159.2001.00046.x. [DOI] [PubMed] [Google Scholar]
- 731.Helbecque N., Abderrahamani A., Meylan L., Riederer B., Mooser V., Miklossy J., Delplanque J., Boutin P., Nicod P., Haefliger J.A., Cottel D., Amouyel P., Froguel P., Waeber G. Islet-brain1/C-Jun N-terminal kinase interacting protein-1 (IB1/JIP-1) promoter variant is associated with Alzheimer’s disease. Mol. Psychiatry. 2003;8(4):413–422. doi: 10.1038/sj.mp.4001344. [DOI] [PubMed] [Google Scholar]
- 732.Hashioka S., Klegeris A., Schwab C., Yu S., McGeer P.L. Differential expression of interferon-gamma receptor on human glial cells in vivo and in vitro. J. Neuroimmunol. 2010;225(1-2):91–99. doi: 10.1016/j.jneuroim.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 733.Fiala M., Mahanian M., Rosenthal M., Mizwicki M.T., Tse E., Cho T., Sayre J., Weitzman R., Porter V. MGAT3 mRNA: A biomarker for prognosis and therapy of Alzheimer’s disease by vitamin D and curcuminoids. J. Alzheimers Dis. 2011;25(1):135–144. doi: 10.3233/JAD-2011-101950. [DOI] [PubMed] [Google Scholar]
- 734.Frenkel D., Wilkinson K., Zhao L., Hickman S.E., Means T.K., Puckett L., Farfara D., Kingery N.D., Weiner H.L., El Khoury J. Scara1 deficiency impairs clearance of soluble amyloid-beta by mononuclear phagocytes and accelerates Alzheimer’s-like disease progression. Nat. Commun. 2013;4:2030. doi: 10.1038/ncomms3030. https://www.nature.com/articles/ncomms3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 735.Rai V. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and alzheimer disease risk: A meta-analysis. Mol. Neurobiol. 2017;54(2):1173–1186. doi: 10.1007/s12035-016-9722-8. [DOI] [PubMed] [Google Scholar]
- 736.Ferrer I. Differential expression of phosphorylated translation initiation factor 2 alpha in Alzheimer’s disease and Creutzfeldt-Jakob’s disease. Neuropathol. Appl. Neurobiol. 2002;28(6):441–451. doi: 10.1046/j.1365-2990.2002.t01-1-00410.x. [DOI] [PubMed] [Google Scholar]
- 737.Szot P., Leverenz J.B., Peskind E.R., Kiyasu E., Rohde K., Miller M.A., Raskind M.A. Tyrosine hydroxylase and norepinephrine transporter mRNA expression in the locus coeruleus in Alzheimer’s disease. Brain Res. Mol. Brain Res. 2000;84(1-2):135–140. doi: 10.1016/s0169-328x(00)00168-6. [DOI] [PubMed] [Google Scholar]
- 738.Tejani-Butt S.M., Yang J., Zaffar H. Norepinephrine transporter sites are decreased in the locus coeruleus in Alzheimer’s disease. Brain Res. 1993;631(1):147–150. doi: 10.1016/0006-8993(93)91201-3. [DOI] [PubMed] [Google Scholar]
- 739.Uhrig M., Ittrich C., Wiedmann V., Knyazev Y., Weninger A., Riemenschneider M., Hartmann T. New Alzheimer amyloid beta responsive genes identified in human neuroblastoma cells by hierarchical clustering. PLoS One. 2009;4(8):e6779. doi: 10.1371/journal.pone.0006779. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0006779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Huang Y., Liu F., Grundke-Iqbal I., Iqbal K., Gong C.X. NF-kappaB precursor, p105, and NF-kappaB inhibitor, IkappaBgamma, are both elevated in Alzheimer disease brain. Neurosci. Lett. 2005;373(2):115–118. doi: 10.1016/j.neulet.2004.09.074. [DOI] [PubMed] [Google Scholar]
- 741.Li X., Long J., He T., Belshaw R., Scott J. Integrated genomic approaches identify major pathways and upstream regulators in late onset Alzheimer’s disease. Sci. Rep. 2015;5:12393. doi: 10.1038/srep12393. https://www.nature.com/articles/srep12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 742.Guttler B.H., Cynis H., Seifert F., Ludwig H.H., Porzel A., Schilling S. A quantitative analysis of spontaneous isoaspartate formation from N-terminal asparaginyl and aspartyl residues. Amino Acids. 2013;44(4):1205–1214. doi: 10.1007/s00726-012-1454-0. [DOI] [PubMed] [Google Scholar]
- 743.Pontillo A., Catamo E., Arosio B., Mari D., Crovella S. NALP1/NLRP1 genetic variants are associated with Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2012;26(3):277–281. doi: 10.1097/WAD.0b013e318231a8ac. [DOI] [PubMed] [Google Scholar]
- 744.Saresella M., La Rosa F., Piancone F., Zoppis M., Marventano I., Calabrese E., Rainone V., Nemni R., Mancuso R., Clerici M. The NLRP3 and NLRP1 inflammasomes are activated in Alzheimer’s disease. 2016 doi: 10.1186/s13024-016-0088-1. https://molecularneurodegeneration.biomedcentral.com/articles/10 [DOI] [PMC free article] [PubMed]
