Abstract
Background
Brain glioma is a type of common primary intracranial malignant tumor, the prognosis of which is frequently unfavorable. Enhancer of zeste homolog 2 (EZH2) belongs to poly-sulfur protein family and can mediate cell proliferation and differentiation via the modulation of various genes expressions. In addition, it is further related with occurrence and metastasis of malignant tumors. This study investigated the effect of EZH2 expression on proliferation and tumorigenesis of brain glioma cells.
Material/Methods
Glioma tumor tissues were collected from 3 patients who received surgery, and the glioma stem cells were then separated, cultured, and identified by flow cytometry. RNA interference approach was used to suppress EZH2 expression, which was confirmed by quantitative real-time polymerase chain reaction (qRT-PCR). Clonal formation assay analyzed the change of cell proliferation potency. The effect on tumorigenesis potency of glioma stem cells was determined by mouse transplantation assay. Western blot investigated the effect of EZH2 on levels of oncogenes such as HER-2, c-myc, PI3K, and Akt.
Results
Flow cytometry revealed cancer stem cells in glioma tissues took up 39.4%, and qRT-PCR showed that EZH2 expression was decreased by 72% after the treatment of RNA interference in glioma cells (P<0.05). Both cell clonal formation and xenograft assays showed that the downregulation of EZH2 inhibited glioma cell proliferation (P<0.05) and weakened tumorigenesis potency (P<0.05). Western blot results showed that the reduction of EZH2 also suppressed expressions of oncogenes including c-myc and Akt (P<0.05).
Conclusions
Our data demonstrated that in brain glioma cells, the decrease of EZH2 level could suppress cell proliferation and tumorigenesis potency, and meanwhile inhibit the expressions of oncogenes including c-myc and Akt.
MeSH Keywords: Cell Proliferation, Cells, Glioma
Background
Brain glioma is the most popular primary intracranial malignant brain tumor, which is prominently featured with high malignancy, rapid growth, and heterogeneous. Brain glioma usually manifests as diffused and infiltrated patterns in human brain tissues, as well as increases the difficulty for the treatment, leading to high recurrent rate and short survival time [1]. Besides other types of malignant tumors, the pathogenesis of brain glioma remains unclear, although widely accepted opinions suggest congenital susceptibility and environmental oncogenic factors [2]. Multiple therapeutic strategies have been deployed including the combined treatment of surgery, radio-chemotherapy or immunotherapy. Recent progress has been made in clinics, but no radical therapy has been developed for treatment of brain glioma [3,4].
Enhancer of zeste homolog 2 (EZH2) belongs to PcG (polycomb group) gene family [5]. Previous study assigned EZH2 as the core protein in PcG family, as its can maintain silent status of homologous genes via chromosomal modification. It has been found that abnormal expression of EZH2 was closely correlated with the development of multiple human malignant tumors [6]. Previous finding unraveled significant elevation of EZH2 expression in human prostate carcinoma cells, and its level had close correlation with cancer cell proliferation and disease progression [7]. In an analysis of brain glioma, in vitro application of EZH2 inhibitor 3′-denitrification adenine A could weaken proliferation potency of glioma tumor stem cells [8]. Moreover, the suppression of EZH2 gene expression could reverse temozolomide resistance in patients with brain glioma. Recent evidence speculated that EZH2 may contribute to regulate oncogenic gene expression via mediating DNA methylation level [9]. This study thus aimed to study the effect of EZH2 expression on proliferation of brain glioma cell and tumorigenesis potency.
Material and Methods
Identification of research patients
Three brain glioblastoma patients (2 males and 1 female, average age=39.1±6.5 years, all were primary cases) from the First Affiliated Hospital, Dalian Medical University were recruited. No chemo-, radio- or immuno-therapy was performed on study patients. After admission, magnetic resonance imaging examination was conducted to confirm tumor lesion. All patients received confirmed diagnosis based on the guideline of World Health Organization. Glioma tissues were obtained by intracranial surgery, without electrical coagulation, necrosis, or cystic lesion. Tissues were stored in sterile phosphate-buffered saline (PSB) on ice for further experiments. This study was pre-approved by the ethical committee of First Affiliated Hospital, Dalian Medical University with informed consents from all participants.
