SUMMARY
We investigate the survival distribution of the patients who have survived over a certain time period. This is called a conditional survival distribution. In this paper, we show that one-sample estimation, two-sample comparison and regression analysis of conditional survival distributions can be conducted using the regular methods for unconditional survival distributions that are provided by the standard statistical software, such as SAS and SPSS. We conduct extensive simulations to evaluate the finite sample property of these conditional survival analysis methods. We illustrate these methods with real clinical data.
Keywords: Delta method, Fieller method, Kaplan-Meier estimator, Log-rank test, Martingale central limit theorem, Proportional hazards model
1. Introduction
Traditionally, survival analysis in clinical researches has been to investigate the distribution of patients’ survival times measured from the diagnosis of a disease or the start of a treatment (i.e., baseline). This type of analysis provides the survival probability of patients expected at the start of treatment that will be useful to predict their prognosis before starting the treatment. However, the survival probability evolves over time and usually decrease with increased survivorship, so that both patients and clinicians are interested in the change in survival probability over the progress of treatment and disease. Residual lifetime of individuals when they have survived over a relevant landmark of time can serve towards this end. For example, when comparing the efficacy of an intensive treatment with a standard treatment, patients receiving the prior may have a higher risk due to treatment-related mortality during treatment period, but may have a much lower risk once they survive over the treatment period with the disease cured.
While researches on residual lifetime theory have been very active in reliability area (e.g. Bryson and Siddiqui, 1969; Hollander and Proschan, 1975; Muth, 1977; Ruiz and Navarro, 1994), those in biostatistics field have been sparse. Among some biostatistical examples are Jeong and Jung (2008) on two-sample comparison of median residual lifetime, and Jung, Jeong and Bandos (2009) extending the two-sample problem to regression analysis. More recently, residual lifetime analysis has been very popularly used in clinical trials to analyze the change in survival distribution of patients as a progress of disease (e.g. Zamboni et al. 2010; Zabor et al. 2013; Bischof et al. 2015; Mertens et al. 2015) under the name of conditional survival analysis. Let T be the survival variable of a population with survivor function S(t) = P (T ≥ t). The t-year conditional survival distribution for patients who have survived for t0 years, P (T ≥ t + t0|T ≥ t0), is denoted as S(t|t0) = S(t + t0)/S(t0) for t ≥ 0. In the clinical literatures (e.g. Zabor et al. 2013; Mertens et al. 2015), investigators estimate the conditional survival distributions by replacing the survivor functions with their Kaplan-Meier (1958) estimates, but they do not provide a formal statistical testing to compare them between patient groups.
This paper can be regarded as a review paper supporting the analysis methods that are popularly used in medical field without theoretical justification. In this paper, we present analysis methods for conditional survival distributions including confidence interval of conditional survivor function for 1-sample problem, the conditional log-rank test for 2-sample test, and the conditional Cox (1972) proportional hazards model for regression analysis. We present simulation results to evaluate the performance of these methods. The proposed methods are demonstrated with real clinical data.
2. Analysis of Conditional Survival Distributions
2.1. One-Sample Problem
Suppose the lifetimes from n patients T1, …,Tn are independent and identically distributed with survivor function S(t) = P(Ti ≥ t) and cumulative hazard function ⋀(t) = — log S(t). From patient i, we observe (Xi,δi), where Xi is the minimum of Ti and censoring time Ci, and δi is an event indicator taking 1 if patient i had an event and 0 otherwise. We assume that censoring times are independent of the survival times.
For patients who have survived for at least t0(≥ 0) years, the probability that they live additional t years, S(t|t0) = P(Ti ≥ t + t0|Ti ≥ t0), is given as
| (1) |
by the definition of conditional probabilities. S(t|t0) is called the conditional survivor function for patients who have survived for t0 years.
The conditional cumulative hazard function ⋀(t|t0) = – log S(t|t0) is given as ⋀(t|t0) = ⋀(t + t0) – ⋀(t0) from (1). Hence, for t ≥ 0, the conditional hazard function λ(t|t0) = ∂⋀(t|t0)/∂t is identical to the unconditional (or marginal) hazard function λ(t + t0) = ∂⋀(t + t0)/∂t. Jeong, Jung and Costantino (2008) propose a nonparametric inference method on conditional median residual lifetime θ that satisfies S(θ|t0) = 1/2.