- 745.Liu S., Zeng F., Wang C., Chen Z., Zhao B., Li K. The nitric oxide synthase 3 G894T polymorphism associated with Alzheimer’s disease risk: A meta-analysis. Sci. Rep. 2015;5:13598. doi: 10.1038/srep13598. https://www.nature.com/articles/srep13598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 746.Argentieri M.A., Nagarajan S., Seddighzadeh B., Baccarelli A.A., Shields A.E. Epigenetic pathways in human disease: The impact of DNA methylation on stress-related pathogenesis and current challenges in biomarker development. EBioMedicine. 2017;18:327–350. doi: 10.1016/j.ebiom.2017.03.044. https://www.ebiomedicine.com/article/S2352-3964 (17)30140-8/fulltext [DOI] [PMC free article] [PubMed] [Google Scholar]
- 747.Leal M.C., Magnani N., Villordo S., Buslje C.M., Evelson P., Castano E.M., Morelli L. Transcriptional regulation of insulin-degrading enzyme modulates mitochondrial amyloid beta (Abeta) peptide catabolism and functionality. J. Biol. Chem. 2013;288(18):12920–12931. doi: 10.1074/jbc.M112.424820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 748.von Otter M., Landgren S., Nilsson S., Zetterberg M., Celojevic D., Bergstrom P., Minthon L., Bogdanovic N., Andreasen N., Gustafson D.R., Skoog I., Wallin A., Tasa G., Blennow K., Nilsson M., Hammarsten O., Zetterberg H. Nrf2-encoding NFE2L2 haplotypes influence disease progression but not risk in Alzheimer’s disease and age-related cataract. Mech. Ageing Dev. 2010;131(2):105–110. doi: 10.1016/j.mad.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 749.Chen Z., Simmons M.S., Perry R.T., Wiener H.W., Harrell L.E., Go R.C. Genetic association of neurotrophic tyrosine kinase receptor type 2 (NTRK2) With Alzheimer’s disease. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147(3):363–369. doi: 10.1002/ajmg.b.30607. [DOI] [PubMed] [Google Scholar]
- 750.Arlt S., Demiralay C., Tharun B., Geisel O., Storm N., Eichenlaub M., Lehmbeck J.T., Wiedemann K., Leuenberger B., Jahn H. Genetic risk factors for depression in Alzheimer’s disease patients. Curr. Alzheimer Res. 2013;10(1):72–81. [PubMed] [Google Scholar]
- 751.Hamilton G., Proitsi P., Jehu L., Morgan A., Williams J., O’Donovan M.C., Owen M.J., Powell J.F., Lovestone S. Candidate gene association study of insulin signaling genes and Alzheimer’s disease: Evidence for SOS2, PCK1, and PPARgamma as susceptibility loci. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B(4):508–516. doi: 10.1002/ajmg.b.30503. [DOI] [PubMed] [Google Scholar]
- 752.Satoh J., Tabunoki H., Ishida T., Saito Y., Arima K. Immunohistochemical characterization of gamma-secretase activating protein expression in Alzheimer’s disease brains. Neuropathol. Appl. Neurobiol. 2012;38(2):132–141. doi: 10.1111/j.1365-2990.2011.01206.x. [DOI] [PubMed] [Google Scholar]
- 753.Kawamata T., Taniguchi T., Mukai H., Kitagawa M., Hashimoto T., Maeda K., Ono Y., Tanaka C. A protein kinase, PKN, accumulates in Alzheimer neurofibrillary tangles and associated endoplasmic reticulum-derived vesicles and phosphorylates tau protein. J. Neurosci. 1998;18(18):7402–7410. doi: 10.1523/JNEUROSCI.18-18-07402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 754.Rahman A., Ting K., Cullen K.M., Braidy N., Brew B.J., Guillemin G.J. The excitotoxin quinolinic acid induces tau phosphorylation in human neurons. 2009 doi: 10.1371/journal.pone.0006344. http://journals.plos.org/plosone/article?id= 10.1371/ [DOI] [PMC free article] [PubMed]
- 755.Silva D.F., Selfridge J.E., Lu J., Roy N., Hutfles L., Burns J.M., Michaelis E.K., Yan S., Cardoso S.M., Swerdlow R.H. Bioenergetic flux, mitochondrial mass and mitochondrial morphology dynamics in AD and MCI cybrid cell lines. Hum. Mol. Genet. 2013;22(19):3931–3946. doi: 10.1093/hmg/ddt247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 756.Shinohara M., Fujioka S., Murray M.E., Wojtas A., Baker M., Rovelet-Lecrux A., Rademakers R., Das P., Parisi J.E., Graff-Radford N.R., Petersen R.C., Dickson D.W., Bu G. Regional distribution of synaptic markers and APP correlate with distinct clinicopathological features in sporadic and familial Alzheimer’s disease. Brain. 2014;137(Pt 5):1533–1549. doi: 10.1093/brain/awu046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 757.Cimino P.J., Sokal I., Leverenz J., Fukui Y., Montine T.J. DOCK2 is a microglial specific regulator of central nervous system innate immunity found in normal and Alzheimer’s disease brain. Am. J. Pathol. 2009;175(4):1622–1630. doi: 10.2353/ajpath.2009.090443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 758.Hoshino T., Namba T., Takehara M., Nakaya T., Sugimoto Y., Araki W., Narumiya S., Suzuki T., Mizushima T. Prostaglandin E2 stimulates the production of amyloid-beta peptides through internalization of the EP4 receptor. J. Biol. Chem. 2009;284(27):18493–18502. doi: 10.1074/jbc.M109.003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 759.Yermakova A.V., Rollins J., Callahan L.M., Rogers J., O’Banion M.K. Cyclooxygenase-1 in human Alzheimer and control brain: Quantitative analysis of expression by microglia and CA3 hippocampal neurons. J. Neuropathol. Exp. Neurol. 1999;58(11):1135–1146. doi: 10.1097/00005072-199911000-00003. [DOI] [PubMed] [Google Scholar]