Separation and identification of brain glioma stem cells
Glioma tissue samples collected from the surgery were cut into small pieces and were digested in trypsin for 10 min. Tissue lysate was filtered and centrifuged to remove the supernatant. Glioma stem cells were kept in DMEM/F12 medium containing 20 μg/L basic fibroblast growth factor (bFGF) and 20 μg/L epidermal growth factor (EGF). Cells were kept at 37°C with 5% CO2 for 14 days, during which fresh culture medium was replenished every 5 days.
Cells after passage were digested by trypsin and re-suspended. After cells were blocked in 1% bovine serum albumin, mouse anti-human CD133 primary antibody was added for 2 hours incubation, and excess antibody was removed by centrifugation. Rhodamine-labeled goat anti-mouse secondary antibody was added for 2 hours incubation at room temperature, and excess antibody was also removed by centrifugation. Ratio of CD133+ glioma tumor stem cells was measured by flow cytometry.
RNA interference
Based on mRNA sequence of EZH2 gene (Genebank access ID: NM_004456), siRNA targeting EZH2 and scramble control were synthesized and methyl-modified on all residues by Sangon (China) as shown in Table 1. Liposome transfection kit INTERFERin (Polypus transfection) was used for RNA interference (RNAi) assay. In brief, cells were cultured until log-growth phase, and were digested in trypsin. Cells were then counted and diluted into 3×105 per mL for further culture into 96-well plate. After 24-hour culture, transfection assay was performed following the manual instruction of test kit [10]. All experiments were performed in 3 groups: anti-EZH2 group, scramble group, and blank control group (PBS for transfection). During transfection, 1 μL Lipofectamine 2000 (Invitrogen) was diluted in 50 μL antibiotic-free and serum-free DMEM medium for 5 min. All cells were then mixed with anti-EZH2 siRNA, scramble siRNA, or PBS to prepare transfection mixture. 100 μL of working mixture was added into each well. The 96-well plate was then cultured for 6 hours at 37°C with 5% CO2. Antibiotics and serum-free medium was then switched to DMEM medium containing 10% fetal bovine serum and gentamycin for 48~72 hours incubation, followed by further assays [11].
Table 1.
Chemically synthesized nucleic acids sequence for cell transfection.
| Name | Sequence |
|---|---|
| Anti-EZH2 | GACTCTGAATGCAGTTGCTTCAGTA AACTGAGACTTACGTCAACGAAGTC |
| Scramble | CTTCATGGACAGTATGCTAGAGT GAGAAGTACCTGTCATACGATCT |
Quantitative real-time polymerase chain reaction (qRT-PCR)
Those cells with transfection were tested for EZH2 expression by quantitative real-time polymerase chain reaction (qRT-PCR), using primers as shown in Table 2. One-step total RNA extraction kit TRIzol (Invitrogen, USA) was used to extract total RNA. Reverse transcription was first performed at 37°C for 2 hours. Using cDNA by reverse transcription as the template qRT was performed with SYBR commercial kit from TianGen (China). In brief, qRT-PCR was conducted under the following conditions: 95°C for 5 min, followed by 45 cycles each containing 95°C 15 sec, 60°C 30 sec. Relative gene expression was semiquantitative analyzed by 2−ΔΔCt method with β-actin as internal reference. 2−ΔΔCt=gene copy number in test group/gene copy number in control. Experiments were carried out in triplicates.
Table 2.
qRT-PCR primer sequence.
| Primer | Sequence | |
|---|---|---|
| β-actin | F | 5′-TGTCCCGATGGCGAGTGTTT-3′ |
| R | 5′-CCTGTTGGCCATAGTACTGC-3′ | |
| EZH2 | F | 5′-ACGGCAGCCTTGTGACAGTTC-3′ |
| R | 5′-ACACTCTCGGACAGCCAGGTAG-3′ | |
Western blot
Cultured cells were collected and mixed with 100 μL cell lysis buffer. Tissues were homogenized, and the lysate was centrifuged at 13 000 g for 10 min. The supernatant was separated by protein electrophoresis for western blot analysis. In brief, SDS-PAGE was performed using 15% separating gel and 5% condensing gel. After electrophoresis, proteins were transferred to PVDF membrane. The membrane was first blocked at 37°C for 1 hour using 5% defatted milk powder. After the membrane was rinsed by TBST, primary antibody working solution (mouse anti-human HER-2, c-myc, PI3K, Akt and β-actin, all at 1: 2000) was added for 4°C overnight incubation. Excess primary antibody was removed by TBST washing. Goat anti-mouse IgG secondary antibody with horseradish peroxidase conjugation (1: 1000) was added for 1 hour of incubation at room temperature. The membrane was rinsed in TBST and mixed with freshly prepared 3,3′-diaminobenzidine chromogenic substrate under 10 min dark incubation. Then the film was developed. The integrated gray values of images were tested by gel imaging system. Using β-actin as the internal reference, relative expression of EZH2 was calibrated.
