Abstract
Historically, it has been difficult to propagate cells in vitro that are derived directly from human tumors or healthy tissue. However, in vitro preclinical models are essential tools for both the study of basic cancer biology and the promotion of translational research, including drug discovery and drug target identification. This protocol describes conditional reprogramming (CR), which involves coculture of irradiated mouse fibroblast feeder cells with normal and tumor human epithelial cells in the presence of a Rho kinase inhibitor (Y-27632). CR cells can be used for various applications, including regenerative medicine, drug sensitivity testing, gene expression profiling and xenograft studies. The method requires a pathologist to differentiate healthy tissue from tumor tissue, and basic tissue culture skills. The protocol can be used with cells derived from both fresh and cryopreserved tissue samples. As approximately 1 million cells can be generated in 7 d, the technique is directly applicable to diagnostic and predictive medicine. Moreover, the epithelial cells can be propagated indefinitely in vitro, yet retain the capacity to become fully differentiated when placed into conditions that mimic their natural environment.
INTRODUCTION
For decades, scientists have endeavored to develop methods for propagating and studying primary tumors and normal cells outside of the human body1,2. To date, traditional established cell lines have been the mainstay of cell, molecular and cancer biology. However, the establishment of tumor cell lines is hindered by the low rate of success (1–10%, depending on the tissue of origin and state of disease progression)3,52. More importantly, the complex heterogeneity of primary tumors is often lacking in these cell lines, and normal cell lines from many tissues do not exist, thereby preventing the use of such cultures for predicting tumor cell responses4. Tumor heterogeneity is recognizable at both the microscopic level (immunohistochemistry) and at the genetic level (DNA mutations and/or changes in RNA expression). Genetic analysis of primary tumor samples has further identified critical variations within the tumor groups and even the relatedness of tumors from different tissues5. Thus, genetic analysis is the core principle of predictive tumor biology and response to therapies. However, despite the great potential for using genomic analysis as the basis for patient care, it is clear that the interplay of genetics, epigenetics, signaling alterations and cell–cell interactions is probably best evaluated by in vitro and/or rapid ex vivo models. In this article, we present a protocol on how to establish primary epithelial cell cultures in vitro from healthy human tissue and human cancer samples. We compare the utility of the CR method with other models, including patient-derived xenografts (PDXs) and organoid cultures, for advancing diagnostic and regenerative medicine. In addition, we discuss the exciting potential for the CR method to be used as a complementary platform for basic, translational and clinical research.
Development and applications of the protocol
Unlike other model systems, this protocol has been proven to easily establish patient-derived CR cell cultures from both normal and cancer tissues that have the capacity to grow indefinitely without genetic manipulation6–8. The technique uses irradiated mouse fibroblast cells and the Rho-associated kinase (ROCK) inhibitor (Y-27632) to propagate epithelial cells. Originally, Y-27632 was identified in a caspase/kinase inhibitor library as being capable of increasing the cloning efficiency of human embryonic stem (ES) cells9 and, in later studies, as being capable of increasing the viability of human keratinocyte stem cells10. When added to keratinocyte/feeder cocultures developed by Green11, Y-27632 induced indefinite cell proliferation6. Unexpectedly, we observed that feeders and Y-27632 could be used to establish both normal and tumor cell cultures from nonkeratinocyte tissues7. We also noted that the ability of feeders and Y-27632 to induce unlimited cell proliferation is similar to the ability of the HPV-16 E6 and E7 oncogenes to immortalize cells. Both E6 and feeder cells activate telomerase7, whereas both E7 and Y-27632 disrupt the actin cytoskeleton12 and inactivate Rho13. The effects of Y-27632 are completely reversible8, in that CR cultures stop proliferating or terminally differentiate after its removal, depending on culture conditions.
The induction of CR is rapid (within 2 d) and results from reprogramming of the cell population rather than clonal selection8, as is the case with conventional cell lines. Unlike ES cells and induced pluripotent stem (iPS) cells ,CR cells from normal tissue do not express high levels of Sox2, Oct4, Nanog or Klf4 (ref. 8) and do not form teratomas in mice7. Moreover, CR cells maintain developmental potential and do not require intricate manipulation to differentiate into the tissue of origin6–8. In tumor CR cell cultures, phenotypic and genotypic features of the primary tumor are maintained7 and the technique has recently been used to identify an appropriate therapy for respiratory papillomatosis14. Therefore, it is clear that potential applications of the CR method are far-reaching, and, as described by independent laboratories, it can be adapted for live biobanking7 and basic research15–21, as well as for diagnostic22,23, therapeutic21,24,25 and regenerative medicine21,26,27
Overview of the procedure
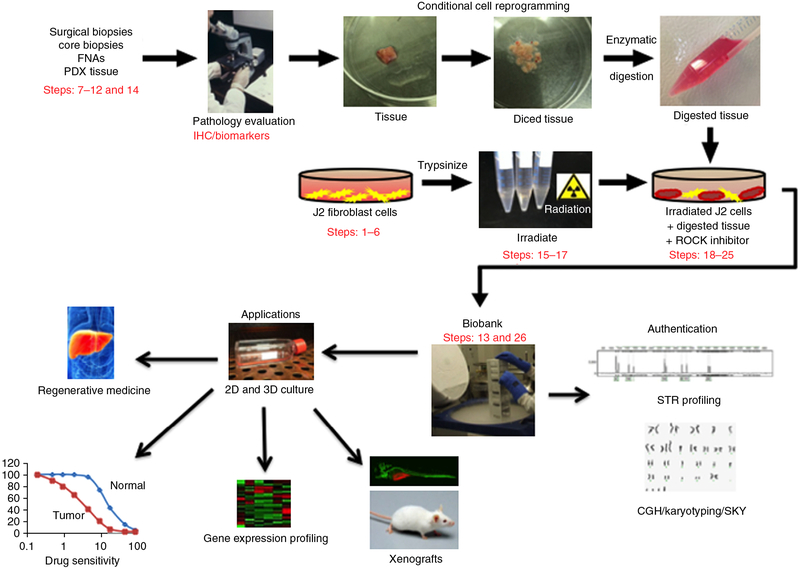
Figure 1 shows an overview of the protocol beginning with a pathologist evaluating tissue biopsies grossly and microscopically. The biopsies are then bisected: one half is fixed in formalin, processed, paraffin-embedded and examined histologically. The pathologist evaluates the viability of the cells and the percentage of the biopsy sample that is composed of tumor cells. The other half of the specimen can be frozen at this stage, or dispersed into single cells by enzymatic digestion and plated in medium containing irradiated Swiss 3T3 J2 mouse fibroblasts (feeder cells) and Y-27632 (ROCK inhibitor). Epithelial cell colonies are readily visible after 2 d, and cultures usually reach confluence in ~5 d. During this time, results of the histological analysis refine the initial determination of the tissue as normal, malignant or mixed. Subsequently, short tandem repeat (STR) profiling, comparative genomic hybridization and spectral karyotyping can be performed on both the original tissue and the corresponding CR cells to authenticate the derivation of the cultured cells.
Figure 1 |.
Overview of the CR method for collection of specimens, establishment of cultures and potential applications of CR technology. Briefly, human tissue samples are obtained from surgical and core biopsies, fine-needle aspiration (FNA) or patient-derived xenograft (PDX) tissue. The samples are thoroughly evaluated by a pathologist using immunohistochemistry (IHC), and specific biomarkers are identified to ensure their normal/tumor status. CR cell lines are generated from the tissue samples using coculture with irradiated J2 feeder cells and ROCK inhibitor, and then are banked in liquid nitrogen. Subsequently, the authenticity of the cell lines should be validated by short tandem repeat (STR) profiling, comparative genomic hybridization (CGH) and spectral karyotyping (SKY). The CR cells can then be used for various applications, including regenerative medicine, drug sensitivity testing, gene expression profiling and xenograft studies. ‘Pathology evaluation’ image courtesy of V.R. Dowell (CDC; Public Health Image Library ID no. 14677). ‘Regenerative medicine’ image reproduced with permission from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. Human tissue samples were collected with the informed consent of patients, according to Georgetown University Institutional Review Board protocols.