- 760.Miyashita A., Hatsuta H., Kikuchi M., Nakaya A., Saito Y., Tsukie T., Hara N., Ogishima S., Kitamura N., Akazawa K., Kakita A., Takahashi H., Murayama S., Ihara Y., Ikeuchi T., Kuwano R., Japanese Alzheimer’s Disease Neuroimaging Initiative Genes associated with the progression of neurofibrillary tangles in Alzheimer’s disease. Transl. Psychiatry. 2014;4:e396. doi: 10.1038/tp.2014.35. https://www.nature.com/articles/tp201435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 761.Luo D., Long Y., Chen G.J. Cyclooxygenase-2 gene polymorphisms and risk of Alzheimer’s disease: A meta-analysis. J. Neurol. Sci. 2015;359(1-2):100–105. doi: 10.1016/j.jns.2015.10.053. [DOI] [PubMed] [Google Scholar]
- 762.Madhusoodanan K.S., Murad F. NO-cGMP signaling and regenerative medicine involving stem cells. Neurochem. Res. 2007;32(4-5):681–694. doi: 10.1007/s11064-006-9167-y. [DOI] [PubMed] [Google Scholar]
- 763.Couturier J., Stancu I.C., Schakman O., Pierrot N., Huaux F., Kienlen-Campard P., Dewachter I., Octave J.N. Activation of phagocytic activity in astrocytes by reduced expression of the inflammasome component ASC and its implication in a mouse model of Alzheimer disease. J. Neuroinflammation. 2016;13:20. doi: 10.1186/s12974-016-0477-y. https://jneuroinflammation.biomedcentral.com/articles/ 10.1186/s12974-016-0477-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 764.Lakshmana M.K., Chung J.Y., Wickramarachchi S., Tak E., Bianchi E., Koo E.H., Kang D.E. A fragment of the scaffolding protein RanBP9 is increased in Alzheimer’s disease brains and strongly potentiates amyloid-beta peptide generation. FASEB J. 2010;24(1):119–127. doi: 10.1096/fj.09-136457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 765.Lin K.G., Tang M., Guo Y.B., Han H.Y., Lin Y.H. Two polymorphisms of RCAN1 gene associated with Alzheimer’s disease in the Chinese Han population. East Asian Arch. Psychiatry. 2011;21(2):79–84. [PubMed] [Google Scholar]
- 766.Seripa D., Matera M.G., Franceschi M., Daniele A., Bizzarro A., Rinaldi M., Panza F., Fazio V.M., Gravina C., D’Onofrio G., Solfrizzi V., Masullo C., Pilotto A. The RELN locus in Alzheimer’s disease. J. Alzheimers Dis. 2008;14(3):335–344. doi: 10.3233/jad-2008-14308. [DOI] [PubMed] [Google Scholar]
- 767.Taguchi K., Yamagata H.D., Zhong W., Kamino K., Akatsu H., Hata R., Yamamoto T., Kosaka K., Takeda M., Kondo I., Miki T. Identification of hippocampus-related candidate genes for Alzheimer’s disease. Ann. Neurol. 2005;57(4):585–588. doi: 10.1002/ana.20433. [DOI] [PubMed] [Google Scholar]
- 768.Kamboh M.I., Minster R.L., Kenney M., Ozturk A., Desai P.P., Kammerer C.M., DeKosky S.T. Alpha-1-antichymotrypsin (ACT or SERPINA3) polymorphism may affect age-at-onset and disease duration of Alzheimer’s disease. Neurobiol. Aging. 2006;27(10):1435–1439. doi: 10.1016/j.neurobiolaging.2005.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Sahin P., McCaig C., Jeevahan J., Murray J.T., Hainsworth A.H. The cell survival kinase SGK1 and its targets FOXO3a and NDRG1 in aged human brain. Neuropathol. Appl. Neurobiol. 2013;39(6):623–633. doi: 10.1111/nan.12023. [DOI] [PubMed] [Google Scholar]
- 770.Xie Z., Dong Y., Maeda U., Xia W., Tanzi R.E. RNA interference silencing of the adaptor molecules ShcC and Fe65 differentially affect amyloid precursor protein processing and Abeta generation. J. Biol. Chem. 2007;282(7):4318–4325. doi: 10.1074/jbc.M609293200. [DOI] [PubMed] [Google Scholar]