Let denote the Kaplan-Meier estimator of S(t). Then, S(t|t0) is consistently estimated by
From Corollary 3.2.1 of Fleming and Harrington (1991), we have
| (2) |
where is a 0-mean martingale, and and denote the event and the at-risk processes, respectively. Let y(t) denote the uniform limit of n−1Y (t) for t ≤ τ, where τ denotes the minimum of the upper limits of the supports of censoring and survival distributions. By the martingale central limit theorem, converges to N (0, Σ) in distribution, where
which can be consistently estimated by replacing y(t), S(t) and d⋀(t) with their consistent estimators Y(t)/n, and Y(t)−1dN(t), respectively. That is, a consistent estimator of Σ is given as
where is a consistent estimator of that is given as
by using the Nelson-Aalen (Nelson, 1969; Aalen, 1978) estimator
The partial differentiation of θ1/θ2 with θ1 = S(t + t0) and θ2 = S(t0) is given as
Hence, by the delta-method, converges to N (0,σ2) in distribution, where
A consistent estimator can be estimated by replacing S(t), y(t) and d⋀(t) with their consistent estimators , n−1Y(t) and Y(t)−1dN(t), respectively. That is,
A 100(1 — α)% confidence interval for the conditional survival probability S(t|t0) can be calculated using this asymptotic result, i.e.
where z1-α denote the 100(1 — α) percentile of the standard normal distribution.
These inferences are available only when there are patients who are at risk at t0 in the data set. In fact, these inferences can be much simplified. Suppose that P(Ti ≥ t0)P (C ≥ t0) > 0, i.e. the maximum survival time is longer than t0 and some patients are followed for longer than t0. Then, for t > 0, we have
| (3) |
The right hand side of (3) equals
since Xi = min(Ti,Ci), and Ti and Ci are independent. Hence, we have S(t|t0) = P(Ti ≥ t + t0|Xi ≥ t0), denoting the survival probability at t + t0 for patients who are at risk at t0. This implies that the conditional survival probability S(t|t0) can be estimated by calculating the Kaplan-Meier estimator at t + t0 from the patients who are at risk at t0.
This relationship becomes clearer by the definition of Kaplan-Meier estimator. For simplicity of notation, suppose that there are no ties among Xi, …,Xn. Then, by the definition of Kaplan-Meier estimator, (2) is expressed as
which is the Kaplan-Meier estimator at t + t0 calculated from the data set consisting of patients who are at risk at time t0, and its variance is consistently estimated by . Hence, one-sample inference of conditional survival distribution using the delta-method will be identical to that based on the standard (or, unconditional) survival distribution using Kaplan-Meier estimator to the subset of data consisting of the patients who are at risk at t0. These results hold with tied survival data too.
An alternative confidence interval for S(t|t0) can be obtained using the Fieller’s (1954) method. Let . By using the asymptotic result for Kaplan-Meier estimator, is asymptotically normal with mean 0, and its variance can be consistently estimated by , where is the (i, j)-component of . Hence, we have
| (4) |
We can obtain a 100(1 – α)% confidence interval of S(t|t0) by solving the equation within the probability of (4) with respect to p, i.e.
where , and . This formula gives an appropriate confidence interval when
2.2. Two-Sample Log-Rank Test
Suppose that nk patients are randomized to arm k(= 1, 2), and the survival time from the nk patients of arm k are independent and identically distributed with survivor function Sk(t) = P(Tki ≥ t) and cumulative hazard function Λk(t) = – log Sk(t). From patient i(= 1,–, nk) in arm k(= 1, 2), we observe (Xki, δki), where Xki is the minimum of Tki and censoring time Cki, and δki is an event indicator taking 1 if the patient had an event and 0 otherwise. We assume that the censoring times are independent of the survival times within each arm. Let and denote the event and the at-risk processes for arm k, respectively. Also, let n = n1 + n2, N(t) = N1(t) + N2(t) and Y(t) = Y1 (t) + Y2 (t).
For conditional survivor function Sk(t|t0) = Sk (t + t0)/Sk (t0), we want to derive a log-rank test to test H0 : S1(t|t0) = S2(t|t0) for all t ≥ 0 against H1 : S1(t|t0) ≠ S2(t|t0) for some t ≥ 0. From the previous section, d⋀(t|t0) = d⋀(t + t0) for t ≥ 0, so that we can consider a log-rank test for comparing conditional survival distributions for patients who have survived over t0,
which is identical to
where H(t) is a predictable function that is uniformly convergent to h(t) over [t0,τ] and τ is the minimum of the supports of the censoring and survival distributions. The logrank statistic (Peto and Peto, 1972) uses H(t) = n−1Y1(t)Y2(t)/Y(t), the Gehan-Wilcoxon test (Gehan, 1965) uses H(t) = n−2Y1(t)Y2(t), and the Prentice-Wilcoxon test (Prentice, 1978) uses , where is the left-continuous version of the Kaplan-Meier (1958) estimate from the pooled data.