Cell clonal formation assay
Those glioma cells after transfection were cultured until log-growth phase. After trypsin digestion, cells were re-suspended into fresh culture medium, and were inoculated into culture dish containing 10 mL fresh medium with 100 cells per dish. The dish was gently swirled to evenly distribute cells. Triplicated experiments were performed for each group of cells. Cells were cultured in a chamber for 10 days. The supernatant was discarded, and cells were rinsed in PBS followed by fixation in 2% paraformaldehyde for 15 min, and 5% crystal violet staining for 15 min. After staining buffer was washed off, the dish was inverted and stacked onto a transparent film with grids. Clonal formation number was counted to calculate clonal formation rate, which was equal to clonal formation number/100×100%.
Xenograft transplantation of human glioma
Xenograft transplantation of cancer cells was performed to measure the effect of EZH2 expression on tumorigenesis potency of glioma cells. In brief, cells were cultured at 37°C for 2 hours and were re-suspended in PBS for counting. 12 adult NOD/SCID mice (6 weeks age, 6 males and 6 females, body weight=32.6±2.5 g) were divided into two groups. Each mouse received 105 cells via intraperitoneal injection and were provided with normal diet at 25°C with 60±10% relative humidity. All animal experimental protocols followed the guideline stipulated by Animal Care Committee of NIH.
Statistical analysis
All experimental data were processed by SPSS 20.0 software. Results were presented as mean ± standard deviation (SD). Statistical comparisons of multiple groups were made via an ANOVA, followed by Bonferroni post hoc test. The differences between 2 groups using Student’s t-tests were 2-sided. A statistical significance was defined when P<0.05, and extreme significance was identified when P<0.01.
Results
Separation and passage culture of brain glioma stem cells
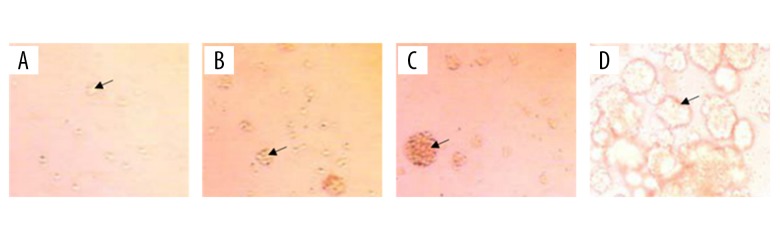
Brain glioma stem cells were separated and cultured from tissues of patients with glioma tumor and were passaged every 1 week. As shown in Figure 1, we observed cell death at the inner side of cell sphere with elongated incubation time, whilst peripheral cells gradually attached the flask, grew and differentiated. General morphology of cells after passage manifested as regular sphere.
Figure 1.
Morphology of cultured brain glioma stem cells by passage (100×). (A) Glioma cells at 24 hours of passage. (B) Glioma cells at 48 hours with passage culture. (C) Glioma cells at 72 hours with passage culture. (D) Glioma cells at 7 days with passage incubation.
Identification of brain glioma stem cells
Glioma stem cells separated previously were firstly labeled with antibody and were sorted by flow cytometry. 2.6×107 CD133+ cells were selected from a total of 6.6×107 cells, which occupies 39.4% of all cells (Figure 2).
Figure 2.