- 771.Wang Q., Tian Q., Song X., Liu Y., Li W. SNCA Gene polymorphism may contribute to an increased risk of Alzheimer’s disease. J. Clin. Lab. Anal. 2016;30(6):1092–1099. doi: 10.1002/jcla.21986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.Spisak K., Klimkowicz-Mrowiec A., Pera J., Dziedzic T., Aleksandra G., Slowik A. rs2070424 of the SOD1 gene is associated with risk of Alzheimer’s disease. Neurol. Neurochir. Pol. 2014;48(5):342–345. doi: 10.1016/j.pjnns.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 773.Reitz C., Tosto G., Vardarajan B., Rogaeva E., Ghani M., Rogers R.S., Conrad C., Haines J.L., Pericak-Vance M.A., Fallin M.D., Foroud T., Farrer L.A., Schellenberg G.D., George-Hyslop P.S., Mayeux R., Alzheimer’s Disease Genetics Consortium Independent and epistatic effects of variants in VPS10-d receptors on Alzheimer disease risk and processing of the amyloid precursor protein (APP). Transl. Psychiatry. 2013;3:e256. doi: 10.1038/tp.2013.13. https://www.nature.com/articles/tp201313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 774.Reid S.J., Whittaker D.J., Greenwood D., Snell R.G. A splice variant of the TATA-box binding protein encoding the polyglutamine-containing N-terminal domain that accumulates in Alzheimer’s disease. Brain Res. 2009;1268:190–199. doi: 10.1016/j.brainres.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 775.Bertram L., McQueen M.B., Mullin K., Blacker D., Tanzi R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat. Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 776.Randall C.N., Strasburger D., Prozonic J., Morris S.N., Winkie A.D., Parker G.R., Cheng D., Fennell E.M., Lanham I., Vakil N., Huang J., Cathcart H., Huang R., Poduslo S.E. Cluster analysis of risk factor genetic polymorphisms in Alzheimer’s disease. Neurochem. Res. 2009;34(1):23–28. doi: 10.1007/s11064-008-9626-8. [DOI] [PubMed] [Google Scholar]
- 777.Yang Q., Wang E.Y., Jia H.W., Wang Y.P. Association between polymorphisms in transforming growth factor-beta1 and sporadic Alzheimer’s disease in a Chinese population. Int. J. Neurosci. 2016;126(11):979–984. doi: 10.3109/00207454.2015.1088849. [DOI] [PubMed] [Google Scholar]
- 778.Minoretti P., Gazzaruso C., Vito C.D., Emanuele E., Bianchi M., Coen E., Reino M., Geroldi D. Effect of the functional toll-like receptor 4 Asp299Gly polymorphism on susceptibility to late-onset Alzheimer’s disease. Neurosci. Lett. 2006;391(3):147–149. doi: 10.1016/j.neulet.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 779.Alvarez V., Mata I.F., Gonzalez P., Lahoz C.H., Martinez C., Pena J., Guisasola L.M., Coto E. Association between the TNFalpha-308 A/G polymorphism and the onset-age of Alzheimer disease. Am. J. Med. Genet. 2002;114(5):574–577. doi: 10.1002/ajmg.10515. [DOI] [PubMed] [Google Scholar]
- 780.Fossati S., Ghiso J., Rostagno A. TRAIL death receptors DR4 and DR5 mediate cerebral microvascular endothelial cell apoptosis induced by oligomeric Alzheimer’s Abeta. 2012 doi: 10.1038/cddis.2012.55. https://www.nature.com/articles/cddis [DOI] [PMC free article] [PubMed]
- 781.Shang Z., Lv H., Zhang M., Duan L., Wang S., Li J., Liu G., Ruijie Z., Jiang Y. Genome-wide haplotype association study identify TNFRSF1A, CASP7, LRP1B, CDH1 and TG genes associated with Alzheimer’s disease in Caribbean Hispanic individuals. Oncotarget. 2015;6(40):42504–42514. doi: 10.18632/oncotarget.6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 782.Luckhaus C., Mahabadi B., Grass-Kapanke B., Janner M., Willenberg H., Jager M., Supprian T., Fehsel K. Blood biomarkers of osteoporosis in mild cognitive impairment and Alzheimer’s disease. J. Neural Transm. (Vienna) 2009;116(7):905–911. doi: 10.1007/s00702-009-0241-x. [DOI] [PubMed] [Google Scholar]
- 783.Manczak M., Reddy P.H. RNA silencing of genes involved in Alzheimer’s disease enhances mitochondrial function and synaptic activity. Biochim. Biophys. Acta. 2013;1832(12):2368–2378. doi: 10.1016/j.bbadis.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Chiappelli M., Borroni B., Archetti S., Calabrese E., Corsi M.M., Franceschi M., Padovani A., Licastro F. VEGF gene and phenotype relation with Alzheimer’s disease and mild cognitive impairment. Rejuvenation Res. 2006;9(4):485–493. doi: 10.1089/rej.2006.9.485. [DOI] [PubMed] [Google Scholar]
- 785.Okuizumi K., Onodera O., Namba Y., Ikeda K., Yamamoto T., Seki K., Ueki A., Nanko S., Tanaka H., Takahashi H., Oyanagi K., Mizusawa H., Kanazawa I., Tsuji S. Genetic association of the very low density lipoprotein (VLDL) receptor gene with sporadic Alzheimer’s disease. Nat. Genet. 1995;11(2):207–209. doi: 10.1038/ng1095-207. [DOI] [PubMed] [Google Scholar]