Note that has the same expression as the standard rank tests W0 except that the range of the integration is restricted to [t0, ∞). Using the same arguments as those used for the standard rank tests (e.g. Gill, 1980; Fleming and Harrington, 1991), we can show that is asymptotically standard normal with
under H0. Hence, we reject H0 in favor of H1, if with two-sided type I error rate α.
For example, for the conditional log-rank test, we have
and
From the expression of and , it is obvious that the conditional log-rank test at t0 can be carried out by applying the standard log-rank test to the data set consisting of patients who are at risk at t0, . The 2-sample conditional log-rank test can be easily extended to the log-rank test for K-sample cases with K > 2. These results holds for other types of conditional rank tests.
2.3. Regression Method
From patient i = 1,…, n, we observe covariates zi = (z1i, …, zmi)T together with the minimum of the survival and censoring times Xi and event indicator δi. We assume that, given zi, the survival and censoring times are independent. Suppose that the conditional survival distribution for patients who have survived over t0 has a proportional hazards model
| (5) |
for t ≥ 0, where λi(t|t0) denotes the baseline conditional hazard function. As was shown in the previous sections, the conditional hazard function λi(t|t0) is identical to the unconditional hazard function λi(t + t0), so that (5) can be expressed as the regular proportional hazards model
for t ≥ t0. Hence, if the (unconditional) survival distribution has a proportional hazards model with constant covariate effect over the whole time span, then the conditional survival distribution for any t0(> 0) has the same proportional hazards model. However, if the covariate effect changes over time, then the regression model for conditional survival distribution changes in t0.
The partial score and information matrix (Cox 1972) are given as
and
respectively, where z⊗2 = zzT for a column vector z. Using the same asymptotic theory for the Cox regression method, we can show that is approximately normal with mean 0 and its variance-covariance matrix can be consistently approximated by .
For a univariate proportional hazards model with a dichotomous covariate, it is easy to show that the partial score test under , is identical to the conditional log-rank test, , that was discussed in the previous section. From the expression of the partial score and information, it is obvious that the conditional Cox regression model (5) can be fitted by applying the standard Cox regression method to the data set consisting of the patients who are at risk at t0, i.e. D (t0) = {(Xi,δi,zi) : Xi ≥ t0,i = 1, …,n}. Kurta et al. (2014) proposed this analysis method without any theoretical justification.
3. Numerical Studies
3.1. Simulations
We want to show that the standard inference methods using the subset of data appropriately reflect the conditional survival distribution concept. At first, we conduct simulations on one-sample problems using a piecewise exponential distribution with survivor function
This distribution has a hazard function of λ1 for 0 ≤ t ≤ 2 and λ2 for t ≥ 2. By choosing λ1 = 0.3466 and λ2 = λ1/2, the median survival for the whole patients is 2 years at the baseline, while that for those who have survived the first t0 = 2 years is 4 years starting from the 2-year time point. Survival times are generated from S(t) and censoring times from U(0,a), where a is chosen for 30% of censoring rate. With a fixed at this value, we consider censoring distribution U(b, a + b) with b chosen for 15% of censoring. We generate 10,000 simulation samples of size n = 200. From each sample, we estimate S(t|t0) and its 95% confidence interval by the delta-method and Fieller’s method for t0 = 1, 2, 3, and 4, and t = 1, 2,…, 5 – t0.
Table 1 reports mean bias and the sample standard deviation over the simulation samples (SSD) of the estimator, , and empirical coverage probabilities of the two confidence interval methods. Table 1 also reports the mean of the standard deviation (MSD) of estimated by the delta method over the simulation samples. We observe that the estimated conditional probabilities have very small bias and the bias tends to increase in censoring proportion. As expected, SSD and MSD tend to increase in censoring proportion. They are very close for each simulation setting, but MSD is slightly smaller than SSD. This may result in slightly anti-conservative empirical coverage probability of the confidence intervals by the delta method. As t0 and t increase, the number of subjects at risk decreases and the bias tends to be negative. The two confidence interval methods have empirical coverage probabilities close to the nominal 95% overall, but Fieller’s method always have slightly larger average length and more accurate coverage probability than the delta method. It is known that Fieller’s method usually provides better large sample approximation for a ratio of parameters than the delta method, e.g. Herson (1975). For each method, the average length increases in censoring proportion.
