Abstract
Anion channelrhodopsin-2 (ACR2), a light-gated channel recently identified from the cryptophyte alga Guillardia theta, exhibits anion channel activity with exclusive selectivity. In addition to its novel function, ACR2 has become a focus of interest as a powerful tool for optogenetics. Here we combined experimental and computational approaches to investigate the roles of conserved carboxylates on the anion transport activity of ACR2 in Escherichia coli membrane. First, we replaced six conserved carboxylates with a neutral residue (i.e. E9Q, E56Q, E64Q, E159Q, E219Q and D230N), and measured anion transport activity using E. coli expression system. E159Q and D230N exhibited significantly lower anion transport activity compared with wild-type ACR2 (1/12~1/3.4), which suggests that E159 and D230 play important roles in the anion transport. Second, to explain its molecular aspects, we constructed a homology model of ACR2 based on the crystal structure of a cation channelrhodopsin (ChR). The model structure showed a cavity formed by four transmembrane helices (TM1, TM2, TM3 and TM7) similar to ChRs, as a putative anion conducting pathway. Although E159 is not located in the putative pathway, the model structure showed hydrogen bonds between E159 and R129 with a water molecule. D230 is located in the pathway near the protonated Schiff base (PSB) of the chromophore retinal, which suggests that there is an interaction between D230 and the PSB. Thus, we demonstrated the functional importance and the hypothetical roles of two conserved carboxylates, E159 and D230, in the anion transport activity of ACR2 in E. coli membrane.
Keywords: microbial rhodopsin, anion channel, retinal, ion transport
Significance.
Anion channelrhodopsin-2 (ACR2) was recently identified from the eukaryotic alga Guillardia theta as a light-gated anion channel that consists of seven-transmembrane helices and a retinal chromophore. Here we investigated the roles of six conserved carboxylates in the anion transport activity of ACR2 using an Escherichia coli expression system. Two of those carboxylates, E159 and D230, were identified as functionally important residues. A homology model structure of ACR2 showed that E159 and D230 have significant interactions with R129 and the retinal chromophore, respectively. These combined experimental and theoretical approaches provide molecular insights into the anion transport mechanism of ACR2.
Organisms such as archaea, eubacteria and eukarya receive light to capture energy and information from their environments. Rhodopsin, which is a photoreceptive protein family consisting of a 7-transmembrane helical apoprotein called opsin and vitamin-A aldehyde retinal as a chromophore, plays central roles in the light reception of those organisms [1,2]. The chromophore retinal binds to a conserved Lys residue of opsin through a protonated Schiff base (PSB) linkage, and trans-cis or cis-trans photoisomerization of the retinal triggers the sequential conformational changes of the protein moiety [1,2].
Based on their amino acid sequences, rhodopsins are classified into two types, microbial and animal rhodopsins. Animal rhodopsins are widely distributed in the animal world (including humans) and generally work as G protein coupled receptors (GPCRs) and as retinal photoisomerases in the retina and in light-sensitive organs [2,3]. In contrast, microbial rhodopsins are widely distributed in the microbial world, including archaea, eubacteria and eukarya, and show a variety of biological functions, such as light-driven ion pumps, light-gated ion channels and light sensors [1,4]. In 1971, Drs. Oesterhelt and Stoeckenius identified the first microbial rhodopsin, bacteriorhodopsin (BR), from the archaeon Halobacterium salinarum as a light-driven outward proton pump that produces adenosine triphosphate (ATP) [5]. Since then, BR has become a focus of interest as a model for photoactive proteins, ion transporters and membrane proteins [1,6]. Discoveries of novel microbial rhodopsins showing a variety of biological functions were started in 1999 by the advances in genomics and bioinformatics [7,8]. Among them, in 2002 and 2003, two novel microbial rhodopsins, channelrhodopsin-1 (CrChR1) and channelrhodopsin-2 (CrChR2), were identified from the eukaryotic alga Chlamydomonas reinhardtii as light-gated cation-selective channels [9,10]. Both of those cation channelrhodopsins (ChRs) absorb blue light (~480 nm for CrChR1 and ~465 nm for CrChR2) and are responsible for the phototactic behavior of the alga [10,11]. In 2005, it was reported that ChRs ectopically expressed in mammalian neurons control the action potential firing in high time- and spatial-resolution upon blue-light irradiation [12]. This is due to the depolarization of the cell membrane upon light irradiation. Thus, this finding indicates that microbial rhodopsins are useful as powerful optogenetic tools for controlling neural activity and animal behavior with light by changing the ion gradient between the extracellular and intracellular sides of cell membranes [13,14].