- 786.Helbecque N., Richard F., Cottel D., Neuman E., Guez D., Amouyel P. The very low density lipoprotein (VLDL) receptor is a genetic susceptibility factor for Alzheimer disease in a European Caucasian population. Alzheimer Dis. Assoc. Disord. 1998;12(4):368–371. doi: 10.1097/00002093-199812000-00019. [DOI] [PubMed] [Google Scholar]
- 787.Chibnik L.B., Yu L., Eaton M.L., Srivastava G., Schneider J.A., Kellis M., Bennett D.A., De Jager P.L. Alzheimer’s loci: Epigenetic associations and interaction with genetic factors. Ann. Clin. Transl. Neurol. 2015;2(6):636–647. doi: 10.1002/acn3.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 788.Di Francesco A., Arosio B., Gussago C., Dainese E., Mari D., D’Addario C., Maccarrone M. Involvement of 5-lipoxygenase in Alzheimer’s disease: A role for DNA methylation. J. Alzheimers Dis. 2013;37(1):3–8. doi: 10.3233/JAD-130506. [DOI] [PubMed] [Google Scholar]
- 789.Foraker J., Millard S.P., Leong L., Thomson Z., Chen S., Keene C.D., Bekris L.M., Yu C.E. The APOE gene is differentially methylated in Alzheimer’s disease. J. Alzheimers Dis. 2015;48(3):745–755. doi: 10.3233/JAD-143060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 790.Chang L., Wang Y., Ji H., Dai D., Xu X., Jiang D., Hong Q., Ye H., Zhang X., Zhou X., Liu Y., Li J., Chen Z., Li Y., Zhou D., Zhuo R., Zhang Y., Yin H., Mao C., Duan S., Wang Q. Elevation of peripheral BDNF promoter methylation links to the risk of Alzheimer’s disease. 2014 doi: 10.1371/journal.pone.0110773. http://journals.plos.org/plosone/article?id= 10.1371/ [DOI] [PMC free article] [PubMed]
- 791.Wang B.Y., Zhong Y., Zhao Z., Miao Y. Epigenetic suppression of hippocampal BDNF mediates the memory deficiency induced by amyloid fibrils. Pharmacol. Biochem. Behav. 2014;126:83–89. doi: 10.1016/j.pbb.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 792.Sen A., Nelson T.J., Alkon D.L. ApoE4 and abeta oligomers reduce BDNF expression via HDAC nuclear translocation. J. Neurosci. 2015;35(19):7538–7551. doi: 10.1523/JNEUROSCI.0260-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 793.Rao J.S., Keleshian V.L., Klein S., Rapoport S.I. Epigenetic modifications in frontal cortex from Alzheimer’s disease and bipolar disorder patients. Transl. Psychiatry. 2012;2:e132. doi: 10.1038/tp.2012.55. https://www.nature.com/articles/tp201255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 794.Humphries C., Kohli M.A., Whitehead P., Mash D.C., Pericak-Vance M.A., Gilbert J. Alzheimer disease (AD) specific transcription, DNA methylation and splicing in twenty AD associated loci. Mol. Cell. Neurosci. 2015;67:37–45. doi: 10.1016/j.mcn.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 795.Sanchez-Mut J.V., Aso E., Heyn H., Matsuda T., Bock C., Ferrer I., Esteller M. Promoter hypermethylation of the phosphatase DUSP22 mediates PKA-dependent TAU phosphorylation and CREB activation in Alzheimer’s disease. Hippocampus. 2014;24(4):363–368. doi: 10.1002/hipo.22245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 796.Drzewinska J., Walczak-Drzewiecka A., Ratajewski M. Identification and analysis of the promoter region of the human DHCR24 gene: involvement of DNA methylation and histone acetylation. Mol. Biol. Rep. 2011;38(2):1091–1101. doi: 10.1007/s11033-010-0206-z. [DOI] [PubMed] [Google Scholar]
- 797.Siegmund K.D., Connor C.M., Campan M., Long T.I., Weisenberger D.J., Biniszkiewicz D., Jaenisch R., Laird P.W., Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2(9):e895. doi: 10.1371/journal.pone.0000895. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0000895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 798.Hendrickx A., Pierrot N., Tasiaux B., Schakman O., Kienlen-Campard P., De Smet C., Octave J.N. Epigenetic regulations of immediate early genes expression involved in memory formation by the amyloid precursor protein of Alzheimer disease. PLoS One. 2014;9(6):e99467. doi: 10.1371/journal.pone.0099467. http://journals.plos.org/plosone/article? id=10.1371/journal.pone.0099467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 799.Yao X.Q., Li X.C., Zhang X.X., Yin Y.Y., Liu B., Luo D.J., Wang Q., Wang J.Z., Liu G.P. Glycogen synthase kinase-3beta regulates leucine-309 demethylation of protein phosphatase-2A via PPMT1 and PME-1. FEBS Lett. 2012;586(16):2522–2528. doi: 10.1016/j.febslet.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 800.Nicolia V., Fuso A., Cavallaro R.A., Di Luzio A., Scarpa S. B vitamin deficiency promotes tau phosphorylation through regulation of GSK3beta and PP2A. J. Alzheimers Dis. 2010;19(3):895–907. doi: 10.3233/JAD-2010-1284. [DOI] [PubMed] [Google Scholar]