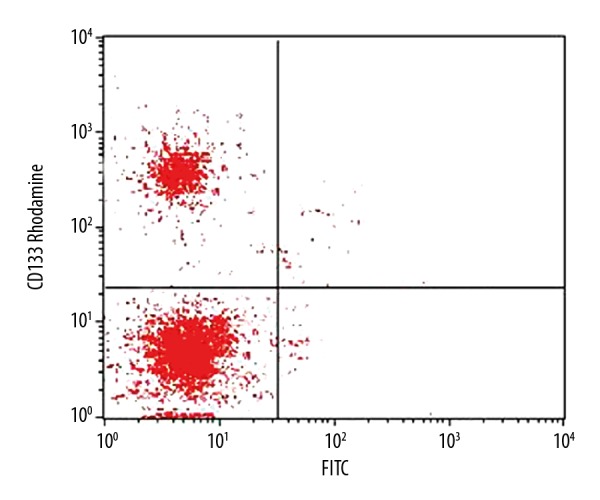
Flow cytometry identification of brain glioma stem cells.
EZH2 expression level after RNA interference
Compared to the control group, EZH2 expression in glioma cells was significantly decreased by 72% after siRNA transfection (P<0.05), indicating RNAi assay was successfully inhibited the level of EZH2 (Figure 3).
Figure 3.
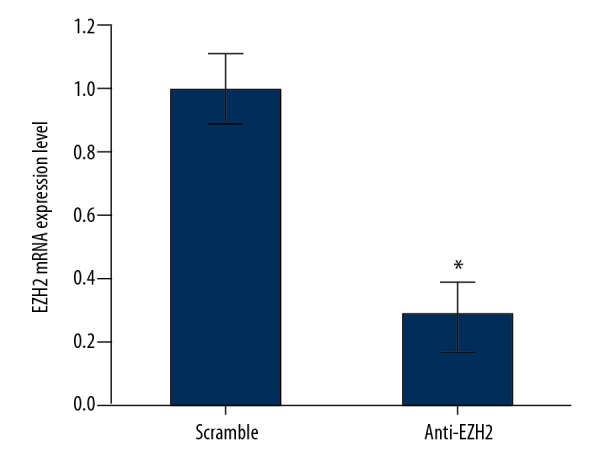
Expression profile of enhancer of zeste homolog 2 (EZH2) by qRT-PCR. * P<0.05 compared to scramble group.
Effects of EZH2 expression on glioma cell proliferation
The proliferation potency of glioma cells was then analyzed by clonal formation assay. Results showed that 73.5±8.2% clonal formation rate in scramble group, whilst the rate of glioma cells in anti-EZH2 group was merely 27.3±7.3%, indicating proliferation potency of glioma cells was reduced after EZH2 expression was inhibited.
Effects of EZH2 expression of tumorigenesis potency of glioma cells
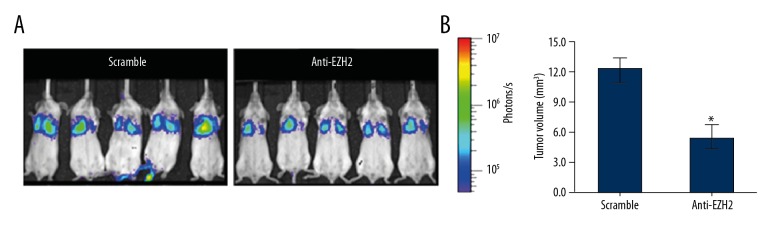
After the expression of EZH2 was downregulated by RNAi, xenograft transplantation assay was performed to analyze the effect of EZH2 expression on tumorigenesis potency of glioma cells. Of note, significantly small tumor size was found in anti-EZH2 group compared to those in control group in Figure 4, suggesting that low EZH2 expression could efficiently decrease tumorigenesis potency of glioma cells.
Figure 4.
Effects of enhancer of zeste homolog 2 (EZH2) expression on glioma cell tumorigenesis potency. (A) Results of glioma cell xenograft transplantation; (B) Effects of EZH2 expression on glioma cell tumorigenesis potency. * P<0.05 compared to scramble group.