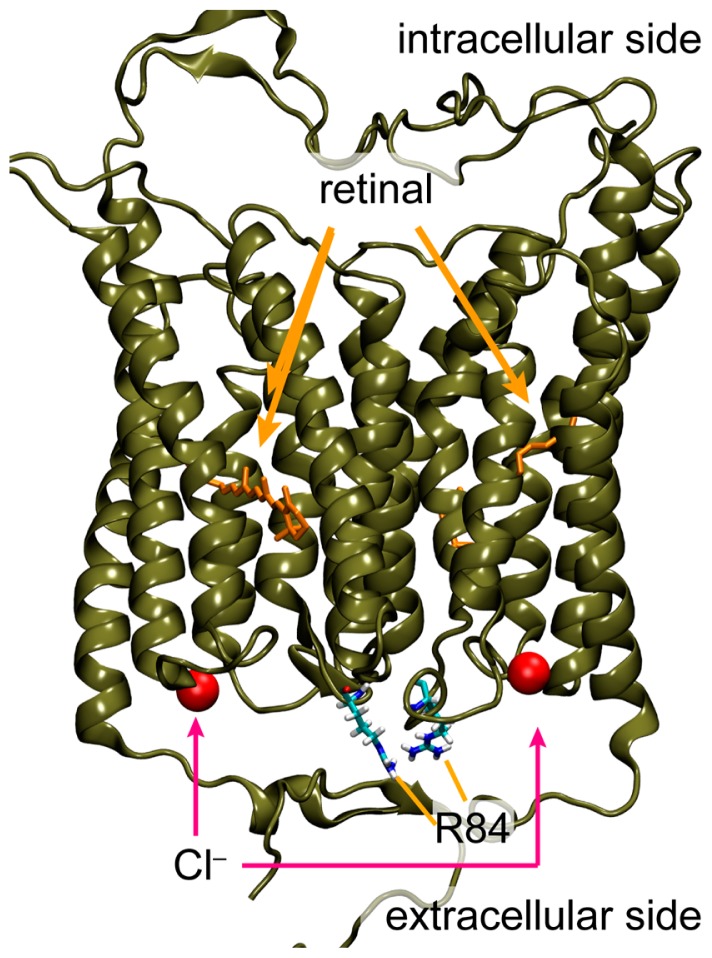
As opposed to ChRs, an outward proton pumping rhodopsin, Archaerhodopsin-3 (AR3), and an inward chloride pumping rhodopsin, Natronomonas pharaonis Halorhodopsin (NpHR) have been used to suppress the action potential firing in mammalian neurons [15–17]. This is due to the hyperpolarization of the cell membrane upon light irradiation. Recently, anion channelrhodopsin-1 (ACR1) and anion channelrhodopsin-2 (ACR2) were identified from the cryptophyte alga Guillardia theta, as light-gated anion-selective channels, after which other related homologues have been identified in different cryptophyte species [18,19]. When ACRs are expressed in mammalian cells in culture, they transport monovalent anions (chloride and bromide) upon light irradiation and exhibit exclusive anion channel selectivity. ACRs also generate ~1000-fold higher hyperpolarizing photocurrents than the conventional neural silencer AR3, suggesting that ACRs can be used as a powerful neural silencer for optogenetics [18]. So far, many electrophysiological, spectroscopic, structural and computational studies of ChRs have been performed to elucidate the molecular mechanism of the efficient cation conduction in a light-dependent manner [9,10,20–24]. Especially, high-resolution crystal structure of a chimeric protein named C1C2 has provided valuable structural information at the atomic level [22]. During the preparation of our manuscript, the crystal structure of CrChR2 has been published [23]. Of note, the overall structure is similar to that of the chimeric ChR (root mean square deviation, RMSD = 0.9 angstrom). As opposed to ChRs, little is known about the molecular mechanism of the anion transport in ACRs (Fig. 1A). Judging from the sequence analysis, the functionally important amino acid residues and structural modules of ChRs are not conserved in ACRs. Therefore, the channel mechanisms of ACRs, such as their gating-kinetics, ion selectivity and color-tuning, are seemingly different from those of ChRs and an analogy-based expectation about their molecular functions may not be feasible.
Figure 1.
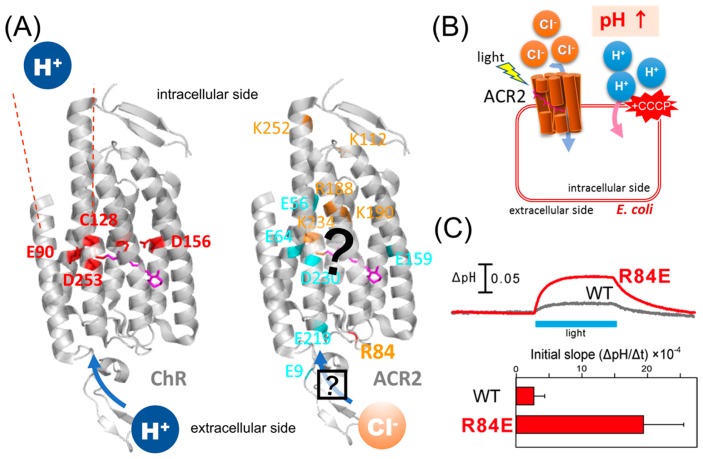
Characteristics of ChRs and ACRs. (A) (Left) Crystal structure of ChR (PDB;3UG9) showing that the cation conducting pathway is formed by TM1, TM2, TM3 and TM7. Key residues for the cation transport in ChRs are colored red. (Right) High resolution structure and the anion conducting pathway of ACR2 are still unclear. Conserved basic and acidic amino acid residues are colored orange and cyan, respectively. (B) Light-induced secondary proton movement across the cell membrane by the inward chloride channeling activity of ACR2 expressed in E. coli cells, which is facilitated by the addition of CCCP. The proton movement results in increases in extracellular pH. (C) (Upper) Light-induced pH changes of E. coli cells expressing wild-type ACR2 or the R84E mutant in a solution containing 300 mM NaCl in the presence of CCCP. The cell suspensions were illuminated with blue light (480±10 nm) for 3 min (blue stripe). (Lower) Comparison of the anion transport activity of wild-type ACR2 and the R84E mutant. These data are taken from our previous study [25].