- 801.Wang Y., Yang R., Gu J., Yin X., Jin N., Xie S., Wang Y., Chang H., Qian W., Shi J., Iqbal K., Gong C.X., Cheng C., Liu F. Cross talk between PI3K-AKT-GSK-3beta and PP2A pathways determines tau hyperphosphorylation. Neurobiol. Aging. 2015;36(1):188–200. doi: 10.1016/j.neurobiolaging.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 802.Bollati V., Galimberti D., Pergoli L., Dalla Valle E., Barretta F., Cortini F., Scarpini E., Bertazzi P.A., Baccarelli A. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav. Immun. 2011;25(6):1078–1083. doi: 10.1016/j.bbi.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 803.Cook C., Carlomagno Y., Gendron T.F., Dunmore J., Scheffel K., Stetler C., Davis M., Dickson D., Jarpe M., DeTure M., Petrucelli L. Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 2014;23(1):104–116. doi: 10.1093/hmg/ddt402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 804.Narayan P.J., Lill C., Faull R., Curtis M.A., Dragunow M. Increased acetyl and total histone levels in post-mortem Alzheimer’s disease brain. Neurobiol. Dis. 2015;74:281–294. doi: 10.1016/j.nbd.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 805.Mastroeni D., Grover A., Delvaux E., Whiteside C., Coleman P.D., Rogers J. Epigenetic changes in Alzheimer’s disease: Decrements in DNA methylation. Neurobiol. Aging. 2010;31(12):2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 806.Tannorella P., Stoccoro A., Tognoni G., Petrozzi L., Salluzzo M.G., Ragalmuto A., Siciliano G., Haslberger A., Bosco P., Bonuccelli U., Migliore L., Coppede F. Methylation analysis of multiple genes in blood DNA of Alzheimer’s disease and healthy individuals. Neurosci. Lett. 2015;600:143–147. doi: 10.1016/j.neulet.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 807.Piaceri I., Raspanti B., Tedde A., Bagnoli S., Sorbi S., Nacmias B. Epigenetic modifications in Alzheimer’s disease: Cause or effect? J. Alzheimers Dis. 2015;43(4):1169–1173. doi: 10.3233/JAD-141452. [DOI] [PubMed] [Google Scholar]
- 808.Yoshino Y., Mori T., Yoshida T., Yamazaki K., Ozaki Y., Sao T., Funahashi Y., Iga J.I., Ueno S.I. Elevated mRNA expression and low methylation of SNCA in japanese Alzheimer’s disease subjects. J. Alzheimers Dis. 2016;54(4):1349–1357. doi: 10.3233/JAD-160430. [DOI] [PubMed] [Google Scholar]
- 809.Silva P.N., Gigek C.O., Leal M.F., Bertolucci P.H., de Labio R.W., Payao S.L., Smith Mde A. Promoter methylation analysis of SIRT3, SMARCA5, HTERT and CDH1 genes in aging and Alzheimer’s disease. J. Alzheimers Dis. 2008;13(2):173–176. doi: 10.3233/jad-2008-13207. [DOI] [PubMed] [Google Scholar]
- 810.Kaut O., Ramirez A., Pieper H., Schmitt I., Jessen F., Wullner U. DNA methylation of the TNF-alpha promoter region in peripheral blood monocytes and the cortex of human Alzheimer’s disease patients. Dement. Geriatr. Cogn. Disord. 2014;38(1-2):10–15. doi: 10.1159/000357126. [DOI] [PubMed] [Google Scholar]
- 811.Wang T., Chen K., Li H., Dong S., Su N., Liu Y., Cheng Y., Dai J., Yang C., Xiao S. The feasibility of utilizing plasma MiRNA107 and BACE1 messenger RNA gene expression for clinical diagnosis of amnestic mild cognitive impairment. J. Clin. Psychiatry. 2015;76(2):135–141. doi: 10.4088/JCP.13m08812. [DOI] [PubMed] [Google Scholar]
- 812.Banzhaf-Strathmann J., Benito E., May S., Arzberger T., Tahirovic S., Kretzschmar H., Fischer A., Edbauer D. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014;33(15):1667–1680. doi: 10.15252/embj.201387576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 813.Tan L., Yu J.T., Liu Q.Y., Tan M.S., Zhang W., Hu N., Wang Y.L., Sun L., Jiang T., Tan L. Circulating miR-125b as a biomarker of Alzheimer’s disease. J. Neurol. Sci. 2014;336(1-2):52–56. doi: 10.1016/j.jns.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 814.Galimberti D., Villa C., Fenoglio C., Serpente M., Ghezzi L., Cioffi S.M., Arighi A., Fumagalli G., Scarpini E. Circulating miRNAs as potential biomarkers in Alzheimer’s disease. J. Alzheimers Dis. 2014;42(4):1261–1267. doi: 10.3233/JAD-140756. [DOI] [PubMed] [Google Scholar]