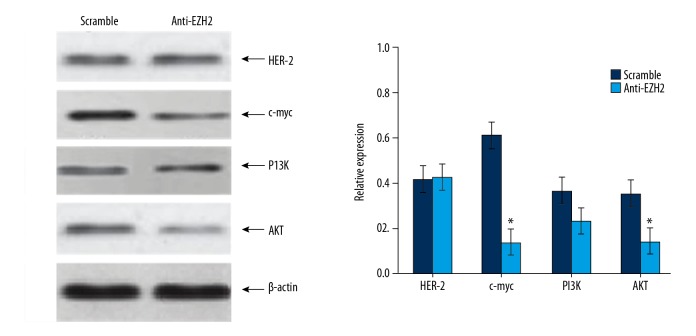
Effects of EZH2 expression on oncogene expression for glioma
To illustrate the effect of EZh2 expression on the levels of oncogenes in brain glioma cells, western blot was performed to analyze expression of oncogenes including HER-2, c-myc, PI3K, and Akt in glioma cells (Figure 5). After EZH2 gene expression was inhibited by RNAi, significantly downregulation was found in c-myc (P<0.01) and Akt (P<0.05), whilst no significant change of expression of HER-2 or PI3K was observed.
Figure 5.
Relative protein expression levels in glioma cells after RNA interference assay. * P<0.05 compared to scramble group.
Discussion
EZH2 represents a kind of histone-lysine-N-methyltransferase, and contains catalytic subunits including polycomb repressive complex 1 (PRC1) and PRC2 [12]. EZH2 participates in mediating DNA methylation thus affecting gene expression [13]. Previous study showed the involvement of EZH2 in the formation of heterochromatin. Biological studies also revealed the close correlation between EZH2 and occurrence/progression of multiple human malignant tumors including breast cancer, prostate cancer, and brain glioma [9,10,14]. In this study, we first separated and identified glioma stem cells. Further assays showed that when EZH2 gene was downregulated, cell proliferation potency was suppressed, along with weakened tumorigenesis potency.
Studies have shown that within PRC2 complex, EZH2 can bind with other DNA methyltransferase, enhance the binding affinity between DNA methyltransferase and target DNA sequence, and eventually facilitate DNA methylation modification [15]. Analysis for expressional profiles of various tumor related genes showed EZH2 gene expression was significantly elevated in various malignant tumors. In breast cancer cells, evidence indicated that EZH2 expression was correlated with tumor size, lymph node metastasis, pathological grade and estrogen receptor (ER) levels. Follow-up studies revealed relatively worse prognosis in those patients with comparably high level of EZH2 [16]. This study found that the inhibition of EZH2 expression significantly suppressed proliferation and tumorigenesis potency of glioma cells. Suva et al. cultured brain glioma cells in vitro and found that when EZH2 activity was inhibited by specific inhibitor, proliferation potency of glioma cells was remarkably suppressed [8]. Fan et al. also demonstrated that downregulation of EZH2 in glioma cells by RNA interference and found decreased cell proliferation potency and weakened resistance toward temozolomide, thus speculating the close correlation between EZH2 expression and proliferation or drug resistance mechanism of brain glioma cells [9], as was consistent with our study.
Currently, it is believed that EZH2 is under the regulation of multiple mechanisms in tumor cells. Varambally et al. reported that miR-101 could target EZH2 gene and suppress the expression, while miR-101 expression deficit was found in cancer cells with EZH2 overexpression, manifesting that EZH2 expression was under regulation of miRNA expression [17]. More evidences showed that EZH2 was involved at the downstream of p53-RB-E2F signal pathway. The activation of tumor suppressor gene p53 deactivated RB-E2F pathway via the inhibition of EZH2 promoter activity by p21 [18]. Chen et al. [19] and Wilson et al. [20] presented that EZH2 expression was also modulated of cell cycle protein CDK and transcriptional regulatory factor ETS.
We found that in glioma cells, EZH2 expression inhibition can downregulate the levels of several oncogenes including c-myc and Akt. The correlation between EZH2 and tumor proliferation or metastasis has also exhibited [13]. Rao et al. showed that EZH2 could modulate H3K27m3 activity in favor of somatic cell transformation, via interaction between c-myc and Sox2 [21]. Gonzalez et al. showed that BRCA12 downregulation is closely correlated with EX, probably due to PI3K-Aktt signal pathway and EZH2 up-regulation [22,23].
