Abstract
Clear links between human genes and influenza disease susceptibility are scarce. A recent study uncovered a gene variant coupled to severe influenza and showed how it hampers expression of an antiviral gene key to immune cell survival.
Influenza viruses can cause severe and sometimes fatal disease in humans, owing to differences in the pathogenicity of individual virus strains and variation in human susceptibility. Risk factors—including age, underlying co-morbidities, and pregnancy—influence susceptibility, but do not explain all of the circumstances under which serious influenza-associated complications occur1. Host genetic factors have emerged as potential regulators of human influenza disease susceptibility, and may explain severe disease in otherwise apparently healthy individuals. In this issue of Nature Medicine, Allen et al. reveal a new single nucleotide polymorphism (SNP) risk allele in the IFITM3 gene that is strongly associated with severe influenza disease, and further detail a novel mechanism through which this SNP regulates IFITM3 expression to influence survival of CD8+ T lymphocytes (CTLs) (Figure 1)2.
Figure 1. The rs34481144 allele regulates IFITM3 expression and influenza severity.
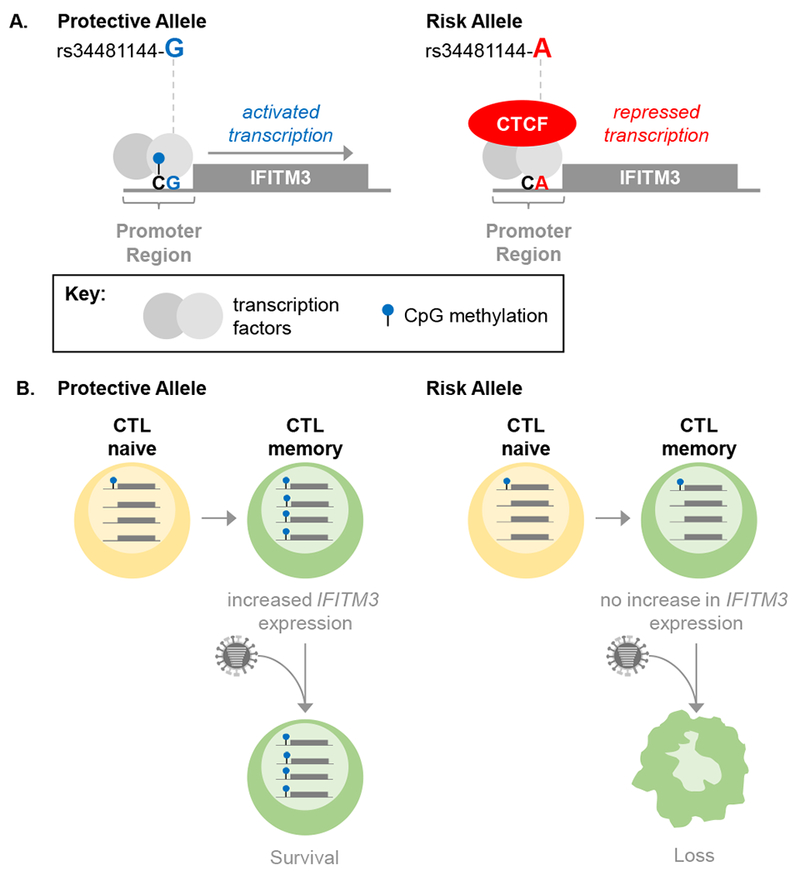
(A) Shown is the IFITM3 promoter region and open reading frame. The G (protective) allele promotes methylation of an adjacent nucleotide, while the A (risk) allele ablates methylation. Methylation blocks binding of the CTCF transcriptional repressor, allowing transcription to occur, while in the absence of methylation, CTCF binds to the IFITM3 promoter and represses IFITM3 transcription. A key is shown at the bottom of the panel. (B) IFITM3 promoter methylation promotes IFITM3 protein expression in memory CTL, which protects CTL survival after influenza virus challenge. CTL, CD8+ T lymphocyte.
IFITM3 is a gene induced by interferons and is a strong candidate for genetic regulation of human influenza susceptibility because of its ability to inhibit influenza virus replication in vitro and restrict influenza virus pathogenicity in mice. It also shapes adaptive immune responses to influenza by protecting memory CTL survival during secondary influenza virus challenge3. A SNP in the coding region of the IFITM3 gene (rs12252-C) was previously shown to increase influenza disease severity in some patients4–8, but the mechanism remains ambiguous.
To identify new IFITM3-associated SNPs with the potential to influence influenza disease severity, Allen et al. first assembled candidate SNPs located in the IFITM3 core promoter and coding regions, and then prioritized those candidates based on common allele frequency in European populations and putative functional attributes. A single SNP (rs34481144), located in the IFITM3 promoter region, satisfied their prioritization criteria. Genotyping analysis of three different patient cohorts revealed statistically significant associations of the rs34481144-A (risk) allele with more severe disease symptoms, earlier virus replication in nasal tissue, and pediatric mortality. Combining data from all three patient cohorts, the authors estimated that individuals carrying the rs34481144-A (risk) allele are ~2.6 times more likely to experience a severe outcome upon influenza virus infection.
Given rs34481144’s location in the IFITM3 core promoter and the rs34481144-A (risk) allele’s association with lower IFITM3 expression in blood cells (based on the authors’ query of expression quantitative trait loci data available through the Genotype-Tissue Expression project, www.gtexportal.org), it seems likely that rs34481144 controls influenza disease risk by regulating IFITM3 expression levels. Allen et al. provide substantial data supporting an association between the rs34481144-A (risk) allele and reduced IFITM3 expression in immune cells responding to influenza virus infection. They further demonstrate that the rs34481144 genotype directly impacts IFITM3 promoter function (with the rs34481144-A (risk) allele linked to diminished promoter activity), and identify rs34481144 genotype-specific differences in transcription factor binding to the IFITM3 promoter. Of particular importance, the rs34481144-A (risk) allele increases binding of the methylation-sensitive CTCF transcriptional repressor, which has not previously been linked to the regulation of IFITM3 expression.