Table 1.
Bias, SSD, and MSD of , and average length and empirical coverage probability of 95% confidence intervals by Delta and Fieller methods
| Delta Method | Fieller Method | |||||||
|---|---|---|---|---|---|---|---|---|
| t0 | t | Bias | SSD | MSD | ECP | Length | ECP | Length |
| Under 15% Censoring | ||||||||
| 1 | 1 | 0.0004 | 0.0308 | 0.0299 | 0.9407 | 0.1096 | 0.9411 | 0.1098 |
| 2 | 0.0005 | 0.0381 | 0.0379 | 0.9471 | 0.1486 | 0.9476 | 0.1489 | |
| 3 | 0.0001 | 0.0396 | 0.0390 | 0.9465 | 0.1528 | 0.9468 | 0.1530 | |
| 4 | 0.0000 | 0.0379 | 0.0371 | 0.9430 | 0.1453 | 0.9434 | 0.1456 | |
| 2 | 1 | 0.0003 | 0.0386 | 0.0384 | 0.9470 | 0.1505 | 0.9477 | 0.1511 |
| 2 | −0.0001 | 0.0440 | 0.0432 | 0.9442 | 0.1694 | 0.9451 | 0.1701 | |
| 3 | −0.0001 | 0.0434 | 0.0425 | 0.9433 | 0.1665 | 0.9435 | 0.1672 | |
| 3 | 1 | −0.0004 | 0.0487 | 0.0475 | 0.9387 | 0.1860 | 0.9425 | 0.1879 |
| 2 | −0.0004 | 0.0548 | 0.0536 | 0.9404 | 0.2099 | 0.9433 | 0.2120 | |
| 4 | 1 | −0.0002 | 0.0607 | 0.0589 | 0.9350 | 0.2307 | 0.9399 | 0.2352 |
| Under 30% Censoring | ||||||||
| 1 | 1 | 0.0003 | 0.0308 | 0.0299 | 0.9457 | 0.1170 | 0.9460 | 0.1172 |
| 2 | 0.0006 | 0.0425 | 0.0416 | 0.9458 | 0.1631 | 0.9462 | 0.1634 | |
| 3 | 0.0000 | 0.0438 | 0.0432 | 0.9427 | 0.1696 | 0.9428 | 0.1699 | |
| 4 | −0.0001 | 0.0422 | 0.0415 | 0.9398 | 0.1629 | 0.9402 | 0.1632 | |
| 2 | 1 | 0.0004 | 0.0434 | 0.0425 | 0.9438 | 0.1668 | 0.9451 | 0.1875 |
| 2 | −0.0002 | 0.0488 | 0.0482 | 0.9420 | 0.1891 | 0.9433 | 0.1899 | |
| 3 | −0.0002 | 0.0486 | 0.0477 | 0.9398 | 0.1872 | 0.9406 | 0.1881 | |
| 3 | 1 | −0.0006 | 0.0542 | 0.0531 | 0.9349 | 0.2085 | 0.9388 | 0.2109 |
| 2 | −0.0007 | 0.0620 | 0.0604 | 0.9376 | 0.2369 | 0.9418 | 0.2397 | |
| 4 | 1 | −0.0003 | 0.0686 | 0.0666 | 0.9320 | 0.2614 | 0.9374 | 0.2677 |
Now, we investigate the finite sample properties of the two-sample log-rank test on conditional distributions. Suppose that arm 1 has an exponential distribution with Si (t) = exp(–λ1t) for t ≥ 0, and arm 2 has a piecewise exponential distribution with survivor function
Note that, if λ1 ≠ λ2, then the two arms have different survival distributions, but with t0 ≥ 2, their conditional distributions are identical with Sk(t|t0) = exp(–λ1t) and λk(t|t0) = λk(t + t0) = λ1 for t ≥ 0. So, the log-rank test will have some power for t0 < 2, but not for t0 ≥ 2. We set λ1 = 0.3466, λ2 = λ1/2, and n1 = n2 = 100, 150 or 200. We consider 15% and 30% censoring by uniform censoring variables as in the previous simulations. We generate 10,000 samples, apply the 2-sample log-rank test with 2-sided α = 0.05 for t0 = 0,1, 2, 3, 4 to each sample, and estimate the empirical power as the proportion of samples that the log-rank test rejects the null hypothesis that two arms have the same conditional survival distributions. Note that the test with t0 = 0 corresponds to the standard log-rank test to compare two unconditional distributions.