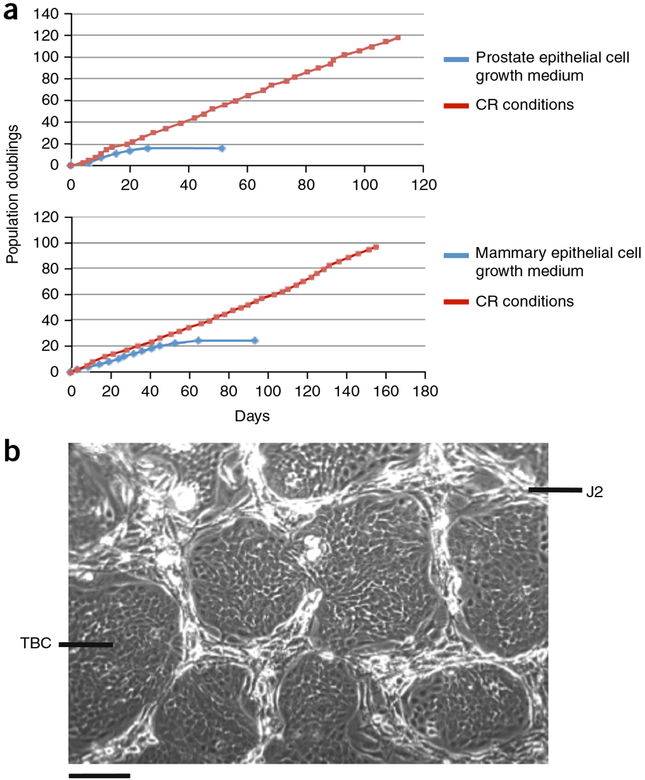
Beyond supporting the rapid expansion of primary epithelial cells in vitro, the CR method conditionally induces long-term proliferation. For example, as shown in Figure 2a, primary prostate and mammary cells were passaged every 3 d in CR coculture at 1:4–1:32 dilutions and continued to proliferate at the same rate for at least 110 d and 100 population doublings until the experiment was terminated. STR analysis of the prostate cells at passage 33 confirmed genetic stability of the CR cells as compared with the initial population7. By contrast, only 20 population doublings were supported by traditional cell culture media (Fig. 2a). The morphology of epithelial cell colonies growing in CR coculture with feeders at passage 4 (eight population doublings) is shown in Figure 2b.
Figure 2 |.
CR cells are conditionally immortalized. (a) Normal prostate and mammary epithelial cells were propagated in both CR culture and synthetic media (prostate epithelial cell growth medium and mammary epithelial cell growth medium). Growth curves were plotted using cumulative population doubling time over time. a adapted with permission from ref. 7, Elsevier.(b) Morphology of primary tracheal–bronchial cells (TBCs) growing in CR coculture with irradiated J2 feeder cells (J2s) at passage 4 (eight population doublings). Scale bar, 100 mm. Human tissue specimens were collected with the informed consent of patients according to Georgetown University Institutional Review Board protocols.
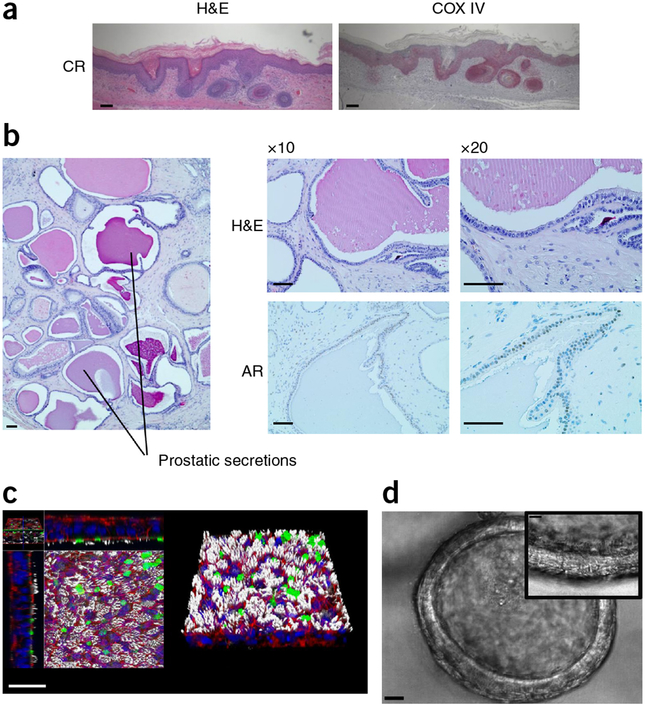
Importantly, CR cells retain lineage commitment and differentiate into the tissue of origin when removed from CR culture6–8. For example, Figure 3a shows that CR keratinocytes form follicular and epidermal lineages when grafted as dermal–epidermal composites, as described earlier for non-CR keratinocytes28,29. In addition, a well-differentiated prostate epithelium that expresses the androgen receptor forms when CR prostate cells are injected below the renal capsule of an immunodeficient mouse30 (Fig. 3b). Finally, CR tracheal–bronchial cells differentiate into both ciliated cells and mucocytes in vitro27,31 (Fig. 3c,d, Supplementary Video 1).
Figure 3 |.
CR cells maintain lineage commitment in vitro and in vivo. (a) Dermal–epidermal composites were constructed using CR human neonatal foreskin keratinocytes and human dermal papilla cells (both passage 3) grafted to nude mice. H&E staining of sections of grafts obtained after 8 weeks show hair follicle structures and interfollicular stratified squamous epithelium. The epithelial cells stain positively with a human-specific antibody (COX IV), confirming their identity as human (anti-COX IV antibody from Cell Signaling, cat. no. 4850, 1:500 dilution). Scale bars, 100 μm. (b) Well-differentiated prostate epithelium formed from normal human prostate CR cells co-injected into the renal capsule of an immunodeficient mouse along with Matrigel and urogenital mesenchyme cells (these specifics are essential)39. H&E staining shows normal prostate morphology and prostatic secretions. The prostate epithelium expresses the androgen receptor (AR; Santa Cruz Biotechnology antibody, cat. no. sc-816, 1:500 dilution). Scale bars, 100 μm. (c) Tracheal–bronchial CR cells were differentiated in air–liquid interface culture. Confocal microscopy demonstrates the presence of cilia (white: α-tubulin; Millipore antibody, cat. no. MAB1854, 1:500 dilution), mucins (green: Muc 5AC/Muc 5B; Thermo/Lab Vision antibody, cat. no. MS-551, 1 mg/ml), F-actin (red: AlexaFluor 488-phalloidin; Molecular Probes, cat. no. A12379, 200 units/ml) and DNA (blue: Hoechst 33258, Molecular Probes, cat. no. H3569, 1:1,000 dilution). Scale bar, 30 μm. c adapted with permission from ref. 8, National Academy of Sciences. (d) Low-magnification view of differentiated tracheal–bronchial CR cells showing bronchosphere. Scale bar, 20 μm. Inset is a high-magnification frame of the bronchosphere from time-lapse photography (Supplementary Video 1) showing cilia. Scale bar, 10 mm. Tissue specimens were collected with the informed consent of patients, according to Georgetown University and The University of North Carolina School of Medicine Institutional Review Board protocols.
Comparison with other methods
The most common method of immortalizing cells has been transformation with SV40 virus large T antigen or overexpression of hTERT32,33. However, these genetic manipulations lead to genomic instability, such that after a few passages there is an irreversible loss of critical biological and genetic characteristics as compared with those of the primary tissues from which they were derived34. Most primary cell cultures, regardless of the method used to generate them, are difficult to maintain, as they have a limited life span because of a gradual decrease in proliferation, eventually leading to senescence34,35.
Both ES and iPS cells can be propagated in vitro and retain a normal karyotype. They can differentiate into several germ layers and show great potential in regenerative medicine36,37. However, with limited exceptions, it is not feasible to precisely direct their differentiation into specific adult tissues. In addition, the use of human ES cells can, in certain cases, be prohibited for ethical reasons38. Generation of iPS cells from adult tissues usually requires transduction with exogenous genes that can alter cellular growth and differentiation properties37, unlike the generation of CR cells, which maintain lineage commitment. More recently, a defined chemical method has been used to generate iPS cells and, although promising, further testing and verification are required before this reprogramming method can be used in regenerative medicine39.
