ABSTRACT
Objectives
Mutations in mgrB, phoP/phoQ, pmrA, pmrB, pmrC, and crrABC regulatory systems have been found responsible for colistin resistance. The aim of our study was to investigate the role of alteration in mgrB gene and plasmid mediate mcr-1 and mcr-2 genes as a source of colistin resistance in 17 non duplicate Klebsiella pneumoniae clinical isolates.
Methods
All isolates classified as resistant to colistin by VITEK 2 system (BioMerieux, Marcy I’ Etoile, France) were included. Susceptibility to colistin was also determined by broth microdilution using breakpoints recommended by EUCAST (>2mg/L resistant; and ≤2mg/L susceptible). PCR amplification of mgrB gene was performed and sequenced using specific primers. Presence of mcr-1 and mcr-2 was also investigated using PCR.
Results
PCR amplification of the mgrB gene of the 17 K.pneumoniae isolates revealed a larger (~1000bp) amplicon in three isolates when compared with the wild type mgrB ampiclon (250 bp). Sequencing of these amplicons showed that mgrB was disrupted by the insertion of ISKpn14, a IS element belonging to the IS1 family. Sequencing, of the 250 bp mgrB gene in the remaining 14 isolates revealed frame shift mutation after the second codon leading to a premature stop codon in only one isolate.
Conclusions
The study showed that colistin resistance in 20% of the K. pneumoniae isolates was due to loss of function of mgrB. We describe for the first-time from India, insertional inactivation of mgrB by ISKpn14 inserted at different sites, responsible for colistin resistance.
Key words: mgrB, colistin, insertion sequence, polymyxins, Klebsiella pneumoniae
RESUMEN
Objetivos
Las mutaciones en los sistemas de regulación mgrB, phoP/phoQ, pmrA, pmrB, pmrC y crrABC se han asociado a la resistencia a colistina. El objetivo del estudio fue investigar el papel de la alteración en el gen mgrB y los genes mediados por plásmidos mcr-1 y mcr-2 como fuente de la resistencia a colistina en 17 aislados clínicos de Klebsiella pneumoniae.
Material y métodos
Todos los aislados que fueron clasificados como resistentes a colistina por el sistema VITEK 2 system (BioMerieux, Marcy I’ Etoile, France) fueron incluidos. La sensibilidad a colistina fue también determinada por microdilución en caldo empleando los puntos de corte recomendados por el EUCAST (> 2mg/L resistente y ≤ 2mg/L sensible). Se realizó la amplificación del gen mgrB por PCR empleando cebadores específicos. La presencia de los genes mcr-1 y mcr-2 fue también realizada empleando la PCR.
Resultados
La amplificación por PCR del gen mgrB de 17 aislados clínicos de K. pneumoniae mostró un amplicón más grande (~1000pb) en 3 cepas cuando se comparó con el amplicón salvaje (250 pb). La secuenciación de estos 3 amplicones mostró que el gen mgrB estaba alterado por la inserción de ISKpn14, un elemento IS que pertenece a la familia IS1. La secuenciación de los 250 pb del gen mgrB en el resto de los 14 aislados reveló mutaciones con desplazamiento del marco de lectura después del segundo codón que conducía a una interrupción de la lectura en sólo un aislado.
Conclusiones
Este estudio mostró que la resistencia a colistina en el 20% de los aislados de K. pneumoniae fue debida a la pérdida de la función del gen mgrB. Describimos por primera vez en India que la inactivación insercional en el gen mgrB por ISKpn14 es responsable de la resistencia a colistina.