Table 2 reports the empirical power of the log-rank tests. As expected, the empirical power of the conditional log-rank test is close to the nominal level α = 0.05 with t0 ≥ 2, for which two conditional distributions are identical. However, with t0 = 0 and 1, it has some power, and the power becomes higher with a smaller t0(= 0) since the time interval over which the two conditional distributions are different is wider in this case. The empirical power for t0 = 0 and 1 also increases in n(= n1 + n2), while that for t0 ≥ 2 is close to the nominal α = 5% regardless of the sample size. We observe that the power with t0 < 2 does not much depend on the censoring proportion under the simulation setting.
Table 2.
Empirical power of the conditional log-rank tests between S1(t) and S2(t) for t0 = 0,1, 2, 3, 4
| n | Censoring | t0 = 0 | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|---|
| 200 | 15% | 0.5490 | 0.1971 | 0.0522 | 0.0510 | 0.0558 |
| 30% | 0.5608 | 0.1853 | 0.0476 | 0.0549 | 0.0551 | |
| 300 | 15% | 0.7202 | 0.2601 | 0.0508 | 0.0480 | 0.0485 |
| 30% | 0.7324 | 0.2597 | 0.0515 | 0.0509 | 0.0516 | |
| 400 | 15% | 0.8451 | 0.3345 | 0.0501 | 0.0535 | 0.0546 |
| 30% | 0.8490 | 0.3206 | 0.0504 | 0.0502 | 0.0505 |
We consider two regression models for simulations on Cox regression analysis of conditional survival distributions. In Model 1, given covariate value zi, the hazard function is given as
Since the hazard function for t ≥ 2 does not depend on zi, the conditional Cox regression with t0 > 2 will be free of the covariate. The cumulative hazard function is given as
and the survivor function is given as
For a U(0,1) random variable Ui, we generate Ti by solving Si(Ti) = Ui. We set β= 0.3 and λ0 = 0.3 for Model 1.
In Model 2, we consider a piecewise exponential distribution with a time-dependent covariate effect: for patient i with covariate value zi, λi(0) = 0,
for j = 1, 2,.… For this model, the covariate effect β/j decreases in t. The cumulative hazard function given zi is,
Since ⋀i(t) = – log Si(t) and Si(Ti) ~ U(0,1) for Ti with cumulative survivor function Si(t), we generate Ti by solving equation ⋀i(Ti) = – log Ui for Ui ~ U(0,1), i.e.
We set λ0 = 0.3 and β = 0.4 for Model 2.
For each of the survival models, we generate 10,000 simulation samples of size n = 500 and generate 15% and 30% censoring from uniform distributions U(b, a + b) as in the previous simulations. For each subject, covariate zi is generated from the standard normal distribution. From each sample, conditioning on (Ti ≥ t0) with t0 = 0, 1, 2, 3, or 4, we fit a proportional hazards model with a time-independent regression coefficient, estimate the regression coefficient, and test on H0 : β= 0 with 2-sided α = 0.05.
Table 3 report the mean regression estimate and empirical power under the two models. For Model 1, with t0 ≥ 2, the mean regression estimate is close to 0 and the empirical power is close to the nominal 0.05 level as expected. But, for t0 < 2, the regression estimate is smaller than β= 0.3 since the covariate effect is diluted over the time interval t ≥ 2 which has no covariate effect. The regression estimate is smaller with t0 = 1 than with t0 = 0 since the former case has a narrower time interval with non-zero regression coefficient. With 30% of censoring, the regression estimate is larger since the additional censoring over 15% censoring occurs over t ≥ 2 (b = 2.2 for Model 1) for which the covariate has no effect. For Model 2, we observe that the regression estimate decays in t0. And the decaying trend is more prominent with 30% censoring since the additional censoring occurs after b = 2.1 where the covariate effect is smaller than that over the earlier time interval. The empirical power quickly decreases in t0 since both the mean covariate effect and the number of observations used in analysis decrease. However, the decrease of power in censoring proportion is smaller since the additional censoring occurs over the time interval with smaller covariate effect.
Table 3.