In recent years, PDXs have emerged as a system that in some respects serves as a better model for cancer studies than cultured cells40. They often preserve the molecular and cellular basis of tumor heterogeneity41 and have been shown in certain cases to predict therapeutic responses42. However, as PDX models are established from tumors as subcutaneous xenografts in mice, the normal tissue counterpart and surrounding interacting stroma of each patient is missing. Humanized PDX models are costly, difficult to develop and not amenable to high-throughput platforms; in addition, their success rates frequently fall below 30%42,43.
Clevers44–46, Guo47,48 and colleagues have successfully established 3D organoid cultures of epithelial cells using Matrigel or collagen. The organoid culture method supports the growth of both normal and cancer tissue, and cultures can be manipulated at the genetic level by transfection49. Although organoids are also useful for evaluating drug responses, they are not readily adaptable to high-throughput screening.
Limitations of the CR method
A limitation of the basic CR method is that the outgrowth of human stromal cells in biopsies is inhibited. This phenomenon was observed decades ago and was attributed to the J2 feeder cells11. Thus, although overgrowth of tumor-associated fibroblasts is inhibited, which allows selected propagation of epithelial carcinomas, it is difficult to assay the influence of stromal cells on tumor cell growth and their effect on the response of the tumor cells to therapeutic agents. However, previously unpublished modifications of the CR method have allowed some stromal tumors to be propagated, opening new possibilities for the examination of direct epithelial/stromal interactions (Table 1).
TABLE 1 |.
CR cell growth conditions.
| Cell culture conditions | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Standard CR coculture | Conditioned Medium |
2% O2 |
Collagen coating |
Poly L-ornithine coating |
B-27 | R-spondin | N-2 Supplement |
|||
| Tissue type | F medium | J2 cells | Y-27632 | |||||||
| Breast (N, T) | + | + | + | |||||||
| Prostate (N, T) | + | + | + | |||||||
| Lung (N, T) | + | + | + | |||||||
| Cervix (N, T) | + | + | + | |||||||
| Oral (N, T) | + | + | + | |||||||
| Ovary (N, T) | + | + | + | |||||||
| Skin (N, T) | + | + | + | |||||||
| Salivary gland (N,T) | + | + | + | |||||||
| Kidney (N, T) | + | + | + | |||||||
| Colon (T) | + | + | + | + | + | |||||
| Colon (N) | + | + | + | + | + | |||||
| Pancreas (N, T) | + | + | + | + | + | |||||
| Thyroid (N, T) | + | + | + | + | ||||||
| Schwannomas | + | + | + | + | + | |||||
| Neuroendocrine cells (T) | + | + | + | |||||||
| GIST cells (T) | + | + | + | |||||||
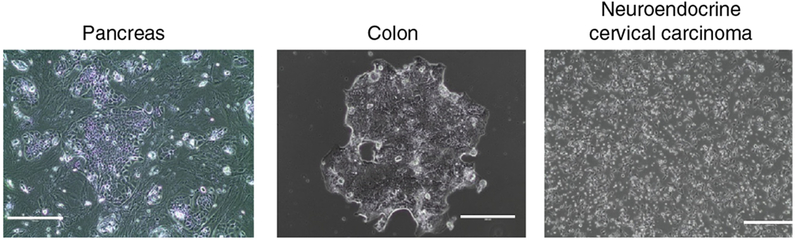
Normal (N) and tumor (T) cells from various tissues, including pancreas, colon and neuroendocrine carcinoma (Fig. 4) require modifications of standard CR culture conditions for optimal proliferation. The concentrations of all components are listed in the ‘REAGENTS’ and ‘REAGENT SETUP’ sections, except for that for R-spondin, which is 1 μg/ml.
Another limitation of the method is that the use of the Y-27632 might potentially interfere with migration and invasion assays of tumor cells, as it alters the actin cytoskeleton. Interestingly, however, we have been able to distinguish normal and tumor cells in invasion assays even in the presence of Y-27632, suggesting that this biological activity is not blocked under CR conditions (Supplementary Fig. 1).
Finally, the CR method allows the outgrowth of both normal and tumor epithelial cells, which means that cultures of biopsies will often contain such mixtures. Indeed, the replication of normal cells can sometimes exceed that of tumor cells, and thereby the normal cells eventually overtake the culture. However, previously unpublished, minor modifications of the standard CR method (such as eliminating the feeder cell component) can select for certain tumor cells in a mixed population (Table 1, Fig. 4).
Figure 4 |.
Modification of standard CR conditions for certain tissue-derived cells. CR cell cultures from pancreas, colon and neuroendocrine cervical carcinoma exhibited unique morphologies and required modifications of standard CR culture conditions for optimal proliferation (Table 1). The CR neuroendocrine cervical carcinoma cells form tumors in nude mice equivalent to the primary tumor (data not shown). The differentiation of pancreas and colon CR cells has not been evaluated. Scale bars, 200 μm (colon), 400 μm (pancreas and neuroendocrine cervical carcinoma). Tissue specimens were collected with the informed consent of patients according to Georgetown University Institutional Review Board protocols.
Experimental design
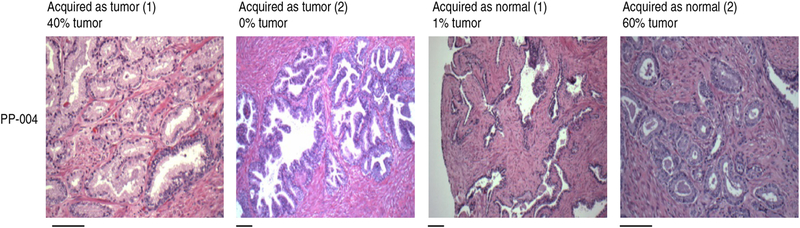
It is important to examine the biopsy material histologically, as in our experience ~25% of the tumor biopsies of primary prostate carcinomas from whole prostatectomy specimens contained no tumor cells. Similarly, 50% of the prostate carcinoma biopsies contained fewer than 10% tumor cells. Biopsies of normal regions of the prostate from the same patient usually contained mostly normal cells, but there were samples in which tumor cells were present (Fig. 5, Table 2).
Figure 5 |.
Evaluation of paired tumor and normal biopsies. Thirty-seven cases of primary prostate cancer were enrolled in the present study. A board-certified pathologist at Georgetown University Medical Center (B.K.) collected biopsies from four sites for each case: two that were deemed malignant and two that were deemed normal. For each biopsy, slices were made for CR culture and for histological evaluation. Using hematoxylin to stain proteins, filaments and intracellular membranes (pink) and eosin to stain nuclei and nucleic acids (blue), the percentages of tumor cells in the biopsies were determined according to standard histopathological criteria (Table 2). Representative histological images are shown for patient PP-004. Scale bars, 100 μm; scale bars apply to images directly above. Tissue specimens were collected with the informed consent of patients, according to Georgetown University Institutional Review Board protocols.
TABLE 2 |.
Evaluation of paired tumor and normal patient prostate biopsies.