As a first step toward characterizing the anion channeling mechanism, in this study, we combined mutagenesis and computational analysis of ACR2 to complement the respective advantages. In an earlier study, we established a functional expression system of ACR2 using Escherichia coli cells (Fig. 1B). The E. coli expression system has great advantages with respect to the rapid growth of those cells and the ease of genetic and proteomic modifications. By taking advantage of the expression system, we performed mutational analysis on the conserved “basic” amino acid residues among ACRs, and found that the positive charge of R84 located at the extracellular side plays an inhibitory role in its anion transport activity (Fig. 1C) [25], although the precise position and the interaction remain unknown. The prediction of protein structure by molecular simulation and informatics is still developing. However, the effective combination of abundant experience and knowledge can lead to useful insights for detailed molecular structures and functions. Indeed, we proposed a model structure of ChR2 using computational approaches, and predicted the key residues involved in channel function [26], which were confirmed by the crystal structure of the chimeric ChR [22].
Here we focused on six conserved “acidic” amino acid residues in ACR2 identified by sequence analysis and investigated their roles in anion transport. First, we replaced each of the six conserved carboxylates in ACR2 with a neutral residue (i.e. E9Q, E56Q, E64Q, E159Q, E219Q and D230N), and measured the anion transport activity of the mutant proteins using our newly developed E. coli expression system. Second, we then built a homology model of ACR2 from the crystal structure of the chimeric ChR, followed by molecular dynamics (MD) simulations for structural refinement and evaluation of stability. Finally, we discuss the roles of the carboxylates on anion transport in ACR2 from experimental and structural aspects.
Materials and Methods
Gene preparation and protein expression
A cDNA encoding the 7-transmembrane domain of ACR2 (Genbank accession no. KP171709, amino acid residues from the first to the 266th position) was inserted into the pET22b plasmid vector (Novagen, USA) with NdeI and XhoI restriction enzyme sites as previously described [25]. Consequently, the plasmid encoded a cDNA with a hexahistidine-tag at the C-terminus. Mutant genes were constructed using the QuickChange site-directed mutagenesis method, the SLiCE method or an In-Fusion Cloning Kit according, to the manufacturer’s instructions as previously described [25,27,28]. Proteins were expressed in E. coli BL21 (DE3) cells using the same experimental procedures as previously described [25]. Briefly, freshly colonized cells harboring the expression plasmids were incubated in 100 mL LB medium containing 50 μg/mL ampicillin and were grown at 30°C for 6 hr. After the pre-culture, the growth medium was directly transferred to 1.9 L LB medium containing 50 μg/mL ampicillin, and the cells were grown at 30°C until the optical density at 660 nm reached 0.2–0.4. The cells were kept on ice for 15 min, then isopropyl -D-thiogalactopyranoside (IPTG, final concentration = 0.5 mM) and all-trans retinal (final concentration = 10 μM) were added to the culture medium and incubated at 18°C for 12–14 hr to induce protein expression. The cells were then collected by centrifugation (7500×g for 10 min at 4°C).
SDS-PAGE and western blotting analysis
SDS-PAGE and western blotting analysis were performed according to the standard method as previously described [25]. Briefly, E. coli cells expressing wild-type ACR2 or its mutants were suspended in a buffer containing 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl. The optical density at 660 nm of all samples was initially set at 0.6 and the cell suspensions were then diluted 10-times with the same buffer. The diluted samples (5 μL) were suspended in SDS-PAGE loading buffer (5 μL) containing 5% 2-mercaptoethanol, and were heated at 95°C for 5 min. After that, the samples (10 μL) were separated by 12% acrylamide SDS-PAGE. Immunoblotting analysis was then performed using an anti His-tag HRP conjugate antibody (Novagen, USA) according to the manufacturer’s instructions. E. coli cells harboring the pET22b vector plasmid alone were simultaneously analyzed as a negative control. To assess the protein expression level of the wild-type ACR2 and its mutants, we quantitatively evaluated the band intensities using ImageJ public domain software.
Light-driven anion transport measurements
Anion transport activity was measured by light-induced pH changes using essentially the same method as previously described [25,29]. Briefly, E. coli BL21(DE3) cells harboring expression plasmids were suspended and washed 3 times in a solution containing 300 mM NaCl to remove LB culture medium. After that, cells were resuspended in the same solutions and kept on ice until they were used for measurements. For the measurements, the optical density at 660 nm of all samples was set at 6 and the samples were kept in the dark until the pH of the solution became stable. The samples were then illuminated with a Xenon lamp (Asahi Spectra Co. Ltd., Tokyo, Japan) through a band-pass filter (480±10 nm), where the light intensity was measured and adjusted to 8–10 mW/cm2 using an optical power meter (Hioki, Ueda, Japan) with an optical sensor (Hioki, Ueda, Japan). If necessary, the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP, Sigma-Aldrich, USA) was added to the medium to a final concentration of 10 μM. E. coli cells harboring the pET22b vector plasmid alone were simultaneously analyzed as a negative control. The temperature of the samples was maintained at 25°C. Statistical significance was evaluated by the Dunnett’s test for two group comparisons.