Palabras claves: mgrB, colistina, secuencia inserción, polimixinas, Klebsiella pneumoniae
INTRODUCTION
Due to high rates of infections due to ESBL producing Enterobacteriaceae in India, carbapenems are extensively used for their treatment [1]. This lead to the emergence of carbapenem-resistant Enterobacteriaceae (CRE) due to plasmid mediated NDM-1 metallo-betalactamase in 2010 [2]. Soon after, NDM-1 producing CRE were reported from all parts of India, including the remote islands of Andaman & Nicobar. Subsequently polymyxins (colistin & polymyxin B) were launched as an effective option to treat CRE. Polymyxins are cationic compounds that bind to the negatively charged phosphate group of the lipopolysaccharide (LPS) causing cell death by disruption and loss of integrity of cell membrane [3]. They are active against a wide variety of Gram-negative pathogens but has no activity against Gram-positive and anaerobic pathogens. For the past few years, colistin and polymyxin B have been used mostly in combination with a broad spectrum betalactam to treat infections due to CRE [4]. Though colistin was used extensively, the dosing, most often would have been sub-therapeutic due to reasons like lack of clarity on the optimum dose, high cost, absence of loading dose and poor renal function leading to emergence of colistin resistance. Colistin resistance is primarily due decrease in the negative charge of the outer membrane due to addition of positively charged L-Ara-N and PEtN molecules thereby decreasing the affinity between colistin and its target [5]. This modification is mediated by the pmrHFIJKLM operon which in turn is regulated by the,phoP/phoQ two component system [5]. A small transmembrane protein mgrB negatively regulates the phoP/phoQsystem preventing activation of pmrHFIJKLM operon in K. pneumoniae. Previous studies have reported that insertional inactivation of mgrB gene in K.pneumoniae lead to upregulation of the phoP/phoQ system, causing overexpression of the pmrHFIJKLM operon, resulting in colistin resistance [6]. It was also found that insertion of different types of insertion sequence (IS) at different locations into the mgrB gene lead to its inactivation [5,6]. Mutations in mgrB, phoP/phoQ, pmrA, pmrB, pmrC, and crrABC regulatory systems have also been found responsible for colistin resistance [7]. Plasmid mediated colistin resistance due to mcr-1 to mcr-5 encoding for phosphoethanolamine transferase are also being increasingly reported from all over the world [8]. Though colistin resistant K. pneumoniae have often been reported in India there are only two recent articles characterizing the underlying mechanism [9,10].
MATERIALS AND METHODS
The study was conducted from January to June 2017 in a 1,200 bedded tertiary care teaching hospital in South India. Colistin-resistant non-duplicate (one isolate per patient) K. pneumoniae isolates encountered during routine susceptibility testing using VITEK 2 (bioMerieux) automated system were included in the study. Out of 932 isolates, 17 were found to be resistant to colistin using breakpoints recommended by EUCAST (> 2mg/L resistant; and ≤ 2mg/L susceptible) [11]. Resistance to colistin in these 17 isolates was confirmed by CLSI recommended broth microdilution method [12]. Colistin-susceptible K. pneumoniae ATCC 70063 was used as control. They were also tested for bla CTX-M (detects CTX-M-1-, CTX-M-2-, and CTX-M-9-like-encoding genes) and bla NDM-1 genes by PCR [2,13]. To determine the mechanism for colistin resistance PCR amplification and sequencing of the specific for mgrB gene was performed using specific primers (mgrB-extF:5′-TTAAGAAGGCCGTGCTATCC-3’ and mgrB-extR:5′-AAGGCGTTCATTCTACCACC-3’) [7]. Plasmid mediated colistin resistance due mcr-1 and mcr-2 to was investigated by PCR [8]. The study was approved by the Institutional Ethics Committee of our institute. (IEC-AIMS 2017-MICROB-117)
RESULTS
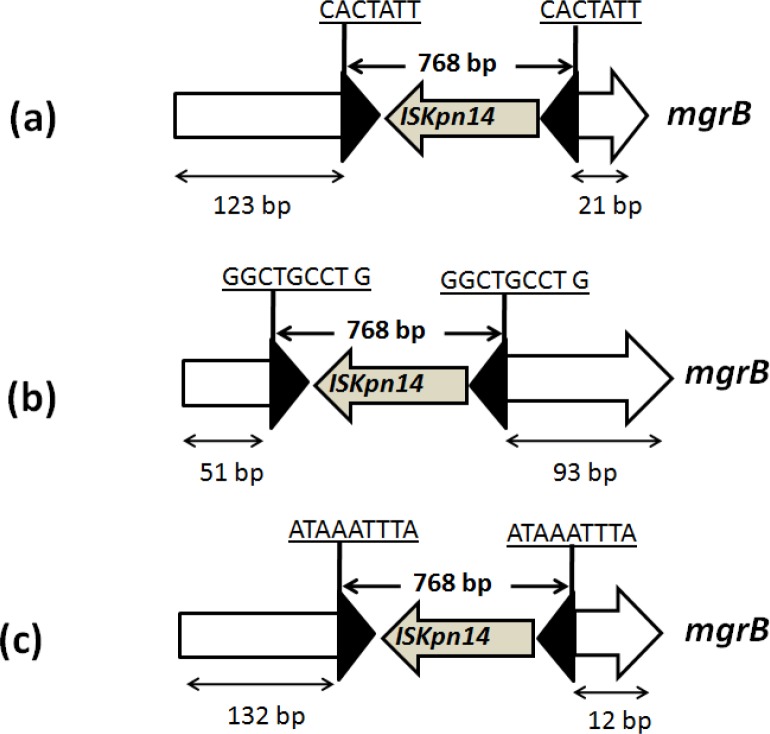
PCR amplification of the mgrB gene of the 17 K. pneumoniae isolates revealed a larger~1000bp) amplicon in three (CR8, 12, & 28) when compared with the wild type mgrB ampiclon (250 bp). Sequencing of these amplicons showed that mgrB was disrupted by the insertion of ISKpn14, a IS element belonging to the IS1 family (figure 1). In CR8 the insertion occurred between the nucleotides 123 and 124 and was bracketed by 7 bp target site duplication (CACTATT). In the CR28 the insertion occurred between the nucleotides 51 and 92 and was bracketed by 9 bp target site duplication (GGCTGCCTG) while in CR12 insertion was observed between the nucleotides 132 and 133 and was bracketed by 9 bp target site duplication (ATAAATTTA). In all the three cases transposon was inserted in the reverse orientation. Sequencing of the remaining 14 isolates with wild type mgrB ampiclon (250 bp) showed mutation only in one isolate (CR6). The CR6 isolate had a frame-shift mutation after the second codon leading to six alternative amino acids followed by a premature stop codon.