| Patient no. |
Tumor no. 1 location |
Slide results |
Tumor no. 2 location |
Slide results | Benign no. 1 location |
Slide results |
Benign no. 2 location |
Slide results |
|---|---|---|---|---|---|---|---|---|
| PP-001 | Left mid | 5% tumor | Left posterior | 70% tumor | Apex | 0% tumor | Mid | 0% tumor |
| PP-002 | Right apex | 100% tumor | Left apex | 20% tumor | Right mid anterior | 0% tumor | Left mid anterior | 5% tumor |
| PP-003 | Right mid | 1% tumor | Right anterior mid | 1% tumor | Left mid | 5% tumor | Left mid anterior | 0% tumor |
| PP-004 | Left mid | 40% tumor | Left base | 0% tumor | Right mid/base | 1% tumor | Right base | 60% tumor |
| PP-005 | Left apex/mid | 0% tumor | Left mid/base | 5% tumor | Right mid | 0% tumor | Right mid | 0% tumor |
| PP-006 | Right base | 60% tumor | Right apex | 90% tumor | Left apex/mid | 1% tumor | Left mid | 0% tumor |
| PP-007 | Left apex | 5% tumor | NA | NA | Right apex | 0% tumor | Right mid | 0% tumor |
| PP-008 | Right mid | 80% tumor | ? | NA | Left mid | 0% tumor | ? | NA |
| PP-009 | Left base | 0% tumor | Left mid/base | 60% tumor | Right mid | 0% tumor | Right mid/apex | 0% tumor |
| PP-010 | Right | 0% tumor | NA | NA | Right | 0% tumor | Left | 0% tumor |
| PP-011 | Left base Lateral | 1% tumor | Left base | 5% tumor | Right apex | 1% tumor | ? | NA |
| PP-012 | Left apex | 80% tumor | Right base | 0% tumor | Right mid | 0% tumor | Left mid | 50% tumor |
| PP-013 | Right apex | 20% tumor | Right mid | 1% tumor | Left mid | 5% tumor | Left apex | 0% tumor |
| PP-014 | Right base | 0% tumor | Right base | 0% tumor | Left mid | 0% tumor | Left mid | 0% tumor |
| PP-015 | Left apex | 0% tumor | Left mid | 0% tumor | Right apex | 0% tumor | Right mid | 0% tumor |
| PP-016 | Right base/mid | 80% tumor | Left mid | 80% tumor | Right mid/apex | 100% tumor | Right base | 1% tumor |
| PP-017 | Right mid | 80% tumor | Left mid/apex | 0% tumor | Right mid/base | 0% tumor | Left apex | 0% tumor |
| PP-018 | Left mid/apex | 0% tumor | Left mid | 0% tumor | Right mid | 0% tumor | Right base | 0% tumor |
| PP-019 | Right apex no. 1 | 1% tumor | Right apex no. 2 | 0% tumor | Left apex no. 1 | 0% tumor | Left apex no. 2 | 0% tumor |
| PP-020 | Left mid/apex | 5% tumor | Left/right apex | 90% tumor | Right base | 0% tumor | Left base | 0% tumor |
| PP-021 | Left mid anterior | 90% tumor | Left mid posterior | 30% tumor | Right mid | 100% tumor | Right mid/base | 0% tumor |
| PP-022 | Right mid | 20% tumor | Right mid/base | 10% tumor | Left mid | 0% tumor | Left mid/base | 0% tumor |
| PP-023 | Left mid/base | 0% tumor | Left Lateral mid | 100% tumor | Left mid | 0% tumor | Right apex/mid | 0% tumor |
| PP-024 | Left apex/mid no. 1 | 70% tumor | Left apex/mid no. 2 | 10% tumor | Right mid | 0% tumor | Right mid/base | 0% tumor |
| PP-025 | Left mid | 90% tumor | Right apex/mid | 90% tumor | Left mid | 0% tumor | Right apex/mid | 30% tumor |
| PP-026 | Right mid | 80% tumor | Left mid | 70% tumor | Right mid/base | 0% tumor | Left mid/base | 0% tumor |
| PP-027 | Left mid anterior | 20% tumor | Left base | 5% tumor | Right mid | 5% tumor | Right base | 10% tumor |
| PP-028 | Left base | 80% tumor | Left mid/base | No slide | Right mid/base | 0% tumor | Right mid | 0% tumor |
Twenty-eight out of thirty-seven cases of primary prostate cancer enrolled in the present study are shown; representative histological images are shown for patient PP-004 in Figure 5. Mid, middle; NA, not available.
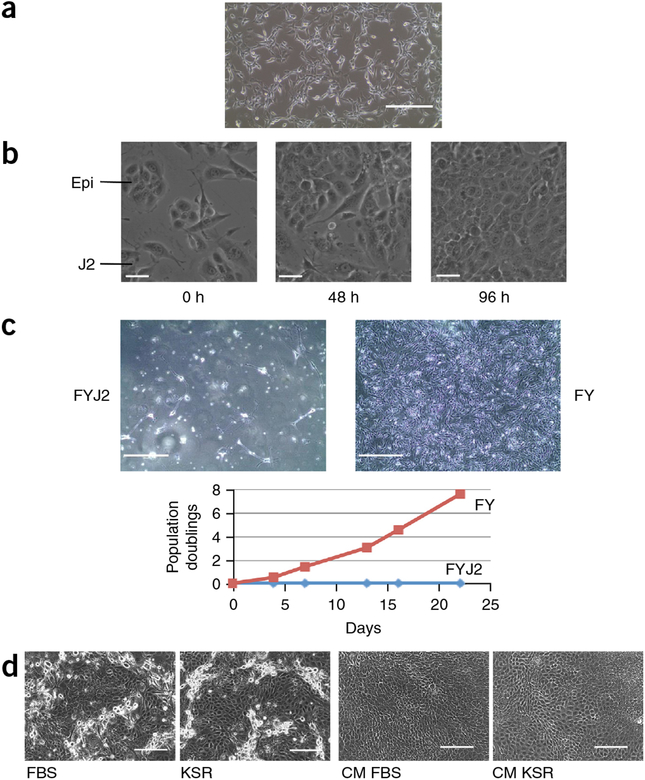
Irradiated J2 feeder cells should be plated at a confluence of 70–80% for optimal CR coculture (Fig. 6a). Feeder cells support epithelial cell growth, and colonies form rapidly as a consequence of proliferation and subsequent migration and merging of small colonies into larger colonies (Fig. 6b, Supplementary Video 2). By contrast, feeder cells inhibit the growth of fibroblast and stromal cells, as shown for gastrointestinal stromal tumor (GIST) cells in Figure 6c. It is clear from these results that GIST cells proliferate in CR medium only in the absence of feeders. Removal of feeder cells is also optimal for the growth of cells from the pancreas and colon (Fig. 4).
Figure 6 |.
Irradiated feeder cells, conditioned medium and serum replacement. (a) Irradiated J2 feeder cells plated at 70–80% confluence. Scale bar, 400 μm. (b) Colonies of primary epithelial cells (Epi) form between irradiated feeder cells (J2) by proliferation and rapid merging of small colonies (also see Supplementary Video 2). Scale bars, 30 mm. (c) Proliferation of fibroblast/stromal cells is inhibited by irradiated feeder cells. Gastrointestinal stromal tumor (GIST) cells proliferate in F medium containing Y-27632 (FY) only when feeder cells (FYJ2) are not present (compare left and right images). Scale bars, 400 μm. The graph shows proliferation of GIST cells (measured as population doublings) in FY in the presence (blue line) or absence (red line) of irradiated J2 feeder cells. (d) Both FBS and knockout serum replacement (KSR) similarly support the proliferation of primary human keratinocytes in CR culture, or in culture with conditioned medium (CM FBS, CM KSR). Scale bars, 200 μm. Human tissue specimens were collected with the informed consent of patients, according to Georgetown University Institutional Review Board protocols.
In addition to these changes, we found that it is possible to replace FBS with a defined serum replacement (knockout serum replacement; KSR) in all CR cocultures containing J2 cells (Fig. 6d). Furthermore, J2 cells can be replaced by medium conditioned by irradiated J2 cells (conditioned medium; CM) for established CR cultures (i.e., after three passages using standard coculture conditions), thereby eliminating the need to constantly maintain and irradiate J2 cells50. CM is critical for growing some cells in low O2 and makes high-throughput assays much simpler, as any background that could be attributed to feeder cells is eliminated.
However, unless CM is specifically listed for a cell type in Table 1, standard coculture conditions should be used.
MATERIALS REAGENTS
Amphotericin B, 250 μg/ml (Fisher Scientific, cat. no. BP264550)
B-27 Supplement (50×) (Gibco, cat. no. 17504044)
Cholera toxin, 11.7 μM (Sigma-Aldrich, cat. no. C-3012) ! CAUTION Cholera toxin is toxic. Handle it with gloves.