Generation of a homology model structure of ACR2
We generated a homology model of ACR2 using the chimeric ChR crystal structure as a template [22] with the SWISS-MODEL web interface [30]. The crystal structure corresponds to the residues 1 to 279 of ACR2. Therefore the length of ACR2 for the theoretical simulation (amino acids from 1 to 279) is different from that for the experimental studies (amino acids from 1 to 266). The alignment used for homology modeling is shown in Supplementary Figure S1A. In the homology model, all aspartic and glutamic acid residues except for E64 were negatively charged, while all lysine and arginine residues were positively charged. On the other hand, we assumed that all histidine residues were electrostatically neutral with a hydrogen bound to Nδ atom. The internal water molecules were placed at the same positions as in the chimeric ChR crystal structure. The model structure was dimerized and embedded in a POPC lipid bilayer because the chimeric ChR was solved as a homo-dimer in the lipidic cubic phase [22]. The membrane assembly was solvated and electrostatically neutralized by the addition of ions to the bulk water phase. The system was then structurally optimized, which was followed by a 10 ns-equilibrating MD simulation. The production MD run was conducted for 100 ns with a Nose-Hoover thermostat [31] at a time constant tt = 0.5 ps and 300 K reference temperature. A pressure of 1.0 bar was maintained using the Parrinello-Rahman barostat [32] at a time constant τp = 5.0 ps and a compressibility parameter of β = 4.5×10−6 bar−1 for the x, y direction and β = 4.5×10−5 bar−1 for the z direction. All molecular simulations were carried out using GROMACS 5.1.2 [33] with an adopted CHARMM27 force-field [34].
Results and Discussion
It is well known that charged residues in membrane-embedded ion transporters, such as Lys, Arg, Glu and Asp, play crucial roles in their function. In our recent study, we performed mutational analysis of conserved “basic” amino acid residues (Fig. 1) taking advantage of our newly developed E. coli expression system of ACR2 [25]. In viable cellular conditions, ACR2 conducts chloride ions from the extracellular to the intracellular side. Thus, R84 in the extracellular side of ACR2 intuitively seems to be related to the anion uptake. Notably, however, we found that R84 plays an inhibitory role [25], which implied the presence of other relevant residues to the anion uptake.
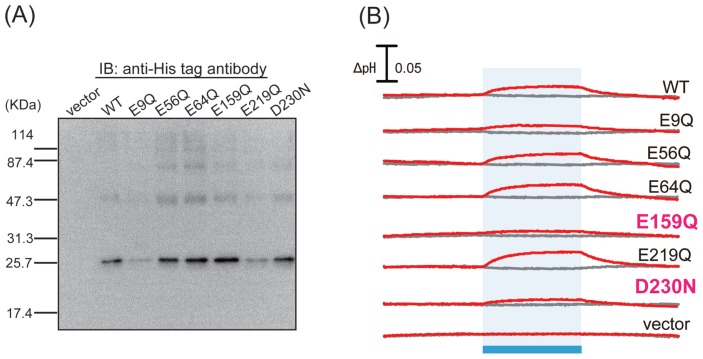
On the basis of that background, as the next step to identify key residues involved in the anion transport of ACR2, we focused on the conserved “acidic” amino acid residues in ACRs. From the alignment of the amino acid sequences of ACR1 and ACR2 (Supplementary Fig. S1A), we identified six conserved carboxylates, E9, E56, E64, E159, E219 and D230. We then constructed mutants in which those carboxylates are replaced with a neutral residue, Gln or Asn (i.e. E9Q, E56Q, E64Q, E159Q, E219Q and D230N), and expressed those mutant proteins using our E. coli expression system. To assess protein expression levels, we performed western blotting analysis and compared them with wild-type ACR2 (Fig. 2A). As a result, the relative band intensity of the mutant protein to the wild-type one was estimated to be 0.3, 1.2, 1.4, 1.5, 0.6 and 1.3 for E9Q, E56Q, E64Q, E159Q, E219Q and D230N, respectively. Thus, the band intensity of E9Q was remarkably lower than the wild-type and the other mutants, suggesting a significantly lower expression level of E9Q. In contrast, the other 5 mutants were successfully expressed in E. coli cells with roughly comparable expression levels to the wild-type.
Figure 2.
Expression and measurement of anion transport of wild-type ACR2 and its mutants. (A) Western blotting analysis of wild-type ACR2 and its mutants. Cells harboring the pET22b vector alone were used as a negative control. (B) Light-induced pH changes of E. coli cells expressing wild-type ACR2 or its mutants in a solution containing 300 mM NaCl in the absence or presence of CCCP (gray and red lines, respectively). The cell suspensions were illuminated with blue light (480±10 nm) for 3 min (blue stripe).
In order to analyze the anion transport activity, we then measured the light-induced pH changes in E. coli cells expressing the wild-type or mutant ACR2 proteins in a solution containing 300 mM NaCl in the presence or absence of the protonophore, CCCP, which can conduct protons across lipid bilayers (Fig. 2B). In the wild-type ACR2, a weak decrease or an increase in pH was observed upon blue light illumination in the presence or absence of CCCP, respectively, as was observed in our previous study [25]. The light-induced increase in extracellular pH reflects the secondary inward proton transport across the cell membrane, which is facilitated by the inward chloride ion transport of ACR2 in the presence of CCCP. On the other hand, the weak decrease in pH observed in the absence of CCCP reflects an outward proton pumping activity of ACR2 expressed in E. coli cells [25]. The mutants also had the same tendency as the wild-type, although the signal amplitudes were different.