- 815.Tiribuzi R., Crispoltoni L., Porcellati S., Di Lullo M., Florenzano F., Pirro M., Bagaglia F., Kawarai T., Zampolini M., Orlacchio A., Orlacchio A. miR128 up-regulation correlates with impaired amyloid beta(1-42) degradation in monocytes from patients with sporadic Alzheimer’s disease. Neurobiol. Aging. 2014;35(2):345–356. doi: 10.1016/j.neurobiolaging.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 816.Smith P.Y., Hernandez-Rapp J., Jolivette F., Lecours C., Bisht K., Goupil C., Dorval V., Parsi S., Morin F., Planel E., Bennett D.A., Fernandez-Gomez F.J., Sergeant N., Buee L., Tremblay M.E., Calon F., Hebert S.S. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 2015;24(23):6721–6735. doi: 10.1093/hmg/ddv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 817.Liu C.G., Wang J.L., Li L., Xue L.X., Zhang Y.Q., Wang P.C. MicroRNA-135a and -200b, potential biomarkers for Alzheimers disease, regulate beta secretase and amyloid precursor protein. Brain Res. 2014;1583:55–64. doi: 10.1016/j.brainres.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 818.Liu C.G., Wang J.L., Li L., Wang P.C. MicroRNA-384 regulates both amyloid precursor protein and beta-secretase expression and is a potential biomarker for Alzheimer’s disease. Int. J. Mol. Med. 2014;34(1):160–166. doi: 10.3892/ijmm.2014.1780. [DOI] [PubMed] [Google Scholar]
- 819.Liu C.G., Song J., Zhang Y.Q., Wang P.C. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol. Med. Rep. 2014;10(5):2395–2400. doi: 10.3892/mmr.2014.2484. [DOI] [PubMed] [Google Scholar]
- 820.Zhang Y., Xing H., Guo S., Zheng Z., Wang H., Xu D. MicroRNA-135b has a neuroprotective role via targeting of beta-site APP-cleaving enzyme 1. Exp. Ther. Med. 2016;12(2):809–814. doi: 10.3892/etm.2016.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 821.Cheng C., Li W., Zhang Z., Yoshimura S., Hao Q., Zhang C., Wang Z. MicroRNA-144 is regulated by activator protein-1 (AP-1) and decreases expression of Alzheimer disease-related a disintegrin and metalloprotease 10 (ADAM10). J. Biol. Chem. 2013;288(19):13748–13761. doi: 10.1074/jbc.M112.381392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 822.An Y.W., Jhang K.A., Woo S.Y., Kang J.L., Chong Y.H. Sulforaphane exerts its anti-inflammatory effect against amyloid-beta peptide via STAT-1 dephosphorylation and activation of Nrf2/HO-1 cascade in human THP-1 macrophages. Neurobiol. Aging. 2016;38:1–10. doi: 10.1016/j.neurobiolaging.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 823.Long J.M., Ray B., Lahiri D.K. MicroRNA-153 physiologically inhibits expression of amyloid-beta precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J. Biol. Chem. 2012;287(37):31298–31310. doi: 10.1074/jbc.M112.366336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 824.Kim J., Yoon H., Chung D.E., Brown J.L., Belmonte K.C., Kim J. miR-186 is decreased in aged brain and suppresses BACE1 expression. J. Neurochem. 2016;137(3):436–445. doi: 10.1111/jnc.13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 825.Zhang J., Hu M., Teng Z., Tang Y.P., Chen C. Synaptic and cognitive improvements by inhibition of 2-AG metabolism are through upregulation of microRNA-188-3p in a mouse model of Alzheimer’s disease. J. Neurosci. 2014;34(45):14919–14933. doi: 10.1523/JNEUROSCI.1165-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 826.Zhu H.C., Wang L.M., Wang M., Song B., Tan S., Teng J.F., Duan D.X. MicroRNA-195 downregulates Alzheimer’s disease amyloid-beta production by targeting BACE1. Brain Res. Bull. 2012;88(6):596–601. doi: 10.1016/j.brainresbull.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 827.Xing H., Guo S., Zhang Y., Zheng Z., Wang H. Upregulation of microRNA-206 enhances lipopolysaccharide-induced inflammation and release of amyloid-beta by targeting insulin-like growth factor 1 in microglia. Mol. Med. Rep. 2016;14(2):1357–1364. doi: 10.3892/mmr.2016.5369. [DOI] [PubMed] [Google Scholar]
- 828.Lee S.T., Chu K., Jung K.H., Kim J.H., Huh J.Y., Yoon H., Park D.K., Lim J.Y., Kim J.M., Jeon D., Ryu H., Lee S.K., Kim M., Roh J.K. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 2012;72(2):269–277. doi: 10.1002/ana.23588. [DOI] [PubMed] [Google Scholar]
- 829.Liu H., Chu W., Gong L., Gao X., Wang W. MicroRNA-26b is upregulated in a double transgenic mouse model of Alzheimer’s disease and promotes the expression of amyloid-beta by targeting insulin-like growth factor 1. Mol. Med. Rep. 2016;13(3):2809–2814. doi: 10.3892/mmr.2016.4860. [DOI] [PubMed] [Google Scholar]