Collagen I, rat tail, 1 mg/ml (Corning, cat. no. 354236)
Collagenase IV, 1 mg/ml (Stemcell Technologies, cat. no. 07909)
Collagenase/hyaluronidase solution (10×; Fisher Scientific, cat. no. NC-9694308)
Deionized water
Dispase (Fischer Scientific, cat. no. 354235)
DMEM (Gibco, cat. no. 11965–092)
DMSO (Fisher Scientific, cat. no. BP-231–100)
Epidermal growth factor (EGF), human recombinant, 0.125 μg/ml (Life Technologies, cat. no. PHG0313)
Endothelial Cell Growth Medium (EGM-2, Lonza, cat. no. CC-3162)
Ethanol 200 proof, anhydrous (Fisher Scientific, cat. no. AC 61510–0010)
Ethanol, 70% (vol/vol) (Fisher Scientific, cat. no. R40135)
F-12 nutrient mix (Gibco, cat. no. 11765–054)
FBS (Gibco, cat. no.16140–071)
Gelatin, 0.1% solution (EMD Millipore, cat. no. ES-006-B or equivalent)
Gentamicin, 10 mg/ml (Gibco, cat. no. 15710–064)
Human patient tissue (We have used this protocol on prostate, breast, lung, salivary gland, cervix, liver, colon and pancreas tissue samples.) ! CAUTION Normal or tumorous human specimens must be collected with the informed consent of the donors. Example data shown here were obtained according to Georgetown University Institutional Review Board protocols. ! CAUTION Every unfixed human tissue should be treated as if it is known to be infectious for HIV, HBV or other blood-borne pathogens. Handle tissues with gloves. ! CAUTION The cell lines generated should be regularly checked to ensure that they are authentic and are not infected with mycoplasma.
Human umbilical vein endothelial cells (HUVECs, Lonza cat. no. C2519AS)
Hydrocortisone, 25 μg/ml (Sigma-Aldrich, cat. no. H-0888)
Insulin, 5 mg/ml (Sigma-Aldrich, cat. no. I-5500)
Knockout serum replacement (Gibco, cat. no. 10828–028)
l-Glutamine, 200 mM (Gibco, cat. no. 25030–081)
Mammary epithelial cell growth medium (MEGM; Lonza, cat. no. CC-3051)
N-2 Supplement, 100× (Thermo Fisher Scientific, cat. no. 17502048)
Nystatin, 50 mg/ml (Sigma-Aldrich, cat. no. N6261)
PBS (Gibco, cat. no. 14190250)
Penicillin/streptomycin (pen/strep) mix, 10,000 U/ml (penicillin), 10,000 mg/ml (streptomycin) (Gibco, cat. no. 15140–122)
Poly-l-ornithine solution, 0.01% (Sigma-Aldrich, cat. no. P4957)
Prostate epithelial cell growth medium (PrEGM; Lonza, cat. no. CC-3166)
ROCK inhibitor (Y-27632), 10 mM in sterile water (Enzo, cat. no. 270–333M025)
R-Spondin-1, human recombinant, 5 μg/ml (PeproTech, cat. no. 120–38)
Trypsin/EDTA 0.05% (Gibco, cat. no. 25300120)
Trypsin/EDTA 0.25% (Gibco, cat. no. 25200114)
EQUIPMENT
Aluminum foil (Fisher Scientific, cat. no. 01-213-100 or equivalent)
Biopsy processing/embedding cassettes (Simport, cat. no. M506–2)
Biopsy foam pads (Simport, cat. no. M476–1)
Cell culture flasks, CELLSTAR, 25 cm2, 75 cm2, 175 cm2 (Greiner Bio-One, cat. nos. 690160, 658170 and 660 or equivalent)
Cell culture incubator (37 °C, 95% humidity, 5% CO2; HERACELL 150i; cat. no. 51026282 or equivalent)
Cell culture variable oxygen control incubator (37 °C, 95% humidity, 5% CO2, 2% O2; HERACELL 150i, cat. no. 51026408 or equivalent)
Cell strainer, 100 μm (Corning, cat. no. 431752 or equivalent)
CryoTube vials, 1.8 ml (Thermo Scientific, cat. no. 375418 or equivalent)
Disposable sterile plastic pipettes, CELLSTAR, 5 ml, 10 ml, 25 ml (cat. nos. EK-33107, EK-34107, EK-53107, or equivalent)
Dissecting forceps (Fisher Scientific, cat. no. 08875 or equivalent)
E-plates 16 (ACEA Biosciences, cat. no. 05469830001)
EVOS XL digital inverted microscope (×4, ×10, ×20, ×40 objectives; Advanced Microscopy Group, cat. no. AMEX1000)
Examination gloves, powder-free, nitrile (VWR, cat. no. 82026 or equivalent)
Freezer, −80 °C (Thermo Scientific, cat. no. TSX600D or equivalent)
Inverted phase-contrast microscope (×4, ×10, ×20 objectives; Zeiss Primovert, cat. no. 4155101100000000 or equivalent)
Laminar flow hood with vacuum (Thermo Electron Corporation, cat. no. 1323TS or equivalent)
Liquid nitrogen freezer (Kryos, K-series Cryostage Systems; Taylor-Wharton, cat. no. 6900–62 or equivalent)
Microcentrifuge, accuSpin Micro 17 (Fisher Scientific, cat. no. 13-100-675 or equivalent)
Microcentrifuge tubes, Costar, 1.7 ml (Sigma-Aldrich, cat. no. CLS3620 or equivalent)
P1000, P200, P20 Rainin automatic pipettes with sterile tips with filter (Rainin, cat. no. 17014406 or equivalent)
Pipet-Aid (Drummond, cat. no. 4-000-105)
Refrigerated centrifuge, Beckman Allegra 6R with CH-3.8 swinging bucket rotor (Beckman Coulter, cat. no. 366816 or equivalent)
Rocking double-tier platform (VWR, cat. no. 40000–304 or equivalent)
Scalpels, stainless steel surgical blade (Bard-Parker, cat. no. 08-915-18 or equivalent)
Scepter handheld automated cell counter (Millipore, cat. no. PHCC20060), with disposable Scepter Sensors (cat. no. PHCC60050 or equivalent)
Sealing sterilization pouch (Fisher Scientific, cat. no. 01-812-54 or equivalent)
Time-lapse microscope (Nikon Eclipse TE-300 Spinning Disk Timelapse Microscope System) with Volocity 6.2.1 computer software (PerkinElmer or equivalent)
Tissue culture plates, 6 wells, 12 wells and 96 wells (VWR, cat. nos. 10062–892, 10062–894, 10062–900 or equivalent)
Tubes, conical bottom, CELLSTAR, 15 ml and 50 ml (cat. nos. 82050–278, 82050–350 or equivalent)
Vacuum filtration unit, 500 ml (VWR, cat. no. 10040–436 or equivalent)
Water bath, 37 °C (VWR, cat. no. 89501–460 or equivalent)
xCELLigence real-time cell analysis (RTCA) dual-purpose (DP) complete system (Acea Biosciences, cat. no. 00380601050)
REAGENT SETUP
Complete DMEM (1×) Mix 500 ml of DMEM, 50 ml of FBS, 5.5 ml of l-glutamine and 5.5 ml of pen/strep. Store the mixture at 4 °C for up to 4 months.
Hydrocortisone/EGF mix (1,000×) Dissolve hydrocortisone in 100% ethanol at a concentration of 0.5 mg/ml. Mix 1 ml with 19 ml of DMEM containing 2.5 μg of EGF. Store sterile 1.1-ml aliquots at −20 °C for up to 1 year (hydrocortisone = 25 μg/ml; EGF = 0.125 μg/ml).
Complete F medium (1×) For 500 ml, mix 373 ml of complete DMEM, 125 ml of F12 nutrient mix, 0.5 ml of hydrocortisone/ EGF mix, 0.5 ml of insulin, 0.5 ml of amphotericin B, 0.5 ml of gentamicin and 4.3 μl of cholera toxin. Filter-sterilize the solution using a 0.2-μm sterile filter and store it at 4 °Cfor up to 2 weeks (final concentrations: insulin, 5 μg/ml; amphotericin B, 250 ng/ml; gentamicin, 10 mg/ml; cholera toxin, 0.1 nM; EGF, 0.125 ng/ml; hydro-cortisone, 25 ng/ml). After adding the ROCK inhibitor Y-27632 at a final concentration of 10 mM, the medium may be stored at 4 °C for up to 2 months.
Sample collection (transport medium) Mix 500 ml of DMEM, 20 ml of pen/strep mix (100×), 2 ml of gentamicin (10 μg/ml), 1 ml of amphotericin B (250 μg/ml) and 1 ml of nystatin (50 mg/ml). Store the medium at 4 °C for up to 1 year. Add 10 μM Y-27632 immediately before use.