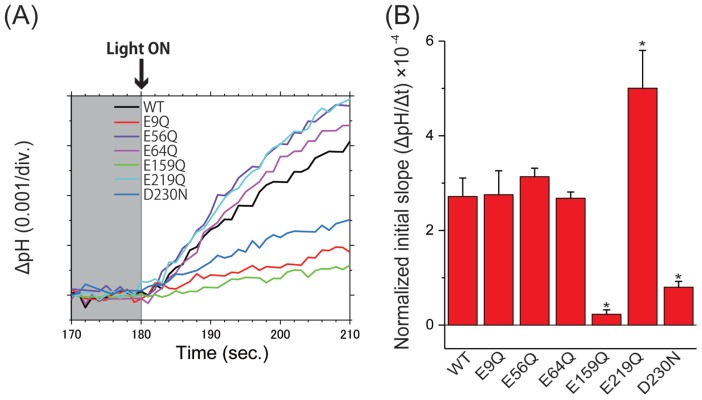
We then evaluated the initial slope amplitudes of the light-induced pH increase from 0 to 10 s upon blue light illumination in the presence of CCCP as chloride ion transport activity of the wild-type and mutant ACR2 proteins (Fig. 3). The initial slope amplitudes were normalized with the band intensities of the western blotting analysis (Fig. 2A) as an index of the protein expression level. The amplitudes of some mutants (E9Q, E56Q and E64Q) were similar to that of the wild-type, suggesting that the mutants maintain comparable chloride ion transport activity to the wild-type. On the other hand, the amplitudes of E159Q and D230N were 12- and 3.4-fold lower than the wild-type, respectively, indicating that E159 and D230 play important roles in the anion transport activity of ACR2. In order to exclude the possibility that the decrease in amplitudes was due to changes of spectral sensitivity caused by the E159Q and D230N mutations, we compared the signal amplitudes of the mutants at varying wavelengths of light (Supplementary Fig. S2). The amplitudes in all wavelength conditions were equivalent with or lower than those observed in blue light illumination (480±10 nm). These results strongly suggested that the decrease in signal amplitudes reflected the anion transport activity. The mean values of E159Q irradiated at 420 and 520 nm were slightly negative, suggesting outward proton transport. However, the absolute values were much less (1/10) than those of the wild-type (Fig. 3B), and therefore it is difficult to have some conclusion. In addition, the normalized amplitude of E219Q was significantly (1.9-fold) higher than that of the wild-type, suggesting that E219 plays an inhibitory role in the anion transport activity of ACR2.
Figure 3.
Comparison of anion transport activity of wild-type ACR2 and its mutants. (A) Light-induced pH changes of wild-type ACR2 and its mutants in the presence of CCCP. The pH values are offset by the initial pH values. (B) Comparison of the anion transport activity of wild-type ACR2 and its mutants. The initial slope amplitudes of the light-induced pH changes, which were obtained from the data in Panel A, were normalized with the band intensities of the western blotting analysis (Fig. 2A) as an index of the protein expression level. All error bars represent the SEM of more than three independent measurements. Asterisks (*) indicate a significant difference from the wild-type (P<0.05; Dunnett’s test).
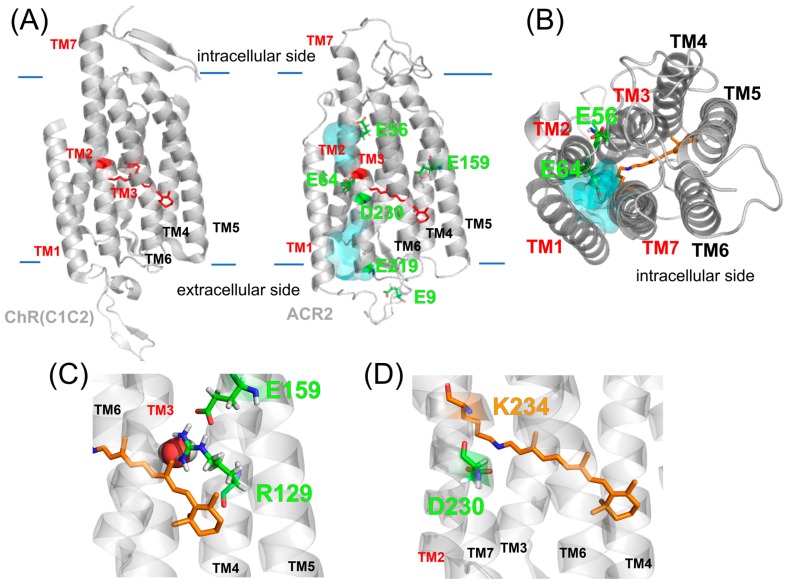
Since we successfully identified the key carboxylates involved in the anion transport of ACR2, we next constructed a homology model of ACR2 from the crystal structure of the chimeric ChR [22] to understand the functional mechanism of the carboxylates in anion transport at the atomic level. The crystal structures of ChRs revealed an electronegative pore formed by TM1, TM2, TM3 and TM7 [22,23]. In addition, the previous electrophysiological analysis indicated that some mutations of residues within the pore reduced the peak amplitudes of the photocurrent [22,26], which strongly suggested that the cation conducting pathway is formed by TM1, TM2, TM3 and TM7 in ChRs. The overall structure of the homology model of ACR2 is shown in Figures 4A and 4B. In that model structure, we found a cavity consisting of TM1, TM2, TM3 and TM7, and water molecules were distributed in that cavity, suggesting that the anion conducting pathway is formed by TM1, TM2, TM3 and TM7 in ACR2, which is similar to the cation conducting pathway in ChRs. Indeed, we observed that a chloride ion comes into the cavity of the respective monomers of a dimeric ACR2 during these MD simulations (Supplementary Movie S1). The previous MD simulations for CrChR2 suggested that cations might be transported along some polar and/or negatively charged residues, N53, Q56 E97 and E101. In particular, it is likely that the negative charge of E97 and E101 are related to the cation uptake [35]. On the other hand, ACR2 does not have any counterion residues (positively charged residues) in the corresponding sites. Instead, chloride ions are coordinated by several backbone NH groups around the edge of TM2 and the loop region between TM2 and TM3 (BC-loop) (Fig. 5). Note that the BC-loop in the ACR model was shorter and showed low sequence homology compared to CrChR2, which suggested that the homology model was not necessarily accurate for the BC-loop. To assess the model dependency of chloride binding, we built an alternative homology model from a different sequence alignment where the BC-loop firmly forms a β-sheet as occurs in ChRs (Supplementary Fig. S1B), and then conducted MD simulations for 200 ns. However, this model did not show rigid chloride binding, which thus implied the sensitivity of chloride binding and modeling difficulty. In other words, the low similarity between ChRs and ACRs implies that the BC-loop may be closely involved with the anion channel function. Indeed, mutation of R84 in the BC-loop affects the anion conductance (Fig. 1C) [25]. In the present models, R84 is located at the dimer interface interacting with some negatively charged amino acids in its own or in the other monomer loop regions. We suppose that the attractive interaction of R84 may inhibit the channel opening and thus a mutation of that residue can substantially enhance the anion transport activity as experimentally reported (Fig. 1C). A similar inhibitory role in the anion transport (i.e., enhancement of the anion transport activity by the mutation) was also observed for E219Q mutant (Fig. 3B). In the model structure, E219 was located in TM7 forming the putative anion conductance pathway (Figs. 4A and 4B) and was close to R84 (Fig. 1A). Therefore we hypothesized that E219 interacts with R84 and hampers the anion transport.