Mean regression estimate and empirical power of the conditional regression method
| Model 1 | Model 2 | ||||
|---|---|---|---|---|---|
| Censoring | t0 | Mean Est | Power | Mean Est | Power |
| 15% | 0 | 0.2022 | 0.8866 | 0.2610 | 0.9817 |
| 1 | 0.1327 | 0.3706 | 0.1456 | 0.3912 | |
| 2 | 0.0036 | 0.0522 | 0.1076 | 0.1724 | |
| 3 | 0.0035 | 0.0530 | 0.0862 | 0.1043 | |
| 4 | 0.0026 | 0.0565 | 0.0717 | 0.0768 | |
| 30% | 0 | 0.2191 | 0.8824 | 0.2787 | 0.9793 |
| 1 | 0.1470 | 0.3538 | 0.1514 | 0.3409 | |
| 2 | 0.0021 | 0.0545 | 0.1102 | 0.1475 | |
| 3 | 0.0004 | 0.0560 | 0.0869 | 0.0951 | |
| 4 | −0.0033 | 0.0553 | 0.0692 | 0.0741 | |
3.2. Real Data Analysis
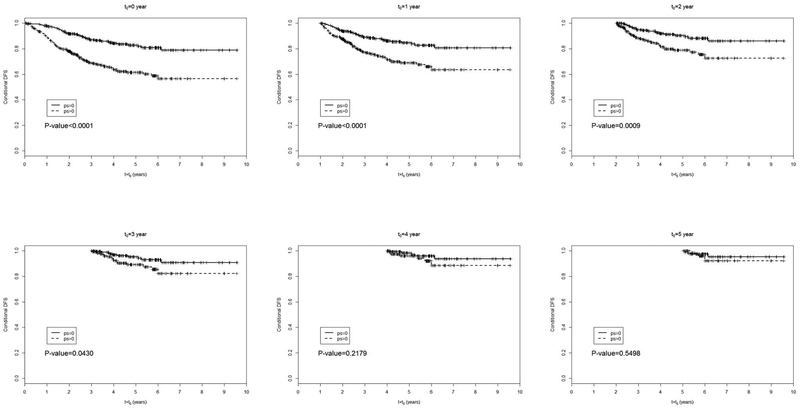
Kim et al. (2016) report analysis results of a retrospective record study on 723 lung adenocarcinoma patients. All the patients underwent complete resection and mediastinal lymph node dissection with or without postsurgical adjuvant therapy. From each patient, overall survival (OS), time to death of any cause from surgery for tumor resection, and progression-free survival (PFS), time to tumor progression, were observed as outcomes together with risk factors including ECOG performance score (PS), with or without adjuvant chemotherapy (adj), tumor-shadow disappearance ratio (TDR) on CT value, and maximum standardized uptake value (SUV) on 18F-uoro-2-deoxyglucose (FDG)-PET/CT. SUV is log-transformed to lower the effect of outliers. The objectives of the study is to associate OS and PFS with the latter four clinical and image predictors using conditional survival analysis. We report the analysis results on OS to illustrate the conditional survival analysis methods. At first, patients are partitioned into two groups by PS = 0 and PS ≥ 1. Figure 1 displays the conditional survivor functions of the two PS groups and conditional log-rank p-value for t0 = 0,1,…, 6. We observe that the effect of PS decays as time passes from surgery and becomes insignificant for patients who have survived for t0 = 4 years or longer. For each t0 value, we regress the conditional survival at t0 = 0, 1, …, 6 on these four covariates using a multivariate proportional hazards model. Figure 2 displays the regression estimate of each covariate and its 95% confidence interval against t0. The covariate effect diminishes for longer survivors except history of adjuvant therapy which has a strong and consistent negative effect on OS. A high log-SUV tends to be associated with shorter OS, but it is not so significant for the t0 values considered. Poor PS is significantly associated with poor OS until t0 = 2 years and its effect becomes weak after t0 = 4 years. High TDR is associated with longer until about t0 = 4 years, but its effect diminishes among survivors over t0 = 5 years.