Cryopreservation medium Mix 90% FBS, 10% DMSO and 10 mM Y-27632. Store the medium at −80 °C for up to 6 months.
Collagen-coated flasks/wells Prepare 1 mg/ml collagen I solution using sterile deionized water, and store it at 4 °C overnight. Cover the bottom of a flask/dish with diluted collagen I (1 ml for a T25 flask; 0.5 ml per well of a six-well plate) and keep it at room temperature (18–22 °C) for 30 min. Transfer the flask/dish to 4 °C for another 30 min. Aspirate leftover collagen I, rinse once with PBS and aspirate it. Store the flask/dish at room temperature for up to 4 weeks.
Poly-L-ornithine-coated flasks/wells Cover the bottom of a flask/dish with 0.01% poly-l-ornithine solution (2 ml for a T25 flask; 1 ml per well of a 6-well plate) and keep it at room temperature for 1 h. Aspirate the poly-l-ornithine solution and rinse the flask/dish once with sterile deionized water. Aspirate the water and repeat this step twice more. Let the flask/dish dry completely (lid on) in a cell culture hood. Store the flask/dish at 4 °C, wrapped in aluminum foil, for up to 4 weeks.
PROCEDURE
Feeder cell culture ● TIMING 1 h
1| Maintain Swiss-3T3-J2 mouse fibroblasts at 37 °C and 5% CO2 in complete DMEM and passage two to three times per week.
▲ CRITICAL STEP Use only the J2 strain of Swiss-3T3 mouse fibroblasts as feeder cells for CR cocultures. The J2 cell line should be regularly checked to ensure that it is not infected with mycoplasma.
? TROUBLESHOOTING
2| To passage J2 cells, rinse with PBS and incubate with 0.05% trypsin/EDTA solution at room temperature for 20 s. Monitor closely using a phase-contrast microscope. When the cells look round and begin to detach from the substrate, tap the cultures gently to detach the cells.
3| Stop the trypsin/EDTA incubation by adding four volumes of complete DMEM to the cells.
4| Centrifuge the cells at 300g for 5 min at 4 °C.
5| Resuspend the cell pellet in 10 ml of complete DMEM. Plate 1 × 103 cells/cm2 if the next passage will occur after 3 d. If you require the next passage to occur after 2 d, plate 2.3 × 103 cells/cm2, and if you are passaging after 1 d, plate 4 × 103 cells/cm2. Incubate the plate at 37 °C and 5% CO2 in complete DMEM.
Fresh tissue handling, transportation and shipping ● TIMING 0.5 h
▲ CRITICAL Only de-identified, coded, consented and institutional review board–approved specimens should be used in this protocol. Undertake the following steps in sterile conditions within 12 h after surgery.
6| Quickly rinse resected tissue sample(s) with 95–100% ethanol (up to 3 s) and then with cold (4 °C) PBS, and transfer to a sterile Petri dish. Using a set of forceps and a scalpel, remove residual fat tissue.! CAUTION Do not extend ethanol rinse time longer than 3 s.
7| Cut fresh specimen/s into small (<10 mm) pieces.
▲ CRITICAL STEP Every unfixed human tissue should be handled with caution to prevent risk of infection from blood-borne pathogens. Wear a protective coat and handle human tissue specimens with gloves.
8| Dissect the tumor lesions from surrounding normal tissues as thoroughly as possible. Use at least one section from each sample for pathology.
9| If you are handling paired normal and tumor samples from the same subject, place each sample in a separate, clearly labeled container. The matching normal tissue should be taken from a site that is as far from the tumor lesion as possible.
10| Place all specimens (in separate containers) with 4 ml of sample collection (transport) medium. Close the lids of the containers tightly.
11| Package the samples with wet ice bags (4 °C) and ship using priority overnight delivery.
▲ CRITICAL STEP Never snap-freeze or fix the samples before shipping.
12| If you are unable to process the specimen immediately upon receipt (optimal), place it in cryopreservation medium (Re-agent Setup). Freeze it on dry ice and store it at −80 °C for up to 1 week, or freeze in liquid nitrogen for long-term storage.
■ PAUSE POINT Cryopreserved tissue specimens may be kept indefinitely.
13| To isolate primary cells from human prostate, breast, lung, salivary gland, cervix and liver tissue, follow option A; to isolate primary cells from human colon and pancreas tissue, follow option B; and to isolate primary cells from cryopreserved human tissue, follow option C.
-
(A)
Isolation of primary cells from human prostate, breast, lung, salivary gland, cervix and liver tissue ● TIMING 2–4 h
-
(i)
Dilute one part of 10× collagenase/hyaluronidase solution with nine parts of F medium and place the solution into a 15-ml tube.
-
(ii)
Mix 3 ml of F medium/collagenase/hyaluronidase with 1 ml of dispase.
-
(iii)
Quickly rinse resected tissue sample(s) with 95–100% ethanol (up to 3 s) and then with cold (4 °C) PBS.
? TROUBLESHOOTING
-
(iv)
Mince the tissue with a scalpel into small (<10 mm) pieces.
-
(v)
Transfer the tissue fragments to the 15-ml tube containing 4 ml of F medium/collagenase/hyaluronidase/dispase and incubate for 1–3 h at 37 °C on a rocking platform.
-
(vi)
After dissociation, centrifuge the cells at 500g for 5 min at 4 °C and discard the supernatant.
-
(vii)
Resuspend the cell pellet in 10 ml of complete DMEM. Filter the cell suspension through a 100-mm cell strainer into a new 50-ml centrifuge tube. Centrifuge at 300g for 5 min at 4 °C and discard the supernatant.
-
(B)
Isolation of primary cells from human colon and pancreas tissue ● TIMING 2–4 h
-
(i)
Rinse resected tissue sample(s) with 95–100% ethanol (up to 3 s) and then with cold (4 °C) PBS (as in Step 13A(iii), and transfer the samples to a sterile Petri dish.
-
(ii)
Mince the tissue with a scalpel into small (<10 mm) pieces.
-
(iii)
Transfer the tissue fragments to the tube containing collagenase IV (4 ml) and incubate for 15 min at 37 °C, on a rocking platform.
▲ CRITICAL STEP Do not extend collagenase IV treatment longer than 20 min.
-
(iv)
Add 7 ml of DMEM to the tube and mix gently.
-
(v)
Filter the cell suspension through a 100-μm cell strainer into a new 50-ml centrifuge tube. Centrifuge at 300g for 5 min at 4 °C and discard the supernatant.
-
(vi)
Resuspend the cell pellet in complete F medium (or CM, for normal colon tissue samples, as shown in Table 1) and plate in a collagen-I-coated T25 flask (Reagent Setup). Total media volume is 5 ml. Change the medium every 7 d.
! CAUTION The cell lines generated should be regularly checked to ensure that they are authentic and are not infected with mycoplasma.
▲ CRITICAL STEP Maintain cell cultures in a variable oxygen control incubator with a low oxygen level (2% O2).
? TROUBLESHOOTING
-
(C)
Isolation of primary cells from cryopreserved human tissue ● TIMING 2–4 h
-
(i)
Thaw the container of tissue (in cryopreservation medium) in a 37 °C water bath.
-
(ii)
Remove tissue from the container using sterile forceps.
-
(iii)
Place the tissue on a sterile Petri dish containing sufficient sterile PBS to rinse.
-
(iv)
Move the piece of tissue onto another sterile Petri dish and proceed as in Step 13A(iv–vii) (for prostate, breast, lung, salivary gland, cervix and liver tissue) or Step 13B(ii–vi) (for colon and pancreas tissue).
Feeder cell irradiation and coculture ● TIMING 1 h
14| For the irradiation procedure, start with a T175 flask of J2 cells (2 × 104 cells/cm2) in DMEM.
▲ CRITICAL STEP Never grow J2 cells above 90% confluence. Overgrowth changes their sensitivity to irradiation.
15| Detach the cells as described in Steps 3–5 and resuspend the cell pellet in 10 ml of DMEM.
16| Irradiate the cell suspension with an appropriate Cesium Irradiator at 30 Gy (=3,000 rad).