Figure 4.
Homology model structure of ACR2. (A) Crystal structure of the chimeric ChR (PDB; 3UG9) and the homology model of ACR2. In both models, the cavity (cyan colored) seems to be formed by TM1, TM2, TM3 and TM7. (B) Intracellular side of the homology model of ACR2. The cavity is colored cyan. (C, D) Detailed structures around E159 (C) and D230 (D).
Figure 5.
Hypothetical chloride binding site identified in the MD simulations. Overview of the hypothetical binding site in the dimeric structure of the homology model of ACR2.
In the model structure, although E159 was located in the center of TM5 apart from the putative anion conducting pathway, the E159Q mutation decreased the chloride ion transport activity (Fig. 3A). It is notable that in the present model, E159 formed a characteristic structural hydrogen bonding network with C98, S126 and R129 (Figs. 4B and 4C). For ChRs, D156 (in CrChR2) in the center of TM4 is probably protonated and forms a hydrogen bond with C128 (in CrChR2) in TM3 via a water molecule, where D156 is a proton donor to the deprotonated Schiff base [21,23,35]. Prof. Heberle and coworkers demonstrated that mutations of D156 and C128 significantly slow the photocycle kinetics in ChRs and thus proposed the hydrogen bond network as a key structural module (called the DC-gate) in the control of channel kinetics in ChRs [36,37]. In contrast, in ACR2, the Asp residue (D156 in CrChR2) is replaced with Ser (S126 in ACR2), suggesting the lack of the DC-gate. Instead, the interactions of C98 with S126 and E159 are mediated by R129. Therefore, we hypothesized that this remarkable hydrogen bond network plays an important role in the control of channel kinetics, such as the equilibrium between the conductive and non-conductive state during the photoreaction, in ACR2 instead of the DC-gate.
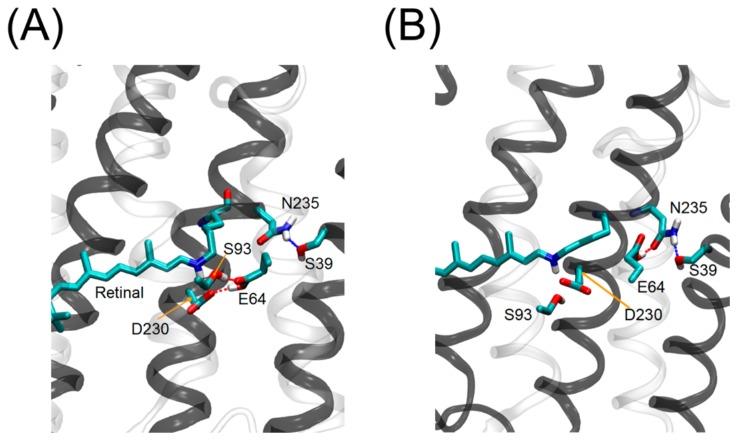
D230 in TM7 was located in the vicinity of the protonated retinal Schiff base. The present study demonstrated that the D230N mutation in ACR2 decreased but did not completely abolish the anion transport activity (Fig. 3). It is well known that the corresponding residue, D253 in CrChR2, (Supplementary Fig. S1A), works as a counterion which stabilizes the PSB in the original state and receives a proton from the PSB during the photocycle [21,22]. Mutations of D253 to an Ala or Asn residue in ChRs almost completely abolished the photocurrent [21,22]. In contrast, it is reported that the mutation of the corresponding residue (D234N) in ACR1 did not substantially influence the amplitude of the photocurrent [38,39]. Alternatively, they found that the E68Q mutation slowed the formation of the M intermediate where the Schiff base is deprotonated. Since deprotonation of the Schiff base is a primary factor for the photocycle presumably both for ChRs and ACRs, E68 has been assumed to be the proton acceptor in ACR1 (i.e., deprotonated residue) [40]. On the other hand, Yi et al. claimed that both E68 (E64 in ACR2) and D234 (D230 in ACR2) are protonated and there is a negatively charged residue that stabilizes the positively charged Schiff base nitrogen [41]. Thus, the protonation states both of E68 and D234 in ACR1 are still ongoing debate. As described, ACR2 has a blue-shifted absorption maximum (460 nm). The visible colors of microbial rhodopsins originate from an energy gap between the electronic ground and excited states. When both E64 and D230 are protonated in the dark state, the absorption maximum of ACR2 should be shifted to the longer wavelength, because of decreases in energy gap between electronic ground and excited states. Therefore, we presume that either E64 or D230 is deprotonated in ACR2. As a possibility, we suppose that D230 is more likely to be deprotonated on the basis of a number of experiences for microbial rhodopsins [1], although the present experiments are not decisive to identify which residue is negatively charged. E64 in ACR2 is located in the center of TM2. Regardless of the high sequential homology between ACR1 and ACR2, the E64Q mutation in ACR2 did not show a distinct difference from the wild-type regarding the chloride ion transport activity. This is inconsistent with a previous report for the same mutation of the corresponding residue E68Q in ACR1, which led to a substantial suppression of the gating mechanism. Therefore, although we assumed that D230 is not the primary proton acceptor in ACR2, D230 seems to play some role in the photocycle, such as the turnover rate and the equilibrium