- 830.Absalon S., Kochanek D.M., Raghavan V., Krichevsky A.M. MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J. Neurosci. 2013;33(37):14645–14659. doi: 10.1523/JNEUROSCI.1327-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 831.Jennewein C., von Knethen A., Schmid T., Brune B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J. Biol. Chem. 2010;285(16):11846–11853. doi: 10.1074/jbc.M109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 832.Zhang Y., Liu C., Wang J., Li Q., Ping H., Gao S., Wang P. 2016 https://www.nature.com/articles/
- 833.Pereira P.A., Tomas J.F., Queiroz J.A., Figueiras A.R., Sousa F. Recombinant pre-miR-29b for Alzheimer’s disease therapeutics. Sci. Rep. 2016;6:19946. doi: 10.1038/srep19946. https://www.nature.com/ articles/srep19946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 834.Yang G., Song Y., Zhou X., Deng Y., Liu T., Weng G., Yu D., Pan S. DNA methyltransferase 3, a target of microRNA-29c, contributes to neuronal proliferation by regulating the expression of brain-derived neurotrophic factor. Mol. Med. Rep. 2015;12(1):1435–1442. doi: 10.3892/mmr.2015.3531. [DOI] [PubMed] [Google Scholar]
- 835.Lei X., Lei L., Zhang Z., Zhang Z., Cheng Y. Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 2015;8(2):1565–1574. [PMC free article] [PubMed] [Google Scholar]
- 836.Yang G., Song Y., Zhou X., Deng Y., Liu T., Weng G., Yu D., Pan S. MicroRNA-29c targets beta-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol. Med. Rep. 2015;12(2):3081–3088. doi: 10.3892/mmr.2015.3728. [DOI] [PubMed] [Google Scholar]
- 837.Kim J., Yoon H., Horie T., Burchett J.M., Restivo J.L., Rotllan N., Ramirez C.M., Verghese P.B., Ihara M., Hoe H.S., Esau C., Fernandez-Hernando C., Holtzman D.M., Cirrito J.R., Ono K., Kim J. MicroRNA-33 regulates ApoE lipidation and amyloid-beta metabolism in the brain. J. Neurosci. 2015;35(44):14717–14726. doi: 10.1523/JNEUROSCI.2053-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 838.Long J.M., Ray B., Lahiri D.K. MicroRNA-339-5p down-regulates protein expression of beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J. Biol. Chem. 2014;289(8):5184–5198. doi: 10.1074/jbc.M113.518241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 839.Zhao Y., Bhattacharjee S., Jones B.M., Dua P., Alexandrov P.N., Hill J.M., Lukiw W.J. Regulation of TREM2 expression by an NF-small ka, CyrillicB-sensitive miRNA-34a. Neuroreport. 2013;24(6):318–323. doi: 10.1097/WNR.0b013e32835fb6b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 840.Zheng D., Sabbagh J.J., Blair L.J., Darling A.L., Wen X., Dickey C.A. MicroRNA-511 binds to FKBP5 mRNA, which encodes a chaperone protein, and regulates neuronal differentiation. J. Biol. Chem. 2016;291(34):17897–17906. doi: 10.1074/jbc.M116.727941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 841.Mezache L., Mikhail M., Garofalo M., Nuovo G.J. Reduced miR-512 and the elevated expression of its targets cFLIP and MCL1 localize to neurons with hyperphosphorylated tau protein in Alzheimer disease. Appl. Immunohistochem. Mol. Morphol. 2015;23(9):615–623. doi: 10.1097/PAI.0000000000000147. [DOI] [PubMed] [Google Scholar]
- 842.Li F., Wei G., Bai Y., Li Y., Huang F., Lin J., Hou Q., Deng R., Zhou J.H., Zhang S.X., Chen D.F. MicroRNA-574 is involved in cognitive impairment in 5-month-old APP/PS1 mice through regulation of neuritin. Brain Res. 2015;1627:177–188. doi: 10.1016/j.brainres.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 843.Zhang C., Lu J., Liu B., Cui Q., Wang Y. Primate-specific miR-603 is implicated in the risk and pathogenesis of Alzheimer’s disease. Aging (Albany N.Y.) 2016;8(2):272–290. doi: 10.18632/aging.100887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 844.Li W., Li X., Xin X., Kan P.C., Yan Y. MicroRNA-613 regulates the expression of brain-derived neurotrophic factor in Alzheimer’s disease. Biosci. Trends. 2016;10(5):372–377. doi: 10.5582/bst.2016.01127. [DOI] [PubMed] [Google Scholar]
- 845.Zhao Z.B., Wu L., Xiong R., Wang L.L., Zhang B., Wang C., Li H., Liang L., Chen S.D. MicroRNA-922 promotes tau phosphorylation by downregulating ubiquitin carboxy-terminal hydrolase L1 (UCHL1) expression in the pathogenesis of Alzheimer’s disease. Neuroscience. 2014;275:232–237. doi: 10.1016/j.neuroscience.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 846.Hu Y.K., Wang X., Li L., Du Y.H., Ye H.T., Li C.Y. MicroRNA-98 induces an Alzheimer’s disease-like disturbance by targeting insulin-like growth factor 1. Neurosci. Bull. 2013;29(6):745–751. doi: 10.1007/s12264-013-1348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]