▲ CRITICAL STEP Using a parallel culture, ensure that there is no proliferation of J2 cells after irradiation. (This is necessary only until it is clear that the routine irradiation is sufficient to stop proliferation.)
? TROUBLESHOOTING
17| Immediately after irradiation, plate the J2 cells in a T25 flask (1 × 104 cells/cm2) together with processed tissue or desired epithelial cells in complete F medium. Alternatively, prepare CM and plate epithelial cells using CM following the steps in Box 1. Store excess irradiated J2 feeder cells at 4 °C in DMEM in a sterile 15-ml tube. Use them within 4 d or freeze the cells in cryopreservation medium at −80 °C for up to 1 week or place in liquid nitrogen for long-term storage as described in Step 12.
Box 1 | Procedure for preparation of CM and plating of cells using CM
● TIMING 3 d and 1 h, respectively
-
Plate 7 × 106 irradiated J2 cells in a T175 flask in 30 ml of F medium.
▲ CRITICAL STEP If complete serum-free CM is necessary, replace FBS with 20% KSR before conditioning by irradiated J2 cells.
-
Incubate the cells for 72 h at 37 °C.
▲ CRITICAL STEP To obtain the optimal concentration of factors released by the irradiated J2 cells, allow the F medium to be conditioned for 72 h.
? TROUBLESHOOTING
Transfer the medium to 50-ml conical tubes and centrifuge at 300g for 5 min at 4 °C.
Collect the supernatant and filter using a 0.22-μm vacuum filtration system.
-
Aliquot into 15-ml conical tubes. For immediate use, mix three parts of the filtered supernatant with one part of fresh F medium and supplement with 10 mM Y-27632. The resulting complete CM can be stored at 4 °C up to 1 week.
■ PAUSE POINT For longer storage, freeze complete CM on dry ice and store at −80 °C for up to 6 months.
For epithelial cells that require culturing in CM (Table 1), or for all epithelial cells after three passages in standard CR coculture, plate in complete CM (if desired). (Use all cells when plating from dissociated tissue; split 1:4–1:20 for existing cultures, depending on the growth rate.)
Change the medium every 2–3 d; passage when confluent.
! CAUTION Swiss-3T3-J2 mouse fibroblasts should be regularly checked to ensure that they are not infected with mycoplasma.
? TROUBLESHOOTING
18| Maintain coculture/s at 37 °C, 95% humidity and 5% CO2 until confluent.
! CAUTION The cell lines generated should be regularly checked to ensure that they are authentic and are not infected with mycoplasma.
Differential trypsin treatment for passage of cocultured cells ● TIMING 0.5 h
19| Rinse coculture with PBS and incubate with 1 ml of 0.05% trypsin/EDTA solution (for a T25 flask) at room temperature for 20 s.
20| Monitor closely using a phase-contrast microscope. When the feeder cells become round and begin to detach from the substrate, tap the flask gently and remove the detached cells by aspiration. Colonies of epithelial cells should remain tightly adherent.
▲ CRITICAL STEP Do not extend trypsin/EDTA incubation time longer than 1 min.
? TROUBLESHOOTING
21|Rinse the epithelial cells with PBS and re-treat them with 1 ml of 0.05% trypsin/EDTA at 37 °C for 5 min.
22|Stop the trypsin/EDTA incubation by adding four volumes of complete DMEM to the cells and transfer the mixture to a 15-ml centrifuge tube.
23|Centrifuge the cells at 300g for 5 min at 4 °C.
24|After centrifugation, resuspend the cell pellet in complete F medium.
25|During passaging, surplus epithelial cells can be cryopreserved (Step 12).
? TROUBLESHOOTING
26| To assess the cells, follow option A for time-lapse microscopy and cell motility testing, or option B to perform a cell invasion assay.
■ PAUSE POINT Cryopreserved cells can be stored indefinitely.
-
(A)
Time-lapse microscopy and cell motility testing ● TIMING 1–5 d
-
(i)
Plate 103 epithelial cells in a six-well plate, together with irradiated feeder cells (104 cells/cm2) or CM. Place the coculture in a tissue culture incubator overnight.
-
(ii)
Check cell confluence the next morning.
? TROUBLESHOOTING
-
(iii)
Aspirate the medium from the cells and add 7 ml of fresh complete F medium to each well.
-
(iv)
Place the six-well plate in an incubator (37 °C, 5% CO2) equipped with a microscope, camera and time-lapse recording system.
▲ CRITICAL STEP Prepare the incubator by setting an appropriate temperature, CO2 concentration and humidity level. Prewarm the plate to 37 °C before placing it in the incubator in order to eliminate water condensation, which degrades visibility.
? TROUBLESHOOTING
-
(v)
Check the visibility and condition of the cells and choose fixed points as areas the pictures will be taken from (as required by the software).
-
(vi)
Set up the software to take a picture of each area every 15 min.
-
(vii)
Record cell motility (1–4 d).
▲ CRITICAL STEP Each day check the temperature, CO2 concentration and humidity level in the incubator and adjust if necessary.
? TROUBLESHOOTING
-
(viii)
Stop recording. Make a video document using the Volocity 6.2.1 software (or equivalent).
-
(B)
Cell invasion assay ● TIMING 2.5 d
-
(i)
Place the xCELLigence RTCA DP instrument in a standard tissue culture incubator (37 °C, 5% CO2).
-
(ii)
Coat 16-well E-plates with 100 μl per well of 0.1% gelatin for 1 h at 37 °C and rinse three times with PBS.
-
(iii)
Add 200 μl of EGM-2 per well.
-
(iv)
Add of 3.5 × 104 HUVECs per well and incubate them for 21 h to allow a monolayer to form.
-
(v)
Aspirate the EGM-2 medium and replace it with 200 ml per well of complete F medium containing 104 normal or tumor CR cells originating from the head and neck region. Incubate the cells for 10 h.
-
(vi)
Return the plate to the xCELLingence RTCA instrument and set the background to zero. Record data for 11 h.
-
(vii)
Analyze the data using Acea RT-CA software, which is included with the xCELLigence RTCA DP system.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
TABLE 3 |.
Troubleshooting table.
| Step | Problem | Possible reason | Possible solution |
|---|---|---|---|
| 1 | No growth or slow growth of epithelial cells in coculture | Use of a nonrecommended feeder cell line | Use only the J2 strain of mouse Swiss-3T3 fibroblasts |
| 13A(iii) | No live cells after tissue processing | Extended ethanol rinse time | Rinse no longer than 3 s |
| 13B(vi) | No growth of colon or pancreas cell lines | Incubation in an incubator with an atmospheric gas mixture | Incubate in low oxygen (2% O2) |
| 16 | Observed J2 cell proliferation after irradiation | Irradiation dose is too low | Increase irradiation dose (up to 60 Gy) |
| 17 | Low cell viability after freeze-thaw procedure | Storage of cells at −80 °C | Store cells in a liquid nitrogen freezer |
| 20 | Epithelial cells detach together with J2 feeder cells during first step of differential trypsin treatment |
Trypsin treatment was too long | Do not extend trypsin treatment beyond 1 min |
| 25 | Low cell viability after freeze-thaw procedure | Storage of cells at −80 °C | Store cells in a liquid nitrogen freezer |
| 26A(ii) | No colonies visible when recording starts | Cell confluence is too low | Increase the number of cells added to the wells |
| 26A(iv) | No cells visible | Condensation on the plate surfaces | Let the plate adapt to incubator conditions for at least 30 min |
| 26A(vii) | No motility; wrong cell morphology | Temperature, C02 concentration and/or humidity not correct | Make sure that temperature, C02 level and humidity level are within the range for cell growth |
| Box 1 | Low quality of conditioned medium-does not support cell growth | Suboptimal concentration of growth factors | Use the correct number of cells. Condition the medium for no less than 72 h |
● TIMING
Steps 1–5, feeder cell culture: 1 h
Steps 6–12, fresh tissue handling, transportation and shipping: 0.5 h
Step 13A, isolation of primary cells from human prostate, breast, lung, salivary gland, cervix and liver tissue: 2–4 h Step 13B, isolation of primary cells from human colon and pancreas tissue: 2–4 h
Step 13C, isolation of primary cells from cryopreserved human tissue: 2–4 h
Steps 14–18, feeder cell irradiation and coculture: 1 h
Steps 19–25, differential trypsin treatment for passage of cocultured cells: 0.5 h
Step 26A, time-lapse microscopy and cell motility testing: 1–5 d
Step 26B, cell invasion assay: 2.5 d
Box 1, conditioned medium preparation and plating of cells using conditioned medium: 3 d and 1 h, respectively
ANTICIPATED RESULTS
This procedure describes how to generate approximately a million cells from a variety of fresh or cryopreserved human tissue biopsies in as little as 7 d. ‘Fast-growing’ tissues (e.g., prostate, lung, cervix, skin and salivary gland tissues) can yield up to 2 million cells after 6–7 d. Tissues with an intermediate growth rate (breast and kidney tissues) yield 1–2 million cells after ~2 weeks. ‘Slow-growing’ tissues (colon, pancreas, ovary and thyroid tissues) yield ~10,000 cells after 4 weeks. We have not observed any consistent differences between the growth rates of normal and tumor cells from the same tissue. The viability of cells at the initial plating from dissociated tissue is 80–90% (using trypan blue staining).