Figure 1.
Schematic representation of different insertion events identified in the mgrB gene. (a) isolate CR8, (b) isolates CR 28,(c) isolate CR 12
The clinical and demographic details and the susceptibility patterns are given in table 1. All isolates were resistant to colistin (> 16 mg/L) by Vitek 2 system. MICs for colistin by broth-microdilution ranged from 4-1,024 mg/L. None of the isolates were positive for NDM-1 by PCR (table 1). Isolate CR12 was a non-ESBL susceptible to all antibiotics except for colistin. Isolate CR28 and CR8 were ESBL producer. None of the isolates were positive for plasmid mediated colistin resistance genes mcr-1 and mcr-2 by PCR .
Table 1.
Demographics, susceptibility, and molecular characteristics of colistin-resistant K. pneumoniae isolates.
| Sample ID | CR28 | CR12 | CR8 | CR6 |
|---|---|---|---|---|
| Age/Sex | 58/M | 44/M | 68/M | 49/M |
| Underlying condition | DM,CLD, Child B, Left above knee amputation | Asymptomatic bacterinuria | CLD, Child B, subdural haematoma, 2 months hospitalization | Motor neuron disease, bronchopneumonia, sepsis. |
| Outcome | Survived | Asymptomatic | Expired | Survived |
| Sample | Tissue | Urine | Urine | Sputum |
| Ticarcillin/clavulanic acid | R | S | R | R |
| Piperacillin/tazobactam | R | S | R | R |
| Ceftazidime | R | S | R | R |
| Cefoperazone/sulbactam | R | S | R | R |
| Cefepime | R | S | R | R |
| Aztreonam | R | S | R | R |
| Doripenem | S | S | S | R |
| Imipenem | S | S | S | R |
| Meropenem | S | S | S | R |
| Amikacin | R | S | R | R |
| Gentamicin | R | S | R | R |
| Ciprofloxacin | R | S | R | R |
| Minocycline | R | S | R | R |
| Tigecycline | I | S | R | I |
| Colistin MIC broth microdilution (VITEK) | 32 mg/L (16 mg/L) | 16 mg/L (8 mg/L) | 1,024 mg/L (16 mg/L) | 4 mg/L (8 mg/L) |
| Cotrimoxazole | S | S | S | R |
| PCR | ||||
| NDM-1 | Neg | Neg | Neg | Neg |
| CTX-M | Pos | Neg | Pos | Neg |
| mcr-1 | Neg | Neg | Neg | Neg |
| mcr-2 | Neg | Neg | Neg | Neg |
| mgrB truncated by IS | Kpn14 | Kpn14 | Kpn14 | Premature stop codon |
| Insertion site between | +51 and +92 | +132 and +133 | +123 and +124 | NA |
S = susceptible; I = intermediate; R= resistant; Neg = negative; Pos = positive
DISCUSSION
Reports on molecular characterization of colistin resistant isolates from India are scarce. Only two publications from India reported ten K. pneumoniae isolates with mutations in the mgrB, phoP/phoQ, pmrA, pmrB, pmrC, and crrABC regulatory systems [10]. To our knowledge there have been no previous reports of colistin resistance in K. pneumoniae due to insertional inactivation of the mgrB from India. Here we report three cases of infection with K. pneumoniae resistant to colistin due to insertional inactivation of mgrB gene.
Isolate CR8 was surprisingly susceptible to most of the antibiotics and was resistant only to colistin. Further it was isolated from a patient with asymptomatic bacterinuria. Similarly, Kieffer et al. had also reported a susceptible K. pneumoniae isolate recovered from a case of bovine mastitis with resistance to colistin due to insertional inactivation by IS 903B element belonging to IS5 family [14]. The most common IS causing truncation of the mgrB gene belong to the IS5 family [15]. ISKpn14 is a 768 bp IS belonging to the IS1 family. Insertional inactivation by ISKpn14 has been previously reported from a solitary isolate from Colombia and France, three isolates from Turkey and two from Italy [7,14-16]. In all our three isolates the ISKpn14 was inserted at different sites.