between the conductive and non-conductive states during the photoreaction. MD simulations have provided useful insights as follows: although E64 forms a hydrogen bond with N235 at the beginning of the MD simulation as it appears in the template chimeric ChR crystal structure, we observed that E64 swaps the hydrogen bond partner from N235 to D230 and S93 from time to time (Fig. 6). It is notable that the orientation of E90 (E64 in ACR2) is inconsistent between the crystal structures of the chimeric ChR and CrChR2. In the chimeric ChR crystal structure, E64 forms a hydrogen bond with N235, while it is oriented to the active site in the CrChR2 crystal structure. Therefore, the two observed orientations of E64 in the present ACR2 simulations correspond to those in the chimeric ChR and CrChR2 crystal structures, respectively. In addition, the previous simulations suggested that E90 is probably protonated in the chimeric ChR crystal structure [35]. The orientation of E64 interacting with D230 (Fig. 6A) corresponds with that of E90 in the CrChR2 crystal structure [23]. Therefore, we assume that the E64 orientation is sensitive to the electrostatic conditions of the active site, in particular the protonation state of the counterion, and E64 can work as an alternative proton acceptor when D230 is replaced by a neutral residue (or the other way around). However, in order to identify which basic residue works as a primary counterion in ACR2, further spectroscopic experiments using purified mutant proteins are needed.
Figure 6.
Sidechain orientations of E64 in ACR2. Two sidechain orientations of E64 were observed in the MD simulations. E64 interacts with D230 (A) and N235 (B) in the model structure of ACR2.
We then focused on mutations of the three conserved carboxylates, E56, E64 and E219. In the model structure, two of them, E56 and E64, were located in TM2 while the other one, E219, was located in TM7 forming the putative anion conductance pathway (Figs. 4A and 4B). E56 and E64 in ACR2 correspond to E82 and E90 in CrChR2, respectively (Supplementary Fig. S1A). It has been reported that the E82A and E90Q mutations significantly reduce the amplitude of the photocurrent in CrChR2 [20,42]. In addition, the E90K and E90R mutations convert the cation channel activity into an anion channel activity [20]. Thus, E82 plays an important role in cation conductance and selectivity in CrChR2. However, the E56Q mutation in ACR2 did not substantially influence the chloride ion transport activity, which is consistent with a previous report for the same mutation of the corresponding residue in ACR1 (E60Q) [39]. E56 is not oriented to the putative anion conducting pathway in the model structure of ACR2 (Fig. 4B). Thus, the carboxylates would play different roles in ChRs and ACRs. The sum of our experimental and computational results suggest that interactions of two conserved carboxylates (E159-R129 and D230-PSB) control the channel kinetics and anion transport activity in ACR2.
Conclusion
In this study, we performed experimental and computational analysis on six conserved carboxylates of ACR2 to investigate their roles in anion transport. We demonstrated that E159 and D230 form important interactions with R129 and the PSB, respectively, to play important roles in the anion transport activity of ACR2.
Supplementary Information
Acknowledgements
This work was financially supported by JSPS KAKENHI Grant Numbers JP17K15101 to HW, JP18H05155 to HI, and JP15H04363, JP15H00878, JP25104005 and JP17H05726 to YS. This research was partially supported by JST PRESTO (JPMJPR17GC) to HW; JST CREST (JPMJCR1656) to HI and YS, AMED to HI and YS, the Materials Integration for Engineering Polymers of Cross-Ministerial Strategic Innovation Promotion Program (SIP), the Interdisciplinary Computational Science Program in Center for Computational Sciences at the University of Tsukuba, the TSUBAME Encouragement Program for Young/Female Users of Global Scientific Information and Computing Center at the Tokyo Institute of Technology, and the Joint Usage/Research Center for Interdisciplinary Large-Scale Information Infrastructures in Japan.
Footnotes
Conflict of Interest
There are no conflicts of interest to declare. The authors declare no competing financial interests.
Author contributions
K. K. and S. D. performed experimental studies. H. C. W., M. K. and H. I performed theoretical studies. N. M. analyzed data. H. I. and Y. S. designed research. K. K., H. C. W., H. I. and Y. S. wrote the paper.