Figure 1:
Conditional survivor functions for t0 = 0,1, 2, 3, 4, 5. The p-values are from the conditional log-rank test between PS=0 and PS> 0 groups
Figure 2:
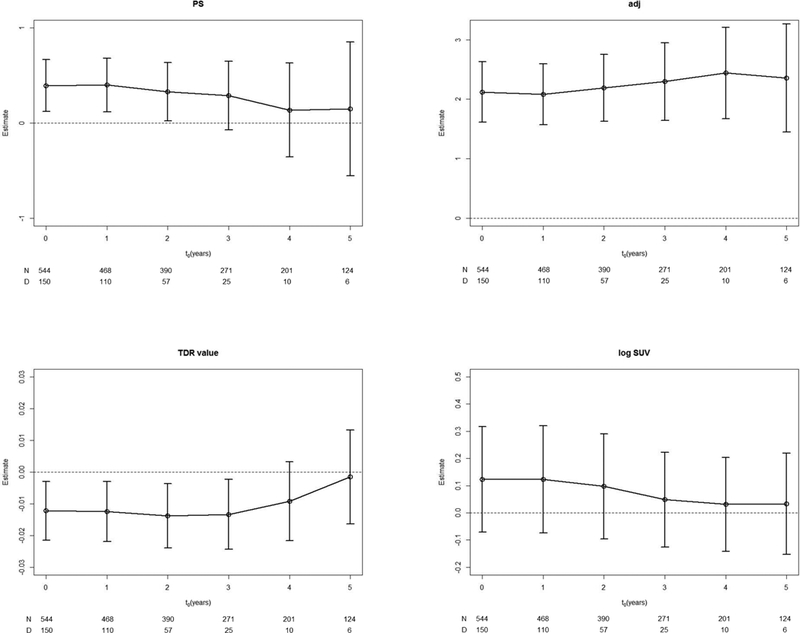
Regression estimate of each covariate and its 95% confidence interval against t0
4. Conclusions
Conditional survival analysis has been popularly used to investigate the long term effect of treatment and baseline characteristics on the prognosis in clinical researches. As a reviewer points out, this analysis provides some new insight on the difference and effect of non-proportional hazards, early and late treatment and baseline patient characteristics. One meaningful scenario is that an aggressive surgical treatment may have a high early mortality, but leads to much higher survival or even cure after the treatment period, while a chemotherapy does not have a severe treatment-related mortality, but leads to a moderate treatment effect over a long time span. The conditional survival analysis would be particularly useful to compare this kind of early and late survival benefit between treatments.
Without any theoretical justification, investigators have applied the standard survival analysis methods, such as log-rank test and Cox regression, to the data removing the patients whose censoring or event times are shorter than t0 claiming that this results in the survival distributions of the survivors over t0. This paper is to theoretically justify this claim. We have reviewed inference methods of one-sample, two-sample and regression analysis for conditional survival distributions. For a reliable estimation of S(t|t0), we need enough number of patients who are at risk at t0 and enough number of patients followed for at least t + t0 unless all patients have events before this time point. Hence, a conditional survival analysis will not be available unless the follow-up period is long enough. Since the conditional survival analyses among the patients who survived over t0 are identical to the standard survival analyses using the data set consisting of patients who are at risk at time t0. Hence, we can conduct any conditional survival analysis using existing statistical softwares, such as SAS or SPSS, with the standard survival analysis procedures. These methods are based on large sample theory. Through simulations, we find that these methods accurately reflect the change in risk function over time and have good finite sample properties.
If the marginal survival distribution satisfies a proportional hazards model (PHM) assumption with time-fixed covariate effect, then the regression estimates from conditional survival analysis will give similar regression estimates for various t0 values. We may be able to develop a goodness of fit test for PHM assumption of a marginal survival distribution using this concept. Jung and Wieand (1999) propose a goodness of fit test for PHM using a similar approach. By plotting the trend of regression estimates of conditional survival analysis over t0, we can also model the time trend of covariates with time-varying effect, refer to Therneau and Grambsch (2000).
Acknowledgments
Funding
This research was supported by a grant from the National Cancer Institute (CA142538–01).