It is important to note that normal and tumor CR cell cultures can be generated from the same patient, including for rare tumors such as adenocystic carcinoma of the salivary gland and neuroendocrine cervical cancer, for which few or no cell lines exist. CR cultures are suitable for early-stage, late-stage, low-grade, high-grade and metastatic tumors, which should aid tremendously in investigating progression from pre-neoplastic to neoplastic to metastatic states. Furthermore, CR cultures make an in-depth analysis of tumor heterogeneity possible, as they preserve the cellular diversity of tumors. Finally, CR cell cultures can be generated from xenograft and organoid tissue, suggesting that these three platforms can work in synergy to advance research into both normal and cancer cell biology using primary human-tissue-derived cells51.
As CR cells maintain developmental potential and differentiate into 3D cultures representative of their tissue of origin in vitro or in xenografts upon removal of CR conditions, developmental abnormalities associated with specific tumor cells can be analyzed by immunohistochemical analysis of frozen or paraffin-embedded sections.
Supplementary Material
ACKNOWLEDGMENTS
Studies of conditional cell reprogramming were funded predominantly by internal funds from the Center for Cell Reprogramming at Georgetown University Medical Center and grants R33CA177466, R21CA180524 and R01RR032315 from the National Institutes of Health. We thank A. Wellstein, P. Furth, A. Riegel and B. Haddad for valuable discussions during the course of this study.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Cunderlíková B Issues to be considered when studying cancer in vitro. Crit. Rev. Oncol. Hematol 85, 95–111 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Kahn J et al. Preclinical models in radiation oncology. J. Natl. Cancer Inst 51, 1417–1423 (1973).4357758 [Google Scholar]
- 3.Giard DJ et al. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst 51, 1417–1423 (1973). [DOI] [PubMed] [Google Scholar]
- 4.Herrmann D et al. Three-dimensional cancer models mimic cell-matrix interactions in the tumour microenvironment. Carcinogenesis 35, 1671–1679 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Schmidt F & Efferth T Tumor heterogeneity, single-cell sequencing, and drug resistance. Pharmaceuticals (Basel) 9, 33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman S et al. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Invest 120, 2619–2626 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol 180, 599–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suprynowicz FA et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. USA 109, 20035–20040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe K et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol 25, 681–686 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Terunuma A et al. Efficient procurement of epithelial stem cells from human tissue specimens using a Rho-associated protein kinase inhibitor Y-27632. Tissue Eng. Part A 16, 1363–1368 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rheinwald JG & Green H Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6, 331–343 (1975). [DOI] [PubMed] [Google Scholar]
- 12.Yue J et al. Cutaneous human papillomavirus type 38 E7 regulates actin cytoskeleton structure for increasing cell proliferation through CK2 and the eukaryotic elongation factor 1A. J. Virol 85, 8477–8494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charette ST & McCance DJ The E7 protein from human papillomavirus type 16 enhances keratinocyte migration in an Akt-dependent manner. Oncogene 26, 7386–7390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan H et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N. Engl. J. Med 367, 1220–1227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prater MD et al. Mammary stem cells have myoepithelial cell properties. Nat. Cell Biol 16, 942–950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walters BJ et al. Pseudo-immortalization of postnatal cochlear progenitor cells yields a scalable cell line capable of transcriptionally regulating mature hair cell genes. Sci. Rep 5, 17792 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hata AN et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med 22, 262–269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garraway LA Lander ES Lessons from the cancer genome. Cell 153, 17–37 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Brown DD et al. Developing in vitro models of human ductal carcinoma in situ from primary tissue explants. Breast Cancer Res. Treat 153, 311–21 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Boehm JS & Golub TR An ecosystem of cancer cell line factories to support a cancer dependency map. Nat. Rev. Genet 16, 373–4 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Chu HW et al. CRISPR-Cas9-mediated gene knockout in primary human airway epithelial cells reveals a proinflammatory role for MUC18. Gene Ther 22, 822–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman AA et al. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer 15, 747–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crystal AS et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 346, 1480–1486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beglyarova N et al. Screening of conditionally reprogrammed patient-derived carcinoma cells identifies ERCC3-MYC interactions as a target in pancreatic cancer. Clin. Cancer Res 22, 6153–6163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeed K et al. Comprehensive drug testing of patient-derived conditionally reprogrammed cells from castration-resistant prostate cancer. Eur. Urol 10.1016/j.eururo.2016.04.019 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Veit G et al. Ribosomal stalk protein silencing partially corrects the DF508-CFTR functional expression defect. PLoS Biol 14, e1002462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler CR et al. Rapid expansion of human epithelial stem cells suitable for airway tissue engineering. Am. J. Respir. Crit. Care Med 194, 156–168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thangapazham RL et al. Dissociated human dermal papilla cells induce hair follicle neogenesis in grafted dermal-epidermal composites. J. Invest. Dermatol 134, 538–540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thangapazham RL et al. A model system to analyse the ability of human keratinocytes to form hair follicles. Exp. Dermatol 23, 443–446 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Jin K et al. Patient-derived human tumour tissue xenografts in immunodeficient mice: a systematic review. Clin. Transl. Oncol 12, 473–480 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Fulcher ML & Randell SH Human nasal and tracheo-bronchial respiratory cell culture. Methods Mol. Biol 945, 109–121 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Ali SH & DeCaprio JA Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol 11, 15–23 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Shay JW & Wright WE Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis 26, 867–874 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Wilding JL & Bodmer WF Cancer cell lines for drug discovery and development. Cancer Res 74, 2377–2384 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Wright WE & Shay JW The two-stage mechanism controlling cellular senescence and immortalization. Exp. Gerontol 27, 383–389 (1992). [DOI] [PubMed] [Google Scholar]
- 36.Curry EL et al. Using induced pluripotent stem cells as a tool for modelling carcinogenesis. World J. Stem Cells 7, 461–469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seki T & Fukuda K Methods of induced pluripotent stem cells for clinical application. World J. Stem Cells 7, 116–125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Miguel-Beriain I The ethics of stem cells revisited. Adv. Drug Deliv. Rev 82.–, 176–180 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Chen G et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8, 424–429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams SA et al. Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Lab. Invest 93, 970–982 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Tentler JJ et al. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol 9, 338–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boone JD et al. Ovarian and cervical cancer patient derived xenografts: the past, present, and future. Gynecol. Oncol 138, 486–491 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Bernardo C et al. Patient-derived bladder cancer xenografts: a systematic review. Transl. Res 166, 324–331 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Boj SF et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van de Wetering M et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sachs N & Clevers H Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev 24, 68–73 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Li X et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med 20, 769–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nadauld LD et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol 15, 428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drost J et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 52, 43–47 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Palechor-Ceron N et al. Radiation induces diffusible feeder cell factor(s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. Am. J. Pathol 183, 1862–1870 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McAuliffe PF et al. Cancer xenografts is enhanced in chemoresistant disease and predicts poor patient outcomes. PLoS One 10, e0136851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gillet JP, Varma S & Gottesman MM The clinical relevance of cancer cell lines. J. Natl. Cancer Inst 105, 452–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.