References
- 1.Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shichida Y, Matsuyama T. Evolution of opsins and photo-transduction. Philos Trans R Soc Lond, B, Biol Sci. 2009;364:2881–2895. doi: 10.1098/rstb.2009.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yau KW, Hardie RC. Photo-transduction motifs and variations. Cell. 2009;139:246–264. doi: 10.1016/j.cell.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurihara M, Sudo Y. Microbial rhodopsins: wide distribution, rich diversity and great potential. Biophys Physicobiol. 2015;12:121–129. doi: 10.2142/biophysico.12.0_121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature New Biol. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 6.Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 7.Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 8.Sharma AK, Spudich JL, Doolittle WF. Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol. 2006;14:463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 10.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudo Y, Okazaki A, Ono H, Yagasaki J, Sugo S, Kamiya M, et al. A blue-shifted light-driven proton pump for neural silencing. J Biol Chem. 2013;288:20624–20632. doi: 10.1074/jbc.M113.475533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science. 2015;349:647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govorunova EG, Sineshchekov OA, Rodarte EM, Janz R, Morelle O, Melkonian M, et al. The Expanding Family of Natural Anion Channelrhodopsins Reveals Large Variations in Kinetics, Conductance, and Spectral Sensitivity. Sci Rep. 2017;7:43358. doi: 10.1038/srep43358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenhauer K, Kuhne J, Ritter E, Berndt A, Wolf S, Freier E, et al. In channelrhodopsin-2 Glu-90 is crucial for ion selectivity and is deprotonated during the photocycle. J Biol Chem. 2012;287:6904–6911. doi: 10.1074/jbc.M111.327700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lórenz-Fonfría VA, Resler T, Krause N, Nack M, Gossing M, Fischer von Mollard G, et al. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc Natl Acad Sci USA. 2013;110:E1273–1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, Hirata K, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volkov O, Kovalev K, Polovinkin V, Borshchevskiy V, Bamann C, Astashkin R, et al. Structural insights into ion conduction by channelrhodopsin 2. Science. 2017;358 doi: 10.1126/science.aan8862. [DOI] [PubMed] [Google Scholar]
- 24.Wietek J, Wiegert JS, Adeishvili N, Schneider F, Watanabe H, Tsunoda SP, et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 25.Doi S, Tsukamoto T, Yoshizawa S, Sudo Y. An inhibitory role of Arg-84 in anion channelrhodopsin-2 expressed in Escherichia coli. Sci Rep. 2017;7:41879. doi: 10.1038/srep41879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe HC, Welke K, Schneider F, Tsunoda S, Zhang F, Deisseroth K, et al. Structural model of channelrhodopsin. J Biol Chem. 2012;287:7456–7466. doi: 10.1074/jbc.M111.320309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Werling U, Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40:e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kojima K, Yamashita T, Imamoto Y, Kusakabe TG, Tsuda M, Shichida Y. Evolutionary steps involving counterion displacement in a tunicate opsin. Proc Natl Acad Sci USA. 2017;114:6028–6033. doi: 10.1073/pnas.1701088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doi S, Mori A, Tsukamoto T, Reissig L, Ihara K, Sudo Y. Structural and functional roles of the N- and C-terminal extended modules in channelrhodopsin-1. Photochem Photobiol Sci. 2015;14:1628–1636. doi: 10.1039/c5pp00213c. [DOI] [PubMed] [Google Scholar]
- 30.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoover WG, Holian BL. Kinetic moments method for the canonical ensemble distribution. Phys Lett A. 1996;211:253–257. [Google Scholar]
- 32.Parrinello M, Rahman A. Polymorphic Transitions in Single-Crystals— a New Molecular-Dynamics Method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 33.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 34.Feller SE, Gawrisch K, MacKerell AD., Jr Polyunsaturated fatty acids in lipid bilayers: intrinsic and environmental contributions to their unique physical properties. J Am Chem Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe HC, Welke K, Sindhikara DJ, Hegemann P, Elstner M. Towards an understanding of channelrhodopsin function: simulations lead to novel insights of the channel mechanism. J Mol Biol. 2013;425:1795–1814. doi: 10.1016/j.jmb.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 36.Radu I, Bamann C, Nack M, Nagel G, Bamberg E, Heberle J. Conformational changes of channelrhodopsin-2. J Am Chem Soc. 2009;131:7313–7319. doi: 10.1021/ja8084274. [DOI] [PubMed] [Google Scholar]
- 37.Nack M, Radu I, Gossing M, Bamann C, Bamberg E, von Mollard GF, et al. The DC gate in Channelrhodopsin-2: crucial hydrogen bonding interaction between C128 and D156. Photochem Photobiol Sci. 2010;9:194–198. doi: 10.1039/b9pp00157c. [DOI] [PubMed] [Google Scholar]
- 38.Sineshchekov OA, Govorunova EG, Li H, Spudich JL. Gating mechanisms of a natural anion channelrhodopsin. Proc Natl Acad Sci USA. 2015;112:14236–14241. doi: 10.1073/pnas.1513602112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sineshchekov OA, Li H, Govorunova EG, Spudich JL. Photochemical reaction cycle transitions during anion channelrhodopsin gating. Proc Natl Acad Sci USA. 2016;113:E1993–2000. doi: 10.1073/pnas.1525269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sineshchekov OA, Li H, Govorunova EG, Spudich JL. Photochemical reaction cycle transitions during anion channelrhodopsin gating. Proc Natl Acad Sci USA. 2016;113:E1993–2000. doi: 10.1073/pnas.1525269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi A, Mamaeva N, Li H, Spudich JL, Rothschild KJ. Resonance Raman Study of an Anion Channelrhodopsin: Effects of Mutations near the Retinylidene Schiff Base. Biochemistry. 2016;55:2371–2380. doi: 10.1021/acs.biochem.6b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugiyama Y, Wang H, Hikima T, Sato M, Kuroda J, Takahashi T, et al. Photocurrent attenuation by a single polar-to-nonpolar point mutation of channelrhodopsin-2. Photochem Photobiol Sci. 2009;8:328–336. doi: 10.1039/b815762f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.