REFERENCES
- Aalen OO (1978). Nonparametric estimation of partial transition probabilities in multiple decrement models. Annals of Statistics 6:534–545. [Google Scholar]
- Bischof DA, Kim Y, Dodson R, Jimenez MC, Behman R, Cocieru A, Fisher SB,Groeschl RT, Squires MH, Maithel SK, Blazer DG, Kooby DA, Gamblin TC, Bauer TW, Quereshy FA, Karanicolas PJ, Law CH, Pawlik TM (2015). Conditional disease-free survival after surgical resection of gastrointestinal stromal tumors: A multi-institutional analysis of 502 patients. JAMA Surgery 150:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson C, Siddiqui MM (1969). Some criteria for aging. Journal of the American Statistical Association 64:1472–1483. [Google Scholar]
- Cox DR (1972). Regression models and life tables. Journal of the Royal Statistical Society, Ser. B 34:187–220. [Google Scholar]
- Fieller EC (1954). Some problems in interval estimation. Journal of the Royal Statistical Society, Ser. B bf 16:175185. [Google Scholar]
- Fleming TR, Harrington DP (1991). Counting Processes and Survival Analysis. New York: Wiley. [Google Scholar]
- Gehan EA (1965). A generalized Wilcoxon test for comparing arbitrarily single censored samples. Biometrika 52:203–223. [PubMed] [Google Scholar]
- Gill RD (1980). Censoring and Stochastic Integrals Mathematical Centre Tracts 124, Mathematisch Centrum, Amsterdam. [Google Scholar]
- Herson J (1975). Fieller’s theorem versus the delta method for significance intervals for ratios. Journal of Statistical Computing and Simulation 3:265–274. [Google Scholar]
- Hollander E, Proschan F (1975). Tests for mean residual life. Biom etrika 62:585–593. [Google Scholar]
- Jeong JH, Jung SH, Costantino JP (2008). Nonparametric inference on median residual life function. Biometrics 64:157–163. [DOI] [PubMed] [Google Scholar]
- Jung SH, Jeong JH, Bandos H (2009). Regression on median residual life. Biometrics 65:1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SH, Wieand S (1999). Analysis of Goodness-of-Fit for Cox Regression Model. Statistics and Probability Letters 41:379–82. [Google Scholar]
- Kaplan EL, Meier P (1958). Nonparametric estimation from incomplete observations. Journal of American Statistical Association 53:457481. [Google Scholar]
- Kim W, Lee HY, Jung SH, Kim HK, Choi YS, Kim J, Zo J, Shim YM, Han J, Jeong JY, Choi JY, Lee KS (2016). Dynamic prognostication using conditional survival analysis for patients with operable lung adenocarcinoma. Oncotarget DOI: 10.18632/oncotarget.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurta ML, Edwards RP, Moysich KB, McDonough K, Bertolet M, Weissfeld JL, Catov JM, Modugno F, Bunker CH, Ness RB, Diergaarde B (2014). Prognosis and conditional disease-free survival among patients with ovarian cancer. Journal of Clinical Oncology 32:4102–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens AC, Yong J, Dietz AC, Kreiter E, Yasui Y, Bleyer A, Armstrong GT, Robison LL, Wasilewski-Masker K (2015). Conditional survival in pediatric malignancies: Analysis of data from the childhood cancer survivor study and the surveillance, epidemiology, and end results program. Cancer 121:1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth EJ (1977). Reliability models with positive memory derived from the mean residual life function, In Theory and Applications of Reliability, (Edited by Tsokos CP and Shimi IN), PP 401–434, Academic Press. [Google Scholar]
- Nelson W (1969). Hazard plotting for incomplete failure data. Journal of Quality Technology 1:27–52. [Google Scholar]
- Peto R, Peto J (1972). Asymptotically efficient rank invariant test procedures (with discussion). Journal of the Royal Statistical Society, Ser. A 135:185–206. [Google Scholar]
- Prentice RL (1978). Linear rank tests with right censored data. Biometrika 65:167–179. [Google Scholar]
- Ruiz JM, Navarro J (1994). Characterization of distributions by relationships between failure rate and mean residual life. IEEE Transactions on Reliability 43:640–644. [Google Scholar]
- Therneau TM, Grambsch PM (2000). Modeling Survival Data: Extending the Cox Model. Springer, New York, NY, USA. [Google Scholar]
- Zabor EC, Gonen M, Chapman PB, Panageas KS (2013). Dynamic prognostication using conditional survival estimates. Cancer 119:3589–3592. [DOI] [PubMed] [Google Scholar]
- Zamboni BA, Yothers G, Choi M, Fuller CD, Dignam JJ, Raich PC, Thomas CR, OConnell MJ, Wolmark N, Wang SJ (2010). Conditional survival and the choice of conditioning set for patients with colon cancer: An analysis of NSABP trials C-03 through C-07. Journal of Clinical Oncology 28:2544–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabor EC, Gonen M, Chapman PB, Panageas KS (2013). Dynamic prognostication using conditional survival estimates. Cancer 119:3589–3592. [DOI] [PubMed] [Google Scholar]