CTCF promoter binding is disrupted by DNA methylation, and interestingly, the rs34481144-A (risk) allele ablates a CpG methylation site in the IFITM3 promoter. In their final set of experiments, Allen et al. examined the relationship between the rs34481144 genotype, IFITM3 promoter methylation, CTCF promoter binding, and IFITM3 expression in cell types relevant to influenza disease in humans. Their observations are consistent with the notion that the rs34481144 genotype controls IFITM3 promoter methylation in memory CTLs, and support a model in which methylation at the rs34481144 locus reduces CTCF binding to increase IFITM3 expression, leading to increased memory CTL survival and more efficient viral clearance from infected airways (Figure 1). Therefore, while a previous study suggested that IFITM3 expression is important for memory CTL survival during secondary influenza virus challenge3, the work presented by Allen et al. provides the first evidence that a human allele associated with influenza disease severity directly regulates IFITM3 expression in this cell type.
The identification of host genetic polymorphisms that regulate risk for severe influenza disease is imperative for reducing influenza-associated morbidity and mortality. Until now, no studies have confidently linked a human genetic variant with influenza disease risk and directly established its mechanism of action. Previously, targeted genetic analyses have associated the IFITM3 rs12252-C allele with severe influenza disease, particularly in populations with Asian ancestry5,6, but mechanistic information is lacking. Other attempts at targeted genetic analysis have suggested potential risk factor alleles in the CCR59, TNF9 and ST3GAL110 genes, but evidence for association with influenza disease severity is modest. Several groups have attempted genome-wide association studies, and while some SNPs have been identified (e.g., in CD5511 and FCGR2A12), no candidates have reached genome-wide significance, none have been authenticated through multiple studies or in multiple patient cohorts, and few attempts to identify mechanisms have been performed. In their work, Allen et al. used a targeted approach to identify a SNP (rs34481144) that regulates the expression of IFITM3. The association of the rs34481144-A (risk) allele with higher influenza disease severity across three different patient cohorts, as well as detailed mechanistic studies, provide substantial credibility to their finding, which clearly surpasses the limitations of previous work.
Future studies based on the current work should address several open questions to fully clarify how rs34481144 contributes to human influenza disease. IFITM3 protein limits influenza virus infection in cell types other than CTL that are central to influenza pathogenicity (e.g., epithelial and endothelial cells), so potential contributions of these cell types to rs34481144-regulated disease severity need to be examined carefully. At the molecular level, mechanistic insights may be garnered by evaluating rs34481144 effects on global IFITM3 promoter occupancy, since Allen et al. described effects for only a small subset of transcription factors capable of binding IFITM3 on sites overlapping the rs34481144 locus. Most critically, future efforts must focus on the development of large, well-powered cohort(s) including patients with various human ancestries, both for the wider validation of the rs34481144 allele and identification of additional SNPs that regulate human influenza disease. Ideally, these patient cohorts should include individuals that have been infected with different influenza viruses, since susceptibility may vary depending on the type of influenza virus strain (e.g., seasonal human H1N1 versus avian H5N1 viruses). Moreover, the identification and inclusion of patients that have experienced asymptomatic infections could assist in identification of resistance alleles. Given the time and expense required for the development of such cohort(s), emphasis should also be placed on new animal models that more closely represent human influenza disease (e.g., the Collaborative Cross or Diversity Outbred mice) as a means for identification of potential susceptibility alleles that can be further examined in limited patient cohorts. Ultimately, the identification of genetic risk factors that regulate influenza disease susceptibility may improve clinical case management by assisting in the identification of patients with a greater risk for severe complications, and further, may reveal critical insights into influenza disease pathophysiology. The work reported by Allen et al. is an important step toward achieving this goal.
ACKNOWLEDGEMENTS
This study was funded by the Leading Advanced Projects for medical innovation (LEAP) from the Japan Agency for Medical Research and Development (AMED); and grant U19AI106772, provided by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (USA). The authors thank Susan Watson for editing the manuscript.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors do not have any competing financial interests to declare.
REFERENCES
- 1.Writing Committee of the, W.H.O.C.o.C.A.o.P.I., et al. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N Engl J Med 362, 1708–1719 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Allen EK, et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat Med 23, 975–983 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey CC, Zhong G, Huang IC & Farzan M IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu Rev Virol 1, 261–283 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everitt AR, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci U S A 111, 769–774 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang YH, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun 4, 1418 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Rodriguez M, et al. IFITM3 and severe influenza virus infection. No evidence of genetic association. Eur J Clin Microbiol Infect Dis 35, 1811–1817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills TC, et al. IFITM3 and susceptibility to respiratory viral infections in the community. J Infect Dis 209, 1028–1031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horby P, Nguyen NY, Dunstan SJ & Baillie JK An updated systematic review of the role of host genetics in susceptibility to influenza. Influenza Other Respir Viruses 7 Suppl 2, 37–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maestri A, et al. Siaalpha2-3Galbeta1- Receptor Genetic Variants Are Associated with Influenza A(H1N1)pdm09 Severity. PLoS One 10, e0139681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, et al. A functional variation in CD55 increases the severity of 2009 pandemic H1N1 influenza A virus infection. J Infect Dis 206, 495–503 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Zuniga J, et al. Genetic variants associated with severe pneumonia in A/H1N1 influenza infection. Eur Respir J 39, 604–610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]