Due to rapid growth and high heterogeneity, brain glioma is characterized as a type of human intracranial tumors with the highest malignancy. Despite the treatment, the 5-year old survival rate is still less than 10%. Therefore, further studies about cellular and molecular mechanism of brain growth are required to develop treatment strategy for brain glioma. This study observed the effect of EZH2 in glioma cell proliferation and tumorigenesis, and meanwhile possible biological mechanism of EZh2 on brain glioma cells. The limitation in this study, however, still exists that requires prospective investigation of EZH2 on DNA methylation in various tumor gene promoter regions, and further analysis between EZH2 and cellular behavior of glioma cells, such as cell apoptosis.
Conclusions
Our data demonstrated that, within brain glioma cells, reducing levels of EHZ2 can decrease cell proliferation or tumorigenesis potency, and meanwhile suppress expressions of c-myc and Akt, which provides fundamental insights for the further development of novel therapy against glioma tumor by targeting EHZ2 in clinical practice.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: A clinical review. JAMA. 2013;310:1842–50. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 2.Oberoi RK, Parrish KE, Sio TT, et al. Strategies to improve delivery of anticancer drugs across the blood-brain barrier to treat glioblastoma. Neuro Oncol. 2016;18:27–36. doi: 10.1093/neuonc/nov164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizoguchi M, Hata N, Suzuki SO, et al. Pediatric glioblastoma with oligodendroglioma component: Aggressive clinical phenotype with distinct molecular characteristics. Neuropathology. 2013;33:652–57. doi: 10.1111/neup.12029. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–26. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciarapica R, De Salvo M, Carcarino E, et al. The polycomb group (PcG) protein EZH2 supports the survival of PAX3-FOXO1 alveolar rhabdomyosarcoma by repressing FBXO32 (Atrogin1/MAFbx) Oncogene. 2014;33:4173–84. doi: 10.1038/onc.2013.471. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi H, Hung MC. Regulation and role of EZH2 in cancer. Cancer Res Treat. 2014;46:209–22. doi: 10.4143/crt.2014.46.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–29. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 8.Suva ML, Riggi N, Janiszewska M, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–18. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 9.Fan TY, Wang H, Xiang P, et al. Inhibition of EZH2 reverses chemotherapeutic drug TMZ chemosensitivity in glioblastoma. Int J Clin Exp Pathol. 2014;7:6662–70. [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez ME, Moore HM, Li X, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci USA. 2014;111:3098–103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lotan TL, Gurel B, Sutcliffe S, et al. PTEN protein loss by immunostaining: Analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17:6563–73. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–35. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae WK, Hennighausen L. Canonical and non-canonical roles of the histone methyltransferase EZH2 in mammary development and cancer. Mol Cell Endocrinol. 2014;382:593–97. doi: 10.1016/j.mce.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim E, Kim M, Woo DH, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–52. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bitler BG, Aird KM, Garipov A, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–38. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reijm EA, Timmermans AM, Look MP, et al. High protein expression of EZH2 is related to unfavorable outcome to tamoxifen in metastatic breast cancer. Ann Oncol. 2014;25:2185–90. doi: 10.1093/annonc/mdu391. [DOI] [PubMed] [Google Scholar]
- 17.Varambally S, Cao Q, Mani RS, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–99. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiogama S, Yoshiba S, Soga D, et al. Aberrant expression of EZH2 is associated with pathological findings and P53 alteration. Anticancer Res. 2013;33:4309–17. [PubMed] [Google Scholar]
- 19.Chen S, Bohrer LR, Rai AN, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–14. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson BG, Wang X, Shen X, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–28. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao RA, Dhele N, Cheemadan S, et al. Ezh2 mediated H3K27me3 activity facilitates somatic transition during human pluripotent reprogramming. Sci Rep. 2015;5:8229. doi: 10.1038/srep08229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez ME, DuPrie ML, Krueger H, et al. Histone methyltransferase EZH2 induces Akt-dependent genomic instability and BRCA1 inhibition in breast cancer. Cancer Res. 2011;71:2360–70. doi: 10.1158/0008-5472.CAN-10-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puppe J, van de Ven M, van der Burg E, et al. EZH2 inhibition sensitizes BRCA1-deficient breast tumors to the PI3K inhibitor BKM120. Geburtshilfe und Frauenheilkunde. 2016;76:FV032. [Google Scholar]