The study showed that colistin resistance in 23.5% (4/17) K. pneumoniae isolates was due to loss of function of mgrB which is not in agreement with a recent study from India which reported a rate of 50% [9]. We describe for the first-time from India, insertional inactivation of mgrB by ISKpn14 inserted at different sites, responsible for colistin resistance. Frame shift mutation of mgrB resulting in colistin resistance has been described earlier in a solitary study from India [10]. The resistance in the remaining 13 isolates may be due to mutations in phoP/phoQ, pmrA, pmrB, pmrC, and crrABC regulatory systems as reported in a previous study from India. As opposed to previous reports of pan resistant isolates from India, two of our isolates were not carbapenemase producer among them one was also susceptible to most of the antibiotics. Our study was limited by the fact that mutations in phoP/phoQ, pmrA, pmrB, pmrC, and crrABC regulatory systems which are also responsible for colistin resistance were not investigated and K. pneumoniae strain typing was also not done to determine clonality.
The study showed that colistin resistance in 20% of the K. pneumoniae isolates was due to loss of function of mgrB. We describe for the first-time from India, insertional inactivation of mgrB by ISKpn14 inserted at different sites, responsible for colistin resistance. Plasmid mediated colistin resistance due to mcr-1 and mcr-2 was not identified in K. pneumoniae from India.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest
FUNDING
None to declare
REFERENCES
- 1.Ah Y, Kum AJ, Lee JY. Colistin resistance in Klebsiella pneumoniae. Int.J.Antimicrob.Agents. 2014; 44:8–15. doi: 10.1128/AAC.04763-14 [DOI] [PubMed] [Google Scholar]
- 2.Kumarasamy KK, Toleman MA, Walsh TR, Bagari`a J, Butt F et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010; 10(9):597-602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmeli Y, Akova M, Cornaglia G, Daikos GL, Garau J, et al. Controlling the spread of carbapenemase-producing Gram-negatives: therapeutic approach and infection control. Clin.Microbiol.Infect. 2010; 16:102–111. doi: 10.1111/j.1469-0691.2009.03115.x. [DOI] [PubMed] [Google Scholar]
- 4.Zusman O, Avni T, Leibovici L, Adler A, Friberg L, Stergiopoulou T, Carmeli Y, Paul M. Systematic review and meta-analysis of in vitro synergy of polymyxins and carbapenems. Antimicrob Agents Chemother. 2013; 57(10):5104-11. doi: 10.1128/AAC.01230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M et al. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 6.Cannatelli A, D’Andrea MM, Giani T, Di Pilato V, Arena F et al. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother. 2013; 57: 5521–6. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study.Int J Antimicrob Agents. 2014; 44(6):500-7. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium. Euro Surveill 2016; 7;21(27). doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 9.Pragasam AK, Shankar C, Veeraraghavan B, Biswas I, Nabarro LE et al. Molecular Mechanisms of Colistin Resistance in Klebsiella pneumoniae Causing Bacteremia from India—A First Report. Front. Microbiol. 2017;7:2135. doi: 10.3389/fmicb.2016.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veeraraghavan B, Perumalla SK, Devanga Ragupathi NK, Pragasam AK, Muthuirulandi Sethuvel DP, Inian S, et al. Coexistence of Fosfomycin and Colistin Resistance in Klebsiella pneumoniae: Whole-Genome Shotgun Sequencing. Genome Announc. 2016;23:4(6). doi: 10.1128/genomeA.01303-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing 2015. Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0. http://www.eucast.org/.
- 12.Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow. Aerobically; Approved Standard–Tenth Edition. CLSI document M7-A10 Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 13.Mathai D, Kumar VA, Paul B, Sugumar M, John KR, Manoharan A, et al. Fecal carriage rates of extended-spectrum b-lactamase-producing Escherichia coli among antibiotic naive healthy human volunteers. Microb Drug Resist 2015;21:59–64. doi: 10.1089/mdr.2014.0031. [DOI] [PubMed] [Google Scholar]
- 14.Kieffer N, Poirel L, Nordmann P, Madec JY, Haenni M. Emergence of colistin resistance in Klebsiella pneumoniae from veterinary medicine. J Antimicrob Chemother. 2015; 70(4):1265-7. doi: 10.1093/jac/dku485. [DOI] [PubMed] [Google Scholar]
- 15.Cannatelli A, Giani T, D’Andrea MM, Di Pilato V, Arena F et al. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. 2014; 58(10):5696-703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordmann P, Jayol A, Poirel L. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis. 2016; 22:1038-1043. doi: 10.3201/eid2206.151840. [DOI] [PMC free article] [PubMed] [Google Scholar]